Abstract
The retroviral Gag protein is capable of directing the production and release of virus-like particles in the absence of all other viral components. Budding normally occurs after Gag is transported to the plasma membrane by its membrane-targeting and -binding (M) domain. In the Rous sarcoma virus (RSV) Gag protein, the M domain is contained within the first 86 amino acids. When M is deleted, membrane association and budding fail to occur. Budding is restored when M is replaced with foreign membrane-binding sequences, such as that of the Src oncoprotein. Moreover, the RSV M domain is capable of targeting heterologous proteins to the plasma membrane. Although the solution structure of the RSV M domain has been determined, the mechanism by which M specifically targets Gag to the plasma membrane rather than to one or more of the large number of internal membrane surfaces (e.g., the Golgi apparatus, endoplasmic reticulum, and nuclear, mitochondrial, or lysosomal membranes) is unknown. To further investigate the requirements for targeting proteins to discrete cellular locations, we have replaced the M domain of RSV with the product of the unique long region 11 (UL11) gene of herpes simplex virus type 1. This 96-amino-acid myristylated protein is thought to be involved in virion transport and envelopment at internal membrane sites. When the first 100 amino acids of RSV Gag (including the M domain) were replaced by the entire UL11 sequence, the chimeric protein localized at and budded into the Golgi apparatus rather than being targeted to the plasma membrane. Myristate was found to be required for this specific targeting, as were the first 49 amino acids of UL11, which contain an acidic cluster motif. In addition to shedding new light on UL11, these experiments demonstrate that RSV Gag can be directed to internal cellular membranes and suggest that regions outside of the M domain do not contain a dominant plasma membrane-targeting motif.
Although viruses use a variety of different replication strategies, one step common to all of these pathways is the accumulation of virion components at a specific location within the cell to form new infectious particles. To efficiently arrive at the designated site of assembly, viral proteins contain specific targeting signals. Retroviruses are useful for investigating these targeting signals because budding is directed by a single, well-studied viral protein named Gag (Fig. 1).
FIG. 1.
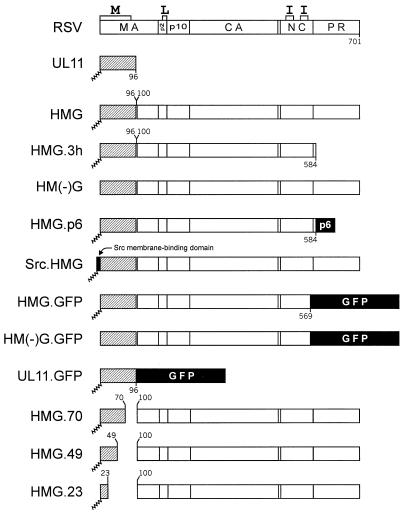
UL11-Gag chimeras. The wild-type RSV Gag (unshaded) and UL11 (hatched) proteins are aligned at their N termini. The positions of the assembly domains are marked along the top of Gag. Sites cleaved by the viral protease are marked by vertical lines through Gag. N-terminal myristylation is indicated by a squiggled line. Numbers indicate the amino acids included in each construct. The p6 sequence of HIV, the first 10 amino acids of the Src oncoprotein, and the GFP sequences are indicated by black boxes.
Gag proteins are synthesized on free ribosomes and subsequently transported and assembled into particles on the cytoplasmic face of the plasma membrane. Only three small modular regions of Gag (termed assembly domains) are required for particle formation (39). The interaction (I) domain within the nucleocapsid (NC) region facilitates the formation of the Gag-Gag contacts necessary for the production of dense particles (3, 6, 43). The membrane-targeting and -binding (M) domain is responsible for directing Gag from the cytoplasm to the plasma membrane (21, 40, 47). Once at the plasma membrane, the late (L) domain coordinates the final release of the particle from the cell surface (14, 22, 45). At this time, Gag is cleaved by the viral protease to generate the mature viral proteins (MA, p2, p10, CA, NC, PR; J. W. Wills and R. C. Craven, Editorial, AIDS 5:639–654, 1991) (Fig. 1).
Although Gag is normally directed to the plasma membrane, proteins from some other viruses concentrate at different membrane sites. For example, herpes simplex virus type 1 (HSV-1) expresses a 96-amino-acid protein from the UL11 gene (18) (here referred to as the UL11 protein) (Fig. 1) that appears to be targeted to perinuclear membranes (1). Because Gag and UL11 localize to distinct cellular locations, it is likely that they use different targeting signals.
The targeting signals of Gag have been extensively studied. The retroviral M domain is contained within the matrix (MA) sequence at the amino terminus of Gag (Fig. 1). Positively charged amino acids are important features of the Rous sarcoma virus (RSV) and human immunodeficiency virus (HIV) M domains (23, 50), as is myristate for the M domains of HIV, murine leukemia virus, and Mason-Pfizer monkey virus (7, 15, 29, 30, 38). M is required for plasma membrane localization, and disruption of its amino terminus abrogates budding and results in Gag remaining cytoplasmic (7, 38, 47). In addition, M alone is sufficient to target heterologous cytoplasmic proteins to the plasma membrane (40, 50).
Despite its importance for plasma membrane localization, it is not entirely clear that the M domain is the sole targeting determinant within Gag. For example, cleavage of Gag during retroviral particle maturation releases MA from the rest of the protein. Upon entry of the mature particle into a host cell, some of this free MA no longer associates with the plasma membrane, even though it contains the intact M domain (8, 16, 28). Why is the M domain inactive in this context? It is possible that cleavage of MA from the remainder of Gag allows conformational rearrangements that disrupt the M domain and prevent plasma membrane binding (37, 50). An alternate explanation is that sequences in regions of Gag other than MA affect localization in some way, either by providing additional targeting information that is required for transport to the plasma membrane or by providing cooperativity to strengthen the avidity of the membrane-binding domain. After cleavage of MA, these secondary signals would no longer be available to promote plasma membrane association. To address this issue, we replaced the M domain of RSV with the UL11 protein from HSV-1. Our results show that UL11-Gag chimeras localized to internal membranes (like UL11 alone) rather than to the plasma membrane, suggesting that the C terminus of Gag does not contain dominant plasma membrane-targeting signals. These experiments also revealed some of the sequence elements required for UL11-directed membrane binding and provided reagents for further investigation of the role of UL11 in herpesvirus assembly.
MATERIALS AND METHODS
Previously constructed gag alleles.
The RSV gag gene was obtained from pATV-8, an infectious molecular clone of the Prague C genome (34). The HSV-1 UL11 gene was amplified from the KOS strain (35). The plasmid pSV.Myr1 used in these experiments has been described previously (46). Standard protocols were used for all DNA manipulations (31), and all plasmids were propagated in Escherichia coli DH-1 cells in 2× YT medium containing 25 μg of ampicillin per ml, with the exception of the green fluorescent protein (GFP) vectors, which were propagated in the presence of 50 μg of kanamycin per ml.
Newly constructed gag alleles.
Several Gag chimeras were made to replace the RSV M domain with various portions of the UL11 protein of HSV-1. For the herpesvirus-myristylated Gag (HMG) construct, the entire UL11 coding sequence was obtained by PCR amplification from the KOS genome with a forward primer that was complementary to the HSV sequence located 100 bases upstream of the initiator ATG and a reverse primer complementary to the sequence coding for the final five amino acids of UL11. The 5′ ends of the primers also contained recognition sites for the SstI (forward) and SpeI (reverse) restriction endonucleases (underlined). After amplification with these primers (forward, 5′-ACCAGGCCGTTGAGCTCGCCCTGATCATTA-3′; reverse, 5′-CATTGTTTTGGTACTAGTTCGCTATCGGACAT-3′), the product was digested with SstI and SpeI and ligated to the large fragment generated by digesting pSV.ΔMA6S (21) with SstI and SpeI. The resulting plasmid was named pSV.HMG. The constructs in which N-terminal fragments of UL11 were fused to Gag were also made by PCR with the forward primer listed above in combination with reverse primers that contained an SpeI site (underlined) and that were complementary to internal UL11 sequences (ending at codon 70 [5′-TGAGTGTGGCGCACTAGTGGGTCCGAT-3′], codon 49 [5′-CCCGCGCATATCCACTAGTACGTAGAAAT-3′], or codon 23 [5′-GCGAGACGACCACTAGTTCGTCGGTGAT-3′). The resulting plasmids were named pSV.HMG.70, pSV.HMG.49, and pSV.HMG.23, respectively.
A mutant encoding a myristate-minus form of HMG was produced by combining the products of two PCR fragments before cloning with the SstI and SpeI sites. The “left” fragment was amplified by using the HMG forward primer and a reverse primer that was complementary to the region flanking the UL11 initiator ATG. This reverse primer (5′-GAGAGGCCCATATGTCGGCGAGCGT-3′) retains the ATG, but contains two changes that create an overlapping NdeI site (underlined). The PCR mixture for the “right” fragment used the original HMG reverse primer and a forward primer that contains two mismatches to create an NdeI site (underlined) at the same position mentioned above. This forward primer (5′-GCTCGCCGACATATGGCCCTCTCGTTCT-3′) also contains an additional mismatch (double underlined) that changes the second UL11 codon (glycine) to one for alanine. The two amplified products were digested with NdeI, ligated together, and reamplified with the forward primer used to make the left half and the reverse primer used to make the right half. This product was digested with SstI and SpeI and cloned into pSV.ΔMA6S to create pSV.HM(−)G.
pSV.Src.HMG was made by a similar strategy. For this construct, the left fragment was amplified with pSV.Myr1 as the template and upstream and downstream primers completely complementary to pSV.Myr1 sequences flanking the SstI (nucleotide [nt] 255) and MluI (nt 408) sites, respectively. The right fragment was amplified from pSV.HMG template DNA with a forward primer (5′-ACGCTCGCCACGCGTTGGGCCTCTCGTT-3′) containing an MluI site which overlaps a mismatch (double underlined) that changes the first base of the start codon from an A to a T. The left and right fragments were digested with MluI, ligated together, and reamplified as described above. The resulting fragment was digested with SstI and SpeI and ligated to the large fragment generated by digesting pSV.ΔMA6S with SstI and SpeI.
The protease (PR)-coding region was deleted from pSV.HMG by digesting pSV.Myr1.3h (42) with KpnI and SstII and ligating the large fragment to the small fragment produced by digesting pSV.HMG with KpnI and SstII. The resulting PR mutant was named pSV.HMG.3h. To create pSV.HMG.p6, the large fragment generated by digesting pSV.T10C.p6 (22) with SstI and BlpI was ligated to the small fragment generated by digesting pSV.HMG with the same enzymes.
GFP-containing constructs were made after cloning Gag-coding sequences into the Clontech pEGFP-N2 vector. To do this, oligonucleotide-directed M13 mutagenesis of the wild-type gag gene (46) was used to create an ApaI site (underlined) near the junction of the NC- and PR-coding sequences (5′-TCGGGGCCGTGGCCCGGGCCCGAGCCACCTGCCGTCTCG-3′). Gag sequences were removed from replicative form DNA by SstI-ApaI digestion and placed in frame into the Clontech vector to form pGag.GFP. To create pHMG.GFP, which encoded a GFP-tagged form of the UL11-Gag chimera, the small fragment from an SstI-EspI digest of pSV.HMG was purified and ligated to the large fragment from an SstI-EspI digest of pGag.GFP. pHM(−)G.GFP was generated by the same procedure, but the small fragment was obtained from pSV.HM(−)G. To create pUL11.GFP, the UL11 coding sequence was amplified with the same forward primer that was used to create HMG. The reverse primer (5′-TCAGGAATTCGCTATCGGA-3′) was complementary to the coding sequence of the C terminus of UL11 and had a noncomplementary region that contained an EcoRI site (underlined). This product was digested with SstI and EcoRI and ligated to the large fragment generated by digestion of pEGFP-N2 with the same enzymes.
To enable transient and stable expression in QT6 avian cells, UL11-coding sequences were transferred into an RSV proviral vector that contains the hygromycin resistance gene. pRC.HMG was constructed by ligating the large fragment generated by digesting the BHRCAN vector (12) with SstI and HpaI to the small fragment generated by digesting pSV.HMG with BssHII, treating it, with Klenow fragment, and digesting it with SstI.
Transfection and labeling of cells and immunoprecipitation of Gag proteins.
COS-1 cells were transfected by the DEAE-dextran-chloroquine method, as previously described (46). Approximately 48 h after transfection, the cells were labeled for either 5 min or 2.5 h with 50 μCi (>1,000 Ci/mmol) of l-[35S]methionine. The cells and growth medium from each labeled culture were separated and mixed with lysis buffer containing protease inhibitors (46). The Gag proteins were immunoprecipitated with polyclonal rabbit serum against whole RSV (reacts with MA, CA, NC, and PR [42]). The immunoprecipitated proteins were resolved by electrophoresis in sodium dodecyl sulfate–12% polyacrylamide gels and detected by fluorograpy (46). QT6 cells were transfected by the calcium phosphate transfection method as previously described (12).
Budding efficiency was determined by first calculating, for each construct, the ratio of protein released into the medium during a 2.5-h labeling to the amount made in the lysates during a 5-min pulse. The ratio of each mutant was divided by the ratio of the wild type to yield its relative budding efficiency.
Immunofluorescence and confocal and electron microscopy.
Cells were transfected as described above for all microscopic analyses. For immunofluorescence, cells were fixed with 5% acetic acid–95% ethanol at −20°C (48), blocked with 0.1% rabbit serum albumin in phosphate-buffered saline, and incubated with rabbit anti-RSV polyclonal primary antibodies (42) and goat anti-rabbit secondary antibodies conjugated to either fluorescein isothiocyanate (FITC) or tetramethyl rhodamine isothiocyanate (TRITC) (Sigma). The procedure for double-label immunofluorescence was the same as that described above, except that the primary antibody was a mixture of the anti-RSV polyclonal antibody and a mouse anti-Golgi 58-kDa protein (Sigma) and the secondary antibody was a mixture of goat anti-rabbit immunoglobulin G conjugated to TRITC and goat anti-mouse immunoglobulin G conjugated to FITC (Sigma). Cells were visualized by light microscopy and a filter set appropriate for the fluorescent label, and images were captured on Kodak T400 black and white photographic film. Confocal microscopy was done with a Zeiss LSM microscope, and the captured images were colored and digitally combined in Adobe Photoshop. For electron microscopy, transfected cells were grown in Permanox plates, fixed with glutaraldehyde-paraformaldehyde, postfixed in osmium-potassium ferrocyanide, dehydrated through increasing concentrations of ethanol, and embedded in Epon 812 (11). Thin sections were stained with uranyl acetate-lead citrate and viewed on a Phillips 400 electron microscope.
RESULTS
To investigate the role of sequences other than the M domain in plasma membrane targeting, we replaced the first 100 amino acids of RSV Gag with a heterologous membrane-binding sequence. Because its size approximates that of the M domain (96 versus 86 amino acids, respectively) and it is localized to internal membranes, the myristylated herpesvirus protein UL11 was used (Fig. 1). If the M domain is the only source of targeting information within wild-type Gag, then the localization of the UL11-Gag chimera (HMG; Fig. 1) should be similar to that of UL11 alone (i.e., at internal membranes). If Gag sequences outside the M domain promote plasma membrane targeting, then the localization of HMG might be similar to that of full-length Gag or at least different from that of UL11.
The UL11-RSV Gag chimera HMG is budding deficient.
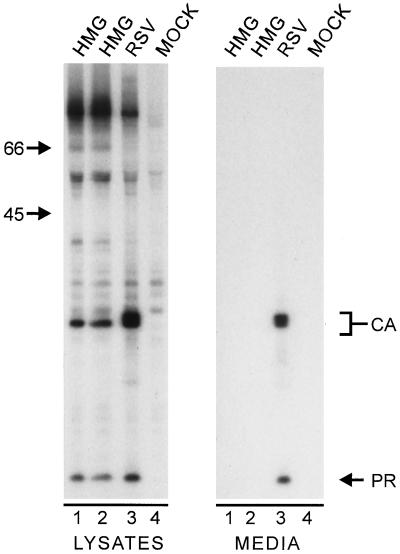
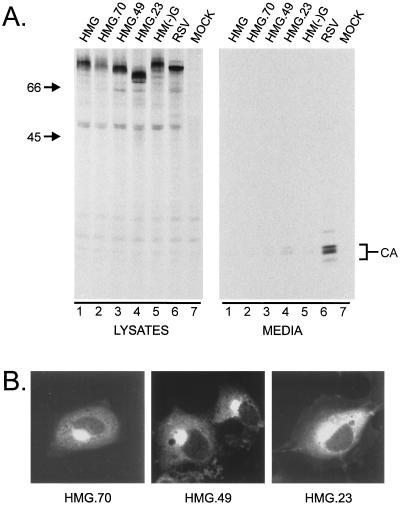
Since Gag can mediate budding when it is localized to the plasma membrane by a heterologous signal (47), particle release was initially monitored as a marker for plasma membrane targeting. When two clones of HMG were expressed in COS-1 cells (Fig. 2), a precursor protein of the expected size (∼76 kDa) was observed in the cell lysates along with the CA and PR Gag cleavage products (4, 11, 46). Although the efficient proteolytic processing of the Gag precursor seen here is indicative of membrane binding (46, 47), it was also noted that the CA in the HMG lanes was not further processed to form the characteristic triplet of bands. This phenotype has previously been associated with RSV Gag mutants that do not bud (47), so it was not surprising that no Gag products (and hence no virus-like particles) from either of the two HMG clones were visible in the extracellular medium (Fig. 2, compare lanes 1 and 2 with lane 3). It is important to note that the lack of budding is not caused by the lack of CA processing, since PR-deficient mutants as well as Gag proteins engineered to produce only the lower CA species bud efficiently (46, 49).
FIG. 2.
Expression of HMG. COS-1 cells transfected with the indicated constructs were labeled for 2.5 h with l-[35S]methionine, and the Gag proteins from the media and cell lysates were immunoprecipitated with anti-RSV antibodies, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and visualized by autoradiography. The numbers to the left are the positions (in kilodaltons) of molecular mass markers. The positions of the CA and PR Gag cleavage products are also indicated. Gag products in the medium are indicative of budding.
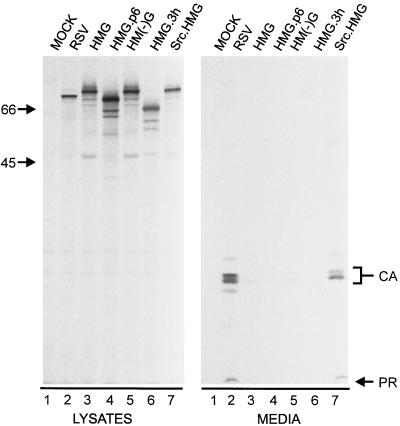
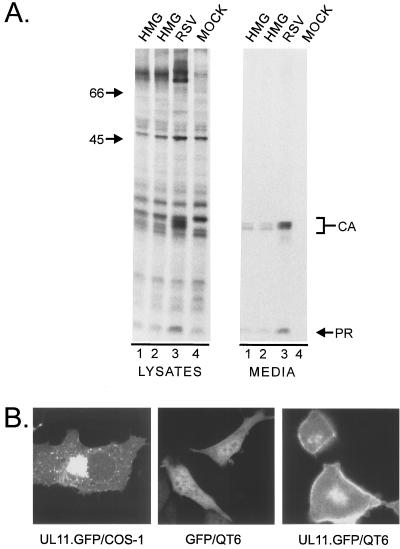
Since these findings suggest that HMG is membrane bound, the lack of budding is consistent with HMG being localized to a membrane other than the plasma membrane. However, the lack of budding is a negative result, and there are ways that particle release could be inhibited even if HMG was located at the plasma membrane. For example, if the viral PR is more active than usual in the context of HMG, then the UL11-Gag molecules would be cleaved before they are able to complete the budding process (9). This would cause the nascent buds to collapse back into the cytoplasm and result in the observed lack-of-budding phenotype. Since PR is not required for budding, 90% of it was deleted to form HMG.3h (Fig. 1). This chimera was neither cleaved nor released into the medium (Fig. 3, lanes 6), indicating that HMG is not budding deficient because of enhanced PR activity.
FIG. 3.
Release properties of modified forms of HMG. The indicated constructs were transfected into duplicate plates of COS-1 cells. The right panel (media) was from one set of plates that were treated as described in the legend to Fig. 2. Gag proteins in the left panel (lysates) were collected and visualized as for Fig. 2, but were from the second set of plates that had been labeled for only 5 min.
Another way that HMG might be blocked for budding even if it had been directed to the plasma membrane is by interference with the nearby L domain (Fig. 1), which is required for the virus-cell separation step (45). L domain mutants are targeted to the plasma membrane and, like HMG, undergo proteolytic processing, but do not bud. Previous experiments have shown that budding can be restored to RSV L domain mutants by the addition of the HIV p6 sequence, which also contains an L domain (22), and with this in mind, HMG.p6 was constructed (Fig. 1). Although it was expressed well (Fig. 3, lane 4, lysates), none of the protein was released into the medium. Thus, it appears unlikely that an L domain defect caused the retention of HMG inside the cell.
A third possibility to explain the failure of HMG to be released after being targeted to the plasma membrane is that the UL11 sequences are bulky and inhibit the budding process. To test this idea, the strong plasma membrane-binding domain of the Src oncoprotein was attached to the amino terminus of HMG to create Src.HMG (Fig. 1). If HMG is already at the plasma membrane and is prevented from budding by the UL11 sequences, then this modification should have no effect. However, if HMG is directed to an internal membrane, then release into the culture medium might be restored. We found that Src.HMG was released into the medium (Fig. 3, lanes 7; 40% efficiency relative to the wild type), indicating that the UL11 sequences are not incompatible with budding and further supporting the idea that the L domain within HMG is intact.
An additional possibility to explain the failure of HMG to be released is that it is not bound to any membrane at all. If HMG is partitioned into the cytoplasm, then it might be packaged into particles when coexpressed with wild-type Gag molecules. However, unlike membrane-binding mutants of RSV (5, 47), HMG was not detectably rescued (data not shown). Although this is a negative result, it is consistent with a rapid transport of HMG to an internal-membrane location.
HMG is targeted to the Golgi apparatus.
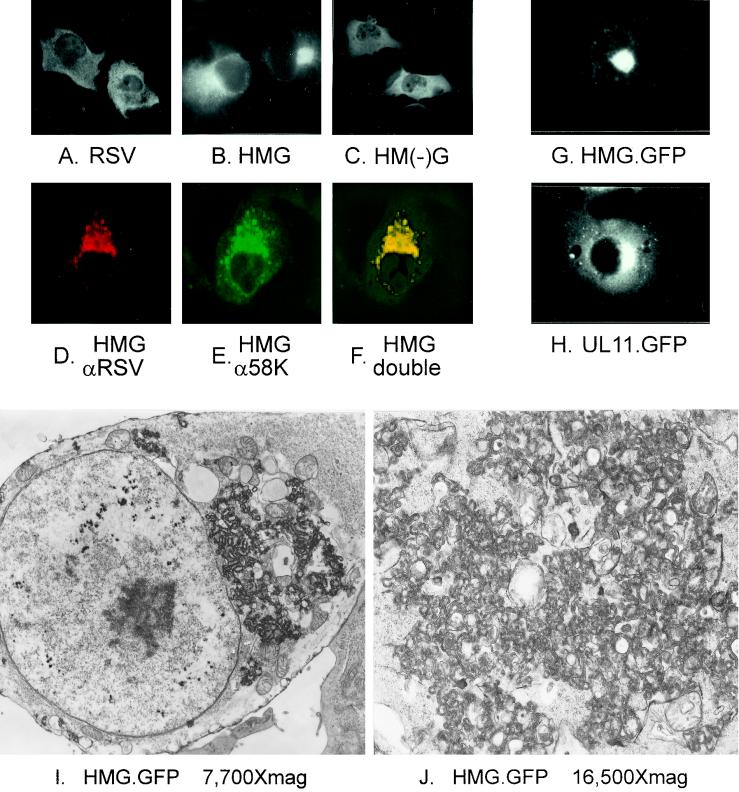
In an attempt to gather direct evidence that HMG is localized to a membrane other than the plasma membrane, immunofluorescence analysis was performed. Expression of full-length RSV Gag in COS cells, followed by fixation and staining, revealed a diffuse fluorescence throughout the cell (Fig. 4A). In contrast, similar analyses with HMG showed a distinct staining pattern in a tight juxtanuclear location (Fig. 4B). To identify the cellular compartment containing HMG, double-label immunofluorescence and confocal microscopy were used. A 1-μm optical slice of a single transfected cell labeled with antibodies against RSV (Fig. 4D) and against the Golgi 58-kDa protein (58K) (Fig. 4E) shows colocalization of HMG and the Golgi apparatus-specific marker (Fig. 4F).
FIG. 4.
Microscopic analyses of HMG-expressing cells. COS-1 cells were transfected with the indicated constructs. Cells in panels A to F were analyzed by immunofluorescence. In panels A to C, antibodies specific for RSV Gag were used. Cells in panels D to F were double labeled with a mixture of rabbit antibodies against Gag and mouse antibodies against the Golgi 58K protein, which were detected by using a mixture of goat anti-rabbit antibodies conjugated to TRITC and goat anti-mouse antibodies conjugated to FITC. Panels D and E are the same field viewed by confocal microscopy with the appropriate wavelength to excite TRITC (D) or FITC (E) and were digitally combined to provide the image in panel F. Cells transfected with GFP constructs in panels G and H were viewed by light microscopy without fixing or staining. Cells in panels I and J were analyzed by standard electron microscopy.
Although it is clear that HMG is localized to the Golgi apparatus within fixed cells, it was possible that the chemical permeabilization and fixation processes might have altered the normal cellular distribution of HMG (20). To completely rule out this possibility, GFP was fused to the C terminus of HMG, and the localization of the resulting chimera (HMG.GFP; Fig. 1) was visualized in living cells. HMG.GFP appeared to be exclusively perinuclear (Fig. 4G), indicating that the Golgi apparatus targeting observed by immunofluorescence was not a fixation artifact. To assess whether the Gag sequences of HMG contribute to the Golgi apparatus targeting, UL11 was fused directly to either GFP (UL11.GFP; Fig. 1) or the nine-amino-acid hemagglutinin (HA) tag. The localization of UL11.GFP (Fig. 4H) and UL11.HA (data not shown) was identical to that of HMG.GFP, suggesting that UL11 alone is able to travel to and bind Golgi membranes.
To determine whether HMG merely binds to the cytoplasmic face of the Golgi membranes or whether interactions among the Gag portion of the chimera also mediate budding into the Golgi cisternae, electron microscopy was employed to achieve increased resolution. In contrast to untransfected COS-1 cells (not shown), normal Golgi stacks were not observed (Fig. 4I and J). Instead, thickened, electron-dense membranes similar in appearance to budding retroviruses were observed in the same perinuclear location that was fluorescently labeled, but they were not seen at the plasma membrane. Due to the substantial disruption of the Golgi compartment, it is not clear whether virus-like particles are released into the cisternae. However, because membrane distortion is required during Gag-mediated budding and because HMG is capable of directing budding when targeted to the plasma membrane with the Src membrane-binding domain, this result is consistent with internal budding.
Requirements for Golgi targeting by UL11.
Many (but not all) myristylated proteins require the addition of myristate for proper targeting and/or membrane binding. To determine whether this requirement exists for HMG, we constructed a myristate-negative form of HMG (Fig. 1). Although HM(−)G was expressed well, it was not released into the medium (Fig. 3, lanes 5), and immunofluorescence analysis showed diffuse cytoplasmic staining (Fig. 4C). The lack of perinuclear staining indicates that myristylation is required for Golgi localization.
To determine which amino acids are required for Golgi localization, three C-terminal deletions of the UL11 portion of HMG were made (Fig. 1). Metabolic labeling of the deletion mutants showed protein production and processing, but little (HMG.23; Fig. 5A, lane 4; 7% relative to RSV Gag) or no (HMG.70 and HMG.49; Fig. 5A, lanes 2 and 3, respectively) release into the medium. By immunofluorescence, HMG.70 and HMG.49 show strong Golgi staining, while HMG.23 shows cytoplasmic staining in addition to weak Golgi staining (Fig. 5B). The significant intracellular cleavage of HMG.23, as well as the increased amount of budding by this protein (Fig. 5A, lanes 4), indicates that it is directed to the plasma membrane to some extent. Thus, the Golgi apparatus-specific targeting signal is contained entirely within the first 49 but not the first 23 amino acids of UL11. Taken together, these data suggest that the first 23 residues of UL11 contain a nonspecific membrane-binding function that is affected by the information contained within residues 24 to 49.
FIG. 5.
Deletion analysis of UL11. (A) The level of particle release of the indicated mutants was measured by labeling the transfected cells as in Fig. 3. (B) COS-1 cells were transfected, fixed, and stained with RSV Gag-specific antibodies and visualized by confocal microscopy.
The efficiency of Golgi targeting was found to be cell type dependent. In contrast to the lack of budding from COS-1 cells (Fig. 2 and 3), HMG is able to escape from QT6 avian cells with 40% the efficiency of RSV Gag (Fig. 6A, lanes 1 and 2 versus lanes 3), suggesting some degree of plasma membrane targeting. This differential targeting is not due to the Gag portion of the chimera, since the localization of UL11.GFP (which contains no Gag sequences) is exclusively perinuclear in mammalian cells (COS-1; Fig. 4 and 6B; BHK, Vero, and human melanoma cells [data not shown]), but shows both perinuclear and plasma membrane staining in QT6 cells (Fig. 6B). These data raise the possibility that membrane localization is influenced by cellular factors that are divergent between mammalian and avian cells and that these factors may play a role in herpesvirus infections.
FIG. 6.
Expression of HMG in avian cells. The HMG and RSV Gag genes were transferred into the BHRCAN (12) vector for expression in avian cells. (A) Plasmids were transfected by the calcium-phosphate precipitation method into QT6 cells, and the proteins were labeled and collected as described in the legend to Fig. 2. (B) COS-1 or QT6 cells were transfected as for panel A with either GFP or UL11.GFP and viewed by confocal microscopy.
DISCUSSION
Role of RSV Gag C terminus in plasma membrane localization.
By redirecting a UL11-RSV Gag chimera to the Golgi apparatus, our current experiments have provided evidence that the C terminus of Gag does not contain dominant plasma membrane targeting sequences. However, the efficiency of membrane localization of both HIV and RSV Gag is affected by sequences downstream of the amino-terminal M domain (27, 47). These downstream sequences may influence membrane localization by enhancing the ability of Gag to remain bound at its destination after arrival. One way to increase the avidity of the M domain-membrane interaction is by linking large numbers of Gag proteins together. Since the I domains are found within NC and are involved in Gag multimerization (39), they are likely mediators of the C-terminal sequence contribution to Gag localization (32).
Another way in which the Gag C terminus may increase the efficiency of membrane localization is by facilitating the transport of Gag from the cytoplasm to the destination specified by the M domain. Trafficking of both cellular (10) and viral (13, 36) components can be accomplished by interaction with the microtubule and microfilament elements of the cytoskeletal transport network. Therefore, the colocalization of Gag with actin in infected cells (24, 25) and the binding of NC within the virion to actin (17, 44) suggest that an interaction of the C terminus of Gag with the cytoskeleton may also play a role in Gag transport.
Mechanism of UL11 Golgi apparatus targeting.
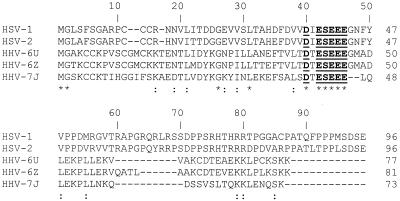
Although it is less than 100 amino acids long, UL11 must contain signals to direct it to and retain it specifically at the Golgi apparatus. Evidence presented here indicates that these targeting sequences are contained within the myristylated amino-terminal 49 residues. What information is contained in the first 49 residues? When we aligned the UL11 sequences of several herpesviruses, a striking acidic cluster (33) was revealed (Fig. 7, residues 37 to 43). Since certain acidic clusters are known to direct proteins to the trans-Golgi network (TGN) via an association with the PACS-1 protein (41), it is possible that UL11 is targeted to the Golgi apparatus in a similar manner. Indeed, the peripheral membrane protein Nef from HIV-1 has recently been shown to recycle from the plasma membrane to the TGN via a PACS-1-dependent mechanism (26).
FIG. 7.
Alignment of UL11 homologs. The amino acid sequences of the UL11 homologs from several herpesviruses are shown. HHV, human herpesvirus. Identical amino acids are indicated by an asterisk, while similar amino acids are marked by a colon. A conserved acidic cluster motif is underlined.
The role of myristylation in UL11 localization is also unclear. The loss of Golgi apparatus targeting of HM(−)G is presumably due to the loss of membrane binding, rather than a loss of targeting, since nonspecific binding (as was seen for HMG.23) was not observed. However, we cannot rule out the possibility that myristate is required for UL11 to associate with a PACS-1-like protein and therefore is needed for Golgi targeting as well as membrane binding.
Role of UL11 in herpesvirus replication.
UL11 is targeted to internal cellular membranes (1) that are identified here as those of the Golgi apparatus. We have also shown that this specific localization is independent of all other viral proteins. However, the role of UL11 at these membranes during HSV infection remains unclear. UL11 is required for efficient envelopment and particle release (2, 19), but it is not known whether UL11 itself is the primary mediator of the budding event. Our microscopy data indicated severe involution of the internal membranes. We attributed these effects to the Gag portion of the HMG.GFP protein, but we cannot discount the possibility of a contribution from UL11.
Another possibility is that UL11 does not drive envelopment but instead recruits other viral proteins to the site of assembly. As a membrane-bound virion protein, UL11 would be ideal as a “bridge” linking the viral envelope to other components of the tegument. These additional tegument proteins, alone or in combination, could provide the budding machinery. The amino-terminal halves of UL11 homologs from several herpesviruses, which include the targeting information, have significant sequence similarity (Fig. 7). This is expected for domains that must interface with conserved cellular structures, such as those of a trafficking pathway. The C termini of the homologs are more divergent, presumably to accommodate binding to other viral proteins not conserved between herpesviruses. Experiments to test these models are currently under way.
The assembly pathways of retroviruses and herpesviruses are very different, but they do share the common requirement of targeting the virion proteins to the correct site of assembly. We have now demonstrated that RSV Gag sequences downstream of the M domain do not contain dominant plasma membrane-specific localization signals. We have also identified and partially characterized the membrane-binding and -targeting motif of the HSV-1 UL11 protein. Although further experiments will be needed to dissect the functional components of the first half of UL11, the information provided here will be useful in designing these and other investigations into the mechanisms of protein targeting and herpesvirus assembly.
ACKNOWLEDGMENTS
We thank Roland Meyers for performing the thin sectioning and assisting with all aspects of the electron microscopy.
This research was sponsored by National Institutes of Health (NIH) grants to R.J.C. (CA42460 and CA60395) and J.W.W. (CA47482). J.B.B. was partially supported by NIH training grant CA60395, and J.S.L. was partially supported by an LSC fellowship from Pennsylvania State University.
REFERENCES
- 1.Baines J D, Jacob R J, Simmerman L, Roizman B. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J Virol. 1995;69:825–833. doi: 10.1128/jvi.69.2.825-833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1992;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett R P, Rhee S, Craven R C, Hunter E, Wills J W. Amino acids encoded downstream of gag are not required by Rous sarcoma virus protease during Gag-mediated assembly. J Virol. 1991;65:272–280. doi: 10.1128/jvi.65.1.272-280.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett R P, Wills J W. Conditions for copackaging Rous sarcoma virus and murine leukemia virus Gag proteins during retroviral budding. J Virol. 1999;73:2045–2051. doi: 10.1128/jvi.73.3.2045-2051.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowzard J B, Bennett R P, Krishna N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burstein H, Bizub D, Skalka A M. Assembly and processing of avian retroviral gag polyproteins containing linked protease dimers. J Virol. 1991;65:6165–6172. doi: 10.1128/jvi.65.11.6165-6172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carson J H, Kwon S, Barbarese E. RNA trafficking in myelinating cells. Curr Opin Neurobiol. 1998;8:607–612. doi: 10.1016/s0959-4388(98)80088-3. [DOI] [PubMed] [Google Scholar]
- 11.Craven R C, Leure-duPree A E, Erdie C R, Wilson C B, Wills J W. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus Gag protein. J Virol. 1993;67:6246–6252. doi: 10.1128/jvi.67.10.6246-6252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craven R C, Leure-duPree A E, Weldon R A, Jr, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 14.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retrovir. 1993;9:817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 17.Liu B, Dai R, Tian C-J, Dawson L, Gorelick R J, Yu X-F. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J Virol. 1999;73:2901–2908. doi: 10.1128/jvi.73.4.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLean C A, Clark B, McGeoch D J. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J Gen Virol. 1989;70:3147–3157. doi: 10.1099/0022-1317-70-12-3147. [DOI] [PubMed] [Google Scholar]
- 19.MacLean C A, Dolan A, Jamieson F E, McGeoch D J. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J Gen Virol. 1992;73:539–547. doi: 10.1099/0022-1317-73-3-539. [DOI] [PubMed] [Google Scholar]
- 20.Melan M A, Sluder G. Redistribution and differential extraction of soluble proteins in permeabilized cultured cells. Implications for immunofluorescence microscopy. J Cell Sci. 1992;101:731–743. doi: 10.1242/jcs.101.4.731. [DOI] [PubMed] [Google Scholar]
- 21.Nelle T D, Wills J W. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J Virol. 1996;70:2269–2276. doi: 10.1128/jvi.70.4.2269-2276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parent L J, Wilson C B, Resh M D, Wills J W. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J Virol. 1996;70:1016–1026. doi: 10.1128/jvi.70.2.1016-1026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearce-Pratt R, Malamud D, Phillips D M. Role of the cytoskeleton in cell-to-cell transmission of human immunodeficiency virus. J Virol. 1994;68:2898–2905. doi: 10.1128/jvi.68.5.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perotti M-E, Tan X, Phillips D M. Directional budding of human immunodeficiency virus from monocytes. J Virol. 1996;70:5916–5921. doi: 10.1128/jvi.70.9.5916-5921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piguet V, Wan L, Borel C, Mangasarian A, Demaurex N, Thomas G, Trono D. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat Cell Biol. 2000;2:163–167. doi: 10.1038/35004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Platt E J, Haffar O K. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: requirement for a C-terminal domain. Proc Natl Acad Sci USA. 1994;91:4594–4598. doi: 10.1073/pnas.91.10.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popov S, Rexach M, Zybarth G, Reiling N, Lee M A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M I. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rein A, McClure M R, Rice N R, Luftig R B, Schultz A M. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schafer W, Stroh A, Berghofer S, Seiler J, Vey M, Kruse M-L, Kern H F, Klenk H-D, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 35.Smith K O. Relationship between the envelope and the infectivity of herpes simplex virus. Proc Soc Exp Biol Med. 1964;115:814–816. doi: 10.3181/00379727-115-29045. [DOI] [PubMed] [Google Scholar]
- 36.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spearman P, Wang J-J, Vander Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanstrom R, Wills J W. Synthesis, processing, and assembly of viral proteins. In: Weiss R, Teich N, Varmus H E, Coffin J M, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 40.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan L, Molloy S S, Thomas L, Liu G, Xiang Y, Rybak S L, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 42.Weldon R A, Jr, Erdie C R, Oliver M G, Wills J W. Incorporation of chimeric Gag protein into retroviral particles. J Virol. 1990;64:4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilk T, Gowen B, Fuller S D. Actin associates with the nucleocapsid domain of the human immunodeficiency virus Gag polyprotein. J Virol. 1999;73:1931–1940. doi: 10.1128/jvi.73.3.1931-1940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wills J W, Srinivas R V, Hunter E. Mutations of the Rous sarcoma virus env gene that affect the transport and subcellular location of the glycoprotein products. J Cell Biol. 1984;99:2011–2023. doi: 10.1083/jcb.99.6.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiang Y, Ridky T W, Krishna N K, Leis J. Altered Rous sarcoma virus Gag polyprotein processing and its effects on particle formation. J Virol. 1997;71:2083–2091. doi: 10.1128/jvi.71.3.2083-2091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]