Abstract
In cytopathic bovine viral diarrhea virus genotype 1 (BVDV1) isolates, insertions are reported at position A (amino acid [aa] 1535) and position B (aa 1589). Insertions at position B predominate. In this survey it was found that in BVDV2, insertions at position A predominate. Possible reasons for this difference in relative frequency are discussed.
Bovine viral diarrhea viruses (BVDV) are segregated into two genotypes, BVDV1 and BVDV2 (18, 26). Viruses from either genotype may be characterized as cytopathic or noncytopathic (6, 11, 23) based on their activity in cell culture. Noncytopathic BVDV code for an NS2-3 protein that in cytopathic BVDV is processed to an NS3 protein (5). Processing of the NS2-3 protein in cytopathic BVDV1 occurs by several different strategies depending on the viral strain (3, 9, 10, 13, 15, 20, 21, 27, 28). Most commonly it is associated with the insertion of sequences into the viral genome. In BVDV1, the amino acid sequences flanking the carboxy-terminal end of inserted sequences correspond to either amino acid position 1535 or position 1589 (numbering is based on BVDV-1 strain SD-1, GenBank accession number M96751), referred to as positions A and B, respectively (13). Most reported insertions into BVDV1 have been at position B and consist of duplicated viral genomic sequences and/or ubiquitin coding sequences (8, 21), although insertions of other cellular coding sequences have been reported (2, 13, 20). In contrast, limited information is available on the molecular characterization of cytopathic BVDV2 insertions.
In this study, nine cytopathic BVDV2 were examined for type and location of insertions. All viruses were isolated in North America, between 1990 and 1998, from tissues submitted to diagnostic laboratories. Isolation and propagation of viruses were performed as described earlier (25). All cytopathic viruses, except BVDV2-96B3491c, were coisolated with noncytopathic BVDV. Viruses were cloned at limiting dilution and genotyped based on PCR amplification and comparison of the 5′ untranslated region (26). Biotype was determined by activity in cell culture (6, 11) and production of NS3 (5, 19), as determined by radioimmunoprecipitation using bovine polyclonal antisera (25). Viruses were found to be free of defective interfering particles by Northern blot analysis, which was performed as previously described (17) using a BVDV-specific probe prepared from sequences from the NS3 coding region (data not shown).
Total RNA was prepared from cultured cells 24 h after inoculation (24). Primer design and PCR amplification were performed as described previously (26). Three PCR products were generated for each virus using the primers listed in Table 1. Both strands of each PCR product were sequenced in duplicate (22). Phylogenetic analysis was performed using the Align Plus (Scientific and Educational Software, State Line, Pa.), GeneWorks (Intelligenetics Inc., Mountain View, Calif.), and DNASIS (Hitachi Software, San Bruno, Calif.) software packages.
TABLE 1.
PCR primers for detection and sequencing of insertions in NS2-3
| Primer set | Forward primer
|
Reverse primer
|
||
|---|---|---|---|---|
| Sequence | Positiona | Sequence | Positiona | |
| A | ATG-ACC-ACC-ATG-TGG-GC | 4244 | GAG-TTG-TCC-TCT-GGC-TAT-GT | 5029 |
| B | GTG-TGG-AAG-ACA-AGG-AAA-GC | 4894 | TTG-GTA-GGT-TCC-TCA-GGA-AG | 5069 |
| C | AGT-GAG-AAA-TGA-AGC-CGT-CC | 4730 | TGT-ACC-TCT-AGG-CAT-GAT-CC | 5234 |
Nucleotide position numbers correspond to the sequence of BVDV1-SD-1 (GenBank accession number M96751).
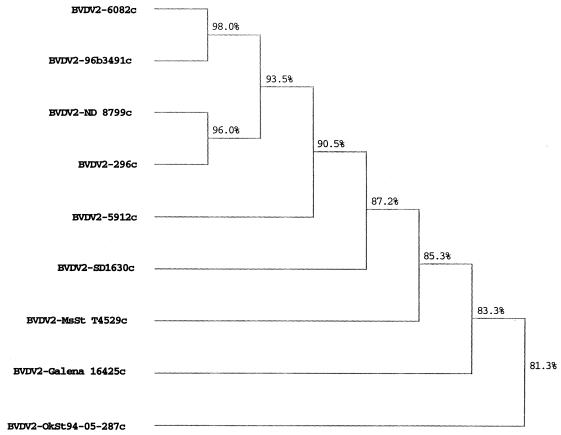
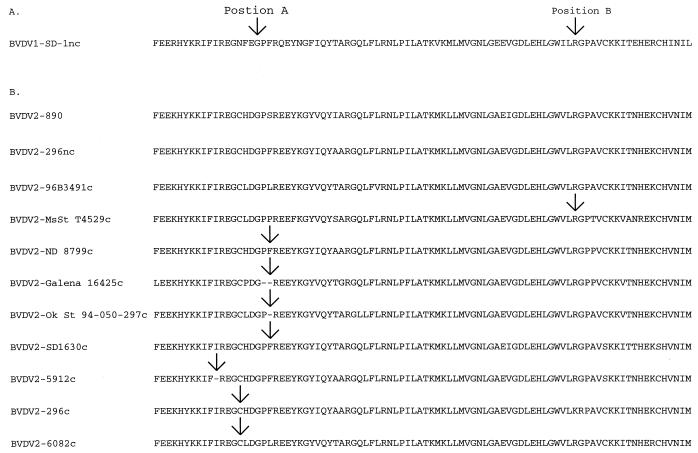
The viruses represented a genetically diverse sample, with derived NS2-3 amino acid sequence similarities ranging between 81.3 and 98% (Fig. 1). No correlation was seen between amino acid sequence similarity and size and type of genomic insertion. No insertion was detected in BVDV2-96B3491c; the remaining eight viruses had insertions (Fig. 2). Only one virus, BVDV2-MsSt T4529c, had an insertion at position B. This insertion consisted of two full-length ubiquitin genes and one partial ubiquitin gene (Fig. 3A). The partial ubiquitin gene lacked 34 amino acid residues from the amino terminus. Four viruses, BVDV2-ND 8799c, BVDV2-Galena 16425c, BVDV2-Ok St 94-050-297c, and BVDV2-SD1630c, had insertions at position A. Three viruses, BVDV2-296c, BVDV2-5912c, and BVDV2-6082c, had insertions within 10 amino acid positions of position A. In all seven viruses the insertion contained a portion of cellular sequence coding for a DnaJ-like protein (J. D. Neill and J. F. Ridpath, Abstr. 17th Annu. Meet. Am. Soc. Virol. 1998, p. W3–W10, 1998; unpublished data). This sequence had previously been identified in BVDV1-NADL (12) and in a cytopathic border disease isolate (1) (Fig. 3). For three of these viruses, BVDV2-Galena 16452c, BVDV2-OkSt 94-050-297c, and BVDV2-6082c, the DnaJ-like sequence was preceded by 5, 22, and 28 amino acids residues, respectively; while in a fourth virus, BVDV2-ND 8799c, the DnaJ-like sequences were followed by a string of 10 amino acid residues. No significant sequence similarity was found between any of these flanking amino acid sequences or the nucleotide sequence coding for them and sequences in the GenBank.
FIG. 1.
Phylogenetic analysis of deduced amino acid sequences flanking the NS2/NS3 junction. Consensus amino acid sequences of the NS2/NS3 junction region minus insertion sequences were deduced from sequences generated by using primers shown in Table 1. Sequences compared corresponded to amino acid residues 1290 to 1610 in BVDV1-SD-1. The dendrogram was produced by applying the unpaired geometric mean analysis method using the Higgins-Sharp algorithm (CLUSTAL4) supplied in the MacDNASIS software package (Hitachi Software). Calculated matching percentages are indicated at each branch point of the dendrogram.
FIG. 2.
Insertion sites in the NS2-3 coding region. (A) Arrows show the sites of reported insertions into cytopathic BVDV1. Position A corresponds to amino acid residue 1535 and position B corresponds to amino acid residue 1589 of BVDV-1 strain SD-1 (GenBank accession number M96751). (B) The NS2-3 coding regions derived from nine cytopathic BVDV2 minus inserted sequences are shown. One noncytopathic virus coisolated with a cytopathic virus (BVDV2-296nc) and one noncytopathic BVDV2 sequence from GenBank (BVDV2-890, GenBank accession number BVU18059) were included for comparison purposes. Arrows indicate sites of insertions. No arrow indicates that there was no insertion. Unlike their noncytopathic counterparts, eight of these viruses had genomic insertions. No insertion was detected in BVDV2-96B3491c. One virus, BVDV2-MsSt T4529c, had an insertion at position B. Four viruses, BVDV2-ND 8799c, BVDV2-Galena 16425c, BVDV2-Ok St 94-050-297c, and BVDV2-SD1630c, had insertions at position A. Three viruses, BVDV2-296c, BVDV2-5912c, and BVDV2-6082c, had insertions within 10 amino acid residues of position A.
FIG. 3.
Inserted sequences detected in cytopathic BVDV2. (A) The insertion in BVDV2-MsSt T4529c at position B consisted of two full-length ubiquitin genes and a partial ubiquitin gene (34 amino acid residues from the amino acid terminus are missing). (B) Seven viruses had insertions at or within 10 amino acid residues of position A. Each of these insertions contained a portion of a cellular gene that codes for a novel DnaJ-like protein (underlined sequences). For three of these viruses, BVDV2-Galena 16452c, BVDV2-OkSt 94-050-297c, and BVDV2-6082c, the DnaJ-like sequence was proceeded by 5, 22, and 28 amino acid residues, respectively. For virus BVDV2-ND 8799c, the DnaJ-like sequence was followed by a string of 10 amino acid residues.
Regardless of the existence, identity, or position of insertion, all cytopathic viruses produced an NS3 with the same molecular weight based on mobility in polyacrylamide gel electrophoresis (data not shown). It is theorized that insertions at position B result in processing of NS2-3 by introducing either a new cleavage site at the carboxy terminus of the insertion or by introducing sequences with autocatalytic activity that act at the carboxy terminus of the insertion (for a review, see reference 16). The result of either mechanism is cleavage at position B, making Gly1590 the N-terminal amino acid of NS3. The amino acid sequences flanking the 3′ end of the insertions in the vicinity of position A were approximately 50 amino acid residues upstream of Gly1590 and were not highly conserved. However, as stated above, the NS3 proteins produced by each of the cytopathic BVDV2 were the same size, regardless of the insertion position. This suggests that processing of the NS2-3 by cytopathic viruses with insertions upstream from position B occurs by a different mechanism. It has been proposed that insertions upstream of position B induce a conformational change that allows cleavage via a cryptic mechanism at Gly1590 (16), but no experimental data are available.
Previously, BVDV2 cytopathology has been associated with recombination between a noncytopathic BVDV2 and a cytopathic BVDV1 at position A (23) or the presence of viral subgenomic RNAs with ubiquitin inserts at position B (2). In this study we show that cytopathic BVDV2, like cytopathic BVDV1, may have insertions at positions A or B or may have no insertions at all. Unlike cytopathic BVDV1, in which insertions are most often seen at position B and contain ubiquitin coding sequences, insertions in BVDV2 were most frequently observed in the vicinity of position A and contained portions of a gene coding for a DnaJ-like protein (Neill et al., Abstr. 17th Annu. Meet. Am. Soc. Virol. 1998). Three possible explanations for the differences in relative frequency of position and type of insertion are (i) differences in recombination frequencies due to sequence variation, (ii) differences in the stability of recombinations, or (iii) differences in the relative amounts of mRNA coding for ubiquitin and DnaJ-like protein in BVDV1- and BVDV2-infected cells.
Nucleotide sequence accession numbers.
GenBank accession numbers for sequences derived in this study are AF268171 through AF268180.
Acknowledgments
We thank Margaret Walker, Tracy Waltz, Becky Zaworski, Sharon Stark, and Bernadette Hackbart for excellent technical support on this project.
REFERENCES
- 1.Becher P, Meyers G, Shannon A D, Thiel H J. Cytopathogenicity of border disease virus is correlated with integration of cellular sequences into the viral genome. J Virol. 1996;70:2992–2998. doi: 10.1128/jvi.70.5.2992-2998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becher P, Orlich M, König M, Thiel H J. Nonhomologous RNA recombination in bovine viral diarrhea virus: molecular characterization of a variety of subgenomic RNAs isolated during an outbreak of fatal mucosal disease. J Virol. 1999;73:5646–5653. doi: 10.1128/jvi.73.7.5646-5653.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becher P, Orlich M, Thiel H J. Ribosomal S27a coding sequences upstream of ubiquitin coding sequences in the genome of a pestivirus. J Virol. 1998;72:8697–8704. doi: 10.1128/jvi.72.11.8697-8704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collett M S, Larson R, Belzer S K, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genomic organization of a pestivirus. Virology. 1988;165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 5.Donis R O, Dubovi E J. Differences in virus-induced polypeptides in cells infected by cytopathic and noncytopathic biotypes of bovine virus diarrhea-mucosal disease virus. Virology. 1987;158:168–173. doi: 10.1016/0042-6822(87)90250-9. [DOI] [PubMed] [Google Scholar]
- 6.Gillespie J, Baker J, McEntee K. A cytopathogenic strain of virus diarrhea virus. Cornell Vet. 1960;50:73–79. [PubMed] [Google Scholar]
- 7.Greiser-Wilke I, Dittmar K E, Liess B, Moennig V. Heterogeneous expression of the non-structural protein p80/p125 in cells infected with different pestiviruses. J Gen Virol. 1992;73:47–52. doi: 10.1099/0022-1317-73-1-47. [DOI] [PubMed] [Google Scholar]
- 8.Greiser-Wilke I, Haas L, Dittmar K, Liess B, Moennig V. RNA insertions and gene duplications in the nonstructural protein p125 region of pestivirus strains and isolates in vitro and in vivo. Virology. 1993;193:977–980. doi: 10.1006/viro.1993.1209. [DOI] [PubMed] [Google Scholar]
- 9.Kümmerer B M, Stoll D, Meyers G. Bovine viral diarrhea virus strain Oregon: a novel mechanism for processing of NS2-3 based on point mutations. J Virol. 1998;72:4127–4138. doi: 10.1128/jvi.72.5.4127-4138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kupfermann H, Thiel H J, Dubovi E J, Meyers G. Bovine viral diarrhea virus: characterization of a cytopathogenic defective interfering particle with two internal deletions. J Virol. 1996;70:8175–8181. doi: 10.1128/jvi.70.11.8175-8181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee K, Gillespie J. Propagation of virus diarrhea virus of cattle in tissue culture. Am J Vet Res. 1957;18:952–955. [PubMed] [Google Scholar]
- 12.Meyers G, Rumenapf T, Tautz N, Dubovi E J, Thiel H J. Insertion of cellular sequences in the genome of bovine viral diarrhea virus. Arch Virol Suppl. 1991;3:133–142. doi: 10.1007/978-3-7091-9153-8_15. [DOI] [PubMed] [Google Scholar]
- 13.Meyers G, Stoll D, Gunn M. Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle. J Virol. 1998;72:4139–4148. doi: 10.1128/jvi.72.5.4139-4148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyers G, Tautz N, Dubovi E J, Thiel H J. Viral cytopathogenicity correlated with integration of ubiquitin-coding sequences. Virology. 1991;180:602–616. doi: 10.1016/0042-6822(91)90074-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyers G, Thiel H J. Cytopathogenicity of classical swine fever virus caused by defective interfering particles. J Virol. 1995;69:3683–3689. doi: 10.1128/jvi.69.6.3683-3689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyers G, Thiel H J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 17.Neill J D, Mengeling W L. Further characterization of the virus-specific RNAs in feline calicivirus infected cells. Virus Res. 1988;11:59–72. doi: 10.1016/0168-1702(88)90067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellerin C, van den Hurk J, Lecomte J, Tussen P. Identification of a new group of bovine viral diarrhea virus strains associated with severe outbreaks and high mortalities. Virology. 1994;203:260–268. doi: 10.1006/viro.1994.1483. [DOI] [PubMed] [Google Scholar]
- 19.Pocock D H, Howard C J, Clarke M C, Brownlie J. Variation in the intracellular polypeptide profiles from different isolates of bovine virus diarrhoea virus. Arch Virol. 1987;94:43–53. doi: 10.1007/BF01313724. [DOI] [PubMed] [Google Scholar]
- 20.Qi F, Ridpath J F, Berry E S. Insertion of a bovine SMT3B gene in NS4B and duplication of NS3 in a bovine viral diarrhea virus genome correlate with the cytopathogenicity of the virus. Virus Res. 1998;57:1–9. doi: 10.1016/s0168-1702(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 21.Qi F, Ridpath J F, Lewis T, Bolin S R, Berry E S. Analysis of the bovine viral diarrhea virus genome for possible cellular insertions. Virology. 1992;189:285–292. doi: 10.1016/0042-6822(92)90704-s. [DOI] [PubMed] [Google Scholar]
- 22.Ridpath J F, Bolin S R. Comparison of the complete genomic sequence of the border disease virus, BD31, to other pestiviruses. Virus Res. 1997;50:237–243. doi: 10.1016/s0168-1702(97)00064-6. [DOI] [PubMed] [Google Scholar]
- 23.Ridpath J F, Bolin S R. Delayed onset postvaccinal mucosal disease as a result of genetic recombination between genotype 1 and genotype 2 BVDV. Virology. 1995;212:259–262. doi: 10.1006/viro.1995.1480. [DOI] [PubMed] [Google Scholar]
- 24.Ridpath J F, Bolin S R. Differentiation of types 1a, 1b and 2 bovine viral diarrhoea virus (BVDV) by PCR. Mol Cell Probes. 1998;12:101–106. doi: 10.1006/mcpr.1998.0158. [DOI] [PubMed] [Google Scholar]
- 25.Ridpath J F, Bolin S R. Viral protein production in homogeneous and mixed infections of cytopathic and noncytopathic BVD virus. Arch Virol. 1990;111:247–256. doi: 10.1007/BF01311058. [DOI] [PubMed] [Google Scholar]
- 26.Ridpath J F, Bolin S R, Dubovi E J. Segregation of bovine viral diarrhea virus into genotypes. Virology. 1994;205:66–74. doi: 10.1006/viro.1994.1620. [DOI] [PubMed] [Google Scholar]
- 27.Tautz N, Meyers G, Stark R, Dubovi E J, Thiel H J. Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion. J Virol. 1996;70:7851–7858. doi: 10.1128/jvi.70.11.7851-7858.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tautz N, Meyers G, Thiel H J. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology. 1993;197:74–85. doi: 10.1006/viro.1993.1568. [DOI] [PubMed] [Google Scholar]
- 29.Wengler G, Bradley D, Collett M. Family Flaviviridae. In: Murphy F, Fauquet C, Bishop D, editors. Virus taxonomy. 6th ed. New York, N.Y: Springer-Verlag; 1995. pp. 415–427. [Google Scholar]