ABSTRACT
The canonical arsRBC genes of the ars1 operon in Pseudomonas putida KT2440, which confer tolerance to arsenate and arsenite, are followed by a series of additional ORFs culminating in phoN1. The phoN1 gene encodes an acetyltransferase that imparts resistance to the glutamine synthetase inhibitor herbicide phosphinothricin (PPT). The co‐expression of phoN1 and ars genes in response to environmental arsenic, along with the physiological effects, was analysed through transcriptomics of cells exposed to the oxyanion and phenotypic characterization of P. putida strains deficient in different components of the bifan motif governing arsenic resistance in this bacterium. Genetic separation of arsRBC and phoN1 revealed that their associated phenotypes operate independently, indicating that their natural co‐regulation is not functionally required for simultaneous response to the same signal. The data suggest a scenario of associative evolution, akin to Pavlovian conditioning, where two unrelated but frequently co‐occurring signals result in one regulating the other's response – even if there is no functional link between the signal and the response. Such surrogate regulatory events may provide an efficient solution to complex regulatory challenges and serve as a genetic patch to address transient gaps in evolving regulatory networks.
Keywords: arsenic, herbicide, phosphinothricin, Pseudomonas putida , surrogate regulation
Similar to Pavlov's dog experiment, where two different stimuli can elicit the same response, environmental bacteria exposed to one of two simultaneous and unrelated stimuli may eventually trigger responses to both.
1. Introduction
The default view of how expression of bacterial operons is regulated by physicochemical and nutritional cues includes the action of transcriptional factors (TFs, either activators or repressors) that recognize specifically such signals and in turn either enable or curb transcription of often clustered genes whose products build the physiological response to the upstream inputs (Janga, Salgado, and Martínez‐Antonio 2009; Dudek and Jahn 2021). This general scenario then splits in a large number of particular cases that diverge in significant ways from the rule. For instance, many TFs show a degree of side‐promiscuity towards, for example, gratuitous inducers which make promoters to be turned on/off by a physiologically wrong signal (Abril et al. 1989; de Lorenzo and Pérez‐Martín 1996). In other cases, the evolutionary history of the system makes regulation of a new operon to keep the TF of the earlier precursor and thus maintaining a faulty signal‐response profile (de Las Heras, Chavarría, and de Lorenzo 2011). Finally, some genes are expressed upon signals which proxy the environment where the responses are to unfold but do not produce a direct response to the trigger input. This last scenario is most typical of virulence genes, which often recruit iron‐starvation promoters controlled by the Fur repressor for effective expression in characteristically Fe‐deprived target niches, for example, animal tissues (Mekalanos 1992; Porcheron and Dozois 2015).
The growing availability of bacterial genomes and transcriptomes has revealed a wealth of naturally occurring regulatory architectures whose evolutionary logic is sometimes difficult to grasp (Stormo and Tan 2002). One conspicuous example is the regulation of the two co‐existing As‐resistance operons found in the genome of the Gram‐negative soil bacterium and plant colonizer Pseudomonas putida KT2440 (Figure 1). Ars operons, which are widespread through the bacterial realm (Páez‐Espino et al. 2009), enable tolerance to arsenate (AsV) through reduction to arsenite (AsIII) by a cytoplasmic arsenate reductase ArsC, followed by extrusion of the thereby reduced oxyanion by a cognate efflux pump (see Kruger et al. 2013, for a review). The first oddity of As resistance in P. putida is the redundance of the ars operons: both gene clusters separately deliver high levels of As tolerance, their duplication resulting in only a minor advantage in terms of enduring exposure to the oxyanion at different temperatures (Fernández et al. 2014; Páez‐Espino, Durante‐Rodríguez, and de Lorenzo 2015). The second is the cross‐regulation between their promoters Pars1 and Pars2 by their corresponding regulators ArsR1 and ArsR2 (Fernández et al. 2016), which originate an intriguing bifan motif (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021) of uncertain functionality (Figure 1). Finally, each of the core arsRDC resistance genes of the two clusters is followed by and co‐transcribed with additional ORFs that may or may not have any relationship with arsenic (Páez‐Espino et al. 2009; Páez‐Espino, Durante‐Rodríguez, and de Lorenzo 2015).
FIGURE 1.
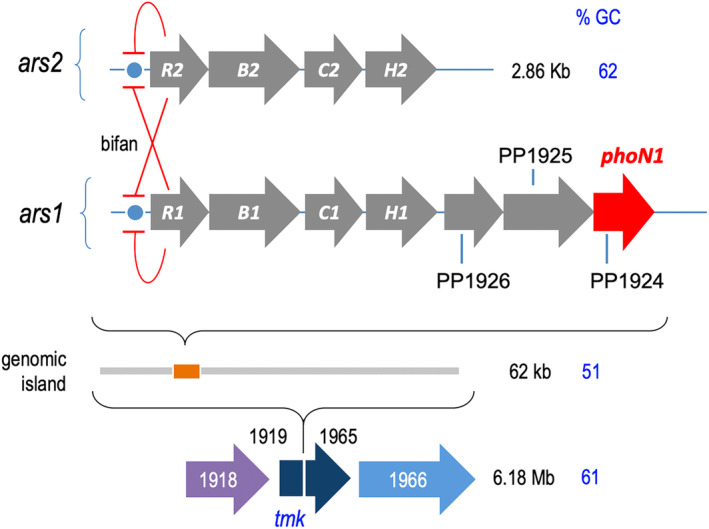
Organization of the two arsenic resistance operons of P. putida KT2440 in respect to phoN1 (PPTR) and regulatory interplay between them. The upper part of the figure shows the arrangement of the 2.86 kbp chromosomal segment of P. putida KT2440 with the As‐resistance ars2 operon with canonical genes R (repressor), B (transporter), C (reductase) and H (NADPH‐dependent flavin mononucleotide reductase). The GC contents accredit its complete assimilation to the rest of the genomic DNA of this bacterium. The arrangement of the ecoparalogous ars2 cluster (Páez‐Espino, Durante‐Rodríguez, and de Lorenzo 2015) is shown below aligned with ars1. The high amino acid sequence similarity through the arsRBCH genes (Páez‐Espino, Durante‐Rodríguez, and de Lorenzo 2015) stops after the H cistron. In the case of ars1, H is followed by 3 co‐transcribed genes, the last of which (phoN1) encodes an acetylase that endows resistance to the herbicide phosphinothricin (Páez‐Espino, Chavarría, and de Lorenzo 2015). The ars1 operon is included in a 62 kbp genomic island inserted in the tmk gene with a clearly different origin (GC content 51%). The distinct bifan motif that rules the regulatory interplay of ars1 and ars2 is sketched between the two schemes.
One of such genes is phoN1 which is located 4 ORFs downstream of the last arsC in the canonical operon (Páez‐Espino, Chavarría, and de Lorenzo 2015), but forming part of the same transcriptional unit (Fernández et al. 2016). phoN1 encodes an acetylase which inactivates the commercial herbicide Glufosinate [phosphinothricin (PPT)], a structural analogue of glutamate that, similarly to antibiotic arsinothricin (Kuramata et al. 2016; Nadar et al. 2019), inhibits glutamine synthetase (Hoerlein 1994). PTT is naturally produced by several species of Streptomyces (Schwartz et al. 2004), but it started to be massively produced by chemical synthesis for selection of genetically modified crops (McElroy and Brettell 1994; Duke and Cerdeira 2010) implanted with bacterial bar or pat genes which endow resistance to the herbicide (Herouet et al. 2005). The location of phoN1 downstream of an arsenic‐response promoter and its plausible induction by the oxyanion is intriguing, as the only known effector of ArsR1 (and ArsR2) is arsenite (Fernández et al. 2014, 2016; Páez‐Espino, Durante‐Rodríguez, and de Lorenzo 2015; Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021). Under this arrangement, one signal triggers responses to two separate and quite distinct environmental challenges, one directly related to the inducing signal (arsenic) and the other not (PPT). Note also that the regulatory cross‐talk between the two As‐resistance operons of P. putida makes mutual control of their respective promoters to shape a distinct bifan motif which is believed to provide a better temporal regulation of signal propagation and filtering noisy signal inputs (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021). Still, the agency of this specific regulatory arrangement, in particular the predicted surrogate regulation of phoN1 expression by environmental arsenic, remains puzzling.
In this work, we have genetically and physiologically dissected the interplay between arsenic tolerance and herbicide resistance in P. putida KT2440 and investigated how the two traits ended up being manifested simultaneously in response to only one of the two signals. For this, we document below the transcriptional association between their cognate genes and its consequences in vivo when having them expressed together or by separate under a heterologous expression system. The results hint at a case of heritable evolutionary memory (Casadesús and D'Ari 2002; Mitchell et al. 2009) embodied in the ars regulatory device. The resulting genetic patch is reminiscent of a Pavlovian‐like learning process (Pearce and Hall 1980; Tagkopoulos, Liu, and Tavazoie 2008; Zhang et al. 2014) in which two simultaneous signals can end up eliciting a response to both of them even if one of the initial stimuli is later missing. The consequences of such patches in the evolution of regulatory networks are discussed.
2. Results and Discussion
2.1. Regulatory Architecture of the Ars Operons Clusters in P. putida KT2440
The high tolerance of P. putida KT2440 to arsenic oxyanions can be traced to the combined action of the products of two separate ars operons encoded in separate locations of the genome of this bacterium (Figure 1). The gene cluster named ars2 consists of a streamlined canonical arsenic resistance module that includes the genes encoded by an arsenite transporter (arsB2) and an arsenate reductase (arsC2). These are preceded of arsR2, which encodes the As (III)‐responsive repressor ArsR2. No other ORFs directly connected to arsenic can be spotted upstream or downstream of such a basic arsRBC cluster. Inspection of the GC contents of the region and tetranucleotide distribution (Páez‐Espino, Durante‐Rodríguez, and de Lorenzo 2015) suggest that such a DNA segment has been assimilated to the core genetic complement of this bacterium for a long time (Figure 1). The genomic context and evolutionary roadmap of the other arsenic resistance gene cluster (ars1) looks however very different. Not only it maps in a location quire distant from ars2 but also its sequence signatures indicate that it was acquired more recently. The ars1 cluster forms part of a large (62 kb) genomic island inserted in the midst of the tmk gene, an occurrence absent in other P. putida isolates (Belda et al. 2016; Wirth et al. 2023). Furthermore, inspection of the genomic region nearby the core arsR1B1C1 cluster reveals the presence of a number of other ORFs which could be co‐expressed with the ars genes and thus suspect of being functionally related. Some of these have been annotated and their role predicted and experimentally verified – for example, arsH (Chen, Bhattacharjee, and Rosen 2015; Páez‐Espino et al. 2020). The most intriguing of them is the one called phoN1 also called arsN1 (Nadar et al. 2019) which encodes an acetyltransferase able to inactivate the glutamine synthetase inhibitor PTT, commercially known as glufosinate. This is a synthetic, simplified derivative of the antibiotic bialaphos, which is naturally produced by some Streptomyces strains. PPT has been widely used as herbicide for the selection of transgenic plants engineered to express the cognate resistance gene bar. Note also that PPT has structural similarity to yet another unrelated antibiotic, arsinothricin, which is produced also by some rare Burkholderia isolates (Kuramata et al. 2016; Nadar et al. 2019). The question thus arises on how and why resistance to a modern herbicide appears together with an otherwise classical operon for arsenic resistance. Furthermore, the fact that both ArsR variants can regulate each other's promoters (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021) could enter a physiological connection between two‐tiered arsenic tolerance and herbicide resistance.
To clarify the regulatory scenario above, we started by inspecting in detail the expression of the ars gene clusters of wild‐type P. putida in the presence or absence of their canonical inducer, arsenite. To this end, we blew up the regions of interest out of the Ars (III)‐responsive transcriptome of P. putida (Figure A1). The data clearly show that the oxyanion induced arsR2B2C2H2 transcription as an apparent stand‐alone single multicistronic operon, with no significant effects on flanking or nearby genes (Figure A2a). In contrast, as shown in detail in Figure 2, a number of genes located downstream of the ars1 core were also co‐induced in response to As (III) in what looked like a single transcript encompassing arsR1B1C1H1‐PP1926‐PP1925‐phoN. The predicted functions of the genes preceding phoN include a phosphatase and a mono‐oxygenase, respectively (Yoshinaga, Cai, and Rosen 2011), but their actual encoded function in As resistance (or other tasks) remains uncertain. In any case, the data indicated that the mRNA that starts at the Pars1 promoter reaches out and finishes at the end of the phoN1 cistron (Figure 2a).
FIGURE 2.
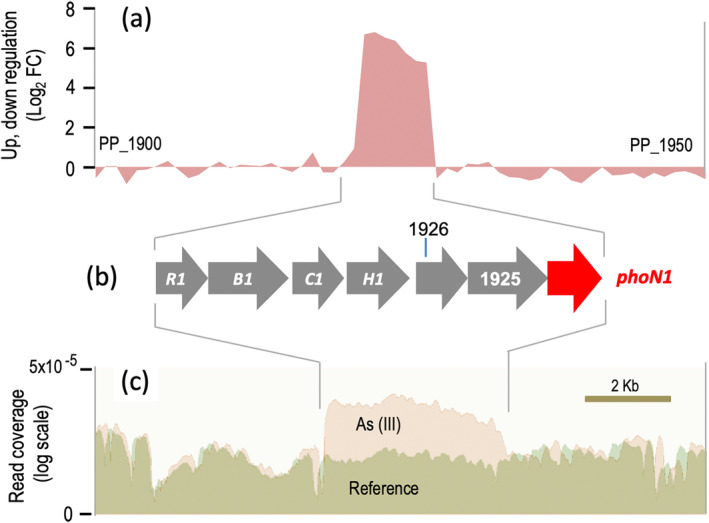
Induction of phoN1 by As (III) along with the rest of the ars1 operon of P. putida as revealed by transcriptomic analysis of genomic region PP_1900‐PP_1950. (a) Sequence coverage plots from transcriptome profile of area between PP_1900 and PP_1950 of P. putida KT2440 genome, that encompasses the ars1 operon. The plot shows relative gene expression intensity of ars1 genes in P. putida KT2440 grown in LB and supplemented with 1 mM arsenite, using cells grown in the absence of the oxyanion as reference conditions. (b) Genomic organization of the ars1 operon in P. putida KT2440 constituted by arsR1 (R1), arsB1 (B1), arsC1 (C1), arsH1 (H1), two genes of unknown function (1925 and 1926) and phoN1 (highlighted in red). (c) RNA sequencing‐based transcriptome profile of ars1 genes in P. putida KT2440. Total number of cDNA reads is annotated responding to the ars genes in the reference (pale green) or arsenite‐exposed cells (pale orange). Geneious software was used for visualization.
To test whether the transcriptional regulatory scenario shown in Figure 1 had its corresponding physiological counterpart, we also inspected resistance of P. putida to PPT in the presence or absence of arsenite. To this end, we grew cultures of the reference strain P. putida (wild‐type, Table 1) in minimal M9 + citrate medium (supplemented with uracil) and added with combinations of the oxyanion and the herbicide ranging 0–10 mM As (III) and 0–10 mM PPT. As shown in Figure 3, bacteria hardly grew in the presence of the herbicide unless the culture was added with subinhibitory concentrations of As (III), maximum growth happening at 2–3 mM of the inducer. Note that higher levels of arsenite are toxic in any case and cells stop growing by As (III) concentrations of 6–7 mM. In contrast, an isogenic strain with a complete deletion of the ars1 operon was sensitive to both PTT and concentrations of As (III) above 2 mM. The residual resistance to the oxyanion could be trailed to the action of the second As‐resistance operon ars2, since deletion of both clusters made P. putida altogether very sensitive to arsenite concentrations above 0.2 mM (Páez‐Espino, Durante‐Rodríguez, and de Lorenzo 2015).
TABLE 1.
Strains and plasmids.
| Relevant characteristics | Reference | |
|---|---|---|
| Strains | ||
| P. putida KT2440 | Prototrophic, TOL plasmid‐cured P. putida mt‐2 | (Belda et al. 2016) |
| P. putida TEC1 | RifR, P. putida KT2440 with an internal deletion of pyrF | (Galvão and de Lorenzo 2005) |
| P. putida ∆ars1 | TEC1 with a seamless deletion of the whole ars1 gene cluster | (Páez‐Espino, Chavarría, and de Lorenzo 2015) |
| P. putida ∆phoN1 | TEC1 with a seamless ∆phoN1 deletion | (Páez‐Espino, Chavarría, and de Lorenzo 2015) |
| P. putida ∆arsR1 | TEC1 with a seamless ∆arsR1 deletion | This work |
| P. putida ∆arsR2 | TEC1 with a seamless ∆arsR2 deletion | This work |
| E. coli CC118λpir | Δ(ara‐leu) araD ΔlacX74 galE galK phoA20 thi‐1 rpsE rpoB argE recA1 λpir lysogen | Laboratory collection |
| E. coli HB101 | Smr; rpsL recA thi pro leu hsdR − M + ( E. coli K‐12/ E. coli B. hybrid) | Laboratory collection |
| E. coli DH5α | F− F80d [lacZ∆M15], ∆(lacZYA‐argF), ∆U169, recA1, endA1, hsdR17, R − M +, supE44, thil, gyrA, relA | Laboratory collection |
| E. coli M15 | K12, ∆M15 lacZ deletion for α complementation | Laboratory collection |
| Plasmids | ||
| pTEC | KmR, FOAS, Ura+, MCS‐Km‐MCS, oriR6K/origin of transfer mobRK2 and pyrF + | (Galvão and de Lorenzo 2005) |
| pTU•DR1 | pTEC inserted with a NotI‐SacI 1.23 kb DNA segment composed of a 0.69 kb fragment upstream of the arsR1 gene and 0.54 kb downstream, composed upon assembly of PCR products resulting from amplification of genomic DNA with oligonucleotide primers FWDR1Up/RVSR1Up and FWDR1Down/RVSR1Down (Table A1). | This work |
| pTU•DR2 | pTEC inserted with a NotI‐SacI 1.15 kb DNA segment composed of a 0.54 kb fragment upstream of the arsR1 gene and 0.61 kb downstream, composed upon assembly of PCR products resulting from amplification of genomic DNA with oligonucleotide primers FWDR2Up/RVSR2Up and FWDR2Down/RVSR2Down (Table A1). |
This work |
| pVLT33 | KmR, RSF1010‐lacI q/P tac hybrid broad‐host‐range expression vector, MCS of pUC18 | (de Lorenzo et al. 1993) |
| pVPPT1 | pVLT33 inserted with a 0.55‐kb EcoRI‐HindIII PCR fragment containing the phoN1 gene of P. putida | (Páez‐Espino, Chavarría, and de Lorenzo 2015) |
| pRK600 | CmR, oriColE1, mobRK2, traRK2 | (Kessler, de Lorenzo, and Timmis 1992) |
| pQE32 | ApR, CmR His6‐tagging, T 5 lac‐driven expression vector. | (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021) |
| pQR32‐ArsR1 | pQE32 inserted with structural arsR1 gene | (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021) |
| pREP4 | KmR, oriV p15A, lacI q , partner of pQE32 for IPTG‐inducible expression | (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021) |
| pSEVA225T | KmR, low copy number vector for ‘lacZ translational fusions | (Martínez‐García et al. 2023) |
| pSEVA225T•R1 | pSEVA225T inserted with a 297 bp EcoRI‐BamHI fragment spanning −150 pb upstream of the transcription initiation site of P ars1 and 78 bp (26 leading amino acids) of the arsR1 structural gene, amplified with PCR primers 5Pars1Eco and 3Pars1Bam RVSR2Down (Table A1). | This work |
FIGURE 3.
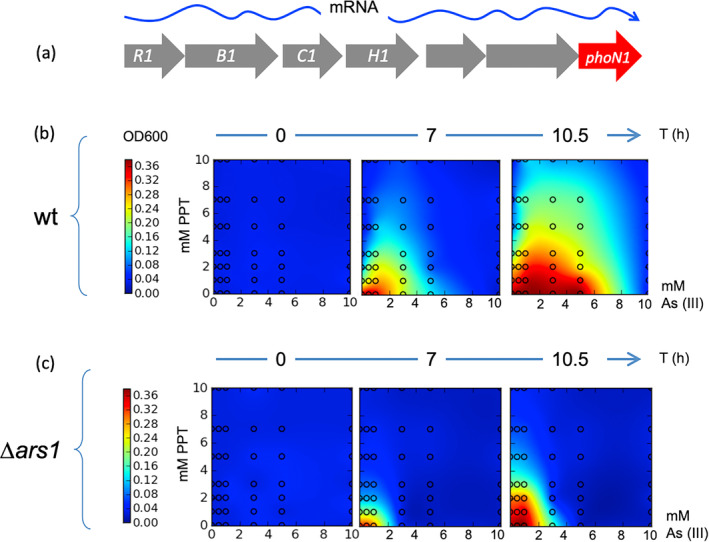
The PPTR phenotype of P. putida relies on induction of the ars1 operon by As (III). (a) Sketch of the polycistronic mRNA starting in arsR1 upon exposure of the cells to As (III) and reaching out the end of the transcript with the phoN1 sequence. (b) Wild‐type P. putida cells were inoculated in M9‐citrate medium (supplemented with uracil, see Experimental Procedures) in microtitre plates and cultures at 30°C for the times indicated in the presence of the PPT and As (III) concentrations shown in each case. Growth was recorded as optical density (OD) at 600 nm and the values were interpolated using the function stat_density2d to generate density heat maps from the R package ggplot2. (c) Same analyses using a complete deletion of the ars1 operon (Δars1 strain).
2.2. PPT Resistance and As Tolerance Are Biologically Independent Traits
To clarify the functional dependence (or lack of it thereof) between herbicide resistance and tolerance to As oxyanions, the genomic region encompassing the phoN1 gene of P. putida wild type was deleted seamlessly from its site in the ars1 operon, thereby generating strain P. putida ∆phoN1 (Table 1 and Páez‐Espino, Chavarría, and de Lorenzo 2015). As shown in Figure 4, using as a reference the concentrations of PPT and As (III) where clearer effects were seen in the experiments of Figure 3, it became clear that phoN1 was responsible for resistance to the herbicide and that its expression to an effective level was elicited by As (III) by means of readthrough transcription from the upstream promoter Pars1. These results not only verified the known function of phoN1 as a PPT resistance gene but also that its efficacy in vivo was dependent on arsenic and that this could be traced to its co‐transcription under the same signals that elicited tolerance to arsenic. The ensuing question was whether the two resistances were physiologically linked, that is, phenotypic manifestation of PPT resistance inherently depends on the presence of arsenic and/or co‐occurrence of ars products. To examine this, we used plasmid pVPTT1, in which transcription of the phoN gene was engineered under the control of an IPTG‐inducible promoter (Table 1). pVPPT1 was then placed in P. putida ∆phoN1 and the resulting strain grown in the presence of different As and PPT concentrations. As evidenced in the heat maps shown in Figure 5, the two phenotypes could be manifested separately by just splitting their regulatory devices. Furthermore, overexpression of PPT resistance upon addition of IPTG to the ∆phoN1 (pVPTT1) strain regardless of As (III) led to cell overgrowth (Figure 5c), thereby ruling out participation of either arsenic itself or any of the other gene products of the ars1 operon otherwise induced by the oxyanion.
FIGURE 4.
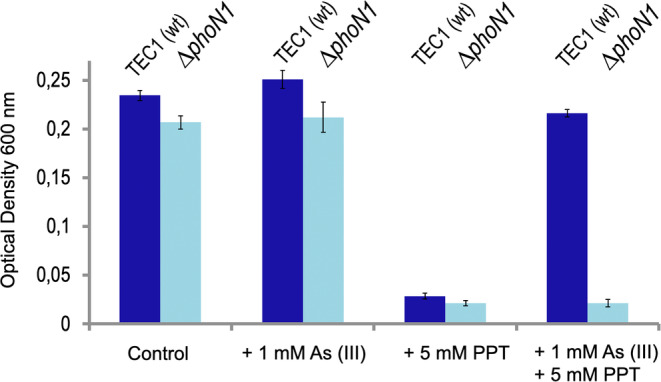
Resistance of wild‐type P. putida TEC1 and its ∆phoN1 derivative to PPT in the presence or absence of As (III). The growth of each strain was followed in 96‐well plates with M9‐citrate (with uracil) and added with As (III) and PPT at the concentrations indicated in each case. The experiments were carried out with biological triplicates and technical duplicates at 30°C. OD600 measurements after 12 h of incubation are shown. Note that manifestation of the PPTR phenotype requires phoN1 and As (III).
FIGURE 5.
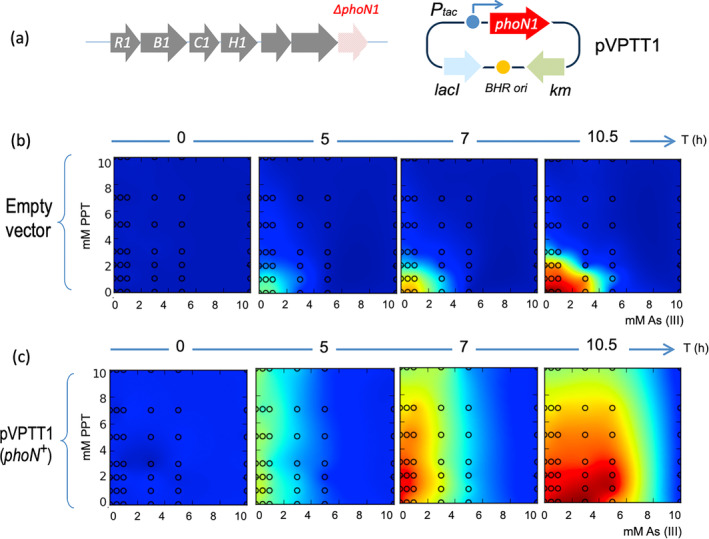
PPT resistance and As (III) tolerance in strains with arsR1B1C1H1 and phoN1 expressed separately in trans. (a) Relevant genetic constructs in test strains. P. putida ∆phoN1 is altogether identical to wild‐type P. putida TEC1 except for the seamless deletion of the phoN1 gene at the end of the ars1 operon. This strain was endowed with either an empty IPTG‐inducible expression vector pVLT33 (Table 1) or the same vector containing the inserted structural phoN1 gene (plasmid pVPTT1 sketched to the right). (b) Growth of P. putida ∆phoN1 (pVLT33) under various PPT and As (III) concentrations. The experiment was run as indicated in the legend to Figure 3, although the medium was added with 1 mM IPTG. (c) Same with strain P. putida ∆phoN1 (pVTPP1). OD600 scale is the same as in Figure 3.
The scenario that emerges from these data is that expression of the herbicide resistance has evolutionarily become subject to a regulatory mechanism that responds to a signal alien to the function of the phoN gene proper, that is, arsenic induces not only resistance to the oxyanion but also to an herbicide whose mechanism of action is completely different and thus lack any cross‐protection or functional synergy. To shed some light on this apparent paradox, we inspected the in vivo consequences of such unusual regulatory setup.
2.3. Arsenic Resistance and Herbicide Tolerance Are Physiologically Intermingled in P. putida
That As tolerance can be genetically and phenotypically separated from PPT resistance says little of the logic behind the regulatory takeover of phoN1 expression by an As‐dependent regulatory device. In this sense, it is worth noting that Pars1 activity (which reaches out phoN1) is controlled not only by its cognate repressor ArsR1 but also cross‐regulated by ArsR2, the equivalent regulator of the paralogous operon ars2 (Figure 1). To sort out the role and plausible interplay of the two regulatory systems with the manifestation of the herbicide resistance phenotype, we built P. putida strains seamlessly deleting arsR1 and arsR2—either together or separated—but keeping intact the Pars1 and Pars2 promoter sequences and their ArsR operators.
The resulting strains were then subject to separate sensitivity tests to either PPT or arsenite. The results of these assays are shown in Figure 6. Expectedly, in the absence of As (III), wild‐type cells and those which had lost only one of the ArsR variants were sensitive to PPT (Figure 6a). However, the double mutant ∆arsR1 ∆arsR2 was tolerant to the herbicide, plausibly due to the unrestrained expression of the whole ars1 operon in the absence of any repressor (Figure 6a). However, the tests for assessing the resistance to As (III) of the same strains revealed that the loss of arsR1 caused a considerable sensitivity to the oxyanion, which was only reverted in the double mutant ∆arsR1 ∆arsR2. This was counterintuitive, as one would expect the loss of just one of the repressors to maintain the resistance level of the wild‐type strain. Instead, the experiment of Figure 6b hinted at a separate role of the ArsR1 protein in As resistance different from its mere action as a transcriptional repressor. But what could that be?
FIGURE 6.
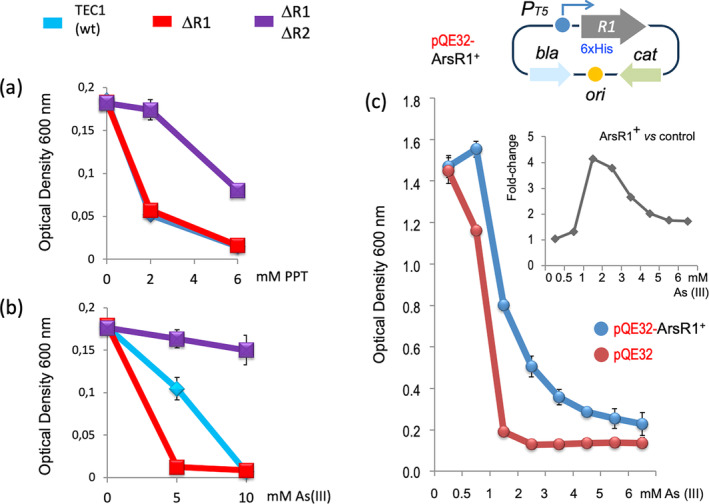
Physiological effects of phoN1 expression regulation by ArsR repressors. (a) Differential sensitivity to PPT of P. putida with or without repressors ArsR1, ArsR2 or both. Cultures inoculated with the strains indicated were grown for 7 h in M9 citrate + uracil medium in the presence of the PPT concentrations indicated. Growth was then recorded as OD600 in duplicated technical and biological replicas. (b) Same strains tested for sensitivity to growing concentrations of As (III). Cultures inoculated with the strains indicated were grown for 7 h in M9 citrate + uracil medium in the presence of the As (III) concentrations indicated. Growth was then recorded as OD600 in duplicated technical and biological replicas. (c) ArsR1 acts as an As metallothionein‐like protein in E. coli . Triplicate cultures of E. coli M15 (pREP4) in LB carrying plasmids pQE32‐Control (red line) or pQE32‐ArsR1 (sketched on top blue line) were grown for 24 h at 37°C in the presence of 0.5 mM IPTG and the specified concentrations of arsenite from 0 to 6 mM. The OD600 was recorded at the end of the period. The insert shows the fold change between the growth of control versus the ArsR1+ strains at the specified arsenite concentrations.
2.4. ArsR1 Contributes to As Tolerance Independently of arsBCH Genes
It has been reported before that overexpression of ArsR of various origins in E. coli allows intracellular accumulation of As and thus increased tolerance to the oxyanion (Kostal et al. 2004; Maleki and Shahpiri 2022). Since the fully‐derepressed Pars1 promoter is strong (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021) and the ribosomal‐binding site (RBS) sequence of the arsR1 gene is also good (Páez‐Espino, Durante‐Rodríguez, and de Lorenzo 2015), chances are that ArsR1 is produced at high levels in cells exposed to the oxyanion. To verify this, we built a translational fusion between the leading 26 triplets of arsR1 and lacZ, including Pars1 and preceded by −150 nt upstream of the transcription start site. The resulting plasmid (pSEVA225T•R1, Table 1) was then placed in P. putida cells. These were grown in LB with or without added 0–20 mM As and β‐galactosidase measured after 5 h. (Figure A3), we hypothesized that the abundance of the regulator could provide an extra layer of protection due to its inherent ability to bind arsenite and thus act as trap for otherwise loose intracellular ions (Kostal et al. 2004; Maleki and Shahpiri 2022). To test this possibility as an inherent, host‐independent biological activity, arsR1 was cloned in vector pQE32, originating pQE32‐ArsR1 (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021), where a LacIq‐controlled T5 lac promoter drives expression of a His6‐tagged ArsR1 protein. Plasmids were then separately transformed in E. coli M15 strain, bearing compatible LacIq+ plasmid pREP4 (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021). Transformants were grown in LB medium with antibiotics for securing plasmid retention, induced with IPTG and then with various concentrations of As (III). As shown in Figure 6c, at low mM As (III), cells expectedly loaded with ArsR1 were more tolerant to the chemical stressor than the control without the expression device. ArsR1 thus endowed E. coli with a superior endurance to arsenic which was independent of its role as a transcriptional regulator. Therefore, it appears that ArsR1 can functionally behave as a sort of As metallothionein‐like protein likely by binding the arsenite in the cytoplasm of the cell and improving tolerance to the oxyanion. Taken together, these results provide a rationale to the observation that the arsR1 deletion of P. putida is more sensitive to arsenic than the wild‐type strain while still exhibiting resistance to PPT. That the double mutant ∆arsR1 ∆arsR2 shows resistance to the oxyanion (Figure 6b) could be due to the unrestrained expression of the second operon ars2 in cells lacking any regulation. One way or the other, the core arsRBCH modules of each cluster seems to have evolved for synergically merging the potential of each of the activities encoded, whereas phoN1 appears as an add‐on which benefits from the upstream transcriptional flow but without any apparent functional connection to the ars genes. But how could this come about?
2.5. Surrogate Regulation: A Case of Pavlovian‐Reminiscent Evolutionary Process
The genome of P. putida contains two recognizable operators for binding ArsR repressors (Figure A4), which correspond to gene clusters ars1 and ars2 (Figure 1). Also, the chromosome encodes two distinct genes for dealing each with two known and widely used herbicides. These are PPT (resistance encoded by phoN1) and methionine sulfoximine (MetSox), another glutamine synthetase inhibitor resistance encoded by phoN2 (Páez‐Espino, Chavarría, and de Lorenzo 2015). The evolutionary phylogeny of each of them is likely to stem from acetylases active on structurally related natural compounds produced by soil bacteria. Specifically, PPT is a structural analogue of yet another As‐containing antibiotic (arsinothricin) produced by a rice rhizosphere‐associated Burkholderia strain (Kuramata et al. 2016; Nadar et al. 2019), which may have acted as a primary driver of evolutionary phoN1 emergence. Yet, while phoN2 is expressed constitutively (Páez‐Espino, Chavarría, and de Lorenzo 2015), phoN1 is transcribed only upon exposure of cells to arsenic salts. It is also intriguing that phoN1‐type PPT acetyl transferases may or may not appear in bacterial genomes associated with As‐related genes (VanDrisse, Hentchel, and Escalante‐Semerena 2016), thereby suggesting autonomous evolutionary trajectories. On these bases, we entertain that the connection between As resistance and PPT resistance observed in P. putida may stem from an earlier evolutionary contingency. The scenario could be one in which soil Pseudomonads bearing the arsRBCH cluster were simultaneously exposed to inorganic arsenic salts and either PPT (Schwartz et al. 2004) or arsinothricin (Kuramata et al. 2016; Nadar et al. 2019) in sites inhabited by Streptomyces and Burkholderia strains naturally producing one or both herbicides. Such a niche‐specific pressure may have caused that the signal for triggering transcription of ars genes ends up being co‐opted for phoN1 expression. Once this association emerged, it is conceivable that it later propagated and further evolved upon intensive exposure to synthetic counterparts of the herbicides. This is not a mere speculation, as inorganic arsenicals have not only been used in agriculture as pesticides or defoliants for many years (Bencko and Yan Li Foong, 2017) but also contaminated soils as a result of fallout in mining operations (Walsh, Sumner, and Keeney 1977). It is plausible that such soils were later treated with chemically synthesized PPT, thereby creating an extra pressure for stabilization of the extant regulatory architecture that we see in P. putida , which could be later propagated through its incorporation to a mobile DNA segment (Figure 1).
The scenario for the development of the hereby described surrogate regulation phenomenon is reminiscent of an associative learning process in which a simultaneous response to two unrelated challenges are triggered by just one of the stimuli – if that happen to co‐occur with the other. Following Pavlov's terminology, arsenic would be both an unconditional stimulus (because the logic reaction to it would be transcription of the As resistance genes) as well as a conditional stimulus for expression of PPT resistance, because response to herbicide is conditional upon the association between the herbicide and the As salt. Under this frame (Figure 7c,d), conditioning herbicide resistance to arsenic is entertained to result of a contingent concurrence in time and space between the As salts and PPT, thereby resulting in an evolutionary case of positive conditioning. Figure 7 summarizes this scenario, which by no means may be exclusive of the particular case at stake. It is plausible that new genes systematically leverage heterologous transcriptional signals for expression and can benefit from co‐opting other triggering signals that act as proxies for the genuine target of the corresponding activity. If such co‐occurrence persists, the regulatory device may be fixed as a surrogate control. In contrast, if they diverge, each function might evolve different regulatory architectures. Transcriptional networks, for example, those involved in virulence (Mekalanos 1992; Porcheron and Dozois 2015), reveal abundant control schemes in which responses to an environmental cue trigger expression of a very specific virulence factor. Surrogate regulation of the sort hereby described is thus one more evolutionary mechanism for solving multi‐objective optimization challenges.
FIGURE 7.
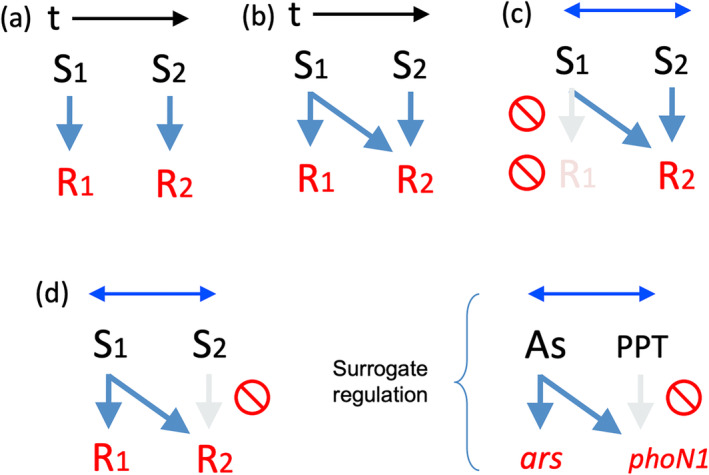
Evolutionary scenarios for emergence of surrogate Pavlovian‐type regulatory nodes. (a) Independent stimuli (S1 S2) at subsequent times generate unrelated responses (R1 R2). (b) Anticipatory regulation: The earlier stimulus activates simultaneously an early response for dealing with S1 as well as another response R2 for a stimulus that will come later, that is, the system embodies a memory device that anticipates S2, even before S2 materializes (Mitchell et al. 2009). (c) Associative Pavlovian response: Two simultaneous but unrelated stimuli trigger the same response to just one of them. (d) Surrogate Pavlovian‐type response: one of the two simultaneous and unrelated stimuli trigger responses to both. This is the scenario that we entertain for the As (III)‐dependent expression of phoN1 in P. putida.
3. Experimental Procedures
3.1. Materials, Culture Conditions and General Procedures
The bacterial strains and the plasmids used in this work are listed in Table 1. Pseudomonas putida was grown at 30°C with vigorous shaking (170 rpm) in rich LB or M9 minimal medium (Sambrook and Rusell 2001) with 2 mM MgSO4 and 0.2% (w/v) citrate as the sole carbon source. E. coli was grown in LB medium at 37°C. When required, antibiotics were added to the medium: kanamycin (Km, 50 μg/mL), ampicillin (100 μg/mL) and chloramphenicol (Cm, 30 μg/mL). Isopropyl‐1‐thio‐β‐galactopyranoside (IPTG) was added at a concentration of 1 mM where indicated for activating the lacI q /P tac expression system of plasmids derived from vector pVLT33 or pQE32 (Table 1). For growing the ∆pyrF reference strain P. putida TEC1 strain and its derivatives, uracil (Sigma Aldrich) was added to all media at 20 μg/mL. Where required, plasmids were transferred from E. coli donors to P. putida recipients as explained (Páez‐Espino, Durante‐Rodríguez, and de Lorenzo 2015). β‐Galactosidase levels of P. putida cells bearing an arsR1’‐‘lacZ translational fusion were measured as indicated (Durante‐Rodríguez, Páez‐Espino, and de Lorenzo 2021).
3.2. Directed Genomic Deletions
Seamless deletion mutants of the arsR1 and arsR2 genes of P. putida TEC1 were made with a reported method (Galvão and de Lorenzo 2005) with delivery plasmids pTU•DR1 and pTU•DR2, respectively the inserts of which bear the boundaries of the desired deletions. These plasmids were separately transferred from donor E. coli CC118λpir to P. putida TEC1 strain (Table 1) by tripartite conjugation. Genomic co‐integration, was followed by resolution of the co‐integrates with fluoroorotic acid (FOA) as described (Galvão and de Lorenzo 2005). The accuracy of the resulting deletions was verified by PCR of the FOAR P. putida clones with the upstream and downstream primers indicated in Appendix A. The double deleted strain P. putida ∆arsR1 ∆arsR2 was built by successive deletion of one gene after the other with the same method (Galvão and de Lorenzo 2005).
3.3. Sensitivity Tests
Herbicide resistance experiments were done in M9 medium with citrate as sole carbon source, supplemented with uracil as mentioned above and added with the concentration the inhibitory compound indicated in each case. Tests were performed by growing strains under examination in 96‐well plates and with biological triplicates and technical duplicates for each condition tested. Similarly, sodium arsenite (NaAsO₂ from Sigma Aldrich Chemicals) was added to the cultures in the same microtitre plate format at the levels marked for each experiment.
3.4. Transcriptomics
P. putida KT2440 cells were grown at 30°C in LB medium (Sambrook and Rusell 2001). Overnight cultures were diluted with fresh media to OD600 of 0.05, in triplicate, and supplemented with 1 mM NaAsO₂(treated) or without arsenite (control) and incubated at 30°C with aeration until mid‐exponential phase (OD600 of 0.6). Cells were harvested by centrifugation at 13000 × g for 5 min at 4°C. Total RNA was extracted from the cell pellets by using the High Pure Isolation Kit (Roche) following the protocol established by the manufacturer. RNA concentration was determined spectrophotometrically with a Nanodrop and genomic contamination was analysed by using the RNA as template in a PCR reaction. RNA final concentration and purity was confirmed by gel electrophoresis and by using an RNA 6000 Nano kit in an Agilent 2100 Bioanalyzer (Agilent Technologies), respectively. The integrity of the RNA was estimated at the Genomic Service of Complutense University (Madrid). After the optimal quality was determined as optimum (RNA integrity number [RIN] > 7), the triplicates of each conditions (control and treatment) were mixed and RNAseq was performed with total RNA obtained (2.3 μg for control sample and 2.7 μg for treatment sample) by Illumina platform at Macrogen (Seoul, South Korea), using their pipeline protocols that follow previously described techniques (Martin and Wang 2011). Briefly, after PCR‐amplifying fragments from the obtained cDNA fragments with insert sizes between 200 and 400 bp were selected for paired‐end sequencing. The cDNA sequences fragments obtained from were mapped using P. putida KT2440 genome as a reference (GCA_000007565.2).
3.5. Data Analyses
Density heat maps of Figures 3 and 5 were generated using the ggplot2 package in R. The stat_density2d function was used to visualize the density distribution of data points. Expression profiles were calculated for each sample and gene as read count. DEG (Differentially Expressed Genes) analysis was performed on four comparisons pairs as requested using edgeR software (Robinson, McCarthy, and Smyth 2010). The results showed 442 genes which satisfied |FC| ≥ 2 and exactTest raw p value < 0.05 conditions in at least one of comparison pairs. To reduce biases in analysis, artefacts such as low‐quality reads, adaptor sequence, contaminant DNA or PCR duplicates are removed. Transcript abundances were measured with mapped read count within gene region. In case of strand specific library, sense or anti‐sense read counts are extracted by strand. Expression profiles are represented as read count and normalization value which is based on the transcript length and depth of coverage. The FPKM (fragments per kilobase of transcript per million mapped reads) values were used as a normalization value.
Author Contributions
David Paez‐Espino: conceptualization, investigation, methodology, formal analysis, software. Gonzalo Durante‐Rodríguez: methodology, investigation. Elena Alonso Fernandes: investigation, methodology. Manuel Carmona: investigation, methodology, software. Victor de Lorenzo: conceptualization, funding acquisition, investigation, writing – original draft, writing – review and editing, formal analysis, supervision, resources.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgements
This work was funded by the NYMPHE (HORIZON‐CL6‐2021‐UE 101060625) Contract of the European Union, the BIOSINT‐CM (Y2020/TCS‐6555) Project of the Comunidad de Madrid‐European Structural and Investment Funds (FSE, FECER) and Project PID2022‐142540OB‐100 of the Spanish Ministerio de Ciencia, Educación y Universidades.
Appendix A.
FIGURE A1.
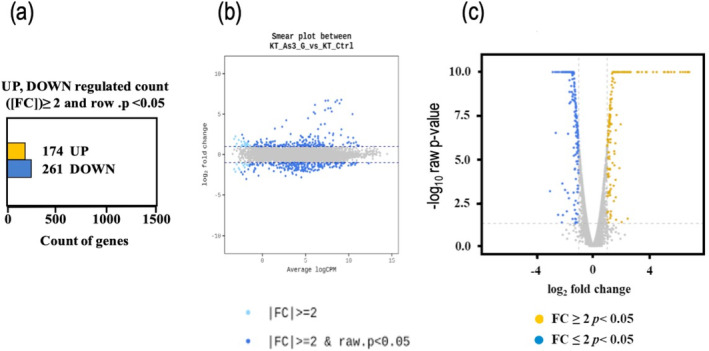
Changes in the P. putida KT2440 transcriptome upon exposure to As (III). Representations of the number of genes regulated by arsenite in P. putida KT2440. (a) Overall number of up and down regulated genes based on fold change. (b) Smear plot of transcriptomic changes of P. putida KT2440 in response to arsenite. (c) Log2 fold change and p‐value obtained from the comparison between two groups plotted as volcano representation. BioProject ID Reference: PRJNA1101689. BioSample accessions SAMN41000924, SAMN41000925.
FIGURE A2.
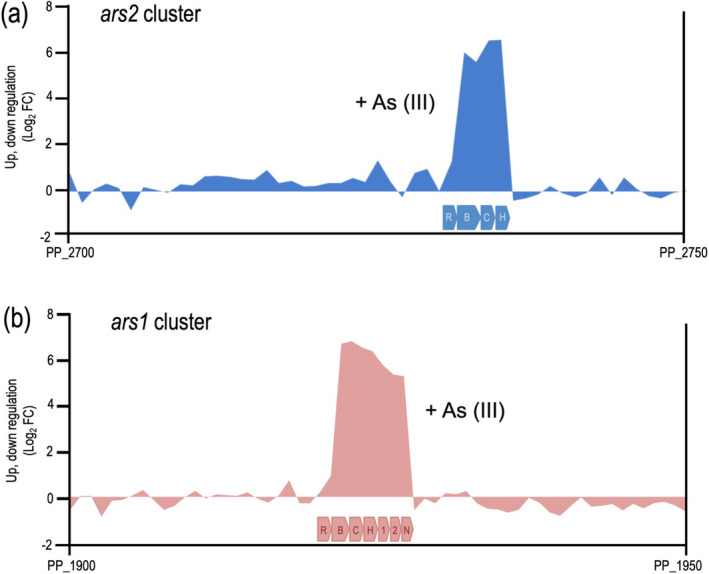
Detail of the gene expression profile through the genomic segments of P. putida KT2440 encompassing the ars2 and ars1 gene clusters. Sequence coverage plots from transcriptome profile of P. putida KT2440 ars genes. (a) Relative gene expression intensity of ars2 genes in P. putida KT2440 grown in LB and supplemented with 1 mM As (III), using cells grown in the absence of As (III) as reference conditions. Blue boxes represent the ars2 operon represented by arsR2 (R), arsB2 (B), arsC2 (C) and arsH2 (H) genes. (b) Relative gene expression intensity of ars1 genes in P. putida KT2440 grown in LB and supplemented with 1 mM As (III), using cells grown in the absence of As (III) as reference conditions. Pale orange boxes represent the ars1 operon represented by arsR1 (R), arsB1 (B), arsC1 (C) and arsH1 (H), two genes with unknown function (1 and 2), and phoN (N). Data replotted for comparison from Figure 2 of main text.
FIGURE A3.
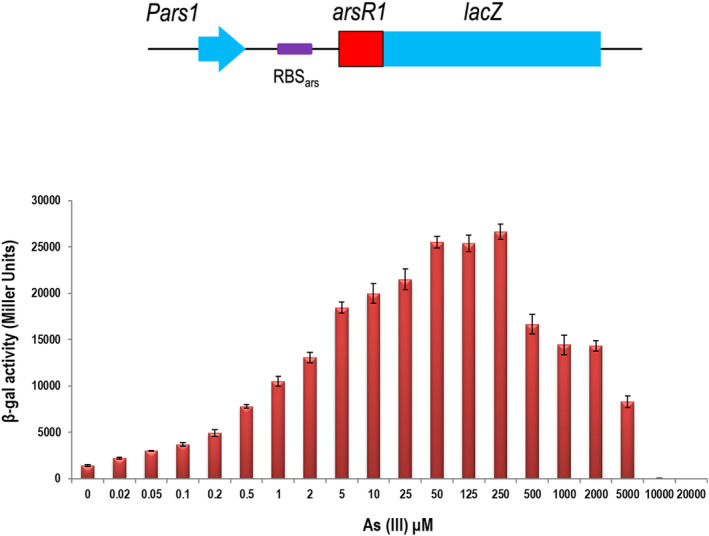
Responsiveness of a low copy plasmid‐borne translational arsR1’‐‘lacZ to As (III). P. putida KT2440 transformed with pSEVA225T•R1 (Table 1) bearing the insert sketched above was grown at 30°C in LB medium with increasing concentrations of arsenite as indicated until the mid‐ exponential culture phase, at which point β‐galactosidase activity was measured and expressed in Miller units.
FIGURE A4.
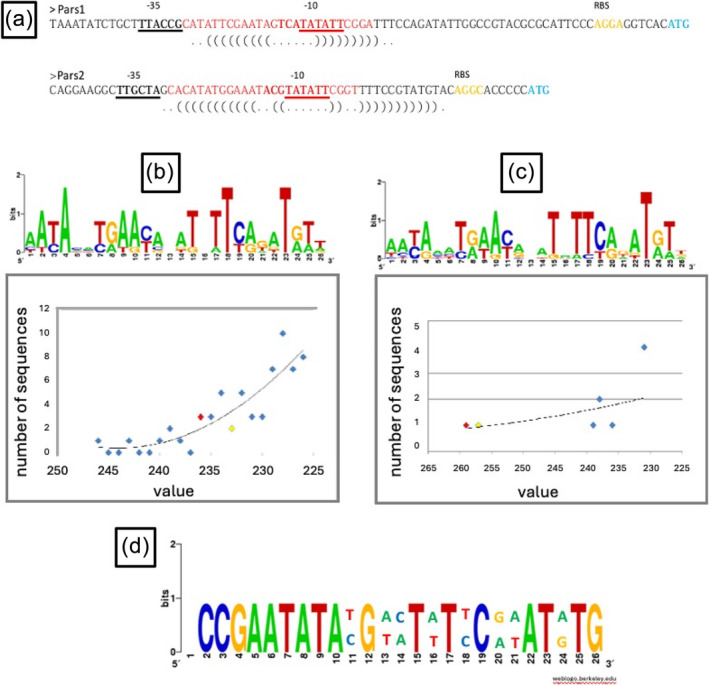
In silico analysis of ArsR1‐ and ArsR2‐binding sites to intergenic regions of the P. putida KT2440 genome. (a) Putative promoters Pars1 and Pars2. The regions corresponding to the −35 and −10 boxes (sigma‐70 transcription factor‐dependent promoters) for each case are underlined. The first amino acid (Met) of the arsR regulators is indicated in blue. The RBS regions are indicated in orange, and the operator region for each case, based on the consensus for the SmtB/ArsR family (tgtgATTTAATCATATG CG TTTTTGGTTATGtgtt) is shown in red. Predictions were made using the BPROM program from the SoftBerry package. Parentheses indicate the palindromic region using the RNAfold program. (b) DNA‐binding region consensus using the 7 homologs of the SmtB/ArsR family. Consensus sequence representations were generated using WebLogo program (http://weblogo.berkeley.edu/). The graph below indicates the number of intergenic sequences (12.5% of the total genome) in P. putida KT2440 relative to the identical number of bases pairs to the consensus (in arbitrary units). The higher the value, the greater the number of bases identical to the consensus. The intergenic regions were extracted using the coderet application and the similarity matrix of the characterized homologs used to search in the non‐ coding genome of P. putida was generated using the prophecy application. The group including Pars1 is shown in red, and Pars2 in yellow. (c) Pars1 and Pars2 are included along with the 7 previous homologs from panel (b). The graph shows that both Pars1 and Pars2 have, by far, the highest values, indicating their specificity for their promoter regions. (d) Identity between ars operons in P. putida KT2440, consistent with the observed cross‐regulation between the two ars systems.
TABLE A1.
Oligonucleotides used in this work.
| Name | Sequence 5′‐3′ |
|---|---|
| FWDR1Up | CCACCAGCGGCCGCTCCTGGGACACCTGAGAACGAACTC |
| RVSR1Up | ACTACAACTCAATCAGCGAAGGGAAGTCGTGACCTCCTGGGAATGCGCGTA |
| FWDR1Down | GACTTCCCTTCGCTGATTGAGTTGTAGT |
| RVSR1Down | GTCCCGAGCTCCGTACTCGCTGAAACCGATGCCGAA |
| FWDR2Up | CCACCTGCGGCCGCACCGAATACACGGGTGAACTGCCG |
| RVSR2Up | GCAGCATGAAAATCTCGCTTGGTGATGAGGGGGTGCCTGTACATACGGAAAAC |
| FWDR2Down | TCATCACCAAGCGAGATTTTCATGCTGC |
| RVSR2Down | GTCCCGAGCTCTACAGGAATAGCACCAGCAGGGTGG |
| 5 Pars1 Eco | GCGAATTCTGATCGGTACCAAGCAATCGG |
| 3 Pars1 Bam | AGAGGATCCATCAGCAGGGTCATCCGGGC |
Funding: This study was funded by the NYMPHE (HORIZON‐CL6‐2021‐UE 101060625) Contract of the European Union, the BIOSINT‐CM (Y2020/TCS‐6555) Project of the Comunidad de Madrid‐European Structural and Investment Funds (FSE, FECER) and Project PID2022‐142540OB‐100 of the Spanish Ministerio de Ciencia, Educación y Universidades.
Data Availability Statement
Transcriptomic data presented in the study are deposited in the NCBI repository, accession numbers SAMN41000924 and SAMN41000925; BioProject PRJNA1101689: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1101689.
References
- Abril, M. A. , Michan C., Timmis K. N., and Ramos J. L.. 1989. “Regulator and Enzyme Specificities of the TOL Plasmid‐Encoded Upper Pathway for Degradation of Aromatic Hydrocarbons and Expansion of the Substrate Range of the Pathway.” Journal of Bacteriology 171: 6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda, E. , van Heck R. G., José Lopez‐Sanchez M., et al. 2016. “The Revisited Genome of Pseudomonas putida KT2440 Enlightens Its Value as a Robust Metabolic Chassis.” Environmental Microbiology 18: 3403–3424. [DOI] [PubMed] [Google Scholar]
- Bencko, V. , and Yan Li Foong F.. 2017. “The History of Arsenical Pesticides and Health Risks Related to the Use of Agent Blue.” Annals of Agricultural and Environmental Medicine: AAEM 24: 312–316. [DOI] [PubMed] [Google Scholar]
- Casadesús, J. , and D'Ari R.. 2002. “Memory in Bacteria and Phage.” BioEssays 24: 512–518. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Bhattacharjee H., and Rosen B. P.. 2015. “ArsH is an Organoarsenical Oxidase That Confers Resistance to Trivalent Forms of the Herbicide Monosodium Methylarsenate and the Poultry Growth Promoter Roxarsone.” Molecular Microbiology 96: 1042–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Las Heras, A. , Chavarría M., and de Lorenzo V.. 2011. “Association of DNT Genes of Burkholderia sp. DNT with the Substrate‐Blind Regulator DntR Draws the Evolutionary Itinerary of 2,4‐Dinitrotoluene Biodegradation.” Molecular Microbiology 82: 287–299. [DOI] [PubMed] [Google Scholar]
- de Lorenzo, V. , Eltis L., Kessler B., and Timmis K. N.. 1993. “Analysis of Pseudomonas Gene Products Using laciq/Ptrp‐Lac Plasmids and Transposons That Confer Conditional Phenotypes.” Gene 123: 17–24. [DOI] [PubMed] [Google Scholar]
- de Lorenzo, V. , and Pérez‐Martín J.. 1996. “Regulatory Noise in Prokaryotic Promoters: How Bacteria Learn to Respond to Novel Environmental Signals.” Molecular Microbiology 19: 1177–1184. [DOI] [PubMed] [Google Scholar]
- Dudek, C.‐A. , and Jahn D.. 2021. “PRODORIC: State‐of‐the‐Art Database of Prokaryotic Gene Regulation.” Nucleic Acids Research 50: D295–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke, S. O. , and Cerdeira A. L.. 2010. “Transgenic Crops for Herbicide Resistance.” In Transgenic Crop Plants, Berlin‐Heidelberg: Springer Verlag, vol. 10, 133–166. [Google Scholar]
- Durante‐Rodríguez, G. , Páez‐Espino D., and de Lorenzo V.. 2021. “A Bifan Motif Shaped by ArsR1, ArsR2, and Their Cognate Promoters Frames Arsenic Tolerance of Pseudomonas putida .” Frontiers in Microbiology 12: 641440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, M. , Morel B., Ramos J. L., and Krell T.. 2016. “Paralogous Regulators ArsR1 and ArsR2 of Pseudomonas putida KT2440 as a Basis for Arsenic Biosensor Development.” Applied and Environmental Microbiology 82: 4133–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, M. , Udaondo Z., Niqui J. L., Duque E., and Ramos J. L.. 2014. “Synergic Role of the Two Ars Operons in Arsenic Tolerance in Pseudomonas putida KT2440.” Environmental Microbiology Reports 6: 483–489. [DOI] [PubMed] [Google Scholar]
- Galvão, T. C. , and de Lorenzo V.. 2005. “Adaptation of the Yeast URA3 Selection System to Gram‐Negative Bacteria and Generation of a ∆betCDE Pseudomonas putida Strain.” Applied and Environmental Microbiology 71: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herouet, C. , Esdaile D. J., Mallyon B. A., et al. 2005. “Safety Evaluation of the Phosphinothricin Acetyltransferase Proteins Encoded by the Pat and Bar Sequences That Confer Tolerance to Glufosinate‐Ammonium Herbicide in Transgenic Plants.” Regulatory Toxicology and Pharmacology 41: 134–149. [DOI] [PubMed] [Google Scholar]
- Hoerlein, G. 1994. “Glufosinate (Phosphinothricin), a Natural Amino Acid With Unexpected Herbicidal Properties.” Reviews of Environmental Contamination and Toxicology 138: 73–145. [DOI] [PubMed] [Google Scholar]
- Janga, S. C. , Salgado H., and Martínez‐Antonio A.. 2009. “Transcriptional Regulation Shapes the Organization of Genes on Bacterial Chromosomes.” Nucleic Acids Research 37: 3680–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, B. , de Lorenzo V., and Timmis K. N.. 1992. “A General System to Integrate lacZ Fusions Into the Chromosomes of Gram‐Negative Eubacteria: Regulation of the Pm Promoter of the TOL Plasmid Studied With All Controlling Elements in Monocopy.” Molecular and General Genetics 233: 293–301. [DOI] [PubMed] [Google Scholar]
- Kostal, J. , Yang R., Wu C. H., Mulchandani A., and Chen W.. 2004. “Enhanced Arsenic Accumulation in Engineered Bacterial Cells Expressing ArsR.” Applied and Environmental Microbiology 70: 4582–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger, M. C. , Bertin P. N., Heipieper H. J., and Arsène‐Ploetze F.. 2013. “Bacterial Metabolism of Environmental Arsenic—Mechanisms and Biotechnological Applications.” Applied Microbiology and Biotechnology 97: 3827–3841. [DOI] [PubMed] [Google Scholar]
- Kuramata, M. , Sakakibara F., Kataoka R., et al. 2016. “Arsinothricin, a Novel Organoarsenic Species Produced by a Rice Rhizosphere Bacterium.” Environmental Chemistry 13: 723–731. [Google Scholar]
- Maleki, F. , and Shahpiri A.. 2022. “Efficient and Specific Bioaccumulation of Arsenic in the Transgenic Escherichia coli Expressing ArsR1 From Corynebacterium glutamicum .” Biometals 35: 889–901. [DOI] [PubMed] [Google Scholar]
- Martin, J. A. , and Wang Z.. 2011. “Next‐Generation Transcriptome Assembly.” Nature Reviews. Genetics 12: 671–682. [DOI] [PubMed] [Google Scholar]
- Martínez‐García, E. , Fraile S., Algar E., et al. 2023. “SEVA 4.0: an Update of the Standard European Vector Architecture Database for Advanced Analysis and Programming of Bacterial Phenotypes.” Nucleic Acids Research 51: D1558–d1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy, D. , and Brettell R. I.. 1994. “Foreign Gene Expression in Transgenic Cereals.” Trends in Biotechnology 12: 62–68. [Google Scholar]
- Mekalanos, J. J. 1992. “Environmental Signals Controlling Expression of Virulence Determinants in Bacteria.” Journal of Bacteriology 174: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, A. , Romano G. H., Groisman B., et al. 2009. “Adaptive Prediction of Environmental Changes by Microorganisms.” Nature 460: 220–224. [DOI] [PubMed] [Google Scholar]
- Nadar, V. S. , Chen J., Dheeman D. S., et al. 2019. “Arsinothricin, an Arsenic‐Containing Non‐Proteinogenic Amino Acid Analog of Glutamate, is a Broad‐Spectrum Antibiotic.” Communications Biology 2: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Páez‐Espino, A. D. , Chavarría M., and de Lorenzo V.. 2015. “The Two Paralogue phoN (Phosphinothricin Acetyl Transferase) Genes of Pseudomonas putida Encode Functionally Different Proteins.” Environmental Microbiology 17: 3330–3340. [DOI] [PubMed] [Google Scholar]
- Páez‐Espino, A. D. , Durante‐Rodríguez G., and de Lorenzo V.. 2015. “Functional Coexistence of Twin Arsenic Resistance Systems in Pseudomonas putida KT2440.” Environmental Microbiology 17: 229–238. [DOI] [PubMed] [Google Scholar]
- Páez‐Espino, A. D. , Nikel P. I., Chavarría M., and de Lorenzo V.. 2020. “ArsH Protects Pseudomonas putida From Oxidative Damage Caused by Exposure to Arsenic.” Environmental Microbiology 22: 2230–2242. [DOI] [PubMed] [Google Scholar]
- Páez‐Espino, D. , Tamames J., Lorenzo V., and Cánovas D.. 2009. “Microbial Responses to Environmental Arsenic.” Biometals 22: 117–130. [DOI] [PubMed] [Google Scholar]
- Pearce, J. M. , and Hall G.. 1980. “A Model for Pavlovian Learning: Variations in the Effectiveness of Conditioned But Not of Unconditioned Stimuli.” Psychological Review 87: 532–552. [PubMed] [Google Scholar]
- Porcheron, G. , and Dozois C. M.. 2015. “Interplay Between Iron Homeostasis and Virulence: Fur and RyhB as Major Regulators of Bacterial Pathogenicity.” Veterinary Microbiology 179: 2–14. [DOI] [PubMed] [Google Scholar]
- Robinson, M. D. , McCarthy D. J., and Smyth G. K.. 2010. “edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data.” Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , and Rusell D. W.. 2001. Molecular Cloning: A Laboratory Manual (3rd ed). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schwartz, D. , Berger S., Heinzelmann E., Muschko K., Welzel K., and Wohlleben W.. 2004. “Biosynthetic Gene Cluster of the Herbicide Phosphinothricin Tripeptide From Streptomyces viridochromogenes Tu494.” Journal of Applied and Environmental Microbiology 70: 7093–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo, G. D. , and Tan K.. 2002. “Mining Genome Databases to Identify and Understand New Gene Regulatory Systems.” Current Opinion in Microbiology 5: 149–153. [DOI] [PubMed] [Google Scholar]
- Tagkopoulos, I. , Liu Y. C., and Tavazoie S.. 2008. “Predictive Behavior Within Microbial Genetic Networks.” Science 320: 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDrisse, C. , Hentchel K., and Escalante‐Semerena J.. 2016. “Identification of Phosphinothricin Acetyltransferases Using In Vivo, In Vitro and Bioinformatics Analyses.” Applied and Environmental Microbiology 82: 7041–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, L. M. , Sumner M. E., and Keeney D. R.. 1977. “Occurrence and Distribution of Arsenic in Soils and Plants.” Environmental Health Perspectives 19: 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth, N. T. , Rohr K., Danchin A., and Nikel P. I.. 2023. “Recursive Genome Engineering Decodes the Evolutionary Origin of an Essential Thymidylate Kinase Activity in Pseudomonas putida KT2440.” mBio 14: e0108123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga, M. , Cai Y., and Rosen B. P.. 2011. “Demethylation of Methylarsonic Acid by a Microbial Community.” Environmental Microbiology 13: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Lin M., Shi H., et al. 2014. “Programming a Pavlovian‐Like Conditioning Circuit in Escherichia coli .” Nature Communications 5: 3102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Transcriptomic data presented in the study are deposited in the NCBI repository, accession numbers SAMN41000924 and SAMN41000925; BioProject PRJNA1101689: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1101689.


