Abstract
The role of valine aminoacylation of the two genomic RNAs of Peanut clump virus (PCV) was studied by comparing the amplification in vivo of RNAs with GAC, GΔC, or CCA anticodons in the tRNA-like structure (TLS) present at the 3′ end of each viral RNA. The PCV RNA1 TLS of isolate PCV2 possesses a GAC anticodon and is capable of highly efficient valylation, whereas the RNA2 TLS has a GΔC anticodon that does not support valylation. The presence in RNA1 of GΔC or CCA anticodons that conferred nonvalylatability resulted in about 2- to 4-fold and a 14- to 24-fold reduction, respectively, in RNA accumulations in tobacco BY-2 protoplasts inoculated with the RNA1 variants together with wild-type RNA2(GΔC). No differences in RNA levels were observed among protoplasts inoculated with the three variant RNA2s in the presence of wild-type RNA1(GAC). All combinations of valylatable and nonvalylatable RNAs 1 and 2 were similarly infectious in Nicotiana benthamiana plants, and viral RNAs accumulated to similar levels; all input TLS sequences were present unchanged in apical leaves. In direct competition experiments in N. benthamiana plants, however, both RNA1 and RNA2 with GAC valylatable anticodons outcompeted the nonvalylatable variants. We conclude that valylation provides a small but significant replicational advantage to both PCV RNAs. Sequence analysis of the TLS from RNA2 of a second PCV isolate, PO2A, revealed the presence of an intact GAC valine anticodon, suggesting that the differential valylation of the genomic RNAs of isolate PCV2 is not a general characteristic of PCV.
The recent sequencing of the genomes of several fungus-transmitted rod-shaped viruses (furoviruses and furo-like viruses) has revealed members of the Pecluvirus, Furovirus, and Pomovirus genera to possess genomic RNAs with a tRNA-like structure (TLS) at the 3′ end of the 3′-untranslated region (7, 17). All of these TLSs can be specifically and efficiently aminoacylated with valine and thus are functionally related in their in vitro properties to the TLSs of tymoviruses (7). The TLSs of the furoviruses and pomoviruses are indeed closely related in structure to the TLSs of Turnip yellow mosaic virus (TYMV) and other tymoviruses, while the TLSs of Peanut clump and Indian peanut clump pecluvirus RNAs have a 42-nucleotide (nt) insert between the aminoacyl acceptor/TΨ and anticodon/ D halves of a TYMV-like TLS (see Fig. 1) (7). A unique aspect of the TLSs of the two peanut clump virus genomic RNAs is the association of a valine anticodon and efficient valine acceptance with the TLS of RNA1 but the absence of both from the TLS of RNA2 due to a single nucleotide deletion (see Fig. 1) (7, 8, 12). Such differential aminoacylation properties have not been previously described for a plant virus and suggest the existence of RNA component-specific regulation of some aspect of the virus replication cycle in a way that is sensitive to the aminoacylation status of the RNA.
FIG. 1.
Sequences and proposed secondary structures of TYMV and PCV TLSs. The main structural features of the TLSs are depicted on the TYMV structure: T and D loops analogous to those of tRNAs, the acceptor/T arm pseudoknot indicated by helical segments S1 and S2 and connecting loops L1 and L2, and the major valine identity nucleotides (arrowheads) in the anticodon loop (anticodon underlined), which are recognized by valyl-tRNA synthetase. The nucleotide in reverse shading in the PCV RNA2 TLS is one of the two nucleotides that differ from the PCV RNA2 TLS sequence. The other difference, a single nucleotide deletion in the anticodon of PCV RNA2, is shown with a dash. The stem-loop (5′-SL) structure shown beside the PCV RNA1 TLS is present at the 5′ end of the PCV RNA1 and -2 TLS RNAs used for in vitro valylation assays and is represented by “SL” in each structure. Nucleotides shown in lowercase at the 3′ ends of the PCV RNA1 and -2 TLSs are of nonviral origin, derived from the MluI and HindIII linearization, respectively, of plasmid templates used to make the infectious transcripts used in in vivo replication experiments. Those nucleotides are not present in the SLTLS transcripts used for valylation assays. During plant cell inoculation, the additional nucleotides are thought to be removed by exonuclease action and repair of the 3′ CCA by CCA nucleotidyltransferase or by internal initiation during minus-strand synthesis. The mutations introduced into the RNA1 and RNA2 anticodons are indicated below the structures. TLS nucleotides are numbered from the 3′ A of the viral sequences.
Apart from the tymoviruses and furo-like viruses mentioned above, aminoacylatable TLSs are also found in the genomes of bromoviruses, cucumoviruses, hordeiviruses and tobamoviruses (13). The functions of TLSs in viral biology have been studied in the TYMV and Brome mosaic virus (BMV) systems by investigating the effects of mutations that decrease or effectively abolish the aminoacylation capacity of the viral RNA. Point mutations in the CAC valine anticodon of the genomic RNA of TYMV (a monopartite virus) resulted in parallel losses of in vitro valine binding and replication in protoplasts (18). Because valine identity is strongly centered on the middle and 3′ anticodon nucleotides (4), single or double mutations at these positions decrease valine charging to background levels. Such mutant genomic RNAs are noninfectious to Chinese cabbage plants and replicate to trace levels (≤0.2% of wild type) in protoplasts (18), demonstrating the importance of an intact valine anticodon. Conversion of aminoacylation identity from valine to methionine, in part by mutation of the anticodon, further demonstrated that efficient aminoacylation rather than the presence of a valine anticodon is required for infectivity and amplification in protoplasts (5). Although we have found means to circumvent the requirement for aminoacylation with chimeric TYMV genomes bearing heterologous 3′ termini (7), wild-type TYMV RNA clearly has a strong requirement for efficient aminoacylation.
Experiments with BMV, whose three genomic RNAs have near-identical TLSs that accept tyrosine, have also utilized mutations designed to specifically decrease the RNAs' aminoacylation efficiency (2). However, since the tyrosine identity elements within the BMV TLS have not been mapped, strong point mutations of the type used with TYMV RNA were not available and the role of aminoacylation has not been so clearly identified. The presence of aminoacylation mutations on RNA3, which does not encode essential replication proteins, has little if any influence on RNA amplification (3, 16). On the other hand, marked decreases in viral RNA amplification in protoplasts result when these mutations are present on RNA1 (6; A. L. N. Rao, personal communication) and RNA2 (16), which encode essential RNA replication proteins. These observations suggest that the role of aminoacylation in a multipartite virus can differ among the genomic RNAs. This could be a consequence of the increased debilitation of a replication defect in an RNA component that itself encodes an essential replication protein (an RNA replicated in cis) compared to the same defect in an RNA not encoding an essential protein (an RNA replicated in trans).
Peanut clump virus (PCV) was chosen as an excellent system for further studying the role of aminoacylation in a multipartite virus, with likely differential effects of aminoacylation on RNAs replicated in cis and in trans. Because of the similarity of the PCV TLS to that from TYMV, strong point mutations with known effects on valylation were available. A differential role for aminoacylation of RNA1 and RNA2 in the bipartite PCV genome was suggested by the existence of an intact GAC valine anticodon in RNA1 (8) but an incomplete GΔC anticodon in RNA2 (see Fig. 1) (12). The PCV RNA1 TLS can be valylated as efficiently as the TYMV TLS and tRNAVal, but virtually no valylation was detected for the RNA2 TLS (7). PCV RNA1 (5,897 nt) encodes all the essential viral replication proteins and is able to replicate independently in protoplasts (9). PCV RNA2 (4,503 nt) encodes proteins involved in viral spread through the plant and probably in fungus transmission (9).
In order to address the role of valylation in the bipartite genome of PCV, we have compared the amplification of viral RNAs in protoplasts and plants after inoculation with valylation-proficient and -deficient variants of both genomic RNAs. We find that valylatability is not essential for infectivity, although valylatable RNAs 1 and 2 are able to outcompete nonvalylatable RNAs during virus amplification in plants. These results differ sharply from the effects of similar mutations in TYMV RNA. Our results also indicate that the differential valylatability of PCV RNAs 1 and 2 does not serve a critical regulatory function in plants and is not characteristic of all Pecluvirus isolates.
MATERIALS AND METHODS
Preparation of wild-type and mutant genomic RNAs.
A 380-bp fragment containing the 3′ TLS of PCV RNA1 was subcloned from pPC1, which contains full-length infectious RNA1 sequences (9), into the phagemid pLITMUS39 (New England Biolabs) using NheI and MluI restriction enzymes. The corresponding 358-bp RNA2 fragment from pPC2 (9) was subcloned using SpeI and HindIII. Single-stranded deoxyuridine-containing encapsidated DNA was prepared using M13K07 helper phage, and oligonucleotide-directed mutagenesis was performed as described before (11). The following mutagenic deoxyoligomers, with anticodon sequences underlined, were used (note that the RNA1 and RNA2 sequences were cloned in opposite orientations): AAGTGTCGTGCTTGACACTCCCGA and AAGTGTCGTTGGTTGACACTCCCGA for mutation of the RNA1 TLS to GΔC and CCA anticodons, respectively; GAGTGTCAAGACACGACACTTAGTGGCT and GAGTGTCAACCAACGACACTTAGTGGCT for mutation of the RNA2 TLS to GAC and CCA anticodons, respectively. After sequencing of the entire subcloned fragments, each mutated sequence was returned to the full-length genomic clones using the restriction sites mentioned above.
After linearization of pPC1 and its derivatives with MluI, and pPC2 and its derivatives with HindIII, 5′-capped genomic transcripts were made using T7 RNA polymerase (9).
Aminoacylation analysis.
To test the valine acceptance after mutation of the anticodon, 136- or 135-nt-long RNAs comprising the TLS adjacent to a heterologous stem-loop (PCV1-SLTLS and PCV2-SLTLS and their variants; see Fig. 1) were prepared by T7 transcription from PCR-generated template DNA as described previously (7). Valine acceptance was assessed by incubation with partially purified wheat germ valyl-tRNA synthetase in TM buffer (25 mM Tris-HCl [pH 8.0], 2 mM MgCl2, 1 mM ATP, and 0.1 mM spermidine) and 10 μM [3H]valine at 30°C (7).
Inoculation and analysis of protoplasts and plants.
Protoplasts were prepared from Nicotiana tabacum suspension cell line BY-2 (10), and 106 protoplasts were inoculated by electroporation with 5 μg each of capped RNA1 and RNA2 transcripts as described before (9). Total nucleic acids were extracted 48 h postinoculation.
Nicotiana benthamiana plants were grown to two or three fully expanded leaves, and each plant was inoculated with 200 μl containing 5 μg each of capped RNA1 and RNA2 transcripts in 5 mM sodium phosphate (pH 7.5) and 0.03% macaloid.
At 11 days postinoculation (dpi), apical and inoculated leaves were harvested and frozen in liquid nitrogen. RNA was extracted from 200 mg of frozen, ground leaf powder. To prepare total RNA, the powder was extracted with 600 μl of ice-cold buffer A (200 mM Tris-HCl [pH 9], 400 mM KCl, 35 mM MgCl2, 25 mM EGTA, and 200 mM sucrose), and the recovered RNA was extracted with phenol-chloroform and ethanol precipitated.
Positive- and negative-sense genomic RNAs were detected in Northern blots after electrophoresis through 1% agarose gels. The strand-specific 32P-labeled RNA probes represented the 3′-most 124 nt of RNA1 for detecting the negative strands of both RNA1 and -2 and the complementary sequence for detecting the positive strands of both RNA1 and -2. The riboprobes were made by transcription with T7 RNA polymerase from PCR-generated products. Northern blots were analyzed (9) and quantitated with a phosphorimager (Fuji BAS1000).
Sequence analysis of progeny RNA isolated from infected plants.
Total RNA was extracted from 200 mg of N. benthamiana leaves as described above. The RNAs were 3′ polyadenylated using Escherichia coli poly(A) polymerase (Gibco-BRL), and subjected to reverse transcription-PCR (RT-PCR), using T35GG to prime reverse transcription and as the downstream PCR primer. To amplify RNA1 and RNA2 3′ sequences, the upstream primers AGCGAGAAACTCTGTTGGCT and CATAGCTTTTGCATCCTACTAC, respectively, were used. This resulted in amplification of a 436- or 435-bp product from RNA1 and a 373/372-bp product from RNA2. The specificity of these amplifications was verified with test template mixtures.
Gel-purified PCR products were sequenced by automated fluorescent dye terminator sequencing (model 373, Applied Biosystems) using the primer GCGAGCCATAGAGCACGGTT for both RNA1 and RNA2 products; this oligonucleotide primes 247 or 246 nt from the 3′ terminus.
When RNA from plants inoculated with mixed RNA sequences was analyzed, the relative amounts of the competing sequences were determined by comparing the areas of peaks in the fluorescent dye trace. For competition involving a GΔC anticodon, in which there is a 1-nt shift between the two sequences downstream of the anticodon, three or four well-isolated downstream peaks were used for area comparison. For competition between sequences with GAC and CCA anticodons, the first nucleotide of the anticodon was compared.
Nucleotide sequence accession number.
The 3′-untranslated region of RNA2 from strain PO2A has been deposited in GenBank (accession no. AJ277545).
RESULTS
Valylation mutants of PCV RNAs 1 and 2.
In order to investigate the role of valylation in PCV, mutations were introduced into the anticodons of the RNA1 and RNA2 TLSs, permitting the influence of GAC, GΔC, and CCA anticodons (Fig. 1) to be tested for each genomic RNA. A GAC anticodon, as present in the wild-type sequence of RNA1 of PCV (isolate PCV2; reference 8), results in highly efficient valylation. A GΔC anticodon, present in the wild-type sequence of PCV RNA2 (isolate PCV2; reference 12), lacks the central nucleotide of the anticodon and directs virtually no valylation: the Vmax/Km ratio for valylation of the PCV RNA2 TLS is 7.6 × 10−4 relative to that for the RNA1 TLS (7). A CCA anticodon in the structurally related TYMV TLS results in a similar loss of valylatability (18). This anticodon sequence was chosen because of its strong effect on viral replication in the TYMV system. Inoculation of Chinese cabbage protoplasts with genomic RNA bearing a CCA anticodon led to ≤0.002 times the levels of coat protein accumulated after inoculation with wild-type TYMV RNA, and the mutant RNA was not infectious in plants (18). Interconversion between the GAC, GΔC, and CCA anticodons by mutation during plant inoculation experiments is unlikely in view of the deletion or triple substitutions distinguishing these anticodons.
Before observing the effects of mutated anticodons of the RNAs in planta, the in vitro valylation properties of each mutant RNA were examined. As previously (7), valylation was tested in the context of the short SLTLS RNAs with 3′ CCA termini shown in Fig. 1; these RNAs are transcribed from PCR-amplified templates and have a stable stem-loop at the 5′ end of the TLS that includes sequences conducive to T7 transcription. Both the RNA1 and RNA2 TLSs with a GAC anticodon were efficiently valylated by wheat germ valyl-tRNA synthetase (Fig. 2), confirming the dominant role of the anticodon in specifying valylation. The RNA2 nucleotide substitution in the D stem-loop (Fig. 1) evidently does not interfere with the folding of a TLS conformation able to support efficient valylation.
FIG. 2.
Comparison of the in vitro valylation profiles of PCV RNA1 and -2 TLSs with GAC, GΔC, and CCA anticodons. RNAs comprising the TLS and the additional 5′ stem-loop (SLTLS RNAs as shown in Fig. 1) were valylated with wheat germ valyl-tRNA synthetase in TM buffer. The valylation profiles of SLTLS RNAs 1GΔC, 1CCA, and 2CCA coincided with that of the water control and are omitted from the graph for reasons of clarity.
None of the SLTLS RNAs with GΔC and CCA anticodons showed valylation above background (Fig. 2), consistent with previous experience with PCV RNA2 SLTLS (GΔC anticodon) (7) and TYMV RNA mutants (CCA anticodon) (18).
RNA accumulation in protoplasts is affected by anticodon mutations in RNA1 but not in RNA2.
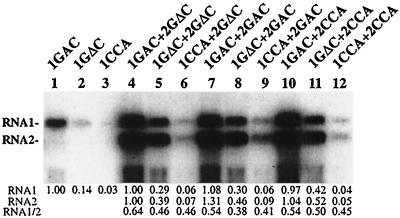
BY-2 tobacco protoplasts were inoculated with all combinations of the valylatable and nonvalylatable capped genomic transcripts. Since RNA1 is able to replicate independently in protoplasts, replication of the RNA1 mutants was also studied without RNA2 coinoculation. Total RNA was extracted from protoplasts 48 h postinoculation, and RNA1 and RNA2 were detected by Northern blot hybridization using a 124-nt riboprobe complementary to the PCV TLSs. The use of a probe that hybridizes equally to both genomic RNAs enabled a direct comparison of RNA1 and RNA2 levels.
When RNA1 was inoculated alone (Fig. 3, lanes 1 to 3) or when RNAs 1 and 2 were coinoculated (Fig. 3, lanes 4 to 12), the viral RNA accumulations were determined by the anticodon sequence of RNA1, decreasing in the order GAC>GΔC>CCA. Thus, inoculations with the nonvalylatable RNA1 mutants resulted in lower levels of RNA accumulation than inoculations with the valylatable RNA1 (compare lanes 1, 4, 7, and 10 with the rest). Interestingly, RNA1 + RNA2 inoculations, including RNA 1CCA, resulted in substantially lower viral RNA accumulations (a 14- to 24-fold reduction relative to RNA 1GAC) than inoculations including RNA 1GΔC (a 2.0- to 3.6-fold reduction) (Fig. 3, lanes 4 to 12). When RNA1 was inoculated alone, the nonvalylatable mutants (especially RNA 1GΔC) accumulated to even lower levels relative to RNA 1GAC (Fig. 3, lanes 1 to 3). Perhaps the encapsidation function provided by RNA2 plays a role in preventing the degradation of genomic RNA, especially when the levels are subnormal.
FIG. 3.
Northern blot analysis of PCV RNAs with GAC, GΔC, and CCA anticodons amplified in tobacco BY-2 protoplasts. Protoplasts (106) were inoculated with the RNA1 variants or with all possible combinations of RNA1 and -2 transcripts as indicated above each lane. At 48 h postinfection, total RNAs were extracted and separated on a 1% agarose gel, and the viral RNAs were detected by Northern hybridization using a riboprobe complementary to the 3′-most 124 nt, whose sequences are almost identical in RNA1 and 2. The positions of RNA1 and -2 are indicated at the left. The relative accumulations of RNA1 and RNA2, as well as the RNA1/RNA2 ratios, are shown beneath each lane (averages of three independent inoculations).
In contrast to the RNA1 mutations, there was no apparent influence of the RNA2 anticodon sequence on the accumulation of viral RNAs in protoplasts. Similar accumulations were observed for all three RNA1 + RNA2 inoculations involving RNA 1GAC (Fig. 3, lanes 4, 7, and 10), RNA 1GΔC (Fig. 3, lanes 5, 8, and 11), or RNA 1CCA (Fig. 3, lanes 6, 9, and 12). Thus, the role of valylation appears to differ between RNA1 and RNA2, reminiscent of the differential effects of aminoacylation mutations in the three BMV RNAs (6).
The ratio of RNA1 to RNA2 did not vary significantly (95% confidence) among the permuted RNA1 + RNA2 inoculations, although this ratio tended to be higher in infections with RNA 1GAC than in infections with the other RNA1 variants (Fig. 3, lanes 4, 7, and 10; cf. lanes 5 and 6, 8 and 9, and 11 and 12). The valylatability of the PCV RNAs is clearly not an important determinant of the RNA1/RNA2 ratio. Likewise, no differences in the ratios of minus to plus strands were observed for either RNA1 or RNA2 among the various inoculations (data not shown). In summary, the protoplast inoculations indicate that the valylatability of RNA1 enhances the overall amplification of PCV RNAs in BY-2 cells, with no apparent specific effects on plus or minus RNA1 or RNA2 accumulation.
All anticodon mutants are similarly infectious in whole plants.
N. benthamiana plants, systemic hosts for PCV, were inoculated with the same combinations of RNA1 + RNA2 used above. Surprisingly, in view of the protoplast results, symptoms developed at similar times (6 to 8 dpi) and to similar severities in all inoculated plants. At 11 dpi, total RNA was extracted from apical leaves, and the viral genomic RNAs were detected by Northern blotting (Fig. 4). Similar levels of viral RNA were present in all plants, with at most only a slight reduction in RNA1 accumulations for infections including nonvalylatable RNA1 relative to those with RNA 1GAC (Fig. 4, lanes 2 and 3, 5 and 6, and 8 and 9; cf. lanes 1, 4, and 7). No systematic differences were observed in RNA2 accumulations among the various infections. Note that, for unknown reasons, RNA1/RNA2 ratios are typically lower in whole plant extracts than in protoplast extracts (9).
FIG. 4.
Northern blot analysis of PCV RNAs with GAC, GΔC, and CCA anticodons amplified in N. benthamiana plants. The indicated RNA1 + RNA2 combinations (as in Fig. 3) were inoculated onto N. benthamiana plants. Total RNAs were extracted at 11 dpi and separated on a 1% agarose gel, and the positive strands of the viral RNA genome were detected by Northern hybridization as in Fig. 3. The positions of RNAs 1 and 2 are indicated at left. The relative accumulations of RNA1 and RNA2, as well as the RNA1/RNA2 ratios, are shown beneath each lane.
To determine whether any mutations were introduced into the TLSs of the viral RNAs amplified in plants, the sequences of the 3′-terminal 234 nt (233 nt for GΔC RNAs) were derived by sequencing PCR products amplified after polyadenylation and reverse transcription. RNA1 and RNA2 sequences were specifically amplified by this procedure. All amplified RNA1 and RNA2 sequences perfectly matched the inoculated sequences, both in the anticodon and throughout the analyzed 3′ region (data not shown). These results clearly demonstrate that the valylation properties do not measurably affect PCV infectivity and the amplification of viral RNAs in N. benthamiana plants. The reasons for the differential effects produced by inoculation of protoplasts and plants with nonvalylatable RNA1 are not clear.
Competition experiments in plants indicate an advantage for valylation of both RNA1 and RNA2.
In order to explore further a possible role of valylation in N. benthamiana plants and the difference from the results obtained with protoplasts, coinoculations involving two RNA1 or two RNA2 variants (together with the other wild-type genomic RNA) were conducted, permitting direct competition between the two genotypes to occur in planta. At 11 dpi, both apical and inoculated leaves were collected separately for total RNA extraction and 3′ sequencing of RNA1 and RNA2 as described above. Mixed sequences involving the GΔC anticodon were readily evident in the fluorescent dye trace generated by the automated sequencer, with a clear transition from a single sequence to a mixed sequence occurring at the anticodon (Fig. 5). By sequencing known mixtures of GAC and GΔC or CCA and GΔC DNAs in the ratios 5:1, 2:1, and 1:1, we verified that isolated nucleotides downstream of the anticodon could be used to estimate the allele ratio by determining the extent of peak splitting into a doublet; the relative peak areas of each peak in the doublet were proportional to the input GAC/GΔC ratios. Thus, three or four peaks in the fluorescence profile (Fig. 5), which became doublets when both 3-nt (GAC or CCA) and 2-nt (GΔC) anticodons were present, were used to estimate the ratio of alleles present in the viral RNAs extracted from the plants. The identification of mixed sequences containing GAC and CCA anticodons relied on the sequence of the anticodon triplet alone. Test mixtures verified that both alleles in a mixture with a GAC/CCA ratio of 1:1 could be detected.
FIG. 5.
Sequence profile of the RNA1 from apical leaves of the mixed inoculations of wild-type RNA 1GAC and RNA 1GΔC. The sequence profile was obtained from the RT-PCR products whose template RNAs had been extracted from the inoculated and apical leaves of an N. benthamiana plant 11 dpi with an equal amount of the 5′-capped RNA 1GAC and RNA 1GΔC transcripts. No mixed nucleotide peak was observed in the profile upstream of the second nucleotide of the anticodon (boxed, upper panel), where the signal of A overlaps with that of C. The 1-nt-shifted peaks (examples of such red and blue doublets are indicated on the right of the upper panel) were evident throughout the remaining viral sequence. The doublet peak areas representing G6, C14, C19, and T24 shown in the lower panel (refer to Fig. 1) were measured and used to estimate the ratio of wild-type to mutant RNA in the extract.
Analysis of the progeny from inoculations with RNAs 1GAC+1GΔC+2GΔC and RNAs 1GAC+1CCA+2GΔC revealed the valylatable RNA 1GAC to be at a competitive advantage over nonvalylatable RNA1 (Table 1). The advantage over RNA 1GΔC was very slight, with a 1GAC/1GΔC ratio of 1.4 in inoculated leaves and 2.4 in apical leaves. The increased dominance of RNA 1GAC in apical compared with inoculated leaves suggests that true selection is occurring, rather than that the 1GAC/1GΔC ratio is perhaps being determined by a slight excess of RNA 1GAC in the inoculum. Marked preference for the amplification of RNA 1GAC over RNA 1CCA was observed, however, with selective amplification of RNA 1GAC in inoculated leaves and no RNA 1CCA evident in apical leaves. In both cases, the RNA2 sequences were verified to be unchanged for the inoculated GΔC anticodon. These experiments have revealed the same GAC>GΔC>CCA ranking of amplification capacity among RNA1 variants as observed in protoplasts.
TABLE 1.
Direct competition between RNA1 and RNA2 variants with different anticodon sequences in N. benthamiana plantsa
| RNA in competitionb | Anticodon sequence
|
RNA ratio (WT/mutant)
|
||
|---|---|---|---|---|
| WT | Mutant | Inoculated leaves | Apical leaves | |
| RNA1 | 1GAC | 1GΔC | 1.4 | 2.4 |
| 1GAC | 1CCA | 8.1 | Only GACc | |
| RNA2 | 2GΔC | 2GAC | 0.14 | Only GACc |
| 2GΔC | 2CCA | 0.70 | 0.45 | |
RNAs were extracted 11 dpi, polyadenylated, and subjected to RT-PCR, amplifying the 3′ 234 nt, which were sequenced by the fluorescent dye terminator method (see Fig. 5). The ratios of anticodon sequences were determined as described in the text. WT, wild type.
The indicated variants of RNA1 were coinoculated with wild-type RNA2 (GΔC). The indicated variants of RNA2 were coinoculated with wild-type RNA1 (GAC).
The detection limit for a minor sequence is about 10% of the predominant sequence.
Competition between RNA 2GΔC and RNA 2GAC or RNA 2CCA (presence of RNA 1GAC) was also monitored. RNA 2GAC was strongly favored over the wild-type RNA 2GΔC, which accumulated seven times less than RNA 2GAC in inoculated leaves and was not detected in apical leaves (Table 1). This result clearly indicates that RNA 2GAC has a replicative advantage over the natural RNA 2GΔC of PCV isolate PCV2. After coinoculation of the two nonvalylatable variants of RNA2 (2GΔC and 2CCA), RNA 2CCA was moderately more abundant than RNA 2GΔC in both inoculated and apical leaves (Table 1).
No mutations in the TLS outside the anticodon were observed in any of the progeny RNAs isolated from plants.
A GAC anticodon is present in the RNA2 TLS of PCV isolate PO2A.
The indication from our competition experiments that valylatability of RNA2 should be a selection criterion in the evolution and maintenance of PCV suggests that isolate PCV2 is not an optimized strain. In order to see whether RNA2 in another PCV strain has the expected GAC anticodon, we sequenced the 3′-untranslated region of RNA2 from strain PO2A. The origin of strain PO2A has been described by Manohar et al. (12). The only difference from RNA2 of strain PCV2 in the 3′ 200 nt (i.e., including the entire TLS) is the presence in strain PO2A of a GAC instead of a GΔC anticodon. This result makes it clear that there is no fundamental requirement for a GΔC anticodon in the RNA2 TLS of PCV.
DISCUSSION
Protoplast and plant experiments with valylatable and nonvalylatable PCV RNAs 1 and 2 have clearly demonstrated that the GAC valine anticodon and the capacity for efficient valylation are not essential properties for the amplification of either RNA1 or RNA2, nor for PCV infectivity in general. This result contrasts sharply with results from the TYMV system, in which the loss of valylatability through mutation of the valine anticodon—including mutation to the same CCA anticodon as studied in this paper with PCV—abrogated infectivity (18). Nevertheless, while nonvalylatable PCV mutants supported efficient infections in plants (Fig. 4), definite replicational advantages were observed for the valylatable RNA1 (RNA 1GAC) over the two nonvalylatable variants, both in N. tabacum BY-2 protoplasts (Fig. 3) and in competition experiments in N. benthamiana plants (Fig. 5 and Table 1). A similar replicational advantage of the RNA 2GAC allele was also observed in the plant competition experiments (Table 1), even though the natural sequence of RNA2 of the PCV2 isolate of PCV is a GΔC anticodon (12). Differential effects were observed between the GΔC and CCA mutations in both protoplasts (Fig. 3) and in the whole plant competition experiments (Table 1), but the reasons for these differences are not understood.
We conclude that an intact valine anticodon and the resultant valylatability are mildly advantageous attributes to both of the PCV genomic RNAs, perhaps more so for RNA1, although the protoplast and whole plant experiments differ on this point, for unknown reasons that may be related to the specific host cell. Our protoplast results indicating a greater role for valylation in RNA1 than in RNA2 are reminiscent of results from the BMV system, in which nonaminoacylation mutations are more critical in RNA1 and in RNA2 than in RNA3 (6). BMV RNA1 and PCV RNA1 both encode essential replication proteins whose action may be at least partially cis-limited, acting preferentially (presumably at the earliest stages of an infection) on the replication of RNA1 itself. Aminoacylation may play a role in this process and so be more crucial for those RNAs of a multipartite virus that are replicated in cis than for those replicated in trans (BMV RNA3 and PCV RNA2). There is no information on what mechanistic role valylation plays in the replication of PCV, nor on whether that role is similar to, though less crucial than, the postulated role of valylation in the regulated suppression of minus-strand synthesis in the TYMV system (1).
Curiosity about the significance of the differential valylation of RNA1 and -2 from the PCV2 isolate was a major incentive in conducting these studies. Protoplast inoculations failed to detect any effect of RNA valylatability on the accumulation of positive- relative to negative-sense RNA1 or RNA2, and thus on RNA1/RNA2 ratios (Fig. 3 and data not shown). We had imagined that differential valylatability might have evolved because of an involvement in the fine regulation of viral RNA synthesis. However, the plant competition experiments make it clear that a valylatable RNA2 with a GAC anticodon is at a competitive advantage over the GΔC anticodon of RNA2 of isolate PCV2, at least in N. benthamiana plants. In this regard, the sequences of other PCV or Indian PCV isolates should be informative. Indeed, we find that RNA2 from PCV isolate PO2A possesses a GAC instead of GΔC anticodon. The same is true of RNA2 from the closely related Indian peanut clump virus, L serotype (15), whose RNA1 contains a valylatable TLS very similar to that of PCV RNA (7, 14).
It thus appears that a GΔC anticodon has become fixed in RNA2 of the PCV2 isolate by a replicational error and that differential valylation is not an important biological trait nor a defining characteristic of the Pecluviridae. The survival of this mutation is indicative of the minor role played by valylation in the PCV system, especially in the case of RNA2. Nevertheless, sufficient advantage is conferred during infections in N. benthamiana plants for the selection of RNAs 1 and 2 with intact GAC valine anticodons that are capable of efficient valylation. Further studies are needed to determine whether this selective advantage is mechanistically related to the crucial role of valylation in TYMV.
ACKNOWLEDGMENTS
D. Matsuda and P. Dunoyer contributed equally to these studies.
We thank the Central Services Facility of the Oregon State University Center for Gene Research for DNA sequencing.
These studies were supported by NIH grant GM-54610 (T.W.D.).
Footnotes
Technical report no. 11655 of the Oregon Agricultural Experiment Station.
REFERENCES
- 1.Dreher T W. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu Rev Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- 2.Dreher T W, Hall T C. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA. Sequence and structural requirements for aminoacylation and 3′-adenylation. J Mol Biol. 1988;201:41–55. doi: 10.1016/0022-2836(88)90437-8. [DOI] [PubMed] [Google Scholar]
- 3.Dreher T W, Rao A L N, Hall T C. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J Mol Biol. 1989;206:425–438. doi: 10.1016/0022-2836(89)90491-9. [DOI] [PubMed] [Google Scholar]
- 4.Dreher T W, Tsai C H, Florentz C, Giege R. Specific valylation of turnip yellow mosaic virus RNA by wheat germ valyl-tRNA synthetase determined by three anticodon loop nucleotides. Biochemistry. 1992;31:9183–9189. doi: 10.1021/bi00153a010. [DOI] [PubMed] [Google Scholar]
- 5.Dreher T W, Tsai C H, Skuzeski J M. Aminoacylation identity switch of turnip yellow mosaic virus RNA from valine to methionine results in an infectious virus. Proc Natl Acad Sci USA. 1996;93:12212–12216. doi: 10.1073/pnas.93.22.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duggal R, Lahser F C, Hall T C. Cis-acting sequences in the replication of plant viruses with plus-sense RNA genomes. Annu Rev Phytopathol. 1994;32:287–309. [Google Scholar]
- 7.Goodwin J B, Dreher T W. Transfer RNA mimicry in a new group of positive-strand RNA plant viruses, the furoviruses: differential aminoacylation between the RNA components of one genome. Virology. 1998;246:170–178. doi: 10.1006/viro.1998.9193. [DOI] [PubMed] [Google Scholar]
- 8.Herzog E, Guilley H, Manohar S K, Dollet M, Richards K, Fritsch C, Jonard G. Complete nucleotide sequence of peanut clump virus RNA 1 and relationships with other fungus-transmitted rod-shaped viruses. J Gen Virol. 1994;75:3147–3155. doi: 10.1099/0022-1317-75-11-3147. [DOI] [PubMed] [Google Scholar]
- 9.Herzog E, Hemmer O, Hauser S, Meyer G, Bouzoubaa S, Fritsch C. Identification of genes involved in replication and movement of peanut clump virus. Virology. 1998;248:312–322. doi: 10.1006/viro.1998.9287. [DOI] [PubMed] [Google Scholar]
- 10.Koiwai A, Noguchi M, Tamaki E. Changes in the amino acid composition of tobacco cells in suspension culture. Phytochemistry. 1971;10:561–566. [Google Scholar]
- 11.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 12.Manohar S K, Guilley H, Dollet M, Richards K, Jonard G. Nucleotide sequence and genetic organization of peanut clump virus RNA 2 and partial characterization of deleted forms. Virology. 1993;195:33–41. doi: 10.1006/viro.1993.1343. [DOI] [PubMed] [Google Scholar]
- 13.Mans R M, Pleij C W, Bosch L. tRNA-like structures. Structure, function and evolutionary significance. Eur J Biochem. 1991;201:303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- 14.Miller J S, Wesley S V, Naidu R A, Reddy D V, Mayo M A. The nucleotide sequence of RNA-1 of Indian peanut clump furovirus. Arch Virol. 1996;141:2301–2312. doi: 10.1007/BF01718632. [DOI] [PubMed] [Google Scholar]
- 15.Naidu, R. A., J. S. Miller, M. A. Mayo, S. V. Wesley, and A. S. Reddy. The nucleotide sequence of Indian peanut clump virus RNA 2: sequence comparisons among pecluviruses. Arch. Virol., in press. [DOI] [PubMed]
- 16.Rao A L N, Hall T C. Interference in trans with brome mosaic virus replication by RNA-2 bearing aminoacylation-deficient mutants. Virology. 1991;180:16–22. doi: 10.1016/0042-6822(91)90004-u. [DOI] [PubMed] [Google Scholar]
- 17.Shirako Y, Wilson T M A. Furoviruses. In: Granoff A, Webster R, editors. Encyclopedia of virology. San Diego, Calif: Academic Press; 1999. pp. 587–596. [Google Scholar]
- 18.Tsai C H, Dreher T W. Turnip yellow mosaic virus RNAs with anticodon loop substitutions that result in decreased valylation fail to replicate efficiently. J Virol. 1991;65:3060–3067. doi: 10.1128/jvi.65.6.3060-3067.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]