Abstract
Nonsense mutations, often resulting from single-nucleotide substitutions, produce mRNA harboring a premature termination codon (PTC), which causes the premature termination of protein synthesis. This produces truncated and non-functional proteins, which cause different genetic diseases, including cystic fibrosis (CF). This work aims to investigate the ability of NV848 (N-(5-methyl-1,2,4-oxadiazol-3-yl)acetamide), a translational readthrough-inducing drug (TRID), to rescue CF transmembrane conductance regulator (CFTR) protein expression in a murine model characterized by the G542X nonsense mutation in the CFTR gene. In vitro experiments assessed the drug’s stability in human hepatic metabolism, and in vivo investigations on wild-type mice allowed us to clarify the distribution of the drug to the target organs. Moreover, its efficacy in recovering the CFTR protein after chronic treatment was assessed in G542X homozygous mice. Our results provide valuable insights into the biodistribution and therapeutic attributes of NV848, representing a promising therapeutic tool for enhanced clinical outcomes in individuals affected by CF with nonsense mutations.
Keywords: nonsense, cystic fibrosis, oxadiazole, stop codons, Mendelian diseases
Graphical abstract
Fiduccia and collaborators investigate the potential of NV848, a translational readthrough-inducing drug (TRID), to restore CFTR protein expression in a murine model with the G542X nonsense mutation linked to cystic fibrosis. The study evaluates the drug’s metabolism, biodistribution, and efficacy, offering promising therapeutic insights for CF treatment.
Introduction
Cystic fibrosis (CF) is an autosomal recessive multiorgan disorder caused by loss-of-function mutations in the CF transmembrane conductance regulator (CFTR) gene. Approximately 10% of the CF population worldwide have at least one CFTR allele with a nonsense mutation that generates a premature termination codon (PTC) in the mRNA. This causes protein synthesis to end prematurely, forming shortened, non-functional polypeptides.1,2 In this investigation, we focused our attention on a small molecule that has recently been demonstrated in vitro to show potential in rescuing the effects of nonsense mutations causing CF.3
Physiologically, a cell surveillance mechanism known as nonsense-mediated mRNA decay (NMD) is activated, attempting to mitigate the presence of aberrant mRNAs. Nevertheless, a notable fraction of mRNAs, ranging from 5% to 25%, evade this regulatory mechanism and are subjected to a process known as translational readthrough.4,5 This process involves the incorporation of a near-cognate tRNA (nc-tRNA) misreading the stop signal. As a result, translation continues until reaching the natural termination codon (NTC), producing a full-length protein.6,7 Consequently, scientists started exploring molecules to enhance the rescue of protein expression, and among these compounds, aminoglycosides were studied, mainly due to their readthrough-inducing activity of nonsense mutations.8,9 However, these compounds have not found clinical application for chronic treatments due to their severe side effects, such as renal, auditory, and vestibular toxicities.10,11
To overcome these issues, the synthetic aminoglycoside ELX-02 has been designed. It lacks the characteristic aminoglycosides’ toxicity in chronic treatment, and it went through a phase 1 clinical trial to verify the drug’s tolerability and safety12,13 but failed the phase 2 trial due to inconsistent results.14
However, the translational readthrough-inducing drug (TRID) family is not only limited to aminoglycosides and their derivatives. Indeed, several non-aminoglycoside compounds have drawn attention as readthrough-inducing drugs. For example, a medication known for anti-allergic and anti-inflammatory properties that has been used for more than 30 years to treat asthma and aphthous ulcers15,16 has, in the last decade, been studied for its ability to inhibit NMD and boost the readthrough of PTCs. However, to date, results have shown that it is not effective enough in restoring a full-length protein either alone or in combination with other compounds.17
Another molecule worthy of being mentioned for its ability to promote the readthrough mechanism is a purine derivative, 2,6-diamino purine (DAP), that holds a prominent place because it can selectively promote the readthrough for the UGA stop codon in the TP53 gene in cancer cells. It works by inhibiting the activity of the RNA 2′-O-methyltransferase 1 (FTSJ1), and it has been tested, showing very low toxicity and high efficacy also in vivo.18,19
Despite exhaustive efforts, scientists have not yet discovered a cure for treating nonsense mutations. Notably, the only medication that has been granted conditional marketing authorization for Duchenne muscular dystrophy (DMD) caused by nonsense mutations was PTC124 in July 2014, also known as ataluren and commercially available as Translarna.20,21
However, the development of this drug has encountered numerous challenges. Upon its initial authorization, the Committee for Medicinal Products for Human Use (CHMP) acknowledged uncertainties about Translarna’s beneficial effects. Consequently, further tests have been requested for PTC Therapeutics to assess the drug’s effectiveness. The subsequent outcomes prompted the CHMP to conclude in January 2024 that Translarna’s benefit-risk balance is negative. Consequently, the committee questioned the medicine’s marketing authorization in the European Union.22
Despite facing setbacks and the intricate challenges of conducting proper clinical trials, ataluren has captured the attention of numerous research groups throughout the years. This intrigue arises from its promising results as a readthrough promoter, its proven low toxicity in both in vitro and in vivo tests, and the desire to learn about its mechanism of action.23,24,25,26
Based on ataluren’s structure, many analogs have been synthesized and tested over the years,27,28,29 and among them, N-(5-methyl-1,2,4-oxadiazol-3-yl)acetamide, designated as NV848, stood out.3 Identified through a virtual high-throughput screening (VHTS) process, NV848 displayed promising preliminary results, prompting an in-depth exploration of its molecular activity. Experimental phases involved Fisher rat thyroid (FRT) cells transfected with a vector expressing nonsense-mutated CFTR mRNA, with validation through immunofluorescence, western blot analyses, and functional assessments.3
Furthermore, NV848 demonstrated good tolerability in vivo and exhibited specificity for PTCs without interfering with NTCs.30,31 The potential of NV848 as a candidate for treating nonsense-related diseases (NRDs) has led us to conduct further investigations across various genetic contexts.32 In this study, to further evaluate the potential of this compound, we characterized NV848 in vitro metabolism on human liver microsomes (HLMs), performed an in vivo study to explore biodistribution in wild-type (WT) mice, and ultimately, investigated its ability to restore CFTR expression in the lungs of a CF mouse model carrying the G542X nonsense mutation.
Results
Evaluation of NV848 metabolic stability in human liver microsomes
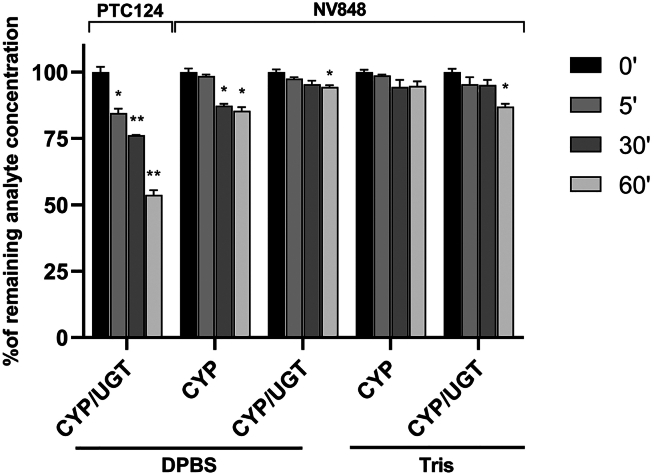
Human liver microsomes (HLMs) were used to investigate cytochrome-P450 (CYP) and UDP-glucuronyl-transferase (UGT) activity to evaluate the consumption of the NV848 molecule, mimicking the liver biotransformation ability in vitro. The human liver vesicles used in this study were derived from 50 different donors to collect more enzyme isoforms in a single microsome mix. In this system, all donors were equally represented for a valid population sample (25 female and 25 male donors; mixed gender). HLMs were incubated with PTC124 or NV848 in two conditions (DPBS and Tris). The phosphate buffer (DPBS) and the other one without phosphate components (Tris) were used to understand possible differences in HLM enzyme efficiency.
The HML mix contained the necessary cofactors (NADPH) and substrates to evaluate the metabolic activity of either cytochrome P450 (CYP) alone or a combination of both UGT (UDP-glucuronyltransferase) and CYP activities (CYP/UGT).
We used PTC124 (ataluren) as a control to verify the compound consumption observed by Kong et al.33 PTC124 incubation with HLMs confirmed the compound’s metabolic consumption, which reached 46.25% after 60 min. The glucuronate derivative was the primary metabolite detected after HLM incubation, proving the correct biotransformation in the experimental system (Figure 1).
Figure 1.
Percentage of tested compounds after 0, 5, 30, and 60 min incubated with human liver microsomes
Samples concerning enzymatic activity (CYP or CYP/UGT) and phosphate buffers (DPBS and TRIS) have been distinguished. Data are expressed as mean values ± SEM. ∗p < 0.05 and ∗∗p < 0.01 (one-way ANOVA test) compared to the percentage of remaining analyte evaluated at time 0 (black bars) for every sample condition.
On the other hand, NV848 showed higher metabolic stability compared to PTC124, with its significant consumption reaching 14.62% and 13.04% after 60 min, respectively, in CYP-DPBS and CYP/UGT-Tris conditions. In the other buffer conditions (CYP/UGT-DPBS and CYP-Tris), the NV848 consumption percentages were very low (5.65% and 5.27%, respectively), confirming the HLM metabolic stability of this compound.
In general, NV848 is more stable against biotransformation mediated by CYP, UGT, and other liver microsome enzymes than PTC124. Interestingly, no glucuronate metabolites were detected after incubation of NV848 in the presence of the cofactors needed for evaluating UGT activity (also in two different buffers; Figure 1).
Biodistribution evaluation of NV848
After establishing NV848’s stability in hepatic metabolism, in vivo studies were performed to assess its biodistribution and efficacy in animal models.
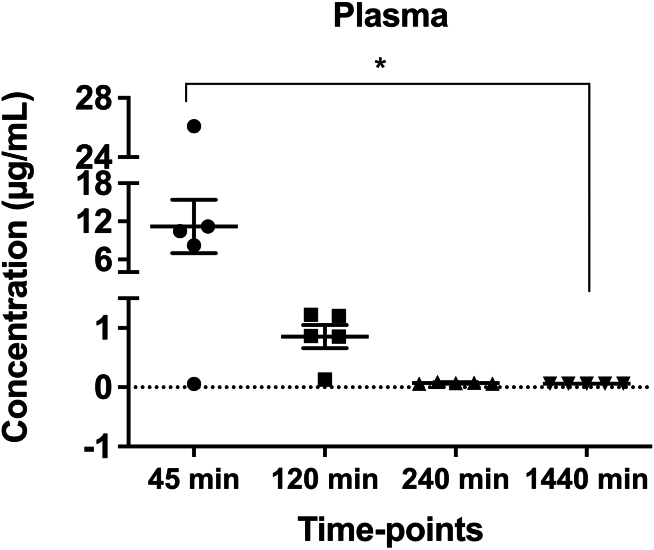
WT mice received a single oral dose of NV848 at a concentration of 60 mg/kg. Distribution was followed up to 24 h after administration in plasma and organs (lungs, pancreas, kidney, intestines, and brain).
The quantification of NV848 revealed organ-specific variations in measured concentrations. Notably, in specific organs, the analyte was found within the initial 45 min, while in others, NV848 was still present up to 120 min post-administration. We observed that the NV848 plasma concentration ranged from 0.05 to 26 g/mL, with an average plasma concentration of 11.21 g/mL, 45 min after the oral administration, which dropped to an average of 0.06 g/mL by the 24 h (Figure 2).
Figure 2.
Plasma concentration of NV848 at established time points
Data are expressed as mean values ± SEM. N = 5. ∗p < 0.05 (one-way ANOVA test) compared to NV848 levels at 45 min.
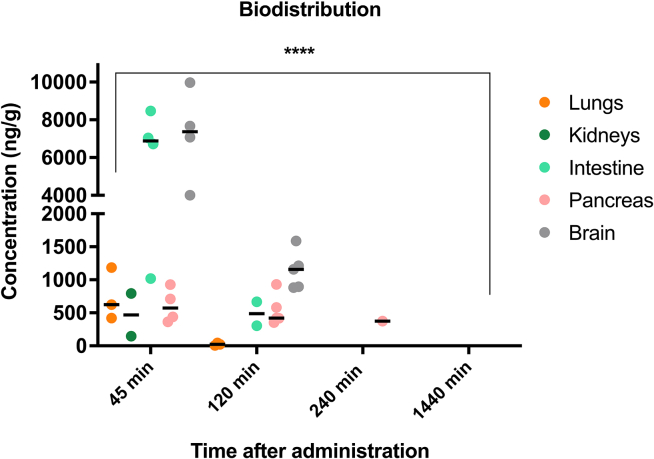
In the lungs, NV848 concentrations ranged from 421 to 1,184 ng/g at 45 min, dropping down after 2 h, marking its last detection. The pancreas exhibited stable concentrations from 45 min to 2 h, then lowered in the following hours. In the intestine, concentrations ranged from 1018 to 8,465 ng/g after 45 min, decreasing after 2 h.
Within the brain, concentrations spanned from 4,003 to 9,969 ng/g at 45 min and lingered between 883 and 1,587 ng/g 2 h later. In the kidneys, NV848 appeared in two mice at 145 and 794 ng/g after 45 min (Figure 3). It is important to note that some measurements fell below the limit of quantification (LoQ); thus, these lower concentrations are not represented in the graph.
Figure 3.
Concentration of NV848 in all organs for each interval (N = 5)
∗∗∗∗p < 0.0001 (two-way ANOVA test) compared to NV848 levels at 45 min.
Intriguingly, the recovery test, uniformly applied across all samples, posed challenges in aligning with specific organs, ranging from 0% to 55%.
Characterization of the CFTRG542X mutant colony and evaluation of the NV848 activity in CFTRG542X/G542X mice
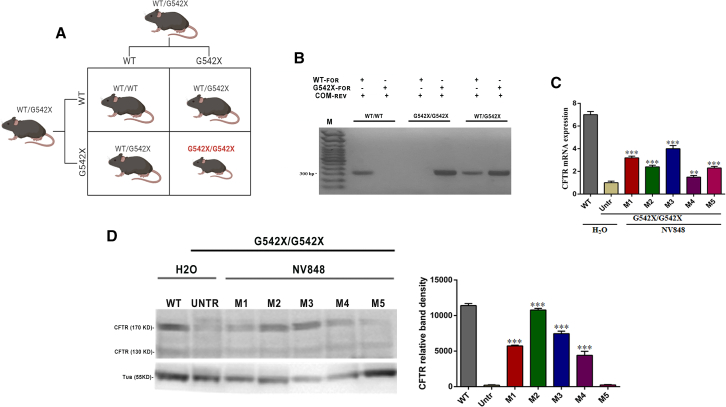
To study the rescue of the CFTR protein expression in the nonsense animal model for CF, we performed a 15-day chronic treatment of NV848 in homozygous CFTRG542X/G542X mice obtained by breeding heterozygous CFTRWT/G542X mice (Figure 4A). As expected, CFTRG542X/G542X homozygous mice displayed, as reported in the literature,34 reduced weight and significantly reduced length compared to sex-matched WT littermates (Table S1).
Figure 4.
CFTRG542X/G542X mice express CFTR protein after 15 days of treatment with NV848 molecule. (A) Punnet’s mutant CFTR mouse colony production scheme shows animal matings to obtain G542X mice. (B) Example of genotyping and expected results: for each mouse in the gel, the first lane shows DNA amplified with primers recognizing the WT allele and the second lane with primers recognizing the G542X allele. (C) Real-time RT-PCR evaluated lung CFTR mRNA level expression in mutant mice treated for 15 days with H2O and NV848; data are means ± SEM (in triplicate for each mouse), p < 0.05 (two-way ANOVA test) compared to the untreated (untr) mutant mouse. (D) CFTR protein expression was evaluated by western blot assay of lung protein extracted from mutant mice treated for 15 days with H2O and NV848 and a histogram showing CFTR protein quantification normalized by tubulin and negative control; data are means ± SEM (in triplicate for each mouse), p < 0.05 (two-way ANOVA test) compared to the untr mutant mouse.
Specific primers for the two CFTR variant alleles were used to genotype offspring and identify CFTRG542X/G542X mice (Figure 4B).
After daily oral administration of 60 mg/kg NV848 for 15 days, mice were sacrificed. CFTR protein rescue evaluation in lung tissues was performed comparing tissues from CFTRG542X/G542X to tissues from CFTRWT and CFTRG542X/G542X mice treated with vehicle (positive and negative controls, respectively). Then, CFTR mRNA and protein expression after the chronic treatment were analyzed.
As shown in Figure 4C, an increase in CFTR mRNA expression was observed in the treated mice’s lungs, and protein CFTR expression was observed in the lungs of 4 out of 5 treated mutant mice. There was a striking increase in CFTR protein expression in treated mice compared to the negative control (Figure 4D).
IHC assessed the safety of the chronic treatment and the recovery of CFTR expression
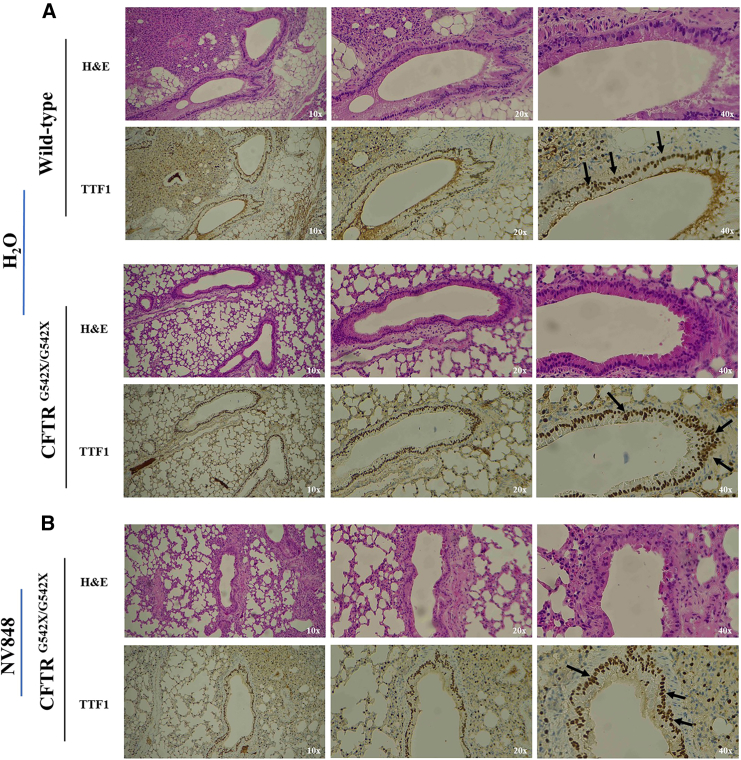
Lung sections of CFTRWT and CFTRG542X/G542X mice were analyzed by immunohistochemistry (IHC) to visualize the correct localization of the CFTR protein in the apical surface of the pulmonary ducts. To this aim, sections of the lungs of one of the CFTR-positive mutant mice (M2) and the control mice (WT and CFTRG542X/G542X H2O treated) were analyzed. Firstly, the possible occurrence of tissue damage after chronic administration was assayed by hematoxylin and eosin (H&E) staining performed on isolated tissues of mice from each group. The analyses of all samples revealed no signs of tissue damage, confirming the safety of the chronic administration (Figures 5A and 5B).
Figure 5.
CFTRG542X/G542X mice express CFTR protein( in lung sections after 15 days of treatment with NV848 molecule. (A) H&E staining and TTF1 expression in CFTRWT and untreated (H2O) CFTRG542X/G542× lung sections (magnifications: 10×, 20×, 40×). (B) H&E staining and TTF1 expression in NV848-treated CFTRG542X/G542× mouse lung sections (magnifications: 10×, 20×, 40×).
The thyroid transcription factor 1 (TTF1) protein signal in the nuclei was used as a positive control for the staining reaction.
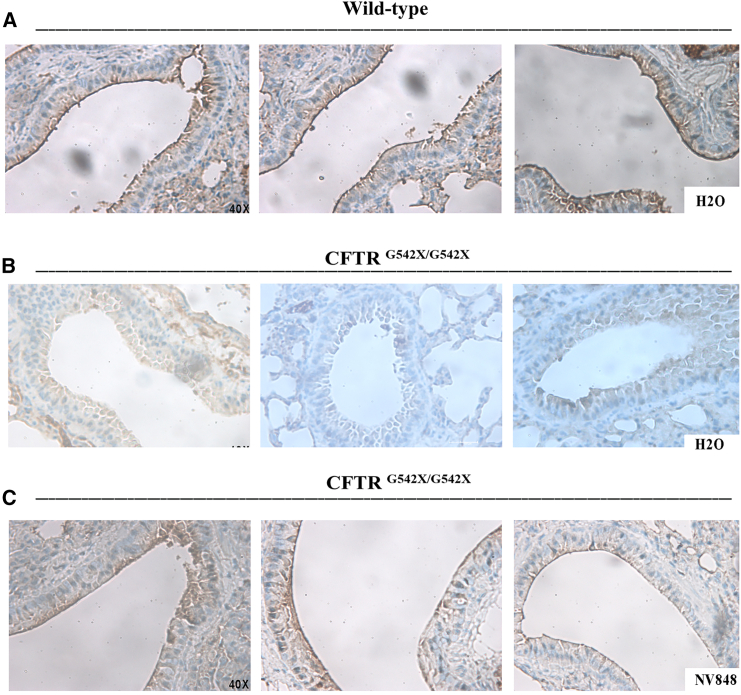
CFTR protein expression was detected in the WT lung sections, particularly on the apical membranes of the pulmonary ducts, which displayed an intense and rich brown coloration (Figure 6A). Conversely, no detection of CFTR was observed in the lung of the negative control, where the staining was not uniform (Figure 6B).
Figure 6.
Expression of CFTR protein in different lung regions of CFTRG542X/G542X mice. (A) Different regions of the CFTR staining (brown) in the CFTRWT lung sections and (B) CFTRG542X/G542X untreated (H2O) control. (C) NV848-treated CFTRG542X/G542X mouse lung sections show CFTR expression of the apical surface of the lung ducts (brown). All the images are shown with a 40× magnification.
Lastly, in homozygous CFTRG542X/G542X NV848 sections, a solid and uniform CFTR detection, compared to the negative control, was observed, mainly in the apical membranes of the pulmonary ducts, with a well-defined staining pattern exhibiting an intensity similar to CFTRWT (Figure 6C). Some brown spots were also observed in the intracellular areas, though fewer than in the negative control.
These findings suggest that NV848 treatment can rescue CFTR expression and localization in the CFTRG542X/G542X treated mice.
Discussion
Our exploration delved into the metabolic stability, biodistribution, and efficacy of NV848 in a murine model carrying the UGA-G542X mutation in the CFTR gene, providing comprehensive insights into its therapeutic perspective.
So far, among the approved therapies for modulating the CFTR gene in patients, a conspicuous void exists regarding treatments that address nonsense mutations. Nonsense mutations typically result in the synthesis of truncated protein and, depending on the severity of the truncation, lead to a significant reduction or total absence of protein function. Current evidence suggests that increasing mRNA levels by inhibiting the NMD pathway could boost the readthrough effect achieved by TRIDs.
We focused on a new patented molecule, NV848, to perform a biodistribution and efficacy study. This study demonstrated, for the first time, NV848’s ability to reach different target organs in vivo and restore the expression of CFTR in an animal model of CF.
In the first step, we performed a metabolic investigation to study the stability of NV848 during hepatic metabolism. Our findings highlighted the higher stability of NV848 compared to PTC124, a benchmark compound in the field.33
Moving forward, the biodistribution study was conducted on C57BL/6 male mice by administering NV848 once, in alignment with the dosage regimen utilized in the Du study35 for Translarna at 60 mg/kg, and the evaluation was carried out, covering 24 h with four main time points (45, 120, 240, and 1,440 min). Interestingly, the results uncovered organ-specific variations with a distinct preference for brain, intestine, lungs, and pancreas localization. Interestingly, our compound seems to be resistant to factors that can affect the designing of an orally viable formulation of the drug, such as pH stability in the stomach and the likely ability of the compound to be absorbed from the intestinal epithelium, as suggested by the recovery of NV848 in blood and the organs.
A consistent recovery test protocol ensured methodological uniformity, highlighting the experimental rigor. However, the variation in suitability across specific organs prompted us to interpret the obtained data with a qualitative lens, enriching our insights into the diverse dynamics of NV848 distribution.
Moreover, the natural inter-individual variation could be an essential factor to consider. Nonetheless, seeing the high concentration achieved in the brain, we could speculate that it could be worth exploring the effective ability of NV848 to cross the blood-brain barrier and imagine a possible use in pathologies involving the brain or the central nervous system.
The presence of NV848 in possible organ targets for disease as CF encouraged us to evaluate the rescue of a nonsense mutation in vivo. G542X homozygous mice exhibited reduced growth or failure to thrive, as reported in the literature.34 Oral treatment for 2 weeks in CFTRG542X/G542X, a CF nonsense mouse model, at the same dosage as that utilized in the biodistribution study, was well tolerated in mice and did not cause toxicity, as observed even after repeated administrations.30 In addition, chronic administration enhanced CFTR protein expression in the lungs, as displayed by western blot assays, where the protein expression level was significantly higher in treated mutants compared to the negative control. These results were also confirmed by immunohistochemical analyses on lung tissues of treated mutant CFTRG542X/G542X mice and control subjects (both CFTRWT and CFTRG542X/G542X). The positive and specific staining on plasma membranes of epithelial cells in the pulmonary ducts in the CFTRWT sample was also found in the mutant CFTRG542X/G542X mouse treated with NV848 at the daily dosage of 60 mg/kg, confirming the recovery of CFTR expression. On the contrary, we observed a lack of specificity in the untreated mutant mouse in the staining. Altogether, these results suggest the rescue of CFTR expression after NV848 treatment, proposing this compound as a potential therapeutic avenue for individuals with CF carrying nonsense mutations. Interestingly, H&E staining displayed that the tissue integrity, after 15 days of chronic treatment with 60 mg/kg NV848, was maintained, confirming once more that the chronic treatment is also well tolerated and does not influence tissue integrity or morphology, as no alterations were evidenced.
Limitations of the study
Our study highlights NV848 as a promising candidate identified through VHTS. However, further investigations could be useful, aiming to clarify NV848’s action mechanism and evaluate the possible introduction of TRIDs into clinics.
Materials and methods
Synthesis of NV848
NV848 was prepared as reported in the literature3 (patents: WO2019101709A1 [IT], US20210002238A1 [US], and EP3713934B1 [EU]).
In vitro metabolism on HLMs
To evaluate NV848 metabolism, an assay on HLMs was performed. Specifically, NV848 at a concentration of 1 mM underwent incubation with pooled HLMs of 50 different donors of both genders (Gibco, Thermo Fisher Scientific, USA) (20 mg protein/mL) at 37°C ± 1°C in 0.2 mL incubation mixtures (final volume) in two different conditions, one containing potassium phosphate buffer (100 mM, pH 7.4) and NADPH (20 mM) and the other one containing Tris- HCl buffer (100 mM, pH 7.4) and NADPH (20 mM). Then, for the measurement of UGT activity, MgCl2 (1 mM), alamethicin (50 μg alamethicin/50 mg microsomal protein), and uridine diphosphate glucuronic acid (UDPGA; 5 mM) were added to the mixtures (adjustment of volume buffer to maintain the same final volume). Reactions were initiated by the addition of cofactors and terminated at the designated intervals: (1) T0, (2) T 5 min, (3) T 30 min, and (4) T 60 min (1 h) using a stop reagent (0.2 mL acetonitrile [ACN]). Centrifugation at 3,000 rpm for 5 min allowed removal of the precipitated proteins. High-performance liquid chromatography mass spectrometry (HPLC-MS) analyses of the supernatant fractions enabled the determination of the percentage loss of substrate in the incubations. Ataluren (PTC124) was used as a benchmark.
The concentration of NV848 at specific time points post-incubation was compared with the initial amount at 0 min to calculate the percentage of NV848 remaining.
Animals
All experiments were performed on adult control C57BL/6 mice purchased from ENVIGO Srl (San Pietro al Natisone UD, Italy) and CFTRG542X/G542X homozygous mice obtained by mating heterozygous CFTRWT/G542X mice, kindly gifted by Professor C. Hodges (Case Western Reserve University, Cleveland, USA).34 Animals were maintained in temperature-controlled rooms (22°C–25°C, 50%–60% humidity) in a 12-h light/darkness cycle. All experimental procedures were carried on in conformity with the Italian D.Lgs 26/2014, under the University of Palermo Animal Care and Use Ethics Committee approval and the authorization of the Italian Ministry of Health (authorization no. 1235/2020; authorization no. 2/2022-PR) and following the national and EU Directive 2010/63/EU for animal handling and experimentation.
Biodistribution study
Experiments were conducted on 25 C57BL/6 male mice (4 weeks old, weight: 18.0 ± 4.0 g). Animals received a single dose of NV848 at a concentration of 60 mg/kg by oral gavage, and they were sacrificed to collect blood and tissues at different time points after gavage (n = 5/each point): 45 min, 2 h, 4 h, and 24 h. Blood samples were placed into tubes containing EDTA as an anticoagulant and centrifuged at 3,000 rpm for 10 min. Following blood collection, all internal tissues were harvested. Tissues were washed three times with 0.9% NaCl, wiped with filter paper, weighed, and stored at −80°C until analyses. A further group of 5 mice receiving vehicles constituted the control group (T0).
Preparation of the organs’ samples for HPLC-MS analyses
Samples were prepared for molecule extraction following Kayali’s protocol36 with slight changes. Specifically, organs were homogenized with Omni TH Tissue Homogenizer (Kennesaw GA, USA) in a 10 mL tube, adding 200 μL of 0.9% NaCl solution followed by protein precipitation using 600 μL of ice-cold ACN CHROMASOLV LC-MS (Honeywell, USA). After vortexing for 2 min, the samples were centrifuged (Thermo Scientific, Sorvall RC 6 Plus Centrifuge) at 14,000 rpm at 4°C for 10 min, and the organic phases were preserved at −20°C. Then, samples were dried with a freeze dryer (CoolSafe, SCANVAC, Labogene, Allerød, Denmark) at −100°C under vacuum conditions. Residues were reconstituted with a 100 μL 3:1 solution of ACN CHROMASOLV LC-MS (Honeywell, USA) and high-purity water CHROMASOLV LC-MS (Honeywell, USA), respectively. Then, the samples were vortexed for 5 min and centrifuged at 14,000 rpm at 4°C for 10 min (Thermo Scientific, Sorvall RC 6 Plus Centrifuge). Finally, the supernatant was used for HPLC-MS analysis by HPLC Agilent 1250 Infinity (Agilent Technologies 6540 UHD accurate mass Q-TOF LC/MS).
HPLC-MS analyses
The matrix-matching technique was employed to detect and assess matrix effects. Calibration curve standard solutions were prepared by combining 50 μL of NV848 at concentrations of 0.1, 0.5, 10, 20, and 50 μg/mL with 50 μL of blank matrix, resulting in a final volume of 100 μL. The standard concentrations of NV848 were 0.05, 0.25, 5, 10, and 25 μg/mL. The calibration curve was calculated by performing a linear regression using the least squares method on the calibration standards.
Generation and management of a CFTRG542X/G542X mouse colony
The generation of CFTRG542X/G542X homozygous mice was performed in the animal facility of the ATeNCenter University of Palermo.
At postnatal day 21 (P21), all mice were genotyped by DNA extraction (PureLink Genomic DNA Mini Kit, Invitrogen, Thermo Fisher Scientific, USA, following the manufacturer’s instruction) from ear biopsies, followed by PCR analysis.
The primer sequences used for genotyping each of the mice by PCR are reported in Table S2.34
PCR was performed following the manufacturer’s instructions using the REDTaq DNA Polymerase (Sigma-Aldrich, USA). Two different reaction mixes, one for each allele, were prepared.
The PCR cycling parameters were an initial denaturation of 3 min at 94°C; 40 cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C; and a final extension of 7 min at 72°C (2720 Thermal Cycler, Applied Biosystems, Waltham, MA, USA). PCR products were visualized on a 1% agarose gel in 1× TBE buffer.
Chronic treatment for a drug efficacy study in vivo
Daily oral administration of NV848 at 60 mg/kg for 15 days starting at P21 was performed in 5 CFTRG542X/G542X mice. Moreover, 6 animals, 3 CFTRWT and 3 CFTRG542X/G542X mice, received an oral dose of vehicle (negative control, water). At the end of the treatment, blood and organs were collected, weighed, and stored at −80°C for western blot and IHC. All the experimental mice were maintained with a liquid diet (Peptamen, Nestlè) to avoid the development of intestinal obstructions, displayed in CFTRG542X/G542X homozygous mice.
RNA extraction and real-time RT-PCR
Protocol for RNA extraction
20–25 mg of tissues were weighted for RNA extraction.
The procedure was performed using the RNeasy Mini Kit (Qiagen, Germany) following the manufacturer’s instructions.
Briefly, a mix of RLT (RNeasy Lysis Buffer) buffer and β-mercaptoethanol was added to each sample for mechanical homogenization. Then, 1 vol of 70% DNase-RNase-free ethanol was mixed into samples, which were transferred to the RNaeasy spin column and centrifuged. A first washing step was followed by in-column DNase treatment, and then previously reconstituted buffer RPE (washing membrane-bound RNA) was added twice. Centrifuged samples were washed twice and centrifuged. A further centrifuge was performed to eliminate any residual reagent. Lastly, after centrifuging samples, RNA was collected from spin columns with 30 μL of DNase-RNase-free water elution.
RNA was quantified by a spectrophotometric assay using NanoDrop 2000 (Thermo Scientific, Invitrogen, USA).
RT
Using the manufacturer’s instructions, 1 μg of total RNA extracted was retrotranscribed in cDNA using the High-Capacity Reverse Transcription Kit (Thermo Fisher Scientific, USA).
For each sample, a reaction mix was prepared as follows:
-
•
2 μL buffer RT
-
•
2 μL random primers
-
•
1 μL dNTP
-
•
1 μL reverse transcriptase
-
•
RNase-free water to reach 20 μL of the final volume
cDNA was then used to perform qPCR using the following steps.
Real-time RT-PCR
Specific primer sequences were used to detect CFTR gene expression, as reported in Table S3.
A reaction mix was prepared for each couple of primers, specific for the gene of interest and the related housekeeping, here in detail indicated for each sample:
(1) SYBR MIX (Life Technologies, Thermo Fisher Scientific, USA) 12.5 μL
(2) Primers (forward + reverse 2 μM) 5 μL
(3) H2O DNase/RNase-free 5.5 μL
(4) cDNA 2 μL
Each sample was loaded in triplicate with a final volume of 25 μL each. The plate was centrifuged at 2,500 rpm for 5 min before the experiment started. The real-time RT-PCR machine (7300 Real-Time PCR System, Applied Biosystems) and the 7300 System Software were used to perform the analysis. Data were analyzed using triplicate values of Ct (cycle threshold). RNA levels were determined using the SDS software version (Applied Biosystems) according to the 2−ΔΔCt method, and Ct values were normalized to the internal control actin.
IHC analyses
Immunohistochemical examination was conducted on the lungs of CFTRG542X/G542X mice treated with NV848 in comparison to the lungs of CFTRG542X/G542X mice treated with water (negative control) and the lungs of CFTRWT mice treated with water (positive control). Tissues fixed in buffered formalin (10% [v/v]) were embedded in paraffin, and sections of 3 μm were prepared. These sections were incubated with the antibody anti-TTF1 (SP141, Roche Ventana 749-4756, ready to use) since TTF1 was chosen as an epithelial control marker to check tissue reactivity. CFTR was detected by incubating tissue slides with the antibody anti-CFTR (1:200, SAB4501942, Sigma-Aldrich, Merck, Darmstadt, Germany). Briefly, to unmask the antigen, the buffer ULTRA Cell Conditioning solution (ULTRA CC1, Roche Ventana 950-224) was used for 64 min at 95°C. The ultraView Universal HRP Multimer Detection Kit (Roche Ventana, 760-500) and UltraBenchmark (Roche Ventana) instruments were used as detection systems, according to the manufacturer’s instructions.
Protein extraction and western blot analyses
Total proteins were extracted from 40 mg of samples mechanically homogenized using RIPA (radioimmunoprecipitation assay buffer) lysis solution (Pierce RIPA Buffer, Thermo Fisher Scientific) and 1% protease inhibitors (Halt Protease and Phosphatase Inhibitor Cocktail, Thermo Fisher Scientific). The homogenates were then incubated for 1.5 h on ice at 4°C. Afterward, the samples were centrifuged at 12,000 rpm for 20 min at 4°C, and the supernatants were collected and preserved at −80°C. Finally, the Bradford colorimetric assay used 2 μL of each sample for quantification.
Western blot setup
For the analysis, 30 μg of proteins were loaded onto a 3%–8% SDS-PAGE gel and electrophoresed at 150 V and 100 mA for 1 h. Subsequently, proteins were transferred onto a PVDF (polyvinylidene fluoride or polyvinylidene difluoride) membrane (Invitrogen, Thermo Fisher Scientific, USA) overnight at 4°C at 12 V and 50 mA. Blocking was performed for 1 h at room temperature using 5% non-fat milk in TBS-TWEEN 1× (Pierce TBS-Tween 20×, Invitrogen, Thermo Fisher Scientific, USA).
The primary antibodies used were anti-CFTR (#PA1-935, Invitrogen, Thermo Fisher Scientific, USA) and anti-β-tubulin (Sigma-Aldrich, USA) 1:5,000, and they were incubated overnight. Membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies, anti-rabbit (Promega, USA, 1:3,000), or anti-mouse (Invitrogen, Thermo Fisher Scientific, USA, 1:5,000). Protein detection was carried out by adding West Femto (SuperSignal West Femto Maximum Sensitivity Substrate, Thermo Fisher Scientific), while Chemidoc XRS (BioRad) and Quantity One software (BioRad) were used for analysis. Protein bands were quantified using ImageJ software (NIH).
Statistics
All data are given as means ± SEM. n in the results section refers to the number of animals on which observations were made. The sample size was calculate using G∗Power 3.1 software. Statistically significant differences were calculated by analysis of variance, followed by a post hoc test when appropriate. A probability value of 0.05 was regarded as significant (GraphPad Software Prism v.6.0, La Jolla, CA, USA).
Data and code availability
All data presented in this work are available from the authors upon request.
Acknowledgments
We thank the University of Palermo for the PJ UTILE 2022 VQR Misura B_D15 Pibiri grant, the HEAL ITALIA Foundation, for funding from the European Union - NextGenerationEU through the Italian Ministry of University and Research under PNRR - M4C2-I1.3 Project PE_00000019 "HEAL ITALIA" to I.P. and L.L. (CUP B73C22001250006 Dip. STEBICEF-University of Palermo) and the Italian Cystic Fibrosis Research Foundation with grant FFC#06/2020 to L.L. and I.P. The views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them. We thank Prof. Craig Hodges, who provided us with the heterozygous CFTRG542X/+ mice.
Author contributions
Conceptualization, L.L., A.P., M.G.Z., and I.P.; methodology, I.F., F.C., R.P., M.G.Z., F.G., and R.M.; data curation, I.F., E.V., F.C., L.L., I.P., R.M., and D.R.; investigation, M.T., A.P., and R.M.; formal analysis, I.F., F.C., R.P., E.V., D.R., F.G., and M.G.Z.; validation, I.F., F.C., R.P., L.L., and I.P.; resources, L.L. and I.P.; writing – original draft, I.F., F.C., R.P., L.L., A.P., M.G.Z., M.T., and I.P.; writing – review & editing, I.F., F.C., M.G.Z., R.M., I.P., M.T., and A.P.; supervision, L.L., I.P., M.G.Z., and A.P.; project administration, L.L. and I.P.; funding acquisition, L.L., I.P., and A.P. All authors have read, edited, and approved the final manuscript.
Declaration of interests
I.P. is a scientific advisor of the CCM Bioscience group. I.P., L.L., A.P., M.T., and R.M. have patent licenses to WO2019101709.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2024.10.028.
Contributor Information
Laura Lentini, Email: laura.lentini@unipa.it.
Ivana Pibiri, Email: ivana.pibiri@unipa.it.
Supplemental information
References
- 1.Clarke L.A., Awatade N.T., Felício V.M., Silva I.A., Calucho M., Pereira L., Azevedo P., Cavaco J., Barreto C., Bertuzzo C., et al. The effect of premature termination codon mutations on CFTR mRNA abundance in human nasal epithelium and intestinal organoids: a basis for read-through therapies in cystic fibrosis. Hum. Mutat. 2019;40:326–334. doi: 10.1002/humu.23692. [DOI] [PubMed] [Google Scholar]
- 2.Potapova N.A. Nonsense Mutations in Eukaryotes. Biochemistry. 2022;87:400–412. doi: 10.1134/S0006297922050029. [DOI] [PubMed] [Google Scholar]
- 3.Pibiri I., Melfi R., Tutone M., Di Leonardo A., Pace A., Lentini L. Targeting Nonsense: Optimization of 1,2,4-Oxadiazole TRIDs to Rescue CFTR Expression and Functionality in Cystic Fibrosis Cell Model Systems. Int. J. Mol. Sci. 2020;21:6420. doi: 10.3390/ijms21176420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isken O., Maquat L.E. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 5.Lombardi S., Testa M.F., Pinotti M., Branchini A. Molecular Insights into Determinants of Translational Readthrough and Implications for Nonsense Suppression Approaches. Int. J. Mol. Sci. 2020;21:9449. doi: 10.3390/ijms21249449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy B., Friesen W.J., Tomizawa Y., Leszyk J.D., Zhuo J., Johnson B., Dakka J., Trotta C.R., Xue X., Mutyam V., et al. Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proc. Natl. Acad. Sci. USA. 2016;113:12508–12513. doi: 10.1073/pnas.1605336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palma M., Lejeune F. Deciphering the molecular mechanism of stop codon readthrough. Biol. Rev. Camb. Philos. Soc. 2021;96:310–329. doi: 10.1111/brv.12657. [DOI] [PubMed] [Google Scholar]
- 8.Burke J.F., Mogg A.E. Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin. Nucleic Acids Res. 1985;13:6265–6272. doi: 10.1093/nar/13.17.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manuvakhova M., Keeling K., Bedwell D.M. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huth M.E., Ricci A.J., Cheng A.G. Mechanisms of aminoglycoside ototoxicity and targets of hair cell protection. Int. J. Otolaryngol. 2011;2011 doi: 10.1155/2011/937861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Novoa J.M., Quiros Y., Vicente L., Morales A.I., Lopez-Hernandez F.J. New insights into the mechanism of aminoglycoside nephrotoxicity: an integrative point of view. Kidney Int. 2011;79:33–45. doi: 10.1038/ki.2010.337. [DOI] [PubMed] [Google Scholar]
- 12.Leubitz A., Frydman-Marom A., Sharpe N., van Duzer J., Campbell K.C.M., Vanhoutte F. Safety, Tolerability, and Pharmacokinetics of Single Ascending Doses of ELX-02, a Potential Treatment for Genetic Disorders Caused by Nonsense Mutations, in Healthy Volunteers. Clin. Pharmacol. Drug Dev. 2019;8:984–994. doi: 10.1002/cpdd.647. [DOI] [PubMed] [Google Scholar]
- 13.Leubitz A., Vanhoutte F., Hu M.-Y., Porter K., Gordon E., Tencer K., Campbell K., Banks K., Haverty T. A Randomized, Double-Blind, Placebo-Controlled, Multiple Dose Escalation Study to Evaluate the Safety and Pharmacokinetics of ELX-02 in Healthy Subjects. Clin. Pharmacol. Drug Dev. 2021;10:859–869. doi: 10.1002/cpdd.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eloxx Pharmaceuticals Reports Topline Results from Phase 2 Combination Clinical Trial of ELX-02 in Class 1 Cystic Fibrosis (CF) Patients. https://investors.eloxxpharma.com/news-releases/news-release-details/eloxx-pharmaceuticals-reports-topline-results-phase-2/ Eloxx Pharmaceuticals, Inc.
- 15.Saijo T., Kuriki H., Ashida Y., Makino H., Maki Y. Mechanism of the action of amoxanox (AA-673), an orally active antiallergic agent. Int. Arch. Allergy Appl. Immunol. 1985;78:43–50. doi: 10.1159/000233861. [DOI] [PubMed] [Google Scholar]
- 16.Meng W., Dong Y., Liu J., Wang Z., Zhong X., Chen R., Zhou H., Lin M., Jiang L., Gao F., et al. A clinical evaluation of amlexanox oral adhesive pellicles in the treatment of recurrent aphthous stomatitis and comparison with amlexanox oral tablets: a randomized, placebo controlled, blinded, multicenter clinical trial. Trials. 2009;10:30. doi: 10.1186/1745-6215-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McHugh D.R., Cotton C.U., Hodges C.A. Synergy between Readthrough and Nonsense Mediated Decay Inhibition in a Murine Model of Cystic Fibrosis Nonsense Mutations. Int. J. Mol. Sci. 2020;22:344. doi: 10.3390/ijms22010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trzaska C., Amand S., Bailly C., Leroy C., Marchand V., Duvernois-Berthet E., Saliou J.-M., Benhabiles H., Werkmeister E., Chassat T., et al. 2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations. Nat. Commun. 2020;11:1509. doi: 10.1038/s41467-020-15140-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leroy C., Spelier S., Essonghe N.C., Poix V., Kong R., Gizzi P., Bourban C., Amand S., Bailly C., Guilbert R., et al. Use of 2,6-diaminopurine as a potent suppressor of UGA premature stop codons in cystic fibrosis. Mol. Ther. 2023;31:970–985. doi: 10.1016/j.ymthe.2023.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch E.M., Barton E.R., Zhuo J., Tomizawa Y., Friesen W.J., Trifillis P., Paushkin S., Patel M., Trotta C.R., Hwang S., et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 21.Ryan N.J. Ataluren: first global approval. Drugs. 2014;74:1709–1714. doi: 10.1007/s40265-014-0287-4. [DOI] [PubMed] [Google Scholar]
- 22.Meeting Highlights from the Committee for Medicinal Products for Human Use (CHMP) 22-25 January 2024. https://www.ema.europa.eu/en/news/meeting-highlights-committee-medicinal-products-human-use-chmp-22-25-january-2024 European Medicines Agency.
- 23.Tutone M., Pibiri I., Perriera R., Campofelice A., Culletta G., Melfi R., Pace A., Almerico A.M., Lentini L. Pharmacophore-Based Design of New Chemical Scaffolds as Translational Readthrough-Inducing Drugs (TRIDs) ACS Med. Chem. Lett. 2020;11:747–753. doi: 10.1021/acsmedchemlett.9b00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng M.Y., Li H., Ghelfi M.D., Goldman Y.E., Cooperman B.S. Ataluren and aminoglycosides stimulate read-through of nonsense codons by orthogonal mechanisms. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020599118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S., Bhattacharya A., Ghelfi M.D., Li H., Fritsch C., Chenoweth D.M., Goldman Y.E., Cooperman B.S. Ataluren binds to multiple protein synthesis apparatus sites and competitively inhibits release factor-dependent termination. Nat. Commun. 2022;13:2413. doi: 10.1038/s41467-022-30080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carollo P.S., Tutone M., Culletta G., Fiduccia I., Corrao F., Pibiri I., Di Leonardo A., Zizzo M.G., Melfi R., Pace A., et al. Investigating the Inhibition of FTSJ1, a Tryptophan tRNA-Specific 2’-O-Methyltransferase by NV TRIDs, as a Mechanism of Readthrough in Nonsense Mutated CFTR. Int. J. Mol. Sci. 2023;24:9609. doi: 10.3390/ijms24119609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pibiri I., Lentini L., Melfi R., Gallucci G., Pace A., Spinello A., Barone G., Di Leonardo A. Enhancement of premature stop codon readthrough in the CFTR gene by Ataluren (PTC124) derivatives. Eur. J. Med. Chem. 2015;101:236–244. doi: 10.1016/j.ejmech.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 28.Pibiri I., Lentini L., Tutone M., Melfi R., Pace A., Di Leonardo A. Exploring the readthrough of nonsense mutations by non-acidic Ataluren analogues selected by ligand-based virtual screening. Eur. J. Med. Chem. 2016;122:429–435. doi: 10.1016/j.ejmech.2016.06.048. [DOI] [PubMed] [Google Scholar]
- 29.Pibiri I., Lentini L., Melfi R., Tutone M., Baldassano S., Ricco Galluzzo P., Di Leonardo A., Pace A. Rescuing the CFTR protein function: Introducing 1,3,4-oxadiazoles as translational readthrough inducing drugs. Eur. J. Med. Chem. 2018;159:126–142. doi: 10.1016/j.ejmech.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 30.Corrao F., Zizzo M.G., Tutone M., Melfi R., Fiduccia I., Carollo P.S., Leonardo A.D., Caldara G., Perriera R., Pace A., et al. Nonsense codons suppression. An acute toxicity study of three optimized TRIDs in murine model, safety and tolerability evaluation. Biomed. Pharmacother. 2022;156 doi: 10.1016/j.biopha.2022.113886. [DOI] [PubMed] [Google Scholar]
- 31.Perriera R., Vitale E., Pibiri I., Carollo P.S., Ricci D., Corrao F., Fiduccia I., Melfi R., Zizzo M.G., Tutone M., et al. Readthrough Approach Using NV Translational Readthrough-Inducing Drugs (TRIDs): A Study of the Possible Off-Target Effects on Natural Termination Codons (NTCs) on TP53 and Housekeeping Gene Expression. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms242015084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bezzerri V., Lentini L., Api M., Busilacchi E.M., Cavalieri V., Pomilio A., Diomede F., Pegoraro A., Cesaro S., Poloni A., et al. Novel Translational Read-through-Inducing Drugs as a Therapeutic Option for Shwachman-Diamond Syndrome. Biomedicines. 2022;10:886. doi: 10.3390/biomedicines10040886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong R., Ma J., Hwang S., Moon Y.-C., Welch E.M., Weetall M., Colacino J.M., Almstead N., Babiak J., Goodwin E. In vitro metabolism, reaction phenotyping, enzyme kinetics, CYP inhibition and induction potential of ataluren. Pharmacol. Res. Perspect. 2020;8 doi: 10.1002/prp2.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McHugh D.R., Steele M.S., Valerio D.M., Miron A., Mann R.J., LePage D.F., Conlon R.A., Cotton C.U., Drumm M.L., Hodges C.A. A G542X cystic fibrosis mouse model for examining nonsense mutation directed therapies. PLoS One. 2018;13 doi: 10.1371/journal.pone.0199573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du M., Liu X., Welch E.M., Hirawat S., Peltz S.W., Bedwell D.M. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc. Natl. Acad. Sci. USA. 2008;105:2064–2069. doi: 10.1073/pnas.0711795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kayali R., Ku J.-M., Khitrov G., Jung M.E., Prikhodko O., Bertoni C. Read-through compound 13 restores dystrophin expression and improves muscle function in the mdx mouse model for Duchenne muscular dystrophy. Hum. Mol. Genet. 2012;21:4007–4020. doi: 10.1093/hmg/dds223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this work are available from the authors upon request.