Abstract
Science-informed decisions are best guided by the objective synthesis of the totality of evidence around a particular question and assessing its trustworthiness through systematic processes. However, there are major barriers and challenges that limit science-informed food and nutrition policy, practice, and guidance. First, insufficient evidence, primarily due to acquisition cost of generating high-quality data, and the complexity of the diet-disease relationship. Furthermore, the sheer number of systematic reviews needed across the entire agriculture and food value chain, and the cost and time required to conduct them, can delay the translation of science to policy. Artificial intelligence offers the opportunity to (i) better understand the complex etiology of diet-related chronic diseases, (ii) bring more precision to our understanding of the variation among individuals in the diet-chronic disease relationship, (iii) provide new types of computed data related to the efficacy and effectiveness of nutrition/food interventions in health promotion, and (iv) automate the generation of systematic reviews that support timely decisions. These advances include the acquisition and synthesis of heterogeneous and multimodal datasets. This perspective summarizes a meeting convened at the National Academy of Sciences, Engineering, and Medicine. The purpose of the meeting was to examine the current state and future potential of artificial intelligence in generating new types of computed data as well as automating the generation of systematic reviews to support evidence-based food and nutrition policy, practice, and guidance.
Keywords: nutrition, computed evidence, artificial intelligence, evidence synthesis, systematic reviews
Introduction
Science-informed decisions are guided by the objective synthesis of the totality of evidence around a particular question and assessing its trustworthiness through the process of conducting a systematic review (SR). This approach has become fundamental to evidence-based food and nutrition policy, practice, and guidance (1–3). Evidence synthesis and evaluation considers the strength of all forms of scientific data and is used across medicine, public health, and the social sciences.
SRs guide the process for setting essential nutrient intake recommendations for individuals and populations, such as the Dietary Reference Intakes (DRIs) (4), and for food-based intake recommendations, including the Dietary Guidelines for Americans (3). Guidance on nutrient and other food substances is based on derived normative values and include the Recommended Dietary Allowance, the Estimated Average Requirement, and the Tolerable Upper Intake Level. The DRIs inform food and nutrition policies, including the Dietary Guidelines for Americans (5); food fortification policies (6); food assistance programs (7); food safety, labeling, and other regulatory decisions (8); and nutrition education programs (9), and can influence food production systems (8). The food and agriculture economy contributes $1.53 trillion to the United States gross domestic product (∼5.6% of overall) (10), while food-related health effects due to cardiometabolic diseases including hypertension, stroke, type 2 diabetes, and heart disease, account for $50 billion per year in healthcare costs (11), highlighting the importance of bringing the very best and current science available to policy and other decision makers. However, there are major barriers and bottlenecks that limit the opportunity to achieve science-informed food and nutrition policy. These include a dearth of high-quality scientific data to inform policy decisions, the costs of generating high-quality food and nutrition experimental data, and the vast and rapidly growing literature base; the sheer number of SRs required to address all policy-related questions across the entire agriculture and food value chain; and the cost and time required to conduct SRs, among others. These challenges have been reviewed elsewhere (12, 13).
The landscape is further complicated by the increasing interest in setting food and nutrition guidance and policies to lower rates of diet-related chronic diseases, which are a major driver of healthcare costs in the United States (11). Historically, the DRIs and the Dietary Guidelines for Americans were established to inform food and nutrient intakes in “apparently healthy” individuals to maintain nutritional adequacy and avoid diseases of nutrient deficiencies. Compared with diet-related chronic diseases, nutritional deficiency in otherwise healthy individuals generally has a single cause, which is a lack of dietary intake of a particular essential nutrient. Furthermore, virtually all healthy individuals respond similarly to dietary deficiency of a particular nutrient in terms of the dose-response relationship and resulting clinical manifestations. This is not the case when diet-related chronic disease is the outcome used for setting food and nutrition guidance, programs, and policies. The etiologies of chronic diseases are highly complex, resulting from the interactions among many essential nutrients and nonessential dietary components. In addition, chronic disease etiologies are modified by differences in individual biology as well as by multiple lifestyle factors and exposures including physical activity, sleep, stress, diet, eating behaviors, immune responses, and toxins, among other factors. The contextual factors that modify connections between food and health are even more complex in low- and middle-income country settings. Hence, it is not surprising there is significant population heterogeneity in the diet-chronic disease relationship compared with that between diet and nutrient deficiencies, indicating the need for new approaches to stratify populations to improve the precision of recommendations based on various contexts (14).
Population-based diet, food, and nutrition recommendations have focused on avoiding essential nutrient deficiencies with consideration for “apparently healthy individuals,” because the disease process can alter nutritional requirements (15). However, when considering chronic disease reduction as an endpoint for nutrient intake recommendations, individuals at risk for or who have chronic disease cannot be excluded because diet-related chronic diseases can initiate as early as during embryonic development and manifest over a lifetime. More than 60% of US adults are affected by a chronic disease, and food- and nutrient-based guidance based on avoidance of nutritional deficiency may not apply to them (16). Globally, essential nutrient deficiencies occur in the obese state. Hence, inclusion of chronic disease outcomes for food and nutrition guidance greatly expands the population under consideration and adds additional heterogeneity in response to dietary and nutrient intake.
Consideration of chronic disease endpoints also expands the number of food components under consideration from essential nutrients to any food component that, while not essential, confers a health benefit (17), further increasing the complexity of food and nutrition guideline development. As such, inclusion of chronic disease endpoints in food and nutrition guidance requires expansion of the populations under consideration. Considering these issues, the National Academies of Sciences, Engineering, and Medicine recently expanded the definition of the target population for DRI values to include those with or at risk for chronic disease, with each expert committee being responsible for establishing exceptions that apply specifically to the nutrient(s) under review (18). This expansion of the population under consideration adds to the complexity of data required for establishing recommendations.
Technology terms and applications to nutrition evidence synthesis
Broadly defined, artificial intelligence (AI) refers to technologies capable of mimicking human intelligence, including having the capacity to solve complex problems and inclusive of various terms and types of strategies as it pertains to evidence synthesis (19). Over the past decade, AI has emerged as an important technology that may provide decision support, early on with specialized deep learning architectures and more recently with general, pretrained large language models (LLMs) (20, 21).
Modern LLMs have emerged because of (i) parameter estimation algorithms that make it possible to train models with billions or trillions of parameters; (ii) computing infrastructure such as graphics processing units that make it possible to fit models in days or weeks, rather than in decades, but may be cost prohibitive to most researchers; and (iii) Internet-scale training data corpuses, enabling an arsenal of applications, some of which are deeply embedded in our everyday lives (22, 23). In food and nutrition, AI is now being utilized to guide more precise and accurate food and nutrition guidance to improve health (24). Data science methods offer the opportunity to (i) automate and thereby accelerate the process of synthesizing data and generating SRs, saving cost and providing decision makers with up-to-date and comprehensive scientific information to make timely decisions; (ii) provide new types of computed data with respect to the complex etiology of the diet-disease relationship; and (iii) identify and classify variation in individual responses to diet. As such, AI offers a timely and cost-effective avenue to develop the strong evidence base necessary to establish effective nutrition/food interventions that prevent and/or manage chronic disease, including data such as electronic medical records (EMRs), as well as take advantage of new types of personalized data from wearables. However, the quality and sparsity of data currently available for such AI-based analyses limit its utility.
Purpose of the summary
This perspective summarizes a meeting convened at the National Academy of Sciences, Engineering, and Medicine (Tables S1, S2). The purpose of the meeting was to examine the current state and future potential of AI in generating new types of computed data as well as in automating the generation of SRs to support evidence-based food and nutrition policy, practice, and guidance. Participants included expert computational, data, and nutrition scientists, as well as scientists from federal research and regulatory agencies. The conference agenda was organized into two main areas (i.e. “parts”) as outlined subsequently.
Part I: emerging sources of scientific evidence
Establishing scientific recommendations for chronic disease risk reduction through food and nutrition presents an enormous data challenge. This is due to the complexity of food and food components that individuals are exposed to, the variation in individual response to food and nutrient exposures, the number of chronic diseases that are affected by food, and the latency and cumulative effects of nutrition on the progression of diet-related diseases that manifest over a lifetime, among many other factors that have been described elsewhere (17). This complexity and the associated costs limit the generation of high-quality scientific evidence through randomized controlled trials (RCTs) that are most often short in duration due to the funding structure of research. The availability of large EMR databases and related real-world health and exposure data, coupled with advances in AI models that mine and automate the synthesis of these resources, provides additional inputs into causal inference models that may provide a less expensive approach to understand the diet-disease relationship and its inherent individual variation. However, EMRs currently have limited data on dietary intakes, nutritional biomarkers and other relevant variables at present. With all AI models, a key consideration is the nature of the training or input data. Cross-sectional data, for example, are limited for making causal claims, whereas longitudinal data are logistically challenging to collect, and experience confounding, but represent the longer latency of nutritional exposures and chronic disease risk. Ultimately, the quality of any synthesis of any data relies on the scientific rigor and data available for (i.e. “garbage in, garbage out”). Training AI models to advance evidence synthesis can be coordinated and managed by efforts to collect the optimal combinations of data needed to leverage the potential of and amount of data needed to seed AI models.
Lessons learned from cancer drug response prediction models
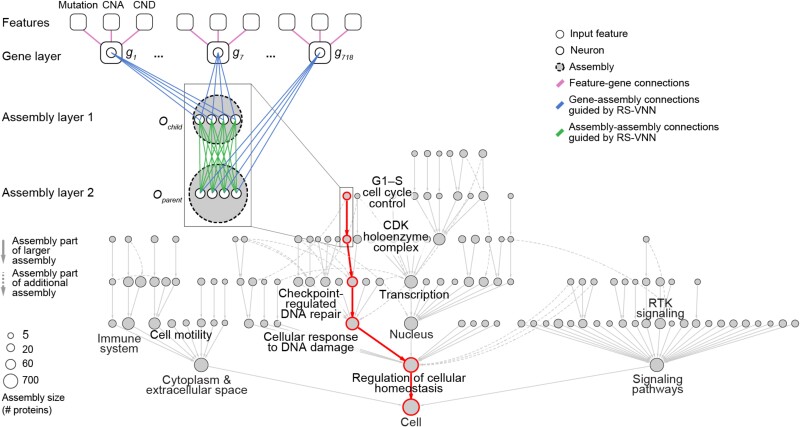
Deep neural networks are actively being deployed in sophisticated models for predicting therapeutic responses in cancer (25). However, two major challenges continue to prevent their integration into broader clinical practice (26). The first is lack of model interpretability. The ability to scrutinize the inner workings of a model is critical to building trustworthy AI tools, especially in high-stakes applications such as precision medicine. Visible neural networks (VNNs) enable direct model interpretation by mapping the neural network architecture to hierarchical knowledge graphs of biological components and functions (Figure 1) (27, 28). A recently published VNN predicted palbociclib efficacy in breast cancer treatment; it captured 8 molecular assemblies integrating rare and common mutations in 90 genes (29). Another recent publication highlighted 41 assemblies involved in modulating response to common chemotherapies (30). These works serve as illustrative proofs of concept to develop robust composite biomarkers.
Fig. 1.
VNN. The first layer of the VNN incorporates gene-level features, including gene mutations, copy number amplifications (CNAs), and copy number deletions (CNDs). Subsequent assembly layers aggregate gene-level features into assembly-level information, guided by the hierarchical relationships defined by a map of protein assemblies. The output state of each gene (g) and assembly (O) is represented by artificial neurons (1 neuron per gene, multiple neurons per assembly). Each node in the hierarchy indicates a protein assembly. An example path of information flow is shown in red. Adapted from Park et al. (29). RS-VNN, random set VNN.
A second challenge is related to generalizability. Drug response prediction models are often trained on preclinical datasets. Transferring information from large preclinical datasets to accurately predict treatment response to smaller patient datasets is particularly challenging, and may require careful causal modeling. For predictive tasks, massive pretrained networks can adapt to new tasks when provided only a handful of examples; this is called “few-shot learning” and was used to perform Translation of Cellular Response Prediction (31). This approach realized better predictive performance across multiple data types, including tumor cell lines, patient-derived tumor cell cultures, and patient-derived tumor xenografts. An independent reusability study was able to apply this approach to 2 patient cohorts and demonstrated its superior performance (32).
VNN models are not yet common in elucidating the role of dietary exposures and their availability on cellular networks and biomarkers of disease etiology. If well executed and validated, these tools can inform biomarker discovery from basic research for clinical utility. This requires the transfer of information across a series of contexts (e.g. from cell lines to patients, from one patient cohort to another, from large populations to small ones or even individuals) with limited data. Similarly, few-shot learning may be applied to transfer biomarkers across contexts.
Knowledge graphs to reveal the etiology of chronic diseases
Mining multidimensional patient data that includes endome-type data (e.g. clinical exams, laboratory data, imaging, genetics) and ectome-type data (e.g. age, demographics, exposures, food, social determinants of health) may allow comprehensive consideration of the risk factors underpinning the etiologies of chronic disease. Extracting trustworthy information from large datasets that is both statistically and biologically meaningful, and that can infer causal factors and their relationships, is yet unrealized (33) but is essential for developing effective interventions that are tailored to the context of an individual's circumstances. Knowledge graphs are a tool to convert large volumes of new data to information and ultimately actionable knowledge but must come from well-established information and include layers of hierarchical organization, their interactions, and relationships across the continuum, including consideration of biological and social complexity. Such bottom-up approaches interconnect layered networks within and across known biological, social, and other domains. The domains can include genes to proteins to pathways (metabolic, signaling, etc.), to cells, organs, the microbiome, and the individual within the social context, including the complexity of spaces and locations related to disease incidence, exposures, and temporal changes that individuals experience.
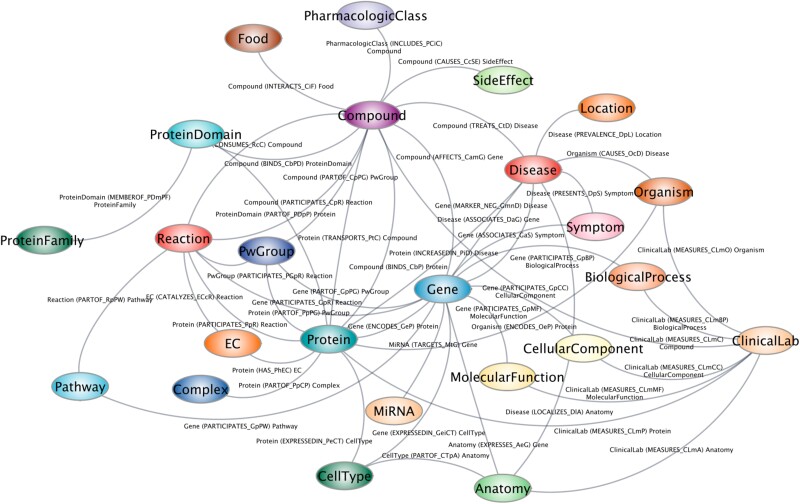
One example knowledge graph is the Scalable Precision Medicine Open Knowledge Engine (SPOKE) (34). It has over 40 million concepts that are connected by over 120 established biologically meaningful relationships gathered from existing knowledge in the scientific literature. SPOKE was built by integrating information from more than 50 public databases and contains experimentally determined information on various biological pathways and their architecture, with every node within a network receiving a weighted score relative to its overall importance explaining to risk or function. SPOKE has recently incorporated more than 1,000 food items and their relationships to biochemical compounds as determined by mass spectrometry (Figure 2) (34). When SPOKE analyzed data from 6 million EMRs, it led to the identification of the nodes of most importance to Parkinson's disease. SPOKE retrospectively predicted individuals who would develop the disease 3 years prior to a diagnosis with 83% accuracy and performed similarly to that of clinical expert predictions (35). While not currently clinical grade/caliber, further development and refinement of SPOKE is expected to support its deployment in medical practice. SPOKE includes more than 10,000 disease states and can be used toward discovery and applications related to food, health, and disease. In nutrition research, SPOKE can be used to generate hypotheses by predicting the immediate biological and long-term health effects of consuming individual nutrients and other bioactive food components through a dietary supplement, the optimal combinations of nutrient intakes, and/or effects of consuming specific foods or dietary patterns.
Fig. 2.
SPOKE. The SPOKE biomedical knowledge graph draws upon and integrates over 45 databases.
Predictive modeling and individual responses
The concept of precision nutrition is founded on the premise that identifiable subgroups of individuals respond differently to nutrients, foods, and dietary patterns when chronic disease endpoints are considered (14, 36). The need for precision nutrition is supported by our understanding of human evolution. Human responses to food and nutrition have been under a strong selective pressure in the face of increasing genetic diversity through adaptation to local food environments, which differed considerably across the globe. Adaptation to local food environments enabled population expansion, as classically seen with genetic variation that enabled lactase persistence (37). However, the degree of meaningful biological variation among individuals that necessitates more precision in food and nutrition interventions and recommendations for chronic disease risk reduction remains unresolved. To fully establish the need for greater precision in food and nutrition guidance, two critical questions need to be addressed. First, are the differences among individuals clinically meaningful? Second, do we have predictive biomarkers for the diet-health response?
Prediction models are widely used to mine massive datasets to explore the complexity underlying the interactions among endogenous biological factors and environmental exposures that define or relate to human health. Importantly, they offer the possibility of identifying causal dietary and other factors and predicting their intervention response (38). Establishing reliable predictive models of intervention responses has proved challenging due to limitations including bias resulting from several sources, such as algorithmic bias (39), data collected for one purpose being used for other purposes, lack of participant diversity, and lack of domain expertise in data selection, among others (40). This, and a dearth of success stories, has led to skepticism for identifying biomarkers that are predictive of medical and nutritional intervention responses. For example, AI prediction models of antipsychotic medications trained on RCT data failed to predict patient outcomes when applied to out-of-sample patients, indicating that treatment outcomes are not generalizable for schizophrenia, emphasizing the strong modifying effects of an individual's contexts (41).
Traditional clinical trials focusing on nutrition and pharmaceuticals are typically designed to determine the average effect of an intervention, which becomes the evidence base for establishing generalized population-based applications. These trial designs give less attention to the variation in response among individuals and in fact may mask positive or negative outcomes among subgroups of participants. In contrast, N-of-1 trials seek to identify and characterize variation in responses to multiple interventions provided to the same individual, often separated by washout periods, and thereby optimize interventions for that individual (42). N-of-1 trials thereby determine which interventions are better suited for individuals with certain characteristics (43). Such studies that seek to identify and quantify variation around an average response, or a more discreet effect revealing overt responders and nonresponders, can be cost-efficient, as the statistical power is optimized when the number of observations is maximized on fewer individuals, compared with fewer observations on more individuals. N-of-1 trials have been used in the fields of psychology and education research, but to a lesser extent in nutrition research.
Predictive model reliability can be improved by combining sparse real-world data in large samples with more rigorously collected and outcome-focused data, including data collected during clinical trials. This approach can be more efficient than using sparse data on large number of individuals or very costly yet plentiful experimental data on fewer individuals. For example, models built on massive, randomly sampled, sparse, real-world data, such as the UK Biobank and the National Institutes of Health–funded All Of Us Study, can be strengthened by calibrating with more sophisticated dense, yet costly, empirical data (44), such as that derived from aggregated N-of-1 studies. In this light, aggregated N-of-1 trials might be efficient and appropriate vehicles for vetting or testing the predictions of population-based AI/LLM analyses. Thus, if a new AI/LLM-based model is designed to determine which individuals are likely to benefit from a nutritional intervention, then more detailed studies of well-chosen data subsets from individuals for whom predictions were made should shed light on their veracity and expose limitations. This approach is essential to advance the concept of precision nutrition.
Other strategies have been employed to strengthen real-world data to understand variation in response (45). AI techniques used to identify factors that are associated with an intervention response are limited by the datasets that they are trained on and cannot be used to infer causation. Training models on more detailed experimental trial data, with limited training on readily available contextual real-world data (e.g. EMRs, large epidemiological datasets), enhances their ability to identify predictive factors and account for variation in individual responses. Such approaches, carefully deployed, have the potential to be more cost-effective and potentially more reliable than conducting large RCTs.
Use of digital twins can also improve predictive models by accounting for the factors that lead to variation in responses. This is achieved by limiting training sets to specified subsets of individuals within a dataset who share similar characteristics. Digital twins may better anticipate the health trajectory of a target individual (i.e. “digital twins” of the target individual) as opposed to using all individuals in the large dataset when making predictions about the target individual's health trajectory. Digital twins may share similar genetic, demographic, microbiome, and other characteristics (46–48).
Addressing the complexity of food systems, diets, and their relationship to health
Food systems, diets, nutrition, and human health exist along a continuum. Dietary patterns differ by context across geography, culture, and socioeconomic status, among other factors (49). Food consumption also has temporal, hedonic, religious, and social dimensions, all of which may relate to health outcomes (14). This has motivated interest in applying AI tools to establish connections across the food value chain and thereby identify opportunities to improve the health-promoting properties of the food system. Traditionally, meta-analyses have been instrumental in understanding the impact of dietary practices and help inform medical decisions. Data science technologies permit a comprehensive approach to addressing food systems and health within these contexts.
One example of a dietary pattern that is used clinically is FODMAP (fermentable oligosaccharides, disaccharides, monosaccharides, and polyols). A recent AI model used data from various studies to correlate the success of low FODMAP diets for the treatment of patients with irritable bowel syndrome, in which only 50% to 70% of the patients respond well to this standard of care treatment. The AI model combined metagenomics and machine learning analysis and provided hypotheses about the mechanism explaining patient segmentation, predicted patient response, and informed treatment decision based on 3 biomarkers (50). Expanding this approach for management of other chronic diseases through diet is an active area of investigation.
Knowledge graphs are also playing a key role in assembling and structuring data related to food composition. Agricultural food products contain tens of thousands of chemicals. The FoodData Central database from the US Department of Agriculture curates compositional information from 236 foods and 400 chemicals that have been validated (51). Recently, there have been advances in streamlining the generation of knowledge graphs with using deep natural language processing (NLP) techniques and LLMs to support decision support and accelerated discovery (52). Food Atlas (53) is an AI-generated knowledge graph that has extracted more than 230,000 food-chemical composition relationships from more than 155,000 scientific papers, and ranked the confidence level of each relationship based on the existing published evidence (54). This analysis estimated that approximately half of the identified relationships were not previously discovered. While false discovery rate is always a caveat to consider, this lends credence to the potential for utilizing such techniques for discovery. By applying knowledge graph completion methods, new hypotheses can be formed and experimentally validated, providing a framework for automated hypothesis generation. The next version of Food Atlas that is under release uses a combination of LLMs and hybrid knowledge graph language models to integrate food, ingredients, chemicals, flavors and health effects.
Part II: accelerating the process of evidence synthesis
The body of unstructured biomedical data is vast and growing rapidly, hindering physicians’ and policymakers’ ability to make the most informed decisions grounded in the totality of the evidence base. SRs and evidence synthesis are key to developing evidence informed decisions whether they are from a medical, research, or policy lens. However, the process of conducting and publishing SRs is time-consuming and expensive, and many of the tasks are highly repetitive but cannot be automated trivially. Consequently, only half of high-quality reviews in biomedical and allied health fields are completed within 2 years of protocol publication (55). SRs can be expensive to produce and can quickly become outdated, sometimes even before they are published (56), lending credence for the need for newer methods that function in real time. Study screening, data extraction, and synthesis are key bottlenecks in generating SRs. There is a need to design, implement, and deploy NLP tasks, corpora, and models to help domain experts navigate and make sense of the vast array of biomedical evidence, ranging from notes in EMRs to published reports of clinical trials, which are generally stored as unstructured text and therefore not readily accessible or mineable.
High-quality evidence synthesis adheres to the principles of transparency, reproducibility, and methodological rigor, following prespecified processes (57, 58). Otherwise, SR findings/conclusions can be highly dependent or influenced by subjective judgments (59, 60). It is these and related challenges that motivated the development of AI tools for SRs, but the uptake has been slow (61). By necessity and logic, the process must include human judgment or oversight in the identification of the relevant literature base from raw search results (based on prespecified search criteria that is then screened) as well as in the rating the risk of bias of individual studies and grading the overall certainty of the available evidence (3). Literature screening, usually conducted manually by human non–content experts (e.g. trainees, students, contractors), is the most time- and resource-intensive stage of the process and can be subject to various types of bias. To mitigate bias and the temporal currency of the SR process, human-AI hybrid approaches have been developed, and evaluated for their effectiveness, in accelerating the generation of high-quality evidence synthesis products that promote timely evidence-based scientific guidance for decision makers.
Goals of including AI applications in the evidence synthesis process include accelerating innovation and time to completion, improving productivity, and cost reduction (62). Title and abstract screening for inclusion in an SR generally reduces the number of studies identified through a literature search by 95%, and hence is a task that is well suited for automated text classification. Early NLP models were frequency-based models, classifying studies by the frequency of individual terms within a document/text. More recent approaches use neural network–based methods, up to and including LLMs. Currently, there are several AI-powered screening tools (both commercial and open source) available to accelerate title and abstract screening but rely on frequency-based models (e.g. EPPI-Reviewer, abstrackr, DistillerSR, RobotReviewer, Rayyan) that represent the industry standard (63–66). As an example, the US Department of Agriculture Nutrition Evidence SR group, which conducts SRs in support of establishing the Dietary Guidelines for Americans, uses AI-powered screening tools (3).
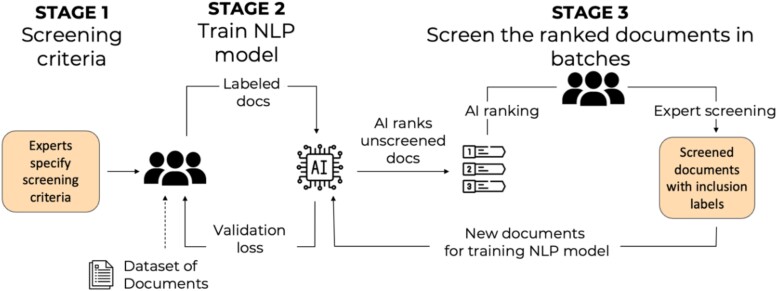
An early and relatively large pretrained neural network was the bidirectional encoder representations from transformers (BERT). BERT is pretrained on a large volume of text, and can be fine-tuned for particular tasks. This model has been incorporated into human-AI hybrid evidence synthesis teams (62). The collaborative screening process involves subject matter experts identifying the screening criteria, followed by the training of the NLP using a limited number of studies screened by subject matter experts. Once the model is judged to function adequately, it ranks new documents never seen by the model (62). A final review of all selected documents is conducted by experts. The approach is iterative as feedback from the experts continuously constrains and improves the model. The approach may incorporate active learning into the human-AI hybrid team by exploring and testing different sampling strategies, including random sampling, least confidence sampling, and highest priority sampling, and evaluating their effectiveness on the collaborative screening process.
Incorporating the BERT-based AI agent into a human team was found to reduce the human screening effort, including the number of documents that humans need to read, by 68.5% compared with the case of no AI assistance, and by 16.8% compared with the industry standard that uses a frequency-based language model and a support vector machine–based classifier (Figure 3). These values are for the human screening effort required to identify 80% of all relevant documents. The process was further improved by applying an high priority sampling strategy to the human screening effort, resulting in 78.3% reduction in human screening effort to identify 80% of all relevant documents compared with no AI assistance. The BERT-based model uniformly outperformed the industry standard NLPs in classification performance.
Fig. 3.
A human-AI workflow for document screening in evidence synthesis. In stage 1, experts specify screening criteria for documents, then screen a subset of the documents-of-interest for inclusion or exclusion in an evidence synthesis product. In stage 2, an AI model is trained on the expert labels of screened documents, and then performs screening of additional documents. In stage 3, expert labelers evaluate the AI's screening decisions. The final validated screening decisions are used to iteratively retrain the AI model.
Key limitations to using an active learning–enhanced human-AI hybrid team workflow process are the time of communication among subject matter experts and computational scientists; the level of measurement error inherent to human labels, which is addressed with additional iterative training; and trust among the experts and the model. Future expansion to full-text screening is expected to improve classification performance but can be limited by inaccessible documents that are not published in open-access format. It is important to note that the field of LLMs is changing rapidly and becoming more powerful with generative models, which could improve accuracy and be able to summarize evidence but will require validation.
Extracting and synthesizing medical evidence with LLMs
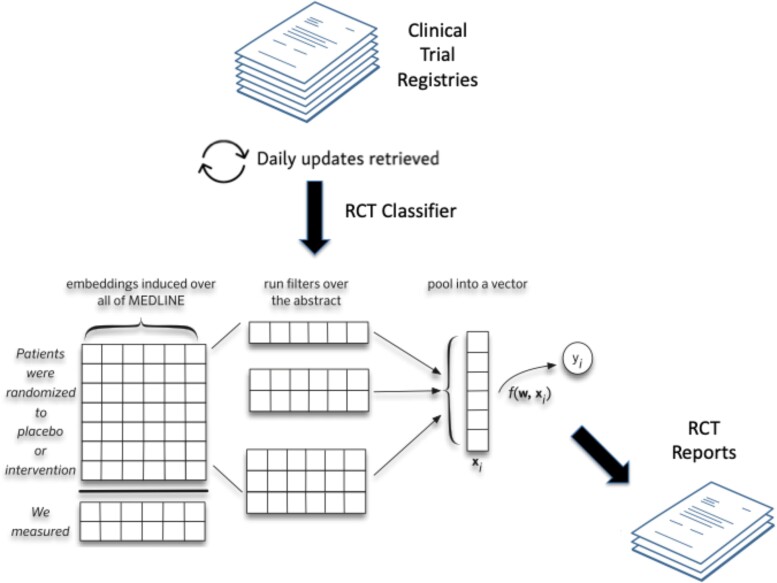
Clinical trial results are disseminated through natural language articles and hence are largely unstructured or semi-structured, including clinical trial databases such as ClinicalTrials.gov. NLP methods in general, and automated summarization in particular, offer a potential means of helping domain experts identify and make better use of the totality of scientific data to inform treatment and other-related decisions. Variants of LLMs are being used to extract and structure findings from clinical trial reports, and to generate automatic summaries of all published evidence pertaining to a particular clinical question. An available prototype, Trialstreamer, is a publicly available living repository of all articles describing RCTs in humans that makes RCT data fully computable (Figure 4) (64, 67). It monitors PubMed and other sources daily, then structures the data using models that extract and tabulate key information including PICO (population, intervention, comparison, outcome) element information and other metrics such as sample sizes. Trialstreamer can conduct aspects of Cochrane-style risk-of-bias assessments, such as whether a trial was randomized or blinded, which otherwise involves subjective judgments by humans. Trialstreamer can infer main findings of a study through a semi-automated process that accelerates human assessment by about 30% (68), and the results are generally in agreement with human assessments. The database can be searched for all studies relevant to a well-formed clinical question if indexed by PubMed (emerging prepublication websites, by lack of peer review, are not incorporated).
Fig. 4.
Trialstreamer: a living automated automatically updated database of clinical trial reports (67).
In development for the next iteration of Trialstreamer is the capability to generate Cochrane-style SRs, including meta-analyses, and a natural language narrative that describes the summary of results. Current technologies permit automatic generation of plausible summaries but may, or even often, include “hallucinations” in the conclusions, which is a real problem that needs to be addressed to ensure “trustworthy” information. Other limitations pertain to the assessment of more nuanced information from studies, such as extraction and critical appraisal of intervention and outcome ascertainment methods given the discipline- and method-specific nature of this kind of data.
Key performance indicators for AI-assisted evidence synthesis
Looking forward, automated evidence synthesis products must be fit for purpose, and the evidence synthesis processes should be robust a predictably changing environment (e.g. the increasing rate at which primary research is published) and rapidly responsive to unpredictable shocks (e.g. health emergencies such as the COVID-19 pandemic). This will require new tools and processes but should also build upon an understanding of three key performance indicators (KPIs): (i) time use and time to completion, (ii) resource use and economic sustainability, and (iii) correctness (69, 70). Shaping the future of evidence synthesis, both technologically and culturally, is essential to ensure that it continues to meet stakeholder needs.
The three KPIs have been assessed in a limited number of cases. A study by Tercero-Hidalgo et al. examined the influence of using AI in the SR process related to COVID-19 (71). The prespecified study included 3,999 SRs, 28 of which used AI. The use of AI was associated with publication in journals with a higher impact factor (8.9 vs. 3.5), more abstracts screened per author (302 vs. 140), and fewer texts screened per author (5.3 vs. 14 full texts) but curiously no effect on time to completion. In another prespecified study, Muller et al. examined the KPIs person-hours and time to completion prior to and following adoption of machine learning in the SR process from August 2020 to January 2023 at the Norwegian Institute of Public Health (70). This study also found using machine learning required more person-hours and other resources, with no effect on time to completion.
The third KPI, correctness, is the most difficult to assess, but could be evaluated by (i) comparing AI outputs with human reviewers, who are assumed to be making correct decisions; (ii) comparing AI outputs with results such as meta-analytical estimates from closed reviews under the assumption that findings in closed reviews are sufficiently close to the truth (reviews are closed if adding additional studies is expected not to change the existing findings); and (iii) a simulation approach in which AI tools for evidence synthesis are applied to bodies of literature using computed data, generated using models such as LLMs, in which the true values of effect measures such as hazard ratios are known by construction, facilitating comparison of AI outputs with known ground truth. To date, only the first approach has been used for analyses, and is biased by the assumption that human reviewers are correct when in reality they can introduce inconsistency due to human judgment.
Looking forward, there are many limitations of AI approaches that must be overcome to achieve correctness. It is recognized that there is a trade-off between accuracy and confidence with time savings and efficiency when automating evidence synthesis. Understanding what type of scientific product is needed for a particular purpose (e.g. guideline development) in which the need for comprehensiveness and accuracy versus expediency can be prespecified and reported transparently, otherwise cheap, fast, and possibly incorrect evidence synthesis may result.
AI tools may also be abused to quickly produce poor-quality “reviews,” which poses new threats to evidence synthesis. LLMs may also facilitate the production of fake, fraudulent, or flawed primary studies (e.g. zombie trials). It is estimated that hundreds of thousands of zombie trials already circulate in the literature, and their inclusion in evidence syntheses is problematic (72). Furthermore, online AI tools are vulnerable to digital attack including denial-of-service attacks and dataset poisoning (73). Other concerns include privacy violations, underrepresentation of studies in minority languages, and the commercial interests of companies marketing AI tools out of alignment with stakeholder needs.
Finally, AI tools are perhaps only necessary because scientific results are not reported using standardized structured data formats that permit accurate and comprehensive automated search and data extraction across the entire literature. While reports for some trials are available in machine-readable formats such as JSON and FHIR from ClinicalTrials.gov, future work could focus on dramatically extending the coverage and depth of scientific reporting, perhaps using fine-grained and federated graph databases and standardized ontologies.
Discussion
Advances in AI are providing decision makers new ways of accessing and making sense of scientific evidence. Although AI tools alone cannot generate evidence de novo, they are capable of processing, daisy-chaining, and/or merging evidence across existing datasets into new formats. They have been used to create synthetic dose-response relationships drawing on pathway data from different datasets, which have aided authoritative organizations in setting food and nutrition policy (74, 75). However, the trustworthiness of computed data, including information from VNN and knowledge graphs, and its relative positioning in the hierarchy of evidence, has not been addressed (76).
The established evidence hierarchy describes the strength of data types based on study design as they relate to causal inference. As one moves up the hierarchy, it is assumed that study quality increases and risk of bias decreases, and thereby the certainty of relationships between interventions/exposures and outcomes is higher (76). Well-designed RCTs, which sit at the top of the hierarchy, can determine causal relationships. As such, SRs, and meta-analyses of these trials are considered the highest level of evidence (76). However, like traditional RCT designs, SRs and meta-analyses generally emphasize average responses across many studies, and often fail to consider variation in response between studies or individuals (77). VNNs and knowledge graphs provide the opportunity to address overall effects of an intervention, as well as address variation in response among individuals, but their potential to determine causality has not been established (78), nor has there been consideration to how computed evidence compares with other traditional types of evidence. The quality of AI-assisted SRs is also dependent on the body of literature available.
Limitations to the established hierarchy of evidence include uncertain generalizability of the findings, even when the evidence for causation is strong. The lack of generalizability is rooted in biological heterogeneity within populations that contributes to variance in the exposure-outcome relationship. Likewise, social, environmental, and other contexts in free-living populations can influence efficacy and effectiveness of interventions or exposures. These effects on context limit the ability to predict nutrition intervention outcomes in low- and middle-income countries based on relationships and contexts established in high-income countries. Furthermore, the strength of evidence does not always inform whether interventions will have a meaningful magnitude of effect that has a clinical and public health value even when causal inference is strong. Knowledge graphs consider the many biological and social dimensions of food, individuals and health. Their application to nutrition questions, especially when combined with LLMs, presents an exciting and transformational opportunity to connect food and health in a way that considers individuals and their contexts.
Ideally, computed data will lead to multiple new types of evidence that will be available to decision makers, yet frameworks and appraisal tools do not exist to guide their use. Rather than a single hierarchy, there is an increasing need for a multidimensional assessment of the totality of the evidence that is fit for purpose and considers the properties of the evidence and how the outcomes are affected in multidimensional situations. Such a framework should consider and potentially rank the properties of different forms of scientific evidence including causality, generalizability, risk of bias, precision, dose-response, and magnitude of effect, and their relative importance for different purposes.
Decision makers emphasize the need to accelerate the synthesis of scientific data in response to emergent and sustained societal needs. This includes outcomes of efficacy, effectiveness, and equity across a population. Understanding the generalizability of even the strongest scientific evidence is also essential, as many policy decisions are made locally and include contextual realities in which research and policy making is done. Automating the SR process and incorporating computed evidence can address many of these concerns. For example, elements of equity can be improved by including data reported in underrepresented languages, which are often excluded, through LLMs.
In the ideal case, automated real-time collection and analyses of data of high relevance to clinical and public health from all sources is the goal. This will allow more rapid science-informed policies and create a continuously learning health system. Learning systems characterized by automated real-time collection and analyses of data in nutrition could facilitate regular updates to both the DGAs and DRIs as new data become available through a semi-automated process that includes expert input and review (79).
AI can also inform future research priorities. AI approaches can assist research funding agencies in identifying gaps in knowledge (identify holes or uncertainty in networks) in real time to guide and prioritize high-impact research needs that have a high societal return on investment, especially concerning both continuing and emerging public health threats, including setting priorities for the Dietary Guidelines for Americans.
Trust in food and nutrition research is essential, otherwise science-informed guidance and recommendations will not achieve or will diminish the impact of their intended health outcomes (80). The inclusion of validated and reliable data science tools into the process of food and nutrition research, and its translation for public benefit, offers the opportunity to increase public trust. This will be challenging, as current LLMs and other tools are essentially “black boxes”; no one knows exactly how they work, or when they will “hallucinate,” rather than provide correct information. While this may be less of a concern when these technologies are used in an analytical mode to screen, identify, or extract straightforward evidence during semi-automated evidence synthesis, applications of the technology that generate computed evidence will have to be carefully validated, replicated, and communicated transparently. On the other hand, data science tools offer the potential for more personalized nutrition guidance in which individuals can access the science and realize the benefit, as opposed to generalized recommendations that may not be optimal for everyone. These tools also offer the opportunity to reduce bias in nutrition. While data from individuals of European ancestry are overrepresented relative to US demographics in many health-related databases, AI tools such as digital twin approaches may allow us to minimize or eliminate biases and data misalignments by moving away from population averages that might poorly reflect underrepresented individuals and toward causal inferences and predictions that address the unique characteristics of individuals.
Finally, meaningful advances in the application of AI to nutrition research, policy, and practice will require the inclusion of more, consistently collected, richer nutrition and diet data in EMRs; greater engagement of data scientists with nutrition scientists; and ensuring the next generation of nutrition scientists are trained in the data sciences.
Supplementary Material
Acknowledgments
The authors are grateful to the Food and Nutrition Board of the National Academies of Sciences, Engineering, and Medicine for hosting the meeting. They are grateful to Mikkel Helding Vembye and Jens Dietrichson at the Danish Center for Social Science Research, Aarhus and Copenhagen, Denmark, for sharing their title and abstract screening results. The authors wish to acknowledge the attendance and contributions of meetings participants listed in Table S1. The views expressed in this paper are those of the authors and should not be construed to represent those of the National Academies of Sciences, Engineering, and Medicine; US Department of Agriculture; US Department of Health and Human Services; or any US government determination or policy.
Contributor Information
Regan L Bailey, Department of Nutrition, Texas A&M University, Cater-Mattil Hall, 373 Olsen Blvd Room 130, College Station, TX 77843, USA; Institute for Advancing Health Through Agriculture, Texas A&M University, Borlaug Building, College Station, TX 77843, USA.
Amanda J MacFarlane, Department of Nutrition, Texas A&M University, Cater-Mattil Hall, 373 Olsen Blvd Room 130, College Station, TX 77843, USA; Texas A&M Agriculture, Food, and Nutrition Evidence Center, 801 Cherry Street, Fort Worth, TX 76102, USA.
Martha S Field, Division of Nutritional Sciences, Cornell University, Savage Hall, Ithaca, NY 14850, USA.
Ilias Tagkopoulos, Department of Computer Science and Genome Center, University of California, Davis, One Shields Avenue, Davis, CA 95616, USA; USDA/NSF AI Institute for Next Generation Food Systems, University of California, Davis, One Shields Avenue, Davis, CA 95616, USA.
Sergio E Baranzini, Department of Neurology, Weill Institute for Neurosciences, University of California, San Francisco, 1651 4th St, San Francisco, CA 94158, USA.
Kristen M Edwards, Department of Mechanical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA.
Christopher J Rose, Cluster for Reviews and Health Technology Assessments, Norwegian Institute of Public Health, PO Box 222 Skøyen, 0213 Oslo, Norway; Centre for Epidemic Interventions Research, Norwegian Institute of Public Health, Lovisenberggata 8 0456, 0213 Oslo, Norway.
Nicholas J Schork, Translational Genomics Research Institute, City of Hope National Medical Center, 445 N. Fifth Street, Phoenix, AZ 85004, USA.
Akshat Singhal, Department of Computer Science and Engineering, University of California San Diego, 9500 Gilman Drive, San Diego, CA 92093, USA.
Byron C Wallace, Khoury College of Computer Sciences, Northeastern University, #202, West Village Residence Complex H, 440 Huntington Ave, Boston, MA 02115, USA.
Kelly P Fisher, Institute for Advancing Health Through Agriculture, Texas A&M University, Borlaug Building, College Station, TX 77843, USA.
Konstantinos Markakis, Department of Computer Science and Genome Center, University of California, Davis, One Shields Avenue, Davis, CA 95616, USA.
Patrick J Stover, Department of Nutrition, Texas A&M University, Cater-Mattil Hall, 373 Olsen Blvd Room 130, College Station, TX 77843, USA.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
This work was funded by a grant from the Bill and Melinda Gates Foundation, MN-Systematic approach to evaluate nutrition biomarkers for MNCH outcomes - INV-047386.
Author Contributions
Conceptualization: R.L.B. and P.J.S.; Writing: R.L.B., A.J.M., M.S.F., I.T., S.E.B., K.M.E., C.J.R., N.J.S., A.S., B.C.W., K.M., and P.J.S. K.P.F. provided project administration.
Data Availability
There are no data underlying this work.
References
- 1. Kelley GA, Kelley KS. 2019. Systematic reviews and meta-analysis in nutrition research. Br J Nutr. 122:1279–1294. [DOI] [PubMed] [Google Scholar]
- 2. Brannon PM, Taylor CL, Coates PM. 2014. Use and applications of systematic reviews in public health nutrition. Annu Rev Nutr. 34:401–419. [DOI] [PubMed] [Google Scholar]
- 3. Spill MK, et al. 2022. Perspective: USDA nutrition evidence systematic review methodology: grading the strength of evidence in nutrition- and public health-related systematic reviews. Adv Nutr. 13:982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trumbo PR, Barr SI, Murphy SP, Yates AA. 2013. Dietary reference intakes: cases of appropriate and inappropriate uses. Nutr Rev. 71:657–664. [DOI] [PubMed] [Google Scholar]
- 5. Murphy SP. 2008. Using DRIs as the basis for dietary guidelines. Asia Pac J Clin Nutr. 17(Suppl 1):52–54. [PubMed] [Google Scholar]
- 6. Institute of Medicine (US) Committee on Use of Dietary Reference Intakes in Nutrition Labeling . 2003. Dietary reference intakes: guiding principles for nutrition labeling and fortification. Washington (DC): National Academies Press. [PubMed] [Google Scholar]
- 7. Murphy SP, Yates AA, Atkinson SA, Barr SI, Dwyer J. 2016. History of nutrition: the long road leading to the dietary reference intakes for the United States and Canada. Adv Nutr. 7:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. National Research Council (US) Subcommittee on the Tenth Edition of the Recommended Dietary Allowances . 1989. Recommended dietary allowances: 10th edition. Washington (DC): National Academies Press. [PubMed] [Google Scholar]
- 9. Institute of Medicine (US) Committee to Review Child and Adult Care Food Program Meal Requirements; Murphy SP, Yaktine AL, Suitor CW, Moats S, eds., 2011. Child and adult care food program: aligning dietary guidance for all. Washington (DC): National Academies Press. [PubMed] [Google Scholar]
- 10.Economic Research Service. What is agriculture's share of the overall U.S. economy? 2023 [accessed 2024 May]. https://www.ers.usda.gov/data-products/chart-gallery/gallery/chart-detail/?chartId=58270.
- 11. Jardim TV, et al. 2019. Cardiometabolic disease costs associated with suboptimal diet in the United States: a cost analysis based on a microsimulation model. PLoS Med. 16:e1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brannon PM, et al. 2016. Scanning for new evidence to prioritize updates to the dietary reference intakes: case studies for thiamin and phosphorus. Am J Clin Nutr. 104:1366–1377. [DOI] [PubMed] [Google Scholar]
- 13. Field MS, et al. 2022. Scanning the evidence: process and lessons learned from an evidence scan of riboflavin to inform decisions on updating the riboflavin dietary reference intakes. Am J Clin Nutr. 116:299–302. [DOI] [PubMed] [Google Scholar]
- 14. Bailey RL, Stover PJ. 2023. Precision nutrition: the hype is exceeding the science and evidentiary standards needed to inform public health recommendations for prevention of chronic disease. Annu Rev Nutr. 43:385–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stover PJ, Garza C, Durga J, Field MS. 2020. Emerging concepts in nutrient needs. J Nutr. 150:2593S–2601S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benavidez GA, Zahnd WE, Hung P, Eberth JM. 2024. Chronic disease prevalence in the US: sociodemographic and geographic variations by zip code tabulation area. Prev Chronic Dis. 21:230267–230277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yetley EA, et al. 2017. Options for basing dietary reference intakes (DRIs) on chronic disease endpoints: report from a joint US-/Canadian-sponsored working group. Am J Clin Nutr. 105:249S–285S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. E. National Academies of Sciences, and Medicine . 2022. Defining populations for dietary reference intake recommendations: a letter report. Washington (DC): The National Academies Press. [Google Scholar]
- 19. Helm JM, et al. 2020. Machine learning and artificial intelligence: definitions, applications, and future directions. Curr Rev Musculoskelet Med. 13:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thirunavukarasu AJ, et al. 2023. Large language models in medicine. Nat Med. 29:1930–1940. [DOI] [PubMed] [Google Scholar]
- 21. Guo E, et al. 2024. Automated paper screening for clinical reviews using large language models: data analysis study. J Med Internet Res. 26:e48996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaswani A, et al. 2017. Attention is all you need. Adv Neural Inf Process Syst. 30:6000–6010. [Google Scholar]
- 23. Wu T, et al. 2023. A brief overview of ChatGPT: the history, Status quo and potential future development. IEEE/CAA J Automat Sinica. 10:1122–1136. [Google Scholar]
- 24. Eetemadi A, et al. 2020. The computational diet: a review of computational methods across diet, microbiome, and health. Front Microbiol. 11:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Partin A, et al. 2023. Deep learning methods for drug response prediction in cancer: Predominant and emerging trends. Front Med (Lausanne). 10:1086097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roscher R, Bohn B, Duarte MF, Garcke J. 2020. Explainable machine learning for scientific insights and discoveries. IEEE Access. 8:42200–42216. [Google Scholar]
- 27. Ma J, et al. 2018. Using deep learning to model the hierarchical structure and function of a cell. Nat Methods. 15:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuenzi BM, et al. 2020. Predicting drug response and synergy using a deep learning model of human cancer cells. Cancer Cell. 38:672–684 e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park S, et al. 2024. A deep learning model of tumor cell architecture elucidates response and resistance to CDK4/6 inhibitors. Nat Cancer. 5(7):996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao X, et al. 2024. Cancer mutations converge on a collection of protein assemblies to predict resistance to replication stress. Cancer Discov. 14:508–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma J, et al. 2021. Few-shot learning creates predictive models of drug response that translate from high-throughput screens to individual patients. Nat Cancer. 2:233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. So E, Yu F, Wang B, Haibe-Kains B. 2023. Reusability report: evaluating reproducibility and reusability of a fine-tuned model to predict drug response in cancer patient samples. Nat Mach Intell. 5:792–798. [Google Scholar]
- 33. Naser MZ. 2024. Causality and causal inference for engineers: beyond correlation, regression, prediction and artificial intelligence. WIREs Data Mining and Knowledge Discovery. 14:e1533. [Google Scholar]
- 34. Morris JH, et al. 2023. The scalable precision medicine open knowledge engine (SPOKE): a massive knowledge graph of biomedical information. Bioinformatics. 39:btad080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soman K, et al. 2023. Early detection of Parkinson's disease through enriching the electronic health record using a biomedical knowledge graph. Front Med (Lausanne). 10:1081087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Food and Nutrition Board; Committee on the Development of Guiding Principles for the Inclusion of Chronic Disease Endpoints in Future Dietary Reference Intakes; Oria MP, Kumanyika S, editors. 2017. Guiding principles for developing dietary reference intakes based on chronic disease. Washington (DC): National Academies Press. [PubMed] [Google Scholar]
- 37. James WPT, et al. 2019. Nutrition and its role in human evolution. J Intern Med. 285:533–549. [DOI] [PubMed] [Google Scholar]
- 38. Dahabreh IJ, Bibbins-Domingo K. 2024. Causal inference about the effects of interventions from observational studies in medical journals. JAMA. 331(21):1845–1853. [DOI] [PubMed] [Google Scholar]
- 39. Messeri L, Crockett MJ. 2024. Artificial intelligence and illusions of understanding in scientific research. Nature. 627:49–58. [DOI] [PubMed] [Google Scholar]
- 40. Lea AS, Jones DS. 2024. Mind the gap - machine learning, dataset shift, and history in the age of clinical algorithms. N Engl J Med. 390:293–295. [DOI] [PubMed] [Google Scholar]
- 41. Chekroud AM, et al. 2024. Illusory generalizability of clinical prediction models. Science. 383:164–167. [DOI] [PubMed] [Google Scholar]
- 42. Schork NJ, Beaulieu-Jones B, Liang WS, Smalley S, Goetz LH. 2023. Exploring human biology with N-of-1 clinical trials. Camb Prism Precis Med. 1:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Potter T, Vieira R, de Roos B. 2021. Perspective: application of N-of-1 methods in personalized nutrition research. Adv Nutr. 12:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Angelopoulos AN, Bates S, Fannjiang C, Jordan MI, Zrnic T. 2023. Prediction-powered inference. Science. 382:669–674. [DOI] [PubMed] [Google Scholar]
- 45. Shah P, et al. 2019. Artificial intelligence and machine learning in clinical development: a translational perspective. NPJ Digit Med. 2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Katsoulakis E, et al. 2024. Digital twins for health: a scoping review. NPJ Digit Med. 7:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun T, He X, Li Z. 2023. Digital twin in healthcare: recent updates and challenges. Digit Health. 9:20552076221149651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. National Academies of Sciences, Engineering, and Medicine; National Academy of Engineering; Division on Earth and Life Studies; Division on Engineering and Physical Sciences; Board on Life Sciences; Board on Atmospheric Sciences and Climate; Computer Science and Telecommunications Board; Board on Mathematical Sciences and Analytics. 2023. Opportunities and challenges for digital twins in biomedical research: proceedings of a workshop—in brief. Washington (DC): National Academies Press. [PubMed]
- 49. Reedy J, et al. 2018. Evaluation of the Healthy Eating Index-2015. J Acad Nutr Diet. 118:1622–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eetemadi A, Tagkopoulos I. 2021. Methane and fatty acid metabolism pathways are predictive of low-FODMAP diet efficacy for patients with irritable bowel syndrome. Clin Nutr. 40:4414–4421. [DOI] [PubMed] [Google Scholar]
- 51. Jennings-Dobbs EM, Forester SM, Drewnowski A. 2023. Visualizing data interoperability for food systems sustainability research-from spider webs to neural networks. Curr Dev Nutr. 7:102006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Youn J, Rai N, Tagkopoulos I. 2022. Knowledge integration and decision support for accelerated discovery of antibiotic resistance genes. Nat Commun. 13:2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. AI Institute for Next Generation Food Systems. 2024. Food Atlas. Davis (CA): University of California at Davis.
- 54. Youn J, Li F, Simmons G, Kim S, Tagkopoulos I. 2024. FoodAtlas: automated knowledge extraction of food and chemicals from literature. Comput Biol Med. 181:109072. [DOI] [PubMed] [Google Scholar]
- 55. Andersen MZ, Gulen S, Fonnes S, Andresen K, Rosenberg J. 2020. Half of Cochrane reviews were published more than 2 years after the protocol. J Clin Epidemiol. 124:85–93. [DOI] [PubMed] [Google Scholar]
- 56. Shojania KG, et al. 2007. How quickly do systematic reviews go out of date? A survival analysis. Ann Intern Med. 147:224–233. [DOI] [PubMed] [Google Scholar]
- 57. Oxman AD, Guyatt GH. 1993. The science of reviewing research. Ann N Y Acad Sci. 703:125–133. [DOI] [PubMed] [Google Scholar]
- 58. Cumpston M, et al. 2019. Updated guidance for trusted systematic reviews: a new edition of the Cochrane handbook for systematic reviews of interventions. Cochrane Database Syst Rev. 10:ED000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Konsgen N, et al. 2020. Inter-review agreement of risk-of-bias judgments varied in Cochrane reviews. J Clin Epidemiol. 120:25–32. [DOI] [PubMed] [Google Scholar]
- 60. Minozzi S, Cinquini M, Gianola S, Gonzalez-Lorenzo M, Banzi R. 2020. The revised Cochrane risk of bias tool for randomized trials (RoB 2) showed low interrater reliability and challenges in its application. J Clin Epidemiol. 126:37–44. [DOI] [PubMed] [Google Scholar]
- 61. O'Connor AM, et al. 2019. Still moving toward automation of the systematic review process: a summary of discussions at the third meeting of the international collaboration for automation of systematic reviews (ICASR). Syst Rev. 8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Edwards KM, et al. 2024. ADVISE: accelerating the creation of evidence synthesis for global development using natural language processing-supported human artificial intelligence collaboration. J. Mech. Des. 146:1–15. [Google Scholar]
- 63. Shemilt I, et al. 2021. Cost-effectiveness of Microsoft academic graph with machine learning for automated study identification in a living map of coronavirus disease 2019 (COVID-19) research. Wellcome Open Res. 6:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ramprasad S, Marshall IJ, McInerney DJ, Wallace BC. 2023. Automatically summarizing evidence from clinical trials: a prototype highlighting current challenges. Proc Conf Assoc Comput Linguist Meet. 2023:236–247. [PMC free article] [PubMed] [Google Scholar]
- 65. Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. 2016. Rayyan-a web and mobile app for systematic reviews. Syst Rev. 5:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Murray CJL, et al. 2020. Five insights from the global burden of disease study 2019. The Lancet. 396:1135–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Marshall IJ, et al. 2020. Trialstreamer: a living, automatically updated database of clinical trial reports. J Am Med Inform Assoc. 27:1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Soboczenski F, et al. 2019. Machine learning to help researchers evaluate biases in clinical trials: a prospective, randomized user study. BMC Med Inform Decis Mak. 19:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Clark J, McFarlane C, Cleo G, Ramos CI, Marshall S. 2021. The impact of systematic review automation tools on methodological quality and time taken to complete systematic review tasks: case study. JMIR Med Educ. 7:e24418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Muller AE, et al. 2023. The effect of machine learning tools for evidence synthesis on resource use and time-to-completion: protocol for a retrospective pilot study. Syst Rev. 12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tercero-Hidalgo JR, et al. 2022. Artificial intelligence in COVID-19 evidence syntheses was underutilized, but impactful: a methodological study. J Clin Epidemiol. 148:124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ioannidis JPA. 2021. Hundreds of thousands of zombie randomised trials circulate among us. Anaesthesia. 76:444–447. [DOI] [PubMed] [Google Scholar]
- 73. Carlini N, et al. 2024. Poisoning web-scale training datasets is practical. arXiv:2302.10149, preprint: not peer reviewed.
- 74. Crider KS, Qi YP, Devine O, Tinker SC, Berry RJ. 2018. Modeling the impact of folic acid fortification and supplementation on red blood cell folate concentrations and predicted neural tube defect risk in the United States: have we reached optimal prevention? Am J Clin Nutr. 107:1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Crider KS, et al. 2014. Population red blood cell folate concentrations for prevention of neural tube defects: Bayesian model. BMJ. 349:g4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Djulbegovic B, Guyatt GH. 2017. Progress in evidence-based medicine: a quarter century on. Lancet. 390:415–423. [DOI] [PubMed] [Google Scholar]
- 77. IntHout J, Ioannidis JP, Rovers MM, Goeman JJ. 2016. Plea for routinely presenting prediction intervals in meta-analysis. BMJ Open. 6:e010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hernan MA. 2018. The C-Word: scientific euphemisms do not improve causal inference from observational data. Am J Public Health. 108:616–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Agency for Healthcare Research and Quality. 2024. About learning health systems. Rockville (MD): AHRQ.
- 80. Garza C, et al. 2019. Best practices in nutrition science to earn and keep the public's trust. Am J Clin Nutr. 109:225–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are no data underlying this work.