Abstract
Naturally colored cotton (NCC; Gossypium spp.) does not require additional chemical dyeing and is an environmentally friendly textile material with great research potential and applications. Our previous study using linkage and association mapping identified TRANSPARENT TESTA 2 (Gh_TT2) as acting on the proanthocyanin synthesis pathway. However, limited information is available about the genetic regulatory network of NCC. Here, we verified the effectiveness of Gh_TT2 and the roles of Gh_TT2 and red foliated mutant gene (Re) in pigment formation and deposition of brown fiber cotton (BFC). Variations in Gh_TT2 derived from interspecific hybridization between Gossypium barbadense acc. Pima 90-53 and Gossypium hirsutum acc. Handan208 resulted in gene expression differences, thereby causing phenotypic variation. Additionally, the MYB-bHLH-WD complex was found to be negatively modulated by TRANSPARENT TESTA 16/ARABIDOPSIS BSISTER (TT16/ABS). RNA-seq suggested that differential expression of homologous genes of key enzymes in the proanthocyanin synthesis pathway strongly contributes to the color rendering of natural dark brown and light brown cotton. Our study proposes a regulatory model in BFC, which will provide theoretical guidance for the genetic improvement of NCC.
TRANSPARENT TESTA 16 and the transcriptional complex MYB-bHLH-WD40 regulate proanthocyanin biosynthesis, leading to brown fiber cotton.
Introduction
Cotton (Gossypium spp.), a major cash crop and an important textile material, has been widely cultivated in the worldwide. Naturally colored cotton (NCC) refers to the varieties in which pigment gradually deposit in the cavity cells of the fiber during the development stage. Its color occurs naturally, without the help of any modern molecular biotechnology, and the varieties currently validated and grown in a large scale are limited to brown and green cotton. Naturally brown fiber has been extensively studied as an excellent model for the cultivation of elite NCC. Researchers (Xiao et al. 2007; Tan et al. 2013) have concluded that the formation and deposition of brown pigments are directly related to the metabolic pathway of flavonoids, and the pigments are most likely to be proanthocyanidins in the flavonoid family by isolating and extracting pigments from brown cotton fiber. In addition, silencing key structural genes of the proanthocyanidin biosynthesis pathway using RNAi technology resulted in a lighter color, demonstrating the contribution of proanthocyanidins to the coloration of brown cotton fiber (Liu et al. 2018; Zheng et al. 2023). Genetically, the gene loci of brown fiber cotton (BFC) have been demonstrated to be Lc1, Lc2, Lc3, Lc4, Lc5, and Lc6 (Kohel 1985), while Lc1 is the best studied and identified via linkage and association mapping.
It was first reported in Arabidopsis thaliana that the complex module MYB-bHLH-WDR (MBW), formed by 3 transcription factors including TT2 (TRANSPARENT TESTA 2, a R2R3-MYB TF), TT8 (TRANSPARENT TESTA 8, a bHLH TF), and TTG1 (TRANSPARENT TESTA GLABROUS1, a WD-repeat protein), could bind to the upstream promoter region of the BAN (BANYULS) to synergistically regulate the proanthocyanin synthesis in the seed coat. While TT8 could be replaced by closely related bHLHs, TT2 was specifically essential for the specific recognition of target DNA and could not be substituted by other MYBs (Baudry et al. 2004). In addition, TT8 expression was controlled by different combinations of MYB (TT2; Production of Anthocyanin Pigment 1, PAP1) and bHLH factors (GLABRA3, GL3; ENHANCER OF GLABRA3, EGL3) in planta (Baudry et al. 2006). Xu et al. (2013) revealed that TT8 simultaneously regulated the synthesis of anthocyanins and proanthocyanidins through promoter analysis, indicating that 6 different MBW complexes were involved in facilitating this regulatory process. Interestingly, the diversity of MBW complexes was also manifested in the fact that R3 MYB (MYBL2) competed with R2R3 MYB to bind bHLH family transcription factors, thereby negatively regulating the proanthocyanidin pathway (Dubos et al. 2008; Matsui et al. 2008; Jiang et al. 2023b). MBW complex co-regulating proanthocyanidins pathway synthesis has also been reported in various plants (Rajput et al. 2022; Jiang et al. 2023a). In cotton, most of the researches on MBW complexes were focused on fiber initiation and determination of fiber cell fate (Walford et al. 2011; Zhu et al. 2018; Tian et al. 2020; Ye et al. 2022). It has been shown that MYB25-like (MIXTA MYB transcription factor) controls fiber development. Wang et al. (2021) reviewed these findings in detail and proposed the hypothesis of sugar signals activating fiber initiation under auxin. Furthermore, researches showed that MYB transcription factors could regulate pigment formation and accumulation in cotton petals, resulting in the phenotype of red spots at the base or a pink corolla (Abid et al. 2022; Cai et al. 2023; Liang et al. 2023).
When exploring the genetic mechanism of brown cotton, GhTT2, as a transcription factor of the R2R3 MYB, was first localized and cloned, but the detail regulation mechanism underlying Gh_TT2 has not been elucidated (Hinchliffe et al. 2016; Wen et al. 2018; Yan et al. 2018). It is currently unclear whether the MBW complex also plays a regulatory role in brown cotton fibers. The function of the MBW complex varies depending on the different core transcription factors MYB and bHLH (Wang et al. 2022a). Therefore, detailed researches on the composition and hierarchical regulation of the MBW complex in brown cotton are of crucial importance for understanding the genetic regulatory network of brown cotton. Colored fiber creation originally depended on traditional hybrid breeding. However, traditional methods struggle to effectively tackle issues like narrow NCC chromatography, pigment accumulation, and the negative relationship between fiber quality and yield. As a result, researchers have increasingly embraced the tools of molecular biology and synthetic biology in recent years (Wen et al. 2024). Wang et al. (2022b) identified that Re can simultaneously activate the anthocyanin and proanthocyanin metabolic pathways, leading to the production of brown fiber when highly expressed specifically in fiber. Ke et al. (2022) created genetic lines of the GhOMT1 and hybridized it with NCC, and developed a series of colored fiber lines by conducting continuous selection and breeding over several years. What's even more impressive is that researchers have successfully introduced the beet red pigment synthesis pathway into cotton, resulting in a successful creation of pink fiber, which provides an approach for the innovation of colored cotton (Ge et al. 2023; Li et al. 2023).
Although substantial progresses have been achieved in understanding proanthocyanidins synthesis pathway, further efforts are needed to close the gaps between genomic studies and practical breeding. It is crucial to explore how to effectively utilize endogenous pigments in cotton, and how to modify and balance anthocyanins and proanthocyanidins through molecular strategies. Furthermore, the exploration and utilization of the different combination of the MBW complex, as well as the internal competition, coordination, and game relationship within the MBW complex, will be a feasible breakthrough solution.
Results
Gh_TT2 is the causal gene for cotton brown fiber
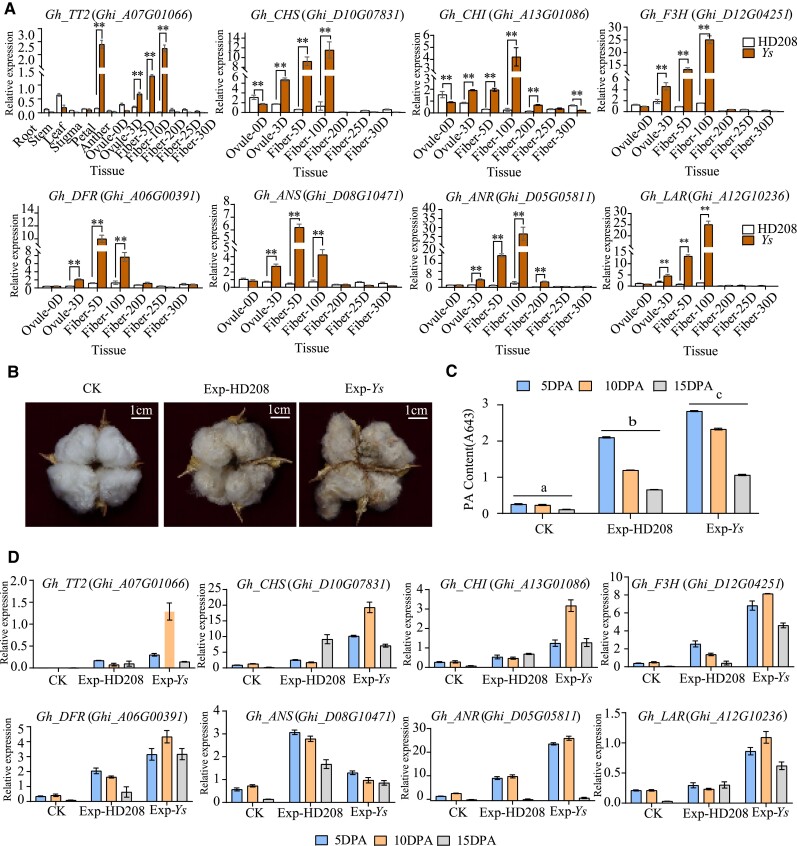
Previously, Gh_TT2 (Ghi_A07G01066) was identified as the candidate gene of BFC in our laboratory (Wen et al. 2018). Expression assays showed that Gh_TT2 specifically expressed in 5 to 15 DPA (days post-anther) fiber and the expression in Ys (dark brown fiber) was significantly higher than that in Handan208 (white fiber) (Fig. 1A). Simultaneously, the expression of related proanthocyanidins biosynthesis genes (PABGs) was tested, which showed the common trend. The expression of PABGs at 5 to 15 DPA during the development of dark brown fiber was significantly higher than that of white fiber (Fig. 1A). We previously found that a single SNP in the coding region of Gh_TT2 resulting in an amino acid variation from proline to threonine (Wen et al. 2018). To verify the influence of the single SNP and function of Gh_TT2, the fiber elongation-specific promoter, promoter of α-expansin 2 (PGbEXPA2) (Li et al. 2015) was used to specifically express Gh_TT2 in 5 to 10 DPA fibers, and cDNAs of genotypes of HD208 and Ys were overexpressed in Gossypium hirsutum transgenic receptor Jin668. Compared with the CK, the mature fibers of transgenic lines of 2 genotypes both showed light brown (Fig. 1B and Supplementary Fig. S1). Correspondingly, the content of proanthocyanidins and expression levels of Gh_TT2 and PABGs were detected. The content of proanthocyanidins gradually decreased in 5 to 15 DPA developing fiber of transgenic lines and exhibited highest amount in PGbEXPA2: Gh_TT2Ys transgenic lines than other materials (Fig. 1C). The PGbEXPA2: Gh_TT2Ys transgenic lines also had the highest expression of Gh_TT2 in 5 to 15 DPA fiber (Fig. 1D). Compared with the CK, the relative expression of PABGs was greatly increased and showed high levels in PGbEXPA2:Gh_TT2Ys transgenic lines. The results indicates that cDNAs of both genotypes have the function of generating a light brown phenotype in fibers, while the single SNP variation in the coding region of Ys has a stronger effect in regulation of downstream genes. Further, in order to explore whether the single SNP in the coding region causes the change of gene expression position, the full length of Gh_TT2Ys and Gh_tt2HD208 was cloned into the vector pMDC43 with GFP tag fused to the N-terminus and transiently expressed in Nicotiana benthamiana epidermal cells following Agrobacterium-mediated transfection, which showed that both Gh_TT2Ys and Gh_tt2HD208 were strongly expressed in the nucleus (Supplementary Fig. S2), suggesting that the expression position of the Gh_TT2 is not altered by the mutation in the exon. In conclusion, the high expression of Gh_TT2 in developing fiber was closely related to the formation and deposition of proanthocyanidins in brown fiber and the SNP in coding regions could affect the expressions of PABGs.
Figure 1.
Expression levels of Gh_TT2 and PABGs, and phenotypes of transgenic lines. The expression of Gh_TT2 and PABGs was significantly higher in Ys than in HD208. PABGs, proanthocyanidins biosynthesis genes. A) The relative expression level was obtained by RT-qPCR. Error bars represent ± SD (3 biological replicates), and the statistical significance was calculated using t-test (**P < 0.01, *P < 0.05). B) The mature fiber of transgenic lines of PGbEXPA2:Gh_tt2HD208 and PGbEXPA2: Gh_TT2Ys. A transgenic negative line named CK was photographed as a control. The fibres of both lines were brown compared with CK. Scale bar, 1 cm. C) Determination of the relative content of PA (Proanthocyanidin) in the 5 to 15 DPA fibers of transgenic lines. Error bars represent ± SD (3 biological replicates), and the statistical significance was calculated using 1-way ANOVA (P < 0.05), and different lowercase letters represent significant differences in pigment content between transgenic lines. DPA, days post-anther. D) The expression of Gh_TT2 and PABGs in the 5 to 15 DPA fibers of transgenic lines. Except ANS, the expression levels of other genes in transgenic lines showed the following trend, with the highest expression in PGbEXPA2: Gh_TT2Ys. Error bars represent ± SD (3 biological replicates).
Variations in the promoter of Gh_TT2 are related to its gain-of-function
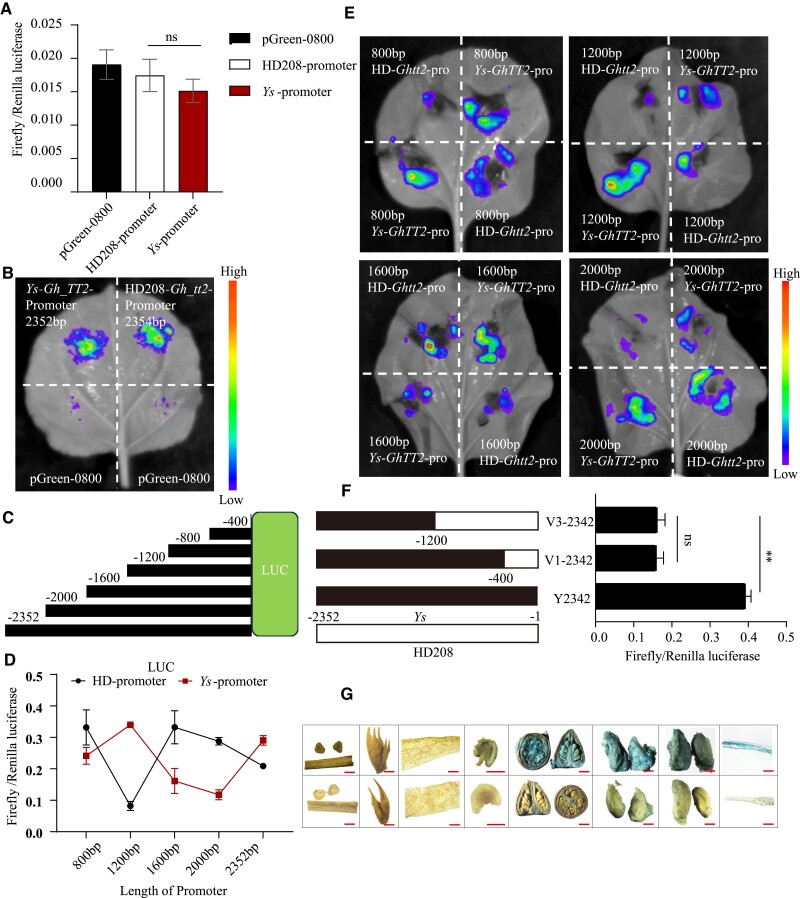
Besides the SNP in the coding region, an Indel was identified in the TATA box in the promoter (Wen et al. 2018), which could be the reason for the induction of gain-of-function (Supplementary Fig. S3). Therefore, we shifted to studying the promoter's own driving ability. The LUC reporter activation assays were used to verify the activity of promoter, and the 2,532 bp sequence of ProYs and the 2,534 bp of ProHD208 were cloned and conducted into the pGreen II 0800-LUC vector. Both N. benthamiana and protoplasts acted as conversion receptors, and there was no significant difference in the fluorescence value of the fireflies (Fig. 2, A and B). We then considered that different lengths might correspond to different levels of driving ability; we intercepted 400, 1,200, 1,600, and 2,000 bp of promoter (Fig. 2C), and conducted the same experiment in protoplasts and N. benthamiana leaves. Surprisingly, the promoters of both genotypes showed significant differences in length at 1,200, 1,600, and 2,000 bp (Fig. 2, D and E). Following this, we attempted to analyze the effect of individual SNPs in the promoter. We first utilized the ProYs as a reference point to substitute the 400 and 1,200 bp segments of ProHD208 with the pGreen 0800 vector serving as an intermediate. Notably, the initial mutation of the TATA insertion in the 400 bp region resulted in a rapid decrease in its driving ability, and SNPs between the 400 and 800 bp segments did not induce any discernible alterations (Fig. 2F and Supplementary Figs. S3 and S4).
Figure 2.
Driving capacity of the promoter region of Gh_TT2. A) LUC assays were performed in cotton protoplasts. The 2,352 bp of ProYs and 2,354 bp of ProHD208 were constructed into pGreen 0800 vector. Error bars represent ± SD (3 biological replicates), and the statistical significance was calculated using t-test (**P < 0.01, *P < 0.05). ns, no significance. B) LUC assays were performed in N. benthamiana. The 2,352 bp of ProYs and 2,354 bp of ProHD208 were constructed into pGreen 0800 vector. C) Schematic diagram of vector construction with different length of promoter fragments fused firefly luciferase reporter gene. D) The intensity of fluorescence in dual-luciferase reporter assays. Different lengths of promoter fused LUC were transformed into protoplasts. Error bars represent ± SD (3 biological replicates), and the statistical significance was calculated using t-test (**P < 0.01, *P < 0.05). E) LUC assays were performed in N. benthamiana for exploring driving activity of different length of promoter. F) LUC performed in protoplasts after mutation introduction. The black part represents ProYs, and the white part indicates the introduced ProHD208. Error bars represent ± SD (3 biological replicates), and the statistical significance was calculated using t-test (**P < 0.01, *P < 0.05). ns, no significance. G) Histochemical assays of GUS activity in cotton. The top line represents ProYs:Gus, and the bottom line represents the negative control. From left to right: stem, leaf, petal, anther, 0 DPA ovule, 5 DPA seed and fiber, 10 DPA seed fiber, and 15 DPA fiber. Scale bar, 1,000 μm.
Qualitative detection experiments for the GUS reporter gene were used to investigate the spatiotemporal expression of Gh_TT2. The results of different tissue staining of A. thaliana showed that blue color was only observed in the 4-leaf stage (Supplementary Fig. S5). We also sampled and stained different tissues of cotton transformation lines fused with GUS reporters, and the expression of the reporter gene was detected in the 0 DPA ovule, 3 DPA ovule, and 5 to 15 DPA fibers (Fig. 2G), which was consistent with its function of producing brown fiber.
The MBW complex is involved in the regulation of proanthocyanidin pathways
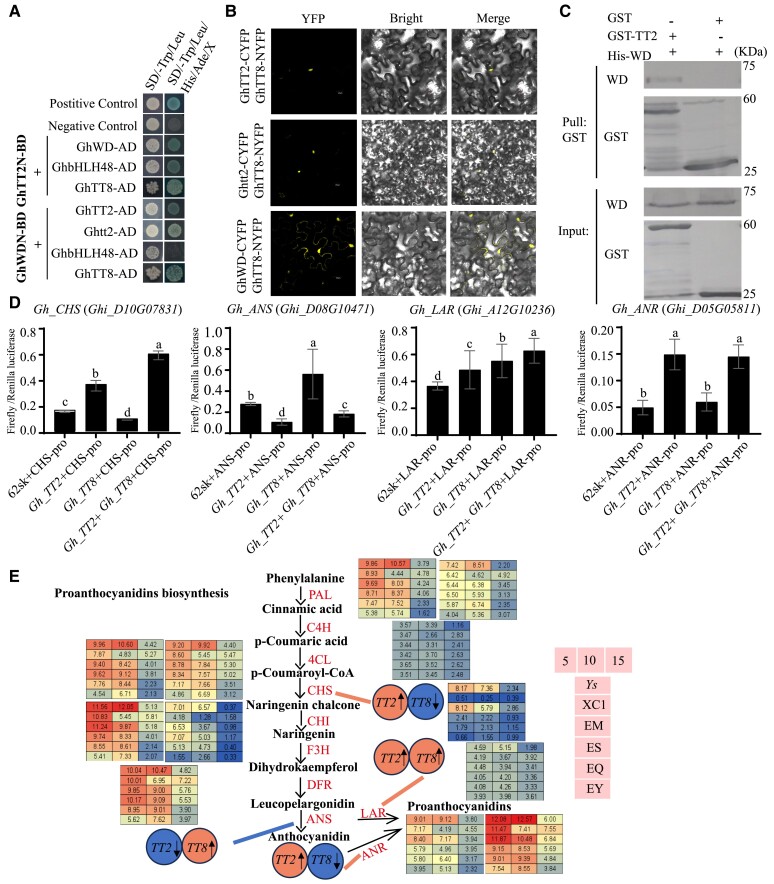
To explore the mechanism of Gh_TT2 regulating brown fiber, the yeast-2-hybrid (Y2H) system was used to screen for the interacting protein of Gh_TT2. In addition to Gh_WD, reported to be involved in the shading of brown cotton (Wen et al. 2018), Gh_TT8 (Ghi_A08G11656) and Gh_bHLH48 (Ghi_A01G11151) were also found (Fig. 3A and Supplementary Fig. S6). Luciferase complementation imaging (LCI) and bimolecular fluorescence complementation (BiFC) were also used to verify the interactions, which showed that Gh_TT2 and Gh_WD individually interacted with Gh_TT8 and Gh_bHLH48, respectively. The interaction was also observed between Gh_TT2 and Gh_WD (Fig. 3B and Supplementary Figs. S7 and S8). GhTT2-GST and GhWD-His proteins were purified to perform pull-down assays. Compared with GST-tag, GhWD-His could be pulled down by GhTT2-GST and successfully detected via immunoblot (Fig. 3C).
Figure 3.
Interactions within the MBW complex and their regulatory role in controlling PA synthesis. A) Yeast-2-hybrid assays showing the pairwise interaction of Gh_TT2, Gh_TT8, GhbHLH48, and Gh_WD. Yeast cells were plated on SD-Trp-Leu and SD-Trp-Leu-His-Ade (with X-α-Gal) media. BD, pGBKT7; AD, pGADT7. B) BiFC assays in N. benthamiana epidermal cells. Bars = 20 μm. C) Pull-down assays for Gh_TT2 and Gh_WD proteins. GhWD-His protein was pulled down by Gh_TT2-GST protein, GST was used as a negative control. D) LUC assays in cotton callus protoplasts. Promoters of CHS, LAR, ANS, and ANR were respectively fused upstream of LUC. Pro35S: REN was used as the internal control. Gh_TT2 and Gh_TT8 were fused downstream of 35Spro in pGreenII 62-SK. Error bars represent ± SD (3 biological replicates), and the statistical significance was calculated using 1-way ANOVA (P < 0.05), and different lowercase letters represent significant differences in different treatment groups. E) Possible regulatory positions of the Gh_TT2 and Gh_TT8 in the PA synthesis pathway. Red represents a potential positive regulatory effect, blue indicates a negative inhibitory effect. Ys: dark brown fiber mutant; XC1: light brown cotton; EM: overexpression transgenic lines of Gh_TT2 with light brown; ES: overexpression transgenic lines of Re, named GbEXP-E1, with reddish brown; EQ: overexpression transgenic lines of Re, named GbEXP-E3, with light brown; EY: transgenic negative control line with white cotton; 5, 10, and 15 represents days post-anther.
In order to explore the specific functions of the interaction genes, the tissue expression patterns of the above 3 genes were tested (Supplementary Fig. S9). Gh_WD was a constitutively expressed gene in roots, stems, leaves, petals, anthers, ovules, and fibers. The expression of Gh_WD in 5 to 10 DPA fiber in BFC was higher than that in white fiber cotton, and the opposite trend was shown in other tissues and the late stage of fiber development. The expression of Gh_TT8 varied significantly at various stages of fiber development except for 25 DPA, and its expression in BFC was significantly higher than that in white fiber cotton. Gh_bHLH48 had a higher expression level in root, stigma, and petal in brown fiber than in white fiber cotton, while its expressions were completely reversed in other tissues.
Based on the tissue expression pattern, Gh_TT8 with the same trend as Gh_TT2 was selected for follow-up research. Yeast 3-hybrid (Y3H) was used to verify MBW complex formation, which showed that Gh_TT8 promoted the interaction between Gh_TT2 and Gh_WD, and Gh_WD had no significant effect on the interaction between Gh_TT2 and Gh_TT8 (Supplementary Fig. S10).
It is unclear how the MBW complex exerts a regulatory role in the proanthocyanidin (PA) pathway. First, we investigated whether Gh_TT2 binds to promoter regions of critical pathway enzyme genes via dual-luciferase reporter system, and then the co-expression of Gh_TT2 and Gh_TT8 was also considered. The co-expression of Gh_TT2 with ProCHS:LUC led to much stronger LUC activity than expressing ProCHS:LUC alone. Furthermore, Gh_TT8 enhanced the positive regulatory effect of Gh_TT2 on CHS (chalcone synthase). When the ProLAR:LUC was co-expressed with Gh_TT2, the LUC activity became more higher compared with the control, and Gh_TT8 produced higher LUC activity than expressing ProLAR:LUC without effector. The combination of Gh_TT8 and Gh_TT2 played a promoting role. Gh_TT2 co-expressed with ProANR:LUC could promote the LUC activity. Lower LUC activity caused by the co-expression of Gh_TT2 with ProANS:LUC can be attenuated by Gh_TT8 (Fig. 3, D and E). These results showed that Gh_TT2 and Gh_TT8 played a regulatory role in PAs pathway, individually or as dimers to target the promoter of CHS, LAR (leucoanthocyanidin reductase), ANR (anthocyanidin reductase), and ANS (anthocyanin synthase).
VIGS (virus induced gene silencing) was taken in Ys for analyzing the role played by Gh_WD in regulating PAs pathway, and RT-qPCR was used to verify silencing efficiency. Leaves from TRV:00, TRV: Gh_WD & GhCEN (CENTRORADIALIS), and TRV: Gh_TT2 & GhCEN were sampled to check the expression of related genes and the content of PAs. The expression of GhCEN, as a precocious gene, was silenced together with the target genes, and Gh_WD was significantly decreased in the silenced lines compared with the TRV:00 lines. Additionally, the expression of Gh_WD was significantly increased in the TRV: GhTT2 & GhCEN lines. The content of PAs in the leaves of the silent lines was significantly lower than that of the control (Supplementary Fig. S11). This result suggested that Gh_WD was correlated with the PA synthesis as a regulator.
TRANSPARENT TESTA16 inhibits Gh_TT2 and influences the PAs pathway
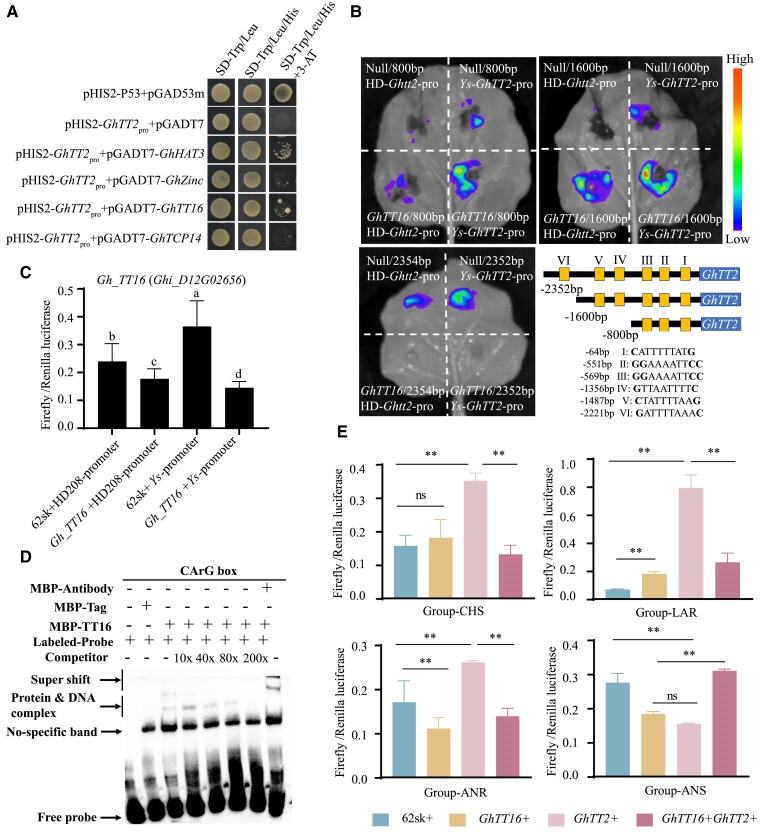
To mine the transcription factors regulating Gh_TT2, we obtained the 1,000 bp upstream fragment of Gh_TT2 through a yeast-1-hybrid (Y1H) experiment. There was no detectable interaction between ProGh_TT2 and the empty pGADT7 vector when cultured on SD/-Trp-Leu-His medium. However, in the presence of 3-amino-1,2,4-triazole (3-AT), ProGh_TT2 interacted with GhHAT3 (Homebox-leucine zipper protein), GhZinc (Zinc finger CCCH domain-containing protein), GhTCP14 (a bHLH TF), and GhTT16 (Fig. 4A), confirming that the above 4 genes could bind to the promoter of Gh_TT2. This combination was also confirmed in protoplasts and N. benthamiana through LUC experiments (Fig. 4, B and C, and Supplementary Fig. S12).
Figure 4.
Verification of the regulatory relationship between GhTT16 and GhTT2. A) Yeast-1-hybrid assays revealed that GhTT16, GhHAT3, GhZinc, and GhTCP14 could bind the Gh_TT2 promoter region. Yeast cells were sequentially screened on the SD-Trp/Leu, SD-Trp/Leu/His, SD-Trp/Leu/His with 3-AT medium. B) LUC assays in N. benthamiana leaves. Different lengths of promoter of Gh_TT2Ys and Gh_tt2HD208 were fused downstream of 35Spro in pGreen II 0800. The pGreenII 62-SK empty vector named Null as a control. And, different length promoter regions contain the number of CArG boxes and their respective positions and sequences. C) LUC assays in cotton callus protoplasts. Promoter of Gh_TT2Ys and Gh_TT2HD208 was fused upstream of LUC, respectively. Pro35S: REN was used as an internal control. Gh_TT16 was fused downstream of 35Spro in pGreenII 62-SK. Error bars represent ± SD (3 biological replicates), and the statistical significance was calculated using 1-way ANOVA (P < 0.05), and different lowercase letters represent significant differences in different treatment groups. D) EMSAs of the DNA binding activity of GhTT16 for the Gh_TT2 promoter. Probes were labeled with biotin as hot probes and incubated with recombinant GhTT16-MBP protein. Unlabeled probes as competitor probes were added to compete with biotin-labeled probes. MBP protein was used as negative control. E) LUC assays in cotton callus protoplasts. Promoters of 4 structural genes were fused upstream of LUC, respectively. Pro35S: REN was used as an internal control. Gh_TT2 and Gh_TT16 were fused downstream of 35Spro in pGreenII 62-SK. Error bars represent ± SD (3 biological replicates), and the statistical significance was calculated using t-test (**P < 0.01, *P < 0.05). ns, no significance.
Given the important role of the ABS-MADS gene AtTT16 in brown seed coat formation in A. thaliana (Nesi et al. 2002), we decided to thoroughly examine the regulatory relationship of GhTT16 with Gh_TT2. Six CArG boxes were detected in the 2,532 bp upstream promoter region of GhTT2 (Fig. 4B). A probe of 50 bp containing 2 CArG boxes (II, Ⅲ) was generated to conduct electrophoretic mobility shift assays (EMSAs). The results showed that MBP alone could not bind to the Gh_TT2 promoter, while GhTT16-MBP was found to directly bind to the biotin-labeled probes, which was gradually diminished upon the addition of unlabeled competitive probes (Fig. 4D). Moreover, after incubation with MBP antibody, the composite bands exhibited a slower migration rate, providing strong evidence for the binding of GhTT16 to Gh_TT2.
To further clarify the regulatory effect of GhTT16 on Gh_TT2 and its impact on PA synthesis, LUC experiments in cotton protoplasts were conducted, which showed that GhTT16 could weaken the activation effect of Gh_TT2 on CHS, DFR (dihydroflavonol 4-reductase), LAR, and ANR. Specifically, Gh_TT2 exhibited higher LUC activity when binding to the promoter regions of the 4 key enzymes, whereas LUC activity significantly decreased after GhTT16 binding (Fig. 4E). Likewise, the attenuating effect of Gh_TT2 on ANS was also counteracted by GhTT16. In addition, GhTT16 could directly bind to the promoter of PABGs to regulate their activities.
The proanthocyanidin metabolic network in BFC
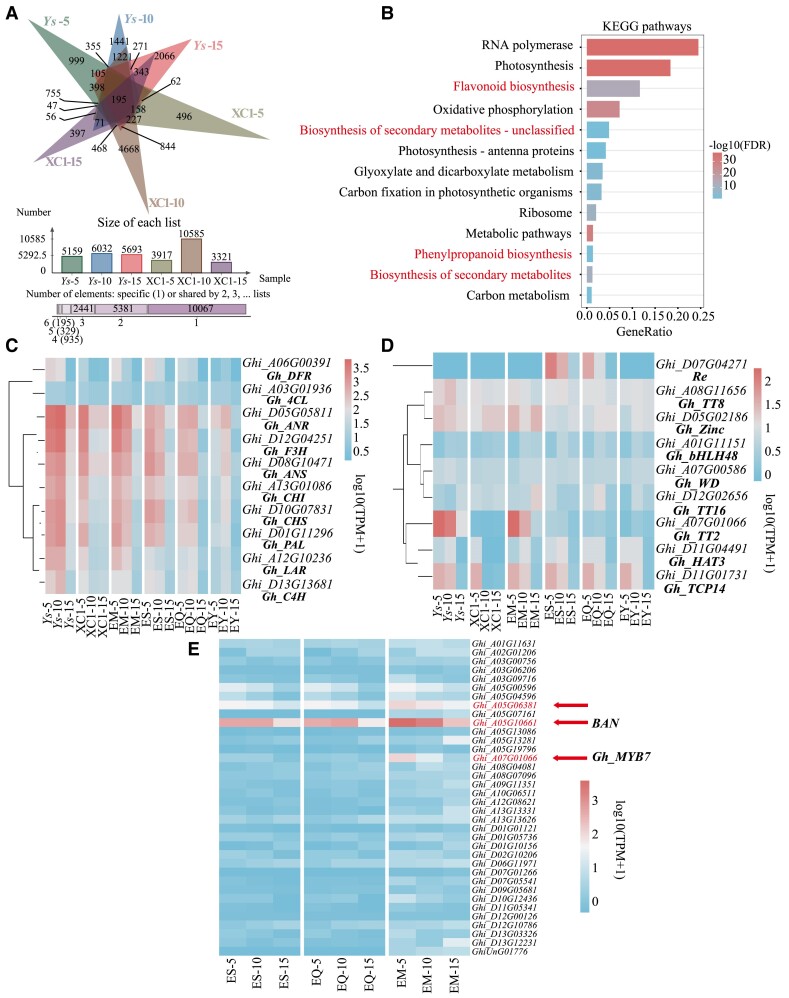
To investigate the regulatory network of pigment formation and accumulation in colored cotton, samples from 3 different stages of fiber development were collected for transcriptome analysis. The transcriptional regulation of natural dark brown cotton Ys and light brown cotton Xincaimian 1 (XC1) showed that the number of differentially expressed genes between XC1 and Ys, at 5 DPA, 10 DPA, and 15 DPA, are 2,711, 12,679, and 5,427, respectively.
Notably, the highest number of differentially expressed genes appeared in 10 DPA fibers, implying its significance as the key period of color alteration (Fig. 5A and Supplementary Fig. S13). Furthermore, enrichment analysis of these genes revealed a significant enrichment in flavonoid biosynthesis, phenylpropanoid biosynthesis, and secondary metabolism biosynthesis (Fig. 5B and Supplementary Fig. S14). Enrichment analysis was then conducted on the differentially expressed genes from the 3 periods and a dynamic bubble map was created to illustrating the genes enriched in flavonoid biosynthesis, phenylpropanoid biosynthesis, and biosynthesis of secondary metabolites (Supplementary Fig. S15). This visual representation showcased the specific situation of these genes in both materials and across the 3 periods. The expression levels of critical pathway enzyme genes exhibited varying trends across the 3 stages in Ys and XC1. Particularly, higher expression levels of these key enzyme genes were observed at 5 DPA and 10 DPA in Ys, while they were only found at 5 DPA in XC1, suggesting that the rapid decay of XC1 at 10 DPA could potentially contribute to the color difference between Ys and XC1 (Supplementary Fig. S16). The expression levels of related regulatory genes and structural genes were detected at 5 DPA, 10 DPA, and 15 DPA. Importantly, there was a noticeable disparity in the expression levels of DFR and LAR between Ys and XC1 (Fig. 5C). Additionally, the expression levels of Gh_TT16 and Gh_TT2 consistently exhibited contrasting trends (Fig. 5D).
Figure 5.
Transcriptome analysis of pigment formation and regulation in brown cotton. A) Number of differentially expressed genes at 5 to 15 DPA between Ys and XC1. DPA, days post-anther. B) KEGG enrichment analysis of differentially expressed genes at 5 to 15 DPA between Ys and XC1. KEGG, Kyoto Encyclopedia of Genes and Genomes. C) Heatmap of critical pathway enzymes expression at 5 to 15 DPA in 6 materials. D) Heatmap of regulated gene expression at 5 to 15 DPA in 6 materials. E) Heatmap of co-downregulated expression genes at 5 to 15 DPA in EM, ES, and EQ.
Our previous research showed that overexpression of Re produced brown cotton fibers (Wang et al. 2022b). Consequently, our efforts were directed toward examining the differential mechanism underlying the alteration in fiber color caused by Re and Gh_TT2. A comparison of the number of differentially expressed genes in the 3 overexpressed lines with that of NCC revealed a deviation from the expected pattern (Supplementary Fig. S17). This observation suggests that the PGbEXPA2 may exert an influence on the expression pattern of genes. Enrichment analysis of these differential genes revealed that some genes were significantly enriched in the photosynthetic pathway, which may be due to the regulation of Re expression by photosynthetic factors, while no similar pattern was found for Gh_TT2 (Supplementary Fig. S18). Compared with EY (transgenic negative line; white fiber), a total of 121 differentially expressed genes were found to be common across the ES, EQ, and EM, which is crucial in understanding the difference in color regulation between Gh_TT2 and Re. Among them, 36 genes were jointly downregulated and 23 were jointly upregulated. The differential expression of these genes was visually depicted in a heat map and 3 genes attracted our particular attention. Of these, Ghi_A07G01066, annotated as MYB7, was found to be downregulated in all 3 materials; however, it exhibited significantly higher expression in EM compared with the ES and EQ (Fig. 5E). Ghi_A05G10661, annotated as Ban, as a negative regulator of flavonoid biosynthesis, showed the same trend as MYB7. Ghi_A05G06381, an unannotated gene, was also detected with high expression only at 5 DPA and 10 DPA in EM, possibly played a role in pigment synthesis (Fig. 5E). These 3 genes were thought to play a role in balancing the pigment ratio of proanthocyanidin and anthocyanidin. Considering the regulation of anthocyanin synthesis by Re and proanthocyanidin synthesis by Gh_TT2, we compared the expressions of LAR, ANS, and ANR among ES, EQ, and EM, and discovered that the levels of LAR and ANR were relatively high, while ANS was relatively low in EM (Supplementary Figs. S19 and S20).
Weighted gene co-expression network analysis (WGCNA) of the core gene set of proanthocyanidins metabolism in BFC
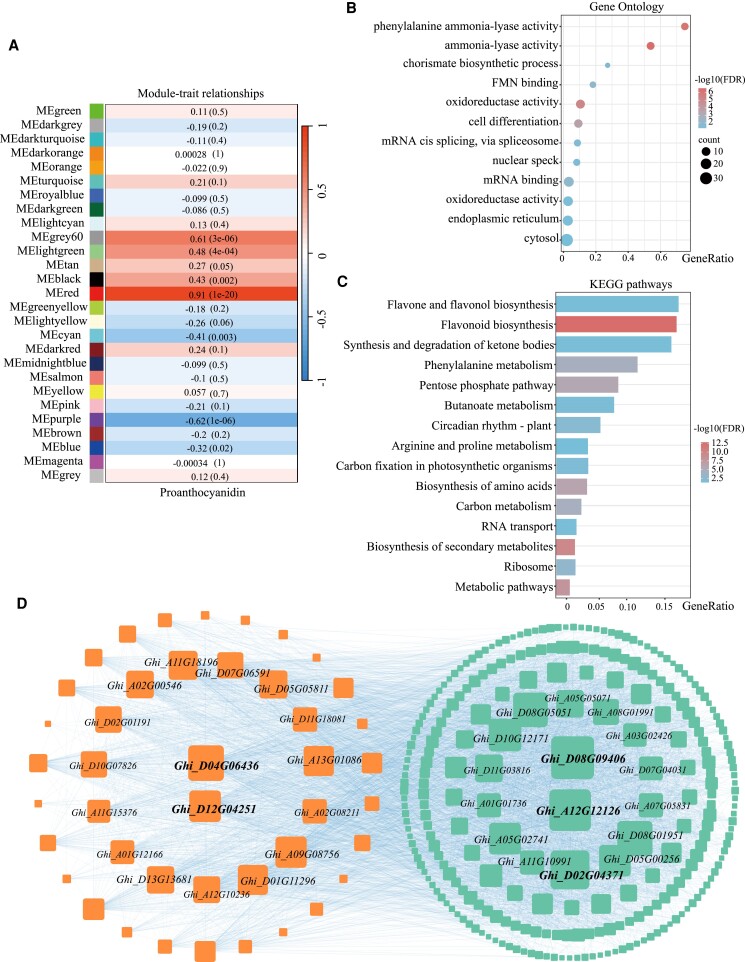
The gene co-expression regulatory network of proanthocyanidins biosynthesis in brown cotton was investigated. We first defined genes with an expression level of TPM > 1 in any period as the expression genes, and filtered the input genes for weighted gene co-expression network analysis (WGCNA) using the median absolute deviation method, resulting in a total of 34,524 genes. A soft threshold of 13 was then determined, leading to the detection of 27 modules. Notably, the red module contained 835 genes, while the grey60 module contained 201 genes, both of which were found to have a significant positive correlation with proanthocyanidin (PA) content. Conversely, the purple module, comprising 494 genes, displayed a significant negative correlation with PA content (Fig. 6A). Remarkably, the red module exhibited the highest correlation of 0.91, and encompassed key structural genes such as CHS, CHI (chalcone isomerase), F3H (flavanone 3-hydroxylase), ANR, LAR, C4H (cinnamate 4-hydroxylase), and PAL (phenylalanine ammonia lyase), along with regulatory genes including Gh_TT2 and Gh_TT8, which are associated with the PA synthesis pathway. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed (Fig. 6, B and C). Simultaneously, the KEGG enrichment analysis of the module genes revealed enrichment in pathways such as “flavonoid biosynthesis”, “secondary metabolites biosynthesis”, “phenylalanine metabolism”, and “flavone and flavonol biosynthesis”, confirming that this is the core gene set for proanthocyanin biosynthesis in cotton (Fig. 6C). The red module contained 835 genes, out of which 507 exhibited differential expression; after filtering with criteria of MM (module membership) > 0.8 and GS (gene significance) > 0.2, a total of 259 genes were retained, including 226 differentially expressed genes. These 259 genes were analyzed using PlantTFDB to predict associated transcription factors, which revealed 19 transcription factors, with MYB exhibiting the greatest number of 7 (Supplementary Table S1). Notably, all MYB transcription factors were differentially expressed, implying their significant involvement in the regulation of pigment synthesis. Specifically, MYB3 has been identified to be involved in phenylalanine synthesis (Zheng et al. 2021; Kim et al. 2022). The visualization of the 259 differentially expressed genes in Cytoscape highlighted 43 genes in the orange portion, enriched in “flavonoid biosynthesis”, “secondary metabolites biosynthesis”, “phenylalanine metabolism”, and “flavone and flavonol biosynthesis”. Within this portion, the critical pathway enzymes exhibited the highest degree (Fig. 6D). Additionally, in the remaining green part, Ghi_D08G09406, showing the highest correlation, is annotated as Cinnamoyl-CoA Reductase; Ghi_A05G02741, encoding NADPH-cytochrome P450 reductase, catalyzes the first oxidative step of the phenylpropane pathway.
Figure 6.
WGCNA of the core gene set of upland cotton proanthocyanidins metabolism in brown cotton. A) Figure depicting the association between traits and modules. Red represents positive correlation, while blue represents negative correlation. The larger the value, the stronger the correlation. B) Gene ontology enrichment analysis of the red module. C) KEGG pathway enrichment analysis of the genes in the red module. D) Visualization of the gene co-expression network.
The grey60 module contained 201 genes enriched in KEGG for “SNARE interactions in vesicular transport” and in GO for “ubiquitin-dependent protein catabolic process” and “ubiquitin conjugating enzyme activity” (Supplementary Fig. S21). The purple module contained 494 genes enriched in KEGG pathways such as “Photosynthesis”, “Oxidative phosphorylation”, and “Metabolic pathways”, and the GO enrichment was mainly in “proton-transporting ATP synthase activity”, “rotational mechanism”, “chloroplast thylakoid membrane”, and “aerobic respiration”, indicating that the purple module was primarily associated with photosynthesis and energy conversion (Supplementary Fig. S22). This suggests that the biosynthesis of proanthocyanins requires a significant amount of energy.
Discussion
Expression differences caused by variations in Gh_TT2 result in the BFC
In this study, we indicated that Gh_TT2, a MYB transcription factor, expressed in a specific high amount in 5 to 15 DPA fibers resulting in the accumulation of PAs to generate brown fiber. Missense mutations in the coding region were the first to be considered for further investigation of its functional variation (Gao et al. 2023; Huang et al. 2023). In our study, cDNAs of both genotypes induced brown fiber when overexpressed, and the Ys genotype having a stronger effect on its own and PABGs' expressions; however, the Ys genotype did not generate dark brown fiber. Therefore, the focus was shifted to the variations in the promoter region. Different lengths of ProYs and ProHD208 showed significant differences in driving ability, which might be due to the variations of active regions caused by SNPs and Indels. We speculated that different active fragments existed in different regions of the upstream promoter of the gene, which might be due to the activation or inhibition effects of different transcription factors binding regulatory elements or the influence of apparent modification (Fan et al. 2018; Liu et al. 2023). In addition, the spatiotemporal specific expression of Gh_TT2 and the presence of different length promoters indeed endow the gene with differential expression. This differential expression has important implications for fine-tuning the expression level of Gh_TT2 and subsequently modifying the proanthocyanin synthesis pathway. Molecular biology techniques can be used to balance the synthesis of proanthocyanins and anthocyanins (Zhou et al. 2019), thus providing avenues for improving colored cotton.
More than 1 MBW complex involves in proanthocyanin biosynthesis
Proanthocyanidins are a natural products unique to vascular plants with a variety of beneficial effects such as anti-ultraviolet, anti-oxidant, anti-disease, and anti-pest (Lu et al. 2021), and their synthetic pathway is relatively conserved. Like anthocyanins, it was regulated by the MBW complex and other transcription factors as an end products of phenylalanine metabolism (Ni et al. 2021; Bi et al. 2023; He et al. 2023; Zhao et al. 2023). In our work, we identified the presence of MYB (TT2)-bHLH (TT8)-WDR (WD40) in BFC and investigated their crucial regulatory roles in proanthocyanin biosynthesis. An interesting finding is that, despite showing a consistent expression with Gh_TT2, Gh_TT8 exhibits divergent regulation on ANS. Gh_TT8 promotes the expression of ANS, while Gh_TT2 inhibits the expression of ANS, thereby preventing more metabolic flux toward anthocyanin biosynthesis. The collaboration between Re and Gh_TT8 promotes the expression of ANS was also mentioned by Wang et al. (2022b).
Considering that the specific overexpression of Re and Gh_TT2 in the fiber can both induce brown coloration, transcriptome sequencing was performed on the transgenic materials (EM, EQ, and ES) to analyze potential differential regulatory mechanisms. The outcome of the competition between Gh_TT2 and Re is the presence of varying proportions of proanthocyanins and anthocyanins in the cells, ultimately resulting in different colors exhibited in the fibers (Fig. 7). Unexpectedly, 2 MYB transcription factors, namely MYB7 and MYB3, were previously known to regulate proanthocyanin synthesis (Wang et al. 2020; Zheng et al. 2021; Kim et al. 2022), were also found to have a negative regulatory effect on BFC production. Another notable finding is that Gh_TT2 expression was not detected in light brown cotton XC1 compared with Gh_TT8 and Gh_WD, suggesting different regulatory genes between XC1 and Ys (Fig. 5D). In summary, there is a complex regulatory network involved in the formation and accumulation of brown cotton pigments, in which the MBW complex composed of transcription factors plays an irreplaceable role through division of labor and cooperation.
Figure 7.
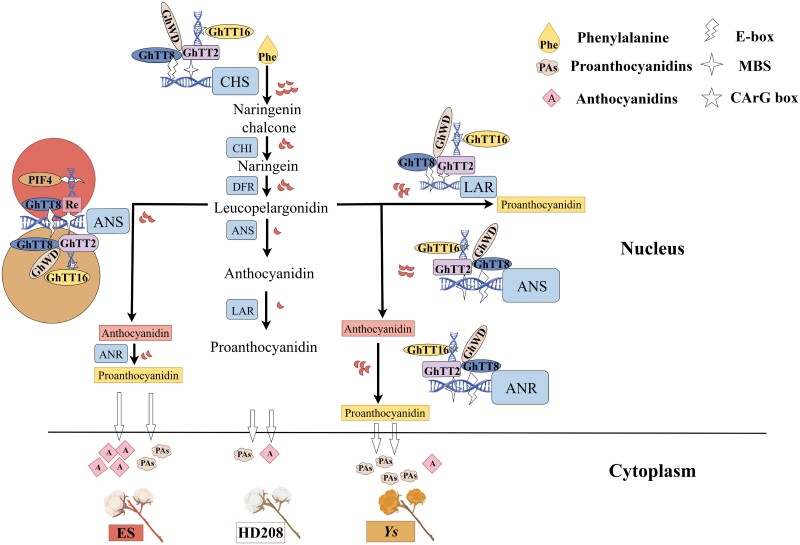
Model outlining the proposed relationship between Gh_TT16, Re, MBW complex, and PABGs in proanthocyanin synthesis by Figdraw. We propose that Gh_TT2, regulated by the upstream transcription factor Gh_TT16, forms a complex with Gh_TT8 and Gh_WD. This complex binds to the promoter regions of pathway structural enzymes CHS, ANR, ANS, and LAR, exerting regulatory effects. During this process, the regulatory element Re involved in anthocyanin synthesis also binds to ANS as part of the complex. The competitive regulation between the complex and Re directs the metabolic flux, ultimately affecting the ratio of proanthocyanins and anthocyanins in cells. Consequently, different colors are manifested in cotton fibers. Phe, phenylalanine; PAs, proanthocyanins; A, anthocyanidins. The red crescent represents metabolic flow, the white wavy shape indicates bHLH transcription factor binding sites (E-box), the white 4-pointed star represents MYB transcription factor binding sites (MBS), and the white 5-pointed star represents ABS-MADS transcription factor binding sites (CArG box).
Multiple transcription factors play a role in regulating the MBW complex
In A. thaliana, TT16 has been reported to regulate the accumulation of proanthocyanidins in the chalaza and micropyle areas, and ectopic expression of TT2 can complement the tt16 mutant phenotype; TT16 acts upstream in the proanthocyanidin biosynthetic pathway, but the ability of cells to accumulate proanthocyanidins is not directly dependent on the activity of TT16 (Nesi et al. 2002). In this study, a similar subtle relationship between GhTT16 and Gh_TT2 was observed in the formation process of proanthocyanidins in BFC. Transcriptome data analysis revealed that the expression levels of GhTT16 and Gh_TT2 showed opposite trends (Fig. 5D), suggesting an inhibitory role of GhTT16 on Gh_TT2 and MBW complex. Differences are that GhTT16 not only depends on Gh_TT2 but also plays a regulatory role on PABGs independently (Fig. 4E).
In addition, GhTCP14, reported to be a crucial regulator in auxin-mediated differentiation and elongation of cotton fiber cells, could also bind to the upstream region of Gh_TT2 (Supplementary Fig. S12). Like GhZinc and GhHAT3, the above 3 genes also exhibited significant expression differences in the 5 to 15 DPA fibers of brown and white cotton (Fig. 5D). Their specific functions await further explorations.
Superior expression pattern of homologous genes has a cumulative effect on phenotype presentation
RNA-seq and WGCNA were first used to investigate the regulatory differences underlying light and dark brown fibers. As expected, the homologous of PABGs was only highly expressed at 5 DPA and rapidly declined between 10 and 15 DPA in light brown cotton XC1, which may lead to reduced accumulation of pigments compared with Ys (Supplementary Fig. S16). The dosage effect of homologous genes is quite common in polyploid species. In polyploid organisms or homologous genomes, duplicated genes or homologous factors coordinate in various ways to mediate interactions between gene dosage effects, gene balance, cis-acting and trans-acting factors, and the reconstruction of gene expression networks (You et al. 2023). Dixon et al. (2018) identified that increased dosage of TEOSINTE BRANCHED1 (TB1) altered inflorescence architecture and growth rate, leading to enhanced wheat yield. Therefore, using a transcriptional activation system to synergistically increase the expression levels of PABGs may be effective to enhance the color of brown cotton fibers.
Complicated regulatory relationships of genes affect pigment proportion and fiber coloration
As described in Fig. 7, Gh_TT2, regulated by the upstream transcription factor Gh_TT16, forms a complex with Gh_TT8 and Gh_WD. This complex binds to the promoter regions of the structural enzyme genes CHS, ANR, ANS, and LAR to exert regulatory effects. During this process, the regulatory element Re involved in anthocyanin synthesis also binds to ANS as part of the complex. The competitive regulation between the complex and Re directs the metabolic flux, ultimately affecting the ratio of proanthocyanins and anthocyanins in cells. Consequently, when both Gh_TT2 and Re are expressed at low levels, there is minimal formation and accumulation of both pigments, resulting in white fibers. When their expression levels are higher, 2 scenarios may occur. If the content of proanthocyanins exceeds that of anthocyanins, the fibers will appear dark brown. Conversely, the fibers appear reddish brown.
Materials and methods
Plant materials
A dark brown fiber mutant named Youse (Ys) resulting from distant hybridization between Gossypium barbadense acc. Pima 90-53 and G. hirsutum acc. Handan208 with these 2 parents were planted in Wuhan, Hubei Province, China (Supplementary Fig. S23). Transgenic plants were created by Agrobacterium-mediated transformation as described previously (Jin et al. 2006). All transgenic plants mentioned in this study were also planted at Huazhong Agricultural University, Wuhan, China.
To investigate the molecular mechanisms and regulatory networks of fiber pigment formation in different brown cotton, the following materials were sampled post-planting and sent for transcriptome sequencing: Ys (dark brown fiber mutant); XC1 (Xincaimian1, light brown cotton) (Wen et al. 2018); EM (overexpression transgenic lines of Gh_TT2Ys; light brown); ES (overexpression transgenic lines of Re, named GbEXP-E1; reddish brown); EQ (overexpression transgenic lines of Re, named GbEXP-E3; light brown) (Wang et al. 2022b); and EY (transgenic negative control line; white cotton). The pigment content data obtained from the above samples (Supplementary Fig. S24) were served as the input data for the WGCNA analysis of module-trait associations.
Cloning and sequencing
The primers of the related genes were designed by Primer Premier 5 (https://www.premierbiosoft.com) based on the TM-1 reference genome (Wang et al. 2019). The genomic DNA of samples used in the test was extracted by CTAB method (Paterson et al. 1993). Amplified target gene fragments were subsequently ligated into the pGEM-T Easy cloning vector (Biotech Co. Ltd, Promega, Beijing), which was kept at 4 ℃, overnight. The inserts were sequenced using M13 primers (5′-CCCAGTCACGACGTTGTAAAACG-3′, 5′-GCGGATAACAATTTCACACAGGA-3′) by Wuhan Qingke Biotechnology Co., Ltd. The final sequences were blasted and analyzed with DNAMAN software (https://www.lynnon.com/dnaman.html).
Vector construction and genetic transformation
The cloned CDS of Gh_TT2Ys and Gh_tt2HD208 was inserted downstream of the fiber-specific expression vector PGbEXPA2 provided by researchers (Li et al. 2015). The resultant vector mentioned was transferred into an Agrobacterium tumefaciens strain (GV3101). The high-efficiency transformation line Jin668 was used as the transformation receptor as described in a previous protocol (Li et al. 2019a, b).
RNA extraction and RT-qPCR
All samples were taken from the experimental field of Huazhong Agricultural University, Wuhan, China. Cotton bolls were tagged on the day of flowering as 0 DPA. Bolls of 5 DPA, 10 DPA, 15 DPA, 20 DPA, 25 DPA, and 30 DPA were taken and frozen with liquid nitrogen immediately after removing the cotton husks, then stored in an ultralow temperature freezer at −80 °C. The fiber peeled off from the ovule was ground into powder in liquid nitrogen. Except fiber, other tissues (root, stem, leaf, petal, stigma, anther, and ovule of 0 DPA, 3 DPA, 5 DPA) were also taken at 9 to 10 AM on sunny days. RNA prep Pure Plant Kit (TIANGEN Biotech, Beijing) was used to extract total RNA. For each sample, 3 μg of RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega). For RT-qPCR analysis, we followed the same steps as Li et al. (2020).
Quantification of anthocyanin and proanthocyanidin (PA) contents
Anthocyanins in fiber of 5 to 15 DPA were extracted and quantified as previously described (Gao et al. 2013). The absorbances at 530 and 657 nm were determined using a multimode plate reader (PerkinElmer), and the relative level of anthocyanin was calculated as A530 − (0.25 ∗ A650) (Rabino and Mancinelli 1986). The relative PA content determination was performed according to the method of Li et al. (2020).
Y2H assays
The full-length sequences of Gh_TT2 and Gh_WD were constructed into pGBKT7 vector and transformed into Y2H strain, and then screened library after self-activation verification. After the transformed Y2H strain was malted with Y187 yeast library at 30 °C for 24 h, the yeast morphology was observed under a microscope. The bacteria were collected by centrifugation and screened sequentially in the deficient medium. The final selected yeast plaque was amplified and sequenced.
After removing the unmatched or frameshift preys, the remaining protein coding preys (designated preyAD) were transferred into the Y187 strain separately and malted with the Y2H strain containing either the pGBKT7 empty vector (BD) or pGBKT7-Gh_TT2/Gh_WD (Gh_TT2-BD/Gh_WD-BD). Both prey-AD/BD and prey-AD/Gh_TT2-BD/Gh_WD-BD combinations showing activated reporter genes were considered as false interactions. Only those plaques with prey-AD + Gh_TT2-BD/Gh_WD-BD activated reporter genes while the prey-AD/BD did not were considered as true interactions.
Subcellular localization
Full length of Gh_TT2 and Gh_WD were cloned into the vector pMDC43 with GFP tag fused to the N-terminus (Cai et al. 2015). The 35S:GFP-Gh_TT2 and 35S:GFP-Gh_WD constructs were transiently expressed in N. benthamiana epidermal cells following Agrobacterium-mediated transfection. The pMDC43 empty vector harboring 35S:GFP was used as control. GFP fluorescence was detected under an Olympus FV1200 confocal microscope (488 nm excitation wavelength, 44% transmissivity, 100 nm collection bandwidth, and gain were 1), and the RFP fluorescence signal of nuclear marker was also detected (559 nm excitation wavelength, 35% transmissivity, 100 nm collection bandwidth, and gain were 1) after 48 h following Agrobacterium-mediated transfection.
LCI and BiFC assays
For Split-LCI assays, the full length of Gh_TT2, Gh_WD, Gh_TT8, and Gh_bHLH48 was respectively constructed into the vectors JW771 and JW772. For BiFC assays, the CDSs of Gh_TT2, Gh_WD, Gh_TT8, and Gh_bHLH48 were respectively constructed into the vectors N-terminal fusion yellow fluorescent protein (YFP) vector pxy106 and C-terminal fusion yellow fluorescent protein (YFP) vector pxy104. The recombinant vectors were transformed into A. tumefaciens GV3101. YFP fluorescence in BiFC assays were observed using an Olympus FV1200 confocal microscope (515 nm excitation wavelength, 29% transmissivity, 100 nm collection bandwidth, and gain was 1). LUC luminescence in LCI assays was observed by CCD camera (Lumazome PyLoN 2048B).
Y3H assays
Using the vector named pBridge as a bridge carrier, Gh_TT2/Gh_TT8 and Gh_TT2/Gh_WD were constructed into the pBridge vector, and then transformed into Y2H strain, while Gh_WD and Gh_TT8 were constructed into the pADKT7 vector and transformed into Y187 strain. Y2H strain and Y187 strain were malted at 30 ℃ for 24 h and cultivated in different medium. Through different configurations and combinations, the influence of each member on the interaction of the other 2 members was verified.
GST pull-down assays
The full-length CDS sequences of Gh_TT2 and Gh_WD were cloned into the vectors PET-28-a (Novagen) and pGEX-4T-1 (Pharmacia), respectively. The constructs His-Gh_WD and GST-Gh_TT2 were transformed into Escherichia coli BL21 (DE3). Empty GST and recombinant GST-Gh_TT2 proteins were used to pull down the His-Gh_WD. The pull-down proteins were purified with Magne GST Protein Purification System (Promega V8603) and Magne His Protein Purification System (Promega V8550). The pull-down assays were performed as described previously (Yang et al. 2017).
Y1H assays and LUC assays
The sequences of ProYs and ProHD208 were cloned and inserted into the pHIS2 vector as baits. After co-transformation of the prey and bait into the yeast strain, Saccharomyces cerevisiae Y187 using the lithium acetate method, the resultant yeast cells were plated onto a selective medium lacking Trp, Leu, and His (SD/-Trp/-Leu/-His). Subsequently, the positive colonies were inoculated on a Trp/-Leu/-His medium supplemented with an appropriate concentration of 3-AT (320 mm) and grown for 3 d at 28 ℃.
For LUC assays, in order to explore the activation capacity of different promoter, the sequences of ProYs and ProHD208 were cloned and inserted into the pGreen II 0800-LUC vector. Pro35S: REN in the same vector was used as an internal control. The promoters of CHS, CHI, F3H, DFR, ANS, ANR, and LAR were also cloned into the pGreen II 0800-LUC vector. And, the full length of Gh_TT2 and Gh_TT8 was cloned and inserted into pGreenII 62-sk-LUC vector. Empty pGreen II 0800-LUC/62-sk was used as control. In brief, vector each of effector (1:1) and reporter constructs (total effectors: reporter = 1:1) were co-transformed into protoplasts using 40% polyethylene glycol 4000 (v/v) (Sigma), then cultured at 25 ℃ in darkness for 16 h. LUC and REN activity were detected for transactivation assays and its candidate targets by using the dual-LUC assay reagents (Promega, Madison, WI) with the Multimode Plate Reader (PerkinElmer).
EMSAs
The CDS of GhTT16 was constructed into the vector pMAL-c4x, and the MBP-tagged protein was induced and expressed in E. coli, and then affinity purified by chromatographic column. MBP protein was used as a negative control. A 50 bp probe containing 2 CArG boxes of the Gh_TT2 promoter was synthesized, and amplified using biotin-labeled primers. Nonlabeled probes were used as competitors. Binding and competition reactions were carried out using the EMSA/Gel-Shift Kit (Beyotime, Shanghai, China). Signals were captured by ECL.
Histochemical assays of GUS activity
GUS activity assays was performed as described previously (Li et al. 2018). In brief, fresh tissues were collected from A. thaliana plants and cotton, which were incubated in staining solution immediately at 37 ℃ overnight and then washed with 75% (v/v) ethanol. The staining solution was composed of 0.9 g L−1 5-bromo-4-chloro-3-indolylglucuronide, 50 mmol L−1 sodium phosphate buffer (pH 7.0), 20% (v/v) methanol, and 100 mg L−1 Chloromycetin. The samples were examined and photographed with a stereomicroscope (Leica Microsystems, Germany). The quantitative analysis of GUS activity in ProYs:GUS and ProHD208:GUS transformants was performed as described previously (Li et al. 2018). Briefly, the quantitative analyses of GUS activity were expressed as pmol 4-methylumbelliferone mg−1 protein min−1.
VIGS assays
VIGS assays were performed as reported previously (Gao et al. 2013). Gene fragments (300 to 500 bp) of Gh_WD and Gh_TT2 from CDS regions were constructed into the vector TRV2. The primers are listed in Supplementary Table S2. The vector constructs were introduced into A. tumefaciens strain GV3101. The recombinant vector was injected into the cotyledons of wildtype cotton plants, Ys (TRV:00; TRV: Gh_WD & GhCEN; TRV: Gh_TT2 & GhCEN). Plants were grown in controlled environment rooms at 25 °C with a 16 h light/8 h dark photoperiod. VIGS efficiency was determined 2 wk after infiltration, the leaves were collected and frozen in liquid nitrogen for target gene expression analysis.
Transcriptome sequencing and WGCNA
Total RNA was extracted from the fiber of 5 to 15 DPA of the materials: Ys, XC1, EM, ES, EQ, and EY and sequenced with Illumina Hiseq 2000 system. Clean RNA-seq reads were mapped to TM-1 reference genome (Yang et al. 2023) using HISAT2.0 (Kim et al. 2019). featureCounts was used to calculate the transcription levels of annotated genes (Liao et al. 2014). DEseq2 in R was used to identify DEGs, and its absolute value was log2(fold change) > 1. The TPM value of gene expression level was calculated by featureCounts. The co-expression network was constructed using WGCNA with default settings (Langfelder and Horvath 2008). And then, the relative content of proanthocyanins was measured and used as phenotypic data, as well, the mean value of TPM was used as input data. The networks were visualized by Cytoscape 3.0.0 (Otasek et al. 2019).
Statistical analysis
Student's t-test was performed using SPSS 25.0. Difference was considered significant at *P < 0.05 and highly significant at **P < 0.01.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: Gh_TT2 (XM_016861402), Re (MH746529), Gh_TT8 (XM_016869231), Gh_TT16 (XM_016824431), Gh_WD (XM_041117089), CHS (XM_016823419), CHI (XM_016810061), ANS (NM_001327309), ANR (XM_016832763), DFR (KF749429), F3H (XM_041083987), and LAR (CP023745).
Supplementary Material
Acknowledgments
The computations in this article were run on the bioinformatics computing platform of the National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University.
Contributor Information
Yuanxue Li, National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Tian Yao, National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Chao Fu, National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Nian Wang, National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Zhiyong Xu, National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Ningyu Yang, National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Xianlong Zhang, National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Tianwang Wen, Key Laboratory of Crop Physiology, Ecology and Genetic Breeding, College of Agronomy, Jiangxi Agricultural University, Nanchang 330045, Jiangxi, China.
Zhongxu Lin, National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Author contributions
Y.L. designed and completed the main experiments, and manuscript writing. T.Y. and T.W. completed the creation of genetic transformation materials and participated in the discussion of experimental design. C.F. was responsible for the analysis of transcriptome data. N.W. provided guidance on experimental design, transcriptome analysis methods, and paper writing. Z.X. and N.Y. performed the experiments. X.Z., T.W., and Z.L. revised the manuscript. Z.L. conceived and designed the project.
Supplementary data
The following materials are available in the online version of this article.
Supplementary Figure S1. Phenotypic photographs in the field of transgenic lines of pGbEXPA2:Gh_tt2HD208 and pGbEXPA2:Gh_TT2Ys.
Supplementary Figure S2. Subcellular localization of Gh_TT2Ys and Gh_tt2HD208.
Supplementary Figure S3. Variation sites in gene coding regions and promoter regions in Handan 208, Pima90-53, and Ys.
Supplementary Figure S4. Sequence differences in 2 genotype promoter regions.
Supplementary Figure S5. Histochemical assays of GUS activity in Arabidopsis thaliana.
Supplementary Figure S6. Construction of pGBKT7 vectors and self-activation detection for Gh_TT2 and Gh_WD.
Supplementary Figure S7. LUC signals detected in N. benthamiana leaves.
Supplementary Figure S8. BiFC assays in N. benthamiana epidermal cells.
Supplementary Figure S9. The expression of Gh_TT8, Gh_bHLH48, and Gh_WD in Ys and HD208.
Supplementary Figure S10. Yeast-3-hybrid assays were used to verify the effect of Gh_TT8 or Gh_WD on the interaction of the other 2 members.
Supplementary Figure S11. VIGS of Gh_WD in Ys causes changes in PA pigment content of leaves.
Supplementary Figure S12. LUC in cotton protoplasts and N. benthamiana verified that GhTCP14, GhHAT3, and GhZinc could bind to the promoter of Gh_TT2.
Supplementary Figure S13. Number of differentially expressed genes at 5 to 15 DPA between Ys and XC1.
Supplementary Figure S14. GO enrichment analysis of differentially expressed genes at 5 to 15 DPA between Ys and XC1.
Supplementary Figure S15. KEGG enrichment analysis of common differentially expressed genes at 5 to 15 DPA between Ys and XC1.
Supplementary Figure S16. The bubble plot represents the dynamic expression of genes enriched in biosynthesis of secondary metabolites, flavonoid biosynthesis, and phenylpropanoid biosynthesis.
Supplementary Figure S17. Number of differentially expressed genes at 5 to 15 DPA in ES and EQ, compared with EM.
Supplementary Figure S18. KEGG and GO enrichment analysis of differentially expressed genes at 10 to 15 DPA in EM, ES, and EQ.
Supplementary Figure S19. Heatmap of ANR and ANS at 5 to 15 DPA in EM, ES, and EQ.
Supplementary Figure S20. Heatmap of LAR at 5 to 15 DPA in EM, ES, and EQ.
Supplementary Figure S21. Gene ontology and KEGG pathway enrichment analysis of the genes in the grey60 module.
Supplementary Figure S22. GO and KEGG pathway enrichment analysis of the genes in the purple module.
Supplementary Figure S23. A brown fiber cotton mutant from interspecific hybridization between Gossypium hirsutum acc. Handan 208 and Gossypium barbadense acc. Pima 90-53.
Supplementary Figure S24. Pigment contents in materials for transcriptomic analysis.
Supplementary Table S1. Prediction of transcription factors for genes in the red module.
Supplementary Table S2. Primers for vector construction and functional verification.
Funding
This work was financially supported by the National Key Research and Development Plan of China (No. 2022YFD1200301) and the Natural Science Foundation of Jiangxi, China (No. 20212BAB215009).
Data availability
The raw RNA-seq data in this study are available in the NCBI data bank (No. PRJNA1084885).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Abid MA, Wei Y, Meng Z, Wang Y, Ye Y, Wang Y, He H, Zhou Q, Li Y, Wang P, et al. Increasing floral visitation and hybrid seed production mediated by beauty mark in Gossypium hirsutum. Plant Biotechnol J. 2022:20(7):1274–1284. 10.1111/pbi.13805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Caboche M, Lepiniec L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006:46(5):768–779. 10.1111/j.1365-313X.2006.02733.x [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004:39(3):366–380. 10.1111/j.1365-313X.2004.02138.x [DOI] [PubMed] [Google Scholar]
- Bi M, Liang R, Wang J, Qu Y, Liu X, Cao Y, He G, Yang Y, Yang P, Xu L, et al. Multifaceted roles of LhWRKY44 in promoting anthocyanin accumulation in Asiatic hybrid lilies (Lilium spp.). Hort Res. 2023:10(9):uhad167. 10.1093/hr/uhad167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Goodman JM, Pyc M, Mullen RT, Dyer JM, Chapman KD. Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell. 2015:27(9):2616–2636. 10.1105/tpc.15.00588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Zhou F, Li W, Yu Y, Guan Z, Zhang B, Guo W. The R2R3-MYB transcription factor GaPC controls petal coloration in cotton. Crop J. 2023:11(5):1319–1330. 10.1016/j.cj.2023.03.013 [DOI] [Google Scholar]
- Dixon LE, Greenwood JR, Bencivenga S, Zhang P, Cockram J, Mellers G, Ramm K, Cavanagh C, Swain SM, Boden SA. TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum). Plant Cell. 2018:30(3):563–581. 10.1105/tpc.17.00961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Gourrierec JL, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul J-M, Alboresi A, Weisshaar B, Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008:55(6):940–953. 10.1111/j.1365-313X.2008.03564.x [DOI] [PubMed] [Google Scholar]
- Fan D, Wang X, Tang X, Ye X, Ren S, Wang D, Luo K. Histone H3K9 demethylase JMJ25 epigenetically modulates anthocyanin biosynthesis in poplar. Plant J. 2018:96(6):1121–1136. 10.1111/tpj.14092 [DOI] [PubMed] [Google Scholar]
- Gao Z, Liu C, Zhang Y, Li Y, Yi K, Zhao X, Cui ML. The promoter structure differentiation of a MYB transcription factor RLC1 causes red leaf coloration in empire red leaf cotton under light. PLoS One. 2013:8(10):e77891. 10.1371/journal.pone.0077891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhao Y, Zhao Z, Liu W, Jiang C, Li J, Zhang Z, Zhang H, Zhang Y, Wang X, et al. RRS1 shapes robust root system to enhance drought resistance in rice. New Phytol. 2023:238(3):1146–1162. 10.1111/nph.18775 [DOI] [PubMed] [Google Scholar]
- Ge X, Wang P, Wang Y, Wei X, Chen Y, Li F. Development of an eco-friendly pink cotton germplasm by engineering betalain biosynthesis pathway. Plant Biotechnol J. 2023:21(4):674–676. 10.1111/pbi.13987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhang R, Jiang S, Wang H, Ming F. The MYB transcription factor RcMYB1 plays a central role in rose anthocyanin biosynthesis. Hortic Res. 2023:10(6):uhad080. 10.1093/hr/uhad080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe DJ, Condon BD, Thyssen G, Naoumkina M, Madison CA, Reynolds M, Delhom CD, Fang DD, Li P, McCarty J. The GhTT2_A07 gene is linked to the brown colour and natural flame retardancy phenotypes of Lc1 cotton (Gossypium hirsutum L.) fibres. J Exp Bot. 2016:67(18):5461–5471. 10.1093/jxb/erw312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Liu L, Yi D, Lu Y, Zhu L, Chen M, Wang Y, Zhou J, Hu X, Wei Y, et al. DlMYB113 mutation affects anthocyanin accumulation in red pericarp longan (Dimocarpus longan Lour.). Hortic Adv. 2023:1(1):11. 10.1007/s44281-023-00014-3 [DOI] [Google Scholar]
- Jiang L, Yue M, Liu Y, Zhang N, Lin Y, Zhang Y, Wang Y, Li M, Luo Y, Zhang Y, et al. A novel R2R3-MYB transcription factor FaMYB5 positively regulates anthocyanin and proanthocyanidin biosynthesis in cultivated strawberries (Fragaria × ananassa). Plant Biotechnol J. 2023a:21(6):1140–1158. 10.1111/pbi.14024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Yin Q, Liu J, Su X, Han X, Li Q, Zhang J, Pang Y. The APETALA2-MYBL2 module represses proanthocyanidin biosynthesis by affecting formation of the MBW complex in seeds of Arabidopsis thaliana. Plant Commun. 2023b:5:2590–3462. 10.1016/j.xplc.2023.100777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zhang X, Nie X, Guo X, Liang S, Zhu H. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plant. 2006:50(4):519–524. 10.1007/s10535-006-0082-5 [DOI] [Google Scholar]
- Ke L, Yu D, Zheng H, Xu Y, Wu Y, Jiao J, Wang X, Mei J, Cai F, Zhao Y, et al. Function deficiency of GhOMT1 causes anthocyanidins over-accumulation and diversifies fibre colours in cotton (Gossypium hirsutum). Plant Biotechnol J. 2022:20(8):1546–1560. 10.1111/pbi.13832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Jeon SJ, Yanders S, Park S-C, Kim HS, Kim S. MYB3 plays an important role in lignin and anthocyanin biosynthesis under salt stress condition in Arabidopsis. Plant Cell Rep. 2022:41(7):1549–1560. 10.1007/s00299-022-02878-7 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019:37(8):907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohel RJ. Genetic analysis of fiber color variants in cotton. Crop Sci. 1985:25(5):793–797. 10.2135/cropsci1985.0011183X0025000500017x [DOI] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008:9(1):559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang M, Li Y, Zhang Q, Lindsey K, Daniell H, Jin S, Zhang X. Multi-omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation process. Plant Biotechnol J. 2019a:17(2):435–450. 10.1111/pbi.12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mitchell M, Rolland V, Allen S, MacMillan C, Pettolino F. ‘Pink cotton candy’—a new dye-free cotton. Plant Biotechnol J. 2023:21(4):677–679. 10.1111/pbi.13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ouyang X, Zhang Z, He L, Wang Y, Li Y, Zhao J, Chen Z, Wang C, Ding L, et al. Over-expression of the red plant gene R1 enhances anthocyanin production and resistance to bollworm and spider mite in cotton. Mol Genet. 2019b:294:469–478. 10.1007/s00438-018-1525-3 [DOI] [PubMed] [Google Scholar]
- Li Y, Min L, Zhang L, Hu Q, Wu Y, Li J, Xie S, Ma Y, Zhang X, Zhu L. Promoters of Arabidopsis Casein kinase I-like 2 and 7 confer specific high-temperature response in anther. Plant Mol Biol. 2018:98(1–2):33–49. 10.1007/s11103-018-0760-7 [DOI] [PubMed] [Google Scholar]
- Li Y, Tu L, Ye Z, Wang M, Gao W, Zhang X. A cotton fiber-preferential promoter, PGbEXPA2, is regulated by GA and ABA in Arabidopsis. Plant Cell Rep. 2015:34(9):1539–1549. 10.1007/s00299-015-1805-x [DOI] [PubMed] [Google Scholar]
- Li Z, Su Q, Xu M, You J, Khan AQ, Li J, Zhang X, Tu L, You C. Phenylpropanoid metabolism and pigmentation show divergent patterns between brown color and green color cottons as revealed by metabolic and gene expression analyses. J Cotton Res. 2020:3(1):27. 10.1186/s42397-020-00069-x [DOI] [Google Scholar]
- Liang Q, Jin Y, Zhu Q, Shao D, Wang X, Ma X, Liu F, Zhang X, Li Y, Sun J, et al. A MYB transcription factor containing fragment introgressed from Gossypium bickii confers pink flower on Gossypium hirsutum L. Ind Crops Prod. 2023:192:116121. 10.1016/j.indcrop.2022.116121 [DOI] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014:30(7):923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu H, Luo C, Song W, Shen H, Li G, He Z, Chen W, Cao Y, Huang F, Tang S, et al. Flavonoid biosynthesis controls fiber color in naturally colored cotton. PeerJ. 2018:18(6):e4537. 10.7717/peerj.4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Shu Q, Lin W, Espley RV, Allan AC, Pei MX, Li X, Su J, Wu J. DNA methylation reprogramming provides insights into light-induced anthocyanin biosynthesis in red pear. Plant Sci. 2023:326:111499. 10.1016/j.plantsci.2022.111499 [DOI] [PubMed] [Google Scholar]
- Lu N, Rao X, Li Y, Jun JH, Dixon RA. Dissecting the transcriptional regulation of proanthocyanidin and anthocyanin biosynthesis in soybean (Glycine max). Plant Biotechnol J. 2021:19(7):1429–1442. 10.1111/pbi.13562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008:55(6):954–967. 10.1111/j.1365-313X.2008.03565.x [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, Lepiniec L. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002:14(10):2463–2479. 10.1105/tpc.004127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Premathilake AT, Gao Y, Yu W, Tao R, Teng Y, Bai S. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2021:105(1):167–181. 10.1111/tpj.15049 [DOI] [PubMed] [Google Scholar]
- Otasek D, Morris JH, Bouças J, Pico AR, Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019:20(1):185. 10.1186/s13059-019-1758-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Brubaker CL, Wendel JF. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Report. 1993:11(2):122–127. 10.1007/BF02670470 [DOI] [Google Scholar]
- Rabino I, Mancinelli AL. Light, temperature, and anthocyanin production. Plant Physiol. 1986:81(3):922–924. 10.1104/pp.81.3.922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput R, Naik J, Stracke R, Pandey A. Interplay between R2R3 MYB-type activators and repressors regulates proanthocyanidin biosynthesis in banana (Musa acuminata). New Phytol. 2022:236(3):1108–1127. 10.1111/nph.18382 [DOI] [PubMed] [Google Scholar]
- Tan J, Tu L, Deng F, Hu H, Nie Y, Zhang X. A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol. 2013:162(1):86–95. 10.1104/pp.112.212142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Du J, Wu H, Guan X, Chen W, Hu Y, Fang L, Ding L, Li M, Yang D, et al. The transcription factor MML4_D12 regulates fiber development through interplay with the WD40-repeat protein WDR in cotton. J Exp Bot. 2020:71(12):3499–3511. 10.1093/jxb/eraa104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walford S-A, Wu Y, Llewellyn DJ, Dennis ES. GhMYB25-like: a key factor in early cotton fibre development. Plant J. 2011:65(5):785–797. 10.1111/j.1365-313X.2010.04464.x [DOI] [PubMed] [Google Scholar]
- Wang L, Kartika D, Ruan Y. Looking into ‘hair tonics’ for cotton fiber initiation. New Phytol. 2021:229(4):1844–1851. 10.1111/nph.16898 [DOI] [PubMed] [Google Scholar]
- Wang B, Luo Q, Li Y, Du K, Wu Z, Li T, Shen W-H, Huang C-H, Gan J, Dong A. Structural insights into partner selection for MYB and bHLH transcription factor complexes. Nat Plants. 2022a:8(9):1108–1117. 10.1038/s41477-022-01223-w [DOI] [PubMed] [Google Scholar]
- Wang M, Tu L, Yuan D, Zhu D, Shen C, Li J, Liu F, Pei L, Wang P, Zhao G, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet. 2019:51(2):224–229. 10.1038/s41588-018-0282-x [DOI] [PubMed] [Google Scholar]
- Wang X, Wu J, Guan M, Zhao C, Geng P, Zhao Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020:101(3):637–652. 10.1111/tpj.14570 [DOI] [PubMed] [Google Scholar]
- Wang N, Zhang B, Yao T, Shen C, Wen T, Zhang R, Li Y, Le Y, Li Z, Zhang X, et al. Re enhances anthocyanin and proanthocyanidin accumulation to produce red foliated cotton and brown fiber. Plant Physiol. 2022b:189(3):1466–1481. 10.1093/plphys/kiac118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T, Luo W, Li Y, Lin Z. Advances and new insights in naturally colored cotton breeding and research. Ind Crops Prod. 2024:211:118252. 10.1016/j.indcrop.2024.118252 [DOI] [Google Scholar]
- Wen T, Wu M, Shen C, Gao B, Zhu D, Zhang X, You C, Lin Z. Linkage and association mapping reveals the genetic basis of brown fibre (Gossypium hirsutum). Plant Biotechnol J. 2018:16(9):1654–1666. 10.1111/pbi.12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Zhang Z, Yin M, Luo M, Li X, Hou L, Pei Y. Cotton flavonoid structural genes related to the pigmentation in brown fibers. Biochem Biophys Res Commun. 2007:358(1):73–78. 10.1016/j.bbrc.2007.04.084 [DOI] [PubMed] [Google Scholar]
- Xu W, Grain D, Gourrierec JL, Harscoët E, Berger A, Jauvion V, Scagnelli A, Berger N, Bidzinski P, Kelemen Z, et al. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 2013:198(1):59–70. 10.1111/nph.12142 [DOI] [PubMed] [Google Scholar]
- Yan Q, Wang Y, Li Q, Zhang Z, Ding H, Zhang Y, Liu H, Luo M, Liu D, Song W, et al. Up-regulation of GhTT2-3A in cotton fibres during secondary wall thickening results in brown fibres with improved quality. Plant Biotechnol J. 2018:16(10):1735–1747. 10.1111/pbi.12910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li C, Cai Z, Hu Y, Nolan T, Yu F, Yin Y, Xie Q, Tang G, Wang X. SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev Cell. 2017:41(1):47–58.e4. 10.1016/j.devcel.2017.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wang J, Huang Y, Wang S, Wei L, Liu D, Weng Y, Xiang J, Zhu Q, Yang Z, et al. CottonMD: a multi-omics database for cotton biological study. Nucleic Acids Res. 2023:51(D1):D1446–D1456. 10.1093/nar/gkac863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Chen Y, Chen C, Yang D, Ding L, Yang Q, Xu C, Chen J, Zhang T, Hu Y. Cotton genes GhMML1 and GhMML2 control trichome branching when ectopically expressed in tobacco. Gene. 2022:820:146308. 10.1016/j.gene.2022.146308 [DOI] [PubMed] [Google Scholar]
- You J, Liu Z, Qi Z, Ma Y, Sun M, Su L, Niu H, Peng Y, Luo X, Zhu M, et al. Regulatory controls of duplicated gene expression during fiber development in allotetraploid cotton. Nat Genet. 2023:55(11):1987–1997. 10.1038/s41588-023-01530-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li P, Zuo H, Peng A, Lin J, Li P, Wang K, Tang Q, Tadege M, Liu Z, et al. CsMYBL2 homologs modulate the light and temperature stress-regulated anthocyanin and catechins biosynthesis in tea plants (Camellia sinensis). Plant J. 2023:115(4):1051–1070. 10.1111/tpj.16279 [DOI] [PubMed] [Google Scholar]
- Zheng H, Duan B, Yuan B, Chen Z, Yu D, Ke L, Zhou W, Liu H, Sun Y. Flavanone and flavonoid hydroxylase genes regulate fiber color formation in naturally colored cotton. Crop J. 2023:11(3):766–773. 10.1016/j.cj.2022.10.004 [DOI] [Google Scholar]
- Zheng J, Wu H, Zhao M, Yang Z, Zhou Z, Guo Y, Lin Y, Chen H. OsMYB3 is a R2R3-MYB gene responsible for anthocyanin biosynthesis in black rice. Mol Breed. 2021:41(8):51. 10.1007/s11032-021-01244-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin-Wang K, Wang F, Espley RV, Ren F, Zhao J, Ogutu C, He H, Jiang Q, Allan AC, et al. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019:221(4):1919–1934. 10.1111/nph.15486 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Yuan Y, Stiller W, Jia Y, Wang P, Pan Z, Du X, Llewellyn D, Wilson I. Genetic dissection of the fuzzless seed trait in Gossypium barbadense. J Exp Bot. 2018:69(5):997–1009. 10.1093/jxb/erx459 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq data in this study are available in the NCBI data bank (No. PRJNA1084885).