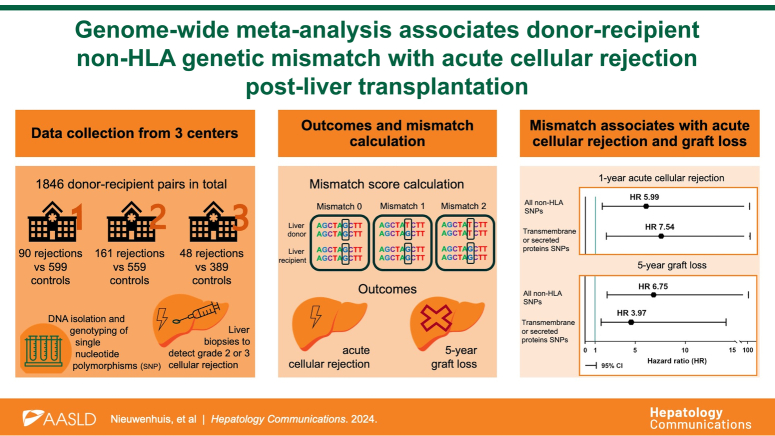
Abstract
Background:
Acute cellular rejection (ACR) remains a common complication causing significant morbidity post-liver transplantation. Non–human leukocyte antigen (non-HLA) mismatches were associated with an increased risk of ACR in kidney transplantation. Therefore, we hypothesized that donor-recipient non-HLA genetic mismatch is associated with increased ACR incidence post-liver transplantation.
Methods:
We conducted an international multicenter case-control genome-wide association study of donor-recipient liver transplant pairs in 3 independent cohorts, totaling 1846 pairs. To assess genetic mismatch burden, we calculated sum scores for single-nucleotide polymorphism (SNP) mismatch based on all non-HLA functional SNPs, specifically SNPs coding for transmembrane or secreted proteins as they more likely affect the immune system. We analyzed the association between the non-HLA mismatch scores and ACR in a multivariable Cox regression model per cohort, followed by a weighted meta-analysis.
Results:
During the first year post-transplantation, 90 of 689 (13%), 161 of 720 (22%), and 48 of 437 (11%) recipients experienced ACR in cohorts 1–3, respectively. Weighted meta-analyses showed that higher mismatch in functional non-HLA SNPs was associated with an increased incidence of ACR (HR 5.99; 95% CI: 1.39–20.08; p=0.011). Moreover, we found a larger effect of mismatch in SNPs coding for transmembrane or secreted proteins on ACR (HR 7.54; 95% CI 1.95–28.79; p=0.003). Sensitivity analyses showed that imputed HLA mismatch did not affect the associations between both non-HLA mismatch scores and ACR.
Conclusions:
Donor-recipient mismatch of functional non-HLA SNPs overall and, especially, of SNPs encoding transmembrane or secreted proteins correlated with 1-year ACR post-liver transplantation. Identifying high-risk immunological burdens between pairs may prevent early graft rejection and aid in personalizing immunosuppressive therapy. Future studies are, however, needed to validate our findings using a genotyped HLA cohort.
Keywords: clinically relevant acute rejection, functional single-nucleotide polymorphisms, genetics, mismatch score, transmembrane and secreted proteins
INTRODUCTION
Despite the inherent tolerogenic capacity of the liver graft, 15%–25% of recipients of liver transplants develop acute T-cell–mediated cellular rejection (ACR), which necessitates additional immunosuppressive treatment and may lead to graft loss.1,2 Several studies have identified risk factors for the development of ACR after liver transplantation. These include primary liver diseases with an autoimmune etiology, high preoperative aspartate transaminase level, low recipient age, high donor age, prolonged cold ischemia time, low tacrolimus trough levels, and donor-recipient mismatches of the human leukocyte antigen (HLA), also known as major histocompatibility complex (MHC).3,4
HLA matching is standard practice in kidney transplantation but is not routinely performed in liver transplantation as the association with rejection is controversial. The impact of non-HLA genetic mismatch on rejection after any solid organ transplant, however, had until recently not been investigated. A study in a kidney transplant cohort found that mismatches in genes coding for transmembrane or secreted proteins were associated with an increased risk of functional graft loss, independently of the degree of HLA compatibility.5 Furthermore, a study reported that the presence of mismatched single-nucleotide polymorphisms (SNPs), determined by exome sequencing, was associated with an increased risk of rejection in kidney transplantation recipients.6 In addition to genome-wide mismatch approaches, studies have investigated the impact of a single variant mismatch between donors and recipients of a kidney transplant, thereby linking mismatch in LIMS1 and CFHR1-3 deletion to ACR.7,8 In the field of liver transplantation, however, the contribution of non-HLA genetic mismatch between donor and recipient to ACR has not been investigated to date.
Based on the profound effect of non-HLA mismatch on graft patency and rejection after kidney transplantation, as shown in recent literature, we aimed to investigate whether non-HLA mismatches between donor and recipient pairs are associated with an increased incidence of biopsy-proven ACR after liver transplantation.
METHODS
Study design and patients
A post hoc multicenter analysis was performed of 3 observational cohort studies consisting of patients who underwent liver transplantation in the University Medical Center Groningen, Groningen, the Netherlands, between 1993 and 2019 (“UMCG” cohort 1), in the Baylor University Medical Center, Dallas, TX, between 1998 and 2010, and were enrolled in the Annette C. and Harold C. Simmons Transplant Institute biorepository and liver transplant research database, between 1998 and 2010 (“Baylor,” cohort 2),9 and in the Penn Medicine Transplant Institute, Philadelphia, PA, between 2012 and 2017, and enrolled in BioTIP (Biorepository of the Transplant Institute at Penn) study (“BioTIP” cohort 3).10 Characteristics of the donor and recipient pairs were collected from institutional databases. All postoperative transplant care, including the immunosuppression regimen, was according to the center standard protocol. Detailed information on immunosuppression regimen per cohort is described in the studies from Li et al11 (cohort 1) and Shaked et al10 (cohorts 2 and 3).
Ethical approval
Cohort 1 was registered in the Netherlands Trial Register (www.trialregister.nl – Trial NL6334) and was conducted within the TransplantLines cohort study,12 which was approved by the institutional research board (METc 2014/077). Cohorts 2 and 3 were approved by the Institutional Review Boards at the respective institutions (Baylor: FWA 00004415/00003358, protocol 013-065; UPenn: FWA 00004028, protocol 820091). The participants signed informed consent prior to transplantation. All study protocols adhered to the Declaration of Helsinki and were in concordance with the principles of the Declaration of Istanbul on Organ Trafficking and Transplant Tourism. STrengthening the REporting of Genetic Association studies (STREGA) guidelines were adhered to.13
Transplantation outcomes
We defined clinically relevant ACR as Banff grade 2 (moderate) or Banff grade 3 (severe) histologically proven rejection within 1-year after liver transplantation. The biopsies were assessed by specifically trained pathologists. As it was clinical practice in all cohorts to not perform biopsies without clinical indication, we cannot with certainty exclude grade 1 cases in all patients when there was no routine biopsy performed. This is because grade 1 rejection can present without or with minimal symptoms and can be missed. We therefore focused on grade 2 and 3 rejections and excluded all recipients with clinically suspected rejection (without biopsy) or grade 1 biopsy-proven ACR. Additionally, we recorded all instances of graft loss in each cohort. Graft loss was defined as the need for retransplantation or the occurrence of patient death.
Sample collection and DNA extraction
Blood for DNA extraction was collected at the time of transplantation for both donors and recipients. Genomic DNA was extracted from whole blood or peripheral blood mononuclear cells using standard procedures and stored at −80 °C. DNA quality was assessed for all samples using agarose gel electrophoresis. DNA concentrations were estimated using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific Inc., Asheville, NC). DNA samples were diluted to 50 ng/μL with nuclease-free water.
Genotyping and imputation
In cohort 1, genotyping was performed using the Infinium Global Screening Array-24 v1.0 (Illumina Inc., San Diego, CA), a genotyping array that measured 494,059 SNPs, including both rare and common SNPs. In cohorts 2 and 3, genotyping was performed using the Affymetrix Axiom Transplant Array, which measured 767,203 SNPs, including both rare and common SNPs. Genotype data were clustered using the GenomeStudio software,14 and quality control was performed using PLINK.15 Normalized intensities for all samples were called using Optical clustering.16
Raw genotypes were imputed using the TOPMed reference panel (version r2, hg38), which is a diverse panel including information from 97.256 human genomes.17,18,19 SNPs in the MHC region were excluded. Postimputation quality control was performed with an estimated imputation accuracy of at least 0.4.5,20 Markers with a call rate <99%, a MAF<5%, or with significant deviation from Hardy-Weinberg equilibrium (p<1×10−6) were also excluded.
SNP selection and mismatch calculation
We accounted for the genetic mismatch between donors and recipients by including sum scores for SNP mismatch based on non-HLA nonsynonymous SNPs as estimated by Ensembl (GRCh38) annotation using SnpEff (version 5.0). Selected nonsynonymous SNPs included frameshift variants, protein-altering variants, and missense variants (ie, stop gained, stop lost, start lost, splice acceptor variant, splice donor variant, splice region variant, missense variant); hereafter called “functional” SNPs. We calculated the amount of SNP mismatches for all donor and recipient pairs. The mismatch score for each SNP was either 0 (no mismatch), 1 (one mismatched nucleotide), or 2 (2 mismatched nucleotides). To address the heterogeneity of the 3 cohorts and possible differences in functional SNPs and to account for varying SNP coverage of the different genotype arrays, we checked in a sensitivity analysis the overlap of the functional non-HLA SNPs between the 3 cohorts and calculated a separate mismatch score for all donor-recipient pairs in all cohorts using only the overlapping SNPs.
Secreted and transmembrane SNPs
To further reduce noise, and in an effort to replicate the results shown in the study by Reindl-Schweighofer et al,5 we selected solely the SNPs coding for transmembrane or secreted proteins, as these have a higher likelihood to affect the immune system. For each cohort, we constructed a separate list of SNPs for mismatch calculation which only included functional SNPs that coded for transmembrane or secreted proteins. Transcripts from UniProt database were used to select SNPs coding for transmembrane or secreted proteins.21
HLA imputation and mismatch calculation
HLA class l and ll genotypes of all donor-recipient pairs were imputed by the HLA-dedicated imputation algorithms CookHLA22 (cohort 1; v1.0.1) and SNP2HLA23 (cohorts 2 and 3; v1.0) against the reference panel from the type 1 diabetes Genetics Consortium.24 For both CookHLA and SNP2HLA, we used unphased plink GSA data in build hg18 as input, specifically selecting SNPs located within the 29–34Mb region of chromosome 6, which encompasses the MHC region. The class l and ll 4-digit alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, HLA-DQA1, HLA-DPB1, and HLA-DPA1 were included in the mismatch calculation, which was done using HLA-matchmaker.25
Statistical analyses
Characteristics of patients were described using medians and interquartile ranges (IQRs) for continuous variables and frequencies and percentages for categorical variables. For each cohort, we individually assessed the association with ACR and 5-year graft loss for (1) total non-HLA mismatch burden, and (2) mismatch burden in SNPs coding for transmembrane or secreted proteins, using a multivariable Cox proportional hazards model. Possible confounders were selected by performing univariable regression analyses, in which we only included variables from donor and recipient pairs with >80% valid data. Variables with a p-value <0.1 in the univariable analysis were, in addition to transplantation indication, included in the multivariable analysis and were accounted for as possible confounders. To ensure sufficient power, we performed a weighted random effects meta-analysis, in which we included the hazard and SE of the association between non-HLA mismatch score and ACR or 5-year graft loss. The non-HLA mismatch scores were log-transformed to allow for better interpretability. Restricted cubic spline models were used to visualize the nonlinear relationship between non-HLA mismatch score and risk of ACR. To account for the fact that there was a varying number of functional SNPs between cohorts, we performed a sensitivity analysis in which we only selected the functional SNPs that overlap between all cohorts. Additionally, we performed a sensitivity analysis in which HLA mismatch was included as a possible confounder in the Cox regression analyses that tested the association between both non-HLA mismatch scores and ACR or 5-year graft loss. Correlation analyses between HLA and non-HLA mismatch were done using the Spearman correlation test. Two-sided p<0.05 were considered statistically significant. All calculations and statistical analysis were done using R (versions 4.1.2 and 4.3.1) and SPSS software (v28; IBM, Chicago, IL).
RESULTS
Patient characteristics
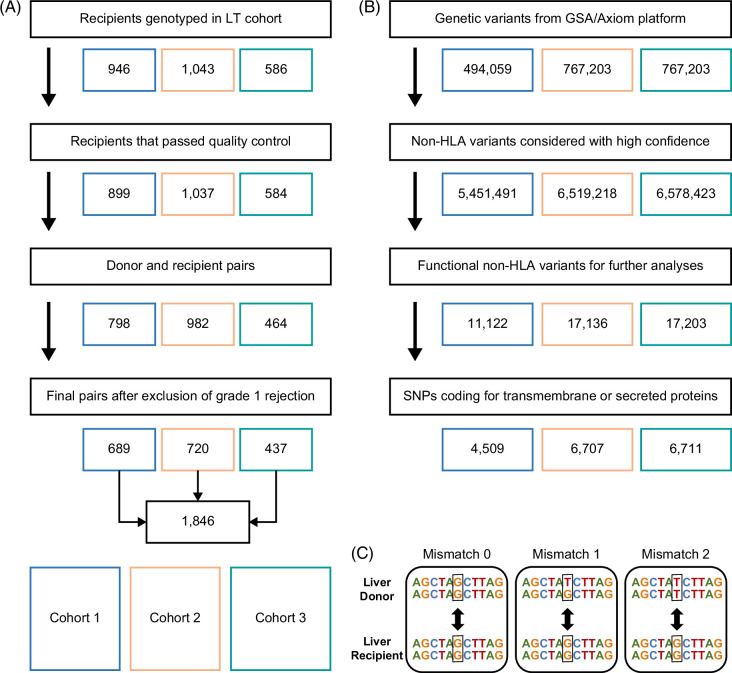
We included phenotypic and genotypic data of 1846 donor-recipient pairs who underwent liver transplantation for a variety of indications from 3 medical centers (Figure 1A). Donor, recipient, and surgical characteristics are described in Table 1. The median follow-up was 10.3 years (IQR, 5.1–17.0) in cohort 1, 7.2 years (IQR, 4.7–10.0) in cohort 2, and 2.3 years (IQR, 1.2–3.8) in cohort 3. Ninety recipients (13%) in cohort 1, 161 recipients (16%) in cohort 2, and 48 recipients (11%) in cohort 3 experienced grade 2 or 3 ACR, which occurred after a median of 10 (IQR, 7–25), 14 (IQR, 6–52), and 38 (IQR, 14–131) days, respectively. In total, 190 (28%), 164 (23%), and 64 (15%) recipients experienced graft loss within the first 5 years after transplantation, which occurred after a median of 90 (IQR, 9–678), 471 (IQR, 44–1090) and 954 (IQR, 543–1470) days in cohorts 1–3, respectively.
FIGURE 1.
Flowchart of data management. (A) Inclusion of donor-recipient pairs after genotyping quality control. (B) Quality control, imputation, and non-HLA functional SNP selection from SNPs genotyped by Global Screening Array-24 v1.0 (Illumina, San Diego, CA) and Affymetrix Axiom Transplant Arrays. (C) The non-HLA SNP mismatch score is calculated by comparing allele overlap between the donor and recipient for each functional variant. Abbreviations: GSA, Global Screening Array; HLA, human leukocyte antigen; LT, liver transplantation; SNP, single-nucleotide polymorphism.
TABLE 1.
Demographics of all donor-recipient pairs
| Cohort 1 | Cohort 2 | Cohort 3 | |
|---|---|---|---|
| N=689 | N=720 | N=437 | |
| N/median (%/IQR) | N/median (%/IQR) | N/median (%/IQR) | |
| Recipient age, y | 46 (16–58) | 52 (47–58) | 59 (52–64) |
| Pediatric recipients | 183 (26.6) | 0 | 0 |
| Recipient sex, male | 391 (56.7) | 465 (65) | 313 (72) |
| Recipient BMI, kg/m2 | 23.3 (19.2–26.9) | 27.9 (24.3–32) | 27.1 (24.2–30.8) |
| Transplantation indication | |||
| PBC/AIH | 69 (10.0) | 68 (9.4) | 33 (7.6) |
| PSC | 102 (14.8) | 33 (4.6) | 26 (5.9) |
| Metabolic | 95 (13.8) | 8 (1.1) | 7 (1.6) |
| Congenital biliary disease | 108 (15.7) | 0 | 0 |
| Alcoholic liver disease | 67 (9.7) | 84 (11.7) | 76 (17.4) |
| Viral hepatitis | 67 (9.7) | 385 (53.5) | 174 (39.8) |
| MASLD | 40 (5.8) | 9 (1.3) | 45 (10.3) |
| Acute liver failure | 32 (4.6) | 19 (2.6) | 6 (1.4) |
| Other | 109 (15.8) | 114 (15.8) | 70 (16.0) |
| Donor age, y | 45 (30–55) | 41 (24–55) | 43 (28–55) |
| Donor sex, male | 362 (52.5) | 423 (59) | 245 (56) |
| Donor BMI, kg/m2 | 24 (22–26) | 25.8 (22.4–29.8) | 27.2 (23.8–32.7) |
| Cold ischemia time, min | 472 (389–589) | 475 (356–595) | 301 (241–382) |
Note: categorical variables are presented as numbers and percent (%), continuous variables are shown in median and interquartile range.
Abbreviations: AIH, autoimmune hepatitis; BMI, body mass index; HLA, human leukocyte antigen; MASLD, metabolic dysfunction–associated steatotic liver disease; PBC, primary biliary cholangitis; PSC, primary sclerosing cholangitis.
Non-HLA mismatch score calculation
To assess the donor-recipient genetic mismatch burden, we calculated sum scores for SNP mismatches based on 11,122, 17,136, and 17,203 non-HLA functional SNPs (Figure 1B) in respectively cohorts 1, 2, and 3, with a mismatch score of 0, 1, or 2 for each SNP (Figure 1C). The median non-HLA mismatch score for all donor-recipient pairs was 3191 (IQR 3111–3265) in cohort 1, 5345 (IQR 5143–5798) in cohort 2, and 5084 (IQR 4965–5741) in cohort 3. Of the 11,122 functional SNPs in cohort 1, 10,528 SNPs (95%) were also present in cohorts 2 and 3. The median mismatch score of overlapping functional non-HLA SNPs was 3082 (IQR 3008–3158) in cohort 1, 3321 (IQR 3226–3434) in cohort 2, and 3312 (IQR 3218–3437) in cohort 3.
We then selected solely the SNPs coding for transmembrane or secreted proteins, which were 4509 in cohort 1, 6707 in cohort 2, and 6711 in cohort 3 (Figure 1B). The median mismatch score of SNPs coding for transmembrane or secreted proteins for all donor-recipient pairs was 1313 (IQR 1266–1360) in cohort 1, 2113 (2027–2262) in cohort 2, and 2088 (1996–2281) in cohort 3.
Risk factors for ACR
We calculated the risk of ACR within the first year after transplantation in relation to donor, recipient, and surgical characteristics. Using univariable regression models, we identified cold ischemia time and transplantation era (time in years since transplantation) as potential risk factors for ACR (p<0.1) in cohort 1, and recipient age, recipient sex, and transplantation era as potential risk factors for ACR (p<0.1) in cohort 2. Subsequently, these factors were included in the multivariable Cox regression analysis in addition to transplantation indication, overall functional non-HLA, and transmembrane or secreted protein mismatch score (Table 2 and Supplemental Table S1, http://links.lww.com/HC9/B784). In the individual cohorts, the association between overall functional non-HLA mismatch score and ACR did not pass the significance threshold after adjusting for possible confounders in a multivariable Cox regression model. However, when looking at the nonlinear association between genetic mismatch and ACR using a restricted cubic spline, we see a trend toward a higher HR for developing ACR for patients with a higher mismatch score in our largest cohort (Supplemental Figure S1, http://links.lww.com/HC9/B785 and Supplemental Table S1, http://links.lww.com/HC9/B784). The association between mismatch in SNPs coding for transmembrane or secreted proteins and ACR was significant in the multivariable Cox regression analysis in cohort 2 (HR 7.67; 95% CI: 1.14–51.8; p=0.036). In cohorts 1 and 3, we observe a higher HR for patients with a higher mismatch in SNPs coding for transmembrane or secreted proteins; however, this association does not reach statistical significance (cohort 1: HR 12.95; 95% CI: 0.60–278.97; p=0.10 and cohort 3: HR 5.24; 95% CI: 0.47–57.9; p=0.18; Supplemental Table S1, http://links.lww.com/HC9/B784).
TABLE 2.
Univariable Cox regression analysis for ACR
| Cohort 1 | Cohort 2 | Cohort 3 | ||||
|---|---|---|---|---|---|---|
| 90 rejection 599 control | 161 rejection 559 control | 48 rejection 389 control | ||||
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
| Recipient age, y | 0.99 (0.99–1.01) | 0.83 | 0.98 (0.97-0.99) | 0.04 | 1.00 (0.97–1.03) | 0.88 |
| Recipient sex, male | 1.25 (0.83–1.89) | 0.30 | 0.73 (0.54–1.00) | 0.05 | 1.20 (0.62–2.30) | 0.59 |
| Recipient BMI, kg/m2 | 0.98 (0.94–1.02) | 0.32 | 0.99 (0.97–1.02) | 0.59 | 0.98 (0.93–1.04) | 0.48 |
| Transplantation indication | ||||||
| Alcoholic liver disease (ref) | 1.00 (1.00–1.00) | NA | 1.00 (1.00–1.00) | NA | 1.00 (1.00–1.00) | NA |
| PBC/AIH | 2.54 (0.98–6.60) | 0.06 | 2.95 (1.34–6.49) | 0.007 | 0.41 (0.09–1.83) | 0.24 |
| PSC | 2.99 (1.23–7.29) | 0.016 | 2.80 (1.11–7.06) | 0.03 | 1.15 (0.36–3.60) | 0.82 |
| Metabolic | 1.10 (0.39–3.08) | 0.86 | 2.52 (0.54–11.66) | 0.24 | 0.00 (0.00–0.00) | NA |
| Congenital biliary disease | 1.16 (0.42–3.19) | 0.78 | NA | NA | NA | NA |
| Viral hepatitis | 2.37 (0.90–6.24) | 0.08 | 2.43 (1.23–4.82) | 0.01 | 0.74 (0.35–1.56) | 0.43 |
| MASLD | 1.10 (0.31–3.91) | 0.88 | 2.16 (0.47–10.01) | 0.32 | 0.72 (0.25–2.07) | 0.54 |
| Acute liver failure | 0.72 (0.15–3.56) | 0.68 | 1.94 (0.52–7.16) | 0.32 | 4.58 (1.28–16.45) | 0.02 |
| Other | 0.78 (0.26–2.31) | 0.65 | 2.20 (1.02–4.74) | 0.04 | 0.39 (0.12–1.21) | 0.10 |
| Donor age, y | 0.99 (0.99–1.01) | 0.45 | 1.01 (1.00–1.01) | 0.20 | 1.01 (0.99–1.03) | 0.44 |
| Donor sex, male | 1.37 (0.90–2.07) | 0.14 | 0.97 (0.71–1.33) | 0.85 | 1.62 (0.89–2.96) | 0.11 |
| Donor BMI, kg/m2 | 0.99 (0.94–1.04) | 0.61 | 1.00 (0.98–1.03) | 0.91 | 1.01 (0.98–1.05) | 0.52 |
| Cold ischemia time, min | 1.001 (1.000–1.002) | 0.08 | 1.000 (0.999–1.002) | 0.25 | 0.998 (0.996–1.001) | 0.12 |
| Transplantation era (time since transplantation in years) | 1.07 (1.04–1.10) | <0.001 | 1.10 (1.04–1.17) | 0.001 | 1.07 (0.89–1.28) | 0.47 |
Note: Univariable Cox regression analyses were performed with ACR as a binary outcome (presence or absence). Data are presented as HRs with confidence ratios and corresponding p-values. Variables in bold will be included as possible confounders in the multivariable Cox regression analyses.
Abbreviations: AIH, autoimmune hepatitis; BMI, body mass index; HLA, human leukocyte antigen; MASLD, metabolic dysfunction–associated steatotic liver disease; PBC, primary biliary cholangitis; PSC, primary sclerosing cholangitis; SNPs, single-nucleotide polymorphisms.
ACR meta-analysis
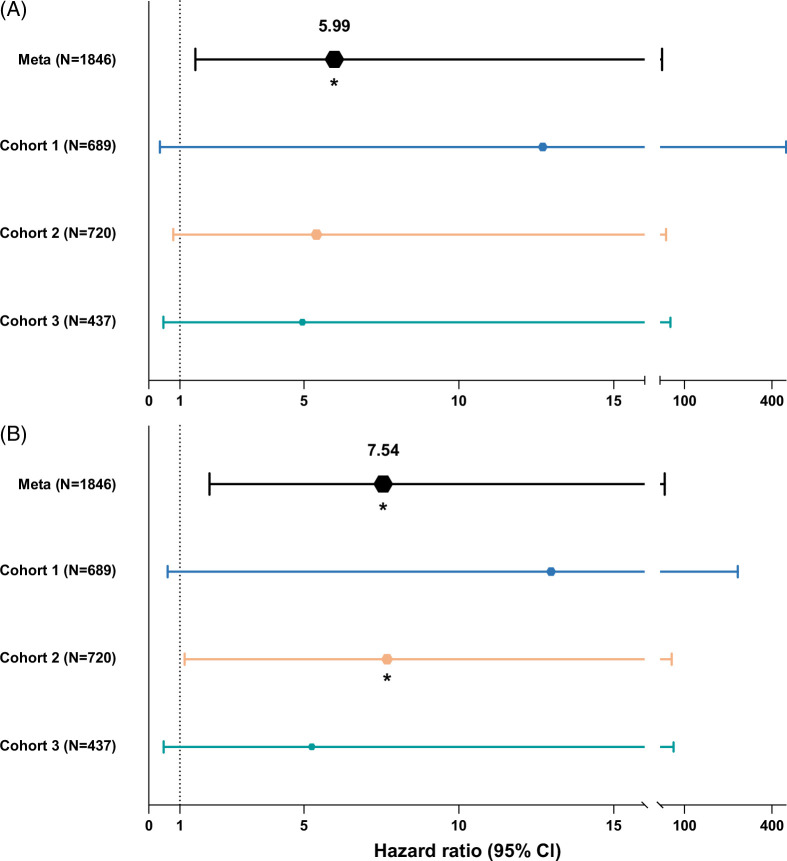
We performed weighted meta-analyses to assess the association with ACR for both overall functional non-HLA mismatch as well as mismatch in SNPs coding for transmembrane or secreted proteins. The meta-analyses combined the individual multivariable Cox regression results of each cohort. Due to the increased power, we show that a higher (log-normalized) overall functional non-HLA mismatch score, as well as higher (log-normalized) mismatch score of SNPs coding for transmembrane or secreted proteins, is significantly associated with an increased incidence of ACR after liver transplantation (HR 5.99; 95% CI: 1.51–23.57; p=0.011) and (HR 7.54; 95% CI: 1.95–28.79; p=0.003), respectively (Figure 2 and Supplemental Table S1, http://links.lww.com/HC9/B784). We performed a sensitivity analysis in which we only included functional SNPs present in all cohorts. The association of mismatch burden of overlapping SNPs with ACR lost its statistical significance in the individual cohorts as well as in the meta-analysis due to loss of power. However, the direction and magnitude of the HR were similar to the analysis with all functional SNPs (HR 4.39, 95% CI: 0.94–20.74, p=0.060; Supplemental Table S2, http://links.lww.com/HC9/B786)
FIGURE 2.
Forest plot depicting the results of regression analyses for acute T-cell-mediated cellular rejection, presented for individual cohorts as well combined using a meta-analysis. This graph depicts for each cohort the results of the associations found with acute T-cell mediated cellular rejection in a proportional hazard Cox regression model for (A) overall functional non–human leukocyte antigen mismatch score and (B) mismatch in single-nucleotide polymorphisms coding for transmembrane or secreted proteins. The top line in both panels shows the results of the meta-analysis of all 3 cohorts, with individual cohorts as random effects.
The association between non-HLA mismatch and graft survival
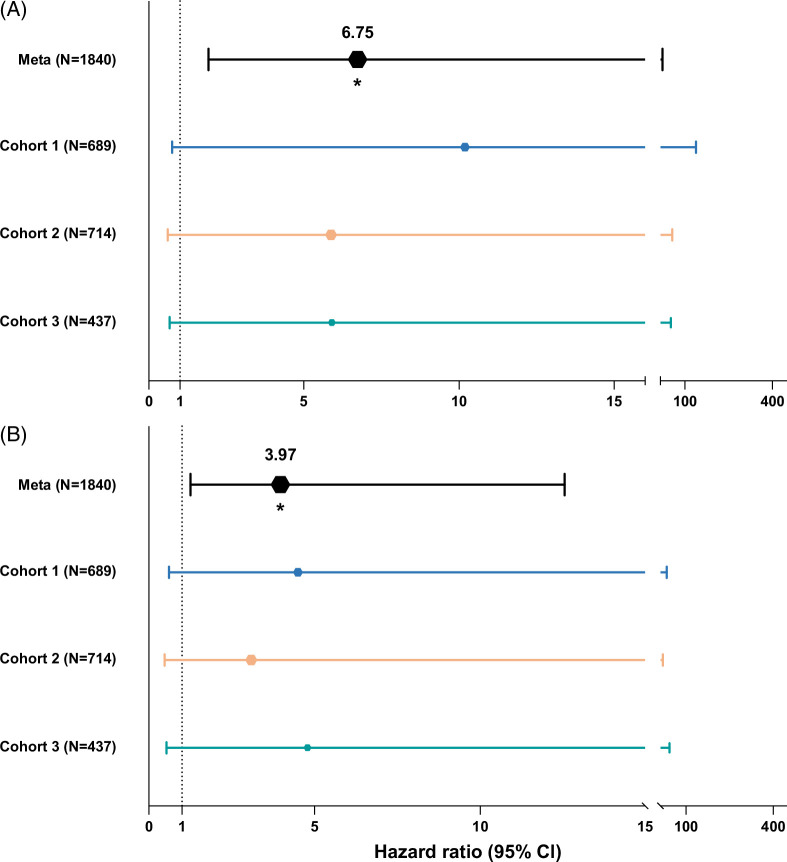
To investigate whether non-HLA mismatch has an effect beyond ACR, we performed the same analyses with 5-year graft survival as the outcome. First, we calculated the risk of 5-year graft loss in relation to donor, recipient- and surgical characteristics. We identified donor age, cold ischemia time, transplantation era, and transplantation indication as potential confounders in cohort 1; donor age and BMI, transplantation era, and transplantation indication in cohort 2; and donor age and transplantation era in cohort 3. In the individual cohorts, the association with 5-year graft survival for both overall functional non-HLA mismatch score and mismatch in SNPs encoding transmembrane or secreted proteins did not pass the significance threshold after adjusting for possible confounders in a multivariable Cox regression model (Supplemental Table S3, http://links.lww.com/HC9/B787). We then performed weighted meta-analyses to assess the association with 5-year graft loss for both overall functional non-HLA mismatch as well as a mismatch in SNPs coding for transmembrane or secreted proteins. The meta-analyses combined the individual multivariable Cox regression results of each cohort. Due to the increased power, we show that a higher (log-normalized) overall functional non-HLA mismatch score, as well as higher (log-normalized) mismatch score of SNPs coding for transmembrane or secreted proteins, is significantly associated with higher rates of graft loss after liver transplantation (HR 6.75; 95% CI: 1.93–23.57; p=0.003) and (HR 3.97; 95% CI: 1.25–12.55; p=0.020), respectively (Figure 3 and Supplemental Table S3, http://links.lww.com/HC9/B787).
FIGURE 3.
Forest plot depicting 5-year graft survival Cox regression analyses results of individual cohorts and all combined using a meta-analysis. This graph depicts for each cohort the results of the associations found with 5-year graft survival in a proportional hazard Cox regression model for (A) overall functional non–human leukocyte antigen mismatch score, and (B) mismatch in single-nucleotide polymorphisms coding for transmembrane or secreted proteins. The top line in both panels shows the results of the meta-analysis of all 3 cohorts, with individual cohorts as random effects.
Adjusting for imputed HLA mismatch
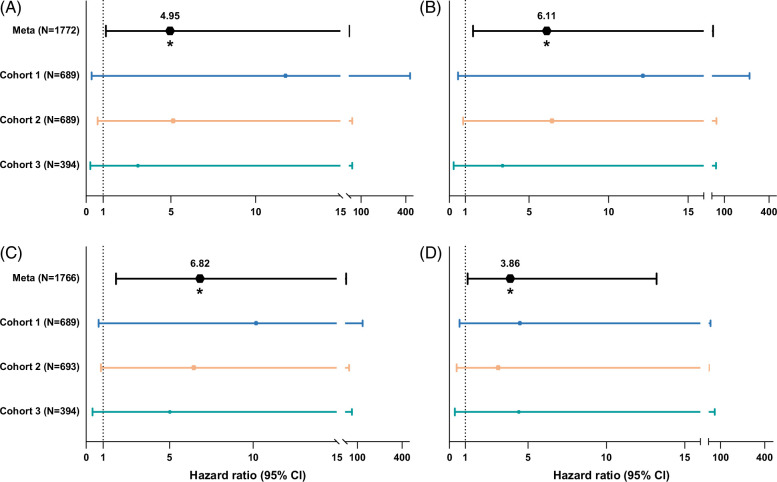
Of 1772 pairs (96% of the total study group) HLA eplet data were available through imputation with either CookHLA or SNP2HLA. For these pairs, we were able to calculate their HLA mismatch and perform the subsequent analyses. The median HLA mismatch score, calculated by HLA-matchmaker, for donor-recipient pairs in cohort 1 was 37 (IQR 26–48), 56 (IQR 44–67) in cohort 2, and 55 (IQR 43–67) in cohort 3. We show, in univariable Cox regression analyses, that there is no significant association between HLA mismatch and ACR (cohort 1: HR: 1.02; 95% CI: 0.90–1.16; p=0.77, cohort 2: HR: 1.00; 95% CI: 0.99–1.01; p=0.52, cohort 3: HR: 1.00; 95% CI: 0.99–1.02; p=0.55). After incorporating HLA mismatch into the meta-analyses of all multivariable Cox regression analyses linking overall functional non-HLA SNP mismatch and mismatch in SNPs coding for transmembrane or secreted proteins to ACR risk, our results remained significant with unchanged direction and magnitude of association (overall functional SNP mismatch: HR: 4.95, 95% CI: 1.16–21.11, p=0.030; transmembrane or secreted SNP mismatch: HR: 6.11, 95% CI: 1.48–25.03, p=0.013; Figure 4 and Supplemental Table S4, http://links.lww.com/HC9/B788). Similarly, correcting for HLA mismatch did not affect the association between non-HLA mismatch and 5-year graft loss (overall functional SNP mismatch: HR: 6.82, 95% CI: 1.79–26.31, p=0.005; transmembrane or secreted SNP mismatch: HR: 3.86, 95% CI: 1.13–13.20, p=0.032; Figure 4 and Supplemental Table S5, http://links.lww.com/HC9/B789).
FIGURE 4.
Forest plot depicting the results of human leukocyte antigen mismatch-corrected Cox regression analyses for acute T-cell mediated cellular rejection and 5-year graft survival, shown for individual cohorts and combined through a meta-analysis. This graph depicts for each cohort the results of the associations found with acute T-cell mediated cellular rejection (A and B) and 5-year graft survival (C and D) in a proportional hazard Cox regression model for overall functional non–human leukocyte antigen mismatch score (A and C), and mismatch in SNPs coding for transmembrane or secreted proteins (B and D). The top line in both panels shows the results of the meta-analysis of all 3 cohorts, with individual cohorts as random effects.
Living donor liver transplantations–related donor-recipient pairs
In cohort 1, there are 27 liver transplant recipients who received their graft from a related living donor. Of these 27 pairs, 22 living donors were first-degree relatives, with the remaining 5 pairs being second-degree related. These 27 related donor-recipient pairs had a median non-HLA mismatch score of 1767 (IQR: 1720–2133), and a median HLA mismatch score of 19 (IQR: 16–27). The median non-HLA and HLA scores of related donor-recipient pairs were significantly lower than those of nonrelated donor-recipient pairs in cohort 1 (non-HLA: 3195 (IQR: 3119–3267, p<0.001); HLA: 38 (IQR: 27–48, p<0.001). In the related group, non-HLA mismatch is significantly correlated to HLA mismatch (r2: 0.602, p<0.001, Supplemental Figure S2, http://links.lww.com/HC9/B785). However, in the nonrelated group, non-HLA mismatch is not correlated to HLA mismatch (r2: 0.056, p=0.15, Supplemental Figure S2, http://links.lww.com/HC9/B785). Interestingly, just 1 recipient of a related donor experienced clinically relevant ACR, with the donor being a second-degree relative. Moreover, none of the recipients of a related donor graft experienced graft loss within the first 5 years after transplantation.
DISCUSSION
In this multicenter, genome-wide meta-analysis, we show that genetic non-HLA mismatch between donors and recipients is an important predictor of ACR after liver transplantation. Our genome-wide association study (GWAS), including data from an unparalleled 1846 donor-recipient pairs, represents the largest genotyped paired-liver transplantation cohort to date. We demonstrate that liver transplant recipients who receive grafts from donors with a higher non-HLA mismatch in crucial functional genetic regions have an increased risk to develop ACR. Furthermore, we show that beyond its impact on ACR risk, a higher non-HLA mismatch is linked to lower 5-year graft survival rates. This study demonstrates the nuanced interplay of genetic factors in transplantation outcomes. By discerning between low-risk and high-risk immunological burden, our findings have the potential to improve current clinical practice by tailoring immune suppression strategy and thereby optimizing patient care.
Graft rejection, subdivided into hyperacute, acute, or chronic has conventionally been understood as a process mediated through the direct and indirect pathways of allorecognition. This intricate mechanism involves the processing and presentation of donor antigens to the cells of the recipient.26 ACR can occur where genetic disparities exist between donors and recipients, which may lead to the presentation of polymorphic peptides that the recipient’s immune system recognizes as abnormal.27 There is a compelling need for identification of immunogenic epitopes that, when mismatched between donor and recipient, can cause an antibody or alloreactive T-cell response.28 GWAS, through the examination of a wide diversity of common genetic variations, have unveiled more than a thousand associations of SNPs with diseases and traits across the genome. A number of GWASs have yielded insight into the genetic architecture of ACR.29,30,31,32 Several genes not located on the HLA, otherwise known as MHC, region were observed to play a role in recipients of kidney transplants with ACR, supporting the hypothesis of immune-accessible epitopes. Sindhi et al33 reported a family-based association study, which identified the minor allele of rs9296068, near HLA class II region, to be significantly associated with enhanced B-lymphocyte participation in pediatric liver transplant rejection. Our team has, however, not been able to link specific SNPs to ACR through GWAS, either because of insufficient sample size or the inability to properly investigate SNP-to-SNP interaction through GWAS. We believe that it is likely that multiple SNPs, both from the donor and recipient, have a combined effect on ACR risk. We therefore focused, as did the team of Reindl-Schwaighofer et al,5 on the combined effect of mismatches in functional SNPs between donor and recipient pairs.
To allow for a more functional interpretation of our results, we furthermore performed an analysis in which we only selected functional SNPs coding for transmembrane or secreted proteins, like was done in the study by Reindl-Schwaighofer et al.5 Transmembrane proteins, embedded within cellular membranes, contribute to vital cellular functions like signal transduction and molecule transport across membranes, influencing immune responses. Secreted proteins, ie, cytokines, chemokines, or complement proteins, produced within cells and released into the extracellular environment, actively engage in intercellular communication, playing essential roles in immune system modulation and various physiological processes. Mismatches in SNPs in a region coding for these proteins are therefore more likely to have a downstream effect on the immune system,34,35,36 For example, there have been studies that investigate the impact on graft patency of polymorphisms in individual secreted proteins. One study identified donor-recipient mismatch of complement factor H-related 3,1 deletion as a risk factor for kidney graft rejection.8 Another study found that specific polymorphisms in IL6 and IL10 are associated with reduced risk of graft loss.37
GWAS in the field of liver transplantation have been notoriously difficult to perform due to a limited availability of genotyped cohorts, complex covariates, and era effects, resulting in poor statistical power for detecting donor-recipient interactions.38 To overcome a lack of statistical power, we performed a weighted meta-analysis, with the cohorts as random effects, in which we included the hazard and SE of the association between non-HLA mismatch score and ACR. The collaboration of 3 independent cohorts has resulted in sufficient power to identify a significant association between functional genetic variant mismatch and higher risk of both ACR and 5-year graft loss (overall functional SNP mismatch, ACR: HR of 5.99, 5-year graft loss: HR of 6.75; mismatch in SNPs coding for transmembrane or secreted proteins, ACR: HR of 7.54, 5-year graft loss: HR of 3.97). We chose to only select functional SNPs to reduce noise, as was done in the study by Reindl-Schwaighofer et al.5
One significant advantage of contemporary genetic testing lies in its capacity to construct a comprehensive genetic risk profile, enabling preemptive screening to predict the likelihood of graft rejection. Leveraging our SNP-based array, we meticulously identified the genetic signals linked with ACR and graft loss across crucial functional genetic regions. However, it is crucial to acknowledge the influence of nongenetic factors, such as highly intricate multifactorial covariates and phenotypes, which have historically impeded investigations into the genetic architecture underlying various morbidities impacting graft rejection. To address this challenge, we corrected for potential confounders in our analyses. For instance, the evolution of immunosuppression management over time may manifest as a gradual decline in the incidence of rejection. The management of immunosuppressive medication is therefore tied to the year of transplantation, which varies significantly in cohorts 1 and 2. Consequently, we have included the transplantation era in all multivariable Cox regression models for these cohorts. By adjusting for transplantation era, we also aim to account for any changes in surgical techniques or improvements in post-surgical hospital care. By accounting for such confounding variables, we aimed to disentangle the genuine genetic associations from outside influences, thereby enhancing the robustness and reliability of our findings.
This study has some limitations, such as its historical design. Due to changes in management related to routine biopsies, we cannot definitively determine if a patient has experienced grade 1 rejection. Therefore, we focused on grade 2 and 3 rejections, which have clear clinical implications, are unlikely to be missed in clinical care, and are therefore consistently biopsied. Another limitation is the different genotyping arrays used for the different cohorts. However, we sought to reduce any further possible bias and differences between cohorts by using identical cutoff values during quality control and the same reference cohort for imputation. Because of this, we believe it is sound to perform a meta-analysis of the regression analysis of the 3 cohorts. In addition, in a sensitivity analysis where we only included the functional non-HLA SNPs present in all cohorts, we show similar results, albeit not significant, compared to the analysis with all functional SNPs.
HLA genes, located on chromosome 6, encode major histocompatibility complex molecules that are expressed on the cell surface and coordinate the adaptive immune system. In kidney transplantation, a higher number of HLA mismatches increases the risk of allograft rejection.39,40 However, the impact of HLA matching on outcomes in liver transplantation remains controversial.41 Liver allografts are believed to exhibit immunologic inertness similar to that of a native liver, resulting in a unique immunological status distinct from other solid organ transplants. Consequently, donor-recipient pairs in liver transplantation are not matched based on their HLA type.
Acknowledging the significant role of HLA in immunity and the fact that some studies suggest an effect of HLA,42,43 we investigated whether HLA mismatch, based on imputed HLA alleles, is associated with ACR and how it influences the association between non-HLA mismatch and ACR. Our findings indicate that the mismatch of imputed HLA alleles is not associated with ACR. After incorporating the mismatch of imputed HLA alleles into our meta-analysis model, the significant associations between overall functional non-HLA SNP mismatch and ACR risk (HR: 4.95, 95% CI: 1.16–21.11, p=0.030) and between SNP mismatch in transmembrane or secreted proteins and ACR risk (HR: 6.11, 95% CI: 1.48–25.03, p=0.013) remained unchanged. Similarly, correcting for imputed HLA mismatch did not affect the association between non-HLA mismatch and 5-year graft loss (overall functional SNP mismatch: HR: 6.82, 95% CI: 1.79–26.31, p=0.005; transmembrane or secreted SNP mismatch: HR: 3.86, 95% CI: 1.13–13.20, p=0.032). However, our study is limited by the use of imputed rather than genotyped HLA types, as genotyped HLA alleles offer greater accuracy. Furthermore, we did not incorporate HLA donor-specific antibodies, which are recognized in the literature as a key player in developing post-transplant rejection. Although genotyped HLA types and HLA donor-specific antibodies would give more precise insights, our analysis using imputed alleles remains informative and provides valuable direction for future research. We, however, recommend that future studies investigate and validate our findings using a high-resolution HLA-genotyped cohort. This follow-up study would be worthwhile to further uncover the immunological pathways involved in post liver transplant rejection.
Of particular interest is the observation that in a subset of cohort 1, living-related donor liver transplantation pairs showed a significantly lower non-HLA mismatch burden. Moreover, no recipients of a graft from a first-degree–related living donor developed clinically relevant ACR. The fact that family members have less genetic mismatch and less ACR cases supports our finding that genetic mismatch is associated with the risk of ACR. Albeit small numbers, this observation merits future investigation. Furthermore, should larger follow-up studies find similar results, this information might be useful when deciding on an immune suppression regimen for pediatric recipients with a related donor.
In conclusion, our study sheds light on the significant association between genetic non-HLA mismatch in liver transplant donor-recipient pairs and clinically relevant ACR following liver transplantation. This finding underscores the importance of considering genetic factors beyond HLA compatibility in predicting transplant outcomes. The insights gleaned from our research pave the way for a more nuanced understanding of the impact of donor-recipient genetic mismatch burden on post-transplantation outcomes. By recognizing and incorporating this crucial factor into clinical decision-making, we hold the potential to tailor immunosuppression regimens to individual patients, thereby optimizing long-term living donor transplant patency. A first step could be to closely monitor recipients whose donor compatibility mismatch falls within the top 5 to 10th percentile.
In essence, our findings not only contribute to the growing body of knowledge on transplantation genetics but also offer a tangible pathway toward personalized medicine in liver transplantation, with the ultimate goal of improving patient outcomes and enhancing the long-term success of liver transplantation.
Supplementary Material
AUTHOR CONTRIBUTIONS
Lianne M. Nieuwenhuis and Yanni Li share the first authorship. The principal investigators (Eleonora A. M. Festen and Vincent E. de Meijer) share senior authorship. Eleonora A. M. Festen and Vincent E. de Meijer conceived and designed the analysis and were responsible for daily supervision, data analysis, and writing of the initial paper draft. Lianne M. Nieuwenhuis, Yanni Li, and Bao-Li Loza analyzed genotype data and completed the statistical analysis. Lianne M. Nieuwenhuis curated the phenotype and genetic data, interpreted the results, and wrote the initial paper draft. Other co-authorship places were awarded in accordance with international (ICMJE) guidelines. All authors have critically read the final paper draft and provided feedback prior to final submission.
CREDIT – CONTRIBUTOR ROLES TAXONOMY
Conceptualization: Vincent E. de Meijer and Eleonora A. M. Festen. Data curation: Lianne M. Nieuwenhuis, Annechien J. A. Lambeck, Bernadien H. Jansen, Marius C. van den Heuvel, and Bouke G. Hepkema. Formal Analysis: Lianne M. Nieuwenhuis, Yanni Li, Bao-Li Loza, Shixian Hu, Ranko Gacesa, and Michiel D. Voskuil. Methodology: Vincent E. de Meijer and Eleonora A. M. Festen. Supervision: Vincent E. de Meijer and Eleonora A. M. Festen Visualization: Lianne M. Nieuwenhuis. Writing—original draft: Lianne M. Nieuwenhuis and Yanni Li. Writing—review and editing: all authors.
ACKNOWLEDGMENTS
The authors thank the patients, study nurses, technicians, laboratory staff, and clinicians for their collaboration. The authors acknowledge Prof Cisca Wijmenga, Prof Alexandra Zhernakova, Prof Jingyuan Fu, and Prof Klaas Nico Faber for their scientific guidance. The authors acknowledge Dr Serena Sanna, Dr Arnau Vich Vila, Iwan Hidding, Dr Alexander Kurilshikov, and Dr Valerie Collij for their bioinformatic and statistical guidance. The authors also acknowledge the support of the International Genetics and Translational Research in Transplantation Network (iGeneTRAiN) consortium.
FUNDING INFORMATION
The work was supported by a grant from Stichting Louise Vehmeijer. This organization had no influence on the design, conduct, or data analysis of the study. Yanni Li was supported by the China Scholarship Council (No. 201706940045).
CONFLICTS OF INTEREST
Vincent E. de Meijer reports a VENI research grant by the Dutch Research Council (NWO; grant #09150161810030), a Research grant from the Dutch Ministry of Economic Affairs (Health~Holland Public Private Partnership grant #PPP-2019-024), and a Research grant from the Dutch Society for Gastroenterology (NVGE #01-2021), all outside the submitted work. Eleonora A. M. Festen is supported by a Clinical Fellow grant from the Dutch Research Council (NWO; grant #90719075) and a Research grant from the Dutch Ministry of Economics. Hans Blokzijl received grants from Abbvie. Henkjan Verkade is on the speaker’s bureau, and received grants from Albireo/ Ipsen. He consults and received grants from Mirum. He consults for Orphalan and Vertex. He is on the speaker’s bureau for Astellas. Rinse Weersma consults and received grants from Takeda Pharmaceuticals. He is on the speaker’s bureau for Abbvie and Ferring. He received grants from Johnson and Johnson. Eleonora Festen received grants from Takeda. The remaining authors have no conflicts to report.
DATA TRANSPARENCY STATEMENT
The deidentified participant data that support the findings of this study are available upon reasonable request from the corresponding author, following institutional review board approval. All data, including raw genomics data, is stored in encrypted environments of all respective institutions.
SUPPORTING INFORMATION STATEMENT
Additional supporting information may be found online in the Supporting Information section at the end of the article.
LIST OF NAMES INCLUDED IN AUTHOR ‘TRANSPLANTLINES INVESTIGATORS’
Coby Annema, Stefan P. Berger, Hans Blokzijl, Frank AJA Bodewes, Marieke T. de Boer, Kevin Damman, Martin H. de Borst, Arjan Diepstra, Gerard Dijkstra, Rianne M. Douwes, Caecilia S. E. Doorenbos, Michele F. Eisenga, Michiel E. Erasmus, C Tji Gan, Antonio W. Gomes Neto, Eelko Hak, Bouke G. Hepkema, Marius C. van den Heuvel, Frank Klont, Tim J. Knobbe, Daan Kremer, Jip Jonker, Anna M. Posthumus, Coretta van Leer-Buter, Henri G. D. Leuvenink, Marco van Londen, Willem S. Lexmond, Vincent E. de Meijer, Hubert G. M. Niesters, Gertrude J. Nieuwenhuis-Moeke, L Joost van Pelt, Robert A. Pol, Adelta V. Ranchor, Jan Stephan F. Sanders, Marion J. Siebelink, Riemer J. H. J. A. Slart, J Cas Swarte, Daan J. Touw, Charlotte A. te Velde-Keyzer, Erik A. M. Verschuuren, Michel J. Vos, Rinse K. Weersma, Stephan J. L. Bakker
Footnotes
Lianne M. Nieuwenhuis, Yanni Li, Eleonora A.M. Festen and Vincent E. de Meijer contributed equally to this study.
Eleonora A.M. Festen and Vincent E. de Meijer accept full responsibility for the conduct of the study. Both authors had access to the data and had control of the decision to publish.
Abbreviations: ACR, acute T-cell–mediated cellular rejection; GWAS, genome-wide association study; HLA, human leukocyte antigen; MHC, major histocompatibility complex; SNP, single-nucleotide polymorphism.
Part of the preliminary results were presented at the International Liver Transplant Society (ILTS) conference in May 2023 in Rotterdam, the Netherlands.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.hepcommjournal.com.
Contributor Information
Lianne M. Nieuwenhuis, Email: l.m.nieuwenhuis@umcg.nl.
Yanni Li, Email: yanni19907300@gmail.com.
Bao-Li Loza, Email: baoli@pennmedicine.upenn.edu.
Annechien J.A. Lambeck, Email: a.j.a.lambeck@umcg.nl.
Shixian Hu, Email: dhu.sxhu@hotmail.com.
Ranko Gacesa, Email: r.gacesa@umcg.nl.
Michiel D. Voskuil, Email: m.d.voskuil@umcg.nl.
Bouke G. Hepkema, Email: b.g.hepkema@umcg.nl.
Bernadien H. Jansen, Email: b.h.jansen@umcg.nl.
Hans Blokzijl, Email: h.blokzijl@umcg.nl.
Henk-Jan Verkade, Email: h.j.verkade@umcg.nl.
Marius C. van den Heuvel, Email: m.c.van.den.heuvel@umcg.nl.
Sumeet Asrani, Email: Sumeet.Asrani@BSWHealth.org.
Giuliano Testa, Email: Giuliano.Testa@BSWHealth.org.
Goran Klintmalm, Email: gklintmalm@gmail.com.
James Trotter, Email: James.Trotter@imail.org.
Kim M. Olthoff, Email: kim.olthoff@uphs.upenn.edu.
Abraham Shaked, Email: abraham.shaked@pennmedicine.upenn.edu.
Brendan J. Keating, Email: bkeating@pennmedicine.upenn.edu.
Rinse K. Weersma, Email: r.k.weersma@umcg.nl.
Eleonora A.M. Festen, Email: e.a.m.festen@umcg.nl.
Vincent E. de Meijer, Email: v.e.de.meijer@umcg.nl.
Collaborators: Coby Annema, Stefan P Berger, Hans Blokzijl, Frank AJA Bodewes, Marieke T. de Boer, Kevin Damman, Martin H. de Borst, Arjan Diepstra, Gerard Dijkstra, Rianne M. Douwes, Caecilia SE Doorenbos, Michele F Eisenga, Michiel E. Erasmus, C Tji Gan, Antonio W. Gomes Neto, Eelko Hak, Bouke G. Hepkema, Marius C. van den Heuvel, Frank Klont, Tim J. Knobbe, Daan Kremer, Coretta van Leer-Buter, Henri G. D. Leuvenink, Marco van Londen, Willem S Lexmond, Vincent E de Meijer, Hubert G. M. Niesters, Gertrude J. Nieuwenhuis-Moeke, L Joost van Pelt, Robert A. Pol, Jip Jonker, Anna M. Posthumus, Adelta V. Ranchor, Jan Stephan F. Sanders, Marion J. Siebelink, Riemer JHJA Slart, J Cas Swarte, Daan J. Touw, Charlotte A. te Velde-Keyzer, Erik A. M. Verschuuren, Michel J. Vos, Rinse K. Weersma, and Stephan J. L. Bakker
REFERENCES
- 1.Burra P. The adolescent and liver transplantation. J Hepatol. 2012;56:714–722. [DOI] [PubMed] [Google Scholar]
- 2.Choudhary NS, Saigal S, Bansal RK, Saraf N, Gautam D, Soin AS. Acute and chronic rejection after liver transplantation: What a clinician needs to know. J Clin Exp Hepatol. 2017;7:358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiesner RH, Demetris JA, Belle SH, Seaberg EC, Lake JR, Zetterman RK, et al. Acute hepatic allograft rejection: Incidence, risk factors, and impact on outcome. Hepatology. 1998;28:638–645. [DOI] [PubMed] [Google Scholar]
- 4.Choi D, Liu M, Guttikonda D, Galen K, Guzman G, Jeon H, et al. Recipient risk factors for acute cellular rejection after orthotopic liver transplant - a single-center, retrospective study. Transpl Int. 2020;33:1779–1787. [DOI] [PubMed] [Google Scholar]
- 5.Reindl-Schwaighofer R, Heinzel A, Kainz A, van Setten J, Jelencsics K, Hu K, et al. Contribution of non-HLA incompatibility between donor and recipient to kidney allograft survival: Genome-wide analysis in a prospective cohort. Lancet. 2019;393:910–917. [DOI] [PubMed] [Google Scholar]
- 6.Pineda S, Sigdel TK, Chen J, Jackson AM, Sirota M, Sarwal MM. Novel non-histocompatibility antigen mismatched variants improve the ability to predict antibody-mediated rejection risk in kidney transplant. Front Immunol. 2017;8. doi: 10.3389/fimmu.2017.01687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steers NJ, Li Y, Drace Z, D’Addario JA, Fischman C, Liu L, et al. Genomic mismatch at LIMS1 locus and kidney allograft rejection. N Engl J Med. 2019;380:1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markkinen S, Helanterä I, Lauronen J, Lempinen M, Partanen J, Hyvärinen K. Mismatches in gene deletions and kidney-related proteins as candidates for histocompatibility factors in kidney transplantation. Kidney Int Rep. 2022;7:2484–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Leary JG, Kaneku H, Banuelos N, Jennings LW, Klintmalm GB, Terasaki PI. Impact of IgG3 subclass and C1q-fixing donor-specific HLA alloantibodies on rejection and survival in liver transplantation. Am J Transplant. 2015;15:1003–1013. [DOI] [PubMed] [Google Scholar]
- 10.Shaked A, Loza BL, Van Loon E, Olthoff KM, Guan W, Jacobson PA, et al. Donor and recipient polygenic risk scores influence the risk of post-transplant diabetes. Nat Med. 2022;28:999–1005. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Nieuwenhuis LM, Voskuil MD, Gacesa R, Hu S, Jansen BH, et al. Donor genetic variants as risk factors for thrombosis after liver transplantation: A genome-wide association study. Am J Transplant. 2021;21:3133–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenga MF, Gomes-Neto AW, van Londen M, Ziengs AL, Douwes RM, Stam SP, et al. Rationale and design of TransplantLines: A prospective cohort study and biobank of solid organ transplant recipients. BMJ Open. 2018;8:e024502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Little J, Higgins JPT, Ioannidis JPA, Moher D, Gagnon F, von Elm E, et al. Strengthening the reporting of genetic association studies (STREGA): An extension of the STROBE Statement. Hum Genet. 2009;125:131–151. [DOI] [PubMed] [Google Scholar]
- 14.Illumina. Infinium ® Genotyping Data Analysis. Tech Note. 2014; (Figure 1):10p.
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah TS, Liu JZ, Floyd JAB, Morris JA, Wirth N, Barrett JC, et al. OptiCall: A robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics. 2012;28:1598–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minimac Imputation -- Fuchsberger C. Abecasis GR, Hinds DA. minimac2: Faster genotype imputation. Bioinformatics. 2014;31:782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.https://www.well.ox.ac.uk/~wrayner/tools/Post-Imputation.html IC. A post-Imputation data checking program. Affymetrix Web site. Accessed August 23, 2022.
- 21.UniProt Consortium T . UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook S, Choi W, Lim H, Luo Y, Kim K, Jia X, et al. CookHLA: Accurate imputation of human leukocyte antigens. Accessed March 26, 2023. https://www.nature.com/articles/s41467-021-21541-5 [DOI] [PMC free article] [PubMed]
- 23.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rich S. Type 1 Diabetes Genetics Consortium (V7) [Dataset]. NIDDK Central Repository. 2023. 10.58020/qrdt-eh40 [DOI] [Google Scholar]
- 25.Duquesnoy RJ, Askar M. HLAMatchmaker: a molecularly based algorithm for histocompatibility determination. V. Eplet matching for HLA-DR, HLA-DQ, and HLA-DP. Hum Immunol. 2007;68:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang JYC, Sarwal MM. Transplant genetics and genomics. Nat Rev Genet. 2017;18:309–326. [DOI] [PubMed] [Google Scholar]
- 27.Loupy A, Lefaucheur C. Antibody-mediated rejection of solid-organ allografts. N Engl J Med. 2018;379:1150–1160. [DOI] [PubMed] [Google Scholar]
- 28.Kohut TJ, Barandiaran JF, Keating BJ. Genomics and liver transplantation: Genomic biomarkers for the diagnosis of acute cellular rejection. Liver Transplant. 2020;26:1337–1350. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien RP, Phelan PJ, Conroy J, O'Kelly P, Green A, Keogan M, et al. A genome-wide association study of recipient genotype and medium-term kidney allograft function. Clin Transplant. 2013;27:379–387. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez-Fuentes M, Stapleton CP, Cavalleri GL, Conlon P, Weale ME, Lord GM. The genetic determinants of renal allograft rejection. Am J Transplant. 2018;18:2100–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez-Fuentes MP, Franklin C, Rebollo-Mesa I, Mollon J, Delaney F, Perucha E, et al. Long- and short-term outcomes in renal allografts with deceased donors: A large recipient and donor genome-wide association study. Am J Transplant. 2018;18:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghisdal L, Baron C, Lebranchu Y, Viklický O, Konarikova A, Naesens M, et al. Genome-wide association study of acute renal graft rejection. Am J Transplant. 2017;17:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sindhi R, Higgs BW, Weeks DE, AshokKumar C, Jaffe R, Kim C, et al. Genetic variants in major histocompatibility complex-linked genes associate with pediatric liver transplant rejection. Gastroenterology. 2008;135:830–839.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, Zhang X, Li Y, Yu J, Chen Z, Niu Y, et al. Interorgan communication with the liver: novel mechanisms and therapeutic targets. Front Immunol. 2023;14:1314123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilgelm AE, Richmond A. Chemokines modulate immune surveillance in tumorigenesis, metastasis, and response to immunotherapy. Front Immunol. 2019;10:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Köberle M, Kaesler S, Kempf W, Wölbing F, Biedermann T. Tetraspanins in mast cells. Front Immunol. 2012;3:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eskandari SK, Gaya da Costa M, Faria B, Petr V, Azzi JR, Berger SP, et al. An interleukin 6-based genetic risk score strengthened with interleukin 10 polymorphisms associated with long-term kidney allograft outcomes. Am J Transplant. 2022;22(suppl 4):45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Nieuwenhuis LM, Keating BJ, Festen EAM, de Meijer VE. The impact of donor and recipient genetic variation on outcomes after solid organ transplantation: A scoping review and future perspectives. Transplantation. 2022;106:1548–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tinckam KJ, Rose C, Hariharan S, Gill J. Re-examining risk of repeated HLA mismatch in kidney transplantation. J Am Soc Nephrol. 2016;27:2833–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiebe C, Kosmoliaptsis V, Pochinco D, Gibson IW, Ho J, Birk PE, et al. HLA-DR/DQ molecular mismatch: A prognostic biomarker for primary alloimmunity. Am J Transplant. 2019;19:1708–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forner D, Liwski R, Alwayn I. Human leukocyte antigen, allele, and eplet mismatches in liver transplantation; observations from a small, single center cohort. Hum Immunol. 2018;79:154–159. [DOI] [PubMed] [Google Scholar]
- 42.Muro M, López-Álvarez MR, Campillo JA, Marin L, Moya-Quiles MR, Bolarín JM, et al. Influence of human leukocyte antigen mismatching on rejection development and allograft survival in liver transplantation: Is the relevance of HLA-A locus matching being underestimated? Transpl Immunol. 2012;26. doi: 10.1016/j.trim.2011.11.006 [DOI] [PubMed] [Google Scholar]
- 43.Na GH, Kim EY, Hong TH, You YK, Kim DG. Effects of preoperative positive cross-match and HLA mismatching on early acute cellular rejection and graft survival in living donor liver transplantation. Ann Transplant. 2015;20:553–560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.