Abstract
Influenza virus matrix protein (M1), a critical protein required for virus assembly and budding, is presumed to interact with viral glycoproteins on the outer side and viral ribonucleoprotein on the inner side. However, because of the inherent membrane-binding ability of M1 protein, it has been difficult to demonstrate the specific interaction of M1 protein with hemagglutinin (HA) or neuraminidase (NA), the influenza virus envelope glycoproteins. Using Triton X-100 (TX-100) detergent treatment of membrane fractions and floatation in sucrose gradients, we observed that the membrane-bound M1 protein expressed alone or coexpressed with heterologous Sendai virus F was totally TX-100 soluble but the membrane-bound M1 protein expressed in the presence of HA and NA was predominantly detergent resistant and floated to the top of the density gradient. Furthermore, both the cytoplasmic tail and the transmembrane domain of HA facilitated binding of M1 to detergent-resistant membranes. Analysis of the membrane association of M1 in the early and late phases of the influenza virus infectious cycle revealed that the interaction of M1 with mature glycoproteins which associated with the detergent-resistant lipid rafts was responsible for the detergent resistance of membrane-bound M1. Immunofluorescence analysis by confocal microscopy also demonstrated that, in influenza virus-infected cells, a fraction of M1 protein colocalized with HA and associated with the HA in transit to the plasma membrane via the exocytic pathway. Similar results for colocalization were obtained when M1 and HA were coexpressed and HA transport was blocked by monensin treatment. These studies indicate that both HA and NA interact with influenza virus M1 and that HA associates with M1 via its cytoplasmic tail and transmembrane domain.
Influenza viruses, enveloped RNA viruses containing single-stranded, segmented RNA of negative polarity, assemble and bud from the plasma membrane of virus-infected cells into the outside environment. Complete virions are usually not observed inside the cell in the productive infectious cycle. Furthermore, in polarized epithelial cells, influenza viruses bud asymmetrically, i.e., predominantly from the apical plasma membrane (30). For virus budding to occur, two processes are obligatory (27). Firstly, all viral structural components, namely, the matrix protein (M1), the viral nucleocapsid (viral ribonucleoprotein [vRNP]) containing vRNA, nucleoprotein (NP), polymerase proteins (PB1, PB2, and PA), and NS2 (NEP) as well as the viral envelope containing the host lipids and three transmembrane proteins (hemagglutinin [HA], neuraminidase [NA], and M2) must be transported and targeted either individually or as complex subviral components to the assembly site at the plasma membrane. Secondly, these viral proteins and/or subviral components must interact with each other to initiate the budding processes leading to morphogenesis of virus particles and release of virions.
Influenza virus M1, the most abundant protein in the virus particle, plays a critical role in the assembly and budding processes of virions (3, 22). Although viral glycoproteins may provide critical determinants in the selection of the assembly site of the virion in virus-infected cells, neither HA nor NA is absolutely required for virus assembly, budding, and release since mature virus particles lacking either HA or NA can be formed and released from the infected cells (21, 28). On the other hand, M1 protein is critically important for viral morphogenesis and budding, as particle formation is drastically reduced in abortively infected cells exhibiting reduced M1 synthesis (22) and in cells infected at the nonpermissive temperature with temperature-sensitive (ts) virus having a defect in M1 protein (20, 29, 40, 41). Because of the presumed juxtaposition of the M1 protein between the viral envelope and the nucleocapsid (vRNP), M1 is proposed to interact with the cytoplasmic tail of transmembrane viral proteins on the outer side and the viral nucleocapsid (vRNP) on the inner side. These interactions are believed to trigger the budding process leading to the formation and release of virus particles.
Although vRNP-M1 complexes have been demonstrated both for virus-infected cells and for mature virus particles after nonionic detergent treatments (42, 44, 45), interactions between M1 and envelope glycoproteins (HA and NA) have been difficult to demonstrate. Experiments to demonstrate the specific interaction of M1 with HA and NA have been inconclusive and yielded conflicting results (5, 18, 44). These studies have used floatation gradient analysis in which membrane-bound proteins float to a lighter density in a sucrose gradient (i.e., top fractions) whereas free proteins not bound to membrane do not float and remain at the bottom of the gradient containing the denser sucrose solution. Two reports using coexpression of M1 with HA, NA, and M2 in various combinations using the vaccinia virus T7 transfection system did not find any significant increase in the membrane association of M1 compared to that of M1 expressed alone (18, 44). However, one report using a recombinant vaccinia virus (RVV) expression system showed a significant increase in membrane binding of M1 when coexpressed with HA and NA compared to that of M1 expressed alone (5). The major problem encountered in all of these experiments was the inherent membrane-binding ability of M1 expressed alone. M1 is a hydrophobic protein which binds to lipids (9), and in addition, there was a great deal of variation in the membrane-binding ability of M1 expressed alone in different studies, e.g., 15% (18), 45 to 60% (44), and 20 to 30% (5). To avoid this problem, in this report we have developed an assay using nonionic detergent (Triton X-100 [TX-100]) treatment to distinguish the membrane-bound M1 in the presence of homologous influenza virus glycoproteins from the membrane-bound M1 alone. Using this assay, we demonstrate that the membrane-bound M1 became detergent resistant in influenza virus-infected cells and in cells coexpressing M1 with HA and NA but not in cells expressing M1 alone or coexpressing M1 with a heterologous protein such as Sendai virus F protein. Furthermore, we show that both the transmembrane domain and the cytoplasmic tail of HA help the membrane-bound M1 protein to acquire its detergent-resistant state.
MATERIALS AND METHODS
Cells, virus, and antibodies.
MDBK, MDCK, and HeLa cells were obtained from the American Type Culture Collection (Manassas, Va.) and maintained in minimal essential medium (MEM) and Dulbecco's modified essential medium (DMEM; GIBCO-BRL, Rockville, Md.) supplemented with 10% fetal bovine serum (FBS), 250 U of penicillin/ml, and 250 μg of streptomycin/ml. Influenza virus A/WSN/33 (H1N1) was plaque purified and grown in MDCK cells. Virus stocks were made from individual plaques as previously described (27) and had titers ranging from 5 × 107 to 5 × 108 PFU/ml. Polyclonal anti-WSN antibodies were made in rabbits by using purified virus. Monoclonal anti-HA antibodies were obtained from W. Gerhard (Wistar Institute, Philadelphia, Pa.), and rabbit polyclonal anti-M1 antibodies were obtained from M. Krystal (Bristol-Myers Squibb, Wallingford, Conn.). Polyclonal antibodies against whole Sendai virus and Sendai virus F were obtained from J. Seto (California State University, Los Angeles). Anti-rabbit immunoglobulin G (IgG) conjugated with fluorescein isothiocyanate (FITC) and anti-mouse IgG conjugated with tetramethyl rhodamine isothiocyanate (TRITC) were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Construction of RVVs.
cDNAs of WSN influenza virus HA, Sendai virus Z strain F, and chimeric constructs were inserted into the multiple cloning site of the vaccinia virus expression vector pSC11, which contains the 7.5 promoter sequence upstream of the multiple cloning site and the thymidine kinase gene. Chimeric constructions were made by swapping domains between WSN HA and Sendai virus F (see Fig. 4A). Chimeric constructs were designated FHH, FFH, and HHF indicating the ectodomain, transmembrane domain, and cytoplasmic tail, respectively, of either Sendai virus F protein (F) or influenza virus HA protein (H). Each construct was sequenced to ensure that PCR mutations were not made and assayed for protein expression and transport before being used in coexpression experiments. RVVs were obtained as described previously (31). The vaccinia virus recombinant VP273-expressing M1 (RVVM1) protein was obtained from E. Paoletti (Virogenetics, Troy, N.Y.). All vaccinia viruses were propagated in HeLa cells, and plaque titers in CV-1 cells were determined as previously described (31). For expression of M1 alone, HeLa cells were infected with RVVM1 at a multiplicity of infection (MOI) of 10. For coexpression of M1 with HA, NA, or chimeric constructs using RVVs, a ratio of 2:1 (i.e., an MOI of RVVM1 of 8 and an MOI of RVVHA or RVVNA of 4) was used.
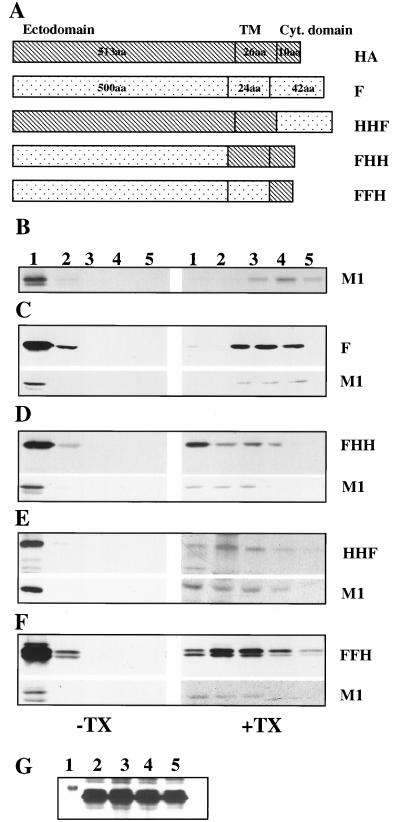
FIG. 4.
Detergent resistance of membrane-associated M1 when coexpressed with chimeric constructs of influenza virus HA and Sendai virus F proteins. (A) Schematic presentation of chimeric constructs between Sendai virus F and influenza virus HA. aa, amino acids; Cyt., cytoplasmic. (B to F) Detergent-resistant membrane fractions of M1 in the presence of heterologous or chimeric proteins. HeLa cells (5 × 106) were infected with RVV expressing M1 alone (B) or M1 with Sendai virus F (C), M1 with FHH (D), M1 with HHF (E), or M1 with FFH (F) as described in Materials and Methods. At 4 hpi, cells were pulse-labeled with 400 μCi of 35S Easy Tag for 30 min and chased for 90 min. Pure membrane fractions were isolated from 4K supernatants with floatation gradients, treated without (−) or with (+) TX-100 (0.05%), and analyzed again with floatation gradients. Fractions were collected, immunoprecipitated, and analyzed by SDS-PAGE. (G) Expression of F and chimeric proteins. HeLa cells (5 × 106) were infected with RVV expressing F, FHH, HHF, and FFH. At 4 hpi, cells were pulse-labeled for 30 min and chased for 90 min. The whole-cell extract was immunoprecipitated and analyzed by SDS-PAGE. Note that similar levels of F and chimeras were expressed. Lanes 1, marker; lanes 2, FHH; lanes 3, FFH; lanes 4, HHF; lanes 5, F.
Radiolabeling.
For influenza viruses, MDBK cells (5 × 106) were infected with WSN virus at an MOI of 10. For RVVs, 5 × 106 HeLa cells were infected with RVVs at an MOI of 10 or 12 as stated above. The infected cells were then incubated at 37°C in DMEM plus 2.5% FBS. At the indicated times (hours postinfection [hpi]), cells were starved with DMEM deficient in methionine and cysteine for 30 min and pulse-labeled with 35S Easy Tag Express Protein labeling mix (NEN Life Science Products Inc., Boston, Mass.). The labeling medium was then replaced with the chase medium (DMEM plus 2.5% FBS supplemented with 10 mM unlabeled cysteine and methionine) and chased for indicated times. The pulse and chase times and the amount of 35S-amino acids varied with different experiments and are stated in the figure legends.
Subcellular fractionation.
Preparative fractionation of influenza virus-infected MDBK cells or RVV-infected HeLa cells was performed as follows. Cell monolayers were washed twice in ice-cold phosphate-buffered saline containing Ca2+ and Mg2+, scraped from dishes, and pelleted by centrifugation. The cell pellet was resuspended in 0.5 ml of hypotonic lysis buffer (10 mM Tris HCl [pH 7.5], 10 mM KCl, 5 mM MgCl2) and incubated on ice for 30 min before disruption of cells by repeated passages (25 times) through a 26-gauge hypodermic needle. Unbroken cells and nuclei were removed by centrifugation at 1,000 × g for 5 min (SW50 rotor at 4,000 rpm) at 4°C, and the resulting postnuclear supernatant (4K supernatant) was then subjected to floatation analysis as described below. For TX-100 (Boehringer, Mannheim, Germany) detergent treatment, 1.0% TX-100 (freshly prepared) was added to the pure membrane fraction to a final concentration as indicated, gently mixed, and kept on ice for 15 min before floatation analysis.
Floatation analysis.
Floatation analysis was performed as described by Sanderson et al. (31) with the following modifications. Aliquots of the 4K postnuclear supernatants (0.4 ml) were dispersed into 2 ml of 75% (wt/wt) sucrose in low-salt buffer (LSB) containing 50 mM Tris-HCl (pH 7.5), 25 mM KCl, and 5 mM MgCl2 and layered on 0.5 ml of 80% (wt/wt) sucrose, overlaid with 2 ml of 55% (wt/wt) sucrose in LSB and approximately 0.6 ml of 5% (wt/wt) sucrose in LSB. Gradients were then centrifuged for 18 h at 38,000 rpm using an SW55 Ti rotor at 4°C, and a 500-μl fraction containing the visible membrane fraction (called the pure membrane fraction) was collected from the top. Four hundred microliters of this pure membrane fraction was treated with or without TX-100 on ice for 15 min and used for a second floatation gradient. Five 1-ml fractions were collected from the top by using a Hacki-Buchler Auto Densiflow II gradient remover (Buchler Instruments, Lenexa, Kans.) and used for immunoprecipitation. Therefore, in all gradients the top fraction is no. 1 and the bottom fraction is no. 5. In these floatation gradients, fractions 1 and 2 contain the membrane fraction and fractions 3, 4, and 5 contain the nonmembrane soluble proteins. To avoid any variation in detergent and membrane concentration, the same number of cells were used in each experiment, the protein concentration in the pure membrane fraction was determined, and the same amounts of membrane fraction were used for detergent treatment and flotation gradient analysis.
Immunoprecipitation.
Prior to immunoprecipitation, all fractions were diluted with 3 ml of LSB before addition of 1 ml of 5× concentrated radioimmunoprecipitation assay (RIPA) buffer (1× RIPA buffer contains 50 mM Tris [pH 7.5], 150 mM NaCl, 1.0% TX-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1.0 mM phenylmethylsulfonyl fluoride [Sigma], and 2% aprotinin [Sigma]). For immunoprecipitation, samples were shaken at 4°C for 2 h before the addition of antibodies. Each fraction was immunoprecipitated with polyclonal anti-WSN or polyclonal anti-Sendai virus rabbit antibodies (AS no. 74). Subsequently, 7 mg of protein A-Sepharose (Pharmacia, Uppsala, Sweden) was added to each sample and the mixture was incubated for 1.5 h at 4°C. Immunoprecipitates bound to Sepharose beads were pelleted by centrifugation and washed three times in RIPA buffer containing 5 mg of bovine serum albumin (BSA) per ml, followed by another wash with RIPA buffer. Immunoprecipitates were then dissolved in SDS sample buffer (50 mM Tris-HCl [pH 6.8], 5% 2-β-mercaptoethanol, 2% SDS, 10% [wt/vol] glycerol, and 0.1% [wt/vol] bromophenol blue) at 95°C for 5 min and analyzed by SDS (0.1%)-polyacrylamide (10%) gel electrophoresis (SDS-PAGE) and autoradiography. Quantifications were done by densitometric scanning of autoradiographs with an LKB 2222-020 Ultrascan-XL laser densitometer (Pharmacia-LKB) using QuanTN software (Molecular Dynamics, Sunnyvale, Calif.). Data from three or more independent experiments were used for quantification analysis.
Western blot analysis.
MDBK cells (5 × 106) were infected at an MOI of 10 with influenza viruses for 1 h at room temperature. The infected cells were then incubated at 37°C in DMEM plus 2.5% FBS for 7.0 h. The 4K supernatant was prepared and fractionated as described above. Each 1-ml fraction was diluted in 3.0 ml of LSB. Proteins from each fraction were precipitated with tricholoroacetic acid followed by washing of the pellet with 100% methanol. Proteins were dissolved in 2× sample buffer and separated under reducing conditions by SDS-PAGE as described above in a minigel apparatus (Bio-Rad Laboratories, Hercules, Calif.) and used for overnight electrotransfer (250 mA) onto nitrocellulose membranes (Bio-Rad Laboratories) in blotting buffer (25 mM Tris-HCl [pH 7.2], 190 mM glycine, 20% methanol). The membranes were then incubated for 30 min in Western blocking buffer (WBB) containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% Tween 20, and 3% (vol/vol) nonfat milk. They were subsequently incubated with either anti-M1 or anti-HA monoclonal antibodies for 1 h and washed three times with WBB. Finally, a WBB solution containing secondary alkaline phosphatase-conjugated goat anti-mouse antibodies (Cappel Laboratories, Durham, N.C.) was applied to the membranes for 1 h. The membranes were developed with 5-bromo-4-chloro-3-indolylphosphate toluidinium nitroblue tetrazolium phosphatase substrate (Kirkegaard & Perry Laboratories Inc., Gaithersburg, Md.) diluted 1:2 in 100 mM Tris-HCl (pH 9.5).
Immunofluorescence by confocal microscopy.
MDBK or HeLa cells (4 × 105) were grown overnight in tissue culture chamber slides (Nunc, Naperville, Ill.) and synchronously infected with WSN virus or RVV, respectively, for 1.0 h at 4°C. Following adsorption, 1.5 ml of prewarmed (37°C) DMEM containing 2.5% FBS was added to the cell monolayers for indicated times. For monensin (Sigma) treatment of the virus-infected cells, prewarmed DMEM containing 2.5% FBS and monensin (10 μM final concentration) was added at 2 hpi and incubated for a further 5 h at 37°C. Infected MDBK cells were then fixed with 100% acetone at −20°C for 20 min. RVV-infected HeLa cells were fixed with 4% formaldehyde for 20 min at room temperature and permeabilized with 1% NP-40 for 30 min at room temperature. To block the nonspecific antibody binding, the cells were incubated in 3% BSA (Sigma) for 30 min. Primary antibodies, anti-M1 rabbit polyclonal antibodies, and anti-HA mouse monoclonal antibodies were diluted in 3% BSA and incubated with cells for 1 h at room temperature as described before (1). Cells were then stained with fluorescein (FITC)-tagged anti-rabbit IgG and rhodamine (TRITC)-tagged anti-mouse IgG (Sigma). Cells were mounted in Vectashield (Vector Laboratories, Burlingame, Calif.). Specimens were imaged on a Leica TCS-SP inverted confocal microscope (Leica Microsystems GmbH, Heidelberg, Germany) equipped with an argon laser for 488-nm blue excitation for FITC and a krypton laser for 568-nm red excitation for TRITC. The thickness of each digital section obtained by the microscope was 0.6 μm, and at least 30 sections throughout the cells were analyzed. Image analysis was performed using the Leica TCS-NT software provided with the microscope. Fluorescent images were superimposed digitally to allow fine comparison. Colocalization by superimposition of green (FITC) and red (TRITC) signals in a single pixel produces yellow or orange, while separated signals remain green and red.
RESULTS
Membrane association of the influenza virus M1 protein in WSN virus-infected cells and in RVV M1-infected cells.
Membrane association of M1 protein in influenza virus-infected cells was investigated by subcellular fractionation and floatation gradient analysis using a modified procedure described previously (31). To determine the membrane association of the M1 protein immediately after synthesis and after chase, WSN virus-infected MDBK cells (MOI of 10) were pulse-labeled at 7 hpi with 300 μCi of 35S Easy Tag for 15 min and chased for 1 h. The 4K supernatants from cells immediately after the pulse and after chase were analyzed by floatation gradient centrifugation, and fractions were collected and immunoprecipitated using rabbit anti-WSN polyclonal antibodies. At 7 hpi immediately after the pulse, approximately 60% of M1 protein in the 4K supernatant was membrane associated, whereas after 1 h of chase the fraction of membrane-associated M1 increased to 75% (Fig. 1A). Similar data on membrane association of M1 immediately after pulse and an increased level of membrane association after chase in WSN virus-infected cells were previously observed by others (5, 13, 44). We have consistently observed that, immediately after pulse, relatively large amounts of immature HA remained in fractions at the bottom half of the gradient. This reflected the association of immature HA with the endoplasmic reticulum membranes which are denser than the plasma or trans-Golgi membranes. We have also observed that less HA was present in MDBK cells after chase (compare HA in Fig. 1A before and after chase). This reduction in HA was due to efficient cleavage of HA into HA1 and HA2 of WSN virus and release of virus particles from MDBK cells. Both virus particles and NP were present in the medium at this stage of infection (data not shown). A significant fraction of NP was membrane associated, and the percentage of membrane association of NP increased with chase (Fig. 1A, before and after chase). Membrane association of NP is likely due to the membrane association of vRNP (or M1-vRNP complex) during the assembly process. We also obtained similar data on the membrane association of M1 in WSN virus-infected MDCK and HeLa cells (data not shown). Using Western blot analysis, we have also observed about 60% membrane-bound M1 and 80% membrane-bound HA in 4K supernatant of WSN-infected MDBK cells at 7.0 hpi (data not shown).
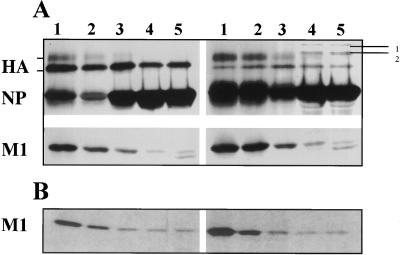
FIG. 1.
Membrane association of M1 protein in WSN virus-infected cells. (A) Pulse-chase analysis of WSN virus-infected MDBK cells. At 7 hpi, WSN virus-infected MDBK cells (5 × 106) were pulse-labeled with 300 μCi of 35S Easy Tag for 15 min and chased for 1 h. Labeled cells were fractionated, and the 4K supernatant was analyzed with floatation gradients (31). Each fraction was immunoprecipitated with an anti-WSN polyclonal antibody and analyzed by SDS–10% PAGE. Fractions are numbered 1 (top) to 5 (bottom). Lines marked 1 and 2 (upper right-hand corner) denote nonspecific cellular proteins. Both panels are from the same gel and same autoradiograph. (B) Pulse-chase analysis of RVVM1-infected HeLa cells. HeLa cells were infected with RVVM1 at an MOI of 10. At 6 hpi, cells were pulse-labeled with 400 μCi of 35S Easy Tag and chased as described above, the 4K supernatant was analyzed with floatation gradients, and fractions were immunoprecipitated and analyzed by SDS-PAGE.
To examine the membrane association of M1 in RVVM1-infected cells, HeLa cells were infected with RVVM1 at an MOI of 10, and at 6 hpi, cells were pulse-labeled with 35S Easy Tag (400 μCi) for 15 min or chased for 60 min. The 4K supernatant was prepared and analyzed with floatation gradients. Fractions were immunoprecipitated and analyzed by PAGE. Results show that, when expressed alone, 60% of M1 in the 4K supernatant was membrane associated immediately after pulse and the membrane-associated fraction of M1 increased to 70% after chase (Fig. 1B, fractions 1 and 2).
All expression studies using RVVs were done in HeLa cells because both MDBK and MDCK cells are poorly infected by vaccinia viruses. Furthermore, although HeLa cells are not highly permissive for influenza virus infection, the transport, glycosylation, and processing of glycoproteins as well as membrane interactions of M1 and glycoproteins including particle formation of WSN virus occurred similarly to the way in which they did in MDBK or MDCK cells (12).
Membrane association of M1 protein after TX-100 detergent treatment.
The above results (Fig. 1B) show that M1 expressed in the absence of other viral proteins became membrane associated, and as indicated earlier, this was due to the inherent membrane-binding ability of M1 expressed alone since the M1 protein possesses hydrophobic and amphipathic regions (10, 18) which bind to lipids and membranes. Therefore, to distinguish the membrane-bound M1 in the presence of influenza virus glycoproteins from the membrane-bound M1 alone and to selectively enrich the membrane-bound fraction of M1 associated with HA and NA, we used TX-100 detergent treatment. We reasoned that, during exocytic transport to the plasma membrane, mature HA and NA specifically associate in the trans-Golgi network with lipid rafts enriched in glycosphingolipids and cholesterol (2, 19, 33, 37, 38), which are relatively resistant to neutral detergents like TX-100. Furthermore, since lipid rafts are formed in both polarized and nonpolarized cells (33, 36, 38, 43), we also reasoned that, if M1 associates with mature HA and NA, it will become resistant to TX-100 due to either direct or indirect association of M1 with such lipid rafts. Therefore, to determine the minimum TX-100 concentration which would render the membrane-associated M1 completely soluble, we expressed M1 alone using the RVV expression system in HeLa cells and prepared the 4K supernatant and the pure membrane fraction was isolated from the top (Fig. 1, fraction 1) of the floatation gradient. The pure membrane fraction was then treated with different concentrations of TX-100 and subjected to a second floatation gradient analysis (Fig. 2). Without any detergent treatment, the membrane-bound M1 floated to the top of the gradient as expected. After treatment of the membrane fraction with different concentrations of TX-100, it was found that an 0.05% or higher concentration of TX-100 completely solubilized the membrane-bound M1. Consequently, M1 could not be detected in the membrane fractions (Fig. 2, 0.05%, fractions 1 and 2) and was present only as soluble protein in the bottom fractions (Fig. 2, fractions 3 to 5) after treatment with 0.05% TX-100.
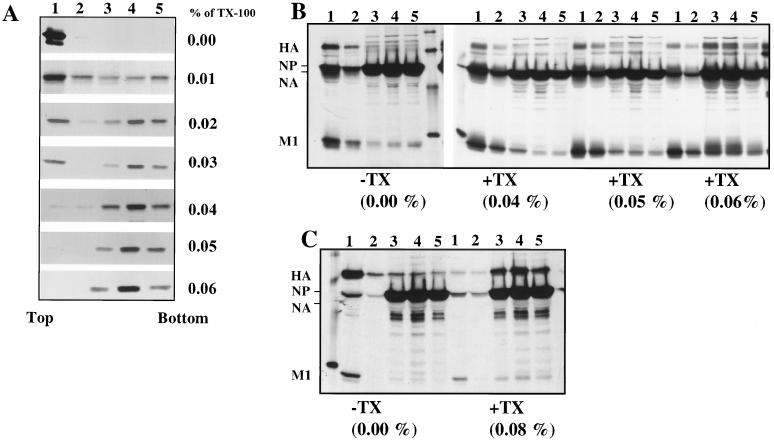
FIG. 2.
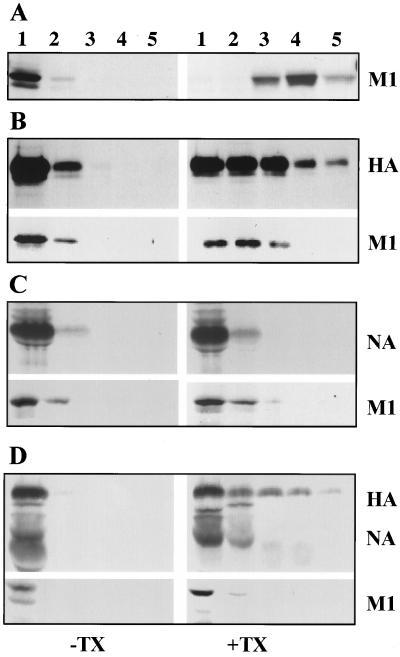
Analysis of membrane-associated M1 expressed alone by RVV and in influenza virus-infected cells after TX-100 detergent treatment. (A) HeLa cells (5 × 106) were infected at an MOI of 10 with RVVM1. At 6.0 hpi, infected cells were labeled for 30 min with 400 μCi of 35S Easy Tag and chased for 90 min. Infected cells were harvested and fractionated for 4K supernatant. The membrane fractions were isolated from the 4K supernatant with a floatation gradient as described in Materials and Methods and were either mock treated or treated with varying concentrations (0.01, 0.02, 0.03, 0.04, 0.05, and 0.06%) of TX-100 detergent for 15 min on ice. Each sample was then analyzed again by floatation in sucrose gradients. Gradient fractions were immunoprecipitated using rabbit anti-WSN polyclonal antibodies and analyzed by SDS–10% PAGE. (B and C) Influenza virus-infected cells were labeled at 6.5 hpi for 30 min and chased for 90 min. The total 4K supernatants were prepared and analyzed with floatation gradients before (−) and after (+) TX-100 treatment at different concentrations. Note that both HA and M1 decreased in the membrane fractions (fractions 1 and 2) after treatment with higher TX-100 concentrations (0.06 and 0.08%). Results in panels B and C are from two separate experiments.
To determine the effect of TX-100 on the M1 and HA proteins in influenza virus-infected cells, virus-infected cells at 6.5 hpi were pulse-labeled (30 min) and chased (90 min). The 4K supernatants were prepared, treated with different concentrations of TX-100, and analyzed with a floatation gradient. As can be seen, TX-100 treatment at 0.04 and 0.05% concentrations did not affect the membrane-bound M1 and HA (Fig. 2B and C, fractions 1 and 2) but higher concentrations (0.06 and 0.08%) of TX-100 reduced the membrane-bound fraction of both HA and M1 in influenza virus-infected cells. Similar results were also obtained using pure membrane fractions from influenza virus-infected cells (data not shown; also see Fig. 5). Since TX-100 at a 0.05% concentration solubilized all membrane-bound M1 in cells expressed alone but not in influenza virus-infected cells, we used TX-100 at a 0.05% concentration for detergent treatment in all subsequent experiments. Our method of detergent treatment of membrane-bound protein was clearly different from the standard methods used for assaying lipid raft association of apical proteins using a higher concentration (usually 1%) of TX-100 in a number of ways. Firstly, in assaying lipid raft-associated proteins, the whole cell rather than the pure membrane is treated with TX-100 and raft-associated proteins are measured directly in the TX-100-insoluble fractions (1). In the floatation assay, only a small fraction of raft-associated apical proteins floats to the top of the floatation gradient after TX-100 treatment, and this fraction does not often correlate with raft association. Secondly, as shown in Fig. 2B and C, raft-associated apical proteins like HA were not resistant to even 0.08% TX-100 under our experimental conditions. Finally, our goal in these experiments was not to analyze and characterize the raft association of M1 but to eliminate the nonspecific M1-membrane complex and assay the specific membrane-M1 complex formed in the presence of HA and NA. Therefore, our method was designed for determining specific membrane binding of M1 protein in the presence of HA and NA rather than protein raft association.
FIG. 5.
Detergent resistance of membrane-associated M1 from WSN virus-infected MDBK cells. (A and B) TX-100 treatment of membrane-associated M1 protein synthesized early (2.5 hpi) in the virus replication cycle. WSN virus-infected (MOI of 10) MDBK cells (5 × 106) were labeled with 300 μCi at 2.5 hpi for 20 min (A) and chased for 3 h (B) in the presence of 1.0 mM cycloheximide. Cells were then harvested and fractionated, and membrane fractions were isolated from the 4K supernatant with a floatation gradient. The membrane fractions were then treated without (−) or with (+) 0.05% TX-100 and analyzed with a second floatation gradient. Fractions were immunoprecipitated and analyzed by SDS-PAGE. (C and D) Analysis of M1 protein synthesized late (6.5 hpi) in the virus replication cycle. WSN virus-infected MDBK cells were labeled with 300 μCi at 6.5 hpi for 20 min (C) and chased for 1 h (D) in the presence of cycloheximide as described above. Membrane fractions were isolated from virus-infected cells as described above, treated without (−) or with (+) 0.05% TX-100, and analyzed with a floatation gradient. Fractions were immunoprecipitated and analyzed by SDS-PAGE.
The effect of HA and NA on the membrane association of M1.
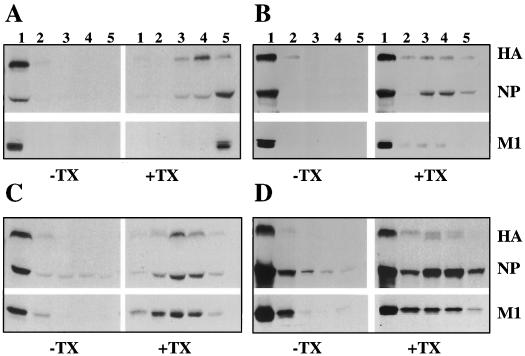
To determine the effect of influenza virus glycoproteins on the membrane association of M1, HeLa cells were infected with RVVs expressing either M1 alone or M1 with influenza virus HA or NA or both HA and NA. Following RVV infection at 6 hpi, HeLa cells were labeled with 35S Easy Tag (400 μCi) for 30 min and chased for 90 min. The membrane fraction was isolated from 4K supernatants, either mock treated or detergent treated (0.05% TX-100), and analyzed by floatation gradient centrifugation (31). Results (Fig. 3) show that major fractions of membrane-bound HA and NA became detergent resistant as expected from their interaction with lipid raft. The membrane-bound M1 from cells expressing M1 alone was completely detergent soluble (Fig. 3A) as expected. On the other hand, in cells coexpressing M1 and HA, 85% of membrane-bound M1 was detergent resistant (Fig. 3B and Table 1). Similarly, 87 and 93% of membrane-bound M1 became detergent resistant when coexpressed with NA and with both HA and NA, respectively (Fig. 3C and D and Table 1). These results demonstrated that both HA and NA rendered the membrane-bound M1 detergent resistant, supporting the interaction of M1 with HA and NA.
FIG. 3.
Detergent resistance of membrane-associated M1 when coexpressed with influenza virus HA and NA. For coexpression studies, HeLa cells (5 × 106) were infected with RVV expressing M1 alone or M1 in combination with HA or NA or with both HA and NA proteins as stated in Materials and Methods. At 4 hpi, cells were labeled with 400 μCi of 35S Easy Tag for 30 min and chased for 90 min. Cells were fractionated, and pure membrane fractions of the 4K supernatant were isolated with a floatation gradient as stated in Materials and Methods. Aliquots of membrane fractions were either untreated (−) or treated (+) with TX-100 (0.05%) and analyzed with a second floatation gradient. Fractions were collected, immunoprecipitated with anti-WSN antibodies, and analyzed by SDS-PAGE. (A) Expression of influenza virus M1 alone. (B) Coexpression of influenza virus M1 with influenza virus HA protein. (C) Coexpression of influenza virus M1 with NA protein. (D) Coexpression of M1 with both influenza virus HA and NA. Left-hand panels are without TX-100 treatment, and right-hand panels are after TX-100 treatment.
TABLE 1.
TX-100 resistance of the membrane-bound M1 in cells coexpressing viral proteinsa
| Coexpressed protein(s) | % of detergent-resistant membrane-bound M1 |
|---|---|
| None | 0 |
| Sendai F | 0 |
| HA | 85 ± 5 |
| NA | 87 ± 3 |
| HA and NA | 93 ± 3 |
| FHH | 75 ± 5 |
| HHF | 57 ± 8 |
| FFH | 70 ± 5 |
Domains of HA involved in rendering membrane-bound M1 TX-100 resistant.
The above results demonstrated that HA affected the membrane interaction of M1 by rendering the membrane-bound M1 resistant to TX-100 detergent. To further examine the domains of HA involved in rendering the membrane-bound M1 resistant to TX-100 detergent, chimeric constructs were made by switching the cytoplasmic tail (HHF), ectodomain (FHH), or transmembrane and ectodomain (FFH) of HA with that of Sendai virus F protein (Fig. 4A). RVVs were made from each chimeric construct, and HeLa cells were coinfected with vaccinia viruses expressing M1 and one of the chimeric proteins. Chimeric proteins used in these experiments were expressed efficiently from RVVs (Fig. 4G) and exhibited similar maturity as evident from their migration as a single band in gels, except for FFH, which was somewhat slow to mature, exhibiting two bands (Fig. 4D and F). Pure membrane fractions were isolated from the 4K supernatant, detergent treated, and analyzed with a floatation gradient. Results (Fig. 4 and Table 1) show that the membrane-bound M1 from cells expressing M1 alone or coexpressing heterologous F protein was completely detergent soluble (Fig. 4B and C). Although F protein was also detergent soluble, the results showed that the expression of any transmembrane protein would not render the membrane-bound M1 detergent insoluble. On the other hand, when both the transmembrane domain and the cytoplasmic tail of HA (FHH) were present, 75% of membrane-bound M1 was TX-100 resistant (Fig. 4D). Furthermore, when either the cytoplasmic tail (FFH) or the transmembrane domain (HHF) of HA was present, the proportion of membrane-bound M1 present after detergent treatment was 70 or 57%, respectively (Fig. 4E and F; Table 1). These results demonstrated that both the cytoplasmic tail and the transmembrane domain of HA played important roles in rendering the membrane-bound M1 resistant to TX-100.
TX-100 resistance of the membrane-bound M1 in virus-infected cells.
The results presented above demonstrated that a significant fraction of membrane-bound M1 when coexpressed with HA or NA became TX-100 resistant. To determine if the membrane-bound M1 in influenza virus-infected cells also became TX-100 resistant, the following experiments were done. WSN virus-infected MDBK cells were pulse-labeled either early (2.5 hpi) or late (6.5 hpi) in the infectious cycle for 20 min and then chased for 3 h (early) or 1 h (late) in the presence of cycloheximide. Membrane fractions were isolated from the 4K cytoplasmic supernatants, treated without (−) or with (+) 0.05% TX-100, and analyzed by floatation gradient centrifugation. Results (Fig. 5A) show that, when influenza virus-infected cells were pulse-labeled early in the infectious cycle (2.5 hpi), all membrane-bound M1 immediately after labeling was completely detergent soluble (Fig. 5A, +TX). However, upon chase for 3 h even in the presence of cycloheximide, 80% of membrane-bound M1 became detergent resistant (Fig. 5B, +TX). This could be explained by maturation of glycoproteins during the chase in the presence of cycloheximide and interaction of M1 with mature glycoproteins. When cells were pulse-labeled late in the infectious cycle (6.5 hpi), a significant fraction (35%) of membrane-bound M1 immediately after labeling became TX-100 resistant (Fig. 5C, +TX), showing that some M1 immediately after synthesis became associated with the preexisting mature glycoproteins. This would be expected because late in the infectious cycle some of the newly synthesized M1 will bind to the Golgi and the plasma membranes containing mature glycoproteins and detergent-resistant lipids. Similarly, the lack of TX-100 insolubility of the newly synthesized membrane-bound M1 at 2.5 hpi (Fig. 5A, +TX) was likely due to the absence of mature glycoproteins in detergent-resistant membranes in the early phase of the infectious cycle. Upon chase for 1 h, 75% of the membrane-bound M1 became TX-100 resistant (Fig. 5D). Early in the infectious cycle, the chase was extended to 3 h to ensure that all glycoproteins became mature and acquired detergent-resistant membrane-bound forms. These results taken together also support the idea that the interaction of M1 with mature glycoproteins rendered the membrane-bound M1 detergent resistant in influenza virus-infected cells.
Colocalization of M1 and HA in virus-infected cells and in cells coexpressing M1 and HA.
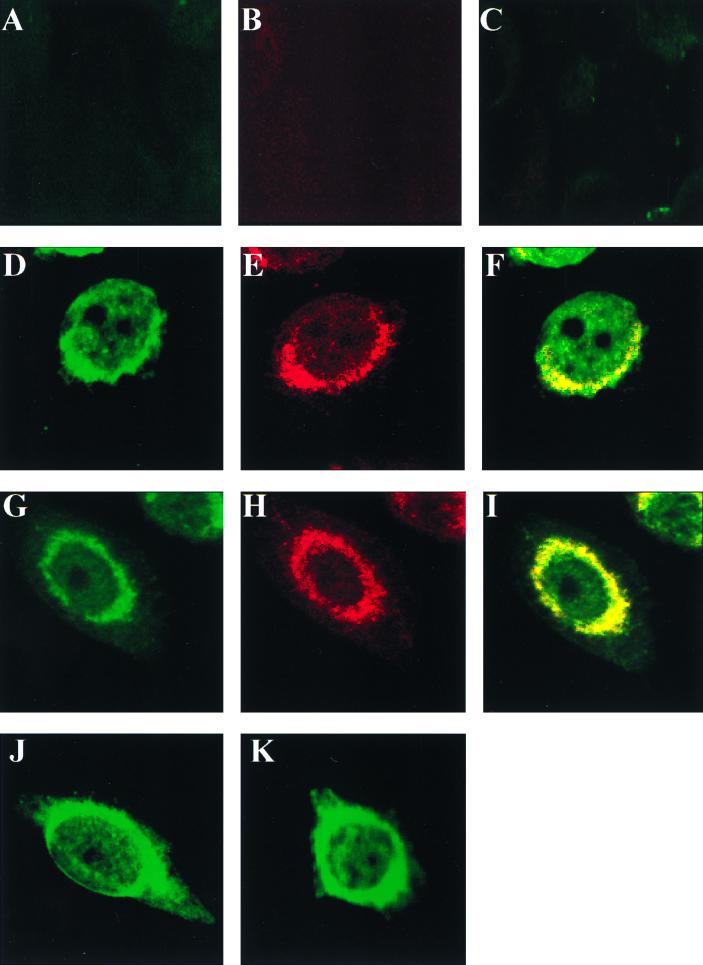
To determine if M1 colocalizes with HA in virus-infected cells, MDBK cells were synchronously infected with WSN virus at an MOI of 10 at 4°C. Cells were then washed and incubated at 37°C for 2 h when monensin (10 μM, final concentration) was added to some cells. Monensin is known to block the transport of glycoproteins from the medial Golgi compartments to plasma membranes (11). Influenza virus-infected cells were then incubated further for 5 h at 37°C in the presence or absence of monensin, and at 7 hpi, the virus-infected cells were fixed, permeabilized, and stained for HA and M1 using monoclonal anti-HA and polyclonal anti-M1 antibodies and analyzed by confocal microscopy. Results show that, in the absence of monensin, HA (red) was present throughout the cell including the cell periphery but concentrated in the perinuclear region and absent in the nucleus as expected (Fig. 6E). M1 (green), on the other hand, was present throughout the cell including the nucleus and cell periphery (Fig. 6D). Superimposition of staining showed the orange and yellow staining indicating colocalization of M1 and HA throughout the cell cytoplasm and the cell periphery. The intensity of the orange and yellow color indicates colocalization with a preponderance of HA and M1, respectively (Fig. 6F). In virus-infected cells treated with monensin, HA was present predominantly in the perinuclear Golgi region and absent in the plasma membrane (Fig. 6K) due to transport block of the exocytic pathway by monensin. In cells treated with monensin, M1 was present both in the nucleus and in the cytoplasm, but the cytoplasmic distribution of M1 was clearly different from that without monensin treatment. M1 was concentrated more in the perinuclear region and less on the cell periphery (Fig. 6J). Again, yellow and discrete orange staining in the perinuclear region of cells treated with monensin indicates colocalization of HA and M1 (Fig. 6L). These results demonstrated that a fraction of M1 colocalized with HA in WSN virus-infected cells, with or without monensin treatment, and that the degree of colocalization varied in the perinuclear region and cell periphery. However, total colocalization of HA and M1 was neither seen nor expected as concentrations of HA and M1 vary in different compartments of the cell.
FIG. 6.
Morphological distribution of M1 and HA in influenza virus-infected MDBK cells by confocal microscopy. MDBK cells (4 × 105) were grown on chamber slides and synchronously infected with WSN virus at an MOI of 10 at 4°C. Cells were then washed and incubated at 37°C. Monensin at a 10 μM final concentration was added to some cells at 2 hpi (G to L), and the cells were incubated for another 5 h at 37°C. At 7 hpi, all virus-infected cells were fixed with ice-cold acetone, incubated with a mixture of anti-M1 rabbit polyclonal and anti-HA mouse monoclonal antibodies, and stained with anti-rabbit IgG (green) and anti-mouse IgG (red). The stained cells were examined by confocal microscopy as described in Materials and Methods. (A to C) Mock-infected cells without monensin; (D to F) virus-infected cells without monensin treatment; (G to I) mock-infected cells with monensin; (J to L) virus-infected cells with monensin. Image analysis was done as follows: panels A, D, G, and J for M1 (green); panels B, E, H, and K for HA (red); and panels C, F, I, and L superimposed for both HA and M1 (original magnification, ×1,000).
Finally, to determine if M1 and HA colocalize when expressed from RVVs, HeLa cells were infected with RVVM1 and RVVHA. At 4 hpi, cells were fixed and stained for HA and M1 proteins and examined by confocal microscopy. Vaccinia virus infection causes a cytopathic effect and often renders the cell round, making it difficult to visualize the distribution of proteins in the cell cytoplasm. However, in some cells coexpressing both HA and M1, colocalization was clearly observed as yellow or orange depending on the level of expression of HA and M1 with (Fig. 7I) or without (Fig. 7F) monensin treatment. Again, as mentioned in Materials and Methods, the entire cell was examined by confocal microscopy at different planes and colocalization of M1 and HA was shown to be specific. Finally, M1 expressed alone exhibited similar subcellular distributions in the presence and in the absence of monensin (Fig. 7J and K). Taken together, these results indicate that fractions of HA and M1 colocalize in cells infected with influenza virus as well as in cells doubly infected with RVVM1 and RVVHA.
FIG. 7.
Morphological distribution of M1 and HA in HeLa cells coinfected with RVVM1 and RVVHA. HeLa cells (4 × 105) were grown on chamber slides and infected with RVVM1 and RVVHA. Cells were incubated with or without monensin, and at 4 hpi, they were double stained for M1 and HA as described in the legend to Fig. 6 and examined by confocal microscopy. (A to C) Cells infected with wild-type vaccinia virus; (D to F) cells without monensin treatment; (G to I) cells with monensin treatment. Image analysis was done as follows: panels A, D, and G for M1; panels B, E, and H for HA; and panels C, F, and I superimposed for both HA and M1. Cells in panels J (without monensin) and K (with monensin) were infected with RVVM1 only and stained for M1 (original magnification, ×600).
DISCUSSION
The critical role of M1, the most abundant protein in the virus particle, in influenza virus assembly and budding is undisputed. Results presented in this report indicate that both viral glycoproteins HA and NA affect the membrane association of M1 proteins, thereby providing evidence for the interaction of M1 with HA and NA. This conclusion was further strengthened by the requirement for the homologous transmembrane domain and cytoplasmic tail of HA in detergent resistance of the membrane-bound M1. Earlier attempts to demonstrate the interaction between M1 and influenza virus glycoproteins showing increased membrane association of M1 protein in the presence of homologous viral glycoproteins yielded conflicting results (5, 18, 44), which can be attributed to significant variation (15 to 60%) in the intrinsic membrane-binding ability of M1 protein expressed alone. Experimental factors including the expression system used and relative ratios of M1 to glycoproteins present in coexpressing cells as well as the process of cell disruption used in releasing membranes and preparing the 4K supernatant may have contributed to these variations. To overcome these difficulties, we designed an assay which would eliminate the membrane association of M1 protein expressed alone without eliminating the membrane association of M1 from the M1-glycoprotein(s) interactions.
Influenza virus transmembrane proteins are sorted to the apical plasma membrane, the budding site of influenza viruses in polarized epithelial cells. Many of these apical proteins including HA and NA have been shown to preferentially cluster on the lipid rafts enriched in cholesterol and glycosphingolipids during their transport from the trans-Golgi membrane to the plasma membrane (2, 19, 33, 37), and this interaction of apical proteins with lipid rafts occurs in both polarized and nonpolarized cells (38). Furthermore, we and others have shown that the transmembrane domains of influenza virus NA and HA provide an apical determinant and associate with TX-100-resistant lipid rafts (2, 19, 33). However, M1, a cytoplasmic protein, which is not transported by the exocytic pathway, is not expected to be raft associated and TX-100 detergent resistant unless it binds to another raft-associated protein, as has been shown for a number of signaling molecules (36). Therefore, TX-100 detergent treatment essentially eliminates all lipid-protein interactions except for those proteins present in cholesterol- and glycosphingolipid-enriched membranes, and these detergent-resistant membrane-bound proteins will float to the top of the gradient. However, such detergent extraction of membranes should not be confused with TX-100 treatment used for assaying cytoskeleton-protein interactions (25, 32) since cytoskeleton-protein interactions are resistant to a higher detergent concentration (1% TX-100) as well as to octylglucoside (1) and the cytoskeletal components and proteins will not float to the top of the gradient following detergent extraction. Furthermore, we and others have shown previously that M1 protein interacts with cytoskeletal components in influenza virus-infected cells but not in cells expressing either M1 alone or M1 with influenza virus NP (1, 44).
Analysis of membrane association of M1 in influenza virus-infected cells (Fig. 5) also supports the idea that mature glycoproteins are required for the association of M1 with detergent-resistant membranes and that the newly synthesized M1 can bind to preexisting mature influenza virus glycoproteins, associated with TX-100-resistant lipid rafts in the trans-Golgi membrane and plasma membrane. However, we cannot rule out additional conformational modification of M1 during chase facilitating further M1-glycoprotein interaction. It is possible that the M1-vRNP complex may have further facilitated M1-glycoprotein interactions in influenza virus-infected cells. However, it is clear that M1 can bind to HA or NA in the absence of other influenza virus proteins.
Immunofluorescence analysis by confocal microscopy also supports the interaction of M1 with HA. In influenza virus-infected cells, colocalization of M1 and HA could be seen both at the plasma membrane and in the perinuclear cell cytoplasm but not in the nucleus. Colocalization of HA and M1 was observed both with and without monensin treatment. However, distribution of M1 and HA differed in monensin-treated cells. Almost all HA was present in the perinuclear region after monensin treatment, whereas M1, being more abundant than HA, was not restricted to the perinuclear region but was present throughout the cell. However, the distribution of M1, particularly at the cell periphery, was much less pronounced and distinctly different after monensin treatment due to lack of HA at the plasma membrane (compare Fig. 6D and J). Essentially similar results were obtained for cells coexpressing HA and M1 from RVVM1- and RVVHA-infected cells. Finally, although biochemical and morphological studies demonstrate interaction of M1 with mature glycoproteins (i.e., glycoproteins present in the mid-Golgi complex and trans-Golgi complex and plasma membrane), we cannot rule out M1-glycoprotein interaction in the cis- or pre-Golgi complex or endoplasmic reticulum.
Coexpression of M1 with heterologous (F) and homologous (HA and NA) glycoproteins showed that homologous glycoproteins were critical for M1 to acquire detergent-resistant membrane association and that heterologous glycoproteins such as Sendai virus F failed to render the membrane-bound M1 detergent resistant (Fig. 4C). These experiments clearly demonstrated that the interaction of M1 with HA was essential for M1 to become associated with detergent-resistant membranes. Analysis with chimeric constructs between F and HA revealed that both the transmembrane domain and the cytoplasmic tail were involved in interacting with M1. The cytoplasmic tail of HA and the transmembrane domain were independently capable of rendering a fraction of M1 detergent resistant; however, the fraction was less than that obtained with HA or with FHH containing both the transmembrane domain and the cytoplasmic tail of HA (Table 1). High-resolution cryoelectron microscopy (6) and the X-ray crystal structure of the N terminus of M1 (35) also support the idea that M1 interacts with the inner leaflet of the lipid bilayer and therefore is likely to interact with the COOH half of the transmembrane domain of HA.
The cytoplasmic tails of HA and NA are highly conserved among virus strains. The role of cytoplasmic tails of NA and HA has been investigated using reverse genetics (8, 15, 16, 17, 24). Since viruses having either tail-minus HA, tail-minus NA, or both tail-minus HA and tail-minus NA could be rescued, it was shown that the cytoplasmic tail of HA and NA individually or together was not an absolute requirement for assembly and particle formation. However, tails of both glycoproteins provided a considerable advantage in efficient budding since the yield of infectious virus in tail-minus mutants was considerably lower and any revertant virus possessing cytoplasmic tail outgrew the mutant viruses (15, 17). In addition, the influenza virus lacking both tail-minus HA and tail-minus NA exhibited bizarre filamentous morphology (17). Earlier studies using ts mutants demonstrated that viral morphogenesis can take place in the absence of either HA or NA (21, 28), suggesting that there is considerable redundancy in the assembly and budding processes and that only one envelope protein may be sufficient for assembly and budding. However, in none of these experiments was foreign cytoplasmic tail replaced in the transfectant viruses. Furthermore, the role of the transmembrane domain of viral proteins in viral morphogenesis is less clear. HA molecules containing foreign cytoplasmic and foreign transmembrane domains failed to be incorporated into virus particles possessing the wild-type HA (25), whereas a foreign protein containing the transmembrane domain and cytoplasmic tail of HA was incorporated into virus particles (9).
In other viruses, the role of the cytoplasmic tail of the envelope protein in the assembly process and incorporation into virus particles appears to vary greatly. With Sindbis viruses, alteration in the cytoplasmic tail of E2 glycoprotein can prevent particle formation (7, 39). With vesicular stomatitis virus, the G protein containing a foreign cytoplasmic tail of specific length can be incorporated efficiently (34), and with rabies virus, budding can take place in the complete absence of spike glycoprotein (23). Similarly, viruslike particles can be formed and released in the absence of envelope protein in retroviruses including human immunodeficiency virus (4, 14).
In conclusion, the data presented here show that a major fraction of influenza virus M1 protein when expressed alone or in virus-infected cells becomes membrane associated immediately after synthesis. Since at this stage M1 protein nonselectively binds to intracellular membranes, the membrane-M1 association is TX-100 detergent soluble. In the presence of homologous viral glycoproteins HA and NA, in either influenza virus-infected cells or cells expressing homologous glycoprotein, M1 interacts with influenza virus glycoproteins and the membrane-M1 interaction becomes TX-100 resistant because of the association of mature HA and NA with lipid rafts enriched in cholesterol and glycosphingolipids. Furthermore, colocalization data reported here indicate that M1 can interact with viral glycoproteins present in the plasma membrane as well as with glycoproteins in transit through the exocytic pathway. M1 interaction with chimeric constructions of glycoproteins demonstrates that both the cytoplasmic tail and the transmembrane domain of influenza virus HA can help membrane-bound M1 to acquire TX-100 resistance, supporting the idea that M1 interacts with both the transmembrane domain and the cytoplasmic tail of HA.
ACKNOWLEDGMENTS
This work was supported by USPHS grants (AI-16348 and AI-41681) from the NIH, NIAID.
We are grateful to Jose Orozco for constructing the RVVs and Eleanor Berlin for typing the manuscript. Confocal microscopy and image analysis were done using the UCLA Brain Research Institute Core Imaging Facility. We thank Matthew J. Schibler for his kind help with confocal microscopy.
REFERENCES
- 1.Avalos R T, Yu Z, Nayak D P. Association of influenza virus NP and M1 proteins with cellular cytoskeletal elements in influenza virus-infected cells. J Virol. 1997;71:2947–2958. doi: 10.1128/jvi.71.4.2947-2958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barman S, Nayak D P. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J Virol. 2000;74:6538–6545. doi: 10.1128/jvi.74.14.6538-6545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Compans R W, Choppin P W. Reproduction of myxoviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. IV. New York, N.Y: Plenum Press; 1975. pp. 179–252. [Google Scholar]
- 4.Deml L, Schirmbeck R, Reimann J, Wolf H, Wagner R. Recombinant human immunodeficiency Pr55gag virus-like particles presenting chimeric envelope glycoproteins induce cytotoxic T-cells and neutralizing antibodies. Virology. 1997;235:26–39. doi: 10.1006/viro.1997.8668. [DOI] [PubMed] [Google Scholar]
- 5.Enami M, Enami K. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J Virol. 1996;70:6653–6657. doi: 10.1128/jvi.70.10.6653-6657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujiyoshi Y, Kume N P, Sakata K, Sato S B. Fine structure of influenza A virus observed by electron cryo-microscopy. EMBO J. 1994;13:318–326. doi: 10.1002/j.1460-2075.1994.tb06264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaedigk-Nitschko K, Schlesinger M J. Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology. 1991;183:206–214. doi: 10.1016/0042-6822(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Sastre A, Palese P. The cytoplasmic tail of the neuraminidase protein of influenza A virus does not play an important role in the packaging of this protein into viral envelopes. Virus Res. 1995;37:37–47. doi: 10.1016/0168-1702(95)00017-k. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Sastre A, Muster T, Barclay W S, Percy N, Palese P. Use of a mammalian internal ribosomal entry site element for expression of a foreign protein by transfectant influenza virus. J Virol. 1994;68:6254–6261. doi: 10.1128/jvi.68.10.6254-6261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregoriades A, Frangione B. Insertion of influenza M protein into the viral lipid bilayer and localization of site of insertion. J Virol. 1981;40:323–328. doi: 10.1128/jvi.40.1.323-328.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths G, Quinn P, Warren G. Dissection of Golgi complex. I. Monensin inhibits the transport of viral membrane proteins from medial to trans-Golgi cisternae in baby hamster kidney cells infected with Semliki Forest virus. J Cell Biol. 1983;96:835–850. doi: 10.1083/jcb.96.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gujuluva C N, Kundu A, Murti K G, Nayak D P. Abortive replication of influenza virus A/WSN/33 in HeLa 229 cells: defective membrane function during entry and budding processes. Virology. 1994;204:491–505. doi: 10.1006/viro.1994.1563. [DOI] [PubMed] [Google Scholar]
- 13.Hay A J. Studies on the formation of the influenza virus envelope. Virology. 1974;60:398–418. doi: 10.1016/0042-6822(74)90335-3. [DOI] [PubMed] [Google Scholar]
- 14.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 15.Jin H, Leser G, Lamb R A. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 1994;13:5504–5515. doi: 10.1002/j.1460-2075.1994.tb06885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin H, Subbarao K, Bagai S, Leser G P, Murphy B R, Lamb R A. Palmitylation of the influenza virus hemagglutinin (H3) is not essential for virus assembly or infectivity. J Virol. 1996;70:1406–1414. doi: 10.1128/jvi.70.3.1406-1414.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin H, Leser G P, Zhang J, Lamb R A. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kretzschmar E, Bui M, Rose J K. Membrane-association of influenza virus matrix protein does not require specific hydrophobic domains or the viral glycoproteins. Virology. 1996;220:37–45. doi: 10.1006/viro.1996.0283. [DOI] [PubMed] [Google Scholar]
- 19.Kundu A, Avalos R T, Sanderson C M, Nayak D P. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J Virol. 1996;70:6508–6515. doi: 10.1128/jvi.70.9.6508-6515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Xu M, Coelingh K. Electroporation of influenza virus ribonucleoprotein complexes for rescue of the nucleoprotein and matrix genes. Virus Res. 1995;37:153–161. doi: 10.1016/0168-1702(95)00031-k. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Eichelberger M C, Compans R W, Air G M. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol. 1995;69:1099–1106. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohmeyer J, Talens L T, Klenk H D. Biosynthesis of the influenza virus envelope in abortive infection. J Gen Virol. 1979;42:73–88. doi: 10.1099/0022-1317-42-1-73. [DOI] [PubMed] [Google Scholar]
- 23.Mebatsion T, Konig M, Conzelmann K-K. Budding of rabies virus particles in absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 24.Mitnaul L J, Castrucci M R, Murti K G, Kawaoka Y. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J Virol. 1996;70:873–879. doi: 10.1128/jvi.70.2.873-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison T G, McGinnes L J. Cytochalasin D accelerates the release of Newcastle disease virus from infected cells. Virus Res. 1985;4:93–106. doi: 10.1016/0168-1702(85)90023-1. [DOI] [PubMed] [Google Scholar]
- 26.Naim H Y, Roth M G. Basis of selective incorporation of glycoproteins into the influenza virus envelope. J Virol. 1993;67:4831–4841. doi: 10.1128/jvi.67.8.4831-4841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nayak D P. A look at assembly and morphogenesis of orthomyxo- and paramyxoviruses. ASM News. 1996;62:411–414. [Google Scholar]
- 28.Pattnaik A K, Brown D J, Nayak D P. Formation of influenza virus particles lacking hemagglutinin on the viral envelope. J Virol. 1986;60:994–1001. doi: 10.1128/jvi.60.3.994-1001.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rey O, Nayak D P. Nuclear retention of M1 protein in a temperature-sensitive mutant of influenza (A/WSN/33) virus does not affect nuclear export of viral ribonucleoproteins. J Virol. 1992;66:5815–5824. doi: 10.1128/jvi.66.10.5815-5824.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Boulan E, Sabatini D D. Asymmetric budding of viruses in epithelial cell monolayers: a model system for study of epithelial cell polarity. Proc Natl Acad Sci USA. 1978;75:5071–5075. doi: 10.1073/pnas.75.10.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanderson C M, Wu H-H, Nayak D P. Sendai virus M protein binds independently to either the F or the HN glycoprotein in vivo. J Virol. 1994;68:69–76. doi: 10.1128/jvi.68.1.69-76.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanderson C M, Avalos R, Kundu A, Nayak D P. Interaction of Sendai viral F, HN, and M protein cytoskeletal and lipid components in Sendai virus-infected BHK cells. Virology. 1995;209:701–707. doi: 10.1006/viro.1995.1308. [DOI] [PubMed] [Google Scholar]
- 33.Scheiffele P, Roth M G, Simons K. Interaction of influenza virus hemagglutinin with sphingolipid-cholesterol membrane domains via transmembrane domain. EMBO J. 1997;16:5501–5508. doi: 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnell M J, Buonocore L, Boritz E, Ghosh H P, Chernis R, Rose J K. Requirement for a nonspecific cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis viruses. EMBO J. 1998;17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sha B, Luo M. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat Struct Biol. 1997;4:239–244. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- 36.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 37.Simons K, van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 38.Skibbens J E, Roth M G, Matlin K S. Differential extractability of influenza virus hemagglutinin during intracellular transport in polarized epithelial cells and nonpolar fibroblasts. J Cell Biol. 1989;108:821–832. doi: 10.1083/jcb.108.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suomalainen M, Liljestrom P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittaker G, Kemler I, Helenius A. Hyperphosphorylation of mutant influenza virus matrix protein M1 causes its retention in the nucleus. J Virol. 1995;69:439–445. doi: 10.1128/jvi.69.1.439-445.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yasuda J, Bucher D J, Ishihama A. Growth control of influenza A virus by M1 protein: analysis of transfectant viruses carrying chimeric M gene. J Virol. 1994;68:8141–8146. doi: 10.1128/jvi.68.12.8141-8146.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye Z, Liu T, Offringa D P, McInnis J, Levandowski R A. Association of influenza virus matrix protein with ribonucleoproteins. J Virol. 1999;73:7467–7473. doi: 10.1128/jvi.73.9.7467-7473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshimori T, Keller P, Roth M G, Simons K. Different biosynthetic transport routes to the plasma membrane in BHK and CHO cells. J Cell Biol. 1996;133:247–256. doi: 10.1083/jcb.133.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Lamb R A. Characterization of the membrane-association of the influenza virus matrix protein in living cells. Virology. 1996;225:255–265. doi: 10.1006/viro.1996.0599. [DOI] [PubMed] [Google Scholar]
- 45.Zhirnov O P. Isolation of matrix protein M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology. 1992;186:324–330. doi: 10.1016/0042-6822(92)90090-c. [DOI] [PubMed] [Google Scholar]