Abstract
We have identified a previously unknown nucleotide sequence important for the packaging of murine leukemia virus. This nucleotide sequence is located downstream from the stop codon of the env gene but does not overlap the polypurine tract. Deletion of 17 bp from this region resulted in a more than 10-fold decrease in viral titer. Consistent with this result, the deletion mutant showed a 20- to 30-fold drop in the amount of virion RNA in the culture supernatant. The total amount of virion protein in the culture supernatant was comparable for the deletion mutant and the parental virus, suggesting that the mutant construct could release the empty viral particles. These results suggested that the packaging signal sequence might be present at the two extreme sites of the viral genome, one in the region around the splice donor sequence downstream from the 5′ long terminal repeat (LTR) and the other immediately upstream from the 3′ LTR. Implications for gene therapy, especially in regard to construction of retroviral vectors and packaging constructs, are discussed.
Murine leukemia virus (MLV) appears to have a complex array of nucleotide sequences that are involved in viral packaging. We recently found at least three regions that influence viral titer (11): the core region A, from +228 to +371, whose deletion completely abolishes viral packaging; region B, downstream from the core region (+377 to +527), which is necessary for optimal packaging; and region C, around the gag coding sequence (+739 to +1016), which inhibits the packaging function. The discovery of region C was somewhat unexpected because the 250-bp N-terminal gag coding region was previously known to contain the so-called extended packaging signal sequence (1, 2, 3). However, another group has recently reported our data confirming that the gag coding region may not be involved in viral packaging, and, on the contrary, deletion of this region may increase viral titer, at least in certain environments (9). These results suggested that the viral nucleotide sequence necessary for optimum packaging has not yet been fully identified in MLV.
MLV is still the most widely used gene delivery system in gene therapy trials (14; The Journal of Gene Medicine website [http://www.wiley.co.uk/genetherapy/clinical/vectors.html]), and one of the major limiting factors hindering successful application of amphotropic MLV in the real world is low viral titer (reviewed in references 6 and 10). Therefore it is vital to understand the possible involvement of any other nucleotide sequence in viral packaging. We have been trying to construct a retroviral vector(s) containing a minimum-length viral sequence, with the aim of constructing retroviral vectors and packaging constructs whose nucleotide sequences do not overlap at all, thus making a retroviral production system free of homologous recombination. During this work, we found that a previously unknown region can influence viral titer minimally by an order of magnitude.
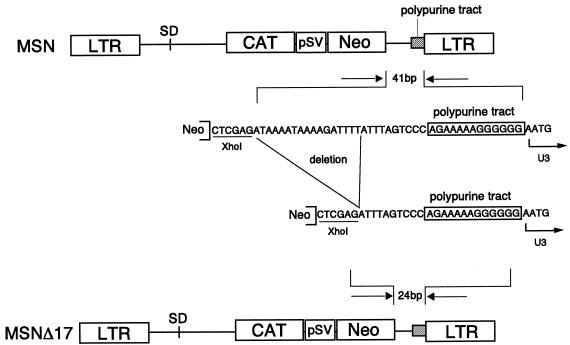
The retroviral vector MSN contains the entire 5′ long terminal repeat (LTR), its downstream sequence (right to before the start codon of the gag gene), and the entire sequence downstream from the stop codon of the env gene, including the polypurine tract and 3′ LTR (Fig. 1). MSNΔ17 is identical to MSN except that the former lacks 17 bp located immediately downstream from the stop codon of the env gene, as shown in Fig. 1. The bacterial chloramphenicol acetyltransferase (CAT) sequence was initially used to compare the levels of gene expression between the vectors, while the neo gene was used to estimate viral titer. In these constructs, CAT is driven from the retroviral LTR, while neo is expressed from the internal simian virus 40 (SV40) early promoter.
FIG. 1.
Schematic representation of retroviral vectors used in this study. MSN contains the nucleotide sequence from the 5′ end of the 5′ LTR to the region right before the start codon of the gag gene. At the 3′ side, MSN has all the nucleotide sequences downstream from the stop codon of the env gene. MSNΔ17 is identical except that it lacks the 17-nucleotide sequence immediately downstream from the stop codon of the env gene. The bacterial CAT sequence was used as a reporter gene and the selectable marker neo is driven by the internal SV40 early promoter. Plasmids used in this study were constructed by PCR using proofreading Pfu DNA polymerase (Stratagene, La Jolla, Calif.). The nucleotide sequences of final constructs were always determined to confirm that there were no mutations introduced by this amplification step. The retroviral vector MSN, which does not have any viral coding sequences, was constructed as follows. pMLV (22) was used for the amplification of the 5′ and 3′ LTR regions. The nucleotide sequences of primers used in amplifying the 5′ LTR region of MLV are as follows (the restriction linkers attached to each primer are underlined): (i) HHIR, AAGCTTATGTGAAAGACCCCTCCTG (the HindIII region is underlined), and (ii) 5LTR3, GGATCCGCGGGCCCACGCGTATTTTCAGACAAATACAGAAAC (the BamHI, SacII, ApaI, and MluI regions are underlined). The amplified product covered the 5′ LTR and 5′ noncoding regions containing packaging signals of MLV. The amplified HindIII-BamHI fragment was cloned into pUC 18, generating p5LTR. To amplify the 3′ LTR region, PCR was performed using primers 3LTR5 and 3LTR3 (3LTR5, GGATCCTCGAGGATAAAATAAAAGATTTTATTTAGTCTCC [the BamHI and XhoI regions are underlined], and 3LTR3, GAATTCAATGAAAGACCCCCGCTGAC [the EcoRI region is underlined]). The amplified product covered the entire 3′ untranslated region downstream from the stop codon of env, containing the polypurine tract and 3′ LTR. The amplified BamHI-EcoRI fragment was then cloned into p5LTR, resulting in pM, retroviral backbone. To insert the SV/Neo cassette into pM, the BamHI-XhoI fragment from pDON-AI (Takara, Shiga, Japan) was inserted into the BamHI-XhoI site of pM, resulting in MSN. To construct MSNΔ17, the 3′ LTR region was amplified using primers M3L52 and 3LTR3 (M3L52, AAAGGATCCATTTAGTCTCC [the BamHI region is underlined], and 3LTR3, GAATTCAATGAAAGACCCCCGCTGAC [the EcoRI region is underlined]). The amplified product covered a 3′ untranslated region 17 bp downstream from the stop codon of env, containing the polypurine tract and 3′ LTR. The amplified BamHI-EcoRI fragment was then cloned in p5LTR, resulting in pMΔ17, retroviral backbone. The BamHI-XhoI fragment from pDON-AI was then inserted into the BamHI-XhoI site of pMΔ17, resulting in MSNΔ17. To construct the retroviral vectors expressing CAT, the BamHI CAT fragment from PCRII-CAT (9) was inserted into the BamHI site of each vector.
The two constructs were transfected to the 293T cells using the three-plasmid transfection method (23). Then, the resulting supernatants were passed through a 0.45-μm-pore-size filter; NIH 3T3 cells were transduced with these cell-free viral supernatants, and the level of CAT activity was measured 2 days after transduction. Viral titer was estimated by counting the number of G418-resistant colonies. All experiments were performed in triplicate more than five times by two different investigators. The transfection efficiency was checked by using another plasmid expressing LacZ or green fluorescent protein and found to be always comparable between transfections.
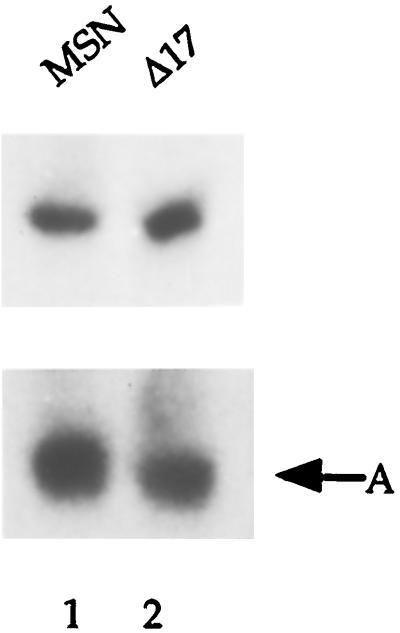
Initially, levels of CAT activity were compared in transiently transfected 293T cells. This level would represent the overall gene expression from a given construct. As summarized in Table 1, levels of CAT activity were comparable for MSN and MSNΔ17, suggesting that the deletion of 17 bp immediately downstream from the stop codon of the env gene did not have any significant effect on the level of gene expression. To be certain, we prepared total RNAs from 293T cells transfected with two retroviral constructs followed by Northern hybridization analysis using a CAT probe. These vectors produce two RNA species, a genomic and a subgenomic transcript. However, the CAT probe recognizes only the former RNA species that produces the CAT protein and is packaged into the virion. As shown in Fig. 2, levels of viral RNAs that specifically hybridized with the CAT probe were comparable for MSN and MSNΔ17, confirming that a 17-bp deletion has no effect on levels of RNA produced from the vector.
TABLE 1.
Effects of deletion of 17 nucleotide sequences on the level of gene expression and viral titera
| Vector | Relative CAT activity
|
Viral titer (105) | ||
|---|---|---|---|---|
| Transient transfection | Transduction
|
|||
| Transient | Stable | |||
| MSN | 100 | 100 | 100 | 17.0 ± 2.0 |
| MSNΔ17 | 106 ± 3 | 8.0 ± 3.2 | 100 ± 0.2 | 1.3 ± 0.4 |
293T cells were transfected with two retroviral constructs, MSN-CAT and MSNΔ17-CAT, using a three-plasmid transfection method. Other packaging lines such as Phoenix and FLYA13 were also used, and similar results were obtained. Therefore, only the results from 293T cells are summarized here. The level of CAT activity and viral titer were determined 2 days after transfection, as described previously (9). Cell-free viral supernatants were prepared using a 0.45-μm-pore-size filter and were used to transduce NIH 3T3 cells. The viral titer was standardized by diluting cell-free supernatants with fresh media or filtered untransfected culture supernatants from 293T cells prior to transduction. A CAT assay was performed either 2 to 3 days after transduction (transient) or after G418-resistant cultures were obtained. In all CAT assays, the same amounts of protein extracts were used. All assays were performed in triplicate by two different investigators more than four different times.
FIG. 2.
RNA blot analysis of 293T cells transfected with MSN-CAT or MSNΔ17-CAT retroviral constructs. Total RNA (20 μg) was subjected to 1% formaldehyde-agarose gel electrophoresis, blotted to nitrocellulose membrane (Hybond-C; Amersham Pharmacia, Piscataway, N.J.), and hybridized with a 32P-labeled CAT probe. A, cellular actin RNA.
However, when NIH 3T3 cells were transduced with cell-free viral supernatants and the level of gene expression was compared after 2 days, the result was dramatically different (Table 1). The level of CAT activity in cells transduced with MSN vector was always a minimum of 10-fold higher than the level in cells transduced with MSNΔ17. We estimated viral titer by counting the number of G418-resistant colonies and found that MSN reproducibly produced a minimum of 10-fold-higher levels of viral titer than MSNΔ17, suggesting that a 10-fold difference in the level of gene expression was probably due to a difference in viral titer.
To be certain, we diluted the MSN virus to the level of the MSNΔ17 virus using filtered untransfected culture supernatants or fresh media. NIH 3T3 cells were transduced with the same titers (multiplicity of infection [MOI] = 0.1) of the two viruses, and the level of gene expression was compared 2 days after transfection. Because the 17-bp region had no effect on the level of gene expression, cells transduced with the same amount of MSN and MSNΔ17 viruses would give comparable levels of gene expression. Indeed, levels of CAT activity were found to be comparable for the two constructs (data not shown). Lastly, transduced cells were selected in the presence of G418, and drug-resistant cultures were compared for the level of gene expression using the same amount of protein. The level of CAT activity was similar for the constructs, once again confirming that the 17-bp region has no significant effect on the level of gene expression.
The above results do not exclude a possible involvement of factors other than viral titer, for example, steps that occur between viral entry and integration. This is because we biologically determined viral titer by using the neo gene in the vector. For example, in this assay system, the above results could also be interpreted to mean that the 17-bp nucleotide sequence might influence reverse transcription. Therefore, our next step was to physically quantitate the amount of virus produced from two constructs. To obtain a consistent amount of virus and to rule out any artifacts derived from the transient transfection system, we constructed producer lines using PG13 packaging cell lines (18). 293T cells were transfected with MSN-CAT or MSNΔ17-CAT using a three-plasmid transfection method (23). The viral titer was determined using HT1080 cells (which are prone to viruses pseudotyped with GaLV env), and PG13 cells were transduced with a low MOI viral titer to ensure less than a single copy of integration in a majority of transduced cells. Transduced PG13 cells were subjected to drug selection, and drug-resistant cells were obtained. These producer cells were plated at 5 × 106 cells per 100-mm dish, and 2 days later viruses were harvested and passed through a 0.45-μm-pore-size filter, at which point protein extracts were also prepared for CAT analysis. Cell-free viral supernatants were used to transduce HT1080 cells and also to isolate virion RNA and protein in order to determine the viral titer, both biologically and physically. As shown in Table 2, levels of CAT activity were always comparable for PG13 producer lines, confirming the above results that deletion of 17 nucleotide sequences did not have any significant effect on gene expression.
TABLE 2.
Transduction using the virus produced by PG13 cells transduced with MSN or MSNΔ17a
| Vector | Relative CAT activity
|
Viral titer (105) | ||
|---|---|---|---|---|
| Producer line | Transduction
|
|||
| Transient | Stable | |||
| MSN | 100 | 100 | 100 | 3.2 ± 1.5 |
| MSNΔ17 | 83 ± 6 | 7.1 ± 2.7 | 85 ± 10 | 0.14 ± 0.06 |
Cell-free viral supernatants were prepared as described in Table 1 and were used to transduce the PG13 line (13) at the same low MOI, around 0.1. All other procedures were identical to those described in Table 1 except that HT1080 cells, instead of NIH 3T3 cells, were used as target cells, as they are prone to viruses pseudotyped with GaLV env.
Next, HT1080 cells transiently or stably transduced with cell-free viral supernatants were analyzed for their levels of CAT activity. Consistent with results obtained from the transient transfection system shown in Table 1, MSN produced an approximately 20-fold-higher level of CAT activity than MSNΔ17 in transiently transduced cells, while the two levels were comparable in cells stably transduced, with a low MOI viral titer. The viral titer from the PG13 producer line containing MSN-CAT was almost 20-fold higher than that from the PG13 line harboring MSNΔ17-CAT.
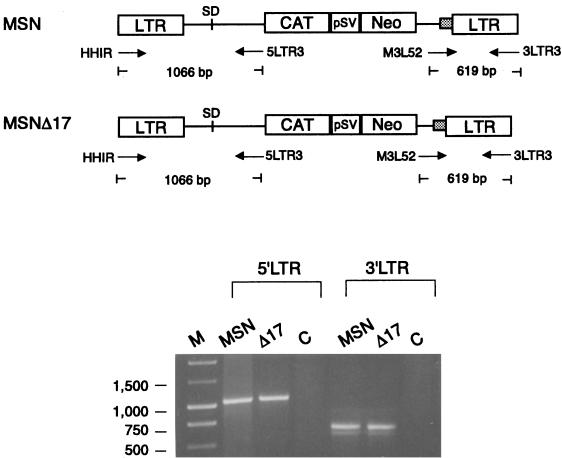
To ensure that the deletion of 17 bp did not affect the stabilities of the retroviral gene after transduction, total DNAs were prepared from PG13 lines producing MSN-CAT or MSNΔ17-CAT, followed by PCR with oligonucleotide primers, as shown in Fig. 3. If retroviral vectors have stably transferred the retroviral sequence to target cells, these primers would amplify 1,066 bp and 619 bp of the 5′ and 3′ LTR regions of the viral genome, respectively. DNA fragments of the expected lengths were present in all cells, suggesting that proviral structures were preserved in PG13 producer lines, and the defect in viral production of MSNΔ17 was not due to the defect of the proviral structure.
FIG. 3.
Test for preservation of retroviral sequences in transduced cells. Total cellular DNAs were prepared from G418-resistant PG13 cells transduced with retroviral vectors as described by Kim et al. (11). PCR was performed with 5 μg of total genomic DNA and oligonucleotide primers specific to various regions of the retroviral vector as indicated. The two pairs of oligonucleotide primers used to amplify the retroviral regions were described in the Fig. 1 legend.
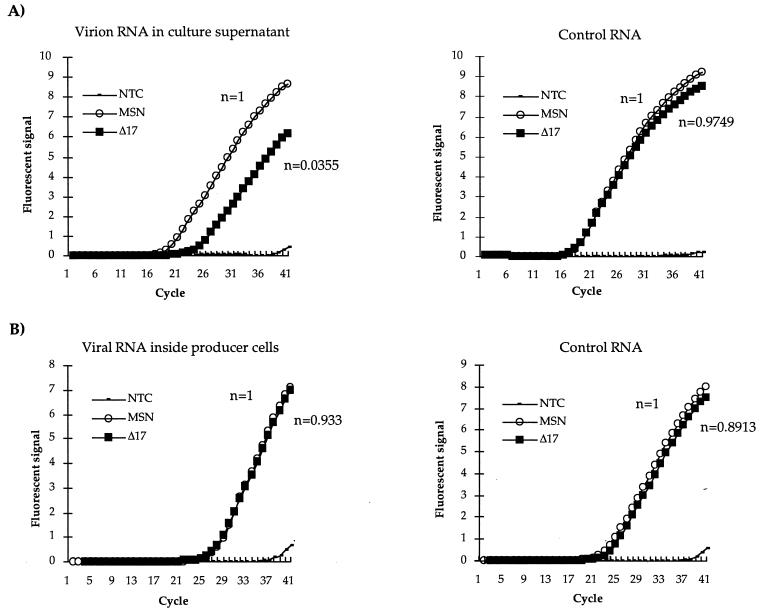
To test whether a 10- to 20-fold difference in the level of CAT activity in transiently transduced cells was due to decreased viral titer in the culture supernatant, virion RNAs and proteins in culture supernatants from PG13 cells were examined by quantitative reverse transcription-PCR (RT-PCR) and Western blot analysis. The same amount of culture supernatant was harvested from PG13 lines producing MSN-CAT or MSNΔ17-CAT viruses, subjected to low-speed centrifugation to remove cells and cellular debris, and then centrifuged at 25,000 rpm for 90 min in an SW41 rotor. Viral pellets were suspended in 50 mM Tris–1 mM EDTA (pH 7.5 to 8). A final concentration of 0.1% sodium dodecyl sulfate and 200 μg of tRNA per ml was added to the viruses, and the mixtures were extracted once with phenol, once with phenol-chloroform, and once with chloroform. A one-tenth volume of 3 M sodium acetate and 2 volumes of 100% ethanol were added to RNAs. RNA pellets were resuspended in 100 μl of diethylpyrocarbonate-treated water. cDNAs were then synthesized by reverse transcription followed by PCR using oligonucleotide primers specific for the CAT gene to analyze the level of virion RNA (Fig. 4A). As an internal control for the RT-PCR procedure, equal amounts of cellular RNAs prepared from PG13 cells were added to virion RNAs prepared from cells producing MSN or MSNΔ17 virions. RNA samples were then subjected to RT-PCR amplification using oligonucleotide primers specific for GaLV env present as an integrated form in the PG13 producer line (Fig. 4A). For accurate quantitative analysis, real-time quantitative PCR was employed using the ABI Prism 5700 sequence detector system (The Perkin-Elmer Corp., Foster City, Calif.). As shown in Fig. 4, typical amplification curves were obtained and the CT value and relative quantity of input RNA target were calculated as previously described (16). The starting copy number of MSNΔ17 virion RNA was on average 28-fold lower than that of MSN, while the amount of GaLV env RNA used as an internal control for the RT-PCR reaction was comparable for MSN and MSNΔ17. As another control experiment, the amounts of both MSN and MSNΔ17 viral RNAs expressed inside the PG13 line were analyzed in the same way. As shown in Fig. 4B, the starting copy numbers of the viral RNAs expressed in MSN and MSNΔ17 producer lines were comparable. The amounts of GaLV env RNAs used as a control were also similar for the two PG13 cell lines. Because both expressed amounts of MSN and MSNΔ17 viral RNAs were comparable within the PG13 cells, these results directly demonstrated that the MSNΔ17 virus was packaged less efficiently.
FIG. 4.
Quantitative analysis of virion RNA by real-time quantitative RT-PCR. (A) Amplification curves of virion RNAs (left) or control cellular RNAs (right). Virion RNAs of MSN-CAT or MSNΔ17-CAT were purified and mixed with equal amounts of total cellular RNA from PG13 cell lines. cDNAs were then synthesized by reverse transcription followed by PCR analysis using specific primers for CAT (viral RNA as it is present in the viral genome) or GaLV env genes (control “cellular” RNA as it is present in the cell as an integrated form), respectively. At each PCR reaction, the CT value was obtained from the amplification curve. The relative copy numbers of starting RNA were calculated as previously described (16) and are indicated as n. NTC, control containing no template. (B) Amplification curves of viral RNAs of retroviral vectors (left) inside PG13 cells or cellular GaLV env gene RNA (right) of PG13 cells are also shown. Total cellular RNAs of PG13 cells containing MSN-CAT or MSNΔ17-CAT proviruses were purified. cDNAs were then synthesized by reverse transcription followed by PCR analysis using specific primers for the CAT (left) or GaLV env genes (right). The result shown here is one representative of three independent assays.
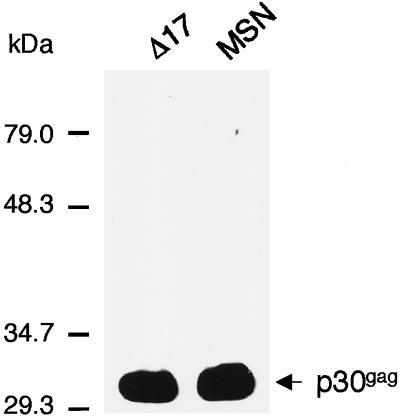
Finally, the amount of virion protein present in the culture supernatants was compared by extracting protein directly from viral pellets. Western blot analysis (Fig. 5) showed that the amounts of p30gag protein in the culture supernatant were comparable for MSN-CAT and MSNΔ17-CAT, indicating that the total amount of viral particles produced by MSNΔ17 was similar to that of the wild type.
FIG. 5.
Analysis of viral protein p30gag by Western blotting. Producer cells were plated at 5 × 106 cells per 100-mm dish, and 2 days later viruses were harvested and filtered through a 0.45-μm-pore-size filter. The same amount of filtered supernatants was concentrated by centrifugation at 25,000 rpm for 90 min in an SW41 roter. Viral pellets were suspended in a Laemmli buffer (13). An equal amount of viral protein lysates was subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with antisera specific for p30gag of MLV. The result shown here is one representative of more than three independent assays using different dilutions of the sample.
These RNA and protein analyses suggested that deletion of the 17 bp did not exert influence on either the level of gene expression in producer cells or the release of viral particles. However, this deletion significantly attenuated the packaging capability of MLV, releasing empty particles. Similar results were obtained using different producer lines based on FLYA13 (5) (data not shown). These results clearly indicated that 17 bp downstream from the MLV env gene are involved in viral packaging.
Viral packaging is an area that remains poorly understood (1, 2, 3, 4). Our study localized a previously unknown nucleotide sequence involved in viral packaging. The sequence is located immediately downstream from the stop codon of the env gene. This sequence does not overlap the polypurine tract needed for reverse transcription (20, 21). It is not yet clear how the sequence defined in this study interacts with the previously characterized packaging signal sequence located downstream from the 5′ LTR. It is possible that the interaction between the viral protein(s) and the MLV nucleotide sequence may require a three-dimensional structure and that the sequence defined in this work may play an important role in determining such a structure.
Our data have implications for designing MLV-based retroviral vectors and packaging constructs. For example, this sequence has to be included in the vector to obtain the highest viral titer. Without understanding the role of this 17-bp region, many previously constructed MLV-based vectors appear to contain this sequence (7, 12, 17, 19). However, some env expression vectors used to construct the packaging plasmids contain this 17-nucleotide sequence (8, 15, 23), increasing the possibility of homologous recombination. Ideally, in order to minimize the chance of homologous recombination, there should be no overlapping sequence between the vector and the packaging constructs. We recently constructed a series of retroviral vectors that contain none of the viral coding sequences, as well as expression plasmids for gag-pol and env, precisely starting from the start and stop codons of each gene. These systems should be safer than any other constructs without compromising viral titer.
Acknowledgments
We thank Hongchan Cho and Eunyoung Han for technical assistance.
This work was supported in part by grants from the Korean Science and Engineering Foundation (S.K.; 96-0403-03-01-3), ViroMed Limited (S.K.), and the Ministry of Science and Technology (S.K.).
REFERENCES
- 1.Adam M A, Miller A D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J Virol. 1988;62:3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armentano D, Yu S F, Kantoff P W, von Ruden T, Anderson W F, Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987;61:1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 5.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crystal R G. Transfer of genes to humans: early lessons and obstacles to success. Science. 1995;270:404–410. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- 7.Danos O, Mulligan R C. Safe and efficient generation of recombinant retroviruses with amphotropic and ecotropic host ranges. Proc Natl Acad Sci USA. 1988;85:6460–6464. doi: 10.1073/pnas.85.17.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finer M H, Dull T J, Qin L, Farson D, Roberts M R. kat: a high-efficiency retroviral transduction system for primary human T lymphocytes. Blood. 1994;83:43–50. [PubMed] [Google Scholar]
- 9.Hildinger M, Abel K L, Ostertag W, Baum C. Design of 5′ untranslated sequences in retroviral vectors developed for medical use. J Virol. 1999;73:4083–4089. doi: 10.1128/jvi.73.5.4083-4089.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay M A, Liu D, Hoogerbrugge P M. Gene therapy. Proc Natl Acad Sci USA. 1997;94:12744–12746. doi: 10.1073/pnas.94.24.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S H, Yu S S, Park J S, Robbins P D, An C S, Kim S. Construction of retroviral vectors with improved safety, gene expression, and versatility. J Virol. 1998;72:994–1004. doi: 10.1128/jvi.72.2.994-1004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kral W J, Skelton D C, Yu X J, Reviere I, Lehn P, Mulligan R C, Kohn D B. Increased levels of spliced RNA account for augmented expression from the MFG retroviral vector in hematopoietic cells. Gene Ther. 1995;3:37–48. [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Marcel T, Grausz J D. The TMC Worldwide Gene Therapy Enrollment Report, end 1996. Hum Gene Ther. 1997;8:775–800. doi: 10.1089/hum.1997.8.6-775. [DOI] [PubMed] [Google Scholar]
- 15.Markowitz D, Goff S, Bank A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martell M, Gomez J, Esteban J I, Sauleda S, Quer J, Cabot B, Esteban R, Guardia J J. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J Clin Microbiol. 1999;37:327–332. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller A D, Roseman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 18.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rattray A J, Champoux J J. Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J Mol Biol. 1989;208:445–456. doi: 10.1016/0022-2836(89)90508-1. [DOI] [PubMed] [Google Scholar]
- 21.Robson N D, Telesnitsky A. Effects of 3′ untranslated region mutations on plus-strand priming during Moloney murine leukemia virus replication. J Virol. 1999;73:948–957. doi: 10.1128/jvi.73.2.948-957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 23.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]