Abstract
Background
Livestock-related emissions have been associated with aggravations of respiratory symptoms in patients with chronic obstructive pulmonary disease (COPD), potentially by altering the respiratory resistome.
Objectives
This study investigates the structure of the acquired oropharyngeal (OP) resistome of patients with COPD and controls, its interplay with the respiratory microbiome and associations with residential livestock exposure.
Methods
In a matched case–control study in the rural Netherlands, we analysed OP swabs from 35 patients with COPD and 34 controls, none of whom had used antibiotics in the preceding 4 weeks. Resistome profiling was performed using ResCap, complemented by prior characterization of the microbiome via 16S rRNA-based sequencing. Residential livestock farm exposure was defined using distance-based variables alongside modelled concentrations of livestock-emitted microbial pollutants. We compared resistome profiles between patients with COPD and controls, examining alpha and beta diversity as well as differential abundance. Additionally, we assessed the interplay between the resistome and microbiome using co-occurrence networks and Procrustes analysis. Variations in resistome profiles were also analysed based on residential livestock exposures.
Results
Patients with COPD exhibited higher resistome diversity than controls (Shannon diversity, P = 0.047), though resistome composition remained similar between groups (PERMANOVA, P = 0.19). Significant correlations were observed between the OP resistome and microbiome compositions, with distinct patterns in co-occurrence networks. Residential exposure to livestock farms was not associated with resistome alterations.
Conclusions
Our findings reveal the COPD airway as a hospitable environment for antimicrobial resistance genes, irrespective of recent antimicrobial usage. Demonstrating the interplay between the resistome and microbiome, our study underscores the importance of a deeper understanding of the resistome in respiratory health.
Introduction
The impact of livestock farm emissions on the respiratory health of neighbouring residents is an important concern within public health. Previous studies have established significant associations between exposure to these emissions and respiratory morbidity and mortality.1–8 Livestock farm emissions constitute a complex mixture of gases and particulate matter,9–11 comprising microbiological components such as endotoxins and antimicrobial resistant (AMR) bacteria,12–16 thereby presenting a multifaceted challenge. Notably, it has been observed that patients with chronic obstructive pulmonary disease (COPD) residing in close proximity to farms demonstrate increased risks of airway inflammation, cough and dyspnoea, compared with those residing further away.17,18
In a recent study, researchers discovered an association between residential exposure to livestock-related air pollution and increased microbial richness in the upper respiratory tract, highlighting the potential influence of the livestock environment on the microbiome.19 The composition of the airway microbiome plays a significant role in the onset and progression of lung diseases. Previous research consistently demonstrates that alterations in the airway microbiome are associated with the development and progression of COPD.20–22 These studies indicate a decline in microbial diversity as COPD progresses and during acute exacerbations.23–27 The microbiome and resistome (the complete collection of acquired bacterial genes potentially responsible for acquired antimicrobial resistance) reciprocally interact.28,29 The microbiome provides a diverse ecosystem that facilitates the acquisition and preservation of antimicrobial resistance genes (ARGs) through gene transfer. Simultaneously, the resistome influences the microbiome by conferring advantages to microorganisms under selective pressures, influencing their abundance and composition based on their resistance potential.
Despite the growing interest in microbiome research, the investigation of the relationship between the respiratory resistome and respiratory health remains limited. Although studies have demonstrated the transmission of AMR bacteria in occupational settings to farmworkers from farm dust,30–32 the impact of residential exposure to livestock emissions on the respiratory resistome remains unexplored. Gaining insight into the resistome among both patients with COPD and control individuals allows for the identification of disease-specific resistome patterns and enhances our understanding of the impacts of livestock farm exposure. Investigating the interplay between the microbiome and resistome unravels intricate dynamics, potentially providing valuable insights into interactions of ARGs within the airways. Given the escalating global health threat posed by AMR,33 and the potential of ARGs within the airway to lead to severe and difficult-to-treat infections, it is important to address this critical knowledge gap by shedding light on the structure of the respiratory tract resistome in health and disease and by investigating the dynamics between residential exposure and the resistome.33
This study characterized the acquired ARGs in the oropharynx of rural residents living in a livestock-dense region. We aimed to investigate associations between the oropharyngeal (OP) resistome composition and (i) COPD status, (ii) the OP microbiome composition and (iii) residential exposure to livestock-related microbial emissions. In this case–control study, we collected OP samples from 35 patients with COPD and 34 matched controls. To characterize the resistome, we employed resistome-enrichment (ResCap) metagenomic shotgun sequencing. We hypothesize that the OP resistome of patients with COPD is inherently different to that of the controls and that the microbiome and resistome mutually shape one another. Furthermore, we anticipate that residential exposure to livestock-related emissions will induce alterations in the load and structure of the OP resistome, potentially contributing to the aggravation of respiratory symptoms in patients with COPD.
Methods
Study design and population
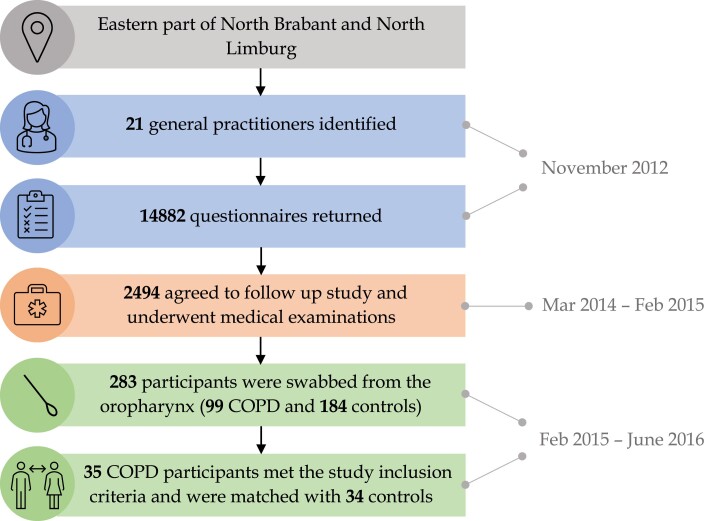
The study population comprises rural residents in the Netherlands, both with and without a diagnosis of COPD. This study is nested within the VGO programme (Dutch acronym for ‘livestock farming and the health of neighbouring residents’) initiated in 2012 to investigate the health effects associated with residential proximity to livestock farms. The initial selection procedure for VGO participants was previously outlined.18,34 Subsequently, participants were selected for a COPD case–control microbiome study,19 from which we selected participants for this resistome study. The inclusion criteria stipulated that patients with COPD must (i) live in the VGO area in 2015–16, (ii) have a COPD diagnosis based on spirometry results and a categorization into Global Initiative for Chronic Obstructive Lung Disease (GOLD) Stages 1–3 and (iii) be aged ≥ 40 years. The exclusion criteria included: (i) any participants with missing data, (ii) farm workers, (iii) residing on a farm, (iv) current smokers and (v) recent use (within the past 4 weeks) of oral antibiotics (Figure 1). Control participants were carefully matched with the patients with COPD based on age category, sex, smoking status (never, ex-smoker) and farm childhood.
Figure 1.
A flow chart of the participant selection procedure from the VGO study, illustrating the procedure for the selection of matched patients with COPD and controls. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Sample collection
OP samples from 35 patients with COPD and 34 control participants were used for this study, collected during home visits in the period February 2015 to July 2016 in addition to three field blanks. Previous research affirms the resemblance between bacterial communities in the oropharynx and lungs, establishing OP samples as a valuable proxy for our research.35 Copan eSwabs were used for sampling, and these were stored in 1 mL liquid Amies Medium (483CE, Copan Diagnostics Inc., CA, USA). During transportation, these samples were stored on ice until transfer to the −80°C freezer on the same day. Blanks were unused swabs treated on-site using the same procedure as participant samples. The DNA extracts were identical to those described in the microbiome analysis protocol outlined by van Kersen et al..19
Sequencing and bioinformatics
Resistome characterization of the OP samples was performed using the ResCap method, a shotgun metagenomic enrichment approach tailored for resistome analyses.36 The ResCap procedure consists of four main steps: (i) total microbial DNA isolation,19 (ii) whole-metagenome shotgun library construction (Roche SeqCap EZ workflow), (iii) hybridization and enrichment and (iv) enriched library deep sequencing (Illumina), as previously described.36 After quality control and trimming of the raw sequences, these were mapped against the ResFinder database of acquired ARGs.37 Mapping data were subsequently normalized for ARG length and microbial load using 16S rRNA qPCR. For additional information on sequencing details and bioinformatics, refer to Text S1.
Residential livestock exposure assessment
Residential exposure for each participant was assessed using their geocoded home address. To investigate potential factors influencing the structure of the OP resistome, we considered both proxies of livestock exposure and predicted concentrations of livestock-related emissions derived from previously established models.38,39 Exposure proxies were computed using Geographic Information System (GIS) software (ArcGIS; version 10.2.2, Esri)40 using geolocated information on livestock farms alongside geocoded residential addresses, as previously described.13 Proxies included general, species-specific and farm type-specific variables. Land-use regression, dispersion and random forest models were applied to estimate residential exposure to generic and more specific livestock-related emissions. These emissions included PM10, endotoxin, two livestock commensals (Escherichia coli and Staphylococcus spp.) and two ARGs (tetW and mecA); all frequently encountered on livestock farms and in their surrounding areas.16,41–43 All models were previously developed and validated for our specific geographic region, and they were employed in our study to predict the annual average residential exposure to these emissions.38,39
Data analysis
All statistical analyses were conducted using R version 4.2.2 (2022-10-31).44 The OP resistome was characterized using established methods, including evaluations of within-sample diversity (alpha diversity), between-sample compositional differences (beta diversity) and in-depth investigations of differences in ARG abundances [differential abundance (DA) analysis]. Firstly, we compared these resistome characteristics between patients with COPD and controls. Secondly, we examined the relationship between the microbiome and resistome using Procrustes analysis and co-occurrence networks. Finally, we examined the potential association between the OP resistome and residential livestock exposure. For a comprehensive explanation of the applied statistical methods, refer to Text S2.
Results
Study population
Characteristics of the study population, stratified by COPD status, are presented in Table 1. The majority of participants were male (59%), with a mean age of 61 years. Among patients with COPD, all were categorized into GOLD Stage 1 or 2 (51% and 49%, respectively), indicating mild and moderate COPD according to the GOLD criteria. For detailed livestock exposure proxy values for each participant, refer to Table S2 (available as Supplementary data at JAC Online).
Table 1.
Baseline characteristics of patients with COPD and control participants included in the study
| Patients with COPD | Controls | P value | |
|---|---|---|---|
| (n = 35) | (n = 34) | ||
| Gender | |||
| Male | 21 (60.0%) | 20 (58.8%) | 1 |
| Female | 14 (40.0%) | 14 (41.2%) | |
| Age | |||
| Mean (SD) | 61.5 (7.36) | 60.2 (7.82) | 0.473 |
| Median (min, max) | 62.6 (43.9, 71.6) | 62.7 (41.8, 70.6) | |
| BMI | |||
| Mean (SD) | 26.7 (4.15) | 27.5 (4.40) | 0.473 |
| Median (min, max) | 25.9 (20.1, 34.1) | 26.3 (21.4, 39.2) | |
| COPD GOLD grade | |||
| 0 | 0 (0%) | 34 (100%) | <0.001 |
| 1 | 18 (51.4%) | 0 (0%) | |
| 2 | 17 (48.6%) | 0 (0%) | |
| Childhood on farm | |||
| No | 22 (62.9%) | 22 (64.7%) | 1 |
| Yes | 13 (37.1%) | 12 (35.3%) | |
| Smoking status | |||
| Former smoker | 27 (77.1%) | 26 (76.5%) | 1 |
| Never | 8 (22.9%) | 8 (23.5%) | |
| Pack-years of cigarettes smoked | |||
| Mean (SD) | 16.8 (15.1) | 13.8 (22.0) | 0.506 |
| Median (min, max) | 16.7 (0, 54.6) | 7.25 (0, 117) | |
| Education level | |||
| No | 22 (62.9%) | 22 (64.7%) | 1 |
| Yes | 13 (37.1%) | 12 (35.3%) | |
| Number of farms within 3 km radius | |||
| Mean (SD) | 82.7 (30.1) | 90.0 (23.1) | 0.265 |
| Median (min, max) | 78.0 (13.0, 137) | 89.5 (20.0, 129) |
For continuous variables, data are presented as mean (SD) and median (min, max), and P values were derived from t tests between COPD and control groups. For categorical variables, data are presented as number (%) per category and P values were derived from the χ2 test of independence.
Oropharyngeal resistome of patients with COPD and controls
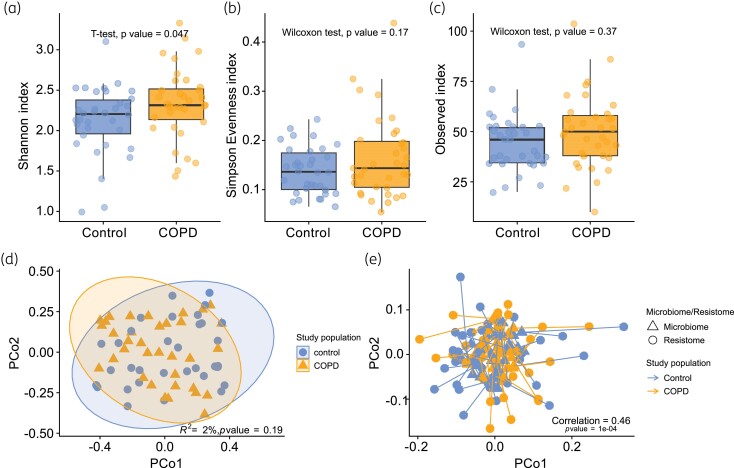
The COPD group exhibited significantly higher Shannon diversity compared with the control group (two-sample t test; P value = 0.047) (Figure 2a). Observed richness and Simpson’s evenness did not reveal significant differences between patients with COPD and controls (Figure 2b and c).
Figure 2.
Comparison of the OP resistomes of patients with COPD and controls. Alpha diversity indices (a) Shannon, (b) Simpson’s evenness and (c) observed richness for COPD and control OP resistome samples. P values were derived from t tests and Wilcoxon’s rank sum tests. (d) PCoA plot based on the Bray–Curtis dissimilarity matrix of the OP resistome of patients with COPD and controls. Ellipses show the 95% CI for the centroid of each group. Points represent the resistance gene communities of one participant. The first two principal coordinates of the PCoA (Axis 1 and Axis 2) explained 28.5% and 17.2% of the variance, respectively. (e) Procrustes superimposition plot showing the association between microbiome and resistome PCoAs. The protest function was used to calculate the correlation coefficient and P value for paired participant samples using 9999 permutations. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
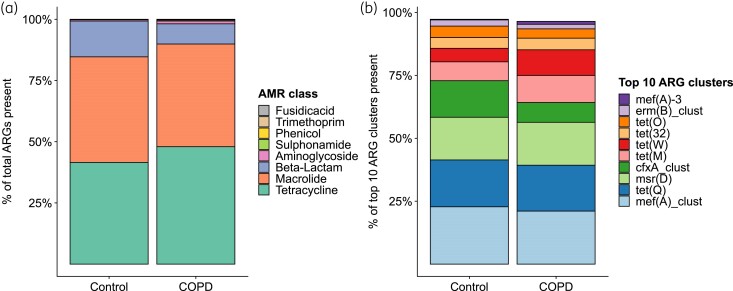
Across the entire study population, we identified 85 distinct ARGs (90% identity clusters). Figure 3(a) displays the relative abundances of ARGs, grouped by their AMR class, while Figure 3(b) presents the relative abundances of the top 10 ARGs, which collectively explain 99.2% of the data from the control population and 96.4% from the COPD population. Within the top 10 ARGs across all samples, 50% conferred tetracycline resistance, 40% macrolide resistance and 10% beta-lactam resistance. The predominant ARG observed in all samples was mef(A)_clust, associated with macrolide resistance. Relative abundance patterns of the eight AMR classes did not appear to differ considerably between COPD and control groups.
Figure 3.
Relative resistome abundances across patients with COPD and controls where (a) shows the relative abundances ARGs grouped by their AMR class and (b) shows the relative abundance of the top 10 ARGs (90% gene identity) within the total resistome. ARG counts have been (relatively) rarefied, gene length-corrected and 16S qPCR-corrected prior to scaling to 100%. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
In each group, we identified eight core ARGs (see Text S2 for definition), conferring resistance potential to tetracyclines, macrolides and beta-lactams. Our analysis revealed an identical core resistome shared between patients with COPD and controls (Figure S1). Notably, the two ARGs that were found in low quantities in the blank samples exhibited no overlap with the resistome of patients with COPD and controls. A heatmap showing the abundance of ARGs across the groups is shown in Figure S2. No distinct clustering patterns were observed between patients with COPD and controls, as depicted in the principal coordinates analyses (PCoAs) (PERMANOVA; P value = 0.19, R2 = 0.0201) (Figure 2d). Similarly, DA analysis using DESeq and ALDEx did not reveal any statistically significant differentially abundant ARGs associated with COPD status, as detailed in Tables S3 and S4.
Association between the oropharyngeal resistome and microbiome
Procrustes analysis demonstrated a moderate yet significant correlation between PCoA ordinations of the resistome and microbiome from the same individuals (correlation = 0.46, P value < 0.001), which was absent when microbiome participant IDs were randomized (correlation = 0.16, P value = 0.32). The paired Procrustes superimposition plot is depicted in Figure 2(e).
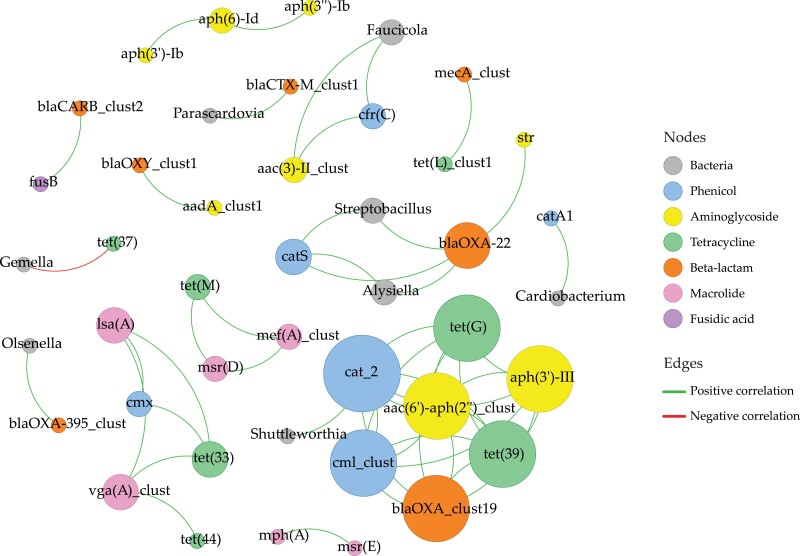
Co-occurrence correlation analysis between bacteria and ARGs uncovered associations among various ARGs and bacterial genera, as well as interactions among the ARGs themselves (Table S5a and b). Seven bacterial genera exhibited positive associations with one or more specific ARGs. Figure 4 illustrates a co-occurrence network depicting these correlations, with nodes representing bacteria and ARGs and edges representing positive and negative correlations. Notably, cat_2 ARG (conferring phenicol resistance) emerged as central in the network, exhibiting the highest degree centrality (number of connections). Co-occurrence patterns were observed within the same AMR class and across different AMR classes, without significant negative correlations among ARGs. Bacteria–ARG correlations revealed a smaller cluster involving Streptobacillus, Alysiella and ARGs cat_2 and blaOXA-22 (phenicol and beta-lactam resistance genes). Gemella exhibited a significant negative correlation with tet(37) (a tetracycline ARG), representing the sole negative correlation identified.
Figure 4.
Bacteria–ARG and ARG–ARG co-occurrence network, based on correlation analysis. Nodes representing bacteria are named on their genus level, and nodes representing ARGs are named by their 90% cluster identity name. Nodes representing bacteria are coloured grey, and the colour of nodes representing ARGs shows their AMR class. Node size is proportional to the nodal degree (number of connections in the network). Edges show strong (Spearman’s |ρ| ≥ 0.6) and significant [P value (BH-adjusted) < 0.01] pairwise correlations. Edges coloured in green represent the positive correlations, while edges in red represent the negative correlations. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Association between the oropharyngeal resistome and livestock exposure
Livestock exposure analysis revealed no significant differences in resistome composition and diversity across individuals with differing levels of exposure, regardless of the specific exposure variable used. Detailed results are given in Tables S6 (PERMANOVA), S7 and S8 (analysis of alpha diversity indices). DA analyses, exploring potential relationships between residential livestock exposure variables and ARG expression, also did not reveal any significant differences (both DESeq and ALDEx methods produced Benjamini–Hochberg (BH)-corrected P values > 0.05).
Discussion
The primary aim of our study was to advance our understanding of the respiratory resistome in rural residents, both with and without COPD. Alongside this, we aimed to conduct a comparative analysis of the resistome and microbiome compositions among the study participants. Additionally, we sought to examine the potential associations between the respiratory resistome and residential exposure to microbial emissions from livestock farms.
The oropharyngeal resistome of patients with COPD versus controls
Consistent with our hypothesis, individuals with COPD demonstrated higher resistome alpha diversity compared with controls, even in the absence of antibiotic usage within the 4 weeks preceding sampling. Compelling evidence suggests that recurrent antibiotic use in individuals with COPD contributes significantly to the increased occurrence of phenotypic antibiotic resistance.45 On average, individuals with COPD tend to use antimicrobial agents more frequently, exerting a selection pressure for ARG expression. Additionally, chronic inflammation within the COPD airway can lead to the production of excess exudate which creates conditions that are more favourable for the survival and growth of microorganisms, including AMR bacteria.46 Consequently, individuals with altered immune function, such as those with COPD, may exhibit higher levels of AMR bacteria, even without increased antimicrobial usage, thus possibly leading to a higher diversity of ARGs. Despite this elevated resistome diversity in patients with COPD compared with controls, no differences in composition or overall resistome load [Fragments Per Kilobase of transcript per Million mapped reads (FPKM)] were observed between the two groups.
Associations between the oropharyngeal resistome and microbiome
Having both resistome and microbiome data for our study participants, we had the unique opportunity to investigate their relationship. A strong correlation was observed between the compositional differences in the resistome and microbiome, suggesting a degree of interdependency. The resistome composition is likely influenced by the presence of specific bacterial species within the microbiome that can be reservoirs for ARGs. Additionally, host-specific factors, such as genetics, immune responses and exposure history, may contribute to the observed correlations. Despite significant overlap between the two compositions shown in the Procrustes analysis, resistome alpha diversity did not correlate with microbial alpha diversity.
Co-occurrence network analysis offered more nuanced insights into the complex relationships among ARGs and bacteria and between ARGs themselves. The distinct clusters of ARGs, which were not directly associated with a specific bacterium, suggest shared genetic elements or co-localization on mobile genetic elements. Positive associations between specific ARGs and bacterial genera imply multi-drug resistance or functional relationships, potentially identifying these bacteria as ARG reservoirs. Alternatively, associations may indicate shared ecological niches. The negative correlation between Gemella and tet(37) suggests that the presence of Gemella may exert an influence on tet(37), or the possibility that another bacterium carrying tet(37) might be more prevalent or involved in competitive interactions with Gemella, thus unveiling intricate and dynamic interactions within this airway microbial ecosystem.
Livestock exposure and its impact on the oropharyngeal resistome
Examining the relationship with livestock exposure, our analyses did not reveal any discernible shifts in resistome composition, relative abundance of ARGs, or alpha diversity associated with livestock exposure. The persistent presence of ARGs in the airways suggests a consistent underlying resistome across all study participants. It is crucial to acknowledge that all participants in our study had some degree of livestock exposure, given the overall high livestock density in the study region, thereby limiting the contrast between exposure levels, and possibly hindering our ability to discern potential differences.
Study strengths and limitations
Strengths of our study include the use of novel ARG-enriched shotgun metagenomics to characterize the OP resistome.36,47 This approach offers a comprehensive view of the resistome along with their relative abundances which surpasses the limitations of conventional methods such as antibiotic susceptibility testing or qPCR, as previously demonstrated.48 Secondly, we assessed exposure to livestock-related emissions through a multifaceted approach, incorporating distance-based variables and predicted microbial exposures based on models developed and validated for our study region.39 Furthermore, the OP microbiota composition of the same study population was previously characterized utilizing 16S rRNA-based sequencing.19 This multi-omics approach enabled us to decipher potential associations between paired microbiome and resistome samples, revealing co-occurrence patterns between specific ARGs and bacteria.
Our study has certain limitations, primarily stemming from its exploratory nature. Notably, the generalizability of our findings to broader populations may be constrained by the homogeneity of the participants as they are all selected from the southern part of the Netherlands with relatively high livestock exposure levels. Additionally, the modest sample size likely compromised our ability to detect significant differences or associations where they may be present. It is also important to note that our study assessed ARGs, offering insights into the antimicrobial potential of the identified genes rather than their actual phenotypic resistance. While ARGs often show a strong correlation with resistance phenotypes, it is crucial to acknowledge the potential for discrepancies.49 Furthermore, our study lacks information on home or work-related exposures other than residential exposure to livestock farming, which could significantly influence the resistome. Additionally, our research exclusively involved participants with mild to moderate COPD, potentially limiting the case–control contrast in our comparisons. Respiratory diseases like COPD are typically more closely associated with microbiota in the lower respiratory tract. However, for practical reasons, we utilized OP samples as a proxy, as studies have demonstrated that the microbiota of these samples bears a closer resemblance to the lung microbiota than their nasopharyngeal counterparts.50 However, it is essential to recognize that OP samples, collected from the upper respiratory tract, may not perfectly mirror the microbiota in the lower respiratory tract.
Study implications
Our findings revealed the pervasive presence of ARGs in both individuals with and without COPD living in a livestock-dense region, raising attention to potential clinical implications. Future research is needed to assess the risks associated with the presence of ARGs in the airways, particularly in the context of respiratory infections and treatment options. Livestock exposure did not emerge as a modifier of the OP resistome composition, consistent with microbiome analysis results from a previous study where no compositional changes were observed.19 To our knowledge, only one prior study has investigated the upper airway resistome in relation to livestock exposure. Conducted in an occupational setting, the study compared OP resistomes of 23 farmworkers with those of 12 nearby villagers. The findings revealed heightened abundance of ARGs associated with farm exposure, influenced by specific farm-related tasks and their duration.51 Another study, specifically focussing on nasal methicillin-resistant Staphylococcus aureus carriage, revealed an increased prevalence associated with residential proximity to livestock farms.52
Comprehensively understanding the impact of different degrees and types of livestock exposure on the resistome and its clinical implications requires further investigation. We recommend for future research that studies encompass diverse geographic and demographic settings to enhance generalizability, including a broader range of exposure levels across participants. The widespread detection of airborne ARGs raises concerns about their contribution to the dissemination of AMR, warranting additional research to evaluate associated risks. While we examined livestock exposure and COPD status as contributing factors to AMR, comprehensive investigations into various host-specific factors are essential for a better understanding of their influence.
In conclusion, this study provides novel insights into the structure of the respiratory resistome and its interplay with the microbiome, residential exposure to livestock farm emissions and respiratory health. Developments in cutting-edge technologies such as metagenomic shotgun sequencing36,47 have enabled the elucidation of the structure of the resistome in OP samples of our rural study population and the investigation of its determinants. These findings make valuable contributions to the emerging field of respiratory microbiome and resistome research, highlighting the importance of comprehending the interplay between the resistome and microbiome in disease dynamics. The identified associations between exposure and respiratory symptoms in individuals with COPD emphasize the need for comprehensive studies that integrate environmental, microbiological and clinical perspectives to unravel these complex interactions. This ‘One Health’ approach aligns with global efforts to both mitigate AMR and enhance our understanding of respiratory diseases by recognizing the interconnectedness of human health, animal health and the environment.
Supplementary Material
Acknowledgements
The authors would like to thank the participating residents who cooperated with us in the study. The authors are grateful to Marieke Oldenwening for performing the fieldwork and Gerdit Greve and Betty Jongerius-Gortemaker for technical assistance (Institute for Risk Assessment Sciences, Utrecht University). The authors thank Frank Harders and Albert de Boer for the deep sequencing and ResCap laboratory work (Wageningen Bioveterinary Research, Lelystad, The Netherlands). The authors acknowledge the provinces of Noord-Brabant and Limburg for their provision of livestock data used in this study.
Contributor Information
Beatrice Cornu Hewitt, Institute for Risk Assessment Sciences (IRAS), Utrecht University, P.O. Box 80178, Utrecht 3508 TD, The Netherlands.
Alex Bossers, Institute for Risk Assessment Sciences (IRAS), Utrecht University, P.O. Box 80178, Utrecht 3508 TD, The Netherlands.
Warner van Kersen, Institute for Risk Assessment Sciences (IRAS), Utrecht University, P.O. Box 80178, Utrecht 3508 TD, The Netherlands.
Myrna M T de Rooij, Institute for Risk Assessment Sciences (IRAS), Utrecht University, P.O. Box 80178, Utrecht 3508 TD, The Netherlands.
Lidwien A M Smit, Institute for Risk Assessment Sciences (IRAS), Utrecht University, P.O. Box 80178, Utrecht 3508 TD, The Netherlands.
Funding
This study was part of the Livestock Farming and Neighbouring Residents’ Health (VGO) that was funded by the Ministry of Health, Welfare and Sports and the Ministry of Agriculture, Nature and Food Quality of the Netherlands. Data collection for this study was supported by a grant from the Lung Foundation Netherlands (Grant No: 3.2.11.022). The current study was funded by a Dutch Research Council (Nederlandse Organisatie voor Wetenschappelijk Onderzoek) Aspasia grant to L.A.M.S. (015.014.067).
Transparency declarations
None to declare.
Data availability
Metagenomic sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA1049329. The phyloseq object used for this project is available at Zenodo (https://zenodo.org/records/10104830). Scripts used for the analyses and figures are available at https://github.com/BeatriceCornuHewitt/VGO_COPDcaco_resistome.
Supplementary data
Figures S1 and S2 and Tables S2, S3, S4, S6, S7 and S8 are available as Supplementary data at JAC Online.
References
- 1. Simões M, Janssen N, Heederik DJJet al. Residential proximity to livestock animals and mortality from respiratory diseases in The Netherlands: a prospective census-based cohort study. Environ Int 2022; 161: 107140. 10.1016/j.envint.2022.107140 [DOI] [PubMed] [Google Scholar]
- 2. Borlée F, Yzermans CJ, Aalders Bet al. Air pollution from livestock farms is associated with airway obstruction in neighboring residents. Am J Respir Crit Care Med 2017; 196: 1152–61. 10.1164/rccm.201701-0021OC [DOI] [PubMed] [Google Scholar]
- 3. Mirabelli MC, Wing S, Marshall SWet al. Asthma symptoms among adolescents who attend public schools that are located near confined swine feeding operations. Pediatrics 2006; 118: e66–75. 10.1542/peds.2005-2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pavilonis BT, Sanderson WT, Merchant JA. Relative exposure to swine animal feeding operations and childhood asthma prevalence in an agricultural cohort. Environ Res 2013; 122: 74–80. 10.1016/j.envres.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Radon K, Schulze A, Ehrenstein Vet al. Environmental exposure to confined animal feeding operations and respiratory health of neighboring residents. Epidemiology 2007; 18: 300–8. 10.1097/01.ede.0000259966.62137.84 [DOI] [PubMed] [Google Scholar]
- 6. Schinasi L, Horton RA, Guidry VTet al. Air pollution, lung function, and physical symptoms in communities near concentrated swine feeding operations. Epidemiology 2011; 22: 208–15. 10.1097/EDE.0b013e3182093c8b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schulze A, Römmelt H, Ehrenstein Vet al. Effects on pulmonary health of neighboring residents of concentrated animal feeding operations: exposure assessed using optimized estimation technique. Arch Environ Occup Health 2011; 66: 146–54. 10.1080/19338244.2010.539635 [DOI] [PubMed] [Google Scholar]
- 8. Sigurdarson ST, Kline JN. School proximity to concentrated animal feeding operations and prevalence of asthma in students. Chest 2006; 129: 1486–91. 10.1378/chest.129.6.1486 [DOI] [PubMed] [Google Scholar]
- 9. Cambra-López M, Aarnink AJA, Zhao Yet al. Airborne particulate matter from livestock production systems: A review of an air pollution problem. Environ Pollut 2010; 158: 1–17. 10.1016/j.envpol.2009.07.011 [DOI] [PubMed] [Google Scholar]
- 10. Hamon L, Andrès Y, Dumont E. Aerial pollutants in swine buildings: a review of their characterization and methods to reduce them. Environ Sci Technol 2012; 46: 12287–301. 10.1021/es3025758 [DOI] [PubMed] [Google Scholar]
- 11. Koerkamp G, Metz PWG, Uenk JHMet al. Concentrations and emissions of ammonia in livestock buildings in Northern Europe. J Agric Eng Res 1998; 70: 79–95. 10.1006/jaer.1998.0275 [DOI] [Google Scholar]
- 12. Davis MF, Pisanic N, Rhodes SMet al. Occurrence of Staphylococcus aureus in swine and swine workplace environments on industrial and antibiotic-free hog operations in North Carolina, USA: a one health pilot study. Environ Res 2018; 163: 88–96. 10.1016/j.envres.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Rooij MMT, Hoek G, Schmitt Het al. Insights into livestock-related microbial concentrations in air at residential level in a livestock dense area. Environ Sci Technol 2019; 53: 7746–58. 10.1021/acs.est.8b07029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Franceschini G, Bottino M, Millet Iet al. Assessment of the exposure of Turkey farmers to antimicrobial resistance associated with working practices. Vet Sci 2019; 6: 13. 10.3390/vetsci6010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao FZ, He LY, Bai Het al. Airborne bacterial community and antibiotic resistome in the swine farming environment: metagenomic insights into livestock relevance, pathogen hosts and public risks. Environ Int 2023; 172: 107751. 10.1016/j.envint.2023.107751 [DOI] [PubMed] [Google Scholar]
- 16. Gibbs SG, Green CF, Tarwater PMet al. Isolation of antibiotic-resistant bacteria from the air plume downwind of a swine confined or concentrated animal feeding operation. Environ Health Perspect 2006; 114: 1032–7. 10.1289/ehp.8910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smit LAM, Hooiveld M, Van Der Sman-de Beer Fet al. Air pollution from livestock farms, and asthma, allergic rhinitis and COPD among neighbouring residents. Occup Environ Med 2014; 71: 134–40. 10.1136/oemed-2013-101485 [DOI] [PubMed] [Google Scholar]
- 18. Borlée F, Yzermans CJ, Van Dijk CEet al. Increased respiratory symptoms in COPD patients living in the vicinity of livestock farms. Eur Resp J 2015; 46: 1605–14. 10.1183/13993003.00265-2015 [DOI] [PubMed] [Google Scholar]
- 19. van Kersen W, Bossers A, de Steenhuijsen Piters WAAet al. Air pollution from livestock farms and the oropharyngeal microbiome of COPD patients and controls. Environ Int 2022; 169: 107497. 10.1016/j.envint.2022.107497 [DOI] [PubMed] [Google Scholar]
- 20. Turek EM, Cox MJ, Hunter Met al. Airway microbial communities, smoking and asthma in a general population sample. EBioMedicine 2021; 71: 103538. 10.1016/j.ebiom.2021.103538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dickson RP, Erb-Downward JR, Huffnagle GB. Homeostasis and its disruption in the lung microbiome. Am J Physiol Lung Cell Mol Physiol 2015; 309: L1047–55. 10.1152/ajplung.00279.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J, Ran Z, Wang Fet al. Role of pulmonary microorganisms in the development of chronic obstructive pulmonary disease. Crit Rev Microbiol 2021; 47: 1–12. 10.1080/1040841X.2020.1830748 [DOI] [PubMed] [Google Scholar]
- 23. Dicker AJ, Huang JTJ, Lonergan Met al. The sputum microbiome, airway inflammation, and mortality in chronic obstructive pulmonary disease. J Allergy Clin Immunol 2021; 147: 158–67. 10.1016/j.jaci.2020.02.040 [DOI] [PubMed] [Google Scholar]
- 24. Galiana A, Aguirre E, Rodriguez JCet al. Sputum microbiota in moderate versus severe patients with COPD. Eur Resp J 2014; 43: 1787–90. 10.1183/09031936.00191513 [DOI] [PubMed] [Google Scholar]
- 25. Garcia-Nuñez M, Millares L, Pomares Xet al. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J Clin Microbiol 2014; 52: 4217–23. 10.1128/JCM.01967-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mayhew D, Devos N, Lambert Cet al. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax 2018; 73: 422–30. 10.1136/thoraxjnl-2017-210408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Z, Maschera B, Lea Set al. Airway host-microbiome interactions in chronic obstructive pulmonary disease. Respir Res 2019; 20: 113. 10.1186/s12931-019-1085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Casals-Pascual C, Vergara A, Vila J. Intestinal microbiota and antibiotic resistance: perspectives and solutions. Hum Microbiome J 2018; 9: 11–5. 10.1016/j.humic.2018.05.002 [DOI] [Google Scholar]
- 29. Schwartz DJ, Langdon AE, Dantas G. Understanding the impact of antibiotic perturbation on the human microbiome. Genome Med 2020; 12: 82. 10.1186/s13073-020-00782-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bos MEH, Verstappen KM, Van Cleef BAGLet al. Transmission through air as a possible route of exposure for MRSA. J Expo Sci Environ Epidemiol 2016; 26: 263–9. 10.1038/jes.2014.85 [DOI] [PubMed] [Google Scholar]
- 31. Dohmen W, Schmitt H, Bonten Met al. Air exposure as a possible route for ESBL in pig farmers. Environ Res 2017; 155: 359–64. 10.1016/j.envres.2017.03.002 [DOI] [PubMed] [Google Scholar]
- 32. Van Gompel L, Luiken REC, Hansen RBet al. Description and determinants of the faecal resistome and microbiome of farmers and slaughterhouse workers: a metagenome-wide cross-sectional study. Environ Int 2020; 143: 105939. 10.1016/j.envint.2020.105939 [DOI] [PubMed] [Google Scholar]
- 33. Murray CJ, Ikuta KS, Sharara Fet al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022; 399: 629–55. 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Borlée F, Yzermans CJ, Krop Eet al. Spirometry, questionnaire and electronic medical record based COPD in a population survey: comparing prevalence, level of agreement and associations with potential risk factors. PLoS One 2017; 12: e0171494. 10.1371/journal.pone.0171494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Charlson ES, Bittinger K, Haas ARet al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 2011; 184: 957–63. 10.1164/rccm.201104-0655OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lanza VF, Baquero F, Martínez JLet al. In-depth resistome analysis by targeted metagenomics. Microbiome 2018; 6: 11. 10.1186/s40168-017-0387-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zankari E, Hasman H, Cosentino Set al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Rooij MMT, Heederik DJJ, van Nunen EJHMet al. Spatial variation of endotoxin concentrations measured in ambient PM10 in a livestock-dense area: implementation of a land-use regression approach. Environ Health Perspect 2018; 126: 017003. 10.1289/EHP2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cornu Hewitt B, Smit LAM, van Kersen Wet al. Residential exposure to microbial emissions from livestock farms: implementation and evaluation of land use regression and random forest spatial models. Environ Pollut 2024; 346: 123590. 10.1016/j.envpol.2024.123590 [DOI] [PubMed] [Google Scholar]
- 40. Esri . ArcGIS Desktop: Release 10.2.2. Environmental Systems Research Institute, 2014. [Google Scholar]
- 41. de Rooij MMT, Smit LAM, Erbrink HJet al. Endotoxin and particulate matter emitted by livestock farms and respiratory health effects in neighboring residents. Environ Int 2019; 132: 105009. 10.1016/j.envint.2019.105009 [DOI] [PubMed] [Google Scholar]
- 42. McEachran AD, Blackwell BR, Hanson JDet al. Antibiotics, bacteria, and antibiotic resistance genes: aerial transport from cattle feed yards via particulate matter. Environ Health Perspect 2015; 123: 337–43. 10.1289/ehp.1408555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nguyen XD, Zhao Y, Evans JDet al. Survival of Escherichia coli in airborne and settled poultry litter particles. Animals (Basel) 2022; 12: 284. 10.3390/ani12030284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, 2022. https://www.R-project.org/ [Google Scholar]
- 45. Brill SE, Law M, El-Emir Eet al. Effects of different antibiotic classes on airway bacteria in stable COPD using culture and molecular techniques: a randomised controlled trial. Thorax 2015; 70: 930–8. 10.1136/thoraxjnl-2015-207194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dickson RP, Huffnagle GB. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog 2015; 11: e1004923. 10.1371/journal.ppat.1004923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Crofts TS, Gasparrini AJ, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol 2017; 15: 422–34. 10.1038/nrmicro.2017.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Macedo G, van Veelen HPJ, Hernandez-Leal Let al. Targeted metagenomics reveals inferior resilience of farm soil resistome compared to soil microbiome after manure application. Sci Total Environ 2021; 770: 145399. 10.1016/j.scitotenv.2021.145399 [DOI] [PubMed] [Google Scholar]
- 49. Urmi UL, Nahar S, Rana Met al. Genotypic to phenotypic resistance discrepancies identified involving β-lactamase genes, blaKPC, blaIMP, blaNDM-1, and blaVIM in uropathogenic Klebsiella pneumoniae. Infec Drug Resist 2020; 13: 2863–75. 10.2147/IDR.S262493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Prevaes SMPJ, de Steenhuijsen Piters WAA, de Winter-de Groot KMet al. Concordance between upper and lower airway microbiota in infants with cystic fibrosis. Eur Respir J 2017; 49: 1602235. 10.1183/13993003.02235-2016 [DOI] [PubMed] [Google Scholar]
- 51. Ding D, Zhu J, Gao Yet al. Effect of cattle farm exposure on oropharyngeal and gut microbial communities and antibiotic resistance genes in workers. Sci Total Environ 2022; 806: 150685. 10.1016/j.scitotenv.2021.150685 [DOI] [PubMed] [Google Scholar]
- 52. Zomer TP, Wielders CCH, Veenman Cet al. MRSA in persons not living or working on a farm in a livestock-dense area: prevalence and risk factors. J Antimicrob Chemother 2017; 72: 893–9. 10.1093/jac/dkw483 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metagenomic sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA1049329. The phyloseq object used for this project is available at Zenodo (https://zenodo.org/records/10104830). Scripts used for the analyses and figures are available at https://github.com/BeatriceCornuHewitt/VGO_COPDcaco_resistome.