Abstract
The tumor suppressor TP53 is frequently mutated in hormone receptor-negative, HER2-positive breast cancer (BC), contributing to tumor aggressiveness. Traditional ancillary methods like immunohistochemistry (IHC) to assess TP53 functionality face pre- and post-analytical challenges. This proof-of-concept study employed a deep learning (DL) algorithm to predict TP53 mutational status from H&E-stained whole slide images (WSIs) of BC tissue. Using a pre-trained convolutional neural network, the model identified tumor areas and predicted TP53 mutations with a Dice coefficient score of 0.82. Predictions were validated through IHC and next-generation sequencing (NGS), confirming TP53 aberrant expression in 92 % of the tumor area, closely matching IHC findings (90 %). The DL model exhibited high accuracy in tissue quantification and TP53 status prediction, outperforming traditional methods in terms of precision and efficiency. DL-based approaches offer significant promise for enhancing biomarker testing and precision oncology by reducing intra- and inter-observer variability, but further validation is required to optimize their integration into real-world clinical workflows. This study underscores the potential of DL algorithms to predict key genetic alterations, such as TP53 mutations, in BC. DL-based histopathological analysis represents a valuable tool for improving patient management and tailoring treatment approaches based on molecular biomarker status.
Keywords: Breast cancer, Artificial intelligence, Deep learning, TP53
1. Introduction
Tumor suppressor TP53 is among the most frequently mutated genes in hormone receptor (HR)-negative breast cancer (BC), significantly contributing to both tumorigenesis and tumor progression [1], [2], [3], [4], [5]. The co-occurrence of TP53 mutations and HER2 gene amplifications further worsen the prognosis of these patients [6], [7]. Notably, most TP53 mutations are missense mutations, often linked to gain-of-function activities that enhance the aggressiveness of cancer cells [8]. Immunohistochemistry (IHC) is a commonly employed method to assess p53 functionality; however, this approach is prone to pre- and post-analytical challenges and provides limited insight into the underlying gene status [9], [10], [11]. Recent advancements in deep learning (DL) systems, particularly convolutional neural networks (CNNs), offer the potential for highly accurate molecular biomarker assessment [12], [13], [14]. DL-based algorithms have been successfully utilized to predict biomarker status from H&E-stained tissue in BC, encompassing markers such as homologous recombination deficiency (HRD), programmed death ligand-1 (PD-L1), and HR [7], [15], [16], [17], [18], [19]. These methods have shown superior effectiveness compared to traditional machine learning (ML) approaches, requiring less human intervention in pattern recognition tasks [20], [21]. DL techniques not only provide a more efficient and accessible alternative to IHC but also show promise in identifying morphological features correlated with biomarker status [22], [23].
Considering these technological advancements, we hypothesized that DL algorithms could predict the mutational status of key cancer-related genes in BC through weakly-supervised analysis of H&E slides. In this proof-of-concept study, we aimed to characterize the TP53 mutational status in HR-/HER2 + BC using a pre-trained H&E-based DL framework.
2. Methods and results
2.1. Data preparation and tissue digitalization
This study was approved by Scientific Direction and the Data Processor of the European Institute of Oncology IRCCS (IEO), Milan, Italy (approval number: UID3472). In accordance with Institutional policy, all patient data were pseudoanonymized prior to analysis. H&E-stained tissue slides from a case of poorly differentiated (G3), early-stage (pT1c, pN0), HR-/HER2 + , invasive BC of no special type (NST) was retrieved from the archives of the Division of Pathology at IEO, Milan, Italy, digitized using a Panoramic 1000 system (3DHISTECH Kft, Budapest, Hungary) and converted to ScanScope Virtual Slide (*.svs) format for subsequent analysis. Two breast pathologists (N.F. and M.I.) reviewed the digitalized images, selected the most representative slide, and annotated both normal and tumor tissue areas.
2.2. WSI pre-processing, model inference and quantification of tissue areas
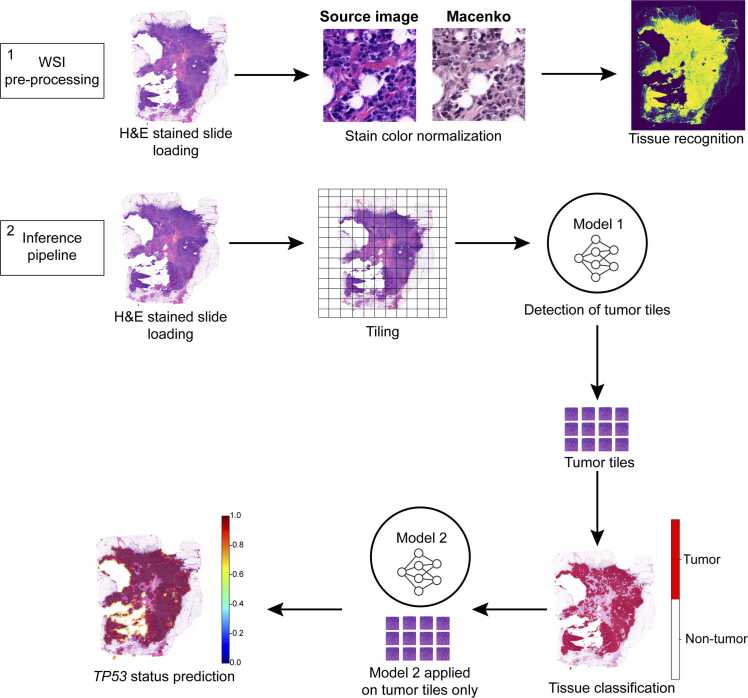
The WSI underwent pre-processing consisting of three steps: image loading, stain-color normalization, and tissue mask generation [24], as shown in Fig. 1. The pre-processing phase facilitated stain-color normalization and enabled the identification of the total tissue area relative to the background. Subsequently, the inference pipeline, which applies image-derived attention regions (IDARs) models, was executed. This pipeline requires slide-level labels and consists of four main steps: tiling and encoding, tile scoring, aggregation, and final decision-making. The algorithm successfully utilized the annotated tumor area to execute each of these four steps, culminating in the prediction of TP53 mutational status. Using the TIAToolbox python library [25], the slides were loaded onto a virtual machine hosted on the GARR Cloud Platform (https://cloud.garr.it/). Stain-color normalization was performed using the Macenko technique [26] to minimize potential artifacts, while tissue segmentation was achieved via Otsu thresholding [27], [28], generating a tissue mask that was subsequently exported in Portable Network Graphics (*.png) format. To identify the tumor areas, a pre-trained fine-tuned convolutional neural network based on ResNet18 [29], [30] was employed as a tumor detection model, allowing for the identification of tumor-specific tiles within the slide. As described by Bilal et al. [30], ResNet18 is a deep neural network model with 18 layers, frequently employed in image analysis tasks. It is initially pre-trained on ImageNet, a large dataset of general images, to learn basic visual features. In their study, the ResNet18 model underwent additional training using a specialized dataset of medical images, which included 35,436 image tiles from the TCGA-CRC-DX cohort, as well as two publicly available datasets from Kather et al. [31] and Shaban et al. [32]. This transfer learning approach enabled the ResNet18 model to accurately differentiate between tumor and non-tumor regions within colorectal cancer samples. The convolutional neural network used, ResNet18, is a robust and well-established model, chosen for its strong performance in image recognition tasks. Its residual learning framework mitigates the vanishing gradient problem, enabling effective training of deeper networks. We employed a pre-trained version of ResNet18, fine-tuned for breast cancer image analysis using transfer learning, thus maximizing performance with limited data. To quantify the normal tissue area predicted by the algorithm, the area of a single patch (4096 pixels) was calculated by multiplying the pixel area (0.0625 µm²) to obtain the total area of each patch (256 µm²). This value was then multiplied by the total number of patches into which the slide had been segmented. The same calculation was applied to determine the tumor tissue area, using only tumor-positive patches. The tumor tiles were aggregated and saved as an independent mask, serving as input for the second stage of analysis. In this stage, a task-specific prediction model was applied to each tumor patch (256 × 256 pixels) to predict the TP53 mutational status. This second pre-trained model generated a digital score reflecting the molecular status of each tile and identified the most predictive visual fields across the entire WSI. Notably, the model employed in this second stage was trained using slide-level labels, thus eliminating the need for detailed annotations at the cellular or regional level. Comparative assessment between the deep learning (DL) model and a human breast pathologist showed a high degree of similarity regarding total tissue area (2.6 cm² vs. 2.7 cm², respectively) and its identification. Following the application of the ResNet18 model, the image was segmented into patches, which served as input for differentiating between tumor and non-tumor tiles. The tumor area calculated by both DL and breast pathologists exhibited absolute concordance (1.0 cm² for both) (Fig. 2).
Fig. 1.
: Deep-learning pipeline workflow. The workflow begins with the pre-processing of whole slide images (WSI), which are first loaded into the Python environment. Once loaded, the images undergo stain-color normalization to standardize variations in staining, followed by the identification of relevant tissue areas for analysis. The inference pipeline then processes these images by segmenting them into smaller tiles. These tiles serve as the input for the first model, which is responsible for distinguishing between tumor and non-tumor regions. The classification results are visualized using a red-and-white mask, where red represents tumor areas and white represents non-tumor tissue. Finally, tiles identified as tumorous are further analyzed by a second model that predicts molecular features, such as TP53 mutation status, based on the whole image.
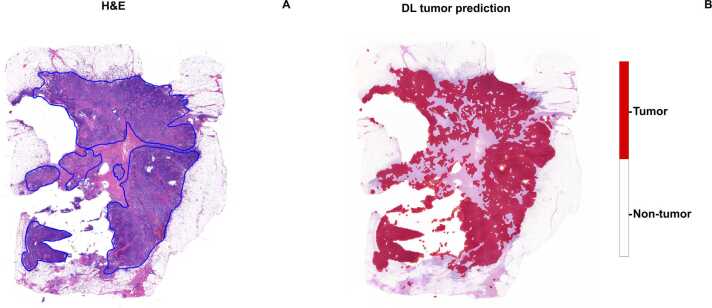
Fig. 2.
: Deep-learning tissue classification. The initial output of the deep-learning pipeline involves classifying tissue tiles as either tumor or non-tumor based on their morphological features. A: annotations made by the pathologist on the H&E slide indicate the tumor area manually identified across the image. B: the algorithm's tumor classification is shown, with red highlighting the regions where the model has detected tumor-containing tiles, visually marking the zones identified by the deep-learning process.
2.3. TP53 prediction pipeline
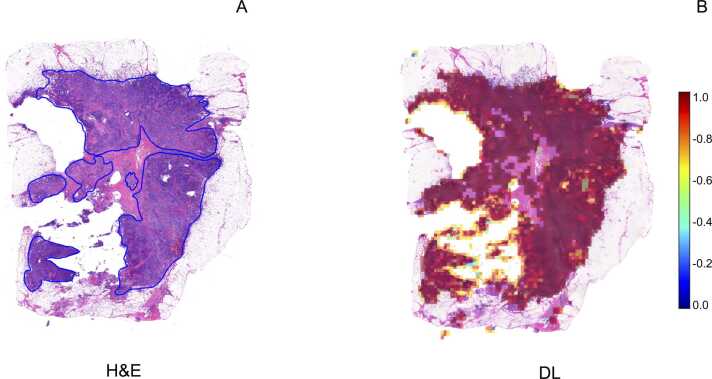
To classify the morphology of the tiles in which the WSI was segmented, a weakly supervised DL Iterative Draw and Rank Sampling (IDaRS) model pre-trained by Bilal et al., was employed [30]. This model was developed using 502 diagnostic slides of primary colorectal tumors from 499 patients in The Cancer Genome Atlas colon and rectal cancer (TCGA-CRC-DX) cohorts [30]. The IDaRS model utilizes a two-stage approach: i) patch-level tumor classification and ii) patch-level WSI classification. Initially, the WSI is segmented, and a ResNet18 network is used to distinguish tumor from non-tumor tiles. Subsequently, the tumor tiles are processed with a ResNet34 network, pre-trained on ImageNet for the prediction of slide labels and using iterative draw and rank sampling to generate prediction scores for high mutation density molecular labels. For deploying these pre-trained models, TIAToolbox, an open-source Python library developed by the TIA Centre, was utilized [25]. This PyTorch-based computational pathology library offers comprehensive tools for WSI processing, stain normalization, tile-based classification, and segmentation of both tissue and nuclei. Finally, the Dice similarity coefficient scores were calculated to evaluate the performance of the algorithm in predicting TP53 aberrant expression. All analyses were performed on a virtual machine equipped with an NVIDIA A30 24 GB GPU and Compute Unified Device Architecture (CUDA) version 12.3. Python v. 3.8 was used, with all scripts executed via Jupyter, a web-based interactive computing platform launched with a remote server connection to a local machine. To evaluate the predictive accuracy of the DL algorithm, ResNet34 was applied to representative tumor area patches. Through iterative draw and rank sampling, the DL analysis generated prediction scores, indicating that TP53 was aberrantly overexpressed in 92 % of the tumor area, with an overall Dice coefficient of 0.82 (Fig. 3).
Fig. 3.
: Deep-learning pipeline prediction ofTP53status and spatial distribution. Graphical output illustrating the spatial distribution of the TP53 mutation as predicted by the algorithm, linking molecular features to specific regions within the tumor. A: pathologist's annotations on the H&E slide, marking the tumor area identified within the tissue. B: deep-learning model's prediction of TP53 status, visually represented in the tumor-containing tiles.
2.4. Validation by immunohistochemistry (IHC)
Tumor samples were subjected to IHC for p53 using the DO-7 antibody clone (1:200 dilution; Agilent DAKO). Two independent blinded pathologists evaluated p53 expression based on the following criteria [33]: Mutant overexpression: Diffuse strong nuclear positivity in > 80 % of tumor cells. Null mutant: Complete absence of p53 expression with a positive internal control. Cytoplasmic expression: Significant p53 cytoplasmic staining in > 80 % of tumor cells. Wild-type p53 expression was defined as variable nuclear staining in 1–80 % of tumor cells. IHC analysis revealed aberrant p53 overexpression in 90 % of the tumor area, with both pathologists in agreement. This result closely matched the deep learning (DL) assessment, which showed p53 overexpression in 92 % of the tumor area, as illustrated in Fig. 4.
Fig. 4.
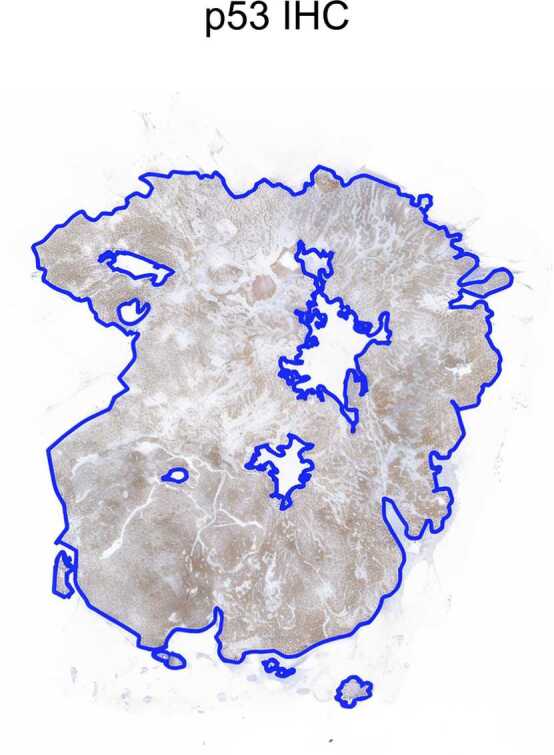
: Immunohistochemistry of p53 expression. IHC of p53 revealed aberrant expression across 90 % of tumor area as it is visible from the strongly positive, brown-stained cells in the image.
2.5. Validation by next-generation sequencing (NGS)
DNA isolation was performed as described previously [34] and subsequently, TP53 mutational status was assessed by next-generation sequencing (NGS) using the SOPHiA DDM™ Solid Tumor Solutions (SOPHiA GENETICS) [35]. To confirm the predictions obtained by DL, IHC and NGS analyses were conducted. TP53 mutational status assessment by NGS showed a splice site alteration (i.e. c.376–1 G>A, exon 5), known for its likely pathogenic effect, further confirming DL and IHC results (Supplementary Figure 1).
3. Discussion
In this study, we predicted the TP53 status in BC from H&E-stained WSIs and analyzed its spatial distribution evaluating the impact of intra-tumor heterogeneity. TP53 is highly mutated in HR-/HER2 + BC and is associated with aggressive clinical behavior [20], [36]. In clinical practice, p53 expression is assesses by IHC and can be troubled by issues in pre- and post-analytical phases [37]. Thus, testing alternative approaches to assist and complement traditional diagnostic methods represents a current need in biomarker testing. Our analyses confirmed that DL algorithms can reliably identify and quantify tumor and non-tumor tissue areas [38], [39]. Of note, TP53 prediction on H&E-stained WSIs was particularly accurate. In particular, the DL-based approach assigned TP53 status with a high Dice coefficient score without being particularly affected by the increased intra-tumor heterogeneity seen in the selected case. We further confirmed this finding both at protein and gene level with IHC and NGS, respectively.
Manual assessment of H&E-stained histology slides suffers from low throughput and is prone to intra- and inter-observer variability [40]. Accurate tissue identification and quantification on WSI are essential steps for biomarker testing [41]. Our DL model and BP assessment exhibited remarkable concordance in total tissue area quantification (2.6 cm² vs. 2.7 cm²) and tumor area calculation (1.0 cm² for both). This high level of agreement underscores the reliability of the DL approach in accurately distinguishing tumor from non-tumor regions. These findings are in line with previous studies [42], [43], in which segmentation and identification of multi-tissue histology images was achieved with high accuracy. The precision in tumor area quantification is critical for subsequent biomarker analysis, ensuring that predictions are based on relevant tissue sections.
The clinical actionability of gene mutations is becoming increasingly important for the management of patients with solid tumors, making the prediction of key genetic alterations based on WSI a significant advancement for tailored molecular biomarkers testing [44]. Several studies have explored the predictive capabilities of DL models in oncology. For instance, Wang et al. developed a model to predict the risk of germline BRCA (gBRCA) mutations by analyzing whole-slide pathology features from breast cancer (BC) H&E-stained images in conjunction with patients' gBRCA mutation status [45]. Furthermore, ongoing clinical trials are evaluating the utility of DL models for predictive biomarker analysis. Notably, the TROPION-Lung01 Phase 3 trial has investigated a novel computational pathology-based TROP2 biomarker to predict clinical outcomes for patients receiving datopotamab deruxtecan in the treatment of non-small cell lung cancer (NSCLC) [46]. TP53 is a tumor suppressor gene that plays a key role in many cellular pathways controlling cell proliferation, cell survival, and genomic integrity [47]. The DL algorithm predicted TP53 aberrant overexpression in 92 % of the tumor area, achieving a high Dice coefficient of 0.82. The predictive performance of the DL model in assessing TP53 status was validated with other diagnostic methods such as IHC and NGS. This prediction was corroborated by IHC, through which BPs identified aberrant p53 expression in 90 % of the tumor area, demonstrating a high degree of alignment between the DL predictions and traditional histopathological assessment. Importantly, these results align with previous findings reported by Qu et al. who managed to predict TP53 point mutations with high accuracy (0.729) [44]. Moreover, in our study, the selected case showed a TP53 splicesite alteration (i.e. c.376–1 G>A, exon 5), which is a type of alteration known to create diverse transcript variants, even if their effects on downstream genes are unclear [48]. Notably, many studies have shown that certain TP53 mutation types are associated with specific p53 IHC patterns where splice site mutations may result in a null pattern [49]. Interestingly, Alsner et al. reported that the clinical outcome for breast cancer patients is significantly different based on diverse TP53 mutation types [50], [51]. With the prediction of TP53 mutation, the tested DL model is able to offer insights for improving patient clinical management.
While DL models demonstrate high accuracy, it is important to acknowledge their limitations, particularly in comparison to traditional methods like IHC and NGS. DL models may underperform in scenarios with limited or highly variable datasets, highlighting the need for comprehensive algorithm training and robust database structuring [52]. In clinical practice, molecular tests are guided by well-established protocols and recommendations that ensure quality and consistency. In contrast, digital pathology approaches, including DL models, have not yet achieved widespread adoption. Consequently, despite their potential advantages, DL models should be viewed as complementary tools that enhance biomarker testing rather than as complete replacements [41]. Testing the IDaRS pre-trained model developed by Bilal et al. [30] exhibited superior performance in distinguishing TP53 mutational status, corroborating previous findings that highlight the robustness and accuracy of DL in complex pattern recognition tasks [53], [54]. The ability of DL algorithms to autonomously learn and extract relevant features from high-dimensional data with minimal human intervention positions them as powerful tools for histopathological analysis [55], [56].
Despite the promising results, our study has several intrinsic limitations. First, the proof-of-concept nature of this work, which focuses on a single case of BC, limits the generalizability of the findings. While our study provides valuable insights into the integration of deep learning (DL) with molecular pathology in breast cancer, it is based on the analysis of a single case and should thus be considered hypothesis-generating. A more extensive and robust analysis is needed to validate these findings and assess their generalizability. Future research should involve larger, more diverse cohorts, encompassing a wide range of breast cancer stages and the full spectrum of HER2 expression (score 0-null, score 0-ultra low, score 1 +, score 2 +) [57]. Additionally, extending the analysis to other heterogeneous cancer types, such as gastric cancer, may provide crucial insights into the performance of DL models across varied tumor microenvironments and genetic landscapes [58]. Such efforts are essential to optimize therapeutic strategies across different cancer types. Additionally, given the heterogeneity in mutant p53 expression (e.g., mutant overexpression or null mutations), a multi-label algorithm should be employed to address this variability and enhance the clinical relevance of the predicted molecular features. Furthermore, as tumors can acquire additional mutations over time, potentially altering the molecular profile and influencing disease progression, it would be beneficial to analyze samples from different time points (e.g., at diagnosis and after therapy) to better understand the evolution of TP53 mutations over the course of treatment [59]. To achieve this, DL models could be adapted for the analysis of longitudinal samples, allowing for the assessment of temporal heterogeneity in breast cancer. By incorporating time-series data, these models can capture dynamic changes in tumor characteristics, potentially providing insights into disease progression, treatment response, and evolving molecular profiles [60].
Integrating DL models into pathology workflows demands planning and substantial infrastructure investment [61]. The time required to analyze a WSI largely depends on the algorithm's complexity and the hardware employed. Modern DL models, when optimized, can analyze a single slide within minutes to an hour. However, achieving this level of performance necessitates high-performance computational resources, such as GPUs or dedicated cloud-based infrastructure, which are critical for both the training and inference stages of the DL pipeline [62]. From a hardware perspective, clinical centers must invest in advanced imaging systems, including high-resolution scanners to digitize tissue slides rapidly and with high quality [63], [64]. Additionally, robust data storage solutions are needed to handle the vast amount of image data generated, often requiring terabytes of space. Efficient data retrieval and scalable storage infrastructure are particularly important when managing large datasets or conducting retrospective analyses [65]. For effective integration of our DL model, medical-grade display monitors with high resolution and refresh rates are essential to ensure accurate and comfortable visualization, both for pathologists and during model validation [66]. The computational setup must include dedicated GPUs to accelerate the processing of large, complex datasets and cloud-based solutions for scalability, especially in centers with limited local GPU resources, though they introduce challenges related to data privacy and transfer speeds [67]. Moreover, deploying DL systems in clinical practice requires compliance with regulatory standards and the acquisition of necessary certifications (e.g., FDA approval, CE marking). Successful implementation also depends on the availability of skilled personnel, including data scientists, bioinformaticians, and IT specialists, to support the integration, maintenance, and troubleshooting of the AI pipeline. Pathologists will need training to interpret AI-assisted results effectively and incorporate these findings into clinical decision-making [66], [68], [69]. This underscores the importance of multidisciplinary collaboration and continuous education to fully leverage DL models for enhanced diagnostic accuracy and efficiency in pathology. By addressing these practical and logistical challenges, hospitals can strategically plan for the adoption of DL technologies, ensuring they are equipped to capitalize on AI-driven advancements in a sustainable and scalable manner [70]. Furthermore, while deep learning can significantly enhance diagnostic accuracy and efficiency, interpretability for pathologists remains a challenge, as these models often function as "black boxes” [71]. The complex nature of DL applications often makes it difficult for pathologists to interpret the decision-making processes behind specific model outputs, potentially diminishing their confidence in employing these tools in clinical practice [72]. Therefore, additional comparative studies between DL and traditional testing methods are needed to emphasize the importance of robust validation frameworks and to inform future recommendations focused on interpretability, standardization, and clinical integration. Although advances in explainability techniques, such as heat maps and gradient-based visualizations [73], have made progress in mitigating this challenge, further refinement of these methods is required. Moreover, these issues extend beyond technical challenges and raise several ethical concerns. Privacy, equity, and trust are key ethical considerations in AI-driven digital pathology, and they remain central to the responsible implementation of these tools in clinical settings [72], [74]. Thus, visualization tools and explainability techniques can help bridge this gap, making the outputs more understandable for clinical use. Despite addressing some issues inherent in manual histopathological assessment, DL models can still be influenced by variability in tissue processing and slide preparation. Differences in staining protocols, scanner settings, and image quality could impact the DL performance. Moreover, this study focused solely on TP53 status. While TP53 is a crucial biomarker, BC prognosis and treatment rely on multiple biomarkers. Lastly, the IDaRS pre-trained model by Bilal et al. [30], was trained and validated on a cohort of colorectal cancer slides, reaching an AUC of 0.7 for TP53 status prediction, thus lacking any further testing on tissue slides coming from other cancer types, such BC. A better understanding on how the algorithm performs on BC slides it would be necessary to give robustness to the model. Since this study focused on the application of a pre-trained DL model, it would be of interest to apply other algorithms or deploy a DL model, to increase reliability for biomarker status prediction [52]. This proof-of-concept study lays the groundwork for more comprehensive evaluations in future research. Future studies should incorporate key diagnostic measures, such as confusion matrices and ROC curves on large and heterogeneous datasets. These tools will provide deeper insights into the model’s strengths and limitations, revealing areas of potential misclassification and offering a clearer picture of its diagnostic accuracy.
4. Conclusions
This study demonstrated that DL algorithms can accurately predict TP53 status in BC from H&E-stained WSIs, even in the presence of intra-tumor heterogeneity. The DL model showed high concordance with traditional methods like IHC and NGS. While promising, the study's single-case focus limits generalizability, highlighting the need for larger cohort studies. Future research should include other biomarkers and cancer types to enhance DL's applicability in clinical practice. DL models can improve diagnostic precision and efficiency but require further validation for broader adoption.
Author statement
The raw data analyzed in this study are available from the authors upon reasonable request. Additionally, all datasets referenced in this study are publicly available and can be accessed using the citations provided in the manuscript.
CRediT authorship contribution statement
Chiara Frascarelli: Writing – original draft, Software, Methodology. Carmen Criscitiello: Writing – review & editing. Annarosa Farina: Writing – review & editing. Alberto Concardi: Writing – review & editing. Francesca Maria Porta: Writing – review & editing. Giulia Cursano: Writing – review & editing. Mariia Ivanova: Writing – review & editing. Eltjona Mane: Writing – review & editing. Arnaud Gerard Michel Ceol: Writing – review & editing. Nicola Fusco: Writing – review & editing, Project administration, Conceptualization. Antonio Marra: Writing – review & editing. Elena Guerini-Rocco: Writing – review & editing. Konstantinos Venetis: Writing – review & editing, Writing – original draft, Supervision. Giuseppe Curigliano: Writing – review & editing.
Declaration of Competing Interest
A. M. has received support from Menarini Group and served on the Speakers' Bureau for Roche and AstraZeneca. C. C. has participated in advisory or consultancy roles and speakers' bureau engagements for Eli Lilly, Pfizer, Novartis, Roche, AstraZeneca, MSD, Daiichi Sankyo, Gilead, and Seagen. G. Curi. has received honoraria for speaker engagements from Roche, Seattle Genetics, Novartis, Lilly, Pfizer, Foundation Medicine, NanoString, Samsung, Celltrion, BMS, and MSD; honoraria for consultancy from Roche, Seattle Genetics, and NanoString; honoraria for participation in advisory boards from Roche, Lilly, Pfizer, Foundation Medicine, Samsung, Celltrion, and Mylan; honoraria for writing engagements from Novartis and BMS; and honoraria for participation in the Ellipsis Scientific Affairs Group. He has also received institutional research funding for conducting phase I and II clinical trials from Pfizer, Roche, Novartis, Sanofi, Celgene, Servier, Orion, AstraZeneca, Seattle Genetics, AbbVie, Tesaro, BMS, Merck Serono, Merck Sharp & Dohme, Janssen-Cilag, Philogen, Bayer, Medivation, and Medimmune. E. G-R. has received advisory fees, honoraria, travel accommodations/expenses, grants, and/or non-financial support from AstraZeneca, Exact Sciences, GSK, Illumina, MSD, Novartis, Roche, and Thermo Fisher Scientific. N.F. has received honoraria for consulting, advisory roles, speakers' bureau participation, travel, and/or research grants from Merck Sharp & Dohme (MSD), Novartis, AstraZeneca, Roche, Menarini, Daiichi Sankyo, GlaxoSmithKline (GSK), Gilead, Diaceutics, Adicet Bio, Sermonix, Reply, and Leica Biosystems. The remaining authors declare no conflicts of interest.
Acknowledgements
This work was partially supported by the Italian Ministry of Health through Ricerca Corrente 5 × 1000 funds; the Italian Ministry of Innovations via the Sustainable Growth Fund – Innovation Agreements under the Ministerial Decree of December 31, 2021, and the Director's Decree of November 14, 2022 (2nd Call), Project No.: F/350104/01–02/X60; and the Italian Ministry of University and Research (MUR) 2023 through the “Future Artificial Intelligence Research – FAIR” program, PE0000013, CUP D53C22002380006, within the National Recovery and Resilience Plan (PNRR), Mission 4, Component 2, Investment 1.3 – funded by the European Union – NextGenerationEU. Project: “AIDH – Adaptive AI Methods for Digital Health.” Konstantinos Venetis was supported by the Fondazione Umberto Veronesi, Mariia Ivanova by the IEO-Monzino Foundation, and Antonio Marra by the ESMO José Baselga Fellowship for Clinician Scientists 2023–2025. The authors acknowledge support from the University of Milan through the APC initiative. The final proofreading of grammar and syntax for the manuscript was conducted using ChatGPT 4 and Grammarly v.6.8.263.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.11.037.
Appendix A. Supplementary material
Supplementary Figure S1: TP53 mutational assessment. NGS analysis revealed the mutational status of TP53, as depicted in the Integrative Genomics Viewer.
.
References
- 1.Feroz W., Sheikh A.M.A. Exploring the multiple roles of guardian of the genome: P53. Egypt J Med Hum Genet. 2020;21(1):49. [Google Scholar]
- 2.Schaafsma E., Takacs E.M., Kaur S., Cheng C., Kurokawa M. Predicting clinical outcomes of cancer patients with a p53 deficiency gene signature. Sci Rep. 2022;12(1):1317. doi: 10.1038/s41598-022-05243-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubrey B.J., Strasser A., Kelly G.L. Tumor-suppressor functions of the TP53 pathway. Cold Spring Harb Perspect Med. 2016;6(5) doi: 10.1101/cshperspect.a026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J., Chang X., Li X., Liu J., Wang N., Wu Y., et al. The heterogeneous impact of targeted therapy on the prognosis of stage III/IV colorectal cancer patients with different subtypes of TP53 mutations. Cancer Med. 2023;12(24):21920–21932. doi: 10.1002/cam4.6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammer L., Rebernick R., McFarlane M., Westbrook T., Hazime M., Hammoud T., et al. Clinical impact of mutations in driver oncogenes and TP53/RB1 in advanced prostate cancer. J Clin Oncol. 2023;41:263. [Google Scholar]
- 6.Russo A., Bazan V., Iacopetta B., Kerr D., Soussi T., Gebbia N. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23(30):7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 7.Børresen-Dale A.L. TP53 and breast cancer. Hum Mutat. 2003;21(3):292–300. doi: 10.1002/humu.10174. [DOI] [PubMed] [Google Scholar]
- 8.Chen X., Zhang T., Su W., Dou Z., Zhao D., Jin X., et al. Mutant p53 in cancer: from molecular mechanism to therapeutic modulation. Cell Death Dis. 2022;13(11):974. doi: 10.1038/s41419-022-05408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vousden K.H., Prives C. P53 and prognosis: new insights and further complexity. Cell. 2005;120(1):7–10. doi: 10.1016/j.cell.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 10.Bailey M.H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173(2):371–385. doi: 10.1016/j.cell.2018.02.060. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olivier M., Hollstein M., Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu G., Pan C., Bei J.X., Li B., Liang C., Xu Y., et al. Mutant p53 in cancer progression and targeted therapies. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.595187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen K., Yang D., Li X., Sun B., Song F., Cao W., et al. Mutational landscape of gastric adenocarcinoma in Chinese: implications for prognosis and therapy. Proc Natl Acad Sci USA. 2015;112(4):1107–1112. doi: 10.1073/pnas.1422640112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robles A.I., Jen J., Harris C.C. Clinical outcomes of TP53 mutations in cancers. Cold Spring Harb Perspect Med. 2016;6(9) doi: 10.1101/cshperspect.a026294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russo A., Bazan V., Iacopetta B., Kerr D., Soussi T., Gebbia N. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23(30):7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W., Edwards A., Flemington E.K., Zhang K. Significant prognostic features and patterns of somatic TP53 mutations in human cancers. Cancer Inf. 2017;16 doi: 10.1177/1176935117691267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueno H., Yoshida K., Shiozawa Y., Nannya Y., Iijima-Yamashita Y., Kiyokawa N., et al. Landscape of driver mutations and their clinical impacts in pediatric B-cell precursor acute lymphoblastic leukemia. Blood Adv. 2020;4(20):5165–5173. doi: 10.1182/bloodadvances.2019001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escudeiro C., Pinto C., Vieira J., Peixoto A., Pinto P., Pinheiro M., et al. The role of TP53 pathogenic variants in early-onset HER2-positive breast cancer. Fam Cancer. 2021;20(3):173–180. doi: 10.1007/s10689-020-00212-2. [DOI] [PubMed] [Google Scholar]
- 19.Fedorova O., Daks A., Shuvalov O., Kizenko A., Petukhov A., Gnennaya Y., et al. Attenuation of p53 mutant as an approach for treatment Her2-positive cancer. Cell Death Discov. 2020;6(1):100. doi: 10.1038/s41420-020-00337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marvalim C., Datta A., Lee S.C. Role of p53 in breast cancer progression: an insight into p53 targeted therapy. Theranostics. 2023;13(4):1421–1442. doi: 10.7150/thno.81847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frascarelli C., Bonizzi G., Musico C.R., Mane E., Cassi C., Guerini Rocco E., et al. Revolutionizing cancer research: the impact of artificial intelligence in digital biobanking. J Pers Med. 2023;13(9) doi: 10.3390/jpm13091390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Haan L.M., de Groen R.A.L., de Groot F.A., Noordenbos T., van Wezel T., van Eijk R., et al. Real-world routine diagnostic molecular analysis for TP53 mutational status is recommended over p53 immunohistochemistry in B-cell lymphomas. Virchows Arch. 2023 doi: 10.1007/s00428-023-03676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanova M., Porta F.M., D’Ercole M., Pescia C., Sajjadi E., Cursano G., et al. Standardized pathology report for HER2 testing in compliance with 2023 ASCO/CAP updates and 2023 ESMO consensus statements on HER2-low breast cancer. Virchows Arch. 2024;484(1):3–14. doi: 10.1007/s00428-023-03656-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoque M.Z., Keskinarkaus A., Nyberg P., Seppänen T. Stain normalization methods for histopathology image analysis: a comprehensive review and experimental comparison. Inf Fusion. 2024;102 [Google Scholar]
- 25.Pocock J., Graham S., Vu Q.D., Jahanifar M., Deshpande S., Hadjigeorghiou G., et al. TIAToolbox as an end-to-end library for advanced tissue image analytics. Commun Med. 2022;2(1):120. doi: 10.1038/s43856-022-00186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macenko M., Niethammer M., Marron J., Borland D., Woosley J., Guan X., et al. A Method for Normalizing Histology Slides for Quantitative Analysis2009. 1107–1110 p.
- 27.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst, Man, Cybern. 1979;9(1):62–66. [Google Scholar]
- 28.Li S., Ye L. Multi-level thresholding image segmentation for rubber tree secant using improved Otsu's method and snake optimizer. Math Biosci Eng. 2023;20(6):9645–9669. doi: 10.3934/mbe.2023423. [DOI] [PubMed] [Google Scholar]
- 29.He K., Zhang X., Ren S., Sun J., editors. Deep residual learning for image recognition2016.
- 30.Bilal M., Raza S.E.A., Azam A., Graham S., Ilyas M., Cree I.A., et al. Development and validation of a weakly supervised deep learning framework to predict the status of molecular pathways and key mutations in colorectal cancer from routine histology images: a retrospective study. Lancet Digit Health. 2021;3(12):e763–e772. doi: 10.1016/S2589-7500(21)00180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kather J.N., Pearson A.T., Halama N., Jäger D., Krause J., Loosen S.H., et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25(7):1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaban M., Awan R., Fraz M.M., Azam A., Tsang Y.W., Snead D., et al. Context-aware convolutional neural network for grading of colorectal cancer histology images. IEEE Trans Med Imaging. 2020;39(7):2395–2405. doi: 10.1109/TMI.2020.2971006. [DOI] [PubMed] [Google Scholar]
- 33.Lee H., Cho Y.A., Kim D.G., Son J.Y., Cho E.Y. Real-world comparison of P53 immunohistochemistry and TP53 mutation analysis using next-generation sequencing. Anticancer Res. 2024;44(9):3983–3994. doi: 10.21873/anticanres.17227. [DOI] [PubMed] [Google Scholar]
- 34.Venetis K., Pepe F., Munzone E., Sajjadi E., Russo G., Pisapia P., et al. Analytical performance of next-generation sequencing and RT-PCR on formalin-fixed paraffin-embedded tumor tissues for PIK3CA testing in HR+/HER2- breast cancer. Cells. 2022;11(22) doi: 10.3390/cells11223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pepe F., Guerini-Rocco E., Fassan M., Fusco N., Vacirca D., Ranghiero A., et al. In-house homologous recombination deficiency testing in ovarian cancer: a multi-institutional Italian pilot study. J Clin Pathol. 2023 doi: 10.1136/jcp-2023-208852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sirvent J.J., Salvadó M.T., Santafé M., Martínez S., Brunet J., Alvaro T., et al. p53 in breast cancer. Its relation to histological grade, lymph-node status, hormone receptors, cell-proliferation fraction (ki-67) and c-erbB-2. Immunohistochemical study of 153 cases. Histol Histopathol. 1995;10(3):531–539. [PubMed] [Google Scholar]
- 37.Caselli E., Pelliccia C., Teti V., Bellezza G., Mandarano M., Ferri I., et al. Looking for more reliable biomarkers in breast cancer: Comparison between routine methods and RT-qPCR. PLoS One. 2021;16(9) doi: 10.1371/journal.pone.0255580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sajjadi E., Frascarelli C., Venetis K., Bonizzi G., Ivanova M., Vago G., et al. Computational pathology to improve biomarker testing in breast cancer: how close are we? Eur J Cancer Prev. 2023 doi: 10.1097/CEJ.0000000000000804. [DOI] [PubMed] [Google Scholar]
- 39.L'Imperio V., Cazzaniga G., Mannino M., Seminati D., Mascadri F., Ceku J., et al. Digital counting of tissue cells for molecular analysis: the QuANTUM pipeline. Virchows Arch. 2024 doi: 10.1007/s00428-024-03794-9. [DOI] [PubMed] [Google Scholar]
- 40.Dunn C., Brettle D., Cockroft M., Keating E., Revie C., Treanor D. Quantitative assessment of H&E staining for pathology: development and clinical evaluation of a novel system. Diagn Pathol. 2024;19(1):42. doi: 10.1186/s13000-024-01461-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baxi V., Edwards R., Montalto M., Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol. 2022;35(1):23–32. doi: 10.1038/s41379-021-00919-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham S., Vu Q.D., Raza S.E.A., Azam A., Tsang Y.W., Kwak J.T., et al. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal. 2019;58 doi: 10.1016/j.media.2019.101563. [DOI] [PubMed] [Google Scholar]
- 43.Vairamoorthy P., Venkatraman S, S.P., Malarvannan S, A.K. Multi-Attention Integrated Deep Learning Frameworks for Enhanced Breast Cancer Segmentation and Identification2024.
- 44.Qu H., Zhou M., Yan Z., Wang H., Rustgi V.K., Zhang S., et al. Genetic mutation and biological pathway prediction based on whole slide images in breast carcinoma using deep learning. npj Precis Oncol. 2021;5(1):87. doi: 10.1038/s41698-021-00225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Zou C., Zhang Y., Li X., Wang C., Ke F., et al. Prediction of BRCA gene mutation in breast cancer based on deep learning and histopathology images. Front Genet. 2021;12 doi: 10.3389/fgene.2021.661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daiichi S. Novel computational pathology-based TROP2 biomarker for datopotamab deruxtecan was predictive of clinical outcomes in patients with non-small cell lung cancer in TROPION-Lung01 Phase III trial Astrazeneca Media Press: Astrazeneca; 2024 [Available from: 〈https://www.astrazeneca.com/media-centre/press-releases/2024/novel-computational-pathology-based-trop2-biomarker-for-dato-dxd-was-predictive-of-clinical-outcomes-in-patients-with-nsclc-in-tropion-lung01-phase-iii-trial.html〉.
- 47.Wang H., Guo M., Wei H., Chen Y. Targeting p53 pathways: mechanisms, structures, and advances in therapy. Signal Transduct Target Ther. 2023;8(1):92. doi: 10.1038/s41392-023-01347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holmila R., Fouquet C., Cadranel J., Zalcman G., Soussi T. Splice mutations in the p53 gene: case report and review of the literature. Hum Mutat. 2003;21(1):101–102. doi: 10.1002/humu.9104. [DOI] [PubMed] [Google Scholar]
- 49.Bellizzi A.M. p53 as exemplar next-generation immunohistochemical marker: a molecularly informed, pattern-based approach, methodological considerations, and pan-cancer diagnostic applications. Appl Immunohistochem Mol Morphol. 2023;31(7):507–530. doi: 10.1097/PAI.0000000000001144. [DOI] [PubMed] [Google Scholar]
- 50.Alsner J., Jensen V., Kyndi M., Offersen B.V., Vu P., Børresen-Dale A.L., et al. A comparison between p53 accumulation determined by immunohistochemistry and TP53 mutations as prognostic variables in tumours from breast cancer patients. Acta Oncol. 2008;47(4):600–607. doi: 10.1080/02841860802047411. [DOI] [PubMed] [Google Scholar]
- 51.Alsner J., Yilmaz M., Guldberg P., Hansen L.L., Overgaard J. Heterogeneity in the clinical phenotype of TP53 mutations in breast cancer patients. Clin Cancer Res. 2000;6(10):3923–3931. doi: 10.1186/bcr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El Nahhas O.S.M., van Treeck M., Wölflein G., Unger M., Ligero M., Lenz T., et al. From whole-slide image to biomarker prediction: end-to-end weakly supervised deep learning in computational pathology. Nat Protoc. 2024 doi: 10.1038/s41596-024-01047-2. [DOI] [PubMed] [Google Scholar]
- 53.Saba T. Recent advancement in cancer detection using machine learning: systematic survey of decades, comparisons and challenges. J Infect Public Health. 2020;13(9):1274–1289. doi: 10.1016/j.jiph.2020.06.033. [DOI] [PubMed] [Google Scholar]
- 54.Nemade V., Pathak S., Dubey A. A systematic literature review of breast cancer diagnosis using machine intelligence techniques. Arch Comput Methods Eng. 2022;29 [Google Scholar]
- 55.Huang X., Qian S., Fang Q., Sang J., Xu C., editors. Csan: Contextual self-attention network for user sequential recommendation2018.
- 56.Jagannath M. Editorial: Feature extraction and deep learning for digital pathology images. Front Signal Process. 2023;3 [Google Scholar]
- 57.Fusco N., Viale G. The "lows": update on ER-low and HER2-low breast cancer. Breast. 2024;78 doi: 10.1016/j.breast.2024.103831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fusco N., Rocco E.G., Del Conte C., Pellegrini C., Bulfamante G., Di Nuovo F., et al. HER2 in gastric cancer: a digital image analysis in pre-neoplastic, primary and metastatic lesions. Mod Pathol. 2013;26(6):816–824. doi: 10.1038/modpathol.2012.228. [DOI] [PubMed] [Google Scholar]
- 59.Ahrenfeldt J., Christensen D.S., Sokač M., Kisistók J., McGranahan N., Birkbak N.J. Computational analysis reveals the temporal acquisition of pathway alterations during the evolution of cancer. Cancers (Basel) 2022;14(23) doi: 10.3390/cancers14235817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dadsetan S., Arefan D., Berg W.A., Zuley M.L., Sumkin J.H., Wu S. Deep learning of longitudinal mammogram examinations for breast cancer risk prediction. Pattern Recognit. 2022;132 doi: 10.1016/j.patcog.2022.108919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dammu H., Ren T., Duong T.Q. Deep learning prediction of pathological complete response, residual cancer burden, and progression-free survival in breast cancer patients. PLoS One. 2023;18(1) doi: 10.1371/journal.pone.0280148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim I., Kang K., Song Y., Kim T.J. Application of artificial intelligence in pathology: trends and challenges. Diagn (Basel) 2022;12(11) doi: 10.3390/diagnostics12112794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang D.Y., Venkat A., Khasawneh H., Sali R., Zhang V., Pei Z. Implementation of digital pathology and artificial intelligence in routine pathology practice. Lab Investig. 2024;104(9) doi: 10.1016/j.labinv.2024.102111. [DOI] [PubMed] [Google Scholar]
- 64.Ardon O., Klein E., Manzo A., Corsale L., England C., Mazzella A., et al. Digital pathology operations at a tertiary cancer center: infrastructure requirements and operational cost. J Pathol Inform. 2023;14 doi: 10.1016/j.jpi.2023.100318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonizzi G., Zattoni L., Fusco N. Biobanking in the digital pathology era. Oncol Res. 2021;29:229–233. doi: 10.32604/or.2022.024892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bruce C., Prassas I., Mokhtar M., Clarke B., Youssef E., Wang C., et al. Transforming diagnostics: the implementation of digital pathology in clinical laboratories. Histopathology. 2024;85(2):207–214. doi: 10.1111/his.15178. [DOI] [PubMed] [Google Scholar]
- 67.Xu H., Usuyama N., Bagga J., Zhang S., Rao R., Naumann T., et al. A whole-slide foundation model for digital pathology from real-world data. Nature. 2024;630(8015):181–188. doi: 10.1038/s41586-024-07441-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinto D.G., Bychkov A., Tsuyama N., Fukuoka J., Eloy C. Real-world implementation of digital pathology: results from an intercontinental survey. Lab Investig. 2023;103(12) doi: 10.1016/j.labinv.2023.100261. [DOI] [PubMed] [Google Scholar]
- 69.Hanna M.G., Reuter V.E., Samboy J., England C., Corsale L., Fine S.W., et al. Implementation of Digital Pathology Offers Clinical and Operational Increase in Efficiency and Cost Savings. (1543–2165 (Electronic)). [DOI] [PMC free article] [PubMed]
- 70.Alowais S.A., Alghamdi S.S., Alsuhebany N., Alqahtani T., Alshaya A.I., Almohareb S.N., et al. Revolutionizing healthcare: the role of artificial intelligence in clinical practice. BMC Med Educ. 2023;23(1):689. doi: 10.1186/s12909-023-04698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petch J., Di S., Nelson W. Opening the black box: the promise and limitations of explainable machine learning in cardiology. Can J Cardiol. 2022;38(2):204–213. doi: 10.1016/j.cjca.2021.09.004. [DOI] [PubMed] [Google Scholar]
- 72.Chauhan C., Gullapalli R.R. Ethics of AI in pathology: current paradigms and emerging issues. Am J Pathol. 2021;191(10):1673–1683. doi: 10.1016/j.ajpath.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hägele M., Seegerer P., Lapuschkin S., Bockmayr M., Samek W., Klauschen F., et al. Resolving challenges in deep learning-based analyses of histopathological images using explanation methods. Sci Rep. 2020;10(1):6423. doi: 10.1038/s41598-020-62724-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McKay F., Williams B.J., Prestwich G., Bansal D., Hallowell N., Treanor D. The ethical challenges of artificial intelligence-driven digital pathology. J Pathol Clin Res. 2022;8(3):209–216. doi: 10.1002/cjp2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: TP53 mutational assessment. NGS analysis revealed the mutational status of TP53, as depicted in the Integrative Genomics Viewer.