Abstract
Budding of lentiviruses occurs at the plasma membrane, but the preceding steps involved in particle assembly are poorly understood. Since the Gag polyprotein mediates virion assembly and budding, studies on the localization of Gag within the cell should provide insight into the mechanism of particle assembly. Here, we utilize biochemical fractionation techniques as well as high-resolution confocal imaging of live cells to demonstrate that Gag is localized at the plasma membrane in a striking punctate pattern. Mutation of the N-terminal myristoylation site results in the formation of large cytosolic complexes, whereas mutation of the N-terminal basic residue cluster in the matrix domain redirects the Gag protein to a region partially overlapping the Golgi apparatus. In addition, we show that Gag and Env colocalize at the plasma membrane and that mistargeting of a mutant Gag to the Golgi apparatus alters the pattern of surface expression of Env.
Morphological studies have established that the budding of type C retroviruses and lentiviruses such as human immunodeficiency virus type 1 (HIV-1) occurs at the plasma membrane (9, 13). However the earlier steps involving assembly of viral particles are poorly understood. Expression of the viral Gag polyprotein is both necessary and sufficient for assembly and budding of virus-like particles (5, 8, 28). Gag is synthesized on cytoplasmic polysomes and is posttranslationally targeted to the plasma membrane, where it oligomerizes into electron-dense structures that can be visualized by electron microscopy. Thus, studies of the localization of Pr55Gag should provide insights into the mechanisms of particle assembly.
HIV-1 Gag has been shown to be localized to the plasma membrane by both biochemical and cytological methods (1, 10). The membrane binding domain (M) of Pr55Gag has previously been mapped within the N-terminal region of the matrix domain (MA), and it includes N-terminal myristate and a cluster of basic amino acids (36, 41). Myristate inserts into the lipid bilayer by hydrophobic interactions, while the positively charged residues associate via electrostatic interactions with negatively charged phospholipids on the inner face of the plasma membrane (20, 27, 32, 41). Although ultrastructural characterization of full-length Pr55Gag is not yet available, nuclear magnetic resonance and X-ray crystal structures for the nonmyristoylated p17MA protein indicate that the basic residues form a β-pleated sheet that is exposed to solvent and potentially available for membrane binding (12, 18, 19). Moreover, cryoelectron microscopy reveals an apparently elongated structure of full-length Gag within viral particles, suggesting close accessibility of the MA region of the Gag precursor to viral membranes (7). These results predict that both the myristate and the basic motif in HIV-1 Gag should contribute significantly to membrane interactions.
Recently, several reports have used subcellular fractionation coupled with sucrose gradient sedimentation to identify large complexes of Gag proteins in the cytoplasm (14, 22, 24). These Gag complexes have been postulated to represent putative virus assembly intermediates that form in the cytosol prior to budding. However, since these methods involve cell disruption with detergent and/or salt addition, they may not accurately reflect the localization of Gag in intact cells. Here, we have applied high-resolution confocal microscopy and quantitative imaging to the study of Gag localization in intact, live cells. The results from confocal imaging as well as biochemical fractionation clearly indicate that Gag is localized at the plasma membrane in a striking punctate pattern. In addition, we provide evidence for colocalization of Gag and Env at the plasma membrane and show that intracellular targeting of a mutant Gag results in altered surface expression of Env.
MATERIALS AND METHODS
Antibodies and reagents.
Anti-p24CA monoclonal antibody was purchased from ABI (Columbia, Md.). Anti-p17MA polyclonal antibody was from Intracel (Bartels Diagnostics, Issaquah, Wash.). Polyclonal anti-p24CA and anti-Env antibodies were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (Rockville, Md.). Anti-Grp78 was from StressGene (Victoria, British Columbia, Canada); anti-GM130 and anti-p115 were from Transduction Labs (Lexington, Ky.). Anti-IL2-R (CD25) was from Chemicon (Temecula, Calif.). Anti-rabbit rhodamine, anti-goat fluorescein isothiocyanate (FITC), anti-mouse Cy 5, and anti-human rhodamine secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, Pa.). β-Glycan was detected with a polyclonal antibody (a gift from J. Massaguè, Sloan-Kettering Institute, New York, N.Y.), and W6/32 HLA was detected with a monoclonal antibody (Monoclonal Core Facility, Memorial Sloan-Kettering Cancer Center, New York, N.Y.). 2-Hydroxymyristic acid, streptolysin O, and brefeldin A were purchased from Sigma (St. Louis, Mo.). All other reagents were analytical grade or better. Brefeldin A was prepared in methanol as a 5-mg/ml stock and used at a final concentration of 5 μg/ml for 1 or 2 h.
DNA plasmids.
Pr55Gag was expressed from the noninfectious vector pHXB2ΔBalID25S, a proviral construct containing a deletion within Pol and an inactivating point mutation at the protease active site. Mutations abrogating myristoylation (G2A) or replacing the eight basic residues with neutral asparagine residues (8N) were introduced into the proviral construct and are described elsewhere (41). Plasmid pGag-EGFP was constructed as follows. Plasmid p55M1234, a Rev-independent full-length gag gene clone (31), was fused to green fluorescent protein in pEGFPN1 (Clontech, Palo Alto, Calif.) by PCR-directed mutagenesis (W. van't Hof, unpublished results). Plasmid pTac-TGN38, which expresses a cytoplasmic domain of CD25 fused to TGN38, was a generous gift from Tim McGraw (Cornell University Medical College, New York, N.Y.).
Cell culture and transfections.
COS-1 and COS-7 cells (American Type Culture Collection) and HeLa S3 cells (a kind gift from Robert G. Roeder, The Rockefeller University, New York, N.Y.) were cultured in 10% fetal bovine serum in Dulbecco modified Eagle medium (DMEM) and subcultured the day prior to transfection. COS cells were transfected with FuGene 6 (Roche Diagnostics, Indianapolis, Ind.) or Lipofectamine 2000 (Life Technologies, Gaithersburg, Md.) and HeLa S3 cells were transfected with Lipofectin with PLUS reagent (Life Technologies) according to the manufacturer's instructions. Cells were processed at 48 h posttransfection as described below. For immunofluorescence, cells were subcultured 18 to 24 h posttransfection into 6-cm-diameter dishes containing microscope coverslips and fixed and stained 48 h posttransfection as described below.
Cell fractionation and sucrose flotation assays.
Cell homogenates were obtained as described previously (11) with minor modifications. Cells were washed twice in ice-cold 10 mM Tris (pH 8.0)–1 mM EDTA–150 mM NaCl and harvested by centrifugation at 1,000 × g for 3 min at 4°C. The cells were swollen on ice in hypotonic buffer (10 mM HEPES [pH 7.5], 1.5 mM MgCl2, 5 mM KCl, 1 mM dithiothreitol). Cell homogenates were obtained by osmotic shock using Dounce homogenization and adjusted to 0.25 M sucrose and 1 mM EDTA, and the nuclei were removed by centrifugation at 1,000 × g for 3 min at 4°C. A P-10 fraction resulted from centrifugation at 10,000 × g for 10 min at 4°C. S-100 and P-100 fractions were obtained by ultracentrifugation of the S-1 and S-10 supernatants, respectively, at 100,000 × g for 1 h at 4°C. Membrane flotation assays were performed as described previously (33). Briefly, 200 μl of cell homogenates or individual pellet fractions from P-10 and P-100 fractionation were adjusted to 70% (wt/vol) sucrose in phosphate-buffered saline (PBS) in a 1-ml volume and were layered successively with 7 ml of 65% sucrose and 3.5 ml of 10% sucrose, respectively, in SW40 tubes. Samples were centrifuged for 14 h at 100,000 × g, and fractions were collected from the top and analyzed by sodium dodecyl sulfate (SDS)-gel electrophoresis and Western blotting using an anti-p24CA antibody. Sometimes, membranes were stripped and reprobed with a sheep anti-Env polyclonal antibody.
Metabolic myristoylation inhibition.
Cells transfected with pGag-EGFP or pHXB2 expressing wild-type (wt) Gag or 8N Gag were treated with 2-hydroxymyristate (2-OH-Myr) 24 h following transfection essentially as described previously (37). Cells were washed twice in serum-free DMEM and incubated overnight in 100 μM 2-OH-Myr in DMEM containing 2% dialyzed fetal bovine serum and 5% delipidated bovine serum albumin. Alternatively, cells were treated for 6 h with 250 μM 2-OH-Myr prior to cell harvesting or cell processing for immunofluorescence.
Streptolysin O treatment.
Treatment of COS-7 cells transfected with pHXB2 expressing wt Gag, pHXB2 expressing 8N Gag, or pGag-EGFP with streptolysin O was performed as described previously (25). Cells grown on coverslips were chilled and treated with 4 U of streptolysin O/ml for 10 min on ice, followed by a 7.5-min incubation at 37°C. Cells were chilled again for 1 h on ice to release the cytosolic fraction. Cells expressing Gag-enhanced green fluorescent protein (EGFP) were stained with the DNA nuclear stain Hoechst 33342 and visualized directly by confocal microscopy. Cells expressing pHXB2-derived constructs were fixed and processed for immunofluorescence as described below.
Immunofluorescence.
Cell immunofluorescence staining was performed as described previously (21) with modifications. COS-7 or HeLa S3 transfected cells were grown on coverslips and processed 48 h posttransfection. All manipulations were carried out at room temperature unless otherwise indicated. Cells were washed with PBS, fixed with 3.7% formalin–PBS for 15 min, incubated for 10 min in 100 mM glycine in Tris-buffered saline (10 mM Tris [pH 7.5], 150 mM NaCl), and blocked for 30 min in 4% donkey normal serum in PBS. Cells were incubated with a (1:200) dilution of a goat anti-p17MA primary antibody in 4% serum for 2 h. Following four 5-min washes in PBS cells were incubated with an FITC-conjugated anti-goat secondary antibody for 1 h. For costaining experiments for the endoplasmic reticulum (ER), Golgi, and trans-Golgi network (TGN), cells were incubated with monoclonal antibodies against Grp78 (ER), p115, and GM130 (cis and medial Golgi); COP-I (cis to medial Golgi); or IL2R (for Tac-TGN38), respectively, along with the anti-Gag antibody at the dilutions suggested by the manufacturer. For multiple labeling experiments, staining with all secondary antibodies was done simultaneously. Monoclonal antibodies were then incubated with either rhodamine red X-conjugated anti-mouse secondary antibodies or Cy 5-conjugated anti-mouse secondary antibodies. For F105 human monoclonal anti-Env antibody, a rhodamine-conjugated anti-human secondary antibody was used. All secondary-antibody incubations were performed at a 1:500 dilution for 1 h. Cell nuclei were then stained for 5 min with 250 ng of Hoechst 33258/ml, washed four times for 5 min each with PBS, and mounted on microscope slides.
Gag-EGFP expression was visualized directly in live transfected cells. COS-7 or HeLa S3 cells grown on coverslips were washed extensively at room temperature with Hanks' buffered balanced salt solution (Life Technologies) containing HEPES and bicarbonate, and cell nuclei were stained with 1 μg of Hoechst 33342/ml. The coverslips were mounted onto microscope slides and visualized directly.
Confocal microscopy.
Laser scanning confocal microscopy was performed on a Zeiss LSM510 confocal microscope equipped with an Axiovert 100M inverted microscope using a ×63, 1.2-numerical-aperture water immersion lens for imaging. Laser beams with 364-, 488-, 543-, and 633-nm excitation wavelengths were used for Hoechst, FITC, rhodamine, and Cy 5 imaging, respectively. Emission filters were 385 to 470 nm band pass (BP) for Hoechst, 505 to 530 nm BP for FITC, 560 line pass (LP) for rhodamine, and 650 BP for Cy 5. For COS-7 cells, which are about 8 μm thick, z stacks of typically 24 0.4-μm slices were taken, each slice being the average of four laser scans. For HeLa S3 cells, which are about 14 μm thick, 28 0.5-μm slices were imaged. Single confocal sections and z-stack images were processed in Adobe Photoshop. Image rendering of confocal data files was done by deconvolution using Metamorph Imaging Systems (Universal Imaging Corp., West Chester, Pa.).
RESULTS
Subcellular fractionation of wt Gag and M domain mutants.
The first set of experiments was designed to study the localization of Pr55Gag using biochemical fractionation techniques. Transfected COS-1 cells expressing Pr55Gag and M domain Gag mutants were homogenized as described in Materials and Methods, and P-10, P-100, and S-100 fractions were prepared by differential ultracentrifugation. Aliquots from each subcellular fraction were subjected to SDS-gel electrophoresis, and Pr55Gag was detected by Western blotting using an anti-p24CA polyclonal antibody. The results are shown in Table 1. Approximately 95% of wt Gag and 8N Gag fractionated in the P-10 and P-100 fractions. Most (75%) of the 8N Gag partitioned into the P-10 fraction, which may reflect formation of large aggregates or the interaction of 8N Gag with denser membranes due to altered subcellular localization (see below). In contrast, at least 40% of G2A Gag was found in the S-100 (cytosolic) fraction. The remaining G2A Gag in the P-10 and P-100 fractions may represent protein that is either transiently membrane bound or present in large cytoplasmic complexes.
TABLE 1.
Subcellular localization of wt and mutant Pr55Gag in COS cells
| Construct | % in fractiona:
|
||
|---|---|---|---|
| S-100 | P-100 | P-10 | |
| wt Pr55Gag | 6 ± 3 | 35 ± 2 | 59 ± 3 |
| 8N Gag | 4 ± 1 | 21 ± 3 | 75 ± 5 |
| G2A Gag | 41 ± 1 | 34 ± 1 | 24 ± 1 |
Values represent the averages of three independent experiments ± standard deviations.
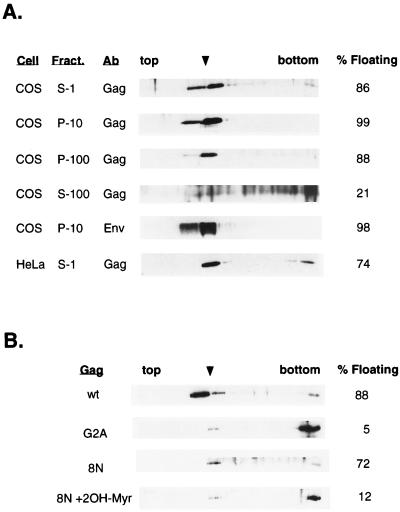
Subcellular fractionation by velocity sedimentation does not resolve macromolecular cytoplasmic complexes or aggregates from membrane-bound protein. Therefore, we performed sucrose flotation analyses of transfected cells expressing wt Gag and Gag mutant proteins (33). As depicted in Fig. 1A, fractionation of S1 homogenates from cells expressing wt Pr55Gag showed that virtually all of the protein floated up to the 10 to 65% sucrose interface. The blots were then stripped and reprobed with an anti-gp120Env polyclonal antibody. Greater than 97% of the HIV-1 Env protein (Fig. 1A) also floated up to the interface. We then subjected the individual subcellular P-10, P-100, and S-100 fractions to flotation analysis. Nearly all wt Gag from both P-10 and P-100 fractions floated up through the sucrose cushion to the 10 to 65% sucrose interface (Fig. 1A). In contrast, the small amount of wt Gag that was found in the S-100 fraction remained predominantly in the bottom part of the gradient after ultracentrifugation, consistent with this fraction representing membrane-free complexes.
FIG. 1.
Fractionation of wt Pr55Gag by sucrose flotation analysis. COS-1 cells transfected with pHXB2ΔBalID25S, which bears the gene encoding wt Pr55Gag, were harvested 48 h posttransfection. Cell homogenates were fractionated and subjected to sucrose flotation centrifugation as described in Materials and Methods. Fractions were collected from the top and analyzed by SDS-gel electrophoresis and Western blotting with an anti-p24CA antibody. (A) Fractionation of wt Pr55Gag S-1 cell homogenates and individual subcellular fractions. Blotting membranes containing wt Pr55Gag in S-1, P-10, and P-100 fractions were stripped and reprobed with a sheep anti-gp160/gp120Env polyclonal antibody (Env). Shown here is the Env fractionation profile for the P-10 fraction. The flotation profile of the HeLa S3 S-1 fraction is shown at the bottom. The distribution of Pr55Gag in the 10 to 65% sucrose interface (arrowhead) over the total protein is indicated at the right. The exposure time for the S-100 fraction was longer than that for the other fractions because very little wt Gag is present in this fraction (see Table 1). Ab, antibody. (B) Fractionation of wt Pr55Gag, G2A Gag, and 8N Gag by sucrose flotation analysis. Transfected COS cells were harvested 48 h posttransfection. Denucleated P-100 fractions were adjusted to 70% sucrose and analyzed by sucrose flotation as described above. For myristoylation inhibition, 8N Gag-transfected cells were incubated with 250 μM 2-OH-Myr for 5 h prior to sucrose flotation analysis.
Flotation analyses were also performed on wt Gag expressed in HeLa S3 cells, where Gag expression levels are about 10-fold lower than in COS cells. Cells were transfected with pHXB2ΔBalID25S. Forty-eight hours posttransfection, S-1 cell homogenates were prepared and membranes were isolated by sucrose flotation assays as described in Materials and Methods. Greater than 74% of the Gag floated to the 10 to 65% sucrose interface (Fig. 1A). Taken together, these results clearly indicate that, at steady state, most of wt Pr55Gag is membrane bound.
Analysis of M domain Gag mutants on sucrose gradients.
We next analyzed the behavior of the M domain Gag mutants by sucrose flotation assays. Greater than 95% of the G2A Gag from the P-100 (Fig. 1B) or P-10 (not shown) fractions remained at the bottom of the gradient. This result is in agreement with that of others (14, 22, 24, 33) and is consistent with the nonmyristoylated protein being present in large cytosolic complexes. The gradient profile obtained with the 8N Gag mutant showed that the majority of the protein in the P-100 fraction (Fig. 1B) and the P-10 fraction (not shown) floated to the 10 to 65% sucrose interface. Since the 8N mutant contains an intact myristoylation site, it was likely that the presence of myristate at the N terminus contributed to membrane association. To verify that myristate was responsible for the membrane binding of 8N Gag, the protein was expressed in the presence of the myristoylation inhibitor 2-OH-Myr. Cells were homogenized as described above, and subcellular fractions were subjected to sucrose flotation. In the presence of 2-OH-Myr, the distribution of 8N Pr55Gag in either the P-100 fraction (Fig. 1B) or the S-1 fraction (not shown) shifted to the bottom of the gradient, with only ∼10 to 15% of the total 8N Gag remaining at the 10 to 65% interface. Likewise, treatment of COS cells expressing wt Gag in the presence of 2-OH-Myr resulted in a shift of nearly all of the protein to the bottom of the tubes (not shown). We conclude that both wt Gag and 8N Gag proteins are membrane associated, whereas nonmyristoylated Gag is largely present in cytosolic complexes. The membrane binding of wt and 8N Gag proteins is dependent on the presence of myristate, implying that the myristate moiety is exposed and available for insertion into the lipid bilayer.
Confocal microscopy reveals the presence of wt Pr55Gag primarily at the plasma membrane.
In order to determine the subcellular localization of Gag more precisely, we analyzed cells expressing wt Gag and Gag mutants by laser scanning confocal microscopy. A Rev-independent construct expressing Pr55Gag fused to green fluorescent protein (pGag-EGFP) was used to observe Gag directly in live cells. This construct expresses Pr55Gag without other viral proteins; viral particles are released at levels similar to those obtained with the proviral pHXB2 expressing wt Pr55Gag (not shown). COS-7 cells transfected with pHXB2-derived constructs expressing wt Pr55Gag, G2A Gag, and 8N Gag were fixed 48 h posttransfection and stained for immunofluorescence.
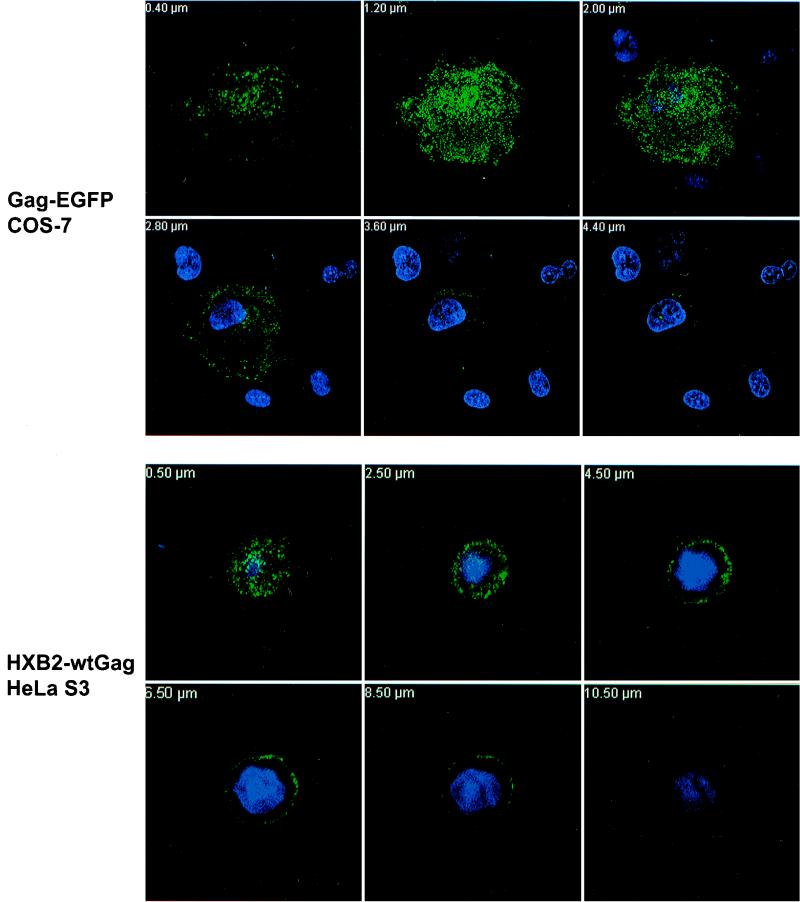
Figure 2 shows a z stack of wt Pr55Gag expressed from live cells transfected with pGag-EGFP (top). The wt Pr55Gag displayed a punctate pattern localized at the cell surface; the strongest FITC fluorescence was observed in the earlier z-stack slices most distal from the coverslip, where the nuclear fluorescence was minimal. Deeper slices inside the cell, where nuclear staining was maximal, showed diminished Gag fluorescence. Surface staining of wt Gag was confirmed by comparison with Nomarski optics (not shown). Identical results were obtained from cells transfected with pHXB2 expressing wt Gag that had been fixed prior to staining (not shown). We also examined HeLa S3 cells, which have a more spherical morphology. HeLa S3 cells transfected with either pHXB2 expressing wt Pr55Gag or pGag-EGFP exhibited distinct plasma membrane fluorescence (Fig. 2, bottom). These results clearly indicate that Pr55Gag is predominantly localized at the plasma membrane.
FIG. 2.
Immunofluorescence of Pr55Gag in live and fixed COS-7 and HeLa S3 cells. (Top) Confocal microscopy analysis of live COS-7 cells transfected with wt Gag-EGFP and stained with Hoechst as described in Materials and Methods. A composite z stack corresponding to 6 confocal sections (from a total of 24) is shown. (Bottom) HeLa S3 cells were transfected with pHXB2ΔBalID25S expressing wt Pr55Gag. Cells were fixed and stained with an anti-p17MA polyclonal antibody and an FITC-conjugated secondary antibody. Nuclei were stained with Hoechst. A composite z stack of 6 confocal sections (out of 28) is shown.
M domain Pr55Gag mutants exhibit altered cellular localization.
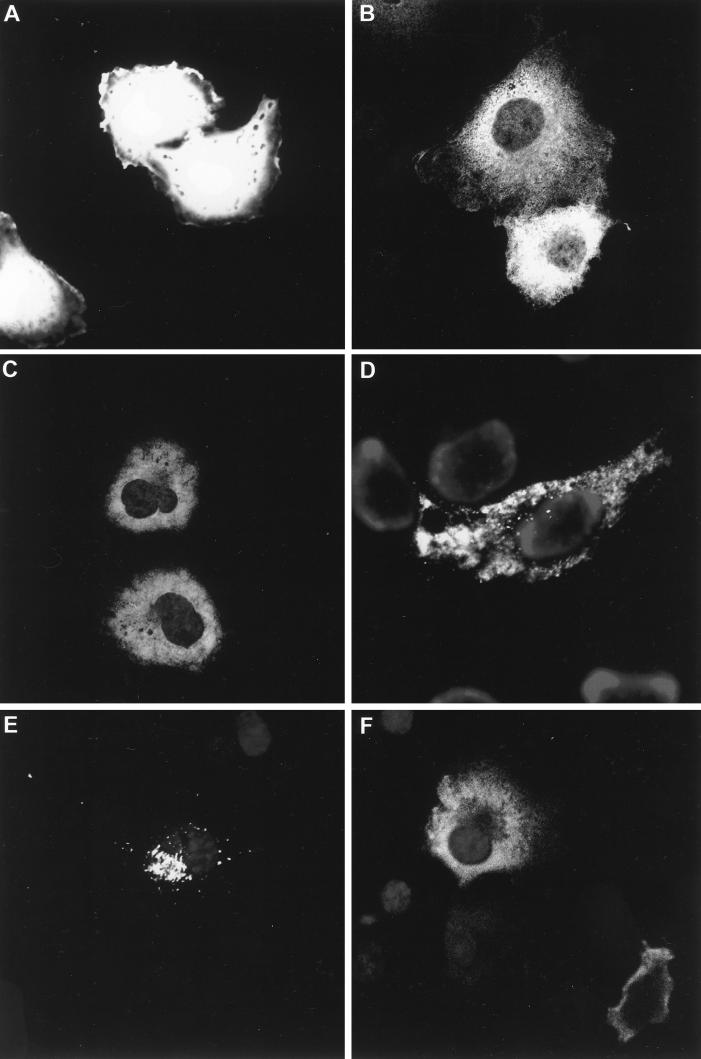
We next examined the cellular localization of the Gag mutants. COS-7 cells were transfected with pHXB2 expressing G2A Gag or 8N Gag and analyzed by confocal microscopy. Single confocal sections are shown in Fig. 3. The nonmyristoylated G2A Gag mutant (B) was dispersed throughout the cell except in the nucleus. The staining pattern of G2A Gag appeared flocculent and differed from the diffuse pattern seen with the soluble green fluorescent protein (Fig. 3A). Superimposition of the FITC images with Nomarski optics confirmed that the G2A staining did not extend to the plasma membrane (not shown). Identical results were obtained with live COS cells transfected with pGag-EGFP and treated with 2-OH-Myr (Fig. 3C) or with 2-OH-Myr-treated COS cells transfected with pHXB2 expressing wt Gag (not shown) or 8N Gag (Fig. 3F). Thus, it is likely that the images in Fig. 2B represent G2A Gag in intracellular cytoplasmic aggregates.
FIG. 3.
Immunofluorescence of Pr55Gag and M domain Gag mutants. Single confocal sections of transfected COS-7 cells are shown. (A) Cells transfected with pEGFP; (B) cells transfected with pHXB2ΔBalID25S expressing G2A Gag; (C) cells transfected with pGag-EGFP and incubated overnight with 100 μM 2-OH-Myr; (D) cells transfected with pGag-EGFP and incubated overnight with 100 μM 2-OH-Myr followed by streptolysin O permeabilization; (E) cells transfected with pHXB2ΔBalID25S expressing 8N Gag; (F) cells transfected with pHXB2ΔBalID25S expressing 8N Gag and incubated overnight with 100 μM 2-OH-Myr to generate nonmyristoylated 8N Gag protein. EGFP confocal imaging was performed on live cells. G2A Gag and 8N Gag cells were fixed and stained as described in Materials and Methods. Cells were fixed and stained with anti-MA antibody and Hoechst as described above.
In cellular fractionations about 40% of G2A Gag is present in the soluble S-100 fraction. If the nonmyristoylated Gag present in the S-100 fraction represents small cytosolic complexes, it should be released by gentle cell permeabilization. pGag-EGFP-transfected COS cells were left untreated or were treated with 2-OH-Myr, followed by incubation with the membrane permeabilizer streptolysin O (25). The untreated wt pGag-EGFP-transfected cells that were permeabilized with streptolysin O maintained the punctate staining at the cell surface (not shown). In contrast, permeabilization of live cells expressing 2-OH-Myr-treated (nonmyristolyated) wt Gag-EGFP with streptolysin O revealed a different pattern from that of the unpermeabilized cells, in that the remaining nonmyristoylated Gag was distributed in very large aggregates throughout the cytoplasm (compare Fig. 3C and D). These data support the notion that nonmyristoylated Gag forms cytosolic complexes, some of which are small enough to be released through the 30-nm pores created by streptolysin O.
Analysis of the cellular distribution of 8N Pr55Gag showed intracellular perinuclear staining polarized on one side of the nucleus (Fig. 3C), which is consistent with the finding that 8N Gag was enriched in the cellular fraction produced by centrifugation at 10,000 × g (Table 1). The morphology of the perinuclear fluorescence in 8N Pr55Gag-transfected COS cells varied considerably. Most cells displayed a crescent shape fluorescence pattern, while some cells displayed a very round, cylindrical structure reminiscent of the Golgi apparatus and other cells presented fragmented fluorescence distributed around the nucleus and included vesicle-like structures dispersed through the cytosol. These observations were consistently seen and cannot be attributed to differences in protein expression levels among the cells. Some minor surface staining was seen as well, perhaps a reflection of the ability of the 8N Gag mutant to produce viral particles at less than 10% of the levels obtained with wt Pr55Gag.
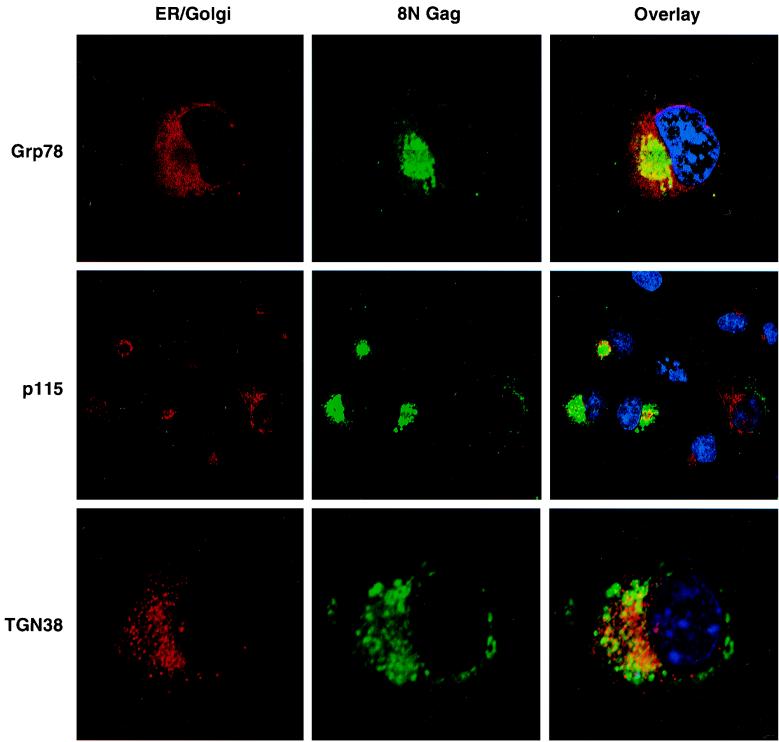
In order to determine the intracellular localization of the 8N Pr55Gag protein more precisely, monoclonal antibodies against cellular markers for ER (Grp78), cis and medial Golgi (p115 and GM130), and coatomer (COP-I) were used. A plasmid encoding TGN38 fused to the cytoplasmic domain of the interleukin-2 receptor (CD25) was used in cotransfection experiments to visualize the TGN. The fusion protein was visualized by staining with an anti-CD25 monoclonal antibody. Comparison of 8N Gag localization with Grp78, a resident ER protein, is shown in Fig. 4. Image rendering of the 8N Gag-Grp78 confocal data files was done through deconvolution methods and showed no overlap between 8N Gag and the ER. The staining pattern of p115, a medial Golgi marker protein, is also shown in Fig. 4 (middle). No costaining of 8N Gag with p115 or any of the other Golgi markers used was observed. It should be noted that cells expressing 8N Pr55Gag from the HXB2 proviral constructs displayed an abnormal Golgi morphology relative to nonexpressing cells. This may be attributed to the expansion of the Golgi to accommodate the processing of other HIV proteins such as Env in the ER and Golgi compartments. The location of 8N Gag relative to the Golgi varied among the three distinct morphologies seen (Fig. 4, middle). In the cells displaying round, compact staining, 8N Gag fluorescence was surrounded by the Golgi; in the cells with the crescent 8N Gag pattern, the Golgi became diffused and expanded and its localization was found to be internal to, but not to overlap, the 8N Gag fluorescence. Finally, in cells where 8N Gag exhibited a scattered, vesicle-like morphology, there was no colocalization between the Golgi and 8N Gag. Incubation of 8N Gag-transfected cells with the Golgi-disrupting agent brefeldin A showed no obvious change in the 8N Gag fluorescence, confirming that the 8N Gag mutant localization does not overlap the Golgi apparatus (not shown). Fluorescence staining of 8N Pr55Gag-transfected cells partially overlapped the TGN; however, the z stacking of these cells revealed that the majority of the TGN was directly below the 8N Gag in the cells analyzed (Fig. 4, bottom). We conclude that membrane-bound 8N Gag localizes at or near the Golgi and TGN-proximal region. Although there is partial overlap of 8N Gag with TGN38 fluorescence, the origin of the membranes to which 8N Gag is bound is unclear.
FIG. 4.
Intracellular localization of 8N Gag and costaining with ER and Golgi markers. Single confocal sections are shown for cells transfected with pHXB2 expressing and 8N Gag stained with cellular markers for ER (rhodamine) and Golgi and TGN (Cy 5) (left column), or anti-p17MA antibodies (FITC; middle column). The overlays are shown on the right. (Top) 8N Gag and Grp78 (ER). (Middle) 8N Gag and p115 (cis to medial Golgi marker). Notice the distinct morphologies of the 8N Gag fluorescence (FITC; center). (Bottom) 8N Gag and TGN38. Notice that the confocal section is focused on the TGN vesicles and that 8N Gag is out of focus.
Colocalization of wt Pr55Gag with Env at the cell surface.
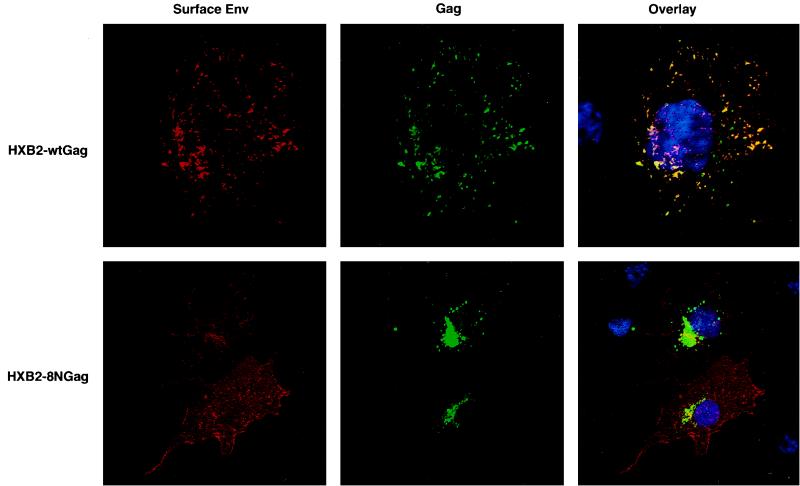
Since Env glycosylation and processing occur in the ER and Golgi compartments, we used double labeling to examine the localization of 8N Gag and Env by confocal microscopy. Partial overlap of 8N Gag with Env was observed in cells permeabilized and stained for 8N Gag and Env (not shown). We then examined the distribution of Env at the cell surface, which represents a small fraction of the total envelope protein expressed in COS cells. Formalin-fixed cells were first incubated with an anti-Env monoclonal antibody and a rhodamine-conjugated secondary antibody to label Env proteins exposed at the cell surface. Cells were then permeabilized and stained with anti-Gag and an FITC-conjugated antibody. Interestingly, results of surface staining of Env differed significantly between cells transfected with pHXB2 expressing wt Gag and 8N Gag. In cells expressing wt Gag, Env surface staining was punctate and overlapped the punctate staining of wt Gag (Fig. 5, top). In contrast, redirection of 8N Gag assembly to intracellular sites resulted in diffuse staining of Env at the surface of the cell (Fig. 5, bottom), with no overlap between 8N Gag and Env staining. Similarly, diffuse surface staining of Env was also observed in cells expressing G2A Gag (not shown).
FIG. 5.
wt Gag directs surface localization of Env to the sites of viral assembly. Cell surface staining of Env in COS cells transfected with pHXB2 expressing wt Gag (top) and 8N Gag (bottom) is shown. Transfected cells were fixed and stained for plasma membrane-associated Env as described in Materials and Methods. Left, Env staining (rhodamine); middle, Gag staining (FITC); right, Gag-Env overlay. Yellow areas indicate wt Gag and Env colocalization at the plasma membrane.
In order to confirm that 8N Gag expression did not alter the integrity of the ER and/or Golgi, cells were fixed and stained with antibodies against endogenous plasma membrane proteins β-glycan (transforming growth factor β RIII) and HLA class I (β2-microglobulin), which traffic through the secretory pathway. Confocal analysis showed no change in expression patterns of either marker in pHXB2-transfected or nontransfected cells, ruling out a functional defect in the Golgi caused by 8N Gag (not shown). These findings strongly suggest that the presence of Gag at the plasma membrane promotes the recruitment of Env to the site of assembly.
DISCUSSION
wt Pr55Gag is predominantly plasma membrane bound.
In this study, we have utilized subcellular fractionation and confocal imaging to determine the localization of Pr55Gag in transfected cells. Biochemical analyses of cell fractions on discontinuous sucrose gradients revealed that greater than 85% of the wt Gag was membrane associated in both COS and HeLa cells (Fig. 1). A similar conclusion was reached by Spearman and coworkers (33) using continuous sucrose gradients. In contrast, several recent reports have found only 30 to 40% of the total Pr55Gag at the 10 to 65% sucrose interface (14, 22, 24). The reasons for the discrepancies in the amount of Gag that floats may be related to the use of sonication and/or salt addition during sample preparation by other investigators or to the use of different cell types and virus strains.
All of the cellular fractions analyzed in Fig. 1 were denucleated by centrifugation at 1,000 × g. Any Gag complexes that are associated with the nuclear pellet would therefore not have been detected. It is difficult to perform gradient analyses with the dense nuclear pellet because many intracellular membranes stick to the nucleus nonspecifically. To circumvent this problem and to obtain a precise view of Gag localization in intact cells, we performed confocal imaging of both fixed and live cells. The fluorescence of wt Gag was maximal during the first one-third of the confocal slices, which corresponded to the region of the cell most distal from the coverslip, and was well resolved from the Hoechst-stained nucleus, which had the strongest intensity in the last one-third of the slices of the confocal z stacks proximal to the coverslips. In addition, maximal wt Gag fluorescence overlapped with the cell surface on the basis of Nomarski optics. The images depicted in Fig. 2, as well as numerous images that were analyzed but not presented here, clearly establish that Gag is predominantly localized at the plasma membrane. This finding is further confirmed by the images of Fig. 5 showing Gag colocalization with surface-stained Env. A small amount of intracellular Gag was also observed in the perinuclear region. This may represent the non-membrane-bound Gag complexes that do not float up through the sucrose gradient (Fig. 1). Punctate staining patterns of wt Gag have also been reported by others (23, 29, 33). Taken together, we conclude that Pr55Gag is predominantly plasma membrane bound. Since Gag is the “particle-forming machine” (8) of the virion, it is likely that virus assembly occurs mainly at the plasma membrane.
Nonmyristoylated Gag forms cytosolic complexes.
Myristoylation has previously been shown to be required for Gag membrane binding and efficient virus production (1, 10). Mutation of the N-myristoylation site produces a mutant protein (G2A Gag) that pellets with the P-100 fraction but that does not float to the top of sucrose gradients (Fig. 1). These observations have led us and others to conclude that G2A Gag is present in large, cytosolic complexes (14, 22, 24). Alternatively, G2A Gag is weakly or transiently associated with membranes. A unique feature of the present study is the use of 2-OH-Myr, a myristoylation inhibitor, to demonstrate that nonmyristoylated wt Gag and nonmyristoylated 8N Gag are present in cytosolic complexes that are indistinguishable from those containing G2A Gag by confocal imaging. Most of these complexes must be fairly large, as they cannot be released from the cell by streptolysin O treatment. Myristoylation is therefore necessary for membrane binding as well as plasma membrane targeting of HIV-1 Gag.
Basic amino acids in MA constitute a plasma membrane targeting motif.
The data in Fig. 2 to 5 clearly establish that the basic motif, in conjunction with N-terminal myristoylation, is required for the specific targeting of Pr55Gag to the plasma membrane. A similar conclusion has been reached by others (4, 23, 40). The finding that the Pr55Gag 8N Gag mutant is membrane bound contrasts with findings concerning the behavior of 8N p17MA matrix chimeras, which display reduced membrane association in the absence of the basic cluster (41). It is likely that, when MA alone is present, the basic motif binds directly to acidic membrane phospholipids via electrostatic interactions. However, in the context of wt Pr55Gag, domains downstream of MA may contribute to membrane binding (17, 26). In the absence of a basic motif, 8N Gag must use the same or additional domains for membrane binding and targeting within perinuclear membranes, since myristate alone is insufficient for a stable membrane association. The I domain within the nucleocapsid (NC), which has been implicated in the membrane association of Gag, may contribute directly or indirectly to membrane binding of 8N Gag (3, 29). In fact, a recent study (1a) has shown that the MA basic cluster and the NC domain both promote recruitment of RNA and subsequent Gag multimerization. Perhaps formation of large Gag-RNA complexes enhances the membrane binding of multimerized Gag molecules via myristate alone.
Gag and Env colocalize at the plasma membrane.
Several lines of evidence support the hypothesis that Gag and Env interact. Mutations within the MA domain of Gag affect the ability of Env to be incorporated into virions (4, 6, 16, 39). In polarized epithelial cells, expression of Env restricts Gag-mediated budding to the basolateral plasma membrane (15). Moreover, expression of Env has been shown to direct Gag localization to the somatic region in neurons (38). In addition, glutathione S-transferase pull-down assays revealed an interaction between Gag and Env in vitro (2). However, no study has shown an interaction between Gag and Env in vivo using immunocytochemistry at the light microscopy level. Here, we provide evidence for Gag-Env interaction by demonstrating that Gag staining coincides with surface-stained Env by confocal microscopy analysis.
Compared to that in wt Gag-expressing cells, the surface staining pattern of Env in 8N Gag-expressing cells is strikingly different (Fig. 5). At least two explanations can be invoked to account for this result. It is possible that the presence of Gag at the plasma membrane restricts Env localization at the surface to discrete, punctate locations. This would be consistent with the hypothesis that Gag-Env interactions are important for virion budding. Alternatively, it is possible that expression of 8N Gag results in disorganization (or in a morphological or functional alteration) of the Golgi apparatus and therefore affects the subsequent surface targeting of proteins trafficking through the secretory pathway. However, no alteration of surface staining of β-glycan or HLA was observed in cells expressing G2A or 8N Gag compared to that for the wt. Moreover, Env surface fluorescence was also diffuse in cells expressing G2A Gag. It is therefore likely that an interaction between Gag and Env occurs at the plasma membrane and serves to incorporate the Env protein into Gag assembly site complexes.
In conclusion, the biochemical and high-resolution cytological analyses presented here clearly establish that the majority of HIV-1 Pr55Gag in intact COS and HeLa cells is localized at the plasma membrane. Our results contrast with several recent biochemical fractionation studies documenting the presence of Pr55Gag in large, “cytosolic” complexes (14, 22, 24). This finding has led others to conclude that Gag multimerization precedes membrane binding and that the non-membrane-bound Gag complexes represent assembly intermediates. Using pulse-chase analysis, our laboratory has recently shown that cytosolic Gag complexes are transiently formed by newly synthesized Gag but that they represent a minor short-lived species and are rapidly degraded intracellularly (35). It is therefore likely that the cytosolic Gag complexes observed by others represent membrane-dissociated protein produced during cell lysis or subcellular fractionation. Our findings are consistent with ultrastructural electron microscopy studies, in which the appearance of electron-dense structures was found only at the plasma membrane in HIV-1-infected cells (9), and with biochemical studies showing that the presence of membranes induces Gag multimerization in vitro (30). The formation of cytosolic Gag assembly complexes has previously been invoked as an explanation for the ability of mutant, nonmyristoylated Gag proteins to be incorporated into wt particles. However, the presence of wt Gag oligomers at the plasma membrane could also serve to recruit nonmyristoylated mutant Gag molecules to the membrane through protein-protein (through the CA dimerization domain) or protein-RNA interactions (between I domains). This mode of recruitment is somewhat analogous to the recruitment of soluble SH2-containing proteins to the membrane by tyrosine-phosphorylated membrane receptors (34). We propose that the assembly of HIV-1 Pr55Gag into oligomers and ultimately into virus like particles primarily occurs at the plasma membrane and that Pr55Gag directs recruitment of Env to sites of virus assembly at the plasma membrane.
ACKNOWLEDGMENTS
We thank George Pavlakis for the Rev-independent Gag construct, Wouter van't Hof for the Gag-EGFP construct and many helpful discussions, James Rothman (Sloan-Kettering Institute) for the generous gift of Golgi and ER cellular markers, and Carl Blobel (Sloan-Kettering Institute) for the anti-CD25 antibody. We also thank Michael Weiden (New York University School of Medicine) and Yael Webb for critical reading of the manuscript, Marc Tritel and Onn Wolf Lindwasser for many helpful discussions, and Raisa Louft-Nisenbaum for expert technical support. We are thankful to Katia Manova-Todorova and Ali McBride at the Molecular Cytology Core Facility for expert assistance with confocal fluorescence microscopy and Debra Alston for secretarial assistance.
This work was supported by NIH grant CA72309.
REFERENCES
- 1.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Burniston M T, Cimarelli A, Colgan J, Curtis S P, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 3.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Facke M, Janetzko A, Shoeman R L, Krausslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freed E O. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 6.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller S D, Wilk T, Gowen B E, Krausslich H G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 8.Garnier L, Bowzard J B, Wills J W. Recent advances and remaining problems in HIV assembly. AIDS. 1998;12:S5–S16. [PubMed] [Google Scholar]
- 9.Gelderblom H R, Hausmann E H, Ozel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 10.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermida-Matsumoto L, Resh M D. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55Gag and p17MA. J Virol. 1999;73:1902–1908. doi: 10.1128/jvi.73.3.1902-1908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krausslich H G, Ochsenbauer C, Traenckner A M, Mergener K, Facke M, Gelderblom H R, Bosch V. Analysis of protein expression and virus-like particle formation in mammalian cell lines stably expressing HIV-1 gag and env gene products with or without active HIV proteinase. Virology. 1993;192:605–617. doi: 10.1006/viro.1993.1077. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y M, Liu B, Yu X F. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J Virol. 1999;73:5654–5662. doi: 10.1128/jvi.73.7.5654-5662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodge R, Gottlinger H, Gabuzda D, Cohen E A, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger H G. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol. 1995;69:3824–3830. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mammano F, Ohagen A, Hoglund S, Gottlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massiah M A, Starich M R, Paschall C, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 19.Massiah M A, Worthylake D, Christensen A M, Sundquist W I, Hill C P, Summers M F. Comparison of the NMR and X-ray structures of the HIV-1 matrix protein: evidence for conformational changes during viral assembly. Protein Sci. 1996;5:2391–2398. doi: 10.1002/pro.5560051202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray D, Hermida-Matsumoto L, Buser C A, Tsang J, Sigal C T, Ben-Tal N, Honig B, Resh M D, McLaughlin S. Electrostatics and the membrane association of Src: theory and experiment. Biochemistry. 1998;37:2145–2159. doi: 10.1021/bi972012b. [DOI] [PubMed] [Google Scholar]
- 21.Okamura H, Resh M D. p80/85 cortactin associates with the Src SH2 domain and colocalizes with v-Src in transformed cells. J Biol Chem. 1995;270:26613–26618. doi: 10.1074/jbc.270.44.26613. [DOI] [PubMed] [Google Scholar]
- 22.Ono A, Freed E O. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono A, Orenstein J M, Freed E O. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J Virol. 2000;74:2855–2866. doi: 10.1128/jvi.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paillart J-C, Göttlinger H G. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J Virol. 1999;73:2604–2612. doi: 10.1128/jvi.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prekeris R, Klumperman J, Chen Y A, Scheller R H. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reil H, Bukovsky A A, Gelderblom H R, Gottlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resh M D. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 28.Sakalian M, Hunter E. Molecular events in the assembly of retrovirus particles. Adv Exp Med Biol. 1998;440:329–339. doi: 10.1007/978-1-4615-5331-1_43. [DOI] [PubMed] [Google Scholar]
- 29.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scarlata S, Ehrlich L S, Carter C A. Membrane-induced alterations in HIV-1 Gag and matrix protein-protein interactions. J Mol Biol. 1998;277:161–169. doi: 10.1006/jmbi.1997.1615. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigal C T, Zhou W, Buser C A, McLaughlin S, Resh M D. Amino-terminal basic residues of Src mediate membrane binding through electrostatic interaction with acidic phospholipids. Proc Natl Acad Sci USA. 1994;91:12253–12257. doi: 10.1073/pnas.91.25.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas S M, Brugge J S. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 35.Tritel M, Resh M D. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J Virol. 2000;74:5845–5855. doi: 10.1128/jvi.74.13.5845-5855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veronese F D, Copeland T D, Oroszlan S, Gallo R C, Sarngadharan M G. Biochemical and immunological analysis of human immunodeficiency virus gag gene products p17 and p24. J Virol. 1988;62:795–801. doi: 10.1128/jvi.62.3.795-801.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webb Y, Hermida-Matsumoto L, Resh M D. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 38.Weclewicz K, Ekstrom M, Kristensson K, Garoff H. Specific interactions between retrovirus Env and Gag proteins in rat neurons. J Virol. 1998;72:2832–2845. doi: 10.1128/jvi.72.4.2832-2845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X, Yuan X, Matsuda Z, Lee T H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan X, Yu X, Lee T H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]