Abstract
Diatoms derive from a secondary endosymbiosis event, which occurred when a eukaryotic host cell engulfed a red alga. This led to the formation of a complex plastid enclosed by 4 membranes: 2 innermost membranes originating from the red alga chloroplast envelope, and 2 additional peri- and epiplastidial membranes (PPM and EpM). The EpM is linked to the endoplasmic reticulum (ER). The most abundant membrane lipid in diatoms is monogalactosyldiacylglycerol (MGDG), synthesized by galactosyltransferases called MGDG synthases (MGDs), conserved in photosynthetic eukaryotes and considered to be specific to chloroplast membranes. Similar to angiosperms, a multigenic family of MGDs has evolved in diatoms, but through an independent process. We characterized MGDα, MGDβ, and MGDγ in Phaeodactylum tricornutum, combining molecular analyses, heterologous expression in Saccharomyces cerevisiae, and studying overexpressing and CRISPR-Cas9-edited lines. MGDα localizes mainly to thylakoids, MGDβ to the PPM, and MGDγ to the ER and EpM. MGDs have distinct specificities for diacylglycerol, consistent with their localization. The results suggest that MGDα is required for thylakoid expansion under optimal conditions, while MGDβ and MGDγ play roles in plastid and nonplastid membranes and in response to environmental stress. Functional compensation among MGDs likely contributes to diatom resilience under adverse conditions and to their ecological success.
A multigenic family of membrane-localized monogalactosyldiacylglycerol synthases is present in diatoms, with specific isoforms for thylakoid expansion and galactolipid production in response to stress.
IN A NUTSHELL.
A multigenic family of membrane-localized monogalactosyldiacylglycerol synthases is present in diatoms, with MGDα a specific isoforms for thylakoid expansion and MGDβ and MGDγ for galactolipid production in plastid and nonplastid membranes in response to stress.
Introduction
Diatoms form a monophyletic phylum of photosynthetic eukaryotes, representing one of the largest groups of stramenopiles (de Vargas et al. 2015; Serôdio and Lavaud 2020). They dominate phytoplankton biodiversity in oceans and freshwater ecosystems (Mann and Droop 1996; Mann 1999; Kooistra et al. 2007; Guiry 2012; de Vargas et al. 2015). Our understanding of their cellular functional organization relies mainly on the model species Phaeodactylum tricornutum, for which intensive efforts have been made to sequence its genome (Bowler et al. 2008), develop molecular tools and resources (Siaut et al. 2007; De Riso et al. 2009; Daboussi et al. 2014; Kroth et al. 2018), and characterize membrane glycerolipids at whole-cell level (Abida et al. 2015) and in organelle-enriched fractions (Lupette et al. 2019; Huang et al. 2024).
They have a cell architecture comprising membrane systems with no equivalent in “simpler’ photosynthetic models, like Chlamydomonas reinhardtii or Arabidopsis (Arabidopsis thaliana), containing primary plastids surrounded by only 2 membranes. This complex membrane compartmentation derives from their evolutionary origin following 2 endosymbiosis events. An initial event at the origin of most photosynthetic eukaryotes involved the engulfment of a cyanobacterium by an unknown heterotrophic cell, 1 to 1.5 b.y.a. (Jensen and Leister 2014; Maréchal 2024). This primary endosymbiosis led to the formation of the classical chloroplast, bounded by a 2-membrane envelope (Sato and Awai 2017; Maréchal 2024). Lineages radiated into chlorophytes (green algae), rhodophytes (red algae), and glaucophytes. Emergence of diatoms has occurred more recently, ∼180 to 250 m.y.a. (Kooistra et al. 2003; Sorhannus 2007), stemming from the engulfment of a red algal symbiont by a secondary eukaryotic host cell (Reyes-Prieto et al. 2007; Bowler et al. 2008; Benoiston et al. 2017).
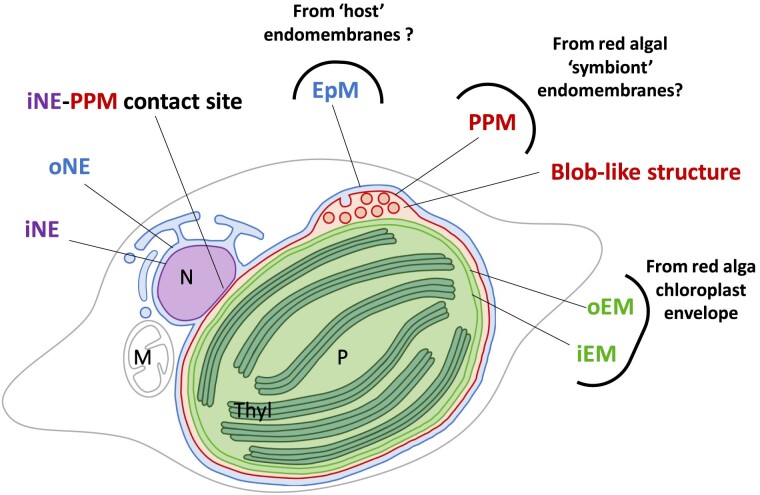
Diatom photosynthetic organelle derives vertically from the symbiont cell, reduced in the form of a “complex” or “secondary plastid,” bounded by 4 membranes (Fig. 1). The 2 innermost membranes, i.e. the “inner” and “outer envelope membranes” (iEM and oEM, respectively; Fig. 1) correspond to the chloroplast envelope of the engulfed alga. The outermost one, named the “epiplastidial membrane” (EpM; Fig. 1), is thought to derive from the host cell phagosome. It is continuous with the outer membrane of the nuclear envelope (oNE) and is connected to the endoplasmic reticulum (ER; Murakami and Hashimoto 2009; Tanaka et al. 2015; Flori et al. 2016). Underneath, the “periplastidial membrane” (PPM; Fig. 1) is usually considered to derive from the symbiont plasma membrane (Grosche et al. 2014). An alternative hypothesis is that the PPM could derive from the host ER, like the EpM (Gould et al. 2015). A “blob-like” structure, or in short a “blob,” containing a vesicular network (VN) visible in confocal and transmission electron microscopy (Fig. 1), has been detected between the PPM and the oEM, but its organization and function remain elusive (Kilian and Kroth 2005; Flori et al. 2016). Cavalier-Smith (2018) postulated that the blob corresponds to a relic of the symbiont ER. The EpM, PPM, and blob are therefore ontogenetically related to the endomembrane system. To picture diatoms’ sophisticated plastid architecture completely, membrane contact sites occur between the inner membrane of the nuclear envelope (iNE) and the PPM (Flori et al. 2016; Fig. 1).
Figure 1.
Chimeric origin of the secondary plastid in diatom. The scheme shows a fusiform cell of P. tricornutum. The plastid is limited by 4 membranes. The EpM is continuous with the oNE. The PPM is in tight contact with the iNE. The iEM and oEM, shown in light green, are tightly apposed. A specific PPC relies on the detection of a “blob”-like structure observed by confocal and transmission electron microscopy. In confocal microscopy, the precursors of blob-residing proteins fused to GFP cross only the EpM and PPM and form fluorescent spots in the middle of the plastid (Lang et al. 1998; Kilian and Kroth 2005; Huang et al. 2024). In electron microscopy, the blob is marked by the development of a VN budding from the PPM (Flori et al. 2016). Each membrane surrounding the plastid derive from distinct evolutionary origin. The oEM and iEM stem from the chloroplast of the engulfed red algae. The PPM potentially derive from the endomembrane systems of the red algal symbiont and the EpM from that of the host cell, respectively. M, mitochondrion; N, nucleus; P, plastid; Thyl: thylakoids.
It is challenging to identify the protein and lipid components of the iEM, oEM, PPM, and EpM, and to unravel the biogenesis, dynamics, and function of each of these membranes, since little knowledge can be transferred from simple eukaryotic models. Concerning proteins, the import of some nuclear-encoded precursors was dissected molecularly in P. tricornutum and Thalassiosira pseudonana. Plastid protein precursors contain a bipartite N-terminal targeting peptide, with a signal peptide (Sp) fused to a chloroplast transit peptide-like sequence (Ctp), and harbor a conserved amino acid motif called “ASAFAP” (Apt et al. 2002; Kilian and Kroth 2005; Gruber et al. 2007; Gruber et al. 2015). Based on the detection of Sp-Ctp sequences and ASAFAP motifs, using the 2 predictions tools ASAFind (Gruber et al. 2015) and HECTAR (Gschloessl et al. 2008) only 1,471 and 561 proteins, respectively, were considered chloroplast-targeted among the >12,000 nuclear-encoded proteins in P. tricornutum. These predictions are likely underestimated. In contrast, our recent proteomic study of P. tricornutum purified plastids, allowed the detection of 2,758 nuclear-encoded and 92 plastid-encoded proteins (Huang et al. 2024), possibly including contaminants, and requesting experimental validation by expressing protein fusions with fluorescent reporters.
Concerning lipids, the composition of the different plastid membranes and the precise sites of their biosynthesis are unknown. Membrane glycerolipids contain 2 fatty acids (FAs) esterified at Positions sn-1 and sn-2 of a glycerol backbone, and harbor a polar head group at Position sn-3. The nature of the head group defines glycerolipid “classes,” while FA chain lengths and unsaturation levels define “molecular species” (Li-Beisson et al. 2010; Petroutsos et al. 2014). It was considered plausible but not demonstrated that the iEM and oEM may contain the lipid classes conserved in cyanobacteria and primary chloroplast, i.e. 2 galactolipids, monogalactosyl- and digalactosyldiacylglycerol (MGDG and DGDG), a sulfolipid, sulfoquinovosyldiacylglycerol (SQDG) and a phospholipid, phosphatidylglycerol (PG; Boudiere et al. 2014; Petroutsos et al. 2014). The current consensus is that this lipid quartet is a marker of plastids in eukaryotes (Boudiere et al. 2014; Petroutsos et al. 2014). The EpM and PPM were hypothesized to have an ER/endomembrane-type lipid composition, with phospholipids, e.g. phosphatidylcholine (PC) and phosphatidylethanolamine (PE), and betaine lipids, e.g. diacylglyceryl hydroxymethyltrimethyl-β-alanine (DGTA; Dolch and Marechal 2015). Extraplastidial membranes were speculated to be either devoid of galactolipids and sulfolipids or to contain very low levels of these lipid classes. Contradicting this view, a quantitative analysis of glycerolipids between whole cells and plastid-enriched subfractions showed similar compositions (Huang et al. 2024), suggesting that MGDG, DGDG, or SQDG presence could expand to the PPM, EpM, and nonplastid membranes in diatom cells.
Here, we addressed the committing step of galactolipid synthesis, i.e. the production of MGDG by MGDG synthase (MGD) enzymes. MGDs catalyze the transfer of a galactosyl residue from a UDP-galactose (UDP-Gal) to the sn-3 position of a diacylglycerol (DAG):
In P. tricornutum, MGDG is the most abundant membrane lipid in whole extracts, representing one-third of all lipid classes, supporting its presence at least in thylakoids, the largest membrane system (Abida et al. 2015). It is the main platform for the production of highly unsaturated 16-carbon FAs deriving from plastid palmitoyl-ACP (16:0-ACP), in particular 16:2, 16:3 and 16:4 (Dolch and Marechal 2015; Smith et al. 2021), mostly esterified at its sn-2 position (Abida et al. 2015). P. tricornutum MGDG molecular species are also enriched in eicosapentaenoic acid (20:5), a very-long-chain polyunsaturated FA. Biosynthesis of 20:5 follows a sequence of acyl-chain-elongations and desaturations, likely occurring at the ER (Moog et al. 2011; Dolch and Marechal 2015; Dolch et al. 2017b; Smith et al. 2021; Billey et al. 2021b) and/or at the EpM (Huang et al. 2024), on acyl-chains esterified on phospholipids and betaine lipids. The route of 20:5 from its biosynthesis site in the ER/EpM to inner compartments of the plastid, tentatively called the “Ω-pathway” (Petroutsos et al. 2014; Dolch and Marechal 2015; Dolch et al. 2017b) is unresolved.
Three MGDs have been predicted in P. tricornutum based on sequence similarity (Petroutsos et al. 2014; Hori et al. 2016; Shang et al. 2022). Since MGD genes were previously assigned confusing numbering schemes, the 3 gene loci, Phatr3_J14125 (on Chromosome 13), Phatr3_J43938 (on Chromosome 3), and Phatr3_J9619 (on Chromosome 1) (Bowler et al. 2008), were renamed here MGDα, MGDβ, and MGDγ, respectively. MGD proteins were detected in P. tricornutum proteome analyses (Lupette et al. 2022; Huang et al. 2024), validating their expression. It would be tempting to assume that multiple isoforms of MGDs may be important for lipid production in the different membrane compartments of the secondary plastid. However, other lineages in stramenopiles, like eustigmatophytes, harbor also a 4-membrane plastid but contain only one MGD in their genome (Vieler et al. 2012; Corteggiani Carpinelli et al. 2014). Thus, the presence of 3 isoforms in diatoms is intriguing.
In this study, we characterized the 3 MGD isoforms in P. tricornutum, addressing the question of their subcellular localization, nature of DAG substrates, involvement in the different MGDG and DGDG species productions, as well as their putative role under environmental stress, taking nitrogen starvation as model stress condition.
Results
Gene structure and molecular phylogeny of P. tricornutum MGDα, MGDβ, and MGDγ
MGD sequences were recovered from genomic data of P. tricornutum CCAP 1055/1 via the EnsemblProtist web portal (Yates et al. 2022) and compared with entries in the National Centre for Biotechnology Information (NCBI) and Uniprot databases (Supplementary Table S1). The Phatr3_J14125/Phatr3_EG02525 gene was previously numbered as either MGD1 (Bullmann et al. 2010) or MGD3 (Dolch et al. 2017a; Shang et al. 2022). To resolve this ambiguity, we named genes with Greek alphabet letters. The presence and length of introns was confirmed by cDNA sequencing and curated manually. The structure of MGDα, MGDβ, and MGDγ coding regions, with 1,615, 1,805, and 2,099 bp lengths, respectively, is summarized in Supplementary Fig. S1.
Molecular phylogeny of MGDs has previously shown that angiosperm and diatom sequences fall in distinct groups (Hori et al. 2016; Shang et al. 2022). We confirmed this finding with a midpoint-rooted molecular phylogeny, updated with sequences of stramenopiles and other secondary endosymbionts available from public databases (Supplementary Table S2 and Fig. S2). Angiosperm MGD encoded sequences form 2 clusters, corresponding to the 2 types identified in A. thaliana, namely Types A and B, reflecting a gene duplication and functional differentiation, at the base of flowering plants’ radiation (Awai et al. 2001). The MGD sequences encoded in bryophyta and lycopodiophyta cluster with Type A, highlighting that this type is the ancestral isoform in plants.
MGDs from stramenopiles are close to rhodophyta (Supplementary Fig. S2), reflecting their origin from the red algal symbiont. In eustigmatophytes, like Microchloropsis gaditana (Fawley et al. 2015), a unique MGD sequence is detected, clustering with diatom MGDα, along with pelagophyceae and phaeophyceae sequences, in a sister clade to the alveolates. This supports the conservation of an ancestral type corresponding to MGDα. The multigenic family of diatom MGDs highlights 2 additional clusters corresponding to β and γ, which have likely stemmed from gene duplication at the base of diatoms’ radiation (Supplementary Fig. S2).
MGDα, MGDβ, and MGDγ predicted targeting sequences and protein structures
P. tricornutum protein sequences were analyzed using A. thaliana MGDs as a reference (Supplementary Figs. S3 and S4). In Arabidopsis, AtMGD1, AtMGD2, and AtMGD3 have distinct subcellular localizations and roles (Awai et al. 2001). AtMGD1 belongs to “Type A,” defined by the presence of an N-terminal Ctp that targets its precursor to the iEM of the chloroplast (Awai et al. 2001). It is the major isoform producing MGDG for thylakoid membranes, and its knockout (KO) mutation is lethal in photoautotrophic conditions (Kobayashi et al. 2007a, 2007b). AtMGD2 and AtMGD3 belong to “Type B,” lacking any N-terminal Ctp. They are dispensable under optimal conditions (Kobayashi et al. 2009b). Type B MGDs localize to the oEM of chloroplasts (Awai et al. 2001) and are crucial for the adaptation of plant cells to nutrient stress (Kobayashi et al. 2009b). A recent work also shows a localization of AtMGD2 in the cytosol of pollen tube (Billey et al. 2021a). This suggested that angiosperm MGDs might have also some unknown function outside of plastids.
In P. tricornutum, nuclear-encoded sequences harboring a Sp-Ctp bipartite presequence and an ASAFAP motif were shown to go across the 4 limiting membranes of the plastid (Apt et al. 2002). ASAFAP-dependent targeting can be tuned by changing some residues, leading to the arrest of preproteins after crossing the EpM, at the level of the PPM and in the so-called periplastidial compartment (PPC), locally forming a blob-like structure (Apt et al. 2002; Kilian and Kroth 2005).
In MGDα protein sequence, an Sp-Ctp could be detected using SignalP (Emanuelsson et al. 2007; Almagro Armenteros et al. 2019) and WoLF PSORT (Horton et al. 2007) software. This prediction was confirmed using the HECTAR method designed for stramenopiles (Gschloessl et al. 2008). An ASAFAP-like motif was tentatively identified in MGDα Sp-Ctp, with a “SAAFSP” sequence (Supplementary Fig. S3). In MGDβ, an Sp was detected using SignalP and HECTAR; a Ctp could only be predicted by ChloroP and WoLF PSORT tools, but not by HECTAR. A low similarity ASAFAP-like motif was tentatively identified with a “GSGFVL” sequence (Supplementary Fig. S3). In MGDγ, no prediction of a bipartite presequence could be obtained with any of these tools, confirming a recent analysis of this isoform (Shang et al. 2022).
Specific domains involved in the binding to the UDP-Gal sugar donor, the DAG acceptor, and the lipid activator PG were characterized structurally and functionally in AtMGD1 (Rocha et al. 2016). Supplementary Fig. S3 shows the sequence alignment of the A. thaliana and P. tricornutum MGDs and the conservation of these domains and key residues. Two residues are particularly critical for catalysis in AtMGD1 (Botte et al. 2005; Dubots et al. 2010; Rocha et al. 2016). Firstly, a histidine in Position 159 (H159) of the alignment (Supplementary Fig. S3) is involved in the deprotonation of the nucleophile hydroxyl (OH) group of DAG. Secondly, a lysine (K576) binds to the sugar donor, stabilizing the phosphates of the departing UDP during the reaction. The presence of anionic lipids such as PA, PG, and SQDG is critical for AtMGD1 activity. A PG-H catalytic dyad was proposed to form an acid/base relay system facilitating the deprotonation of the acceptor substrate by H159 (Makshakova et al. 2020; Nitenberg et al. 2020). It was further shown that PG develops interactions with H159 and the neighboring arginine (R160), thus placing this lipid close to the active site (Makshakova et al. 2020). R160 is also conserved in all P. tricornutum sequences (Supplementary Fig. S3).
MGDs belong to the glycosyltransferase B (GT-B) superfamily (Rocha et al. 2016). The conserved GT-B fold consists of 2 Rossmann-type α/β/α domains of comparable size, named the N- and C-domains, separated by a large cleft, and stabilized by 2 long C-terminal α-helices. The N-domain of AtMGD1 corresponds to Positions 141 to 338 and 655 to 672, the C-domain to Positions 362 to 654. They are connected by a loop at Positions 339 to 361 (Supplementary Fig. S3). Secondary and tertiary structures of P. tricornutum MGDs were predicted using Phyre² (Kelley et al. 2015), based on the 3-dimensional structure generated from the crystal of AtMGD1 in complex with UDP (template 4X1T; Rocha et al. 2016) and AlphaFold (Jumper et al. 2021; Jumper and Hassabis 2022; Mirdita et al. 2022). All P. tricornutum MGDs, starting approximately at Position 145 of the alignment in Supplementary Fig. S3 and ending around Position 670, are typical GT-B fold structures, highly similar to AtMGD1 (Supplementary Fig. S4, B to E). The MGDα C-domain is approximately the same size as that of AtMGD1 (∼170 amino acids), whereas it is longer in MGDβ (∼223 amino acids) and MGDγ (∼290 amino acids). The hinge connecting N- and C-domains is longer in all P. tricornutum MGDs, possibly providing some flexibility in the structure. Additional segments in the C-domain of MGDβ and MGDγ, at the level of an unresolved loop in AtMGD1 structure, were predicted with a lower confidence level (50% and 70%, respectively) than the rest of the structure (100%). The last amino acids of the C-terminal stabilizing helix were also modeled with a lower confidence for MGDα and MGDγ (50%). Overall, these analyses show the prediction of the canonical double-Rossmann fold structure defined in AtMGD1 in all P. tricornutum MGDs, with conserved domains and residues that are critical for enzymatic activity, and highlight some additional features that may provide specific functionalities. These models have allowed designing guide RNA (gRNA) sequences in structured and conserved regions, or within the enzymes active sites for the CRISPR-Cas9 editing experiments.
Functional analysis of MGDG synthase activity by heterologous expression in yeast
Sequences encoding the 3 mature MGDs were cloned under the control of the galactose-inducible promoter pGAL1 and introduced into Saccharomyces cerevisiae for heterologous expression. A transformed yeast expressing AtMGD2 was used as a positive control. The lipid profile of 3 independent expressing lines was analyzed per transgene. The expression of all P. tricornutum MGD isoforms led to the biosynthesis of MGDG (Supplementary Fig. S5), a lipid class which is absent in yeast. It is important to note that DAG substrates produced naturally in yeast are distinct from those found in P. tricornutum. In particular, yeast lacks 20:5, which is abundant in diatom MGDG. Therefore, the observed MGDG profiles do not reflect the enzymatic specificity per se, and lower MGDG level may simply reflect a lack of favored DAG substrate for the considered enzyme. Nevertheless, the pattern of MGDG species produced by the different lines highlighted some divergence among isoforms. MGDα-expressing yeast showed the most distinct MGDG pattern compared with yeast expressing AtMGD2, with higher proportions of MGDG-36:1 and MGDG-36:2 compared with any other line. MGDβ-expressing yeast showed higher proportions of MGDG-32:1 and 32:2, while no traces of MGDG 36:1 and MGDG-36:2 could be detected. Finally, MGDγ was the only isoform able to produce MGDG-28:0 and MGDG-30:0, i.e. with saturated acyl species. Although caution is needed when interpreting heterologous expression, results presented here support different substrate specificities in the 3 MGD isoforms of P. tricornutum.
P. tricornutum MGDα, MGDβ, and MGDγ subcellular localization
P. tricornutum lines overexpressing MGD genes fused to eGFP were generated. For each isoform, 2 independent overexpressing strains were analyzed and gave identical results, namely MGDα–eGFP-A and -B, MGDβ–eGFP-A and -B, and MGDγ–eGFP-A and -B. Identical GFP signals were also obtained with a third series of overexpressing lines, used for colocalization studies.
We first performed an immunoblot analysis on total protein extracts from each strain using anti-GFP–horseradish peroxidase–coupled antibodies to assess the size and expression level of each construct (Supplementary Fig. S6). The molecular weight expected for each MGD are 51, 61, and 66 kDa for MGDα, β, and γ, respectively; therefore, we expected to detect proteins around 78, 89, and 93 kDa for MGDα–, MGDβ–, and MGDγ–eGFP, respectively. Immunoblot analyses revealed a band at ∼70 and ∼80 kDa for MGDα– and MGDβ–eGFP, respectively, which are in the expected range (Supplementary Fig. S6), coherent with the cleavage of a targeting presequence in these proteins. A broad signal was detected for MGDγ–eGFP, around 90 to 100 kDa. These analyses indicate that the transformed lines expressed MGD–eGFP fusions, and could be used for subcellular localization studies as well as phenotypic analyses of MGD overexpression. Overexpressing lines highlighted distinct subcellular localizations of MGD isoforms (Fig. 2).
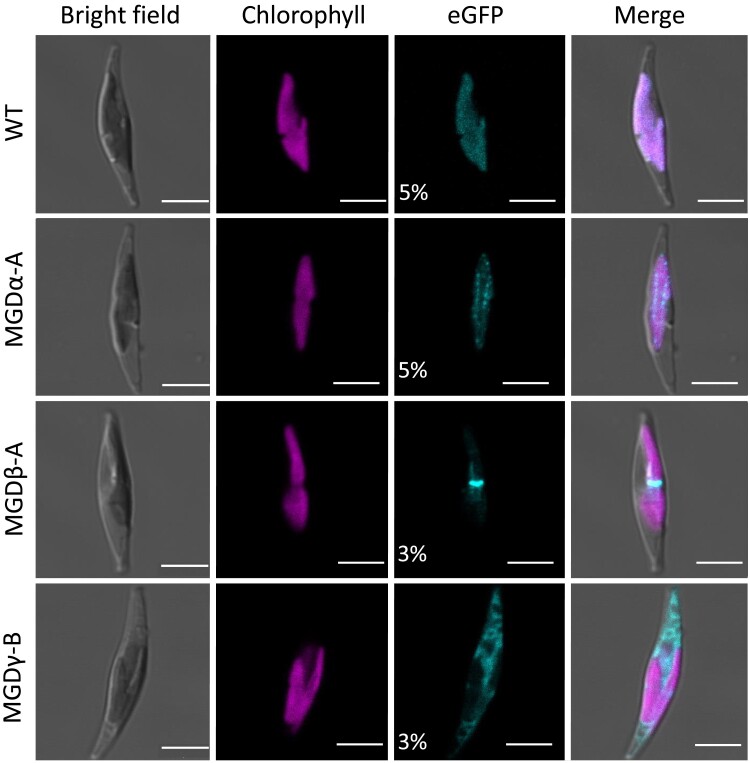
Figure 2.
Subcellular localization of MGD isoforms fused to eGFP in P. tricornutum. Detection of MGD–eGFP expressed in transgenic P. tricornutum lines with WT as a negative control. For each cell, bright field, chlorophyll fluorescence, and eGFP signal are shown, with a merge of all images. eGFP excitation was achieved at either 5% or 3% of laser power, as indicated. The WT control shows low unspecific detection of fluorescence from thylakoid photosystems. An intraplastidial localization was obtained when MGDα–eGFP is expressed. With MGDβ–eGFP, the eGFP signal corresponds to the blob-like structure, indicating a localization of MGDβ at the level of the periplasitidal compartment, and possibly the PPM. MGDγ–eGFP is detected mainly in the ER, and at lower level in the EpM. Scale bar: 5 µm. Similar independent observations are provided in Supplementary Fig. S7. MGD, monogalactosyldiacylglycerol synthase; WT, wild type.
MGDα–eGFP showed an intraplastidial signal visible as dots inside the plastid (Fig. 2, Supplementary Fig. S7A), corresponding to a localization in thylakoids (Huang et al. 2024). This localization is consistent with the predicted bipartite Sp-Ctp, assuming that the detected SAAFSP sequence has the same targeting function as the canonical ASAFAP motif (Apt et al. 2002). In contrast, MGDβ–eGFP fluorescence was detected in a single spot corresponding to the blob-like structure (Fig. 2, Supplementary Fig. S7B). A membrane VN was shown to protrude inside the blob from the PPM but not the oEM (Flori et al. 2016). Therefore, MGDβ localization might correspond to the VN and extend to the PPM. This localization is consistent with the predicted Sp-Ctp, and suggests that the GSGFVL motif may be a specific alteration of the ASAFAP motif leading to a retention in the blob and PPM (Apt et al. 2002). MGDγ–eGFP signal appeared in a reticulated membrane structure corresponding to the ER, and at the periphery of the plastid (EpM; Fig. 2, Supplementary Fig. S7C). This localization is consistent with the absence of any predicted targeting sequence in MGDγ.
We sought to confirm the observed localization of the 3 isoforms. We co-expressed MGDα–eGFP with a gene coding for the photosynthetic electron transport C subunit (PetC), addressed to thylakoids (Liu et al. 2016), fused to a monomeric red fluorescent protein (mRFP). MGDα–eGFP and PetC–mRFP signals showed an overlapping localization in thylakoids, with slightly different distribution patterns (Supplementary Fig. S8A). We co-expressed MGDβ–eGFP with the symbiont Derlin1-2 (sDer1-2), a component of the protein translocation channel located at the PPM (Hempel et al. 2009), fused to mRFP. The MGDβ–eGFP and sDer1-2-mRFP signals showed identical localization in the PPM, concentrated at the level of the blob-like structure (Supplementary Fig. S8B). Finally, we co-expressed MGDγ–eGFP with the mRFP, highlighting the difference between the MGDγ localization at the periphery of the plastid and in the ER network and the soluble form of mRFP in the cytosol (Supplementary Fig. S8C).
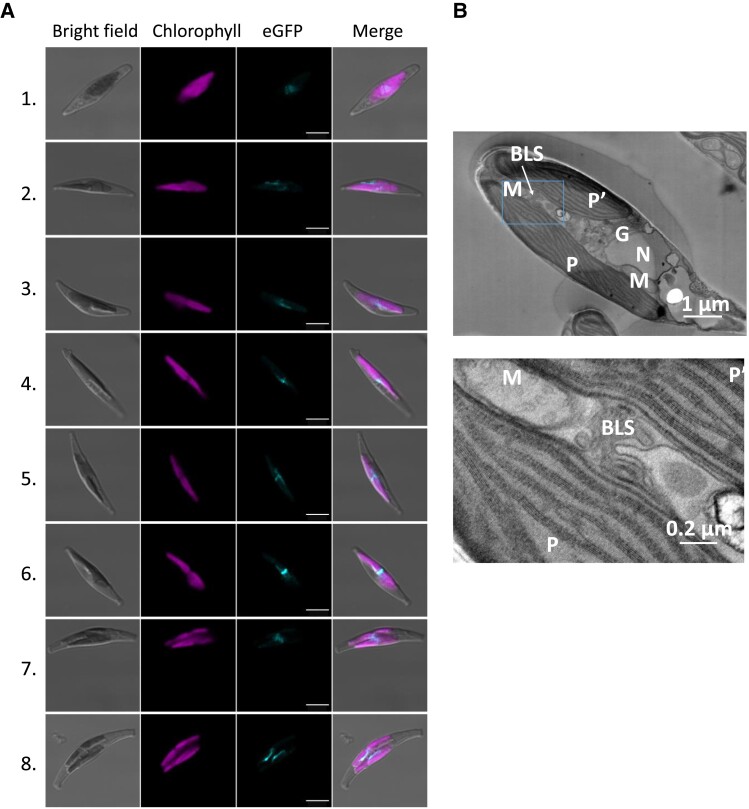
The MGDβ–eGFP signal always appeared at a location corresponding to the center of plastids (Figs. 2 and 3A, Supplementary Figs. S7 and S8). The blob-associated fluorescence was also observed in dividing plastids, which could suggest a role of MGDβ during plastid division. In dividing plastids observed by confocal microscopy, the blob was localized at the interface between the 2 forming plastids (Fig. 3A, Supplementary Fig. S7B). This observation was supported by scanning transmission electron microscopy (STEM) observation of the blob VN in a zone connecting dividing plastids (Fig. 3). After separation of the daughter cells, blobs appeared facing each other. When MGDβ–eGFP was observed in a nondividing plastid, the signal was much more diffuse. The blob-like structure may either become more exposed as plastids undergo divisions, making the signal more visible, or it may become more compacted during this particular time. Thus, it seems that the blob may play an important role during cytokinesis, possibly as a glycerolipid synthesis machinery, and that this machinery might be scattered in the PPM in nondividing cells.
Figure 3.
Subcellular localization of the blob-like structure, associated to MGDβ–eGFP fluorescence. Observation of blob-like structure localization by confocal microscopy (A) and transmission electron microscopy (B). P. tricornutum transgenic lines overexpressing MGDβ–eGFP were used to monitor the appearance of blob-like structures in the cell (A). Cells 1 and 2 show a scattered eGFP signal, Cells 3 to 6 show a blob-like structure at the central constriction of dividing plastids, Cell 7 shows a blob-like structure extending from 1 plastid to the other inside a dividing cell, and Cell 8 shows 2 blob-like structures facing each other on distinct plastids during cytokinesis. Cell 6 was also shown in Fig. 2. Observation of a WT P. tricornutum cell containing 2 plastids following plastid division inside the cell (B). Cleavage furrows are visible at both end of the cell. The blob-like structure (outlined in a rectangle) was observed connecting the 2 plastids at 2 magnifications. BLS, blob-like structure (or blob); G, Golgi apparatus; M, mitochondrion; MGD, monogalactosyldiacylglycerol synthase; N, nucleus; P and P′, plastids.
Generation of MGDα, MGDβ, and MGDγ edited lines
We generated KO lines by CRISPR-Cas9 editing, using single gRNA (sgRNA) sequences selected to interrupt protein function. Since random genomic insertions and off-target mutations can lead to unrelated phenotypes, we selected several independent strains for each gene, obtained from different sgRNAs (distinct potential off-targets), and not originating from the same initial transformed colony (distinct vector insertions in the genome). (Primers and sgRNAs are shown in Supplementary Table S3).
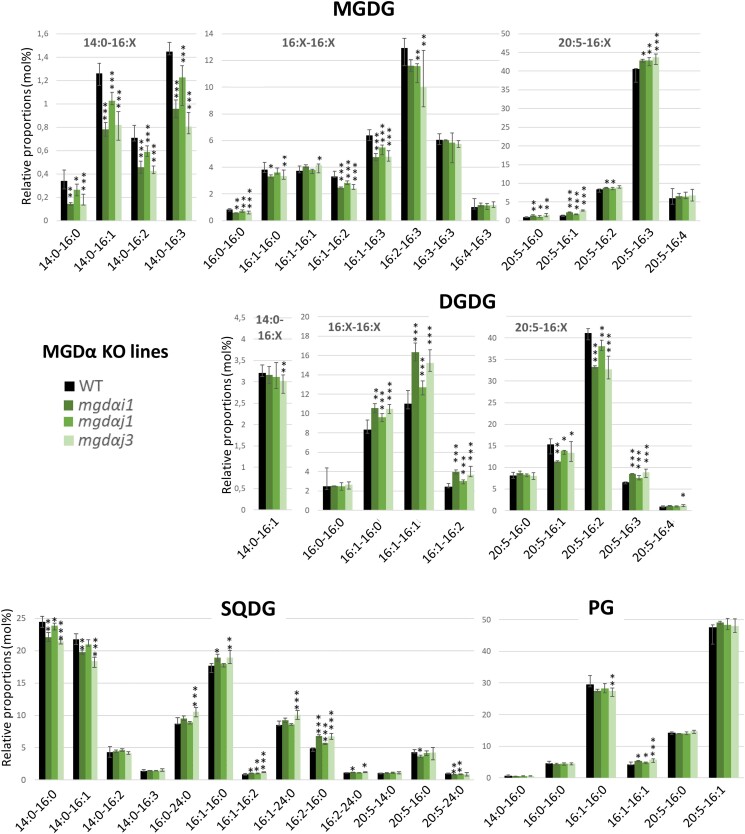
For MGDα, we selected 3 pure KO lines with frameshift disruptions, i.e. 1 KO obtained with sgRNA 125i (mgdαi1), and 2 independent KO lines obtained with sgRNA 125j (mgdαj1 and mgdαj3; Supplementary Fig. S9). Two mgdα mutants conserved for at least 1 yr highlighted homozygous profiles and were stable after repeated rounds of cultivation in liquid medium (Supplementary Fig. S10).
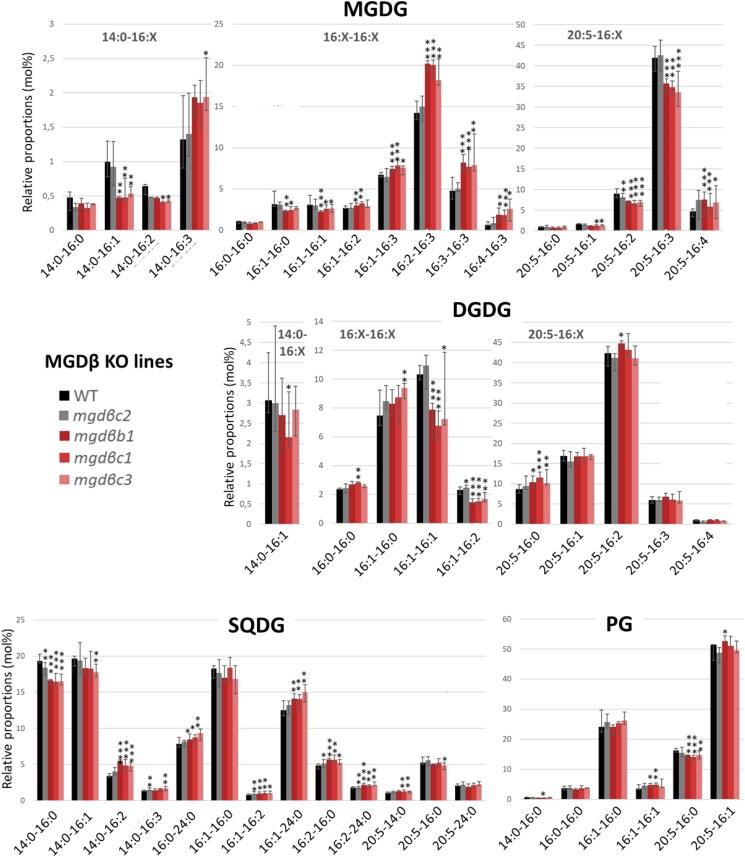
For MGDβ, we selected 1 KO line with sgRNA 168b (mgdβb1), and 3 mutants obtained with sgRNA 168c (mgdβc1, mgdβc2, and mgdβc3; Supplementary Fig. S9). The 3 lines mgdβc1, mgdβc2, and mgdβc3 derive from the same initially transformed cell. They harbor the same vector insertion, but different editions at the targeted locus. The mgdβc2 mutant has a deletion of 6 nucleotides (DEL6), which does not generate a frameshift in the sequence (Supplementary Fig. S9). This mutant lacks 2 amino acids between the predicted Sp and the first N-terminal conserved region. This deletion was not expected to impair the enzyme function and thus mgdβc2 was kept as a possible control for mgdβc1 and mgdβc3 KO lines. All mgdβ mutants highlighted homozygous profiles and were stable after repeated rounds of cultivation in liquid medium (Supplementary Fig. S10).
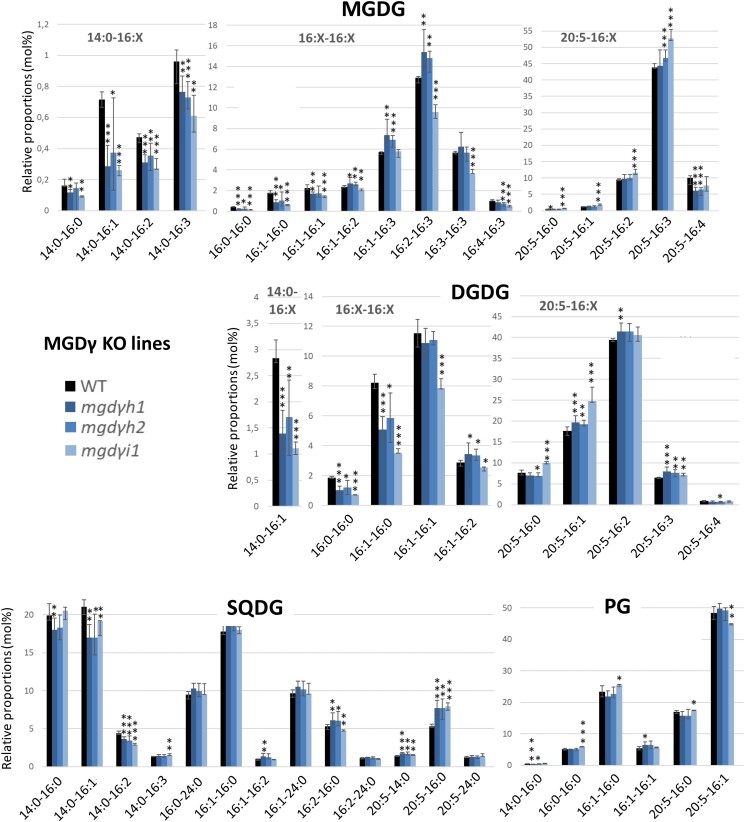
For MGDγ, we selected 2 KO lines obtained with sgRNA 619h (mgdγh1 and mgdγh2) deriving from the same initially transformed cell, and 1 obtained with sgRNA 619i (mgdγi1; Supplementary Fig. S9). For this gene, the generation of mutant colonies was difficult, as transformed lines were greatly affected in their ability to form colonies on plates. Only one mosaic colony was obtained after transformations with 619h and 619i sgRNAs. We managed to isolate 3 strains from the mosaic colony obtained with 619h sgRNA. For the mosaic colony obtained with 619i sgRNA, the mutant profile corresponds to a mixed population, without any wild type (WT); all indels detected led to either a frameshift in the sequence or the deletion of crucial amino acids in the active site region (Supplementary Fig. S9). We used this third heterogeneous KO line, without further purification. The growth phenotype observed on plates may indicate some important role in stressful environmental conditions. In addition, all mgdγ mutants evolved, as observed after 1 yr of repeated rounds of cultivation in liquid medium (Supplementary Fig. S10), without any trace of WT MGDγ sequence, and in all cases, in the direction of larger deletions and functional loss. They were therefore considered as valid KO lines.
Impact of MGD KO and overexpression on Phaeodactylum growth and photosynthesis efficiency in optimal growth conditions
A first key observation is that none of the MGD isoform KO was lethal. The growth of each KO line was monitored in optimal condition, in a rich medium (Supplementary Fig. S11A). Most of the KO mutants had a slightly slower growth than the WT strain. Overexpression of MGDα and MGDγ but not MGDβ led to a slight increase in growth (Supplementary Fig. S11B). The moderate impact of MGD KO on growth in liquid medium suggests possible mutual compensating effects between isoforms.
As MGDG plays an important role in the stabilization and function of photosystems, and in the xanthophyll cycle (Azadi-Chegeni et al. 2022; Garab et al. 2022), we sought whether any effect on photosynthesis and/or photoprotection could be detected. We measured the effective photochemical quantum yield of photosystem II (ΨII) and nonphotochemical quenching (NPQ) during 2 different kinds of light stress: a high light (700 µmol photons m−2 s−1) and a moderate light stress (2 steps increase in light intensity, at 55 then 335 µmol photons m−2 s−1; Supplementary Fig. S12). ΨII recovery and NPQ relaxation were monitored at low light intensity (20 µmol photons m−2 s−1; Ruban et al. 2004; Goss and Jakob 2010). Under all tested conditions, no significant changes in photosynthetic efficiency or NPQ were observed, except in the case of mgdgγi1. This line showed lower photosynthetic efficiency under 55 and 335 µmol photons m−2 s−1 and reduced recovery under low light intensity following high light intensity measurements. Additionally, mgdgγi1 exhibited slower relaxation of NPQ under the same low light conditions (Supplementary Fig. S12C and F).
Impact of MGD KO and overexpression on Phaeodactylum cell morphology and plastid architecture
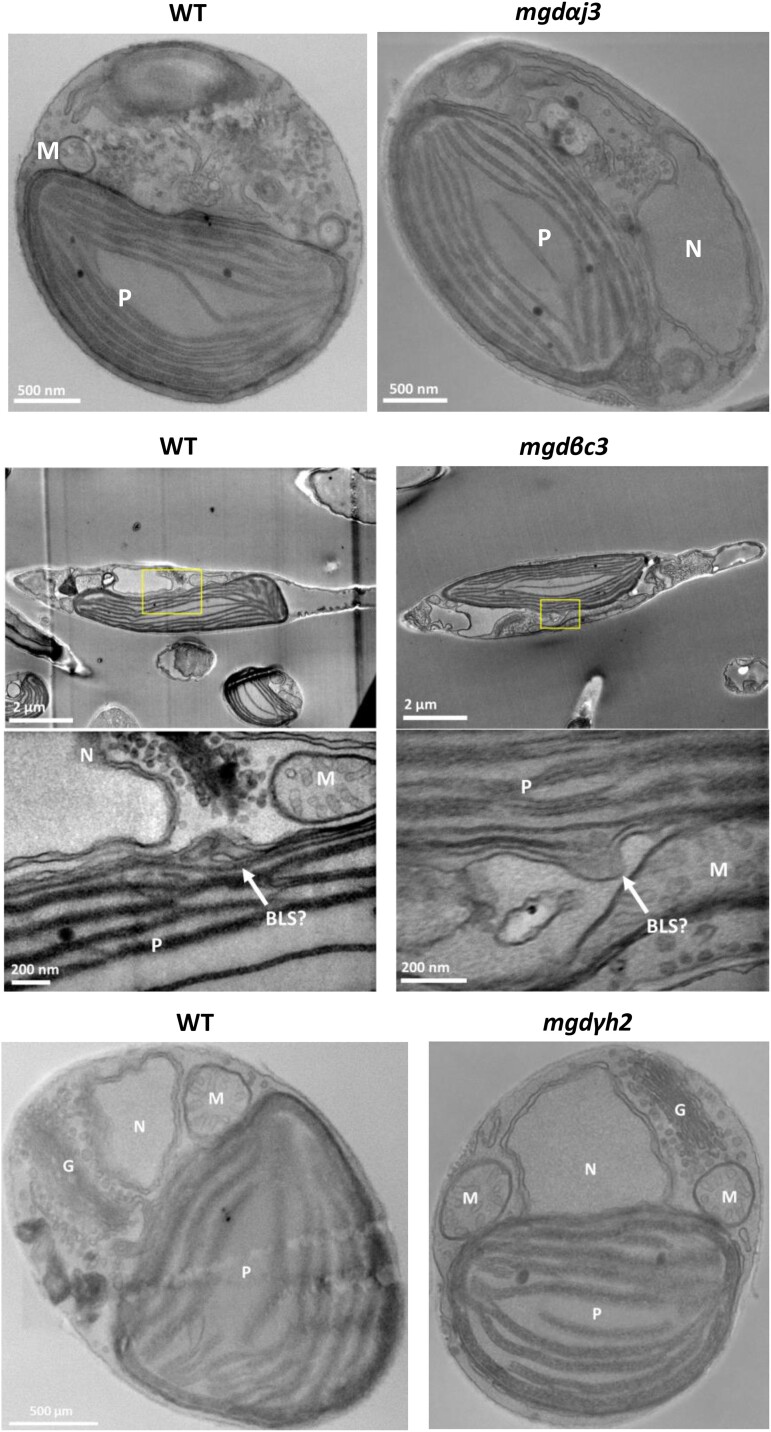
Cell morphology of MGD KO and overexpressing lines was examined by light and epifluorescence microscopy (Supplementary Fig. S13). All mutant cells were fusiform with a normally sized plastid compared with the WT. No change in cell size was detected. Potential impact of MGD KO on plastid membranes was checked by STEM (Fig. 4), with cells from mgdαj3, mgdβc3, and mgdγh2 strains. No modifications could be observed in plastids nor in any other parts of the cell in any MGD KO line. In MGDβ null mutants, blob-like structures were still present (Fig. 4). Likewise, no change in cell ultrastructure could be noticed in overexpressing lines (Supplementary Fig. S14).
Figure 4.
Phaeodactylum tricornutum cell ultrastructure in MGD KO lines determined by STEM. A WT and KO mgdαj3 and mgdγh2 lines were cultured in parallel. In a separate experiment, a WT and a mgdβc3 mutant were cultured in parallel. Cell ultrastructure is shown in each mutant with corresponding WT control on the left. No impact of MGDα, MGDβ, or MGDγ KO could be observed at the level of membrane compartments, including plastids. BLS outlined in rectangles was observed at 2 magnifications. BLS, blob-like structure; G, Golgi; KO, KO; M, mitochondria; MGD, monogalactosyldiacylglycerol synthase; N, nucleus; P, plastid; WT, wild type.
Impact of MGD KO and overexpression on Phaeodactylum total FA and glycerolipid class profiles
We cultured WT, KO, and overexpressing lines in parallel in a nutrient-replete medium (10N10P). The total amount of FAs in all strains was similar to the WT analyzed in parallel (Supplementary Figs. S15A and S16A), and FA distributions were similar to those already reported in the literature in the same conditions (Abida et al. 2015). As observed previously, P. tricornutum is rich in 20:5 (about 25% to 30% of total FAs) and C16 molecular species (mainly 16:0, 16:1, 16:3), while poor in C18 FA. The distribution of the different lipid classes was analyzed by LC–MS/MS (Supplementary Figs. S15B and S16B). In all strains, the glycerolipid profiles were dominated by MGDG, PC, and SQDG, which accounted for about 70% of total glycerolipid content, as previously reported (Abida et al. 2015).
Only minor changes were observed in the relative proportion of membrane lipid classes (Supplementary Fig. S15B). Most importantly, MGD KO had no significant impact on the levels of galactolipids. Concerning the other lipid classes, MGDα KO strains showed slightly lower levels of SQDG (16.0% vs. 19.6% in the WT) and more PC (17.8% vs. 14.10% in the WT; Supplementary Fig. S15B). In MGDβ KO strains, the proportion of PC was higher (14.10%) compared with both WT and silent mgdβc2 controls (12.7%; Supplementary Fig. S15B), consistently with an opposite trend in the MGDβ overexpressing strains (Supplementary Fig. S16B). All MGDγ KO strains had slightly lower proportions of PG (5.1% vs. 6.3% in the WT) and slightly higher proportions of DAG (1.23% vs. 1.05% in the WT; Supplementary Fig. S15B). DAG tended to decrease in MGDγ overexpressing strains (Supplementary Fig. S16B), in line with a more active utilization of this MGD substrate.
Impact of MGD KO and overexpression on acyl profiles in each glycerolipid class
Since KO or overexpression of MGD isoforms had minor impacts on cell division, photosynthesis, morphology, total FAs, and glycerolipid class profiles, one may wonder whether the 3 isoforms might be fully interchangeable. Expression of the different MGD isoforms in yeast suggested that they have distinct substrate specificities (Supplementary Fig. S5). We looked therefore in more detail at the variation of each lipid molecular species in each lipid class (Abida et al. 2015; Jouhet et al. 2017).
We based our analyses on the following assumptions: (i) a decrease in a molecular species should reflect the KO of the considered MGD isoform, and (ii) an opposite trend should be observed in the corresponding overexpressing lines (shown in Supplementary Figs. S17 and S18). An additional principle was to assume that (iii) an increase of a molecular species in a KO line might be determined by a compensation by the other intact isoforms. Finally, (iv) we focused our analyses on the 4 glycerolipids known to be conserved in photosynthetic membranes, i.e. MGDG, DGDG, SQDG, and PG (Boudiere et al. 2014; Figs. 5 to 7), and on other phospho- and betaine lipids, supposed to be related to the endomembrane system, and acting as molecular platforms for the synthesis of 20:5.
Figure 5.
Impact of MGDα mutations on MGDG, DGDG, SQDG, and PG molecular species. Lipid molecular species in MGDG, DGDG, SQDG, and PG were analyzed by LC–MS/MS. Each result is the median of 6 biological replicates ± min and max values. *P < 5 × 10−2, **P < 1 × 10−2, and ***P < 1 × 10−3, based on an unpaired multiple t test. DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol.
Figure 7.
Impact of MGDγ mutations on MGDG, DGDG, SQDG, and PG molecular species. Lipid molecular species were analyzed by LC–MS/MS. Each result is the median of 6 biological replicates ± min and max values. *P < 5 × 10−2, **P < 1 × 10−2, ***P < 1 × 10−3, based on an unpaired multiple t test. DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol.
In MGDα KO lines, most molecular species containing a C16 in Position sn-1 decreased in MGDG (Fig. 5). MGDG 16:1-16:2, 16:1-16:3, and 16:2-16:3 species increased in MGDα–eGFP-B overexpressing strain (Supplementary Fig. S17). In contrast, the proportion of MGDG molecular species containing a 20:5 FA increased, except the 20:5-16:4. MGDG 20:5-16:3 decreased in MGDα–eGFP-B overexpressor. The proportion of all molecular species containing a 14:0 FA also decreased in MGDα KO lines, but did not change in overexpressing lines. Concerning DGDG, the change in molecular species profile did not reflect that of MGDG. An increase in species with a 16:1 in Position sn-1 was observed in the MGDα KO lines, while species containing a 20:5 decreased, except for 20:5-16:0 and 20:5-16:4 (Fig. 5). In the MGDα–eGFP-B overexpressor, DGDG 16:1-16:0 and 16:1-16:1 decreased, while 20:5-16:2 species increased (Supplementary Fig. S17). Concerning SQDG, in the KO lines, the sn-1 C16-SQDG tended to increase, while species containing a 20:5 FA tended to decrease. SQDG 14:0-16:0 and 14:0-16:1 species also decreased. In PG, the proportion of 16:1-16:0 molecular species decreased while that of 16:1-16:1 species increased (Fig. 5).
The acyl profile of other membrane glycerolipids, i.e. PC, PE, DGTA, and DAG changed in MGDα KO and overexpressing lines (Supplementary Figs. S18 and S19). In brief, all 14:0-containing molecular species decreased, and they consistently increased in MGDα–eGFP-B strain (Supplementary Fig. S18). All C18-containing species decreased, with the exception of 18:2-18:1 and 18:2-18:2 in PC, and 20:5-18:4 in PE. All species containing a C16 in Positions sn-1 and sn-2 increased in PC and DGTA. However, 16:0-16:0 and 16:0-16:1 molecular species decreased in DAG while 16:1-16:1 increased. Species containing a 20:5 at Position sn-1 and a C16 at Position sn-2 increased or tended to increase in PC, DGTA, and PE. In MGDα KO, the 20:5-22:6 molecular species increased in PC (Supplementary Fig. S19), while it consistently decreased in MGDα–eGFP-B (Supplementary Fig. S18).
For the analysis of MGDβ KO lines, we used the silent mgdβc2 strain as a negative control in addition to the WT. MGDG molecular species containing a 20:5 in Position sn-1 decreased (Fig. 6), with the exception of 20:5-16:0 and 20:5-16:4. MGDG 16:1-16:0 and MGDG 16:1-16:1 decreased, while other MGDG species containing a C16 FA in Positions sn-1 and sn-2 and 2 or more unsaturations on a C16 increased. The opposite trend was observed in the MGDβ overexpressing strains (Supplementary Fig. S17). By comparison, DGDG molecular species were little affected (Fig. 6). DGDG 14:0-16:1 tended to decrease and 16:1-16:1 strongly decreased, in line with similar changes in MGDG. The DGDG species containing 20:5 were almost not affected, with only a slight increase in 20:5-16:0 compared with the controls. In contrast with MGDα mutants, only a few changes were observed in other glycerolipids in MGDβ KO lines: for PC, DGTA, and PE, 16:1-16:2, 16:1-16:3, and 20:5-16:2 species tended to increase in the mutants, while 20:5-18:1, 20:5-18:2, and 20:5-18:3 tended to decrease (Supplementary Fig. S20). PE 20:5-20:5 species increased in MGDβ KO, while it decreased consistently in the MGDβ overexpressing strains (Supplementary Fig. S18).
Figure 6.
Impact of MGDβ mutations on MGDG, DGDG, SQDG, and PG molecular species. Lipid molecular species were analyzed by LC–MS/MS. The mgdβc2 mutant contains a silent mutation and was used as a control. Each result is the median of 6 biological replicates ± min and max values. *P < 5 × 10−2, **P < 1 × 10−2, ***P < 1 × 10−3, based on an unpaired multiple t test. DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol.
Finally, the levels of 16:0-16:0, 16:1-16:0, and 16:1-16:1 strongly decreased in both MGDG and DGDG in MGDγ KO lines (Fig. 7) and increased in MGDγ overexpressors (Supplementary Fig. S17). The proportion of all 14:0 FA-containing species were decreased compared with the WT. For several molecular species, the mgdγh1 and mgdγh2 mutants behaved slightly differently compared with mgdγi1 (Fig. 7). Nevertheless, the common increase in MGDG 16:1-16:3, 16:2-16:3, and 16:3-16:3 species was coherent with an opposite trend in the overexpressors (Supplementary Fig. S17). No change was observed in 20:5-containing MGDG species, except a decrease in 20:5-16:4 mirrored by an increase in the overexpressing lines. In DGDG and SQDG, 20:5-containing species tended to increase except for DGDG 20:5-16:4. PG molecular species distribution was globally unaffected by MGDγ mutations.
In other glycerolipid classes linked with the endomembrane system, many changes in species proportions were observed in MGDγ mutants (Supplementary Fig. S21). In PC and DGTA, 16:0-16:1, 16:1-16:0, 16:1-16:1, 16:1-18:1, and 16:1-18:2 species increased, while DGTA 16:1-18:3 and PC 16:1-18:4 decreased. There was a striking decrease in 20:5-20:4 and 20:5-20:5 in PC, DGTA, and PE. By comparison with MGDα and MGDβ mutations, 14:0-containing species were not impacted in MGDγ mutants. In DAG, changes were limited to 16:0-16:1 and 20:5-16:0, which tended to increase and decrease, respectively.
Impact of each MGD KO mutation on the expression level of other MGD isoforms
We performed reverse transcription qPCR (RT-qPCR) analyses on MGD KO strains to check for possible compensations at the transcriptional level. In replete conditions, the WT, MGDβ, and MGDγ were expressed at similar levels, while MGDα was ∼4 times more expressed (Supplementary Fig. S22). We observed a decrease in MGDβ gene expression in MGDβ KO lines (2.25-fold decrease on average), which was not observed in the silent mutant mgdβc2. In MGDγ mutants, an increase of MGDα and MGDβ expression (1.37- and 1.69-fold increase, respectively) suggests the possible activation of a compensation mechanism at the transcriptional level, when MGDγ is affected. No apparent transcriptional compensation could be detected when MGDα or MGDβ were affected (Supplementary Fig. S22). This does not exclude some other compensation mechanisms at the posttranscriptional level.
Impact of MGDα, MGDβ, and MGDγ mutations on the response of P. tricornutum to a nitrogen starvation
The diversification of the MGD isoforms in diatoms might play a role in response to environmental variations. We have previously reported that nitrogen deprivation led to a change in MGD protein proportions (Lupette et al. 2022): the protein levels of MGDα and MGDγ were reduced to 6.1% and 26.3% of control level grown in replete conditions, respectively. In contrast, MGDβ protein level showed an increase to 127.0% of its level in control culture.
Two KO mutants per MGD were used for the analysis of the impact of nitrogen deprivation: mgdαi1, mgdαj3, mgdβb1, mgdβc1, mgdγh2, and mgdγi1. Cells grown in nutrient-replete medium (enriched ESAW 10N10P) were shifted to nitrogen-deprived medium (00N10P) at a starting cell concentration of 3 to 3.5 × 106 cells mL−1. Growth was monitored after 3, 4, and 5 d of nitrogen deprivation (Supplementary Fig. S23). Nitrogen deprivation condition led to a slowing down of cell division. Cultures kept growing between Day 3 and Day 5, with WT cells reaching a concentration of ∼9 × 106 cells mL−1 at Day 5, which is about half of the concentration that would be expected in 10N10P condition. The growth of MGDα and MGDβ KO lines were similar to that of the WT, whereas growth was affected in both MGDγ KO lines (Supplementary Fig. S23), suggesting a function of this isoform in this condition.
The triacylglycerol (TAG) accumulation in response to nitrogen deprivation was first monitored by Nile Red staining at Days 3 and 4 following deprivation. We observed that TAG accumulation was faster in MGDβ KO lines compared with the WT (Supplementary Fig. S24). Cells stained with Nile Red at Day 5 of nitrogen deprivation were observed by epifluorescence microscopy (Supplementary Fig. S25). All strains presented a fusiform morphotype and showed large lipid droplets. Plastids appeared shrunken compared with cells grown in 10N10P condition (Supplementary Fig. S13), consistent with the senescence of plastid membranes previously reported during nitrogen starvation (Abida et al. 2015). Besides a few lipid droplets detected in the medium in the mgdγi1 KO mutant, no difference in cell, plastid and lipid droplet size could be noticed between WT and KO lines (Supplementary Fig. S25).
After 5 d of nitrogen deprivation, cells were harvested for glycerolipid analysis. The response of P. tricornutum to nitrogen shortage was consistent with previous reports (Abida et al. 2015) with a total FA profile becoming close to the composition of TAG molecular species (Supplementary Fig. S26A), due to the increase in TAG, reaching 4.10 nmol 10−6 cells after 5 d of nitrogen deprivation, and representing ∼60% of total lipid content (Supplementary Fig. S26B).
Consistent with the very low MGDα protein level in the WT in low nitrogen (Lupette et al. 2022), no change was detected in the proportions of membrane glycerolipids in MGDα KO lines compared with the WT. For MGDβ KO lines, a slight increase in PE and PG, and a very small decrease in PI were observed. The changes of membrane glycerolipid distribution in MGDγ KO lines were more striking, with lower proportions in DGDG (∼22% decrease) and DAG (∼10% decrease), and an increase in PC (∼27% increase) and DGTA (∼57% increase) compared with the WT (Supplementary Fig. S26B). Quantitatively, the decrease in DGDG was roughly equivalent to the increase in PC.
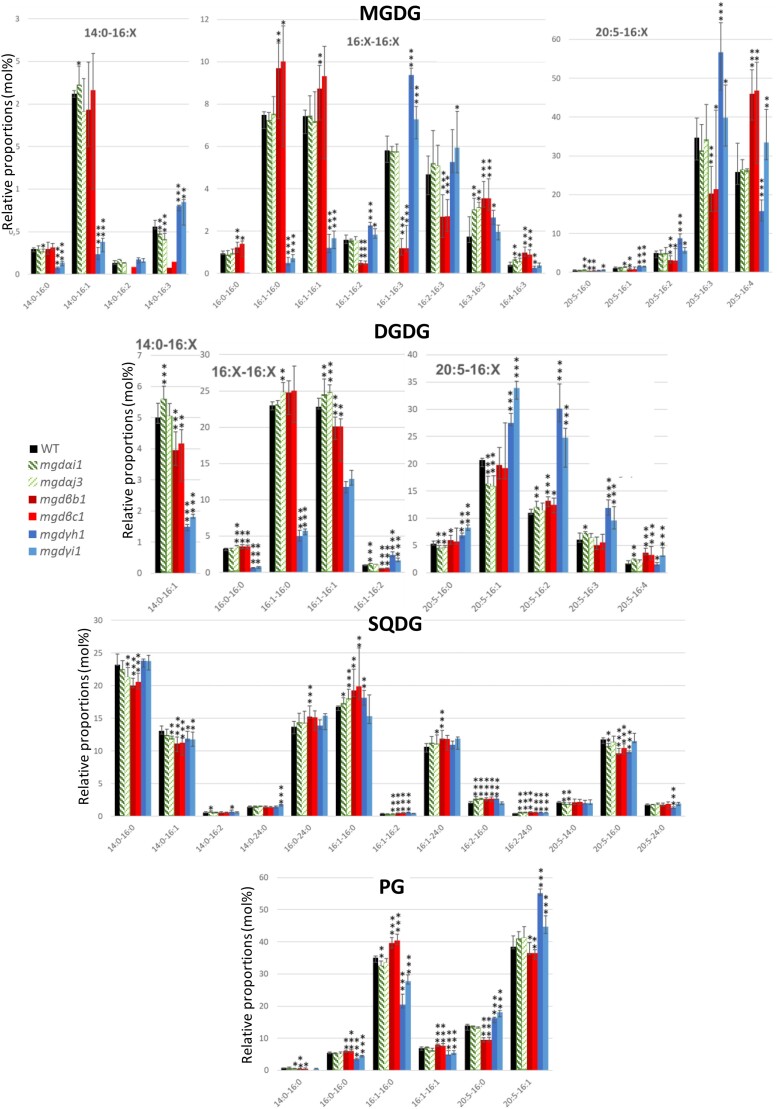
Concerning MGDG acyl composition, the impact was barely detected in MGDα KO mutants (Fig. 8), in line with the expected lack of involvement of this isoform in nitrogen shortage. In contrast, MGDG molecular species composition was severely affected in MGDβ and MGDγ KO lines (Fig. 8) suggesting that these 2 isoforms play a role under nitrogen stress. In MGDβ KO lines, a strong decrease in 14:0-16:2 and 14:0-16:3 was observed (Fig. 8). Although MGDG 14:0-16:1 seemed stable, a decrease in DGDG 14:0-16:1 was observed. MGDG 16:1-16:2, 16:1-16:3, and 16:2-16:3 strongly decreased while 16:0-16:0, 16:1-16:1, 16:3-16:3 and 16:4-16:3 increased. All 20:5-containing MGDG species decreased, except for 20:5-16:4 MGDG that strongly increased. In DGDG, 16:1-16:1, 16:1-16:2, and only 20:5-16:2 and 20:5-16:4 species showed a moderate increase, while other 20:5-containing species were stable.
Figure 8.
Impact of MGD mutations on the molecular species constituting MGDG, DGDG, SQDG, and PG in nitrogen-deprived P. tricornutum cells. Lipid molecular species in MGDG, DGDG, SQDG, and PG were analyzed by LC–MS/MS. Each result is the median of 6 biological replicates ± min and max values. *P < 5 × 10−2, **P < 1 × 10−2, ***P < 1 × 10−3, based on an unpaired multiple t test. DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol.
Changes in galactolipid species distribution were the most spectacular in MGDγ mutants (Fig. 8): MGDG and DGDG species containing a 16:0 or a 16:1 in Position sn-2 and either a 14:0 or a saturated or monounsaturated C16 in Position sn-1 decreased. On the contrary, species with a 16:2 or 16:3 in Position sn-2 increased. Most 20:5-containing galactolipid species were mainly increased, showing that compensatory mechanisms most likely relying on MGDβ, channeled 20:5 to galactolipids.
In other lipids, the impact of MGDα mutation was mild (Fig. 8, Supplementary Fig. S27). In MGDβ KO lines, the proportion of 14:0-16:0, 14:0-16:1, 20:5-16:0 in SQDG and 14:0-16:0, 20:5-16:0 and 20:5-16:1 in PG decreased. The proportion of 16:1-16:0, 16:1-16:2, 16:2-16:0 and 16:2-24:0 in SQDG, and that of 16:0-16:0, 16:1-16:0 and 16:1-16:1 in PG increased. MGDγ mutation had no effect on SQDG, whereas the proportion of PG species containing a C16 FA in Position sn-1 decreased while 20:5-containing PG increased. It seems that PG species were affected similarly to the galactolipid species with the same FA composition, suggesting that they shared some substrates in their biosynthetic pathways.
The MGDγ KO lines were the only ones to show a striking impact on cytosolic TAG upon nitrogen shortage, verified by opposite effects in overexpressing lines. MGDγ inactivation led in particular to an increase of 14:0-16:1-16:1, 16:1:16-1:16-1, and 20:5-16:1-16:1 TAG species that were consistent with an opposite trend in the overexpressing lines in optimal conditions (Supplementary Figs. S18 and S27). Additionally, KO of MGDγ in nitrogen starvation led to a decrease of 14:0-16:0-16:0, 16:0-16:0-16:0, 16:0-16:0-16:1, 16:0-18:1-16:0, 16:1-18:1-16:0, and 16:1-18:1-16:1 TAG species, consistent with an increase of these species in the MGDγ overexpressing lines (Supplementary Figs. S18 and S27).
Considering that MGDα is naturally absent under nitrogen shortage, the KO of either MGDβ or MGDγ could be seen as a double invalidation (mgdα/mgdβ or mgdα/mgdγ, respectively), resulting in stronger phenotypes compared with those observed in replete medium. MGDβ looked important to keep a high level of polyunsaturated galactolipids in the absence of MGDα. MGDγ seems to play a specific role in response to nitrogen shortage, in particular on galactolipids with a low unsaturation level. MGDγ KO had a stronger effect on nonplastidial lipids than MGDβ KO. The increase in DGTA 16:0-16:1 and 16:1-16:1 in MGDγ KO lines was particularly striking and highlights a role for this isoform outside of the plastid, consistent with its ER/EpM localization. Modification of TAG species distribution suggests a coupling between MGDγ products and TAG synthesis.
MGDβ/MGDγ double mutations
To better understand the role of MGDα, 3 MGDβ/MGDγ double KO mutants were generated (2KO4, 2KO6, and 2KO9; Supplementary Fig. S28). Their growth in optimal conditions was slightly affected compared with the WT (Supplementary Fig. S29); nevertheless, the mgdβ/mgdγ KO proved nonlethal. The double KO mutants showed no variation in the MGDG levels compared with the WT, demonstrating that MGDα alone was capable of maintaining MGDG production in the cell at a level equivalent to the WT, at least quantitatively. A mild decrease of DGDG (Supplementary Fig. S30) contrasted with the results of the MGDβ and MGDγ single KO mutants. The weakly unsaturated C16-containing MGDG species showed a decrease whereas the highly unsaturated ones increased drastically (Supplementary Fig. S31), confirming that these MGDG species are more specifically produced by MGDα. The vicinity of plastidial desaturases (Dolch and Marechal 2015) and their strong activity may be at the origin of both variations, unbalancing the ratio between weakly and strongly unsaturated C16-containing MGDG. A reduction of the 20:5-containing MGDG species, but not their elision, demonstrates that MGDα is able to process DAG species containing 20:5 at Position sn-1. The acyl profile of DGDG is peculiar and very informative as well (Supplementary Fig. S31). A dramatic decrease in 14:0-16:1, 16:1-16:0, and double 16:1 was recorded, accompanied with a slight increase in 20:5-16:0 and 20:5-16:1. The lack of reduction in the 16:1-16:2 DGDG may imply that this species is produced by a DGDG synthase unable to process MGDα products. The increase in 20:5-containing DGDG species supports the hypothesis that a specific DGDG synthase uses 20:5-containing MGDG as preferential substrate and is highly active because, although its substrate is reduced in the double KO mutants, its product is either unvaried or increased. Considering all other lipid classes (Supplementary Fig. S31 and S32), acyl profiles show some opposite trends to those observed in MGDα KO lines (Fig. 5 and Supplementary Fig. S19).
Discussion
The emergence of a multigenic family of MGDs likely occurred before the radiation of diatoms and pelagophyceae from other photosynthetic stramenopiles
Phylogenetic analyses of MGD proteins reported earlier (Hori et al. 2016; Shang et al. 2022) and completed here (Supplementary Fig. S2) revealed the conservation of 3 major isoforms in diatoms, originating from gene duplications and specialization, similar but distinct to the process that led to the emergence of Types A and B in angiosperms. Based on parsimonious assumptions, an ancestral MGDα type might have been vertically inherited from a red alga during the secondary endosymbiosis event at the base of all stramenopiles. Then, a first gene duplication occurred, before the radiation of diatoms and phaeophyceae from other stramenopiles. This first gene duplication is likely to have led to the emergence of a common ancestor to MGDβ and MGDγ. The second duplication potentially followed the separation of pelagophyceae from diatoms. Gene or genome duplications are crucial drivers in the evolution of stramenopiles (Martens et al. 2008; Parks et al. 2018). In pennate diatoms for instance, up to 6 whole-genome duplications have been identified (Parks et al. 2018). It is important to stress that the genomes of some nondiatom stramenopiles contain only one MGD, such as the eustigmatophytes Nannochloropsis or Microchloropsis (Vieler et al. 2012; Corteggiani Carpinelli et al. 2014). A single isoform seems, therefore, sufficient for the biogenesis of a secondary plastid bounded by 4 membranes.
P. tricornutum MGDα, MGDβ, and MGDγ are active MGDG synthases and have distinct substrate specificities.
After assessing the accurate sequences of MGD proteins encoded by the genome of P. tricornutum, structural modeling highlighted a typical double-Rossmann fold (Supplementary Fig. S4) characterizing the 3-dimensional structure of MGD determined in angiosperms (Rocha et al. 2016). The critical residues for activity and involved in a PG-H catalytic dyad are conserved supporting that diatom proteins are functional MGDs. A noticeable distinctive feature lied in the relative size of the N- and C-domains among isoforms. Overall, the hinge separating the N- and C-Rossmann folds was longer in P. tricornutum proteins than in plant sequences. Compared with AtMGD1 and MGDα, the C-domain of MGDβ and MGDγ contains additional stretches of 50 and 110 amino acids, respectively. These 2 regions appeared to be essentially hydrophilic and could influence protein function. In addition, the N-domain of MGDβ and MGDγ contains 82 and 81 additional amino acids, respectively. MGDβ and MGDγ mature proteins are therefore bigger; the role of the detected protein extensions is unknown. The enzymatic function was confirmed by (i) heterologous expression of MGD isoforms in yeast and (ii) lipid changes in P. tricornutum MGD KO lines and overexpressors. These 2 sets of experiments showed that substrate specificity was distinct for each isoform, with consistent results.
P. tricornutum MGDα, MGDβ, and MGDγ localize to distinct subcellular compartments inside and outside of the plastid
MGDα is predicted to contain a bipartite Sp-Ctp presequence suggesting its localization inside the plastid. A putative cleavage site is predicted at the level of a SAAFSP sequence, matching partly with the profile of the “ASAFAP motif” characterized in nuclear-encoded plastid proteins (Kilian and Kroth 2005). In P. tricornutum expressing MGDα–eGFP fusion proteins, localization was consistently observed in thylakoid membranes (Fig. 2). The localization was confirmed by co-expression with a thylakoid marker (Supplementary Fig. S8).
In MGDβ, an Sp and a GSGFVL sequence with weak similarity to the ASAFAP motif were predicted. MGDβ–eGFP fusion proteins localize to the blob-like structure (Figs. 2 and 3) and were confirmed by coexpression with a blob/PPM membrane marker (Supplementary Fig. S8). These results indicate that the precursor of MGDβ is targeted to the secondary plastid, successfully crosses the EpM, but, at least in overexpressing system, is not transported further through the oEM and iEM. Arrest in the blob-like structure might be determined by specific changes in the ASAFAP motif, corresponding to a different amino acid pattern than previously characterized (Apt et al. 2002; Kilian and Kroth 2005). This may explain why MGDβ was not detected by bioinformatics methods developed to predict the secondary plastid proteome in P. tricornutum (Gruber et al. 2015). Additionally, MGDβ localization could extend to the PPM, as the VN present in the blob-like structure emerges from the PPM (Flori et al. 2016). Since the oEM and iEM correspond to the chloroplast envelope in primary plastids, in rigorous terms, the suborganellar localization of MGDβ represents a case of MGD outside a membrane system vertically related to the primary chloroplast. This evolution may have been determined by relevant mutations of the ASAFAP motif of an MGDα-like ancestral protein.
Finally, no targeting sequence could be predicted in MGDγ, which was consistent with an MGDγ–eGFP fluorescence detected outside of the secondary plastid, in the ER and possibly the EpM (Fig. 2 and Supplementary Fig. S8). In a recent report, MGDγ–GFP expression in P. tricornutum showed a signal partly overlapping that of chlorophyll, and outside of the plastid (Shang et al. 2022). Authors suggested that such pattern was that of an intraplastidial localization, whereas presented images rather show a localization at the periphery of the plastid consistent with results presented here. Localization of MGDγ in the ER and EpM is consistent with its detection in 2 replicates of the proteome of cytosolic lipid droplets isolated from P. tricornutum (Leyland et al. 2020). Overall, these results support that MGDγ is distributed between the ER and the EpM, and possibly the cytosol. In Arabidopsis, although MGDs are classically considered bound to chloroplast envelope membranes, the initial study of AtMGD2 and AtMGD3 fused to GFP showed some labeling diffusing in the cytosol (Awai et al. 2001), and AtMGD2 was shown to localize to the cytosol of elongating pollen tubes (Billey et al. 2021a). Based on this localization, MGDγ represents an evolutionary innovation, performing MGDG synthesis outside of the plastid, for membrane lipid synthesis involving other cell compartments. This is in line with the lipid phenotype we observed. The results suggest therefore an evolution by gene duplication and acquisition of new cell function, in the following order, MGDα → MGDβ → MGDγ, at the base of diatoms’ radiation.
None of the isoforms seems to localize to the iEM nor the oEM, as in primary chloroplasts of plants and green algae (Petroutsos et al. 2014). The concentric localization (MGDα/thylakoid; MGDβ/PPM; MGDγ/ER-EpM) suggests distinct roles for each of membrane compartment, and some kind of sophisticated interactions among glycerolipid pathways occurring inside and outside the secondary plastid. This localization also contradicts the assumption that MGDG, and its derivative DGDG, would be restricted to thylakoid, iEM and oEM membranes. Multiple pools of MGDG are likely to occur in diatom cells, an important feature to take into account in the interpretation of the lipid phenotypes in MGD KO and overexpressing lines.
The blob-like structure, a lipid synthesis machinery playing a role during cytokinesis?
Observation of MGDβ–eGFP in the blob-like structure between the PPM and oEM also gave insightful information about the positioning of this structure, particularly during plastid division (Fig. 3). Our observations of MGDβ–eGFP fluorescence by confocal microscopy and of the WT by STEM suggest that the VN might converge and take a blob-like structure mostly during cytokinesis, from the moment the plastid starts dividing until the whole cell division has been completed. When the plastid does not show any sign of division, the MGDβ–eGFP fluorescence signal was more diffuse, either suggesting that the VN itself loses the blob structure and become more scattered, or that MGDβ protein relocates to the PPM near the blob. During plastid division, the blob-like structure was located at the central constriction site of the dividing plastid. Following division, the 2 generated plastids seem to remain attached by the blob-like structure, meaning that the PPM and EpM are not fully divided, probably until the cleavage furrow comes through the blob-like structure. This is particularly important as it supports that the oNE–EpM continuum is never broken during plastid division, a point that was not addressed yet in studies of diatoms’ plastid division (Tanaka et al. 2015). These observations suggest that MGDβ could be involved in the provision of MGDG to the growing and dividing plastid membranes. Nevertheless, such possible role appears as nonessential, since MGDβ KO lines grow nearly as well as WT.
Functional compensation mechanisms between MGD isoforms
For phenotypic analyses of KO lines, a minimum of 2 CRISPR-Cas9 KO mutants, each obtained with a different sgRNA, were used. None of the isoforms proved strictly necessary for cell survival. In spite of careful scrutiny, neither cell growth and morphology, nor membrane integrity, nor photosynthetic activity was affected, suggesting either a dispensable role of the targeted isoforms or, more likely, a performant complementation by the action of intact isoforms. Under optimal culture condition, moderate differences could be observed in the relative proportions of lipid classes of all mutant strains. No change in transcript level was observed in MGDα and MGDβ KO lines. In contrast, in MGDγ mutants, an increase in MGDα and MGDβ transcripts is one of the possible compensation mechanisms involved in addition to the control of MGD protein stability and enzymatic regulation (i.e. activation of remaining isoforms via metabolic regulatory loops). In addition, to establish appropriate MGDG levels at the different subcellular compartments, intense fluxes of MGDG between membranes need to be postulated.
Specialization of MGDα in thylakoid membrane synthesis
Expression analysis in P. tricornutum WT strain showed that MGDα was the most expressed isoform under our optimal laboratory conditions, with relative expressions ∼4 times higher than that of MGDβ and MGDγ. In a recent study, MGDγ was reported to be relatively more expressed than other isoforms, when cells were grown in a medium containing less nitrogen (0.88 mm instead of 5.5 mm used here; Shang et al. 2022), a nutrient controlling the level of MGDα expression. MGDα basal role under optimal conditions is consistent with its higher similarity to MGDs from other phylogenetic lineages, and the conserved role in MGDG production for the massive demand of lipids for photosynthetic membranes. The relative protein level of MGDα was shown to drop to 6% in a whole-proteome study of nitrogen starved P. tricornutum (Lupette et al. 2022), coinciding with a decrease of thylakoid membranes observed in this stress condition (Abida et al. 2015). It also reveals that MGDβ and MGDγ might be recruited to sustain the cell in MGDG during nitrogen starvation. The complementary role of MGDβ and MGDγ for an accurate biogenesis of thylakoids is further demonstrated in MGDα KO lines, showing only a moderate phenotype, with apparently well-structured plastids and healthy photosynthesis.
MGDα uses a DAG pool deriving from plastid 16:0- and 16:1-ACP and produces MGDG with 16 carbon FA at Position sn-1, unsaturated up to 16:4 by plastid acyl-desaturases
Through combined observations of MGDα, MGDβ, and MGDγ and MGDβ/MGDγ KO effects, we identified the main DAG substrates used by each isoform and the fate of their products (Fig. 9). It is important to note that this reasoning is based on whole-cell lipidomic changes, considering both the location of an MGD isoform and the DAG substrate at its vicinity. Substrate specificity was further confirmed for medium chain FAs (C16) by the analysis of yeasts after heterologous expression.
Figure 9.
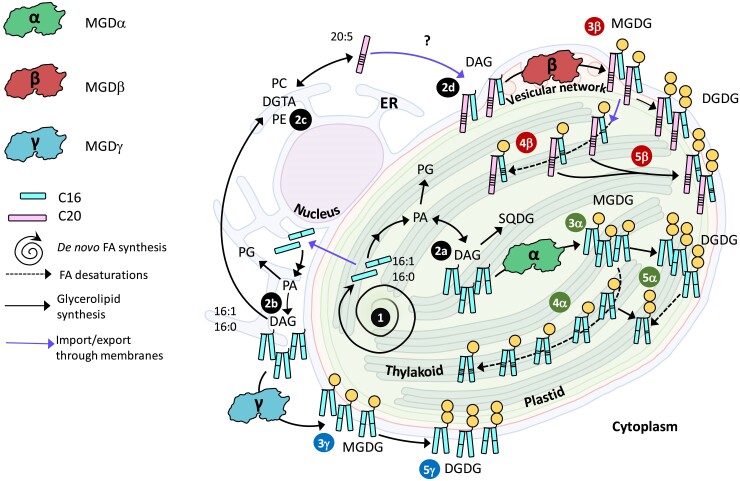
Function of MGDα, MGDβ, and MGDγ in P. tricornutum cell. The model summarizes all compartmentalization, structural and functional data obtained in this study. FAs are synthesized in the stroma as 16:0-ACP and 16:1-ACP (1), where they can be used for the synthesis of DAG (2a). Alternatively, FAs are exported to extraplastidial membranes where they can serve for the synthesis of DAG (2b) and phospholipids such as PC and PE, or the betaine lipid DGTA (2c). PC and DGTA serve as platforms to produce 20:5 (2c), used inside the plastid (at an unknown membrane site) for the production of 20:5-containing DAG (2d). MGDα is localized at the level of thylakoids, and functional studies reported here are consistent with a utilization of DAG available inside the plastid (3α), to form MGDG species rapidly desaturated (16:1 → 16:4) by plastid desaturases (4α). Based on obtained phenotype of KO and overexpressing lines, MGDG produced by MGDα is likely used by a specific DGD enzyme (5α) to form DGDG. MGDβ is localized at the level of the blob-like structure at the PPM, and functional studies reported here are consistent with a utilization of DAG either produced inside the plastid or with 20:5 acyls imported from the ER by a still uncharacterized Ω-pathway (3β). MGDG produced can be desaturated (16:1 → 16:4) by plastid desaturases (4β). Based on obtained phenotype of KO and overexpressing lines, MGDG produced by MGDβ is likely used by a specific DGD enzyme (5β) to form DGDG. MGDγ is localized at the periphery of the plastid, possibly partitioned between the ER and the EpM. Functional studies are consistent with a utilization of DAG containing FA freshly exported from the plastid (3γ), producing MGDG at a location far from plastid desaturases. Based on obtained phenotype of KO and overexpressing lines, MGDG produced by MGDγ is also likely associated to a specific DGD enzyme (5γ). In nitrogen shortage, MGDα protein level decreases drastically, and MGDG production relies on MGDβ and MGDγ. When 1 MGD is knocked out, compensation mechanisms rely on intact isoforms located in distinct membrane systems, requiring rapid inward and outward trafficking of glycerolipids including MGDG, through the 4 membranes limiting the plastid. KO, knockout; DAG, diacylglycerol; DGTA, diacylglyceryl hydroxymethyltrimethyl-β-alanine; DGD, digalactosyldiacylglycerol synthase; DGDG, digalactosyldiacylglycerol; ER, endoplasmic reticulum; FA; fatty acid; MGD, monogalactosyldiacylglycerol synthase; MGDG, monogalactosyldiacylglycerol; PC, phosphatidylcholine.
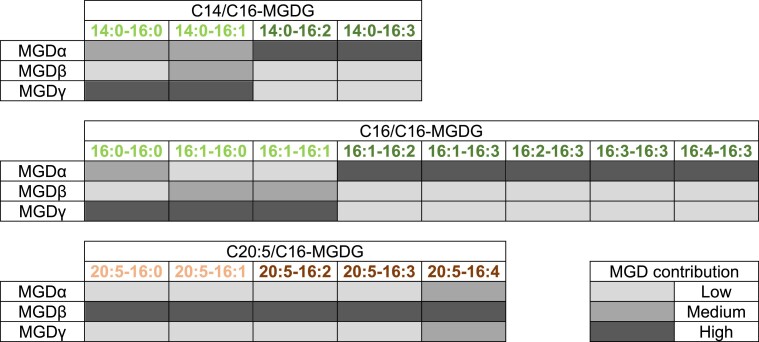
MGDα substrate specificity is consistent with its localization close to de novo FA synthesis, as well as to the plastid desaturases. Under optimal condition, MGDα KO most strongly affected MGDG species containing a C16 FA with 2 or more unsaturations, except those containing a C20:5 in Position sn-1. These species are highly reduced in nitrogen deprivation, when MGDα is not expressed. In the total FA distribution in MGDα mutants, a small increase in 16:1 was observed, while 16:2, 16:3, and 16:4 FAs tended to decrease. Given that in P. tricornutum 16:0-ACP is produced by the FA synthase in the stroma of the plastid, desaturated into 16:1-ACP by a plastid palmitoyl-ACP-desaturase (Smith et al. 2021), the pool of DAG used by MGDα is likely generated inside the plastid. In addition, C16 desaturations are performed by desaturases acting mostly on MGDG, which were predicted to localize to the stromal side of the plastid issued from the detection of Sp-Ctp and ASAFAP motifs (Dolch and Marechal 2015). Based on observed phenotypes, MGDα could mainly use DAG with the following compositions: 14:0-16:0, 14:0-16:1, 16:0-16:0, 16:1-16:0, and 16:1-16:1 (the main DAG species detected by LC–MS), corresponding to a de novo production of FA. The heterologous expression of MGDα in yeast confirms this capacity to use DAG with C16 unsaturated substrates, but MGDα appears to be also able to use C18 species in this nonnative context. Once the corresponding MGDG products are formed, they do not accumulate, as plastid desaturases rapidly generate MGDG species with higher unsaturations, i.e. 16:2, 16:3, and 16:4 (Figs. 9 and 10).
Figure 10.
Contribution of each MGD isoform to the accumulation of different pools of MGDG species. The level of contribution of each MGD to the accumulation of each MGDG species is indicated in 3 different intensities of grey. Diacyl species corresponding to DAG substrates deriving from FAs synthesized de novo in the stroma of the plastid are shown in C14/C16- and C16/C16-MGDG tables. Diacyl species corresponding to DAG substrates containing eicosapentaenoic acid (20:5) imported from the ER or endomembranes are shown in the C20:5/C16 table. Diacyl species corresponding to MGDG molecular species desaturated inside the plastid are shown in dark green and dark brown. This table summarizes information we deduced from (i) the lipidome analysis of P. tricornutum KO and overexpressing lines, and (ii) the range of medium chain MGDG fatty acyl species obtained after heterologous expression of isoforms in yeast. MGD, monogalactosyldiacylglycerol synthase; MGDG, monogalactosyldiacylglycerol.
The contrasting effects on 20:5-containing DGDG molecular species (decrease in the MGDα KO and increase in the MGDβ/MGDγ double KO) along with the corresponding increase or decrease in DGDG species containing 16:1 at Position sn-1, provide additional insight into the fate of the MGDG pool produced by MGDα. It is well established in A. thaliana that AtMGD1 and AtDGD1 operate together for the bulk of thylakoid galactolipids, whereas AtMGD2/3 and AtDGD2 operate together when galactolipids contribute to whole-cell membrane lipid remodeling processes (Benning and Ohta 2005). A reasonable hypothesis is that MGDα operates in close association with a specific DGDG synthase (MGDα-associated DGD), very active on any 20:5-containing MGDG produced. In the absence of MGDα, the unused DAG pool is available for the other MGD isoforms. The MGDG thus produced close to MGDβ and/or MGDγ would fuel the syntheses of distinct DGDG pools (Fig. 9).
MGDβ uses a DAG pool enriched in 20:5 imported from the cytosol and contributes to the production of highly unsaturated MGDG species in nitrogen deprivation
Under optimal conditions, MGDβ mutation strongly decreased the accumulation of 20:5-containing MGDG species, except MGDG 20:5-16:4. Interestingly, MGDG 14:0-16:1, 16:1-16:0, and 16:1-16:1 species also decreased. This firstly suggests that under normal conditions, MGDβ mostly uses 20:5-16:0 and 20:5-16:1 DAG, and that the corresponding MGDG products are rapidly desaturated, leading to the accumulation of 20:5-containing MGDG species with a higher unsaturation level of the C16 at Position sn-2 (Figs. 9 and 10). Given the localization of this isoform in the blob/PPM, the 20:5-containing MGDG it produces need to be rapidly transported to the thylakoid membranes where C16 desaturases are located. The role of MGDβ in the production of 20:5-containing MGDG species is also consistent with its subcellular localization, since 20:5 FA is produced outside or at the periphery of the plastid following elongation and desaturation steps (Fig. 9). For the massive incorporation of this FA into the plastid, 20:5- and/or 20:5-containing precursor need to cross the 4 limiting membranes, via the PPM where MGDβ is strategically localized.
The decrease of MGDG 14:0-16:1, 16:1-16:0, and 16:1-16:1 species suggests that these 3 species are also direct products of MGDβ, but that most of them are not directly available for desaturation. The heterologous expression of MGDβ in yeast highlights clearly 32:1 and 32:2 MGDG as major products. Therefore, these saturated/monounsaturated MGDG species might derive from another pool of DAG containing FAs synthesized de novo in the stroma. MGDG 16:1-16:0 and 16:1-16:1 proportions increase in the MGDβ overexpressing lines, while the MGDG 16:2-16:3 and 16:3-16:3 decreased. By this mean, MGDβ can produce more saturated forms of MGDG, remaining away from stromal C16 desaturases, thus compensating for the loss of MGDγ.
Thus, on the one hand, MGDβ appears able to use a DAG pool deriving from the de novo synthesis of FA in the stroma, but forming MGDG accumulating in nonphotosynthetic membranes. On the other hand, MGDβ uses a DAG pool enriched in 20:5 imported from the ER. This isoform in the blob-like structure seems therefore to facilitate bidirectional fluxes of MGDG molecular species deriving from internal and external DAG pools.
In this case also, changes in the DGDG profile highlight a possible association of MGDβ isoform with a specific DGDG synthase (DGD) enzyme. Under normal conditions, only DGDG 16:1-16:1 and 16:1-16:2 proportions decreased in MGDβ KO lines, while DGDG 14:0-16:1 tended to decrease. This would be consistent with the role of MGDβ in the production of MGDG 14:0-16:1 and MGDG 16:1-16:1 for the formation of a specific pool of DGDG. The decrease in DGDG 16:1-16:2 is likely related to that of DGDG 16:1-16:1 from which it can be produced through desaturation by the plastid FAD6 desaturase (Dolch and Marechal 2015). The stable proportion of 20:5-containing DGDG species might be due to compensation effects by MGDα and MGDγ. In line with this, 20:5-containing MGDG are still produced and could be used in priority to maintain 20:5-containing DGDG levels, most importantly by the MGDα-associated DGD pointed above.
Finally, under nitrogen deprivation, at low MGDα level and in the absence of MGDβ, MGDγ could compensate for the lack of MGDG, and for the provision of substrates for DGDG species.
MGDγ uses a DAG pool enriched in 16:0 and 16:1 exported to the EpM/ER, for the production of MGDG species with low unsaturation level in nonplatidial membranes
Under optimal conditions, MGDγ mutation led to a strong decrease in all 14:0-containing MGDG species and in MGDG 16:0-16:0, 16:1-16:0, 16:1-16:1, and 20:5-16:4. Therefore, the main DAG substrates for MGDγ under normal conditions are likely those directly deriving from de novo FA synthesis in the stroma, i.e. 14:0-16:0, 14:0-16:1, 16:0-16:0, 16:0-16:1, and 16:1-16:1, requiring that these DAG are exported by specific systems, at least to the EpM (Fig. 9). Heterologous expression in yeast is consistent with this substrate preference. Given that MGDγ is located in the ER and at the outermost membrane of the plastid, its MGDG products are not directly available for plastid desaturases. Coherently, MGDγ mutation did not have any strong impact on MGDG species containing a C16 with 2 or more unsaturations, except for 14:0-16:2 and 14:0-16:3 species. The strong impact on MGDG 14:0-16:0, 14:0-16:1, 16:0-16:0, 16:1-16:0, and 16:1-16:1 species indicates that MGDγ is responsible for their production and accumulation (Figs. 9 and 10).
Under nitrogen deprivation, MGDγ also led to a very strong decrease of the same molecular species. Coherently, MGDG 14:0-16:1, 16:0-16:0, 16:1-16:0, and 16:1-16:1 species increased in MGDγ overexpressing lines, while MGDG 16:1-16:3, 16:2-16:3, and 16:3-16:3 decreased. The products of MGDγ are therefore able to accumulate in cell membranes where plastid C16 desaturases are not present.
The nature of the membranes where these MGDG species accumulate is intriguing, because this represents an observation that MGDG synthesis in a nonplastidial membrane system would occur in a photosynthetic eukaryote. The MGDγ subcellular localization suggests that MGDG might be synthesized in the EpM and the ER. By extension, this pool of MGDG could virtually be transferred to any other compartment of the endomembrane system, directly or indirectly connected to the ER via membrane continuity (like the oNE) or via vesicular trafficking. MGDG presence outside of the plastid is further supported by the comparison of lipidomes of purified plastids and whole cells of P. tricornutum (Huang et al. 2024). We have no element to comprehend the role of MGDG in extraplastidial membranes, and this represents therefore a critical question to address in diatoms. The role of MGDG in membranes, a lipid class usually characterized by the high unsaturation level of its esterified FAs, is often associated to its capacity to form Hexagonal II phase. However, it is uncertain whether the highly saturated products of MGDγ would organize in a similar way or not. Therefore, both the localization and the distinct nature of MGDγ products should be studied in the future.
MGDγ KO led to a decrease of DGDG 14:0-16:1, 16:0-16:0, 16:1-16:0, and 16:1-16:1, and to a proportional increase in 20:5-containing DGDG species. These changes were moderate in optimal conditions, and very strong in nitrogen deprivation condition. Therefore, we can suppose that a particular DGD enzyme might be dependent on MGDγ for the production of DGDG species with a 14:0 or a C16 in Position sn-1. This hypothetical DGD enzyme should localize at or close to the EpM. Nevertheless, it must be noted that under nitrogen deprivation tuning down MGDα expression, in the MGDβ KO, MGDγ was able to produce 20:5-containing MGDG species, and maintained a sufficient provision of substrates for the production of 20:5-containing DGDG species. Therefore, MGDγ appears to have flexibility in its roles, able to also use DAG 20:5-16:0 and 20:5-16:1 for the production of MGDG species (Fig. 9). This role of MGDγ in the production of 20:5-containing MGDG is however minor, when other MGDs are active.
MGD isoforms and extraplastidial lipid homeostasis
Mutation and overexpression of the 3 MGD isoforms led to changes in nonplastidial lipids, showing a high level of integration between membrane glycerolipid syntheses. The observed phenotypes support the existence of a flux of 16:0/16:1-rich DAG going outward from the stromal side of the plastid to the EpM and ER, where this pool is available for extraplastidial lipids formation by MGDγ. They also highlighted that 20:5-DAG species could be imported inward or produced at the PPM, forming a DAG pool available for MGDβ, and further transported to the stroma, where it joined the 16:0/16:1-rich DAG pool available for plastidial lipid syntheses by MGDα. The results presented here show that the impact of MGDα and MGDβ on extraplastidial lipids rather reflects the establishment of a new equilibrium compensating alterations in DAG pools. In contrast, alterations of MGDγ reveal more spectacular changes, supporting a specific role of this isoform in whole-cell lipid remodeling occurring in response to environmental stress conditions.
MGDγ mutation had a stronger impact on nonplastidial lipid composition. In particular, 16:1 and 16:0 containing PC and DGTA increased both under optimal and nitrogen-deprived conditions. MGDγ being located at the periphery and outside of the plastid, it is possible that it sits near to PC and DGTA synthesis sites. Therefore, it is coherent that the DAG species not used by MGDγ to produce MGDG are used as substrate for the synthesis of PC and DGTA (Fig. 9). Following MGDγ KO, 16:0 amounts available for elongation into 18:0 by Δ0-elongases (Dolch et al. 2017b), the committing step for further desaturations up to 18:4 and 20:5 in phospholipids and DGTA increased. The amounts of PC, PE, and DGTA containing an 18:1 or an 18:2 increased. It seems however that the rates of 18:2-to-18:3 and 18:3-to-18:4 desaturations and 18:4-to-20:4 elongation were slower than the 16:0 export rate.
In P. tricornutum and other photosynthetic stramenopiles, an excess of 16:0 or 16:1 in the cytosol is known to be “absorbed” by an increased production of the storage lipid TAG (Dolch et al. 2017b; Smith et al. 2021). Under optimal growth conditions, MGDγ KO led to strong differences in the profile of several TAG species, with a decrease in 16:0-16:0-16:0 and 16:0-16:0-16:1, and an increase in 16:1-16:1-16:1. This impact on TAG was more pronounced when cells were subjected to a nitrogen starvation. This coupling between MGDγ activity and TAG production highlights a contribution of this MGD isoform to the intense membrane and storage glycerolipid remodeling occurring in response to a variety of environmental factors. The control of a MGDG/TAG balance is known to occur in other algal systems, such as C. reinhardtii containing only one MGD gene (Li et al. 2012; Gu et al. 2021; Iwai et al. 2021). Diatoms appear to contain an isoform of MGD in their extraplastidial membranes specialized in controlling part of this equilibrium.
Comparison of the emergence of specialized MGD isoforms in diatoms and in angiosperms
The emergence of specialized MGD isoforms in diatoms and angiosperm is a fascinating example of evolutionary convergence, as important similarities can be drawn. In both cases, MGD isoforms derived from an ancestral form dedicated to the production of the bulk of MGDG for photosynthetic membranes, following gene duplications and speciation occurring before the main radiation of the phylum. Whereas in angiosperms, Type B MGD could gain function distinct from Type A bound to the iEM, following a relocation to the oEM after a loss of the Ctp, MGDβ/γ could gain additional function following a similar topological relocation. A plausible mechanism lies in the evolution of the original Sp-Ctp bipartite sequence characterizing MGDα, with mutations of the ASAFAP motif arresting the import of the protein precursor into the PPM (MGDβ), followed by a loss of the targeting sequence, leading to a localization in the EpM/ER (MGDγ). In both angiosperms and diatoms, the topological separation of MGD isoforms correlated with the use of DAG pools with distinct acyl profiles, the production of MGDG with distinct acyl profiles, and the specific association with DGD enzymes, also producing DGDG with distinct acyl profiles. In both cases, MGDG dedicated to photosynthetic membrane highlighted the highest level of unsaturations (16:3 and 18:3 in angiosperms, up to 16:4 and 20:5 in diatoms).
In both cases, the emergence of “novel” MGD isoforms (Type B MGDs in angiosperm and MGDβ/γ in diatoms) strengthens the integration of the organelle within the cell and implies intense trafficking of lipids. On the one hand, these novel isoforms facilitate the incorporation of DAG molecular species deriving from extraplastidial precursors, via the so-called eukaryotic pathway in angiosperms (Browse et al. 1986), and the still unresolved Ω-pathway in diatoms (Petroutsos et al. 2014). The nature of the imported precursor(s) may differ, as well as the mechanism involved. On the other hand, without sparing MGDG required for photosynthesis, novel MGD isoforms could be involved in the specific production of galactolipids, for novel function, like lipid remodeling occurring under environmental stress. An example is the role of AtMGD2/3 in Arabidopsis upon phosphate shortage (Kobayashi et al. 2009a) and that of MGDγ and to a lesser extent, MGDβ, under a lack of nitrogen. In both cases, the emergence of specialized isoforms correlated with a remarkable ecological success of each phylum populating a variety of terrestrial and marine habitats, marked by constant environmental changes and stress conditions.
Among the differences, the roles played by Types A and B do not overlap in angiosperms: a KO of Type A is lethal, and cannot be compensated by Type B MGDs (Kobayashi et al. 2007a, 2007b). Furthermore, Type B MGDs appear as specific to some environmental conditions, like phosphate starvation, performing function that Type A cannot compensate. In contrast, the multigenic family of MGDs in diatoms showed strong overlapping activities, preventing the loss of integrity of photosynthetic membranes. Any defect of 1 isoform is nearly completely compensated by the 2 others. Even under nitrogen shortage, when MGDα level is drastically reduced, MGDγ compensates for the loss of MGDβ and vice versa. This does not make the dissection of respective roles easy, but this highlights the powerful flexibility of membrane glycerolipid metabolism in diatoms, despite their very sophisticated membrane compartmentation.
Eventually, diatoms introduced an important milestone in the evolution of MGD proteins, unreported to date in any other phylum, marked by the targeting in systems ontogenetically unrelated to membranes deriving from a primary chloroplast, i.e. in the PPM (MGDβ) supposed to derive from the red alga endosymbiont plasma membrane, and the EpM and ER (MGDγ). MGDG has been assumed to be the hallmark of chloroplastic membranes, and in textbooks it is referred to as such. We do not have yet the conceptual framework to address MGDG role outside of the plastid, but this work provides genetic tools to address this fascinating question. It is important to note, that whereas the biogenesis of the iEM and oEM could be supposed to follow that of the primary chloroplast envelope in angiosperms and green algae, and whereas the biogenesis of the EpM could be considered to derive from the ER, no lipid-synthesis protein could be located to the PPM until now. This work provided therefore clues on the biogenesis of all membrane systems of a secondary plastid, with biological processes unsuspected until now, paving the way for future works.
Materials and methods
Primary sequence analyses
P. tricornutum MGD genes were identified based on protein sequence homology using BLAST, with Arabidopsis (A. thaliana) main isoform, i.e. AtMGD1 (NP_194906), as query. The identified locus tags of P. tricornutum were named MGDα (Phatr3_J14125, Uniprot; XM_002181649.1, RefSeq; J14125, EnsemblProtists), MGDβ (Phatr3_J43938; XM_002186319; J54168), and MGDγ (Phatr3_J9619; XM_002176764.1; J9619), respectively. Target sequence predictions were performed considering multiple START/Methionine codons present in the N-terminal part of putative reading frames. Sps were predicted with the SignalP-6.0 tool, with default parameters (Emanuelsson et al. 2007; Almagro Armenteros et al. 2019). Ctps were predicted using WolF PSORT (Horton et al. 2007) and ChloroP—1.1 (Emanuelsson et al. 2007), with default parameters. Specific bipartite presequence was predicted using the HECTAR software specifically designed to detect Sp-Ctp in stramenopiles (Gschloessl et al. 2008).
Protein structure predictions
The software Phyre2 (Protein Homology/analogY Recognition Engine V 2.0; Kelley et al. 2015) and AlphaFold (Jumper et al. 2021; Jumper and Hassabis 2022) run from ChimeraX (Mirdita et al. 2022) were used to model P. tricornutum MGD sequences, which were then viewed on PyMol (Janson et al. 2017). AtMGD1 structure (4X1T- wwPDB consortium, 2019) (Rocha et al. 2016) was used to build Phyre2 PtMGDs models. For each P. tricornutum MGD, the peptide sequences used as input were those deduced after cDNA sequencing and Sp prediction, as represented in Fig. 3E. These correspond to the following sequences: for MGDα, the sequence retrieved from Phatr3_J14125.t1 starting from the second methionine (MCKL to TRTS, 460 amino acid); for MGDβ, the whole sequence from Phatr3_J54168.t1 (MVWS to LNNK, 559 amino acid); for MGDγ, the whole sequence from Phatr3_J9619.t1 (MATG to EPSR, 618 amino acids).
MGD phylogenetic analysis
All MGD protein sequences used for phylogenetic analysis were retrieved from the NCBI (Supplementary Table S1) after a protein BLAST search using MGDα (XM_002181649.1), MGDβ (XM_002186319), and MGDγ (XM_002176764.1) as queries. Sequences from different phyla covering the biodiversity of plastid-containing eukaryotes, including Archaeplastida and Stramenopiles were retained, with at least 2 species per phylum. The dataset was manually curated to validate the presence of full-length sequences with conserved MGD domains including residues at the active site. Alignment was performed with a customized pipeline in NGphylogeny.fr (Lemoine et al. 2019) using the MUSCLE v3.8.1551 software (Edgar 2004). The ambiguously aligned regions were curated using the Block Mapping and Gathering with Entropy (BMGE v1.12_1) software (Criscuolo and Gribaldo 2010) implemented in NGphylogeny.fr using default settings. Preliminary phylogenetic trees were inferred using FastME with 2,500 bootstrap pseudoreplicates. MEGA X v10.0.5 software (Kumar et al. 2018) was fed with the aligned and curated dataset. The best evolutionary model was evaluated, and a maximum likelihood phylogenetic analysis was performed. To define the best evolutionary model, MEGA X was used to compare the 56 models implemented. The LG + G model (Le et al. 2008) was chosen (lowest Bayesian Information Criterion score). The phylogeny was inferred by maximum likelihood with 5,000 bootstrap pseudoreplicates. The tree with the highest log likelihood (−12,414.63) was retained. Initial trees for the heuristic search were obtained by applying the BioNJ method (Gascuel 1997) to a matrix of pairwise distances estimated using a JTT model. A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 0.7081)]. The best phylogenetic tree was then retrieved in Newick format and visualized by iToL (v5) (Letunic and Bork 2021).
Cultivation of P. tricornutum cells
P. tricornutum (Pt1) Strain 8.6 CCAP 1055/1 (CCMP2561; Culture Collection of Marine Phytoplankton, now known as NCMA: National Center for Marine Algae and Microbiota) was used to generate overexpressing and KO lines. Cells were maintained and grown in a modified ESAW (Enriched Seawater, Artificial Water) medium [NaCl 362.7 mm; Na2SO4 25 mm; KCl 8.03 mm; NaHCO3 2.067 mm; KBr 0.725 mm; H3BO3 0.372 mm; NaF 0.0657 mm; MgCl2 47.18 mm; CaCl2 9.134 mm; SrCl2 0.082 mm; Na2-glycerophosphate 21.8 µm; Na2SiO3 105.6 µm; disodium ethylenediaminetetraacetate dehydrate (Na2EDTA) 14.86 µm; Fe(NH4)2(SO4)2 5.97 µm; FeCl3 0.592 µm; MnSO4 2.42 µm; ZnSO4 0.254 µm; CoSO4 0.0569 µm; Na2MoO4 0.52 µm; H3BO3 61.46 µm; Na2SeO3 10 nm; biotin (Vitamin H) 8.18 nm; cobalamin (Vitamin B12) 2.94 nm; thiamine (Vitamin B1) 0.594 µm; Falciatore et al. 2000] using 10 times enriched nitrogen and phosphate sources (10N10P containing 5.49 mm NaNO3 and 0.224 mm NaH3PO4; Abida et al. 2015). Culture in liquid media was performed at 20 °C, on a 12:12 light (40 μmol photons m−2 s−1) dark cycle, under gentle agitation, in an INFORS HT Multitron Pro incubator. Cell concentration was kept between 0.5 × 106 and 10 × 106 cell mL−1. For nitrogen starvation, P. tricornutum cells were cultivated in triplicates in 50 mL of ESAW 10N10P in 250 mL flasks until a concentration of 3 to 4 × 106 cell mL−1. After centrifugation at 1,500×g for 10 min and removal of supernatant, cells were washed with 50 mL of ESAW 0N10P medium, and transferred into to 250 mL flaks containing 50 mL of ESAW 0N10P, at a concentration of 2.5 to 3 × 106 cell mL−1. Cultures on solid medium were performed with ESAW 10N10P complemented with Agar (1% m/v), at 20 °C under continuous light (50 µmol photons m−2 s−1), in an MLR-352-PE Climate Chamber. To guarantee axenic conditions, all media were supplemented with carbenicillin disodium salt (0.237 µm). For culture of mutant lines transformed with CRISPR-Cas9 vectors and of overexpressing lines, zeocin (Invitrogen; 0.07 µm) was added to the media. For each analysis, 3 independent lines were cultivated. Growth was evaluated by cell counting using a TECAN infinite M1000Pro plate reader and determined following the equation y = 0.01834 x + 0.03758 with y the absorbance at 730 nm and x the number of cells in million per milliliter (Conte et al. 2018).
DNA and RNA extractions
Genomic DNA extraction from P. tricornutum was performed on a cell pellet corresponding to 100 × 106 cells, suspended in 500 µL of lysis buffer (250 mm Tris pH 8.2, 100 mm EDTA, 2% SDS v/w, 350 mm NaCl) and transferred to a 1.5 mL tube. The sample was incubated 15 s at 60 °C under agitation (100×g) using a thermobloc. Five hundred microliters of cold phenol:chloroform:isoamyl alcohol (25:24:1) were added to the sample and homogenized gently. The sample was centrifuged at 13,000×g at 4 °C for 10 min. Three hundred microliters of the upper aqueous phase was collected. A second extraction was performed by addition of 1 volume of chloroform:isoamyl alcohol (24:1), and 200 µL of the upper aqueous phase was collected. DNA was precipitated with 30 µL of sodium acetate 3 m pH 5 and 3 volumes of ethanol. After a 20 min incubation at −20 °C and a 10 min centrifugation at 13,000×g at 4 °C the precipitated DNA was washed with 700 µL of ethanol 70% v/v, centrifuged at 13,000×g at 4 °C for 5 min and air dried at room temperature. DNA was resuspended in DNAse-free water. Concentration and purity were evaluated using a NanoDrop2000 (Thermo Fisher Scientific). For RNA extraction from P. tricornutum cells, 1.5 mL of TRI Reagent (Sigma-Aldrich) was added to a frozen cell pellet corresponding to 150 × 106 cells. The sample was first vigorously mixed and incubated 5 min at 60 °C, a process repeated twice for a thorough cell lysis. Three hundred microliters of chloroform were added to the sample then vigorously mixed by tube inversion. The sample was incubated at room temperature for 15 min before centrifugation at 11,000×g at 4 °C for 15 min for phase separation. About 600 µL of the upper aqueous phase was transferred to a Phasemaker tube (Thermo Fisher Scientific). A volume of 1-bromo-3-chloropropane corresponding to one-fifth of the transferred aqueous phase was added and mixed vigorously. The sample was incubated for 3 min at room temperature before centrifugation at 16,000×g at 4 °C for 5 min. After centrifugation, 1 mL of the aqueous phase was collected. RNA was precipitated by addition of 1 volume of isopropanol and a 30 min incubation at room temperature. RNA was pelleted down by centrifugation at 11,000×g at 4 °C for 10 min and further washed in 1 mL of ethanol 75% v/v, followed by a centrifugation at 7,500×g at 4 °C for 10 min. The pellet was left to air dry for 30 min at room temperature. RNA was resuspended in 35 µL of RNA-free water. A first DNAse treatment was applied using the Invitrogen Ambion TURBO DNA-free kit following manufacturer's instructions. Cleaning and purification of the sample was then achieved using the RNeasy MinElute Cleanup kit (Qiagen) following manufacturer's instructions. RNA concentration and quality was assessed using a NanoDrop2000 (Thermo Fisher Scientific) and by electrophoresis. RNA was stored at −80 °C until use.
MGD gene expression analysis by RT-qPCR
This method is detailed in Supplementary material.
Heterologous expression of MGD isoforms in yeast
This method is detailed in Supplementary material.
Construction of MGD overexpressing lines
Full-length coding sequence of MGDα, MGDβ, and MGDγ genes were amplified by PCR using cDNA derived from P. tricornutum as template and primers detailed in Supplementary Table S4. The PCR products were cloned into pEASY-T1 simple (TransGen, Beijing) vector for DNA sequencing. Sequence-confirmed MGDα, MGDβ, and MGDγ genes were excised from T-cloning vector with BamHI and XhoI in the case of MGDα, BamHI and SalI for MGDβ, and NheI and SalI for MGDγ. Each gene was in-frame inserted upstream the eGFP coding sequence in the multi cloning site of pPha-CG vector (GenBank AF219942; Zaslavskaia et al. 2000). The obtained pPhaT1 vectors harboring MGDα–eGFP, MGDβ–eGFP, and MGDγ–eGFP were linearized with ScaI and introduced in P. tricornutum cells by electroporation as previously described (Zhang and Hu 2014). P. tricornutum PetC (Liu et al. 2016) and symbiont Derlin1-2 (sDer1-2; Hempel et al. 2009) were used as markers of thylakoid and PPM, respectively. For colocalization studies, 3 pairs of linearized plasmids, pPhaT1–MGDα–eGFP and pPhaT1–PetC–mRFP, pPhaT1–MGDβ–eGFP and pPhaT1–sDer1-2–mRFP, and pPhaT1–MGDγ–eGFP and pPhaT1-mRFP were cotransformed into P. tricornutum, respectively. Fluorescence of eGFP and plastid autofluorescence were excited at 488 nm and were detected with 2 photomultiplier tubes at 500 to 520 and 625 to 720 nm, respectively, using a Leica TCS SP8 laser scanning confocal microscope. Gain was set at 800. The mRFP fluorescence was excited at 552 nm and detected at 580 to 600 nm.
Construction of CRISPR-Cas9 single and double KO lines
Single guide RNAs (sgRNA) were designed using the PhytoCrispex online tool (Rastogi et al. 2016), choosing NGG as Protospacer Adjacent Motif sequence, and allowing CRISPR targets to start with any nucleotide. sgRNA were selected based on their proximity to START codons and active site residues. sgRNA forward and reverse sequences preceded with the short nucleotide sequences TCGA and AAAC, respectively, are shown in Supplementary Table S3. The sgRNA sequences were inserted in the pKSdiaCas9_sgRNA-zeo vector, kindly provided by Cécile Giustini (LPCV, Grenoble), derived from pKS diaCas9_sgRNA vector (Addgene). P. tricornutum cells were transformed with corresponding vectors by particle bombardment as described previously (Falciatore et al. 1999; Dolch et al. 2017a). For MGDβ/MGDγ double KO, mgdβc1 single KO was chosen as genetic background. The sgRNA 619 h (Supplementary Table S3) was cloned between the 2 BsaI sites of the pYS31 episome, derived from PtPuc3_diaCas9_sgRNA episome (Addgene) with a blasticidin resistance cassette instead of a zeocin. The PtPuc3_diaCas9_sgRNA was digested by SalI and StuI in order to excise the zeocin resistance gene. The Blasticidin S deaminase (BSD) coding sequence, conferring the resistance to blasticidin, was amplified using the primers olYS40 (5′-GAAAAATTAACCAAGATGGCCAAGCCTTTGTCTCA-3′) and olYS41 (5′CCCAGATCTCCGAGGTCAGCCCTCCCACACATAAC-3′). Fifty nanograms of the vector were used for Gibson assembly with a 2-fold molar excess of the BSD fragment. The episome was transformed by bacterial conjugation (Karas et al. 2015). Colony PCR (Phire Plant Direct PCR Master Mix, F160; Thermo Fisher Scientific) was conducted to analyze the mutation profile by sequencing. Primers were designed for each sgRNA to amplify a region of about 500 nucleotides surrounding the cutting site of the Cas9 protein (Supplementary Table S3). In some rare cases, no amplification was obtained and primers amplifying a larger region were used to reveal a possible very large deletion. Three different kinds of edition profiles were usually obtained: either no edition (pure WT colony), several different editions (mosaic colony), or 1 edition only (pure mutant colony). For the analysis of mosaic colonies, TIDE (Tracking of Indels by Decomposition v3.3.0; Brinkman et al. 2014) and ICE (Inference of CRISPR Edits v2, Synthego.com) online software were used to decompose the chromatograms and estimate the edition profile of the colony. Whenever a fraction of cells within the colony presented insertions/deletions (INDELs), a strain isolation was attempted. The equivalent of 200 cells from a mosaic colony was spread on a plate with carbenicillin and zeocin. After 3 to 6 wks, the same steps of PCR and sequencing analyses were conducted. If pure edited colonies were obtained, the screening process was stopped. Otherwise, subsequent isolation attempts were performed.
Bacterial conjugation
This method is detailed in Supplementary material.
Glycerolipid analyses
Glycerolipids were extracted according to the Folch method (Folch et al. 1957), as described previously (Abida et al. 2015). One-tenth of total glycerolipid extract was used to quantify the total FA content, after methanolysis. Lipids were solubilized with 1 mL of chloroform and a 100 µL aliquot was transferred to a 10 mL crim cap vial (Gerstel). Methanolysis was performed by a MultiPurpose Sampler (MPS, Gerstel). Briefly, 5 µL of 15:0 FA (an FA with a 15-carbon chain length) at 1 mg mL−1 was added as internal standard, and 3 mL of methanolysis buffer (methanol/sulfuric acid; 40:1 v/v) was added, vigorously mixed, and incubated at 80 °C for 1 h, yielding FA methyl esters (FAMEs). Reaction was stopped with the addition of 3 mL of distilled water and 3 mL of hexane. FAME retrieved from the upper phase were suspended in hexane and analyzed by gas chromatography coupled to Flame Ionization Detection (GC-FID, Perkin Elmer Clarus 580 equipped with a 30 m long cyanopropyl polysilphenesiloxane column, 0.22 mm diameter). FAMEs were identified by comparison of their retention times with those of standards (Sigma) and quantified by the surface peak method using 15:0 for calibration. Extraction and quantification were performed with 3 biological replicates. Glycerolipids were then analyzed and quantified by high-pressure liquid chromatography–tandem MS (HPLC–MS/MS), as previously described (Dolch et al. 2017a), with appropriate standard lipids (Jouhet et al. 2017). Lipid extracts corresponding to 25 nmol of total FAs were dissolved in 100 µL of chloroform/methanol (2/1, v/v) containing 125 pmol of each internal standard. PE 18:0-18:0 and DAG 18:0-22:6 from Avanti Polar Lipid, and SQDG 16:0-18:0 extracted from spinach thylakoid (Deme et al. 2014) and hydrogenated (Buseman et al. 2006) were used as internal standards. Lipids were then separated by HPLC and quantified by MS/MS. Lipid classes were separated using an Agilent 1200 HPLC system using a 150 mm × 3 mm (length × internal diameter) 5 µm diol column (Macherey-Nagel), at 40 °C. The mobile phases consisted of hexane/isopropanol/water/1 m ammonium acetate, pH 5.3 (625/350/24/1, v/v) (A) and isopropanol/water/1 m ammonium acetate, pH 5.3 (850/149/1, v/v) (B). The injection volume was 20 µL. After 5 min, the percentage of B was increased linearly from 0% to 100% in 30 min and kept at 100% for 15 min. This elution sequence was followed by a return to 100% A in 5 min and an equilibration for 20 min with 100% A before the next injection, leading to a total runtime of 70 min. The flow rate of the mobile phase was 200 µL.min−1. The distinct glycerophospholipid classes were eluted successively as a function of the polar head group. Mass spectrometric analysis was performed on a 6460 triple quadrupole mass spectrometer (Agilent) equipped with a Jet stream electrospray ion source under following settings: drying gas heater at 260 °C, drying gas flow at 13 L min−1, sheath gas heater at 300 °C, sheath gas flow at 11 L min−1, nebulizer pressure at 25 psi, capillary voltage at ±5,000 V and nozzle voltage at ±1,000 V. Nitrogen was used as collision gas. The quadrupoles Q1 and Q3 were operated at widest and unit resolution, respectively. PC and DGTA analyses were carried out in positive ion mode by scanning for precursors of m/z 184 and 236, respectively, at a collision energy (CE) of 34 and 52 eV. SQDG analysis was carried out in negative ion mode by scanning for precursors of m/z −225 at a CE of −56 eV. PE, phosphatidylinositol (PI), PG, MGDG, and DGDG measurements were performed in positive ion mode by scanning for neutral losses of 141, 277, 189, 179, and 341 Da at CEs of 20, 12, 16, 8, and 8 eV, respectively. DAG and TAG species were identified and quantified by multiple reaction monitoring (MRM) as singly charged ions [M + NH4]+ at a CE of 16 and 22 eV, respectively. Quantification was done for each lipid species by MRM with 50 ms dwell time with the various transitions previously recorded (Abida et al. 2015). Mass spectra were processed using the MassHunter Workstation software (Agilent) for identification and quantification of lipids. Lipid amounts (pmol) were corrected for response differences between internal standards and endogenous lipids and by comparison with a qualified control (Jouhet et al. 2017).
Epifluorescence microscopy
Cells were observed using an epifluorescence microscope (Zeiss AxioScope A1) equipped with a Zeiss AxioCam MRc. Images were captured using a Zeiss EC Plan Neofluar 100×/1.3 oil immersion objective. Chlorophyll autofluorescence and Nile Red fluorescence in lipid droplets were visualized with Zeiss filter set 16 (BP 485/20, FT510, LP515), as described previously (Dolch et al. 2017a).
Laser scanning confocal microscopy
Confocal microscopy was performed with a microscope Zeiss LSM880 equipped with a 63×/1.4 oil-immersed Plan-Apochromat objective, running Zen 2.3 SP1 acquisition software (Platform µLife, IRIG, LPCV). Chlorophyll autofluorescence and eGFP fluorescence were excited using the 488 nm ray of an Argon Multiline laser, and were detected at 600 to 730 and 500 to 531 nm, respectively, using a GaAsP detector. Laser intensity was set at 5% for visualization of chlorophyll, and at 2%, 3%, and 5% for eGFP visualization according to the eGFP signal intensity in each cell. On all images, gain was set at 800 for chlorophyll and for MGDβ, 850 for MGDγ and 900 for MGDα. “Pseudo brightfield” images were acquired in parallel by differential interference contrast, using the 488 nm laser ray at 0.6% as light source and a photomultiplier tube detector for transmitted light, with gain set at 500. Z-stacks containing consecutive images with a distance of 0.47 nm were obtained for each cell. For colocalization analyses, fluorescence of eGFP and plastid autofluorescence were excited at 488 nm and were detected with 2 photomultiplier tubes at 500 to 520 and 625 to 720 nm, respectively, using a Leica TCS SP8 laser scanning confocal microscope. The mRFP fluorescence was excited at 552 nm and detected at 580 to 600 nm.
Transmission electron microscopy
Samples were prepared as previously described (Flori et al. 2018). Ultrathin sections (50 to 70 nm) were prepared with a diamond knife on a PowerTome ultramicrotome (RMC products, Tucson, AZ, USA) and collected on 200 µm carbon-coated gold grids. Samples were visualized by STEM using a MERLIN microscope (Zeiss, Oberkochen, Germany) set up at 30 kV and 240 pA or using an FEI tecnai OSIRIS microscope (Hillsboro, OR, USA) set up at 200 kV and ∼300 pA.
Accession numbers
Species and associated MGD protein sequences used for phylogeny (NCBI accession numbers): Vitrella brassicaformis (CEM14063.1, CEM34604.1); Gregarina niphandrodes (XP_011130274.5); Pseudo-nitzschia multistriata (VEU37191.1, VEU39505.1); Fragilariopsis cylindrus (OEU16730.1, OEU06991.1); Fistulifera solaris (GAX23913.1, GAX10983.1, GAX09439.1); P. tricornutum (XP_002186355, XP_002176800, XP_002181685), T. pseudonana (XP_002295865, XP_002293576.1, XP_002294242); Ectocarpus siliculosus (CBJ28381.1, CBJ28372.1, CBN79326.1); Aureococcus anophagefferens (XP_009033523.1, XP_009038839.1, XP_009035780.1); Microchloropsis/Nannochloropsis gaditana (XP_005855249.1); Coccomyxa subellipsoidea (XP_005651388.1); Chlorella sorokiniana (PRW45643.1); Ostreococcus tauri (OUS42062.1); C. reinhardtii (PNW74102.1); Monoraphidium neglectum (XP_013903204.1); Raphidocelis subcapitata (GBF88428.1); A. thaliana (NP_194906.1, NP_565352.1, NP_568394.2); Coffea arabica (XP_027069738.1, XP_027084801.1); Spinacia oleracea (XP_021867203.1, XP_021852153.1); Nymphaea colorata (XP_031495579.1, XP_031480803.1); Amborella trichopoda (XP_006852865.1, XP_006845407.1); Brachypodium distachyon (XP_010238179.1, XP_003573675.2, XP_003570331.1); Oryza sativa (XP_015611851.1, XP_015649135.1, XP_015627712.1); Physcomitrium patens (XP_024403181.1, XP_024404076.1); Selaginella moellendorffii (XP_024533920.1); Cyanidioschyzon merolae (XP_005536420.1); Cyanidiococcus yangmingshanensis (KAF6002292.1); Gracilariopsis chorda (PXF49423.1, PXF42956.1, PXF46603.1); and Blastochloris viridis (WP_055036643.1). P. tricornutum MGD gene accession numbers: MGDα (PHATRDRAFT_14125, NCBI; Phatr3_J14125, Ensembl); MGDβ (PHATR_43938, NCBI; Phatr3_J54168, Ensembl); MGDγ (PHATRDRAFT_9619, NCBI; Phatr3_J9619, Ensembl).
Supplementary Material
Acknowledgments
The authors thank Guillaume Allorent, Baptiste Doussot, and Cécile Giustini for their technical assistance and Sylvaine Roy for lipid data analysis software development.
Contributor Information
Nolwenn Guéguen, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Yannick Sérès, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Félix Cicéron, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Valérie Gros, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Grégory Si Larbi, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Denis Falconet, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Etienne Deragon, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Siraba D Gueye, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Damien Le Moigne, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Marion Schilling, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Mathilde Cussac, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Dimitris Petroutsos, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France; Department of Organismal Biology, Uppsala University, 75236 Uppsala, Sweden.
Hanhua Hu, Key Laboratory of Algal Biology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China.
Yangmin Gong, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan 430062, China; Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture, Oil Crops Research Institute of Chinese Academy of Agricultural Sciences, Wuhan 430062, China.
Morgane Michaud, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Juliette Jouhet, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Juliette Salvaing, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Alberto Amato, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Eric Maréchal, Laboratoire de Physiologie Cellulaire et Végétale, Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement, Centre National de la Recherche Scientifique, Commissariat à l’Energie Atomique et aux Energies Alternatives, Université Grenoble Alpes, IRIG, CEA-Grenoble, 17 rue des Martyrs, 38000 Grenoble, France.
Author contributions
N.G. has performed the most experimental works. Y.S. has generated multiple KO lines and contributed to imaging and lipidomic analyses. F.C. has provided guidance on CRISPR-Cas9 strategy. V.G. and G.S.L. have provided technical assistance on lipidomic analyses and STEM, respectively. D.P. has contributed to the design of photosynthetic measurements and their analyses. A.A., E.D., S.D.G., and M.M. have provided assistance on heterologous expression in yeast. H.H. and Y.G. have developed overexpressing lines. D.L.M. and J.S. have contributed to confocal imaging. J.J. has provided guidance in the analysis of lipidomic profiles. M.S and M.C. have performed lipid analyses. J.S., A.A., and E.M. have contributed to the design of experiments and their analyses. All authors have contributed to the writing of the manuscript.
Supplementary data
The following materials are available in the online version of this article.
Supplementary Figure S1. Schematic view of MGD genes in P. tricornutum.
Supplementary Figure S2. Phylogenetic analysis of MGDs from plastid-containing eukaryotes.
Supplementary Figure S3. Multiple sequence alignment of A. thaliana AtMGD1, AtMGD2, and AtMGD3 and P. tricornutum MGDα, MGDβ, and MGDγ proteins.
Supplementary Figure S4. Protein models of P. tricornutum MGDs.
Supplementary Figure S5. Heterologous expression of P. tricornutum MGD isoforms in yeast.
Supplementary Figure S6. Visualization of MGD–eGFP proteins overexpression by immunoblot.
Supplementary Figure S7. Multiple observations of MGD isoforms fused to eGFP in P. tricornutum cells.
Supplementary Figure S8. Colocalization of MGD isoforms fused to eGFP with subcellular protein makers fused to RFP.
Supplementary Figure S9. Selected KO MGDα, MGDβ, and MGDγ mutants, generated by CRISPR-Cas9 editing.
Supplementary Figure S10. The analysis of loss of heterozygosity and stability of MGD KO lines.
Supplementary Figure S11. Growth curves of MGD mutant and overexpressing strains compared with the WT.
Supplementary Figure S12. Effect of MGDs mutations on photosynthetic properties under high and moderate light stresses.
Supplementary Figure S13. Cell morphology of KO and overexpressing lines.
Supplementary Figure S14. P. tricornutum cell ultrastructure in MGD overexpressing lines.
Supplementary Figure S15. Quantitative analysis of FA and glycerolipid content in MGD KO mutants.
Supplementary Figure S16. Quantitative analysis of FA and glycerolipid content in MGD overexpressing lines.
Supplementary Figure S17. Impact of the overexpression of MGD isoforms on the molecular species constituting MGDG, DGDG, SQDG, and PG in P. tricornutum.
Supplementary Figure S18. Impact of the overexpression of MGD isoforms on the molecular species of endomembrane glycerolipids in P. tricornutum.
Supplementary Figure S19. Impact of MGDα mutations on the molecular species constituting PC, PE, DGTA, and DAG in P. tricornutum.
Supplementary Figure S20. Impact of MGDβ mutations on the molecular species constituting PC, PE, DGTA, and DAG in P. tricornutum.
Supplementary Figure S21. Impact of MGDγ mutations on the molecular species constituting PC, PE, DGTA, and DAG in P. tricornutum.
Supplementary Figure S22. MGD relative gene expression in mutant lines compared with the WT, grown 3 d in 10N10P medium.
Supplementary Figure S23. WT and mutant P. tricornutum growth during nitrogen limitation.
Supplementary Figure S24. Accumulation of nonpolar lipids during nitrogen limitation.
Supplementary Figure S25. Observation of cell morphology and TAG accumulation by epifluorescence microscopy.
Supplementary Figure S26. Quantitative analysis of FA and glycerolipid content in MGD mutant lines upon nitrogen starvation.
Supplementary Figure S27. Impact of MGD mutations on the molecular species constituting PC, PE, DGTA, DAG, and TAG in nitrogen-deprived P. tricornutum cells.
Supplementary Figure S28. Selected MGDβ/MGDγ double KO mutants, generated by CRISPR-Cas9 editing.
Supplementary Figure S29. Growth curves of MGDβ/MGDγ double mutants compared with the WT.
Supplementary Figure S30. Quantitative analysis of glycerolipid content in MGDβ/MGDγ double mutant overexpressing lines.
Supplementary Figure S31. Impact of MGDβ/MGDγ double mutations on MGDG, DGDG, SQDG, and PG molecular species.
Supplementary Figure S32. Impact of MGDβ/MGDγ double mutations on PC, DGTA, PE, and TAG molecular species.
Supplementary Table S1. P. tricornutum MGD entries in NCBI, EnsemblProtist, and Uniprot databases.
Supplementary Table S2. Taxonomic classification of the MGD proteins used for phylogenetic analyses.
Supplementary Table S3. Single guide RNA list.
Supplementary Table S4. Primers list.
Funding
N.G. was supported by a PhD grant from INRAE (BAP program). A.A., F.C., E.D., N.G., S.D.G., V.G, J.J., D.P., G.S.L., Y.S., and E.M. were supported by the French National Research Agency (GRAL Labex ANR-10-LABEX-04, EUR CBS ANR-17-EURE-0003, ANR AlpAlga ANR-20-CE02-0020, ANR DIM ANR-21-CE02-0021, PEPR Algadvance A-22-PEBB-0002, Glyco@Alps Cross-Disciplinary Program; Grant ANR-15-IDEX-02). H.H. and Y.G. were supported by the National Natural Science Foundation of China (41976119, 91751117). A.A., E.M., H.H., and Y.G. were supported by a CEA-CAS bilateral program. Imaging was performed at the µLife Platform of IRIG. Lipid analyzes were performed at the LIPANG platform supported by the Rhône-Alpes Region, the FEDER funds, and French National Research Agency (GRAL Labex ANR-10-LABEX-04, EUR CBS ANR-17-EURE-0003).
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplemental material.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Abida H, Dolch LJ, Mei C, Villanova V, Conte M, Block MA, Finazzi G, Bastien O, Tirichine L, Bowler C, et al. Membrane glycerolipid remodeling triggered by nitrogen and phosphorus starvation in Phaeodactylum tricornutum. Plant Physiol. 2015:167(1):118–136. 10.1104/pp.114.252395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. Signalp 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019:37(4):420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- Apt KE, Zaslavkaia L, Lippmeier JC, Lang M, Kilian O, Wetherbee R, Grossman AR, Kroth PG. In vivo characterization of diatom multipartite plastid targeting signals. J Cell Sci. 2002:115(21):4061–4069. 10.1242/jcs.00092 [DOI] [PubMed] [Google Scholar]
- Awai K, Marechal E, Block MA, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J. Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2001:98(19):10960–10965. 10.1073/pnas.181331498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azadi-Chegeni F, Thallmair S, Ward ME, Perin G, Marrink SJ, Baldus M, Morosinotto T, Pandit A. Protein dynamics and lipid affinity of monomeric, zeaxanthin-binding LHCII in thylakoid membranes. Biophys J. 2022:121(3):396–409. 10.1016/j.bpj.2021.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Ohta H. Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem. 2005:280(4):2397–2400. 10.1074/jbc.R400032200 [DOI] [PubMed] [Google Scholar]
- Benoiston AS, Ibarbalz FM, Bittner L, Guidi L, Jahn O, Dutkiewicz S, Bowler C. The evolution of diatoms and their biogeochemical functions. Philos Trans R Soc Lond B Biol Sci. 2017:372(1728):20160397. 10.1098/rstb.2016.0397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billey E, Hafidh S, Cruz-Gallardo I, Litholdo CG, Jean V, Carpentier MC, Picart C, Kumar V, Kulichova K, Marechal E, et al. LARP6C orchestrates posttranscriptional reprogramming of gene expression during hydration to promote pollen tube guidance. Plant Cell. 2021a:33(8):2637–2661. 10.1093/plcell/koab131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billey E, Magneschi L, Leterme S, Bedhomme M, Andres-Robin A, Poulet L, Michaud M, Finazzi G, Dumas R, Crouzy S, et al. Characterization of the bubblegum acyl-CoA synthetase of Microchloropsis gaditana. Plant Physiol. 2021b:185(3):815–835. 10.1093/plphys/kiaa110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botte C, Jeanneau C, Snajdrova L, Bastien O, Imberty A, Breton C, Marechal E. Molecular modeling and site-directed mutagenesis of plant chloroplast monogalactosyldiacylglycerol synthase reveal critical residues for activity. J Biol Chem. 2005:280(41):34691–34701. 10.1074/jbc.M505622200 [DOI] [PubMed] [Google Scholar]
- Boudiere L, Michaud M, Petroutsos D, Rebeille F, Falconet D, Bastien O, Roy S, Finazzi G, Rolland N, Jouhet J, et al. Glycerolipids in photosynthesis: composition, synthesis and trafficking. Biochim Biophys Acta. 2014:1837(4):470–480. 10.1016/j.bbabio.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008:456(7219):239–244. 10.1038/nature07410 [DOI] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014:42(22):e168. 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the 16-3 plant Arabidopsis thaliana. Biochem J. 1986:235(1):25–31. 10.1042/bj2350025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmann L, Haarmann R, Mirus O, Bredemeier R, Hempel F, Maier UG, Schleiff E. Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. J Biol Chem. 2010:285(9):6848–6856. 10.1074/jbc.M109.074807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buseman CM, Tamura P, Sparks AA, Baughman EJ, Maatta S, Zhao J, Roth MR, Esch SW, Shah J, Williams TD, et al. Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol. 2006:142(1):28–39. 10.1104/pp.106.082115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma. 2018:255(1):297–357. 10.1007/s00709-017-1147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte M, Lupette J, Seddiki K, Mei C, Dolch LJ, Gros V, Barette C, Rebeille F, Jouhet J, Marechal E. Screening for biologically annotated drugs that trigger triacylglycerol accumulation in the diatom Phaeodactylum. Plant Physiol. 2018:177(2):532–552. 10.1104/pp.17.01804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corteggiani Carpinelli E, Telatin A, Vitulo N, Forcato C, D'Angelo M, Schiavon R, Vezzi A, Giacometti GM, Morosinotto T, Valle G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol Plant. 2014:7(2):323–335. 10.1093/mp/sst120 [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010:10(1):210. 10.1186/1471-2148-10-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi F, Leduc S, Marechal A, Dubois G, Guyot V, Perez-Michaut C, Amato A, Falciatore A, Juillerat A, Beurdeley M, et al. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat Commun. 2014:5(1):3831. 10.1038/ncomms4831 [DOI] [PubMed] [Google Scholar]
- Deme B, Cataye C, Block MA, Marechal E, Jouhet J. Contribution of galactoglycerolipids to the 3-dimensional architecture of thylakoids. FASEB J. 2014:28(8):3373–3383. 10.1096/fj.13-247395 [DOI] [PubMed] [Google Scholar]
- De Riso V, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 2009:37(14):e96. 10.1093/nar/gkp448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vargas C, Audic S, Henry N, Decelle J, Mahe F, Logares R, Lara E, Berney C, Le Bescot N, Probert I, et al. Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015:348(6237):1261605. 10.1126/science.1261605 [DOI] [PubMed] [Google Scholar]
- Dolch L-J, Lupette J, Tourcier G, Bedhomme M, Collin S, Magneschi L, Conte M, Seddiki K, Richard C, Corre E, et al. Nitric oxide mediates nitrite-sensing and acclimation and triggers a remodeling of lipids. Plant Physiol. 2017a:175(3):1407–1423. 10.1104/pp.17.01042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolch L-J, Marechal E. Inventory of fatty acid desaturases in the pennate diatom Phaeodactylum tricornutum. Mar Drugs. 2015:13(3):1317–1339. 10.3390/md13031317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolch L-J, Rak C, Perin G, Tourcier G, Broughton R, Leterrier M, Morosinotto T, Tellier F, Faure JD, Falconet D, et al. A palmitic acid elongase affects eicosapentaenoic acid and plastidial monogalactosyldiacylglycerol levels in nannochloropsis. Plant Physiol. 2017b:173(1):742–759. 10.1104/pp.16.01420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubots E, Audry M, Yamaryo Y, Bastien O, Ohta H, Breton C, Marechal E, Block MA. Activation of the chloroplast monogalactosyldiacylglycerol synthase MGD1 by phosphatidic acid and phosphatidylglycerol. J Biol Chem. 2010:285(9):6003–6011. 10.1074/jbc.M109.071928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004:32(5):1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007:2(4):953–971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C. Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol (NY). 1999:1(3):239–251. 10.1007/PL00011773 [DOI] [PubMed] [Google Scholar]
- Falciatore A, d'Alcalà MR, Croot P, Bowler C. Perception of environmental signals by a marine diatom. Science. 2000:288(5475):2363–2366. 10.1126/science.288.5475.2363 [DOI] [PubMed] [Google Scholar]
- Fawley MW, Jameson I, Fawley KP. The phylogeny of the genus Nannochloropsis (Monodopsidaceae, Eustigmatophyceae), with descriptions of N. australis sp. nov. and Microchloropsis gen. nov. Phycologia. 2015:54(5):545–552. 10.2216/15-60.1 [DOI] [Google Scholar]
- Flori S, Jouneau PH, Finazzi G, Marechal E, Falconet D. Ultrastructure of the periplastidial compartment of the diatom Phaeodactylum tricornutum. Protist. 2016:167(3):254–267. 10.1016/j.protis.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Flori S, Jouneau PH, Gallet B, Estrozi LF, Moriscot C, Schoehn G, Finazzi G, Falconet D. Imaging plastids in 2D and 3D: confocal and electron microscopy. Methods Mol Biol. 2018:1829:113–122. 10.1007/978-1-4939-8654-5_7 [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957:226(1):497–509. 10.1016/S0021-9258(18)64849-5 [DOI] [PubMed] [Google Scholar]
- Garab G, Yaguzhinsky LS, Dlouhy O, Nesterov SV, Spunda V, Gasanoff ES. Structural and functional roles of non-bilayer lipid phases of chloroplast thylakoid membranes and mitochondrial inner membranes. Prog Lipid Res. 2022:86:101163. 10.1016/j.plipres.2022.101163 [DOI] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997:14(7):685–695. 10.1093/oxfordjournals.molbev.a025808 [DOI] [PubMed] [Google Scholar]
- Goss R, Jakob T. Regulation and function of xanthophyll cycle-dependent photoprotection in algae. Photosynth Res. 2010:106(1-2):103–122. 10.1007/s11120-010-9536-x [DOI] [PubMed] [Google Scholar]
- Gould SB, Maier UG, Martin WF. Protein import and the origin of red complex plastids. Curr Biol. 2015:25(12):R515–R521. 10.1016/j.cub.2015.04.033 [DOI] [PubMed] [Google Scholar]
- Grosche C, Hempel F, Bolte K, Zauner S, Maier UG. The periplastidal compartment: a naturally minimized eukaryotic cytoplasm. Curr Opin Microbiol. 2014:22:88–93. 10.1016/j.mib.2014.09.017 [DOI] [PubMed] [Google Scholar]
- Gruber A, Rocap G, Kroth PG, Armbrust EV, Mock T. Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 2015:81(3):519–528. 10.1111/tpj.12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Vugrinec S, Hempel F, Gould SB, Maier UG, Kroth PG. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol Biol. 2007:64(5):519–530. 10.1007/s11103-007-9171-x [DOI] [PubMed] [Google Scholar]
- Gschloessl B, Guermeur Y, Cock JM. HECTAR: a method to predict subcellular targeting in heterokonts. BMC Bioinformatics. 2008:9:393. 10.1186/1471-2105-9-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Cao L, Wu X, Li Y, Hu Q, Han D. A lipid bodies-associated galactosyl hydrolase is involved in triacylglycerol biosynthesis and galactolipid turnover in the unicellular green alga Chlamydomonas reinhardtii. Plants (Basel). 2021:10(4):675. 10.3390/plants10040675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiry MD. How many species of algae are there? J Phycol. 2012:48(5):1057–1063. 10.1111/j.1529-8817.2012.01222.x [DOI] [PubMed] [Google Scholar]
- Hempel F, Bullmann L, Lau J, Zauner S, Maier UG. ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Mol Biol Evol. 2009:26(8):1781–1790. 10.1093/molbev/msp079 [DOI] [PubMed] [Google Scholar]
- Hori K, Nobusawa T, Watanabe T, Madoka Y, Suzuki H, Shibata D, Shimojima M, Ohta H. Tangled evolutionary processes with commonality and diversity in plastidial glycolipid synthesis in photosynthetic organisms. Biochim Biophys Acta. 2016:1861(9):1294–1308. 10.1016/j.bbalip.2016.04.015 [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007:35(Web Server issue):W585–W587. 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Pan Y, Maréchal E, Hu H. Proteomes reveal the lipid metabolic network in the complex plastid of Phaeodactylum tricornutum. Plant J. 2024:117(2):385–403. 10.1111/tpj.16477 [DOI] [PubMed] [Google Scholar]
- Iwai M, Yamada-Oshima Y, Asami K, Kanamori T, Yuasa H, Shimojima M, Ohta H. Recycling of the major thylakoid lipid MGDG and its role in lipid homeostasis in Chlamydomonas reinhardtii. Plant Physiol. 2021:187(3):1341–1356. 10.1093/plphys/kiab340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson G, Zhang C, Prado MG, Paiardini A. Pymod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics. 2017:33(3):444–446. 10.1093/bioinformatics/btw638 [DOI] [PubMed] [Google Scholar]
- Jensen PE, Leister D. Chloroplast evolution, structure and functions. F1000Prime Rep. 2014:6:40. 10.12703/P6-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Lupette J, Clerc O, Magneschi L, Bedhomme M, Collin S, Roy S, Maréchal E, Rébeillé F. LC-MS/MS versus TLC plus GC methods: consistency of glycerolipid and fatty acid profiles in microalgae and higher plant cells and effect of a nitrogen starvation. PLoS One. 2017:12(8):e0182423. 10.1371/journal.pone.0182423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021:596(7873):583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Hassabis D. Protein structure predictions to atomic accuracy with AlphaFold. Nat Methods. 2022:19(1):11–12. 10.1038/s41592-021-01362-6 [DOI] [PubMed] [Google Scholar]
- Karas BJ, Diner RE, Lefebvre SC, McQuaid J, Phillips AP, Noddings CM, Brunson JK, Valas RE, Deerinck TJ, Jablanovic J, et al. Designer diatom episomes delivered by bacterial conjugation. Nat Commun. 2015:6(1):6925. 10.1038/ncomms7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015:10(6):845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian O, Kroth PG. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005:41(2):175–183. 10.1111/j.1365-313X.2004.02294.x [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H. Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 2009a:57(2):322–331. 10.1111/j.1365-313X.2008.03692.x [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. Galactolipid biosynthesis is essential for proper chloroplast biogenesis and embryogenesis. Photosynth Res. 2007a:91:216–216. https://link.springer.com/article/10.1007/s11120-007-9197-6 [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci U S A. 2007b:104(43):17216–17221. 10.1073/pnas.0704680104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Nakamura Y, Ohta H. Type A and type B monogalactosyldiacylglycerol synthases are spatially and functionally separated in the plastids of higher plants. Plant Physiol Biochem. 2009b:47(6):518–525. 10.1016/j.plaphy.2008.12.012 [DOI] [PubMed] [Google Scholar]
- Kooistra WH, De Stefano M, Mann DG, Medlin LK. The phylogeny of the diatoms. Prog Mol Subcell Biol. 2003:33:59–97. 10.1007/978-3-642-55486-5_3 [DOI] [PubMed] [Google Scholar]
- Kooistra WH, Gersonde R, Medlin LK, Mann DG. The origin and evolution of the diatoms: their adaptation to a planktonic existence. In: Falkowski PG, Knoll AH, editors. Evolution of primary producers in the sea. Cambridge, USA: Academic Press; 2007. p. 207–249. [Google Scholar]
- Kroth PG, Bones AM, Daboussi F, Ferrante MI, Jaubert M, Kolot M, Nymark M, Rio Bartulos C, Ritter A, Russo MT, et al. Genome editing in diatoms: achievements and goals. Plant Cell Rep. 2018:37(10):1401–1408. 10.1007/s00299-018-2334-1 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018:35(6):1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Apt KE, Kroth PG. Protein transport into “complex” diatom plastids utilizes two different targeting signals. J Biol Chem. 1998:273(47):30973–30978. 10.1074/jbc.273.47.30973 [DOI] [PubMed] [Google Scholar]
- Le SQ, Lartillot N, Gascuel O. Phylogenetic mixture models for proteins. Philos Trans R Soc Lond B Biol Sci. 2008:363(1512):3965–3976. 10.1098/rstb.2008.0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F, Correia D, Lefort V, Doppelt-Azeroual O, Mareuil F, Cohen-Boulakia S, Gascuel O. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019:47(W1):W260–W265. 10.1093/nar/gkz303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021:49(W1):W293–W296. 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyland B, Zarka A, Didi-Cohen S, Boussiba S, Khozin-Goldberg I. High resolution proteome of lipid droplets isolated from the pennate diatom Phaeodactylum tricornutum (Bacillariophyceae) strain pt4 provides mechanistic insights into complex intracellular coordination during nitrogen deprivation. J Phycol. 2020:56(6):1642–1663. 10.1111/jpy.13063 [DOI] [PubMed] [Google Scholar]
- Li X, Moellering ER, Liu B, Johnny C, Fedewa M, Sears BB, Kuo MH, Benning C. A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell. 2012:24(11):4670–4686. 10.1105/tpc.112.105106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. Acyl-lipid metabolism. In: The Arabidopsis book. Washington (DC): BioOne Digital; 2010. p. e0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XJ, Hempel F, Stork S, Bolte K, Moog D, Heimerl T, Maier UG, Zauner S. Addressing various compartments of the diatom model organism via sub-cellular marker proteins. Algal Res. 2016:20:249–257. 10.1016/j.algal.2016.10.018 [DOI] [Google Scholar]
- Lupette J, Jaussaud A, Seddiki K, Morabito C, Brugiere S, Schaller H, Kuntz M, Putaux JL, Jouneau PH, Rebellle F, et al. The architecture of lipid droplets in the diatom Phaeodactylum tricornutum. Algal Res. 2019:38:101415. 10.1016/j.algal.2019.101415 [DOI] [Google Scholar]
- Lupette J, Tardif M, Brugiere S, Coute Y, Salvaing J, Marechal E. Quantitative proteomic analyses reveal the impact of nitrogen starvation on the proteome of the model diatom Phaeodactylum tricornutum. Proteomics. 2022:22(22):e2200155. 10.1002/pmic.202200155 [DOI] [PubMed] [Google Scholar]
- Makshakova O, Breton C, Perez S. Unraveling the complex enzymatic machinery making a key galactolipid in chloroplast membrane: a multiscale computer simulation. Sci Rep. 2020:10(1):13514. 10.1038/s41598-020-70425-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DG. The species concept in diatoms. Phycologia. 1999:38(6):437–495. 10.2216/i0031-8884-38-6-437.1 [DOI] [Google Scholar]
- Mann DG, Droop SJM. Biodiversity, biogeography and conservation of diatoms. Hydrobiologia. 1996:336(1-3):19–32. 10.1007/BF00010816 [DOI] [Google Scholar]
- Maréchal E. How did thylakoids emerge in Cyanobacteria, and how were the primary chloroplast and chromatophore acquired? Methods Mol Biol. 2024:2776:3–20. 10.1007/978-1-0716-3726-5_1 [DOI] [PubMed] [Google Scholar]
- Martens C, Vandepoele K, Van de Peer Y. Whole-genome analysis reveals molecular innovations and evolutionary transitions in chromalveolate species. Proc Natl Acad Sci U S A. 2008:105(9):3427–3432. 10.1073/pnas.0712248105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022:19(6):679–682. 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog D, Stork S, Zauner S, Maier UG. In silico and in vivo investigations of proteins of a minimized eukaryotic cytoplasm. Genome Biol Evol. 2011:3:375–382. 10.1093/gbe/evr031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami R, Hashimoto H. Unusual nuclear division in Nannochloropsis oculata (Eustigmatophyceae, Heterokonta) which may ensure faithful transmission of secondary plastids. Protist. 2009:160(1):41–49. 10.1016/j.protis.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Nitenberg M, Makshakova O, Rocha J, Perez S, Marechal E, Block MA, Girard-Egrot A, Breton C. Mechanism of activation of plant monogalactosyldiacylglycerol synthase 1 (MGD1) by phosphatidylglycerol. Glycobiology. 2020:30(6):396–406. 10.1093/glycob/cwz106 [DOI] [PubMed] [Google Scholar]
- Parks MB, Nakov T, Ruck EC, Wickett NJ, Alverson AJ. Phylogenomics reveals an extensive history of genome duplication in diatoms (Bacillariophyta). Am J Bot. 2018:105(3):330–347. 10.1002/ajb2.1056 [DOI] [PubMed] [Google Scholar]
- Petroutsos D, Amiar S, Abida H, Dolch LJ, Bastien O, Rebeille F, Jouhet J, Falconet D, Block MA, McFadden GI, et al. Evolution of galactoglycerolipid biosynthetic pathways–from cyanobacteria to primary plastids and from primary to secondary plastids. Prog Lipid Res. 2014:54:68–85. 10.1016/j.plipres.2014.02.001 [DOI] [PubMed] [Google Scholar]
- Rastogi A, Murik O, Bowler C, Tirichine L. PhytoCRISP-Ex: a web-based and stand-alone application to find specific target sequences for CRISPR/CAS editing. BMC Bioinformatics. 2016:17(1):261. 10.1186/s12859-016-1143-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007:41(1):147–168. 10.1146/annurev.genet.41.110306.130134 [DOI] [PubMed] [Google Scholar]
- Rocha J, Sarkis J, Thomas A, Pitou L, Radzimanowski J, Audry M, Chazalet V, de Sanctis D, Palcic MM, Block MA, et al. Structural insights and membrane binding properties of MGD1, the major galactolipid synthase in plants. Plant J. 2016:85(5):622–633. 10.1111/tpj.13129 [DOI] [PubMed] [Google Scholar]
- Ruban A, Lavaud J, Rousseau B, Guglielmi G, Horton P, Etienne AL. The super-excess energy dissipation in diatom algae: comparative analysis with higher plants. Photosynth Res. 2004:82(2):165–175. 10.1007/s11120-004-1456-1 [DOI] [PubMed] [Google Scholar]
- Sato N, Awai K. “Prokaryotic pathway” is not prokaryotic: noncyanobacterial origin of the chloroplast lipid biosynthetic pathway revealed by comprehensive phylogenomic analysis. Genome Biol Evol. 2017:9(11):3162–3178. 10.1093/gbe/evx238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serôdio J, Lavaud J. Diatoms and their ecological importance. In: Leal Filho W, Azul AM, Brandli L, Lange Salvia A, Wall T, editors. Life below water. Cham: Springer International Publishing; 2020. p. 1–9. [Google Scholar]
- Shang S, Liu R, Luo L, Li X, Zhang S, Zhang Y, Zheng P, Chen Z, Wang B. Functional characterization of the monogalactosyldiacylglycerol synthase gene ptMGD2 in the diatom Phaeodactylum tricornutum. Front Mar Sci. 2022:9:e874448. 10.3389/fmars.2022.874448 [DOI] [Google Scholar]
- Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, Manfredonia A, Falciatore A, Bowler C. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene. 2007:406(1–2):23–35. 10.1016/j.gene.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Smith R, Jouhet J, Gandini C, Nekrasov V, Marechal E, Napier JA, Sayanova O. Plastidial acyl carrier protein Delta9-desaturase modulates eicosapentaenoic acid biosynthesis and triacylglycerol accumulation in Phaeodactylum tricornutum. Plant J. 2021:106(5):1247–1259. 10.1111/tpj.15231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorhannus U. A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Mar Micropaleontol. 2007:65(1–2):1–12. 10.1016/j.marmicro.2007.05.002 [DOI] [Google Scholar]
- Tanaka A, De Martino A, Amato A, Montsant A, Mathieu B, Rostaing P, Tirichine L, Bowler C. Ultrastructure and membrane traffic during cell division in the marine pennate diatom Phaeodactylum tricornutum. Protist. 2015:166(5):506–521. 10.1016/j.protis.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieler A, Wu G, Tsai CH, Bullard B, Cornish AJ, Harvey C, Reca IB, Thornburg C, Achawanantakun R, Buehl CJ, et al. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 2012:8(11):e1003064. 10.1371/journal.pgen.1003064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates AD, Allen J, Amode RM, Azov AG, Barba M, Becerra A, Bhai J, Campbell LI, Martinez MC, Chakiachvili M, et al. Ensembl genomes 2022: an expanding genome resource for non-vertebrates. Nucleic Acids Res. 2022:50(D1):D996–D1003. 10.1093/nar/gkab1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J Phycol. 2000:36(2):379–386. 10.1046/j.1529-8817.2000.99164.x [DOI] [Google Scholar]
- Zhang C, Hu H. High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation. Mar Genomics. 2014:16:63–66. 10.1016/j.margen.2013.10.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplemental material.