Abstract
Background
Prion diseases are irreversible infectious neurodegenerative diseases caused by a contagious form of prion protein (PrPSc). Since chronic wasting disease (CWD)-infected white-tailed deer are strong carriers of the prion seed through corpses via scavenger animals, preemptive control based on genetic information for a culling system is necessary. However, the risk of CWD-related genetic variants has not been fully evaluated. In the present study, we carried out a quantitative estimation of the risk of a G96S single nucleotide polymorphism (SNP) of the PRNP gene to CWD infection in white-tailed deer.
Methods
We carried out a literature search for genetic data of the G96S (c.286G>A) SNP of the PRNP gene from CWD-infected white-tailed deer and matched controls. We performed a meta-analysis using incorporated eligible studies to evaluate the association of the G96S SNP of the PRNP gene with susceptibility to CWD in white-tailed deer.
Results
We identified a strong association between the G96S (c.286G>A) SNP of the PRNP gene and susceptibility to CWD infection in white-tailed deer using meta-analysis. We observed the most significant association in the recessive model (odds ratio = 3.0050, 95% confidence interval: 2.0593; 4.3851, p < 0.0001), followed by the additive model (odds ratio = 2.7222, 95% confidence interval: 1.9028; 3.8945, p < 0.0001) and the heterozygote (AA vs. AG) comparison (odds ratio = 2.7405, 95% confidence interval: 1.9215; 3.9085, p < 0.0001).
Conclusion
To the best of our knowledge, this was the first meta-analysis of the association between the G96S (c.286G>A) SNP of the PRNP gene and susceptibility to CWD infection.
Keywords: prion, PRNP, PrPSc, scrapie, BSE, CWD, CJD, white-tailed deer
1. Introduction
Prion diseases are malignant contagious neurodegenerative diseases caused by an infectious form of prion protein (PrPSc) converted from a benign form of prion protein (PrPC) (1–3). Several types of prion diseases have been reported worldwide including scrapie in sheep and goats; bovine spongiform encephalopathy (BSE) in cattle; Creutzfeldt-Jakob disease (CJD), fatal familial insomnia (FFI), Gerstmann–Straussler–Scheinker syndrome (GSS), and kuru in humans; and chronic wasting disease (CWD) in the Cervidae family (2, 4, 5). PrPSc acts as a conversion seed and changes PrPC to a ß-sheet-rich aggregate. The aggregate accumulates in brain lesions and is accompanied by several neurotoxic symptoms including neuron loss, vacuolation, and astrogliosis (1, 6, 7). During the conversion process, genetic variants of the amino acid sequences of prion protein (PrP), which are encoded by the prion protein gene (PRNP), play a pivotal role in susceptibility to prion diseases (8, 9). In humans, the M129V single nucleotide polymorphism (SNP) provides resistance to CJD (10). In addition, the G127V natural genetic variant blocks kuru (11). In sheep, scrapie susceptibility is modulated by haplotypes located on codons 136, 154, and 171 (12). In goats, codons at 142, 146, 154, 211, and 222 of the PRNP gene contribute to prion disease resistance (12–14). Like other prion diseases, CWD hosts have unique susceptibility-related genetic variants (5, 15). In elk, M132L SNP is associated with CWD propagation (16). In sika deer, a study indicated that S19N is related to CWD vulnerability (17). In white-tailed deer, several studies have suggested that G96S is the most potent candidate for prion-related genetic factors (18–27). However, quantitative studies have not been performed.
Among the several types of prion disease, CWD is the most infectious because it can be vertically transmitted, as through CWD prion-contaminated soil (28). Therefore, preemptive control with a culling system using genetic information is a crucial step to prevent the extensive spread of CWD. Since some CWD-infected white-tailed deer are free-ranging, there is a greater possibility of prion contamination through corpses via scavenger animals and of the emergence of novel prion hosts and strains (29). In addition, older free-ranging deer of indeterminate slaughter age carrying germline and somatic mutations in the PRNP gene are more likely to develop prion disease. Since CWD seed inducible genetic variants have not been fully elucidated, a quantitative evaluation of the connection between G96S of the PRNP gene and CWD susceptibility in white-tailed deer is necessary.
In the present study, we searched the literature for study data on the genotype and allele distributions of the G96S SNP of the PRNP gene from CWD-infected white-tailed deer and matched controls to evaluate the association between the G96S SNP of the PRNP gene and susceptibility to CWD in white-tailed deer. Then, we carried out a meta-analysis using incorporated eligible studies.
2. Materials and methods
2.1. Search strategy
We carried out the meta-analysis according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. A literature search was conducted in the PubMed, Google Scholar, and Web of Science databases to find studies associated with the G96S (c.286G>A) SNP of the PRNP gene from CWD-infected white-tailed deer. The following terms were employed for the search: “PRNP,” “white-tailed deer,” and “CWD” or “Chronic wasting disease” combined with “polymorphism” or “SNP” (last search update: March 12, 2024).
Eligible studies adhered to the subsequent inclusion criteria: (1) relating to and containing the association between the G96S (c.286G>A) SNP and CWD-infected white-tailed deer; (2) a case-control study; (3) full text available; and (4) written in English. The exclusion criteria were as follows: (1) case reports and (2) insufficient genetic information.
2.2. Meta-analysis
The meta-analysis was performed using the meta package of the R program [https://www.r-project.org/ (accessed on March 12, 2024)] and involved calculation of the pooled odds ratios with 95% confidence intervals between the G96S (c.286G>A) SNP of the PRNP gene and susceptibility to CWD. We utilized a total of 7 genetic models: additive (A vs. G), recessive (AA vs. AG + GG), dominant (AA + AG vs. GG), and over-dominant (AG vs. AA + GG) models and homozygote (AA vs. GG) and heterozygote (AA vs. AG and AG vs. GG) comparisons. Heterogeneity was calculated using the p-value and I2 value. The fixed and random effect models were selected according to the value of the I2 test. Publication bias was estimated using Egger’s weighted regression methods.
3. Results
3.1. Study selection
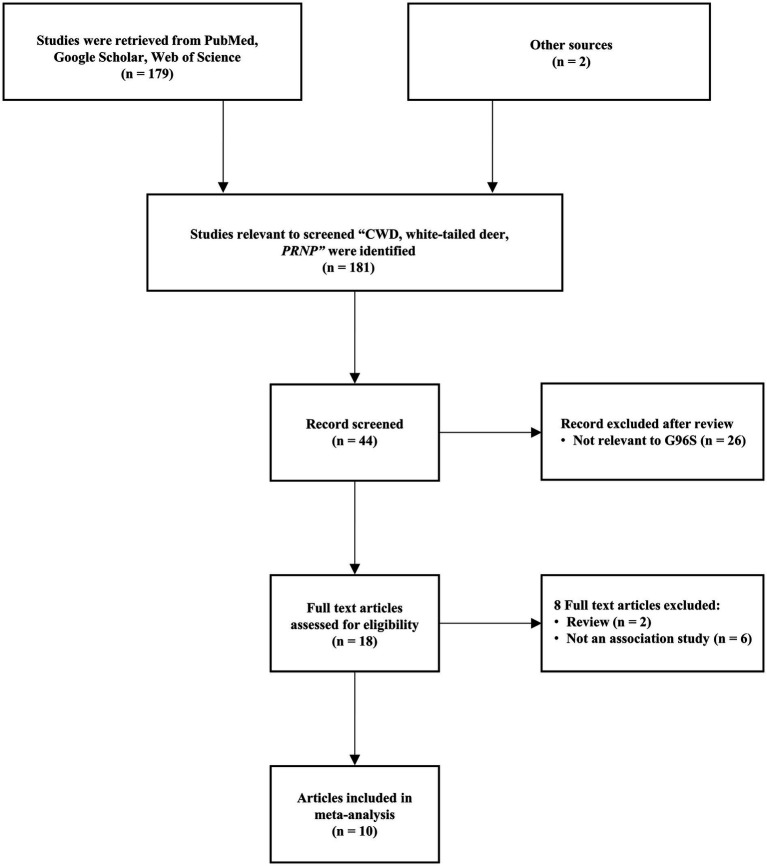
A flow chart of the study selection process is shown in Figure 1. After excluding ineligible articles based on the inclusion and exclusion criteria, 10 relevant studies were extracted from the databases (PubMed, Google Scholar, Web of Science). The meta-analysis comprised 1,003 cases and 1,335 controls (Table 1). All eligible studies utilized a case–control comparison using two genotyping methods. Among the 10 studies, 2 studies were genotyped using polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) methods. The remaining 8 studies were genotyped using PCR-amplicon sequencing methods (Table 1).
Figure 1.
Flow chart of the study selection process.
Table 1.
Characteristics of studies included in the meta-analysis.
| Study | Year | State | Nation | Genotyping methods | Total sample size | Number of controls | Number of cases |
|---|---|---|---|---|---|---|---|
| Chad et al. | 2003 | Wisconsin | USA | RFLP | 120 | 94 | 26 |
| Katherine et al. | 2004 | Nebraska | USA | RFLP | 131 | 64 | 67 |
| Chad et al. | 2006 | Wisconsin | USA | Sequencing | 445 | 153 | 292 |
| Delwyn et al. | 2008 | Wisconsin | USA | Sequencing | 73 | 14 | 59 |
| Amy et al. | 2008 | Illinois | USA | Sequencing | 196 | 120 | 76 |
| Gregory et al. | 2009 | Alberta, Saskatchewan | Canada | Sequencing | 227 | 197 | 30 |
| Julie et al. | 2009 | Wisconsin | USA | Sequencing | 137 | 72 | 65 |
| Adam et al. | 2015 | Illinois, Wisconsin | USA | Sequencing | 240 | 135 | 105 |
| Robert et al. | 2020 | Nebraska | USA | Sequencing | 201 | 103 | 98 |
| Caitlin et al. | 2021 | Michigan | USA | Sequencing | 568 | 383 | 185 |
| Total | — | — | — | — | 2,338 | 1,335 | 1,003 |
3.2. Evaluation of the association between the G96S (c.286G>A) SNP of the PRNP gene and susceptibility to CWD by meta-analysis in white-tailed deer
The pooled, weighted genotyping results were combined according to sample size to evaluate the association between the G96S SNP of the PRNP gene and susceptibility to CWD using the meta package of the R program. The pooled odds ratios with 95% confidence intervals were evaluated based on 7 genetic models: additive (A vs. G), recessive (AA vs. AG + GG), dominant (AA + AG vs. GG), and over-dominant (AG vs. AA + GG) models and homozygote (AA vs. GG) and heterozygote (AA vs. AG and AG vs. GG) comparisons. Heterogeneity was estimated using the p-value and I2 value (Table 2). The dominant model (AA + AG vs. GG) and homozygote (AA vs. GG) and heterozygote (AG vs. GG) comparisons used fixed models according to the I2 value (>0.5). The remaining 4 genetic models used random models according to the I2 value (<0.5).
Table 2.
Meta-analysis of the association between G96S (c.286G>A) of the prion protein gene (PRNP) gene and susceptibility to chronic wasting disease (CWD).
| Genetic model | Association test | Heterogeneity | Publication bias | ||||
|---|---|---|---|---|---|---|---|
| Odds ratio | 95% confidence interval | p-value | Model | p-value | I 2 | Egger’s test p-value | |
| Additive model (A vs. G) | 2.7222 | [1.9028; 3.8945] | 2.963 ×10−7 | Random | <0.01 | 0.70 | 0.2078 |
| Recessive model (AA vs. AG + GG) | 3.0050 | [2.0593; 4.3851] | 8.09 ×10−8 | Random | <0.01 | 0.64 | 0.4141 |
| Dominant model (AA + AG vs. GG) | 2.4081 | [1.3951; 4.1568] | 0.0112199714 | Fixed | 0.18 | 0.29 | 0.0065 |
| Over-dominant model (AG vs. AA + GG) | 0.4067 | [0.2817; 0.5871] | 1.0949 ×10−5 | Random | 0.01 | 0.59 | 0.7684 |
| AA vs. GG | 2.9883 | [1.7221; 5.1856] | 0.0006936882 | Fixed | 0.11 | 0.38 | 0.0043 |
| AA vs. AG | 2.7405 | [1.9215; 3.9085] | 1.827 ×10−7 | Random | 0.02 | 0.55 | 0.6524 |
| AG vs. GG | 1.3313 | [0.7495; 2.3650] | 1 | Fixed | 0.29 | 0.16 | 0.0535 |
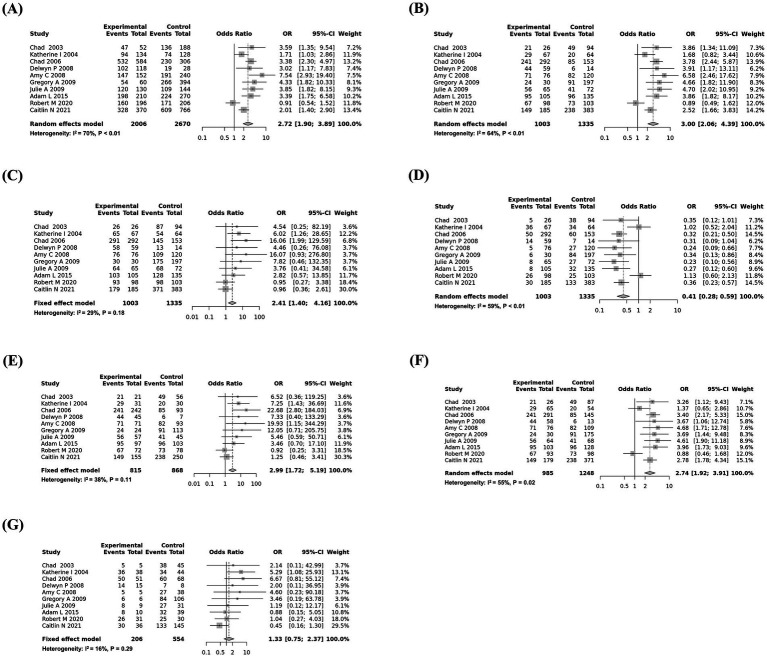
Except for the heterozygote (AG vs. GG) comparison, the data from the 9 remaining genetic models showed a strong association between the risk of CWD and the G96S (c.286G>A) SNP of the PRNP gene in the additive model (odds ratio = 2.7222, 95% confidence interval: 1.9028; 3.8945, p < 0.0001), recessive model (odds ratio = 3.0050, 95% confidence interval: 2.0593; 4.3851, p < 0.0001), dominant model (odds ratio = 2.4081, 95% confidence interval: 1.3951; 4.1568, p < 0.05), over-dominant model (odds ratio = 0.4067, 95% confidence interval: 0.2817; 0.5871, p < 0.001), homozygote comparison (odds ratio = 2.9883, 95% confidence interval: 1.7221; 5.1856, p < 0.001), and heterozygote (AA vs. AG) comparison (odds ratio = 2.7405, 95% confidence interval: 1.9215; 3.9085, p < 0.0001).
To estimate potential publication bias, Egger’s tests were carried out, and publication bias was observed in the dominant model and homozygote (AA vs. GG) and heterozygote comparisons (AG vs. GG). The details of the values are described in Table 2. Except for publication bias that violated genetic models, the most significant association was observed in the recessive model, followed by the additive model and heterozygote (AA vs. AG) comparison. Forest plots are shown in Figure 2.
Figure 2.
Forest plots of the association between the G96S (c.286G>A) single-nucleotide polymorphism (SNP) of the prion protein gene (PRNP) gene and susceptibility to chronic wasting disease (CWD). (A) Additive model (A vs. G). (B) Recessive model (AA vs. AG + GG). (C) Dominant model (AA + AG vs. GG). (D) Over-dominant model (AG vs. AA + GG). (E) Homozygote comparison (AA vs. GG). (F) Heterozygote comparison (AA vs. AG). (G) Heterozygote comparison (AG vs. GG).
4. Discussion
White-tailed deer are raised on farms or live in the wild, and CWD-infected white-tailed deer have been reported in the USA and Canada in North America (23, 30). In previous surveillance for prion diseases in Wisconsin, field cases were reported in several scavenger animals including mink, raccoon, and opossums (29). The results of that study indicated that free-ranging CWD-infected white-tailed deer are strong prion carriers and possible CWD pandemic inducers in the ecosystem. In addition, recent studies have indicated that a sporadic type of animal prion disease, atypical BSE, was identified in old animals (8 years old or older) (31). Since sporadic human prion diseases account for over 85% of the total number (32), and the slaughter age is not determined for free-ranging white-tailed deer, the possibility of sporadic occurrence may be much higher in white-tailed deer than in other farmed animals. Therefore, it is important to investigate CWD-related genetic factors for use in a culling system for preemptive prevention of CWD in white-tailed deer.
Previous genome-wide association analysis (GWAA) suggested that G96S is the large-effect risk locus; however, that study did not provide the quantitative value of the CWD risk (33). In the present study, we quantitatively evaluated the risk of the G96S SNP of the PRNP gene for CWD in white-tailed deer and observed a strong association in the recessive (AA vs. AG + GG) and additive (A vs. G) models and the heterozygote (AA vs. AG) comparison (Table 2). In summary, the 96G allele confers a risk of CWD that is more than 2.5 times higher than that of the 96S allele in these genetic models (Table 2).
Codon 96 is located on the unstructured coil region between the octapeptide repeat region and ß-sheet of cervid PrP (15). Several pathogenic mutations including P102L, P105L, and A117V of human PrP were identified in a similar region (34). Thus, this region seems to play a pivotal role in the conversion of PrPSc. However, although M129V heterozygotes provided potent resistance to CJD, human PrP with the 129M allele showed a similar structure and characteristics to human PrP with the 129V allele. The prion-resistant effect of the 129V allele is related to the homotypic amino acid sequence between the template protein and prion seed (PrPSc) (35). In addition, the results of a recent real-time quaking-induced conversion (RT-QuIC) study indicated that conversion efficiency (PrPC to PrPSc) is significantly related to the correspondence of the amino acid sequence at codon 132 of the elk PrP between the seed (PrPSc) and template protein (36). Thus, further investigation of the effects of codon 96 on seed-template interaction is required to elucidate the pathomechanism of codon 96. In white-tailed deer, several reports have indicated that codon 95 is also associated with susceptibility to CWD (15, 18). In addition, a recent study suggested that codon 116 is related to the stability of CWD prion, sensitivity to proteinase K, and lower prion seeding activity (37). Since the present study did not reflect other codons or the haplotypes of the PRNP gene, further investigation of genetic polymorphisms at other codons and the haplotypes of the PRNP gene are highly desirable in the future. Given the complexity of the pathological mechanisms of CWD, fully understanding them is challenging; therefore, we focused on a single factor (G96S SNP) in this study. However, a comprehensive analysis involving other codons will be necessary in the future to fully capture the phenomenon in real-world settings.
In conclusion, this study is the first meta-analysis to quantitatively assess the G96S SNP’s association with CWD susceptibility in white-tailed deer.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by a Research Grant of Andong National University.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
S-YW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. Y-CK: Conceptualization, Funding acquisition, Investigation, Project administration, Software, Visualization, Writing – original draft, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Johnson RT. Prion diseases. Lancet Neurol. (2005) 4:635–42. doi: 10.1016/S1474-4422(05)70192-7 [DOI] [PubMed] [Google Scholar]
- 2.Imran M, Mahmood S. An overview of human prion diseases. Virol J. (2011) 8:1–9. doi: 10.1186/1743-422X-8-559, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheckel C, Aguzzi A. Prions, prionoids and protein misfolding disorders. Nat Rev Genet. (2018) 19:405–18. doi: 10.1038/s41576-018-0011-4, PMID: [DOI] [PubMed] [Google Scholar]
- 4.Jackson GS, Clarke AR. Mammalian prion proteins. Curr Opin Struct Biol. (2000) 10:69–74. doi: 10.1016/s0959-440x(99)00051-2 [DOI] [PubMed] [Google Scholar]
- 5.Sigurdson C. A prion disease of cervids: chronic wasting disease. Vet Res. (2008) 39:41. doi: 10.1051/vetres:2008018 [DOI] [PubMed] [Google Scholar]
- 6.Ironside JW, Ritchie DL, Head MW. Prion diseases. Handb Clin Neurol. (2018) 145:393–403. doi: 10.1016/B978-0-12-802395-2.00028-6 [DOI] [PubMed] [Google Scholar]
- 7.Cobb NJ, Surewicz WK. Prion diseases and their biochemical mechanisms. Biochemistry. (2009) 48:2574–85. doi: 10.1021/bi900108v, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mead S, Lloyd S, Collinge J. Genetic factors in mammalian prion diseases. Annu Rev Genet. (2019) 53:117–47. doi: 10.1146/annurev-genet-120213-092352 [DOI] [PubMed] [Google Scholar]
- 9.Takada LT, Kim MO, Cleveland RW, Wong K, Forner SA, Gala II, et al. Genetic prion disease: experience of a rapidly progressive dementia center in the United States and a review of the literature. Am J Med Genet B Neuropsychiatr Genet. (2017) 174:36–69. doi: 10.1002/ajmg.b.32505, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YC, Jeong BH. The first Meta-analysis of the M129V single-nucleotide polymorphism (SNP) of the prion protein gene (PRNP) with sporadic Creutzfeldt–Jakob disease. Cells. (2021) 10:3132. doi: 10.3390/cells10113132, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asante EA, Smidak M, Grimshaw A, Houghton R, Tomlinson A, Jeelani A, et al. A naturally occurring variant of the human prion protein completely prevents prion disease. Nature. (2015) 522:478–81. doi: 10.1038/nature14510, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenlee JJ. Update on classical and atypical scrapie in sheep and goats. Vet Pathol. (2019) 56:6–16. doi: 10.1177/0300985818794247, PMID: [DOI] [PubMed] [Google Scholar]
- 13.Torricelli M, Sebastiani C, Ciullo M, Ceccobelli S, Chiappini B, Vaccari G, et al. PRNP polymorphisms in eight local goat populations/breeds from central and southern Italy. Animals. (2021) 11:333. doi: 10.3390/ani11020333, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SK, Kim YC, Won SY, Jeong BH. Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Sci Rep. (2019) 9:15293. doi: 10.1038/s41598-019-51621-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arifin MI, Hannaoui S, Chang SC, Thapa S, Schatzl HM, Gilch S. Cervid prion protein polymorphisms: role in chronic wasting disease pathogenesis. Int J Mol Sci. (2021) 22:2271. doi: 10.3390/ijms22052271, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roh IS, Kim YC, Won SY, Park KJ, Park HC, Hwang JY, et al. Association study of the M132L single nucleotide polymorphism with susceptibility to chronic wasting disease in korean elk: a meta-analysis. Front Vet Sci. (2022) 8:804325. doi: 10.3389/fvets.2021.804325, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roh IS, Kim YC, Won SY, Jeong MJ, Park KJ, Park HC, et al. First report of a strong association between genetic polymorphisms of the prion protein gene (PRNP) and susceptibility to chronic wasting disease in sika deer (Cervus nippon). Transbound Emerg Dis. (2022) 69:e2073–83. doi: 10.1111/tbed.14543 [DOI] [PubMed] [Google Scholar]
- 18.Johnson C, Johnson J, Clayton M, McKenzie D, Aiken J. Prion protein gene heterogeneity in free-ranging white-tailed deer within the chronic wasting disease affected region of Wisconsin. J Wildl Dis. (2003) 39:576–81. doi: 10.7589/0090-3558-39.3.576, PMID: [DOI] [PubMed] [Google Scholar]
- 19.O'Rourke KI, Spraker TR, Hamburg LK, Besser TE, Brayton KA, Knowles DP. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J Gen Virol. (2004) 85:1339–46. doi: 10.1099/vir.0.79785-0 [DOI] [PubMed] [Google Scholar]
- 20.Johnson C, Johnson J, Vanderloo JP, Keane D, Aiken JM, McKenzie D. Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J Gen Virol. (2006) 87:2109–14. doi: 10.1099/vir.0.81615-0, PMID: [DOI] [PubMed] [Google Scholar]
- 21.Keane DP, Barr DJ, Bochsler PN, Hall SM, Gidlewski T, O'Rourke KI, et al. Chronic wasting disease in a Wisconsin white-tailed deer farm. J Vet Diagn Invest. (2008) 20:698–703. doi: 10.1177/104063870802000534, PMID: [DOI] [PubMed] [Google Scholar]
- 22.Kelly AC, Mateus-Pinilla NE, Diffendorfer J, Jewell E, Ruiz MO, Killefer J, et al. Prion sequence polymorphisms and chronic wasting disease resistance in Illinois white-tailed deer (Odocoileus virginianus). Prion. (2008) 2:28–36. doi: 10.4161/pri.2.1.6321, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson GA, Nakada SM, Bollinger TK, Pybus MJ, Merrill EH, Coltman DW. Polymorphisms at the PRNP gene influence susceptibility to chronic wasting disease in two species of deer (Odocoileus Spp.) in western Canada. J Toxicol Environ Health A. (2009) 72:1025–1029. doi: 10.1080/15287390903084264, PMID: [DOI] [PubMed] [Google Scholar]
- 24.Blanchong JA, Heisey DM, Scribner KT, Libants SV, Johnson C, Aiken JM, et al. Genetic susceptibility to chronic wasting disease in free-ranging white-tailed deer: complement component C1q and Prnp polymorphisms. Infect Genet Evol. (2009) 9:1329–35. doi: 10.1016/j.meegid.2009.08.010, PMID: [DOI] [PubMed] [Google Scholar]
- 25.Brandt AL, Kelly AC, Green ML, Shelton P, Novakofski J, Mateus-Pinilla NE. Prion protein gene sequence and chronic wasting disease susceptibility in white-tailed deer (Odocoileus virginianus). Prion. (2015) 9:449–62. doi: 10.1080/19336896.2015.1115179, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zink RM, Najar N, Vázquez-Miranda H, Buchanan BL, Loy D, Brodersen BW. Geographic variation in the PRNP gene and its promoter, and their relationship to chronic wasting disease in North American deer. Prion. (2020) 14:185–92. doi: 10.1080/19336896.2020.1796250, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott-Conn CN, Blanchong JA, Larson WA. Prion protein polymorphisms in Michigan white-tailed deer (Odocoileus virginianus). Prion. (2021) 15:183–90. doi: 10.1080/19336896.2021.1990628, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuznetsova A, McKenzie D, Banser P, Siddique T, Aiken JM. Potential role of soil properties in the spread of CWD in western Canada. Prion. (2014) 8:92–9. doi: 10.4161/pri.28467, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jennelle CS, Samuel MD, Nolden CA, Keane DP, Barr DJ, Johnson C, et al. Surveillance for transmissible spongiform encephalopathy in scavengers of white-tailed deer carcasses in the chronic wasting disease area of Wisconsin. J Toxicol Environ Health A. (2009) 72:1018–24. doi: 10.1080/15287390903084249 [DOI] [PubMed] [Google Scholar]
- 30.Schultze ML, Horn-Delzer A, Glaser L, Hamberg A, Zellner D, Wolf TM, et al. Herd-level risk factors associated with chronic wasting disease-positive herd status in Minnesota, Pennsylvania, and Wisconsin cervid herds. Prev Vet Med. (2023) 218:106000. doi: 10.1016/j.prevetmed.2023.106000, PMID: [DOI] [PubMed] [Google Scholar]
- 31.Dudas S, Czub S. Atypical BSE: current knowledge and knowledge gaps. Food Saf. (2017) 5:10–3. doi: 10.14252/foodsafetyfscj.2016028, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C, Dong X-P. Epidemiological characteristics of human prion diseases. Infect Dis Poverty. (2016) 5:47. doi: 10.1186/s40249-016-0143-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seabury CM, Lockwood MA, Nichols TA. Genotype by environment interactions for chronic wasting disease in farmed US white-tailed deer. G3. (2022) 12:jkac109. doi: 10.1093/g3journal/jkac109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Appleby BS, Shetty S, Elkasaby M. Genetic aspects of human prion diseases. Front Neurol. (2022) 13:1003056. doi: 10.3389/fneur.2022.1003056, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi A, Teruya K, Matsuura Y, Shirai T, Nakamura Y, Yamada M, et al. The influence of PRNP polymorphisms on human prion disease susceptibility: an update. Acta Neuropathol. (2015) 130:159–70. doi: 10.1007/s00401-015-1447-7 [DOI] [PubMed] [Google Scholar]
- 36.Moore SJ, Vrentas CE, Hwang S, West Greenlee MH, Nicholson EM, Greenlee JJ. Pathologic and biochemical characterization of PrPSc from elk with PRNP polymorphisms at codon 132 after experimental infection with the chronic wasting disease agent. BMC Vet Res. (2018) 14:80. doi: 10.1186/s12917-018-1400-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hannaoui S, Triscott E, Duque Velásquez C, Chang SC, Arifin MI, Zemlyankina I, et al. New and distinct chronic wasting disease strains associated with cervid polymorphism at codon 116 of the Prnp gene. PLoS Pathog. (2021) 17:e1009795. doi: 10.1371/journal.ppat.1009795, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.