Abstract
Current immunosuppression regimens for lupus nephritis are incompletely effective, placing patients at risk for poor long-term outcomes. This emphasizes the need to dissect pathogenic mechanisms in lupus nephritis, in order to inform the development of targeted therapies. In this issue of Kidney International, Parikh and colleagues performed transcriptomic analysis of pre- and post-treatment protocol kidney biopsies, segregated into glomerular and tubulointerstitial compartments, to identify candidate molecular pathways distinguishing treatment responders and non-responders.
Commentary body:
After decades of disappointment, investments in translational and clinical science have yielded two new approved agents for the treatment of active lupus nephritis, belimumab and voclosporin. However despite these clinical successes, less than 50% of patients can expect complete renal response despite current optimal treatment. This highlights the critical need for greater understanding of pathogenic mechanism driving kidney inflammation in lupus nephritis. With this goal in mind, the public-private Accelerating Medicines Partnership (AMP) program has funded multicenter projects aimed at generating a single cell atlas of human lupus nephritis. In 2019, companion publications described the cellular complexity of the stromal and infiltrating leukocyte compartments in lupus nephritis1, 2. Despite the value of this resource for future translational studies, important limitations include the lack of spatial resolution of the infiltrating immune populations and analyses focused on initial pre-treatment biopsies with no assessment of transcriptional changes in response to treatment.
In this context, the current study by Parikh et al.3 takes advantage of a cohort of class III or class IV (+/− class V) lupus nephritis patients in whom renal biopsies were obtained during acute nephritis flares and followed by subsequent protocol biopsy after completion of induction therapy. Using laser capture microdissection (LCM) to separately obtain glomeruli and tubulointerstitium from each biopsy, the authors retrospectively probed molecular profiles of each compartment using the Nanostring immunology panel with the goal of identifying immunologic markers and pathways distinguishing complete responders from partial or nonresponders. To the best of our knowledge, this study is the first to utilize a large gene expression panel to characterize the compartment-specific immune landscape of serial renal biopsies to identify molecular signatures of lupus nephritis treatment response.
Important insights gained from this work include: 1) monocyte signatures are prominent in glomerular and peri-glomerular regions of baseline lupus nephritis biopsies, whereas T cell signatures localize to the tubulointerstitial compartment; 2) greater chemokine and proinflammatory cytokine expression at baseline in both compartments portend future non-response; 3) lack of treatment response correlates with enhanced monocyte, interferon, and extracellular matrix signatures in the glomerular compartment, and increased interferon, complement, and T cell gene expression in the tubulointerstitium in repeat biopsies obtained from non-responders. This latter observation provides molecular support for interferon blockade as a leading new potential therapeutic option in lupus nephritis. A unique feature of these studies are the separate analyses of the glomeruli and tubulointerstitium, thereby delineating the immune pathways preferentially localized within each compartment. These findings thus provide insights into mechanisms of treatment resistance or response as well as the identity of potential predictive biomarkers for clinical responsiveness.
The value of repeat kidney biopsy in patients with lupus nephritis has been demonstrated in a variety of contexts over the past several decades. It has frequently been noted that urinary and serum biomarkers correlate poorly with histology in lupus nephritis4, leaving biopsy as the most reliable guide to the institution or maintenance of immunosuppressive therapy. Indeed, as early as 2001, a study of 6 month post-treatment protocol biopsies by Hill et al. demonstrated that glomerular leukocyte infiltration in general and monocyte influx in particular predicted subsequent disease activity5. It is thus notable that a similar glomerular monocyte signature correlated with lack of treatment response in this report. Whether repeat kidney biopsy becomes standard-of-care in the management of lupus nephritis, depends on whether actionable information can be gained from histopathologic findings after initial immunosuppression. In this context, several authors of the current study previously reported that residual histology activity in patients in clinical remission predicted lupus nephritis flares and that, therefore, repeat kidney biopsy could be used to inform immunosuppression withdrawal6, 7. One exciting possibility is that molecular signatures of nephritis remission may be even more predictive of favorable renal outcome than histopathologic findings, allowing tailored therapy for individual patients. In addition, given the importance of previously recognized histopathologic markers, such as monocyte infiltration, endocapillary cell proliferation, and glomerular leukocyte infiltration in predicting lupus nephritis flares, the transcriptional analyses reported here may allow the identification of new urine and serum biomarkers of treatment response.
An important limitation of this study is that, while lupus class and the activity and chronicity of lesions were assessed in biopsy samples, the relative activity and chronicity likely varied across individual tissue compartments undergoing molecular analysis. The assessment of activity and chronicity involves a comprehensive integration of changes across all glomeruli and the tubulointerstitial parenchyma, which is typically sampled in multiple level sections of the biopsy. Since the glomeruli obtained by LCM lacked correlative histology, we cannot ascertain for each biopsy the proportion of captured glomeruli involved by crescents, necrosis, segmental scarring or even the absence of histological alterations. This limits our ability to correlate the molecular findings with histopathology. Indeed, it is surprising that there is no difference in activity indices between first and second biopsies from both complete responders and nonresponders (Table 2 of the current study), despite a significant clinical difference in response. It is also somewhat surprising that that changes in molecular proinflammatory profiles for each parenchymal compartment corresponded to well accepted histopathologic features of active lupus nephritis, but did not translate into corresponding differences in histologic inflammatory activity scores in this specific biopsy cohort.
Biopsy determination of activity and chronicity indices are a current standard for monitoring and guiding therapeutic decisions for lupus nephritis8. This study reported that changes in these indices lacked correlation with measures of clinical response, an observation which requires further study in larger patient cohorts. It is possible that, after validation in additional clinical cohorts, molecular markers of immunologic inflammatory activity may prove a better guide to therapeutic decisions than histopathologic indices in individual patients. Moreover, incorporation of transcriptional signatures may inform future iterations of lupus nephritis pathology classification systems. This study suggests that the renal biopsy of the future may require integration of molecular profiles to accommodate such a paradigm shift.
There are other important limitations to this study. First, the panel of genes used for transcriptional profiling focused on immunologic parameters, such as cytokines, chemokines, interferons and their respective receptors. By definition, the host parenchymal response to both acute and chronic injury cannot be adequately assessed using this panel. However, given the technical feasibility of generating robust transcriptional signatures from FFPE tissue exemplified by this study, future analyses using gene probes of tissue response or even the whole transcriptome should be possible.
Second, as alluded to above, combining individual glomeruli and tubulointerstitial regions for pooled molecular analysis fails to account for the known heterogeneity across individual biopsy samples in lupus nephritis. Fortunately, the development of technologies which enable transcriptomic analysis of biopsy tissue in situ is a fast-moving field. For example, the GeoMx Digital Spatial Profiler (DSP) developed by Nanostring, Inc. allows the interrogation of tissue morphology and spatial transcriptomics on a single tissue section. Using this approach, Smith et al. quantified mRNA from HIV or SARS-CoV-2 infection associated collapsing glomerulopathy at the level of individual glomeruli9. Thus, the limited ability to correlate LCM-dissected glomeruli with histopathologic changes in the present study may be obviated by the use of commercially-available spatial profiling instruments allowing transcriptomic analysis directly on tissue sections while registering the findings to individual structures and preserving their underlying histopathology. These new technologies can now encompass the whole human transcriptome, thereby overcoming the limitations inherent in targeted gene expression panels. Finally, over the next decade, it is likely that molecular analyses of human biopsy samples will achieve true single cell spatial resolution. While not yet commercially available, technologies such as the Xenium platform (10X Genomics, Inc.) or the CosMx™ Spatial Molecular Imager (Nanostring, Inc.) have reported spatially-resolved gene expression at cellular and even subcellular resolution.
In summary, this study brings us one important step closer to the implementation of precision medicine for management of lupus nephritis. The authors demonstrate that current techniques for gene expression profiling can be applied to clinical biopsy tissue to extract meaningful biologic information. One could envision how these data could be used to develop a more precise classification of underlying disease activity at a specific time in order to guide therapeutic management. Importantly, this approach need not be limited to the study of lupus nephritis and could be applied to a wide range of human kidney diseases. While these techniques remain expensive and challenging to perform and validate, and thus unlikely to enter the clinical laboratory soon, one can easily envision a future where targeted molecular profiling becomes a standard component of renal biopsy evaluation. Thus, the future of lupus nephritis management may mirror that of oncology, where gene expression profiles already guide therapeutic decision making in daily clinical practice.
Figure 1. Diagram showing patient cohort and experimental strategy used to determine compartment-specific gene expression.
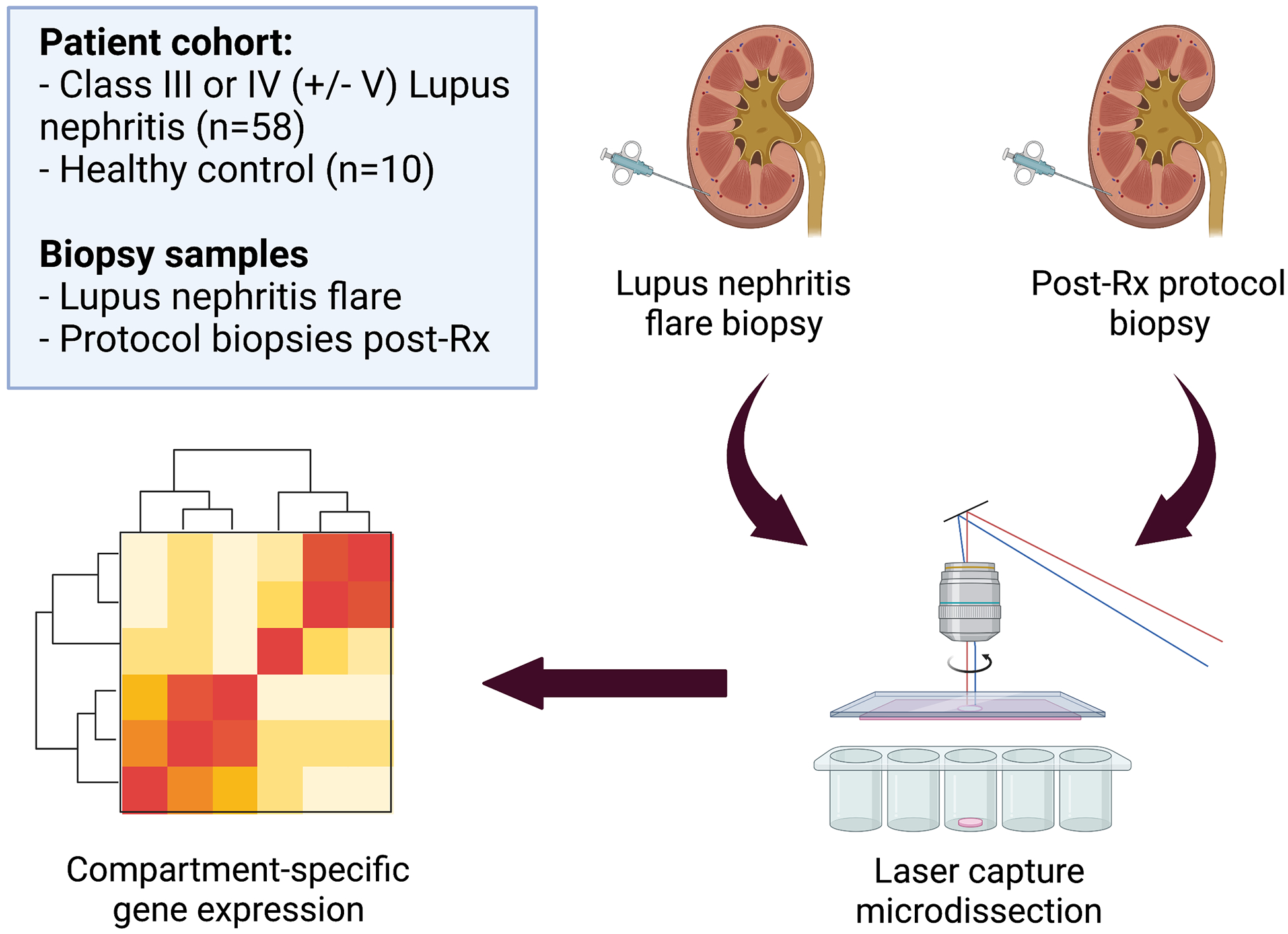
Kidney biopsy tissue from Class III or IV (+/− V) lupus nephritis patients, obtained at flare and following induction immunosuppression, was submitted for laser capture microdissection (LCM) followed by assessment of gene expression using an immunology probe set. Healthy kidney tissue was obtained from pre-implantation living donor kidney biopsies was used as a control. Differential gene expression in renal responders and non-responders was examined to identify molecular pathways of treatment response.
References:
- 1.Der E, Suryawanshi H, Morozov P, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol 2019; 20: 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arazi A, Rao DA, Berthier CC, et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol 2019; 20: 902–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parikh SV. Molecular Profiling of Kidney Compartments from Serial Biopsies to Differentiate Treatment Responders from Non-responders in Lupus Nephritis. Kidney Int 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayne D, Bajema IM. "In my beginning is my end": usefulness of repeat kidney biopsies in lupus nephritis. Kidney Int 2020; 97: 27–29. [DOI] [PubMed] [Google Scholar]
- 5.Hill GS, Delahousse M, Nochy D, et al. Predictive power of the second renal biopsy in lupus nephritis: significance of macrophages. Kidney Int 2001; 59: 304–316. [DOI] [PubMed] [Google Scholar]
- 6.Malvar A, Alberton V, Lococo B, et al. Kidney biopsy-based management of maintenance immunosuppression is safe and may ameliorate flare rate in lupus nephritis. Kidney Int 2020; 97: 156–162. [DOI] [PubMed] [Google Scholar]
- 7.De Rosa M, Azzato F, Toblli JE, et al. A prospective observational cohort study highlights kidney biopsy findings of lupus nephritis patients in remission who flare following withdrawal of maintenance therapy. Kidney Int 2018; 94: 788–794. [DOI] [PubMed] [Google Scholar]
- 8.Bajema IM, Wilhelmus S, Alpers CE, et al. Revision of the International Society of Nephrology/Renal Pathology Society classification for lupus nephritis: clarification of definitions, and modified National Institutes of Health activity and chronicity indices. Kidney Int 2018; 93: 789–796. [DOI] [PubMed] [Google Scholar]
- 9.Smith KD, Prince DK, Henriksen KJ, et al. Digital spatial profiling of collapsing glomerulopathy. Kidney Int 2022; 101: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]


