Abstract
Microbial evolution is typically studied in monocultures or in communities of competing species. But microbes do not always compete and how positive inter-species interactions drive evolution is less clear: Initially facilitative communities may either evolve increased mutualism, increased reliance on certain species according to the Black Queen Hypothesis (BQH), or weaker interactions and resource specialization. To distinguish between these outcomes, we evolved four species for 44 weeks either alone or together in a toxic pollutant. These species initially facilitated each other, promoting each other’s survival and pollutant degradation. After evolution, two species (Microbacterium liquefaciens and Ochrobactrum anthropi) that initially relied fully on others to survive continued to do so, with no evidence for increased mutualism. Instead, Agrobacterium tumefaciens and Comamonas testosteroni (Ct) whose ancestors interacted positively, evolved in community to interact more neutrally and grew less well than when they had evolved alone, suggesting that the community limited their adaptation. We detected several gene loss events in Ct when evolving with others, but these events did not increase its reliance on other species, contrary to expectations under the BQH. We hypothesize instead that these gene loss events are a consequence of resource specialization. Finally, co-evolved communities degraded the pollutant worse than their ancestors. Together, our results support the evolution of weakened interactions and resource specialization, similar to what has been observed in competitive communities.
Keywords: evolution, bacterial community, facilitation, Black Queen Hypothesis, specialization, community function
How natural and engineered microbial communities function depends on ecological interactions between their member species. As species adapt to one another and to their environment, these interactions may change, and as a consequence, the overall functioning of the community (Segar et al., 2020). Being able to predict these evolutionary changes may help to intervene and drive a community toward a desirable function. One could imagine, for example, predicting how the gut microbiome would respond to an intervention against inflammatory bowel disease, or how a community in a microbial bioremediation system could be controlled to evolve toward a more stable, efficient state (Atashgahi et al., 2018; De Roy et al., 2014; Gorter et al., 2020; Widder et al., 2016).
Evolutionary prediction and control relies on understanding how selection acts on interactions between species. One way to study how these inter-species interactions evolve is to perform experimental evolution by passaging multi-species communities over sequential batch cultures or in chemostats over long time periods, and following ecological changes in the relative abundances of different species as well as phenotypic and genotypic changes in each community member. Prior studies using this approach have found that microbes can rapidly adapt to both biotic and abiotic factors (Fiegna et al., 2015; Gravel et al., 2011; Henriksen et al., 2022; Lawrence et al., 2012; Savolainen et al., 2013), but being embedded within a community can limit adaption to abiotic factors (Castledine et al., 2020; Collins, 2011; Gómez and Buckling, 2013; Hall et al., 2018; Lawrence et al., 2012; Runquist et al., 2020).
In terms of inter-species interactions, bacterial communities that initially displayed negative interactions evolved towards neutral (Fiegna et al., 2015; Rivett et al., 2016) or positive interactions (Lawrence et al., 2012). This evolutionary response is intuitive, as species can be expected to reduce resource competition and niche overlap (Hall et al., 2018; Jousset et al., 2016; Liow et al., 2011) and may adapt to use resources generated by other species (Hall et al., 2018; Lawrence et al., 2012; Ridenhour, 2005; Rivett et al., 2016). Accordingly, species evolving in isolation tend to extend their niches in absence of competition and compete when reintroduced into the community context (Castledine et al., 2020; Lawrence et al., 2012).
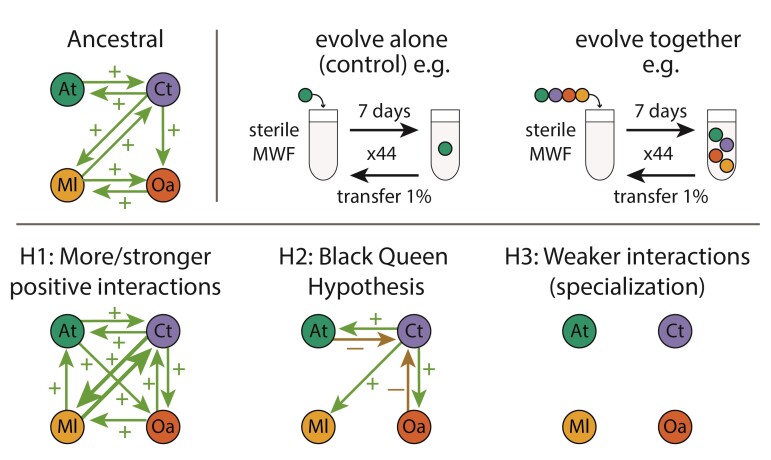
In contrast, studies that have experimentally evolved communities beginning with positive or facilitative interactions mostly contain only two species or two strains of the same species, often with strong dependencies on one another (Fritts et al., 2020; Hillesland et al., 2014; Marchal et al., 2017; Preussger et al., 2020; Summers et al., 2010; Zhang and Reed, 2014) (Henriksen et al., 2022; Turkarslan et al., 2021). This may be because microbial isolates tend to compete with one another when co-cultured in the lab (Foster and Bell, 2012), meaning that a synthetic community assembled in the lab is unlikely to spontaneously display several positive inter-species interactions. We expect three different outcomes compared to initially competitive communities (Figure 1): First, if positive interactions are constant and bi-directional over many generations, this might select for each species to increase its positive effect on the other, resulting in mutualism (Chacón et al., 2021; Sachs and Hollowell, 2012). Second, species evolving together might evolve to exploit resources that are provided by others, resulting in stable co-existence because the providing species itself depends on the resource. As proposed by the Black Queen Hypothesis (Jeffrey Morris et al., 2014; Morris et al., 2012), a common consequence of the reliance on public goods produced by others is that the receiving species are selected to lose genes for costly product pathways (Cordero et al., 2012; Hillesland et al., 2014). Third, positive interactions can weaken, particularly if the cooperative traits are costly, resulting in reduced reliance of species on one another (Sachs and Simms, 2006). If each species grows independently, one might expect species to evolve to each specialize on a different resource, thereby exploiting available niches more efficiently (Lawrence et al., 2012).
Figure 1.
Experiment and hypotheses. Top: An ancestral community with facilitative interactions was evolved in MWF using serial transfers every 7 days for 44 weeks. Different species combinations were explored with species evolving either alone or in a community. Bottom: Hypotheses for how interactions in the community might evolve, assuming that they continue to coexist. H1: More/stronger positive or mutualistic (bi-directional positive) interactions. H2: According to the BQH, few species would provide the public goods for others that would lose the ability to produce them and possibly exploit the producing species. H3: Each species specializes on a different environmental niche, resulting in weaker inter-species interactions.
In our previous work (Piccardi et al., 2019), we studied a community composed of four bacterial species (Agrobacterium tumefaciens (At), Comamonas testosteroni (Ct), Microbacterium liquefaciens (Ml), and Ochrobactrum anthropi (Oa)) and showed that facilitation was more prevalent when the community was grown in a toxic environment, in agreement with the Stress Gradient Hypothesis (Bertness and Callaway, 1994). The toxic environment in question is an emulsion of machine oils used in the manufacturing industry called Metal Working Fluids (MWF), which the four species were capable of degrading when together. They are not known to have a common evolutionary history and were likely isolated from distinct MWF samples (van der Gast and Thompson, 2005, 2014). This community represents a tractable model system for exploring how the abiotic and biotic environment shapes the evolution of positive inter-specific interactions and how they relate to community function, in this case, MWF bioremediation.
In this study, the four bacterial species were grown in MWF and left to evolve either in isolation or together in communities (Figure 1, top) (Castledine et al., 2020; Lawrence et al., 2012). We quantified bacterial growth and MWF degradation efficiency, and identified genomic changes. By the end of the experiment, positive interactions had declined between the two species evolving together that were able to grow on their own, but not for those that still relied on others to survive and grow. The species evolved in isolation were more productive than those evolved in community and tended to compete with one another when co-cultured. We found little evidence to support the Black Queen Hypothesis, as the species that experienced gene loss events did not increase their reliance on others to grow. Gene loss may instead be a signature of resource specialisation. These results suggest that evolving communities that begin with positive interspecies interactions can evolve similarly to those that begin with negative interactions. In our system, interactions weakened whenever dependencies disappeared, possibly due to niche partitioning, and because the evolution of individual species was constrained by coexisting species.
Results
Replicate microcosms for each species combination behaved similarly and converged to even communities
Our central question is how facilitative inter-species interactions drive evolution within a microbial community. We addressed this question using experimental evolution of four species either together in groups of three or four species, or alone as a control. The choice to include this particular three-species combination was based on preliminary data suggesting that Oa may affect community dynamics. While this intuition was confirmed, we do not compare the three- and four-species communities explicitly, but nevertheless include all combinations in our data set.
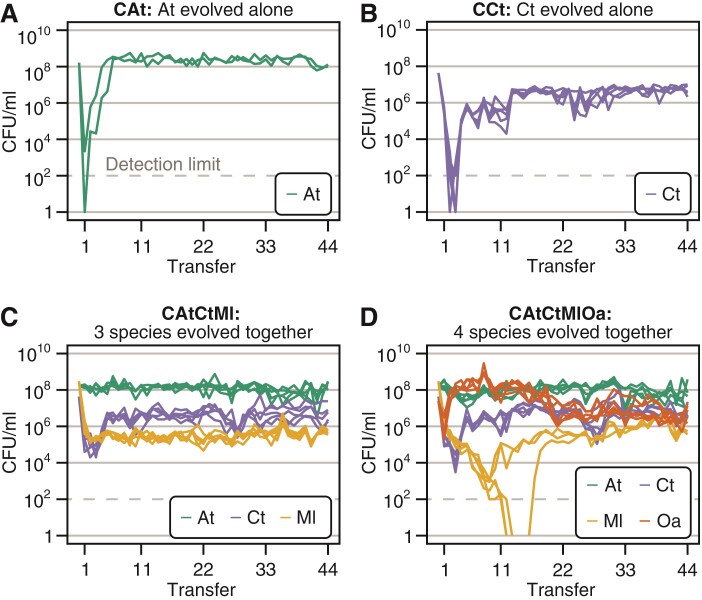
Over the first few weeks, population sizes experienced large fluctuations, which were less pronounced when species were evolving together. When evolving alone, Ml and Oa went extinct after the first transfer (data not shown), which was unsurprising as they do not grow alone in MWF unless the other species are present (Piccardi et al., 2019). When evolving alone, At only persisted in 2 out of 5 lines (henceforth CAt for “combination” At), while Ct survived in all five microcosms (henceforth CCt). The population sizes of both species dropped initially, but stabilized after about 6 and 11 transfers, respectively (Figure 2A and B). When species were evolving together, population sizes stabilized after about four transfers in the three-species community (CAtCtMl, Figure 2C) and 22 transfers in the four-species community (CAtCtMlOa, Figure 2D), with the exception of Ml that went extinct in two out of five microcosms in the four-species community. In the presence of Oa, Ml’s population size dropped dramatically compared to when Oa was absent (Figure 2C, D), indicating a negative interaction. However, since none of them grew alone in MWF, we were unable to quantify this negative interaction in the corresponding pair-wise mono- and co-cultures.
Figure 2.
Population sizes over time. Experiments were started with each species in batch monoculture or co-cultures of three and four species. Every week, we serially transferred cultures by diluting them 100-fold in fresh MWF for 44 weeks. Before each transfer, species abundances were quantified by selective plating. Each species combination (abbreviated as “C” followed by the species combination inoculated at the start, e.g. CAtCtMl) initially consisted of 5 microcosm replicates (culture tubes). CFU/ml counts from selective plates are shown for all combinations: At evolved alone (green), where three microcosms dropped below the detection limit (at CFU/ml and is indicated with a grey horizontal line) and were discontinued (A), Ct evolved alone (blue) (B), At, Ct, and Ml evolved together (C) and At, Ct, Ml, and Oa evolved together (D). In this species combination, Ml dropped below the detection limit in three microcosms, but recovered in one.
In all microcosms where species did not go extinct, the population dynamics in replicate microcosms of the same species combination were similar. By transfer 44, communities were quite even, with relatively small differences in population sizes between species that evolved together (Figure 2C, D), as expected based on similar studies (Rivett et al., 2016). The total population sizes in the two co-evolving communities did not increase over time (as observed by Hillesland and Stahl, 2010; Hillesland et al., 2014), suggesting that species did not evolve to increase their own or other species’ yield (Rivett et al., 2016). In fact, fitting a linear model to the total population size in evolving communities (CAtCtMl and CAtCtMlOa) showed a small yet significant decrease over transfers (slope = , ). Species that evolved alone instead showed no significant change (CAt: ) or increased over time (CCt: slope = , ). Species that survived until the end of the experiment went through approximately 300 generations (Supplementary Table S1).
Positive interspecies interactions weakened when evolving together
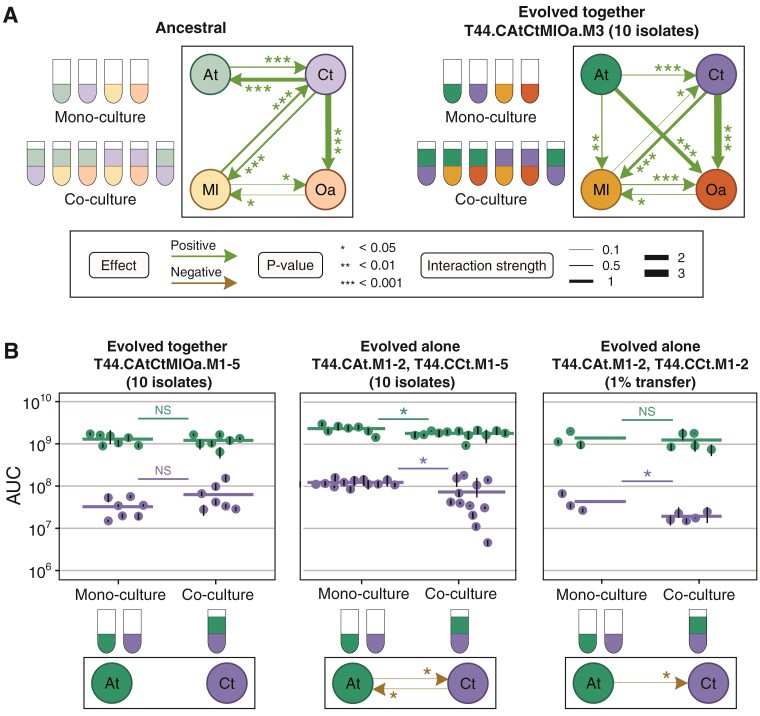
We first explored whether interactions between the evolved species differed from the ancestral ones. We focused on the four species that evolved together (CAtCtMlOa), and to represent the most abundant, genotypically distinct sub-populations of each evolved bacterial species, mixed equal proportions of ten isolates of each species coming from transfer 44 of the same replicate microcosms (see Methods). We used these mixes, as we detected some within-species phenotypic diversity in growth and degradation (Supplementary Figures S1 and S2), but obtained similar results using only one isolate per species, suggesting that growth patterns are consistent across approaches (Supplementary Figures S3 and S5). From now on, when referring to species in these evolved cultures, we mean these isolate mixes.
Using these isolate mixes, we measured the inter-species interactions in one microcosm where four species evolved together (CAtCtMlOa, replicate microcosm 3, arbitrarily selected among microcosms where all four species were present at transfer 44). We incubated each species in monoculture or in pairwise co-cultures with each of the other species from the same microcosm over 12 days (Figure 3A). In monoculture, contrary to its ancestor (Supplementary Figure S4), At was able to survive and grow in MWF alone. Both ancestral and evolved Ct were able to survive and grow in MWF (Supplementary Figure S3C), but the area under the growth curve (AUC) of evolved Ct was significantly lower even when measured with different assays (Supplementary Figures S5 and S6B). Finally, Ml and Oa from all microcosms were still unable to grow alone (Supplementary Figure S8).
Figure 3.
Inter-species interactions. (A) Inter-species interaction network in ancestral species (adapted from Piccardi et al., (2019), Supplementary Figure S4) and between the four species after evolving together (species combination CAtCtMlOa) for 44 transfers (T44) from microcosm 3 only (M3). Each evolved species was represented by a mixture of 10 isolates from T44. To quantify interactions between evolved species, we compared the AUC of each species (mix of 10 isolates) grown for 12 days in mono- versus co-culture (see Methods, Supplementary Figure S3). (B) Interactions between At and Ct evolved together (first column, CAtCtMlOa) or evolved alone (2nd and 3rd column, CAt and CCt) during 8-day growth assays. For growth assays for CAtCtMlOa (first column), we only co-cultured species that had evolved together in the same microcosm, and analyzed all microcosms (M1-5, see Methods for replicate numbers). For the 2nd column, we monocultured At and Ct several times and co-cultured all possible combinations of microcosms that had evolved alone at least once (see Methods for replicate numbers). For the 3rd column, we only tested four combinations using 1% of the population of the corresponding microcosm (see Methods). Dots show means and black bars standard deviations of the AUCs, thick horizontal lines show the means of the dots. Statistical tests outlined in Methods.
By comparing the AUCs of monocultures with pair-wise co-cultures, we were able to reconstruct an interaction network (Figure 3A), as previously done for the ancestral network by Piccardi et al., (2019). Ml and Oa continued to rely on Ct for survival, but we found no evidence for increased mutualism between Ml and Ct (Supplementary Figure S9). Unlike the ancestral community, evolved At promoted the survival and growth of Ml and Oa while it no longer benefited from evolved Ct. The appearance of positive interactions toward the two species that could not grow alone was expected because At could now grow independently (Piccardi et al., 2019). The lack of competition between the two independent species (At and Ct) was however unexpected, as our intuition from previous work was that autonomous species should compete (Piccardi et al., 2019). This motivated us to explore this relationship further.
To understand whether the weakened interaction between At and Ct was consistent across all five microcosms where the species had evolved together (CAtCtMlOa), we compared the growth of evolved At and Ct isolates from the same microcosms in mono- and pair-wise co-cultures. We found that the AUC, the maximal CFU/ml difference between two consecutive days of each species (a proxy for growth rate), and the maximum population size of At, did not differ significantly when co-cultured with Ct from the same evolved microcosm (linear model with biological replicate as a random effect; AUC: , Figure 3B, left column; maximal CFU/ml difference between two consecutive days: , Supplementary Figure S7B, left column; maximum population size: , Supplementary Figure S7C, left column). Instead, Ct had a significantly greater maximal growth rate when co-cultured with At from the same evolved microcosm, but its AUC and its maximum population size did not differ significantly (linear model with biological replicate as a random effect; AUC: , Figure 3B, left column; maximal CFU/mL difference between two consecutive days: , Supplementary Figure S7B, left column; maximum population size: , Supplementary Figure S7C, left column). In other words, taking into account all microcosms and several ways to measure interactions, At and Ct no longer interacted significantly.
Species that evolved alone tended to interact negatively
We wondered whether the reduction in positive interactions between At and Ct when evolved together was simply the result of adaptation to the harsh MWF conditions. We compared the growth of At and Ct that had evolved alone when grown in mono- and pair-wise co-cultures (Figure 3B, middle column). Both species inhibited each other’s growth, where the AUC (linear model with biological replicate as a random effect, CtAt, AtCt) and maximal population size (CtAt, AtCt) (Supplementary Figure S7A, C) of the co-cultures were lower than the monocultures. Although the effect sizes do not appear large on the plot, they are non-negligible (e.g. AUCs of At and Ct were reduced by 22.8% and 40.5% on average, respectively). Overall, this suggests that the evolutionary response of At and Ct is different whether they evolve alone or in the community context.
One explanation for the competitive interactions may be that the isolates we used for these assays had a particularly high fitness within their populations. To test whether our results were biased in this way, we transferred 1% of the entire populations of At and Ct from two microcosms each where they had evolved alone directly into mono- or co-culture assays (see Methods). At still inhibited the growth of Ct (linear model with biological replicate as a random effect, AtCt) (Figure 3B, right column), suggesting that there was likely nothing particular about the 10 isolates. In sum, the positive interactions between At and Ct in the ancestral strains switched toward more neutral interaction when evolving together, and competition when evolving alone.
Species evolved alone were more productive than those evolved together
A possible explanation for why species that evolved alone compete with one another in co-culture, is that evolving alone allows them to increase their niche coverage, resulting in competition with future invaders into its environment. If instead, a focal species is already sharing the environment with other species with which it does not compete, their presence may prevent the focal species from expanding its niche thereby limiting competitive interactions from arising over evolutionary time scales. While niche partitioning is difficult to quantify in a complex chemical environment like MWF, we predicted that if species that evolved alone cover more niche space, they should grow faster or to a larger population size compared to their counterparts that evolved with others. Consistent with this prediction, the AUC of At and Ct that had evolved alone was significantly higher than their counterparts that had evolved in community (linear model with biological replicate as a random effect, At, Ct), even when they were grown in co-culture (linear model with biological replicate as a random effect, At, Ct, Figure 3B, Supplementary Figure S1, S10). While these results do not prove that evolving alone led to greater niche expansion (they may simply have evolved higher yield), they match observations from previous studies (Fiegna et al., 2015; Hall et al., 2018; Lawrence et al., 2012) showing that adaptations to increase productivity are limited when species are evolving with others.
Ecological context influences genomic changes
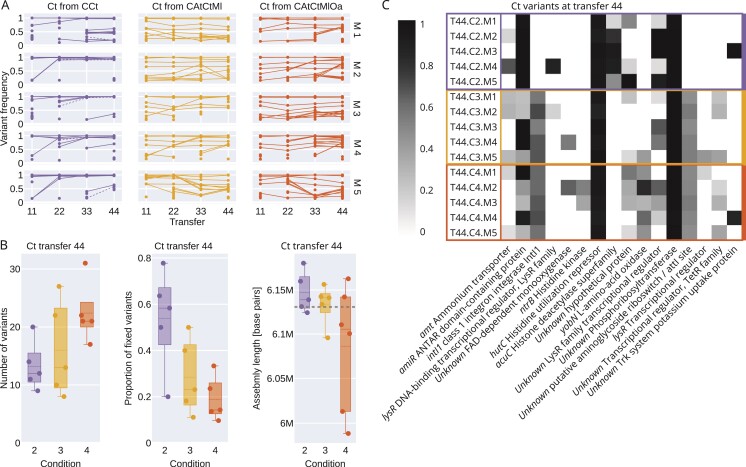
Given the differences between Ct and At that had evolved alone or together, we next wondered whether we could find corresponding genomic variations and determine when they emerged. To this end, we extracted and sequenced the DNA of all microcosm populations every 11 transfers and reconstructed their evolutionary trajectories (see Methods). Because we lack statistical power for At (it only survived in 2 microcosms when evolving alone, analysis in Supplementary Figure S12), we focused on Ct.
We observed distinct patterns for Ct evolved alone or together with other species (Figure 4). When evolved with other species (CAtCtMl and CAtCtMlOa), Ct accumulated a higher number of variants compared to when it was evolving alone (CCt, Kruskal–Wallis chi-squared = 6.818, , Figure 4B left), resulting in a higher total allele frequency (Supplementary Figure S13). But many variants did not fix and remained at intermediate frequencies (Figure 4A center and right). Instead, when evolved alone, a significantly higher number and proportion of variants fixed (number: Kruskal–Wallis chi-squared = 4.165, ; proportion: Kruskal–Wallis chi-squared = 4.810, , Figure 4B, center). This suggests hard sweeps when evolving alone and soft sweeps when evolving in community, which can be explained by the strong drop in population size early on in the experiment when alone compared to in community (Figure 2B vs. C and D).
Figure 4.
Genomic changes. (A) Variant frequency trajectories in all Ct populations. Each dot/line represents a different variant at a different location. Dashed blue lines indicate mutations in the acuC gene (see panel C for a description of acuC). (B) Number of variants found in each Ct population at transfer 44 (left, matches data in panel A), proportion of variants that reached fixation (center, matches data in panel A), and de novo long-read assembly lengths based on PacBio sequencing of selected isolates from transfer 44 (right). The dashed line represents the assembly length of the ancestor. (C) Mutated genes with protein annotation that were found in at least two Ct populations at transfer 44. The grey shade indicates the frequency of the mutated allele.
Given that ecological context affected allele frequencies and fixation rates, we expected variant targets to also depend on the presence or absence of other community members. We annotated the variants and filtered for genes that were mutated in at least two microcosms (Figure 4C). One gene (acuC), which codes for a histone deacetylase, was mutated exclusively in CCt (in all 5 microcosms). Mutations to seven genes were exclusive to combinations CAtCtMl and CAtCtMlOa (two genes were affected in all 10 microcosms), and two genes were mutated and almost completely fixed in all microcosms across all species combinations, likely related to adaptation to MWF.
In Ct coming from one particular microcosm (T44.CAtCtMlOa,M2), we observed that three out of 10 isolates were able to grow alone at twofold higher MWF concentrations and had no measurable lag time, while the remaining seven grew more characteristically for this species (Supplementary Figures S1 and S2). We whole-genome sequenced one isolate from each subpopulation and identified a mutation in ntrB (coding for a Histidine kinase) in the more resistant strain. We confirmed that this variant was present but not fixed in the metagenomic sequencing data of that population. The resistant isolate also had a large deletion (Supplementary Figure S14C), which we discuss below. Why this more resistant variant did not fix, and whether the wildtype-like variant is acting as a cheater is unclear.
No evidence for Black Queen dynamics in our system
The Black Queen Hypothesis (BQH) (Jeffrey Morris et al., 2014; Morris et al., 2012) predicts that if several species in a community are contributing to a public good, all but one species should lose this trait, leading to gene loss in evolving communities. In our system, environmental detoxification can act as a public good. Although we do not know which genes are involved, we explored whether gene loss occurred preferentially for species evolved together compared to alone by long-read sequencing whole genomes of isolates from all microcosms at transfer 44 (see Methods). After assembling full Ct and At genomes, we found that two Ct isolates from CAtCtMlOa were over a 100k base pairs shorter than the reference genome. We mapped these to the reference strain and found an identical deletion of 145k base pairs including 31 genes (see Supplementary Figure S14). We doubt that these deletions are due to increased dependence on other species in the community, as the BQH would predict, as these two isolates grew similarly in isolation to the ones without the deletion (Supplementary Figure S1). Indeed, one of these isolates was the strain that was resistant to higher MWF concentrations described above and grew better than the other isolates (Supplementary Figures S1 and S2). For At we observed a large deletion in one isolate from CAtCtMl, but nothing striking for Ml and Oa (Supplementary Figure S13B). Despite these observations, we lack statistical power to conclude anything general. We also used the assemblies to check if any sequences from other species were integrated in the genomes; however, no transfer events were detected.
As it seemed plausible that 44 weeks were too short for structural changes to occur systematically, we next explored whether point mutations in regulatory regions might have instead led to downregulation in gene expression in evolved communities. We extracted and sequenced RNA from isolates of all microcosms of Ct and At at transfer 44 as well as their ancestors. As the quality of RNA from At samples was low, we focused on Ct. Contrary to the prediction of the BQH, we found no significant difference in the normalized expression levels from the isolates of CAtCtMl and CAtCtMlOa that had evolved in community compared to the ancestor, while several genes in isolates that had evolved alone (CCt) were significantly downregulated when compared to the ancestor (Supplementary Figure S13C–E, Table S2). The only mutation present uniquely and repeatedly in CCt was in the acuC gene, which is expected to affect gene expression.
At degrades MWF better after evolving alone but not in community
Next, we investigated whether the decline in positive inter-specific interactions over the 44 transfers was associated with a shift in MWF degradation efficiency (as in Rivett et al., 2016). If, for example, co-evolved species have indeed reduced their niche overlap and diverged in their resource use, we might expect greater overall MWF degradation. On the other hand, Ct that evolved in the community grew slower than its ancestor, which may lead to worse degradation, as it is one of the main degraders in the community (Piccardi et al., 2019).
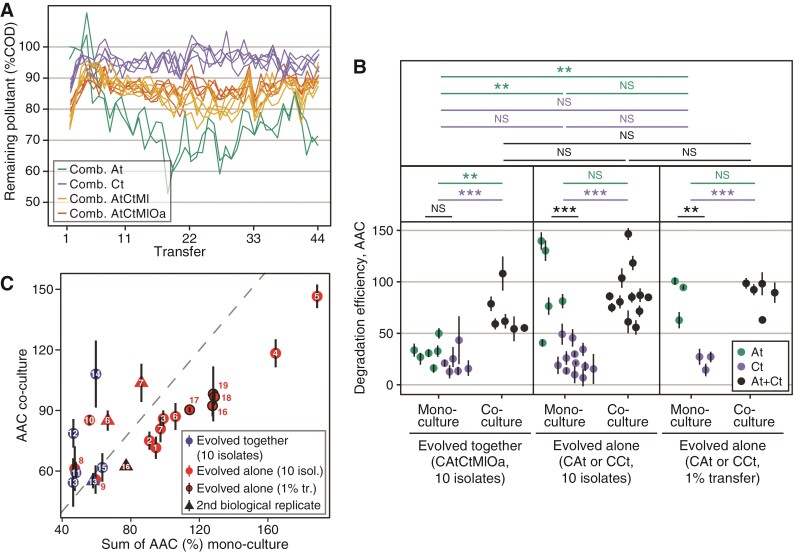
Over the 44 transfers (Figure 5A), Ct and the two evolved communities reduced their degradation efficiency, such that at the end, they degraded less than their ancestral counterparts (%COD on day 7, isolates from transfer 1 vs. 44 in Ct evolved alone, CAtCtMl and CAtCtMlOa, respectively: paired t-tests, , ; , ; , ). In contrast, the two microcosms in which At evolved alone degraded significantly better than their ancestral counterpart (%COD on day 7, isolates from transfer 1 vs. 44 in mono-evolved At: linear model, , , Figure 5A) and even compared to all other microcosms (%COD on transfer 44, day 7, comparing At with Ct and the three- and four-species communities, respectively: linear model, , ; linear model, , ; linear model, , , Figure 5A).
Figure 5.
Degradation efficiency. (A) Remaining pollutant, measured as chemical oxygen demand (COD, g/L) as a percentage of the COD of an abiotic control for each microcosm before each transfer in all four species combinations (lower implies greater degradation). (B) Comparison of the degradation efficiency of At and Ct evolved alone or together with others, in mono- or co-culture, where the area above the degradation curve over 8 days is shown (higher AAC implies greater degradation, unit is sum of percentages). (C) Prediction of an additive model of the sum of degradation efficiencies of individual species is plotted against degradation efficiency of the co-cultures of species evolved alone versus together. Points lying above (or below) the dashed line degrade more (or less) efficiently than predicted by the additive model. Each small number associated with a datapoint indicates a given species combination (e.g. CAt.M1 + CCt.M1). Certain numbers have both a circle and a triangle, which are biological replicates of the same species combination. A list of which number corresponds to which combination can be found in Dataset 1.
Knowing that At was a member of the two evolved communities, we wondered why the degradation efficiency of the communities was worse than At evolved alone. Did the community members inhibit the degradation efficiency of At or did it not evolve improved degradation? We find evidence to support the latter: when grown alone, At from the evolved community degraded less efficiently than when it had evolved alone (%AAC, assays with 10 isolates and 1% transfer, respectively: linear model with biological replicate as random factor, , ; , , Figure 5B). This mirrors our earlier observation that At that had evolved alone grew to greater population sizes than when evolved in community (Figure 3B, Supplementary Figures S1 and S10, compare At monocultures). These data suggest that other species may have constrained the evolution of At, preventing it from evolving greater degradation efficiency by occupying some niches that it could instead fill when evolving alone. If the species that evolved together with At are filling the available niches, might they complement At’s ability to degrade MWF?
Species evolved together degrade MWF synergistically
Following our observation that At evolved in community does not degrade as much as when it evolved alone, we wondered whether the species evolving together with At—notably Ct—could improve its degradation efficiency. By applying an additive null model to degradation efficiency, we compared the combined degradation of the two monocultures of these two species to degradation in their corresponding co-cultures (Fiegna et al., 2015; Foster and Bell, 2012; Piccardi et al., 2019; Rivett et al., 2016) (Figure 5C). Although there were some differences between experimental repeats with different sub-samples of the evolved populations, overall, we found that At and Ct that had evolved in the same microcosms had small positive effects on each other’s degradation efficiency (statistical analysis in Supplementary Figure S11). For the species that had evolved alone, depending on which isolates we used for the assays, we found that in some cases the two species significantly reduced each other’s degradation efficiency (Supplementary Figure S11). Together, this supports the hypothesis that co-evolving species do not overlap much in their niches and can therefore synergistically degrade MWF. Instead, Ct that had evolved alone seems to interfere with the degradation ability of At that had evolved alone, which suggests that the potential of At to expand its niches and increase its own degradation efficiency may have been limited when it evolved in the community context.
Discussion
Our main goal was to establish how interactions within a facilitative community might change as species evolved together. Would interactions become more mutualistic, would one species become parasitized by the others to produce all the public goods (as per the BQH) or would they evolve to specialize or even compete (Figure 1)? Similar experiments with initially competitive communities found that interactions weaken as their members co-evolve to specialize on different resources (Barraclough, 2019; Castledine et al., 2020; Fiegna et al., 2015; Gravel et al., 2011; Lawrence et al., 2012; Rivett et al., 2016). In our 44-week long evolution experiment, species that relied heavily on others to survive in MWF continued to do so, but with no evidence of strengthened mutualism. Instead, one species At, that evolved to grow independently in MWF weakened its positive interaction with the other independent species, Ct. When we evolved each of those two species alone, they competed when put back together.
Our interpretation of this outcome—while well aware that alternative explanations exist—is that in the community, At and Ct experienced weak selection to expand into occupied niches and compete with other residents, driving them to specialize on more available resources (H3 in Figure 1), analogous to character displacement in Darwin’s finches (Grant and Grant, 2006; Pastore et al., 2021; Schluter et al., 1985). Instead, when evolving alone, they may have become generalists by expanding into available niches because no other species were occupying them (Hall et al., 2018). The presence of other species may then have constrained the evolutionary potential of At and Ct (similar to results reported by Hall et al., (2018)). Evidence for this is that after evolving alone, isolates of these two species grew significantly better than those that had evolved in community. Indeed, At evolving alone was the only condition where degradation improved over the course of the experiment and largely surpassed the degradation ability of the community, even though the community includes At. In follow-up experiments reported elsewhere we also found that new, non-resident species were more likely to invade the ancestral compared to the evolved community (Piccardi et al., 2022), suggesting that the community members evolved to cover the available niche space. However, our data may appear to not fully support this interpretation: if the species have really evolved to partition the available niches more efficiently, why would they not degrade the pollutant better? The answer may lie in the COD measure for pollutant degradation, which captures the removal of both nutrients and toxic compounds: while the evolved species may be better at partitioning and consuming the nutrients, their ability to remove toxins may have declined.
An alternative initial hypothesis was that positive interactions might increase, leading to the evolution of mutualism (H1 in Figure 1) because mutants that overproduce public goods should be favored as they promote the growth of species that “help” them (Chacón et al., 2021; Sachs and Hollowell, 2012). This outcome can result in increased community productivity (Harcombe et al., 2018; Zhang and Reed, 2014), increased aggregation between cooperating strains (Marchal et al., 2017; Preussger et al., 2020; Summers et al., 2010) and/or loss of independent growth (Hillesland et al., 2014). By comparing the ancestral and evolved interaction networks (Figure 3), the bi-directional interactions between Ct and Ml were a candidate for this. However, we found no significant increase in the strength of their positive interactions, at least in this one microcosm (Supplementary Figure S9). Second, the number of positive interactions may increase if species generate new niches, which others can evolve to occupy (Fritts et al., 2020; Harcombe, 2010; Lawrence et al., 2012). While it may appear that there are more positive interactions in the evolved community (Figure 3), the positive effects of At on Ml and Oa were already observed in the ancestors growing under conditions where At survives (Piccardi et al., 2019), and are not newly evolved traits. In addition, if stronger positive interactions had evolved, we would expect overall community productivity to go up because resource use becomes more efficient (Hillesland and Stahl, 2010; Lawrence et al., 2012). While we do find some synergy in MWF degradation, the co-evolved co-cultures still degrade less than At evolved alone, and total population sizes even decreased over the evolutionary experiment.
The other question was whether we would find support for the Black Queen Hypothesis (Morris et al., 2012) (H2 in Figure 1): if several species in the ancestral community provide a “service”, others should evolve to lose it, manifesting itself in gene loss for species evolving together (Mas et al., 2016; Morris et al., 2012). What constitutes a “service” in our context is not clear mechanistically, but At and Ct do facilitate the other two species by detoxifying the environment (Piccardi et al., 2019). If they were initially achieving this in overlapping ways, the two species might evolve to specialize on degrading different toxins. This would predict greater gene loss or reduced gene expression in the species evolved together compared to those evolved alone, and greater reliance on one another for survival. We found little evidence in support of this prediction: two Ct isolates that had evolved in community experienced large deletions, but these isolates grew similarly alone to others without the deletion, and at least one of them was even more resistant to MWF compared to a strain that had evolved alone.
These findings made us realize that the evolution of resource specialization within a community predicts similar patterns of gene loss to the BQH, as the ability to use certain resources that are already taken up by others becomes superfluous (Supplementary Figure S15). Given that our data generally support niche specialization rather than increased reliance on other species (at least for the two species we focused on), the deletions we see may be more in line with specialization rather than the BQH, but additional work would be needed to test this idea. In other words, we suggest that the BQH and specialization are two similar processes that we expect to drive genomic changes when species evolve in community. By itself then, gene loss alone should not be taken as evidence supporting the BQH.
Why the bacteria evolved to degrade less in all conditions except for At evolving alone is an important open question when optimizing microbial community function. One possibility is that in the communities and when Ct was alone, selection favored the emergence of cheaters that grew faster but contributed less to MWF degradation, which may have increased the death rate, explaining the lack of increase in total population size (O’Brien et al., 2014; O’Brien and Buckling, 2015). Alternatively, cells might have evolved to resist the toxins without secreting toxin-degrading enzymes, for example by thickening the cell wall or using efflux pumps (Blair et al., 2015; Bottery et al., 2016, 2021). This would make resistance into a “private good” and reduce MWF degradation. Third, the community constrained the evolution of At, explaining why its degradation did not improve when evolving in the community. Regardless of the mechanism, our results suggest that the problem of loss of community function needs to be addressed in future studies. Otherwise, single species like At might be better suited compared to communities, at least for this particular function of MWF degradation.
A final interesting question in community evolution concerns predictability: Do parallel microcosms evolving under the same condition resemble one another? Previous evolutionary experiments using communities found bimodal or trimodal outcomes in final relative abundances (Celiker and Gore, 2014; Hekstra and Leibler, 2012). We observed striking parallel ecological dynamics between microcosms, whereby relative abundances converged by week 44, despite the occasional extinction of Ml. Oa appeared to play a destabilizing role, as population sizes of all species fluctuated more strongly in CAtCtMlOa compared to CAtCtMl before converging (Figure 2C, D). As in other such experiments (Henriksen et al., 2022; Summers et al., 2010; Turkarslan et al., 2021), we also observed some parallelism in genomic evolution, where several mutations and deletions occurred in parallel lines of the same experimental condition, at least in Ct. While it is tempting to speculate on the effects these mutations might have, we prefer to leave mechanistic analyses to future work where we would build the appropriate mutants.
One of the weaknesses of our system is that chemical analysis is challenging, meaning that we lack a mechanistic understanding of pollutant degradation or the interactions between species. We are therefore blind to how resources are being partitioned, what lies behind the positive interactions, the consequences of genomic changes, or why degradation efficiency dropped over time in evolving communities. Another difficulty was our inability to generalize, as the community only includes four species, and each followed a different evolutionary trajectory. Running similar experiments using communities with more species in a simpler chemical environment could help to test our hypotheses further.
Our experiments present a case study of how four species can evolve in a toxic environment, showing that for species pairs whose dependencies were facultative, interactions weakened over time. Positively interacting species are therefore not necessarily expected to evolve towards mutualism (Chacón et al., 2021), and can instead evolve similarly to competitive communities. From an applied perspective, community function dropped over time as the species evolved, suggesting that to maintain function, new strategies are needed. Finally, parallels can be drawn to evolution in other toxic environments, such as those containing antibiotics, a phenomenon that has classically been studied in single species in isolation (De Wit et al., 2022). Being able to predict and control the evolution of microbial communities would be impactful in many such contexts.
Methods and materials
Bacterial species and culture conditions
The ancestral species used in this study were Agrobacterium tumefaciens str. MWF001, Comamonas testosteroni str. MWF001, Microbacterium liquefaciens str. MWF001, and Ochrobactrum anthropi str. MWF001. More details on these strains can be found byPiccardi et al., (2019) and their genome sequences on NCBI (Accession: PRJNA991498). Note that Microbacterium liquefaciens was previously referred to as Microbacterium saperdae but a more recent classification has led us to refer to it differently.
All experiments were performed in 30 ml batch cultures in glass tubes containing 0.5% (vol/vol) Castrol Hysol XF MWF (acquired in 2016) diluted in water with added salts and metal traces (see Piccardi et al., 2019 for detailed recipe). Cultures were incubated at 28 C, shaken at 200 rpm.
Evolution experiment
All the experiments (initially six treatments: four monocultures, one 3-species co-culture, one 4-species co-culture) were conducted simultaneously in five microcosm replicates to give 30 experimental cultures in addition to three sterile controls (Table 1).
Table 1.
Evolved species combinations and microcosm numbers.
| Combination | Species | Number of microcosms Transfer 0 Transfer 44 |
|---|---|---|
| CAt | A. tumefaciens | 5 2 |
| CCt | C. testosteroni | 5 5 |
| CAtCtMl |
A. tumefaciens + C. testosteroni + |
5 5 |
| M. liquefaciens | ||
| CAtCtMlOa |
A. tumefaciens + C. testosteroni + |
5 5 |
|
M. liquefaciens + O. anthropi |
||
| CMl | M. liquefaciens | 5 0 |
| COa | O. anthropi | 5 0 |
All tubes were incubated at C, shaken at 200 rpm for a total of 7 days. Each week, for a total of 44 weeks, 29.7 ml of fresh MWF medium was prepared and 300 l of the week-old culture transferred into it. Before each transfer, population sizes (CFU/ml) were quantified using serial dilution followed by selective plating. CODs (pollution load) were quantified using Macherey Nagel 15 g/L COD tube tests (see Piccardi et al., 2019 for detailed recipe). A sterile tube containing MWF but no bacteria was always used as a control for the COD measurement. Every week, 1 ml of the bacterial cultures was harvested for each treatment, spun down at 10,000 rcf for 5 min, resuspended in glycerol 25% (diluted in PBS) and stocked at C for future analyses (e.g. DNA extraction). All five replicate populations of M. liquifaciens, O. anthropi and three replicate populations of A. tumefaciens in monoculture went extinct, and these microcosms were discarded after 10 weeks.
At the end of the experiments (after transfer 44), we collected 10 individual isolates of each species from each population for further analysis by plating populations on selective media and randomly picking 10 colonies. These colonies were then grown overnight in TSB at C, shaken at 200 rpm, spun down at 10,000 rcf for 5 min, resuspended in glycerol 25% (diluted in PBS) and stocked at C.
Quantifying interspecies interactions
Interactions were quantified by comparing mono- and pair-wise co-cultures following the same protocol as by Piccardi et al., (2019). For the ancestral species, single isolates were picked and grown alone overnight in TSB, diluted to an OD of 0.05 in TSB and grown for 3 hr to exponential phase. To prepare the monocultures, we spun down 200 l of cells at 10,000 rcf for 5 min, and resuspended them in MWF. For co-cultures, we combined 200 l of each species (total of 400 l) before spinning down and re-suspending cells.
To measure interactions between evolved species, we used two different approaches: combining 10 isolates of each species or transferring the whole population. When combining isolates for each species, we plated the frozen stocks of each isolate coming from transfer 44 (described above), randomly picked one colony from each frozen stock and grew it alone overnight in TSB, diluted it to an OD of 0.05 in TSB and grew it again for 3 hr in TSB at 28 C, shaken at 200 rpm, to exponential phase. We then mixed 20 l of each of the 10 monocultures for a given species (total of 200 l, still in TSB). For each species pair, we compared mono- and co-cultures. To prepare the monocultures, we spun down the mixture of 200 l of cells at 10,000 rcf for 5 min, and resuspended them in MWF. For co-cultures, we combined 200 l of each species (total of 400 l) before spinning down and re-suspending cells.
When transferring the whole population, freezer stocks of transfer 43 were thawed entirely, washed and added to 30 ml of fresh MWF. After 1 week, we transferred these (diluted 100-fold in fresh MWF) to recreate microcosms from transfer 44. After an additional week, we inoculated 300 l in fresh MWF either alone or together with the other species (total of 600 l). We only did this for the mono- and co-cultures of CAt and CCt (evolved alone), as we could not separate species from one another in CAtCtMl and CAtCtMlOa.
For all mono- and co-cultures, we collected CFU/ml data over 8 or 12 days (days 0, 1, 2, 3, 6, 8, and sometimes 12), calculated the area under each growth curve (AUC) and took the log of the ratio of the co-culture and monoculture AUCs to estimate the interaction strength. We only consider interactions where the AUCs were significantly different according to generalized linear models with microcosm origin as an explanatory factor and biological replicate as a random factor.
The data-points in Figure 3B were compiled from several experiments involving mono- and co-cultures, such that some microcosms were over-represented. For the first column, we co-cultured At and Ct from microcosm , but every other microcosm once (total ). For the second column, we monocultured CAt.M1 3, CAt.M2 4 (n = 7), CCt.M1-2 3 each and CCt.M3-5 2 each (). We co-cultured all possible combinations of microcosms that had evolved alone (CAt.M1 + CCt.M1, CAt.M1 + CCt.M2, etc.) with CAt.M2 + CCt.M1 and CAt.M2 + CCt.M2 carried out twice (). For the 3rd column, we only tested four combinations: CAt.M1 + CCt.M1 (twice), CAt.M1 + CCt.M2, CAt.M2 + CCt.M1 and CAt.M2 + CCt.M2 (). Matching monocultures were always done in parallel. All data are available at 10.5281/zenodo.10694070.
Bioinformatic analysis
Ancestral lineage sequencing and annotation.
DNA coming from each ancestral species was sequenced using a combination of Illumina (MiSeq) and PacBio (RSII). PacBio raw data for each genome sequencing were assembled using canu v. 2.2 (Koren et al., 2017) and polished with racon v. 1.5.0 (Vaser et al., 2017). The assembly was further corrected using the Illumina data with polypolish v. 0.5.0 (Wick and Holt, 2022). The assemblies were then annotated using bakta v. 1.2.4 (Schwengers et al., 2021).
DNA extraction and sequencing.
To extract DNA from the frozen populations for Illumina sequencing, we defrosted the populations from the T-1 transfer (e.g., to sequence transfer 22, we defrosted transfer 21), washed and resuspended the cells in 1 ml of PBS and inoculated 300 l into 29.7 ml of fresh MWF medium. After 1 week, we collected 15 ml of each sample, split into 1.5 ml Eppendorf tubes and spun down at 10,000 rpm for 10 min. The bi-phasic supernatant was carefully discarded. Pellets coming from the same sample were resuspended in PBS and pooled together into one single 1.5 ml Eppendorf tube. Cells were precipitated and resuspended in PBS twice, to remove any remaining MWF. A negative control was included in the process and followed the same procedure as the samples. To extract DNA from isolates for PacBio sequencing, we grew the previously frozen isolates overnight in TSB at 28 C, shaken at 200 rpm, and spun them down at 10,000 rcf for 5 min.
The resulting pelleted cells were incubated in 150 l of lysozyme solution for 30 min at 37 C. After this incubation period, 5 l of RNAse solution (5 mg/ml) was added. The RNAse treatment was performed for 30 more minutes at the same temperature. The lysozyme action creates pores in the cell wall of the cells, allowing the RNAse to degrade any possible remaining RNA in the sample. After this second incubation period, 600 l of lysis buffer was added to the sample. The lysis buffer solution contains 9.34 ml of TE buffer (pH 8), 600 l of SDS 10%, 60 l of proteinase K and 2 l of -mercaptoethanol. Cell lysis was performed for 1 hr at 56 C. Once the cell suspension became transparent, 700 l (1, vol/vol) of phenol-chlorophorm-isoamylalcohol (PCI, 25:24:1) was added to the tube. Samples were mixed by inversion for 1 min and left to rest on ice to allow phase separation. After the phases were clearly visible, the sample was centrifuged at 13,000 rpm for 15 min at 4 C. The resulting clear supernatant was transferred to a new tube (approximately 600 l of volume). PCI cleaning was performed one more time to purify the DNA, resulting in around 500 l of clear liquid containing the suspended DNA. After the DNA cleaning, 50 l of sodium acetate (5 M) and 500 l of Isopropnol were added to the sample, allowing the DNA to precipitate. Insoluble DNA was incubated at C for 2 hr and centrifuged down at 13,000 rpm for 15 min. The alcoholic supernatant was discarded. The precipitated DNA was washed with 1 ml of ethanol 70% (vol/vol), re-centrifuged at 13,000 rpm for 15 more minutes, and the supernatant removed. The air dried pellet was then redissolved in 50 l nuclease-free water, and the concentration and purity were analyzed using Qubit and Nanodrop.
The obtained DNA was sequenced using the Illumina platform with two different platforms at the Oxford genomics facilities: Samples from transfer 22 were sequenced using HiSeq4000, while transfers 11,33,44 were sequenced using NovaSeq. The reason behind the different platform usage was the discontinuation of the former at the selected facility. Coverage was overall lower for transfer 22 (Supplementary Figure S16). PacBio sequencing was performed on individual isolates of each species from transfer 44 at the Lausanne Genomic Technologies Facility using a Sequel II system (SMRT cell 8M).
RNA extraction and sequencing.
We grew the previously frozen isolates from transfer 44 (see section Evolution Experiment) overnight in TSB, washed them in PBS and then inoculated 300 l into 29.7 ml of fresh MWF medium. After 7 days of growth, the cells were pelleted and the RNA extracted using the RNeasy PowerSoil Total RNA Kit. The extraction yielded a minimum of 30 ng/l in 10 l. The sequencing library was prepared including ribosomal RNA depletion using the Illumina ZeroPlus library perparation kit and sequenced on a NovaSeq 600 sequencer.
Sequence data processing and analysis.
For each Illumina sequencing data-set, an initial quality control was performed using FastQC, to evaluate the overall per-position quality, the k-mer enrichment (which could indicate adapter contamination), and the GC-content (which could indicate origin admixture) (Andrews, 2010). Adapters and low quality sequences were removed using trimmomatic v. 0.36, using the parameters PE, leading 3, trailing 3, slidingwindow 4:15, minlen 60 (Bolger et al., 2014). The resulting cleaned reads were mapped against the ancestral genome references using minimap2 v. 2.22 (Li, 2018). For sequencing data derived from microcosms with multiple species, the reads were aligned against all merged ancestral reference genomes with no secondary mapping in order to avoid cross-mapping. The mapping was filtered to remove distant alignments and low quality alignments using samtools view with the parameters and (Danecek et al., 2021). Based on the filtered alignment files, we identified variants with freebayes version 1.3.6 with the parameters –min-alternate-count 3 –min-alternate-fraction 0.05 –pooled-continuous –haplotype-length 0 –standard-filters (Garrison and Marth, 2012). Variants outputted by freebayes were then filtered by a minimum population frequency of 10% and a minimum Phred quality of 20. A variant was considered fixed if it exceeded a frequency of 95%.
PacBio whole-genome sequencing data were assembled using canu version 2.2 (Koren et al., 2017). The resulting assemblies were polished with racon 1.5.0 (Vaser et al., 2017), and annotated with bakta v. 1.2.4 (Schwengers et al., 2021). To investigate potential intra-species gene transfers, we split theassemblies into 150-mers and taxanomically classified the 150-mers using using krakken 2.1.2 (Vaser et al., 2017; Wood and Salzberg, 2014). RNA sequencing data was analyzed using the RASflow workflow with default parameters wrapping hisat2 2.1.0 as an aligner, htseq-count 0.11.2 for feature counting and edgeR 3.26.0 for differential expression analysis (Zhang and Jonassen, 2020).
Data and code availability
All scripts and data sets are available at the following DOIs: 10.5281/zenodo.10694070 (data) and 10.5281/zenodo.10694150 (code). All sequencing data can be found on NCBI (Accession: PRJNA991498).
Author contributions
P.P. and S.M. conceived the study, P.P., S.M., and M.G.G. designed the experiment, P.P. performed the evolution experiment and all follow-up experiments with the help of S.E.A.T., M.G.G. and P.P. extracted the DNA and sent it for sequencing, E.U. performed bioinformatic analysis after some initial analysis by M.G.G., R.D.M. performed RNA extractions, P.P., E.U., and S.M. wrote thepaper.
Funding
P.P. and E.U. were funded by the University of Lausanne, M.G.G., R.D.M., and S.M. were funded by European Research Council Starting Grant 715097, and S.E.A.T. and S.M. by the NCCR Microbiomes Grant 51NF40_180575 from the Swiss National Science Foundation.
Conflict of interest: The authors declare no conflicts of interest.
Supplementary Material
Acknowledgments
We thank Julien Luneau, Afra Salazar, Oliver Meacock, Massimo Amicone, and Margaret Vogel and two anonymous reviewers for useful and constructive feedback on the manuscript. We thank Christopher van der Gast and Ian Thompson for the four species used. We thank Sabrina Rivera for help with the experiments. We thank the staff of Lausanne Genomic Technologies Facility for DNA and RNA sequencing, and advice on library preparation.
Contributor Information
Philippe Piccardi, Department of Fundamental Microbiology, University of Lausanne, Lausanne 1015, Switzerland.
Eric Ulrich, Department of Fundamental Microbiology, University of Lausanne, Lausanne 1015, Switzerland.
Marc Garcia-Garcerà, Department of Fundamental Microbiology, University of Lausanne, Lausanne 1015, Switzerland; Swiss Institute for Bioinformatics, Lausanne 1015, Switzerland.
Rita Di Martino, Department of Fundamental Microbiology, University of Lausanne, Lausanne 1015, Switzerland; Swiss Institute for Bioinformatics, Lausanne 1015, Switzerland.
Samuele E A Testa, Department of Fundamental Microbiology, University of Lausanne, Lausanne 1015, Switzerland.
Sara Mitri, Department of Fundamental Microbiology, University of Lausanne, Lausanne 1015, Switzerland; Swiss Institute for Bioinformatics, Lausanne 1015, Switzerland.
References
- Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. [Google Scholar]
- Atashgahi, S., Sánchez-Andrea, I., Heipieper, H. J., van der Meer, J. R., Stams, A. J. M., & Smidt, H. (2018). Prospects for harnessing biocide resistance for bioremediation and detoxification. Science, 360(6390):743–746. [DOI] [PubMed] [Google Scholar]
- Barraclough, T. G. (2019). Species matter for predicting the functioning of evolving microbial communities – An eco-evolutionary model. PLoS One, 14(8):e0218692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertness, M. D. & Callaway, R. (1994). Positive interactions in communities. Trends in Ecology & Evolution, 9(5):191–193. [DOI] [PubMed] [Google Scholar]
- Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., & Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nature Reviews Microbiology, 13(1):42–51. [DOI] [PubMed] [Google Scholar]
- Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottery, M. J., Pitchford, J. W., & Friman, V. P. (2021). Ecology and evolution of antimicrobial resistance in bacterial communities. ISME Journal, 15(4):939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottery, M. J., Wood, A. J., & Brockhurst, M. A. (2016). Selective conditions for a multidrug resistance plasmid depend on the sociality of antibiotic resistance. Antimicrobial Agents and Chemotherapy, 60(4):2524–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castledine, M., Padfield, D., & Buckling, A. (2020). Experimental (co)evolution in a multi-species microbial community results in local maladaptation. Ecology Letters, 23(11):1673–1681. [DOI] [PubMed] [Google Scholar]
- Celiker, H. & Gore, J. (2014). Clustering in community structure across replicate ecosystems following a long-term bacterial evolution experiment. Nature Communications, 5:1–8. [DOI] [PubMed] [Google Scholar]
- Chacón, J. M., Hammarlund, S. P., Martinson, J. N., Smith, L. B., & Harcombe, W. R. (2021). The ecology and evolution of model microbial mutualisms. Annual Review of Ecology, Evolution, and Systematics, 52:363–384. [Google Scholar]
- Collins, S. (2011). Competition limits adaptation and productivity in a photosynthetic alga at elevated CO2. Proceedings of the Royal Society B: Biological Sciences, 278(1703):247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero, O. X., Ventouras, L.-A., DeLong, E. F., & Polz, M. F. (2012). Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proceedings of the National Academy of Sciences of the United States of America, 109(49):20059–20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., Whitwham, A., Keane, T., McCarthy, S. A., Davies, R. M., & Li, H. (2021). Twelve years of SAMtools and BCFtools. GigaScience, 10(2):giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roy, K., Marzorati, M., Van den Abbeele, P., Van de Wiele, T., & Boon, N. (2014). Synthetic microbial ecosystems: an exciting tool to understand and apply microbial communities. Environmental Microbiology, 16(6):1472–1481. [DOI] [PubMed] [Google Scholar]
- De Wit, G., Svet, L., Lories, B., & Steenackers, H. P. (2022). Microbial interspecies interactions and their impact on the emergence and spread of antimicrobial resistance. Annual Review of Microbiology, 76(1):179–192. [DOI] [PubMed] [Google Scholar]
- Fiegna, F., Moreno-Letelier, A., Bell, T., & Barraclough, T. G. (2015). Evolution of species interactions determines microbial community productivity in new environments. The ISME Journal, 9(5):1235–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, K. R. & Bell, T. (2012). Competition, not cooperation, dominates interactions among culturable microbial species. Current Biology, 22(19):1845–50. [DOI] [PubMed] [Google Scholar]
- Fritts, R. K., Bird, J. T., Behringer, M. G., Lipzen, A., Martin, J., Lynch, M., & McKinlay, J. B. (2020). Enhanced nutrient uptake is sufficient to drive emergent cross-feeding between bacteria in a synthetic community. The ISME Journal, 14(11):2816–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison, E. & Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio]. [Google Scholar]
- Gómez, P. & Buckling, A. (2013). Real-time microbial adaptive diversification in soil. Ecology Letters, 16(5):650–655. [DOI] [PubMed] [Google Scholar]
- Gorter, F. A., Manhart, M., & Ackermann, M. (2020). Understanding the evolution of interspecies interactions in microbial communities. Philosophical Transactions of the Royal Society B: Biological Sciences, 375(1798):20190256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, P. R. & Grant, B. R. (2006). Evolution of character displacement in Darwin’s finches. Science, 313(5784):224–226. [DOI] [PubMed] [Google Scholar]
- Gravel, D., Bell, T., Barbera, C., Bouvier, T., Pommier, T., Venail, P., & Mouquet, N. (2011). Experimental niche evolution alters the strength of the diversity–productivity relationship. Nature, 469(7328):89–92. [DOI] [PubMed] [Google Scholar]
- Hall, J. P. J., Harrison, E., & Brockhurst, M. A. (2018). Competitive species interactions constrain abiotic adaptation in a bacterial soil community. Evolution Letters, 2(6):580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcombe, W. (2010). Novel cooperation experimentally evolved between species. Evolution, 64(7):2166–72. [DOI] [PubMed] [Google Scholar]
- Harcombe, W. R., Chacón, J. M., Adamowicz, E. M., Chubiz, L. M., & Marx, C. J. (2018). Evolution of bidirectional costly mutualism from byproduct consumption. Proceedings of the National Academy of Sciences of the United States of America, 115(47):12000–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekstra, D. R. & Leibler, S. (2012). Contingency and statistical laws in replicate microbial closed Ecosystems. Cell, 149(5):1164–1173. [DOI] [PubMed] [Google Scholar]
- Henriksen, N. N. S. E., Hansen, M. F., Kiesewalter, H. T., Russel, J., Nesme, J., Foster, K. R., Svensson, B., Øregaard, G., Herschend, J., & Burmølle, M. (2022). Biofilm cultivation facilitates coexistence and adaptive evolution in an industrial bacterial community. NPJ Biofilms and Microbiomes, 8(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillesland, K. L., Lim, S., Flowers, J. J., Turkarslan, S., Pinel, N., Zane, G. M., Elliott, N., Qin, Y., Wu, L., Baliga, N. S., Zhou, J., Wall, J. D., & Stahl, D. A. (2014). Erosion of functional independence early in the evolution of a microbial mutualism. Proceedings of the National Academy of Sciences of the United States of America, 111(41):14822–14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillesland, K. L. & Stahl, D. A. (2010). Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proceedings of the National Academy of Sciences of the United States of America, 107(5):2124–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey Morris, J., Papoulis, S. E., & Lenski, R. E. (2014). Coexistence of evolving bacteria stabilized by a shared Black Queen function. Evolution, 68(10):2960–2971. [DOI] [PubMed] [Google Scholar]
- Jousset, A., Eisenhauer, N., Merker, M., Mouquet, N., & Scheu, S. (2016). High functional diversity stimulates diversification in experimental microbial communities. Science Advances, 2(6):e1600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., & Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Research, 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, D., Fiegna, F., Behrends, V., Bundy, J. G., Phillimore, A. B., Bell, T., & Barraclough, T. G. (2012). Species interactions alter evolutionary responses to a novel environment. PLoS Biology, 10(5):e1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics, 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liow, L. H., Van Valen, L., & Stenseth, N. C. (2011). Red Queen: from populations to taxa and communities. Trends in Ecology and Evolution, 26(7):349–358. [DOI] [PubMed] [Google Scholar]
- Marchal, M., Goldschmidt, F., Derksen-Müller, S. N., Panke, S., Ackermann, M., & Johnson, D. R. (2017). A passive mutualistic interaction promotes the evolution of spatial structure within microbial populations. BMC Evolutionary Biology, 17(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas, A., Jamshidi, S., Lagadeuc, Y., Eveillard, D., & Vandenkoornhuyse, P. (2016). Beyond the Black Queen Hypothesis. ISME Journal, 10(9):2085–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J. J., Lenski, R. E., & Zinser, E. R. (2012). The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio, 3(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien, S. & Buckling, A. (2015). The sociality of bioremediation. EMBO Reports, 16(10):1241–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien, S., Hodgson, D. J., & Buckling, A. (2014). Social evolution of toxic metal bioremediation in Pseudomonas aeruginosa. Proceedings of the Royal Society B: Biological Sciences, 281(1787):20140858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore, A. I., Barabás, G., Bimler, M. D., Mayfield, M. M., & Miller, T. E. (2021). The evolution of niche overlap and competitive differences. Nature Ecology & Evolution, 5(3):330–337. [DOI] [PubMed] [Google Scholar]
- Piccardi, P., Alberti, G., Alexander, J. M., & Mitri, S. (2022). Microbial invasion of a toxic medium is facilitated by a resident community but inhibited as the community co-evolves. The ISME Journal 202216:12,16(12):2644–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccardi, P., Vessman, B., & Mitri, S. (2019). Toxicity drives facilitation between 4 bacterial species. Proceedings of the National Academy of Sciences of the United States of America, 116(32):15979–15984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preussger, D., Giri, S., Muhsal, L. K., Oña, L., & Kost, C. (2020). Reciprocal fitness feedbacks promote the evolution of mutualistic Cooperation. Current Biology, 30(18):3580–3590.e7. [DOI] [PubMed] [Google Scholar]
- Ridenhour, B. J. (2005). Identification of selective sources: partitioning selection based on interactions. The American Naturalist, 166(1):12–25. [DOI] [PubMed] [Google Scholar]
- Rivett, D. W., Scheuerl, T., Culbert, C. T., Mombrikotb, S. B., Johnstone, E., Barraclough, T. G., & Bell, T. (2016). Resource-dependent attenuation of species interactions during bacterial succession. The ISME Journal, 10(9):2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runquist, R. D., Gorton, A. J., Yoder, J. B., Deacon, N. J., Grossman, J. J., Kothari, S., Lyons, M. P., Sheth, S. N., Tiffin, P., & Moeller, D. A. (2020). Context dependence of local adaptation to abiotic and biotic environments: A quantitative and qualitative synthesis. American Naturalist, 195(3):412–431. [DOI] [PubMed] [Google Scholar]
- Sachs, J. & Simms, E. (2006). Pathways to mutualism breakdown. Trends in Ecology & Evolution, 21(10):585–592. [DOI] [PubMed] [Google Scholar]
- Sachs, J. L. & Hollowell, A. C. (2012). The origins of cooperative bacterial communities. mBio, 3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen, O., Lascoux, M., & Merilä, J. (2013). Ecological genomics of local adaptation. Nature Reviews Genetics, 14(11):807–820. [DOI] [PubMed] [Google Scholar]
- Schluter, D., Price, T. D., & Grant, P. R. (1985). Ecological character displacement in Darwin’s finches. Science, 227(4690):1056–1059. [DOI] [PubMed] [Google Scholar]
- Schwengers, O., Jelonek, L., Dieckmann, M. A., Beyvers, S., Blom, J., & Goesmann, A. (2021). Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microbial Genomics, 7(11):000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segar, S. T., Fayle, T. M., Srivastava, D. S., Lewinsohn, T. M., Lewis, O. T., Novotny, V., Kitching, R. L., & Maunsell, S. C. (2020). The role of evolution in shaping ecological Networks. Trends in Ecology & Evolution, 35(5):454–466. [DOI] [PubMed] [Google Scholar]
- Summers, Z. M., Fogarty, H. E., Leang, C., Franks, A. E., Malvankar, N. S., & Lovley, D. R. (2010). Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science, 330(6009):1413–1415. [DOI] [PubMed] [Google Scholar]
- Turkarslan, S., Stopnisek, N., Thompson, A. W., Arens, C. E., Valenzuela, J. J., Wilson, J., Hunt, K. A., Hardwicke, J., de Lomana, A. L. G., Lim, S., Seah, Y. M., Fu, Y., Wu, L., Zhou, J., Hillesland, K. L., Stahl, D. A., & Baliga, N. S. (2021). Synergistic epistasis enhances the co-operativity of mutualistic interspecies interactions. The ISME Journal, 15(8):2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Gast, C. J. & Thompson, I. P. (2005). Effects of pH amendment on metal working fluid wastewater biological treatment using a defined bacterial consortium. Biotechnology and Bioengineering, 89(3):357–66. [DOI] [PubMed] [Google Scholar]
- van der Gast, C. J. & Thompson, I. P. (2014). US patent 8,703,475 B2. [Google Scholar]
- Vaser, R., Sović, I., Nagarajan, N., & Šikić, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Research, 27(5):737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, R. R. & Holt, K. E. (2022). Polypolish: Short-read polishing of long-read bacterial genome assemblies. PLOS Computational Biology, 18(1):e1009802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widder, S., Allen, R. J., Pfeiffer, T., Curtis, T. P., Wiuf, C., Sloan, W. T., Cordero, O. X., Brown, S. P., Momeni, B., Shou, W., Kettle, H., Flint, H. J., Haas, A. F., Laroche, B., Kreft, J.-U. U., Rainey, P. B., Freilich, S., Schuster, S., Milferstedt, K., …Free, A. (2016). Challenges in microbial ecology: building predictive understanding of community function and dynamics. The ISME Journal, 10(11):2557–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, D. E. & Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology, 15(3):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. & Jonassen, I. (2020). RASflow: an RNA-Seq analysis workflow with Snakemake. BMC Bioinformatics, 21(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. & Reed, J. L. (2014). Adaptive evolution of synthetic cooperating communities improves growth performance. PLoS One, 9(10):e108297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts and data sets are available at the following DOIs: 10.5281/zenodo.10694070 (data) and 10.5281/zenodo.10694150 (code). All sequencing data can be found on NCBI (Accession: PRJNA991498).