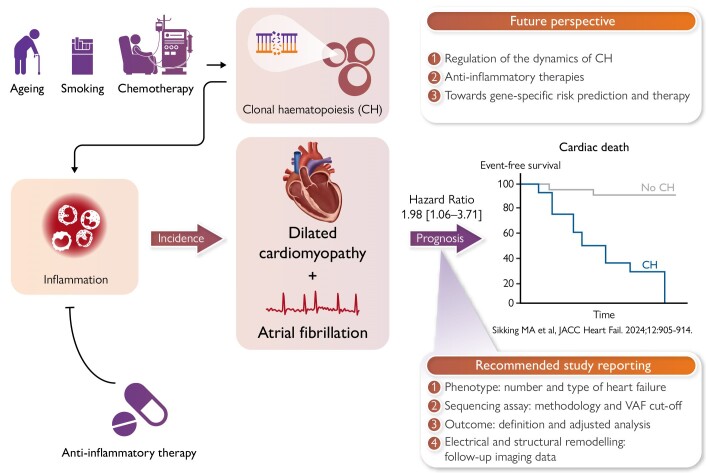
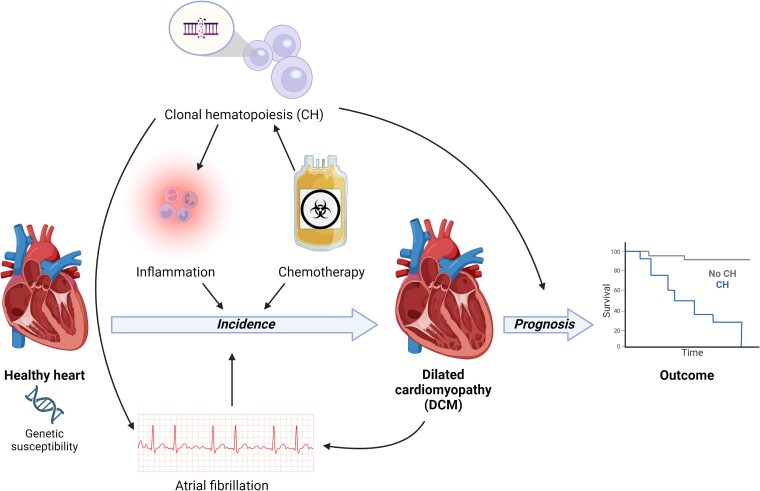
Graphical Abstract
Graphical Abstract.
Clonal haematopoiesis in the pathogenesis of dilated cardiomyopathy. CH, clonal haematopoiesis; VAF, variant allele frequency.
Keywords: Dilated cardiomyopathy, Clonal haematopoiesis, Heart failure, Sequencing, Somatic mutations, CHIP
Abstract
The increased sensitivity of novel DNA sequencing techniques has made it possible to identify somatic mutations in small circulating clones of haematopoietic stem cells. When the mutation affects a ‘driver’ gene, the mutant clone gains a competitive advantage and has the potential to expand over time, a phenomenon referred to as clonal haematopoiesis (CH), which is emerging as a new risk factor for various non-haematological conditions, most notably cardiovascular disease (e.g. heart failure). Dilated cardiomyopathy (DCM) is a form of non-ischaemic heart failure that is characterized by a heterogeneous aetiology. The first evidence is arising that CH plays an important role in the disease course in patients with DCM, and a strong association of CH with multiple aetiologies of DCM has been described (e.g. inflammation, chemotherapy, and atrial fibrillation). The myocardial inflammation induced by CH may be an important trigger for DCM development for an already susceptible heart, e.g. in the presence of genetic variants, environmental triggers, and comorbidities. Studies investigating the role of CH in the pathogenesis of DCM are expected to increase rapidly. To move the field forward, it will be important to report the methodology and results in a standardized manner, so results can be combined and compared. The accurate measurement of CH in patients with DCM can provide guidance of specific (anti-inflammatory) therapies, as mutations in the CH driver genes prime the inflammasome pathway.
Introduction
Dilated cardiomyopathy (DCM) is defined by the presence of left ventricular dilatation and systolic dysfunction unexplained solely by abnormal loading conditions or coronary artery disease.1 The causes of DCM are heterogeneous, including both genetic and environmental factors, also determining the disease progression and outcome.2,3 Chronic myocardial inflammation is often present in patients with DCM, and can be either the cause or consequence of disease progression.4 A broad and aggressive immunosuppressive therapy may be beneficial in DCM patients with increased myocardial inflammation,5,6 but more targeted immunomodulation strategies are lacking.7 Given the connection between clonal haematopoiesis (CH) and inflammation, CH may be a marker and target for immunomodulatory therapy in patients with DCM.8 Recent studies indicate an age-independent effect of CH on the prognosis of DCM, suggesting that CH may also be involved in its pathogenesis. The current review elaborates on (i) the current evidence of CH involvement in DCM, (ii) the knowledge gaps in the road towards clinical impact, and (iii) the future prospects (Graphical Abstract).
General background on clonal haematopoiesis
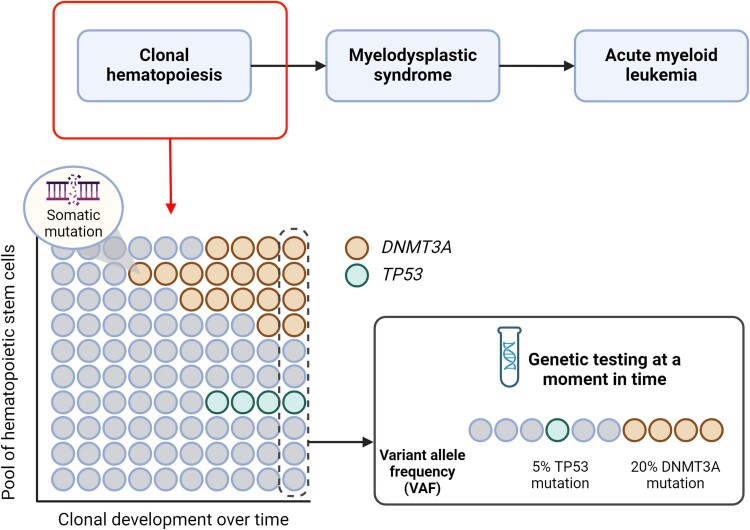
The advent of next-generation DNA sequencing methodologies and their increasing utilization in large human cohorts have led to a new era in human genetics and genomics for clinical and research purposes. Within the specific setting of cardiovascular disease (CVD), investigations into the influence of human genetics have predominantly concentrated on germline variants, which are heritable and therefore present uniformly across all cells within an organism. Numerous inherited genetic variants clearly contribute to a diverse set of different cardiovascular pathologies.9–12 However, research during the last decade has exposed a similarly important role of certain somatic variants. These variants are non-inherited and accumulate in a mosaic manner within an individual from conception onwards as a result of DNA damage or random errors in DNA replication and repair.13 The clinical implications of this somatic genome mosaicism are particularly relevant in the haematopoietic system. Haematopoietic stem cells (HSCs) accumulate random mutations continuously as an individual grows older.14–16 While most of these mutations are deemed neutral ‘passenger’ mutations, a select subset will affect a ‘driver’ gene, providing a competitive advantage to the mutant hematopoietic stem cells (HSC) by promoting its proliferation, survival or self-renewal. Consequently, these mutations drive the progressive expansion of the mutant cell population over time, a phenomenon commonly referred to as somatic mutation-driven CH (Figure 1).17,18
Figure 1.
Clonal haematopoiesis in relation to secondary haematological diseases and genetic testing of clonal expansion. When clonal haematopoiesis is followed by cytopenia and dysplasia it results in myelodysplastic syndrome that can lead to acute myeloid leukaemia. Clonal haematopoiesis is characterized by expansion of a clone that gained a selective advantage due to a somatic mutation (in this example in the DNMT3A and TP53 gene). Genetic testing at a certain time point will detect the variant allele frequency (VAF): the percentage of sequencing read with the specific mutation, reflecting a somatic mosaicism which is different from germline mutations. Clonal haematopoiesis is defined as the presence of a clone with a mutation in a known gene associated with haematological malignancies and a VAF of at least 2%
Clonal haematopoiesis-driving mutations are typically detected through next-generation DNA sequencing of blood samples, which allows the detection of expanded somatic mutations by calculating variant allele frequency (VAF), representing the proportion of reads that support a mutant allele out of the total sequencing reads (Figure 1). Such sequencing analyses have unveiled a diverse array of mutations detectable in blood, including base substitutions [known as single-nucleotide variants (SNVs)], small insertions and deletions (indels), cytogenetic aneuploidies, and structural chromosomal variants. Consequently, it is possible to define different forms of CH based on the detected mutation type. In this context, the type of CH that is gaining more clinical relevance, particularly in the cardiology and haematology fields, is a condition referred to as CH of indeterminate potential (CHIP). CHIP is defined as the presence in blood or bone marrow of an expanded SNV or indel in a known gene associated with haematological malignancies, typically myeloid-biased, with a VAF of at least 2%, and in the absence of overt haematological abnormalities.19 Although a diverse range of cancer-related genes have been identified as potential drivers of CHIP, most mutations occur in a limited subset of genes, most frequently in those encoding the epigenetic regulators DNMT3A, TET2, and ASXL1. Additionally, other frequently mutated CHIP genes encode for DNA damage response (DDR) proteins (e.g. TP53 and PPM1D), splicing factors (e.g. SF3B1, SRSF2, and U2AF1) and signalling mediators (e.g. JAK2).
CHIP is strongly associated with aging20–23 and with age-related diseases.24–38 Unsurprisingly, CHIP mutations increase the risk of incident haematological malignancy.20,22,39 However, CHIP is also emerging as a new risk factor for various non-haematological conditions, most notably CVD.13,24,28,29,31,34,36,37 First, a robust correlation between CHIP mutations and an increased risk of developing atherosclerotic conditions, such as coronary artery disease and peripheral artery disease, independent of age, sex, or traditional cardiovascular risk factors have been described.24,28,31,34 Furthermore, recent studies suggest that CHIP may also be associated with cardiac dysfunction and disease beyond its effects on coronary arteries, as further discussed below (Table 1, Figure 2).
Table 1.
Overview of clinical studies investigating the association of clonal haematopoiesis with the development or prognosis of heart failure
| Study | Patient population | Size population cohort | Size cohort | Age (years)d | CH | Cut-off VAF (%) | Outcome definition | Association (hazard ratio) | Dominant genes |
|---|---|---|---|---|---|---|---|---|---|
| Incidence | |||||||||
| Shi et al.29 | HFrEF + HFpEF | 8592 | 374 (4.3%) | 65 [58–70] | 66 (18%) | 2 | NA | 1.2 [0.9–1.7]a | DNMT3A, TET2 |
| Yu et al.30 | HF | 56 597 | 4.694 (8.3%) | NR | 414 (9%) | 2 | NA | 1.3 [1.1–1.4] | ASXL1, TET2, JAK2 |
| Schuermans et al.40 | HFpEF | 8090 | 459 (5.7%) | NR | NR | 2 | NA | 1.3 [0.9–1.8] | TET2 b |
| HFrEF | 339 (4.2%) | 0.8 [0.5–1.3] | |||||||
| Reiner et al.41 | HFpEF | 5214 | 301 (5.8%) | NR | NR | 2 | NA | 1.4 [1.1–1.9] | TET2 |
| HFrEF | 213 (4.1%) | NR | |||||||
| Prognosis | |||||||||
| Sikking et al.42 | DCM | NA | 520 | 58 [53–66] | 109 (21%) | 0.01 | Cardiac death | 2.0 [1.1–3.7] | DNMT3A |
| Pascual-Figal et al.43 | NICMP | NA | 32 | 74 [69–79] | 12 (38%) | 2 | HF mortality + hospitalization | 2.0 [1.1–3.7] | DNMT3A, TET2 |
| IHF | 30 | 12 (40%) | |||||||
| Wu et al.44 | DCM | NA | 52 | 62 (26–94) | 10 (19%) | 5 | All-cause mortality + hospitalization | 1.7 [0.6–4.9] | DNMT3A, TET2 |
| IHF | 48 | 69 (26–94) | 6 (13%) | 1.4 [0.4–4.7] | DNMT3A, CUX | ||||
| Scolari et al.45 | NICMPc | NA | 446 | 55 ± 15 | 149 (22%) | 2 | All-cause mortality | 2.7 [1.3–5.7] | TET2, ASXL1 |
| IHFc | 213 | ||||||||
| Assmus et al.46 | IHF | NA | 404 | 63 (25–87) | 227 (56%) | 0.5 | All-cause mortality | 1.8 [1.1–2.9] | DNMT3A, TET2 |
| Dorsheimer et al.47 | IHF | NA | 200 | 65 [56–72] | 38 (19%) | 2 | All-cause mortality + hospitalization | 2.1 [1.1–4.0] | DNMT3A, TET2 |
| Cochran et al.48 | HFpEF | NA | 81 | 77 ± 7 | 36 (44%) | 0.5 | Hospitalization | 5.1 [1.1–24.2] | TET2 |
| Cremer et al.49 | IHF | NA | 419 | 63 | 154 (37%) | 0.5 | All-cause mortality | 2.8 [1.5–5.2] | DNMT3A, TET2 |
| Amancherla et al.50 | HTx | NA | 479 | NR | 77 (16%) | 2 | Cardiac allograft vasculopathy | 1.4 [0.9–2.3] | DNMT3A |
CH, clonal haematopoiesis; DCM, dilated cardiomyopathy; HF, heart failure; HFpEF, heart failure with preserved ejection fraction; HFrEF, heart failure with reduced ejection fraction; HTx, heart transplant; IHF, ischaemic heart failure; NA, not applicable; NR, not reported; NICMP, non-ischaemic cardiomyopathy; VAF, variant allele frequency.
aAssociation was significant in the sub-cohort of patients younger than 65 years: HR 2.1 [1.3–3.3].
bAssociation was significant for TET2: HR 2.4 [1.3–4.1].
cStudy including patients with cardiogenic shock with different heart failure aetiologies.
dThe age of included patients with heart failure reported either as mean ± standard deviation, median [inter-quartile range], or median (absolute range).
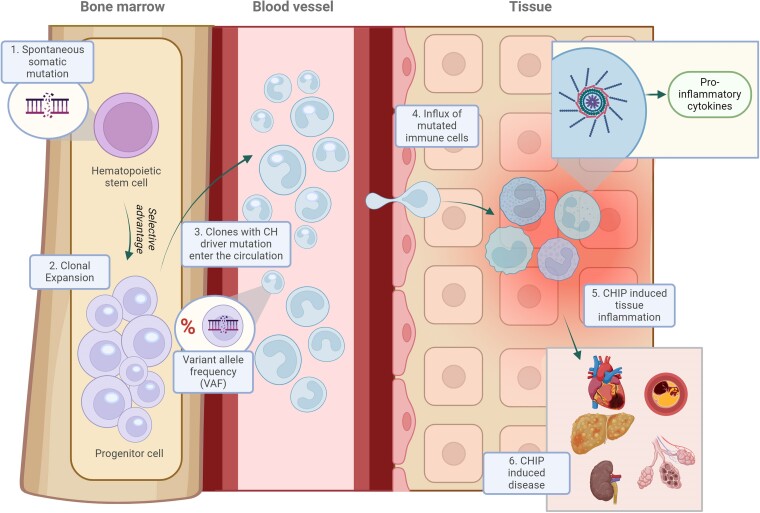
Figure 2.
The association between clonal haematopoiesis, inflammation, and disease development. Somatic mutations origin in haematopoietic stem cells in the bone marrow due to environmental triggers or as a result of ageing (1). When the mutation affects a (driver) gene that provides the cell a competitive advantage, that cell can expand, leading to a mutant cell population (2). The size of the clone can be measured by the variant allele frequency (VAF). The mutated cell progeny enter the circulation (3) where they may infiltrate organ tissue (4) resulting in elevated levels inflammation (5) and consequently leading to tissue damage and organ failure (6)
From a mechanistic standpoint, it is important to note that the clonal expansion of CHIP mutations primarily occurs among the HSC population in the bone marrow, resulting in a variable proportion of progeny immune cells carrying the CHIP mutation. Consequently, CHIP has the potential to significantly influence inflammatory responses, which play a central role in CVD and heart failure (HF). Experimental studies in animal models in various contexts are elucidating the specific immunomodulatory pathways and mechanisms dysregulated in CHIP.28,51–54 While some common mechanisms are emerging, the links between CHIP and CVD appear to be dependent on the specific mutated gene, offering avenues for the development of precision medicine approaches to prevent or treat CVD by targeting the pro-inflammatory effects of specific CHIP mutations.
Clonal haematopoiesis in patients with heart failure
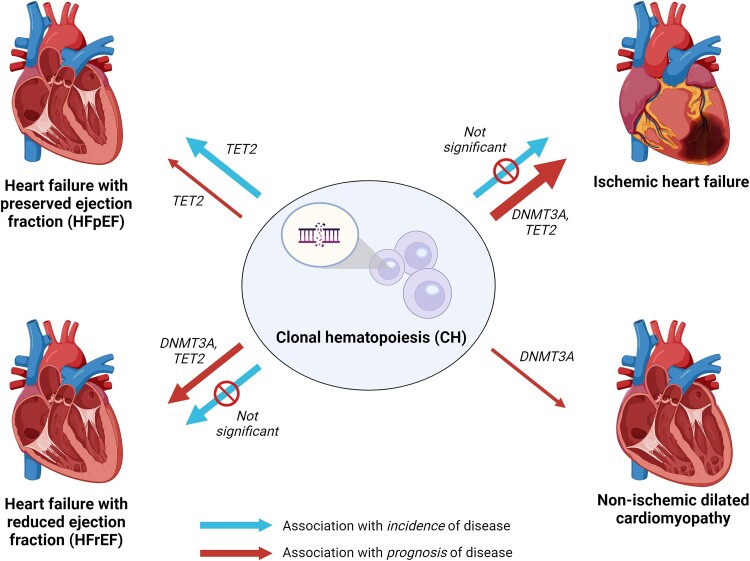
The association between CH and various forms of HF have been extensively described,55 either in relation to the prognosis of patients with HF, or the incidence of HF in the general population (Figure 3). The methodology and cohort characteristics (e.g. the number of individuals and types of HF included) significantly differ between studies, which is important to take into account when interpreting the results.
Figure 3.
Association between clonal haematopoiesis on the development and prognosis of heart failure. The width of the arrow indicates the number of independent studies that investigated the association. Further details on the studies can be found in Table 1
The association of CH with the incidence of HF remains uncertain (Table 1),29,30,40,41 although several predictors and modifiers have been identified. The association is modified by age: CH is mainly associated with new-onset HF in younger individuals (<65 years).29 The largest study with the longest follow-up showed a significant association of CH with incident HF, but mainly due to mutations in ASXL1, TET2, and JAK2 independent of age.30 Most studies distinguish between HF with preserved (HFpEF) or with reduced ejection fraction (HFrEF), creating the opportunity to compare the effect of CH between these types. Two population studies identified TET2 mutations associated with incident HFpEF, but not in HFrEF.40,41 Concluding, CH is predominantly associated with the incidence of HFpEF and not HFrEF, which is mainly attributable to mutations in TET2, with the effect being modified by age. However, we require more studies investigating (gene-specific) CH and HF development to draw definite conclusions.
Most studies investigating the association between CH and prognosis were conducted in patients with ischaemic HF, where CH was a predictor of worse outcome, independent of age (defined as cardiac death and HF hospitalization).43–50,56 As such, the presence of a CH driver mutation is an independent prognostic marker for a worse outcome in patients with an ischaemic form of HF (Figure 3).
Impact of clonal haematopoiesis in patients with dilated cardiomyopathy
Studies investigating the role of CH in the prognosis of patients with DCM are scarce, and thus far only one dedicated DCM cohort.42 Two cohorts included a low number of non-ischaemic (and ischaemic) HF patients,43,44 and one study reported on non-ischaemic HF in patients in cardiogenic shock45 (Table 1). The constitution of the patients with non-ischaemic patients was not further specified in the latter studies, it remains therefore unknown how many patients with DCM were included. The largest cohort including 520 patients with DCM, detected CH in 109 patients (21%) using a VAF of 0.01% to detect small clones.42 Interestingly, relatively large clones (>2%) were already detected in patients below the age of 30, which is higher compared to that observed in the general population.57 CH significantly increased the risk of cardiac death (hazard ratio of 1.98) independent of the clone size, where even a VAF threshold of 0.36% predicted worse outcome. Notably, these clone sizes are smaller than the 2% threshold that is used by the DNA sequence technologies in many of the other studies. In another study, 10 out of 52 patients (19%) with DCM had CH using a VAF threshold of 5%, to detect only relatively large clones.44 The statistical power was too low to draw any conclusions on the prognostic impact of CH. Noteworthy, large CH clones were already detected at a young age in patients with DCM in contrast to the included patients with ischaemic HF. In a cohort of 32 patients with HF of non-ischaemic origin, 12 patients (38%) had CH using a VAF threshold of 2%.43 In the overall cohort (also including patients with ischaemic HF), CH was associated with HF hospitalization and HF-related death, which was independent of ischaemic or non-ischaemic aetiology. A study investigating the role of CH as predictor of mortality in patients with cardiogenic shock included 686 patients, of which 65% had a non-ischaemic cardiomyopathy.45 Patients with cardiogenic shock had a higher prevalence of CH mutations compared to ambulatory HF patients, which was also associated with a decreased survival. However, there was no further distinction in aetiology in the analysis.
Overall, there is a large variation in (i) the sensitivity of DNA sequencing methodologies, (ii) cohort sizes providing well-powered studies, (iii) outcomes reported, and (iv) patient selection. With regard to the latter, the definition of the HF aetiology remains often unclear (i.e. non-ischaemic could indicate hypertrophic, dilated, arrhythmogenic, or even valvular cardiomyopathy). Comparing or aggregating study results, therefore, remain difficult to interpret. We propose the following reporting criteria that should be included in future studies investigating the prognostic impact of CH in patients with DCM (and by extension different cardiac diseases) to better interpret the current evidence and possibilities for future translation:
Definition of HF aetiology (e.g. definition of DCM), and the exact number of patients included with this diagnosis.
-
Sequencing methodology used, required to interpret the number of genes and their coverage, including the sensitivity of the assay to detect clones.
If a high sensitivity assay is used (VAF below 2%), still report a separate analysis using a cut-off value of 2% allowing to compare with earlier studies.
Outcome parameters, especially all-cause mortality and further differentiate into cardiac, or HF-related outcome. We would propose to always include all-cause mortality as an outcome, as this information will be available in all cohorts. Adding more details to the cause of death is recommended in subsequent analysis.
Impact on structural, functional, and/or electrical remodelling is to be determined. We appreciate including any data on the association of CH with follow-up information on echocardiography and/or magnetic resonance imaging.
Second hit model in patients with dilated cardiomyopathy
The exact role or relative contribution of CH in the pathogenesis of DCM remains unclear, as current studies are only associative. The fact that large clones are already present at a younger age in DCM patients44 might suggest a direct role of CH in the development of DCM. Still, the environmental triggers causing DCM, could also be the ones driving CH mutations such as cardiotoxic chemotherapy and inflammation (Figure 4). Although these associations are described indirectly, they render the blueprint of the design for future studies.
Figure 4.
Role of environmental factors in the development of dilated cardiomyopathy, and the association with clonal haematopoiesis. Although there are no studies that have directly investigated the association between clonal haematopoiesis and the incidence of dilated cardiomyopathy, there are associations between drivers of dilated cardiomyopathy development and clonal haematopoiesis. Additionally, clonal haematopoiesis directly impacts the prognosis of patients with dilated cardiomyopathy
Cardiac arrhythmias and dilated cardiomyopathy
Arrhythmias such as atrial fibrillation (AF) occur commonly in patients with DCM, but can also be involved in the development of DCM (e.g. tachycardiomyopathy).4,58,59 The presence of CH is also associated with supraventricular arrhythmias, bradyarrhythmias, and ventricular arrhythmias independent of the presence of HF or coronary artery disease.36 In this study, the increased risk was mainly observed in carriers of TET2, ASXL1, PPM1D, or TP53 mutations. In a separate study using an East Asian cohort, CH was independently associated with AF, with carriers of DNMT3A, TET2, or ASXL1 being most commonly mutated.37 CH driver mutations were associated with a more progressive nature of AF (longer AF duration, larger atrial volume, and elevated E/E′) and unfavourable clinical outcomes defined as HF, ischaemic stroke, or death. Overall, CH seems to play a role throughout the disease course of (supra)ventricular arrhythmias, and potentially the evolution towards HF. Therefore, the interplay between arrhythmias, DCM, and CH in disease progression seems interesting, but remains uninvestigated.
Myocardial inflammation as driver of progression of dilated cardiomyopathy
The presence of leucocyte infiltrates and subsequent inflammation in the myocardium is detected in ∼20% of patients with DCM, and it is often termed as inflammatory DCM or chronic myocarditis.59,60 Whether the inflammation is causal or secondary to the disease progress is in a chronic situation often unknown, but studies with immunosuppressive therapies have shown beneficial effects in a selection of patients with chronic DCM.5,6 Additionally, there is ample evidence that CH driver mutations induce inflammation. Loss of function or deletion of TET2 leads to a higher expression of interleukin (IL)-6 and IL-1β, as well as heightened secretion of IL-1β via the NLRP3-inflammasome in mouse models.51,61–63 Mutations in DNMT3A have been associated with myeloid up-regulation of NLRP3, IL-1, and IL-6,64 and studies suggest that the inflammatory environment can subsequently be beneficial for clonal expansion of DNMT3A clones.65 It could be worthwhile to determine CH in patients with inflammatory DCM to investigate whether the increased myocardial inflammation could be a consequence of CH driver mutations. Elucidation of the role of CH in the disease progression of (inflammatory) DCM could provide a better patient stratification for novel and specific immunomodulatory treatment regimens. As an example, an ongoing phase II study investigates the beneficial effects of colchicine in patients with CH and ischaemic HFrEF (2021-001508-13 in the European Union Clinical Trials Register). The question remains if colchicine is the medication of choice to go forward, as is it a broad-spectrum anti-inflammatory agent that acts by inhibiting microtubule polymerization. Among many other actions, resulting in partial inhibition of the NLRP3-inflammasome although this might depend on the dosage of colchicine.66 The primary outcome of this study is endothelial function assessed by flow-mediated dilation after 60 days of treatment, which is not an outcome relevant for patients with DCM. Specific studies and trials to investigate CH-guided immunomodulatory therapy for patients with (inflammatory) DCM are still absent.
Inflammation and (cardiac) fibrosis are intertwined by multiple pathways.67 Fibrosis in the heart is a well-known risk factor for malignant ventricular arrhythmias and sudden cardiac death in patients with DCM.68 However, studies including patients with DCM investigating the association of CH with cardiac fibrosis are absent. A recent study identified that monocytes isolated from patients with HF and mutations in DNMT3A stimulate the release of heparin-binding epidermal growth factor-like growth factor, thereby facilitating activation of cardiac fibroblasts and subsequent cardiac fibrosis.69 Large CH clones in patients are associated with increased myocardial fibrosis as measured with cardiac magnetic resonance imaging.36 Additionally, a murine model of HFpEF, Tet2-mediated CH led to greater cardiac fibrosis.63 Thus, CH might also prime cardiac fibrosis, independent of the association with inflammation.
Chemotherapy-induced cardiomyopathy
Cardiomyopathies can arise from patient exposure to anthracyclines and other cytotoxic therapies. Some cytotoxic therapies promote the formation of a distinct form of CH that results from mutations in the DDR pathway genes that include TP53, PPM1D, CHK1, CHK2, and ATM.70–73 This type of CH has been referred to as therapy-related CH (t-CH). Unlike the driver genes that give rise to age-related CH by promoting the proliferation and self-renewal of HSCs (e.g. DNMT3A, TET2, and ASXL1), t-CH driver genes confer a selective advantage to HSC by promoting their survival under conditions of genotoxic stress,73,74 which has been previously reviewed.75 This form of CH is of potential interest because patients with cancer and cancer survivors exhibit an increased risk of CVD.75,76 While cytotoxic agents can directly damage the heart by acting on cells of the myocardium, it has been proposed that the cardiac toxicity associated with long latency periods could be due in part to the effects of t-CH that develops in this patient population.77 Of particular interest is the t-CH found in childhood cancer survivors, as it would be assumed that, due to their youth, they would be largely void of age-associated CH and hence exhibit a simplified mutational landscape compared to elderly cancer survivors. Notably, childhood cancer survivors display accelerated biological aging78,79 and a markedly increased risk of CVD.80,81 Although conflicting data on the prevalence of CH among the survivors of childhood cancer has been presented,82,83 Novetsky Friedman et al.84 recently reported that there is a nearly two-fold increase in the frequency of CH in childhood cancer survivors compared to healthy controls when they were assessed by ultradeep, error-corrected DNA sequencing. As expected, childhood cancer survivors showed a significant enrichment of DDR gene-mutant clones compared with clonal mutations in DNMT3A, TET2, or ASXL1 genes. Furthermore, the enrichment of CH in the survivor cohort was also observed relative to a treatment-naïve cohort with solid tumours suggesting that the overrepresentation of CH in the childhood cancer survivors resulted from exposure to some cancer therapies.
Could t-CH be contributing to this increased risk of DCM in this patient population? Recent experimental studies have shown that Trp53 and Ppm1d driver genes can contribute to HF and/or atherosclerotic CVD.28,85,86 An experimental study examined the effects of t-CH on cardiac function by transplanting mice with Trp53 heterozygous-knockout bone marrow cells or bone marrow cells harbouring a common TP53 missense mutation, Trp53R270H.86 To establish a model of t-CH, mice were treated with a course of the chemotherapeutic agent doxorubicin. Doxorubicin accelerated the expansion of haematopoietic Trp53-mutant cells, and these mice displayed worse doxorubicin-induced cardiotoxicity compared with mice transplanted with wild-type bone marrow. Mechanistic studies revealed that doxorubicin promoted greater Trp53-mutant neutrophil infiltration of the myocardium, leading to greater reactive oxygen species production and greater inflammatory cytokine production.86 While these experimental findings suggest that t-CH can contribute to the development of DCM in cancer survivors, clinical evidence in support of this hypothesis is lacking and could potentially be addressed by examining the associations between CH and CVD outcomes in cancer survivors. If validated by further studies, these data would suggest that t-CH is predictive of DCM in cancer survivors and that this subset of patients could have heightened therapeutic responses to anti-inflammatory medications.
Future perspectives
Regulation of the dynamics of clonal haematopoiesis
As our understanding of the mechanisms linking CH to CVD deepens, it becomes essential to also identify the factors regulating mutant cell expansion. Highly sensitive DNA sequencing suggests that very low levels of blood cells carrying CH-related mutations can be found in virtually every middle-aged individual.73,87 However, only a fraction of individuals develop a marked clonal expansion of those mutant cells. Since such expansion is likely a prerequisite for the pathophysiological effects of CH, it is of high clinical value to identify the factors determining whether a mutant HSC remains quiescent and indolent or instead expands to a substantial clone size. To date, our understanding of the regulation of the dynamics of mutant cell expansion is limited, as most previous CH studies were based on cross-sectional sequencing analysis at a single timepoint. An important advancement in this context is the development of a mathematical approach for inferring the fitness advantage conferred by a given somatic mutation based on a single whole genome sequencing time point.88 Applying this tool to a large sequencing dataset led to the identification of inherited genetic variance in the TCL1A gene as an important modulator of the fitness advantage of several commonly mutated driver genes in CH.88 Similar analyses in other large sequencing datasets could deepen our understanding of the biology of CH. Additionally, an increasing number of human cohorts with serially sampled blood over years are allowing for longitudinal sequencing analyses of the dynamics of CH.72,89–93 The data available to date lead to two major conclusions. First, the expansion rates of mutant haematopoietic clones is substantially different among different driver genes. Although further research is needed, available evidence suggest that mutations in epigenetic regulatory genes expand slower than those in genes encoding splicing regulators or involved in the DDR.89–91 Second, even when considering the same mutated gene or the same hotspot mutation, the expansion rates of mutant clones vary considerably between individuals, suggesting that the dynamics of CH are markedly influenced by non-mutational factors that remain to be determined. The clinical implications of identifying the factors or mechanisms controlling clonal outgrowth are manifold. For example, it may aid the development of interventions to slow down or even reverse the expansion of mutant clones, blunting their adverse effects on health. Furthermore, it may lead to new algorithms that enhance the predictive and prognostic value of CH in the setting of CVD, as a small mutant clone identified at a given timepoint may have a substantial long-term impact on health if it undergoes rapid expansion.
Anti-inflammatory therapies
Therapeutic targeting of CH in HF is an emerging area of research with several potential approaches. Understanding the mechanistic link between CH and HF may lead to new drug targets that can specifically modulate the detrimental effects of mutated blood cells on the heart. One can inhibit the enhanced activation of inflammatory pathways related to CH, including IL-1β, IL-6, TNF, NLRP3, or the JAK pathway,64 helping to reduce the inflammatory burden caused by CH. Whereas this approach is not specific for CH itself, it opens the way for tailored medicine in HF and DCM patients. In murine models of HF, CH associated with TET2 loss-of-function leads to worsened cardiac remodelling and function, through an IL-1β-mediated mechanism.62 Thus, individuals with TET2-mediated CH might respond better to IL-1β-NLRP3-inflammasome inhibition, an example of personalized medicine. An illustration of this is the finding that the IL-1β neutralizing antibody, canakinumab, reduced the relative risk of major adverse cardiovascular events by 62% in high-risk CVD patients with TET2 mutations compared with 7% in those without CH.8 Furthermore, it is conceivable that DNA methyltransferase inhibitors could potentially be used to prevent the hypomethylation related to DNMT3A mutations, but these can still have widespread effects on healthy cells. Moreover, histone deacetylase inhibitors, which could alter chromatin structure to activate the transcription of genes that are silenced in the presence of DNMT3A mutations, may potentially counteract some of the negative effects on gene expression.
Eliminating the mutated blood cells themselves by selectively killing these cells is another therapeutic option. The precise identification of cell surface antigens specific to CH cells is crucial in this regard. Chimeric antigen receptor T-cell therapy, which would work by modifying a patient’s T-cells to recognize and attack CH cells expressing specific antigens, is one approach to eliminate these cells.94 Another approach is bispecific T-cell engagers, which are engineered proteins that can simultaneously bind to a T-cell and a target cell. These T-cell engagers would bring T-cells and CH cells into close proximity leading to the destruction of CH cells.95 Since the mutations driving CH predominantly occur in genes that regulate DNA methylation, histone modification, and chromatin organization (e.g. DNMT3A, TET2, and ASXL1), rather than genes that encode cell surface proteins, identifying specific antigens remains a major challenge. However, another more invasive approach in severe cases could include gene editing with CRISPR–Cas9 to correct the mutations in HSCs responsible for CH.96 This approach would involve collecting HSCs from the patient, correcting the mutation ex vivo, and then re-infusing the corrected cells back into the patient, a quite challenging but potentially impactful therapeutic approach in HF patients.
In summary, targeting CH in cardiomyopathies could involve a multifaceted approach, including targeting CH specific inflammatory, metabolic and methylation pathways, developing therapies to eliminate mutated cells, and even considering stem cell transplantation in severe cases. Developing therapies that effectively target mutated cells without harming healthy cells is a significant challenge, and it will require more research to identify CH specific cell surface antigens and CH mediated intracellular pathways that can be therapeutically exploited.
Conclusion
The increased sensitivity of novel DNA sequencing techniques has significantly increased the accessibility of CH sequencing. CH might be the consequence of aetiologies of DCM (e.g. chemotherapy) but can also lead to triggers that subsequently contribute to DCM development and progression (e.g. inflammation, AF). The tissue inflammation triggered by CH provides a specific treatment target. The exact involved inflammatory pathways differ per mutated CH driver gene, thus the benefit of immunomodulatory therapy might differ per individual patient with DCM and CH. However, the number of studies that are currently investigating the role of CH in DCM are low, and the individual studies strongly differ in their set-up and definitions. It will be important for future studies to systematically report on the specification of the DNA sequencing technique, accurate phenotyping of the cohort, and the statistical analysis in order to compare the results of different studies towards developing clinical trials investigating CH-based treatment stratification.
Supplementary data
Supplementary data are not available at European Heart Journal online.
Contributor Information
Job A J Verdonschot, Department of Clinical Genetics, Maastricht University Medical Center+, Maastricht, the Netherlands; Department of Cardiology, Maastricht University, Cardiovascular Research Institute Maastricht (CARIM), P.O. Box 616, 6200 MD Maastricht, the Netherlands; European Reference Network for Rare, Low Prevalence and Complex Diseases of the Heart (ERN GUARD-Heart).
Jose J Fuster, Centro Nacional de Investigaciones Cardiovasculares (CNIC), C. de Melchor Fernández Almagro, 3, Fuencarral-El Pardo, 28029 Madrid, Spain; CIBER en Enfermedades Cardiovasculares (CIBER-CV), Av. Monforte de Lemos, 3-5. Pabellón 11, Planta 0, 28029 Madrid, Spain.
Kenneth Walsh, Division of Cardiovascular Medicine and Robert M. Berne Cardiovascular Research Center, Hematovascular Biology Center, University of Virginia School of Medicine, 415 Lane Rd, Suite 1010, PO Box 801394, Charlottesville, VA, USA.
Stephane R B Heymans, Department of Cardiology, Maastricht University, Cardiovascular Research Institute Maastricht (CARIM), P.O. Box 616, 6200 MD Maastricht, the Netherlands; European Reference Network for Rare, Low Prevalence and Complex Diseases of the Heart (ERN GUARD-Heart); Department of Cardiovascular Science, Katholieke Universiteit Leuven, Herestraat 49, 3000 Leuven, Belgium.
Declarations
Disclosure of Interest
S.R.B.H. receives personal fees for independent scientific advice on early development in the field of heart failure from AstraZeneca, Ribocure, and CSL Behring, and receives research support for early clinical development from AstraZeneca and CSL Behring. The other authors have nothing to declare.
Data Availability
No data were generated or analysed for or in support of this article.
Funding
J.A.J.V. was supported by a Dekker clinical scientist grant from the Dutch Heart Foundation, and the Academic Funds of the Maastricht University Medical Center+. J.J.F. was supported by grant PLEC2021-008194, funded by MICIU/AEI/10.13039/501100011033 and by ‘European Union Next Generation EU/PRTR’; grant PID2021-126580OB-I00, funded by MICIU/AEI/10.13039/501100011033; and grant 202314-31, funded by Fundació ‘La Marató TV3’. He also received research funding from ‘la Caixa’ Foundation under the project code LCF/PR/HR22/52420011, and from Instituto de Salud Carlos III (ISCIII), co-funded by the European Union Next Generation EU/PRTR under the umbrella of the Partnership Fostering a European Research Area for Health (ERA4Health) (GA No 101095426 of the EU Horizon Europe Research and Innovation Programme). The CNIC was supported by ISCIII, the Ministerio de Ciencia e Innovación and the Pro CNIC Foundation, and is a Severo Ochoa Center of Excellence (grant CEX2020-001041-S funded by MICIN/AEI/10.13039/501100011033). K.W. receives funding from the National Institutes of Health (NIH)) grants AG073249, HL142650, and HL152174 and NASA grant 80NSSC21K0549. S.R.B.H. received funding from the European Union Commission’s Seventh Framework programme under grant agreement no. 305507 (HOMAGE); the IMI2-CARDIATEAM, from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement no. 821508, where the JU receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA; support by funding from the Pathfinder Cardiogenomics programme of the European Innovation Council of the European Union (DCM-NEXT project); and further support from the Netherlands Cardiovascular Research Initiative, an initiative with support of the Dutch Heart Foundation, Dutch Cardiovascular Alliance Double Dosis, 2020-B005; ZonMW-Metacor.
References
- 1. Arbelo E, Protonotarios A, Gimeno JR, Arbustini E, Barriales-Villa R, Basso C, et al. 2023 ESC Guidelines for the management of cardiomyopathies. Eur Heart J 2023;44:3503–626. 10.1093/eurheartj/ehad194 [DOI] [PubMed] [Google Scholar]
- 2. Verdonschot JAJ, Heymans SRB. Dilated cardiomyopathy: second hits knock-down the heart. Eur Heart J 2024;45:500–1. 10.1093/eurheartj/ehad778 [DOI] [PubMed] [Google Scholar]
- 3. Hazebroek MR, Moors S, Dennert R, van den Wijngaard A, Krapels I, Hoos M, et al. Prognostic relevance of gene-environment interactions in patients with dilated cardiomyopathy: applying the MOGE(S) classification. J Am Coll Cardiol 2015;66:1313–23. 10.1016/j.jacc.2015.07.023 [DOI] [PubMed] [Google Scholar]
- 4. Verdonschot JAJ, Hazebroek MR, Ware JS, Prasad SK, Heymans SRB. Role of targeted therapy in dilated cardiomyopathy: the challenging road toward a personalized approach. J Am Heart Assoc 2019;8:e012514. 10.1161/JAHA.119.012514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Merken J, Hazebroek M, Van Paassen P, Verdonschot J, Van Empel V, Knackstedt C, et al. Immunosuppressive therapy improves both short- and long-term prognosis in patients with virus-negative nonfulminant inflammatory cardiomyopathy. Circ Heart Fail 2018;11:e004228. 10.1161/CIRCHEARTFAILURE.117.004228 [DOI] [PubMed] [Google Scholar]
- 6. Chimenti C, Russo MA, Frustaci A. Immunosuppressive therapy in virus-negative inflammatory cardiomyopathy: 20-year follow-up of the TIMIC trial. Eur Heart J 2022;43:3463–73. 10.1093/eurheartj/ehac348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hazebroek MR, Henkens M, Raafs AG, Verdonschot JAJ, Merken JJ, Dennert RM, et al. Intravenous immunoglobulin therapy in adult patients with idiopathic chronic cardiomyopathy and cardiac parvovirus B19 persistence: a prospective, double-blind, randomized, placebo-controlled clinical trial. Eur J Heart Fail 2021;23:302–9. 10.1002/ejhf.2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Svensson EC, Madar A, Campbell CD, He Y, Sultan M, Healey ML, et al. TET2-driven clonal hematopoiesis and response to canakinumab: an exploratory analysis of the CANTOS randomized clinical trial. JAMA Cardiol 2022;7:521–8. 10.1001/jamacardio.2022.0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet 2017;18:331–44. 10.1038/nrg.2016.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaidi S, Brueckner M. Genetics and genomics of congenital heart disease. Circ Res 2017;120:923–40. 10.1161/circresaha.116.309140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morrell NW, Aldred MA, Chung WK, Elliott CG, Nichols WC, Soubrier F, et al. Genetics and genomics of pulmonary arterial hypertension. Eur Respir J 2019;53:1801899. 10.1183/13993003.01899-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tayal U, Prasad S, Cook SA. Genetics and genomics of dilated cardiomyopathy and systolic heart failure. Genome Med 2017;9:20. 10.1186/s13073-017-0410-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu Z, Coorens THH, Uddin MM, Ardlie KG, Lennon N, Natarajan P. Genetic variation across and within individuals. Nat Rev Genet 2024:25;548–62. 10.1038/s41576-024-00709-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee-Six H, Øbro NF, Shepherd MS, Grossmann S, Dawson K, Belmonte M, et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 2018;561:473–8. 10.1038/s41586-018-0497-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Osorio FG, Rosendahl Huber A, Oka R, Verheul M, Patel SH, Hasaart K, et al. Somatic mutations reveal lineage relationships and age-related mutagenesis in human hematopoiesis. Cell Rep 2018;25:2308–16.e4. 10.1016/j.celrep.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012;150:264–78. 10.1016/j.cell.2012.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuster JJ, Walsh K. Somatic mutations and clonal hematopoiesis: unexpected potential new drivers of age-related cardiovascular disease. Circ Res 2018;122:523–32. 10.1161/circresaha.117.312115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jaiswal S, Libby P. Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol 2020;17:137–44. 10.1038/s41569-019-0247-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16. 10.1182/blood-2015-03-631747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–98. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 2020;586:763–8. 10.1038/s41586-020-2819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477–87. 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kessler MD, Damask A, O'Keeffe S, Banerjee N, Li D, Watanabe K, et al. Common and rare variant associations with clonal haematopoiesis phenotypes. Nature 2022;612:301–9. 10.1038/s41586-022-05448-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–21. 10.1056/NEJMoa1701719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller PG, Qiao D, Rojas-Quintero J, Honigberg MC, Sperling AS, Gibson CJ, et al. Association of clonal hematopoiesis with chronic obstructive pulmonary disease. Blood 2022;139:357–68. 10.1182/blood.2021013531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agrawal M, Niroula A, Cunin P, McConkey M, Shkolnik V, Kim PG, et al. TET2-mutant clonal hematopoiesis and risk of gout. Blood 2022;140:1094–103. 10.1182/blood.2022015384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim PG, Niroula A, Shkolnik V, McConkey M, Lin AE, Słabicki M, et al. Dnmt3a-mutated clonal hematopoiesis promotes osteoporosis. J Exp Med 2021:218:e20211872. 10.1084/jem.20211872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zekavat SM, Viana-Huete V, Matesanz N, Jorshery SD, Zuriaga MA, Uddin MM, et al. TP53-mediated clonal hematopoiesis confers increased risk for incident atherosclerotic disease. Nat Cardiovasc Res 2023;2:144–58. 10.1038/s44161-022-00206-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi C, Aboumsallem JP, Suthahar N, de Graaf AO, Jansen JH, van Zeventer IA, et al. Clonal haematopoiesis of indeterminate potential: associations with heart failure incidence, clinical parameters and biomarkers. Eur J Heart Fail 2023;25:4–13. 10.1002/ejhf.2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu B, Roberts MB, Raffield LM, Zekavat SM, Nguyen NQH, Biggs ML, et al. Supplemental association of clonal hematopoiesis with incident heart failure. J Am Coll Cardiol 2021;78:42–52. 10.1016/j.jacc.2021.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vlasschaert C, Heimlich JB, Rauh MJ, Natarajan P, Bick AG. Interleukin-6 receptor polymorphism attenuates clonal hematopoiesis-mediated coronary artery disease risk among 451 180 individuals in the UK Biobank. Circulation 2023;147:358–60. 10.1161/circulationaha.122.062126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kestenbaum B, Bick AG, Vlasschaert C, Rauh MJ, Lanktree MB, Franceschini N, et al. Clonal hematopoiesis of indeterminate potential and kidney function decline in the general population. Am J Kidney Dis 2023;81:329–35. 10.1053/j.ajkd.2022.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wong WJ, Emdin C, Bick AG, Zekavat SM, Niroula A, Pirruccello JP, et al. Clonal haematopoiesis and risk of chronic liver disease. Nature 2023;616:747–54. 10.1038/s41586-023-05857-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao K, Shen X, Liu H, Lin Z, Li J, Chen S, et al. Somatic and germline variants and coronary heart disease in a Chinese population. JAMA Cardiol 2024;9:233–42. 10.1001/jamacardio.2023.5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vlasschaert C, Robinson-Cohen C, Chen J, Akwo E, Parker AC, Silver SA, et al. Clonal hematopoiesis of indeterminate potential is associated with acute kidney injury. Nat Med 2024;30:810–7. 10.1038/s41591-024-02854-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schuermans A, Vlasschaert C, Nauffal V, Cho SMJ, Uddin MM, Nakao T, et al. Clonal haematopoiesis of indeterminate potential predicts incident cardiac arrhythmias. Eur Heart J 2024;45:791–805. 10.1093/eurheartj/ehad670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahn HJ, An HY, Ryu G, Lim J, Sun C, Song H, et al. Clonal haematopoiesis of indeterminate potential and atrial fibrillation: an east Asian cohort study. Eur Heart J 2024;45:778–90. 10.1093/eurheartj/ehad869 [DOI] [PubMed] [Google Scholar]
- 38. Tobias DK, Manning AK, Wessel J, Raghavan S, Westerman KE, Bick AG, et al. Clonal Hematopoiesis of Indeterminate Potential (CHIP) and incident type 2 diabetes risk. Diabetes Care 2023;46:1978–85. 10.2337/dc23-0805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Warren JT, Link DC. Clonal hematopoiesis and risk for hematologic malignancy. Blood 2020;136:1599–605. 10.1182/blood.2019000991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schuermans A, Honigberg MC, Raffield LM, Yu B, Roberts MB, Kooperberg C, et al. Clonal hematopoiesis and incident heart failure with preserved ejection fraction. JAMA Netw Open 2024;7:e2353244. 10.1001/jamanetworkopen.2023.53244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reiner AP, Roberts MB, Honigberg MC, Kooperberg C, Desai P, Bick AG, et al. Association of Clonal Hematopoiesis of Indeterminate Potential with Incident Heart Failure with Preserved Ejection Fraction. medRxiv, 10.1101/2023.06.07.23291038, 10 June 2023, preprint: not peer reviewed. [DOI]
- 42. Sikking MA, Stroeks S, Henkens M, Raafs AG, Cossins B, van Deuren RC, et al. Clonal hematopoiesis has prognostic value in dilated cardiomyopathy independent of age and clone size. JACC Heart Fail 2023;12:905–14. 10.1016/j.jchf.2023.06.037 [DOI] [PubMed] [Google Scholar]
- 43. Pascual-Figal DA, Bayes-Genis A, Díez-Díez M, Hernández-Vicente Á, Vázquez-Andrés D, de la Barrera J, et al. Clonal hematopoiesis and risk of progression of heart failure with reduced left ventricular ejection fraction. J Am Coll Cardiol 2021;77:1747–59. 10.1016/j.jacc.2021.02.028 [DOI] [PubMed] [Google Scholar]
- 44. Wu JMF, Bekfani T, Hinze A, Westphal JG, Steinacker B, Zeller M, et al. Clonal haematopoiesis of indeterminate potential-related mutations and outcome in dilated and ischaemic cardiomyopathy. ESC Heart Fail 2022;9:3954–60. 10.1002/ehf2.14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scolari FL, Brahmbhatt DH, Abelson S, Medeiros JJF, Anker MS, Fung NL, et al. Clonal hematopoiesis confers an increased mortality risk in orthotopic heart transplant recipients. Am J Transplant 2022;22:3078–86. 10.1111/ajt.17172 [DOI] [PubMed] [Google Scholar]
- 46. Assmus B, Cremer S, Kirschbaum K, Dorsheimer L, Rasper T, Abou-El-Ardat K, et al. Clonal haematopoiesis in chronic ischaemic heart failure: prognostic role of clone size for DNMT3A- and TET2-driver gene mutations. Eur Heart J 2021;42:257–65. 10.1093/eurheartj/ehaa845 [DOI] [PubMed] [Google Scholar]
- 47. Dorsheimer L, Assmus B, Rasper T, Ortmann CA, Ecke A, Abou-El-Ardat K, et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol 2019;4:25–33. 10.1001/jamacardio.2018.3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cochran JD, Yura Y, Thel MC, Doviak H, Polizio AH, Arai Y, et al. Clonal hematopoiesis in clinical and experimental heart failure with preserved ejection fraction. Circulation 2023;148:1165–78. 10.1161/circulationaha.123.064170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cremer S, Kirschbaum K, Berkowitsch A, John D, Kiefer K, Dorsheimer L, et al. Multiple somatic mutations for clonal hematopoiesis are associated with increased mortality in patients with chronic heart failure. Circ Genom Precis Med 2020;13:e003003. 10.1161/circgen.120.003003 [DOI] [PubMed] [Google Scholar]
- 50. Amancherla K, Schlendorf KH, Vlasschaert C, Lowery BD, Wells QS, See SB, et al. Clonal hematopoiesis of indeterminate potential and outcomes after heart transplantation: a multicenter study. Am J Transplant 2023;23:1256–63. 10.1016/j.ajt.2023.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science (1979) 2017;355:842–47. 10.1126/science.aag1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fuster JJ, Zuriaga MA, Zorita V, MacLauchlan S, Polackal MN, Viana-Huete V, et al. TET2-loss-of-function-driven clonal hematopoiesis exacerbates experimental insulin resistance in aging and obesity. Cell Rep 2020;33:108326. 10.1016/j.celrep.2020.108326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fidler TP, Xue C, Yalcinkaya M, Hardaway B, Abramowicz S, Xiao T, et al. The AIM2 inflammasome exacerbates atherosclerosis in clonal haematopoiesis. Nature 2021;592:296–301. 10.1038/s41586-021-03341-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rauch PJ, Gopakumar J, Silver AJ, Nachun D, Ahmad H, McConkey M, et al. Loss-of-function mutations in Dnmt3a and Tet2 lead to accelerated atherosclerosis and concordant macrophage phenotypes. Nat Cardiovasc Res 2023;2:805–18. 10.1038/s44161-023-00326-7 [DOI] [PubMed] [Google Scholar]
- 55. Sikking MA, Stroeks S, Waring OJ, Henkens MTHM, Riksen NP, Hoischen A, et al. Clonal hematopoiesis of indeterminate potential from a heart failure specialist's point of view. J Am Heart Assoc 2023;12:e030603. 10.1161/jaha.123.030603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Böhme M, Desch S, Rosolowski M, Scholz M, Krohn K, Büttner P, et al. Impact of clonal hematopoiesis in patients with cardiogenic shock complicating acute myocardial infarction. J Am Coll Cardiol 2022;80:1545–56. 10.1016/j.jacc.2022.08.740 [DOI] [PubMed] [Google Scholar]
- 57. Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science (1979) 2019;366:eaan4673. 10.1126/science.aan4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gopinathannair R, Etheridge SP, Marchlinski FE, Spinale FG, Lakkireddy D, Olshansky B. Arrhythmia-induced cardiomyopathies: mechanisms, recognition, and management. J Am Coll Cardiol 2015;66:1714–28. 10.1016/j.jacc.2015.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Verdonschot JAJ, Hazebroek MR, Krapels IPC, Henkens MTHM, Raafs A, Wang P, et al. Implications of genetic testing in dilated cardiomyopathy. Circ Genom Precis Med 2020:13;476–87. 10.1161/CIRCGEN.120.003031 [DOI] [PubMed] [Google Scholar]
- 60. Caforio AL, Pankuweit S, Arbustini E, Basso C, Gimeno-Blanes J, Felix SB, et al. Current state of knowledge on aetiology, diagnosis, management, and therapy of myocarditis: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 2013;34:2636–48. 10.1093/eurheartj/eht210 [DOI] [PubMed] [Google Scholar]
- 61. Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 2015;525:389–93. 10.1038/nature15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sano S, Oshima K, Wang Y, MacLauchlan S, Katanasaka Y, Sano M, et al. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1β/NLRP3 inflammasome. J Am Coll Cardiol 2018;71:875–86. 10.1016/j.jacc.2017.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y, Sano S, Yura Y, Ke Z, Sano M, Oshima K, et al. Tet2-mediated clonal hematopoiesis in nonconditioned mice accelerates age-associated cardiac dysfunction. JCI Insight 2020;5:e135204. 10.1172/jci.insight.135204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Abplanalp WT, Cremer S, John D, Hoffmann J, Schuhmacher B, Merten M, et al. Clonal hematopoiesis-driver DNMT3A mutations alter immune cells in heart failure. Circ Res 2021;128:216–28. 10.1161/circresaha.120.317104 [DOI] [PubMed] [Google Scholar]
- 65. Liao M, Chen R, Yang Y, He H, Xu L, Jiang Y, et al. Aging-elevated inflammation promotes DNMT3A R878H-driven clonal hematopoiesis. Acta Pharm Sin B 2022;12:678–91. 10.1016/j.apsb.2021.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Deftereos SG, Beerkens FJ, Shah B, Giannopoulos G, Vrachatis DA, Giotaki SG, et al. Colchicine in cardiovascular disease. In-depth review. Circulation 2022;145:61–78. 10.1161/circulationaha.121.056171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sikking MA, Stroeks S, Marelli-Berg F, Heymans SRB, Ludewig B, Verdonschot JAJ. Immunomodulation of myocardial fibrosis. JACC Basic Transl Sci 2023;8:1477–88. 10.1016/j.jacbts.2023.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Halliday BP, Gulati A, Ali A, Guha K, Newsome S, Arzanauskaite M, et al. Association between midwall late gadolinium enhancement and sudden cardiac death in patients with dilated cardiomyopathy and mild and moderate left ventricular systolic dysfunction. Circulation 2017;135:2106–15. 10.1161/CIRCULATIONAHA.116.026910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shumliakivska M, Luxán G, Hemmerling I, Scheller M, Li X, Müller-Tidow C, et al. DNMT3A clonal hematopoiesis-driver mutations induce cardiac fibrosis by paracrine activation of fibroblasts. Nat Commun 2024;15:606. 10.1038/s41467-023-43003-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gibson CJ, Lindsley RC, Tchekmedyian V, Mar BG, Shi J, Jaiswal S, et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol 2017;35:1598–605. 10.1200/jco.2016.71.6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 2017;21:374–82.e4. 10.1016/j.stem.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bolton KL, Ptashkin RN, Gao T, Braunstein L, Devlin SM, Kelly D, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet 2020;52:1219–26. 10.1038/s41588-020-00710-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015;518:552–5. 10.1038/nature13968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hsu JI, Dayaram T, Tovy A, De Braekeleer E, Jeong M, Wang F, et al. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell 2018;23:700–13.e6. 10.1016/j.stem.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fuster JJ. Clonal hematopoiesis and cardiovascular disease in cancer patients and survivors. Thromb Res 2022;213:S107–12. 10.1016/j.thromres.2021.12.009 [DOI] [PubMed] [Google Scholar]
- 76. Zamorano JL, Gottfridsson C, Asteggiano R, Atar D, Badimon L, Bax JJ, et al. The cancer patient and cardiology. Eur J Heart Fail 2020;22:2290–309. 10.1002/ejhf.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yura Y, Cochran JD, Walsh K. Therapy-related clonal hematopoiesis: a new link between cancer and cardiovascular disease. Heart Fail Clin 2022;18:349–59. 10.1016/j.hfc.2022.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ness KK, Kirkland JL, Gramatges MM, Wang Z, Kundu M, McCastlain K, et al. Premature physiologic aging as a paradigm for understanding increased risk of adverse health across the lifespan of survivors of childhood cancer. J Clin Oncol 2018;36:2206–15. 10.1200/jco.2017.76.7467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ness KK, Krull KR, Jones KE, Mulrooney DA, Armstrong GT, Green DM, et al. Physiologic frailty as a sign of accelerated aging among adult survivors of childhood cancer: a report from the St Jude Lifetime cohort study. J Clin Oncol 2013;31:4496–503. 10.1200/jco.2013.52.2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Armstrong GT, Liu Q, Yasui Y, Neglia JP, Leisenring W, Robison LL, et al. Late mortality among 5-year survivors of childhood cancer: a summary from the childhood cancer survivor study. J Clin Oncol 2009;27:2328–38. 10.1200/jco.2008.21.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mertens AC, Liu Q, Neglia JP, Wasilewski K, Leisenring W, Armstrong GT, et al. Cause-specific late mortality among 5-year survivors of childhood cancer: the childhood cancer survivor study. J Natl Cancer Inst 2008;100:1368–79. 10.1093/jnci/djn310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Collord G, Park N, Podestà M, Dagnino M, Cilloni D, Jones D, et al. Clonal haematopoiesis is not prevalent in survivors of childhood cancer. Br J Haematol 2018;181:537–9. 10.1111/bjh.14630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hagiwara K, Natarajan S, Wang Z, Zubair H, Mulder HL, Dong L, et al. Dynamics of age- versus therapy-related clonal hematopoiesis in long-term survivors of pediatric cancer. Cancer Discov 2023;13:844–57. 10.1158/2159-8290.Cd-22-0956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Novetsky Friedman D, Chan ICC, Moskowitz CS, Li S, Turner K, Liu J, et al. Clonal hematopoiesis in survivors of childhood cancer. Blood Adv 2023;7:4102–6. 10.1182/bloodadvances.2023009817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yura Y, Miura-Yura E, Katanasaka Y, Min K-D, Chavkin N, Polizio AH, et al. The cancer therapy-related clonal hematopoiesis driver gene ppm1d promotes inflammation and non-ischemic heart failure in mice. Circ Res 2021;129:684–98. 10.1161/circresaha.121.319314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sano S, Wang Y, Ogawa H, Horitani K, Sano M, Polizio AH, et al. TP53-mediated therapy-related clonal hematopoiesis contributes to doxorubicin-induced cardiomyopathy by augmenting a neutrophil-mediated cytotoxic response. JCI Insight 2021;6:e146076. 10.1172/jci.insight.146076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 2016;7:12484. 10.1038/ncomms12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Weinstock JS, Gopakumar J, Burugula BB, Uddin MM, Jahn N, Belk JA, et al. Aberrant activation of TCL1A promotes stem cell expansion in clonal haematopoiesis. Nature 2023;616:755–63. 10.1038/s41586-023-05806-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Robertson NA, Latorre-Crespo E, Terradas-Terradas M, Lemos-Portela J, Purcell AC, Livesey BJ, et al. Longitudinal dynamics of clonal hematopoiesis identifies gene-specific fitness effects. Nat Med 2022;28:1439–46. 10.1038/s41591-022-01883-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fabre MA, de Almeida JG, Fiorillo E, Mitchell E, Damaskou A, Rak J, et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature 2022;606:335–42. 10.1038/s41586-022-04785-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. van Zeventer IA, de Graaf AO, Salzbrunn JB, Nolte IM, Kamphuis P, Dinmohamed A, et al. Evolutionary landscape of clonal hematopoiesis in 3,359 individuals from the general population. Cancer Cell 2023;41:1017–31.e4. 10.1016/j.ccell.2023.04.006 [DOI] [PubMed] [Google Scholar]
- 92. Mack T, Vlasschaert C, von Beck K, Silver AJ, Heimlich JB, Poisner H, et al. Cost-effective and scalable clonal hematopoiesis assay provides insight into clonal dynamics. J Mol Diagn 2024;26:563–73. 10.1016/j.jmoldx.2024.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Andersson-Assarsson JC, van Deuren RC, Kristensson FM, Steehouwer M, Sjöholm K, Svensson P-A, et al. Evolution of age-related mutation-driven clonal haematopoiesis over 20 years is associated with metabolic dysfunction in obesity. EBioMed 2023;92:104621. 10.1016/j.ebiom.2023.104621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Uslu U, June CH. CAR T-cell therapy meets clonal hematopoiesis. Blood Cancer Discov 2022;3:382–4. 10.1158/2643-3230.BCD-22-0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tian Z, Liu M, Zhang Y, Wang X. Bispecific T cell engagers: an emerging therapy for management of hematologic malignancies. J Hematol Oncol 2021;14:75. 10.1186/s13045-021-01084-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ogawa H, Sano S, Walsh K. Employing the CRISPR-Cas system for clonal hematopoiesis research. Int J Phys Med Rehabil 2021;9:582. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were generated or analysed for or in support of this article.