Simple Summary
Carcinoembryonic antigen is a cell-surface glycoprotein and target for anti-cancer drugs. In this study, more than 15,000 samples from 120 different tumor types were analyzed by immunohistochemistry. CEA expression was found at least occasionally in 65 tumor types, most frequently in colorectal cancers and other gastrointestinal tumors, thyroid gland cancers, and pulmonary adenocarcinomas. Reduced CEA expression was linked to colon cancer aggressiveness. In contrast, aggressiveness cancers of the urinary bladder and breast cancers were characterized by CEA overexpression. We present a comprehensive catalog of tumor types that might benefit from anti-CEA therapies.
Keywords: CEA, diagnostic marker, tissue microarray, immunohistochemistry, human tumors
Abstract
Background/Objectives: Carcinoembryonic antigen (CEA) is a cell-surface glycoprotein serving as a drug target, diagnostic marker, and serum marker for cancer monitoring. However, prevalence data on CEA expression in cancer tissues vary considerably. This study was designed to determine CEA expression in normal and neoplastic tissues. Methods: A tissue microarray containing 13,725 samples from 120 different tumor types, as well as 76 different normal tissue types, was analyzed by immunohistochemistry (IHC). Results: CEA was detectable in 65 (54.2%) of 120 tumor categories, including 49 (40.8%) tumor types with at least one strongly positive case. CEA positivity was most common in colorectal adenomas (100%) and carcinomas (98.7%), other gastrointestinal adenocarcinomas (61.1–80.3%), medullary carcinomas of the thyroid (96.3%), pulmonary adenocarcinoma (73.7%), mucinous carcinomas of the ovary (79.8%) and the breast (43.2%), small-cell carcinomas of the lung (64.3%), and urinary bladder (38.9%). CEA overexpression was linked to high tumor grade and invasive growth (p < 0.0001 each) in urinary bladder cancer, and estrogen and HER2 receptor positivity (p ≤ 0.0158) in invasive breast cancer of no special type. In colorectal adenocarcinomas, reduced CEA expression was associated with mismatch repair deficiency (p < 0.0001). Conclusions: The comprehensive list of CEA-positive human tumor types demonstrates that CEA is expressed in a broad range of epithelial neoplasms, many of which might benefit from CEA serum monitoring and anti-CEA therapies.
1. Introduction
Carcinoembryonic antigen (CEA; CEACAM5) is a cell-surface glycoprotein with a role in cell adhesion [1]. CEA is extensively produced in many tissues during fetal development, but even before birth, its expression becomes limited to a limited number of different normal cell types [2]. CEA is overexpressed in various cancers (summarized in [3,4,5,6]). Because CEA is also shed into the blood stream, CEA measurement in the serum is used as a tool for early detection and recurrence monitoring of cancer [7,8]. There are several therapeutic approaches targeting CEA. For example, an ongoing phase I study is investigating the CEA-CD3 bispecific antibody cibisatamab on metastatic CEA-positive colorectal carcinomas. In all, 11% (4/36) of patients treated with cibisatamab monotherapy and 50% (5/10) of patients receiving cibisatamab in combination with a PD-L1-inhibitor showed radiological shrinkage, reflecting its antitumor activity [9,10]. Labetuzumab govitecan, an SN-38 CEA-antibody–drug conjugate, resulted in stable disease in 49% (42/86) of patients with metastatic colorectal cancer in a phase I/II study [11]. A phase I pretargeted radioimmunotherapy trial using anti-CEA-HSG-TF2 showed promising dose optimization for the treatment of CEA-positive metastasized lung carcinomas [12]. Three phase I studies showed that CEA is an auto-antigen that can be safely targeted by CAR T cells, resulting in stable disease in 7/10 patients with metastatic colorectal carcinomas (NCT02349724), 7/14 patients with metastatic gastrointestinal carcinomas (NCT01212887), and 1/6 patients with liver metastases of different adenocarcinomas (NCT01373047) [13]. In addition, vaccination with Ad5 [E1-, E2b-]-CEA(6D)—encoding CEA—induced CEA-specific cell-mediated immune response with antitumor activity in a phase I/II colorectal cancer study [14].
More than 700 studies analyzed the expression of CEA in cancer by immunohistochemistry (IHC) and described CEA positivity in at least 62 different tumor types and subtypes. CEA is considered to occur at a particularly high frequency in colorectal cancers, other gastrointestinal carcinomas, and in pulmonary adenocarcinomas (summarized in [3,4,5,6]). Because CEA expression is rare in hepatocellular carcinoma and in malignant mesothelioma, CEA IHC has been suggested as a tool to distinguish mesothelioma from pulmonary adenocarcinoma and hepatocellular carcinoma from liver metastasis [15,16]. However, the results of previous studies on CEA expression in cancer vary considerably for many tumor types. For example, reported CEA positivity rates ranged from 17% to 100% of pulmonary adenocarcinoma [17,18], 0% to 54% of malignant mesothelioma [15,19], 0% to 100% of hepatocellular carcinoma [20,21], 29% to 100% of colorectal adenocarcinoma [22,23], 25% to 100% of different subtypes of gastric adenocarcinoma [24,25,26,27,28,29], 0% to 100% of cholangiocarcinoma [20,30], 0% to 71% of endometrioid endometrial carcinoma [31,32], 9% to 58% of small-cell carcinoma of the lung [33,34], 0% to 100% of mucinous carcinoma of the ovary [35,36], and 14% to 94% of invasive breast carcinoma of no special type [37,38]. Technical factors, including staining protocols and antibodies used and differences in the definition of thresholds determining positivity, as well as possible selection bias with respect to the analyzed tumors, may have caused these discrepancies. A comprehensive study analyzing as many tumor types as possible under standardized experimental conditions and analysis criteria is, therefore, highly warranted.
Using IHC in a tissue microarray (TMA) format, more than 15,000 tissue samples from 120 different tumor types and subtypes, and 76 non-neoplastic tissues were analyzed, allowing us to better understand the relative importance of CEA expression in various cancers and normal tissues.
2. Materials and Methods
2.1. Tissue Microarrays (TMAs)
The normal tissue TMA was composed of 8 samples from 8 different donors for each of 76 different normal tissue types (608 samples on one slide). The cancer TMAs contained a total of 15,413 primary tumors from 120 tumor types and subtypes. Detailed histopathological and molecular data on grade, pathological tumor stage (pT), pathological lymph node status (pN), HER2, estrogen receptor (ER),progesterone receptor (PR) status, and mismatch repair protein status were available from subsets of adenocarcinomas of the colon (n = 2351), the pancreas (n = 598), and the stomach (n = 327); invasive breast carcinomas of no special type (n = 1208); urothelial bladder carcinomas (n = 1663); endometrioid endometrial carcinomas (n = 182); and endometrioid (n = 40) and serous carcinoma of the ovary (n = 369). Clinical follow-up data (overall survival) were available from 877 breast cancer patients, with a median follow-up time of 49 months (range 1–88 months). The composition of both normal and cancer TMAs is described in detail in the Section 3. All samples were from the archives of the Institutes of Pathology, University Hospital of Hamburg, Germany; the Institute of Pathology, Clinical Center Osnabrueck, Germany; or the Department of Pathology, Academic Hospital, Fuerth, Germany. Tissues were fixed in 4% buffered formalin and then embedded in paraffin. The TMA manufacturing process was described earlier in detail [39,40]. In brief, one tissue spot (diameter: 0.6 mm) was transmitted from a cancer-containing donor block in an empty recipient paraffin block. The use of archived remnants of diagnostic tissues for manufacturing of TMAs, and their analysis for research purposes, as well as patient data analyses, have been approved by local laws (HmbKHG, §12) and by the local ethics committee (Ethics commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration.
2.2. Immunohistochemistry (IHC)
Freshly prepared TMA sections were immunostained on one day in one experiment. Slides were deparaffinized with xylol, rehydrated through a graded alcohol series and exposed to heat-induced antigen retrieval for 5 min in an autoclave at 121 °C in pH 9.0 DakoTarget Retrieval SolutionTM (Agilent, Santa Clara, CA, USA; #S2367). Endogenous peroxidase activity was blocked with Dako Peroxidase Blocking SolutionTM (Agilent #52023) for 10 min. Primary antibody specific against CEA protein (rabbit recombinant, MSVA-465R, MS Validated Antibodies, Hamburg, Germany; #2563-465R) was applied at 37 °C for 60 min at a dilution of 1:150. Bound antibody was visualized using the EnVision KitTM (Agilent #K5007) according to the manufacturer’s directions. For the purpose of antibody validation, the normal tissue TMA was also analyzed by an additional CEA antibody (monoclonal mouse, II-7, Agilent #M7072) on a Agilent DAKO autostainer Link48 according to a protocol suggested by Agilent DAKO. The sections were counterstained with hemalaun. For tumor tissues, the percentage of CEA-positive tumor cells was estimated and the staining intensity was semi-quantitatively recorded (0, 1+, 2+, 3+). For statistical analyses, the staining results were categorized into four groups as follows: negative, no staining at all; weak staining, staining intensity of 1+ in ≤70% or staining intensity of 2+ in ≤30% of tumor cells; moderate staining, staining intensity of 1+ in >70%, staining intensity of 2+ in >30% but in ≤70%, or staining intensity of 3+ in ≤30% of tumor cells; and strong staining: staining intensity of 2+ in >70% or staining intensity of 3+ in >30% of tumor cells.
2.3. Statistics
Statistical calculations were performed with JMP 16 software (SAS Institute Inc., Cary, NC, USA). Contingency tables and the chi2 test were performed to search for associations between CEA immunostaining and tumor phenotype. Overall survival curves were calculated according to Kaplan–Meier. The Log-Rank test was applied to detect significant differences between groups. A p-value of ≤0.05 was considered to be statistically significant.
3. Results
3.1. Technical Issues
In our TMA analysis, a total of 13,725 (89.0%) of 15,413 tumor samples and over three samples for each normal tissue category were analyzable. Non-analyzable samples showed absence of tissue or lack of indisputable tumor cells in their respective TMA spots.
3.2. CEA in Normal Tissues
For MSVA-465R, a moderate-to-strong CEA immunostaining was seen, particularly in the upper layers of the non-keratinizing (but not keratinizing) squamous epithelium, irrespective of the location. In the skin, eccrine glands showed a luminal membrane staining. Strong CEA staining occurred in the transitional epithelium of the anal canal and in most cells of Hassall’s corpuscles in the thymus. The strongest CEA staining in the gastrointestinal tract was seen in the colorectal mucosa, where the staining was strongest in the surface cells and the upper half of crypts. Stomach mucosa surface cell layers showed a moderate-to-strong CEA positivity, while glands were either negative or much less stained. Duodenum and small intestine showed a moderate staining of a subset of goblet cells, primarily at the surface epithelium, while deeper glands were mostly negative. A weak-to-moderate staining occurred at the apical membranes of a fraction of (mainly) mucinous cells in salivary glands. A variable CEA staining occurred in respiratory epithelium, primarily in goblet cells. A significant fraction of pneumocytes also showed an apical membrane positivity. Urothelium was usually CEA-negative but occasionally showed a variable weak staining of umbrella cells. Representative images are shown in Figure 1. All of these cell types were also found to be CEA-positive by using the monoclonal mouse antibody IL-7 (Supplementary Figure S1). CEA staining was absent in mesenchymal, lymphatic, and hematopoietic cells; skin; liver; adrenal gland; kidney; gall bladder epithelium; Brunner glands; prostate; seminal vesicle; epididymis; testis; breast; placenta; endocervix; endometrium; ovary with corpus luteum and follicular cysts; thyroid; hypophysis; cerebrum; and in the cerebellum (Supplementary Table S1).
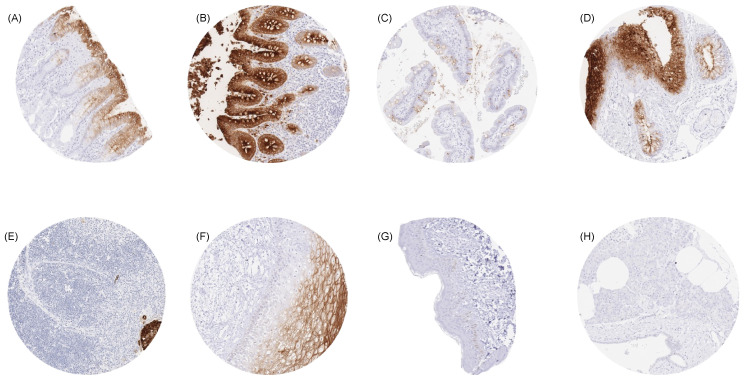
Figure 1.
CEA immunostaining of normal tissues. A membranous and cytoplasmic CEA staining of variable intensity is seen in surface epithelial cells of the stomach (A), epithelial cells (predominantly at the surface) of the colon (B), goblet cells of the small intestine (C), respiratory epithelial cells (D), Hassal’s corpuscles of the thymus (E), and in superficial cell layers of the squamous epithelium of the cervix uteri (F). CEA staining is absent in tissues from the epidermis of the skin (G) and in the pancreas (H). Original magnifications 10×, spot size 600 μm.
3.3. CEA in Cancer
All cancers were analyzed with MSVA-465R. CEA immunostaining was detectable in 4323 (31.5%) of the 13,725 analyzable tumors. Weak CEA immunostaining was seen in 1076 tumors (7.8%), moderate in 425 (3.1%), and strong in 2822 (20.6%). Of the 120 tumor categories included, CEA positivity was found in 65 (54.2%) while 49 (40.8%) tumor categories showed at least one case of strong positivity (Table 1).
Table 1.
CEA immunostaining in human tumors. Abbreviations: int. = number of interpretable samples, neg. = negative, mod. = moderate, str. = strong.
| CEA Immunostaining Result | |||||||
|---|---|---|---|---|---|---|---|
| Tumor Entity | On TMA (n) | Int. (n) | Neg. (%) |
Weak (%) | Mod. (%) | Str. (%) | |
| Tumors of the skin | Pilomatricoma | 35 | 23 | 100.0 | 0.0 | 0.0 | 0.0 |
| Basal cell carcinoma of the skin | 88 | 80 | 96.3 | 3.8 | 0.0 | 0.0 | |
| Benign nevus | 29 | 29 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the skin | 90 | 90 | 65.6 | 31.1 | 2.2 | 1.1 | |
| Malignant melanoma | 46 | 43 | 86.0 | 9.3 | 4.7 | 0.0 | |
| Merkel cell carcinoma | 46 | 42 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the head and neck | Squamous cell carcinoma of the larynx | 109 | 103 | 46.6 | 30.1 | 7.8 | 15.5 |
| Squamous cell carcinoma of the pharynx | 60 | 58 | 60.3 | 22.4 | 5.2 | 12.1 | |
| Oral squamous cell carcinoma (floor of the mouth) | 130 | 126 | 69.8 | 17.5 | 7.1 | 5.6 | |
| Pleomorphic adenoma of the parotid gland | 50 | 39 | 82.1 | 15.4 | 2.6 | 0.0 | |
| Warthin tumor of the parotid gland | 49 | 49 | 91.8 | 8.2 | 0.0 | 0.0 | |
| Basal cell adenoma of the salivary gland | 15 | 15 | 93.3 | 6.7 | 0.0 | 0.0 | |
| Tumors of the lung, pleura, and thymus | Adenocarcinoma of the lung | 196 | 179 | 26.3 | 19.6 | 8.9 | 45.3 |
| Squamous cell carcinoma of the lung | 80 | 71 | 46.5 | 28.2 | 5.6 | 19.7 | |
| Small-cell carcinoma of the lung | 16 | 14 | 35.7 | 0.0 | 7.1 | 57.1 | |
| Mesothelioma, epithelioid | 39 | 28 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mesothelioma, biphasic | 76 | 62 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Thymoma | 29 | 28 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Lung, neuroendocrine tumor (NET) | 19 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the female genital tract | Squamous cell carcinoma of the vagina | 78 | 71 | 57.7 | 25.4 | 9.9 | 7.0 |
| Squamous cell carcinoma of the vulva | 130 | 119 | 60.5 | 30.3 | 2.5 | 6.7 | |
| Squamous cell carcinoma of the cervix | 129 | 123 | 30.9 | 23.6 | 13.8 | 31.7 | |
| Endometrioid endometrial carcinoma | 236 | 227 | 80.6 | 18.1 | 0.4 | 0.9 | |
| Endometrial serous carcinoma | 82 | 68 | 82.4 | 17.6 | 0.0 | 0.0 | |
| Carcinosarcoma of the uterus | 48 | 46 | 91.3 | 4.3 | 2.2 | 2.2 | |
| Endometrial carcinoma, high grade, G3 | 13 | 13 | 92.3 | 7.7 | 0.0 | 0.0 | |
| Endometrial clear cell carcinoma | 8 | 8 | 87.5 | 12.5 | 0.0 | 0.0 | |
| Endometrioid carcinoma of the ovary | 110 | 95 | 76.8 | 15.8 | 2.1 | 5.3 | |
| Serous carcinoma of the ovary | 559 | 499 | 98.0 | 1.8 | 0.2 | 0.0 | |
| Mucinous carcinoma of the ovary | 96 | 84 | 20.2 | 28.6 | 17.9 | 33.3 | |
| Clear cell carcinoma of the ovary | 50 | 43 | 93.0 | 4.7 | 0.0 | 2.3 | |
| Carcinosarcoma of the ovary | 47 | 44 | 95.5 | 4.5 | 0.0 | 0.0 | |
| Brenner tumor | 9 | 9 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the breast | Invasive breast carcinoma of no special type | 1345 | 1042 | 72.2 | 13.1 | 5.4 | 9.4 |
| Lobular carcinoma of the breast | 293 | 193 | 79.3 | 8.8 | 5.7 | 6.2 | |
| Medullary carcinoma of the breast | 26 | 19 | 94.7 | 5.3 | 0.0 | 0.0 | |
| Tubular carcinoma of the breast | 27 | 19 | 89.5 | 0.0 | 0.0 | 10.5 | |
| Mucinous carcinoma of the breast | 58 | 44 | 56.8 | 11.4 | 6.8 | 25.0 | |
| Phyllodes tumor of the breast | 50 | 43 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the digestive system | Adenomatous polyp, low-grade dysplasia | 50 | 50 | 0.0 | 0.0 | 8.0 | 92.0 |
| Adenomatous polyp, high-grade dysplasia | 50 | 47 | 0.0 | 0.0 | 8.5 | 91.5 | |
| Adenocarcinoma of the colon | 1882 | 1773 | 1.3 | 2.1 | 3.3 | 93.3 | |
| Gastric adenocarcinoma, diffuse type | 176 | 149 | 32.2 | 12.1 | 8.1 | 47.7 | |
| Gastric adenocarcinoma, intestinal type | 174 | 161 | 25.5 | 15.5 | 14.9 | 44.1 | |
| Gastric adenocarcinoma, mixed type | 62 | 54 | 38.9 | 13.0 | 9.3 | 38.9 | |
| Adenocarcinoma of the esophagus | 83 | 79 | 29.1 | 16.5 | 6.3 | 48.1 | |
| Squamous cell carcinoma of the esophagus | 76 | 69 | 65.2 | 14.5 | 10.1 | 10.1 | |
| Squamous cell carcinoma of the anal canal | 89 | 80 | 52.5 | 23.8 | 3.8 | 20.0 | |
| Cholangiocarcinoma | 113 | 107 | 72.9 | 12.1 | 6.5 | 8.4 | |
| Hepatocellular carcinoma | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ductal adenocarcinoma of the pancreas | 612 | 553 | 19.7 | 20.3 | 10.8 | 49.2 | |
| Pancreatic/Ampullary adenocarcinoma | 89 | 81 | 22.2 | 18.5 | 11.1 | 48.1 | |
| Acinar cell carcinoma of the pancreas | 16 | 15 | 80.0 | 6.7 | 6.7 | 6.7 | |
| Gastrointestinal stromal tumor (GIST) | 50 | 46 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Appendix, neuroendocrine tumor (NET) | 22 | 15 | 80.0 | 6.7 | 0.0 | 13.3 | |
| Colorectal, neuroendocrine tumor (NET) | 12 | 12 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ileum, neuroendocrine tumor (NET) | 49 | 45 | 97.8 | 2.2 | 0.0 | 0.0 | |
| Pancreas, neuroendocrine tumor (NET) | 97 | 89 | 91.0 | 2.2 | 2.2 | 4.5 | |
| Colorectal, neuroendocrine carcinoma (NEC) | 12 | 10 | 70.0 | 10.0 | 0.0 | 20.0 | |
| Gallbladder, neuroendocrine carcinoma (NEC) | 4 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pancreas, neuroendocrine carcinoma (NEC) | 14 | 14 | 92.9 | 0.0 | 0.0 | 7.1 | |
| Tumors of the urinary system | Non-invasive papillary urothelial ca., pTa G2 low grade | 177 | 141 | 94.3 | 5.0 | 0.0 | 0.7 |
| Non-invasive papillary urothelial ca., pTa G2 high grade | 141 | 116 | 75.0 | 20.7 | 0.9 | 3.4 | |
| Non-invasive papillary urothelial carcinoma, pTa G3 | 187 | 116 | 66.4 | 26.7 | 3.4 | 3.4 | |
| Urothelial carcinoma, pT2-4 G3 | 1206 | 835 | 70.1 | 19.2 | 4.7 | 6.1 | |
| Small-cell neuroendocrine carcinoma of the bladder | 20 | 18 | 61.1 | 16.7 | 5.6 | 16.7 | |
| Sarcomatoid urothelial carcinoma | 25 | 21 | 85.7 | 9.5 | 0.0 | 4.8 | |
| Clear cell renal cell carcinoma | 857 | 835 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Papillary renal cell carcinoma | 255 | 242 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Clear cell (tubulo) papillary renal cell carcinoma | 21 | 21 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Chromophobe renal cell carcinoma | 131 | 127 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Oncocytoma of the kidney | 177 | 165 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the male genital organs | Adenocarcinoma of the prostate, Gleason 3 + 3 | 83 | 83 | 100.0 | 0.0 | 0.0 | 0.0 |
| Adenocarcinoma of the prostate, Gleason 4 + 4 | 80 | 78 | 91.0 | 7.7 | 0.0 | 1.3 | |
| Adenocarcinoma of the prostate, Gleason 5 + 5 | 85 | 85 | 98.8 | 1.2 | 0.0 | 0.0 | |
| Adenocarcinoma of the prostate (recurrence) | 258 | 248 | 87.9 | 10.1 | 0.8 | 1.2 | |
| Small-cell neuroendocrine carcinoma of the prostate | 19 | 16 | 50.0 | 18.8 | 6.3 | 25.0 | |
| Seminoma | 621 | 613 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Embryonal carcinoma of the testis | 50 | 44 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Yolk sac tumor | 50 | 37 | 94.6 | 5.4 | 0.0 | 0.0 | |
| Teratoma | 50 | 45 | 88.9 | 4.4 | 4.4 | 2.2 | |
| Squamous cell carcinoma of the penis | 80 | 76 | 67.1 | 19.7 | 7.9 | 5.3 | |
| Tumors of endocrine organs | Adenoma of the thyroid gland | 113 | 107 | 100.0 | 0.0 | 0.0 | 0.0 |
| Papillary thyroid carcinoma | 391 | 384 | 99.7 | 0.3 | 0.0 | 0.0 | |
| Follicular thyroid carcinoma | 154 | 152 | 98.0 | 0.0 | 0.0 | 2.0 | |
| Medullary thyroid carcinoma | 111 | 107 | 3.7 | 0.9 | 3.7 | 91.6 | |
| Anaplastic thyroid carcinoma | 45 | 42 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adrenal cortical adenoma | 50 | 45 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adrenal cortical carcinoma | 26 | 25 | 96.0 | 0.0 | 4.0 | 0.0 | |
| Pheochromocytoma | 50 | 49 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of hematopoetic and lymphoid tissues | Hodgkin lymphoma | 103 | 98 | 100.0 | 0.0 | 0.0 | 0.0 |
| Small lymphocytic lymphoma, B-cell type (B-SLL/B-CLL) | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Diffuse large B cell lymphoma (DLBCL) | 113 | 113 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Follicular lymphoma | 88 | 88 | 100.0 | 0.0 | 0.0 | 0.0 | |
| T-cell non-Hodgkin lymphoma | 25 | 25 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mantle cell lymphoma | 18 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Marginal zone lymphoma | 16 | 16 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Diffuse large B-cell lymphoma (DLBCL) in the testis | 16 | 16 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Burkitt lymphoma | 5 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of soft tissue and bone | Tenosynovial giant cell tumor | 45 | 44 | 100.0 | 0.0 | 0.0 | 0.0 |
| Granular cell tumor | 53 | 45 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leiomyoma | 50 | 49 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leiomyosarcoma | 87 | 87 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Liposarcoma | 132 | 123 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Malignant peripheral nerve sheath tumor (MPNST) | 13 | 13 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Myofibrosarcoma | 26 | 26 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Angiosarcoma | 73 | 68 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Angiomyolipoma | 91 | 89 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Dermatofibrosarcoma protuberans | 21 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ganglioneuroma | 14 | 14 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Kaposi sarcoma | 8 | 5 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Neurofibroma | 117 | 114 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sarcoma, not otherwise specified (NOS) | 74 | 71 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Paraganglioma | 41 | 41 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ewing sarcoma | 23 | 16 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Rhabdomyosarcoma | 6 | 6 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Schwannoma | 121 | 118 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Synovial sarcoma | 12 | 11 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Osteosarcoma | 43 | 37 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Chondrosarcoma | 38 | 21 | 100.0 | 0.0 | 0.0 | 0.0 | |
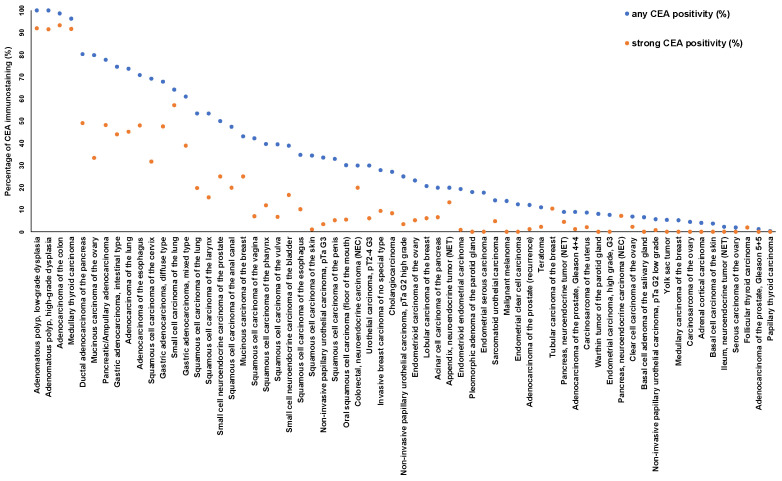
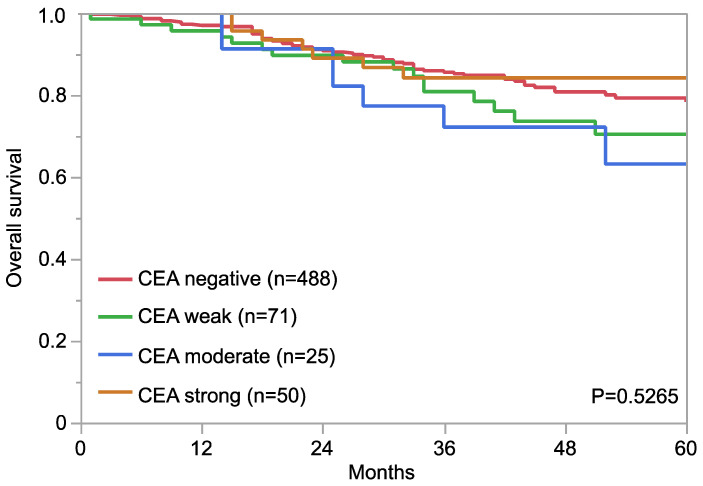
Examples for particularly high positivity rates and high levels of expression were colorectal adenomas (100% positive) and adenocarcinomas (98.7%), other gastrointestinal adenocarcinomas (61.1–80.3%), medullary thyroid carcinomas (96.3%), adenocarcinoma of the lung (73.7%), mucinous carcinomas of the ovary (79.8%) and the breast (43.2%), squamous cell carcinomas of different sites of origin (30.2–69.1%) as well as small-cell carcinomas of the lung (64.3%), the prostate (50.0%), and the urinary bladder (38.9%). In many of these tumor types, CEA expression was often stronger than in the corresponding normal tissues. Figure 2 shows representative images. Figure 3 gives a graphical representation of the ranking order of CEA-positive and strongly positive tumors. Associations between CEA immunostaining and histopathological features are shown in Table 2. High CEA expression was associated with advanced pT stage (p < 0.0001) and muscle-invasive growth (p < 0.0001) in urinary bladder cancer. In invasive breast carcinomas of no special type, high CEA expression was linked to ER positivity (p = 0.0005) and HER2 positivity (p < 0.0001) but was unrelated to grade, pT, pN, and overall survival (p = 0.2520, Figure 4). Reduced CEA expression was associated with defective mismatch repair status (dMMR; p < 0.0001), BRAF V600E mutations (p = 0.0498), and tumor localization in the right colon (p = 0.0024) in colorectal adenocarcinoma. There was no association of the CEA expression level to histopathological, molecular, or clinical tumor characteristics in pancreatic and gastric adenocarcinomas, or endometroid endometrium carcinoma. A combined analysis of 524 squamous cell carcinomas from nine different sites showed a link between HPV infection and CEA positivity (p = 0.0281; Supplementary Table S2) but subgroup analysis by organs of origins did not show significant associations between HPV status and CEA expression levels.
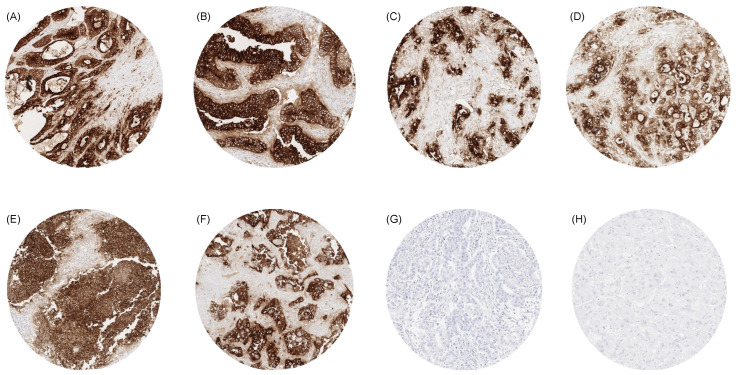
Figure 2.
CEA immunostaining in cancer. The panels show a strong CEA staining in an adenocarcinoma of the colon (A), an adenocarcinoma of the esophagus (B), a ductal adenocarcinoma of the pancreas (C), a cholangiocellular carcinoma of the liver (D), a small-cell neuroendocrine carcinoma of the lung (E), and an adenocarcinoma of the lung (F). CEA staining is lacking in a malignant mesothelioma of the pleura (H) and a hepatocellular carcinoma in the liver (G). Original magnifications 10×, spot size 600 μm.
Figure 3.
Ranking order of CEA immunostaining in cancers. Both the percentage of positive cases (blue dots) and the percentage of strongly positive cases (orange dots) are shown.
Table 2.
CEA immunostaining and cancer phenotype.
| CEA Immunostaining Result | |||||||
|---|---|---|---|---|---|---|---|
| n | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | p | ||
| Invasive breast carcinoma of no special type | pT1 | 491 | 75.2 | 12.0 | 5.3 | 7.5 | 0.5044 |
| pT2 | 369 | 70.5 | 13.6 | 5.1 | 10.8 | ||
| pT3–4 | 80 | 68.8 | 15.0 | 3.8 | 12.5 | ||
| G1 | 153 | 78.4 | 10.5 | 5.2 | 5.9 | 0.1972 | |
| G2 | 472 | 69.9 | 14.0 | 6.6 | 9.5 | ||
| G3 | 345 | 74.2 | 12.5 | 3.5 | 9.9 | ||
| pN0 | 269 | 71.4 | 18.2 | 5.6 | 4.8 | 0.6430 | |
| pN+ | 171 | 66.7 | 20.5 | 8.2 | 4.7 | ||
| pM0 | 137 | 70.1 | 16.8 | 5.1 | 8.0 | 0.8864 | |
| pM1 | 93 | 66.7 | 17.2 | 7.5 | 8.6 | ||
| HER2 negative | 771 | 74.2 | 11.9 | 5.1 | 8.8 | 0.0158 | |
| HER2 positive | 113 | 62.8 | 23.0 | 3.5 | 10.6 | ||
| ER negative | 184 | 83.7 | 8.7 | 2.2 | 5.4 | 0.0005 | |
| ER positive | 662 | 68.9 | 15.4 | 6.0 | 9.7 | ||
| PR negative | 351 | 74.6 | 12.8 | 4.0 | 8.5 | 0.6741 | |
| PR positive | 527 | 71.5 | 13.7 | 5.5 | 9.3 | ||
| Non-triple negative | 701 | 68.9 | 15.4 | 5.6 | 10.1 | <0.0001 | |
| Triple negative | 123 | 91.1 | 4.1 | 2.4 | 2.4 | ||
| Urothelial bladder carcinoma | pTa G2 low | 143 | 93.0 | 5.6 | 0.0 | 1.4 | <0.0001 |
| pTa G2 high | 116 | 75.0 | 20.7 | 0.9 | 3.4 | ||
| pTa G3 | 116 | 66.4 | 26.7 | 3.4 | 3.4 | ||
| pT2 | 136 | 69.1 | 20.6 | 2.9 | 7.4 | 0.0167 | |
| pT3 | 216 | 69.0 | 16.2 | 10.6 | 4.2 | ||
| pT4 | 97 | 76.3 | 17.5 | 2.1 | 4.1 | ||
| G2 | 22 | 68.2 | 22.7 | 4.5 | 4.5 | 0.9255 * | |
| G3 | 427 | 70.7 | 17.6 | 6.6 | 5.2 | ||
| pN0 | 253 | 72.7 | 17.0 | 5.5 | 4.7 | 0.4985 * | |
| pN+ | 170 | 66.5 | 20.6 | 8.2 | 4.7 | ||
| Adenocarcinoma of the pancreas | pT1 | 14 | 21.4 | 14.3 | 7.1 | 57.1 | 0.4277 |
| pT2 | 70 | 10.0 | 27.1 | 11.4 | 51.4 | ||
| pT3 | 373 | 20.6 | 19.0 | 10.7 | 49.6 | ||
| pT4 | 29 | 17.2 | 27.6 | 17.2 | 37.9 | ||
| G1 | 16 | 18.8 | 18.8 | 12.5 | 50.0 | 0.9969 | |
| G2 | 344 | 18.9 | 19.5 | 11.3 | 50.3 | ||
| G3 | 104 | 19.2 | 22.1 | 9.6 | 49.0 | ||
| pN0 | 106 | 19.8 | 17.9 | 13.2 | 49.1 | 0.7820 | |
| pN+ | 379 | 18.5 | 21.4 | 10.6 | 49.6 | ||
| R0 | 249 | 18.5 | 20.9 | 11.6 | 49.0 | 0.8254 | |
| R1 | 198 | 18.2 | 20.7 | 9.1 | 52.0 | ||
| MMR proficient | 440 | 19.3 | 20.7 | 11.8 | 48.2 | 0.2974 | |
| MMR deficient | 4 | 0.0 | 50.0 | 25.0 | 25.0 | ||
| Adenocarcinoma of the stomach | pT1–2 | 57 | 31.6 | 8.8 | 10.5 | 49.1 | 0.6383 |
| pT3 | 110 | 33.6 | 11.8 | 9.1 | 45.5 | ||
| pT4 | 106 | 24.5 | 16.0 | 13.2 | 46.2 | ||
| pN0 | 68 | 27.9 | 8.8 | 19.1 | 44.1 | 0.1081 | |
| pN+ | 204 | 30.9 | 13.7 | 8.3 | 47.1 | ||
| MMR proficient | 232 | 30.6 | 13.8 | 9.9 | 45.7 | 0.0899 | |
| MMR deficient | 37 | 18.9 | 16.2 | 24.3 | 40.5 | ||
| Endometrioid endometrial carcinoma | pT1 | 115 | 79.1 | 20.0 | 0.0 | 0.9 | 0.3498 |
| pT2 | 24 | 83.3 | 12.5 | 4.2 | 0.0 | ||
| pT3–4 | 35 | 88.6 | 11.4 | 0.0 | 0.0 | ||
| pN0 | 50 | 80.0 | 20.0 | 0.0 | 0.0 | 0.7254 | |
| pN+ | 30 | 76.7 | 23.3 | 0.0 | 0.0 | ||
| Endometrioid carcinoma of the ovary | pT1 | 23 | 82.6 | 17.4 | 0.0 | 0.0 | 0.2037 |
| pT2 | 5 | 60.0 | 20.0 | 0.0 | 20.0 | ||
| pT3 | 6 | 50.0 | 33.3 | 0.0 | 16.7 | ||
| pN0 | 21 | 71.4 | 28.6 | 0.0 | 0.0 | 0.0516 | |
| pN1 | 8 | 62.5 | 12.5 | 0.0 | 25.0 | ||
| Serous carcinoma of the ovary | pT1 | 32 | 100.0 | 0.0 | 0.0 | 0.0 | 0.2000 |
| pT2 | 44 | 95.5 | 4.5 | 0.0 | 0.0 | ||
| pT3 | 255 | 99.2 | 0.8 | 0.0 | 0.0 | ||
| pN0 | 83 | 100.0 | 0.0 | 0.0 | 0.0 | 0.1147 | |
| pN1 | 163 | 98.2 | 1.8 | 0.0 | 0.0 | ||
| Adenocarcinoma of the colon | pT1 | 66 | 3.0 | 0.0 | 3.0 | 93.9 | 0.5369 |
| pT2 | 336 | 1.2 | 2.4 | 2.7 | 93.8 | ||
| pT3 | 936 | 1.3 | 1.7 | 3.4 | 93.6 | ||
| pT4 | 345 | 1.2 | 3.5 | 2.9 | 92.5 | ||
| pN0 | 877 | 1.0 | 1.5 | 3.2 | 94.3 | 0.1474 | |
| pN+ | 790 | 1.6 | 2.9 | 3.0 | 92.4 | ||
| V0 | 1225 | 1.1 | 2.2 | 3.1 | 93.6 | 0.6785 | |
| V1 | 432 | 1.9 | 2.1 | 3.2 | 92.8 | ||
| L0 | 629 | 1.1 | 1.3 | 2.7 | 94.9 | 0.1296 | |
| L1 | 1013 | 1.5 | 2.8 | 3.5 | 92.3 | ||
| Right side | 454 | 2.4 | 3.3 | 4.6 | 89.6 | 0.0024 | |
| Left side | 1237 | 0.8 | 1.8 | 2.6 | 94.8 | ||
| MMR proficient | 1162 | 0.6 | 1.3 | 2.0 | 96.1 | <0.0001 | |
| MMR deficient | 88 | 5.7 | 11.4 | 13.6 | 69.3 | ||
| RAS wildtype | 468 | 0.9 | 2.8 | 3.0 | 93.4 | 0.7409 | |
| RAS mutation | 355 | 1.1 | 2.3 | 2.0 | 94.6 | ||
| BRAF wildtype | 124 | 0.8 | 3.2 | 1.6 | 94.4 | 0.0498 | |
| BRAF V600E mutation | 21 | 0.0 | 9.5 | 14.3 | 76.2 | ||
* only in pT2–4 urothelial bladder carcinomas. Abbreviations: pT, pathological tumor stage; G, Grade; pN, pathological lymph node status; pM, pathological status of distant metastasis; R, resection margin status; V, venous invasion; L, lymphatic invasion; PR, progesterone receptor; ER, estrogen receptor; MMR, mismatch repair.
Figure 4.
CEA immunostaining and patient prognosis in invasive breast carcinoma of no special type.
4. Discussion
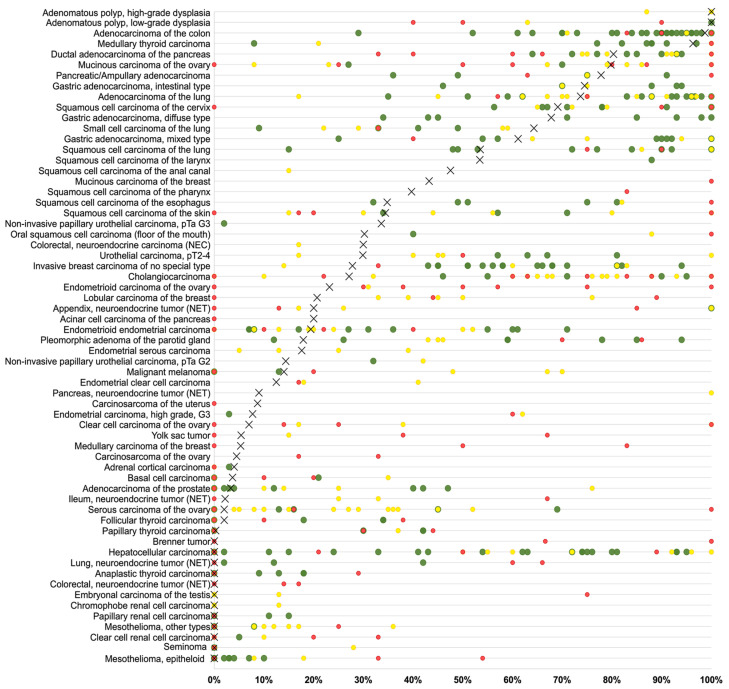
The importance of CEA as a cancer-related protein is reflected by more than 3000 Medline articles meeting the search terms “CEA + immunohistochemistry + cancer” (pubmed_January 2023). The articles generally agree on colorectal adenocarcinoma being the cancer entity most commonly CEA-positive, but data on the prevalence of CEA expression vary considerably in other tumor types. Based on our highly standardized analysis of 120 tumor types and subtypes, we were able to create a ranking order of human tumors according to the prevalence of CEA expression (Figure 3), which allows for a direct comparison of CEA positivity obtained under identical experimental conditions. A collection of data from the literature (Figure 5, Supplementary Table S3) demonstrates that such information could not easily be retrieved from the literature.
Figure 5.
Comparison of CEA expression with previous works in the literature. An “X” represents the proportion of CEA-positive cancers in the present study, dots indicate the frequencies reported in the literature for comparison: studies with ≤10 tumors analyzed are marked with red dots, studies with 11 to 25 tumors analyzed are marked with yellow dots, and green dots mark studies with >25 tumors analyzed.
Although—in line with our data—multiple studies showed high rates of CEA positivity in tumor types arising from epithelial tissues, including gastrointestinal adenocarcinomas, medullary carcinomas of the thyroid, adenocarcinomas of the lung, and squamous cell carcinomas of various origins, as well as in small-cell carcinomas, there were also numerous studies suggesting much lower positivity rates in these tumor types. Moreover, our data identified low CEA positivity rates in several tumor entities for which high CEA expression levels were suggested in multiple earlier studies, such as, for example, in high-grade serous carcinomas of the ovary [41], or prostatic adenocarcinomas [42]. That high levels of CEA expression can be observed in many different tumor entities is consistent with data summarized in The Cancer Genome Atlas (TCGA) database (www.cancer.gov, data available from https://www.proteinatlas.org/ENSG00000105388-CEACAM5/summary/rna, accessed on 21 November 2024).
Considering the high expression levels of CEA in various tumor entities and the relatively low CEA levels in normal tissues, CEA represents an attractive therapeutic target. However, previous attempts at targeting CEA by using humanized anti-CEA antibodies showed disappointing results. It is assumed that in cancers with high serum CEA levels, the therapeutic antibodies are prevented from reaching the tumor cells, as they bind to circulating CEA (summarized in [43]). Promising recent approaches include developing a vaccination (summarized in [44]) and the generation of CAR-T cells (summarized in [45]) targeting CEA. CEA-targeting drugs are currently studied in more than 200 phase I and II clinical trials. Included in these studies are colorectal, breast, esophageal, stomach, lung, gastric, and pancreatic carcinomas as well as CEA-positive tumors regardless of their sites of origin (www.clinicaltrials.gov). If one of these treatment approaches should become clinically available, our ranking order of tumors based on their CEA positivity rates could help to determine the tumor entities for which such approaches would be most beneficial.
Our data support two important diagnostic applications of CEA IHC. The complete CEA-negativity of all our 90 malignant mesotheliomas while 74% of the 179 pulmonary adenocarcinomas were CEA-positive supports the use of CEA IHC for the distinction of malignant mesothelioma from pulmonary adenocarcinoma. Panels that have been proposed for this application also include calretinin, D2-40, WT1, cytokeratin 5/6, D2-40, EpCAM, TTF-1, Ber-EP4/MOC31, and Napsin A [46]. Total absence of CEA staining in all 50 hepatocellular carcinomas analyzed in this study supports the concept of using CEA as a marker for the distinction of primary tumors from metastases in the liver [16]. This is all the more useful, as carcinomas from the entire gastrointestinal tract—the most common source of liver metastases—were often CEA-positive. The high rate of CEA-positive hepatocellular carcinomas described in several previous studies is likely due to the use of less specific and/or polyclonal CEA antibodies, which leads to positivity rates between 15–100% [30,47,48,49]. Previous studies with monoclonal CEA antibodies identified a CEA positivity in 0–55% of hepatocellular carcinomas [35,48,49,50].
For several of our tumor categories, the large number of tumors analyzed allowed an analysis of the potential clinical significance of CEA expression. The strong link between a reduced CEA expression and dMMR in colorectal cancer, in which 96% of mismatch repair-proficient (pMMR) but only 69% of dMMR cancers showed strong CEA staining is in agreement with data from Schiemann et al. [51] finding lower CEA serum levels in colorectal cancer patients with hereditary non-polyposis colorectal carcinomas than found in those with sporadic carcinomas. However, another study showed only a marginal difference between dMMR (67% negative) and pMMR (58%) colorectal carcinomas [52] while three further studies found no difference in CEA immunostaining between dMMR and pMMR carcinomas [53,54,55]. Given the abundance of DNA mutations in microsatellite instable/dMMR cancers, it appears possible that functionally relevant DNA alterations of genes that directly or indirectly regulate CEA expression may cause the reduced CEA expression in a subset of dMMR cancers. The absence of clear-cut associations with histopathological parameters of tumor aggressiveness in several tumor entities and the lack of a prognostic significance in invasive breast cancers of no special type strongly argues against a strong and clinically relevant prognostic role of CEA expression in cancer and suggests that CEA upregulation may parallel tumor development in various cancer types. This interpretation is supported by controversial results of previous studies. Although there were more than 18 studies suggesting an unfavorable tumor phenotype or poor prognosis for tumors with high CEA expression, there were at least 24 studies denying such a role of CEA expression (Supplementary Table S4). Similarly controversial data were seen for specific tumor entities. For example, in colorectal adenocarcinomas, eight studies linked high CEA expression to unfavorable tumor features and/or prognosis while six studies found no association between CEA immunostaining and tumor phenotype and/or prognosis (Supplementary Table S4).
CEA mainly functions as a serum marker for colorectal cancer. Serial CEA serum measurements were recommended in a 2014 update of the European group on tumor marker guidelines for postoperative surveillance of UICC stage II and III patients considered for surgical resection or systemic therapy in case of distant metastasis and for monitoring response to treatment in advanced disease [56]. High preoperative [57] and postoperative [58] serum CEA levels have also been suggested as a prognostic tool in colorectal cancer. Given the abundant and homogeneous expression of CEA in most colorectal cancers, this is likely due to a link between CEA serum levels, tumor burden, and residual disease. Our findings demonstrate that similarly high levels of tumoral CEA expression can occur in many other tumor types, at least in a significant fraction of cases. Given the close to 100% prevalence of CEA expression in colorectal cancer, serological CEA monitoring obviously does not require CEA analysis in tumor tissue. It appears possible, however, that an immunohistochemical tumor tissue analysis documenting high-level CEA expression in a non-colorectal cancer could identify individual patients for whom CEA serum levels would also be suited for monitoring response to therapy and early detection of tumor relapse. Various studies suggested a clinical utility of CEA serum assessment in gastric [59], pancreatic [60], non-small-cell lung [61], and breast cancer [62], although the majority of these studies did not improve patient selection by immunohistochemical CEA analysis of the tumor tissue.
Regarding the large scale of our study, a particular emphasis was placed on the validation of our staining procedure, which was conducted according to the methods proposed by the International Working Group for Antibody Validation (IWGAV). In these guidelines, a comparison with expression data obtained by an additional independent method or a second independent antibody are suggested [63]. Both methods were applied in this study. CEA IHC in 76 different normal tissues was first compared with RNA expression data compiled from the Human Protein Atlas RNA-seq tissue dataset [64], the Functional Annotation of the Mammalian Genome (FANTOM5) project [65,66], and the Genotype-Tissue Expression (GTEx) project [67]. Our normal tissue analysis revealed CEA immunostaining in 10 of 12 organs for which RNA expression had been described (rectum, colon, appendix, small intestine, duodenum, stomach, salivary gland, tonsil, cervix, and lung). Only two organs with documented RNA expression did not show CEA immunostaining (esophagus, smooth muscle). As esophagus and smooth muscle can both be adjacent to CEA-expressing epithelium, these RNA findings may reflect contaminations. In the—CEA RNA negative—thymus and the skin, CEA immunostaining was limited to very small subpopulations that might have been missed in RNA analyses of entire organs. For two other CEA IHC positive tissues (bronchus, anal canal), RNA data were unavailable. The comparison with an independent second antibody (IL-7) confirmed CEA protein expression in bronchus, anal canal, thymus, and the skin, as well as in all other cell types identified as CEA-positive by MSVA-465R.
5. Conclusions
The comprehensive list of CEA positivity across human tumor types shows that CEA is abundantly expressed in a broad range of epithelial neoplasms and serves as a basis for potential future research. Our data identify these tumor entities where most CEA-positive cancers might benefit from CEA serum monitoring and anti-CEA therapies. However, the level of CEA expression does not appear to be largely related to cancer aggressiveness.
Acknowledgments
We are grateful to Melanie Steurer, Laura Behm, Inge Brandt, and Sünje Seekamp for excellent technical assistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers16234052/s1. Supplementary Figure S1: IHC validation by comparison of antibodies. The panels show a concordance of immunostaining results obtained by two independent CEA antibodies. Using MSVA-465R, a cytoplasmic or membranous CEA positivity was seen in a fraction of squamous epithelial cells (A), a fraction of cells in Hassal’s corpuscles (black arrow) of the thymus (B), superficial epithelial cells of the colon (C), respiratory epithelial cells (D), a fraction of alveolar cells in the lung (E), eccrine glands of the skin (F), a fraction of glandular cells in the submandibular gland (G), and some goblet cells of the small intestine (H). Using II-7, a comparable staining was seen in squamous epithelium (I), Hassal’s corpuscles (K), colon (L), respiratory epithelium (M), the lung (N), eccrine glands (O), the submandibular gland (P), and the small intestine (Q). The images A–H and I–Q are from consecutive tissue sections. Supplementary Table S1: CEA immunostaining in normal tissue. Supplementary Table S2: CEA immunostaining and HPV status in squamous cell carcinomas of different sites of origin. Supplementary Table S3: Summary of previous CEA immunostaining studies in tumors. Supplementary Table S4: Summary of previous CEA prognosis studies in tumors.
Author Contributions
K.J., L.K., M.K., R.S., G.S. and C.B.: contributed to conception, design, data collection, data analysis, and manuscript writing. M.L., S.D.R., S.K., V.R., F.V., A.A.B., C.F., N.G., A.M.L., A.M., R.U., T.K., A.H., F.J., E.B., S.S., A.H.M., T.S.C., D.D., P.L. and S.M.: participated in pathology data analysis, data interpretation, and collection of samples. R.S., M.K. and C.H.-M.: data analysis. C.B., R.S. and G.S.: study supervision. All authors agree to be accountable for the content of the work. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The usage of archived diagnostic leftover tissues for manufacturing of TMAs and their analysis for research purposes, as well as patient data analyses, have been approved by local laws (HmbKHG, §12,1) and by the local ethics committee (Ethics commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration.
Informed Consent Statement
Patient consent was waived due to local laws (HmbKHG, §12.1) that permit research with anonymized diagnostic leftover tissue samples.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
The rabbit recombinant antibody, clone MSVA-465R was provided from MS Validated Antibodies GmbH (owned by a family member of GS). All other authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Taheri M., Saragovi U., Fuks A., Makkerh J., Mort J., Stanners C.P. Self recognition in the Ig superfamily. Identification of precise subdomains in carcinoembryonic antigen required for intercellular adhesion. J. Biol. Chem. 2000;275:26935–26943. doi: 10.1016/S0021-9258(19)61463-8. [DOI] [PubMed] [Google Scholar]
- 2.Nap M., Mollgard K., Burtin P., Fleuren G.J. Immunohistochemistry of carcino-embryonic antigen in the embryo, fetus and adult. Tumor Biol. 1988;9:145–153. doi: 10.1159/000217555. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y., Yang J., Li H., Wu Y., Zhang H., Chen W. Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: A meta-analysis. Int. J. Clin. Exp. Med. 2015;8:11683–11691. [PMC free article] [PubMed] [Google Scholar]
- 4.Grunnet M., Sorensen J.B. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer. 2012;76:138–143. doi: 10.1016/j.lungcan.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Fakih M.G., Padmanabhan A. CEA monitoring in colorectal cancer. What you should know. Oncology. 2006;20:579–587; discussion 588, 594, 596. [PubMed] [Google Scholar]
- 6.Chevinsky A.H. CEA in tumors of other than colorectal origin. Semin. Surg. Oncol. 1991;7:162–166. doi: 10.1002/ssu.2980070309. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson B.D., Shinkins B., Pathiraja I., Roberts N.W., James T.J., Mallett S., Perera R., Primrose J.N., Mant D. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst. Rev. 2015;2015:CD011134. doi: 10.1002/14651858.CD011134.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai M., He H., Hong S., Weng J. Synergistic diagnostic value of circulating tumor cells and tumor markers CEA/CA19-9 in colorectal cancer. Scand. J. Gastroenterol. 2023;58:54–60. doi: 10.1080/00365521.2022.2106152. [DOI] [PubMed] [Google Scholar]
- 9.Argiles G., Saro J., Segal N.H., Melero I., Ros W., Marabelle A., Rodriguez M.E., Albanell J., Calvo E., Moreno V., et al. Novel carcinoembryonic antigen T-cell bispecific (CEA-TCB) antibody: Preliminary clinical data as a single agent and in combination with atezolizumab in patients with metastatic colorectal cancer (mCRC) Ann. Oncol. 2017;28:iii151. doi: 10.1093/annonc/mdx302.003. [DOI] [Google Scholar]
- 10.Tabernero J., Melero I., Ros W., Argiles G., Marabelle A., Rodriguez-Ruiz M.E., Albanell J., Calvo E., Moreno V., Cleary J.M., et al. Phase Ia and lb studies of the novel carcinoembryonic antigen (CEA) 1-cell bispecific (CEA CD3 TCB) antibody as a single agent and in combination with atezolizumab: Preliminary efficacy and safety in patients with metastatic colorectal cancer (mCRC) J. Clin. Oncol. 2017;35:3002. doi: 10.1200/JCO.2017.35.15_suppl.3002. [DOI] [Google Scholar]
- 11.Dotan E., Cohen S.J., Starodub A.N., Lieu C.H., Messersmith W.A., Simpson P.S., Guarino M.J., Marshall J.L., Goldberg R.M., Hecht J.R., et al. Phase I/II Trial of Labetuzumab Govitecan (Anti-CEACAM5/SN-38 Antibody-Drug Conjugate) in Patients With Refractory or Relapsing Metastatic Colorectal Cancer. J. Clin. Oncol. 2017;35:3338–3346. doi: 10.1200/JCO.2017.73.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodet-Milin C., Ferrer L., Rauscher A., Masson D., Rbah-Vidal L., Faivre-Chauvet A., Cerato E., Rousseau C., Hureaux J., Couturier O., et al. Pharmacokinetics and Dosimetry Studies for Optimization of Pretargeted Radioimmunotherapy in CEA-Expressing Advanced Lung Cancer Patients. Front. Med. 2015;2:84. doi: 10.3389/fmed.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holzinger A., Abken H. CAR T cells targeting solid tumors: Carcinoembryonic antigen (CEA) proves to be a safe target. Cancer Immunol. Immunother. 2017;66:1505–1507. doi: 10.1007/s00262-017-2045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morse M.A., Chaudhry A., Gabitzsch E.S., Hobeika A.C., Osada T., Clay T.M., Amalfitano A., Burnett B.K., Devi G.R., Hsu D.S., et al. Novel adenoviral vector induces T-cell responses despite anti-adenoviral neutralizing antibodies in colorectal cancer patients. Cancer Immunol. Immunother. 2013;62:1293–1301. doi: 10.1007/s00262-013-1400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halimi M., BeheshtiRouy S., Salehi D., Rasihashemi S.Z. The Role of Immunohistochemistry Studies in Distinguishing Malignant Mesothelioma from Metastatic Lung Carcinoma in Malignant Pleural Effusion. Iran. J. Pathol. 2019;14:122–126. doi: 10.30699/ijp.14.2.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdel-Hamid N.M., Abouzied M.M., Nazmy M.H., Fawzy M.A., Gerges A.S. A suggested guiding panel of seromarkers for efficient discrimination between primary and secondary human hepatocarcinoma. Tumor Biol. 2016;37:2539–2546. doi: 10.1007/s13277-015-4025-7. [DOI] [PubMed] [Google Scholar]
- 17.Pavelic Z.P., Petrelli N.J., Herrera L., Vaughan M.M., Paecock J.S., Pavelic L. D-14 monoclonal antibody to carcinoembryonic antigen: Immunohistochemical analysis of formalin-fixed, paraffin-embedded human colorectal carcinoma, tumors of non-colorectal origin and normal tissues. J. Cancer Res. Clin. Oncol. 1990;116:51–56. doi: 10.1007/BF01612640. [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Ma Y., Zhu Z.H., Situ D.R., Hu Y., Rong T.H. Expression and prognostic relevance of tumor carcinoembryonic antigen in stage IB non-small cell lung cancer. J. Thorac. Dis. 2012;4:490–496. doi: 10.3978/j.issn.2072-1439.2012.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ordonez N.G. The value of antibodies 44-3A6, SM3, HBME-1, and thrombomodulin in differentiating epithelial pleural mesothelioma from lung adenocarcinoma: A comparative study with other commonly used antibodies. Am. J. Surg. Pathol. 1997;21:1399–1408. doi: 10.1097/00000478-199712000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Hurlimann J., Gardiol D. Immunohistochemistry in the differential diagnosis of liver carcinomas. Am. J. Surg. Pathol. 1991;15:280–288. doi: 10.1097/00000478-199103000-00008. [DOI] [PubMed] [Google Scholar]
- 21.Morrison C., Marsh W., Jr., Frankel W.L. A comparison of CD10 to pCEA, MOC-31, and hepatocyte for the distinction of malignant tumors in the liver. Mod. Pathol. 2002;15:1279–1287. doi: 10.1097/01.MP.0000037312.69565.24. [DOI] [PubMed] [Google Scholar]
- 22.Cai J., Ma H., Huang F., Zhu D., Zhao L., Yang Y., Bi J., Zhang T. Low expression of RSK4 predicts poor prognosis in patients with colorectal cancer. Int. J. Clin. Exp. Pathol. 2014;7:4959–4970. [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L., Xie G., Duan G., Yan X., Li Q. High expression of testes-specific protease 50 is associated with poor prognosis in colorectal carcinoma. PLoS ONE. 2011;6:e22203. doi: 10.1371/journal.pone.0022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santeusanio G., Peronace L., Castagna G., De Muro G., Santi D., D’Orazio A., Amanti C., Midiri G., Campisi C., D’Ambra G., et al. Immunohistochemical study of carcinoembryonic antigen (CEA) in gastric tumors: Correlation with preoperative serum levels, histologic type, and grade of anaplasia of the tumor. J. Surg. Oncol. 1988;37:13–19. doi: 10.1002/jso.2930370105. [DOI] [PubMed] [Google Scholar]
- 25.Terada T. An immunohistochemical study of primary signet-ring cell carcinoma of the stomach and colorectum: III. Expressions of EMA, CEA, CA19-9, CDX-2, p53, Ki-67 antigen, TTF-1, vimentin, and p63 in normal mucosa and in 42 cases. Int. J. Clin. Exp. Pathol. 2013;6:630–638. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J., Zhang L., Gu Y., Li K., Nie Y., Fan D., Feng Y. Dynamic expression of CEACAM7 in precursor lesions of gastric carcinoma and its prognostic value in combination with CEA. World J. Surg. Oncol. 2011;9:172. doi: 10.1186/1477-7819-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park S.H., Ku K.B., Chung H.Y., Yu W. Prognostic significance of serum and tissue carcinoembryonic antigen in patients with gastric adenocarcinomas. Cancer Res. Treat. 2008;40:16–21. doi: 10.4143/crt.2008.40.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umeda T., Sakamoto J., Watanabe T., Ito K., Akiyama S., Yasue M., Takagi H. Immunohistochemical analysis of the poorly differentiated stomach adenocarcinoma with medullary growth pattern. J. Surg. Oncol. 1996;62:34–39. doi: 10.1002/(SICI)1096-9098(199605)62:1<34::AID-JSO8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 29.Margaritescu C., Mogoanta L., Manescu P., Simionescu C., Pirici D., Streba L., Mercut D. The immunohistochemical profile of the adenocarcinoma of upper gastric pole. Rom. J. Morphol. Embryol. 2007;48:215–235. [PubMed] [Google Scholar]
- 30.Xiao S.Y., Wang H.L., Hart J., Fleming D., Beard M.R. cDNA arrays and immunohistochemistry identification of CD10/CALLA expression in hepatocellular carcinoma. Am. J. Pathol. 2001;159:1415–1421. doi: 10.1016/S0002-9440(10)62528-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueda S., Tsubura A., Izumi H., Sasaki M., Morii S. Immunohistochemical studies on carcinoembryonic antigen in adenocarcinomas of the uterus. Acta Pathol. Jpn. 1983;33:59–69. doi: 10.1111/j.1440-1827.1983.tb02100.x. [DOI] [PubMed] [Google Scholar]
- 32.Toki T., Oikawa N., Tase T., Wada Y., Yajima A., Suzuki M. Immunohistochemical localization of carcinoembryonic antigen in adenocarcinoma of the endocervix and the endometrium. Tohoku J. Exp. Med. 1985;146:27–32. doi: 10.1620/tjem.146.27. [DOI] [PubMed] [Google Scholar]
- 33.Segawa Y., Ohnoshi T., Hiraki S., Ueoka H., Kiura K., Kamei H., Tabata M., Shibayama T., Miyatake K., Genba K., et al. Immunohistochemical detection of P-glycoprotein and carcinoembryonic antigen in small cell lung cancer: With reference to predictability of response to chemotherapy. Acta Med. Okayama. 1993;47:181–189. doi: 10.18926/AMO/31590. [DOI] [PubMed] [Google Scholar]
- 34.Kimura N., Ghandur-Mnaymen L. An immunohistochemical study of keratin, carcinoembryonic antigen, human chorionic gonadotropin and alpha-fetoprotein in lung cancer. Tohoku J. Exp. Med. 1985;145:23–38. doi: 10.1620/tjem.145.23. [DOI] [PubMed] [Google Scholar]
- 35.Sheahan K., O’Brien M.J., Burke B., Dervan P.A., O’Keane J.C., Gottlieb L.S., Zamcheck N. Differential reactivities of carcinoembryonic antigen (CEA) and CEA-related monoclonal and polyclonal antibodies in common epithelial malignancies. Am. J. Clin. Pathol. 1990;94:157–164. doi: 10.1093/ajcp/94.2.157. [DOI] [PubMed] [Google Scholar]
- 36.Ronnett B.M., Shmookler B.M., Diener-West M., Sugarbaker P.H., Kurman R.J. Immunohistochemical evidence supporting the appendiceal origin of pseudomyxoma peritonei in women. Int. J. Gynecol. Pathol. 1997;16:1–9. doi: 10.1097/00004347-199701000-00001. [DOI] [PubMed] [Google Scholar]
- 37.Croce M.V., Colussi A.G., Price M.R., Segal-Eiras A. Expression of tumour associated antigens in normal, benign and malignant human mammary epithelial tissue: A comparative immunohistochemical study. Anticancer Res. 1997;17:4287–4292. [PubMed] [Google Scholar]
- 38.Saadatmand S., de Kruijf E.M., Sajet A., Dekker-Ensink N.G., van Nes J.G., Putter H., Smit V.T., van de Velde C.J., Liefers G.J., Kuppen P.J. Expression of cell adhesion molecules and prognosis in breast cancer. Br. J. Surg. 2013;100:252–260. doi: 10.1002/bjs.8980. [DOI] [PubMed] [Google Scholar]
- 39.Bubendorf L., Nocito A., Moch H., Sauter G. Tissue microarray (TMA) technology: Miniaturized pathology archives for high-throughput in situ studies. J. Pathol. 2001;195:72–79. doi: 10.1002/path.893. [DOI] [PubMed] [Google Scholar]
- 40.Kononen J., Bubendorf L., Kallioniemi A., Barlund M., Schraml P., Leighton S., Torhorst J., Mihatsch M.J., Sauter G., Kallioniemi O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 41.Silcocks P.B., Herbert A., Wright D.H. Evaluation of PAS-diastase and carcinoembryonic antigen staining in the differential diagnosis of malignant mesothelioma and papillary serous carcinoma of the ovary. J. Pathol. 1986;149:133–141. doi: 10.1002/path.1711490208. [DOI] [PubMed] [Google Scholar]
- 42.Pan C.C., Chen P.C., Tsay S.H., Ho D.M. Differential immunoprofiles of hepatocellular carcinoma, renal cell carcinoma, and adrenocortical carcinoma: A systemic immunohistochemical survey using tissue array technique. Appl. Immunohistochem. Mol. Morphol. 2005;13:347–352. doi: 10.1097/01.pai.0000146525.72531.19. [DOI] [PubMed] [Google Scholar]
- 43.Gold P., Goldenberg N.A. The Carcinoembryonic Antigen (CEA): Past, Presnt, and Future. McGill J. Med. 1997;3:1. doi: 10.26443/mjm.v3i1.472. [DOI] [Google Scholar]
- 44.Turriziani M., Fantini M., Benvenuto M., Izzi V., Masuelli L., Sacchetti P., Modesti A., Bei R. Carcinoembryonic antigen (CEA)-based cancer vaccines: Recent patents and antitumor effects from experimental models to clinical trials. Recent Pat. Anti-Cancer Drug Discov. 2012;7:265–296. doi: 10.2174/157489212801820020. [DOI] [PubMed] [Google Scholar]
- 45.Lei W., Xie M., Jiang Q., Xu N., Li P., Liang A., Young K.H., Qian W. Treatment-Related Adverse Events of Chimeric Antigen Receptor T-Cell (CAR T) in Clinical Trials: A Systematic Review and Meta-Analysis. Cancers. 2021;13:3912. doi: 10.3390/cancers13153912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Husain A.N., Colby T.V., Ordonez N.G., Allen T.C., Attanoos R.L., Beasley M.B., Butnor K.J., Chirieac L.R., Churg A.M., Dacic S., et al. Guidelines for Pathologic Diagnosis of Malignant Mesothelioma 2017 Update of the Consensus Statement From the International Mesothelioma Interest Group. Arch. Pathol. Lab. Med. 2018;142:89–108. doi: 10.5858/arpa.2017-0124-RA. [DOI] [PubMed] [Google Scholar]
- 47.Ferrandez-Izquierdo A., Llombart-Bosch A. Immunohistochemical characterization of 130 cases of primary hepatic carcinomas. Pathol. Res. Pract. 1987;182:783–791. doi: 10.1016/S0344-0338(87)80043-2. [DOI] [PubMed] [Google Scholar]
- 48.Wang L., Vuolo M., Suhrland M.J., Schlesinger K. HepPar1, MOC-31, pCEA, mCEA and CD10 for distinguishing hepatocellular carcinoma vs. metastatic adenocarcinoma in liver fine needle aspirates. Acta Cytol. 2006;50:257–262. doi: 10.1159/000325951. [DOI] [PubMed] [Google Scholar]
- 49.Rishi M., Kovatich A., Ehya H. Utility of polyclonal and monoclonal antibodies against carcinoembryonic antigen in hepatic fine-needle aspirates. Diagn. Cytopathol. 1994;11:358–361; discussion 361–362. doi: 10.1002/dc.2840110409. [DOI] [PubMed] [Google Scholar]
- 50.Lau S.K., Prakash S., Geller S.A., Alsabeh R. Comparative immunohistochemical profile of hepatocellular carcinoma, cholangiocarcinoma, and metastatic adenocarcinoma. Hum. Pathol. 2002;33:1175–1181. doi: 10.1053/hupa.2002.130104. [DOI] [PubMed] [Google Scholar]
- 51.Schiemann U., Gunther S., Gross M., Henke G., Muller-Koch Y., Konig A., Muders M., Folwaczny C., Mussack T., Holinski-Feder E. Preoperative serum levels of the carcinoembryonic antigen in hereditary non-polyposis colorectal cancer compared to levels in sporadic colorectal cancer. Cancer Detect. Prev. 2005;29:356–360. doi: 10.1016/j.cdp.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 52.Zhao N., Cao Y., Yang J., Li H., Wu K., Wang J., Peng T., Cai K. Serum Tumor Markers Combined With Clinicopathological Characteristics for Predicting MMR and KRAS Status in 2279 Chinese Colorectal Cancer Patients: A Retrospective Analysis. Front. Oncol. 2021;11:582244. doi: 10.3389/fonc.2021.582244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yunlong W., Tongtong L., Hua Z. The efficiency of neoadjuvant chemotherapy in colon cancer with mismatch repair deficiency. Cancer Med. 2022;12:2440–2452. doi: 10.1002/cam4.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y., Li Y., Zhu Z., Yang J., Tan Y., Wang Y., Xu X. Development of novel models for predicting mismatch repair protein deficiency and relevant disease-free survival in colorectal cancer patients. Int. J. Color. Dis. 2022;37:1449–1464. doi: 10.1007/s00384-022-04150-6. [DOI] [PubMed] [Google Scholar]
- 55.Ghanipour L., Jirstrom K., Sundstrom M., Glimelius B., Birgisson H. Associations of defect mismatch repair genes with prognosis and heredity in sporadic colorectal cancer. Eur. J. Surg. Oncol. 2017;43:311–321. doi: 10.1016/j.ejso.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 56.Erasmus G., Mostert W.P. Factors associated with the perception of side-effects relating to the use of contraceptive methods. Curationis. 1985;8:45–47. doi: 10.4102/curationis.v8i3.619. [DOI] [PubMed] [Google Scholar]
- 57.Compton C., Fenoglio-Preiser C.M., Pettigrew N., Fielding L.P. American Joint Committee on Cancer Prognostic Factors Consensus Conference: Colorectal Working Group. Cancer. 2000;88:1739–1757. doi: 10.1002/(SICI)1097-0142(20000401)88:7<1739::AID-CNCR30>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 58.Tomita M., Ayabe T., Chosa E., Nakamura K. Postoperative Serum CEA Level is a More Significant Prognostic Factor than Post/Preoperative Serum CEA Ratio in Non-small Cell Cancer Patients. Asian Pac. J. Cancer Prev. 2015;16:7809–7812. doi: 10.7314/APJCP.2015.16.17.7809. [DOI] [PubMed] [Google Scholar]
- 59.Shimada H., Noie T., Ohashi M., Oba K., Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: A systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17:26–33. doi: 10.1007/s10120-013-0259-5. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y., Gao S.G., Chen J.M., Wang G.P., Wang Z.F., Zhou B., Jin C.H., Yang Y.T., Feng X.S. Serum CA242, CA199, CA125, CEA, and TSGF are Biomarkers for the Efficacy and Prognosis of Cryoablation in Pancreatic Cancer Patients. Cell Biochem. Biophys. 2015;71:1287–1291. doi: 10.1007/s12013-014-0345-2. [DOI] [PubMed] [Google Scholar]
- 61.Dal Bello M.G., Filiberti R.A., Alama A., Orengo A.M., Mussap M., Coco S., Vanni I., Boccardo S., Rijavec E., Genova C., et al. The role of CEA, CYFRA21-1 and NSE in monitoring tumor response to Nivolumab in advanced non-small cell lung cancer (NSCLC) patients. J. Transl. Med. 2019;17:74. doi: 10.1186/s12967-019-1828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park B.J., Cha M.K., Kim I.H. Thioredoxin 1 as a serum marker for breast cancer and its use in combination with CEA or CA15-3 for improving the sensitivity of breast cancer diagnoses. BMC Res. Notes. 2014;7:7. doi: 10.1186/1756-0500-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhlen M., Bandrowski A., Carr S., Edwards A., Ellenberg J., Lundberg E., Rimm D.L., Rodriguez H., Hiltke T., Snyder M., et al. A proposal for validation of antibodies. Nat. Methods. 2016;13:823–827. doi: 10.1038/nmeth.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L.M., et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 65.Lizio M., Harshbarger J., Shimoji H., Severin J., Kasukawa T., Sahin S., Abugessaisa I., Fukuda S., Hori F., Ishikawa-Kato S., et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lizio M., Abugessaisa I., Noguchi S., Kondo A., Hasegawa A., Hon C.C., de Hoon M., Severin J., Oki S., Hayashizaki Y., et al. Update of the FANTOM web resource: Expansion to provide additional transcriptome atlases. Nucleic Acids Res. 2019;47:D752–D758. doi: 10.1093/nar/gky1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Consortium G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.