Abstract
Poliovirus binding to its receptor (PVR) on the cell surface induces a conformational transition which generates an altered particle with a sedimentation value of 135S versus the 160S of the native virion. A number of lines of evidence suggest that the 135S particle is a cell entry intermediate. However, the low infection efficiencies of the 135S particle and the absence of detectable 135S particles during infection at 26°C by the cold-adapted mutants argue against a role for the 135S particle during the cell entry process. We show here that binding of 135S-antibody complexes to the Fc receptor (CDw32) increases the infectivity of these particles by 2 to 3 orders of magnitude. Thus, the low efficiency of infection by 135S particles is due in part to the low binding affinity of these particles. In addition, we show that there is an additional stage in the entry process that is associated with RNA release. This stage occurs after formation of the 135S particle, is rate limiting during infection at 37°C, but not at 26°C, and is PVR independent. The data also demonstrate that during infection at 26°C, the rate-limiting step is the PVR-mediated conversion of wild-type 160S particles to 135S particles. This suggests that during infection at 26°C by the cold-adapted viruses, 135S particles are formed, but they fail to accumulate to detectable levels because the subsequent post-135S particle events occur at a significantly faster rate than the initial conversion of 160S to 135S particles. These data support a model in which the 135S particle is an intermediate during poliovirus entry.
Poliovirus is a picornavirus whose 7,400-nucleotide positive-sense RNA genome is encapsidated in an icosahedral protein shell formed by 60 copies of four capsid proteins (VP1, VP2, VP3, and VP4). Virus entry into cells is initiated by binding to the poliovirus receptor (PVR) on the cell surface. PVR binding induces conformational rearrangements within the virus particle which leads to the formation of an altered particle sedimenting at 135S (versus the 160S sedimentation value of the native particle) and the coordinate relocation of capsid protein VP4 and VP1 N termini from the interior of the virion to the outer surface of the particle (9).
A number of lines of evidence suggest that the 135S particles (also known as “A particles”) are intermediates in the cell entry pathway. Thus, the 135S particles are able to bind to liposomes and insert into lipid bilayers to form ion channels (9, 17). In addition, it has been shown that 135S particles infect cells in a receptor-independent fashion (6) and that drugs (for example, Win 51711) that inhibit the conversion of 160S particles to the 135S form by binding to the poliovirus capsid prevent infection (3, 8, 12, 13, 20). However, two observations raise serious questions in some people's minds about the role of the 135S particle as a true intermediate in entry. First, although the 135S particle can infect cells in the absence of PVR expression, infection is highly inefficient (6). Thus, it has been proposed that the infectivity of the 135S particle resulted from the presence of small amounts of the infectious RNA genome that are randomly released due to breakage or degradation of the 135S particles. Second, the 135S particles are not detected during infection at 26°C with virus mutants which have been selected to grow at this lower temperature (7).
Here, we present data to counter these arguments. Specifically, we show that efficiency can be significantly improved when an alternative mechanism is provided for high-affinity binding of 135S particles to the cell surface. Thus, the low efficiency of infection of the 135S particle is due in part to the low binding affinity of this particle to cells. Furthermore, we show that there exists an additional stage in the entry process, which occurs after formation of the 135S particle, which is rate limiting at 37°C. Detectable levels of 135S particles accumulate during infection at 37°C because the rate of production of 135S is faster than this rate-limiting downstream stage. Data also demonstrate that genome entry by wild-type virus at 26°C is significantly more rapid when initiated with the wild-type 135S particles than with the 160S particles, indicating that it is the PVR-mediated conversion of 160S to 135S particles and not the downstream stage that is rate limiting at 26°C. Thus, an alternative interpretation of the failure to detect the 135S particles during infection at 26°C is that at the lower temperature, the 135S particles do not accumulate to detectable concentrations because subsequent downstream (post-135S particle) events during virus entry occur at a significantly faster rate than the initial conversion of 160S to 135S particles.
Antibody enhancement of 135S particle infection.
The 135S particles bind to cells via a receptor-independent, nonsaturable, low-affinity interaction that may simply reflect 135S particles binding to cell membranes (6). This suggested that the efficiency of 135S particle infections might be greater if these particles could bind to cells at higher affinity and that this higher-affinity interaction could potentially be achieved by binding 135S particle-antibody complexes to Fc receptors. Previous studies had shown that foot-and-mouth disease virus–antibody complexes could mediate infection in normally nonsusceptible cells by binding to the Fc receptor (11). In that study, however, parallel experiments with poliovirus-antibody complexes were not infectious. Similar studies with the C3 monoclonal antibody with poliovirus 160S and 135S particles also failed to show antibody-mediated infection with Fc receptor (S. Curry, unpublished observations). More recently, Arita et al. have shown with different antibodies that binding of 160S-antibody complexes to Fc receptors can inefficiently compensate for PVR binding to initiate infection (1).
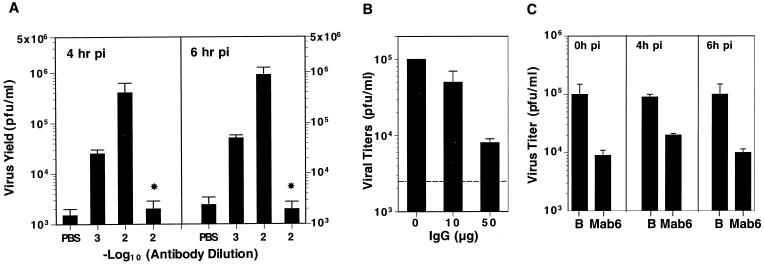
We have investigated a specific monoclonal antibody (MAb6) that binds to neutralizing antigenic site 2 of 160S particles and also binds to 135S particles with slightly lower affinities (9). The 135S-MAb6 complexes were used to infect mouse L cells (L/CD32+), a cell line which stably expresses Fc gamma receptors (CDw32+) (2). The resultant titers were measured over the course of infection (Fig. 1A). Viral titers increased over time, indicating that the titers resulted from active replication rather than titers from input virus particles. The infectivities of the 135S-antibody complexes were 2 to 3 orders of magnitude higher than that of 135S particles alone and were dependent on antibody concentrations. No enhancement was observed when 135S was incubated with a nonpolio-virus-specific antibody, OKT3 (Fig. 1A). The increased efficiency was dependent on Fc-receptor binding of the 135S-antibody complexes, because antibody enhancement of 135S infection did not occur in CDw32− L cells or in Rat-2 cells (data not shown). In addition, coincubation or preincubation of the CDw32+ L cells with the nonspecific immunoglobulin G (IgG) inhibited binding of the 135S-MAb6 complex to Fc receptor and prevented the antibody-mediated increases in titers (Fig. 1B). At very high dilutions (10−5), MAb6 also mediated infection of 160S particles on these cells. However, at the antibody concentrations used for the 135S experiments, MAb6 neutralizes the infectivity of 160S particles rather than enhances it (Fig. 1C). This provides further evidence that the infectivity observed is not due to trace amounts of unconverted 160S particles.
FIG. 1.
Antibody enhancement of infections by 135S particles. (A) Poliovirus 135S particles were generated in vitro by preincubating purified 160S virus in 20 mM Tris-HCl (pH 7.5)–2 mM CaCl2–0.1% Tween 20 at 55°C for 2 min and subsequently chilling the mixture on ice. The levels of residual 160S particles present in these samples of 135S particles were shown to be negligible by the equivalent titers observed when assayed on Rat-2 cells (PVR−) and on CV-1 cells (PVR+). Dilutions of MAb6 (■) or of nonspecific IgG (*) ascites fluids were made in PBS and incubated with the in vitro-generated 135S particles (108 particles) for 1 h at 0°C. The antibody-virus complexes were added to L/CD32+ cells (5 × 105) for 1 h at 20°C, the cells were washed with PBS to remove unbound virus or antibody complexes, and infection at 37°C was initiated with addition of prewarmed medium (Dulbecco's modified medium–5% fetal calf serum). Samples were harvested at various hours p.i. and lysed by freeze-thaw three times, and the resultant virus titers were measured in triplicate by plaque assay on HeLa cells. The average titers at 4 and 6 h p.i. are shown. Titers at 0 h p.i. were below the detectable levels (<50 PFU/ml) for all samples. (B) L/CD32+ cells were incubated with 135S-MAb6 complexes (10−2 dilution of ascites fluid) in the presence of various concentrations of purified nonspecific IgG (0, 10, or 50 μg) and washed with PBS, and infection at 37°C was initiated by addition of prewarmed medium. The resultant titers were measured by plaque assay. The titer obtained upon infection with 135S particles alone is indicated by the dotted line. (C) L/CD32+ cells were incubated with 160S particles in PBS (B) or with 160S-MAb6 complexes (10−2 dilution of ascites fluid) (MAb6) and washed with PBS, and infection at 37°C was initiated by addition of prewarmed medium. Samples were harvested at various hours p.i. and lysed by freeze-thaw three times. Virus titers were measured by plaque assay on HeLa cells. The titers at 0, 4, and 6 h p.i. are shown.
Although the increased infectivity of the 135S-MAb6 complex is not equivalent to that of the wild-type 160S particle, the resultant titers from the 135S-MAb6 infections indicate that the infectivity of the 135S particle is reduced at most 1 to 2 orders of magnitude from that of the 160S particle and is comparable with that observed with several previously characterized viable poliovirus mutants (15, 16). Thus, the low infectivity of the 135S particle is explained in part by the absence of a high-affinity interaction with the cell surface which can be partially compensated for by the affinity of the antibody-Fc receptor interaction. Moreover, the MAb-dependent increase in titers complements previous observations which showed that the infectivity of 135S particles is dependent on the N-terminal 31 amino acids of VP1 and is resistant to digestion with RNase (6). These data reinforce the conclusion that 135S particles are infectious and this infectivity is not dependent on PVR expression on the cell surface.
NR sensitivity of 135S particle infection.
Previous studies have demonstrated that the infectivity of poliovirus grown in the presence of neutral red (NR) is photosensitive (5, 19). This photosensitivity results from the incorporation of the dye within the interior of the virion in close proximity to the RNA and subsequent modification of the RNA genome by the locally high concentrations of the photoactivated dye after exposure to visible light. During infection with NR-poliovirus, the period of photosensitivity has been correlated with the kinetics of RNA release and is thought to be due to diffusion of this chromogenic dye away from the RNA upon release of the genome into the cytoplasm. Infection by NR-virus is light sensitive only at initial stages of infection, and this period of photosensitivity has been used to characterize viable mutants which are defective in uncoating (10). It was of interest to determine whether infection by NR-135S particles was photosensitive and, if so, to compare the periods of photosensitivity as a measure of RNA release. If the 135S particle is an intermediate in the entry process, then one would predict that a similar (perhaps even shorter) period of photosensitivity would be observed during infection by NR-135S particles.
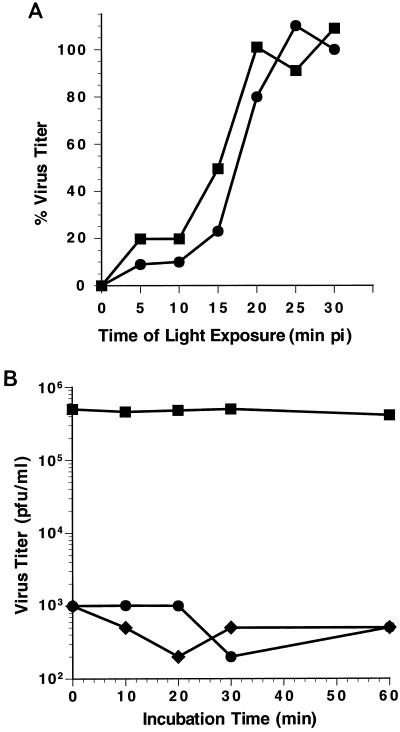
NR-135S particles were generated in vitro from NR-160S particles. To determine the optimal time of light irradiation, the NR-135S and NR-160S particles were exposed to increasing periods of light irradiation, and the residual viral titers were measured by plaque assay (10). The decay curves for NR-135S and -160S particles were very similar. Viral titers for both the 160S and 135S forms of viral particles dropped 300- to 400-fold within a 5-min period of light exposure, indicating that the infectivities of 135S and 160S particles were equivalently sensitive to light (data not shown). The period during which infection by NR-135S particles was light sensitive was measured by exposing cell monolayers to light at different times postinfection (p.i.). This photosensitive period was compared with that observed for infections with NR-160S particles (Fig. 2A). As observed previously, infection by NR-160S particles becomes light resistant within 20 to 30 min p.i. (half-time for acquiring photoresistance for 160S [T1/2-160S] = 17 min p.i.) (10). Similarly, infection by NR-135S particles becomes photoresistant within 20 to 25 min (T1/2-135S = 15 min p.i.).
FIG. 2.
Photosensitivity of infection with NR-135S particles. (A) [35S]methionine-labeled virus was grown in the presence of NR (5, 19). NR-135S particles were converted in vitro from NR-160S particles and diluted to the appropriate titers with PBS. HeLa cell monolayers were incubated in the dark at 4°C for 1 h with 300 to 400 PFU of NR-135S (■) or NR-160S (●) particles. The cells were washed with PBS, and infection was initiated in the dark with the addition of a Dulbecco's modified medium–5% fetal calf serum–0.8% agarose overlay. At the indicated times p.i. the monolayers were exposed to white light for 10 min at 20°C and then subsequently returned to 37°C for 48 h to allow the plaque assay to develop. The resistant titers from duplicate infectious center assays were averaged, and the percentage of light-resistant viral infections was calculated based on the 100% value representing the average titers obtained when infections were constantly maintained in the dark. Representative results for a set of parallel infections with 160S and 135S particles are shown. (B) In vitro-converted NR-135S particles were incubated in PBS at 26°C (⧫) or at 37°C (■, ●). At various times, aliquots were taken to infect Rat-2 cell monolayers, the monolayers were exposed to light (⧫, ●) or maintained in the dark (■), and then infection was initiated by the addition of prewarmed medium. Cells were harvested at 6 h, and the resultant titers were determined by plaque assay.
Photosensitivity is dependent on the concentration of NR present in the particle. It is possible that the conformational changes associated with formation of the 135S particle result in a particle with increased permeability, which leads to diffusion of NR out of the particle. If this were the case, then the period of photosensitivity observed during infection for NR-135S particles could represent the kinetics of NR diffusion out of the particle rather than the kinetics of RNA release into the cell. To test for this, NR-135S particles were incubated in phosphate-buffered saline (PBS) at 26 or 37°C. At various times of incubation, samples were exposed to light and the proportion of viral titer that was photosensitive was measured (Fig. 2B). The intrinsic infectivity of the 135S particles remains unchanged over the incubation period and is reflected by the minimal change in viral titers when the samples were maintained in the dark. However, when the 135S virus particle was exposed to light, virus titers significantly decreased at all times examined. These data indicate that the appearance of photoresistant viral titers at 20 to 25 min p.i. is not an artifact of passive loss of NR, but rather is dependent on downstream events in the entry pathway which are initiated upon interaction of 135S particles with cells.
Interestingly, the transition from photosensitivity to photoresistance began, for infections by either 135S or 160S particles, at approximately 10 to 15 min p.i. The exact periods of light sensitivity for infections with 160S and 135S particles varied from experiment to experiment, with values for T1/2-160S ranging between 16 to 19 min p.i. and T1/2-135S ranging between 14 and 17 min p.i. The variation in half-times likely reflects the several different preparations of virus and passages of HeLa cells used over the course of these studies. Thus, when reviewed over the entire range of experiments, the differences between the T1/2-160S and T1/2-135S are of marginal statistical significance. However, when measured for infections by 135S and 160S particles done in parallel, the period of photosensitivity (as quantitated by T1/2 values) was always shorter for 135S particles than for 160S particles. The steep rise in photoresistant viral titers demonstrates that infections with either 135S particles or 160S particles are highly synchronous (Fig. 2A). Moreover, the appearance of photoresistant virus at 10 to 25 min p.i. identifies an additional stage which occurs downstream from the initial receptor-mediated conformational rearrangements that form 135S particles. The length of this NR-sensitive interval is similar, irrespective of whether infection is initiated by 160S or 135S particles. This suggests that the stage (demarcated by NR sensitivity) is rate limiting during infection at 37°C.
Poliovirus entry during infection at 26°C.
Previous studies indicated that infection at 26°C by wild-type virus is nonproductive because of a block in RNA synthesis (7). Consistent with viral RNA synthesis being inhibited during infections with wild-type virus, the cold-adapted mutants contain mutations in 2C, a nonstructural protein involved in viral RNA replication, which enable these mutants to overcome this block and allow productive replication at 26°C (7). To look for the presence of an NR-sensitive stage in virus entry during infection at 26°C with wild-type virus, it was necessary to separate the events associated with virus entry and uncoating from the process of RNA transcription and replication. This is possible if replication of wild-type virus can occur after the infected cells were initially incubated at 26°C for a period of time and subsequently shifted to 37°C. To test for this, cells were initially infected with wild-type virus at 26°C. At various times p.i. at 26°C, the cells were shifted to 37°C, and the resultant viral titers were measured. Consistent with the presence of a block in RNA synthesis, no viral titers were detected when the infections with wild-type virus were maintained at 26°C over a 16-h period. However, for up to 6 h at 26°C, if the infections subsequently were shifted to 37°C, the titers recovered from these temperature shift infections were 50 to 80% of the titers obtained from infections occurring completely at 37°C (data not shown). These data indicate that subsequent poliovirus replication is not significantly compromised by the initial incubation period at 26°C. Thus, after light exposure to block infection by RNA genomes remaining within the viral particles, RNA genomes released from NR-virus particles during virus entry at 26°C can be detected by the replication of these genomes at 37°C.
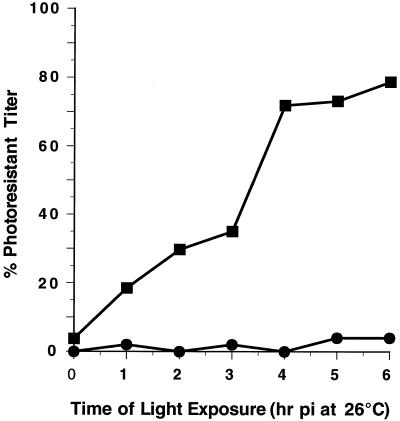
Cell monolayers were infected with wild-type NR-135S or -160S particles at 26°C in the dark. At various times p.i., the monolayers were exposed to light at room temperature and then returned to 26°C. After the last time point at 6 h, all samples (including samples which were maintained in the dark) were shifted to 37°C to allow completion of the replication cycle, and the resultant viral titers were measured (Fig. 3). Infections with NR-160S particles remain highly photosensitive over the 6-h period at 26°C, indicating that very few particles had uncoated their genomes. In striking contrast, photoresistant viral titers are detected in NR-135S infections within 1 h p.i. and continue to increase until infection with these particles becomes largely photoresistant at 4 h p.i. at 26°C. Moreover, there is an interval (between 3 and 4 h p.i.) during the infection at 26°C in which a sharp increase in photoresistant titers occurs. This indicates that a similar NR-sensitive stage exists during infection at 26°C and suggests that release of RNA from 135S particles during infection at 26°C is relatively synchronous. The half-time for acquiring photoresistance (T1/2-135S) is approximately 3.5 h. In contrast, T1/2-160S has not been measurable, but it is significantly longer than 6 h for 160S at 26°C. This is consistent with the conversion of 160S to 135S particles at 26°C being slow and rate limiting in these infections. Thus, the failure to observe 135S particles during infection at 26°C is expected, because the 135S particles are being consumed more rapidly than they are being produced.
FIG. 3.
Photosensitivity of 135S infection at 26°C. NR-160S (●) and in vitro-converted NR-135S (■) particles (300 to 400 PFU) were bound to HeLa cell monolayers for 1 h at 4°C. In the dark, cells were washed with PBS at 4°C, and infection was initiated by the addition of medium prewarmed to 26°C and maintained in a 5% CO2 incubator at 26°C. At various hours p.i., the monolayers were exposed to white light for 10 min and returned to the incubator at 26°C. One set of infected monolayers were maintained constantly in the dark at 26°C. At 6 h p.i., an agarose overlay was added to all monolayers, and the cells were shifted to a 5% CO2 incubator at 37°C. The photoresistant titers were measured after 48 h p.i. The percentage of light-resistant viral infections was calculated based on the 100% value representing the titers obtained for infections maintained in the dark, which were initiated at 26°C and subsequently shifted to 37°C.
Roles of PVR during poliovirus entry.
The data presented here counter several arguments that have been raised against a role for 135S particles during poliovirus entry into cells and are consistent with a model in which 135S particles are an intermediate of the viral entry pathway. First, enhancement of 135S particle infection by antibody-Fc receptor interactions (Fig. 1) demonstrates that the ability to infect cells in a PVR-independent manner is an inherent property of 135S particles. Thus, the infection inefficiencies of 135S particles at 37°C are due in part to the low binding affinity of these particles to the cell. Second, there is at least an additional stage during virus entry which occurs during infection at 37°C at a significantly later time after the PVR-mediated 160S-to-135S conformational transition (Fig. 2). This stage appears to be rate limiting during infection at 37°C, thus allowing accumulation of detectable levels of 135S particles. In contrast, it appears that at 26°C, the conversion of 160S to 135S particles by PVR is rate limiting. Infection with 135S particles bypasses the requirement for PVR-mediated conversions at 26°C, and, consequently, genome release during infection with 135S particles is synchronous and more rapid at this lower temperature (Fig. 3).
If the 135S particle is an intermediate during virus entry, then the differences observed in infection efficiencies of the 160S versus 135S particles at 37 and 26°C confirm that PVR serves at least two roles during virus entry (1, 4, 14). First, PVR provides a high-affinity binding site that the virus uses to “dock” with the cell surface. Second, PVR binding induces the 160S-to-135S conformational transition to occur at physiological temperatures. In vitro, these conformational rearrangements can also be triggered at elevated temperatures in the absence of PVR, indicating that this conformational transition is an energy-requiring event. Indeed, recent measurements suggest that the energy of activation (Ea) of this transition is quite large—on the order of 100 to 140 kcal—and that the rate of this transition in the absence of PVR is very slow at 37°C (18). Thus, in addition to its docking function, PVR interaction alters the energetics of the 160S-135S transition such that it can readily occur under physiological conditions.
Finally, the interval of NR sensitivity defines a temporal window during which an additional stage in the entry pathway occurs which is associated with RNA release. This stage also occurs during infection of Rat-2 cells with 135S particles, indicating that this event is independent of PVR expression. The steep transition from photosensitivity to photoresistance suggests that this stage occurs synchronously during infection. This synchrony suggests that additional unknown factors may be required to trigger RNA release.
Acknowledgments
We are grateful to M. Tosteson for valuable discussions and critiques. We thank M. Cannon and A. Khanolkar for the gifts of CDw32+ L cells and purified OKT3 IgG.
This work was supported by Public Health Service grants AI22627 and AI42390 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Arita M, Horie H, Arita M, Nomoto A. Interaction of poliovirus with its receptor affords a high level of infectivity to the virion in poliovirus infections mediated by the Fc receptor. J Virol. 1999;73:1066–1074. doi: 10.1128/jvi.73.2.1066-1074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banchereau J, de Paoli P, Valle A, Garcia E, Rousset F. Long-term human B cell lines dependent on interleukin-4 and antibody to CD40. Science. 1991;251:70–72. doi: 10.1126/science.1702555. [DOI] [PubMed] [Google Scholar]
- 3.Caliguiri L A, McSharry J J, Lawrence G W. Effect of arildone on modifications of poliovirus in vitro. Virology. 1980;105:86–93. doi: 10.1016/0042-6822(80)90158-0. [DOI] [PubMed] [Google Scholar]
- 4.Chow M, Basavappa R, Hogle J M. The role of conformational transitions in poliovirus pathogenesis. In: Chiu W, Garcea R, Burnette R, editors. Structural biology of viruses. Oxford, United Kingdom: Oxford University Press; 1997. pp. 157–186. [Google Scholar]
- 5.Crowther D, Melnick J L. The incorporation of neutral red and acridine orange into developing poliovirus particles making them photosensitive. Virology. 1962;14:11–21. doi: 10.1016/0042-6822(61)90127-1. [DOI] [PubMed] [Google Scholar]
- 6.Curry S, Chow M, Hogle J M. The poliovirus 135S particle is infectious. J Virol. 1996;70:7125–7131. doi: 10.1128/jvi.70.10.7125-7131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dove A W, Racaniello V R. Cold-adapted poliovirus mutants bypass a postentry replication block. J Virol. 1997;71:4728–4735. doi: 10.1128/jvi.71.6.4728-4735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everaert L, Vrijsen R, Boeyé A. Eclipse products of poliovirus after cold-synchronized infection of HeLa cells. Virology. 1989;171:76–82. doi: 10.1016/0042-6822(89)90512-6. [DOI] [PubMed] [Google Scholar]
- 9.Fricks C E, Hogle J M. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkegaard K. Mutations in VP1 of poliovirus specifically affect both encapsidation and release of viral RNA. J Virol. 1990;64:195–206. doi: 10.1128/jvi.64.1.195-206.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason P W, Baxt B, Brown F, Harber J, Murdin A, Wimmer E. Antibody-complexed foot-and mouth disease virus, but not poliovirus, can infect normal insusceptible cells via the Fc receptor. Virology. 1993;192:568–577. doi: 10.1006/viro.1993.1073. [DOI] [PubMed] [Google Scholar]
- 12.McSharry J J, Caliguiri L A, Eggers H J. Inhibition of uncoating of poliovirus by arildone, a new antiviral drug. Virology. 1979;97:307–315. doi: 10.1016/0042-6822(79)90342-8. [DOI] [PubMed] [Google Scholar]
- 13.Ofori-Anyinam O, Vrijsen R, Kronenberger P, Boeyé A. Effect of a capsid-stabilizing pyridazinamine, R 78206, on the eclipse and intracellular location of poliovirus. J Virol. 1993;67:2367–2369. doi: 10.1128/jvi.67.4.2367-2369.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Racaniello V R. The poliovirus receptor: a hook, or an unzipper? Structure. 1996;4:769–773. doi: 10.1016/s0969-2126(96)00083-4. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds C, Birnby D, Chow M. Folding and processing of the capsid protein precursor P1 is kinetically retarded in neutralization site 3B mutants of poliovirus. J Virol. 1992;66:1641–1648. doi: 10.1128/jvi.66.3.1641-1648.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds C, Page G, Zhou H, Chow M. Identification of residues in VP2 that contribute to poliovirus neutralization antigenic site 3B. Virology. 1991;184:391–396. doi: 10.1016/0042-6822(91)90856-7. [DOI] [PubMed] [Google Scholar]
- 17.Tosteson M T, Chow M. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J Virol. 1997;71:507–511. doi: 10.1128/jvi.71.1.507-511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsang S K, Danthi P, Chow M, Hogle J M. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. J Mol Biol. 2000;296:335–340. doi: 10.1006/jmbi.1999.3483. [DOI] [PubMed] [Google Scholar]
- 19.Wilson J N, Cooper P D. Aspects of the growth of poliovirus as revealed by the photodynamic effects of neutral red and acridine orange. Virology. 1963;21:135–145. doi: 10.1016/0042-6822(63)90249-6. [DOI] [PubMed] [Google Scholar]
- 20.Zeichhardt H, Otto M J, McKinlay M A, Willingmann P, Habermehl K O. Inhibition of poliovirus uncoating by disoxaril (WIN 51711) Virology. 1987;160:281–285. doi: 10.1016/0042-6822(87)90075-4. [DOI] [PubMed] [Google Scholar]