Abstract
Introduction
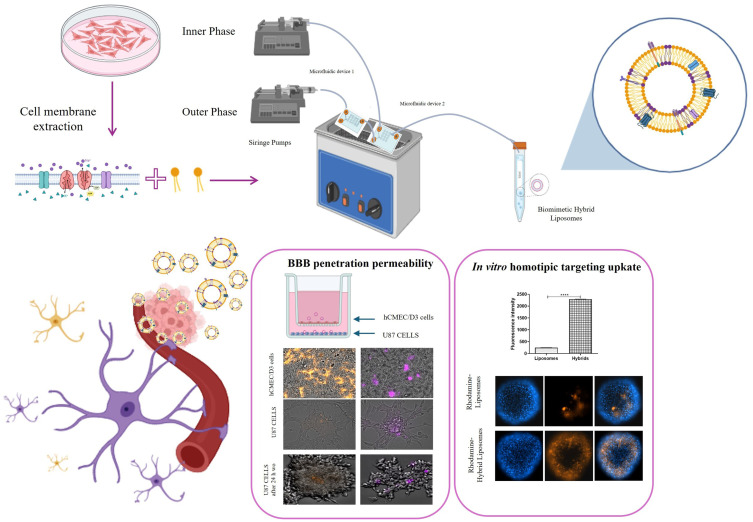
The treatment of glioblastoma is hindered by the blood–brain barrier (BBB) and rapid drug clearance by the immune system. To address these challenges, we propose a novel drug delivery system using liposomes modified with cell membrane fragments. These modified liposomes can evade the immune system, cross the BBB, and accumulate in tumor tissue through homotypic targeting, thereby delivering drugs like paclitaxel and carboplatin more effectively.
Methods
In this work, the hybrid liposomes were synthesized using microfluidics and integrating 3D printing to produce the microfluidic devices. In vitro, we explored the homotypic targeting capability, BBB passing ability, and therapeutic efficacy of paclitaxel and carboplatin.
Results
The production of hybrid liposomes by microfluidics has been key to creating high-quality biomimetic nanoparticles, and the integration of 3D printing has simplified the production of microfluidic devices, making the process more efficient and economical. In vitro experiments have shown that these drug-loaded biomimetic hybrid liposomes are able to reach the homotypic target, cross the BBB, and maintain the efficacy of paclitaxel and carboplatin.
Conclusions
The development of biomimetic hybrid liposomes represents a promising approach for the treatment of glioblastoma. By combining the advantages of liposomal drug delivery with the stealth properties and targeting capabilities of cell membrane fragments, these nanoparticles can potentially overcome the challenges associated with traditional therapies.
Keywords: bioinspired materials, biomimetic nanoparticles, glioblastoma cells, emerging technology, drug delivery system, microfluidics
Graphical Abstract
Introduction
Glioma presents significant challenges due to its unique location and complex etiology. Currently, two major issues hinder glioma treatment. First, chemotherapeutic drugs are rapidly and thoroughly cleared from the bloodstream by the immune system. Second, these drugs cannot effectively cross the blood–brain barrier (BBB), preventing them from accumulating sufficiently in the tumor.1 Therefore, there is a critical need to develop advanced drug delivery systems that can bypass the immune system and enable drugs to penetrate the BBB. Nanomaterials possess properties that make them highly beneficial for drug delivery systems. However, their effectiveness is often compromised as the immune system identifies and clears these foreign nanoparticles (NPs), resulting in decreased targeting efficiency.2 Over the past decade, liposomes have represented NP-based drug delivery systems widely used for their ability to efficiently load, control the release of drugs, and offer excellent modifiability. Although unmodified liposomes are among the most traditional and widely commercialized nanoformulations, they still struggle to effectively cross the BBB effectively. Therefore, rational modifications are crucial to improving the brain-targeting abilities of liposomes.
A recent innovative strategy in nanomedicine involves modifying the surface of NPs with cell membranes (CMs). This approach has gained significant attention due to the natural biological molecules in CMs that provide “stealth” properties and active targeting functions. Leveraging these characteristics, biomimetic CM-coated or hybrid NPs can evade the immune system and accumulate at tumor sites. Cancer cells offer unique advantages for this purpose, such as easy large scale in vitro production and homotypic targeting capabilities, making them ideal candidates for NP camouflage in anti-tumor therapy.3,4 The preparation of CMs functionalized biomimetic NPs is an important aspect for further applications. Among the traditional methods of preparation, we have physical co-extraction and sonication, while more advanced methods include microfluidics.1
In recent years, microfluidic systems have emerged as a promising solution for the precise control of discrete volumes and continuous fluids at the submillimeter scale. These systems have revolutionized nanomedicine and significantly advanced industrial research in this field. The miniaturization of the manufacturing process using microfluidic channels overcomes the limitations of top-down approaches and bulk mixing due to their microscale size, precise flow control, particle size tuning and reproducibility.3,5–7 The definition of biomimetic hybrid NPs includes both NPs coated with CMs and those in which lipids or polymers fuse with CMs. In the literature, examples of biomimetic hybrid coated NPs prepared by microfluidics are numerous and have proven promising with successful applications in gene silencing at the cellular level and in targeted therapy of subcutaneous tumors at the animal level.8 However, regarding the production of biomimetic hybrid fused NPs using microfluidics, there are few examples. In our previous work, we designed a customized, flexible and efficient microfluidic platform to produce biomimetic hybrid fused liposomes. Using the intricate internal geometric architecture of two different 3D printed microfluidic devices combined with active mixing promoted by ultrasound, we produced high-quality monodisperse biomimetic hybrid fused liposomes.3 In the present study, to demonstrate the versatility of the microfluidic platform in preparing biomimetic liposomes, we tested microfluidic devices with different geometries arranged in series and coupled with external force fields, thereby combining active and passive mixing. Once again, microfluidic devices are produced using 3D printing.9,10 The ability to manufacture a microfluidic device in a single step from a CAD model is advantageous in the field of microfluidics. 3D printing techniques offer an effective solution for microfluidic prototyping that is inexpensive, easy, and fast.
The aim of this work was to further demonstrate that the customization of microfluidic chips enabled by 3D printing technology, combined with the versatility of biomimetic hybrid drug delivery systems, can represent a successful strategy for the treatment of cancer. In our previous work, we demonstrated that the biomimetic hybrid liposomes fused with CMs derived from melanoma biopsies were capable of selectively recognizing the original tumor tissue. In this study, we aim to show that hybrid liposomes fused with the CMs of U87 glioblastoma cells are capable of selectively recognizing target tumor cells, being internalized, and subsequently releasing their cargo of anticancer drugs, which are active in the treatment of glioblastoma. Moreover, we have also demonstrated that these systems can cross the blood–brain barrier (BBB) using an in vitro model. Specifically, the anticancer drugs loaded into the hybrid liposomes as a model therapy for glioblastoma were Paclitaxel (PCX) and Carboplatin (CBP). The choice of drugs was based on literature data suggesting the use of paclitaxel and carboplatin in the treatment of glioblastoma. The activity of these drugs has been studied both individually and in combination with glioblastoma animal models11 and directly in humans through surgical intracavitary application of paclitaxel and carboplatin encapsulated in liquid crystalline cubic phases.12 Recently, evidence supported their use in a clinical trial on patients with glioblastoma, where brain access was achieved using an ultrasound technique that induced BBB opening with microbubbles.13 However, since this process of BBB crossing is transient, we considered a more stable drug delivery system for this study.
Thus, we conducted in vitro combination therapy studies, simulating the preliminary crossing of the BBB before reaching the target tumor cell. Moreover, the hybrid liposomes were comprehensively characterized in terms of their physicochemical and morphological properties, including the validation of the effective fusion of CMs with the lipids. Subsequently, they were characterized in in vitro biological experiments to validate homotypic targeting, the ability to cross the BBB, and the capacity to maintain the effect of chemotherapeutic drugs, CBP and PCX, even when loaded in the hybrid liposomes, for the treatment of in vitro glioblastoma cells.
Materials and Methods
Materials
All chemicals were purchased at the highest available purity and used as received without further purification or distillation. Polypropylene (PP) filament was purchased from BASF (Germany). PCX, CBP, Cholesterol, PBS (Dulbecco’s Phosphate Buffered Saline), Tween 80, PKH67 Fluorescent Cell Linker Kits (green, fluorescent dye), PKH26 Red Fluorescent, Ethanol 99% (v/v) (EtOH) and Dimethylsulfoxide (DMSO) were purchased from Sigma Aldrich. Dulbecco’s Modified Eagle’s medium (DMEM), heat inactivated fetal bovine serum (FBS), L-glutamine (200 mm), non-essential amino acids (NEAA), penicillin (100 IU/mL), streptomycin (100 mg/mL) and trypsin (2.5%) were acquired from HyClone Waltham, USA. 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (ammonium salt) (NBD-PE) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Liss Rhod PE) were bought from Avanti Polar Lipids, ab8245 Anti-GAPDH antibody [6C5] - Loading Control (Abcam), and Ripa buffer was purchased from Merck Life Science srl (Milano, Italy).
Cell Membrane Isolation from U87-Glioblastoma Cell Lines
U-87 MG cell membranes (CMs) were extracted following the previously described protocol.3 In brief, the cellular pellet was lysed by adding MilliQ water and then was exposed to two cycles of freezing and thawing, which allowed the cellular structure to disrupt. The resulting dispersion was sonicated with an ultrasound probe tip (9 W) for three minutes in an ice bath, then it was centrifuged at 4°C for 15 minutes at 15,000 RCF. The supernatant was collected and ultra-centrifuged at 100,000 RCF for 60 min. Following, the CMs were dispersed using an adequate amount of water for injectable preparations and stored at –20° C. The amount of CMs extracted was quantified by analyzing the total protein content using the Bradford method.14
Design and Manufacturing of 3D Printing Microfluidic Devices
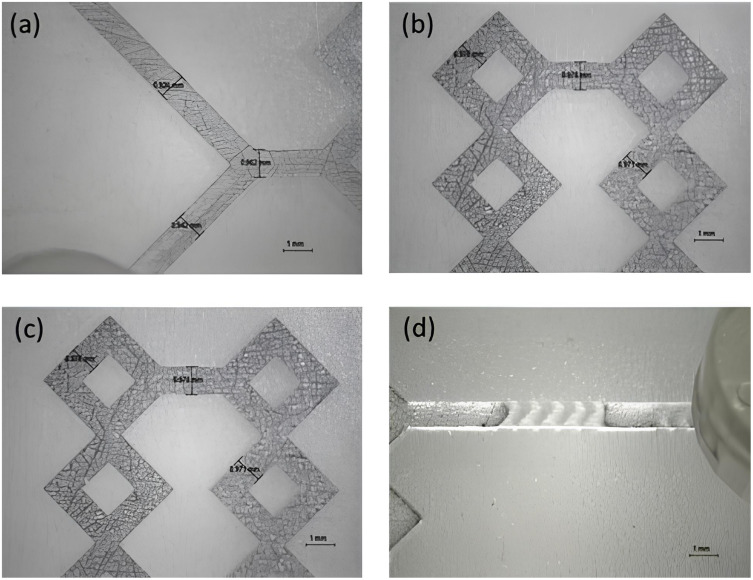
According to a previous research, 3D-printed microfluidic devices were used for the production of liposomes and hybrid liposomes.10 Two different T-shaped devices were applied, device 1 and device 2, respectively. Both chips are characterized by the presence of 90° contact angle at the meeting point of inner and outer phases and are composed by squared channels; as reported in the Figure 1, device 1 consists in a long-circumvolved micromixer, while device 2 is composed by a shorter micromixing path followed by a main channel decorated with herringbone modified structure. Briefly, the designs were generated using an open-source software (TinkerCAD, USA) and they were built using the digital-light processing AsigaMax UV (Asiga, Alexandria, Australia) printer. Following, uploading to Asiga Composer (Version 1.2) slicing software, the devices were printed using PlasCLEAR resin (Asiga, Alexandria, Australia) at a resolution of 0.0025 mm. After manufacturing, they were cleaned according to the printer’s instructions, which included soaking them in isopropyl alcohol (IPA) and sonicating them for eight minutes. After that, the devices were air dried and exposed to a 385 nm UV chamber for 20 minutes to cure (Asiga Flash, Melbourne).
Figure 1.
Representative images of device 1 (a and b), and device 2 (c and d). Both devices have rectangular shape (28.00 × 50.00×2.70 mm) and same dimensions as the micromixing path (1.0 × 1.0×2.30 mm). Additionally, the device 2 is decorated with a modified herringbone geometry (0.8 × 1.94×1.22 mm).
Preparation of Fused Hybrid Liposomes by Microfluidic Technique
The hybrid liposomes were prepared by a nanoprecipitation process using two different 3D-printed microfluidics devices as reported above.10 The miscible fluids were introduced into the microfluidic device through polyethylene syringes attached to syringe pumps, ensuring a steady flow rate.
The organic phase was composed of a mixture containing 4.2 mg/mL of phosphatidylcholine from soybean and 0.8 mg/mL of cholesterol in ethanol, along with 1 mg/mL of PCX and CBP. The aqueous phase consisted solely of PBS for liposomes, while for hybrid liposomes, it included PBS and CMs (equivalent to 0.6 mg of CM protein content). The organic and aqueous phases represented the inner and outer phases, respectively. The preparation of NPs was tailored by adjusting the Total Flow Rate (TFR) (ie, the sum of the flow rates for the aqueous and organic solutions) and to obtain monodispersed liposome and hybrid liposome, the optimization of the process demanded multiple experimental repetitions. Nonetheless, following optimization of the procedure, the two fluids were introduced through distinct inlets into the device at TFR ranging from 6 to 9. The liquids flowed from their respective syringes into the devices through small tubes. While preparing all formulations, the microfluidic devices were subjected to sonication using a bath sonicator (CP102 Ultrasonic Cleaner, France) at 60 kHz and a power of 400 W. The resulting preparations were then purified by ultracentrifugation at 45,000 rpm at 4°C for 120 minutes. The formulation included a fluorescent lipid, specifically 17 µg of Liss Rhod PE, which was added to the organic phase to determine the cellular uptake of liposomes and hybrid liposomes.
Membrane-Lipids Fusion Validation by FRET, Flow Cytometry Co-Localization Study and Western Blotting
To perform the Förster resonance energy transfer (FRET) study, two fluorescent lipids were utilized in the production of biomimetic nanoparticles: 1.2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (ammonium salt) (NBD-PE, excitation/emission = 463/536 nm) and 1.2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Liss Rhod PE, excitation/emission = 560/580 nm). Ethanol (inner phase) was used to dissolve both fluorescent lipids. In short, the same method was used to prepare fluorescent liposomes and hybrid liposomes, but with 1.7 μg of NBD-PE and 17 μg of Liss Rhod PE dissolved in the lipid phase. Each sample’s fluorescence spectrum was read between 500 and 650 nm using a Tecan Infinite M200 plate reader with an excitation wavelength of 470 nm. The fluorescence recovery of the donor (NBD-PE) at the lower emission peak (534 nm) indicated increasing amounts of fusion. FRET efficiency, defined as the proportion of donor molecules (NBD) that have transferred their excess energy to the acceptor molecules (rhodamine), was calculated using the following equation as previously described.
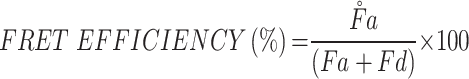 |
where Fa = emission fluorescence of acceptor (rhodamine) and Fd = emission fluorescence of donor (NBD).
As previously described,3 the membrane colocalization study was conducted. PKH67 (excitation/emission = 490/504 nm) was used to label CMs, while 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Liss Rhod PE, excitation/emission = 560/580 nm) was used to label lipid membranes. These dye-labelled membranes were used to prepare hybrid liposomes, which were then analyzed by FCM. Controls included non-labelled CMs and hybrid-liposomes.
Another technique used by us to confirm successful membrane fusion and removal of intracellular components was Western Blot. Gel electrophoresis followed by protein staining was used for the determination of the protein content in samples: membrane extracted from U87 cells, liposomes and hybrid liposomes.
The collected cancer cell membrane, bare liposome, and the hybrid liposome were separately incubated with Radioimmunoprecipitation assay (RIPA) buffer for 30 minutes on the ice with soft agitation and mild sonication for complete lysis to extract the protein. The lysate of the samples was then centrifuged at 16000 g for 20 minutes to remove the impurities, and the supernatant was collected for the protein quantification and Western Blot analysis. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies were used as the main marker.
Particle Size, Size Distribution, Surface Electrostatic Charge and Nanoparticle Tracking Analysis
Using a Zetasizer Nano ZS (Malvern Instruments Ltd., Worcestershire, UK), the size distribution and ζ-potential of all preparations were assessed. Each sample, diluted 1:50 in double-distilled water, was analyzed in disposable polystyrene cuvettes (Sarstedt AG & Co., Germany) at 25 ± 0.1 °C [28]. The surface ζ-potential was measured with 750 μL of the 1:50 dilution in demineralized water of the NPs suspension in a disposable folded capillary cell (DTS1070, Malvern, UK). NanoSight NS300 (Malvern Panalytical) was used to characterize hybrid nanoparticles in terms of size and concentration as hybrid nanoparticles/mL, following manufacturer guidelines (NanoSight NS300 User Manual, MAN0541-02-EN, 2018). The samples were diluted 1:1000 in Milli-Q pre-filtered water and injected with a steady syringe flow rate of 50 μL. Data were obtained from three 60-second movies and reported as mean ± standard deviation for each triplicate.3,15
Transmission Electron Microscopy (TEM) Imaging
Following a well-established procedure,6 the size, shape, and morphology of the NPs were examined.
Ten microliter liposome droplets were dropped on the copper grid (200 mesh), and after 10 minutes, filter paper was used to remove the excessive samples on the grid. Then, 5 μL 0.5% uranyl acetate solution was added to the grid for 2 minutes of negative staining. The TEM (PHILIPS CM120 working at 120 keV) was used for the morphology observation.
In vitro Biological Studies
Cell Culture
Human glioblastoma U87-MG cells were purchased from the American Type Culture Collection (ATCC, VA, USA) and were cultured as previously reported.5,6 For U87-MG tumour spheroid formation, cells were seeded at a density of 5000 cells per well on 96-well plate Corning®Spheroid Microplate and incubated at 37◦C under 5% CO2 for 5 days before performing experiments. Spheroid formation was assessed using the Celldiscoverer 7 Live Cell Imaging System (Zeiss, Jena, Germany). Human brain endothelial cells (hCMEC/D3) were kindly provided by Pierre-Olivier Couraud (Universitè Paris Descartes, Paris, France). As reported in literature,15 cells between passages 25 and 35 were grown for 15 days in EndoGRO medium nourished with EndoGRO MV supplement kit, 10% FBS, basal FGF (200 ng/mL), penicillin–streptomycin (1%), lithium chloride (10 mm) and resveratrol (10 µM) and kept at 37 °C with 5% CO2.
Homotypic Targeting Studies on 2D and 3D Cell Models
In vitro uptake studies were carried out using both 2D and 3D cell models through flow cytometry analysis (FCM) and fluorescence imaging (FI) as detailed by Arduino et al.3 The 2D uptake studies involved U87 cells, from which cell membranes were extracted, after a 2h incubation with liposomes or hybrid liposomes (0.3 μM Liss Rhod PE). These experiments were conducted at both 37°C and 4°C to evaluate the total fluorescence and fluorescence associated solely with the cellular membrane. The analysis utilized the AttuneNxT acoustic focusing cytometer (Thermo Fisher Scientific, Waltham, MA, USA), with 10,000 events counted in the viable gate. The geometric mean of the viable cell population exposed to nanoparticles (NPS) determined internalization, adjusted for cell autofluorescence and NPS compound autofluorescence. The internalized fluorescence data, shown as the difference between the values at 37°C and 4°C, were processed using Attune NxT Analysis Software and CytExpert software v.1.2, presented as mean ± SD, n = 3. For the 3D model, U87 spheroids were incubated with rhodamine-hybrid liposomes and rhodamine-liposomes for 2 h and 24 h and stained with Hoechst 33,342 dye (2 μg/mL, InvitrogenTM) for 30 minutes for nuclei visualization. Spheroids were washed and resuspended in PBS without Ca2+ and Mg2+ for imaging on the Celldiscoverer 7 Cell Imaging System (Zeiss, Jena, Germany) using a Plan-Apochromat 20×/0.95 objective and optovar 0.5x tube lens. Co-localization of the two fluorophores was quantified by circumscribing the internal area of the spheroid and measuring fluorescence intensity using Zen 3.2 software (Carl Zeiss Microscopy, Germany).
In vitro Cytotoxicity Studies
The 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazoliumbromide (MTT) assay was performed in order to evaluate the cytotoxic effect of carboplatin, paclitaxel and drug(s) loaded hybrid liposomes, as described in Di Fonte et al 16. Untreated cells were used as a negative control. Briefly, cells were seeded in 96-well plates at a density of 7500 cells/well and incubated for 24 h in the culture medium to allow attachment. The cells were treated with 0.5 µM paclitaxel/hybrid liposomes–PCX, 50 µM carboplatin/hybrid liposomes–CBP for 24 h. Afterwards, 10 µL of 0.5% MTT was added to each well, and the plates were incubated until the medium was removed and replaced with DMSO (100 µL). The absorbance was measured using a microplate reader. The results are reported in a histogram plot showing the viability of tumor cells as mean (%) ±SD of two different experiments.
BBB Penetration Ability of Hybrid Liposomes
Human brain endothelial cells (hCMEC/D3) were plated at a density of 21,000 cells per well in the upper chamber of a 24-well Transwell™ apparatus (0.4 μm diameter pores-size, Transwell insert surface: 0.33 cm2; Corning Life Sciences) according to the protocol described in Sommonte et al.15 Human U87MG cells were cultured in the lower chamber at a density of 50,000 cells per well. After 21 days, which allowed for the formation of a blood–brain barrier (BBB) indicated by the trans-endothelial electrical resistance (TEER) reaching approximately 150 Ω•cm², a permeability test was conducted. This involved incubating hCMEC/D3 cells with equal amounts of fluorescent hybrid liposomes and liposomes in the upper chamber of the Transwells and then analyzing the BBB penetration ability and the internalized fluorescence in U87 cells in the lower chamber after 2 h and 24 h post-washout by FI.16,17 Fluorescence intensity was quantified by using Celldiscoverer 7 Cell Imaging System (Zeiss, Jena, Germany) and reported as the mean ± SD, n = 3.
Adapting a standard protocol for the determination of BBB penetration of drugs,16,18 the apparent permeability (Papp), in units of cm/second for the Liss Rhod PE labelled Nps, was calculated using the following equation:
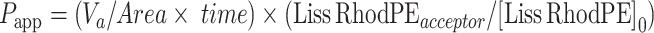 |
where “Va” is the volume in the acceptor well, “Area” is the surface area of the membrane, “time” is the total transport time, “[Liss Rhod PE]acceptor” is the concentration of the Liss Rhod PE measured in the cell culture medium in the bottom chamber using TECAN INFINITE M200 plate reader, and “[Liss Rhod PE]0” is the Liss Rhod PE concentration of the Nps tested in the AP chamber. The flux of fluorescein isothiocyanate-dextran (FD4, Sigma-Aldrich, Italy, 200 ug/mL) and diazepam (75 uM) was used to verify cell barrier function and integrity. The absorbance of Diazepam (tested at 75 uM) samples at wavelengths 254 nm was analyzed on the TECAN INFINITE M200 plate reader, while FD4 samples were evaluated at excitation and emission wavelengths of 485 and 535 nm. Data are reported as the apparent permeability (Papp), calculated in units of cm/s.
Statistical Analysis
The statistical significance was calculated using two-tailed t-tests, while the analysis of variance was performed by using a two-way ANOVA, followed by Bonferroni post hoc tests (GraphPad Prism version. 5.0). Results were reported as the mean ± SD of three independent experiments (**p < 0.01, ***p < 0.001, ****p < 0.0001).
Results and Discussion
Microfluidic technology represents a significant innovation in pharmaceutical technology and nanomedicine. This technique has grown tremendously in recent years, becoming a preferred approach for large-scale nanomedicine applications.19 Among the numerous benefits of microfluidic production are the reduction of time and costs, the elimination of toxic solvents, and a decrease in batch-to-batch variability15,20. This reduction in variability is crucial in drug delivery due to the significant impact of technological properties on the fate of the formulation once administered.
Microfluidics relies on laminar flow in microchannels, where chemical interactions occur at fluid interfaces via diffusion. However, to ensure adequate-phase interaction, systems often require additional elements to promote mixing, typically through passive chaotic advection. This approach uses structured paths and obstacles to introduce controlled disorder, enhancing mixing while maintaining laminar flow. Despite its benefits, microfluidic technology is limited by the high cost and perishability of devices.21,22 Many traditional microfluidic devices are fabricated from glass or silicon using labor-intensive, expensive techniques, which are not only costly but also make the devices fragile and prone to breakage, especially when handling high-throughput production. Additionally, the materials and processes used are often incompatible with certain solvents or chemicals, limiting the range of formulations possible. Another significant drawback is the difficulty in rapidly prototyping and customizing device designs, which is essential for optimizing nanoparticle production processes. 3D printing offers a promising solution to these limitations. By allowing for the rapid, cost-effective fabrication of complex microfluidic structures, 3D printing enables the creation of custom, durable devices tailored to specific production needs. This method also expands the range of compatible materials, allowing for devices made of resilient polymers or other substances that better withstand solvents and physical wear. Moreover, 3D printing allows iterative prototyping, where designs can be quickly adjusted and tested, accelerating optimization of the nanoparticle production process. Thus, 3D printing can make microfluidic technology more accessible, customizable, and durable, addressing many of its current challenges in nanoparticle production.
The present work leverages the beneficial synergy between microfluidics and 3D printing techniques.9 Moreover, it is interesting to note that one of the most important problems to solve when creating biomimetic nanoparticles is the fragmentation of cell membranes (CMs), which may be ineffective using passive microfluidic devices. Among the most effective strategies, in fact, is the use of ultrasound, which facilitates the rupture of membranes and assists in the formation of hybrid biomimetic nanosystems.15 In addition, ultra-rapid and homogeneous mixing is crucial for the fusion of CMs with lipids and to produce NPs with a narrow size distribution. The literature lacks extensive research on biomimetic hybrid liposomes produced via microfluidic techniques. While many studies have focused on coating nanoparticles with cell membranes, few have explored the fusion of cell membranes with synthetic phospholipids. The novelty of our work lies primarily in the microfluidic production technique, which enables high reproducibility and potential for industrial-scale manufacturing. Additionally, the integration of 3D printing with microfluidics allows us to create fully customized devices, adding another layer of innovation to our approach. To our knowledge, there are no examples in the literature of biohybrid fused liposomes produced by microfluidics specifically applied to glioma treatment.
Preparation and Characterization of Biomimetic Hybrid Liposomes
Inspired by a previous study,10 two T-shaped microfluidic devices, namely device 1 and device 2, were applied due to the peculiarity of their internal geometry. In fact, devices 1 and 2 (Figure 2) share the same basic characteristics; in fact, both are T-shaped passive microfluidic devices with an inlet angle of 90°C, and both have squared channels characterized by the presence of angles that could facilitate mixing between phases and CM fragmentation.
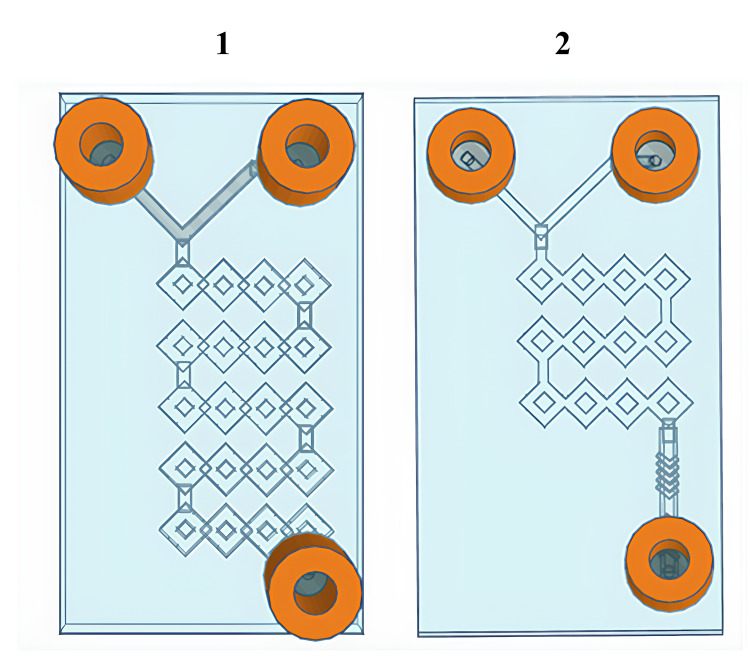
Figure 2.
Graphical representation of 3D-printed microfluidic devices:10 Microfluidic device 1 and microfluidic device 2.
In a more detailed way, the device 1 consists in a long and circumvolved micromixing channel that begins at the point where the two phases meet and continues until the outlet, whereas the device 2 has a shorter micromixer path that is injected into a main channel decorated with a herringbone-modified structure. Both devices were purposely engineered to dramatically increase the micromixing between phases by exploiting the passive chaotic advection phenomenon while keeping the flow over control. Thus, both devices take advantage of the split-and-recombine (SAR) model, allowing to significantly increase the area/volume ratio and thus the number and efficiency of phase-to-phase exchanges without increasing the length of the channel.21,23
Before describing the methodological approach followed for production, it was important to present the tests conducted on the individual devices to evaluate the influence of device geometries on the production of hybrid liposomes. First, the device 2 was applied by testing different TFRs, given the presence of the herringbone structure inside, which, as widely in literature, represents one of the most suitable geometries for producing nanosized and monodispersed lipid nanoparticles.24 Thus, liposomes and hybrid liposomes were generated by immersing device 2 in a sonicator bathtub to aid in the fragmentation and dispersion of CMs in the outer phase, while various TFRs were tested to determine the optimal operating conditions. Based on the results shown in Table 1, it is possible to observe that as the TFR increases, nanosystems become smaller and more monodisperse due to the phenomena of mixing and passive chaotic advection induced by the micromixer and the herringbone geometry, in accordance with the data in the literature.24,25
Table 1.
Brief Overview of the Formulations Produced Using Device 2 Investigating Several TFRs
| Nanoformulations | Total Flow Rate | dmean (nm) | Polydispersity Index (PdI) | ζ-potential (mV) |
|---|---|---|---|---|
| Liposomes | 6 | 260.4 ± 1.42 | 0.192 ± 0.149 | − 32.3 ± 1.85 |
| Hybrid Liposome | 6 | 270.7 ± 9.19 | 0.120 ± 0.040 | − 27.8 ± 0.91 |
| Liposome | 7 | 212.5 ± 6.72 | 0.216 ± 0.199 | − 35.9 ± 3.83 |
| Hybrid Liposome | 7 | 230.5 ± 8.59 | 0.190 ± 0.030 | − 29.1 ± 2.20 |
| Liposome | 8 | 201.9 ± 87.95 | 0.201 ± 0.156 | − 28.1 ± 3.17 |
| Hybrid Liposome | 8 | 211.7 ± 6.71 | 0.180 ± 0.060 | − 23.1 ± 2.07 |
| Liposome | 9 | 189.5 ± 26.13 | 0.092 ± 0.075 | − 29.5 ± 0.92 |
| Hybrid Liposome | 9 | 197.7 ± 13.65 | 0.270 ± 0.030 | − 27.2 ± 2.54 |
Then, a more articulated approach was attempted, taking inspiration from the need to implement the degree of mixing and fragmentation of the CMs to facilitate hybridization with the phospholipids and the formation of a more complex nanosystem. Thus, device 1 and device 2 were connected in series, and both devices were placed inside a sonicator bathtub while producing the formulation. The idea was based on the evidence that by connecting the two devices in series, hybrids were able to form at two critical stages. Device 1 was used to amplify the process of fragmenting cell membranes (CMs) before their encounter with lipids, allowing the two phases to meet more efficiently. The outlet of device 1 serves as an inlet for the aqueous phase containing the CMs, while the organic phase containing dissolved lipids is pumped through the second inlet.
The actual fragmentation of the CMs flowing inside device 1 was evaluated to validate the method by testing various total flow rates (TFRs). Surprisingly, as shown in Table 2, a flow rate of 6 mL/min resulted in the highest degree of CM fragmentation while maintaining a satisfactory level of monodispersity.
Table 2.
Data Resulting from Fragmenting Process of CMs Using Device 1 and the Sonicator Bathtub
| CMs at Different Flows (mL/min) | dmean (nm) | Polydispersity Index (PdI) | ζ-Potential (mV) |
|---|---|---|---|
| 5 | 248.2 ± 12.47 | 0.389 ± 0.105 | − 24.3 ± 2.80 |
| 6 | 122.4 ± 4.18 | 0.308 ± 0.176 | − 26.9 ± 1.65 |
| 7 | 668.8 ± 55.15 | 0.533 ± 0.123 | − 29.0 ± 0.52 |
| 8 | 544.0 ± 19.59 | 0.651 ± 0.108 | − 30.5 ± 0.94 |
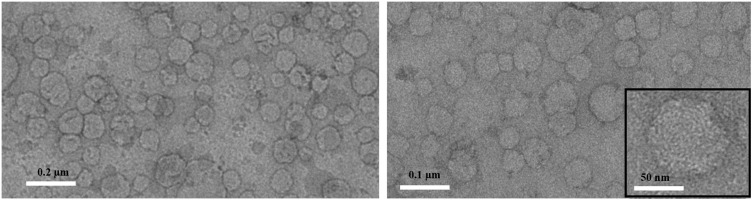
Once the optimal condition was determined, it was tested in the entire microfluidic system consisting of devices 1 and 2 in series, demonstrating extremely high efficiency and robustness. In fact, the TFR of 7 mL/min and an FRR (inner: outer) of 1:6, monodispersed and nanosized liposomes and hybrids liposomes were produced (Table 3). Moreover, a proof of the hybridization could be found evaluating the ζ-potential values which resulted like CMs values and showed a slight increase compared to liposomes. In our previous study,3 we have conducted analyses on the influencing factors in the fabrication of biomimetic hybrid liposomes, including the concentration and mixing ratio of liposomes to CMs. These analyses were aimed at optimizing average diameter, size uniformity, and stability. We believe the insights from that study are applicable here as well. PCX and CBP, two anticancer drugs used as model therapies for glioblastoma, were loaded separately into biomimetic hybrid NPs. High EE values were obtained for the drug PCX, with values exceeding 70%. For CBP, the EE is around 20% due to its hydrophilic nature (Table 3). The hybrid liposomes were analyzed further using transmission electron microscopy (TEM) to examine their morphological properties. TEM images (Figure 3) displayed a general distribution of NPs with spherical shapes. The images (Figure 3) indicate that the NPs are roughly 80 nm in diameter when dry.
Table 3.
Intensity-Average Hydrodynamic Diameter and Corresponding Polydispersity Index (PdI) Determined by DLS, ζ-Potential and Encapsulation Efficiency (%) of Liposome and Hybrid Liposome Produced Through the Chosen Microfluidic Condition (TFR 7 mL/min, FRR (Inner:Outer) 1:6)
| Nanoformulations | DLS | NTA* | Encapsulation Efficiency (%) | |||
|---|---|---|---|---|---|---|
| dmean (nm) | Polydispersity Index (PdI) | ζ-potential (mV) | Mode (nm) | Particle/mL | ||
| Liposomes | 130.1 ± 1.24 | 0.050 ± 0.020 | −26.8 ± 1.93 | 103.5±2.4 | 1.0x1011 | / |
| Hybrid liposomes | 143.9 ± 2.28 | 0.090 ± 0.012 | −21.4 ± 0.50 | 135.6±2.7 | 1.3x1011 | / |
| Hybrid liposomes-PCX | 139.3 ± 2.67 | 0.102 ± 0.044 | −25.7 ± 0.94 | 143.4±3.5 | 8.7x108 | 72.28 ± 1.49 |
| Hybrid liposomes-CBP | 144.3 ± 1.58 | 0.082 ± 0.062 | −23.0 ± 1.58 | 142.3±2.8 | 2.8x109 | 22.45 ± 2.55 |
Notes: *Concentration (particle/mL and size distribution (mode nm) of liposomes and hybrid liposomes determined by NTA.
Figure 3.
Representative TEM micrographs obtained with staining for hybrid liposomes. Scale bar: 0.2 µm (left panel), 0.1 µm (right panel), 50 nm (inset in the right panel).
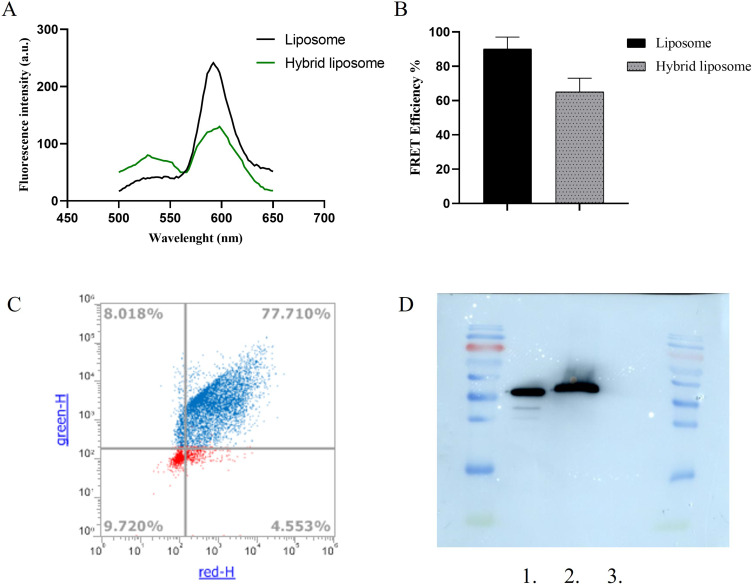
FRET analysis was conducted to verify the hybridization of liposomes and CMs. This technique is commonly utilized to investigate membrane fusion. FRET liposomes were created using the fluorescent pairs PE-NB (donor, λem = 534 nm) and PE-Rh-B (acceptor, λem = 595 nm) in a 1:7 molar ratio. The energy transfer within the FRET liposomes was monitored both before and after hybridization, as depicted in Figure 4A. The spectra for liposomes depicted the stage prior to hybridization, whereas the spectra for hybrid liposomes showed the stage following hybridization. The appearance of the FRET effect post-hybridization indicated an increased separation between FRET pairs. This finding suggested the incorporation of CM content into the liposome’s lipid bilayer, thereby confirming successful hybridization. To measure the reduction in the FRET effect, FRET efficiency was determined. Figure 4B showed a decline in FRET efficiency in the hybrid liposomes. Notably, a reduction of approximately 32% was observed. Furthermore, FCM analysis was used to quantify the fusion of the lipid membrane and CMs in the hybrid liposomes. The lipid membrane was labeled with the lipid Liss Rhod PE (red), and CMs were marked with the green, fluorescent dye PKH67. Figure 4C shows that approximately 77% of NPs in the hybrid liposomes were co-localized.
Figure 4.
Validation of hybrid liposome formation. (A) FRET analysis illustrating the successful fusion of CMs and liposomes. The FRET analysis was performed using the fluorescent donor NBD (λem = 525 nm) and the fluorescent acceptor RhB (λem = 595 nm) at an excitation wavelength of 470 nm. (B) Measurement of FRET efficiency and the decrease in FRET efficiency following the hybridization of CMs. All bars represent mean values ± standard deviation; n = 3. (C) Confirmation of liposome-CMs fusion via FCM by determining the percentage of nanoparticles with overlapping red and green fluorescence in hybrid liposomes. (D) Western blot protein analysis of: 1. cancer cell membrane, 2. hybrid liposomes, 3. bare liposomes.
Moreover, western blot analysis was conducted to assess that the proteins extracted from cancer cells were present and remained intact in hybrid nanoparticles. The protein investigated in this case was the housekeeping protein Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). As illustrated in Figure 4D, both the cancer cell membranes and the liposomes functionalized with the same cancer cell membranes exhibited the presence of the same protein. This protein was not detected in bare liposomes, which consist solely of phosphatidylcholine and cholesterol. The Western blot analysis thus demonstrated that in the hybrid liposome sample, the proteins extracted from cancer cells were present and remained intact.
In vitro Uptake and Homotypic Targeting Studies
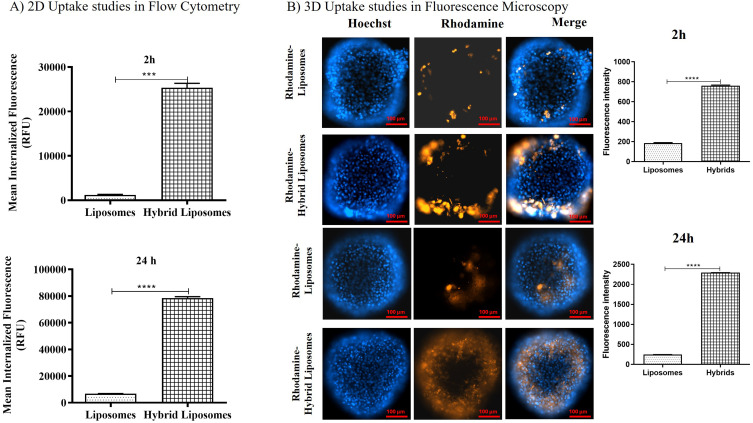
In vitro biological internalization studies allowed us to evaluate the homotypic targeting of hybrid liposomes. The uptake study conducted on the 2D model of U87MG, the cell line from which we extracted the cell membranes, confirmed a higher internalization for hybrid liposomes compared to conventional liposomes. As shown in the histograms in Figure 5A, the amount of internalized fluorescence after 2 h was 25,327 ± 980 for hybrid liposomes and 1,307 ± 100 for liposomes. After 24 h, it was 78,513 ± 1,000 for hybrid liposomes and 6,802 ± 90 for liposomes. The ratio of internalized hybrids to liposomes after 2 h and 24 h was 19.4 ± 2.3 and 11.5 ± 1.2, respectively, highlighting a significant improvement in the internalization of the hybrid formulation into source cells and confirming homotypic targeting.
Figure 5.
2D and 3D in vitro uptake studies (A) Uptake experiment results by flow cytometry analysis after 2 hours and 24 hours of incubation. The histograms represent average internalized fluorescence values (37°C - 4°C values). The tested concentration of liposomes or hybrid liposomes, in terms of fluorescent lipids, was 0.3 μM. Differences were considered very significant with **** p<0.0001, *** p<0.0005. (B) Representative images showing cell uptake of fluorescent hybrid liposomes and liposomes in a 3D U87 model after 2 hours and 24 hours of incubation. The upper panel shows the fluorescence intensities of Rhodamine-Liposomes (orange), Hoechst (blue, nuclei), and their merge after 2 hours; the bottom panel shows the fluorescence intensities of Rhodamine-Hybrid Liposomes (orange), Hoechst (blue, nuclei), and their merge after 24 hours. Scale bar: 100 μm. The histograms show the quantified fluorescence intensities detected in U87 spheroids after 2 hours and 24 hours of incubation with liposomes and hybrid liposomes. Differences were considered very significant with **** p<0.0001.
Moreover, this result was corroborated by uptake studies conducted on 3D models of U87 cells (Figure 5B). As observed in the merged images, there was higher internalized fluorescence for the hybrid formulation. Specifically, the colocalized rhodamine/Hoechst fluorescence, measured in pixel count, was 764 ± 25 for hybrid liposomes and 191 ± 9 for liposomes after 2 h. After 24 h, it was 2,290 ± 22 for hybrid liposomes and 245 ± 13 for liposomes. Thus, the fluorescence intensity (FI) investigation was consistent with flow cytometry (FCM) analysis, demonstrating the efficacy of homotypic targeting in promoting increased cell internalization.
The data obtained in this study regarding hybrid liposomes fused with U87 membranes are even more promising than those obtained for hybrids with melanoma cell membranes reported in Arduino et al.3 This could likely be attributed to a better integration of U87 membranes with the lipids, thanks to the different microfluidic production process. In addition, based on the results of uptake experiments conducted at 4 °C (data not shown), which demonstrated significant inhibition of hybrid liposome and liposome internalization in U87 cells, we suggest that an energy-dependent endocytosis process is the primary route for nanoparticle cellular entry. This finding aligns well with the results of our previous work, in which we also conducted uptake experiments in the presence of specific pathway inhibitors.3 Therefore, given that the composition of these hybrid liposomes and liposomes is similar, we conclude that no specific endocytosis mechanism is involved for either hybrid liposomes or liposomes.
BBB Penetration Ability of Hybrid Liposomes
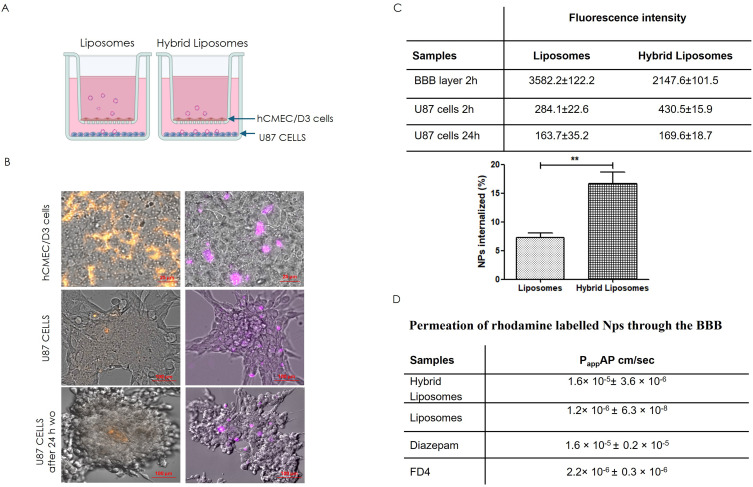
In addition to the improved uptake of hybrid liposomes by their parent cells, the objective of the present study was to investigate the ability of NPs to cross the BBB, focusing on nanoparticle integrity. Thus, we assessed the BBB penetration capacity of hybrid liposomes compared to liposomes, using a standardized in vitro BBB model16,17 (Figure 6A). As reported in the literature, the targeted delivery of therapies for glioblastoma, a heterogeneous brain tumor, presents significant challenges. Namely, the different molecular and histological composition of the tumor results in varying degrees of BBB permeability in different regions. While some areas exhibit disruption of the BBB, others remain intact. In addition, invasive glioblastoma margins, composed of normal brain tissue infiltrated by tumor cells, maintain an intact BBB.26 For these reasons, a study of the nanosystem’s ability to cross the BBB was necessary. Fluorescence imaging of the hCMEC/D3 layer (Figure 6B, top panel) showed that the addition of the liposome samples led to a higher fluorescence signal in the human brain endothelial cells comp26 ared to the hybrid liposome samples, in 2 hours. The lower signal for the hybrids suggests that hybrid liposomes could more easily cross the BBB via transcytosis than liposomes, facilitated by the specific recognition between U87 cell membranes and hCMEC/D3 cells. In the lower chamber, the detection of rhodamine fluorescence in the U87 cells by FI (Figure 6B, second panel) confirmed that hybrid NPs were able to cross the BBB intact and exhibited higher uptake efficiency by U87 cells compared to liposomes, after 2 h of incubation. Moreover, we showed the persistence of hybrid NPs into cells also 24 h post-washout (Figure 6B). The fluorescence intensity quantified and reported in Figure 6C showed that the 16.7 ± 2% of hybrid liposomes added in the upper side of the transwell crossed the BBB and were internalized in U87 cells, as respect to the 7.3 ± 0.8% of liposomes. These in vitro penetration assays highlighted the strong potential of hybrid liposomes to cross the BBB and subsequently target glioblastoma cells. Moreover, the higher apparent permeability (Papp) value (Figure 6D) for hybrid liposomes, calculated by adapting a standard protocol for the determination of BBB penetration of drugs,16,18 highlighted higher BBB permeation for hybrids than liposomes. Additionally, the Papp value for hybrid liposomes was very similar to that of diazepam, used as a marker of the transcellular pathway, confirming that hybrid liposomes could pass through the BBB by transcytosis.
Figure 6.
In vitro BBB penetration ability of hybrid liposome and liposomes and homotypic U87 tumor cell uptake. (A) Schematic illustration of the in vitro BBB model (Transwell™) for evaluating the potential BBB penetration ability of the hybrid liposomes and liposomes. (B) Representative FI images of the hCMEC/D3 BBB layer (top panel) and U87 cells (second and third panels) showing the penetration and targeting ability of liposomes and hybrid liposomes after 2h and 24 h after washing out. Orange: rhodamine-labelled liposomes; Violet: rhodamine-labelled hybrid liposomes. Scale bar: 25μm/100μm for hCMEC/D3 and U87, respectively. (C) Fluorescence intensity quantification of liposomes and hybrid liposomes internalized by U87 cells after crossing the BBB. Data in C were presented as the mean ± SD, n = 3 independent experiment. (D) Apparent permeability (Papp), in units of cm/second for the Liss Rhod PE labelled liposomes and hybrid liposomes.
Antitumor Activity of Drug(s) Loaded-Hybrid Liposomes
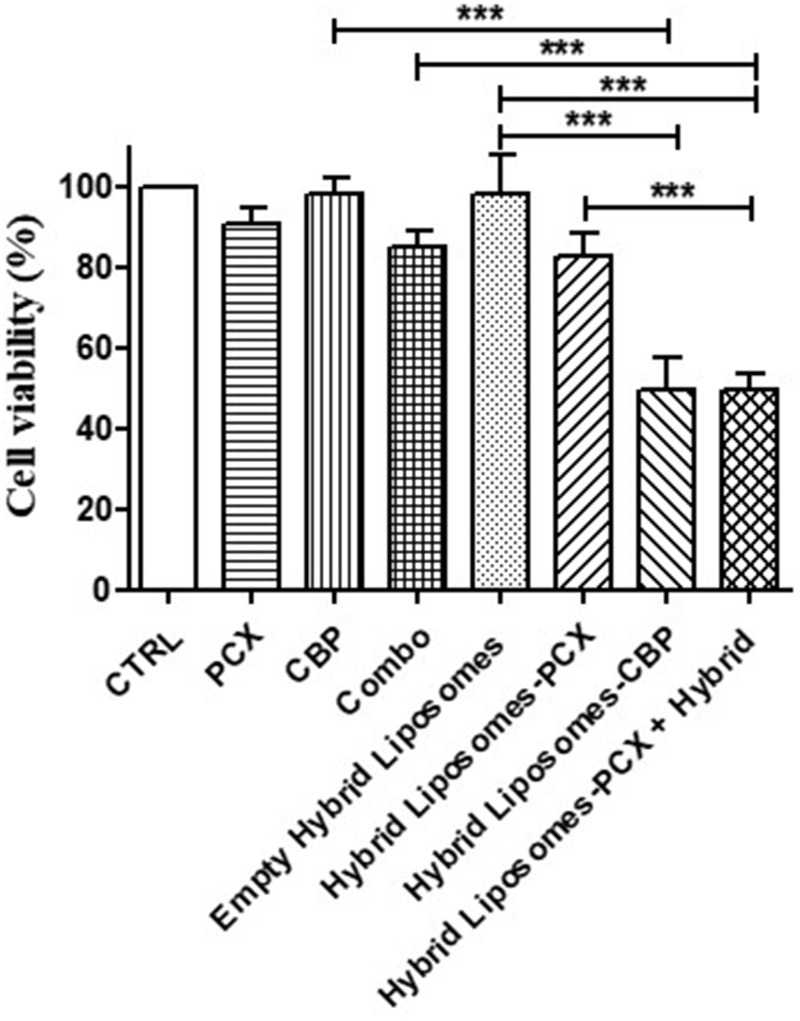
We have demonstrated that hybrid NPs cross the BBB, but the last step was to evaluate whether the drugs chosen and transported by these hybrid nanodevices would show cytotoxic activity at least comparable to that of free drugs. We first assessed the activity of the two free drugs in U87 cells and determined their IC50 values: 152.5 µM for paclitaxel and 492.4 µM for carboplatin. To optimize the effectiveness of the nanosystem, we chose to use low concentrations corresponding to the IC10 of each drug. This approach was based on the expectation that encapsulating the drugs in the nanosystem would enhance their efficacy while also minimizing the potential impact of administering large amounts of hybrid NPs, which, despite lacking intrinsic cytotoxic activity, could still influence various cellular pathways. Consequently, we conducted the experiments with 0.5 µM of paclitaxel and 50 µM of carboplatin. The viability assay, performed after treating tumor cells with paclitaxel, carboplatin, free and transported by hybrid liposomes for 24 h, revealed that the antitumor efficacy of the drugs was retained even after encapsulation. In detail, both paclitaxel and carboplatin, in monotherapy, slightly reduced tumor cell viability by 10%, while their combination was slightly more effective, reducing viability by 15% (Figure 7). Hybrid liposomes-PCX reduced tumor cell viability by about 18% compared to cells treated with empty hybrid liposomes, which is slightly more than the reduction achieved by the free drug (Figure 7). Notably, the hybrid liposomes-CBP significantly increased tumor cell cytotoxicity compared to control cells with empty hybrid liposomes, achieving the strongest antitumor efficacy (49.6%), that was retained in the cells treated with the combination of the two NP formulations (49.5%) (Figure 7). Noteworthy, no cytotoxicity was found in the cells treated with empty hybrid liposomes, demonstrating their safety. These results demonstrate that the drugs loaded in hybrid liposomes maintain at least the same efficacy as free drugs. In addition, the absence of cytotoxicity of the empty hybrid liposomes on U87 cells (Figure 7) and on hCMEC/D3 cells (data not shown), as well as their very low hemolytic activity demonstrated in a previous study,3 could support a prediction of non-cytotoxicity in vivo as well.
Figure 7.
In vitro antitumor efficacy. Histogram plot reporting the results of MTT assay performed in U87 cell line treated with paclitaxel, carboplatin, hybrid liposomes-PCX and hybrid liposomes-CBP for 24 h, showed as mean±SD of three experiments (***p<0.001).
Conclusion
In conclusion, this work has demonstrated that the customization of microfluidic chips enabled by 3D printing technology, combined with the versatility of biomimetic hybrid drug delivery systems, represents a promising strategy for the treatment of cancer. Our previous research showed that biomimetic hybrid liposomes fused with cell membranes (CMs) derived from melanoma biopsies could selectively recognize the original tumor tissue. This study extended that concept to glioblastoma, demonstrating that hybrid liposomes fused with CMs from U87 glioblastoma cells can selectively recognize target tumor cells, internalize, and release their cargo of anticancer drugs effectively. The comprehensive characterization of these hybrid liposomes, in terms of their physicochemical and morphological properties, confirmed the effective fusion of CMs with lipids. Furthermore, biological experiments have unequivocally validated the homotypic targeting capabilities of hybrid NPs and their ability to cross the blood–brain barrier, thus demonstrating that they are a valid delivery strategy for paclitaxel and carboplatin in the treatment of glioblastoma. The absence of cytotoxicity in the empty hybrid liposomes on U87 cells and on hCMEC/D3 cells, as well as their very low hemolytic activity, could support a prediction of non-cytotoxicity in vivo as well. This could be the starting point for the definition of a new therapeutic strategy in this pathology. Furthermore, from a purely technological point of view, these findings underline the potential of customized microfluidic platforms and biomimetic hybrid drug delivery systems as effective strategies to overcome the challenges associated with the treatment of brain tumors that require BBB crossing.
Acknowledgments
We thank Mr. Domenico Cellamare of the Department of Pharmacy–Pharmaceutical Sciences, University of Bari Aldo Moro, 70125 Bari, Italy, for the technical support. We thank M.I.U.R.—Programma Operativo Nazionale (PON) “Ricerca e Innovazione” 2014–2020 Tematica IV.4 “Dottorati e Contratti di ricerca su tematiche dell’innovazione”, and Ministry of University and Research (MUR) Next Generation EU: NRRP Initiative, Mission 4, Component 2, Investment 1.3 – Partnerships extended to universities, research centres, companies and research D.D. MUR n. 341 15.03.2022 – Next Generation EU (PE00000006 - ”MNESYS”).
The authors affiliated to the IRCCS Istituto Tumori Giovanni Paolo II are responsible for the views expressed in this article, which do not necessarily represent the ones of the Institute.
Funding Statement
This work was supported by “European Research Council (ERC) STARTING GRANT” University of Bari, project code: 2023-UNBACLE-0243725, title: “Microfluidic development of biomimetic lipid nanoparticles for homologous-targeting and enhanced therapy against glioma”, and the Ministry of Health, Italy, under grant: RC 2022–2024: Ottimizzazione di trattamenti farmacologici mediante creazione di nanodelivery system per il rilascio selettivo di farmaci nei siti tumorali e analisi di metaboliti cellulari (metabolomica) e di farmaci e loro metaboliti nei fluidi biologici.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors of this paper declare no competing financial or other interests that could affect the work they describe here.
References
- 1.Zhang G, Yao M, Ma S, et al. Application of cell membrane-functionalized biomimetic nanoparticles in the treatment of glioma. J Mat Chem B. 2023;11(30):7055–7068. doi: 10.1039/D3TB00605K [DOI] [PubMed] [Google Scholar]
- 2.Kang W, Xu Z, Lu H, et al. Advances in biomimetic nanomaterial delivery systems: harnessing nature’s inspiration for targeted drug delivery. J Mat Chem B. 2024;12(29):7001–7019. doi: 10.1039/D4TB00565A [DOI] [PubMed] [Google Scholar]
- 3.Arduino I, Di FR, Tiboni M, et al. Microfluidic development and biological evaluation of targeted therapy-loaded biomimetic nano system to improve the metastatic melanoma treatment. Int J Pharm. 2024;650:123697. doi: 10.1016/j.ijpharm.2023.123697 [DOI] [PubMed] [Google Scholar]
- 4.Li H, Zhu L, Zhang Y, Yang L, Wu W, Yang D. Biomimetic nanotherapeutics for homotypic-targeting photothermal/chemotherapy of oral cancer. J Control Release. 2024;366:28–43. doi: 10.1016/j.jconrel.2023.12.039 [DOI] [PubMed] [Google Scholar]
- 5.Arduino I, Liu Z, Iacobazzi RM, et al. Microfluidic preparation and in vitro evaluation of iRGD-functionalized solid lipid nanoparticles for targeted delivery of paclitaxel to tumor cells. Int J Pharm. 2021;610:121246. doi: 10.1016/j.ijpharm.2021.121246 [DOI] [PubMed] [Google Scholar]
- 6.Arduino I, Liu Z, Rahikkala A, et al. Preparation of cetyl palmitate-based PEGylated solid lipid nanoparticles by microfluidic technique. Acta Biomater. 2021;121:566–578. doi: 10.1016/j.actbio.2020.12.024 [DOI] [PubMed] [Google Scholar]
- 7.Fondaj D, Arduino I, Lopedota AA, Denora N, Iacobazzi RM. Exploring the microfluidic production of biomimetic hybrid nanoparticles and their pharmaceutical applications. Pharmaceutics. 2023;15(7):1953. doi: 10.3390/pharmaceutics15071953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Ma X, Wu Z, et al. Microfluidics-prepared ultra-small biomimetic nanovesicles for brain tumor targeting. Adv Healthcare Mater. 2024;13(5):2302302. doi: 10.1002/adhm.202302302 [DOI] [PubMed] [Google Scholar]
- 9.Sommonte F, Denora N, Lamprou DA. Combining 3D printing and microfluidic techniques: a powerful synergy for nanomedicine. Pharmaceuticals. 2023;16(1):69. doi: 10.3390/ph16010069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sommonte F, Weaver E, Mathew E, Denora N, Lamprou DA. In-house innovative “diamond shaped” 3D printed microfluidic devices for lysozyme-loaded liposomes. Pharmaceutics. 2022;14(11):2484. doi: 10.3390/pharmaceutics14112484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.KLv E, Patt S, Kratzel C, Kiwit JCW, Reszka R. Local chemotherapy of F98 rat glioblastoma with paclitaxel and carboplatin embedded in liquid crystalline cubic phases. J Neuro-Oncol. 2005;72(3):209–215. doi: 10.1007/s11060-004-3010-6 [DOI] [PubMed] [Google Scholar]
- 12.Eckardstein KL, Reszka R, Kiwit JC. Intracavitary chemotherapy (paclitaxel/carboplatin liquid crystalline cubic phases) for recurrent glioblastoma -- clinical observations. J Neurooncol. 2005;74(3):305–309. doi: 10.1007/s11060-004-7559-x [DOI] [PubMed] [Google Scholar]
- 13.Habashy KJ, Dmello C, Chen L, et al. Paclitaxel and carboplatin in combination with low-intensity pulsed ultrasound for glioblastoma. Clin Cancer Res. 2024;30(8):1619–1629. doi: 10.1158/1078-0432.CCR-23-2367 [DOI] [PubMed] [Google Scholar]
- 14.Sommonte F, Arduino I, Iacobazzi RM, et al. Microfluidic assembly of “Turtle-Like” shaped solid lipid nanoparticles for lysozyme delivery. Int J pharm. 2023;631:122479. doi: 10.1016/j.ijpharm.2022.122479 [DOI] [PubMed] [Google Scholar]
- 15.Sommonte F, Arduino I, Iacobazzi RM, et al. Microfluidic development of brain-derived neurotrophic factor loaded solid lipid nanoparticles: an in vitro evaluation in the post-traumatic brain injury neuroinflammation model. J Drug Delivery Sci Technol. 2024;96:105699. doi: 10.1016/j.jddst.2024.105699 [DOI] [Google Scholar]
- 16.Arduino I, Iacobazzi RM, Riganti C, et al. Induced expression of P-gp and BCRP transporters on brain endothelial cells using transferrin functionalized nanostructured lipid carriers: a first step of a potential strategy for the treatment of Alzheimer’s disease. Int J Pharm. 2020;591:120011. doi: 10.1016/j.ijpharm.2020.120011 [DOI] [PubMed] [Google Scholar]
- 17.Lu G, Wang X, Li F, et al. Engineered biomimetic nanoparticles achieve targeted delivery and efficient metabolism-based synergistic therapy against glioblastoma. Nat Commun. 2022;13(1):4214. doi: 10.1038/s41467-022-31799-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pisani L, Iacobazzi RM, Catto M, et al. Investigating alkyl nitrates as nitric oxide releasing precursors of multitarget acetylcholinesterase-monoamine oxidase B inhibitors. Eur J Med Chem. 2019;161:292–309. doi: 10.1016/j.ejmech.2018.10.016 [DOI] [PubMed] [Google Scholar]
- 19.Webb C, Forbes N, Roces CB, et al. Using microfluidics for scalable manufacturing of nanomedicines from bench to GMP: a case study using protein-loaded liposomes. Int J pharm. 2020;582:119266. doi: 10.1016/j.ijpharm.2020.119266 [DOI] [PubMed] [Google Scholar]
- 20.Weaver E, O’Hagan C, Lamprou DA. The sustainability of emerging technologies for use in pharmaceutical manufacturing. Expert Opin Drug Delivery. 2022;19(7):861–872. doi: 10.1080/17425247.2022.2093857 [DOI] [PubMed] [Google Scholar]
- 21.Bayareh M, Ashani MN, Usefian A. Active and passive micromixers: a comprehensive review. Chem Eng Process Process Intensif. 2020;147:107771. doi: 10.1016/j.cep.2019.107771 [DOI] [Google Scholar]
- 22.Demello AJ. Control and detection of chemical reactions in microfluidic systems. Nature. 2006;442(7101):394–402. doi: 10.1038/nature05062 [DOI] [PubMed] [Google Scholar]
- 23.Jain V, Patel VB, Singh B, Varade D. Microfluidic device based molecular self-assembly structures. J Mol Liq. 2022;362:119760. doi: 10.1016/j.molliq.2022.119760 [DOI] [Google Scholar]
- 24.Belliveau NM, Huft J, Lin PJ, et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol Ther Nucleic Acids. 2012;1(8):e37. doi: 10.1038/mtna.2012.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhigaltsev IV, Belliveau N, Hafez I, et al. Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing. Langmuir. 2012;28(7):3633–3640. doi: 10.1021/la204833h [DOI] [PubMed] [Google Scholar]
- 26.Noorani I, de la Rosa J. Breaking barriers for glioblastoma with a path to enhanced drug delivery. Nat Commun. 2023;14(1):5909. doi: 10.1038/s41467-023-41694-9 [DOI] [PMC free article] [PubMed] [Google Scholar]