Abstract
We recently described our finding that recombinant baculovirus-produced virus-like particles (VLPs) can induce cell-cell fusion similar to that induced by intact rotavirus in our assay for viral entry into tissue culture cells (J. M. Gilbert and H. B. Greenberg, J. Virol. 71:4555–4563, 1997). The conditions required for syncytium formation are similar to those for viral penetration of the plasma membrane during the course of viral infection. This VLP-mediated fusion activity was dependent on the presence of the outer-layer proteins, viral protein 4 (VP4) and VP7, and on the trypsinization of VP4. Fusion activity occurred only with cells that are permissive for rotavirus infection. Here we begin to dissect the role of VP4 in rotavirus entry by examining the importance of the precise trypsin cleavage of VP4 and the activation of VP4 function related to viral entry. We present evidence that the elimination of the three trypsin-susceptible arginine residues of VP4 by specific site-directed mutagenesis prevents syncytium formation. Two of the three arginine residues in VP4 are dispensable for syncytium formation, and only the arginine residue at site 247 appears to be required for activation of VP4 functions and cell-cell fusion. Using the recombinant VLPs in our syncytium assay will aid in understanding the conformational changes that occur in VP4 involved in rotavirus penetration into host cells.
Rotaviruses are the leading cause of severe dehydrating gastroenteritis in children worldwide (17, 24). Rotavirus, a member of the reovirus family, is a nonenveloped icosahedral virus consisting of three concentric protein layers surrounding a segmented, double-stranded RNA genome. The outer-layer proteins, viral protein 4 (VP4; 88 kDa) and VP7 (34 kDa), are required for viral penetration (6, 13, 16). VP7, a glycoprotein, is the major component of the outer layer, whereas VP4 is much less abundant and forms dimeric spikes that project out from the viral surface (31, 33). VP4 has been shown to be a determinant of host range and virulence and is directly involved in cell attachment and rotaviral entry into cells (19, 22, 30, 32).
Proteolytic cleavage of the precursor VP4 to two noncovalently associated subunits, VP8* (28 kDa) and VP5* (60 kDa) (10, 12, 26), significantly enhances viral infectivity (2, 4, 8). In vivo processing occurs in the lumen of the intestine, while in vitro, cleavage is accomplished by trypsin, a protease with specificity for cleavage after arginine and lysine residues. VP8*, the amino-terminal fragment of VP4, is the subunit involved in binding to specific cell surface receptors (15, 22, 32). The carboxyl-terminal portion of VP4, VP5*, contains two sequence motifs that are hypothesized to be involved in viral penetration of host cells. These motifs are a putative internal fusion peptide sequence and a putative alpha-helical coiled-coil domain (11, 27). It is thought that specific binding of VP4 to the host cell surface receptors must occur in order to initiate viral entry. This binding is hypothesized to trigger entry-related conformational changes in the outer-layer proteins, predominantly in VP4, leading to cellular membrane penetration and viral replication. Whereas viral attachment to the cell occurs regardless of VP4 cleavage, it appears that the conformational changes and productive viral entry are dependent upon the VP4 cleavage event (5, 8, 18, 23).
We described previously an assay that measures the ability of rotavirus to induce syncytia when added to cholesterol-supplemented MA104 cells (14). Syncytium production occurs only with cells that are permissive for rotavirus infection (16). Like rotavirus entry, syncytium production also requires cleavage of VP4 by trypsin. Since molecular analysis of rotavirus functions has been impeded by the fact that a method to alter a specific rotavirus gene product and recover it in infectious virus is not yet available, we have employed recombinant virus-like particles (VLPs) (9) as an alternative to intact rotavirus particles. The rotavirus VLPs are expressed in Spodoptera frugiperda 9 (Sf-9) cells from four different recombinant baculoviruses, each of which expresses one of the four main structural proteins of rotavirus (VP2, VP4, VP6, or VP7). We have recently demonstrated that these recombinant rotavirus VLPs can induce polykaryon formation similarly to intact rotavirus (16). Here we demonstrate the usefulness of these recombinant particles for dissecting the entry of rotavirus into host cells on a molecular level.
In order to understand the mechanism by which rotavirus enters host cells, it is clearly important to precisely define the requirement for trypsinization of VP4 in viral penetration. Arias et al. (1) examined patterns of VP4 trypsin digestion and its correlation with rotavirus infectivity. Within a putative exposed loop of most strains of VP4, three trypsin-susceptible arginine residues, R231, R241, and R247, reside in the trypsin cleavage region (TCR; the sequence between amino acids 231 and 247 [1]). The biochemical analyses of Arias et al. (1) indicated that these three sites have different susceptibilities to trypsinization. When the highest concentration of trypsin required for maximal infectivity was employed, cleavage after residues R231 and R241 was complete. Cleavage after residue R247 occurred in approximately 80% of the molecules. Examination of the infectivity of rotavirus upon digestion with increasing concentrations of trypsin seemed to indicate a correlation between cleavage after R247, rather than after R241, and the induction of infectivity. However, since all VP4 molecules were also cleaved after R231, the importance of the individual cleavage events could not be precisely analyzed.
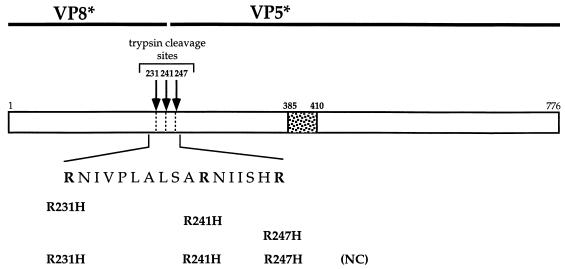
To examine the contribution to rotavirus entry of the individual arginine residues within the TCR, specific arginine residues were changed to histidine residues by site-directed mutagenesis (Fig. 1) (25). The residues were altered either individually (R231H, R241H, and R247H) or together such that all three arginine residues were mutated in combination to create a triple mutant designated NC. The mutant cDNAs were sequenced to confirm that they contained only the appropriate changes. These rhesus rotavirus (RRV) VP4 cDNAs were individually subcloned into the pFASTBAC vector (Gibco/BRL, Gaithersburg, Md.), and this vector was used to create recombinant baculoviruses expressing the mutant VP4 molecules (BAC-to-BAC; Gibco/BRL).
FIG. 1.
Primary structure of VP4 and location of amino acid changes. The cDNA of RRV VP4 encodes a protein containing a predicted 776 amino acids (88 kDa). Proteolytic cleavage of the precursor VP4 to two noncovalently associated subunits, VP8* (28 kDa) and VP5* (60 kDa), is required for infectivity. Cleavage has been shown to occur after three different arginine residues (arginine 231, arginine 241, and arginine 247) within a predicted exposed loop, the TCR (1). These residues were changed to histidine residues by site-directed mutagenesis (25) either individually (to create R231H, R241H, and R247H, respectively) or as a group (to create NC). The mutations were confirmed by sequencing on an ABI automated DNA sequencer. The putative fusion peptide sequence from sites 385 to 410 is shown.
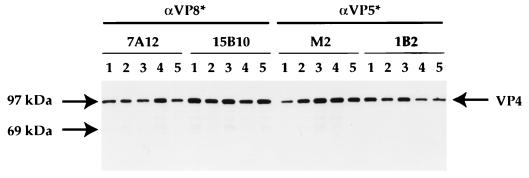
After selection of positive, recombinant baculoviruses, viruses were plaque purified, amplified, and analyzed to assess expression of the VP4 proteins. To ensure that the mutant VP4 proteins retained the antigenic integrity of wild-type RRV VP4, proteins were examined by immunoprecipitation with monoclonal antibodies (MAbs) directed against neutralizing epitopes on VP4. The mutant VP4 proteins were recognized as efficiently as the wild-type VP4 by MAbs directed at both the VP8* and VP5* subunits (MAb 7A12 and 15B10 and MAbs M2 and 1B2, respectively) (Fig. 2). These data indicate that the mutations in the TCR from arginine to histidine residues do not alter the overall structure of the mutant VP4 proteins. Examination of wild-type and mutant VP4-expressing baculovirus-infected Sf-9 cells by immunostaining with additional neutralizing MAbs to both the VP8* and VP5* subunits (1A9 and 2G4, respectively) also demonstrated that there was no detectable difference between mutant and wild-type VP4 in this assay (data not shown) (21).
FIG. 2.
Immunoprecipitation of mutant VP4 proteins. The wild-type and mutated VP4 cDNAs were excised as an NcoI/XhoI fragment from pBluescript KS+ (Stratagene, La Jolla, Calif.) and subcloned into a modified pFASTBAC vector (Gibco/BRL). Recombinant baculoviruses were produced in Sf-9 cells (Invitrogen, San Diego, Calif.) with the BAC-to-BAC system (Gibco/BRL) according to the manufacturer’s instructions. To produce recombinant proteins, Sf-9 cells were infected at a multiplicity of infection of 1. At 24 h, the medium was removed and cells were labelled with [35S]methionine (Amersham, Arlington Heights, Ill.), in methionine-free medium (Ex-Cell 401; JRH Biosciences, Lenexa, Kans.) for an additional 48 h. Cells were harvested and lysed, and the proteins were examined by immunoprecipitation (16). The VP4 antibodies, neutralizing MAbs to VP8* (7A12 and 15B10), and MAbs to VP5* (M2 and 1B2) (references 32 and 34 and this paper) were precoupled to Protein A-Sepharose (Sigma Chemical Company) for 2 h at 4°C. The antibody-bead complexes were washed extensively and added to the 35S-labelled lysates, and the samples were immunoprecipitated overnight at 4°C. The samples were washed and then subjected to SDS-PAGE (12% acrylamide; NOVEX, San Diego, Calif.) as described previously (16). The figure shows immunoprecipitation of wild-type VP4 (lane 1), R231H VP4 (lane 2), R241H VP4 (lane 3), R247H VP4 (lane 4), and NC VP4 (lane 5).
VLPs containing VP2, VP6, VP7, and either wild-type or mutant VP4 were produced and purified as described by Gilbert and Greenberg (16). The CsCl-purified wild-type and mutant VLPs were then examined to confirm that the changes to the TCR did not affect the binding function of the mutant VP4 molecules. Since the background strain of the VP4 molecules is RRV, a sialic acid-dependent strain, we examined whether the changes in the cleavage site of VP4 would affect VLP binding to sialic acid residues. The ability of wild-type and mutant VLPs to hemagglutinate human type O erythrocytes was compared with the ability of wild-type RRV to do so (Table 1). The wild-type and the mutant VLPs hemagglutinate to the same extent, albeit at a slightly lower level than RRV, indicating that the modifications to the TCR did not alter the sialic acid binding functions of the VP8* subunit.
TABLE 1.
Hemagglutination by RRV and several VLPsa
| Virus or particle | HA units/ml |
|---|---|
| RRV | 2,560 |
| Wild-type VLP | 1,280 |
| R231H VLP | 1,280 |
| R241H VLP | 1,280 |
| R247H VLP | 1,280 |
| NC VLP | 1,280 |
RRV, wild-type RRV VLPs, and the mutant VLPs at equivalent protein concentrations in phosphate-buffered saline containing 1% bovine serum albumin (fraction V; Sigma) were serially diluted twofold into the wells of a 96-well dish containing human type O erythrocytes at a concentration of 0.5% in a mixture of phosphate-buffered saline and bovine serum albumin. Samples were incubated at room temperature for 2 h and hemagglutination was assessed. HA units are as described in Hirst et al. (20).
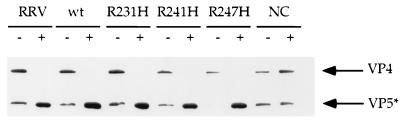
To verify (i) the alterations to the individual arginine residues within the TCR did not affect overall trypsinization of VP4 and (ii) that the elimination of all three arginine residues abrogated trypsin cleavage, trypsinization of the wild-type and mutant VLPs was compared to that of native RRV. Samples were treated with trypsin or mock-treated for 30 min at 37°C, and then the trypsin was inactivated with the inhibitor TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone). The proteins were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with HS-2, a MAb directed to the VP5* subunit (Fig. 3) (32). In all the untreated samples, including RRV, some VP4 appears to have been cleaved to a protein that migrated similarly to VP5*. The observed amounts of this nonspecific digestion varied among VLP preparations. This small amount of preexisting cleavage was not associated with the ability of the preparations to form syncytia (see below) and may be mediated by cellular proteases. The wild-type VLPs and mutant VLPs with single arginine-to-histidine changes were all cleaved by trypsin treatment similarly to intact RRV, as demonstrated by the increase in the amount of VP5* detected by the HS-2 MAb. This indicates that the removal of trypsin sites by changing individual arginine residues to histidine residues had no apparent effect on the ability of the remaining trypsin sites to be accessed and proteolyzed. However, the triple mutant, NC, showed no detectable VP5* production following trypsin treatment, demonstrating that changing all three arginine residues to histidine eliminated the ability of trypsin to process VP4 to VP8* and VP5*.
FIG. 3.
Trypsinization of RRV virus and wild-type and mutant VLPs. Wild-type (wt) and mutant VLPs were treated with 10 μg of trypsin per ml (type XIII, N-tosyl-l-phenylalanine chloromethyl ketone [TPCK] treated; Sigma) for 30 min at 37°C (+) or left untreated (−). The reactions were quenched by treatment with an equimolar amount of TLCK (Sigma). Samples were diluted into protein sample buffer, boiled, and subjected to SDS-PAGE as described in the text. The electrophoresed proteins were transferred to nitrocellulose (Schleicher and Schuell, Keene, N.H.), and the VP4 and VP5* proteins were detected with HS-2 (a carboxyl-terminus-specific VP4 monoclonal antibody). The bound antibodies were detected with goat anti-mouse immunoglobulin G coupled to peroxidase (Kirkegaard and Perry, Gaithersburg, Md.). The peroxidase signal was detected by enhanced chemiluminescence (ECL reagent; Pierce Chemical Company, Rockford, Ill.).
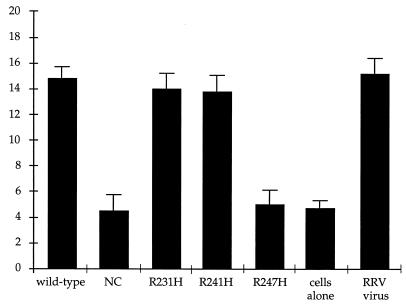
Since the mutant VLPs appeared to be virus-like according to the above biochemical and serologic criteria, we next examined whether these particles could induce syncytia in cholesterol-supplemented MA104 cells in a manner similar to that of native RRV and wild-type VLP particles (14, 16). Trypsinized RRV and wild-type and mutant VLPs were incubated with MA104 cells as described in the legend to Fig. 4. Cells were examined microscopically and the number of nuclei in syncytia as a function of the total number of nuclei in each sample was determined. Cells that were not incubated with virus or VLPs had very few polykaryons. RRV, wild-type VLPs, and the R231H and R241H mutant VLPs all induced syncytia at approximately the same rate and to an equivalent extent (Fig. 4), whereas the NC mutant VLPs could not induce cell-cell fusion. This indicates that the substitution of histidine residues for the three arginine residues completely abrogates the trypsin activation of VP4, resulting in a blockage to viral entry. Interestingly, the R247H mutant VLPs were also unable to induce polykaryon formation. This implies that the arginine residue at site 247 is the only trypsin cleavage site within the TCR whose cleavage is required to promote viral entry.
FIG. 4.
Syncytium formation of RRV and wild-type and mutant VLPs. RRV virus (75 focus-forming units per cell) and wild-type and mutant VLPs (at the protein equivalent of 75 focus-forming units per cell) were treated with trypsin for 30 min at 37°C under conditions previously established and described by Gilbert and Greenberg (16). Samples were incubated with MA104 cells at 4°C for 15 min and at 37°C for 15 min, plated onto six-well tissue culture dishes, and incubated for 2 h at 37°C in a CO2 incubator. Cells were examined microscopically and the number of syncytia per 100 nuclei was counted as described by Gilbert and Greenberg (16).
By employing recombinant rotavirus VLPs, we have been able to conduct a more precise examination of the role in the activation of VP4 of the three arginine residues within the TCR. Our studies demonstrate that, within the scope of our analysis (immunogenicity, hemagglutination, and trypsinization), changing these specific arginine residues to histidine has no detectable effect on VP4. It is possible that within the mutant TCRs, very subtle changes that were not detectable by our assays occurred, but only higher-resolution structural analysis would reveal this. The ability of the VP4 mutants to interact with sialic acid, the primary receptor for VP4 of the RRV strain, is not altered by these amino acid changes, compared with the wild-type VLPs. This indicates that the binding functions of VP8* remain intact. The mutant proteins are also efficiently recognized by a panel of neutralizing MAbs to VP4, affirming that multiple biologically significant epitopes of both VP8* and VP5* are retained in the mutant proteins. It is probable that these arginine residues are contained within an exposed loop domain of VP4 which easily tolerates amino acid changes.
Investigating the susceptibility to trypsinization of the individual arginine-to-histidine mutant VLPs demonstrated that all were equivalently cleaved to VP5*, similarly to wild-type VLPs. Although the analyses of Arias et al. (1) indicated that with intact SA11 rotavirus particles there may be increased accessibility to trypsinization of the arginine residues at sites 231 and 241 compared with site 247, the alteration of either of these individual arginine residues did not result in a diminution of overall VP4 cleavage. Similarly, changing the arginine residue at site 247 had no apparent effect on the cleavage efficiency after arginine residues 231 and 241. Not surprisingly, replacement of all three arginine residues with histidine residues completely abolished any trypsin cleavage of VP4. Although multiple preparations of both RRV and the VLPs contained small portions of inadvertently cleaved VP4 molecules, these particles did not appear to function in our fusion assay because trypsin activation is still required for syncytium formation, as discussed below (data not shown). It is probable that these preexisting cleavages occurred by the action of nonspecific cellular proteases within the putative TCR loop.
The syncytial phenotype of the arginine-to-histidine mutants indicates that substituting histidine for all of the three arginine residues simultaneously (to form NC VLPs) eliminated the ability of these particles to form syncytia with MA104 cells. Alteration of the single residues at arginine 231 or 241 (to form R231H and R241H VLPs, respectively) had no effect on the rate or extent of polykaryon formation, suggesting that the trypsin cleavages seen after these residues on intact viral particles (1) are not strictly required for VP4 activation. Only the mutant with a single change of an arginine to histidine at residue 247 (R247H VLPs) had a syncytium-negative phenotype. This result indicates that arginine 247 is the essential site for VP4 trypsin enhancement of cell-cell fusion. The ramifications of the necessity for cleavage to occur at this site are twofold. First, the newly generated carboxyl terminus of VP8* can be heterogeneous. VP8*, with carboxyl termini at either arginines 231 and 247 or arginines 241 and 247, can still induce syncytia as well as wild-type VP4-containing particles. The unimportance of the VP8* carboxyl terminus is not unexpected since the primary function of VP8*, cell binding, occurs regardless of the trypsinization of VP4. Second, the trypsin-activated amino terminus of VP5*, which is thought to mediate cell entry, must be homogeneous. Unlike the VP8* cell binding function, the VP5* entry-related functions, most probably membrane penetration, do not occur in the absence of trypsinization. Apparently, the amino terminus required for a functional VP5* begins with alanine 248. The requirement for precise cleavage after residue 247 is not surprising. For example, proteases which do not cleave specifically after arginine 247, chymotrypsin (which cleaves after residue 246), and AspN (which cleaves after residue 241) do not activate viral infectivity or syncytium formation (1, 12) (data not shown). Additionally, RRV and wild-type and mutant VLPs will not induce syncytia without trypsin treatment, indicating that the observed nonspecific cleavage of VP4 does not activate VP4 entry-related functions. Finally, it appears that stepwise cleavage, first after arginine 241 and then after arginine 231 (1), is not a prerequisite for the cleavage after arginine 247 required for infectivity, since R231H and R241H VLPs can induce syncytium formation. This result indicates that there are no specific structural constraints on the TCR for activation except that cleavage must occur after arginine 247.
The requirement of cleavage of VP4 to VP8* and to VP5* for infectivity, and presumably membrane penetration, mirrors what is seen for the orthomyxo-, paramyxo-, toga-, and retrovirus envelope glycoproteins (28, 29, 35, 36). These proteins are expressed as inactive precursor molecules in which a proteolytic cleavage event is required for function. For influenza virus hemagglutinin (HA), the paradigm for viral envelope glycoproteins, the cleavage of the precursor HA0 to HA1 and HA2 allows the protein to undergo dramatic conformational changes when the trigger, an acidic pH, is encountered. The synthesis of the precursor HA0 results in a folded protein with a stable conformation. After cleavage of HA0 to HA1 and HA2, however, the protein is no longer in its most stable form but is instead in a state termed metastable. The low-pH-induced conformational changes convert HA from this metastable state to its most stable conformation, which is the conformation capable of membrane penetration as a result of fusion peptide exposure and presentation (3, 7). This transformation in the structure of HA cannot occur without the proteolytic processing of the precursor. Similarly, the conformational changes that presumably must occur within VP4 to allow viral entry also seem to be dependent on the processing of the VP4 precursor to VP8* and VP5*. Without appropriate cleavage, as seen with the R247H and NC VLPs, the putative structural changes required to activate VP4 to a fusogenic conformation upon encountering the appropriate cellular receptor(s) may be blocked. In this scenario, uncleaved VP4 would be unable to carry out penetration-related functions. Elucidating the conformational changes that the proteolytically processed VP4 undergoes will assist in understanding the mechanism of rotavirus entry into cells.
Summary.
Using recombinant baculovirus-produced VLPs, we have examined the requirement that arginine residues (R231, R241, and R247) be present within the TCR for trypsin activation of VP4. By site-directed mutagenesis we have demonstrated that changing the individual arginine residues to histidine residues does not appear to alter VP4 immunogenicity, hemagglutination, or trypsinization. Substituting histidine for all three arginine residues results in a protein that appears to be wild type by serological and hemagglutination criteria but that cannot be cleaved by trypsin. Examination of the cell-cell fusion of the VLPs containing the mutant VP4 proteins indicates that only arginine residue 247 is required for VP4 activation.
Acknowledgments
We thank P. O’Hanley’s lab for the use of their incubators, J.-H. Yu for the 1B2 and 15B10 MAbs, R. Tabtiang for assisting with the figures, N. Madhani for DNA sequencing, and M. Falconer for helpful discussions.
This work was supported by a V.A. Merit Review grant and by NIH grants R37AI21632 and DK38707. H. B. Greenberg was a V.A. Medical Investigator during these studies. J. M. Gilbert is supported by grant T32AI07328-08 from the NIH and by a Bank of America-Giannini Foundation Fellowship in Medical Research.
REFERENCES
- 1.Arias C F, Romero P, Alvarez V, López S. Trypsin activation pathway of rotavirus infectivity. J Virol. 1996;70:5832–5839. doi: 10.1128/jvi.70.9.5832-5839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiuk L A, Mohammed K, Spence L, Fauvel M, Petro R. Rotavirus isolation and cultivation in the presence of trypsin. J Clin Microbiol. 1977;6:610–617. doi: 10.1128/jcm.6.6.610-617.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker D, Agard D A. Influenza hemagglutinin: kinetic control of protein function. Structure. 1994;2:907–910. doi: 10.1016/s0969-2126(94)00091-3. [DOI] [PubMed] [Google Scholar]
- 4.Barnett B B, Spendlove R S, Clark M L. Effect of enzymes on rotavirus infectivity. J Clin Microbiol. 1979;10:111–113. doi: 10.1128/jcm.10.1.111-113.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bass D M, Baylor M R, Chen C, Mackow E M, Bremont M, Greenberg H B. Liposome-mediated transfection of intact viral particles reveals that plasma membrane penetration determines permissivity of tissue culture cells to rotavirus. J Clin Invest. 1992;90:2313–2320. doi: 10.1172/JCI116119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridger J C, Woode G N. Characterization of two particle types of calf rotavirus. J Gen Virol. 1976;31:245–250. doi: 10.1099/0022-1317-31-2-245. [DOI] [PubMed] [Google Scholar]
- 7.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 8.Clark S M, Roth J R, Clark M L, Barnett B B, Spendlove R S. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J Virol. 1981;39:816–822. doi: 10.1128/jvi.39.3.816-822.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford S E, Labbé M, Cohen J, Burroughs M H, Zhou Y-J, Estes M K. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5945–5952. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espejo R T, López S, Arias C. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J Virol. 1981;37:156–160. doi: 10.1128/jvi.37.1.156-160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estes M K. Rotaviruses and their replication. In: Fields B N, editor. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1625–1656. [Google Scholar]
- 12.Estes M K, Graham D Y, Mason B B. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estes M K, Graham D Y, Smith E M, Gerba C P. Rotavirus stability and inactivation. J Gen Virol. 1979;43:403–409. doi: 10.1099/0022-1317-43-2-403. [DOI] [PubMed] [Google Scholar]
- 14.Falconer M M, Gilbert J M, Roper A M, Greenberg H B, Gavora J S. Rotavirus-induced fusion from without in tissue culture cells. J Virol. 1995;69:5582–5591. doi: 10.1128/jvi.69.9.5582-5591.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiore L, Greenberg H B, Mackow E R. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology. 1991;181:553–563. doi: 10.1016/0042-6822(91)90888-i. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert J M, Greenberg H B. Virus-like particle-induced fusion from without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J Virol. 1997;71:4555–4563. doi: 10.1128/jvi.71.6.4555-4563.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glass R I, Gentsch J R, Ivanoff B. New lessons for rotavirus vaccines. Science. 1996;272:46–48. doi: 10.1126/science.272.5258.46. [DOI] [PubMed] [Google Scholar]
- 18.Graham D Y, Estes M K. Proteolytic enhancement of rotavirus infectivity: biology mechanism. Virology. 1980;101:432–439. doi: 10.1016/0042-6822(80)90456-0. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg H B, Flores J, Kalica A R, Wyatt R G, Jones R. Gene coding assignments for growth restriction, neutralization and subgroup specificities of the W and DS-1 strains of human rotavirus. J Gen Virol. 1983;64:313–320. doi: 10.1099/0022-1317-64-2-313. [DOI] [PubMed] [Google Scholar]
- 20.Hirst G K. Agglutination of red cells by allantoic fluid by chick embryos infected by influenza virus. Science. 1941;94:22–23. doi: 10.1126/science.94.2427.22. [DOI] [PubMed] [Google Scholar]
- 21.Ishida S-I, Feng N, Tang B, Gilbert J M, Greenberg H B. Quantification of systemic and local immune responses to individual rotavirus proteins during rotavirus infection in mice. J Clin Microbiol. 1996;34:1694–1700. doi: 10.1128/jcm.34.7.1694-1700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalica A R, Flores J, Greenberg H B. Identification of the rotaviral gene that codes for hemagglutination and protease-enhanced plaque formation. Virology. 1983;125:194–205. doi: 10.1016/0042-6822(83)90073-9. [DOI] [PubMed] [Google Scholar]
- 23.Kaljot K T, Shaw R D, Rubin D H, Greenberg H B. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J Virol. 1988;62:1136–1144. doi: 10.1128/jvi.62.4.1136-1144.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, editor. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1657–1708. [Google Scholar]
- 25.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 26.López S, Arias C F, Bell J R, Strauss J H, Espejo R T. Primary structure of the cleavage site associated with trypsin enhancement of rotavirus SA11 infectivity. Virology. 1985;144:11–19. doi: 10.1016/0042-6822(85)90300-9. [DOI] [PubMed] [Google Scholar]
- 27.Mackow E R, Shaw R D, Matsui S M, Vo P T, Dang M-N, Greenberg H B. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci USA. 1988;85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCune J M, Rabin L B, Feinberg M B, Lieberman M, Kosek J C, Reyes G R, Weissman I L. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 29.Morrison T. Structure, function, and intracellular processing of paramyxovirus membrane proteins. Virus Res. 1988;10:113–136. doi: 10.1016/0168-1702(88)90010-x. [DOI] [PubMed] [Google Scholar]
- 30.Offit P A, Clark H F, Blavat G, Greenberg H B. Reassortant rotaviruses containing structural proteins vp3 and vp7 from different parents induce antibodies protective against each parental serotype. J Virol. 1986;60:491–496. doi: 10.1128/jvi.60.2.491-496.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad B V V, Burns J W, Marietta E, Estes M K, Chiu W. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature. 1990;343:476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- 32.Ruggeri F M, Greenberg H B. Antibodies to the trypsin cleavage peptide VP8* neutralize rotavirus by inhibiting binding of virions to target cells in culture. J Virol. 1991;65:2211–2219. doi: 10.1128/jvi.65.5.2211-2219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw A L, Rothnagel R, Chen D, Ramig R F, Chiu W, Prasad B V V. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell. 1993;74:693–701. doi: 10.1016/0092-8674(93)90516-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw R D, Vo P T, Offit P A, Coulson B S, Greenberg H B. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology. 1986;155:434–451. doi: 10.1016/0042-6822(86)90205-9. [DOI] [PubMed] [Google Scholar]
- 35.Sturman L S, Ricard C S, Holmes K V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J Virol. 1985;56:904–911. doi: 10.1128/jvi.56.3.904-911.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White J, Kielian M, Helenius A. Membrane fusion proteins of enveloped animal viruses. Q Rev Biophys. 1983;16:151–195. doi: 10.1017/s0033583500005072. [DOI] [PubMed] [Google Scholar]