Abstract
A lattice of VP7 trimers forms the surface of the icosahedral bluetongue virus (BTV) core. To investigate the role of VP7 oligomerization in core assembly, a series of residues for substitution were predicted based on crystal structures of BTV type 10 VP7 molecule targeting the monomer-monomer contacts within the trimer. Seven site-specific substitution mutations of VP7 have been created using cDNA clones and were employed to produce seven recombinant baculoviruses. The effects of these mutations on VP7 solubility, ability to trimerize and formation of core-like particles (CLPs) in the presence of the scaffolding VP3 protein, were investigated. Of the seven VP7 mutants examined, three severely affected the stability of CLP, while two other mutants had lesser effect on CLP stability. Only one mutant had no apparent effect on the formation of the stable capsid. One mutant in which the conserved tyrosine at residue 271 (lower domain helix 6) was replaced by arginine formed insoluble aggregates, implying an effect in the folding of the molecule despite the prediction that such a change would be accommodated. All six soluble VP7 mutants were purified, and their ability to trimerize was examined. All mutants, including those that did not form stable CLPs, assembled into stable trimers, implying that single substitution may not be sufficient to perturb the complex monomer-monomer contacts, although subtle changes within the VP7 trimer could destabilize the core. The study highlights some of the key residues that are crucial for BTV core assembly and illustrates how the structure of VP7 in isolation underrepresents the dynamic nature of the assembly process at the biological level.
Orbiviruses, members of the Reoviridae family, possess a large (86 nm in diameter) nonenveloped virus particle, encapsidating 10 segments of double-stranded RNA genome (31, 32, 35). Orbivirus virions are composed of a number of discrete proteins arranged in a specific but nonequimolar ratio. Overall, these viruses are icosahedral, with two protein layers that have radically different geometries, and provide a complex subject to study, both in terms of protein-protein interactions, and protein-RNA interactions. Bluetongue virus (BTV), the prototype orbivirus, consists of seven structural proteins (VP1 to VP7), four of which are major (VP2, VP3, VP5, and VP7) and include proteins that interact with cellular receptors and others that form the underlying framework of the virion (30, 31, 32). The three minor proteins (VP1, VP4, and VP6), which are present in low molar ratios within the virion, have RNA transcriptase- and RNA-modifying properties (33). In its mature form the virus exhibits no transcriptase activity until it is activated upon infection with the modification of the outer capsid to create channels in the core architecture that allow metabolites to enter the capsid and the viral mRNA species to be formed and extruded (10, 12, 13, 29).
A considerable amount of data has recently been accumulated on the transcriptionally active BTV core architecture. A combination of three-dimensional cryo-electron microscopic analysis of the BTV core at a 25-Å resolution and X-ray crystallographic structure of the BTV core at a 3.5-Å resolution has revealed the complexity in the arrangements of the core protein, in particular, how the two major core proteins, VP7 (Mr 38,000) and VP3 (Mr, 103,000) are organized within the core (12, 13). The bristly surface of the 70-nm icosahedral core is made up of 13 icosahedrally independent copies of the VP7 molecule in the form of four trimers (P, Q, R, and S) in general positions and one (T) located with its threefold axis aligned with the icosahedral threefold axis (12, 13, 29). A total of 780 VP7 molecules form the entire core surface. The local threefold axes of these 260 trimers (85 Å long) are perpendicular to the core surface, and the broad, flat base of each of the trimers (65 Å) contacts the inner subcore (11–13). The underlying scaffold is a thin shell, made up of 120 copies of VP3 molecule that are arranged at T=2 icosahedral symmetry within the subcore layer. The VP7 and VP3 layers interact through flat, predominantly hydrophobic surfaces. There are 13 different sites of contact for the VP7 subunit on the VP3 subcore. Since VP7 forms trimers in solution (4), it is likely that trimers are the driving force for VP7 and VP3 interactions during core assembly. However, as yet there have been no studies delineating trimer formation from core assembly.
The crystal structure of VP7 at a 2.6-Å resolution has revealed that three molecules interact extensively to form VP7 trimers (13). Each VP7 subunit or monomer consists of two distinct domains, the upper and lower, and are twisted such that the top domain of one monomer rests on the lower domain of a threefold related subunit. The interactions between monomers involve both domains and are extensive. However, there are significant cavities at the center of the trimer (along the threefold axis) surrounded by predominantly uncharged residues (11, 13).
To develop an understanding of the assembly process of BTV particles and the role played by VP7 in capsid assembly, we have focused on the lower domain of the VP7 molecule, as not only are the lower domains directly in contact with VP3 layer, but also interactions between the lower domains within the trimers are intensive (11, 13). A series of substitutions of amino acid residues have been designed which, based on the X-ray structure, appeared to be involved in intramolecular (within the VP7 subunit) and intermolecular (between the VP7 subunits) interactions within the molecule. Their role in trimer-trimer interaction was, however, unclear. Each mutant VP7 molecule was expressed in insect cells infected with recombinant baculoviruses, and the effects of these mutations on the overall VP7 structure (i.e., correct folding) and on core assembly were analyzed using previously established biological assay systems (9, 25). The formation of the lattice of VP7 trimers on the VP3 layer was examined by an in vivo expression system in which BTV core-like particles (CLPs) are assembled upon coexpression of VP7 and VP3 in insect cells (9). The interaction of VP7 with VP3 within the CLP was also determined by electron microscopy (EM). Trimerization of VP7 was assessed by two different experimental approaches using purified VP7 mutants. It was clear that a single substitution at a key position could destabilize BTV core assembly without drastically disrupting the trimer formation. From these analyses, key residues within the lower domain of VP7 that are essential for core formation were identified.
MATERIALS AND METHODS
Viruses and cells.
Spodoptera frugiperda (Sf) cells were grown in suspension or monolayer cultures at 28°C in TC100 medium supplemented with 10% fetal calf serum. Derivatives of Autographa californica nuclear polyhedrosis virus (AcNPV) containing the wild-type BTV type 10 (BTV-10) VP7 gene (Ac10BTV7) and the VP7 mutants were plaque purified and propagated as described elsewhere (6).
Site-directed mutagenesis, construction of recombinant transfer vectors, and isolation of recombinant baculoviruses expressing mutant VP7 proteins.
Using the single-strand capacity of the baculovirus transfer vector pAcCL29 (24), synthetic oligonucleotides were employed to prepare VP7 mutants by the method described by Kunkel and associates (22). Wild-type BTV-10 VP7 DNA was recovered from the transfer vector pAcYM1.10BTV7 (27) by excision with BamHI and subcloned into the BamHI site of pAcCL29. The oligonucleotides used for mutagenesis and the resulting amino acid changes are shown in Table 1. All the oligonucleotides represent the complement of the coding strand of the BTV-10 gene. The mutated sequences in the recombinant plasmids were identified by sequence analyses (36).
TABLE 1.
Oligonucleotide primers used in site-directed mutagenesisa
| Mutant | Nucleotide sequence |
|---|---|
| E104W | 5′-ATTCGCCGCCCACGTTGTAAA-3′ |
| R111F | 5′-CCCAGTCAGAAAAGCTATTTC-3′ |
| W119D | 5′-CGCTGGCCCGTCTGTCGAAGT-3′ |
| F268R | 5′-GTAAACATTGCGAATCTCAGC-3′ |
| Y271R | 5′-CCTGAAGCTGCGAACATTGAA-3′ |
| D318N | 5′-AGTGTAAACATTAGCAAGTGT-3′ |
| T321R | 5′-CCTTAAAACTCTGTAAACATC-3′ |
The oligonucleotides used for site-directed mutagenesis represent the complement of the coding strand of the VP7 gene. The underlined triplets are the altered anticodons.
The lipofection technique (8) was used to cotransfect monolayers of Sf cells with recombinant transfer vectors and Bsu36I triple-cut AcNPV DNA (17). Recombinant baculoviruses were selected on the basis of their LacZ-negative phenotypes, plaque purified, and propagated as described elsewhere (16).
Purification of CLPs.
Baculovirus-expressed CLPs were purified as described previously (9). In brief, Sf cells were coinfected in suspension culture with recombinant baculovirus, Ac17BTV3 expressing VP3 together with the various mutant VP7 baculoviruses or recombinant baculovirus Ac10BTVP7 expressing wild-type VP7, using a multiplicity of infection of 5 to 10 PFU per cell. After incubation at 28°C for 48 h, cells were harvested, washed with phosphate-buffered saline (PBS), resuspended in TNN buffer (200 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% [vol/vol] Nonidet P-40) and lysed by Dounce homogenization. The lysate was clarified by centrifugation (10 min at 10,000 rpm using a JA-12 rotor) and the CLPs were purified from the supernatant by centrifugation on a (35%) CsCl gradient for 18 h at 35,000 rpm (Beckman SW41 rotor). Alternatively, CLPs were concentrated by a discontinuous sucrose step gradient (66% [wt/wt] and 40% [wt/vol] sucrose in 200 mM Tris-HCl [pH 8.0], 150 mM NaCl) and centrifuged for 3 h at 26,000 rpm using an SW28 rotor. CLPs were collected from the interface as described previously (9). The presence of VP3 and VP7 proteins was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting using anti-BTV-10 polyclonal antibodies, and by EM.
SDS-PAGE and Western blot analyses.
Sf cell monolayers were infected with each recombinant baculovirus using a multiplicity of infection of 10 (9). Cells were harvested at 48 h postinfection, washed with PBS and lysed at 4°C in TNN buffer. Samples were then boiled in protein dissociation buffer (10% [vol/vol] β-mercaptoethanol, 10% [wt/vol] SDS, 25% [vol/vol] glycerol, 10 mM Tris-HCl [pH 6.8], and 0.02% [wt/vol] bromophenol blue) at 100°C or at room temperature (for identification of VP7 trimers) for 10 min and resolved by SDS–10% PAGE followed by staining with Coomassie brilliant blue.
For Western blot analyses proteins were transferred from gels onto a polyvinylidene difluoride membrane by standard blotting procedures. Membranes were incubated with the anti BTV-10 antiserum diluted 1:1,000 in blocking buffer (containing 5% [wt/vol] skim milk and 0.05% [vol/vol] Tween 20 in PBS), followed by incubation with the secondary antibody conjugated with alkaline phosphatase, and developed with the alkaline phosphatase substrate (NBT-BCIP [GIBCO-BRL] in 0.1 M Tris-HCl [pH 8.5], 0.1 M NaCl, 0.05 M MgCl2).
Purification of recombinant VP7 protein by ammonium sulfate precipitation.
The wild-type and mutant BTV-10 VP7 proteins were purified as described previously (4). The infected Sf cells were harvested 48 h postinfection, washed in PBS, resuspended in cold TNN buffer, and homogenized at 4°C as described above. Cell debris and nuclei were pelleted by centrifugation (10 min at 16,000 × g). Ice-cold saturated ammonium sulfate in 100 mM Tris-HCl, pH 7.5 was added to the cytoplasmic cell extracts to a final concentration of 20%. The precipitated protein was pelleted by centrifugation and resuspended in 10 mM Tris-HCl, pH 8.5. The insoluble material was removed by pelleting at 9,000 × g for 10 min and then dialyzed against the same buffer at 4°C overnight. The extract of VP7 was stored at −20°C prior to analysis.
Velocity sedimentation analysis.
Purified VP7 protein was applied onto 15 to 30% (vol/vol) glycerol gradients made in 20 mM Tris (pH 8.0)–100 mM NaCl–1 mM dithiothreitol–0.5 mM EDTA and sedimented at 60,000 rpm in a Beckman VTi 65 rotor at 4°C for 90 min. Molecular mass markers in the gradients were provided by high-molecular-mass calibration kits (Pharmacia). The molecular mass standard consisted of the following proteins: catalase, 4 × 58 kDa = 232 kDa; lactate dehydrogenase, 4 × 36 kDa = 140 kDa; and serum albumin, 67 kDa. Gradients were fractionated from the bottom, and VP7 molecules were detected by SDS-PAGE and Western blotting.
EM.
Purified wild-type and mutant CLPs were resuspended in water, and 10 μl of CLPs were absorbed onto carbon-coated copper 400-mesh EM grids for 15 min, washed with water, and negatively stained with 1% (wt/vol) uranyl acetate. Grids were examined in a Hitachi H-7000 electron microscope at 75 kV.
RESULTS
Rationale for selection of residues within VP7 for site-specific mutagenesis.
The amino acid sequence of VP7 protein is highly conserved among BTV serotypes (up to 99% identity) and other closely related orbiviruses, such as epizootic hemorrhagic disease virus (EHDV) (64% identity) and African horse sickness virus (AHSV) (45% identity) (7, 10, 18, 20, 30–35, 37). Several amino acid residues were selected for mutagenesis studies on the basis of their positions in the X-ray structure of the BTV VP7 molecule as well as their conservation between BTV, EHDV, and AHSV (Fig. 1). The lower domain of VP7, which contains both the N terminus (amino acids 1 to 120) and the C terminus (aa 250 to 349) of the molecule, is composed entirely of α-helices (nine in total) and long extended loops (11). The N-terminal portion provides five of these helices, with the remaining four helices formed by the C-terminal region (Fig. 1 and 2). The helices of the lower domain are packed tightly together (Fig. 2). The interactions between the three VP7 lower domains in the trimer are complex and mostly involve helices 5 and 6 and associated loops (11). Helix 5 is located inside the trimer, interacting with helices 1, 2, 6, and 8. Helix 6 runs across the top of the lower domain, interacting with both the clockwise and anticlockwise related subunits. Our aims were to substitute key residues in these helices and loops with alternative residues and to assess their effect in VP7 trimer formation and core assembly. These mutants would allow us to assess to what extent these specific contact points could be varied. Some of these residues were selected since they are highly conserved among VP7 molecules of three related orbiviruses and to replace these with residues that should be accommodated within the molecule without too drastic effect. Based on the structural analysis, we selected two residues of helix 5 (E104 and R111), one residue of a loop at the end of helix 5 (W119), two residues of helix 6 (F268 and Y271), and two residues of helix 8 (D318 and T321), respectively, for mutational studies (Table 2).
FIG. 1.
Important features relative to VP7 BTV-10 sequence. Residues conserved (1) in VP7 of BTV-10, AHSV type 4 (AHSV-4), and EHDV type 1 (EHDV-1) the three gnat-transmitted orbiviruses (1, 15, 26, 34), are shown in black squares. The seven mutated residues are indicated by stars on the bottom line; vertical arrows show the beginning and the end of the top domain. Secondary elements of VP7 BTV-10 (2, 13) are shown on the top line. α- and 310-helices are indicated by spirals; β-strands are indicated by horizontal arrows.
FIG. 2.
Ribbon diagram of BTV-10 VP7 trimer. Sites of mutation are indicated with the three subunits, a, b, and c, which are related by a threefold molecular axis. Subunit a (cyan), contains helices 5, 6, and 8, in which mutations have been introduced. The b and c subunits are colored yellow and grey, respectively. Details of the mutation are highlighted by ball-and-stick representation. Oxygen is shown in red and nitrogen is shown in blue. Hydrogen bonds on side chains are shown by a dashed line. The Figure was drawn with MOLSCRIPT (21) as modified by R. Esnouf.
TABLE 2.
VP7 mutants and their allocated chemical and functional features
| VP7 mutant | Location of lower domain | Substitution | Charge | Side chain | Chemical descriptiona | Hydropathicityb |
|---|---|---|---|---|---|---|
| E104W | Helix 5 | Glu104 to Trp | Acidic to nonpolar | Aromatic | −→0 | Phil to Phob |
| R111F | Helix 5 | Arg111 to Phe | Basic to nonpolar | Aromatic | +→0 | Phil to Phob |
| W119D | Loop between helices 5 and 6 | Trp119 to Asp | Nonpolar | Aromatic to acidic | 0→− | Phob to Phil |
| F268D | Helix 6 | Phe268 to Arg | Nonpolar | Aromatic to basic | 0→+ | Phob to Phil |
| Y271R | Helix 6 | Tyr271 to Arg | Unchc-polar | Aromatic to basic | −→0 | Phob to Phil |
| D318N | Helix 8 | Asp318 to Asn | Acidic to unch-polar | Amide | −→0 | Phil to Phob |
| T321R | Helix 8 | Thr321 to Arg | Unch-polar | Hydroxyl to basic | 0→+ | Phob to Phil |
Data show that mutations resulted in changes from a negatively (−) or positively (+) charged or neutral (0) state.
Phil, hydrophilic; Phob, hydrophobic.
Unch, uncharged.
Residues on helix 5.
Helix 5 of the lower domain consists of 15 residues, amino acids 103 to 118, (TEAANEIARVTGETS). The first glutamate at position 104 (E104) of helix 5 is located at an interesting site in the molecule as shown in Fig. 2 (also see Fig. 9). It is located at the bottom of a small cavity in the middle of the VP7 trimer; it does not make close contact with any other residue within the subunit. However, it has weak interactions (3.9-Å gap), with the residues of the threefold related subunits close to the molecular threefold axis (Fig. 2). Therefore, we predicted that substitution of this residue with a bulky residue such as W, or F, or Y can be accommodated without perturbing the protein folding, although it may affect the shape of the trimer. We chose to replace E104 with tryptophan (W). The arginine at position 111 (R111) is conserved among BTV serotypes and in AHSV and EHDV (Fig. 1). The R111 is surface accessible and surrounds the small cavity (Fig. 2). Therefore, its substitution by a bulky hydrophobic residue, such as phenylalanine (F), should not interfere in trimerization. However, unlike E104, it is in contact with E266 (see Fig. 9) of the neighboring subunit via a side chain hydrogen bond (11). Therefore, there is a possibility that this substitution may affect the stable monomer-monomer contacts.
FIG. 9.
Position of each residue mutated and the surrounding environment within a trimer. The amino acids charged are highlighted as ball-and-stick models on a wire frame background. Hydrogen bonding between residues is indicated by dashed lines, and the relevant spacing is shown in angstroms. The environment of each mutation is shown for one monomer-monomer interface only, with the residues derived from each monomer designated by a, b, or c. The residues mutated are E104 (A), R111 (B), W119 (C), F268 (D), Y271 (E), and T321 (F).
The tryptophan at 119 (W119) is part of the loop at the end of helix 5, is located on the surface of the molecule, weakly interacting with R341 of the partner subunit, and is far away from the VP7-VP3 interface. From the atomic structure and our previous deletion mutation studies, it appears that the residues 334 to 349 at the carboxy terminus of VP7 are essential for trimer-trimer interactions in the core (13, 23). W119 is also near K255, which is critical for the correct folding of the VP7 molecule. It was, therefore, of interest to determine whether replacement of W119 by a charged residue, such as an aspartate, would affect the trimerization of VP7.
Residues on helix 6.
Helix 6 is the major element that lines up with the internal cavity. Most residues appear not to be involved in close interactions with the partner subunits. For example, as shown in Fig. 2, none of the residues appear to be near the side chain of phenylalanine at position 268 (F268) of helix 6. However, it packs against a lysine residue at residue 255, which is located in the hinge region between the bottom domain and the top domain. Replacement of F268 by a large charged side chain, such as arginine, should bring residue 268 close to a residue in the vicinity (such as the proline at 121 [P121]), since arginine is longer (by at least ∼20 Å) than phenyalanine. This substitution may or may not influence the trimerization. F268 was therefore replaced with arginine. A neighboring tyrosine at position 271 (Y271) of the helix 6 is highly conserved among other orbiviruses. There are strong nonpolar interactions between Y271 and P292 (see Fig. 9). Therefore, Y271 was also selected for replacement by an arginine (Y271R), which may interfere with the trimer formation due to incorporation of charged residues in the nonpolar pocket.
Residues on helix 8.
Helix 8 is directly in contact with helix 5, packing closely against it. The two residues that were selected for mutagenesis are involved in contacts between the two helices. The aspartic acid at position 318 (D318) was replaced by asparagine (D318N) (Table 2), which is an isosteric mutation, in which the size of the side chain remains unchanged. However, the loss of charge on the side chain would only weaken the strong hydrogen bond (or the salt link) between the Asp and Arg residues, since this residue is in contact with R111 in the partner subunit and thus might affect VP7 trimer formation due to repulsion force and, consequently, CLP assembly. T321 was selected for replacement by arginine (T321R) (Table 2) as it is at the end of helix 8 and close to the surface of the trimer (Fig. 2). The latter should not affect the trimer formation.
Synthesis of recombinant VP7 mutants and influence of substitution on VP7 solubility.
We prepared the seven mutations on VP7 DNA as described in Materials and Methods, each having a single substitution (e.g., E104W, R111F, W119D, F268R, Y271R, D318N, and T321R) in the lower domain of the molecule. Each VP7 derivative DNA was then cloned into the baculovirus single-stranded transfer vector, pAcCL29, as described in Materials and Methods. Seven recombinant baculoviruses were isolated, each expressing a single mutant VP7 molecule. The supernatants recovered from the recombinant virus-infected insect cell lysates were examined for the presence of VP7 mutants. Except one, Y271R, synthesis of VP7 protein was present in all six mutants, when analyzed by SDS-PAGE and Western analysis, indicating that the expressed products were in soluble form. Therefore, none of the six remaining mutations had any apparent drastic effects on VP7 solubility. As expected, when compared with the native VP7 protein, both the levels of expression and sizes of the mutant proteins in SDS-PAGE were similar to those of the wild-type VP7, as shown in Fig. 3 (27). It was therefore considered that each of the six mutants probably had characteristics that were overall similar to those of the wild-type VP7.
FIG. 3.
Expression of recombinant VP7 mutants. Sf cells were infected with each recombinant virus, and the infected-cell lysates were analyzed by SDS–10% PAGE and stained with Coomassie brilliant blue. Each mutant protein is indicated. The sizes of the expressed proteins are compared with the purified wild-type VP7 protein as indicated (WT). The position of VP7 is shown by an arrow.
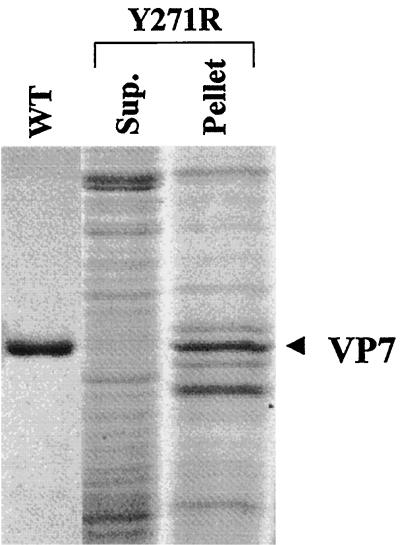
Since the recombinant Y271R mutant protein was not detectable from the solubilized supernatant fraction of the infected-cell lysate, it was assumed that Y271R protein was precipitated with the cell debris upon low-speed centrifugation. Indeed, high-level expression of mutant Y271R protein could be detected in the cell pellet when analyzed by SDS-PAGE (Fig. 4, lane 3). The expressed protein was clearly insoluble and formed aggregated inclusion bodies in infected cells. Due to this it was impossible to recover it in a native soluble form. It was therefore concluded that the arginine for phenylalanine substitution disrupted the packing of the N- and C-terminal helical domains within the VP7 subunit. No further characterization was performed with this mutant.
FIG. 4.
Solubility assay of the 10BTV7-Y271R mutant. Following lysis with nonionic detergent, extracts of the baculovirus-infected Sf cells were separated by low-speed centrifugation. The supernatant and the pellet were resolved by SDS–10% PAGE and Coomassie brilliant blue staining. The lanes shown contain purified wild-type VP7 protein (WT), supernatant after a low-speed centrifugation (Sup.), and the pellet after low-speed centrifugation.
VP7-VP7 interaction at certain specific contact points dictates the assembly of stable capsid.
The X-ray structure of the BTV core revealed that although various VP7 trimers interact with VP3 layers differently, the VP7-VP7 interaction is relatively well fitting and the various VP7 trimers remain virtually uniform in the complex core structure (13). Of the five different trimers, the T trimers seem to be the most tightly attached and are probably the first set of trimers that attach to the VP3 layer, and the P trimer is the most loosely attached and probably the last to assemble (12, 13). Although the substitution mutations were designed to influence the oligomerization of VP7, they may also be able to influence the stability of the VP7-VP3 interaction. An attempt was, therefore, made to investigate whether mutant derivatives of the VP7 molecule could indeed assemble into the BTV capsid. This was performed by employing a biological assembly assay that we had developed previously (9). Each recombinant virus (but not Y271R) was coexpressed together with VP3 protein in insect cells as described in Materials and Methods. Two days postinfection, cells were lysed with nonionic detergent, and the assembled CLPs in the soluble supernatant fraction were purified through a cesium chloride gradient. Only three mutants (E104W, D318N, and F268R) of the six recombinant proteins were able to form the particulate structures that could easily be visualized as bands on the CsCl gradients upon centrifugation and at the same position as the native CLPs. Evidently, either the other three mutants, R111F, W119D, and T32R, did not assemble at all, or the assembled particles were too weak to tolerate the cesium chloride gradient purification (Table 3).
TABLE 3.
Mutations in the BTV-10 VP7 gene and their effect on trimer and CLP formation
| BTV-10 VP7 mutant | Trimer formation | CLP assembly | CLP stability in CsCl gradient |
|---|---|---|---|
| E104W | Yes | Yes | Yes |
| R111F | Yes | Yes | No |
| W119D | Yes | Yes | No |
| F268R | Yes | Yes | Yes |
| Y271Ra | No | No | No |
| D318N | Yes | Yes | Yes |
| T321R | Yes | Yes | No |
Note that this mutant failed to form trimers or assemble CLPs.
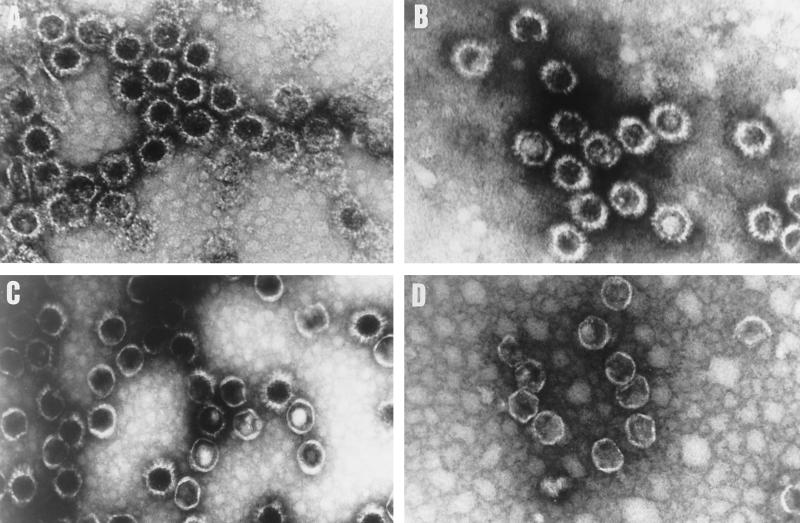
The morphology for purified CLPs formed by E104W, D318N, and F265R VP7 were characterized by EM. The arrangement of the VP7 capsomers on the outer layers of these purified particles appeared more or less dense and regular, similar to the wild-type CLPs. In all three cases, the VP3 inner layer of the CLPs was conserved, exhibiting a thin layer of an icosahedral configuration. However, while in the case of E104W the EM revealed empty CLPs (Fig. 5B) similar in size and appearance to wild-type CLPs (Fig. 5A), the CLPs formed by mutants D318N and F268R were more heterogeneous. Many particles with only VP3 subcore could be seen in the case of mutant F268R (Fig. 5D), and a few were seen with D318N (Fig. 5C). This is probably due to loss of VP7 trimers from these particles during purification and the fixation treatments required for EM.
FIG. 5.
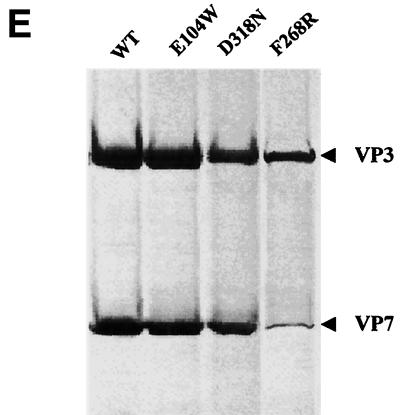
CLP formation by BTV-17 VP3 and mutated VP7 proteins. Sf cells were coinfected with Ac17BTV3 and either Ac10BTV7, expressing the wild-type VP7, or recombinant baculoviruses expressing various VP7 mutants. The CLPs were purified on CsCl gradients as described in Materials and Methods. Shown are the results of EM of CLPs formed by native VP3 protein and native VP7 (A) or different mutant VP7 E104W (B), D318N (C), and F268F (D) proteins. (E) CLPs were analyzed by SDS-PAGE (each CLP indicated at the top). The positions of VP3 and VP7 are indicated.
The efficiency of these VP7 mutants to assemble with the VP3 subcore and form CLPs was further assessed by biochemical analysis. When analyzed by SDS-PAGE (Fig. 5E), it was obvious that E104W formed CLPs in the presence of VP3 that were indistinguishable from the wild-type CLPs. CLPs formed by the other two mutants, D318R and F268R, had fewer VP7 proteins than did native CLPs and the difference could easily be visualized upon SDS-PAGE. In particular, CLPs formed with mutant F268R VP7 had very few VP7 molecules, indicating that most VP7 molecules were lost during purification.
The numbers of VP7 molecules present in each type of CLPs were subsequently estimated. The Coomassie-stained protein bands of each preparation were scanned, and the molar ratio of VP3 to VP7 in each CLP preparation was estimated. Since purified wild-type CLPs usually lack the P trimers (i.e., only 200 trimers/CLP are generally present), 600 VP7 molecules per 120 molecules of VP3 in each CLP were considered the full complement (14; B. V. V. Prasad, personal communication). As shown in Fig. 6, while all of the VP7 trimers were accounted for in CLPs formed by E104W, only approximately one-third of VP7 molecules were present in CLPs formed by D318N and only a very few VP7 trimers (<20) remained attached to the VP3 layer.
FIG. 6.
Number of VP7 molecules per CLP. CsCl gradient-purified CLPs were resolved by SDS-PAGE, and the image of the gel was captured and analyzed by AlphaImager 2000 v.3.2 software (Alpha Innotech Co.). The number of VP7 molecules per CLP was calculated by the ratio of the density of VP7 to VP3, with VP3 as the denominator. From left to right, the bars represent wild-type VP7, VP7-E104W, VP7-D318N, and VP7-F268R, respectively.
It is possible that the remaining three VP7 mutants, R111F, W119D, and T321R, were able to assemble into CLPs but were too fragile to tolerate the purification by the CsCl gradient centrifugation. In order to verify this, an alternative method of CLP isolation was used. CLPs were concentrated from the infected cell's supernatants on a discontinuous step gradient of sucrose instead of CsCl gradient. The material after centrifugation was recovered from the interface at the top of the 66% sucrose cushion and examined by EM. As shown in Fig. 7, a few particles were clearly visible in each case, indicating that all three mutants interacted with VP3 and assembled as CLPs, albeit the assembled particles appeared to be much less abundant than native capsomers. Therefore, it was concluded that these mutants have the ability to assemble, although the assembled particles were highly unstable. It is noteworthy that in similar analyses with the CLPs formed by the other three mutants (E104W, D318N, and F268R), the CLPs appeared to have much more VP7 trimer on the surface (data not shown).
FIG. 7.
EM of unpurified CLPs formed by VP7 mutants. Sf cells were coinfected with Ac17BTV-3 and one of the mutant viruses expressing either R111F, T321R, or W1119D. The infected-cell lysates were loaded onto a discontinuous sucrose step gradient (66% [wt/wt]) and 40% [wt/vol]) in 200 mM Tris-HCl (pH 8.0)–150 mM NaCl and centrifuged for 3 h on an SW28 rotor at 26,000 rpm. The interface above the 66% sucrose was collected and visualized by EM as described in Materials and Methods.
Substitution mutations at key residues in VP7 do not perturb trimerization.
Of the six soluble VP7 proteins, most were able to assemble in vivo with the VP3 subcore layer, albeit at various degrees of stability. These variations were likely due to different extents of intermolecular contact within the VP7 trimers. Weakening of these contacts would lead to the formation of unstable oligomers. Therefore, the effects of substitution mutations on VP7 trimer formation were analyzed on each of these recombinant proteins. Since it is not possible to investigate the oligomerization of the expressed proteins in crude extracts, each expressed protein was therefore partially purified from soluble supernatant fractions. Purification of each VP7 mutant was successfully achieved using the same purification regimen previously established for wild-type recombinant VP7 (4). It was assumed, therefore, that the overall biochemical features of each mutant protein were similar to that of the wild-type VP7. Each purified preparation was used for trimerization assay using two different methods, first, SDS-PAGE analysis of unboiled samples that have previously been established to detect VP7 oligomers (25), and second, glycerol gradient sedimentation of the oligomers. Much to our surprise, all VP7 mutants were able to form trimers that could easily be detected by SDS-PAGE, similar to those of wild-type recombinant protein (Table 3). As shown in Fig. 8, not only did E104W, D318N, and F268R form trimers (Fig. 8A) as expected, but also the other three VP7 mutants, R111F, W119D, and T321R, showed dimeric and trimeric bands and at an easily detectable level. However, dimers were not detectable for the first three mutants, indicating that the stable trimers were formed easily. The ability of these mutant VP7 proteins to form oligomers was also confirmed by an alternative glycerol gradient sedimentation. As expected, when analyzed by the gradient fractions and SDS-PAGE, the trimers were detected in each mutant sample, including R111F, W119D, and T321R (Fig. 8B and C). However, dimers and trimers were not segregated on the gradient used. It was therefore assumed that even though the assembled particles formed by these mutants were not stable, these mutants could still form oligomers that were reasonably stable. The data clearly indicate that these substitution mutants were able to form the trimers in solution.
FIG. 8.
Trimerization assay of the wild-type and mutated BTV-10 VP7. Recombinant wild-type BTV-10 VP7 and VP7 mutants were purified and assayed for trimer formation as described in Materials and Methods. (A and B) Coomassie blue-stained SDS-PAGE profiles of boiled and unboiled (RT) VP7 samples. The mutants and wild type (WT) are indicated. The positions of VP7 monomer, dimer, and trimer are indicated. (C) Sedimentation profiles of soluble VP7 protein on glycerol gradients. Following purification, soluble VP7 mutants were subjected to velocity sedimentation analysis on 15-to-30% glycerol gradients to separate VP7 trimers. Fractions from the bottom to the top (left to right) were subjected to SDS-PAGE. An arrow marks the sedimented position of VP7. (a) High-molecular-weight calibration markers (in thousands [K]) consisting of catalase (4 × 58 K = 232 K), lactate dehydrogenase (4 × 36 K = 140 K), and serum albumin (67 K); (b) wild-type VP7; (c) W119D, stained with Coomassie brilliant blue. Lane M shows molecular weight markers (Bio-Rad) for SDS-PAGE. (D) Western analysis of SDS-PAGE of wild-type VP7 and three VP7 mutants as indicated.
The stability of each mutant in comparison to the wild-type VP7 was further assessed by Western blot analysis. By such analysis, it was possible to visualize differences in relative amounts of VP7 oligomers (Fig. 8D). Clearly each of the three mutant proteins exhibited fewer trimeric forms than monomeric and dimeric forms. In contrast, the native VP7 shows much higher amount of trimers. This is probably due to the fact that these mutant VP7 trimers are not as stable as the trimers formed by the wild-type VP7 and that the mutant oligomers disassemble easily.
Data obtained from trimerization studies together with the data obtained from CLP assembly indicate that the formation of trimers by these VP7 derivatives is not sufficient to generate stable VP3-VP7 interaction, probably due to unstable trimer-trimer interaction and CLP assembly. It is therefore likely that the nature (e.g., shape) of VP7 trimers may also play a crucial role in core assembly.
DISCUSSION
The self-assembly of the BTV VP3 and VP7 proteins into empty CLPs, observed after coexpression of their respective genes in insect cells using recombinant baculoviruses (9), provides a useful model for studying some of the BTV protein-protein interactions that may occur during virus assembly. Before the structure of VP7 was determined, a number of regions essential for the interaction of VP7 and VP3 in CLP formation were mapped by site-directed and deletion-insertion mutagenesis (5, 23). It was demonstrated that replacement of the conserved K255 by L or removal of 14 amino acids from the carboxy terminus of BTV-10 VP7 abrogated CLP formation, possibly by interfering with protein folding and/or trimerization.
The crystal structure of VP7 has revealed a highly unusual molecular organization (11). The β-sandwich structure of the top domain of VP7 resembles a jellyroll, similar to that of majority of other viral capsid proteins (11). In contrast, the α-helical bottom domain is not commonly observed in viral capsid proteins. The crystallographic data suggested that the lower domain of VP7 is responsible for interactions with the VP3 layer in the core via the flat hydrophobic area at the base of the VP7 trimers (11). This has recently been confirmed by the X-ray structure of the BTV core, which also revealed how the VP7 layer in the core avoids the need for different protein conformations for trimer-trimer interaction by creating a thin band around the α-helical bottom domain (13).
Site-directed mutagenesis was used to investigate the role of the VP7 lower domain, specifically, helices 5, 6, and 8 and associated loops in CLP assembly. Several amino acid residues in these helices were selected on the basis of their position and conserved nature among the three orbivirus VP7 proteins. Four residues (E104, R111, Y271, and D318), two from helix 5 and one each from helix 6 and 8, respectively, that are conserved among the related orbiviruses were chosen and exchanged for either more bulky residues (E104W or R111F) or uncharged to charged hydrophilic residues (Y271R) or vice versa (D318N). While it was predicted that a mutation at E104 would not affect either the oligomerization or CLP formation due to its position in the molecule, the other three substitution mutations were likely to have some affect on CLP assembly, as these substitutions should weaken the contacts between the helices. Indeed replacing E104 with tryptophan did not affect the formation of VP7 trimers and their interaction with VP3. In the atomic structure (Fig. 9) there are not many close interactions between E104 and the neighboring arginine residues and there was a lot of space to fit the bulky W residue; hence, there was no apparent effect on CLP assembly. On the other hand, loss of a charge at residue 318, i.e., replacement of aspartate by asparagine, had a greater effect on CLP assembly, yielding only CLPs with one-third of the trimer intact. Mutation at R111 had more severe effect on capsid assembly. It is likely that changing the arginine to phenyalanine (Fig. 9) led to loss of two salt links between the subunits and thus resulted in weaker contacts within the trimer. It might have affected the overall nature (e.g., shape) of the trimer, which would have had an indirect effect on the assembly of the VP7 trimers on the VP3 subcore. VP3 subcores are highly unstable when assembled in the absence of VP7 molecules. Therefore, it is not surprising that no band could be visualized on CsCl gradient centrifugation for R111F CLPs. Nevertheless, when the crude soluble products were concentrated on a sucrose cushion, a few assembled CLPs could still be detected by EM, implying that the contact between VP3 and VP7 did occur, although it appeared to be extremely unstable. These three mutant VP7 proteins were all highly soluble and could trimerize in solution. In contrast, the replacement of the conserved tyrosine at residue 271 (helix 6) by arginine resulted in aggregation of the mutated protein. Tyrosine 271, as a part of a hydrophobic cluster (Fig. 9), contacts the neighboring proline (aa 292). The replacement by arginine might have disturbed this hydrophobic region and interfered with the proper folding of VP7 and its interaction with proline (aa 292), a residue that is known to be crucial for folding and stability (28).
Of the second set of mutants, the residue F268 located on helix 6 maintains a close contact with neighboring residues. The residue F268 also rests against a lysine residue at 255 of the hinge region. Similar structural features are associated with residue W119, which is located just beyond helix 5 and in contact with helix 9, although, while both F268R and W119D could still form stable trimers and could be detected by SDS-PAGE, only F268R assembled with VP3 subcore and could be isolated in purified forms. However, the W119D failed to attach strongly onto VP3 subcore, and consequently resultant CLPs could not tolerate the CsCl gradient purification. The long side chain of arginine in F268R probably accommodated this by avoiding the unfavorable interactions with the nearby basic residues, such as K255 or R274, and interacting with Glu115 (Fig. 9) on the neighboring subunit and allowed the monomer-monomer interaction. However, only some, but not all, trimers which could remain attached to VP3 subcores after purification were detectable in CLP preparations. In contrast, W119D was not able to form stable multimers of trimers, indicating that the mutations in this site, although targeted to monomer-monomer contacts, had a profound effect on the stability of CLPs, possibly due to an indirect effect on multimerization of trimers. By changing the tryptophan to aspartate at the interface, the local nonpolar environment (Fig. 9), which is surrounded by methionine (M253) and alanine (A341), would have been disrupted. This could result in the interference on correct oligomerization of VP7, highlighting the role of the VP7 trimer in core assembly. In addition, aspartate could also interact with the nearby lysine 255 via a side chain, causing some alteration in oligomerization. For the last VP7 mutant, a charged hydrophilic arginine residue was substituted for the threonine at position 321, near the C terminus of the molecule. The substitution at this position also affected the CLP stability. This was probably caused by electrostatic repulsion of the two arginine residues (aa 111 and aa 274) (Fig. 9) in close proximity and probably also influenced the trimer-trimer interaction.
The effects of these VP7 mutations on CLP formation clearly indicate that VP7 trimer formation could not accommodate most of the designed point mutations without any functional effect on capsid assembly. The crystallization data have demonstrated that the VP7 molecule, although rigid in part, is yet capable of significant structural rearrangement (3). From our biochemical and biological studies, it appears that the interactions between subunits are highly specific and mostly inflexible, which is not apparent from the VP7 atomic structural analysis. Our studies have clearly identified a series of key residues that are responsible for correct VP7 trimer formation and have revealed their influence on VP7-VP3 interactions during core assembly. The structural analysis revealed that the interactions between the various VP7 trimers are relatively nonspecific, involving a set of hydrophobic residues forming a thin band around the lower domains. There is only a limited contact area in the interfaces, indicating that the trimer-trimer interactions are relatively weak. The instability of the CLPs in these experiments were likely due to the unstable interactions between the various mutant trimers, indicating that subtle alterations (such as bonding angle or subunit conformation) within a trimer could have a profound effect on trimer-trimer interaction, although formation of stable trimers was still achieved. Thus, the formation of VP7 lattice on the core surface requires the exact fitting of 260 VP7 trimers. In addition, the mutations might also have influenced indirectly the interactions between the VP7 trimers and the VP3 subcore by slightly changing the flat surface of the bottom of the trimer. The results obtained in this report have supplemented the atomic structure of VP7 and have generated biochemical and biological information revealing the significance of correct oligomerization of VP7 in core assembly.
ACKNOWLEDGMENTS
We thank D. Stuart and J. Grimes (University of Oxford) for designing the mutations. We are very grateful to Ian M. Jones (IVEM), Peter E. Prevelige, Jr. (UAB), and Vijay Reddy (The Scripps Research Institute, La Jolla, Calif.) for very fruitful discussions during the preparation of the manuscript.
This work was partly funded by grants from the NIH (United States) and the BBSRC (United Kingdom).
REFERENCES
- 1.Bairoch A, Boeckmann B. The SWISSPROT protein sequence bank. Nucleic Acids Res. 1990;19:2247–2248. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basak A K, Gouet P, Grimes J, Roy P, Stuart D I. Crystal structure of the top domain of African horsesickness virus VP7: comparisons with bluetongue virus VP7. J Virol. 1996;70:3797–3806. doi: 10.1128/jvi.70.6.3797-3806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basak A K, Grimes J, Gouet P, Roy P, Stuart D I. Structures of orbivirus VP7: implications for the role of this protein in the viral life cycle. Structure. 1997;5:871–883. doi: 10.1016/s0969-2126(97)00242-6. [DOI] [PubMed] [Google Scholar]
- 4.Basak A K, Stuart D I, Roy P. Preliminary crystallographic study of bluetongue virus capsid protein, VP7. J Mol Biol. 1992;228:687–689. doi: 10.1016/0022-2836(92)90850-j. [DOI] [PubMed] [Google Scholar]
- 5.Belyaev A S, Roy P. Presentation of hepatitis B virus preS2 epitope on bluetongue virus core-like particles. Virology. 1992;190:840–844. doi: 10.1016/0042-6822(92)90922-c. [DOI] [PubMed] [Google Scholar]
- 6.Brown M, Faulkner P. A plaque assay for nuclear polyhedrosis viruses using a solid overlay. J Gen Virol. 1977;36:361–364. [Google Scholar]
- 7.Eaton B T, Gould A R, Hyatt A D, Coupar B E, Martyn J C, White J R. A bluetongue serogroup-reactive epitope in the amino terminal half of the major core protein VP7 is accessible on the surface of bluetongue virus particles. Virology. 1991;180:687–696. doi: 10.1016/0042-6822(91)90082-m. [DOI] [PubMed] [Google Scholar]
- 8.Felgner P L, Gadek T R, Holm M, Roman R, Chan H W, Wenz H, Northrop J P, Ringold G M, Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French T J, Roy P. Synthesis of bluetongue virus (BTV) core-like particles by a recombinant baculovirus expressing the two major structural core proteins of BTV. J Virol. 1990;64:1530–1536. doi: 10.1128/jvi.64.4.1530-1536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouet P, Diprose J M, Grimes J M, Malby R, Burroughs J N, Zientara S, Stuart D I, Mertens P P C. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97:481–490. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 11.Grimes J, Basak A K, Roy P, Stuart D I. The crystal structure of bluetongue virus VP7. Nature. 1995;373:167–170. doi: 10.1038/373167a0. [DOI] [PubMed] [Google Scholar]
- 12.Grimes J M, Jakana J, Ghosh M, Basak A K, Roy P, Chiu W, Stuart D I, Prasad B V V. An atomic model of the outer layer of the bluetongue virus core derived from X-ray crystallography and electron cryomicroscopy. Structure. 1997;5:885–893. doi: 10.1016/s0969-2126(97)00243-8. [DOI] [PubMed] [Google Scholar]
- 13.Grimes J M, Burroughs J N, Gouet P, Diprose J M, Malby R, Zientara S, Mertens P P C, Stuart D I. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 14.Hewat E A, Booth T F, Loudon P T, Roy P. 3-D reconstruction of bluetongue virus core-like particles by cryo-electron microscopy. Virology. 1992;189:10–20. doi: 10.1016/0042-6822(92)90676-g. [DOI] [PubMed] [Google Scholar]
- 15.Iwata H, Chuma T, Roy P. Characterization of the genes encoding two of the major capsid proteins of epizootic haemorrhagic disease virus indicates a close genetic relationship to bluetongue virus. J Gen Virol. 1992;73:915–924. doi: 10.1099/0022-1317-73-4-915. [DOI] [PubMed] [Google Scholar]
- 16.King L A, Possee R D. The baculovirus expression system. A laboratory guide. London, United Kingdom: Chapman & Hall; 1992. [Google Scholar]
- 17.Kitts P A, Possee R D. A method for producing recombinant baculovirus expression vectors at high efficiency. BioTechniques. 1993;14:810–817. [PubMed] [Google Scholar]
- 18.Kowalik T F, Li J K. Sequence analyses and structural predictions of double-stranded RNA segment S1 and VP7 from United States prototype bluetongue virus serotypes 13 and 10. Virology. 1989;172:189–195. doi: 10.1016/0042-6822(89)90120-7. [DOI] [PubMed] [Google Scholar]
- 19.Kowalik T F, Li J K. Bluetongue virus evolution: sequence analyses of the genomic S1 segments and major core protein VP7. Virology. 1991;181:749–755. doi: 10.1016/0042-6822(91)90911-t. [DOI] [PubMed] [Google Scholar]
- 20.Kowalik T F, Li J K, Chuang R Y, Doi R H, Osburn B I. The complete nucleotide and deduced amino acid sequence of the gene encoding the major inner capsid protein, VP7 of US bluetongue virus serotype 17. Nucleic Acids Res. 1990;18:5302. doi: 10.1093/nar/18.17.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 22.Kunkel T A, Roberts J D, Zakuor R A. Rapid and efficient method of site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 23.LeBlois H, Roy P. A single point mutation in the VP7 major core protein of bluetongue virus prevents the formation of core-like particles. J Virol. 1993;67:353–359. doi: 10.1128/jvi.67.1.353-359.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livingstone C, Jones I M. Baculovirus expression vector with single-strand capability. Nucleic Acids Res. 1989;17:2366. doi: 10.1093/nar/17.6.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monastyrskaya K, Staeuber N, Roy P. Effects of domain-switching and site-directed mutagenesis on the properties and functions of the VP7 proteins of two orbiviruses. Virology. 1997;237:217–227. doi: 10.1006/viro.1997.8776. [DOI] [PubMed] [Google Scholar]
- 26.Moss S R, Jones L D, Nuttall P A. Comparison of the major structural core proteins of tick-borne and Culicoides-borne orbiviruses. J Gen Virol. 1992;73:2585–2590. doi: 10.1099/0022-1317-73-10-2585. [DOI] [PubMed] [Google Scholar]
- 27.Oldfield S, Adachi A, Urakawa T, Hirasawa T, Roy P. Purification and characterization of the major group-specific core antigen VP7 of bluetongue virus synthesized by a recombinant baculovirus. J Gen Virol. 1990;71:2649–2656. doi: 10.1099/0022-1317-71-11-2649. [DOI] [PubMed] [Google Scholar]
- 28.Plaxco K W, Spitzfaden C, Campbell I D, Dobson C M. Rapid refolding of a proline-rich all-β-sheet fibronectin type III module. Proc Natl Acad Sci USA. 1996;93:10703–10706. doi: 10.1073/pnas.93.20.10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasad B V V, Yamaguchi S, Roy P. Three-dimensional structure of single-shelled BTV. J Virol. 1992;66:2135–2142. doi: 10.1128/jvi.66.4.2135-2142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy P. Bluetongue virus proteins. J Gen Virol. 1992;73:3051–3064. doi: 10.1099/0022-1317-73-12-3051. [DOI] [PubMed] [Google Scholar]
- 31.Roy P. Orbiviruses and their replication. In: Fields B N, editor. Fields' virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1995. pp. 1709–1734. [Google Scholar]
- 32.Roy P. Orbivirus structure and assembly. Virology. 1996;216:1–11. doi: 10.1006/viro.1996.0028. [DOI] [PubMed] [Google Scholar]
- 33.Roy P, Mertens P P C. Orbiviruses and coltiviruses: molecular biology. In: Granoff A, Webster R G, editors. Encyclopaedia of virology. 2nd ed. London, United Kingdom: Academic Press; 1999. pp. 1062–1074. [Google Scholar]
- 34.Roy P, Hirasawa T, Fernandez M, Blinov V M, Sanchez-Vizcaino Rodriquez J M. The complete sequence of the group-specific antigen, VP7, of African horsesickness virus serotype 4 reveals a close relationship to bluetongue virus. J Gen Virol. 1991;72:1237–1241. doi: 10.1099/0022-1317-72-6-1237. [DOI] [PubMed] [Google Scholar]
- 35.Roy P, Marshall J J A, French T J. Structure of bluetongue virus genome and its encoded proteins. Curr Top Microbiol Immunol. 1990;162:43–87. doi: 10.1007/978-3-642-75247-6_3. [DOI] [PubMed] [Google Scholar]
- 36.Sanger J A, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wade-Evans A M. The complete nucleotide sequence of genome segment 7 of bluetongue virus, serotype 1 from South Africa. Nucleic Acids Res. 1990;18:4919. doi: 10.1093/nar/18.16.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]