Abstract
Soil salinity adversely affects plant growth and development, reducing the yield of most crops, including wheat. The highly salt-tolerant wheat germplasm lines W4909 and W4910 were derived from a cross between two moderately salt-tolerant lines, the Chinese Spring (CS)/Thinopyrum junceum disomic addition line AJDAj5 (AJ) and the Ph-inhibitor line (Ph-I) derived from CS/Aegilops speltoides. Molecular markers for gene introgressions in W4909 and W4910 were not reported. Four sequence-tagged site (STS) molecular markers of Ph-I were developed and tested in the above-mentioned lines and the F2 progenies of the two crosses, Anza (AZ) × 4740 (sib of W4910) and Yecora Rojo (YR) × 4728 (sib of W4909). Additionally, homogeneity was assessed in several derivatives of W4909, 4728, W4910, and 4740 using the four markers. The four STS markers are not associated with salt tolerance, but they provide an indication of the transfer of chromatin in 3B chromosome of Ae. speltoides via Ph-I. Moreover, salt tolerance and leaf sodium concentration were determined in CS, AJ, Ph-I, 7151 (progeny of W4909), 7157 (progeny of W4910), AZ, and YR under salt treatment and control. Surprisingly, AJ had the lowest leaf sodium concentration under the control and salt treatment, indicating greater sodium exclusion than that in CS, AZ, and YR. This low level of leaf sodium concentration was heritable from 4740 to its hybrid progenies. On the other hand, the higher leaf sodium concentration, indicative of the tissue tolerance to salinity in Ph-I, had been inherited by both W4909 and W4910 and then transmitted to their hybrid progenies. One offspring line each in both W4909 and W4910 (7762 and 7159, respectively) were homozygous for the three molecular markers and lacked the marker psr1205 of Su1-Ph1 gene, making them better materials than the original lines for future research on, for example, whole-genome sequencing and gene mining. The implications of these findings for the utilization of W4909 and W4910 in breeding salt-tolerant wheat cultivars are discussed.
Keywords: hexaploid wheat, Triticum aestivum, salt tolerance, sodium exclusion, tissue tolerance, specific locus-amplified fragment (SLAF), sequence-tagged sites (STS), molecular marker
1. Introduction
Due to a global change in climate, increasing soil salinization that affects soil health and constrains agricultural production has become a major land-degradation problem [1,2]. Worldwide, over 1 billion hectares of land suffer from salinization [3]. Soil salinity greatly affects bread wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) yield and quality; thus, enhancing the salt tolerance of wheat is a vital task to sustain wheat production for human consumption [4]. However, bread wheat has limited genetic variability in salt tolerance that can be broadened by introducing genes from species in the genus Thinopyrum of the Triticeae tribe, which are tolerant to salinity and can easily be hybridized with wheat.
Useful genes from wild Triticeae species can be transferred into tetraploid wheat (Triticum durum Desf., 2n = 4x = 28, AABB) and hexaploid wheat by inducing gene introgression through homoeologous chromosome pairing and suppressing or inhibiting the effect of the homologous pairing (Ph) gene pairing homoeologous 1 (Ph1) on the long arm of 5B chromosome [5]. Other methods, including irradiation and tissue culture, entail several disadvantages, such as a genetic imbalance in translocation lines involving recombination between non-homoeologous chromosomes [6].
Data on chromosome pairing between wheat and Ae. speltoides Tausch. (2n = 2x = 14, SS) revealed natural variation in homoeologous pairing, resulting in low-, intermediate- and high-pairing hybrids [7,8,9,10]. The high-pairing hybrids between wheat containing Ph1 and Ae. speltoides accessions indicated the presence of inhibitors or suppressors of Ph1 in those Ae. speltoides accessions. Consequently, the genes inhibiting and suppressing the Ph1 gene were transferred to hexaploid wheat [11,12,13]. The high-pairing Ph-inhibitor line (Ph-I) carrying the genes PhI was tentatively identified to be a translocation line involving 4D/4S, based on chromosome C-banding, and another unidentified chromosome pair [9]. Alternatively, the suppressor genes Su1-Ph1 and Su2-Ph1 were mapped to the 3S and 7S chromosome of Ae. speltoides, respectively [12]. The genome symbol for Ae. speltoides was recently changed from S to B [14,15], as suggested nearly 30 years ago [10], such that the chromosomes of Ae. speltoides will be written hereafter as 1B to 7B of Ae. speltoides.
The Ph-I line was crossed with AJDAj5, the Chinese Spring (CS)/Thinopyrum junceum disomic addition line, to produce translocation lines W4909 and its sib line 4728, and W4910 and its sib line 4740, which were tolerant of salinity up to EC = 42 dS/m [16]. These lines inherited salt tolerance from both the parental lines AJDAj5 and Ph-I, characterized by having an extremely high leaf sodium concentration, in contrast to the sodium exclusion mechanism that results in low leaf sodium concentrations. Thus, W4909 and W4910 were released as wheat germplasm lines [17]. The tissue tolerance to salinity of W4909 was substantiated and used to develop a high-yielding germplasm line MW#293 tolerant of both salinity and sodicity [18]. It was advocated that tissue tolerance of salinity would be the basis for breeding salt-tolerant wheat cultivars in the future [16]. Other than Ph-I, W4909, W4910, and MW#293, only one Portuguese landrace, Mocho de Espiga Branca, accumulates up to sixfold greater leaf and sheath sodium concentrations than two Australian cultivars, Gladius and Scout [19].
The State of California in the U.S. has a vast acreage of saline soil [3]. Wheat cultivars Yecora Rojo (YR) and Anza (AZ) are well adapted to California wheat production. Therefore, before the release of W4909 and W4910, their sib lines 4728 and 4740 were used to cross with YR and AZ, respectively.
Because the tissue tolerance of salinity was contributed from the Ph-I, it is logical to assume that gene (or genes) conferring this tolerance mechanism is (or are) located on the chromosomes transferred from Ae. speltoides accession TA1786 into Ph-I [11]. Xpsr1205, a molecular marker 0.4 cM distal to Su1-Ph1 [12] on 3B of Ae. speltoides, was used to test whether Xpsr1205 is present in Ph-I, W4909 or W4910. Additionally, three specific locus-amplified fragment sequencing (SLAF) sequences were converted into sequence-tagged sites (STS) markers and tested on parental lines and F2 hybrid derivatives of 4728 (sib of W4909) and 4740 (sib of W4910). STS markers would be useful in the marker-assisted selection (MAS) of breeding materials generated via crosses involving W4909 and/or W4910. This study aimed to achieve the following: (1) the development of STS molecular markers that are present in AJDAj5 or Ph-I and W4909 or W4910 but absent from Chinese Spring, and testing these STS markers to identify newer lines that lack the marker psr1205 for the Su1-Ph1 gene on the 3B chromosome of Ae. speltoides; and (2) the determination of the leaf sodium concentration in parental lines and F2 populations of the YR × 4728 and AZ × 4740, to elucidate the mechanisms of salinity tolerance. The results of this study reveal genetic lines without the potential for chromosome instability in hybrid derivatives and show that the two target lines have differing mechanisms for salt response.
2. Results
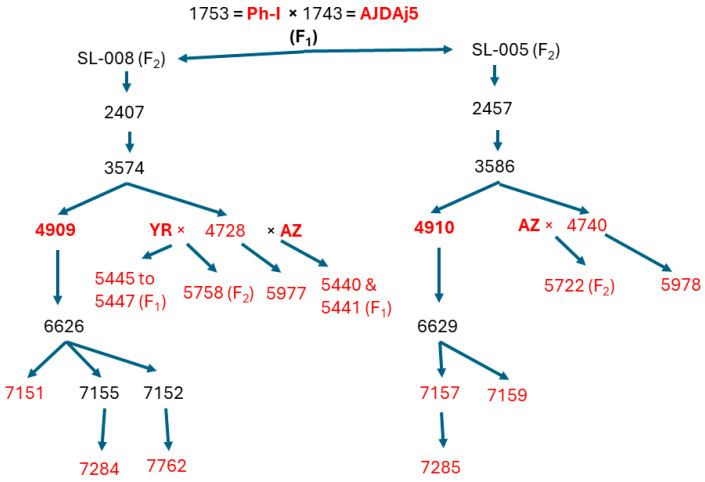
The plant materials used in this study are shown in Figure 1. Because AJDAj5 and the Ph-inhibitor line Ph-I were both developed in the CS background [11,20], W4909 and W4910 also shared the CS background. These five lines were used to generate specific locus-amplified fragment (SLAF) sequences.
Figure 1.
Pedigree of plant materials used in this study (in red color).
2.1. Development of SLAF-Derived STS Markers for W4909 and W4910
SLAF sequences were generated from DNA samples of CS, AJ, Ph-I, W4909, and W4910 and provided by Dr. Xingfeng Li of Shandong Agricultural University, Tai’an, China. An analysis of SLAF sequences yielded some potential markers of W4909 and/or W4910 (Table 1)
Table 1.
Analysis of SLAF sequences in Chinese Spring, AJDAj5, Ph-I, W4909, and W4910.
| SNP Type | Number of SNPs | Number of SLAF Markers with ≥6 SNPs |
|---|---|---|
| Different between AJDAj5 and Ph-I | 125,984 | 2030 |
| Common between W4909 and W4910 | 73,326 | 218 |
| Only in W4909 | 16,244 | 318 |
| Only in W4910 | 19,464 | 456 |
| Common between W4909 and W4910, and tracible to AJDAj5 and/or Ph-I | 20,531 | 40 |
| Total | 255,549 | 3062 |
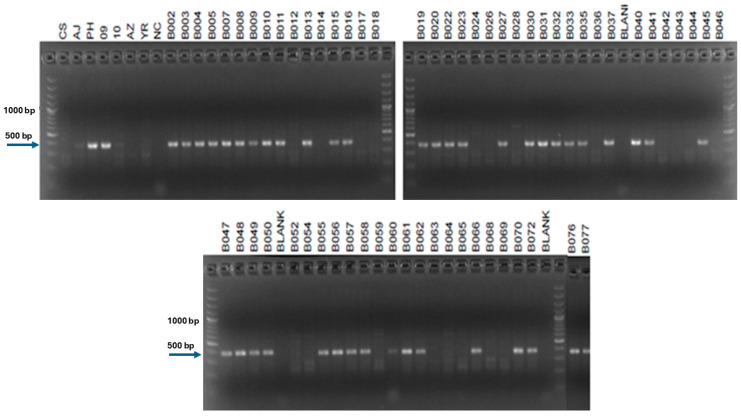
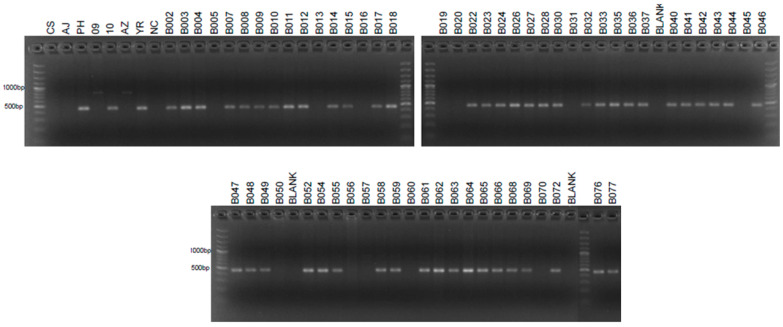
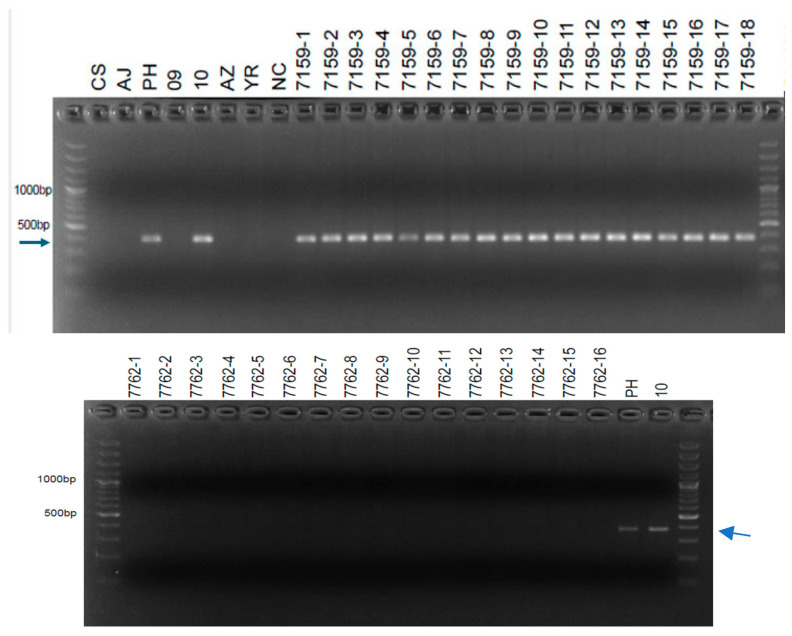
Despite testing 218 primer pairs designed from 156 SLAF sequences on CS, AJ, Ph-I, 7151 (progeny of W4909), 7157 (progeny of W4710), YR, and AZ, only four STS markers were successfully developed for W4909 and W4910 (Table 2). Additional molecular markers could be developed from other sequences identified as polymorphic between CS and W4909/W4910 (Table S1). The results of PCR amplification for Marker4607154 in parental lines and hybrid progenies were the same as those of Marker264410. Therefore, only the results of Marker264410 were presented. The random fragment length polymorphism (RFLP)-derived STS marker psr1205 was previously reported as a marker for the Ph1 suppressor gene, Su1-Ph1 [12]. Now, it is shown that this marker also exists in the Ph1 inhibitor line Ph-I, 7151 (progeny of W4909) and F2 segregants of the YR × 4728 cross (Figure 2). The SLAF-derived STS Marker264410 was present in Ph-I, W4910, YR, and many F2 segregants of YR × 4728 (Figure 3) but absent from F2 segregants of AZ × 4740 (Table 3). The genotype of markers psr1205 and Marker264410 in the 62 out of 66 F2 plants from the YR × 4728 cross suggests that the two molecular markers had a genetic distance of (4 + 21)/(4 + 21 + 17 + 20) = 40.33 centimorgan (cM) (Table 3) and a physical distance of 17.22 Mb (Table 2). The SLAF-derived STS Marker6805321 showed an identical pattern to Marker264410 in Ph-I and line 7157 (progeny of W4910) but was absent in F2 populations (lines 5722 and 5758) of both AZ × 4740 and YR × 4728 crosses (Table 3). Marker6805321 was present in F1 of YR × 4728-y (line 5445 in Table 4) and progenies of W4909 and W4910 (lines 7284, and 5978, 7159, 7285 in Table 5; 7159 in Figure 4).
Figure 2.
PCR amplification of RFLP-derived STS marker psr1205 (362 bp, arrowed) with the template DNA of Chinese Spring (CS), AJDAj5 (AJ), Ph1 inhibitor line (PH), 7151 = progeny of W4909 (09), 7157 = progeny of W4910 (10), Anza (AZ), Yecora Rojo (YR), negative control (NC), and F2 individuals (B numbers) of the cross YR × 4728-x (a sib of W4909).
Figure 3.
PCR amplification of SLAF-derived STS Marker264410 (462 bp) with template DNA of Chinese Spring (CS), AJDAj5 (AJ), Ph1 inhibitor line (PH), 7151 = progeny of W4909 (09), 7157 = progeny of W4910 (10), Anza (AZ), Yecora Rojo (YR), negative control (NC), and F2 individuals (B numbers) of the YR × 4728-x (a sib of W4909) cross.
Figure 4.
PCR amplification of SLAF-derived STS Marker6805321 (377 bp, arrows) with template DNA of Chinese Spring (CS), AJDAj5 (AJ), Ph1 inhibitor line (PH), 7151 = progeny of W4909 (09), 7157 = progeny of W4910 (10), Anza (AZ), Yecora Rojo (YR), negative control (NC), and individuals of lines 7159 (top) and 7762 (bottom), progeny lines from the lineage of W4910 and W4909, respectively.
As 4728 and 4740 were sibs of W4909 and W4910, respectively, the results for Marker264410 and Marker6805321 were puzzling; i.e., these two markers were present in 7157 (progeny of W4910) but absent from all F2 individuals of the cross AZ × 4740 (sib of W4910) (Table 3). Because seeds of lines 4728 and 4740 lost germinability due to long seed storage, we could only check the molecular marker profiles in F1 hybrids of 4728 crossed with Anza and Yecora Rojo (Table 4), as well as other progeny lines of W4909 and W4910 (Table 5), to infer their molecular genotypes.
Table 2.
Primer sequences of working STS markers derived from RFLP marker psr1205 and SLAF sequences 264410, 4607154, and 6805321.
| Marker | Chromosome Location | Primer Name | Primer Sequence 5′ --> 3′ | TA °C | Amplicon bp |
|---|---|---|---|---|---|
| Psr1205 | chr3B:541288907..541289382 | Forward | CGGCAATGATGAGTGTGTCAT | 59 to 56 | 362 |
| Reverse | CAACTCCCAGTTTGCTGACA | ||||
| Marker264410 | chr3B:558511365..558511884 | Forward | ACACTACTCATACGGAACCATCG | 55 | 462 |
| Reverse | TCTTGGCTGACTTGGCATTCA | ||||
| Marker4607154 | chr3B:558833142..558833625 | Forward | ACAAGCAACTAACAGAGCCA | 55 | 486 |
| Reverse | CTGTCGATGCAGGGTTCTACT | ||||
| Marker6805321 | chr2E:49152430..49152911 | Forward | AATGTGAACAATCAACGAGATGT | 52 | 377 |
| Reverse | GTGCACAACACACAGTGGTC |
Apparently, plants in line 4728 used in the crosses with Anza and Yecora Rojo could be classified as two genotypes. Type 4728-x was heterozygous for psr1205 and homozygous-negative for Marker264410 and Marker6805321, while the type 4728-y plants were probably negative for psr1205 and homozygous positive for Marker264410 and Marker6805321 (Table 4; Table S2). The plant 4728-x was the parent in the crosses AZ × 4728 (lines 5440 and 5441) and YR × 4728 (lines 5446 and 5447) (Table 4). The F1 hybrid of Yecora Rojo × 4728-x, that gave rise to the F2 population 5758 (Table S2) would be heterozygous for psr1205, heterozygous for M264410 that came from YR, and homozygous-negative for M6805321 (Table 3). Only the segregation ratios of 38:24 for Marker264410 (Table 3) deviated from the ratio of 3:1 in this F2 population.
Table 3.
Molecular markers in grandparents, parents, and F2 populations of Anza × 4740 and Yecora Rojo × 4728. Green + indicates Yecora Rojo as the source of Marker264410 in the F2 population.
| ID 1 | Description | No. Plants | psr1205 | Marker264410 | Marker6805321 |
|---|---|---|---|---|---|
| 6687 | Chinese Spring | 8 | - | - | - |
| 6598 | AJDAj5 | 7 | - | - | - |
| 6621 | Ph inhibitor | 8 | + | + | + |
| 7151 | Progeny of W4909 | 8 | + | - | - |
| 7157 | Progeny of W4910 | 9 | - | + | + |
| 5976 | Yecora Rojo | 9 | - | + | - |
| 5975 | Anza | 9 | - | - | - |
| 5722 | (Anza × 4740) F2 | 91 | - | - | - |
| Total | 91 | ||||
| 5758 | (Yecora Rojo × 4728-x) F2 | 4 | - | - | - |
| 21 | + | + | - | ||
| 17 | - | + | - | ||
| 20 | + | - | - | ||
| Total | 62 |
1 ID numbers are seed packet numbers, each of them contains the seed harvested from one spike of a plant.
Table 4.
Molecular markers in parental lines and F1 hybrids of 4728 (a sib of W4909). Green + indicates Yecora Rojo as the source of Marker264410 in the F2 population. Yellow + indicates Ph-I and 7151 or 7157 as the source of markers.
| ID 1 | Kind | psr1205 | Marker264410 | Marker6805321 |
|---|---|---|---|---|
| 6687 | Chinese Spring | - | - | - |
| 6598 | AJDAj5 | - | - | - |
| 6621 | Ph inhibitor | + | + | + |
| 7151 | Progeny of W4909 | + | - | - |
| 7157 | Progeny of W4910 | - | + | + |
| 5975 | Anza | - | - | - |
| 5976 | Yecora Rojo | - | + | - |
| 5440-1 | (Anza × 4728-x) F1 | - | - | - |
| 5440-2 | (Anza × 4728-x) F1 | + | - | - |
| 5440-3 | (Anza × 4728-x) F1 | - | - | - |
| 5440-4 | (Anza × 4728-x) F1 | - | - | - |
| 5440-5 | (Anza × 4728-x) F1 | + | - | - |
| 5440-6 | (Anza × 4728-x) F1 | + | - | - |
| 5440-7 | (Anza × 4728-x) F1 | + | - | - |
| 5440-8 | (Anza × 4728-x) F1 | + | - | - |
| 5441-1 | (Anza × 4728-x) F1 | + | - | - |
| 5441-2 | (Anza × 4728-x) F1 | - | - | - |
| 5441-3 | (Anza × 4728-x) F1 | + | - | - |
| 5441-4 | (Anza × 4728-x) F1 | - | - | - |
| 5441-5 | (Anza × 4728-x) F1 | - | - | - |
| 5441-6 | (Anza × 4728-x) F1 | + | - | - |
| 5445-1 | (Yecora Rojo × 4728-y) F1 | - | + | + |
| 5445-2 | (Yecora Rojo × 4728-y) F1 | - | + | + |
| 5445-3 | (Yecora Rojo × 4728-y) F1 | - | + | + |
| 5445-4 | (Yecora Rojo × 4728-y) F1 | - | + | + |
| 5445-5 | (Yecora Rojo × 4728-y) F1 | - | + | + |
| 5446-1 | (Yecora Rojo × 4728-x) F1 | + | + | - |
| 5446-2 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5446-3 | (Yecora Rojo × 4728-x) F1 | + | + | - |
| 5446-4 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5446-5 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5446-6 | (Yecora Rojo × 4728-x) F1 | + | + | - |
| 5446-7 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5446-8 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5447-1 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5447-2 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5447-3 | (Yecora Rojo × 4728-x) F1 | + | + | - |
| 5447-4 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5447-5 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5447-6 | (Yecora Rojo × 4728-x) F1 | - | + | - |
| 5447-7 | (Yecora Rojo × 4728-x) F1 | - | + | - |
1 ID numbers are seed packet numbers, each of them contains the seed harvested from one spike of a plant.
2.2. Homozygosity in Progenies of W4909 and W4910
The molecular-marker profiles of six progeny lines of W4909 and W4910, two to three generations of self-pollination from the uniformly salt-tolerant W4909 and W4910, were unknown. Therefore, they were assessed for homozygosity of the three STS markers in 16 to 20 plants per line. Lines 7762 and 7159 are homozygous for the three STS markers (Table 5). Line 7762 lacked markers M264410 and M6805321, while line 7159 included both. Both lines lacked the psr1205, which is the marker for the Su1-Ph1 gene that could cause the chromosome instability of hybrids of W4909 and W4910, arising from homoeologous chromosome pairing. Therefore, lines 7762 and 7159 are more desirable than the original W4909 and W4910 for crossing with wheat cultivars to breed salt-tolerant wheat.
Table 5.
Molecular markers in different progeny lines of W4909 and W4910. ID numbers are seed packet numbers; each contains seed harvested from a single plant. Red + indicate markers originate from Ph-I and 7151 or 7157; yellow blocks are homozygous markers in progeny lines of W4909 and W4910.
| ID | Kind | psr1205 | Marker264410 | Marker6805321 |
|---|---|---|---|---|
| CS | Chinese Spring | - | - | - |
| AJ | AJDAj5 | - | - | - |
| Ph-I | Ph inhibitor | + | + | + |
| 7151 | Progeny of W4909 | + | - | - |
| 7157 | Progeny of W4910 | - | + | + |
| AZ | Anza | - | - | - |
| YR | Yecora Rojo | - | + | - |
| NC | Negative control | - | - | - |
| 5977 | Progeny of W4909 | 8 : 12 + : - | 0 : 20 + : - | 0 : 20 + : - |
| 7284 | Progeny of W4909 | 0 : 20 + : - | 1 : 19 +: - | 1 : 19 + : - |
| 7762 | Progeny of W4909 | 0 : 16 + : - | 0 : 16 + : - | 0 : 16 + : - |
| 5978 | Progeny of W4910 | 0 : 20 + : - | 17 : 3 + : - | 18 : 2 + : - |
| 7159 | Progeny of W4910 | 0 : 18 + : - | 18 : 0 + : - | 18 : 0 + : - |
| 7285 | Progeny of W4910 | 0 : 20 + : - | 18 : 2 + : - | 20 : 0 + : - |
2.3. Survival, Leaf Sodium Concentration in Parents, and Progenies of W4909 and W4910
AJDAj5 had the lowest leaf sodium ion concentration in both control and salt-treated plants, although that in salt-treated plants was not statistically significantly different from CS, YR, and AZ (Table 6). Ph-I, W4909, and W4910 had significantly higher leaf sodium concentrations than the other four lines under both the control and salt-stressed conditions. The survival rate, leaf sodium concentrations, and marker profiles are presented in Table S3. When percentage survival at a 52-day cutoff between tissue tolerance and susceptibility to salinity was used, AJDAj5, Ph-I, W4909, and W4910 were all more tolerant to salt stress than CS, AZ, and YR.
Table 6.
Mean and 95% confidence interval (CI) of leaf sodium concentration of parental lines. Means with the same letter are not significantly different at p = 0.05 level.
| Plant Materials | Control at EC = 0.6 dS/m | Salt-Treated at EC = 24 dS/m | ||
|---|---|---|---|---|
| Name (ID Number 1) | Mean | 95% CI | Mean | 95% CI |
| Chinese Spring | 1858 b | 1170 to 2546 | 6262 a | 4623 to 7901 |
| (6687) | ||||
| AJDAj5 | 475 a | 222 to 728 | 4801 a | 3002 to 6600 |
| (6598) | ||||
| Ph inhibitor | 4287 c | 2717 to 5857 | 13,016 b | 9821 to 16,211 |
| (6621) | ||||
| W4909 | 5818 c | 5275 to 6561 | 11,943 b | 10,647 to 13,239 |
| (7151) | ||||
| W4910 | 5450 c | 3911 to 6989 | 12,030 b | 10,613 to 13,447 |
| (7157) | ||||
| Anza | 2083 b | 1513 to 2653 | 6610 a | 4746 to 8474 |
| (5975) | ||||
| Yecora Rojo | 1479 b | 1258 to 1700 | 5793 a | 4198 to 7388 |
| (5976) | ||||
1 ID numbers are seed packet numbers.
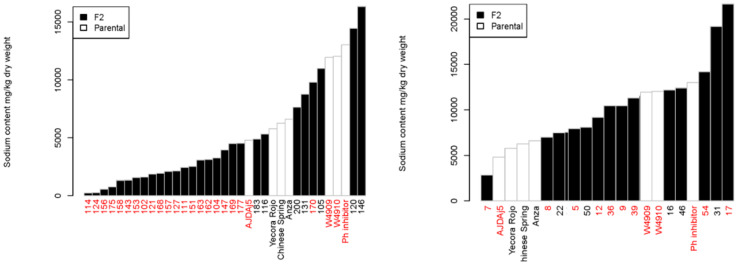
In addition to parental materials, the survival rate in terms of survival days since treatment (SDST) was measured on F2 plants from AZ × 4740 and YR × 4728 crosses (Table S4), and the leaf sodium ion concentration was measured in randomly selected F2 plants (24 of 95 and 14 of 66, respectively) of those two crosses (Figure 5). In the AZ × 4740 cross, 20 salt-tolerant F2 plants out of 24 measured plants had low leaf sodium concentrations, a trait apparently inherited from AJDAj5. On the other hand, 6 out of 10 salt-tolerant F2 plants in the YR × 4728 cross had a high sodium concentration in leaves, a trait attributable to Ph-I. However, a high leaf sodium concentration alone could not confer or account for salt tolerance in F2 segregants in both crosses, such as plants 16, 31, and 46 in YR × 4728 and plants 120, 146, and 170 in AZ × 4740. There is, however, strong evidence (p-value of the Welch two-sample t-test = 0.003529) in the AZ × 4740 cross that salt-tolerant F2 individuals had lower sodium ion concentrations. This supports the conclusion that the mechanism from 4740 (sib of W4910) is sodium exclusion, but tissue tolerance was contributed from 4728 (sib of W4909) in the YR × 4728 cross.
Figure 5.
Leaf sodium concentration in F2 individuals of the crosses Anza × 4740 (left) and Yecora Rojo × 4728 (right). Plants with red identifications are salt-tolerant. Parental lines are represented by white bars.
2.4. Segregation of Salt Tolerance in F2 Populations of the Two Crosses, Anza × 4740 and Yecora Rojo × 4728
F2 individuals of the crosses AZ × 4740 and YR × 4728 segregated in a 3:1 and 7:9 ratio, respectively (Table 7). Thus, a single dominant gene conferred salt tolerance in the hybrid derivatives of AZ × 4740, and two supplementary recessive genes controlled the trait in YR × 4728. STS markers in these two F2 populations (Table S4) did not segregate at these ratios, indicating that these molecular markers were not associated with genes for salt tolerance. Again, these markers are merely indications of the presence of chromatin from the 3B chromosome of Ae. speltoides.
Table 7.
Segregation of salt tolerance in F2 populations of Anza × 4740 and Yecora Rojo × 4728-x.
| ID 1 | Cross | No. Plants | SDST 2 | No. Plants | Segregation Ratio | Chi-Square | p |
|---|---|---|---|---|---|---|---|
| 5722 | Anza × 4740 | 95 | >52 | 67 | 3 : 1 | 1.0140 | 0.3139 |
| 51.8 (50 to 52) | 28 | ||||||
| 5758 | Yecora Rojo × 4728-x | 66 | >52 | 29 | 7 : 9 | 0.0011 | 0.9735 |
| 49.0 (42 to 52) | 37 |
1 ID numbers are seed packet numbers. 2 SDST = survival days since treatment with NaCl salt; >52 = salt-tolerant; ≤52 = salt-sensitive.
3. Discussion
Both AJDAj5 [20] and Ph-I [9] were developed in the CS background with different alien chromosomes of Th. junceum and Ae. speltoides, respectively. The strong band of STS marker psr1205 was present in Ph-I and 7151 (progeny of W4909), as well as some F2 segregants in the YR × 4728 cross (Figure 2). Thus, the intense band possibly originated from the 3B chromosome of Ae. speltoides. This result leads to both implications and applications. Firstly, the presence of marker psr1205 in Ph-I line suggests that Ph-I contains the 3B chromosome of Ae. speltoides, as reported previously [21]. If the PhI gene is indeed located on 4B involving in the 4D/4S (=4D/4B) translocation chromosome, as reported earlier [11], then the unidentified chromosome in the Ph-I line would be the 3B chromosome of Ae. speltoides. Secondly, if both Su1-Ph1 and PhI are located on the 3B chromosome of Ae. speltoides, shown in a previous study [21] and confirmed in the current study, it raises the possibility that PhI and Su1-Ph1 are either different alleles of the same gene or two different genes located on the same chromosome. These possibilities call for future experiments, such as testcrossing them with the same plant material that has alien chromosomes and Ph1 to determine any differences between the progenies. Also, the genetic and physical distances between PhI and Su1-Ph1 should be assessed to determine whether they are different genes located on the same chromosome. The data in the previous study [21] indicated that wmc674 and wmc505 were on the short arm of Ae. speltoides 3B chromosome, whereas the three markers in the present study were near the distal end of 3B long arm. Despite the two studies confirming that the Ph-I line contained segments of both 3BS and 3BL of Ae. speltoides, the precise position of PhI gene is still unknown.
An annotation of the 3B chromosome of Ae. speltoides spanning 13 Mb revealed many candidate genes for salt tolerance near markers psr1205 and Marker264410 reported in this study (Table S5). Some of them had been implicated in the transcriptome study of Chinese Spring, AJDAj5, Ph-I, W4909, and W4910 [22]. These include genes encoding peroxidase, Ser-Thr protein kinase, Myb transcription factor, late embryogenesis abundant protein LEA_2, glutathione S-transferase, calmodulin-binding domain, calcium-dependent vacuole membrane protein, and MIP aquaporin, etc. Although the STS markers identified in this study are not associated with salt tolerance, DNA sequences on 3B chromosome of Ae. speltoides flanking the candidate salt-tolerance genes could be developed as molecular markers useful in MAS for breeding salt-tolerant wheat cultivars.
Na+ exclusion from leaves is associated with salt tolerance in cereal crops, including durum wheat [23,24], bread wheat [25,26,27], and wild relatives such as Hordeum species [28], tall wheatgrass [29], and Triticum tauschii [30]. The bread wheat cultivars ‘Berkut’ and ‘Krichauff’ had Na+ concentration (mg kg−1 DW) of 6308 ± 296 and 5942 ± 442, respectively, while the double haploid lines derived from the hybrid of these two parents had a corresponding value ranging from 2850 to 9733 [31]. Of the five QTL identified for Na+ exclusion, two were co-located with seedling biomass on chromosomes 2A and 6A. The 2A QTL appears to coincide with the previously reported Na+ exclusion locus in durum wheat that hosts one active HKT1;4 (Nax1) and one inactive HKT1;4 gene. Their measurements were comparable to those of CS, YR, and AZ in this study. Fourteen of the twenty salt-tolerant plants in the F2 population of AZ × 4740 had leaf sodium concentrations less than the lowest value of 2850 mg kg−1 DW observed in the doubled-haploid population [31]. The strong sodium-exclusion gene was inherited from AJDAj5 that can be traced to Thinopyrum junceum [16,20]. This gene has not been identified or mapped; thus, no new gene name is given here. Future research is needed to identify and isolate this gene.
Genes controlling sodium exclusion in wheat, Nax1 and Nax2, played a significant role in wheat breeding for salt tolerance [32]. Nax1, which accounted for 38% of the phenotypic variation for a low Na+ concentration in leaf blades, was mapped to the long arm of chromosome 2A via a quantitative trait locus (QTL) analysis [33]. It was identified through fine mapping as an Na+ transporter of the HKT gene family HKT7 (HKT1;4) [34]. Nax2 was previously located at chromosome 5A of T. monococcum and identified as HKT1;5 [35]. However, it was not present in 4A of T. uratu and T. aestivum but mapped to 4B and 4D [36].
The plasma membrane sodium/proton exchanger salt-overly-sensitive 1 (SOS1) is a critical Na+ efflux protein in plants. Gao et al. (2023) cloned three homologues of the TaSOS1 gene in bread wheat, designated as TaSOS1-A1, TaSOS1-B1, and TaSOS1-D1, respectively, according to the location on group 3 chromosomes 3A, 3B, and 3D [37]. Other transporters that may allow the influx of sodium ions have been reported, such as AtPIP2;1 (aquaporin) [37]. In the wheat microarray study of Mott and Wang (2007) [22], the tonoplast aquaporin (Ta.21042; TC205156) was expressed at a higher level in the leaf of Ph-I, W4909, and W4910 than in CS and AJDAj5 under both the control and salt treatments. Another one, aquaporin (TaAffx.8804), appeared to be common to AJDAj5, W4909, and W4910. A potassium-channel protein (Ta.25613) was expressed intermediately in AJDAj5, W4909, and W4910 between CS and Ph-I. Another potassium-channel protein (TaAffx.56132) was lower in AJDAj5, W4909, and W4910 than CS and Ph-I.
Using the normalized salt stress-specific expression datasets developed by Mott and Wang (2007) [22], Mehta et al. (2021) studied the shoot and root tissue-specific expression of the identified genes during the tillering stage [38]. K+/Na+ selectivity in wheat under salt stress was enhanced via Lophopyrum elongatum chromosome arms 1ES, 7ES, and 7EL [39]. Recently, both 7E from Th. elongatum (2n = 14; EE) and 7E1L of Th. ponticum (2n = 70; EEEEEEEEEE) were shown to greatly mitigate the effects of salt stress on root and leaf growth [40]. 7E and 7E1L also enhanced the ability of plants to neutralize ROS and limit their harmful effects via the presence of efficient scavenging systems, involving enzymatic and non-enzymatic antioxidants, including superoxide dismutase (SOD), catalase (CAT), peroxidase (POD), and ascorbate peroxidase (APX) enzymes, as well as ascorbate [40]. Some of these enzymes had been implicated in the microarray study of W4909 and W4910 [22]. These reports support our observation that both W4909 and W4910 had SLAF sequences traceable to the 7E chromosome that carries many candidate genes for salt tolerance (Table S6). Unfortunately, those SLAF sequences were not successfully converted to STS markers in this study. In the future, those SLAF sequences might be converted to other SNP molecular markers.
The genetic distance between molecular markers psr1205 and Marker264410 was fairly large, 40.33 cM. This fact could explain why the marker psr1205 was easily eliminated from the offspring of W4909 (Table 5). The Su1-Ph1 gene linked to psr1205 can lead to chromosome instability, resulting in a low seed set. By selecting a high seed yield in the hybrid progenies of W4909, psr1205 and the Su1-Ph1 gene could be eliminated.
4. Materials and Methods
4.1. Plant Materials
The plant materials used in this study (Figure 1) included bread wheat cultivars Chinese Spring (CS), Anza (AZ), and Yecora Rojo (YR), as well as germplasm lines AJDAj5 (AJ), Ph-inhibitor line (Ph-I), W4909, and W4910. Sibs and offspring of W4909 and W4910, which had uniform salinity tolerance for two self-pollination generations from lines 2407 and 2457, respectively [17], were also analyzed for the three STS markers to select lines lacking psr1205. F1 and F2 of YR and AZ crossed by 4728 (sib of W4909) and F2 of AZ × 4740 (sib of W4910) were analyzed to ascertain their molecular marker profiles. Seeds of the above wheat lines were stored in a refrigerator at 2 °C prior to various studies.
4.2. Greenhouse Study of Salt Tolerance
This study was conducted at a greenhouse located at the USDA-ARS, FRR, on the campus of Utah State University, Logan. The experiment was conducted using a completely randomized split-plot design with two parts to test the analyzed materials’ salt tolerance. The first part involved testing CS, AJ, Ph-I, W4909, W4910, YR, and AZ under control and salt treatment. The second entailed testing the F2 progenies of YR × 4728 and AZ × 4740 for salt tolerance under salt treatment. One hundred and forty plastic stadium cups (900 mL capacity; without drainage) were used for the first testing, which consisted of 10 cups each for the control and salt treatment of seven lines. Within the control and salt-treatment plot, the 70 cups were randomly arranged. In the second part, up to one hundred cups each were used to accommodate the F2 progenies of YR × 4728 and AZ × 4740. A single seed of the plant materials was directly placed into the silica sand-filled cup in this study. Seventy-seven and one hundred seeds were planted, but 66 and 95 seedlings were established for the YR × 4728 and AZ × 4740 F2 populations, respectively. All seedlings of the F2 populations were treated with salt solution the same way as the parental lines in the first part of the experiment.
Plants were grown for 80 days under ambient solar radiation, while the air temperature and relative humidity remained relatively stable at 27 °C and 35%, respectively. Each cup received 50 mL of water-soluble nutrient solution (20-20-20 NPK with micronutrients; Scotts Miracle-Gro Products Inc., Marysville, OH, USA) and was irrigated with deionized water daily to maintain field capacity, i.e., 11.5% soil-water content. Gravimetric soil-water content was determined by weighing individual containers on an electronic microbalance and adding water as needed to reach field capacity. Salinity treatments were imposed via watering with a saline solution of EC = 3 dS/m once a week for 8 weeks, starting when the plants’ fourth leaves had developed. Salt tolerance was determined using survival days since the treatment (SDST) of each plant [41]. Plants with SDST greater than 52 were classified as salt-tolerant. Salt-sensitive plants died between 42 and 52 days after the first salt treatment.
4.3. Molecular Characterization of Parental Lines and Hybrid Progenies
DNA was extracted from the third leaf of these plants using the QIAGEN DNeasy kit, following the manufacturer’s protocol (Germantown, MD, USA. Three DNA samples each from CS, AJ, Ph-I, W4909, and W4910 were used to generate SLAF sequences. Each SLAF sequence is composed of two 100-bp DNA sequences flanking unknown nucleotides of a varying length, which was represented as (N)10. The sequence of each SLAF marker was analyzed using the BLAST function against the whole-genome sequences of Chinese Spring v2.1 [42], Thinopyrum elongatum v1.0 [43], and Aegilops speltoides TS01 [15] using WheatOmics JBrowse (http://wheatomics.sdau.edu.cn) [44]. If marker sequences revealed an identity closer to the alien sequences than to the CS sequences, they were classified as either Th. Junceum-originated or Ae. speltoides-originated. These markers were aligned with their homoeologous wheat sequences to find variable sites that could be used for primer design at the website https://www.ncbi.nlm.nih.gov/tools/primer-blast/, 27 November 2024.
The primer sequences for Xpsr1205 and SLAF-derived STS markers are listed in Table 2. The complex PCR condition for Xpsr1205 was provided by Dr. Karin Deal, UC-Davis, as follow: 96° 5 m, 8× (94 °C 30 s 59 °C 30 s 68 °C 1 min), 8× (94 °C 30 s 57 °C 30 s 68 °C 1 min), 20× (94 °C 30 s 56 °C 30 s 68 °C 1 min), 68 °C 5 min, and 10 °C indefinite. The PCR reagent (25 µL) contained NEB (New England Biolabs, Ipswich, MA, USA) standard Taq polymerase 5 µL, primer pair 1 µL, and template DNA 1 µL, and 18 µL of double-distilled water (ddH2O) was used in the PCR of Xpsr1205. PCR for the Marker264410 and Marker6407154 was carried out using the reagent containing 5X GoTaq flexi 2 µL, GoTaq Flexi G2 Hot Start Polymerase 0.05 µL, primer pair 1 µL, template DNA 1 µL, dNTPs (25 mM) 1 µL, MgCl2 1 µL, and ddH2O with the program set for initial denaturing at 95 °C for 2 min, 30 cycles of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min, with the final extension at 72 °C for 5 min and the plate held at 4 °C for infinity. PCR products were separated in 1% agarose gels and photographed.
4.4. Molecular Markers in Offspring of W4909 and W4910
Sixteen to twenty plants of lines 5977, 7151, 7284, and 7762 from the lineage of W4909, as well as 5978, 7157, 7159, and 7285 from the lineage of W4910, were grown in pots. Plants of these lines were analyzed for the uniformity of the three STS markers, psr1205, Marker264410, and Marker6805321. DNA extraction and PCR conditions were the same as previously described.
4.5. Determination of Sodium Ion Concentration in Leaves
Three randomly selected plants of CS. AJ, Ph-I. W4909, W4910, YR, and AZ, as well as fourteen and twenty-four randomly selected plants from sixty-six and ninety-five established plants in F2 of YR × 4728 and AZ × 4740, respectively, were used for leaf sodium ion analysis. The penultimate leaf (the leaf just below the flag leaf) was collected from three each of the control and salt-treated plants of CS, AJ, Ph-I, W4909, W4910, YR, and AZ. The leaf sodium ion (Na+) concentration (mg Kg−1 DW) was determined in three dried leaf samples per line per treatment via inductively coupled plasma mass spectrometry (ICP-MS) at the Utah State University Analytical Laboratory. The mean sodium ion concentration of parental wheat lines was statistically tested using a one-way ANOVA at the p = 0.05 level.
5. Conclusions
In this study, we presented data showing that genes for sodium exclusion and tissue tolerance were traceable to AJDAj5 and Ph-I via lines 4740 and 4728, sib lines of W4910 and W4909, respectively. A strong band of the STS marker psr1205 located to the 3B chromosome of Aegilops speltoides was attributed to the Ph-I line. Three additional SLAF sequence-derived STS markers were also identified in Ph-I and some progenies of W4909 and W4910. The heterogeneity of molecular markers was observed in the early generations of W4909 and W4910, resulting in variations in molecular marker profiles among F1 hybrids. One homozygous line each (line 7762 and line 7159) in the progenies of W4909 and W4910, respectively, were identified as desirable plant materials for future research, such as whole-genome sequencing and mining for genes conferring salt tolerance, due to their lacking the marker psr1205 of Su1-Ph1 gene.
Acknowledgments
We thank the following people: Xingfeng Li, Shandong Agricultural University, Tai’an, Shandong, China, for providing SLAF-seq data on the five parental lines (CS, AJ, Ph-I, W4909, and W4910); Karin Deal and Jan Dvořák, UC-Davis, CA, for providing advice on the PCR amplification of Xpsr1205; Steve Larson, USDA-ARS-FRRL, Logan, UT, for a helpful discussion on the content of this manuscript; and Anna Hafele and Ramsey Buffham, USDA-ARS-FRRL, Logan, UT, for technical assistance and greenhouse help, respectively.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms252312892/s1.
Author Contributions
Conceptualization, R.R.-C.W.; data curation, R.R.-C.W.; funding acquisition, S.S.X.; investigation, R.R.-C.W.; methodology, T.A.M.; resources, S.S.X.; software, M.D.R.; validation, M.D.R.; visualization, M.D.R.; writing—original draft, R.R.-C.W.; writing—review and editing, S.S.X., T.A.M., and M.D.R. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary materials; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by USDA-ARS CRIS 2080-21000-014-00D.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Tarolli P., Luo J., Park E., Barcaccia G., Masin R. Soil salinization in agriculture: Mitigation and adaptation strategies combining nature-based solutions and bioengineering. iScience. 2024;27:108830. doi: 10.1016/j.isci.2024.108830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamran M., Parveen A., Ahmar S., Malik Z., Hussain S., Chattha M.S., Saleem M.H., Adil M., Heidari P., Chen J.T. An Overview of Hazardous Impacts of Soil Salinity in Crops, Tolerance Mechanisms, and Amelioration through Selenium Supplementation. Int. J. Mol. Sci. 2019;21:148. doi: 10.3390/ijms21010148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang N., Chen S., Huang J., Frappart F., Taghizadeh R., Zhang X., Wigneron J.P., Xue J., Xiao Y., Peng J., et al. Global Soil Salinity Estimation at 10 m Using Multi-Source Remote Sensing. J. Remote Sens. 2024;4:0130. doi: 10.34133/remotesensing.0130. [DOI] [Google Scholar]
- 4.Mujeeb-Kazi A., Munns R., Rasheed A., Ogbonnaya F.C., Ali N., Hollington P., Dundas I., Saeed N., Wang R.R.C., Rengasamy P., et al. Breeding strategies for structuring salinity tolerance in wheat. In: Sparks D., editor. Advances in Agronomy. Volume 155. Academic Press; London, UK: 2019. pp. 121–187. Chapter 5. [Google Scholar]
- 5.Koo D.-H., Friebe B., Gill B.S. Homoeologous Recombination: A Novel and Efficient System for Broadening the Genetic Variability in Wheat. Agronomy. 2020;10:1059. doi: 10.3390/agronomy10081059. [DOI] [Google Scholar]
- 6.Jiang J.M., Friebe B., Gill B.S. Recent advances in alien gene transfer in wheat. Euphytica. 1994;73:199–212. doi: 10.1007/BF00036700. [DOI] [Google Scholar]
- 7.Riley R., Kimber G., Chapman V. Origin of genetic control of diploid-like behavior of polyploid wheat. J. Hered. 1961;52:22–25. doi: 10.1093/oxfordjournals.jhered.a107015. [DOI] [Google Scholar]
- 8.Dvořák J. Genetic variability in Aegilops speltoides affecting homoeologous pairing in wheat. Can. J. Genet. Cytol. 1972;14:371–380. doi: 10.1139/g72-046. [DOI] [Google Scholar]
- 9.Kimber G., Athwal R.S. A reassessment of the course of evolution of wheat. Proc. Natl. Acad. Sci. USA. 1972;69:912–915. doi: 10.1073/pnas.69.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R.R.-C. Understanding the effect of the Ph gene of wheat on chromosome pairing and its implications. In: Kimber G., editor. Proceedings 2nd International Symposium of Chromosome Engineering in Plants. University of Missouri; Columbia, MO, USA: 1990. pp. 259–263. [Google Scholar]
- 11.Chen P.-D., Tsujimoto H., Gill B.S. Transfer of PhI genes promoting homoeologous pairing from Triticum speltoides to common wheat. Theor. Appl. Genet. 1994;88:97–101. doi: 10.1007/BF00222400. [DOI] [PubMed] [Google Scholar]
- 12.Dvořák J., Deal K.R., Luo M.-C. Discovery and mapping of wheat Ph1 suppressors. Genetics. 2006;174:17–27. doi: 10.1534/genetics.106.058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Deal K.R., Luo M.-C., Ji W., Distelfeld A., Dvorak J. Introgression of the Aegilops speltoides Su1-Ph1 Suppressor into Wheat. Front. Plant Sci. 2017;8:2163. doi: 10.3389/fpls.2017.02163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avni R., Lux T., Minz-Dub A., Millet E., Sela H., Distelfeld A., Deek J., Yu G., Steuernagel B., Pozniak C., et al. Genome sequences of three Aegilops species of the section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. Plant J. 2022;110:179–192. doi: 10.1111/tpj.15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L.-F., Zhang Z.-B., Wang Z.-H., Li N., Sha Y., Wang X.-F., Ding N., Li Y., Zhao J., Wu Y., et al. Genome sequences of five Sitopsis species of Aegilops and the origin of polyploid wheat B subgenome. Mol. Plant. 2022;15:488–503. doi: 10.1016/j.molp.2021.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Wang R.R.-C., Li X.-M., Hu Z.-M., Zhang J.-Y., Larson S.R., Zhang X.-Y., Grieve C.M., Shannon M.C. Development of salinity-tolerant wheat recombinant lines from a wheat disomic addition line carrying a Thinopyrum junceum chromosome. Int. J. Plant Sci. 2003;164:25–32. doi: 10.1086/344556. [DOI] [Google Scholar]
- 17.Wang R.R.-C., Larson S.R., Horton W.H., Chatterton N.J. Registration of W4909 and W4910 bread wheat germplasm lines with high salinity tolerance. Crop. Sci. 2003;43:746. doi: 10.2135/cropsci2003.0746. [DOI] [Google Scholar]
- 18.Genc Y., Taylor J., Lyons G., Li Y., Cheong J., Appelbee M., Oldach K., Sutton T. Bread Wheat with High Salinity and Sodicity Tolerance. Front. Plant Sci. 2019;10:1280. doi: 10.3389/fpls.2019.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borjigin C., Schilling R.K., Bose J., Hrmova M., Qiu J., Wege S., Situmorang A., Byrt C., Brien C., Berger B., et al. A single nucleotide substitution in TaHKT15-D controls shoot Na+ accumulation in bread wheat. Plant Cell Environ. 2020;43:2158–2171. doi: 10.1111/pce.13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charpentier A. Production of disomic addition lines and partial amphiploids of Thinopyrum junceum on wheat. C R Acad. Sci. Paris. 1992;315:551–557. [Google Scholar]
- 21.Li H., Gill B.S., Wang X., Chen P.-D. A Ta1-PhI wheat genetic stock facilitates efficient alien introgression. Genet. Resour. Crop Evol. 2011;58:667–678. doi: 10.1007/s10722-010-9609-x. [DOI] [Google Scholar]
- 22.Mott I.W., Wang R.R.-C. Comparative transcriptome analysis of salt-tolerant wheat germplasm lines using wheat genome arrays. Plant Sci. 2007;173:327–339. doi: 10.1016/j.plantsci.2007.06.005. [DOI] [Google Scholar]
- 23.Munns R., James R.A. Screening Methods for Salinity Tolerance: A Case Study with Tetraploid Wheat. Plant Soil. 2003;253:201–218. doi: 10.1023/A:1024553303144. [DOI] [Google Scholar]
- 24.James R.A., Blake C., Zwart A.B., Hare R.A., Rathjen A.J., Munns R. Impact of ancestral wheat sodium exclusion genes Nax1 and Nax2 on grain yield of durum wheat on saline soils. Funct. Plant Biol. 2012;39:609–618. doi: 10.1071/FP12121. [DOI] [PubMed] [Google Scholar]
- 25.Cuin T.A., Tian Y., Betts S.A., Chalmandrier R., Shabala S. Ionic relations and osmotic adjustment in durum and bread wheat under saline conditions. Funct. Plant Biol. 2009;36:110–119. doi: 10.1071/FP09051. [DOI] [PubMed] [Google Scholar]
- 26.James R.A., Blake C., Byrt C.S., Munns R. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J. Exp. Bot. 2011;62:2939–2947. doi: 10.1093/jxb/err003. [DOI] [PubMed] [Google Scholar]
- 27.Cuin T.A., Parsons D., Shabala S. Wheat cultivars can be screened for salinity tolerance by measuring leaf chlorophyll content and shoot sap potassium. Funct. Plant Biol. 2010;37:255–263. doi: 10.1071/FP09229. [DOI] [Google Scholar]
- 28.Garthwaite A.J., von Bothmer R., Colmer T.D. Salt tolerance in wild Hordeum species is associated with restricted entry of Na+ and Cl− into the shoots. J. Exp. Bot. 2005;56:2365–2378. doi: 10.1093/jxb/eri229. [DOI] [PubMed] [Google Scholar]
- 29.Colmer T.D., Flowers T.J., Munns R. Use of wild relatives to improve salt tolerance in wheat. J. Exper. Bot. 2006;57:1059–1078. doi: 10.1093/jxb/erj124. [DOI] [PubMed] [Google Scholar]
- 30.Schachtman D.P., Munns R., Whitecross M.I. Variation in Sodium Exclusion and Salt Tolerance in Triticum tauschii. Crop Sci. 1991;1:992–997. doi: 10.2135/cropsci1991.0011183X003100040030x. [DOI] [Google Scholar]
- 31.Genc Y., Oldach K., Verbyla A.P., Lott G., Hassan M., Tester M., Wallwork H., McDonald G.K. Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor. Appl. Genet. 2010;121:877–894. doi: 10.1007/s00122-010-1357-y. [DOI] [PubMed] [Google Scholar]
- 32.James R.A., Khan M.M., Neogi M.G., Zwart A.B., Munns R., Kabir M.R., Akhond M.A.Y. Impact of Nax. genes for Na+ exclusion from leaves on bread wheat yield on saline soils. J. Agron. Crop Sci. 2023;209:459–474. doi: 10.1111/jac.12643. [DOI] [Google Scholar]
- 33.Lindsay M.P., Lagudah E.S., Hare R.A., Munns R. A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct. Plant Biol. 2004;31:1105–1114. doi: 10.1071/FP04111. [DOI] [PubMed] [Google Scholar]
- 34.Huang S., Spielmeyer W., Lagudah E.S., James R.A., Platten J.D., Dennis E.S., Munns R.A. Sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006;142:1718–1727. doi: 10.1104/pp.106.088864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byrt C.S., Zhao M., Kourghi M., Bose J., Henderson S.W., Qiu J., Gilliham M., Schultz C., Schwarz M., Ramesh S.A., et al. Non-selective cation channel activity of aquaporin AtPIP2;1 regulated by Ca2+ and pH. Plant Cell Environ. 2017;40:802–815. doi: 10.1111/pce.12832. [DOI] [PubMed] [Google Scholar]
- 36.Huang S.B., Spielmeyer W., Lagudah E.S., Munns R. Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J. Exper. Bot. 2008;59:927–937. doi: 10.1093/jxb/ern033. [DOI] [PubMed] [Google Scholar]
- 37.Gao Z., Yang W., Cao Q., Gao X., Liu D., Zhang A. Diversification of three TaSOS1 genes and their roles in sodium exclusion in bread wheat (Triticum aestivum L.) J. Genet. 2023;102:32. doi: 10.1007/s12041-023-01429-7. [DOI] [PubMed] [Google Scholar]
- 38.Mehta G., Muthusamy S.K., Singh G.P., Sharma P. Identification and development of novel salt-responsive candidate gene based SSRs (cg-SSRs) and MIR gene based SSRs (mir-SSRs) in bread wheat (Triticum aestivum) Sci. Rep. 2021;11:2210. doi: 10.1038/s41598-021-81698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deal K.R., Goyal S., Dvorak J. Arm location of Lophopyrum elongatum genes affecting K+/Na+ selectivity under salt stress. Euphytica. 1999;108:193–198. doi: 10.1023/A:1003685032674. [DOI] [Google Scholar]
- 40.Tounsi S., Giorgi D., Kuzmanovic L., Jrad O., Farina A., Capoccioni A., Ben A.R., Brini F., Ceoloni C. Coping with salinity stress: Segmental group 7 chromosome introgressions from halophytic Thinopyrum species greatly enhance tolerance of recipient durum wheat. Front. Plant Sci. 2024;15:1378186. doi: 10.3389/fpls.2024.1378186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen T., Niu Y., Yang C., Liang Y., Xu J. Screening of Rice (Oryza sativa L.) Genotypes for Salinity Tolerance and Dissecting Determinants of Tolerance Mechanism. Plants. 2024;13:1036. doi: 10.3390/plants13071036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu T., Wang L., Rimbert H., Rodriguez J.C., Deal K.R., De Oliveira R., Choulet F., Keeble-Gagnère G., Tibbits J., Rogers J., et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021;107:303–314. doi: 10.1111/tpj.15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., Sun S., Ge W., Zhao L., Hou B., Wang K., Lyu Z., Chen L., Xu S., Guo J., et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;368:eaba5435. doi: 10.1126/science.aba5435. [DOI] [PubMed] [Google Scholar]
- 44.Ma S., Wang M., Wu J., Guo W., Chen Y., Li G., Wang Y., Shi W., Xia G., Fu D., et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant. 2021;14:1965–1968. doi: 10.1016/j.molp.2021.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary materials; further inquiries can be directed to the corresponding author.