Abstract
The hepadnavirus reverse transcriptase binds cotranslationally to the viral pregenomic RNA. This ribonucleoprotein complex is then encapsidated into nascent viral core particles, where the reverse transcriptase copies the viral RNA into DNA. Here we report that 75% of the duck hepatitis B virus reverse transcriptase present in transfected LMH cells does not follow this well-known pathway but rather exists in the cell separate from the core protein or nucleocapsids. The nonencapsidated reverse transcriptase is also abundant in infected duck liver. The nonencapsidated reverse transcriptase exists as a complex set of isoforms that are most likely produced by posttranslational modification. Interestingly, only the smallest of these isoforms is encapsidated into viral core particles. The nonencapsidated reverse transcriptase is bound to a large cellular cytoplasmic structure(s) in a detergent-sensitive complex. The cellular distribution of the reverse transcriptase only partially overlaps that of the core protein, and this distribution is unaffected by blocking encapsidation. These observations raise the possibilities that the metabolic fate of the reverse transcriptase may be posttranscriptionally regulated and that the reverse transcriptase may have roles in the viral replication cycle beyond its well-known function in copying the viral genome.
Hepatitis B virus (HBV) is the type member of the hepadnaviruses, a group of small DNA-containing viruses that replicate by reverse transcription and are highly hepatotropic (8). These viruses have a lipid envelope surrounding an icosahedral protein core particle. Within the core particle, the partially double-stranded viral DNA genome is covalently linked to the viral reverse transcriptase. Other hepadnaviruses infect woolly monkeys, woodchucks, ground squirrels, ducks, and herons (18, 30). Although significant differences exist between various hepadnaviruses, they all share a high degree of hepatotropism, follow the same replication cycle, and are nearly identical in genetic organization.
The hepadnavirus replication cycle starts with binding of the virus to the hepatocyte (8). Fusion of the viral envelope with a cellular membrane liberates the subviral core particle into the cell, where the core particle releases the partially double-stranded viral DNA. In the nucleus the DNA is repaired to a covalently closed circular episome, which is the template for transcription via host RNA polymerase II. The viral mRNAs are transported to the cytoplasm and translated to produce the viral proteins. One of the largest RNAs (the pregenomic RNA [pgRNA]) is packaged into nascent viral core particles as a nucleoprotein complex with the viral polymerase. The pgRNA carries the genetic information of the virus and is also the mRNA for the core and polymerase proteins. Reverse transcription is primed by the reverse transcriptase itself, and hence the viral DNA is covalently attached to the viral polymerase. DNA synthesis is catalyzed by the reverse transcriptase within immature core particles in the cytoplasm. The mature core particles containing DNA either are transported back into the nucleus to maintain the pool of transcriptional templates or bud into the endoplasmic reticulum (ER), where they pick up the envelope and viral surface glycoproteins. The virions are then secreted from the cell noncytolytically.
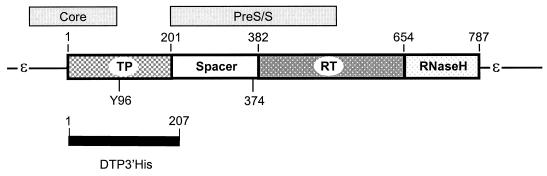
The hepadnavirus reverse transcriptase (polymerase) contains four domains (Fig. 1) (5, 27). The terminal protein and spacer domains are unique to the hepadnavirus polymerases. The terminal protein domain contains the tyrosine residue that primes DNA synthesis and covalently links the polymerase to the viral DNA (41, 45). The spacer domain has no known function other than to link the terminal protein to the rest of the molecule, and the reverse transcriptase and RNase H domains contain the two known enzymatic active sites. These latter two domains are related to the corresponding domains of the polymerases from retroviruses and other retroelements (19, 23, 25).
FIG. 1.
DHBV polymerase structure in the context of the pgRNA. The domain structure of the polymerase is shown in its relative position on the pgRNA (thin line). The positions of the core and pre-S/S surface glycoprotein open reading frames are indicated above, and sequences included in DTP3′His (to which antibodies were raised) are indicated below. TP, terminal protein domain; RT, reverse transcriptase domain; RNaseH, RNase H domain; Y96, tyrosine 96 to which DNA is covalently bound; 374, the amino acid position at which the KOF mutation truncates the polymerase. The approximate amino acid boundaries of the domains are indicated above the polymerase.
The polymerase has two roles in the formation of virions. The first role is structural, because the polymerase must bind to a stem-loop (ɛ) at the 5′ end of the pgRNA to form the ribonucleoprotein complex that is encapsidated into the nascent core particle (1, 12, 15). If this complex does not form, neither the polymerase nor pgRNA is encapsidated. The second role of the polymerase is enzymatic, as the polymerase synthesizes the viral DNA. ɛ is essential for the enzymatic role of the polymerase for two reasons: binding of ɛ to the polymerase promotes the maturation of the polymerase to an enzymatically active form (36, 37), and ɛ contains the origin of reverse transcription (24, 35, 38, 39).
The polymerase has been assumed to be present in low levels in cells and to be located almost exclusively within subviral core particles (13, 14, 20) for four reasons. First, hepadnavirus reverse transcriptase activity cannot be detected outside core particles. Second, the polymerase open reading frame is downstream from the core protein open reading frame in the bicistronic pgRNA, and synthesis of downstream open reading frames in eukaryotic polycistronic mRNAs is typically inefficient. Third, the polymerase is believed to bind to ɛ on the pgRNA cotranslationally (12), and no stable cellular intermediates in core particle formation have been found in cells above the level of core protein dimers (31). Finally, DNA synthesis takes place inside the nascent viral core particles, and hence there is no apparent need for the polymerase outside of viral capsids once it has bound to the pgRNA.
Here we report detection of a large pool of nonencapsidated duck hepatitis B virus (DHBV) polymerase in cultured cells and in infected liver tissue. The nonencapsidated polymerase is found in the cytoplasm in a large detergent-sensitive complex. Its distribution within cells only partially overlaps that of the core protein and is not affected by blocking encapsidation.
MATERIALS AND METHODS
Viruses, cells, and plasmids.
DHBV strain 3 (32) was used in all experiments. LMH cells are a chicken hepatoma cell line (6). D1.5G is a plasmid that contains 1.5 copies of the viral genome cloned into pBS(−) (Stratagene). When transfected into LMH cells, D1.5G directs production of infectious DHBV; D1.5G and its derivatives were used whenever transfections employing the complete DHBV genome were performed. D1.5G(KOF) contains a deletion (nucleotides [nt] 1291 to 4) that causes a frameshift that truncates the polymerase after amino acid 374. D1.5G(Y96F) contains a point mutation in the polymerase open reading frame altering Y96 to F; this mutation prevents covalent attachment of DNA to the polymerase. D1.5G(ɛ-dlBulge), D1.5G(ɛ-LowerL/R), and DHBV(ɛ-Loop5,6) contain mutations in the 5′ copy of the coding sequences for ɛ and have been described elsewhere (26). D1.5G(DR1/SL) contains mutations that flank the 5′ copy of the ɛ coding sequences (C2555A, T2556A, and insertion of GTCGACACCTTTGGTA between nt 2616 and 2617); these mutations have minimal effects on reverse transcription in LMH cells. pCMV-DPol contains DHBV nt 170 to 3021 cloned downstream of the cytomegalovirus (CMV) promoter in pCDNA3.1-Zeo+ (Invitrogen). pCMV-DPol expresses polymerase containing a P2A mutation resulting from optimization of the polymerase Kozak sequence (17). pDTP3′His contains DHBV nt 170 to 791 cloned into the NdeI-EcoRI sites of pRSET-C (Invitrogen); this plasmid directs production of DHBV polymerase amino acids 1 to 207 followed by LGHHHHHH.
Tissue culture, transfection, and isolation of intracellular DHBV cores.
LMH cells were maintained in Dulbecco's modified Eagle's medium–F-12 medium with 10% fetal bovine serum. Transfections employed FuGENE (Roche) according to the manufacturer's instructions. Cores were isolated from cytoplasmic lysates of transfected LMH cells by sucrose sedimentation as described elsewhere (37).
Immunoprecipitation.
Transfected LMH cells were lysed in radioimmunoprecipitation assay buffer (RIPA; 20 mM Tris [pH 7.2], 1% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 150 mM NaCl) for 10 min on ice, and the lysates were clarified at 14,000 × g for 10 min at 4°C. Antibody was bound to fixed Staphylococcus aureus cells (Sigma) and incubated with the lysate on ice for 3 to 4 h. Immunocomplexes were washed three times with 1 ml of RIPA, and the polymerase was released with Laemmli buffer. In some experiments, cells were lysed in CPLB (10 mM Tris [pH 7.5], 1 mM EDTA, 0.25% NP-40, 50 mM NaCl, 8% sucrose) on ice for 10 min, and the lysate was clarified as above. The CPLB lysates were used directly for immunoprecipitation, or detergent was added to bring the concentrations to 1% sodium deoxycholate, 1% Triton X-100, and 0.1% SDS prior to immunoprecipitation. Cells were fractionated into nuclear and cytoplasmic fractions as described previously (33); nuclei were extracted with RIPA, and the cytoplasmic extracts were brought to high detergent concentrations as above prior to immunoprecipitation of the polymerase. Duck liver extracts were prepared by homogenizing liver in RIPA (5%, wt/vol) with a Dounce homogenizer and incubating the tissue on ice for 10 min prior to clarification as above. All lysates were prepared in the presence of 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml, and 3 μg of leupeptin per ml.
Western analysis.
The polymerase was resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to Immobilon P membranes (Millipore) under standard conditions (22). Polymerase was detected with anti-DTP3′His (see Results) monoclonal antibody (MAb) 11 or MAb 9 and anti-mouse immunoglobulin G (IgG)-alkaline phosphatase conjugate (Promega) followed by incubation with nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (Promega) according to the manufacturer's instructions. Core protein and ERP72 were detected with rabbit polyclonal anticore or anti-ERP72 sera and anti-rabbit IgG-alkaline phosphatase conjugate (Roche).
Cytoplasmic fractionation.
The cytoplasmic contents of transfected LMH cells were fractionated as described in reference 9. Briefly, five 100-mm-diameter plates of LMH cells were transfected with D1.5G or pCMV-DPol; 3 days later, cytoplasmic extracts were prepared by harvesting the cells, disrupting them in a Parr bomb, and clarifying the extract at 800 × g for 10 min. Membranes and particulate matter were collected from the lysate by ultracentrifugation at 150,000 × g. The pellet was suspended in Tris-EDTA, homogenized with a Dounce homogenizer, brought to 50 or 65% sucrose, and layered over a saturated sucrose cushion. A 50 to 15% or a 65 to 30% sucrose gradient was poured over the sample, and the sample was centrifuged at 113,000 × g overnight. The tube was pierced, and fractions were taken from the bottom. Sucrose concentration was determined for each fraction by refractometry, the Golgi apparatus was located by measuring UDP-galactose:N-acetylglucossamine galactosyltransferase activity (9), and the ER was located by detecting ERP72 by Western blot analysis (10). Mature cores were tested by the endogenous polymerase assay using conditions described for recombinant DHBV polymerase (34). The presence of DHBV core protein was detected by Western analysis. Nonencapsidated polymerase was detected by diluting the fractions into RIPA and immunoprecipitating the polymerase.
Immunofluorescence.
LMH cells were grown on glass coverslips and transfected with D1.5G, D1.5G(ɛ-dlBulge), pCMV-DPol, or pBS(−). Two days posttransfection, cells were fixed with 3.7% paraformaldehyde and permeabilized with methanol. Cells were blocked by incubation with phosphate-buffered saline (PBS) containing 1% bovine serum albumin and 2% fetal bovine serum at 37°C for 30 min. Primary and secondary antibodies were diluted in PBS–1% bovine serum albumin–2% fetal bovine serum; each incubation was at 37°C for 1 to 1.5 h. Coverslips were washed four times in PBS. Standard immunofluorescence was performed at a magnification of ×1,000, and images were captured digitally with a SPOT camera attached to an Olympus fluorescence microscope. Confocal microscopy was performed at an magnification of ×600 on a Bio-Rad MRC 1024 confocal system attached to a Nikon Optiphot microscope.
RESULTS
Generation of antibodies and immunological detection of the polymerase.
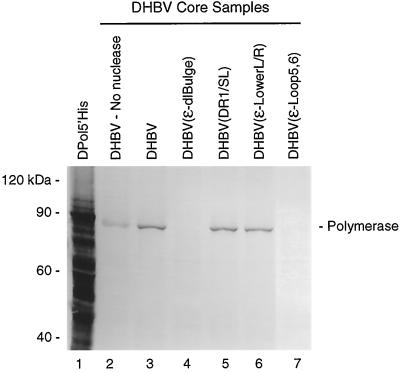
We generated mouse MAbs 9 and 11 and rabbit polyclonal antibodies against the DHBV terminal protein domain expressed in Escherichia coli (DTP3′His; amino acids 1 to 207). MAb 9 and MAb 11 can detect DHBV polymerase encapsidated in viral cores in cytoplasmic extracts from transfected LMH cells by Western analysis (Fig. 2). Detection of the polymerase is enhanced by degradation of the viral DNA prior to electrophoresis (compare lanes 2 and 3), as would be expected for a chimeric DNA-protein molecule. Since polymerase molecules linked to large DNA strands migrate slowly and do not transfer well from gels, the polymerase detected without nuclease digestion (lane 2) is likely to be molecules that have not yet initiated DNA synthesis or those that have synthesized only small amounts of DNA. Mutating ɛ to prevent encapsidation of the pgRNA eliminated detection of the polymerase in core preparations (Fig. 2, lanes 4 and 7), whereas mutations in and near ɛ that have no effect on encapsidation of the pgRNA have no effect on detection of core-associated polymerase (lanes 5 and 6). The level of DNA polymerase activity in these preparations correlated perfectly with the protein level detected by Western analysis (data not shown). This pattern is consistent with the known encapsidation mechanism of the hepadnaviruses in which the polymerase and pgRNA are encapsidated together as a ribonucleoprotein complex (26). This experiment demonstrates that our antibodies are sufficiently sensitive to detect the low levels of polymerase within core particles. It also demonstrates that the antibodies are specific for the polymerase because the large majority of the protein in the extracts is cellular, yet the antibodies recognize only the polymerase.
FIG. 2.
Detection of polymerase within intracellular cores. Cytoplasmic extracts enriched for intracellular core particles were prepared from transfected LMH cells, disrupted in Laemmli buffer, and resolved by SDS-PAGE; the polymerase was detected by Western analysis with MAb 11. Core-associated nucleic acids were destroyed in samples for lanes 3 to 7 by permeabilization at low pH (28) and digestion with micrococcal nuclease. The DHBV genome transfected into cells is indicated at top; DPol5′His is a whole-cell lysate containing recombinant histidine-tagged full-length DHBV polymerase expressed via vaccinia virus.
Detection of nonencapsidated polymerase in transfected cells.
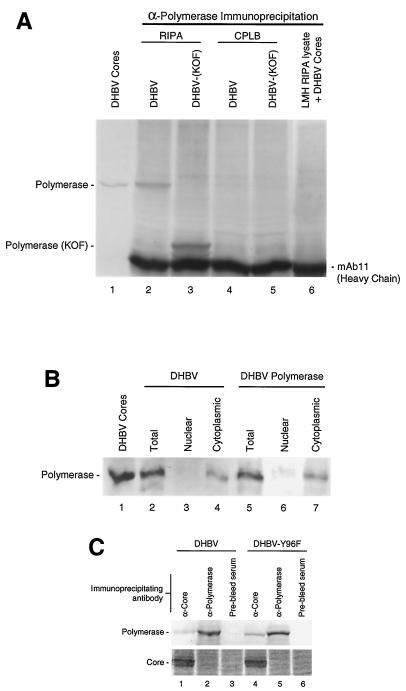
We used our antipolymerase antibodies to determine if the polymerase could be detected in transfected LMH cells, a chicken hepatoma cell line that produces infectious DHBV upon transfection with the viral genome. DHBV polymerase was immunoprecipitated from cells transfected with the complete DHBV genome (Fig. 3A, lane 2). Introduction of a frameshift mutation at amino acid 374 of the polymerase open reading frame reduced the mobility of the polymerase fragment to its predicted position just above that of the MAb 11 IgG heavy chain [DHBV(KOF); lane 3]. The polymerase could readily be detected from whole-cell lysates when the cells were disrupted with a harsh buffer (RIPA) but was not detectable when cells were lysed with CPLB, the mild buffer used to make cytoplasmic extracts from which cytoplasmic viral cores are isolated (compare lanes 2 and 3 to lanes 4 and 5). These results are consistent with three possibilities: (i) the polymerase is bound to a large cellular structure in a detergent-sensitive complex, (ii) the polymerase is in the nucleus, or (iii) the polymerase is from intracellular cores dissociated by RIPA.
FIG. 3.
Transfected LMH cells contain cytoplasmic nonencapsidated polymerase. DHBV Cores, intracellular DHBV cores lacking covalently attached DNA isolated from LMH cells transfected with D1.5G(Y96F). (A) Immunoprecipitation of nonencapsidated polymerase. LMH cells were transfected with D1.5G or D1.5G(KOF) and lysed in RIPA or CPLB; the polymerase was immunoprecipitated and detected by Western analysis with MAb 11. Lane 6 contains an untransfected LMH lysate spiked with purified DHBV core particles prior to immunoprecipitation. (B) The nonencapsidated polymerase is primarily cytoplasmic. Transfected cells were lysed in RIPA (lanes 2 and 5) or were fractionated into nuclear and cytoplasmic fractions and then diluted into RIPA prior to immunoprecipitation of the polymerase and Western analysis with MAb 11. DHBV, D1.5G-transfected cells; DHBV Polymerase, pCMV-DPol-transfected cells. (C) The majority of the polymerase is nonencapsidated. LMH cells transfected with either DHBV or DHBV(Y96F) were lysed, the lysate was divided in three equal portions, and each portion was immunoprecipitated with the indicated polyclonal antibodies. Core and polymerase were detected by Western analysis.
The possibility that the polymerase came from disrupted cores was disproved by adding purified cores to a RIPA lysate of nontransfected LMH cells and attempting to immunoprecipitate the polymerase. Lane 6 in Fig. 3A shows that the polymerase could not be detected by immunoprecipitation in lysates spiked with fivefold more cores than were used as a marker in lane 1, indicating that RIPA was unable to dissociate cores sufficiently to expose the polymerase for immunoprecipitation. High levels of detergent were also needed to immunoprecipitate the polymerase when it was expressed in the absence of the core protein (data not shown), eliminating the possibility that polymerase detected by immunoprecipitation was derived from immature cores that were detergent sensitive. The possibility that the polymerase was in the nucleus was disproved by separating transfected LMH cells into nuclear and cytoplasmic fractions by differential detergent lysis, diluting the nuclear and cytoplasmic fractions into RIPA, and immunoprecipitating the polymerase (Fig. 3B). The polymerase was found primarily in the cytoplasmic fraction, but it was also found at low levels in the nuclear fraction in some experiments.
To determine the relative levels of polymerase associated with core protein versus polymerase not associated with core protein, LMH cells were transfected with the wild-type DHBV genome and a DHBV genome that expressed mutant polymerase (Y96F) unable to be covalently bound to DNA. The cells were lysed with RIPA, and the lysate was clarified. One-third of the lysate was immunoprecipitated with polyclonal anticore serum, one-third was immunoprecipitated with polyclonal anti-polymerase serum, and one-third was immunoprecipitated with preimmune serum. Each immunoprecipitate was assayed for both the core and polymerase proteins by Western analysis. Two observations are apparent in Fig. 3C. First, no core protein was coimmunoprecipitated when antipolymerase antibodies were used for the precipitation, but a significant amount of polymerase was coimmunoprecipitated when anticore antibodies were used. Second, a large majority of the polymerase within cells was not associated with the core protein. Quantitation of these data indicates that 84% of the total polymerase in cells is not associated with the core protein in wild-type DHBV-transfected cells, and 75% was not associated with core protein in DHBV(Y96F)-transfected cells. The encapsidated polymerase is underrepresented in the wild-type DHBV sample because polymerase molecules attached to large DNAs barely enter SDS-polyacrylamide gels and do not transfer well from gels during Western analysis. Therefore, the DHBV(Y96F) sample yields the best estimate of the relative proportions of core-associated and non-core-associated polymerase in cells. Subtraction of the DHBV value from that of DHBV(Y96F) implies that approximately 9% of the total polymerase in LMH cells is covalently bound to long DNA strands and hence is in the latter stages of reverse transcription.
These results indicate that the polymerase we detect by immunoprecipitation of transfected LMH cells is nonencapsidated. These experiments further indicate that the failure to immunoprecipitate the polymerase directly from cytoplasmic extracts without additional detergent (Fig. 3A, lanes 4 and 5) was due to a failure to solubilize the polymerase rather than either a failure to disrupt intracellular viral cores or a nuclear localization of the polymerase. These data therefore indicate that the nonencapsidated polymerase is bound to a large insoluble cytoplasmic structure in a detergent-sensitive complex.
Nonencapsidated polymerase is found in infected duck liver.
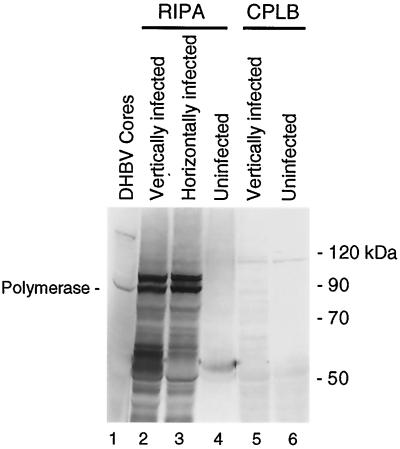
To determine if the nonencapsidated polymerase detected in transfected LMH cells is a normal feature of DHBV replication, we assayed for the polymerase in liver tissue by immunoprecipitation in congenitally DHBV-infected ducks or ducks that had been infected 1 day posthatching with DHBV. Infected or noninfected duck liver was suspended in RIPA (5%, wt/vol) and dissociated with a Dounce homogenizer. The mixture was clarified, and the polymerase was immunoprecipitated with either monoclonal or polyclonal anti-DTP3′His antibodies. The polymerase was easily detected in RIPA extracts from ducks infected either vertically or horizontally (Fig. 4, lanes 2 and 3) but was not found in uninfected liver (lane 3). Because encapsidated polymerase is inaccessible to antibodies and RIPA does not disrupt core particles (Fig. 3A, lane 6), the ability to immunoprecipitate the polymerase indicates that it is nonencapsidated. The polymerase could not be immunoprecipitated from infected duck livers when they were extracted with CPLB (Fig. 4, lane 5). This indicates that the polymerase in infected duck livers is bound to a cellular structure in a detergent-sensitive complex, similar to the situation observed in transfected LMH cells.
FIG. 4.
Nonencapsidated polymerase is found in infected duck liver. Infected or uninfected duck liver was extracted with RIPA or CPLB. The polymerase was immunoprecipitated from the extracts with polyclonal anti-DTP3′His antibodies and detected by Western analysis with MAb 9.
Nonencapsidated polymerase exists in multiple electrophoretic forms.
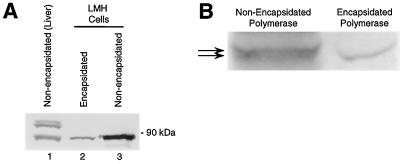
Polymerase from cytoplasmic core particles migrates on SDS-polyacrylamide gels as a single band of about 89 kDa, in agreement with its predicted mobility (Fig. 2). The nonencapsidated polymerase immunoprecipitated from infected liver migrates as two different bands in both vertically and horizontally infected ducks (Fig. 4, lanes 2 and 3). The slower-migrating form of the polymerase is not found within cores (isolated from infected duck liver or transfected LMH cells) or in nonencapsidated polymerase immunoprecipitated from transfected LMH cells (Fig. 5A). It is not known why the upper form of the polymerase is found only in infected duck liver, but the retarded mobility of the upper form is probably not due to attachment of DNA because exhaustive DNase treatment does not alter its mobility (data not shown).
FIG. 5.
The polymerase exists in multiple electrophoretic forms. (A) Nonencapsidated polymerase was immunoprecipitated from liver or LMH cells with polyclonal anti-DTP3′His antibodies, and encapsidated polymerase was immunoprecipitated from LMH cells with anticore antibodies prior to detection of the polymerase and core protein by Western analysis. (B) High-resolution comparison of nonencapsidated polymerase immunoprecipitated from LMH cells and encapsidated polymerase (lacking DNA due to the Y96F mutation). Arrows indicate positions of the barely resolved bands in the nonencapsidated polymerase.
Both the upper and lower electrophoretic forms of the nonencapsidated polymerase resolve into sets of very closely spaced bands, as seen upon careful examination (Fig. 5). These bands are very difficult to resolve clearly (for example, only the largest form in Fig. 5A, lane 1, resolved on that gel). The molecular masses of the upper set of bands found only in liver tissue range from about 95 to 100 kDa, and the molecular masses of the lower set of bands found in liver tissue and LMH cells range from about 88 to 90 kDa. The encapsidated polymerase migrates similarly to the fastest-migrating isoform of the nonencapsidated polymerase immunoprecipitated from either liver or LMH cells. In some experiments we detected a third set of polymerase-specific bands that migrated from 55 to 65 kDa (data not shown). Because these lowest bands were detected at various levels in different immunoprecipitations from the same infected liver sample, we suspect that they may be proteolytic cleavage products of the polymerase generated during extraction.
Nonencapsidated polymerase is found in at least two different forms in the cytoplasm.
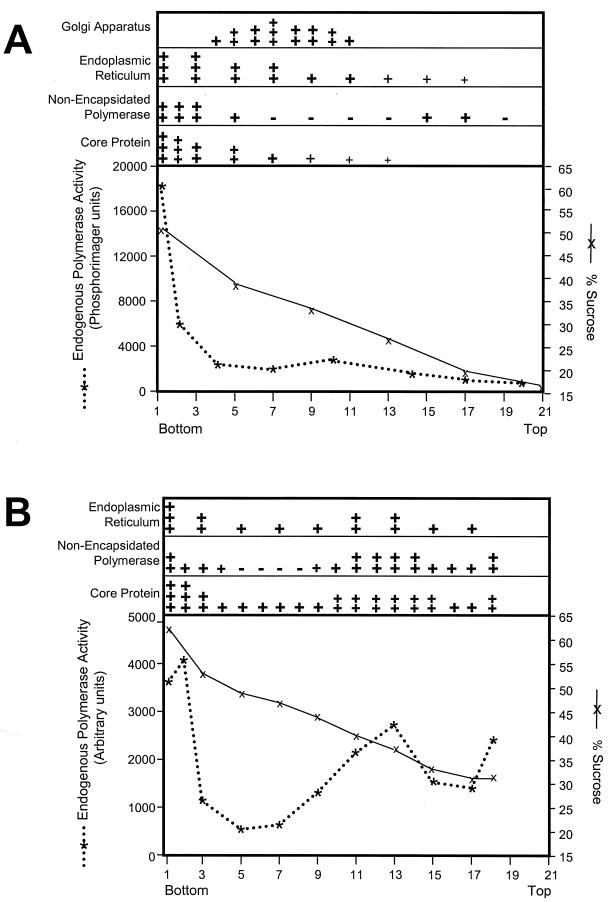
As a first step to identify the structure(s) to which the polymerase is bound, we fractionated the cytoplasm of LMH cells transfected with the DHBV genome (9). Cytoplasmic lysates were prepared by disrupting cells in a Parr bomb in the absence of detergent followed by low-speed centrifugation to remove the nuclei. Membranes and large components of the cytoplasm were collected by ultracentrifugation. The pellet was suspended in 50% sucrose, and a 15 to 50% sucrose gradient was poured over the sample. The cytoplasmic extract was ultracentrifuged overnight, and components with densities less than that of 50% sucrose (∼1.22 g/cm3) floated upward into the gradient. The gradient was split into 21 fractions, and each fraction was assayed for six parameters: (i) sucrose concentration, (ii) nonencapsidated polymerase (by diluting the fractions into RIPA and immunoprecipitating the polymerase), (iii) core protein (by Western analysis), (iv) intact core particles (by the endogenous polymerase assay, an assay that measures DNA synthesis by the polymerase within viral cores), (v) ERP72, an ER-resident chaperone (by Western analysis), and (vi) UDP-galactose:N-acetylglucosamine galactosyltransferase activity, a marker of the Golgi apparatus.
Intact viral core particles were detected at the bottom of the gradient as indicated by endogenous polymerase activity and Western analysis of the core protein (Fig. 6A). This was expected because core particles are large cytoplasmic macromolecular complexes (and hence would be in the sample loaded onto the gradient) that are too dense to float upward during centrifugation. Core protein was also concentrated at the bottom of the gradient, as would be expected for the major protein component of the core particle. The ER (ERP72) was found primarily in fractions 1 to 5, with lower levels extending to the top of the gradient. Galactosyltransferase activity indicative of the Golgi apparatus was found as a broad peak from fractions 4 to 11.
FIG. 6.
The nonencapsidated polymerase is tightly bound to a large cytoplasmic structure. Particulate and membrane-associated material from cytoplasmic lysates of LMH cells transfected with the wild-type DHBV genome were fractionated through sucrose gradients. Sucrose concentration and endogenous polymerase activity are shown in the graphs. The relative levels and position in the gradient of the Golgi apparatus, ER, nonencapsidated polymerase, and core protein are indicated by the number and size of the + symbols. (A) Fractionation of cytoplasmic components through a 50 to 15% sucrose gradient; (B) fractionation through a 65 to 30% gradient.
Nonencapsidated polymerase was detected primarily in fractions 1 to 5 and at a lower level in fractions 15 to 17 (Fig. 6A). Although the bulk of the nonencapsidated polymerase did not resolve from the encapsidated polymerase in this experiment, the encapsidated and nonencapsidated forms of the polymerase are clearly separate because polymerase encapsidated within core particles cannot be immunoprecipitated with antipolymerase antibodies (Fig. 3A, lane 6) and because core particles are released from cells at low detergent concentrations, but high levels of detergent are needed to release the nonencapsidated polymerase (Fig. 3). A portion of the nonencapsidated polymerase was also found at a lighter position in the gradient where there was no detectable core protein (fractions 15 to 17). The nonencapsidated polymerase cosedimented with the ER and was clearly separated from the Golgi apparatus.
To obtain better resolution of the denser fractions, cytoplasmic fractionation of DHBV-transfected cells was repeated using a 65 to 30% sucrose gradient (Fig. 6B). Again, nonencapsidated polymerase was found at the bottom of the gradient. Some of the polymerase floated upward and was detected between fractions 11 and 14. The distribution of core protein, intact core particles (as measured by the endogenous polymerase assay), and ER closely mirrored that of the nonencapsidated polymerase. Cytoplasmic lysates of transfected cells expressing the polymerase under control of the CMV promoter were also fractionated through a 65 to 30% sucrose gradient. Under these conditions, the polymerase was detected as a single broad peak similar to the lighter fraction of the nonencapsidated polymerase from DHBV-transfected cells (data not shown). These data indicate that the nonencapsidated polymerase is found in at least two separate states that are separable by sucrose sedimentation and that some of the nonencapsidated polymerase is in a complex with a density considerably less than that typical for monomeric or aggregated proteins.
The cellular distribution of the polymerase only partially overlaps that of the core protein and is unaffected by blocking encapsidation.
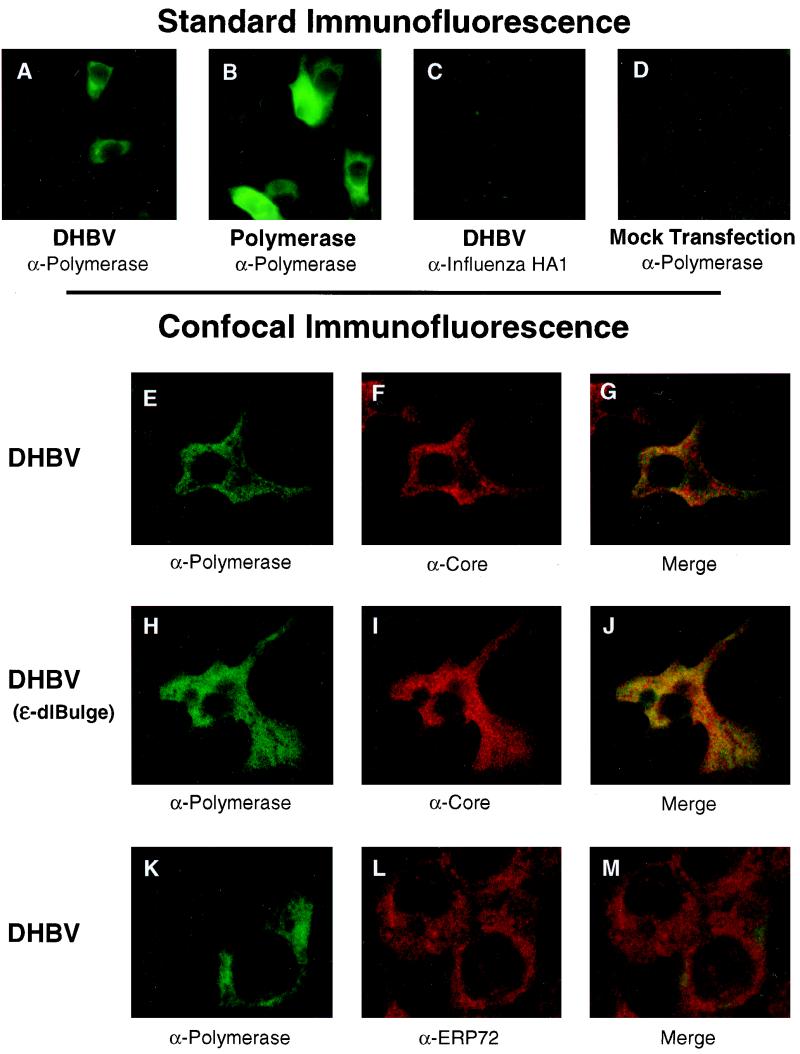
The intracellular distribution of the nonencapsidated polymerase was assessed by immunofluorescence of transfected LMH cells with MAb 9. The polymerase was readily detectable in the cytoplasm of transfected LMH cells in a grainy, uneven pattern (Fig. 7A) that was reminiscent of the distribution of the human HBV polymerase overexpressed in Huh7 cells (46). The majority of the transfected cells were weakly positive, but occasional cells stained extremely brightly for the polymerase. Expression of the polymerase in the absence of the pgRNA or core protein (Fig. 7B) yielded the same staining pattern as was found in cells replicating the virus, but the signal was brighter, presumably due to the strong CMV promoter directing expression of the polymerase. Polymerase was not detected when cells transfected with DHBV were stained with an irrelevant MAb (Fig. 7C) or when nontransfected cells were stained with MAb 9 (Fig. 7D).
FIG. 7.
The cellular distribution of the polymerase only partially overlaps that of the core protein and is unaffected by blocking encapsidation. Transfected LMH cells were fixed and permeabilized, and the polymerase, core, and ERP72 proteins were detected by immunofluorescence microscopy. DHBV, D1.5G-transfected cells; DHBV(ɛ-dlBulge), D1.5G(ɛ-dlBulge)-transfected cells; Polymerase, pCMV-DPol-transfected cells. Antipolymerase antibody was MAb 9; anti-influenza virus HA1 was MAb 12CA5; anticore and anti-ERP72 were rabbit polyclonal antibodies. MAbs were detected with an anti-mouse fluorescein isothiocyanate-labeled secondary antibody, and anticore and anti-ERP72 were detected with an anti-rabbit rhodamine-labeled secondary antibody. (A to D) Standard immunofluorescence; (E to M) confocal immunofluorescence microscopy. Yellow and orange in panels G, J, and M indicate colocalization of the stained proteins.
LMH cells transfected with complete DHBV or DHBV (ɛ-dlBulge) genomes were stained simultaneously with MAb 9 and rabbit anti-DHBV core polyclonal antibodies to determine (i) if the nonencapsidated polymerase was produced in cells that were also producing the core protein, (ii) if the two proteins colocalized, and (iii) if the intracellular distribution of the polymerase or core proteins was altered when encapsidation was blocked by the ɛ-dlBulge mutation. The cells were analyzed by confocal immunofluorescence microscopy to obtain higher resolution than available by standard microscopy. Both the polymerase (Fig. 7E and H) and core protein (Fig. 7F and I) were easily detectable in the cytoplasm of transfected cells in an uneven, grainy pattern. Low levels of polymerase were detected in the nuclei of some cells, but core protein was rarely found in the nucleus (data not shown). The core signal was stronger than that of the polymerase in the large majority of cells, and the level of the polymerase protein varied widely among the transfected cells. Polymerase could be detected in 87% of the cells in which core was detected, and core could be detected in all cells in which polymerase was detected (data not shown). Merging the polymerase and core signals (Fig. 7G and J) revealed a complex, grainy pattern with extensive overlap between the core and polymerase signals (overlap is indicated by yellow). However, significant areas of the cytoplasm contained only core protein (red), only polymerase protein (green), or neither protein (black). There was no noticeable difference in the distribution or overlap of the core or polymerase proteins when encapsidation was blocked by the ɛ-dlBulge mutation (compare Fig. 7G and J).
This experiment excludes the possibility that the nonencapsidated polymerase is found only in a subset of cells in which core protein synthesis (and hence encapsidation) was somehow blocked. These data further indicate that the core and polymerase proteins are found in complex patterns that are only partially overlapping and that these patterns are unchanged when encapsidation is blocked. This observation excludes the possibility that all of the polymerase detectable by immunofluorescence is in association with the core protein and hence confirms our biochemical characterization of at least this subset of the polymerase as nonencapsidated.
Confocal microscopy was also performed on cells transfected with the DHBV or DHBV(ɛ-dlBulge) genomes and stained with antipolymerase MAb 9 and rabbit polyclonal anti-ERP72 antibodies because sucrose sedimentation implied an association between the polymerase and the ER. The polymerase (Fig. 7K) and ERP72 (Fig. 7L) were widely distributed in the cytoplasm, but the merge of the data sets (Fig. 7M) revealed little overlap between the two signals. The distribution and overlap of the polymerase and ERP72 proteins were unaffected by blocking encapsidation with the ɛ-dlBulge mutation (data not shown).
DISCUSSION
Here we report that the large majority of the DHBV polymerase within LMH cells is nonencapsidated and that nonencapsidated polymerase is also easily detectable in infected duck liver. The nonencapsidated polymerase is found in multiple isoforms, only one of which is found within viral core particles. The nonencapsidated polymerase is bound to a cytoplasmic structure in a large detergent-sensitive complex. Finally, the intracellular distribution of the polymerase is highly complex, only partially overlaps that of the core protein, and is not dependent on the encapsidation reaction.
The nonencapsidated DHBV polymerase may have been observed before (40). In this experiment, LMH cells were transfected with a DHBV genome carrying mutations that inserted a protein kinase A phosphorylation site into the polymerase. A cellular lysate was prepared, and core particles were removed by ultracentrifugation. The polymerase was then immunoprecipitated from the lysate, phosphorylated in vitro, and reimmunoprecipitated prior to resolution by SDS-PAGE (3). These experiments revealed a protein of about 90 kDa and a number of smaller bands. Because the lysate had been cleared of cores and as there was no DNA attached to the phosphorylated protein, it was concluded that the 90-kDa band was probably nonencapsidated DHBV polymerase.
We have been unable to demonstrate DNA polymerase activity by the nonencapsidated polymerase (data not shown). This failure may be due to technical reasons. However, no hepadnavirus-specific DNA polymerase activity has been found outside of viral cores in infected cells or tissue despite diligent attempts by multiple groups. This supports the possibility that the nonencapsidated polymerase is truly enzymatically inactive. If the encapsidated pool of the polymerase diverges from the nonencapsidated pool of polymerase at the stage of binding of the polymerase to ɛ on the pgRNA, we expect that the nonencapsidated polymerase would be enzymatically inactive because binding to ɛ promotes enzymatic maturation of the polymerase (36, 37).
Nonencapsidated polymerase from liver tissue migrates as two major bands in SDS-PAGE, whereas nonencapsidated polymerase from LMH cells migrates as only a single major band, equivalent to the lower form observed in liver tissue. It is unknown why the upper form present in liver is not found in LMH cells. The lower form that is found in both liver and LMH cells migrates as predicted from the primary sequence of the gene. Upon close examination, both the upper and lower forms of the nonencapsidated polymerase are seen to resolve into multiple species (Fig. 4). These species are very closely related in size and are difficult to resolve clearly. The slower-migrating isoforms are probably produced by differential posttranslational modification of the polymerase, because all of the polymerase is produced from a single viral gene and there is no evidence for differential splicing in the generation of the polymerase mRNA. Not all isoforms of the polymerase are competent for encapsidation because only the very fastest migrating (and presumably least modified) isoform is found within core particles produced in either LMH cells or duck liver. This raises the possibility that the metabolism of the polymerase within cells may be regulated by posttranslational modification, with the more highly modified molecules remaining nonencapsidated.
The insoluble cytoplasmic component(s) to which the nonencapsidated polymerase is bound appears to be of cellular origin. This is because the only viral components that have not been excluded from being associated with the nonencapsidated polymerase are the RNA sequences encoding the polymerase and the viral surface glycoproteins (which are encoded in a different reading frame within the polymerase gene [Fig. 1]). Because there is no evidence for an interaction between the polymerase and the surface glycoproteins and as association with the DHBV RNA would increase rather than decrease the polymerase's density, these viral components cannot account for the cellular association of the polymerase. Therefore, the polymerase must be either bound to a large cellular component or aggregated. Although aggregation cannot be rigorously excluded, it is insufficient to account for the sedimentation behavior of the nonencapsidated polymerase because the samples in Fig. 6B were dissolved in sucrose at a concentration of about 1.28 g/cm3 and overlaid with a gradient whose density declined to about 1.15 g/cm3. Proteins typically have a density of 1.3 g/cm3 or higher, and they certainly would be denser than the 1.16 g/cm3 at which the lighter fraction of the nonencapsidated polymerase is found. The only way the polymerase could have reached this position in the gradient would have been to float upward during centrifugation due to association with a cellular component of lower density, probably a membrane-containing component. Despite the provocative cosedimentation of the nonencapsidated polymerase with the ER, confocal microscopy indicates there is little colocalization of the polymerase with the ER in cells. Therefore, the identity of putative cellular partner(s) is unknown.
It is not known if nonencapsidated HBV polymerase exists in infected human cells, but one observation suggests that it may. Antibodies to the polymerase can be found in people infected with HBV and in woodchucks infected with woodchuck hepatitis virus (4, 7, 16, 42, 43). If the polymerase were made only in the trace amounts needed to provide one or two molecules per virion, it would be hard to imagine how antibodies could routinely develop against it, especially as the polymerase is not exposed on the outside of the virus. The detection of DHBV polymerase in the cytoplasm of cells producing infectious virus indicates that much more polymerase is made than had been previously assumed. We therefore predict that nonencapsidated HBV polymerase also exists. MAbs against the human HBV polymerase that can detect recombinant HBV polymerase in Western analysis, immunoprecipitation, and immunofluorescence have recently been generated (46), but detection of nonrecombinant HBV polymerase has not yet been reported. However, this inability to detect the polymerase in transfected cells may stem from the low levels of HBV replication supported by hepatoma cell lines (R. Lanford, personal communication).
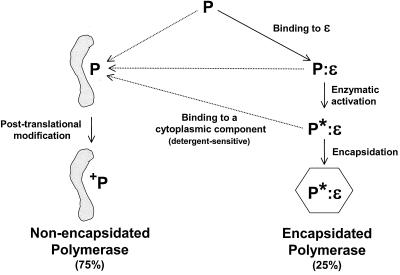
The data presented here reveal that the DHBV polymerase has two possible major metabolic fates that result in either encapsidated or nonencapsidated polymerase (Fig. 8). The encapsidation pathway is well known; it involves cotranslational binding of the polymerase to ɛ (12), followed by a structural alteration to the polymerase that results in its enzymatic activation (36, 37). This ribonucleoprotein complex is then encapsidated into core particles (1, 12, 15). The second possible fate of the polymerase is to bind to a cytoplasmic component and become posttranslationally modified. It is unknown if the polymerase enters this pathway directly following translation, or if polymerase at other stages of encapsidation may enter the nonencapsidated pathway (these possibilities are not mutually exclusive). Figure 8 illustrates the polymerase binding to its cytoplasmic partner(s) prior to posttranslational modification because some of the nonencapsidated polymerase is apparently not modified (Fig. 5). However, it is possible that the polymerase is modified first and then binds to its cytoplasmic partner(s) or that different isoforms of the nonencapsidated polymerase follow different pathways. It is not known what regulates the partitioning of the polymerase into these two pathways.
FIG. 8.
Metabolism of the DHBV polymerase. Two fates of the polymerase are indicated, with the encapsidated pool on the right and the nonencapsidated pool on the left. P, primary translation product of the polymerase gene; P:ɛ, polymerase bound to ɛ on the pgRNA; P∗:ɛ, polymerase (bound to ɛ) that has undergone the structural alteration leading to enzymatic maturation; +P, posttranslationally modified polymerase corresponding to any of the slower-migrating isoforms detectable by immunoprecipitation. The grey shape represents the cytoplasmic component(s) to which the polymerase is bound. Dotted arrows indicate nonexclusive possibilities.
There is no obvious role for the polymerase outside of viral particles in hepadnaviral replication, but we envision three possibilities. First, the nonencapsidated polymerase may be a metabolic by-product of an inefficient encapsidation process. Second, the nonencapsidated polymerase may be an intermediate in the encapsidation reaction. Finally, the nonencapsidated polymerase may regulate cellular or viral processes. These possibilities are not mutually exclusive.
Three observations argue that the nonencapsidated polymerase is not just a metabolic by-product. First, a remarkably large amount of nonencapsidated polymerase accumulates in cells, despite its translation from the downstream open reading frame of a bicistronic mRNA. This accumulation is even more remarkable when the short half-life of the polymerase in cells is considered (the human HBV polymerase has a half-life of ∼40 min [2], and our preliminary data indicate the DHBV nonencapsidated polymerase half-life to be less than 40 min). This implies that the polymerase is synthesized rapidly to maintain the high steady-state level detected by immunoprecipitation. Second, the nonencapsidated polymerase is tightly associated with a cellular structure in what appears to be a specific complex. Third, the nonencapsidated polymerase exists as multiple isoforms, but only one of these forms is encapsidated.
Some of the nonencapsidated polymerase molecules may be encapsidation intermediates. Results of in vitro studies using translation of the HBV core protein in wheat germ extract imply the existence of high-molecular-weight intermediates in the assembly of core particles (21). These studies did not include the polymerase, and other studies in cells have failed to find stable intermediates in core particle assembly larger than core protein dimers (31). However, it is reasonable to suggest that such complexes may exist in cells during the assembly of bona fide core particles containing the polymerase and pgRNA. If these putative intermediates exist, some of the nonencapsidated polymerase could be in detergent-sensitive complexes with partially formed capsids. The denser fraction of immunoprecipitable polymerase detected by sedimentation (Fig. 6B, fractions 1 to 3) may contain polymerase molecules from such intermediates because this portion of the polymerase is found only in cells transfected with the complete DHBV genome. However, much of the nonencapsidated polymerase in the cell must have a fate other than encapsidation for four reasons. First, the detergent extraction characteristics of the polymerase are the same in the presence or absence of the core protein, or in the presence or absence of a functional encapsidation signal. This indicates that the polymerase-containing complexes which are disrupted by RIPA do not require any of the other viral components of the encapsidation reaction. Second, confocal microscopy reveals that some of the polymerase in cells transfected with wild-type DHBV does not colocalize with the core protein (Fig. 7G). Third, the cellular distribution of the polymerase in cells (as measured by confocal microscopy or nuclear/cytoplasmic fractionation) is the same in the presence or absence of the core protein or in the presence or absence of a functional encapsidation signal. Finally, it is difficult to propose that all isoforms of the polymerase are encapsidation intermediates when only one isoform is found in cores.
A role for the polymerase as a regulator of a viral or cellular process is speculative but reasonable given the hepadnaviruses' unusual replication cycle. The hepadnavirus genome is very small, and all nucleotides code for protein, most of them in more than one frame simultaneously. This places severe constraints on the number of proteins that the virus can produce. Three of the four hepadnavirus open reading frames are known to regulate cellular or viral functions. The X open reading frame encodes a regulatory protein with transcriptional transactivation activity (44). The C open reading frame encodes both the major capsid protein and the e antigen, a secreted protein believed to function in immune evasion (8). The S open reading frame encodes the viral surface glycoproteins. The smallest of these glycoproteins is secreted at high levels into the blood and is probably involved in immune evasion, and the largest surface glycoprotein can be a transcriptional regulator (11, 29). Even the polymerase itself has been proposed to be a suppressor of core protein translation (13). Therefore, there is ample precedence for a role of the hepadnavirus polymerase as a regulator of cellular or viral processes beyond its obvious role in genomic replication. Further research into this intriguing possibility is needed to clarify the role of the nonencapsidated polymerase in hepadnavirus biology.
ACKNOWLEDGMENTS
This work was supported by grants AI38447 from the National Institutes of Health and JFRA 616 from the American Cancer Society.
We thank William Mason for the gift of anti-DHBV core antibody and Michael Green for the gift of anti-ERP72 antibody. We are grateful to Cathal O'Sullivan and Michael Green for helpful discussions and to Amy Ruff and Mark Gerber for able technical assistance.
REFERENCES
- 1.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Kuhn C, Schaller H. Expression of the P-protein of the human hepatitis B virus in a vaccinia virus system and detection of the nucleocapsid-associated P-gene product by radiolabelling at newly introduced phosphorylation sites. Nucleic Acids Res. 1992;20:195–202. doi: 10.1093/nar/20.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlater R, Weber M, Schaller H. In vitro phosphorylation of hepatitis B virus P gene product: a general method for radiolabeling of proteins. In: Adolph K W, editor. Methods in molecular genetics. Vol. 4. New York, N.Y: Academic Press; 1994. pp. 391–401. [Google Scholar]
- 4.Chang L J, Dienstag J, Ganem D, Varmus H. Detection of antibodies against hepatitis B virus polymerase antigen in hepatitis B virus-infected patients. Hepatology. 1989;10:332–335. doi: 10.1002/hep.1840100314. [DOI] [PubMed] [Google Scholar]
- 5.Chang L-J, Hirsch R C, Ganem D, Varmus H E. Effects of insertional and point mutations on the functions of the duck hepatitis B virus polymerase. J Virol. 1990;64:5553–5558. doi: 10.1128/jvi.64.11.5553-5558.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condreay L D, Aldrich C E, Coates L, Mason W S, Wu T-T. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J Virol. 1990;64:3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feitelson M A, Millman I, Duncan G D, Bumberg B S. Presence of antibodies to the polymerase gene product(s) of hepatitis B and woodchuck hepatitis virus in natural and experimental infections. J Med Virol. 1988;24:121–136. doi: 10.1002/jmv.1890240202. [DOI] [PubMed] [Google Scholar]
- 8.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2703–2737. [Google Scholar]
- 9.Goldberg D E, Kornfeld S. Evidence for extensive subcellular organization of asparagine-linked oligosaccharide processing and lysosomal enzyme phosphorylation. J Biol Chem. 1983;258:3159–3165. [PubMed] [Google Scholar]
- 10.Haugejorden S, Srinivasan M, Green M. Analysis of the retention signals of two resident luminal endoplasmic reticulum proteins by in vitro mutagenesis. J Biol Chem. 1991;266:6015–6018. [PubMed] [Google Scholar]
- 11.Hildt E, Saher G, Bruss V, Hofschneider P H. The hepatitis B virus large surface protein (LHBs) is a transcriptional activator. Virology. 1996;225:235–239. doi: 10.1006/viro.1996.0594. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch R C, Lavine J E, Chang L J, Varmus H E, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 13.Howe A Y M, Tyrrell D L J. Duck hepatitis b virus polymerase acts as a suppressor of core protein translation. J Virol. 1996;70:5035–5042. doi: 10.1128/jvi.70.8.5035-5042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jean-Jean O, Weimer T, de Recondo A M, Will H, Rossignol J M. Internal entry of ribosomes and ribosomal scanning involved in hepatitis B virus P gene expression. J Virol. 1989;63:5451–5454. doi: 10.1128/jvi.63.12.5451-5454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Junker-Niepmann M, Bartenschlager R, Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kann M, Kochel H G, Uy A, Thomssen R. Diagnostic significance of antibodies to hepatitis B virus polymerase in acutely and chronically HBV-infected individuals. J Med Virol. 1993;40:285–290. doi: 10.1002/jmv.1890400406. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. Structural features in Eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 18.Lanford R E, Chavez D, Brasky K M, Burns III R B, Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci USA. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M D, Bronson D L, Lemke T D, Faras A J. Phylogenetic analyses of 55 retroelements on the basis of the nucleotide and product amino acid sequences of the pol gene. Mol Biol Evol. 1995;12:657–670. doi: 10.1093/oxfordjournals.molbev.a040231. [DOI] [PubMed] [Google Scholar]
- 20.Lin C-G, Lo S J. Evidence for involvement of a ribosomal leaky scanning mechanism in the translation of the hepatitis B virus Pol gene from the viral pregenome RNA. Virology. 1992;188:342–352. doi: 10.1016/0042-6822(92)90763-f. [DOI] [PubMed] [Google Scholar]
- 21.Lingappa J R, Martin R L, Wong M L, Ganem D, Welch W J, Lingappa V R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.McClure M A. Evolutionary history of reverse transcriptase. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 425–444. [Google Scholar]
- 24.Nassal M, Rieger A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J Virol. 1996;70:2764–2773. doi: 10.1128/jvi.70.5.2764-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollack J R, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radziwill G, Zentgraf H, Schaller H, Bosch V. The duck hepatitis B virus DNA polymerase is tightly associated with the viral core structure and unable to switch to an exogenous template. Virology. 1988;163:123–132. doi: 10.1016/0042-6822(88)90239-5. [DOI] [PubMed] [Google Scholar]
- 29.Rothmann K, Schnolzer M, Radziwill G, Hildt E, Moelling K, Schaller H. Host cell-virus cross talk: phosphorylation of a hepatitis B virus envelope protein mediates intracellular signalling. J Virol. 1998;72:10138–10147. doi: 10.1128/jvi.72.12.10138-10147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoedel F, Sprengel R, Weimer T, Fernholz D, Schneider R, Will H. Animal hepatitis B viruses. Adv Viral Oncol. 1989;8:73–102. [Google Scholar]
- 31.Seifer M, Zhou S, Standring D N. A micromolar pool of antigenically distinct precursors is required to initiate cooperative assembly of hepatitis B virus capsids in Xenopus oocytes. J Virol. 1993;67:249–257. doi: 10.1128/jvi.67.1.249-257.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprengel R, Kuhn C, Will H, Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 33.Tavis J E, Frisque R J. Altered DNA binding and replication activities of JC virus T-antigen mutants. Virology. 1991;183:239–250. doi: 10.1016/0042-6822(91)90136-y. [DOI] [PubMed] [Google Scholar]
- 34.Tavis J E, Ganem D. Expression of functional hepatitis B virus polymerase in yeast reveals it to be the sole viral protein required for correct initiation of reverse transcription. Proc Natl Acad Sci USA. 1993;90:4107–4111. doi: 10.1073/pnas.90.9.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavis J E, Ganem D. RNA sequences controlling the initiation and transfer of duck hepatitis B virus minus-strand DNA. J Virol. 1995;69:4283–4291. doi: 10.1128/jvi.69.7.4283-4291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavis J E, Ganem D. Evidence for the activation of the hepatitis B virus polymerase by binding of its RNA template. J Virol. 1996;70:5741–5750. doi: 10.1128/jvi.70.9.5741-5750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavis J E, Massey B, Gong Y. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J Virol. 1998;72:5789–5796. doi: 10.1128/jvi.72.7.5789-5796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavis J E, Perri S, Ganem D. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J Virol. 1994;68:3536–3543. doi: 10.1128/jvi.68.6.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G-H, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber M. Das P-protein des enten hepatitis B virus: Untersuchungen zur struktur und funktion in der hepadnaviralen replikation. Ph.D. dissertation. Heidelberg, Germany: Ruprecht-Karls-Universitaet; 1994. [Google Scholar]
- 41.Weber M, Bronsema V, Bartos H, Bosserhoff A, Bartenschlager R, Schaller H. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J Virol. 1994;68:2994–2999. doi: 10.1128/jvi.68.5.2994-2999.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weimer T, Schoedel F, Jung M C, Pape G R, Alberti A, Fattovich G, Beljaars H, van Eerd P M, Will H. Antibodies to the RNAse H domain of hepatitis B virus P protein are associated with ongoing viral replication. J Virol. 1990;64:5665–5668. doi: 10.1128/jvi.64.11.5665-5668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weimer T, Weimer K, Tu Z X, Jung M C, Pape G R, Will H. Immunogenicity of human hepatitis B virus P-gene derived proteins. J Immunol. 1989;143:3750–3756. [PubMed] [Google Scholar]
- 44.Yen T S B. Hepadnaviral X protein: review of recent progress. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 45.Zoulim F, Seeger C. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J Virol. 1994;68:6–13. doi: 10.1128/jvi.68.1.6-13.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.zu Putlitz J, Lanford R E, Carlson R I, Notvall L, De la Monte S M, Wands J R. Properties of monoclonal antibodies directed against hepatitis B virus polymerase protein. J Virol. 1999;73:4188–4196. doi: 10.1128/jvi.73.5.4188-4196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]