Abstract
Objective
Sjögren disease (SjD) is an autoimmune condition characterized by the dysfunction of the salivary and lacrimal glands. The study aimed to decipher the pathogenic cell populations and their immunologic pathways in the salivary glands. We further determined the therapeutic effect of inhibiting dedicator of cytokinesis 2 (DOCK2) shared by novel clusters of CD8+ T cells in an SjD mouse model.
Methods
This study employed single‐cell RNA sequencing to examine the composition and dynamics of immune cells in the salivary glands of SjD mice. By analyzing the transcriptomic data and employing clustering analysis, a specific target was identified, leading to the treatment of mice with a targeted inhibitor.
Results
The results showed diverse immune cell types, including B cells, CD4+ T cells, CD8+ T cells, macrophages, and natural killer cells. We identified specific clusters possessing phenotypic characteristics of immune cell subpopulations, thereby showing specific genes/pathways associated with the disease. The most striking finding was the elevated expression of DOCK2 in CD8+ T cells in the SjD model. This discovery is significant because subsequent treatment with a DOCK2 inhibitor 4‐[3‐(2‐Chlorophenyl)‐2‐propen‐1‐ylidene]‐1‐phenyl‐3,5‐pyrazolidinedione (CPYPP) led to a marked amelioration of SjD signs.
Conclusion
The effectiveness of DOCK2 inhibition in alleviating SjD signs highlights the potential of DOCK2 as a therapeutic target, opening new avenues for treatment strategies that could modulate the immune response more effectively in SjD.
INTRODUCTION
Sjögren disease (SjD) is a chronic autoimmune disorder in which the body's immune system mistakenly attacks the exocrine glands, primarily the salivary and lacrimal glands. This leads to hallmark symptoms such as dry mouth and eyes, but the pathologic reach of SjD extends to other organ systems, negatively affecting the overall quality of life. 1 , 2 The etiology of SjD remains elusive, although it is widely believed to result from an interplay between genetic predispositions and environmental triggers such as viral or bacterial infections. 3 , 4 These factors initiate an immune response that becomes abnormally self‐directed. T cells are central to this immune dysregulation, infiltrating the glands and disrupting their normal function by releasing cytokines and cell signaling molecules that are involved in recruiting and activating other immune cells, including B cells. 5 , 6 , 7 , 8 The downstream effects of this cytokine signaling lead to the hallmark symptoms of the disease and cause further glandular damage.
Various subtypes of Teff cells contribute differently to the pathogenesis of SjD. Th1 cells, for instance, secrete inflammatory cytokines such as interferon‐gamma (IFN‐γ) and tumor necrosis factor–alpha (TNF‐α), which regulate cell‐mediated immunity and activate other immune cells such as macrophages, natural killer (NK) cells, and CD8+ T cells. 9 Exposure to IFN‐γ has been shown to alter the tight junction components in salivary glands, leading to epithelial cell damage and diminished saliva production. 10 On the other hand, Th2 cells produce cytokines that drive B cell differentiation, eosinophil recruitment, and mucus production. 11 , 12 Genetic ablation of interleukin‐4 (IL‐4) of Th2 in the nonobese diabetic (NOD) mice restored normal levels of secretory function. 13 IL‐4 has also been found to play a crucial function during the clinical manifestation of SjD while having a limited effect on the pathology associated with the preclinical disease. 14 Th17 cells are the primary producers of IL‐17A and IL‐17F as well as other cytokines such as transforming growth factor β (TGFβ), IL‐6, and IL‐12, which have been detected at elevated levels in the plasma and saliva of patients with SjD. 15 , 16 It has been observed that the elimination of IL‐17 reduces sialadenitis more drastically in female mice than in male mice in an SjD mouse model. 17 Furthermore, it has been observed that transferring Th17 cells in IL‐17–deficient mice restores the SjD disease phenotype, highlighting the key role of Th17 cells in the inflammatory cascade and subsequent disease progression. 18 Recent studies have also highlighted the role of CD8+ T cells within the labial salivary gland infiltrates in patients with SjD. 19 , 20 These cells tend to localize with salivary duct epithelial cells and acinar cells, and their depletion protects against the disease's pathologic manifestations. 20
One intriguing molecule that could be central to the migration of immune cells into affected glands is DOCK2 (dedicator of cytokinesis 2); this molecule is a critical regulator of cell migration, particularly in immune cells. 21 It facilitates actin polymerization by activating small GTPases such as Rac, which are essential for cytoskeleton reorganization. 22 Although the specific role of DOCK2 in SjD remains uncertain, transcriptomic data suggests its unique up‐regulation coincides with the initial immune cell invasion into salivary glands in the SjD mouse model. Recent data indicated that DOCK2 had a distinct short‐term up‐regulated profile around 16 weeks of age in the SjD mice, coinciding with the first appearance of leukocytes in the salivary glands. 23 A recent study has shown that DOCK2 controls CD8+ T cell infiltration or migration in the tumors of hepatocellular carcinoma. 24 DOCK2 is involved in the clearance of herpes simplex virus type 1 infections by modulating the initial expansion of CD8+ T cells. 25 Mahajan et al showed that DOCK2 played a role in the conversion of naive T cells into virtual memory CD8+ T cells by lowering the threshold of self‐peptide response. 26 However, the role of DOCK2 in CD8+ T cells has never been explored in SjD. Therefore, in this study, we aimed to investigate the transcriptomic profiles of lymphocytes infiltrating the salivary glands in an SjD mouse model using single‐cell RNA sequencing. In addition, we examined the possible role of DOCK2 in mediating the activity of CD8+ T cells in SjD.
The Gene Expression & Genotyping core performed quantification and qualification, and next‐generation sequencing was provided by the NextGen DNA Sequencing core of the University of Florida Interdisciplinary Center for Biotechnology Research. The single‐cell RNA sequencing data have been deposited in the GEO database (accession number GSE268532).
Methods
Mice
SjD‐susceptible (SjDs) C57BL/6 J.NOD‐Aec1/2 and non‐SjDs C57BL/6 J (B6) control mice were housed under specific pathogen‐free conditions in the animal facilities of the University of Florida. Animal Care Services. The breeding and use of animals described herein were approved by and conducted under the direction of the University of Florida Institutional Animal Care and Use Committee. All methods were performed per the relevant guidelines and regulations. The development of C57BL/6.NOD‐Aec1/2 mouse and its SjD‐like disease phenotype are described previously (1,2). Briefly, the SjDs mouse was developed by introducing two genetic regions, one on chromosome 1 (designated Aec2) and the second on chromosome 3 (designated Aec1) derived from the NOD/LtJ mouse into the B6 mouse. All animals were maintained on a 12‐hour light‐dark schedule, and food and acidified water were provided ad libitum. At times indicated in the study, mice were euthanized by cervical dislocation after deep anesthetization with isoflurane, and their organs and tissues were freshly harvested for analyses. Utilizing the therapeutic approach, mice aged 28 weeks were treated with a DOCK2 inhibitor, CPYPP (4‐[3‐(2‐Chlorophenyl)‐2‐propen‐1‐ylidene]‐1‐phenyl‐3,5‐pyrazolidinedione, TOCRIS, Minneapolis, MN). CPYPP blocks DOCK2 by binding to DOCK2 DHR‐2 (DOCK homology region 2) domain and inhibits its catalytic activity (1). The mice were chosen at 28 weeks of age due the fact that at this age, mice have developed advanced clinical signs of SjD. Mice were given an initial dose of 100 uL of either 50 mg/mL CPYPP in DMSO or DMSO alone as control via intraperitoneal (IP) injection. Three more DMSO or CPYPP IP injections were given on days 3, 9, and 12 before euthanasia on day 14.
Human samples
Immunofluorescent staining for CD8 and DOCK2 was performed on five sicca control and six SjD patients. Sicca control patients were defined as those with xerostomia but without meeting the criteria for an SjD diagnosis; they were referred to the Oral Medicine Clinic at the University of Florida. Biopsies were obtained as reviewed and approved by the University of Florida's Institutional Review Board. SjD patients were identified by a rheumatologist, having met the criteria outlined by the 2016 American College of Rheumatology/European League Against Rheumatism (3). In brief, the classification criteria are based on the weighted sum of 5 items: anti‐SSA(Ro) antibody positivity and focal lymphocytic sialadenitis with a focus score ≥ 1 foci/mm2, each scoring 3; an abnormal ocular staining score ≥ 5 (or van Bijsterveld score ≥ 4), a Schirmer test ≤ 5 mm/5 min, and an unstimulated salivary flow rate ≤ 0.1 mL/min, each scoring 1. Individuals with a total score ≥ 4 for 5 items meet the criteria for primary SjD. Paraffin‐embedded labial salivary gland slides of primary SjD patients were generously provided by The SICCA Biorepository and Data Registry. Available clinical profiles were presented in Table S1.
Immunofluorescent staining
Salivary glands from DMSO and CPYPP‐treated SjDs mice were extracted and fixed in 10% phosphate‐buffered formalin in a histology cassette for 24 hours. Glands were paraffin‐embedded and sectioned at 10μm (Histology Tech Services, Gainesville, FL). Paraffin‐embedded biopsy samples were pressure‐cooked in Trilogy (Cell Marque, Rocklin, CA) for 5 minutes and 10 minutes for mouse salivary glands. After blocking with donkey sera (1 hour, room temperature), primary staining for human CD8 (Abcam, Cambridge, UK) or mouse CD8 (Santa Cruz Biotechnology, Dallas, TX) with DOCK2 (Bioss, Woburn, MA) was performed (4°C, overnight). The following secondary antibodies (Invitrogen, Waltham, MA) were used for humans: donkey anti‐mouse AF594 and donkey anti‐rabbit AF488. For mice, these secondary antibodies were used: donkey anti‐rat AF594 and donkey anti‐rabbit AF 488. Secondary antibodies were each incubated at room temperature for 1 hour. Images were captured with a Nikon Ti‐E fluorescent microscope at 400x magnification. Deconvolution was performed in Nikon NIS Elements. For the enumeration of CD8 T cells, a manual count was performed on a 100x magnification of the field containing an infiltrate, and then ROI intersectional thresholding was used to identify CD8+DOCK2+ cells.
10x Genomics single‐cell sample processing and cDNA library preparation
Samples were prepared using the Chromium Next GEM Single Cell V(D)J Kit v1.1, Mouse (10x Genomics, Pleasanton, CA) following the manufacturer's instructions. In brief, sorted single cells of the salivary glands for each sample were resuspended in RPMI containing 10% FBS to a final concentration of 700‐1200 cells/μl. A total of 8 samples were loaded onto a Chromium Next GEM Chip G, analyzed by the Chromium Controller (10x Genomics, Pleasanton, CA) for Gel Beads‐in‐ emulsion (GEMs) generation and reverse transcription. The generated cDNA was purified with SPRIselect (Beckman Coulter Inc, Indianapolis, IN) and used for 5′ gene expression library construction. The cDNAs and libraries were examined for quality control using D5000 ScreenTape (Agilent Technologies, Waldbronn, Germany), and Qubit (Thermo Fisher Scientific, Waltham, MA) was used for quantification. To achieve 20,000 reads per cell for 5′ gene expression libraries, the libraries were sequenced using Illumina NovaSeq6000 system (Illumina, San Diego, CA).
Measurement of saliva flow
Saliva flow rate (SFR) was recorded prior to drug injection (baseline), then every seven days. Briefly, mice were weighed and given an IP injection of 100 μl isoproterenol (0.2 mg/1 ml of PBS) and pilocarpine (0.05 mg/1 ml of PBS) to stimulate saliva production. Saliva was collected from the oral cavity with a pipet for ten minutes, with a one‐minute break at the midpoint. Saliva was briefly centrifuged, and the SFR was calculated as the volume of saliva (uL) per gram (weight of mouse).
Pathological examination of the mouse salivary glands
Salivary glands were fixed in 10% phosphate‐buffered formalin for 24 hours. The tissues were paraffin‐embedded; sections were cut at a 5‐um thickness and mounted onto slides, followed by hematoxylin and eosin (H&E) staining. Stained sections were observed at 200x magnification by using a Nikon Eclipse Ti‐E inverted microscope (Nikon, Tokyo, Japan). Focus score were determined by enumerating lymphocytic aggregates of ≥ 50 leukocytes for a single whole salivary gland per mouse.
Detection of antinuclear antibodies
Sera of mice was analyzed for the presence of antinuclear antibodies (ANAs) per the manufacturer's instructions (Immuno Concepts, Sacramento, CA). Briefly, sera were evaluated at 1:40 in PBS and incubated on HEP‐2 ANA slides for 30 minutes at room temperature. The secondary antibody, goat anti‐mouse IgG AF488 (Invitrogen, Waltham, MA, A11001), was incubated at room temperature on the slide before sealing with Vectashield DAPI medium (Vector Laboratories, Burlingame, CA) and adding a glass coverslip. ANA staining pattern was observed at 400x with a Nikon Ti‐E fluorescent microscope with an exposure of 200 ms (Nikon, Tokyo, Japan). Samples positive at 1:40 dilution were further titered for ANA analysis.
Analysis of tissues via flow cytometry
Salivary glands were excised, and single cells were isolated as previously described (4). Cells were rinsed, resuspended in FACS buffer, and stained (30 minutes, on ice) with Live/Dead Fixable Aqua Dead Cell Stain Kit, for 405 emission (Life Technologies, Carlsbad, CA) with either a B or T cell panel as follows: B cells: BV650 rat anti‐mouse/human CD45R/B220 (Biolegend, Cat # 103241, San Diego, CA), FITC rat anti‐mouse CD23 (Biolegend, Cat # 101605, San Diego, CA), PE rat anti‐mouse CD21/CD35 (CR2/CR1) (Biolegend, Cat # 123419, San Diego, CA), AF700 rat anti‐mouse IgD (Biolegend, Cat # 405729, San Diego, CA), BV421 rat anti‐mouse IgM (Biolegend, Cat # 406517, San Diego, CA); T cells: BV 785 rat anti‐mouse CD3 (Biolegend, Cat # 100355, San Diego, CA), FITC Rat Anti‐Mouse CD4 (Biolegend, Cat # 116004, San Diego, CA), APC rat anti‐mouse IFN‐γ (Biolegend, Cat # 505810, San Diego, CA), PE rat anti‐mouse CD8 (BD Pharmingen, Cat # MCD0804, Franklin Lakes, NJ), PE/Cy7 rat anti‐mouse IL‐4 (Biolegend, Cat # 504118, San Diego, CA), BV 421 rat anti‐mouse IL‐17A (Biolegend, Cat # 506926, San Diego, CA), and APC‐eF780 Mouse Anti‐Mouse NK‐1.1 (eBioscience, Cat # 47‐5941‐80, Franklin Lakes, NJ). Samples were run on a BD Fortessa flow cytometer, where 100,000 events were captured; in cases where a full 100,000 events were not available, the entire sample was run. Individual antibody compensations were performed using BD CompBeads (BD Biosciences, Franklin Lakes, NJ); in addition to negative compensation bead control, unstained salivary glands were also utilized to confirm gating strategy. Likewise, paired lymph nodes were used as a lymphocyte pure control to also confirm gating with a more robust cell density. Results were analyzed on FlowJo (FlowJo, Ashland, OR) prior to data processing with GraphPad Prism. For all samples, live lymphocyte populations were first selected. Then T cells were selected for either CD4+ or CD8+ for Th1 and Th17 (CD4+) or Tc1 and Tc17 (CD8+) subsets. FO I were IgM‐ IgD+CD23+, FO II were IgM+IgD+CD23+, and MZB were IgM+IgD‐CD21‐CD23+.
Tissue isolation and cell preparation
Salivary glands of C57BL/6.NOD‐Aec1/2 (51 weeks old, n=2 female, 2 male) and B6 (64 weeks old, n=2 female, 2 male) mice were explanted and digested in a buffer containing 1 mg/ml DNase (Sigma‐Aldrich, St. Louis, MO) and 1 mg/ml Collagenase Type 4 (Worthington, Lakewood, NJ, USA) in RPMI (Lonza, Allendale, NJ) complete media (10% FBS, 2 mM L‐glutamine, 0.05 mM β‐ mercaptoethanol). Tissues were placed in a MACS C tube (Miltenyi Biotec, San Diego, CA) for desiccation on GentleMACS V1.02 for a pulse of 38 seconds. After a 10‐minute incubation at 37°C, the digest buffer was removed and placed into 4°C RPMI complete media. The process was repeated twice. Single‐cell suspensions were centrifuged (2500 rpm, 10 min, 4°C) and resuspended in PBS for filtration through a 70‐μm sterile cell strainer (Fisher, Pittsburgh, PA). After a wash with PBS, cells were resuspended again in PBS for lymphocyte isolation with Lympholyte‐M cell separation media (Cedar Lane, Burlington, Canada) per the manufacturer's instructions. Single‐cell suspensions were stained for DAPI, and live cells were sorted with a sorter (SH800S, Sony, San Jose, CA) into RPMI containing 10% FBS on ice for single‐cell sequencing library preparation.
Single‐cell data preprocessing, gene expression quantification, and cell‐type determination
The raw data from each sample were demultiplexed and aligned to the GRCm38 reference genome, and the UMI counts were quantified using the 10x Genomics Cell Ranger pipeline (v 7.0.1). Data analysis continued with the filtered barcode matrix files using the Seurat package (v 4.3.0.1). Cells with >200 detected features and <10% mitochondrial reads were considered valid. LogNormalize in Seurat was used for individual samples before merging for downstream analysis to prevent clusters from being biased by differential sequencing depth. FindVariableFeatures was applied to normalize and find variable features within the single‐cell gene expression data, with ‘vst’ as method to choose top 2000 variable features. Clustering and differential expression analyses were performed using the R package Seurat with default parameters. Based on the ElbowPlot, the first 20 principal components (PCs) (1:20) were selected for the clustering analysis when that number reached the baseline of the standard deviation of the PCs. FindNeighbors uses the previously identified PCs was applied to calculate the distance between cells in the high‐dimensional space. A resolution of 0.6 was applied in FindClusters function to obtain a meaningful number of subclusters within the major cell types, which was identified through the clustree function. Cell clusters were visualized using Uniform Manifold Approximation and Projection (UMAP). Cells were represented in a 2D UMAP plane with 23 distinct clusters, cells that are in the same large population and share the vast majority of classical immune cell signatures are categorized into five major (T cells (Cd3d, Cd3e, CD4, CD8a), Cd68 (macrophage), Ncr1 (NK), and Cd19 (B cells)) and one minor cluster, of which 23 sub‐populations were identified and annotated within the macro‐population according to known biological cell types using canonical marker genes or published reference gene signatures (5–9).
Differential gene expression was performed using model‐based analysis of the single‐cell transcriptomics (MAST) test (10) (log fold‐change ≥0.25, minimum percentage 0.1, and minimum differential percentage >0.15) to select genes with an adjusted P value <0.01. UpSet (v 1.4.0) was used to make UpSet plots for showing matrix layout of all intersections of the comparison datasets. scRNAtoolVis (v 0.08) was used to make the volcano plot show the differentially expressed gene in certain subclusters of each sex and mouse strain. Pathway enrichment analysis was conducted using Metascape (http://metascape.org) for gene function annotation, and enrichment pathway analysis was used under the default setting. T‐like cells (CD3+) were extracted from the global data for downstream analyses to identify T cell subtypes. In the single‐cluster enrichment analysis, the FindAllMarkers function in the Seurat package was performed to obtain the rank of all genes (‘wilcox’, log fold‐change ≥0.25, minimum percentage 0.1, and minimum differential percentage >0.15). Then, the fgsea package (v1.17.1) was used to calculate GSEA enrichment scores and P values for each collection of gene sets. Signatures used for subset identity determination or phenotyping already published are referenced in each figure and were converted to corresponding mouse genes for analysis. All the analyses were conducted in the R environment (v.4.3.1).
Gene set variation analysis (GSVA)
Pathway analyses were performed on the 50 hallmark pathways annotated in the molecular signature database (11). The gene sets we used were from the database MSigDB (v7.4). The expression mean matrix between different subclusters was counted using the AverageExpression function as the input for package GSVA (v1.48.2) to calculate the enrich score. To compare the significance of each group, package limma (v 3.56.2) was used to construct a differential contract matrix and to analyze differences in the enrichment scores for specific groups versus the remaining groups (e.g., group CD8_C4 vs. other CD8 cells).
Trajectory analysis
Package monocle3 (v 1.3.1) was used to estimate the pseudotime path of T cell differentiation. T cells were extracted for Trajectory analysis, and the subset of the data set obtained by Seurat analysis was imported to create a Monocle object. Pseudotime values are assigned to cells using order_cells based on the cell projection on the main graph learned by the learn_graph function and the location of the selected root state. Genes that vary over a trajectory between clusters were identified through graph‐autocorrelation analysis [graph_test()] and genes expression trends of the top 5 differential genes were plotted using plot_genes_in_pseudotime, which is colored by subclusters in CD4 and CD8 T cells.
Statistical analysis
Statistical analyses were performed using Prism 8 software (GraphPad, La Jolla, CA). 2‐way ANOVA, Welch's t‐tests, or Mann‐Whitney U tests were performed where indicated. In all cases, p values < 0.05 were considered significant. For the ANA staining, a Chi‐squared test was performed.
RESULTS
Single‐cell profiling of immune cells in the salivary glands of SjD‐susceptible B6.NOD‐Aec1/2 mice
To elucidate the cellular heterogeneity during the development of SjD within the salivary glands, we employed the SjD‐susceptible (SjDS) B6.NOD‐Aec1/2 mice. The mouse strain was generated previously by breeding the combination of insulin‐dependent diabetes (Idd3) and Idd5 loci derived from the NOD mouse strain on the C57BL/6 background, where autoimmune exocrinopathy 1 (Aec1) corresponds to Idd3 and Aec2 corresponds to Idd5. 27 The strain developed SjD spontaneously and fully recapitulated the SjD phenotype. It exhibits lymphocytic infiltrations of the salivary and lacrimal glands at 12 to 16 weeks of age and production of autoantibodies. By 20 weeks of age, loss in the production of saliva and tears can be observed. 28 , 29 The mouse model develops clinical signs similar to those of human patients; therefore, it is an appropriate model for examining the cellular composition in the salivary glands. To achieve this, we isolated viable lymphocytes from the salivary glands of control B6 mice and SjDS mice that were age and sex matched. Isolated cells were subjected to single‐cell RNA sequencing (scRNA‐seq). A total of 32,796 cellular transcriptomes were retained for comprehensive analysis (Table S2). Unsupervised clustering using the Seurat software package identified five major cell types based on gene expression (B, CD4+ T cells, CD8+ T cells, macrophage, and NK cells) with an unknown or undefined population of cells (Figure 1A). Uniform Manifold Approximation and Projection (UMAP) plot of expression levels of selected genes in different clusters indicated the cell populations identified based on their signature markers (T cells [Cd3d, Cd3e, Cd4, Cd8a], Cd68 [macrophage], Ncr1 [NK], and Cd19 [B cells]) (Figure 1B and Figure S1A and B). Within the analyzed lymphocytes, 28.38% were identified as B cells, 55.55% as T cells, 6.3% as macrophages, and 8.4% as NK cells. T cell populations were further delineated into CD4+ and CD8+ T cells based on their surface markers, constituting 42.37% and 57.63% of the total T cell count, respectively (Figure 1C). A minor subset of cells (1.37%) remained unidentified, potentially arising from incidental epithelial and endothelial cell contamination. The unidentified cell population appeared to express specific markers associated with epithelium, such as Krtcap2 (keratinocyte‐associated protein 2) and Epb41 (erythrocyte membrane protein band 4.1). The presence of these markers and the lack of gene expression for immune cell markers suggest that this population may indeed contain contaminating epithelial/endothelial cells (Figure S1). The gland cell compositions of female and male SjDS mice were distinct, with the proportions of the major cell types being similar in mice of the same sex. The major differences for males were the B cells: 17% in B6 mice and 4% in SjDS mice. In females, 27% of CD8+ T cells were found in the B6 mice versus 17% identified in SjDS mice (Figure 1C). In summary, scRNA‐seq of salivary lymphocytes revealed prototypic cell types that have been shown to associate with SjD. Furthermore, an in‐depth analysis illustrated substantial cellular composition differences in the sex of SjDS mice.
Figure 1.
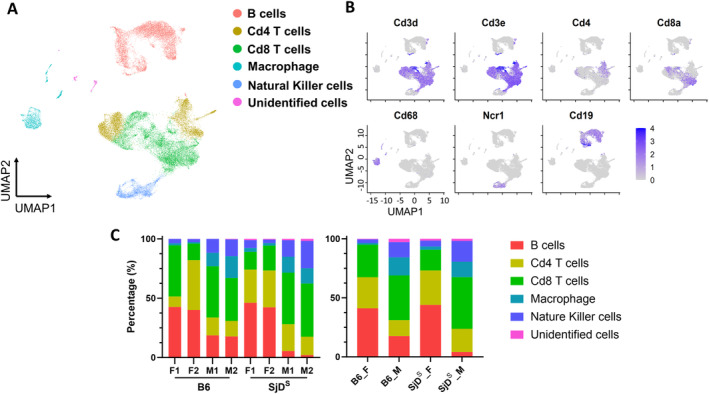
Single‐cell transcriptome of salivary gland immune cells. (A) UMAP (Uniform Manifold Approximation and Projection) plots of immune cells with different colors show the five major cell types in eight mice (B6 mice, n = 2 of each sex; SjDS B6.NOD‐Aec1/2 mice, n = 2 of each sex). (B) UMAP plots show the expression levels of selected marker genes in different clusters, with colors representing clusters expressing the genes and color densities representing different levels of selected gene expression. (C) Proportion of each major cell type in individual mice (left) and groups of mice (right) is shown. B6, C57BL/6; F, female; M, male; NOD, nonobese diabetic; SjDS, Sjögren disease–susceptible; UMAP, XXXX.
Clustering analysis of infiltrating cells to resolve the cellular heterogeneity
As presented previously, we have identified five major cell populations by unsupervised clustering. After further clustering, an analysis revealed 20 distinct immune‐related clusters composed of 6 B cell subsets, 4 subsets of CD4+ T cells, 5 subsets of CD8+ T cells, 3 macrophage subsets, and 2 NK cell subsets, in addition to the 3 subclusters from unidentified cells (Figure 2A and Figure S1C). Each subcluster displayed unique transcription patterns and was accordingly assigned numerical identifiers to differentiate their transcriptional profiles (Table S3). B cell subclusters B_C1, B_C2, B_C5, and B_C6 express genes associated with B cell development, activation, and differentiation, including Cd22 (B_C1), Egr1 (B_C2), Foxo1 and Malt1 (B_C5), and Bach2 and Pax5 (B_C6). Among those subclusters, Ighd and Fcrl1 (B_C1) are directly involved in immunoglobulin production and function, Bank1 (B_C2) is linked to B cell receptor (BCR) signaling, and Lrrk2 (B_C6) suggests roles in cellular signaling and potentially neuroimmune interactions. Other B cell subsets, such as B_C3, show distinct expression patterns of genes involved in B cell signaling and migration through the expression of Cxcr5, and Ighm and Igkc are immunoglobulin genes, indicating that B_C4 cells are heavily involved in antibody production. 30 , 31 , 32 , 33 , 34
Figure 2.
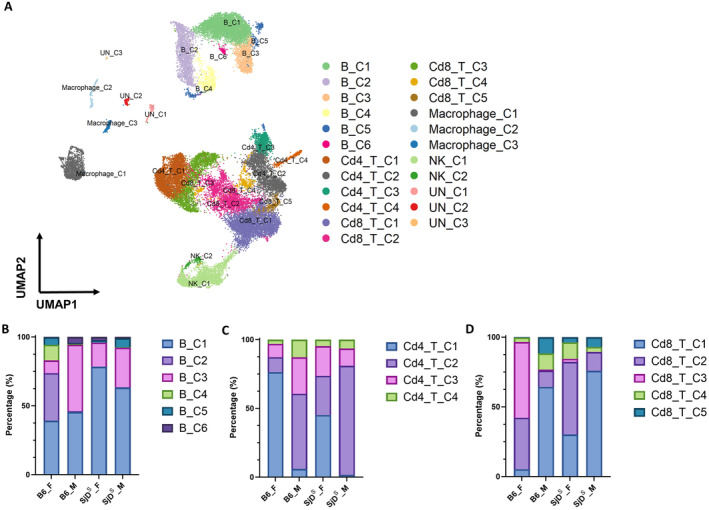
Cellular heterogeneity of salivary gland immune cells. (A) UMAP plots of immune cells show associated cell subpopulations. (B) Proportion of B cell subpopulations of male and female B6 and SjDS mice is shown. Six subpopulations were found in B cells and are indicated by different colors. (C) Proportion of CD4+ T cells subpopulations of male and female B6 and SjDS mice is shown. Four subpopulations were found in CD4+ T cells and are indicated by different colors. (D) Proportion of CD8+ T cell subpopulations of male and female B6 and SjDS mice is shown. Five subpopulations were found in CD8+ T cells and are indicated by different colors. B6, C57BL/6; F, female; M, male; NK, natural killer; SjDS, Sjögren disease–susceptible; UMAP: Uniform Manifold Approximation and Projection; UN: Unknown.
Subclustering analysis of Cd4+ T cells showed the functional heterogeneity and specificity of T cells. The Cd4+ T cell subsets express genes associated with T cell signaling and regulation. Cd4_T_C1 and Cd4_T_C3 express genes vital for T cell proliferation and survival, specifically Stat5a (Cd4_T_C1) and Tnfrsf4/OX40 (Cd4_T_C3). Ifngr2 in Cd4_T_C1 suggests responsiveness to IFN signaling, and Lgals1 (Cd4_T_C3) suggests roles in immune regulation and apoptosis. For other Cd4 T cell subsets, Cd4_T_C2 expresses inducible T‐cell costimulator, which is important for T cell activation and function, and Cd4_T_C4 expresses Rora and Nfkb1, which are involved in T cell differentiation and immune responses. 35 , 36 , 37 , 38 Among the Cd8+ T cell subsets, Cd8_T_C1 expresses genes associated with T cell effector functions and migration, such as Fasl and Cxcr6, whereas Cd8_T_C2 up‐regulates Ccl5, Gzmk, and Eomes, which are related to T cell activation and cytotoxicity. Cd8_T_C3 shows high expression of ribosomal protein genes, suggesting active protein synthesis; particularly, Camk1d expressed in this subcluster has been reported to interact with T cell–mediated cytotoxicity and negatively affect antitumor responses. Cd8_T_C4 expresses genes involved in T cell receptor signaling, such as Themis and Fyn, and Cd8_T_C5 up‐regulates IFN‐stimulated genes (Ifit1, Isg15, and Stat1). 39 , 40 , 41 , 42 , 43 , 44 , 45
Macrophage subpopulations also display unique gene expression profiles, with Macrophage_C1 and Macrophage_C2 expressing genes related to immune regulation and lipid metabolism such as Apoe (Macrophage_C1) and Ifitm3 and Fabp5 (Macrophage_C2), Macrophage_C2 up‐regulating Ifi30 and H2‐Aa involved in antigen presentation, and Macrophage_C3 expressing genes associated with macrophage activation (Aif1 and Trem2). 46 , 47 , 48 , 49 , 50 NK cell subpopulations overall express genes related to cell cytotoxicity, such as Klrk1/NKG2D (NK_C1) and Gzma and Gzmb (NK_C2); Ifngr1 up‐regulated in NK_C1 indicates responsiveness to IFN signaling, and Xcl1 expressed in NK_C2 is associated with dendritic cell recruitment and stimulation. 51 , 52
Salivary gland cellular heterogeneity was attributed by sex dimorphism and disease phenotype
The B, CD4+, and CD8+ T cell subsets showed variations based on sex and disease phenotype (Figure S1). 30 , 31 , 32 , 33 Specifically, a notable presence of B_C2 (34%) and B_C4 (11%) was observed in female B6 mice, and female SjDS mice demonstrated a lower proportion of B_C5 (1.7%) than female B6 (5.4%). In contrast, male SjDS mice exhibited an increase in B_C5: 6.5% compared with 0.7% in B6 (Figure 2B). Analysis of Cd4+ T cell clusters indicated the highest percentage of Cd4_C2 in male SjDS mice and an increased Cd4_C3 in female SjDS mice compared with female B6 mice. The Cd4_T_C1 fraction was more prevalent in female B6 mice (76%) than in female SjDS mice (45%), but the Cd4_T_C2 fraction was greater in male SjDS mice (79%) compared with B6 (54%) (Figure 2C). Examination of the Cd8+ T cell clusters revealed a higher Cd8_T_C1 fraction in male SjDS mice (76%) compared with male B6 mice (64%). A similar trend was observed in the Cd8_T_C2 fraction, where female SjDS mice had 52%, in contrast to 36% in B6 mice. A distinct Cd8_T_C3 (54%) was present in female B6 mice, with a minor Cd8_T_C5 fraction (0.3%) (Figure 2D). In summary, there is a distinct and dynamic change in the B, Cd4+, and Cd8+ T cell subsets between SjDS mice and the healthy controls, and it is most profound when examined by sex.
Recent data suggest that modulating the T cell response has a direct impact on the clinical manifestations of SjD. Additionally, other recent studies highlight the increasingly recognized critical role of CD8+ T cells. We aimed to confirm and further characterize the T cell subclusters, exploring their biologic associations with the disease process. For a comprehensive annotation of the Cd4+ and Cd8+ T cell subpopulations, we referenced marker‐gene signatures from extant literature to determine the cell type or functionality. 53 , 54 , 55 , 56 , 57 Employing these published cellular markers, we discerned a naive Cd4+ T cell subset, termed Cd4_T_C1, that exhibited heightened expression levels of genes such as Sell, Lef1, and Igfbp4 and was enriched in a naive population. The Cd4_T_C2 subset bore a Th1 phenotype, evidenced by the high expression of Tbx21, Ccl5, and Cxcr3 markers. Furthermore, the Cd4_T_C3 subset, characterized by Foxp3 expression, mirrored a regulatory T cell (Treg cell) signature. In contrast, Cd4_T_C4, manifesting elevated Il17 and Rorc expression, presented a Th17 phenotype (Figure 3A and Table S3). Furthermore, using individual cell enrichment of indicated selected signatures from the published dataset, 53 , 54 , 55 , 56 , 57 it has been validated that Cd4_T_C1 shared signature genes with naive Cd4, as well as Cd4_T_C2 with stress CD4 and effector memory T cells re‐expressing Cd45RA (TEMRA) CD4 and Cd4_T_C3 with Treg CD4 (Figure 3B).
Figure 3.
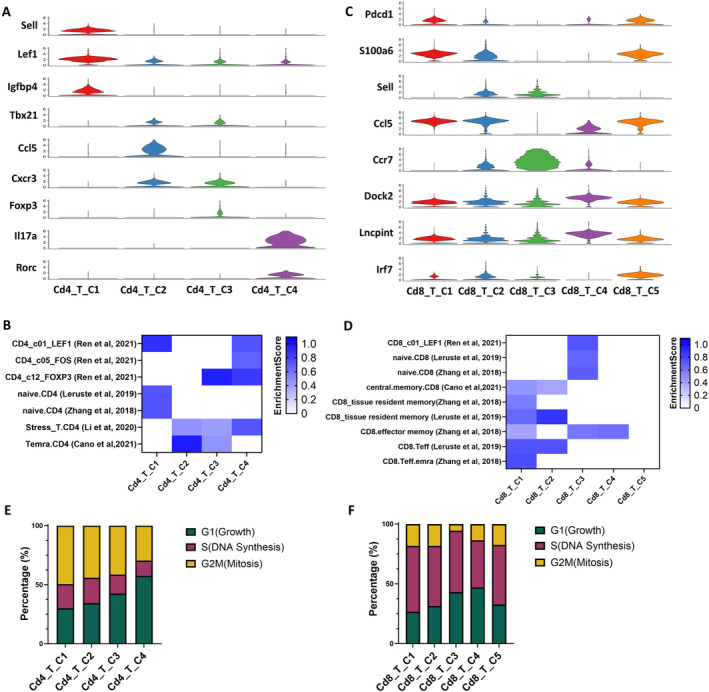
Cluster features of CD4+ and CD8+ T cell subsets in the salivary glands of SjD mice. (A) Violin plots show expression levels of selected marker genes in four different subpopulations of CD4+ T cells: naive T cells (Sell, Lef1, Igfbp4), Th1 (Tbx21, Ccl5, Cxcr3), Treg (Foxp3), and Th17 (Il17a, Rorc). (B) Gene enrichment of functional CD4+ T cell subsets in subpopulations of CD4+ T cells converted from published datasets is shown. 53 , 54 , 55 , 56 , 57 The enrichment scores are shown in heatmap with different color densities. (C) Violin plots show expression levels of selected marker genes in five different subpopulations of CD8+ T cells. Sell was selected as a naive T cell marker, with Ccl5 and Ccr7 expressed differently in naive Cd8_T_C2 and naive Cd8_T_C3 clusters. Pdcd1 was selected as an exhausted T cell marker, with Isg15 expressed in exhausted Cd8_T_C1 (low expression) and exhausted Cd8_T_C5 cluster (high expression). Cd8_T_C4 was a unique Cd8 T cell subcluster with Dock2 and Lncpint highly expressed. (D) Gene enrichment of functional CD8+ T cell subsets in subpopulations of CD8+ T cells converted from published datasets is shown 53 , 54 , 56 , 57 ; enrichment scores are shown in heatmap with different color densities. (E) Proportion of cells in CD4+ T cell subpopulations undergoing each cell cycle phase is shown. Cd4_T_C4 has the least fraction of cells undergoing mitosis (29.6%). (F) Proportion of cells in CD8+ T cell subpopulations undergoing each cell cycle phase is shown. SjD, Sjögren disease; Teff, T effector; TEMRA, effector memory T cells re‐expressing Cd45RA; Treg, regulatory T.
Two naive classifications were identified in the Cd8+ T cell subset, which prominently expressed Sell and epitomized a naive Cd8+ T cell phenotype. Delving deeper, we observed that Cd8_T_C2 exhibited Ccl5 expression, whereas Cd8_T_C3 had heightened Ccr7 expression. Two distinct Cd8+ T cell subsets, Cd8_T_C1 and Cd8_T_C5, manifested the exhaustion marker Pdcd1. However, they diverged in their Isg15 expression, with Cd8_T_C1 being Isg15 negative and Cd8_T_C5 being Isg15 positive; notably, their expression profiles varied subtly. Cd8_T_C1 manifested elevated S100a6 expression, aligning more with a central memory/effective phenotype, whereas Cd8_T_C5 exhibited pronounced Irf7 expression and did not align with the previously mentioned phenotypes. Cd8_T_C4 emerged as a distinctive subset with amplified Dock2 expression and various non–protein‐coding RNAs, such as Lncpint. This subset did not closely align with any pre‐existing clusters (Figure 3C and Table S3). Although Dock2 is most strongly expressed in the Cd8_T_C4 population, it is indeed transcribed at varying levels across all immune cell subsets (Figure S2A and B). Dock2 is also expressed in Cd4+ T cells subpopulations, albeit at lower levels compared to Cd8+ T cells (Figure S2C). This corroborates our previous studies proposing a role for Dock2 in in immune cell migration, activation, and effector functions, beyond its specific association with the Cd8_T_C4 subset.
Similarly, Cd8_T_C1 is shared with central memory (cm) CD8, tissue‐resident memory CD8, and TEMRA CD8; Cd8_T_C2 with effector and tissue‐resident memory CD8; Cd8_T_C3 with naive CD8; and Cd8_T_C4 with effector memory CD8 (Figure 3D). We subsequently assessed the cell cycle dynamics of various T cell subsets. Within the Cd4+ T cell subsets, Cd4_T_C4 (Th17) exhibited the smallest proportion of cells in the mitotic phase with the largest percentage in the growth phase. Cd4_T_C2 (Th1 cells) and Cd4_T_C3 (Treg cells) have a higher percentage of G1 phase but lower G2M phase compared with the Cd4_T_C1 naive cluster (Figure 3E). Within the CD8+ T cell subsets, Cd8_T_C4 possessed the highest percentage of cells in G1 and the least in S phase (Figure 3F). In summary, the identification of these subpopulations and their associated biomarkers provides valuable insights into the functional specialization and activation states of immune cells; further clustering analysis with gene enrichment of infiltrating cells in the salivary glands allowed us to categorize the distinct cell subpopulations and resolved the cellular heterogeneity that has not been observed, specifically the Cd8_T_C4 expressing high levels of Dock2.
Single‐cell transcriptomics uncovers complex biologic pathways of salivary gland CD4 + and CD8 + Teff cells in SjD
Unique patterns were discerned with Cd4+ T cell subsets. The Cd4_T_C1 and Cd4_T_C3 subsets showcased converging gene enrichment profiles, prominently engaging in the regulation of the mitogen‐activated protein kinase cascade, cytokine signaling and production, negative regulation of leukocyte genesis, Th17 lineage differentiation, and inflammatory cascades. The Cd4_T_C2 and Cd4_T_C4 subsets presented alternate regulatory modalities, introducing distinct manners of cytokine production and adaptive immune interactions. Examining gene ontology biologic processes, these subsets again showed divergence: Cd4_T_C1 and Cd4_T_C3 leaned toward regulating biologic processes, immune system processes, and multicellular organismal processes. These two subsets were explicitly enriched in growth, reproductive process, and viral process. Cd4_T_C1 exhibited marked involvement in response to stimulus and developmental processes and was explicitly enriched in the rhythmic process compared to Cd4_T_C3 (Figure 4A and B). Interestingly, gene set variation analysis (GSVA) indicated that Cd4_T_C3 and Cd4_T_C4 overexpress both immunosuppressive hallmark pathways, including TGFβ and reactive oxygen species, as well as multiple proinflammatory pathways, including IFN‐γ and TNF‐α, compared with Cd4_T_C1 and Cd4_T_C2 (Figure S1A). However, an overall protein–protein interaction networks (PPI) analysis of genes enriched in each cluster indicated that various subpopulations of CD4+ T cells were involved in different regulatory pathways (Figure S1B).
Figure 4.
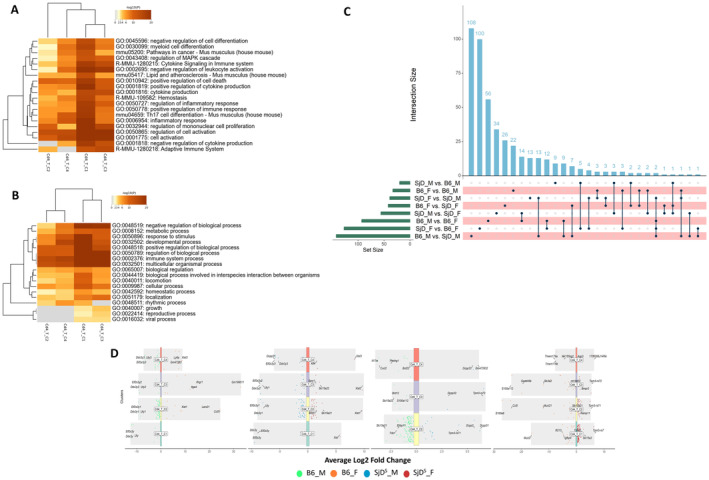
Biologic pathways of CD4+ T cells in the salivary glands of SjD mice. (A and B) Significantly enriched pathway (A) and biologic process of individual T cell subset of CD4+ T cell (B) are shown. (C) UpSet plots show a matrix layout of all intersections of the eight comparison datasets in CD4+ T cells (by each genotype and sex, separately) and sorted by size. The size of each comparison dataset was indicated as the green bar on the left, showing the number of up‐regulated genes in the right dataset compared with the left dataset. Dark circles in the matrix indicate sets of genes with corresponding intersections, with the number of genes in each set shown above the blue bar graph. The connecting lines indicate the comparison sets that share this gene set (two sets or more). (D) Volcano plot of DEGs between paired experimental groups of individual CD4+ T cell subpopulations is given. Different colored dots represent DEGs in each group and are indicated in the legend below the figure. The top three genes in each group (if more than three genes were identified) were labeled in the figure. B6, C57BL/6; DEG, differentially expressed gene; F, female; M, male; MAPK, mitogen‐activated protein kinase; SjD, Sjögren disease.
Cd8+ T cell subsets each exhibit unique functions. The Cd8_T_C5 subset notably regulated cytokine‐responsive stimuli, IFN signaling, and response to the virus. In contrast, a majority of the Cd8+ T cell subsets centralized their functional dynamics around lymphocyte activation. Specifically, the Cd8_T_C2 and Cd8_T_C3 subsets were found to be close regarding functional roles, albeit the Cd8_T_C3 subpopulation plays a dominant role in the negative regulation of cytokine production, response to lipopolysaccharide, positive regulation of immune response, and cytokine signaling in the immune system. Furthermore, although both the Cd8_T_C4 and Cd8_T_C1 subsets were actively involved in the epidermal growth factor receptor 1 signaling cascade, they exhibited distinct characteristics; Cd8_T_C4 was found to be aligned with hepatitis B pathways and demonstrated minimal involvement in type I IFN production, especially compared to Cd8_T_C5. A broader examination into their biologic engagements highlighted that Cd8_T_C5 played an active role in several processes, especially viral processes; Cd8_T_C4 was engaged in intraspecies interactions; Cd8_T_C2 was not involved in biologic processes in interspecies interactions and metabolic processes; and Cd8_T_C4 was not enriched for locomotion (Figure S2A and B). GSVA analysis further corroborated that Cd8_T_C5 overexpressed multiple proinflammatory pathways, including IFN‐γ and TNF‐α (Figure S3A). Meanwhile, each CD8+ T cell subpopulation is relatively independent of signaling regulation with unique functions (Figure S3B).
Furthermore, scRNA‐seq allows us to analyze the gene expression signature of different subclusters’ sample‐specific gene expression signatures. For the four different subsets of CD4+ T cells, we found 112 differentially expressed genes (DEGs) in males compared with 189 DEGs in females in B6 mice, and 52 DEGs in males and 67 DEGs in females in SjDS mice. When comparing the SjDS mice with the B6 mice, we detected 95 DEGs in SjDS mice and 263 DEGs in B6 mice for the females and 20 DEGs in SjDS mice and 76 DEGs in B6 mice for the males (Figure 4C). Using the functionality of scRNA‐seq, we delved into the gene expression profiles of different subgroups characterized by sample‐specific genes. This analytical approach facilitates double comparisons, delineating differences in gene expression according to phenotype (B6 vs SjDS) and sex (female vs male). In the context of the four identified CD4+ T cell subsets, the B6 phenotype exhibited sex‐specific disparities: 112 DEGs were discerned in males, whereas females presented with 189 DEGs. Contrasting this, the SjDS phenotype manifested a subdued differential expression: 52 DEGs in males compared with 67 DEGs in females. Notably, when contrasting the SjDS phenotype against the B6, females exhibited a pronounced difference, with 95 DEGs identified in SjDS mice and 263 DEGs in B6 mice. In males, however, the differential expression was less dramatic, with 20 DEGs in the SjDS mice and 76 DEGs in their B6 counterparts (Figure 4D).
When studying the five CD8+ T cell subsets, the B6 phenotype again displayed sex‐biased gene expression. A substantial 296 DEGs were identified in males, in contrast to the 108 DEGs in females. The SjDS phenotype, akin to the CD4+ T cell subsets, demonstrated a diminished differential gene expression: 59 DEGs in males as opposed to 29 DEGs in females. A phenotype comparison, however, showcased an intriguing pattern. In females, 50 DEGs were noted in the SjDS mice, almost symmetrically countered by 36 DEGs in the B6 mice. Meanwhile, the male cohort presented with 16 DEGs in the SjDS phenotype and a considerably larger 59 DEGs in the B6 phenotype (Figure S2C and D).
Pseudotime trajectories predict the sexual dimorphic developmental and transitional differences
To decipher the developmental trajectory and differentiation context of various T cell subtypes, we utilized Monocle 3 to perform single‐cell trajectories. This approach localizes each T cell on a developmental trajectory based on the changes of the transcriptomic expression. In either CD4+ (Figure 5A) or CD8+ T cells (Figure 5B), we postulated a predictive cell developmental timeline, initiating with the naive phenotype at pseudotime 0 and culminating with the effector cell phenotype until the predicted end of pseudotime. The trajectory matched with T cell subpopulations identified earlier (Figures 2 and 3) and demonstrates a unidirectional developmental trajectory from one cluster to another. In addition, the top five genes that changed over the trajectory in the CD4+ and CD8+ T cell populations were selected and illustrated to change in their expression levels over time (Table S4).
Figure 5.
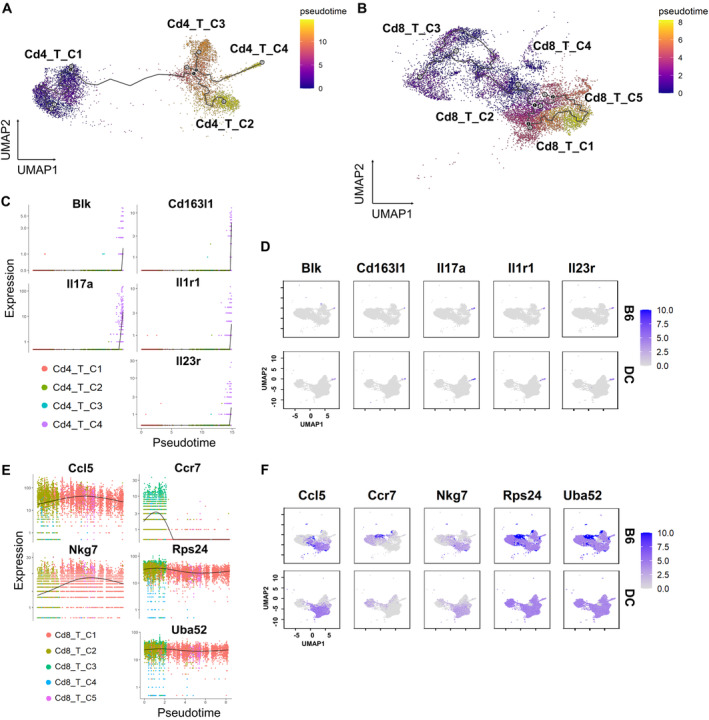
Pseudotime trajectory analysis reveals distinct CD4+ and CD8+ T cell developmental pathways. Pseudotime analysis of CD4+ (A) and CD8+ (B) T cells is shown. The lines represent the developmental trajectories computed by Monocle 3. Each gray plot point is a leaf node at the end of the trajectory, and each black plot point is a developmentally derived branch point. Pseudotime was indicated by color. The dynamics of top five differential genes under pseudotime trajectory and the overall gene expression in T cell population for CD4+ T cells (C and D) and CD8+ T cells (E and F) are shown. Different colors indicate subclusters in CD4+ and CD8+ T cells. B6, C57BL/6; Blk, B‐lymphoid tyrosine kinase; DC, Double Congenic or SjDs mice; IL, interleukin; UMAP, Uniform Manifold Approximation and Projection.
As illustrated, the differential gene expression in CD4+ T cells was all in Cd4_T_C4, in a late developmental mode, and Il17r has an extremely high expression level among the subclusters (Figure 5C). The percentages of Cd4_T_C4 were 3.2%, 12.9%, 4.8%, and 6.5% in female B6, male B6, female SjDS, and male SjDS mice, respectively. Differences in gene expression in Cd4 T cells during differentiation lead to differences in the overall proportion of Cd4_T_C4 cells (Th17 phenotype), which ultimately produces varying degrees of sex‐related interventions in the onset and progression of autoimmune diseases. We mapped the overall expression levels of these five selected genes (Figure 5D) to validate their concentrated expression in Cd4_T_C4, which is involved in cytokine signaling, T cell differentiation, and immune cell signaling. However, in Cd8+ T cells, Ccr7 was activated earlier than other genes and was highly expressed at the beginning of the trajectory (Figure 5E) and was also expressed overall in Cd8_T_C2 and Cd8_T_C3. In female B6, male B6, female SjDS, and male SjDS mice, the percentages of Cd8_T_C3 were 54.2%, 0.8%, 2.1%, and 0.2%, respectively, whereas Ccr7 was highly expressed only in B6 mice (Figure 5F). Ccr7 is a chemokine receptor expressed on naive and central memory T cells, which binds the chemokines Ccl19 and Ccl21 and is critical for T cell homing to secondary lymphoid organs.
Notably, Cd4+ T cells also express some level of Ccr7 but are not sexually distinguishable compared with Cd8+ T cell expression. Ccl5 and Nkg7 have similar expression patterns, in which both genes are activated after Ccr7 expression. Nkg7 is expressed at a lower level than Ccl5 and activated after Ccl5. Nkg7 and Ccl5 are mainly restricted to Cd8 T cells, with Ccl5 being activated in the Cd8_T_C1 and Cd8_T_C2 were expressed and higher in some parts of Cd8_T_C2. The percentages of Cd8_T_C2 in female B6, male B6, female SjDS, and male SjDS mice were 40%, 11.6%, 52%, and 13.5%, respectively, which were more related to sex differences. Rps24 and Uba52, both genes encoding ribosomal proteins, maintained constant expression levels throughout the time period and had overall high expression in parts of Cd8_T_C2 and Cd8_T_C3. This analysis further reveals distinct sexual dimorphic developmental and transitional differences in CD4+ and CD8+ T cell subpopulations, providing insights into the molecular mechanisms underlying sex‐ and genetic background–related variations in autoimmune disease susceptibility and progression.
Inhibition of DOCK2 improves SjD signs in mice
As presented, Cd8_T_C4 expressed high levels of Dock2 in mice, consistent with our previous study as essential mediator for cell trafficking and migration. 23 GSVA analysis also reveals a unique role for Cd8_T_C4 in the maintenance of cellular functions, especially mitochondrial assembly and functioning (Figure S4). To verify DOCK2's elevated expression in CD8d+ T cells across patients and mouse model of SjD, we stained for DOCK2 protein in the salivary glands. As presented in Figure 6A, DOCK2 is highly expressed in the glands of male and female SjDS mice, specifically by CD8d+ T cells. We observed minimal expression by CD4d+ T cells (data not shown). Similar results were found in the labial salivary glands of patients with SjD (Figure 6B). Given DOCK2's potential impact on clinical pathology, we investigated its inhibitory effects using CPYPP, a DOCK2 inhibitor. As presented in Figure 6D, SjDS mice treated with CPYPP showed a marked decrease of CD8+DOCK2+ population (Figure 6C), confirming the effectiveness of CPYPP as a DOCK2 inhibitor. More importantly, SjDS mice treated with CPYPP showed improvement in antinuclear antibody (ANA) (Figure 6D) and increased saliva flow at 12 weeks postinjection (Figure 6E). The most drastic change was the diminished focus score in the glands (Figure 6F). We further explored the impact of DOCK2 inhibition on various cell populations. As shown in Figure S7, treatment with CPYPP led to a substantial reduction in CD8d+ T cells compared with CD4d+ T cells (Figure S7A and B, Table S5). Among the T cell subsets, only cytotoxic T cell 1 (Tc1) was significantly diminished, whereas Th1, Th17, and Tc17 populations remained unaffected (Figure S7C–G and Table S5). CPYPP treatment negatively affected marginal zone B cells (MZBs) and follicular B cell (FOB) I, FOB II, and FOB populations. These findings suggest that DOCK2 plays a crucial role in SjD development, with its inhibition markedly affecting cytotoxic CD8+ T cells and major B cell populations.
Figure 6.
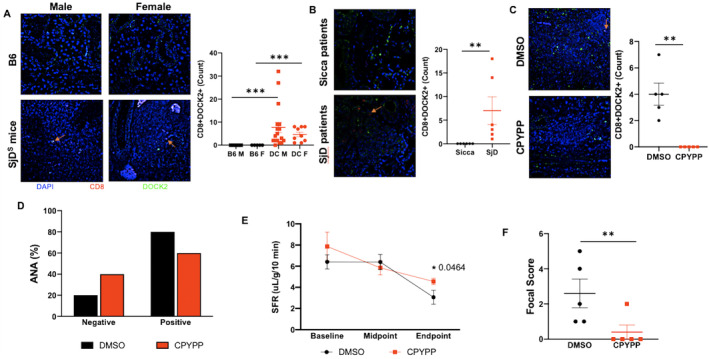
DOCK2 expression and clinical impact of DOCK2 inhibitor in SjD. (A) Immunofluorescent staining was performed for CD8 (red) and DOCK2 (green) with a DAPI counterstain (blue). Images include salivary glands of B6 and SjDS mice. (B and C) Labial salivary gland biopsies from Sjögren's International Collaborative Clinical Alliance control and patients with SjD (B) and DMSO‐ or CPYPP‐treated mice (C) are shown, with representative 400× images provided for each group. A single arrow per relevant image is provided to indicate the CD8+DOCK2+ cells. Enumeration of CD8+DOCK2+ cells in the infiltrates was determined using ROI thresholding in NIS Elements (Nikon), for which Mann‐Whitney t‐test was performed. (D) Percentage of mice positive or negative for ANA is given, with black indicating DMSO‐treated and red CPYPP‐treated mice (n = 5 per group). Chi‐squared test was performed, and although P = 0.490153, there is a shift toward improvement. (E) SFRs at baseline, midpoint, and endpoint are given, with black indicating DMSO‐treated and red CPYPP‐treated mice (n = 5 per group). Welch's one‐tailed t‐test was performed with *P = 0.0464. (F) Focal scores were counted if there were ≥50 lymphocytes in the whole salivary gland. Mann‐Whitney one‐tailed t‐test indicate *P = 0.0159. ANA, antinuclear antibody; B6, C57BL/6; CPYPP, 4‐[3‐(2‐Chlorophenyl)‐2‐propen‐1‐phenyl‐3,5‐pyrazolidinedione; DOCK2, dedicator of cytokinesis 2; F, female; M, male; ROI, region of interest; SFR, salivary flow rate; SjD, Sjögren disease; SjDS, Sjögren disease–susceptible.
DISCUSSION
This study employed scRNA‐seq to profile immune cells in the salivary glands of SjD mice and has offered important insights into the intricate immunologic mechanisms underlying SjD. The detailed analysis of various immune cell types, including B cells, CD4+ and CD8+ T cells, macrophages, NK cells, and their subpopulations, has illuminated the complex cellular milieu associated with SjD. Notably, the study revealed pronounced sex‐specific differences in the immune cell composition of the salivary glands, underscoring the influence of sex on the immune response in SjD and potentially explaining the higher prevalence of the disease in females. Identifying distinct cell subpopulations and their gene expression profiles provides a better understanding of the major immune cells and their cellular clusters in SjD pathogenesis. The study's exploration of gene function annotation and pathway analysis has provided a comprehensive view of the biologic processes and pathways active in different T cell subsets. This analysis enhances our understanding of these cells’ functional roles and interactions in the context of SjD. Lastly, the elevated level of Dock2, high expression of DOCK2 in CD8+ T cells, and the significant amelioration of SjD symptoms after treatment with the DOCK2 inhibitor highlight the potential of targeted therapies. This finding positions DOCK2 as a promising therapeutic target, opening new avenues for treatment strategies that address the underlying immune dysregulation in SjD.
CD4+ T cells are pivotal in initiating and perpetuating the autoimmune response. Once within the affected glands, CD4+ T cells become activated upon encountering their specific antigens and possibly differentiate into Teff cell subsets, which release key cytokines such as IFN‐γ, IL‐4, and IL‐17. 4 The supporting evidence from the animal models suggests that the development of SjD progresses through three distinct but continuous phases. In the innate immune stage, which initiates glandular pathology, aberrant physiologic changes and increased acinar cell apoptosis. 58 During the adaptive immune stage, the unregulated acinar cell apoptosis evokes migration of leukocytes expressing proinflammatory cytokines to the exocrine glands, thereafter establishing lymphocytic foci, first composed of T cell clusters and followed by the recruitment of B lymphocytes. 59 At the clinical stage, loss of salivary and lacrimal gland secretory functions occurs, establishing the clinical phenotypes of SjD. 14 , 59 , 60 Similarly, patients with SjD express elevated Th1 and Th17 cells. 9 , 61 A recent study has found that gamma delta T cells, CD4 naive T cells, and activated memory CD4+ T cells significantly increased in patients with SjD, whereas CD4+ T cells decreased in the mouse model. 62 Another recent study showed that effector memory (em) CD4+ T cells were slightly decreased in patients who were SSA− versus healthy controls and patients who were SSA+. A significant decrease in CD4+ TEMRA frequency was found when comparing healthy controls and SSA+ patients with primary SjD. 63 Our data indicated that Cd4_T_C2, which exhibits a similar profile as CD4+ TEMRA and Th1 phenotype, tends to be elevated in both male and female SjDS mice. Cd4_T_C3 mirrored a Treg phenotype, which is lower in male SjDS mice than male B6 mice but higher in female SjDS mice than female B6 mice. In patients with SjD, Foxp3+ cell frequency was lower in mild gland lesions and high in the intermediate lesion 64 ; fewer Treg cells were detected in the peripheral blood of patients. 65 Our data suggest that the role of Treg cells in SjD may depend on both quantity and functional activity, and it could be sex dependent. A similar trend regarding sex differences was observed in the B cell population. An intriguing finding was that a small subset of Cd19+ cells exhibited transcriptional expression of CD3e and CD3d. Although this could be attributed to a technical artifact in RNA sequencing, 66 a recent study has identified biphenotypic (T/B) lymphocytes in mouse bone marrow, lymph nodes, spleen, peripheral blood, and human peripheral blood mononuclear cells (PBMCs). 67 These cells are involved in both humoral and cellular immune responses triggered by vaccination and have been implicated in autoimmune diseases such as rheumatoid arthritis and multiple sclerosis. 68 The role of biphenotypic lymphocytes in SjD remains unexplored, prompting further investigations to elucidate their function.
Consistent with the literature, CD8+ T cells were identified as a significantly lower proportion of the T cell population compared with CD4+ T cells. The frequency of CD8+ T cells can vary among patients and is influenced by several factors, including disease severity, stage, and individual immune response variability. Their function in SjD has not been fully investigated; however, recent data suggest their important role in the pathogenesis, 69 especially when considered coupled with blood granzyme K+ (GZMK+) and/or CXC chemokine receptors (CXCRs), the latter of which is upstream of DOCK2 activation. Patients were found to have CD8+ T cells with a tissue‐resident memory phenotype. In mice, infiltrating CD8+ T cells showed a significant elevation of IFN‐γ production in the salivary and lacrimal glands. Notably, depletion of CD8+ T cells fully protected mice against the pathologic manifestations of SjD, even after the onset of disease. 70 CD8+ T cells displayed significant clonal expansion in patients. 19 scRNA‐seq on PBMCs and labial gland biopsies revealed that peripheral blood GZMK+CXCR6+CD8+ T cells had an elevated proportion of clones shared with CD69+CD103−CD8+ tissue‐resident memory T cells (Trm) cells in labial glands. Blood samples of patients with SjD showed reduced cytotoxic Tc1 CD8+ T cells, whereas the frequencies of regulatory cytokine‐producing Tc2 and Tc17 CD8+ T cells were significantly elevated. 71 Analyses of T cells in the labial salivary glands of patients with SjD revealed that activated CD8+ Tc cells were the most prominent T cells in these infiltrates with Fas ligand–expressing activated CD8+ T cells accumulating around Fas‐expressing apoptotic epithelial cells. 20 The lacrimal and salivary glands of NOD mice showed proliferating CD8+ T cells that expressed an activated phenotype and produced inflammatory cytokines. 72 Meanwhile, NOD mice with anti‐CXCR3 restored saliva production but not infiltrates; however, CXCR3+CD8+ T cells were reduced, indicating that they may play a role in suppressing saliva production. These studies highlight the critical role of CD8+ T cells in various aspects of the autoimmune process, particularly in its pathogenic effects on glandular tissues. Our findings reveal that the Cd8_T_C1 cluster is significantly elevated in male and female SjD mice. This cluster is characterized by phenotypes of effector memory CD45RA re‐expressing CD8, emCD8, trmCD8, and cmCD8, all of which have been implicated in SjD and other autoimmune disorders. 70 , 73 , 74 Furthermore, we noted an increase in the Cd8_T_C2 cluster, which is similar to tissue‐resident and cmCD8 cells. These clusters exhibited enhanced expression of genes related to lymphocyte activation and the regulation of type I and type II IFN production, processes that are crucial in the pathogenesis of SjD.
Pseudotime trajectories of T cells have yet to be determined in SjD. Our data are the first to suggest Cd4 and Cd8 T cell clusters developed differently based on the sex and genetic‐susceptibility to SjD. Further studies are needed to determine the origin and developmental stages of these cells, how they differentiate into different subsets temporally in the salivary glands. The most striking aspect of the study was the identification of the Cd8_T_C4 cluster, which showed the highest expression of Dock2 compared with other clusters. This cluster exhibits several notable biological pathways, such as lymphocyte activation, hydrolase activity regulation, leukocyte activation, and type I IFN production regulation. Furthermore, its expression of DOCK2 protein functions in leukocyte migration and chemotaxis, which enable immune cells to navigate and reach the glands. DOCK2 is crucial in transducing the chemotactic signals from chemokine receptors on the leukocyte surface to the cell's internal machinery. Upon binding a chemokine to its receptor, DOCK2 becomes activated and subsequently activates Rac, a small GTPase essential for actin cytoskeleton remodeling. Activation of Rac by DOCK2 leads to actin polymerization, assembling actin monomers into filaments. This remodeling of the actin cytoskeleton is a key step in cell migration. 75 , 76 , 77 This functionality of DOCK2 is crucial for migrating immune cells to inflammation and tissue damage sites, facilitating appropriate immune responses. To leverage these critical properties of DOCK2, we treated SjD mice with its inhibitor. As demonstrated, inhibition of DOCK2 alleviated many clinical signs of SjD in the mouse model. As expected, CD8+ T and Tc1 were directly impacted by DOCK2 inhibition.
In conclusion, this comprehensive study using scRNA‐seq to profile immune cells in SjD mice has significantly advanced our understanding of the immunologic landscape of SjD. The identification and characterization of various immune cell types and their subpopulations in the salivary glands, including B cells, CD4+ and CD8+ T cells, macrophages, and NK cells, have illuminated the complex interplay of these cells in the pathogenesis of SjD. The study's revelation of sex‐specific differences in immune cell composition further underscores the impact of sex on the disease's prevalence and progression. The current focus on gene function and biologic pathways by glandular T cell subsets provided a better understanding of the biologic processes driving SjD and the functional roles of these cells. The discovery of the high expression of Dock2 in CD8+ T cells and the significant alleviation of SjD symptoms after treatment with a DOCK2 inhibitor was remarkable. This finding positions DOCK2 as a promising therapeutic target, offering new avenues for treatment strategies to address the underlying immune dysregulation in SjD.
AUTHOR CONTRIBUTIONS
All authors contributed to at least one of the following manuscript preparation roles: conceptualization AND/OR methodology, software, investigation, formal analysis, data curation, visualization, and validation AND drafting or reviewing/editing the final draft. As corresponding author, Dr Nguyen confirms that all authors have provided the final approval of the version to be published, and takes responsibility for the affirmations regarding article submission (eg, not under consideration by another journal), the integrity of the data presented, and the statements regarding compliance with institutional review board/Helsinki Declaration requirements.
Supporting information
Disclosure form
Appendix S1: Supplementary Information
ACKNOWLEDGMENTS
Data and specimens used in this article are from the Sjögren's International Collaborative Clinical Alliance, funded under contract N01‐DE‐32636 by the National Institute of Dental and Craniofacial Research, with funding support from the National Eye Institute and Office for Research in Women's Health.
Supported by the National Institute of Dental and Craniofacial Research, NIH, grants DE‐028544 and DE‐028544‐02S1 (principal investigator: Dr Nguyen).
1Yiran Shen, PhD, Alexandria Voigt, BS: University of Florida College of Veterinary Medicine, Gainesville; 2Indraneel Bhattacharyya, DDS, MSD: University of Florida College of Dentistry, Gainesville; 3Cuong Q. Nguyen, PhD: University of Florida College of Veterinary Medicine and University of Florida College of Dentistry and University of Florida Center for Orphaned Autoimmune Diseases, Gainesville.
Additional supplementary information cited in this article can be found online in the Supporting Information section (http://onlinelibrary.wiley.com/doi/10.1002/acr2.11738).
Author disclosures are available at https://onlinelibrary.wiley.com/doi/10.1002/acr2.11738.
DATA AVAILABILITY STATEMENT
Data available on request from the authors.
REFERENCES
- 1. Mariette X, Criswell LA. Primary Sjögren's syndrome. N Engl J Med 2018;378:931–939. [DOI] [PubMed] [Google Scholar]
- 2. Goules AV, Kapsogeorgou EK, Tzioufas AG. Insight into pathogenesis of Sjögren's syndrome: dissection on autoimmune infiltrates and epithelial cells. Clin Immunol 2017;182:30–40. [DOI] [PubMed] [Google Scholar]
- 3. Teos LY, Alevizos I. Genetics of Sjögren's syndrome. Clin Immunol 2017;182:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Witas R, Gupta S, Nguyen CQ. Contributions of major cell populations to Sjögren's syndrome. J Clin Med 2020;9:3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen W, Yang F, Xu G, et al. Follicular helper T cells and follicular regulatory T cells in the immunopathology of primary Sjögren's syndrome. J Leukoc Biol 2021;109:437–447. [DOI] [PubMed] [Google Scholar]
- 6. Tuzlak S, Dejean AS, Iannacone M, et al. Repositioning TH cell polarization from single cytokines to complex help. Nat Immunol 2021;22:1210–1217. [DOI] [PubMed] [Google Scholar]
- 7. Gupta S, Witas R, Voigt A, et al. Single‐cell sequencing of T cell receptors: a perspective on the technological development and translational application. Adv Exp Med Biol 2020;1255:29–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schnell A, Littman DR, Kuchroo VK. TH17 cell heterogeneity and its role in tissue inflammation. Nat Immunol 2023;24:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Del Papa N, Minniti A, Lorini M, et al. The role of interferons in the pathogenesis of Sjögren's syndrome and future therapeutic perspectives. Biomolecules 2021;11:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Verstappen GM, Pringle S, Bootsma H, et al. Epithelial‐immune cell interplay in primary Sjögren syndrome salivary gland pathogenesis. Nat Rev Rheumatol 2021;17:333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raphael I, Nalawade S, Eagar TN, et al. T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 2015;74:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruterbusch M, Pruner KB, Shehata L, et al. In vivo CD4+ T cell differentiation and function: revisiting the Th1/Th2 paradigm. Annu Rev Immunol 2020;38:705–725. [DOI] [PubMed] [Google Scholar]
- 13. Nguyen CQ, Gao J, Kim H, et al. IL‐4‐STAT6 signal transduction‐dependent induction of the clinical phase of Sjögren's syndrome‐like disease of the nonobese diabetic mouse. J Immunol 2007;179:382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brayer JB, Cha S, Nagashima H, et al. IL‐4‐dependent effector phase in autoimmune exocrinopathy as defined by the NOD.IL‐4‐gene knockout mouse model of Sjögren's syndrome. Scand J Immunol 2001;54:133–140. [DOI] [PubMed] [Google Scholar]
- 15. Verstappen GM, Corneth OBJ, Bootsma H, et al. Th17 cells in primary Sjögren's syndrome: pathogenicity and plasticity. J Autoimmun 2018;87:16–25. [DOI] [PubMed] [Google Scholar]
- 16. Pfeifle R, Rothe T, Ipseiz N, et al. Regulation of autoantibody activity by the IL‐23‐TH17 axis determines the onset of autoimmune disease. Nat Immunol 2017;18:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nguyen CQ, Yin H, Lee BH, et al. Pathogenic effect of interleukin‐17A in induction of Sjögren's syndrome‐like disease using adenovirus‐mediated gene transfer. Arthritis Res Ther 2010;12:R220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen CQ, Yin H, Lee BH, et al. IL17: potential therapeutic target in Sjögren's syndrome using adenovirus‐mediated gene transfer. Lab Invest 2011;91:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu T, Zhu HX, You X, et al. Single‐cell profiling reveals pathogenic role and differentiation trajectory of granzyme K+CD8+ T cells in primary Sjögren's syndrome. JCI Insight 2023;8:e167490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaneko N, Chen H, Perugino CA, et al. Cytotoxic CD8+ T cells may be drivers of tissue destruction in Sjögren's syndrome. Sci Rep 2022;12:15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stevenson C, de la Rosa G, Anderson CS, et al. Essential role of Elmo1 in Dock2‐dependent lymphocyte migration. J Immunol 2014;192:6062–6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanui T, Inayoshi A, Noda M, et al. DOCK2 is essential for antigen‐induced translocation of TCR and lipid rafts, but not PKC‐theta and LFA‐1, in T cells. Immunity 2003;19:119–129. [DOI] [PubMed] [Google Scholar]
- 23. Peck AB, Nguyen CQ, Ambrus JL. Upregulated chemokine and Rho‐GTPase genes define immune cell emigration into salivary glands of Sjögren's syndrome‐susceptible C57BL/6.NOD‐Aec1Aec2 mice. Int J Mol Sci 2021;22:7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang S, Wang R, Xu N, et al. SULT2B1‐CS‐DOCK2 axis regulates effector T‐cell exhaustion in HCC microenvironment. Hepatology 2023;78:1064–1078. [DOI] [PubMed] [Google Scholar]
- 25. Randall KL, Flesch IEA, Mei Y, et al. DOCK2‐deficiency causes defects in anti‐viral T cell responses and impaired control of herpes simplex virus infection. J Infect Dis Published online February 15, 2024. doi: 10.1093/infdis/jiae077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mahajan VS, Demissie E, Alsufyani F, et al. DOCK2 sets the threshold for entry into the virtual memory CD8+ T cell compartment by negatively regulating tonic TCR triggering. J Immunol 2020;204:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brayer J, Lowry J, Cha S, et al. Alleles from chromosomes 1 and 3 of NOD mice combine to influence Sjögren's syndrome‐like autoimmune exocrinopathy. J Rheumatol 2000;27:1896–1904. [PubMed] [Google Scholar]
- 28. Cha S, Nagashima H, Peck AB, et al. IDD3 and IDD5 alleles from NOD mice mediate Sjögren's syndrome‐like autoimmunity. Adv Exp Med Biol 2002;506:1035–1039. [DOI] [PubMed] [Google Scholar]
- 29. Donate A, Voigt A, Nguyen CQ. The value of animal models to study immunopathology of primary human Sjögren's syndrome symptoms. Expert Rev Clin Immunol 2014;10:469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu T, Jiang Y, Shi W, et al. Endoplasmic reticulum stress: a novel targeted approach to repair bone defects by regulating osteogenesis and angiogenesis. J Transl Med 2023;21:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Honjo K, Won WJ, King RG, et al. Fc receptor‐like 6 (FCRL6) discloses progenitor B cell heterogeneity that correlates with pre‐BCR dependent and independent pathways of natural antibody selection. Front Immunol 2020;11:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gururajan M, Simmons A, Dasu T, et al. Early growth response genes regulate B cell development, proliferation, and immune response. J Immunol 2008;181:4590–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Liu J, Burrows PD, et al. B cell development and maturation. Adv Exp Med Biol 2020;1254:1–22. [DOI] [PubMed] [Google Scholar]
- 34. Cook DA, Kannarkat GT, Cintron AF, et al. LRRK2 levels in immune cells are increased in Parkinson's disease. NPJ Parkinsons Dis 2017;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune‐mediated disease. Immunity 2012;36:542–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 2011;29:621–663. [DOI] [PubMed] [Google Scholar]
- 37. Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol 2011;12:21–27. [DOI] [PubMed] [Google Scholar]
- 38. Cedeno‐Laurent F, Dimitroff CJ. Galectin‐1 research in T cell immunity: past, present and future. Clin Immunol 2012;142:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suzuki I, Fink PJ. The dual functions of Fas ligand in the regulation of peripheral CD8+ and CD4+ T cells. Proc Natl Acad Sci USA 2000;97:1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 2012;12:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hinrichs AC, Blokland SLM, Kruize AA, et al. CCL5 release by CCR9+ CD8 T cells: a potential contributor to immunopathology of primary Sjögren's syndrome. Front Immunol 2022;13:887972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Volpin V, Michels T, Sorrentino A, et al. CAMK1D triggers immune resistance of human tumor cells refractory to anti‐PD‐L1 treatment. Cancer Immunol Res 2020;8:1163–1179. [DOI] [PubMed] [Google Scholar]
- 43. Platanias LC. Mechanisms of type‐I‐ and type‐II‐interferon‐mediated signalling. Nat Rev Immunol 2005;5:375–386. [DOI] [PubMed] [Google Scholar]
- 44. Prasad M, Brzostek J, Gautam N, et al. Themis regulates metabolic signaling and effector functions in CD4+ T cells by controlling NFAT nuclear translocation. Cell Mol Immunol 2021;18:2249–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Filby A, Seddon B, Kleczkowska J, et al. Fyn regulates the duration of TCR engagement needed for commitment to effector function. J Immunol 2007;179:4635–4644. [DOI] [PubMed] [Google Scholar]
- 46. Klein S, Golani G, Lolicato F, et al. IFITM3 blocks influenza virus entry by sorting lipids and stabilizing hemifusion. Cell Host Microbe 2023;31:616–633.e20. [DOI] [PubMed] [Google Scholar]
- 47. Stroncek DF, Basil C, Nagorsen D, et al. Delayed polarization of mononuclear phagocyte transcriptional program by type I interferon isoforms. J Transl Med 2005;3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science 1988;240:622–630. [DOI] [PubMed] [Google Scholar]
- 49. De Leon‐Oliva D, Garcia‐Montero C, Fraile‐Martinez O, et al. AIF1: function and connection with inflammatory diseases. Biology (Basel) 2023;12:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Turnbull IR, Gilfillan S, Cella M, et al. Cutting edge: TREM‐2 attenuates macrophage activation. J Immunol 2006;177:3520–3524. [DOI] [PubMed] [Google Scholar]
- 51. Caligiuri MA. Human natural killer cells. Blood 2008;112:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cursons J, Souza‐Fonseca‐Guimaraes F, Foroutan M, et al. A gene signature predicting natural killer cell infiltration and improved survival in melanoma patients. Cancer Immunol Res 2019;7:1162–1174. [DOI] [PubMed] [Google Scholar]
- 53. Zhang L, Yu X, Zheng L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268–272. [DOI] [PubMed] [Google Scholar]
- 54. Leruste A, Tosello J, Ramos RN, et al. Clonally expanded T cells reveal immunogenicity of rhabdoid tumors. Cancer Cell 2019;36:597–612.e8. [DOI] [PubMed] [Google Scholar]
- 55. Li H, van der Leun AM, Yofe I, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 2019;176:775–789.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cano‐Gamez E, Soskic B, Roumeliotis TI, et al. Single‐cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. Nat Commun 2020;11:1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ren X, Wen W, Fan X, et al. COVID‐19 immune features revealed by a large‐scale single‐cell transcriptome atlas. Cell 2021;184:1895–1913.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci 2001;114:4143–4151. [DOI] [PubMed] [Google Scholar]
- 59. Kong L, Robinson CP, Peck AB, et al. Inappropriate apoptosis of salivary and lacrimal gland epithelium of immunodeficient NOD‐scid mice. Clin Exp Rheumatol 1998;16:675–681. [PubMed] [Google Scholar]
- 60. Nguyen KH, Brayer J, Cha S, et al. Evidence for antimuscarinic acetylcholine receptor antibody‐mediated secretory dysfunction in NOD mice. Arthritis Rheum 2000;43:2297–2306. [DOI] [PubMed] [Google Scholar]
- 61. Nguyen CQ, Hu MH, Li Y, et al. Salivary gland tissue expression of interleukin‐23 and interleukin‐17 in Sjögren's syndrome: findings in humans and mice. Arthritis Rheum 2008;58:734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gong H, Qiu X, Li P, et al. Immune infiltration analysis reveals immune cell signatures in salivary gland tissue of primary Sjögren's syndrome. Front Med (Lausanne) 2023;10:1033232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sarkar I, Davies R, Aarebrot AK, et al. Aberrant signaling of immune cells in Sjögren's syndrome patient subgroups upon interferon stimulation. Front Immunol 2022;13:854183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Christodoulou MI, Kapsogeorgou EK, Moutsopoulos NM, et al. Foxp3+ T‐regulatory cells in Sjogren's syndrome: correlation with the grade of the autoimmune lesion and certain adverse prognostic factors. Am J Pathol 2008;173:1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu MF, Lin LH, Weng CT, et al. Decreased CD4+CD25+bright T cells in peripheral blood of patients with primary Sjogren's syndrome. Lupus 2008;17:34–39. [DOI] [PubMed] [Google Scholar]
- 66. Xin H, Lian Q, Jiang Y, et al. GMM‐Demux: sample demultiplexing, multiplet detection, experiment planning, and novel cell‐type verification in single cell sequencing. Genome Biol 2020;21:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Y, Guo C, Zhou Y, et al. A biphenotypic lymphocyte subset displays both T‐ and B‐cell functionalities. Commun Biol 2024;7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen Q, Yuan S, Sun H, et al. CD3+CD20+ T cells and their roles in human diseases. Hum Immunol 2019;80:191–194. [DOI] [PubMed] [Google Scholar]
- 69. Zhou H, Yang J, Tian J, et al. CD8+ T lymphocytes: crucial players in Sjögren's syndrome. Front Immunol 2020;11:602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gao CY, Yao Y, Li L, et al. Tissue‐resident memory CD8+ T cells acting as mediators of salivary gland damage in a murine model of Sjögren's syndrome. Arthritis Rheumatol 2019;71:121–132. [DOI] [PubMed] [Google Scholar]
- 71. Kudryavtsev I, Benevolenskaya S, Serebriakova M, et al. Circulating CD8+ T cell subsets in primary Sjögren's syndrome. Biomedicines 2023;11:2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Barr JY, Wang X, Meyerholz DK, et al. CD8 T cells contribute to lacrimal gland pathology in the nonobese diabetic mouse model of Sjögren syndrome. Immunol Cell Biol 2017;95:684–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Boldison J, Chu CJ, Copland DA, et al. Tissue‐resident exhausted effector memory CD8+ T cells accumulate in the retina during chronic experimental autoimmune uveoretinitis. J Immunol 2014;192:4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Konjar Š, Ficht X, Iannacone M, et al. Heterogeneity of tissue resident memory T cells. Immunol Lett 2022;245:1–7. [DOI] [PubMed] [Google Scholar]
- 75. Nombela‐Arrieta C, Lacalle RA, Montoya MC, et al. Differential requirements for DOCK2 and phosphoinositide‐3‐kinase gamma during T and B lymphocyte homing. Immunity 2004;21:429–441. [DOI] [PubMed] [Google Scholar]
- 76. Nombela‐Arrieta C, Mempel TR, Soriano SF, et al. A central role for DOCK2 during interstitial lymphocyte motility and sphingosine‐1‐phosphate‐mediated egress. J Exp Med 2007;204:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jiang H, Pan F, Erickson LM, et al. Deletion of DOCK2, a regulator of the actin cytoskeleton in lymphocytes, suppresses cardiac allograft rejection. J Exp Med 2005;202:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disclosure form
Appendix S1: Supplementary Information
Data Availability Statement
Data available on request from the authors.


