Abstract
Spinal muscular atrophy (SMA) is a neurodegenerative disease caused by deletions or mutations of survival of motor neuron 1 (SMN1) gene. To date, the mechanism of selective cell death of motor neurons as a hallmark of SMA is still unclear. The severity of SMA is dependent on the amount of survival motor neuron (SMN) protein, which is an essential and ubiquitously expressed protein involved in various cellular processes including regulation of cytoskeletal dynamics. In this review, we discuss the effect of SMN ablation on cytoskeleton organization including actin dynamics, growth cone formation, axonal stability, neurite outgrowth, microtubule stability, synaptic vesicle dynamics and neurofilament protein release in SMA. We also summarized a list of critical proteins such as profilin-2 (PFN2), plastin-3 (PLS3), stathmin-1 (STMN1), microtubule-associated protein 1B (MAP1B) and neurofilament which play an important role in modulating cytoskeleton in SMA. Our aim is to highlight how cytoskeletal defects contribute to motor neuron degeneration in SMA disease progression and concentrating on cytoskeleton dynamics may be a promising approach to develop new therapy or biomarker.
Keywords: Spinal muscular atrophy, Cytoskeleton defect, Actin, Microtubule, Profilin-2, Plastin-3, Microtubule-associated protein
Introduction
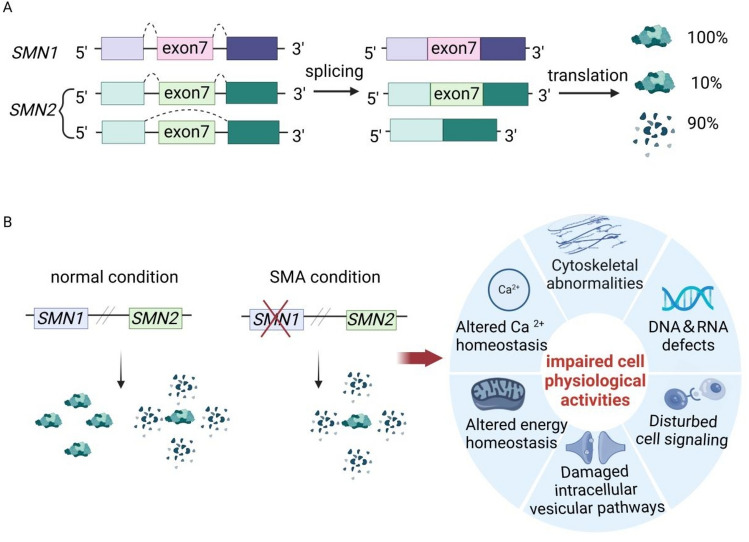
Spinal muscular atrophy (SMA) is an autosomal recessive inheritance disease, characterized by progressive muscle weakness due to the degeneration of spinal α motor neurons. Symmetrical motor function impairment initially happens in proximal skeletal muscles and subsequently spreads to distal muscles [1, 2]. In addition, other tissues in SMA are also affected with the increasing severity of the disease [3]. According to onset time, survival and motor function, SMA is clinically divided into five subtypes from severe type 0 to mild type IV [4, 5]. It is caused by mutations or deletions of survival of motor neuron 1 (SMN1) gene, while the severity of SMA is negatively regulated by the copy number of survival of motor neuron 2 (SMN2) as a paralogous backup gene [6–8]. Despite encoding SMN protein and being highly similar, SMN2 and SMN1 exhibit differences at five bases [9]. One of these differences is C-to-T transition in exon 7 of SMN2 contributing to predominant skipping of exon 7 during pre-mRNA splicing [9–11]. Consequently, 90% SMN2 products are truncated protein called SMNΔ7, whereas 10% transcripts are translated into full-length SMN protein which is insufficient to compensate for the loss of SMN1 [6, 7, 9, 12, 13](Fig. 1A).
Fig. 1.
A Both SMN1 and SMN2 genes are located close together on chromosome 5q13, encoding for SMN protein. Normally, SMN1 transcripts are correctly spliced and translated into a full-length SMN protein, while 90% SMN2 are translated into a truncated and non-functional SMN protein due to the exon 7 skipping caused by alternative splicing. B Under normal condition, the physiological behaviors of healthy individual are supported by the abundant SMN proteins which mainly derive from SMN1 gene. In SMA, the limited SMN protein encoded by SMN2 gene is insufficient to counterbalance the loss of SMN1, impairing various cell physiological activities such as cytoskeleton regulation, Ca2+ and energy homeostasis, intracellular vesicular pathways, cell signaling, RNA metabolism and DNA recombination
Therefore, increasing SMN level is a mainstream treatment for SMA, especially concentrating on modulating SMN2 alternative splicing. In 2016, U.S. Food and Drug Administration approved Nusinersen (Spinraza®), an antisense oligonucleotide (ASO) to correct splicing by blocking an intronic silencer in intron 7 of SMN2. In addition, Risdiplam (Evrysdi®) as an SMN2 pre-mRNA splicing modifier was approved to treat SMA by promoting the recruitment of U1 small nuclear ribonucleoprotein particles (U1-snRNPs). Moreover, onasemnogene abeparvovec (Zolgensma®) as a gene therapy for SMA patient delivers a functional copy of human SMN with adeno-associated virus vector [14–19]. Although this therapeutics made impressive strides in extending patient lifespan and improving neurodevelopmental outcome and motor function, current treatments do not allow to thrive and live a normal life for most SMA patients. Defects of motor axon development in SMA begin prenatally, associated with postnatal degeneration of motor neurons [20]. Thus, as suggested in clinic trials, the earlier treatment can be initiated, the better to attain stabilization of motor function [21]. However, delays in treatment are widespread and some therapies may be currently limited to a certain age population [22]. Even for the patients treated early, the disease burden remains due to irreversible neuronal cell death [23].
Therefore, to understand the mechanisms of selective cell death of motor neurons as a hallmark of SMA is important for SMA patient treatment and diagnosis. SMN is an essential and ubiquitously protein in all cell and tissue types, not just in motor neurons. SMN protein plays a housekeeping role in the regulation of snRNP biogenesis, as well as intracellular homeostasis including cytoskeletal dynamics, RNA metabolism, DNA recombination, cell signaling, Ca2+ homeostasis, intracellular vesicular pathways, ubiquitin–proteasome pathway and mitochondrial activity (Fig. 1B) [24–27]. Hence, SMN plays a major role in SMA pathology and specific vulnerability to motor neurons in this disease. Due to extraordinary extended axon length and their dependency on the cytoskeleton for its stability, signaling, and axonal transport, spinal motor neurons are particularly susceptible to selective and early degeneration compared with other neurons in SMA [28–30]. Moreover, Sumner et al. identified impaired radial growth and Schwann cell ensheathment of motor axons initiated during embryogenesis and caused reduced acquisition of myelinated axons that impeded motor axon function neonatally in SMA [20]. Cytoskeletal proteins have been demonstrated to fulfill crucial functions in various signaling pathways [31–35]. Accumulated evidence suggest that integrity of cytoskeleton is closely linked to cell death [35–43]. Hence, therapeutic strategies targeting cytoskeletal disorders have the potential to be valuable supplements to current treatment of SMA.
In this review, the altered dynamics of cytoskeleton and its mechanisms in SMA were comprehensively described, which explained how potential modifiers might improve phenotypes of SMA. The aim was to provide new insights into SMA pathogenesis and emerging therapies by summarizing the latest advances in the cytoskeletal role of SMA.
Role of cytoskeletal dynamics in neurons
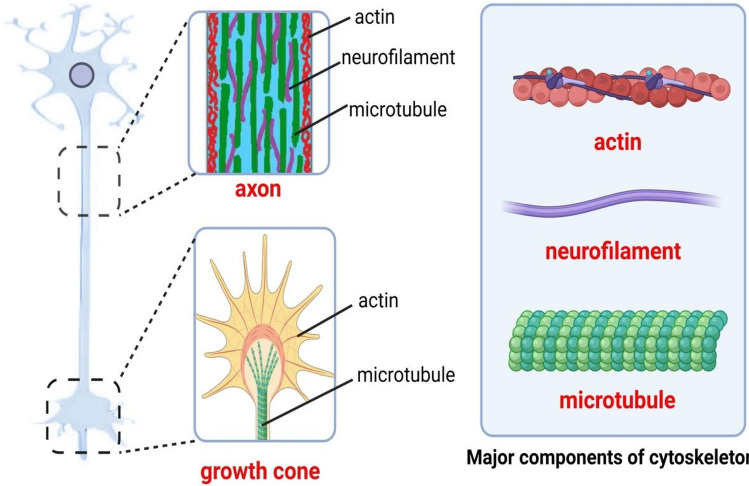
The neuronal cytoskeleton is an adaptive and dynamic structure consisting of three major components: actin-based microfilaments, microtubules and intermediate filaments (neurofilaments) (Fig. 2) [44–47]. Filamentous actin (F-actin) is a heterodimer structure and is composed of α‑ and β‑actin. β‑Actin is known as globular actin (G-actin). Microtubule is a cylinder comprising 13 protofilaments which are heterodimers formed by the alternating arrangement of α‑ and β‑tubulin. Actin and microtubule proteins are indispensable for regulating neuron structure and function by influencing cellular motility processes such as growth cone formation, axonal stability, neurite outgrowth and synaptic vesicle dynamics [44, 48]. Actin cytoskeleton plays a vital role in pathfinding of neuronal axon and control of microtubule dynamics [49–51]. The assembly of microtubule in growth cone promotes the axon extension, influencing neuronal polarization and regeneration [52–54]. Moreover, both actin-based microfilaments and microtubules support the dynamic function and high energy demands of pre- and postsynaptic structures by binding with mitochondria and driving plastic changes [47, 55–57].
Fig. 2.
The neuronal cytoskeleton is an adaptive and dynamic structure consisting of three major components: actin-based microfilaments, microtubules and intermediate filaments (neurofilaments). Actin and microtubule proteins are indispensable for regulating neuron structure and function by influencing cellular motility processes such as growth cone formation, axonal stability, neurite outgrowth and synaptic vesicle dynamics. Neurofilaments are a class of intermediate filaments found primarily in nerve cells, especially in axons. They are essential for the structure of neuronal axons and play a crucial role in maintaining the shape and function of neurons
Therefore, it is perhaps not surprising that cytoskeleton defects may be a common characteristic among neurodegenerative diseases such as Alzheimer’s disease (AD), Huntington’s disease (HD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS) and SMA. In some neurodegenerative diseases, the alteration of cytoskeleton is considered as a potential etiological factor in part [58–60]. However, some cytoskeletal proteins abnormalities may not be causative factors of neurodegenerative disorders because that interventions targeting these proteins fail to rescue the survival of diseased mice [61]. Although the causal link between neurodegenerative diseases and cytoskeletal dysfunctions is unclear, the overwhelming majority of cytoskeleton defects related to neurodegenerative diseases contain the impairments of microtubule stability and actin dynamics [57, 62–69]. In addition, the aggregation of neurofilaments, a class of intermediate filaments found primarily in axons of nerve cells, can be observed in several neurodegenerative diseases [46, 70] and may be related to the hyperphosphorylation of intermediate filaments [46, 70, 71]. In recent years, neurofilament concentrations in biofluids have emerged as a promising clinical biomarker for neurodegeneration [71]. However, compared to actin-based microfilaments and microtubules, the mechanism of neurofilament changes in neurodegenerative diseases remains largely unknown.
SMN protein is not only localized primarily in actin-abundant neurites and growth cones but also regulates the distribution of β-actin within growth cones, which suggests the possible regulatory effects of SMN on cytoskeleton [72–77]. In SMA mouse model, morphological defects of nerve cell, which contributes to neurologic disorder, are often accompanied by cytoskeletal impairments such as destabilization of microtubules and imbalance of F/G-actin levels [45, 78]. In most instances, the depletion of SMN indirectly affects the cytoskeleton through a number of proteins that play a regulatory role in the cytoskeleton. Here, we discuss cytoskeleton-related proteins including PFN2, PLS3 and microtubule-associated proteins (MAPs) that were affected by SMA (Table 1) and highlight the more extensively studied proteins that regulate cytoskeleton in the following sections.
Table 1.
The list of cytoskeleton-related proteins that were affected by SMA
| Protein | Roles in cytoskeleton | Changes in SMA |
|---|---|---|
| PFN2 | Regulates F-actin polymerization by binding with G-actin [82–86] | With the reduction of binding to SMN, available PFN2 is increased and hyperphosphorylated by ROCK [87–90] |
| PLS3 | Binds and bundle actin filaments, and offset the actin depolymerization activity of cofilin and PFN [128–130] | Compared to SMA-affected siblings, some asymptomatic female siblings show high PLS3 level, while PLS3 is decreased in some SMA models [113–125] |
| MAP1B | Serves as a constituent of crossbridge between microtubules in neuron [157–160] | In SMN-depleted cells, increased MAP1B induces down-regulation of tubulin tyrosine ligase that subsequently reduces α-tubulin detyrosination [149] |
| STMN1 | Binds or releases tubulin dimers in a phosphorylation-dependent manner [154] | STMN1 level is increased in SMA-like mouse models [148] |
| MAP2 | Promotes the assembly and stability of microtubules, and have overlapping functions with MAP1B [170–172] | In SMN-deficient motor neuron cells, the expression of MAP2 exhibits a down-regulation independent of MAP1B [150, 173] |
| TAU | Promotes the assembly and stability of microtubules [170] | The hyperphosphorylation of TAU mediated by cyclin-dependent kinase 5 degenerates motor neurons in SMA patients and mouse model [151] |
| EB3 | Promotes the growth and stability of microtubules, and regulate the polarization of microtubules [179–181] | In SMN-depleted cells, EB3 exhibits a significant reduction in a MAP1B-dependent way [173] |
The list of cytoskeleton-related proteins that were affected by SMA. PFN2: profilin-2; PLS3: plastin-3; MAP1B: microtubule-associated protein 1B; STMN1: Stathmin1; MAP2: microtubule-associated protein 2; TAU: tau protein; EB3: end-binding protein 3
PFN2, an important regulator of actin dynamics in SMA
Encoded by four genes PFN1–PFN4, profilins have a variety of expression profiles in mammals. Profilins possess a binding site for actin, two binding sites for phosphatidylinositol 4,5-bisphosphate (PIP2) [79] as well as poly-l-proline (PLP) binding domains, allowing for localization to membranes, interaction with PLP-containing proteins and regulation of actin dynamics [80, 81]. Therefore, profilins are of importance to regulate cellular processes related to actin cytoskeleton, such as cytokinesis, neuronal differentiation, synaptic plasticity, membrane trafficking and nuclear transport [82–86].
PFN2, as a major splice variant of PFN2 gene, is primarily restricted in neuronal cells [82] and is a critical regulator of neuronal growth, development, and dendritic spine formation. PFN2 primarily inhibits actin polymerization by binding with actin monomers alone, whereas SMN protects F-actin from destabilization in the presence of urea, showing positive effect on actin polymerization [87]. In motoneurons, PFN2 is highly concentrated and colocalizes with SMN in the cytoplasm and nuclear gems through binding with PLP domain encoded by exon 5 of SMN1 [88–90], producing more F-actin [87]. Sharma et al. revealed that increased PFN2 caused neurite outgrowth inhibition and axon pathfinding defects by disturbing the normal regulation of dynamic actin cytoskeleton in SMA PC12 cell model [87]. However, decrease of PFN2 via the heterozygous or homozygous knockout (KO) of PFN2 alleles in the intermediate SMA mouse model was not enough to rescue SMA phenotypes [91], indicating that other factors related to actin dynamics may be involved in the development of SMA. Moreover, PFN2 is abundant in postsynaptic structures [72, 76, 92] and actin predominately localizes at presynaptic terminal and postsynaptic density in neurons [47]. These distribution patterns indicate that actin cytoskeleton regulated by PFN2 may play a part in synaptic dysfunction of SMA. The ability of vesicle release was damaged along with a reduction in the size of the readily releasable pool in SMA mouse model [93, 94]. It also has a direct interaction with the PLP domain of Piccolo, a protein regulating neurotransmitter release by promoting F-actin assembly [95, 96]. PFN2 KO mouse exhibited deficiencies in the polymerization of synaptic actin and release of more neurotransmitters, indicating that PFN2 may play a functional role in the exocytosis of vesicles [91, 97]. In SMA Caenorhabditis elegans model, the changes in synaptic endocytic proteins and the deficiencies of endosomal compartments were observed, indicating that SMN depletion impairs synaptic endocytosis [98, 99].
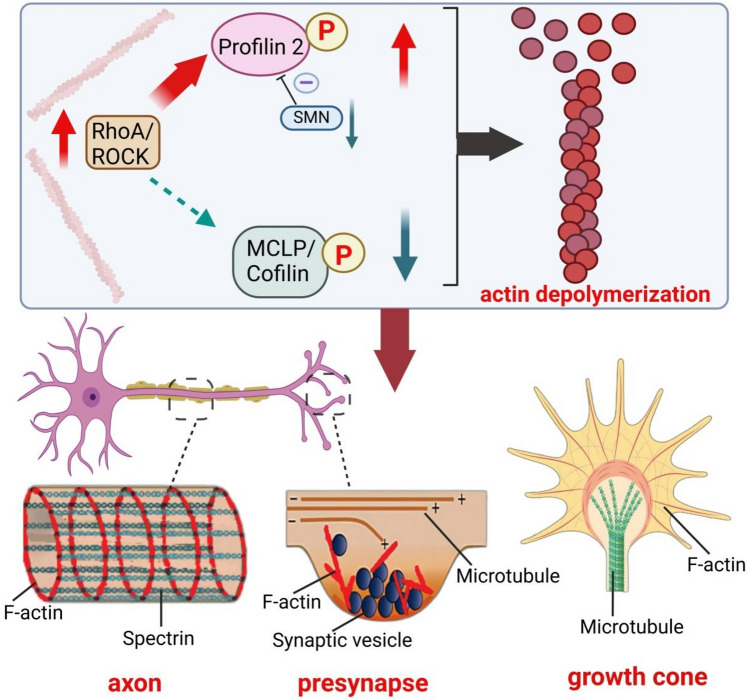
The underlying mechanism of free PFN2 modulating neuronal actin cytoskeleton is through the interaction with ras homolog family member A (RhoA) kinase (ROCK) [100]. PFN2 is phosphorylated at serine 137 (Ser137) close to the PLP-binding domain by ROCK, decreasing combination capacity of PFN2 for some polyproline-rich proteins and inhibiting interaction between G-actin and PFN2 to a lesser extent [101, 102]. In neurons, ROCK interacts with PFN2 in a direct way, which is conducive to increase actin stability and exerts negative effects on neurite outgrowth [98, 103]. Besides, ROCK is capable of directly or indirectly phosphorylating myosin light chain phosphatase (MLCP) and cofilin which are involved in the regulation of actin cytoskeleton and neurite outgrowth [104–106]. However, as shown in PC12 cell with SMN deficiency, the binding of MLCP and cofilin to ROCK is reduced due to the competitive inhibition of PFN2 [88] (Fig. 3).
Fig. 3.
SMN deficiency increases free profilin-2 that is available for combining with ROCK, and subsequently phosphorylated by ROCK. Due to the competitive inhibition, the phosphorylation of MLCP and cofilin is downregulated. Phosphorylated profilin-2 promotes depolymerization of actin filaments, contributing to imbalance of F/G-actin levels in motor neurons, blocked axonal transport, inhibited neurite outgrowth and synaptic dysfunction by influencing actin dynamics in axon, growth cone and synapses
Overactivated ROCK and the changed phosphorylation of its downstream have important impacts on growth cones and neurite outgrowth in SMA. As an initial step, focal F-actin polymerization is an essential process for the onset of axon collaterals [107]. Extending filopodia and lamellipodia from the leading edge of growth cones is crucial in this process. ROCK prevents formation of filopodia and lamellipodia from pre-existing axonal F-actin patches [108]. Moreover, ROCK not only induces growth cone collapse directly to inhibit neurite outgrowth, but also serves as a mediator to affect some signaling processes promoting growth cone collapse [109]. Therefore, ROCK exerted negative regulation on the sprouting of neurites and phosphorylated PFN2 by ROCK is also believed to suppress branching, leading to a severe dysregulation of actin cytoskeleton in PC12 cell with SMN ablation [88, 103].
Under the stimulation of glutamate or electricity, PFN2 is enriched in the head of a dendritic spine and then promote stabilization of dendritic spine morphology via its PLP-binding domain [110]. Therefore, the reduction of PLP-binding capacity of PFN2 may be involved in synapse stripping in SMA PC12 cell model [88]. Moreover, increased available PFN2 and the enhanced ROCK pathway promoted actin rod formation in SMN-lacking cells [111], which induced synaptic loss by blocking axonal transport physically and by disrupting microtubule integrity, leading to the impaired integrity of motoneurons in SMA [111, 112]. Indeed, ROCK inhibitors ameliorated the defect of neurite outgrowth in SMA PC12 cell model [88]. Nevertheless, the inhibition of the RhoA pathway alone is inadequate to fully save neuronal outgrowth and differentiation in SMA mouse model [91]. This may be attributable to the increased availability of PFN2 in SMA also affecting other signaling pathways.
PLS3, a potential protective modifier of SMA
The effect of PLS3 on SMA is controversial. In a study, high PLS3 expression was found in lymphoblastoid cells from asymptomatic female siblings sharing the same SMN genotype with their SMA-affected siblings, indicating that PLS3 may serve as a protective modifier of SMA with a calcium-binding and several actin-binding domains [113, 114]. However, other studies failed to find a relationship between PLS3 expression levels and SMA phenotype [115–117]. As shown in some studies, the effect of PLS3 on SMA is deemed gender-specific and age-dependent, since PLS3 transcript level exhibited a negative relationship with the severity of SMA only in post-pubertal female patients and was associated with SMN2 copy number, gross motor function as well as clinical types [118, 119]. The strong colocalization of PLS3 and SMN was observed in granules throughout motor neuron axons [113], whereas some findings uncovered controversial results about PLS3 expression in SMA models. Ackermann et al. demonstrated that PLS3 did not change with reduction of SMN, suggesting that PLS3 plays a modifying role independent of SMN [120]. By contrast, PLS3 was decreased and dependent on SMN in zebrafish and mouse SMA models [91, 121, 122]. Therefore, overexpression of PLS3 exerts varied therapeutic effects in different SMA models. In zebrafish and intermediate SMA mouse models, overexpression of PLS3 rescued neurite length and axonal outgrowth deficiencies, and promoted survival and motor function [113, 120–123]. However, in severe SMA mice, overexpressed PLS3 did not exhibit a significant beneficial impact on this phenotype [124, 125]. One possible explanation is that PLS3 is able to alleviate the severity of SMA when SMN loss is moderate. Several studies found that human subjects with complete PLS3 protein loss display signs of osteoporosis instead of lower motoneuron degeneration, indicating that PLS3 may act as a protective modifier of SMA rather than an etiological factor [126, 127].
PLS3 can bind and bundle to actin filaments, and offset actin depolymerization activity of cofilin and PFN2 subsequently strengthening actin networks [128–130]. As a result, high expression of PLS3 can elevate F-actin levels in SMA, which rescues axon length and outgrowth defects [113, 131]. Overexpression of PLS3 increased axon input number, muscle fiber and endplate sizes in SMA mice, improving the neuromuscular transmission [120]. Moreover, increased PLS3 restored F-actin intensity, amount of presynapses and synaptic vesicles in SMA, thereby promoting synaptic function [120]. PLS3 restores the intensity and area of Piccolo which is related to F-actin dynamics [95], resulting in the rescue of active zones number in SMA mice. PLS3 also promoted organization of the readily releasable pool in vesicles and rescued vesicle release and electrophysiological defects [120]. In addition, PLS3 plays a pivotal role in cell endocytosis process based the fact that endocytosis of PLS3 KO yeast was impaired [132]. Endocytosis regulated by F-actin is indispensable for replenishing the recycling pool and influences the supply of vesicles to the release of neurotransmitters [133–136]. Overexpressed PLS3 can rescue endocytosis and synaptic vesicle recycling impaired by the depletion of SMN [122]. Moreover, PLS3, SMN and heterogeneous nuclear ribonucleoprotein (hnRNP) F/H act in the same complex that plays a vital role in endocytosis [137]. PLS3 is also important to a well-organized cytoskeleton associated with presynaptic compartment orchestration. The disturbed brain-derived neurotrophic factor/tropomyosin receptor kinase B (BDNF/TrkB)-signaling cascade due to the impaired localization of transmembrane proteins is important in the affected differentiation and maturation of SMA motor neurons [138–141]. PLS3 cooperating with actin-related protein 2/3 (Arp2/3) can improve the localization and cyclic adenosine monophosphate (cAMP)-induced translocation of TrkB in SMA, which strengthens BDNF/TrkB-signaling [141].
In SMA, calcium homeostasis was disturbed and overexpressed PLS3 rescued “cluster-like” formation of Cav2.2 and increased spontaneous calcium ion (Ca2+) transients in motoneurons [141]. Indeed, overexpression of PLS3 without calcium-binding ability is not enough to make up for loss of SMN, while short of actin-binding domains in PLS3, remains capable of rescuing axon morphology [142]. Moreover, PLS3 cooperated with some proteins in a calcium-related way. For example, coronin 1C (CORO1C), as a F-actin-binding protein, can combine with PLS3 directly in a calcium-dependent way. Akin to PLS3, overexpressed CORO1C restores endocytosis in SMA cells by increasing F-actin content [122]. Moreover, calcineurin-like EF-hand protein 1 (CHP1) interacts with PLS3 which exists widely and abundantly at sites related to SMA including growth cones and neuromuscular junctions (NMJs). Together, the effect of PLS3 in SMA is through the cooperation of calcium influx and F-actin dynamics [122, 141, 143].
MAP, the bridge between SMA and microtubule dysfunction
Microtubule dynamics influencing neuronal functions are controlled by MAPs [144, 145], a set of proteins binding to microtubules and regulating their structures [146, 147]. Numerous studies claim that aberrant MAPs disturbing microtubule dynamics are involved in pathomechanism of SMA [148–153].
As a member of stathmin (STMN) phosphoprotein family, STMN 1 is one primary MAP related to SMA. STMN1 has C-terminal “STMN-like domain (SLD)” that can bind or release tubulin dimers in a phosphorylation-dependent manner, participating in regulation of microtubule dynamics [154]. STMN1 identified as a disease modifier for SMA and enhanced in SMN-depleted NSC34 cells and SMA-like mouse models. The aberrant up-regulation of STMN1 is correlated with the severity of SMA and is adverse to microtubule polymerization and mitochondrial transport towards axons [148]. Correspondingly, knockdown of STMN1 ameliorated the deficiencies of microtubule network formation, axonal outgrowth and mitochondrial transport in SMN-depleted NSC34 cells and SMA mouse model [148]. Further study uncovered that heterozygous rather than homozygous STMN1 KO mouse model rescued axonal microtubule density, motor function and NMJ maturation in SMA [61]. In reality, the reduction of STMN1 as a possible pathogenic modifier of motor neuron diseases was reported to be observed in vulnerable motor neurons [155]. By contrast, in intermediate SMA mouse model, overexpressed STMN1 recovered neuromuscular innervation and motor neuron preservation and improved lifespan, weight gain and the righting reflex by promoting microtubule turnover [156]. Contradiction between these findings may be caused by the differences in models. STMN1 is suggested to be a potential therapeutic target for SMA. Despite contributing to the improvement in SMA pathology, the reduction of STMN1 cannot rescue the survival of SMA mice [61]. The potential role of STMN1 on cytoskeleton of motor neuron in SMA needs further investigation.
Microtubule-associated protein 1B (MAP1B), as a member of MAP family, is also related to SMA. It mainly expresses in neurons and serves as a constituent of crossbridge between microtubules in neurons to axon growth, regeneration, growth cone pathfinding and neuronal migration [157–160]. Located on chromosome 5q13, MAP1B locus is close proximity to SMN1 locus [161]. The human MAP1B gene displays close linkage to SMA mutations in a genetic linkage analysis [162]. Moreover, the spatial distribution of MAP1B nearly coincides with SMN granules in axons and presynaptic terminals [163]. These mapping data and colocalization of SMN and MAP1B suggest that MAP1B may be relevant to the pathomechanism of SMA.
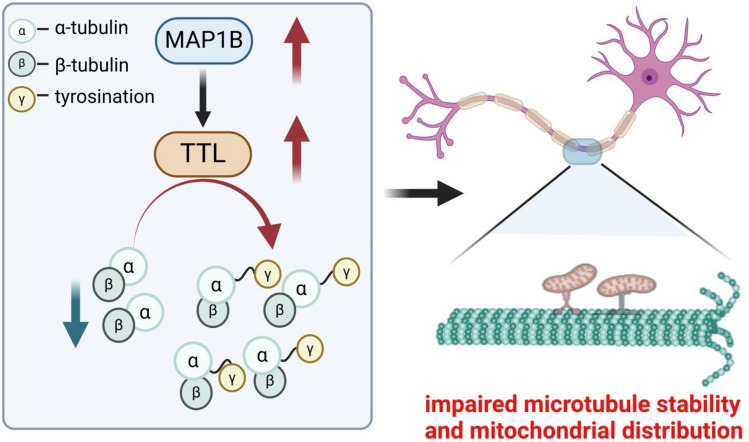
Microtubule stability and dynamics are affected by acetylation and detyrosinated proteins [164–167]. In SMN-depleted cells, increased MAP1B up-regulated tubulin tyrosine ligase (TTL) activity that subsequently decreased α-tubulin detyrosination, impairing microtubule stability [149] (Fig. 4). Down-regulation of MAP1B rescued the aberrant levels of TTL and detyrosinated α-tubulin in SMN-depleted cells [149]. Moreover, damaged microtubule dynamics possibly affect axonal transport, which subsequently influences mitochondrial distribution along neurites in SMA. Decreased MAP1B also ameliorated mitochondrial distribution impaired in SMN-depleted cells, which suggests the restoration of axonal transport [149, 168, 169].
Fig. 4.
In SMA, increased MAP1B up-regulates tubulin tyrosine ligase (TTL) activity which increases tyrosinated α-tubulin. The breakdown of α-tubulin tyrosination balance is harmful to microtubule stability. Moreover, damaged microtubule dynamics possibly affect axonal transport, influencing mitochondrial distribution along neurites
Although studies are relatively limited compared to STMN and MAP1B, several MAPs are also involved in the development of SMA. As a structural MAP abundantly expressed in neurons such as MAP1B and microtubule-associated protein 2 (MAP2) has overlapping functions with MAP1B in terms of neuronal migration and neurite growth, which results in a compensatory mechanism [170, 171]. MAP2 plays a significant role in microtubule nucleation and stabilization, which affects neurite outgrowth and axonal transport by regulating interactions between motor proteins and microtubules [170, 172]. In SMN-deficient motor neuron cells, the expression of MAP2 exhibits a down-regulation independent of MAP1B [150, 173]. In neurons, tau protein is another structural MAP affected by SMA. Similar to other MAP family members in the nervous system, tau maintains the stability of axonal microtubules [170]. In motor neurons of SMA mouse models and spinal cord of SMA patient, the hyperphosphorylation of tau mediated by cyclin-dependent kinase 5 contributes to the dissociation of MAPs from microtubule, subsequently leading to downregulated microtubule stability, impaired axonal transport and neuronal degeneration [151].
As end-binding proteins, plus-end-tracking proteins are regulated by major structural MAPs such as MAP1B, MAP2 and tau, which affects microtubule dynamics later [174–178]. Moreover, MAP1B and MAP2 interplay with microtubule end-binding proteins 1 and 3 (EB1 and EB3) and modulate their actions [174, 176, 178]. Unlike EB1 expressed ubiquitously, EB3 is mainly expressed in neurons and accumulates at the distal ends of freshly polymerized microtubules, and regulates growth of microtubules [179–181]. EB3 rather than EB1 exhibits a significant reduction in SMN-depleted cells. Furthermore, decreased MAP1B can elevate EB3 levels in SMN-depleted cells [173]. Located at both the cytoplasm and microtubule tips, EB3 interplays with proteins at microtubule tips like p150Glued and drebrin, and influences microtubule-dependent transport and interactions between F-actin and microtubules [182–185]. As shown in a study, the decline binding of EB3 to microtubule tips caused by overexpressed MAP1B, indicating that up-regulated MAP1B and decreased EB3, may cooperate in the impaired microtubule dynamics of SMA [176]. On top of that, comets, which are structures formed by agminated EB3 at microtubule ends, serve as markers for newly polymerized microtubules and are regulated by MAP1B and tau [174, 176, 179, 180]. They are up-regulated at proximal neurites in SMN-depleted cells, which indicates more growing microtubules at proximal neurites in SMA [173].
Neurofilament, a potential biomarker of SMA
Neurofilaments, as important structural proteins of neurons, are principally expressed in long axons [46]. As subunits of neurofilaments, neurofilament light chain and neurofilament heavy chain are common detection indicators. In severe SMA patients and mouse models, increased demyelinated axons were observed and degenerated rapidly postnatally, thereby resulting in release of neurofilament light chain [20]. Concentrations of neurofilaments in cerebral spinal fluid (CSF), serum, or plasma indicate neuronal damage and have been proposed as potential prognostic and treatment responsive biomarkers in several neurodegenerative diseases such as ALS, AD, PD and SMA [186].
As a potential biomarker of SMA, the scope of application of neurofilaments is controversial. Neurofilament light chain levels in serum and CSF showed strong correlation with motor function in a pediatric control cohort on SMA patients [187]. In a single-center pilot study, phosphorylated heavy chain and light chain neurofilaments in the CSF of SMA patients correlated with disease severity and activity, indicating that neurofilaments in CSF may serve as marker of neuronal loss and clinical outcome [188]. However, in some studies, although decreased after treatment with nusinersen, levels of neurofilaments and neurofilament light chain in the CSF of SMA type 3 patients did not exhibit correlation with motor functions, suggesting that neurofilaments may be insufficient to serve as an optimal surrogate treatment biomarker [189, 190].
In SMA infants, increased plasma neurofilaments levels correlate with age and several markers of disease severity including Hammersmith Infant Neurological Examination Section 2 motor milestones score, Children’s Hospital of Philadelphia Infant Test of Neuromuscular Disorders score, and peroneal and ulnar nerve compound muscle action potential amplitudes, inversely associated with SMN2 copy number [187, 191]. Moreover, in SMA infants and mouse model, levels of phosphorylated heavy chain and light chain neurofilaments in plasma declined rapidly after nusinersen treatment, suggesting their potential as a peripheral marker reflecting the pathological status of SMA [186, 187, 191, 192]. Nevertheless, SMA patients treated with onasemnogene abeparvovec monotherapy exhibited a significant rise in plasma neurofilaments levels, indicating that neurofilaments may be insufficient to function as the single marker to predict outcomes [186]. Further studies are needed to determine the role of neurofilaments in the pathomechanism of SMA.
Conclusion
Accumulated evidence suggests that cytoskeletal abnormalities may play an important role in motor neuron degeneration in progression of SMA. In SMA condition, cytoskeleton abnormalities induce growth cone formation, neurite extension and microtubule formation defects, thereby amplifying SMN-dependent cellular alterations. SMN deficiency dysregulated actin cytoskeleton by interfering with increased free PFN2 which led to an up-regulation of the ROCK pathway, contributing to neuronal damages such as inhibited neurite outgrowth, growth cone collapse and impaired axon pathfinding. PLS3 regulated calcium influx and F-actin dynamics to promote pathogenesis of SMA. The alterations of microtubule stability which affect axonal transport are associated with the changes of tubulin modifications. The collaboration of several MAPs such as STMN1, MAP1B and tau contributes to downregulated microtubule stability, impaired axonal transport and subsequent neuronal degeneration. Existing clinical studies have shown that the level of neurofilaments in SMA can increase with the demyelination of axon and decrease after nusinersen treatment, suggesting the potential of neurofilaments to serve as prognostic and treatment responsive biomarkers for guiding therapeutic interventions.
The investigation into cytoskeleton holds significant potential for understanding the pathogenesis of SMA and other neurodegenerative disorders. However, the existing studies still have some limitations. First, the trends in the changes of cytoskeleton-associated proteins and influencing degrees of impaired cytoskeleton are controversial among various SMA models. Secondly, the causal link between SMN loss and cytoskeletal dysfunctions is unclear and needs further study. Third, investigations about the effect of cytoskeletal dysfunctions on SMA have focused on animal and cellular models. Although a series of clinical trials have suggested the potential of neurofilaments as biomarkers of SMA, the role of actin disturbance and microtubule instability in the pathomechanism of SMA needs to be further explored in clinical trials. Despite the constraints of current research, comprehending the fundamental mechanisms regulating cytoskeletal proteins is crucial for formulating fresh targeted therapies to treat SMA.
Author contributions
Tianyu Shi: Writing—original draft, writing—review and editing, project administration, conceptualization. Zijie Zhou: writing—review and editing, conceptualization. Taiyang Xiang: writing review and editing, conceptualization. Yinxuan Suo: data curation, visualization. Xiaoyan Shi: visualization, investigation, data curation. Yaoyao Li: investigation, data curation. Peng Zhang: validation, supervision. Jun Dai: data curation, conceptualization. Lei Sheng: validation, supervision, conceptualization.
Funding
This work was supported by the National Nature Science Foundation of China (81902179), the Natural Science Foundation of Jiangsu Province (BK20221241) and the Gusu Talent Program (GSWS2022046).
Data availability
No data were used for the research described in the article.
Declarations
Conflicts of interest
The authors declare that they have no competing interests.
Footnotes
Tianyu Shi and Zijie Zhou have contributed equally to this work.
Contributor Information
Jun Dai, Email: daijun@suda.edu.cn.
Lei Sheng, Email: shenglei510@suda.edu.cn.
References
- 1.Finkel RS, Mercuri E, Meyer OH et al (2018) Diagnosis and management of spinal muscular atrophy: Part 2: Pulmonary and acute care; medications, supplements and immunizations; other organ systems; and ethics. Neuromuscul Disord 28(3):197–207. 10.1016/j.nmd.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 2.Mercuri E, Finkel RS, Muntoni F et al (2018) Diagnosis and management of spinal muscular atrophy: Part 1: Recommendations for diagnosis, rehabilitation, orthopedic and nutritional care. Neuromuscul Disord 28(2):103–115. 10.1016/j.nmd.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 3.Yeo CJJ, Darras BT (2020) Overturning the paradigm of spinal muscular atrophy as just a motor neuron disease. Pediatr Neurol 109:12–19. 10.1016/j.pediatrneurol.2020.01.003 [DOI] [PubMed] [Google Scholar]
- 4.Wang CH, Finkel RS, Bertini ES et al (2007) Consensus statement for standard of care in spinal muscular atrophy. J Child Neurol 22(8):1027–1049. 10.1177/0883073807305788 [DOI] [PubMed] [Google Scholar]
- 5.Zerres K, Rudnik-Schöneborn S (1995) Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch Neurol 52(5):518–523. 10.1001/archneur.1995.00540290108025 [DOI] [PubMed] [Google Scholar]
- 6.Lefebvre S, Bürglen L, Reboullet S et al (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80(1):155–165. 10.1016/0092-8674(95)90460-3 [DOI] [PubMed] [Google Scholar]
- 7.Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B (2002) Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 70(2):358–368. 10.1086/338627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wirth B, Brichta L, Schrank B et al (2006) Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum Genet 119(4):422–428. 10.1007/s00439-006-0156-7 [DOI] [PubMed] [Google Scholar]
- 9.Lorson CL, Hahnen E, Androphy EJ, Wirth B (1999) A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A 96(11):6307–6311. 10.1073/pnas.96.11.6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cartegni L, Krainer AR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30(4):377–384. 10.1038/ng854 [DOI] [PubMed] [Google Scholar]
- 11.Kashima T, Rao N, Manley JL (2007) An intronic element contributes to splicing repression in spinal muscular atrophy. Proc Natl Acad Sci U S A 104(9):3426–3431. 10.1073/pnas.0700343104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorson CL, Strasswimmer J, Yao JM et al (1998) SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet 19(1):63–66. 10.1038/ng0598-63 [DOI] [PubMed] [Google Scholar]
- 13.Lorson CL, Androphy EJ (2000) An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet 9(2):259–265. 10.1093/hmg/9.2.259 [DOI] [PubMed] [Google Scholar]
- 14.Finkel RS, Chiriboga CA, Vajsar J et al (2016) Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 388(10063):3017–3026. 10.1016/S0140-6736(16)31408-8 [DOI] [PubMed] [Google Scholar]
- 15.Mendell JR, Al-Zaidy S, Shell R et al (2017) Single-Dose Gene-replacement therapy for spinal muscular atrophy. N Engl J Med 377(18):1713–1722. 10.1056/NEJMoa1706198 [DOI] [PubMed] [Google Scholar]
- 16.Baranello G, Darras BT, Day JW et al (2021) Risdiplam in type 1 spinal muscular atrophy. N Engl J Med 384(10):915–923. 10.1056/NEJMoa2009965 [DOI] [PubMed] [Google Scholar]
- 17.Ratni H, Ebeling M, Baird J et al (2018) Discovery of risdiplam, a selective survival of motor neuron-2 ( SMN2) gene splicing modifier for the treatment of spinal muscular atrophy (SMA). J Med Chem 61(15):6501–6517. 10.1021/acs.jmedchem.8b00741 [DOI] [PubMed] [Google Scholar]
- 18.Hua Y, Sahashi K, Rigo F et al (2011) Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478(7367):123–126. 10.1038/nature10485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishio H, Niba ETE, Saito T, Okamoto K, Takeshima Y, Awano H (2023) Spinal muscular atrophy: the past, present, and future of diagnosis and treatment. Int J Mol Sci 24(15):11939. 10.3390/ijms241511939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong L, Valdivia DO, Simon CM et al (2021) Impaired prenatal motor axon development necessitates early therapeutic intervention in severe SMA. Sci Transl Med. 13(578):eabb6871. 10.1126/scitranslmed.abb6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dangouloff T, Servais L (2019) Clinical evidence supporting early treatment of patients with spinal muscular atrophy: current perspectives. Ther Clin Risk Manag 15:1153–1161. 10.2147/TCRM.S172291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benini F, Salamon E, Divisic A, Maghini I, Agosto C (2020) Acknowledging limits: statistics and the child’s quality of life in spinal muscular atrophy. J Paediatr Child Health 56(6):995–996. 10.1111/jpc.14959 [DOI] [PubMed] [Google Scholar]
- 23.Day JW, Howell K, Place A et al (2022) Advances and limitations for the treatment of spinal muscular atrophy. BMC Pediatr 22(1):632. 10.1186/s12887-022-03671-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefebvre S, Bürglen L, Frézal J, Munnich A, Melki J (1998) The role of the SMN gene in proximal spinal muscular atrophy. Hum Mol Genet 7(10):1531–1536. 10.1093/hmg/7.10.1531 [DOI] [PubMed] [Google Scholar]
- 25.Coovert DD, Le TT, McAndrew PE et al (1997) The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet 6(8):1205–1214. 10.1093/hmg/6.8.1205 [DOI] [PubMed] [Google Scholar]
- 26.Singh RN, Howell MD, Ottesen EW, Singh NN (2017) Diverse role of survival motor neuron protein. Biochim Biophys Acta Gene Regul Mech 1860(3):299–315. 10.1016/j.bbagrm.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groen EJN, Talbot K, Gillingwater TH (2018) Advances in therapy for spinal muscular atrophy: promises and challenges. Nat Rev Neurol 14(4):214–224. 10.1038/nrneurol.2018.4 [DOI] [PubMed] [Google Scholar]
- 28.Tu WY, Simpson JE, Highley JR, Heath PR (2017) Spinal muscular atrophy: factors that modulate motor neurone vulnerability. Neurobiol Dis 102:11–20. 10.1016/j.nbd.2017.01.011 [DOI] [PubMed] [Google Scholar]
- 29.Gershoni-Emek N, Chein M, Gluska S, Perlson E (2015) Amyotrophic lateral sclerosis as a spatiotemporal mislocalization disease: location, location, location. Int Rev Cell Mol Biol 315:23–71. 10.1016/bs.ircmb.2014.11.003 [DOI] [PubMed] [Google Scholar]
- 30.Baldwin KR, Godena VK, Hewitt VL, Whitworth AJ (2016) Axonal transport defects are a common phenotype in Drosophila models of ALS. Hum Mol Genet 25(12):2378–2392. 10.1093/hmg/ddw105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janmey PA (1998) The cytoskeleton and cell signaling: component localization and mechanical coupling. Physiol Rev 78(3):763–781. 10.1152/physrev.1998.78.3.763 [DOI] [PubMed] [Google Scholar]
- 32.Carpenter CL (2000) Actin cytoskeleton and cell signaling. Crit Care Med 28(4 Suppl):N94-N99. 10.1097/00003246-200004001-00011 [DOI] [PubMed] [Google Scholar]
- 33.Sheng M, Pak DT (2000) Ligand-gated ion channel interactions with cytoskeletal and signaling proteins. Annu Rev Physiol 62:755–778. 10.1146/annurev.physiol.62.1.755 [DOI] [PubMed] [Google Scholar]
- 34.Fletcher DA, Mullins RD (2010) Cell mechanics and the cytoskeleton. Nature 463(7280):485–492. 10.1038/nature08908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laster SM, Mackenzie JM (1996) Bleb formation and F-actin distribution during mitosis and tumor necrosis factor-induced apoptosis. Microsc Res Tech 34(3):272–280. 10.1002/(SICI)1097-0029(19960615)34:3%3c272::AID-JEMT10%3e3.0.CO;2-J [DOI] [PubMed] [Google Scholar]
- 36.Levee MG, Dabrowska MI, Lelli JL, Hinshaw DB (1996) Actin polymerization and depolymerization during apoptosis in HL-60 cells. Am J Physiol 271(6 Pt 1):C1981-C1992. 10.1152/ajpcell.1996.271.6.C1981 [DOI] [PubMed] [Google Scholar]
- 37.van Engeland M, Kuijpers HJ, Ramaekers FC, Reutelingsperger CP, Schutte B (1997) Plasma membrane alterations and cytoskeletal changes in apoptosis. Exp Cell Res 235(2):421–430. 10.1006/excr.1997.3738 [DOI] [PubMed] [Google Scholar]
- 38.Gourlay CW, Carpp LN, Timpson P, Winder SJ, Ayscough KR (2004) A role for the actin cytoskeleton in cell death and aging in yeast. J Cell Biol 164(6):803–809. 10.1083/jcb.200310148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kavanagh E, Vlachos P, Emourgeon V, Rodhe J, Joseph B (2012) p57(KIP2) control of actin cytoskeleton dynamics is responsible for its mitochondrial pro-apoptotic effect. Cell Death Dis 3(5):e311. 10.1038/cddis.2012.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pawlik A, Szczepanski MA, Klimaszewska A, Gackowska L, Zuryn A, Grzanka A (2012) Phenethyl isothiocyanate-induced cytoskeletal changes and cell death in lung cancer cells. Food Chem Toxicol 50(10):3577–3594. 10.1016/j.fct.2012.07.043 [DOI] [PubMed] [Google Scholar]
- 41.van Haelst C, Rothstein TL (1988) Cytochalasin stimulates phosphoinositide metabolism in murine B lymphocytes. J Immunol 140(4):1256–1258 [PubMed] [Google Scholar]
- 42.Rosales C, Brown EJ (1992) Signal transduction by neutrophil immunoglobulin G Fc receptors. Dissociation of intracytoplasmic calcium concentration rise from inositol 1,4,5-trisphosphate. J Biol Chem 267(8):5265–5271 [PubMed] [Google Scholar]
- 43.Lange K, Brandt U (1996) Calcium storage and release properties of F-actin: evidence for the involvement of F-actin in cellular calcium signaling. FEBS Lett 395(2–3):137–142. 10.1016/0014-5793(96)01025-3 [DOI] [PubMed] [Google Scholar]
- 44.Leterrier C, Dubey P, Roy S (2017) The nano-architecture of the axonal cytoskeleton. Nat Rev Neurosci 18(12):713–726. 10.1038/nrn.2017.129 [DOI] [PubMed] [Google Scholar]
- 45.Kapitein LC, Hoogenraad CC (2015) Building the neuronal microtubule cytoskeleton. Neuron 87(3):492–506. 10.1016/j.neuron.2015.05.046 [DOI] [PubMed] [Google Scholar]
- 46.Yuan A, Rao MV, Veeranna, Nixon RA (2017) Neurofilaments and neurofilament proteins in health and disease. Cold Spring Harb Perspect Biol 9(4):a018309. 10.1101/cshperspect.a018309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cingolani LA, Goda Y (2008) Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci 9(5):344–356. 10.1038/nrn2373 [DOI] [PubMed] [Google Scholar]
- 48.Kevenaar JT, Hoogenraad CC (2015) The axonal cytoskeleton: from organization to function. Front Mol Neurosci 8:44. 10.3389/fnmol.2015.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dent EW, Gupton SL, Gertler FB (2011) The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb Perspect Biol 3(3):a001800. 10.1101/cshperspect.a001800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witte H, Bradke F (2008) The role of the cytoskeleton during neuronal polarization. Curr Opin Neurobiol 18(5):479–487. 10.1016/j.conb.2008.09.019 [DOI] [PubMed] [Google Scholar]
- 51.Tahirovic S, Bradke F (2009) Neuronal polarity. Cold Spring Harb Perspect Biol 1(3):a001644. 10.1101/cshperspect.a001644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conde C, Cáceres A (2009) Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci 10(5):319–332. 10.1038/nrn2631 [DOI] [PubMed] [Google Scholar]
- 53.Neukirchen D, Bradke F (2011) Neuronal polarization and the cytoskeleton. Semin Cell Dev Biol 22(8):825–833. 10.1016/j.semcdb.2011.08.007 [DOI] [PubMed] [Google Scholar]
- 54.Hur EM, Saijilafu, Zhou FQ (2012) Growing the growth cone: remodeling the cytoskeleton to promote axon regeneration. Trends Neurosci 35(3):164–174. 10.1016/j.tins.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Sheng ZH (2022) Energy matters: presynaptic metabolism and the maintenance of synaptic transmission. Nat Rev Neurosci 23(1):4–22. 10.1038/s41583-021-00535-8 [DOI] [PubMed] [Google Scholar]
- 56.Cunnane SC, Trushina E, Morland C et al (2020) Brain energy rescue: an emerging therapeutic concept for neurodegenerative disorders of ageing. Nat Rev Drug Discov 19(9):609–633. 10.1038/s41573-020-0072-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheng ZH, Cai Q (2012) Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci 13(2):77–93. 10.1038/nrn3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonini SA, Ferrari-Toninelli G, Montinaro M, Memo M (2013) Notch signalling in adult neurons: a potential target for microtubule stabilization. Ther Adv Neurol Disord 6(6):375–385. 10.1177/1756285613490051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McMurray CT (2000) Neurodegeneration: diseases of the cytoskeleton? Cell Death Differ 7(10):861–865. 10.1038/sj.cdd.4400764 [DOI] [PubMed] [Google Scholar]
- 60.Henriques AG, Oliveira JM, Carvalho LP, da Cruz e Silva OAB (2015) Aβ influences cytoskeletal signaling cascades with consequences to Alzheimer’s disease. Mol Neurobiol 52(3):1391–1407. 10.1007/s12035-014-8913-4 [DOI] [PubMed] [Google Scholar]
- 61.Wen HL, Ting CH, Liu HC, Li H, Lin-Chao S (2013) Decreased stathmin expression ameliorates neuromuscular defects but fails to prolong survival in a mouse model of spinal muscular atrophy. Neurobiol Dis 52:94–103. 10.1016/j.nbd.2012.11.015 [DOI] [PubMed] [Google Scholar]
- 62.Devine MJ, Kittler JT (2018) Mitochondria at the neuronal presynapse in health and disease. Nat Rev Neurosci 19(2):63–80. 10.1038/nrn.2017.170 [DOI] [PubMed] [Google Scholar]
- 63.Millecamps S, Julien JP (2013) Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci 14(3):161–176. 10.1038/nrn3380 [DOI] [PubMed] [Google Scholar]
- 64.Conforti L, Adalbert R, Coleman MP (2007) Neuronal death: where does the end begin? Trends Neurosci 30(4):159–166. 10.1016/j.tins.2007.02.004 [DOI] [PubMed] [Google Scholar]
- 65.Morfini GA, Burns M, Binder LI et al (2009) Axonal transport defects in neurodegenerative diseases. J Neurosci 29(41):12776–12786. 10.1523/JNEUROSCI.3463-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cairns NJ, Lee VMY, Trojanowski JQ (2004) The cytoskeleton in neurodegenerative diseases. J Pathol 204(4):438–449. 10.1002/path.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo W, Stoklund Dittlau K, Van Den Bosch L (2020) Axonal transport defects and neurodegeneration: Molecular mechanisms and therapeutic implications. Semin Cell Dev Biol 99:133–150. 10.1016/j.semcdb.2019.07.010 [DOI] [PubMed] [Google Scholar]
- 68.Nicolas A, Kenna KP, Renton AE et al (2018) Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97(6):1267–1288. 10.1016/j.neuron.2018.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reid E, Kloos M, Ashley-Koch A et al (2002) A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10). Am J Hum Genet 71(5):1189–1194. 10.1086/344210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Didonna A, Opal P (2019) The role of neurofilament aggregation in neurodegeneration: lessons from rare inherited neurological disorders. Mol Neurodegener 14(1):19. 10.1186/s13024-019-0318-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khalil M, Teunissen CE, Lehmann S et al (2024) Neurofilaments as biomarkers in neurological disorders - towards clinical application. Nat Rev Neurol 20(5):269–287. 10.1038/s41582-024-00955-x [DOI] [PubMed] [Google Scholar]
- 72.Pagliardini S, Giavazzi A, Setola V et al (2000) Subcellular localization and axonal transport of the survival motor neuron (SMN) protein in the developing rat spinal cord. Hum Mol Genet 9(1):47–56. 10.1093/hmg/9.1.47 [DOI] [PubMed] [Google Scholar]
- 73.Zhang H, Xing L, Rossoll W, Wichterle H, Singer RH, Bassell GJ (2006) Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J Neurosci 26(33):8622–8632. 10.1523/JNEUROSCI.3967-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rossoll W, Jablonka S, Andreassi C et al (2003) Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol 163(4):801–812. 10.1083/jcb.200304128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Béchade C, Rostaing P, Cisterni C et al (1999) Subcellular distribution of survival motor neuron (SMN) protein: possible involvement in nucleocytoplasmic and dendritic transport. Eur J Neurosci 11(1):293–304. 10.1046/j.1460-9568.1999.00428.x [DOI] [PubMed] [Google Scholar]
- 76.Fan L, Simard LR (2002) Survival motor neuron (SMN) protein: role in neurite outgrowth and neuromuscular maturation during neuronal differentiation and development. Hum Mol Genet 11(14):1605–1614. 10.1093/hmg/11.14.1605 [DOI] [PubMed] [Google Scholar]
- 77.Francis JW, Sandrock AW, Bhide PG, Vonsattel JP, Brown RH (1998) Heterogeneity of subcellular localization and electrophoretic mobility of survival motor neuron (SMN) protein in mammalian neural cells and tissues. Proc Natl Acad Sci U S A 95(11):6492–6497. 10.1073/pnas.95.11.6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baas PW, Rao AN, Matamoros AJ, Leo L (2016) Stability properties of neuronal microtubules. Cytoskeleton (Hoboken) 73(9):442–460. 10.1002/cm.21286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lambrechts A, Jonckheere V, Dewitte D, Vandekerckhove J, Ampe C (2002) Mutational analysis of human profilin I reveals a second PI(4,5)-P2 binding site neighbouring the poly(L-proline) binding site. BMC Biochem 3:12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferron F, Rebowski G, Lee SH, Dominguez R (2007) Structural basis for the recruitment of profilin–actin complexes during filament elongation by Ena/VASP. EMBO J 26(21):4597–4606. 10.1038/sj.emboj.7601874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee SH, Dominguez R (2010) Regulation of actin cytoskeleton dynamics in cells. Mol Cells 29(4):311–325. 10.1007/s10059-010-0053-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jockusch BM, Murk K, Rothkegel M (2007) The profile of profilins. In: Amara SG, Bamberg E, Fleischmann B et al (eds) Reviews of physiology, biochemistry and pharmacology. Springer, Berlin, pp 131–149 [DOI] [PubMed] [Google Scholar]
- 83.Yarmola EG, Bubb MR (2009) How depolymerization can promote polymerization: the case of actin and profilin. BioEssays 31(11):1150–1160. 10.1002/bies.200900049 [DOI] [PubMed] [Google Scholar]
- 84.Birbach A (2008) Profilin, a multi-modal regulator of neuronal plasticity. BioEssays 30(10):994–1002. 10.1002/bies.20822 [DOI] [PubMed] [Google Scholar]
- 85.Witke W (2004) The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol 14(8):461–469. 10.1016/j.tcb.2004.07.003 [DOI] [PubMed] [Google Scholar]
- 86.Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112(4):453–465. 10.1016/s0092-8674(03)00120-x [DOI] [PubMed] [Google Scholar]
- 87.Sharma A, Lambrechts A, Hao LT et al (2005) A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp Cell Res 309(1):185–197. 10.1016/j.yexcr.2005.05.014 [DOI] [PubMed] [Google Scholar]
- 88.Nölle A, Zeug A, Van Bergeijk J et al (2011) The spinal muscular atrophy disease protein SMN is linked to the rho-kinase pathway via profilin. Hum Mol Genet 20(24):4865–4878. 10.1093/hmg/ddr425 [DOI] [PubMed] [Google Scholar]
- 89.Giesemann T, Rathke-Hartlieb S, Rothkegel M et al (1999) A role for polyproline motifs in the spinal muscular atrophy protein SMN. J Biol Chem 274(53):37908–37914. 10.1074/jbc.274.53.37908 [DOI] [PubMed] [Google Scholar]
- 90.Nodelman IM, Bowman GD, Lindberg U, Schutt CE (1999) X-ray structure determination of human profilin II: a comparative structural analysis of human profilins. J Mol Biol 294(5):1271–1285. 10.1006/jmbi.1999.3318 [DOI] [PubMed] [Google Scholar]
- 91.Bowerman M, Anderson CL, Beauvais A, Boyl PP, Witke W, Kothary R (2009) SMN, profilin IIa and plastin 3: a link between the deregulation of actin dynamics and SMA pathogenesis. Mol Cell Neurosci 42(1):66–74. 10.1016/j.mcn.2009.05.009 [DOI] [PubMed] [Google Scholar]
- 92.Murk K, Wittenmayer N, Michaelsen-Preusse K et al (2012) Neuronal profilin isoforms are addressed by different signalling pathways. PLoS ONE. 7(3):e34167. 10.1371/journal.pone.0034167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kong L, Wang X, Choe DW et al (2009) Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci 29(3):842–851. 10.1523/JNEUROSCI.4434-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Torres-Benito L, Neher MF, Cano R, Ruiz R, Tabares L (2011) SMN requirement for synaptic vesicle, active zone and microtubule postnatal organization in motor nerve terminals. PLoS ONE 6(10):e26164. 10.1371/journal.pone.0026164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Waites CL, Leal-Ortiz SA, Andlauer TFM, Sigrist SJ, Garner CC (2011) Piccolo regulates the dynamic assembly of presynaptic F-actin. J Neurosci 31(40):14250–14263. 10.1523/JNEUROSCI.1835-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang X, Kibschull M, Laue MM, Lichte B, Petrasch-Parwez E, Kilimann MW (1999) Aczonin, a 550-Kd putative scaffolding protein of presynaptic active zones, shares homology regions with rim and bassoon and binds profilin. J Cell Biol 147(1):151–162. 10.1083/jcb.147.1.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pilo Boyl P, Di Nardo A, Mulle C et al (2007) Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J 26(12):2991–3002. 10.1038/sj.emboj.7601737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Witke W (1998) In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J 17(4):967–976. 10.1093/emboj/17.4.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dimitriadi M, Derdowski A, Kalloo G et al (2016) Decreased function of survival motor neuron protein impairs endocytic pathways. Proc Natl Acad Sci USA 113(30):E4377–E4386. 10.1073/pnas.1600015113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bowerman M, Shafey D, Kothary R (2007) Smn depletion alters profilin II expression and leads to upregulation of the RhoA/ROCK pathway and defects in neuronal integrity. J Mol Neurosci 32(2):120–131. 10.1007/s12031-007-0024-5 [DOI] [PubMed] [Google Scholar]
- 101.Shao J, Welch WJ, DiProspero NA, Diamond MI (2008) Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol Cell Biol 28(17):5196–5208. 10.1128/MCB.00079-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Diamond MI, Cai S, Boudreau A et al (2015) Subcellular localization and Ser-137 phosphorylation regulate tumor-suppressive activity of profilin-1. J Biol Chem 290(14):9075–9086. 10.1074/jbc.M114.619874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Da Silva JS, Medina M, Zuliani C, Di Nardo A, Witke W, Dotti CG (2003) RhoA/ROCK regulation of neuritogenesis via profilin IIa–mediated control of actin stability. J Cell Biol 162(7):1267–1279. 10.1083/jcb.200304021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meberg PJ (2000) Signal-regulated ADF/cofilin activity and growth cone motility. Mol Neurobiol 21(1–2):97–107. 10.1385/MN:21:1-2:097 [DOI] [PubMed] [Google Scholar]
- 105.Endo M, Ohashi K, Mizuno K (2007) LIM kinase and slingshot are critical for neurite extension. J Biol Chem 282(18):13692–13702. 10.1074/jbc.M610873200 [DOI] [PubMed] [Google Scholar]
- 106.Fujita A, Hattori Y, Takeuchi T, Kamata Y, Hata F (2001) NGF induces neurite outgrowth via a decrease in phosphorylation of myosin light chain in PC12 cells. NeuroReport 12(16):3599–3602. 10.1097/00001756-200111160-00045 [DOI] [PubMed] [Google Scholar]
- 107.Gallo G (2011) The cytoskeletal and signaling mechanisms of axon collateral branching. Dev Neurobiol 71(3):201–220. 10.1002/dneu.20852 [DOI] [PubMed] [Google Scholar]
- 108.Loudon RP, Silver LD, Yee HF, Gallo G (2006) RhoA-kinase and myosin II are required for the maintenance of growth cone polarity and guidance by nerve growth factor. J Neurobiol 66(8):847–867. 10.1002/neu.20258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dontchev VD, Letourneau PC (2003) Growth cones integrate signaling from multiple guidance cues. J Histochem Cytochem 51(4):435–444. 10.1177/002215540305100405 [DOI] [PubMed] [Google Scholar]
- 110.Ackermann M, Matus A (2003) Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci 6(11):1194–1200. 10.1038/nn1135 [DOI] [PubMed] [Google Scholar]
- 111.Walter LM, Rademacher S, Pich A, Claus P (2021) Profilin2 regulates actin rod assembly in neuronal cells. Sci Rep 11(1):10287. 10.1038/s41598-021-89397-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cichon J, Sun C, Chen B et al (2012) Cofilin aggregation blocks intracellular trafficking and induces synaptic loss in hippocampal neurons. J Biol Chem 287(6):3919–3929. 10.1074/jbc.M111.301911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oprea GE, Kröber S, McWhorter ML et al (2008) Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science 320(5875):524–527. 10.1126/science.1155085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Delanote V, Vandekerckhove J, Gettemans J (2005) Plastins: versatile modulators of actin organization in (patho)physiological cellular processes. Acta Pharmacol Sin 26(7):769–779. 10.1111/j.1745-7254.2005.00145.x [DOI] [PubMed] [Google Scholar]
- 115.Yener İH, Topaloglu H, Erdem-Özdamar S, Dayangac-Erden D (2017) Transcript levels of plastin 3 and neuritin 1 modifier genes in spinal muscular atrophy siblings. Pediatr Int 59(1):53–56. 10.1111/ped.13052 [DOI] [PubMed] [Google Scholar]
- 116.Ruhno C, McGovern VL, Avenarius MR et al (2019) Complete sequencing of the SMN2 gene in SMA patients detects SMN gene deletion junctions and variants in SMN2 that modify the SMA phenotype. Hum Genet 138(3):241–256. 10.1007/s00439-019-01983-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wadman RI, Jansen MD, Curial CAD et al (2020) Analysis of FUS, PFN2, TDP-43, and PLS3 as potential disease severity modifiers in spinal muscular atrophy. Neurol Genet 6(1):e386. 10.1212/NXG.0000000000000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stratigopoulos G, Lanzano P, Deng L et al (2010) Association of Plastin 3 expression with disease severity in spinal muscular atrophy only in postpubertal females. Arch Neurol 67(10):1252–1256. 10.1001/archneurol.2010.239 [DOI] [PubMed] [Google Scholar]
- 119.Yanyan C, Yujin Q, Jinli B, Yuwei J, Hong W, Fang S (2014) Correlation of PLS3 expression with disease severity in children with spinal muscular atrophy. J Hum Genet 59(1):24–27. 10.1038/jhg.2013.111 [DOI] [PubMed] [Google Scholar]
- 120.Ackermann B, Kröber S, Torres-Benito L et al (2013) Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum Mol Genet 22(7):1328–1347. 10.1093/hmg/dds540 [DOI] [PubMed] [Google Scholar]
- 121.Hao LT, Wolman M, Granato M, Beattie CE (2012) Survival motor neuron affects plastin 3 protein levels leading to motor defects. J Neurosci 32(15):5074–5084. 10.1523/JNEUROSCI.5808-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hosseinibarkooie S, Peters M, Torres-Benito L et al (2016) The power of human protective modifiers: PLS3 and CORO1C unravel impaired endocytosis in spinal muscular atrophy and rescue SMA phenotype. Am J Hum Genet 99(3):647–665. 10.1016/j.ajhg.2016.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kaifer KA, Villalón E, Osman EY et al (2017) Plastin-3 extends survival and reduces severity in mouse models of spinal muscular atrophy. JCI Insight 2(5):e89970. 10.1172/jci.insight.89970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alrafiah A, Karyka E, Coldicott I et al (2018) Plastin 3 promotes motor neuron axonal growth and extends survival in a mouse model of spinal muscular atrophy. Mol Therapy Methods Clin Dev 9:81–89. 10.1016/j.omtm.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McGovern VL, Massoni-Laporte A, Wang X et al (2015) Plastin 3 Expression does not modify spinal muscular atrophy severity in the ∆7 SMA mouse. PLoS ONE. 10(7):e0132364. 10.1371/journal.pone.0132364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Dijk FS, Zillikens MC, Micha D et al (2013) PLS3 mutations in X-linked osteoporosis with fractures. N Engl J Med 369(16):1529–1536. 10.1056/NEJMoa1308223 [DOI] [PubMed] [Google Scholar]
- 127.Fahiminiya S, Majewski J, Al-Jallad H et al (2014) Osteoporosis caused by mutations in PLS3: clinical and bone tissue characteristics. J Bone Miner Res 29(8):1805–1814. 10.1002/jbmr.2208 [DOI] [PubMed] [Google Scholar]
- 128.Giganti A, Plastino J, Janji B et al (2005) Actin-filament cross-linking protein T-plastin increases Arp2/3-mediated actin-based movement. J Cell Sci 118(6):1255–1265. 10.1242/jcs.01698 [DOI] [PubMed] [Google Scholar]
- 129.Kovar DR, Staiger CJ, Weaver EA, McCurdy DW (2000) AtFim1 is an actin ®lament crosslinking protein from. Plant J 24(5):625–636 [DOI] [PubMed] [Google Scholar]
- 130.Garbett D, Bisaria A, Yang C et al (2020) T-Plastin reinforces membrane protrusions to bridge matrix gaps during cell migration. Nat Commun 11(1):4818. 10.1038/s41467-020-18586-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dent EW, Gertler FB (2003) Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron 40(2):209–227. 10.1016/s0896-6273(03)00633-0 [DOI] [PubMed] [Google Scholar]
- 132.Kübler E, Riezman H (1993) Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J 12(7):2855–2862. 10.1002/j.1460-2075.1993.tb05947.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Doherty GJ, McMahon HT (2009) Mechanisms of endocytosis. Annu Rev Biochem 78:857–902. 10.1146/annurev.biochem.78.081307.110540 [DOI] [PubMed] [Google Scholar]
- 134.Südhof TC (2000) The synaptic vesicle cycle revisited. Neuron 28(2):317–320. 10.1016/s0896-6273(00)00109-4 [DOI] [PubMed] [Google Scholar]
- 135.Rizzoli SO, Betz WJ (2005) Synaptic vesicle pools. Nat Rev Neurosci 6(1):57–69. 10.1038/nrn1583 [DOI] [PubMed] [Google Scholar]
- 136.Mooren OL, Galletta BJ, Cooper JA (2012) Roles for actin assembly in endocytosis. Annu Rev Biochem 81:661–686. 10.1146/annurev-biochem-060910-094416 [DOI] [PubMed] [Google Scholar]
- 137.Walsh MB, Janzen E, Wingrove E et al (2020) Genetic modifiers ameliorate endocytic and neuromuscular defects in a model of spinal muscular atrophy. BMC Biol 18(1):127. 10.1186/s12915-020-00845-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dombert B, Balk S, Lüningschrör P et al (2017) BDNF/trkB induction of calcium transients through Cav2 Calcium channels in motoneurons corresponds to F-actin assembly and growth cone formation on β2-chain laminin (221). Front Mol Neurosci 10:346. 10.3389/fnmol.2017.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tejero R, Balk S, Franco-Espin J et al (2020) R-Roscovitine improves motoneuron function in mouse models for spinal muscular atrophy. iScience 23(2):100826. 10.1016/j.isci.2020.100826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tejero R, Lopez-Manzaneda M, Arumugam S, Tabares L (2016) Synaptotagmin-2, and -1, linked to neurotransmission impairment and vulnerability in Spinal Muscular Atrophy. Hum Mol Genet 25(21):4703–4716. 10.1093/hmg/ddw297 [DOI] [PubMed] [Google Scholar]
- 141.Hennlein L, Ghanawi H, Gerstner F et al (2023) Plastin 3 rescues cell surface translocation and activation of TrkB in spinal muscular atrophy. J Cell Biol 222(3):e202204113. 10.1083/jcb.202204113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lyon AN, Pineda RH, Hao LT, Kudryashova E, Kudryashov DS, Beattie CE (2014) Calcium binding is essential for plastin 3 function in Smn-deficient motoneurons. Hum Mol Genet 23(8):1990–2004. 10.1093/hmg/ddt595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jablonka S, Beck M, Lechner BD, Mayer C, Sendtner M (2007) Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy. J Cell Biol 179(1):139–149. 10.1083/jcb.200703187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mitchison T, Kirschner M (1984) Dynamic instability of microtubule growth. Nature 312(5991):237–242. 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- 145.Desai A, Mitchison TJ (1997) Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13:83–117. 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- 146.Goodson HV, Jonasson EM (2018) Microtubules and microtubule-associated proteins. Cold Spring Harb Perspect Biol 10(6):a022608. 10.1101/cshperspect.a022608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bodakuntla S, Jijumon AS, Villablanca C, Gonzalez-Billault C, Janke C (2019) Microtubule-associated proteins: structuring the cytoskeleton. Trends Cell Biol 29(10):804–819. 10.1016/j.tcb.2019.07.004 [DOI] [PubMed] [Google Scholar]
- 148.Wen HL, Lin YT, Ting CH, Lin-Chao S, Li H, Hsieh-Li HM (2010) Stathmin, a microtubule-destabilizing protein, is dysregulated in spinal muscular atrophy†. Hum Mol Genet 19(9):1766–1778. 10.1093/hmg/ddq058 [DOI] [PubMed] [Google Scholar]
- 149.Bora G, Hensel N, Rademacher S et al (2021) Microtubule-associated protein 1B dysregulates microtubule dynamics and neuronal mitochondrial transport in spinal muscular atrophy. Hum Mol Genet 29(24):3935–3944. 10.1093/hmg/ddaa275 [DOI] [PubMed] [Google Scholar]
- 150.Bora G, Sucularlı C, Hensel N, Claus P, Yurter HE (2019) Investigations of microtubule-associated protein 2 gene expression in spinal muscular atrophy. Jpr 6(2):148–154. 10.4274/jpr.galenos.2019.71473 [Google Scholar]
- 151.Miller N, Feng Z, Edens BM et al (2015) Non-aggregating tau phosphorylation by cyclin-dependent kinase 5 contributes to motor neuron degeneration in spinal muscular atrophy. J Neurosci 35(15):6038–6050. 10.1523/JNEUROSCI.3716-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Farah CA, Nguyen MD, Julien JP, Leclerc N (2003) Altered levels and distribution of microtubule-associated proteins before disease onset in a mouse model of amyotrophic lateral sclerosis. J Neurochem 84(1):77–86. 10.1046/j.1471-4159.2003.01505.x [DOI] [PubMed] [Google Scholar]
- 153.Dubey J, Ratnakaran N, Koushika SP (2015) Neurodegeneration and microtubule dynamics: death by a thousand cuts. Front Cell Neurosci 9:343. 10.3389/fncel.2015.00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chauvin S, Sobel A (2015) Neuronal stathmins: a family of phosphoproteins cooperating for neuronal development, plasticity and regeneration. Prog Neurobiol 126:1–18. 10.1016/j.pneurobio.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 155.Kline RA, Kaifer KA, Osman EY et al (2017) Comparison of independent screens on differentially vulnerable motor neurons reveals alpha-synuclein as a common modifier in motor neuron diseases. PLoS Genet 13(3):e1006680. 10.1371/journal.pgen.1006680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Villalón E, Kline RA, Smith CE et al (2019) AAV9-Stathmin1 gene delivery improves disease phenotype in an intermediate mouse model of spinal muscular atrophy. Hum Mol Genet 28(22):3742–3754. 10.1093/hmg/ddz188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sato-Yoshitake R, Shiomura Y, Miyasaka H, Hirokawa N (1989) Microtubule-associated protein 1B: molecular structure, localization, and phosphorylation-dependent expression in developing neurons. Neuron 3(2):229–238. 10.1016/0896-6273(89)90036-6 [DOI] [PubMed] [Google Scholar]
- 158.Villarroel-Campos D, Gonzalez-Billault C (2014) The MAP1B case: an old MAP that is new again. Dev Neurobiol 74(10):953–971. 10.1002/dneu.22178 [DOI] [PubMed] [Google Scholar]
- 159.Yang M, Wu M, Xia P et al (2012) The role of microtubule-associated protein 1B in axonal growth and neuronal migration in the central nervous system. Neural Regen Res 7(11):842–848. 10.3969/j.issn.1673-5374.2012.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gonzalez-Billault C, Owen R, Gordon-Weeks PR, Avila J (2002) Microtubule-associated protein 1B is involved in the initial stages of axonogenesis in peripheral nervous system cultured neurons. Brain Res 943(1):56–67. 10.1016/s0006-8993(02)02534-9 [DOI] [PubMed] [Google Scholar]
- 161.Brzustowicz LM, Kleyn PW, Boyce FM et al (1992) Fine-mapping of the spinal muscular atrophy locus to a region flanked by MAP1B and D5S6. Genomics 13(4):991–998. 10.1016/0888-7543(92)90012-h [DOI] [PubMed] [Google Scholar]
- 162.Marín I, Fontdevila A (1998) Stable Drosophila buzzatii-Drosophila koepferae hybrids. J Hered 89(4):336–339. 10.1093/jhered/89.4.336 [DOI] [PubMed] [Google Scholar]
- 163.Franco-Espin J, Gatius A, Armengol JÁ et al (2022) SMN is physiologically downregulated at wild-type motor nerve terminals but aggregates together with neurofilaments in SMA mouse models. Biomolecules 12(10):1524. 10.3390/biom12101524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gadadhar S, Bodakuntla S, Natarajan K, Janke C (2017) The tubulin code at a glance. J Cell Sci 130(8):1347–1353. 10.1242/jcs.199471 [DOI] [PubMed] [Google Scholar]
- 165.Schulze E, Asai DJ, Bulinski JC, Kirschner M (1987) Posttranslational modification and microtubule stability. J Cell Biol 105(5):2167–2177. 10.1083/jcb.105.5.2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nieuwenhuis J, Brummelkamp TR (2019) The tubulin detyrosination cycle: function and enzymes. Trends Cell Biol 29(1):80–92. 10.1016/j.tcb.2018.08.003 [DOI] [PubMed] [Google Scholar]
- 167.Janke C, Magiera MM (2020) The tubulin code and its role in controlling microtubule properties and functions. Nat Rev Mol Cell Biol 21(6):307–326. 10.1038/s41580-020-0214-3 [DOI] [PubMed] [Google Scholar]
- 168.Xu CC, Denton KR, Wang ZB, Zhang X, Li XJ (2016) Abnormal mitochondrial transport and morphology as early pathological changes in human models of spinal muscular atrophy. Dis Model Mech 9(1):39–49. 10.1242/dmm.021766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Miller N, Shi H, Zelikovich AS, Ma YC (2016) Motor neuron mitochondrial dysfunction in spinal muscular atrophy. Hum Mol Genet 25(16):3395–3406. 10.1093/hmg/ddw262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dehmelt L, Halpain S (2004) The MAP2/Tau family of microtubule-associated proteins. Genome Biol 6:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Teng J, Takei Y, Harada A, Nakata T, Chen J, Hirokawa N (2001) Synergistic effects of MAP2 and MAP1B knockout in neuronal migration, dendritic outgrowth, and microtubule organization. J Cell Biol 155(1):65–76. 10.1083/jcb.200106025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ramkumar A, Jong BY, Ori-McKenney KM (2018) ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule-associated proteins. Dev Dyn 247(1):138–155. 10.1002/dvdy.24599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zobaroğlu Özer P, Koyunoğlu D, Son ÇD, Erdem-Yurter H, Bora G (2022) SMN loss dysregulates microtubule-associated proteins in spinal muscular atrophy model. Mol Cell Neurosci 120:103725. 10.1016/j.mcn.2022.103725 [DOI] [PubMed] [Google Scholar]
- 174.Sayas CL, Avila J (2014) Regulation of EB1/3 proteins by classical MAPs in neurons. BioArchitecture 4(1):1–5. 10.4161/bioa.27774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sayas CL, Tortosa E, Bollati F, Ramírez-Ríos S, Arnal I, Avila J (2015) Tau regulates the localization and function of End-binding proteins 1 and 3 in developing neuronal cells. J Neurochem 133(5):653–667. 10.1111/jnc.13091 [DOI] [PubMed] [Google Scholar]
- 176.Tortosa E, Galjart N, Avila J, Sayas CL (2013) MAP1B regulates microtubule dynamics by sequestering EB1/3 in the cytosol of developing neuronal cells. EMBO J 32(9):1293–1306. 10.1038/emboj.2013.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ramirez-Rios S, Denarier E, Prezel E et al (2016) Tau antagonizes end-binding protein tracking at microtubule ends through a phosphorylation-dependent mechanism. Mol Biol Cell 27(19):2924–2934. 10.1091/mbc.E16-01-0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kapitein LC, Yau KW, Gouveia SM et al (2011) NMDA receptor activation suppresses microtubule growth and spine entry. J Neurosci 31(22):8194–8209. 10.1523/JNEUROSCI.6215-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Akhmanova A, Hoogenraad CC (2005) Microtubule plus-end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol 17(1):47–54. 10.1016/j.ceb.2004.11.001 [DOI] [PubMed] [Google Scholar]
- 180.Komarova Y, Lansbergen G, Galjart N, Grosveld F, Borisy GG, Akhmanova A (2005) EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol Biol Cell 16(11):5334–5345. 10.1091/mbc.e05-07-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nakagawa H, Koyama K, Murata Y, Morito M, Akiyama T, Nakamura Y (2000) EB3, a novel member of the EB1 family preferentially expressed in the central nervous system, binds to a CNS-specific APC homologue. Oncogene 19(2):210–216. 10.1038/sj.onc.1203308 [DOI] [PubMed] [Google Scholar]
- 182.Poobalasingam T, Bianco F, Oozeer F, Gordon-Weeks PR (2022) The drebrin/EB3 pathway regulates cytoskeletal dynamics to drive neuritogenesis in embryonic cortical neurons. J Neurochem 160(2):185–202. 10.1111/jnc.15502 [DOI] [PubMed] [Google Scholar]
- 183.Geraldo S, Khanzada UK, Parsons M, Chilton JK, Gordon-Weeks PR (2008) Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat Cell Biol 10(10):1181–1189. 10.1038/ncb1778 [DOI] [PubMed] [Google Scholar]
- 184.Bjelić S, De Groot CO, Schärer MA et al (2012) Interaction of mammalian end binding proteins with CAP-Gly domains of CLIP-170 and p150(glued). J Struct Biol 177(1):160–167. 10.1016/j.jsb.2011.11.010 [DOI] [PubMed] [Google Scholar]
- 185.Moughamian AJ, Osborn GE, Lazarus JE, Maday S, Holzbaur ELF (2013) Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J Neurosci 33(32):13190–13203. 10.1523/JNEUROSCI.0935-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Alves CRR, Petrillo M, Spellman R et al (2021) Implications of circulating neurofilaments for spinal muscular atrophy treatment early in life: a case series. Mol Ther Methods Clin Dev 23:524–538. 10.1016/j.omtm.2021.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nitz E, Smitka M, Schallner J et al (2021) Serum neurofilament light chain in pediatric spinal muscular atrophy patients and healthy children. Ann Clin Transl Neurol 8(10):2013–2024. 10.1002/acn3.51449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Johannsen J, Weiss D, Daubmann A, Schmitz L, Denecke J (2021) Evaluation of putative CSF biomarkers in paediatric spinal muscular atrophy (SMA) patients before and during treatment with nusinersen. J Cell Mol Med 25(17):8419–8431. 10.1111/jcmm.16802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Faravelli I, Meneri M, Saccomanno D et al (2020) Nusinersen treatment and cerebrospinal fluid neurofilaments: an explorative study on Spinal Muscular Atrophy type 3 patients. J Cell Mol Med 24(5):3034–3039. 10.1111/jcmm.14939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Musso G, Bello L, Capece G et al (2024) Neurofilament light chain and profilin-1 dynamics in 30 spinal muscular atrophy type 3 patients treated with nusinersen. Eur J Neurol 31(10):e16393. 10.1111/ene.16393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Darras BT, Crawford TO, Finkel RS et al (2019) Neurofilament as a potential biomarker for spinal muscular atrophy. Ann Clin Transl Neurol 6(5):932–944. 10.1002/acn3.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Spicer C, Lu CH, Catapano F et al (2021) The altered expression of neurofilament in mouse models and patients with spinal muscular atrophy. Ann Clin Transl Neurol 8(4):866–876. 10.1002/acn3.51336 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were used for the research described in the article.