Abstract
Combined therapies in cancer treatment aim to enhance antitumor activity. However, delivering multiple small molecules imposes challenges, as different drugs have distinct pharmacokinetic profiles and tumor penetration abilities, affecting their therapeutic efficacy. To circumvent this, poly(lactic-co-glycolic acid) (PLGA)–polyethylene glycol (PEG)–based nanoparticles were developed as a platform for the codelivery of synergistic drug ratios, improving therapeutic efficacy by increasing the percentage of injected dose reaching the tumor. Nonetheless, extravasation-dependent tumor accumulation is susceptible to variations in tumor vasculature; therefore, PLGA-PEG was modified with sulfates to actively target P-selectin–expressing cancers. Here, we show the potential of our platform in unique three-dimensional (3D) in vitro and in vivo models. The P-selectin–targeted nanoparticles showed enhanced accumulation in 3D spheroids and tissues of P-selectin–expressing BRAF-mutated melanomas and BRCA-mutated breast cancers, resulting in superior in vivo efficacy and safety. This nanoplatform could advance the codelivery of a plethora of anticancer drug combinations to various P-selectin–expressing tumors.
A nanoparticle platform codelivers two drugs to P-selectin–expressing cancers, boosting effectiveness in advanced tumor models.
INTRODUCTION
Combination treatment is a fundamental principle in cancer therapy aimed at reducing the required dose of each drug via the enhancement of efficacy through synergistic or additive activity. However, combining drugs may increase the risk of side effects, as each drug may induce diverse adverse events. Moreover, different drugs may display distinct physicochemical properties, thereby presenting substantially varied pharmacokinetics, limiting the potential synergistic effect and making it difficult to optimize the therapeutic window of such drugs combinations. Therefore, there is a need for platforms capable of enhancing the codelivery and maximizing therapeutic benefits, thus advancing the field of drug combinations for cancer treatment strategies.
Here, we propose the use of a poly(lactic-co-glycolic acid)–poly(ethylene glycol) (PLGA-PEG)–based nanosystem as a platform to codeliver a synergistic or additive ratio of drugs, in a single nanoparticle (two-in-one NP). PLGA-PEG–based NPs have attracted attention in drug delivery applications due to their biocompatibility, ease of preparation, and ability to encapsulate hydrophilic/hydrophobic drugs (1). The coencapsulation of a single two-in-one NP is expected to enable the selective and simultaneous delivery of the drugs to the tumor site, thus allowing synergism and enhancement of their t Synthesis of homobifunctional herapeutic efficacy and safety. However, relying on the enhanced permeability and retention (EPR) effect (2) to mediate the extravasation-dependent accumulation of NPs within the tumor site might lead to variation in treatment efficacy due to heterogeneity between tumor types and within the tumor itself (3). Larger tumor volumes and micrometastases were especially shown to have a variable blood vessel permeability, making them susceptible to subtherapeutic drug concentrations (4, 5).
To overcome the limitations of passive targeting, we modified the surface of our PLGA-PEG–based NPs with multivalent sulfates as P-selectin targeting moieties to direct the NPs to these hard-to-reach regions within the tumor microenvironment (6–8). P-selectin was chosen as the targeted receptor as it was shown to be overexpressed on several cancer types, thus enhancing the internalization and accumulation of NPs in melanoma, breast cancer (BC), and glioblastoma mouse models (6, 7). P-selectin is a carbohydrate-binding molecule, physiologically stored in α-granules inside platelets or in Weibel-Palade bodies inside endothelial cells. Upon activation, it is mobilized to the cell surface to enable the recruitment of leukocytes from the bloodstream by mediating their rolling and tethering on the monolayer of the activated endothelial cells (9). P-selectin is not constitutively expressed on platelets or endothelial cells (10), a feature that will prevent the NPs from binding to nonactivated platelets in the bloodstream and affecting their function. Moreover, P-selectin was shown to be expressed on several types of tumor cells (6, 11), which allows them to exploit P-selectin–dependent mechanisms and enables the metastatic process along the tumor endothelium (12, 13). Here, P-selectin was used to ligand-mediate the accumulation of P-selectin–targeted nanomedicines to both compartments: the tumor cells and the activated endothelial cells at the tumor site, which will ultimately improve drug accumulation and, therefore, maximize its efficacy. To test this hypothesis and to demonstrate the versatility and modularity of this platform, we selected two sets of clinically relevant drug combinations to be incorporated in this fully synthetic P-selectin–targeting nanoplatform; the first is composed of V-Raf Murine Sarcoma Viral Oncogene Homolog B (BRAF) and Mitogen-activated protein kinase kinase (MEK) inhibitors (BRAFi and MEKi), while the second includes poly(ADP-ribose) polymerase (PARP) inhibitor (PARPi) and programmed death ligand–1 (PD-L1) inhibitor (PD-L1i). The rationale behind the drug selection was choosing a combination that offers substantial benefits for anticancer therapy for P-selectin cancer tissues.
The combination of BRAFi and MEKi is the standard of care (SoC) for patients with BRAF-mutant melanoma, alongside immunotherapy (14). Dabrafenib (DBF), a BRAFi, and trametinib (TRM), a MEKi, were approved by the Food and Drug Administration (FDA) in 2013 for their notable improvement in response rates, progression-free survival (PFS), and overall survival of patients with melanoma compared to standard chemotherapy and DBF alone, becoming the first line of treatment for patients with melanoma with BRAF V600E or V600K mutations. In 2022, the outcome of three clinical trials involving patients affected by 24 different tumor types supported its accelerated FDA approval for treating adult and pediatric patients with unresectable or metastatic solid tumors with BRAF V600E mutation that progressed after prior treatment (15–17). Yet, the main challenge remained the tumor-acquired resistance, which limits the response duration to about 5 months (18). Efforts to increase treatment efficacy with higher dosing were hampered by dose-limiting toxicities and unaltered response rates (19–21), which point out this combination as an intriguing case study to address the application of our P-selectin–targeted nanoplatform to maximize the therapeutic efficacy of drug combinations.
An additional drug combination was chosen to be encapsulated in the nanoplatform to target a different P-selectin–expressing tumor. Triple-negative breast cancer (TNBC) with Breast Cancer gene 1 and 2 (BRCA) mutations could specifically benefit from a ligand-targeting approach due to their lack of hormone receptors and human epidermal growth factor receptor 2 (HER2) expression (22). Approximately 15% of all BCs are TNBC, and among them, 30% are BRCA1 or BRCA2 mutated, which form highly aggressive and invasive tumors (23). Targeted therapy with PARPi such as talazoparib was shown to prolong the PFS of patients with advanced BC and a germline BRCA 1/2 mutation compared to chemotherapy (8.6 months versus 5.6 months), leading to its FDA approval for the treatment of BRCA-mutated, HER2-negative locally advanced or metastatic BC (24, 25). Inhibiting PARP, an enzyme that repairs DNA single-strand breaks, triggers cell death in BRCA-mutated cells by blocking the base excision repair pathway, causing the accumulation of double DNA strand breaks (26, 27).
Despite their promise, resistance mechanisms to PARPi often develop, compromising the efficacy of these treatments (28). To counteract or delay resistance appearance, the combination of PARPi with immune checkpoint therapy inhibitors has shown promise in clinical trials, particularly in enhancing efficacy in homologous recombination-deficient cancers (29–32). BRCA1-mutated BC has also been associated with high basal expression of PD-L1 and a high abundance of tumor-infiltrating immune cells (33, 34). In addition, PARPi treatment was shown to have an impact on cancer-associated immunity increasing PD-L1 expression on tumor cells, potentially inhibiting T cell activity and thus supporting this drug combination rationale within our P-selectin–targeted platform (30).
Overall, this study reports the design, synthesis, and characterization of two-in-one P-selectin–targeted biodegradable PLGA-PEG NPs. This was achieved by incorporating sulfate groups into PLGA-PEG polymer before NP formation, ensuring the presentation of multiple sulfate groups on the NP surface. The modification did not compromise the physicochemical properties of the NPs, ultimately resulting in superior in vivo activity of both evaluated combinations: BRAF/MEK and PARP/PD-L1.
RESULTS
Design and characterization of drug-loaded PLGA-PEG NPs
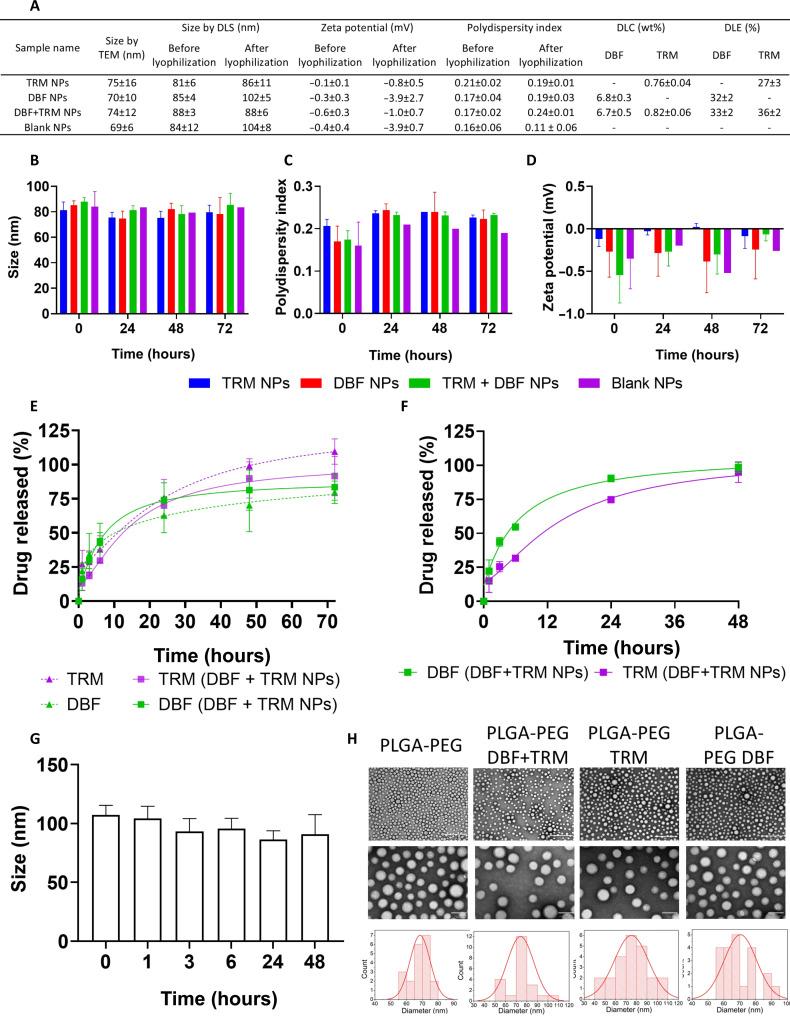
The first step toward the development of a combination therapy platform is to assess the ideal ratio of the two drugs. To achieve that for the DBF and TRM combination, we performed an in vitro toxicity experiment. D4M.3A, 131/4-5B1, and A375 melanoma cell lines were treated with different ratios of DBF and TRM combinations, and the median inhibitory concentration (IC50) was assessed. The combination index (CI) determines the degree of interaction between two drugs, where CI < 1 indicates synergism, CI = 1 is additive activity, and CI > 1 represents antagonism. An average CI of 0.8 and 0.58 was obtained when the cells were treated with the combined drugs at the IC50 and IC70, respectively, compared to each drug separately using the Chou-Talalay method (35). A combination ratio of TRM:DBF at 1:10 was chosen, demonstrating a CI < 1 across all three cell lines (fig. S1). Knowing the ideal drug combination ratio, a prototype of nontargeted NPs was formulated by dissolving PLGA, PLGA-PEG, and the drugs in acetonitrile (ACN), followed by rapid nanoprecipitation in an aqueous solution containing 2.5% polyvinyl alcohol (PVA). The average diameter of the NPs by dynamic light scattering (DLS) was 88 nm with a narrow polydispersity index (PDI) of 0.18, and the average drug loading content (DLC) was 6.7 wt % for DBF and 0.8 wt % for TRM. In addition, the size of the lyophilized NPs was similar to the prelyophilization form, with an average diameter of 95 nm and a PDI of 0.18 (Fig. 1A). The NP size and PDI remained constant for 72 hours of incubation in phosphate-buffered saline (PBS), and the zeta potential (ζ) remained almost neutral. The release profile of the dual-loaded two-in-one NPs closely resembled that of the single-loaded (one-in-two) NPs. Specifically, 50% drug release was recorded for DBF after 10 hours in the DBF-loaded NPs versus 8 hours in the two-in-one NPs, and for TRM, it was 11 hours in the TRM-loaded NPs versus 12.5 hours in the two-in-one NPs (Fig. 1, B to E). Next, the NP stability and drug release profiles were assessed in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). Herein, the NPs’ diameter remained constant during 72 hours of incubation (~95 nm), and 50% of encapsulated TRM was released after 11 hours, and 50% of encapsulated DBF was released after 5 hours (Fig. 1, F and G). The transmission electron microscopy (TEM) images showed a spherical and homogeneous NP structure with an average diameter of 72 nm (Fig. 1H). All in all, these results indicate that PLGA NPs are an ideal platform for the codelivery of drug combinations, and we continued evaluating its anticancer effects in three-dimensional (3D) models of melanoma.
Fig. 1. Single- and dual-drug–loaded NPs demonstrated comparable physicochemical properties.
(A) Summarized physicochemical properties. (B to D) NP stability measurements by DLS in PBS. (E and F) Release profiles of DBF and TRM in (E) PBS and (F) DMEM supplemented with 10% FBS. (G) NP hydrodynamic diameter by DLS (number) after incubation in the DMEM. The graphs show the mean ± SD, N = 3. (H) Representative TEM images and size distribution. Scale bars, 500 nm (top) and 100 nm (bottom). The NPs’ diameter based on at least 20 NPs per formulation was calculated with the ImageJ software.
Combined treatment with DBF and TRM reduced melanoma cell proliferation and 3D tumor spheroid sprouting
The cytotoxic activity of DBF and TRM monotherapy versus combined therapy as drug-loaded NPs or free drugs was evaluated on D4M.3A and A375 melanoma cell lines (Fig. 2, A and B). The IC50 values of the combined free drugs (2.3 nM for D4M.3A and 7.8 nM for A375) were lower than those of the two drugs given as monotherapy (3.3 nM for DBF and 7.7 nM for TRM in D4M.3A, and 9.4 nM for DBF and 12.9 nM for TRM in A375). The dual-loaded NPs surpassed the IC50 values of the combined free drugs in A375 cells (1.1 nM versus 7.8 nM, respectively). However, they remained in the same order of magnitude, as predicted due to the model limitations, which lack the EPR phenomenon and oblivious to the complex pharmacokinetic profiles of the free and entrapped drugs. In D4M.3A cells, the IC50 values of the combined free drugs (2.3 nM) were nearly equal to the dual-loaded NPs (2.9 nM) (fig. S2A). Next, the synergistic activity of the drugs was demonstrated on 3D melanoma spheroids, using 10 nM DBF and 1 nM TRM separately or combined, as free or drug-loaded NPs. In WM115 3D spheroids, the two-in-one NPs reduced the 3D spheroids sprouting to 52%, whereas with the one-in-two NPs, it was reduced to 95% for DBF and 90% for TRM, and to 73% with the combination of free drugs (Fig. 2, C and E). Furthermore, the melanoma cells were combined with brain-resident cells to create a multicellular 3D tumor spheroid (MCTS) model of melanoma brain metastasis (MBM), as melanoma frequently develops secondary lesions in the brain (36, 37), with an incidence of BRAF and NRAS mutations even higher than those observed in the primary melanoma site (38). Here, the combination of free drugs and drug-loaded NPs reduced the sprouting of WM115 spheroids to 40% compared to each drug separately (~50 to 60% for DBF free/NPs and ~70% for TRM free /NPs, respectively), while sparing the brain-resident cells. Blank NPs did not affect the invasiveness of the 3D spheroids (Fig. 2, D and F). In D4M.3A 3D spheroids, the cytotoxic activity of the combined drugs was similar to the monotherapies, resulting in 60% inhibition of the 3D spheroids’ sprouting, possibly due to the rapid growth rates of D4M.3A cells (fig. S2, B and C).
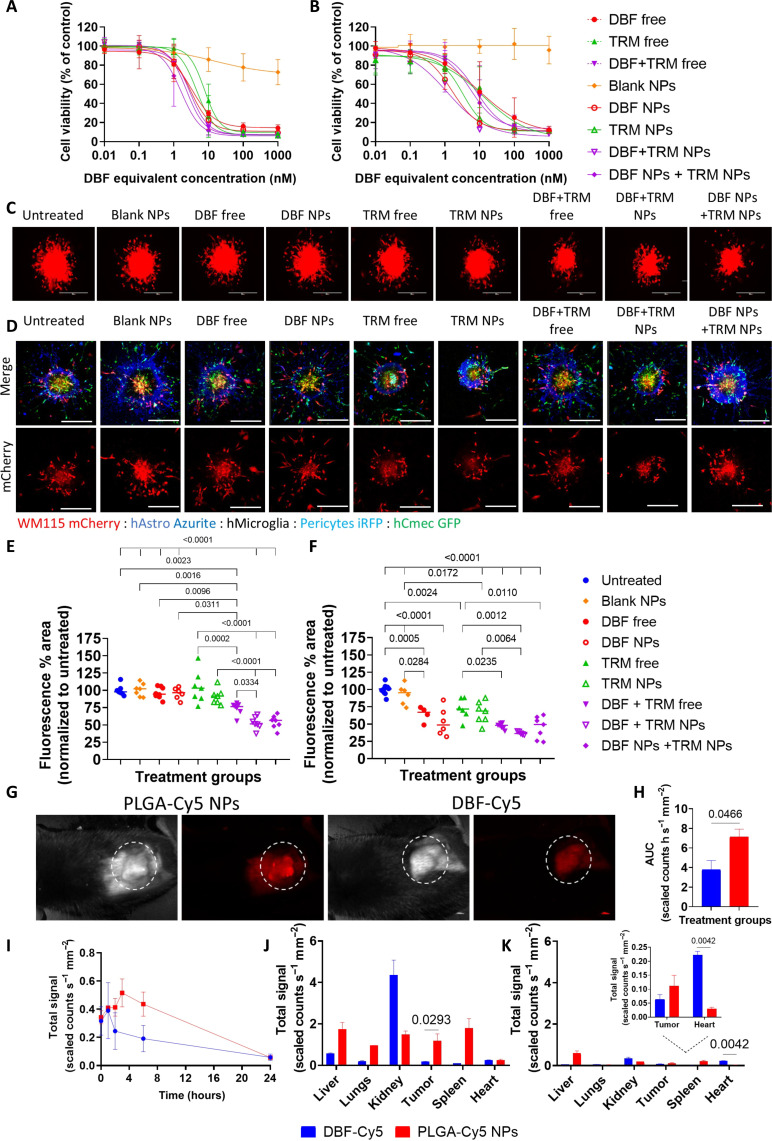
Fig. 2. Drug-loaded NPs inhibited melanoma cell proliferation, reduced 3D spheroids sprouting, and accumulated at the tumor site.
(A) Murine D4M.3A (B) Human A375 melanoma cells were treated with DBF and TRM separately or combined, as drug-loaded NPs or free drugs for 72 hours, then cell viability was evaluated. The data represent mean ± SD, N = 3. (C) Representative images of mCherry-labeled WM115 3D spheroids following 48 hours of incubation with drug-loaded NPs or free drugs at 10 nM DBF and 1 nM TRM, separately or combined. At least three 3D spheroids were imaged for each treatment, N = 3; scale bars, 400 μm. (D) Confocal images of multicellular 3D MBM MCTS after treatments with 10 nM DBF and 1 nM TRM for 72 hours, n > 3, N = 2; scale bar, 400 μm. (E and F) Quantification of mCherry % area of (E) WM115 3D spheroids and (F) WM115 3D MBM MCTS. Statistical analysis was performed using one-way ANOVA and multiple comparisons (P < 0.05) for the mCherry-labeled tumor cells using ImageJ. (G) Representative images of Cy5 signal from D4M.3A tumors 3 hours after intravenous administration of PLGA-Cy5 NPs and DBF-Cy5. (H) Area under the curve (AUC) of Cy5 signal from D4M.3A tumors 24 hours after injection. One-way ANOVA was used for statistical analysis. (I) Quantification of Cy5 signal from D4M.3A tumors, 24 hours after injection. (J) Biodistribution 3 hours and (K) 24 hours after administration. The inset is a magnification of the tumor and heart quantification. Two-way ANOVA was used for statistical analysis. N = 3 mice per group.
Cy5-labeled PLGA-PEG NPs accumulated at the tumor site following systemic administration
Subsequently, the tumor accumulation of Cy5-labeled PLGA-PEG (PLGA-Cy5) (fig. S3, A to C) NPs was compared to Cy5-labeled DBF (Fig. 2G) (39). TRM was not included because it could not be conjugated to Cy5. C57BL/6 mice were subcutaneously inoculated with D4M.3A cells and allowed to establish tumors until the tumor volume reached ~230 mm3. Then, the treatments were injected intravenously, and the Cy5 signal was recorded over 24 hours. Already 3 hours after administration, the Cy5 signal from the tumors in the PLGA-Cy5 NPs group was 6.9 times higher than that from the DBF-Cy5 group (1.18 versus 0.17 scaled counts s−1 mm−2, respectively), and after 24 hours, the cumulative Cy5 signal of the NPs was almost double than the DBF-Cy5 signal (7 versus 4 scaled counts s−1 mm−2, respectively) (Fig. 2, H to J). Of note, according to the release profile, most of the encapsulated drugs are still entrapped inside the NPs after 3 hours (~60 to 70% of DBF and ~75 to 80% of TRM), which will mediate increased drug accumulation at the tumor site. Twenty-four hours after administration, the DBF-Cy5 accumulation in the heart was 7.3 times higher than the PLGA-Cy5 NPs (0.22 versus 0.03 scaled counts s−1 mm−2, respectively) (Fig. 2K).
Drug-loaded PLGA-PEG NPs inhibited the MAPK pathway and prolonged mice survival
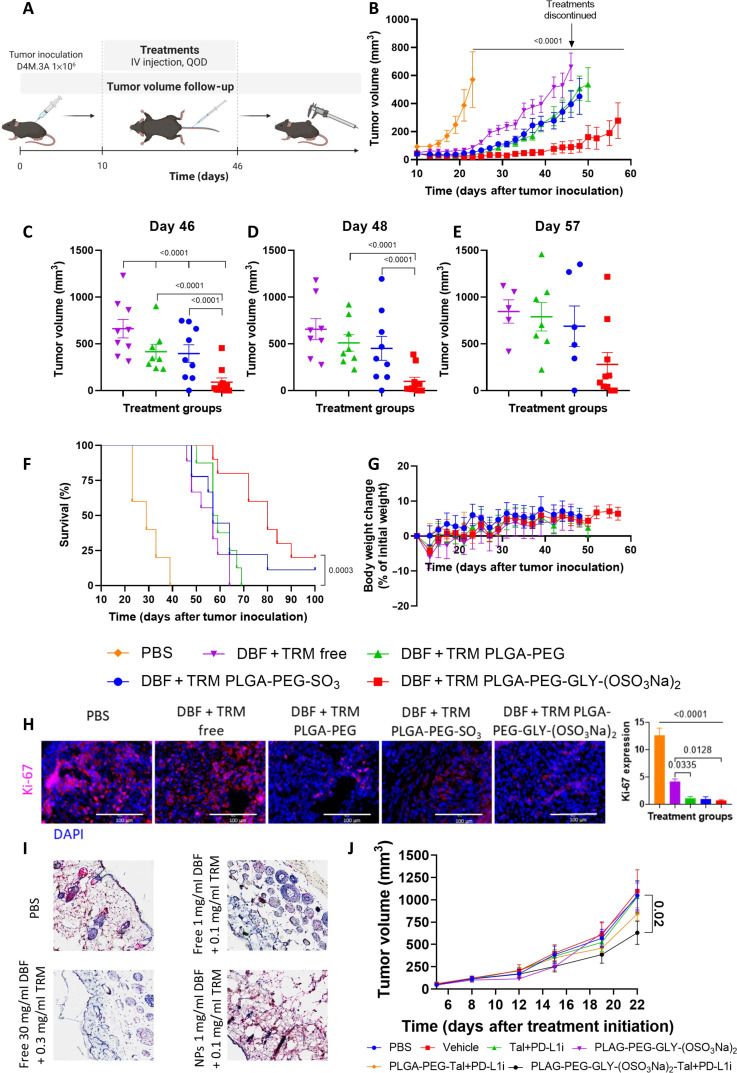
Afterward, we set to assess the antitumor activity of the drug-loaded NPs in vivo. C57BL/6 mice were inoculated subcutaneously with D4M.3A cells and were allowed to establish tumors for 10 days (reaching a tumor size of ~180 mm3). Then, considering the high bioavailability of intravenous dosing and based on previous data (39, 40), mice were treated with DBF and TRM as two-in-one drug-loaded NPs or free drugs, at 3 mg/kg DBF and 0.3 mg/kg TRM, or controls (PBS, vehicle, blank NPs) every other day (QOD). The mice were treated until the first group (PBS) reached the end point of tumor size >1000 mm3. The combined therapy demonstrated increased antitumor activity compared to the controls with no significant body weight change for all treated groups, but no additional efficacy was achieved for the drug-loaded NPs compared to the free drugs (fig. S4, A to C). Yet, the expression of phosphorylated kinases was inhibited by 90-fold for pBRAF and by 80-fold for pMEK and pERK compared to PBS-treated mice (fig. S4, D to G). As a result, another experiment with lower dosing was conducted to leverage the improved tumor accumulation of the NPs. In this regard, C57BL/6 mice with an average tumor volume of ~70 mm3 (day 10) were intravenously treated with a combination of DBF (2 or 1 mg/kg, QOD) and TRM (0.2 or 0.1 mg/kg, QOD) as drug-loaded NPs or free drugs. Both doses of NPs demonstrated superior antitumor efficacy by retaining a small tumor volume (<250 mm3) for 57 (high dose) and 50 days (low dose), compared to the equivalent doses of free drugs (27 days for the high dose and 23 days for the low dose) (Fig. 3, A to E). In addition, the median survival for both NP doses was 71 days, whereas for the free drugs, it was 39 days (low dose) and 52 days (high dose) (Fig. 3F). Neither group suffered from weight loss (Fig. 3G), nor did they present out-of-range parameters in a complete blood count test (fig. S5). The NPs were also hemocompatible, causing minimal hemolysis (8.5%) at high NP concentration (2 mg/ml), indicating their safety even at higher doses from those required for activity (Fig. 3H). In addition, the motor coordination, balance, memory, and learning abilities of the treated mice were not affected as the latency to fall was longer after the second rotarod test in all groups (Fig. 3I).
Fig. 3. DBF + TRM NPs demonstrated substantial antitumor activity in the murine melanoma D4M.3A mouse model.
(A) Timeline of study treatments and follow-up. (B) Tumor volume growth curve. P values correspond to the treatment groups compared to PBS on day 23 and to vehicle on day 25. (C to E) Individual tumor volumes. Statistical significance was calculated only for groups that retained the initial number of mice at the given time point, using two-sided repeated-measures ANOVA (P < 0.05). N = 4 mice for PBS and vehicle groups, N = 6 to 7 mice for the remaining groups. (F) Kaplan-Meier survival curve. The P values in the graph referred to NPs compared to free drugs at equal concentrations. In addition, P = 0.0008 for DBF + TRM NPs 1 mg/kg versus free DBF + TRM 2 mg/kg, and for DBF + TRM NPs 2 mg/kg versus DBF + TRM 1 mg/kg. P = 0.006 and 0.0013 for DBF + TRM NPs 1 mg/kg and 2 mg/kg versus PBS, and P = 0.0005 and 0.0011 versus vehicle, respectively. P = 0.0054 and 0.0006 for free DBF + TRM 1 mg/kg and 2 mg/kg versus PBS, and P = 0.0115 and 0.0005 versus vehicle, respectively. P values were determined using a log-rank test. (G) The body weight change curve is expressed as a percent change from treatment initiation. All graphs represent mean ± SEM. (H) Red blood cells were treated with increasing concentrations of PLGA NPs, dextran (negative control), or sodium dodecyl sulfate (SDS, positive control) for 1 hour, and the percentage of hemoglobin released was measured spectrophotometrically (N = 3). (I) Motor performance of the treated mice. The latency to fall was measured for each mouse individually before treatment initiation and after the seventh treatment. All groups performed better in the later test except the PBS. The data represent mean ± SEM. Individual measurements represent the average of the three longest performances out of five trials.
Talazoparib triggers tumor immunosuppression in BRCA-mutated BC cells by inducing PD-L1 expression
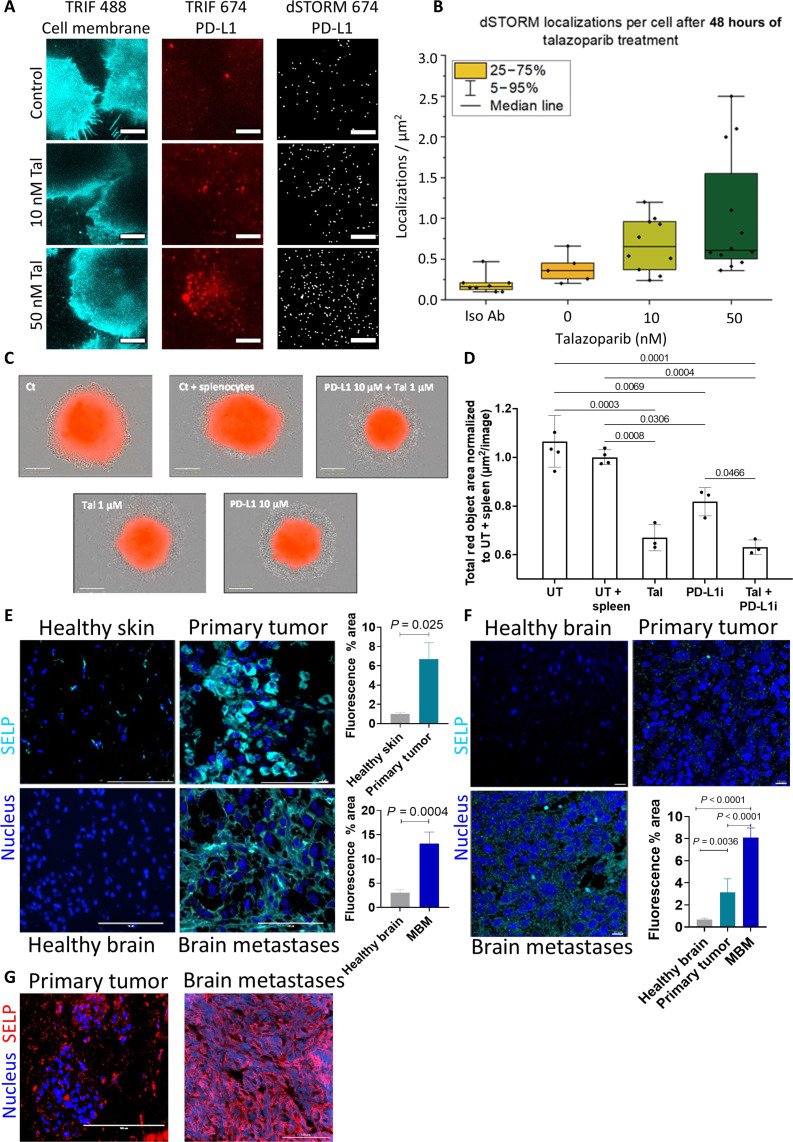
With these results in hand, BRCA-mutated BC was selected as an additional model to prove the versatility of our nanoplatform by coencapsulating another clinically relevant drug combination. Here, the PARPi talazoparib was incubated with four different TNBC cell lines: BRCA-mutated murine EMT6 and human MDA-MB-436 cells, as well as BRCA wild-type murine 4T1 and human MDA-MB-231 cells. Talazoparib exhibited preferred cytotoxic activity in BRCA-mutated cells compared to wild-type cells with IC50 values of 10 and 2 nM for EMT6 and MDA-MB-436 cells, compared to 58 and 411 nM for 4T1 and MDA-MB-231 cells, respectively (fig. S6A). In addition, the cytotoxic activity was demonstrated on 3D spheroids of mCherry-labeled EMT6 cells, where 70% inhibition of the 3D spheroids’ sprouting was observed following treatments with 1 μM talazoparib compared to the control after 72 hours (fig. S6, B and C). Next, we examined whether treatment with PARPi up-regulated PD-L1 expression, as previously reported in other cancer types (30). At this point, treatments with talazoparib induced PD-L1 protein expression in EMT6 cells in a dose-dependent manner, with significantly increased levels at 10 and 100 nM after 48 hours of incubation, resulting in 1.3- and 1.8-fold higher PD-L1 expression, respectively, compared to untreated cells (fig. S6D). In addition, treatments with 10 nM talazoparib for 24 to 72 hours resulted in a time-dependent increase in PD-L1 expression both at the protein and RNA levels (1.5- and ~2.3-fold increase after 72 hours, respectively) (fig. S6, E and F). Moreover, dSTORM superresolution imaging showed an increased number of PD-L1 single-molecule localizations and higher PD-L1 protein density following treatments with increasing concentrations of talazobarib (Fig. 4, A and B). These results confirm that talazoparib treatment can induce tumor immunosuppression, thereby reducing treatment efficacy. This highlights the potential for combination therapy, where codelivery of talazoparib with an immunotherapeutic agent capable of restoring tumor immunogenicity could enhance overall treatment outcomes.
Fig. 4. Talazoparib increased PD-L1 expression levels and evaluation of P-selectin expression in human and murine melanoma, as well as in BRCA-mutated murine BC.
(A and B) dSTORM imaging and analysis showing PD-L1 localizations per square micrometer in EMT6 cells with higher concentrations of talazoparib (Tal) after 48 hours. Blue: TIRF microscopy cell membrane; red: TIRF microscopy PD-L1 and dSTORM microscopy PD-L1 representative images. Data represent mean ± SD (N = 3). Scale bars, 10 μm. (C) Representative images of EMT6 3D spheroids cocultured with activated splenocytes, at a 1:100 ratio, respectively, following treatments with Tal and PD-L1i, separately or combined. Scale bars, 300 μm. (D) Quantification of mCherry % area. Spheroids were either untreated (UT), supplemented with splenocytes (UT + spleen), or treated with 1 μM Tal and 10 μM PD-L1i, separately or combined. Data represent mean ± SD (N = 3, n = 10), and statistical significance was calculated using two-sided repeated-measures ANOVA (P < 0.05). (E and F) Representative images and quantification of immunofluorescence staining of melanoma (E) Murine D4M.3A. Scale bars, 100 μm. (F) Patient-derived specimens. Scale bars, 10 μm. The nucleus was stained with DAPI (blue), and P-selectin was stained with Cy5-labeled antibody (SELP, cyan). Data represent mean ± SD; at least three fields were imaged from each specimen, N = 3. (G) P-selectin expression in BRCA-mutated EMT6 primary (left) and brain metastases (right). Nuclei are stained with DAPI in blue and P-selectin (SELP) in red. Scale bars, 100 μm.
PD-L1 inhibitor reduced PD-L1 expression and 3D spheroid growth when combined with talazoparib
To meet this challenge, a combination treatment of talazoparib and PD-L1i (SM56) (41) was tested in vitro. First, EMT6 cells were incubated with 10 μM PD-L1i for 24 to 72 hours, which caused a reduction in PD-L1 expression in a time-dependent manner (fig. S6G). Then, EMT6 cells were treated with the combination of 10 nM talazoparib and 10 μM PD-L1i to assess their ability to abrogate PD-L1 up-regulation. As seen in the graph, treatment with talazoparib alone up-regulated PD-L1 expression by threefold, while treatments with PD-L1i alone reduced PD-L1 expression by ~40% compared to untreated cells after 48 hours. The addition of PD-L1i to talazoparib reduced PD-L1 expression from 3- to 1.5-fold (fig. S6H). Furthermore, the antiproliferative effect was also assessed in mCherry-labeled EMT6 3D spheroids. The 3D spheroids were established in a U-well–shaped plate for 72 hours, then treated with 1 μM talazoparib and 10 μM PD-L1i separately or combined, and cocultured with activated splenocytes for an additional 72 hours. The activated splenocytes alone inhibited the 3D spheroids growth by 1.1-fold, but the most significant inhibition was achieved for the combined treatment (1.6-fold decrease compared to 1.4 by talazoparib and 1.2 by PD-L1i), confirming the rationale for the combination of these two compounds in our nanoplatform (Fig. 4, C and D). Therefore, this combination could be beneficial in specific subpopulations of patients who bear BRCA-mutated or homologous recombination (HR)–deficient cancers and which express P-selectin.
P-selectin is highly expressed in melanoma and BC tumor tissues and 3D spheroids
The focus was then directed toward enhancing the therapeutic efficacy of NPs through surface modification using P-selectin–binding moieties. Immunostaining for P-selectin on murine D4M.3A primary tumors and brain metastases was higher than on healthy skin and brain tissues (sevenfold and fourfold increase, respectively) (Fig. 4E). In addition, patient-derived human samples showed elevated expression of P-selectin in primary tumors (fivefold increase) and in MBM (12-fold increase) compared with healthy brain samples (11), supporting the clinical relevancy of P-selectin targeting (Fig. 4F). Furthermore, we recently showed that P-selectin is expressed in patient-derived primary and brain metastases of BC (42), and in the current study, we demonstrate P-selectin expression in primary and brain metastases of BRCA-mutated murine EMT6 BC tumors (Fig. 4G). Next, we developed an in vitro model that recapitulates P-selectin expression levels observed in vivo. D4M.3A and WM115 cells grown in 2D cultures expressed low levels of P-selectin, but once grown in 3D spheroids, P-selectin expression was doubled (fig. S7, A and B). When D4M.3A cells were combined with endothelial cells (bEnd.3) to form multicellular 3D spheroids, P-selectin staining was observed in both cell types, tumor and endothelial cells (fig. S7C). In addition, staining for P-selectin in BC EMT6 3D spheroids showed a twofold increase compared to isotype control, verifying the feasibility of P-selectin targeting in this BC model as well (fig. S7D).
Synthetic strategies for the preparation of sulfated PLGA-PEG polymers to target P-selectin
The degree of sulfation was shown to control the binding affinity to L/P-selectin (43, 44). Therefore, we synthesized two polymers: (i) PLGA-PEG-SO3Na (polymer 3) containing one sulfonate group and (ii) PLGA-PEG-GLY-(OSO3Na)2 (polymer 8) containing two sulfate groups (fig. S8). Detailed synthesis is described in Materials and Methods. The introduction of a single sulfonate group and the ratio between PEG and PLGA in polymer 3 were confirmed by 1H nuclear magnetic resonance (NMR) (fig. S9). The coupling of one unit of glycerol in polymer 4 was confirmed by 1H NMR (fig. S10), and the polymers (4 to 8) were characterized by electrospray ionization time-of-flight mass spectrometry (ESI TOF MS; fig. S11), size exclusion chromatography (SEC) (fig. S12), and Fourier transform infrared spectroscopy (fig. S13). Polymers 3 and 8 were then used for encapsulating DBF and TRM for the melanoma model. For the BC model and encapsulating talazoparib and PD-L1i, an optimized polymer was used. Here, to enhance the efficiency of the polymer synthesis—aiming to reduce the number of synthetic and purification steps, increase yields, and improve the structural integrity of the di-block copolymers—we shifted from conjugating PEG to PLGA and then modifying both chain ends, to synthesizing PLGA directly from a PEG macroinitiator. In addition, we used thiol-ene click chemistry (45) to conjugate thio-glycerol, which could be directly transformed into the final di-sulfate functionalized PLGA-PEG polymer. The synthesis procedure is depicted in Materials and Methods as well as in fig. S14. The only difference between the two final polymers is the linker between PEG and glycerol, which changed from carbamate-based in the first version to thio-ether based in the second polymer. This alteration should not substantially affect the overall properties of this large polymer as characterized later.
Twenty percent S content is homogeneously distributed on the surface of PLGA-PEG-GLY-(OSO3Na)2 NPs
The presence of sulfate groups on the surface of the NPs was assessed as they interact with the P-selectin receptors presented on the surface of the cancer cells and the activated endothelial cells. The sulfated polymer was introduced into the polymer mixture, replacing PLGA-PEG polymer used in the nontargeted NP prototype. We conducted a surface-specific quantitative x-ray photoelectron spectroscopy (XPS) analysis to ascertain the elemental analysis of PLGA-PEG-GLY-(OSO3Na)2 NPs and confirm the presence of sulfate groups on their surface. The XPS survey spectrum (fig. S15A) revealed characteristic peaks for carbon, nitrogen, oxygen, and sulfur, indicating the presence of these elements on the NP surface. Additional peaks may suggest the presence of sodium and chlorine. At higher binding energies (above 700 eV), we find peaks for metals or other less common elements. The sulfur peak around 160 to 170 eV suggests sulfate groups, which are relevant for surface interactions that aim to target P-selectin. To accurately pinpoint the peak positions of each element, the C1s (C─C) bond peak was initially located at 284.86 eV, aligning closely with the theoretical expectation of 284.8 eV (fig. S15B). Consequently, no adjustments were needed to align the observed results with theoretical values. The derived peaks are documented in the accompanying table (fig. S15E), which also describes the atomic percentage of each element in relation to the overall sample composition. Figure S15C presents the high-resolution XPS spectra for the sulfate group peak located at 168.48 eV. It resolves the binding energies of the 2p3/2 and 2p1/2 orbitals, confirming the presence of sulfate functional groups on the surface of the PLGA-PEG-GLY-(OSO3Na)2 NPs, which are crucial for targeted interactions. In fig. S15D, the depth profile analysis of the S2p peak indicates a consistent peak shape across different sputtering times, each layer representing 60-s intervals of ion exposure. The accompanying inset corroborates these findings, demonstrating no notable variation in the sulfate content or the sulfur atomic percentage relative to carbon within the top 30 nm of the NP surface.
Sulfation on PLGA-PEG increased the NPs’ internalization and accumulation in 3D spheroids and tumors
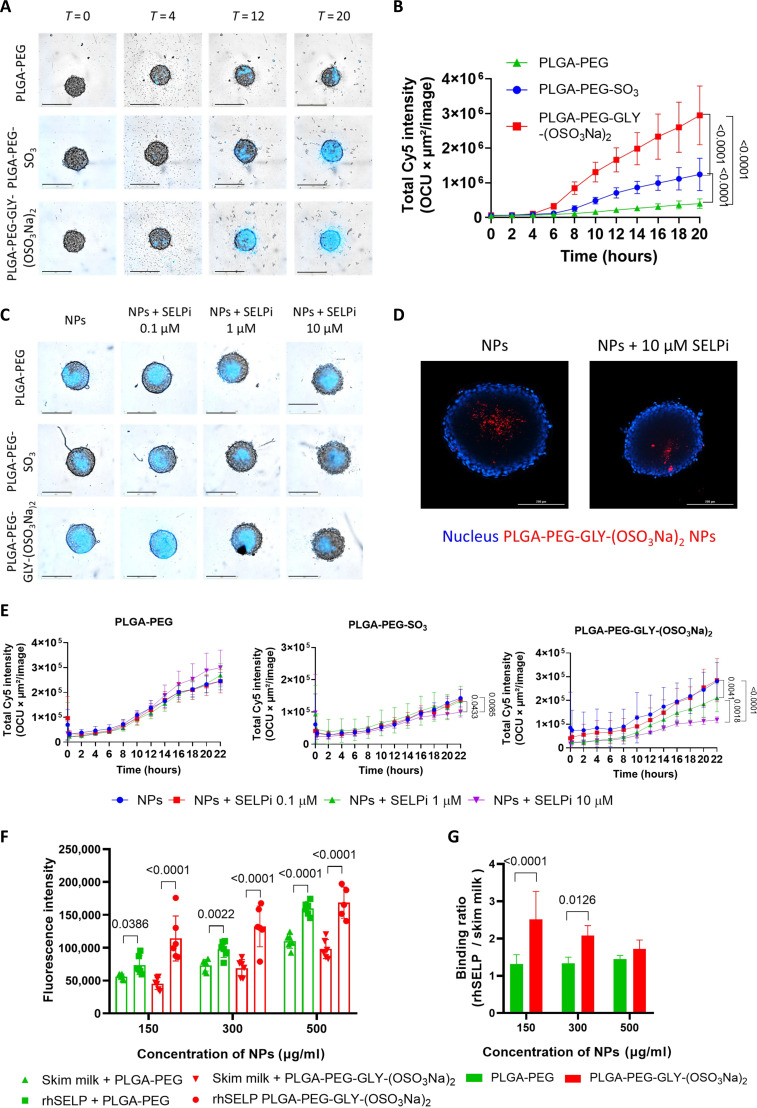
The P-selectin–mediated internalization of the newly synthesized sulfated NPs was assessed on P-selectin–expressing 3D spheroids. Human WM115 and murine D4M.3A melanoma 3D spheroids were treated with Cy5-labeled NPs for 20 hours. PLGA-PEG-GLY-(OSO3Na)2 NPs, followed by PLGA-PEG-SO3Na NPs, exhibited significantly higher internalization rates into the 3D spheroids compared to nontargeted NPs in both cell lines (~6.7- and ~2.8-fold increase, respectively). Furthermore, a 2.3-fold increase was observed for the NPs with bis-sulfates on their surface compared to NPs functionalized with mono-sulfonate (Fig. 5, A and B, and fig. S16, A and C). Upon pretreating the 3D spheroids with 10 μM P-selectin inhibitor (SELPi), the internalization of PLGA-PEG-GLY-(OSO3Na)2 and PLGA-PEG-SO3Na NPs was reduced by 5- and 1.5-fold in D4M.3A 3D spheroids (Fig. 5, C and E), and by 1.5- and 1.3-fold in WM115 3D spheroids, respectively, after 22 hours (fig. S16, B and D to F), while the internalization of nontargeted NPs remained unaffected. A competition assay was also performed, where we compared the internalization of the PLGA-PEG-GLY-(OSO3Na)2 NPs into the core of D4M.3A spheroids in the absence or presence of pretreatment with 10 μM P-selectin inhibitor for 1 hour at 37°C. The spheroids were then incubated with 1 μM Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs for 24 hours. Here, the spheroids were mounted on a slide and imaged from the middle of the Z-stack. As seen in the image, a clear reduction in Cy5 signal was observed after treatment with the P-selectin inhibitor compared to untreated spheroids. The Z-stack images show that the NPs internalized into the core of the spheroid, proving that they did not merely adhere to the spheroids’ surface (Fig. 5D). A similar trend was observed in murine EMT6 3D spheroids, where internalization of the optimized PLGA-PEG-GLY-(OSO3Na)2 NPs was higher than nontargeted NPs and PLGA-PEG-SO3Na NPs (fig. S16, G and H) and was reduced by 1.7-fold in the presence of 10 μM P-selectin inhibitor compared to untreated 3D spheroids after 20 hours (fig. S16, I and J). Together, these results confirm that in both models, the sulfated PLGA-PEG NPs internalization mechanism is primarily mediated by P-selectin. In addition, Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs were incubated with recombinant human P-selectin (rhSELP) or skim milk. The sulfated NPs exhibited increased P-selectin binding compared to nontargeted NPs up to 300 μg/ml. However, at NP concentrations above 300 μg/ml, both NPs showed a high Cy5 signal after incubation with skim milk and rhSELP, suggesting nonspecific binding (Fig. 5, F and G).
Fig. 5. Sulfate conjugation to PLGA-PEG increased the internalization of NPs into 3D spheroids.
(A) Representative images of time course internalization of Cy5-labeled NPs into D4M.3A 3D spheroids. Scale bars, 400 μm. (B) Time course quantification of Cy5 intensity within D4M.3A 3D spheroids. (C) Representative images of Cy5-labeled NPs internalization into D4M.3A 3D spheroids after 22 hours. The spheroids were preincubated with SELPi for 1 hour before NPs addition. Scale bars, 400 μm. (D) Representative Z-stack images of Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs internalization into the core of D4M.3A 3D spheroids after 24 hours, preincubated in the presence or absence of 10 μM SELPi. Scale bars, 200 μm. (E) Quantification of Cy5 intensity over time of nontargeted NPs and P-selectin–targeted NPs, after preincubation of the D4M.3A 3D spheroids with increasing SELPi concentrations. All quantifications are representative of three independent experiments. The data correspond to the mean ± SD of at least eight 3D spheroids per group, and statistical significance was determined using a two-way ANOVA test. (F and G) P-selectin binding assay of PLGA-PEG-GLY-(OSO3Na)2 NPs and PLGA-PEG NPs. Plates were coated with human recombinant P-selectin (rhSELP) or with skim milk (nonspecific binding control) and then incubated with PLGA-PEG-GLY-(OSO3Na)2 NPs or PLGA-PEG NPs for 15 min. (F) Fluorescence intensity of Cy5-labeled NPs after incubation with rhSELP or skim milk. (G) The fluorescence intensity ratio of rhSELP/skim milk. Bars represent mean ± SD, and a two-way ANOVA was used for statistical analysis.
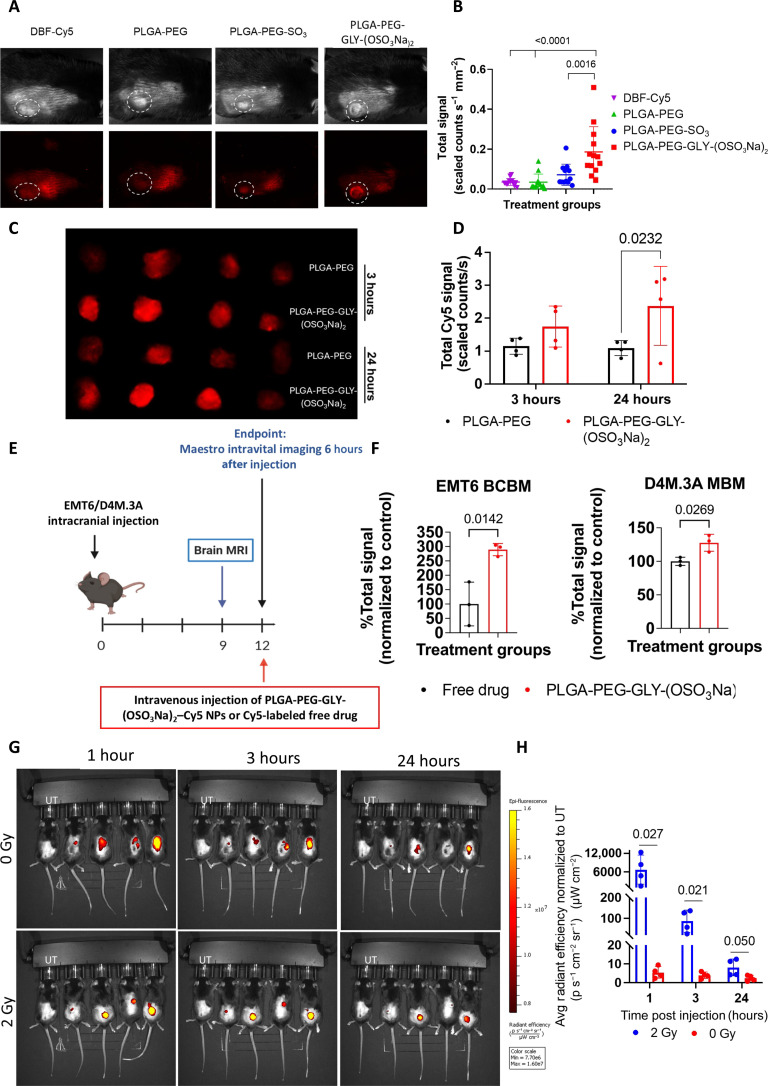
Afterward, P-selectin–targeted NPs were intravenously administered to C57BL/6 mice bearing subcutaneous D4M.3A tumors. The Cy5 signal of PLGA-PEG-GLY-(OSO3Na)2 NPs was 5 times higher than that of DBF-Cy5 and PLGA-PEG NPs, and 2.6 times higher than the signal from PLGA-PEG-SO3Na NPs (Fig. 6, A and B). In addition, the sulfate/sulfonate modification did not alter the distribution of NPs to essential organs, showing reduced accumulation in the liver after 24 hours compared to nontargeted NPs (fig. S17, A to D). We then quantified the drugs’ concentrations in vivo following administration of the P-selectin–targeted two-in-one NP, looking into the early time points until 1 hour, as the NPs start to accumulate at the tumor site at this time point. We intravenously injected the targeted two-in-one NP encapsulating TRM and DBF at a 1:10 ratio, respectively, to non–tumor-bearing mice and collected the blood at the following three time points: 5, 30, and 60 min. Then, TRM and DBF were extracted from the mice’s serum and Liquid Chromatography–Mass Spectrometry (LC-MS/MS) was used to detect the drug concentrations. We found that the initial ratio of TRM and DBF was maintained at these time points, showing that we preserved the effective required concentration of the drugs until the NPs reached the tumor (fig. S17E). Furthermore, the tumor accumulation of Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs was compared to nontargeted Cy5-labeled PLGA-PEG NPs in BALB/C mice that were intramammary fat pad inoculated with EMT6 cells. At 3 and 24 hours after injection, the Cy5 signal from the harvested tumors was significantly higher for the PLGA-PEG-GLY-(OSO3Na)2-Cy5 NPs than for the PLGA-PEG-Cy5 NPs (1.7 versus 1.1 scaled counts s−1 after 3 hours and 2.4 versus 1.1 scaled counts s−1 after 24 hours, respectively). Twenty-four hours after administration, the high Cy5 signal from the tumors indicates the retention of PLGA-PEG-GLY-(OSO3Na)2-Cy5 NPs inside the tumors (Fig. 6, C and D). In addition, 24 hours after administration, the distribution to essential organs was assessed, showing the highest accumulation in the liver, the main elimination pathway, followed by the tumor for PLGA-PEG-GLY-(OSO3Na)2-Cy5 NPs, and the spleen for the PLGA-PEG-Cy5 NPs (fig. S17, F and G). Furthermore, the NP brain accumulation was assessed on BALB/C female mice (BCBM) and C57BL/6 mice (MBM), 12 days after intracranial injection of EMT6 or D4M.3A cells, accordingly (Fig. 6E). Six hours after administration, the NPs’ accumulation in the brain was three times higher than the free drug in the BCBM model, and in the MBM model, there was a 30% increase in the brain Cy5 signal for the NPs compared to the free drug, suggesting that P-selectin–targeted NPs can mediate enhanced accumulation of encapsulated drugs in P-selectin–expressing orthotopic BC and melanoma models as well as brain metastases (Fig. 6F). Nonetheless, based on previous data showing induced P-selectin expression by radiation, which facilitated brain penetration of P-selectin–targeted fucoidan-based NPs (46), we hypothesized that it could also be used to increase the tumor accumulation of our PLGA-PEG-GLY-(OSO3Na)2 NPs. Here, C57BL/6 mice bearing subcutaneous D4M.3A tumors were irradiated [2 gray (Gy)] 4 hours before treatment with PLGA-PEG-GLY-(OSO3Na)2-Cy5 NPs and the Cy5 signal was monitored for 24 hours. Already 1 hour after administration, the irradiated tumors had significantly higher Cy5 signal than the control mice that were not irradiated [6755 versus 5.5 (p s−1 cm−2 sr−1)/(μW cm−2)]. This trend continued throughout the experiment, where 24 hours after administration, the NP tumor accumulation was higher in the irradiated tumors [8.1 versus 2.4 (p s−1 cm−2 sr−1)/(μW cm−2), respectively], strengthening the notion for improved radiation-induced tumor accumulation of P-selectin–targeted NPs (Fig. 6, G and H).
Fig. 6. Sulfate conjugation to PLGA-PEG increased the NPs’ internalization into tumors.
(A) Tumor accumulation of Cy5-labeled NPs or free DBF-Cy5, 2 hours after intravenous administration. N = 6–7 mice per group; the experiment was repeated twice. (B) Quantification of Cy5 intensity within the tumors. The graph represents individual Cy5-intensity values from the tumors and mean ± SEM. Statistical significance was calculated using one-way ANOVA. (C) Representative images of the EMT6 tumor accumulation of Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs or nontargeted PLGA-PEG NPs, 3 and 24 hours after the intravenous administration. (D) Quantification of the Cy5 signal from the EMT6 tumors. N = 3 mice per group. (E) Experimental scheme. Maestro intravital imaging of brains was done 12 days after intracranial injection of EMT6/D4M.3A cells and 6 hours after the injection of Cy5-labeled treatments. (F) Tumor accumulation of Cy5-labeled free drug or Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs in brain metastasis of EMT6 BCBM and D4M.3A MBM. The brains were harvested and imaged 6 hours after the injection of the treatments. N = 3 mice per group. The graphs represent individual Cy5-intensity values from the tumor normalized to the free drug and mean ± SEM. Statistical significance was calculated using Student’s t test. (G) Tumor accumulation of Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs in mice 4 hours after treatment with or without 2 Gy irradiation. N = 4 mice per group. Data was normalized to the untreated mouse on the left side of each image. (H) Quantification of the Cy5 signal from the tumor. The graph represents individual Cy5-intensity values from the tumor and mean ± SEM. Statistical significance was calculated using multiple unpaired t test.
Physicochemical properties of drug-loaded PLGA-PEG-GLY-(OSO3Na)2 NPs
Next, drug-loaded P-selectin–targeted NPs were formulated by dissolving the drug combinations in the polymer’s mixture. The DBF- and TRM-loaded P-selectin–targeted PLGA-PEG NPs exhibited an average diameter of 95 nm and a narrow size distribution (PDI = 0.15), for both the sulfonate and sulfated NPs. The zeta potential was nearly neutral (ζ = −0.95 mV) due to the abundant ester-terminated PLGA and PEG polymers. The DLE and DLC closely resembled that of the nontargeted NPs and the NPs remained stable in PBS for 72 hours (fig. S18, A to F). The 50% drug release of TRM remained unchanged, but that of DBF was reduced to 7 hours in PLGA-PEG-GLY-(OSO3Na)2 NPs and 14 hours in PLGA-PEG-SO3Na NPs. This change resulted from the increased hydrophobicity of DBF (log p = 5.44 for DBF and 3.45 for TRM) and its higher loading ratio (fig. S18G). The NPs showed biocompatibility and safety as they did not affect the viability of NIH/3T3 murine fibroblast cells or cause red blood cell lysis (fig. S18, H and I). For talazoparib- and PD-L1i-loaded NPs, prepared with the optimized PLGA-PEG-GLY-(OSO3Na)2 polymer, the DLC for talazoparib-loaded NPs (Tal NPs) was 18 wt %, and for PD-L1i NPs, it was 10.5 wt %. For the coencapsulation of talazoparib and PD-L1i at a 1:4 ratio, the DLC was 3.5 and 12.5 wt %, respectively. The NPs exhibited an average hydrodynamic diameter of 125 nm with a narrow size distribution (PDI = 0.13) and a nearly neutral zeta potential (fig. S19A). TEM images revealed a homogeneous and spherical morphology with an average diameter of 73 nm (fig. S19B). In addition, the release profile of the dual-loaded NPs resembled that of the single-loaded NPs (fig. S19C). Furthermore, the NPs remained stable in PBS for 72 hours, retaining an average hydrodynamic diameter of ~140 nm and a PDI of 0.18 (fig. S19, D and E). In vitro, talazoparib-loaded NPs demonstrated similar antiproliferative capabilities as free talazoparib, with an average IC50 value of 9.7 nM in EMT6 cells and 5.3 nM in MDA-MB-436 cells, respectively (fig. S19, F and G). Blank NPs (NPs without compounds) did not affect EMT6/MDA-MB-436 cell proliferation, nor did they cause red blood cell lysis up to 5 mg/ml, implying their safety for systemic administration (fig. S19, H to J).
PLGA-PEG-GLY-(OSO3Na)2 NPs show superior in vivo efficacy versus free drugs and nontargeted NPs
Then, the in vivo efficacy of DBF- and TRM-loaded PLGA-PEG-GLY-(OSO3Na)2 NPs was evaluated. C57BL/6 mice bearing D4M.3A tumors of ~50 mm3 (day 10) were intravenously treated with DBF (1 mg/kg) and TRM (0.1 mg/kg), either as free drugs, nontargeted NPs, or P-selectin–targeted NPs, QOD. Treatments continued until the first drug-treated group reached tumor volume > 1000 mm3. Free DBF and TRM only maintained a small tumor volume (<250 mm3) until day 31, while nontargeted NPs and PLGA-PEG-SO3Na NPs retained it until day 39. Surprisingly, 11 days after treatment withdrawal (day 57), the PLGA-PEG-GLY-(OSO3Na)2 NP group had an average tumor volume < 250 mm3, and 7 of 10 mice had tumor volumes <150 mm3 (Fig. 7, B to E). In addition, there was a survival benefit, with a median survival of 80 days for mice treated with the PLGA-PEG-GLY-(OSO3Na)2 NPs compared to 57 to 58 days for the other drug-treated groups and 29 days for the PBS group (Fig. 7F). Immunostaining showed notably decreased cell proliferation (Ki-67), increased apoptosis (cleaved caspase-3), and decreased pERK staining in the PLGA-PEG-GLY-(OSO3Na)2-treated group compared to the other groups. These results supported the better antitumor efficacy obtained in this group (Fig. 7H and fig. S20, A and B). Furthermore, neither group suffered from abnormal weight changes throughout the experiment that could indicate toxicity. In addition, following the eighth treatment, blood was drawn to perform a blood biochemistry analysis, which did not indicate hepatic or renal toxicities (figs. S7G and S21). Notably, because skin toxicity is a known and prevalent adverse event associated with BRAF and MEK inhibitors (47–50), we assessed pERK levels following the administration of low doses of DBF and TRM as free drugs or drug-loaded NPs and compared it to previously reported dosages. Four hours after intravenous administration, reduced levels of pERK were observed in healthy skin tissue for the free drug combination. However, with our NPs, pERK levels were equivalent to the PBS-treated group. On the basis of this observation, we hypothesize that skin toxicity can be mitigated using our P-selectin–targeted NPs (Fig. 7I).
Fig. 7. Drug-loaded PLGA-PEG-GLY-(OSO3Na)2 NPs substantially reduced tumor growth and prolonged mice survival.
(A) Timeline of study treatments and follow-up. (B) Tumor volume growth curve. P values correspond to the tumor volume of the treatment groups compared to PBS at day 23. (C to E) Individual tumor volumes. Statistical significance was calculated for groups that did not reach the first end point using two-sided repeated-measures ANOVA (P < 0.05). N = 5 mice for the PBS group and N = 8 to 10 mice for the remaining groups. (F) Kaplan-Meier survival curve. The statistical significance shown in the graph corresponds to drug-loaded PLGA-PEG-GLY-(OSO3Na)2 NPs compared with free drugs. In addition, P = 0.0006 for PLGA-PEG-GLY-(OSO3Na)2 NPs compared with PLGA-PEG NPs. P < 0.0001 for all groups compared with PBS. P values were determined using a log-rank test. (G) Body weight change graph, expressed as percent change from treatment initiation. (H) Representative immunofluorescence staining of D4M.3A tumors and quantification. Tumor cells are shown in blue (DAPI), and positive immunostaining is shown in magenta. N = 3 mice per group, at least six images per mouse. Statistical significance was determined using a one-way ANOVA test with multiple comparisons adjustment. Scale bars, 100 μm. All graphs show mean ± SEM. (I) Representative images of skin samples from healthy mice, 4 hours after intravenous injection. Red staining for pERK; scale bars, 200 μM. (J) EMT6 tumor volume growth curve. The P values correspond to the tumor volume of the PLGA-PEG-GLY-(OSO3Na)2 NPs group compared to PBS (P = 0.016), vehicle (P = 0.010), and free talazoparib and PD-L1i (P = 0.020) at day 22. Data are shown as mean ± SEM. Statistical significance was determined using two-sided repeated-measures ANOVA (P value <0.05). N = 6 to 9 mice per group.
A proof-of-concept in vivo experiment on the BC model was then conducted after we observed the significant improvement in tumor growth inhibition and survival of the PLGA-PEG-GLY-(OSO3Na)2 NPs in the melanoma model. Here, BALB/C female mice were inoculated in the intramammary fat pad with EMT6 cells and were allowed to establish tumors until the average tumor volume was 50 mm3 (day 15). Then, the mice were randomized into groups and intravenously administered with the combination of PD-L1i (3 mg/kg) and talazoparib (0.3 mg/kg) as free compounds, P-selectin–targeted NPs, nontargeted NPs, or controls [PBS, vehicle, and PLGA-PEG-GLY-(OSO3Na)2 carrier], three times a week. Neither the combination of free talazoparib and PD-L1i nor the nontargeted talazoparib- and PD-L1i-loaded NPs significantly reduced the tumor growth compared to the control groups; however, the P-selectin–targeted NPs substantially reduced the tumor growth compared to the control groups and free compounds (Fig. 7J). Notably, neither group suffered from abnormal weight changes that could indicate toxicity (fig. S22).
DISCUSSION
Combination therapy is at the forefront of cancer therapy, aiming to enhance the efficacy of individual drugs by synergistically inhibiting tumor cell growth. Here, we developed a versatile and effective nanoplatform for the codelivery of multiple drugs. The rationale for selecting two distinct drug combinations was to show the robustness of our nanoplatform, being agnostic to the type of targeted therapies, as long as the cancer tissue expresses P-selectin. The common denominator was the choice of two drugs, the combination of which is advantageous in terms of anticancer therapy. The first duo of BRAFi and MEKi is relevant to BRAF-mutated cancers, and the second duo of PARPi and PD-L1i is relevant to BRCA-mutated or HR-deficient cancers, making a defined subpopulation of patients who will gain benefit from our modular two-in-one NP platform.
BRAFi and MEKi are the SoC for patients with BRAF-mutant melanoma. However, most patients (>80%) discontinued treatments due to disease progression (51). To address this, we designed PLGA-PEG nanocarriers capable of coencapsulating both drugs, ensuring that the optimal ratio for synergism is maintained and that each tumor cell receives equal exposure to both agents. Conversely, coinjection of two single-loaded nanocarriers cannot guarantee the simultaneous and synergistic delivery of combined drugs, resulting in suboptimal efficacy, although it improves the pharmacokinetic profile of each free drug by encapsulating it in an NP (52). Traditionally, nanoparticle production involves manual mixing of organic and aqueous phases, leading to variability and poor batch-to-batch reproducibility. To overcome these challenges, we used microfluidics to achieve homogenous, monodispersed, and reproducible NPs (53). The NP size was tuned to sub-100 nm, with a narrow PDI (0.18) and a slightly negative zeta potential (ζ = −0.35), to prolong blood circulation half-life and allow sufficient tumor accumulation (54, 55). Three hours after injection, the NPs’ tumor accumulation was 6.8 times higher than that of the free drug. In addition, as the MAPK pathway is cardioprotective and essential for normal vascular and cardiac physiology, inhibiting it in the heart with DBF and TRM therapy was expected to lead to cardiotoxicity (56). Although we did not monitor the heart activity and functionality, our NPs did not accumulate in the heart, suggesting a potential reduction in cardiotoxicity compared to free DBF, which exhibited a seven- and fivefold increase in heart accumulation relative to the nanoparticles.
Initially, the two-in-one dual-loaded NPs demonstrated comparable in vivo activity to the combination of the free drugs. However, the ability of the NPs to reduce drug dosing while maintaining high therapeutic efficacy and safety was not manifested at that dose. Therefore, we postulated that the NPs’ improved drug accumulation could allow for lower dosing while still achieving effective antitumor activity. This time, the two lower doses (1 and 2 mg/kg) of NPs resulted in an increased antitumor efficacy compared to free drugs. Cherri et al. showed that intraperitoneally injected dendritic polyglycerol sulfate-bearing poly(caprolactone) (dPGS-PCL) loaded with sunitinib inhibited tumor growth as orally administered free sunitinib at three times higher concentrations (57). In our study, the lower dose of NPs significantly reduced tumor growth (P < 0.0001) compared to free drugs administered even at double the concentration. All treatments were intravenously administered; therefore, factors such as bioavailability did not affect the tumor drug concentrations. In preclinical testing, treatments with 0.3 to 3 mg/kg TRM inhibited tumor growth, while treatments with 0.1 mg/kg resulted in large tumor volumes already after 15 days (58). Similarly, treatments with 10 to 100 mg/kg DBF inhibited tumor growth, whereas treatments with 3 mg/kg did not inhibit tumor growth. In addition, the combination of DBF (30 mg/kg) with TRM (0.3 mg/kg) was superior to each drug alone (40). Our NPs inhibited tumor growth at doses as low as 1/30 of the DBF dose, equivalent to 1 mg/kg DBF and a third of the TRM dose, equivalent to 0.1 mg/kg TRM. Recently, our group published a drug delivery platform based on poly(α, l-glutamic acid) (PGA) conjugated to selumetinib (SLM, MEKi) and modified dabrafenib (mDBF). The combined treatment led to a substantial antitumor effect and prolonged mice survival (39). Nevertheless, SLM, which is indicated for patients with neurofibromatosis type 1, is not the SoC for patients with melanoma (59). On the other hand, TRM lacked a functional group available for covalent conjugation and could not be combined in such a platform. Therefore, herein we developed a drug delivery system based on hydrophobic interactions to encapsulate TRM and DBF within the PLGA matrix. In addition, TRM showed higher potency on D4M.3A cells (IC50 = 7.7 nM) than SLM (IC50 = 200 nM), which facilitated the enhanced antitumor activity of the PLGA-PEG NPs compared to PGA-SLM-mDBF [mDBF (10 mg/kg) and SLM (15 mg/kg)].
To further enhance the NPs’ tumor accumulation, ligand-receptor–mediated targeting was proposed. P-selectin has been demonstrated by us and others to enhance the therapeutic efficacy of targeted NPs against P-selectin–expressing tumors (6, 60, 61). Herein, high levels of P-selectin were observed in patient-derived melanoma samples and D4M.3A melanoma tumor tissues. Furthermore, sections from human MBM exhibited even higher expression levels of P-selectin compared to the primary tumor, suggesting its potential utilization to improve NPs’ brain penetration. In addition, P-selectin was also observed in human and murine primary BC and brain metastasis (BM), which made BC an optimal second model to demonstrate the versatility of our nanoplatform and encapsulate the combination of talazoparib with our PD-L1 small-molecule inhibitor, SM56.
To target P-selectin, we synthesized two modified polymeric amphiphiles, containing either one sulfonate moiety (PLGA-PEG-SO3Na) or two sulfate moieties [PLGA-PEG-GLY-(OSO3Na)2], and used them to prepare targeted NPs, which exhibited greater internalization rates into P-selectin–expressing melanoma and BC 3D spheroids compared to nonmodified NPs. The major driving forces for binding between P-selectin and sulfate/sulfonate groups are charge-charge interactions (62) and counterion release after binding, as demonstrated in molecular dynamics simulations (63). Previously, fucoidan-based NPs were shown to bind P-selectin. Nevertheless, natural polymers lack defined weight-average molecular weight (Mw) and are characterized by broad dispersity (Đ) and variable batch-to-batch degrees of sulfation (64). The use of synthetic polymers that mimic these sulfated natural polymers may improve batch-to-batch reproducibility. However, to date, the existing synthesized amphiphilic polymers that contained sulfates had limited drug loading due to the random distribution of hydrophobic and hydrophilic moieties (57). Polyglycerol sulfate dendrimers (dPGS) were also shown to bind P-selectin. However, the polymers’ hydrophilicity limits their drug-loading content (DLC). The dPGS possessed a 90% degree of sulfation. Hence, drug conjugation to the polymeric backbone was essential to avoid premature drug release (6, 60). Likewise, paclitaxel-loaded fucoidan NPs demonstrated a fast release profile, with more than 70% of the encapsulated drug being released following 8 hours (7). In addition, not all drugs can be covalently bound. Furthermore, Zhang et al. used fucoidan as a coating layer, but the degradation of such charged-based coating could take place already in the blood (65), strengthening the need for a new covalently bound P-selectin–targeting polymer. Our PLGA-PEG self-assembled to form NPs with a hydrophobic core, facilitating the encapsulation of the two hydrophobic drugs. Here, the sulfate groups were conjugated to the PEG block in PLGA-PEG polymer, serving as surface modifiers, so that the hydrophobic core was not affected. Last, the therapeutic advantage of PLGA-PEG-GLY-(OSO3Na)2 was demonstrated by a marked exposure to higher dosing and an optimal drug ratio that enhanced the antitumor activity of the two drugs and delayed tumor escape. Of note, this was achieved by solely altering the pharmacokinetics of the drugs. Together, each sulfated nanocarrier has its advantages and disadvantages, which dictate the drug to be encapsulated/conjugated and the disease indication and its location.
While searching for an effective drug combination to treat TNBS, we observed an induction in PD-L1 expression following the treatment with talazoparib. This gave rise to an additive drug combination with PD-L1i to counteract its induction. In this case, talazoparib was coencapsulated, in P-selectin–targeted NPs with our murine PD-L1 small-molecule inhibitor, SM56, which was developed alongside the human PD-L1 small-molecule inhibitor, SM69 (41). Despite PARPi gaining huge interest in the treatment of cancers with deficient homologous recombination repair, BRCA-mutated BC tumors often develop resistance to PARPi therapy through diverse mechanisms (66–68). Under this scope, combined treatments and drug delivery platforms allow for reducing PARPi dose while increasing tumor-specific delivery can help delay resistance and enhance the overall antitumor effect (69). The accumulating evidence suggests that PARPi can initiate tumor-immune responses, promoting tumor growth (70, 71). Talazoparib demonstrated antiproliferative activity in BRCA-mutated BC cell lines; however, it increased PD-L1 expression, both at the protein and RNA levels in EMT6 cells. This suggested that combining anti–PD-1/PD-L1 therapies with PARPi in BRCA-mutated cancers could mitigate the tumor immunosuppression associated with the PARPi therapy and achieve an additive anticancer effect. Our findings demonstrated that coadministering talazoparib with our PD-L1 inhibitor effectively counteracted PD-L1 up-regulation and significantly reduced EMT6 mCherry tumor 3D spheroid growth in coculture with activated splenocytes. The discovery of P-selectin expression in primary and brain metastases of EMT6 tumors and 3D spheroids prompted us to coencapsulate these drugs in the P-selectin–targeted NPs. In vivo assessments revealed that while free drug combinations or nontargeted NPs did not markedly affect tumor growth, P-selectin–targeted NPs substantially inhibited tumor progression compared to control groups and free drugs, underscoring the potential of this targeted two-in-one nanomedicine approach in enhancing antitumor efficacy in BRCA-mutated BC.
It is now becoming clear that P-selectin is overexpressed in various tumor types compared to healthy tissues, such as glioblastoma versus brain, pancreatic ductal adenocarcinoma versus pancreas, renal cell cancer versus kidney, melanoma versus skin, and BC versus breast tissue and on activated endothelial cells at the tumor site (6, 11). Given that DBF and TRM can now be prescribed to any tumor type with BRAF V600E mutation, and our demonstration of the versatility of our nanoplatform by coencapsulating additional drug combinations, we propose that our P-selectin–targeted platform can be exploited to treat a plethora of P-selectin–expressing tumors bearing BRAF V600E mutation (e.g., primary and metastatic melanoma, TNBC, pediatric low-grade glioma, and rare BRAF-mutated glioblastoma). In addition, this platform could be extended to address HR-deficient tumors, such as BRCA-mutated breast and ovarian cancers, which may benefit from the codelivery of targeted therapies aimed at overcoming resistance and enhancing therapeutic efficacy.
MATERIALS AND METHODS
PLGA (d,l-lactide-co-glycolide 85-15, ester terminated, Mw 50,000 to 70,000 Da), PLGA-PEG [PEG average Mn (number-average molecular weight) 2000 Da, PLGA, actide-co-glycolide 50-50, average Mn 11,500 Da], and PVA (Mw 13,000 to 23,000 Da, 87 to 89% hydrolyzed) were purchased from Sigma-Aldrich (Rehovot, Israel). For P-selectin–targeted polymers: PEG (Mw 2000 Da) and PLGA (d,l-lactide-co-glycolide 50-50, Mw 7000 to 17,000 Da, Resomer 502H, acid terminated) were used. For PLGA-Cy5 synthesis: PLGA (d,l-lactide-co-glycolide 50-50, Mw 7000 to 17,000 Da, Resomer 502, ester terminated) was used. All were purchased from Sigma-Aldrich (Rehovot, Israel). Dialysis tubes—mini-GeBaFlex tube 200 μl, 8 kDa MWCO for release studies, or Mega GeBaFlex tubes 20 ml, 3.5 kDa MWCO for P-selectin–targeted polymers synthesis—were purchased from Gene Bio-Application Ltd. (Yavne, Israel). DBF 98% purity was purchased from Tzamal-Dchem (Petach-Tikva, Israel). TRM 98% purity was purchased from Advanced ChemBlocks Inc. (CA, USA). 2,2-Dimethoxy-2-phenylacetophenone (DMPA, 99%) and 1-thioglycerol were obtained from Sigma-Aldrich. HO-PLGA12k-PEG2k-allyl was obtained from “Creative PEGWorks.” P-selectin inhibitor (KF 38789) was purchased from BioTest (Kfar Saba, Israel). Recombinant human P-selectin (rhSELP; Cat. ADP3) was purchased from R&D Systems (Minneapolis, MN, USA). All chemical reagents, including salts and solvents, were AR grade and purchased from Sigma-Aldrich (Rehovot, Israel). Reactions requiring anhydrous conditions were performed under an Argon atmosphere. Distilled water, filtered using a MilliQ purification system, was used throughout. RPMI 1640, MEM alpha, Advanced DMEM, FBS, and GlutaMAX ×100 were purchased from Gibco (Waltham, MA, USA). Bovine serum albumin (BSA), Dextran, SDS, EDTA, and [2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide] (XTT) reagents were purchased from Sigma-Aldrich (Rehovot, Israel). EndoGRO MV Complete Media Kit was purchased from Merck, Germany. Human astrocyte medium, pericyte medium, or microglia medium were purchased from ScienCell (CA, USA). Penicillin, streptomycin, nystatin, l-glutamine, Hepes buffer, nonessential amino acids solution, sodium pyruvate, β-mercaptoethanol, and the Mycoplasma detection kit were purchased from Biological Industries Ltd. (Kibbutz Beit Haemek, Israel).
Antibodies for flow cytometry: Fluorescein (FITC)–labeled rat anti-mouse CD62P antibody, catalog no. 561923 (1:20 dilution) and FITC rat immunoglobulin G1 (IgG1) Isotype control, catalog no. 553995 (1:20 dilution) were bought from BD Biosciences (Franklin Lakes, NJ, USA). Mouse anti-human E-selectin/P-selectin (CD62E/P) antibody, catalog no. BBA1, lot APB081704 (1:20 dilution) and mouse IgG1 Isotype Control, catalog no. mab002 (1:20 dilution) were bought from R&D Systems (Minneapolis, MN, USA). Secondary antibody: goat anti-mouse Alexa Fluor 647, catalog no. Ab150115 (1:300 dilution) was bought from Abcam (Cambridge, UK). Anti–PD-L1 antibody was purchased from R&D Systems (Mouse PD-L1/B7-H1 Alexa Fluor 647–conjugated antibody; R&D Systems no. FAB90781R) or isotype antibody [rabbit immunoglobulin G (IgG) Alexa Fluor 647–conjugated antibody, R&D Systems no. IC1051R].
Antibodies for immunofluorescence: Rabbit polyclonal anti-human/mouse/rat phospho-p44/42 MAPK (Erk1/2), catalog no. 9101S, lot 1 (1:300 dilution); rabbit mAb anti-human/mouse/rat/monkey phospho-MEK1/2 antibody, catalog no. 2338, lot 9 (1:30 dilution); and rabbit mAb anti-human/mouse/rat/monkey Cleaved Caspase-3, catalog no. 9664L, lot 22 (1:30 dilution) were purchased from Cell Signaling (Danvers, MA, USA). Rabbit anti-human/mouse/rat Phospo B-Raf antibody, catalog no. NBP2-67846 (1:100 dilution), and Rabbit polyclonal anti-human/mouse/rat/porcine Ki67/MKI67 antibody, catalog no. NB500-170, lot G13 (1:50 dilution) were bought from Novus Biologicals (Centennial, CO, USA). Secondary antibodies: goat anti-rabbit IgG H&L (Alexa Fluor 647), catalog no. ab150079, lot GR3176223-2, and goat anti-mouse IgG H&L (Alexa Fluor 647), catalog no. ab150115, lot GR30989-3 were bought from Abcam (Cambridge, UK) (1:350 dilution).
PLGA nanoparticle preparation
PLGA-PEG NPs were prepared using a benchtop NanoAssemblr instrument (Precision NanoSystems Inc., Vancouver, Canada). PLGA and PLGA-PEG at a 2:1 ratio were dissolved in ACN, to a final polymer concentration of 6 mg/ml. For drug-loaded NPs, TRM (2 mg/ml) and DBF (4.5 mg/ml) were dissolved in ACN. Talazoparib (4.5 mg/ml) and PD-L1i SM56 (18 mg/ml) were dissolved in ACN or dimethyl sulfoxide (DMSO), respectively. The compounds were added to the polymer solution. For Cy5-labeled NPs, 2 wt % PLGA-Cy5 was added to the polymer solution. PLGA-PEG NPs were formed by nanoprecipitation achieved by mixing two fluid phases: (i) an organic phase composed of polymers and drug solutions, and (ii) an aqueous phase composed of 2.5% w/v PVA solution. The aqueous and organic phases were mixed at a 12 ml/min flow rate and a 2:1 ratio, respectively. Then, ACN was evaporated under reduced pressure using a rotary evaporator, and the NPs were centrifuged (35,000g, 30 min) and washed twice with 5 ml of MilliQ water. Last, the NPs were resuspended in PBS or MilliQ water. Freeze-dried NPs were resuspended in 5% trehalose, snap frozen, and lyophilized for 48 hours.
Nanoparticle size distribution and zeta potential
Samples were freshly prepared in MilliQ water at 0.1 mg/ml NP concentration. Mean hydrodynamic diameter, PDI, and the zeta potential (ζ) were measured using a DLS Möbius instrument at a 540-nm laser wavelength using a 532-nm long-pass filter (Wyatt Technology Corporation, Santa Barbara, CA 93117 USA). All measurements were performed at 25°C.
Colloidal stability
Freshly prepared PLGA-PEG NP samples were incubated in PBS (pH 7.4) or DMEM containing 10% FBS, and maintained at 37°C. NP size, PDI, and zeta potential (ζ) were monitored by DLS every 24 hours.
Drug loading and encapsulation efficiency
Lyophilized drug-loaded NPs were dissolved in ACN and stirred at 60°C for 1 hour. Samples were filtered through a polyvinylidene fluoride membrane filter with a 0.2-μm pore size and analyzed by HPLC. UltiMate 3000 Nano LC systems (Dionex) was equipped with a 3000 pump, VWD-3000 UV-Vis detector, and Chromeleon 6.80 software. The column used was Jupiter column; particle size, 5 μm; pore size, 300 Å; 4.6 × 250 mm; C18 reversed phase (RP). Chromatographic conditions: flow: 1.0 ml/min, gradient: 30% solution B to 100% solution B [solution A—0.1% trifluoroacetic acid (TFA) in water; solution B—0.1% TFA in ACN]. The injection volume was 20 μl. Drug entrapment efficiency was calculated as both DLC (wt %), Eq. 1, and drug loading efficiency (F %), Eq. 2
| (1) |
| (2) |
Release profile
Drug-loaded NPs were suspended in MilliQ water, aliquoted into several semipermeable dialysis tubes (200 μl), and dialyzed against 1 liter of PBS (pH 7.4) at 37°C. At predetermined time points, an aliquot of the NPs suspension was removed, lyophilized, and dissolved in ACN. The amount of drug released was quantified by HPLC.
Transmission electron microscopy
PLGA-PEG NPs were visualized using 110 keV TEM (JEM-1400Plus Transmission Electron Microscope). TEM ultrathin Formvar-coated 200-mesh copper grids (Ted-pella Inc.) were prepared by depositing the grid over a drop of the sample until drop evaporation on parafilm. To increase the resolution of the images, the samples were stained using the negative staining technique. Briefly, 24 hours after sample preparation, the grids were charged for 5 min with ultraviolet light, then one drop of Uranyl Acetate (staining agent) was deposited over a parafilm strip, and the grid with the sample was deposited over the staining agent drop for 1 min. Then, excess liquid was removed with filter paper, and TEM images were acquired. For size distribution analysis, the size of at least 20 particles was measured and the average size and SD were obtained with the ImageJ software.
Cell culture
Murine primary melanoma D4M.3A cells were kindly provided by D. W. Mullins (Dartmouth College, Hanover) (72). D4M.3A cells were cultured in Advanced DMEM growth media supplemented with 5% FBS, streptomycin (100 μg/ml), penicillin (100 IU/ml), nystatin (12.5 IU/ml), and 1% Glutamax. Human metastatic melanoma 131/4-5B1 cells were kindly provided by R. Kerbel’s laboratory (University of Toronto, Canada) (73) and were cultured in RPMI 1640 supplemented with 10% FBS, streptomycin (100 μg/ml), penicillin (100 IU/ml), nystatin (12.5 IU/ml), and 2 mM l-glutamine. Human primary melanoma A375 cells were purchased from the American Type Culture Collection (ATCC) and were cultured in RPMI 1640 supplemented with 10% FBS, streptomycin (100 μg/ml), penicillin (100 IU/ml), nystatin (12.5 IU/ml), and 2 mM l-glutamine. Human primary melanoma WM115 cells were purchased from the ATCC and cultured in Eagle’s Minimum Essential Medium (EMEM) growth media supplemented with 10% FBS, streptomycin (100 μg/ml), penicillin (100 IU/ml), nystatin (12.5 IU/ml), 1% of nonessential amino acids, 1% of sodium pyruvate, and 2 mM l-glutamine. Mouse fibroblast NIH/3T3 cells were purchased from the ATCC and cultured in DMEM supplemented with 10% FBS, streptomycin (100 μg/ml), penicillin (100 IU/ml), nystatin (12.5 IU/ml), and 2 mM l-glutamine. Human microvascular endothelial hCMEC/D3 cells were purchased from Merck and grown in EndoGRO MV Complete Media Kit (SCM004 - Basal media + supplement kit) supplemented with FGF-2 (1 ng/ml; bFGF). Human astrocytes were purchased from ScienCell and grown in the supplied astrocyte medium complemented with a supplemented kit. Human brain pericytes and human microglia were also purchased from ScienCell and grown in the supplied Pericyte Medium or Microglia medium complemented with a supplement kit. All cells were grown at 37°C and 5% CO2 and were routinely tested for mycoplasma contamination with a mycoplasma detection kit. Human MDA-MB-231 and MDA-MB-436, and murine 4T1 and EMT6 cells were obtained from the ATCC. EMT6, MDA-MB-231, and MDA-MB-436 cells were cultured in DMEM supplemented with 10% FBS, penicillin (100 IU/ml), streptomycin (100 μg/ml), nystatin (12.5 U/ml), and 2 mM l-glutamine. 4T1 cells were cultured in RPMI supplemented with 10% FBS, penicillin (100 IU/ml), streptomycin (100 μg/ml), nystatin (12.5 U/ml), 2 mM l-glutamine, 1 mM sodium pyruvate, and 10 mM Hepes buffer.
Splenocytes were isolated from the spleen of adult Balb/c mice and cultured in RPMI supplemented with 10% FBS, 1% Hepes buffer, 1% sodium pyruvate, and 0.1% β-mercaptoethanol. The splenocytes were activated with anti-CD3 (100 ng/ml; Ultra-LEAF Purified anti-mouse CD3 antibody; BioLegend no. 100340), anti-CD28 (10 ng/ml; Ultra-LEAF Purified anti-mouse CD28; BioLegend no. 102116), and Escherichia coli–derived human IL-2 protein (10 ng/ml; R&D Systems no. 202-IL-500) for 48 hours. All cells were grown at 37°C in 5% CO2.
Proliferation assay
D4M.3A (7.5 × 103 cells per well), 131/4-5B1 (15 × 103 cells per well), and A375 (10 × 103 cells per well) cell lines were plated onto a 24-well plate in growth media as indicated previously for 24 hours. Then, the cultured medium was replaced with a medium containing serial dilutions of DBF, TRM, and their combination as free or drug-loaded NPs. For the combined therapy, TRM concentration was 10 times more diluted than DBF concentration, which is equivalent to the ratio of the drugs in the NPs. Untreated (control) cells were supplemented with fresh drug-free medium 24 hours after seeding. For MDA-MB-436, MDA-MB-23, 4T1, and EMT6, cells were seeded in 24-well plates (2.5 × 104, 1.5 × 104, 5 × 103, and 2 × 103 cells per well, respectively) and incubated for 24 hours. Then, the cultured medium was replaced with a medium containing a serial dilution of talazoparib, talazoparib-loaded NPs, or blank NPs (carrier only) for an additional 72 hours. Untreated (control) cells were supplemented with fresh medium containing 0.1% DMSO. Following 72 hours of incubation, the cells were counted using a Z1 Coulter Counter (Beckman Coulter). The proliferation of cells was normalized to the cell growth in the control group.
Isobolograms and CI
The interactions between DBF and TRM were evaluated by constructing isobolograms for different drug ratios and calculating the CI (35). First, the inhibitory concentration (ICx) values of treatment with DBF, TRM, and their combinations were calculated from the proliferation assays. Then, IC30,50,70 values of free TRM and DBF were marked on the x and y axes, and the lines that represent additive effect were drawn between each IC. Data points in the upper right of the IC line represent an antagonistic effect, while in the lower left, they represent a synergistic effect.
Multicellular 3D tumor spheroids invasion assay
D4M.3A, WM115, and EMT6 mCherry-labeled 3D spheroids were prepared by the “hanging drop” method (74). WM115 3D spheroids contained 1000 cells per spheroid, D4M.3A 3D spheroids contained 700 cells per spheroid, and EMT6 contained 1000 cells per spheroid. For MCTS, a mixture of human WM115 cells and brain-resident cells (astrocytes, microglia, pericytes, and endothelial cells) was prepared at a 1:1:0.5:0.5:2 ratio. Each of the cells was labeled with a different fluorescent label, while the microglia cells were not labeled. The cells were deposited in 25-μl droplets composed of 0.24% w/v methylcellulose in medium and incubated on the inner side of a 20-mm dish for 72 hours at 37°C. The plates were placed upside down to allow the formation of 3D spheroids. Then, the 3D spheroids were seeded in Matrigel (BD, Franklin Lakes NJ, USA), in a 96-well plate (50 μl per well), and incubated for 1 hour at 37°C. Next, the 3D spheroids were treated with free drugs or drug-loaded NPs, and their ability to invade the Matrigel was imaged using the EVOS FL Auto cell imaging system (Thermo Fisher Scientific) and quantified with the ImageJ software.
Quantitative real-time PCR
Total RNA was isolated from cultured cells using the Direct-zol RNA Miniprep Plus kit (Zymo Research) following the manufacturer’s instructions. One microgram of isolated RNA was reverse transcribed into cDNA using the qScript cDNA synthesis kit (Quantabio, MA, USA). The resulting cDNA was then diluted 1:50 in DNase/RNase-Free Water for subsequent quantitative real-time PCR (qPCR) analysis.
PD-L1 mRNA levels were quantified using SYBR green real-time PCR (StepOne Plus; Thermo Fisher Scientific, Waltham, MA, USA). The reaction used PerFecTa SYBR Green FastMix ROX (Quanta BioSciences) and custom primers. The specific primer sequences were as follows:
Murine PD-L1 gene (CD247)
Forward: 5′-TTCAGATCACAGACGTCAAGCTG-3′
Reverse: 5′-ATTCTCTGGTTGATTTTGCGGTA-3′
GAPDH gene
Forward: 5′-ATTCCACCCATGGAATTC-3′
Reverse: 5′-GGATCTCGCTCCTGGAAGATG-3′
In vitro cell viability in the presence of nontargeted NPs or P-selectin–targeted NPs
Cell viability of murine NIH/3T3 was assessed by XTT assay. Briefly, 1 × 103 cells were seeded in a 96-well plate and incubated overnight. Then, the medium was replaced with 200 μl of medium containing different concentrations of NPs. After 72 hours of incubation, 50 μl of XTT reagent was added and incubated for 4 hours. Absorbance was measured at λ = 450 nm using a SpectraMax ELISA plate reader (Molecular Devices LLC, Sunnyvale, CA, USA). Cell culture medium was used as the negative control.
Hemolysis assay
Fresh blood was obtained from male Wistar rats by cardiac puncture and collected in heparinized tubes. The erythrocytes were washed three times with PBS and resuspended to a final 2% w/v solution. The solution was then incubated with serial dilutions of nontargeted NPs or P-selectin–targeted NPs (0.001 to 5 mg/ml) for 1 hour at 37°C. Dextran (Mw 70 kDa) and PBS were used as negative controls, whereas SDS was used as a positive control. Following centrifugation, the supernatants were transferred to a 96-well plate, and the absorbance was measured at 550 nm using a SpectraMax M5e plate reader. The results are normalized to the percentage of hemoglobin released by a 1% w/v of Triton X-100 solution (100% lysis).
In vivo motor coordination behavioral test
DBF- and TRM-related motor coordination was assessed by a Rotarod apparatus (Columbus Instruments, OH, USA). The test measures the mouse’s ability to maintain itself on a rod that turns at an increasing speed. Mice were tested before treatment initiation and following the seventh treatment with DBF (1 or 2 mg/kg) and TRM (0.1 or 0.2 mg/kg) as free or drug-loaded NPs, respectively. The control groups were treated with PBS or vehicle [0.01% DMSO, PEG 200 (83 mg/ml), and Tween 80 (14 mg/ml) in PBS]. The initial speed was 1.6 rpm, with an acceleration rate of 4 rpm per min. Animals were tested five times during each session, with at least 2 min of rest between each test. The three best performances of each mouse were considered, and the results were averaged for the whole group.
In vivo biodistribution and tumor accumulation
For melanoma studies, male C57BL/6 mice were inoculated subcutaneously with 1 × 106 D4M.3A cells at a dorsal site (upper back). Mice bearing ≈230 mm3 tumors were treated intravenously with PLGA-Cy5 NPs, P-selectin–targeted NPs, or DBF-Cy5 at 20 μM Cy5 equivalent concentration. Tumor accumulation of Cy5-labeled DBF or NPs was monitored by CRI Maestro noninvasive intravital fluorescence imaging system. Mice were anesthetized using ketamine (100 mg/kg) and xylazine (12 mg/kg), and shaved, and the accumulation of Cy5 signal was recorded 24 hours after treatment. At 3 and 24 hours after treatment, mice were euthanized, and the NP accumulation in organs and tumors was calculated in ex vivo fluorescence images, in which the total fluorescence was divided by the organ area.
In studies on BRCA-mutated cells, female BALB/c mice were inoculated subcutaneously into the mammary fat pad with 0.2 × 106 EMT6 cells. Mice bearing ≈200 mm3 tumors (10 days) were treated intravenously with Cy5-labeled PLGA-PEG NPs or PLGA-PEG-GLY-(OSO3Na)2 at 20 μM Cy5 concentration. At 3 and 24 hours after treatment, mice were euthanized and the NP accumulation in tumors was calculated ex vivo. The NP distribution to essential organs was monitored 24 hours after injection.
For accumulation in brain tumors, D4M.3A cells (50 × 103 cells/2 μl) or EMT6 cells (15 × 103 cells/2 μl) were stereotactically implanted into the striatum of male C57BL/6 mice (melanoma model) or female BALB/c mice (BC model), respectively. Ten days after implantation, the tumors were visualized by MRI (T1 weighted with a contrast agent, MR Solutions) and, the following day, were treated with DBF-Cy5– or Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs at 20 μM Cy5 equivalent concentration. Three hours (BC model) or 6 hours (melanoma model) after treatment, the mice were euthanized, and the brains were harvested. The brains were imaged by the CRI Maestro noninvasive intravital fluorescence imaging system, and the Cy5 signal was calculated in ex vivo fluorescence images, in which the total fluorescence was divided by the brain area. In all experiments, multispectral image cubes were acquired through the 590- to 750-nm spectral range in 10-nm steps using excitation (635 nm) and emission (675 nm) filter sets. Mice auto-fluorescence and undesired background signals were eliminated by spectral analysis and the Maestro linear unmixing algorithm.
For tumor accumulation in D4M.3A tumors after radiation treatment, male C57BL/6 mice were inoculated subcutaneously with 1 × 106 D4M.3A cells at a dorsal site (upper back). Mice bearing ≈100 mm3 tumors were randomized into two groups; one group of mice was subjected to irradiation treatment with 2 Gy 4 hours before the NP injection, and the other group of mice was not irradiated (BIOBEAM GM, Gamma Service Medical, Germany). Then, the mice were intravenously administered with 20 μM Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs. The Cy5 signal from the tumors was monitored using IVIS instrument and unmixing mode of analysis.
Pharmacokinetic study
Establishment of the LC-MS/MS method
Analysis was performed on LC-Vanquish UHPLC Systems and MS-TSQ Altis. Chromatographic parameters included an ACQUITY UPLCHSS T3 column (2.1 × 100 mm, 1.8 μm) with a mobile phase of ammonium acetate buffer 10 mM and methanol. The flow rate was set at 0.35 ml/min, column temperature was set at 30°C, and injection volume was set at 1 μl. A gradient elution program was applied: 0 to 1 min, 50 to 80% methanol; 1 to 2.5 min, 80% methanol; 2.5 to 3.5 min, 80 to 95% methanol; 3.5 to 5.9 min, 95% methanol; 5.9 to 6.1 min, 95 to 50% methanol; 6.1 to 8 min, 50% methanol. Mass spectrometric parameters included an electrospray ionization (ESI) source with a source voltage of 3.5 kV. The ion transfer tube temperature was 325°C, and the heating module temperature was set at 350°C. Multiple reaction monitoring mode was used to detect ion pairs and collision energies. Omeprazole was used as an internal standard solution.
Sample preparation
C57BL/6 mice were administered intravenously with DBF- and TRM-loaded PLGA-PEG-GLY-(OSO3Na)2 NPs at a dose of 3 mg/kg for DBF and 0.3 mg/kg for TRM. Blood samples (0.5 to 0.8 ml) were collected from the orbital vein into clot activator tubes at 5, 30, and 60 min after administration. The blood samples were centrifuged (10,000 rpm for 5 min at 10°C). After centrifugation, the serum was isolated and stored at −80°C. Then, 100 μl of serum was thawed on an ice-water bath and 10 μl of omeprazole (0.001 mg/ml) was added to the sample following the addition of 1 ml of TBME. Then, the samples were vortex mixed, shaken (10 min at 1400 rpm), and centrifuged (7 min at 20°C and 20,800g). After freezing the samples in a dry ice-ethanol bath, the organic layer was removed and evaporated for 15 min at 40°C under nitrogen. The dry extract was then reconstituted with 100 μl of ACN and 1 μl of the final extract was injected.
P-selectin expression by flow cytometry
P-selectin expression in 2D plated D4M.3A and WM115 cells was measured by flow cytometry. First, the cells were grown to high confluency in a 10-mm petri dish, then scraped with PBS, centrifuged (2000 rpm, 5 min, 4°C), and resuspended in 1 ml of FACS (fluorescence-activated cell sorting) buffer (PBS supplemented with 0.5% BSA and 0.5 mM EDTA). For P-selectin expression in 3D spheroids, tumor spheroids were recovered from the Matrigel using a cell recovery solution (BD, catalog no. 354253). The recovered cells were washed with PBS and centrifuged (2000 rpm, 5 min, 4°C). Then, 0.5 × 106 D4M.3A cells were incubated with FITC rat anti-mouse CD62P antibody or with FITC rat IgG1 Isotype control for 1 hour on ice. For human WM115 cells: 1 × 106 cells were incubated with mouse anti-human CD62E/P antibody or with mouse IgG1 Isotype control for 1 hour on ice. Then, the cells were washed with PBS, centrifuged (2000 rpm, 5 min, 4°C), and incubated with a secondary antibody, goat anti-mouse Alexa Fluor 647, for 1 hour on ice. For EMT6, the cells were washed with PBS, centrifuged (2000 rpm, 5 min), and resuspended in 1 ml FACS buffer (PBS supplemented with 1% FBS, 0.1% NaN3, and 0.5 mM EDTA). A total of 1 × 106 EMT6 cells were then incubated with fluorescent-CD62p antibody (BD, FITC Rat anti-mouse, no. 561923, 1:20 dilution) or with Isotype control [BD, FITC rat immunoglobulin G1 (IgG1), no. 553995, 1:20 dilution] for 1 hour on ice. After incubation, the cells were washed with PBS twice, and the fluorescence intensity was analyzed using an Attune NxT Acoustic Focusing Flow Cytometer (Thermo Fisher Scientific, MA, USA). The results were analyzed by the Kaluza software.
Human formalin-fixed, paraffin-embedded specimens
Formalin-fixed, paraffin-embedded (FFPE) melanoma samples (primary and brain metastasis) were obtained from the Sheba Medical Center following informed consent. The manipulation of the human samples for immunostaining was accepted by the ethics committees of Tel Aviv University and the Sheba Medical Center, under an approved institutional review board (5727-18-SMC).
P-selectin expression by immunohistochemistry
For immunostaining of murine melanoma tumors and brain metastases or murine BC tumors and brain metastases, tissues were embedded in an optimal cutting temperature (OCT) compound and cryo-sectioned into 5-μm-thick sections. Immunostaining was performed using the BOND RX automated immunohistochemistry (IHC) Stainer (Leica Biosystems). Slides were fixated in cold acetone for 20 min and then incubated with 10% goat serum for 30 min to block nonspecific binding sites. Next, the slides were incubated with mouse anti-mouse/rat P-selectin antibody (catalog no. 148301 BioLegend, 1:30 dilution) for 1 hour. Then, the slides were incubated with Mouse-IgG ƙ BP-CFL 488 (catalog no. sc-516176, Santa Cruz Biotechnology, 1:20 dilution) for an additional 1 hour, followed by Hoechst fluorescent dye (1:5000) for an additional 10 min for nuclei counterstaining. Tissues were later mounted with ProLong Gold antifade mountant (Thermo Fisher Scientific) and imaged using the EVOS FL Auto cell imaging system (Thermo Fisher Scientific). For human samples: FFPE slides (10 μm) were re-heated to 60°C for 20 min and then incubated with 10% goat serum for 30 min to block nonspecific binding sites. The slides were subsequently incubated with mouse anti-human E/P selectin (R&D, 1:30 dilution) for 1 hour, and then incubated with mouse-IgG ƙ BP-CFL 488 (catalog no. sc-516176, Santa Cruz Biotechnology, 1:20 dilution) for an additional 1 hour, followed by Hoechst fluorescent dye (1:5000) for an additional 10 min for nuclei counterstaining. Tissues were then mounted with 70% ethanol for 1 min, 95% ethanol for 1 min, 100% ethanol for 1 min, 100% ethanol for 1 min, xylene for 1 min, xylene for 1 min, and xylene-based mounting (Thermo Fisher Scientific).
Synthesis of PLGA-Cy5 polymer
Synthesis of nonsulfated Cy5 dye (Cy5-COOH) was conducted as previously described (75, 76). Then, Cy5-COOH dye was conjugated to PLGA polymer by esterification. Briefly, PLGA (100 mg, 0.008 mmol, 1 equiv), Cy5 (5.8 mg, 0.010 mmol, 1.5 equiv), N,N′-dicyclohexylcarbodiimide (2.6 mg, 0.013 mmol, 1.5 equiv), and 4-dimethylaminopyridine (0.5 mg, 0.004 mmol, 0.5 equiv) were dissolved in 2 ml of dry dichloromethane (DCM) and stirred in the dark for 48 hours, at room temperature (RT). The reaction progress was followed by HPLC. At the end of the reaction, the resulting polymer in DCM (1 ml) was precipitated into 40 ml of hexane and centrifuged. Next, the supernatant was decanted, and the process was repeated thrice. Last, the polymer was dried under a high vacuum (HV) overnight, to yield PLGA-Cy5 as a blue powder (93% yield).
1H NMR (400 MHz; CDCl3; 25°C): δ (ppm) = Cy5: 8.17 (t, J = 6.8 Hz, 2H), 7.46 to 7.35 (m, 4H), 7.26 to 7.22 (m, 2H), 7.18 to 7.06 (m, 2H), 7.01 (s, 1H), 6.73 (d, J = 7.0 Hz, 1H), 6.45 to 6.26 (m, 1H), 2.36 (t, J = 7.48 Hz, 2H). PLGA: 5.4 to 5.1 [m, 92H, CH, PLGA backbone], 5.0 to 4.5 (m, 184H, CH2 PLGA backbone), 1.7 to 1.5 [m, 276H, CH3, PLGA backbone].
P-selectin targeted PLGA-PEG polymer synthesis
Synthesis of PLGA-PEG-SO3Na (polymer 3)
Synthesis of homobifunctional PEG-bis-NH2 (polymer 1)
PEG-bis-OH (Mw 2 kDa, 16.0 g, 16 mmol of OH groups) was melted at 65°C under HV to remove traces of water overnight. After cooling to RT, the polymer was dissolved in 150 ml of anhydrous DCM under argon. Dry triethylamine (11.0 ml, 80 mmol, 5 equiv) was added while stirring and the mixture was cooled with an ice bath, followed by the dropwise addition of methanesulfonyl chloride (4.9 ml, 64 mmol, 4 equiv). The reaction proceeds overnight under argon at RT. The crude mixture was diluted with DCM (200 ml) and washed twice with brine (200 ml). The organic layer was dried with sodium sulfate and concentrated in vacuo. The residue was precipitated in diethyl ether and stirred vigorously for 2 hours, filtered, and washed again with diethyl ether to remove traces of excess triethylamine and methanesulfonyl chloride. Drying in HV yielded the desired PEG-bis-OMs product as a white powder in quantitative yield.
PEG-bis-OMs (15.0 g) was added to 150 ml of 25% aqueous ammonia solution and stirred for 4 days at RT in a sealed flask. The ammonia was evaporated under HV, the pH of the aqueous solution was raised to 13 with 1 N NaOH, and the solution was extracted three times with 100 ml of DCM. The organic layers were combined and dried over Na2SO4, filtered, and evaporated to dryness. Then, the crude was redissolved in DCM and precipitated by the dropwise addition of ether. The solid was filtered, washed with ether, and dried under HV to obtain polymer 1 as a white solid in quantitative yield.
1H NMR (400 MHz; DMSO-d6; 25°C): δ (ppm) = 3.75 to 3.62 (m, 180H, PEG backbone and -OCH2-CH2-NH2), 2.65 (t, J = 5.9 Hz, 4H, -CH2-CH2-NH2).
Synthesis of NH2-PEG-SO3Na (polymer 2)
PEG-bis-NH2 (polymer 1) (1.0 g, 0.5 mmol, 1 equiv) was taken in a round-bottom flask equipped with a magnetic stir bar and dissolved in tetrahydrofuran (THF) (10% w/v). Then, 10% w/v propane sultone (61 mg, 0.5 mmol, 1 equiv) in THF was added dropwise to the PEG solution, and the reaction was stirred at 50°C for 5 hours (77). Then, as the reaction proceeded, the product was precipitated, filtered, washed with cold THF, and dried under HV overnight (yield 52%).
1H NMR (400 MHz; CDCl3; 25°C): δ (ppm) = 3.76 to 3.53 (m, 180 H, PEG backbone, -CH2-SO3H and -CH2-CH2-NH2), 3.132 to 3.14 (m, 2H, -CH2-CH2-NH-CH2- and -CH2-NH2), 3.05 to 2.95 (m, 2H, -CH2-CH2-CH2-SO3H), 2.33 to 2.20 (m, 2H, -CH2-CH2-CH2-SO3H).
Synthesis of PLGA-PEG-SO3Na (polymer 3)
PLGA (Resomer RG 502 H, Sigma-Aldrich) (Mw 7 to 17 kDa, lactide:glycolide 50:50, 1.0 g, 0.143 mmol, 1 equiv) was taken in a round-bottom flask, then NHS (0.083 g, 0.715 mmol, 5.0 equiv) and dry DCM (10 ml) were added and stirred at RT. Next, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) (0.11 g, 0.572 mmol, 4 equiv) was added and the reaction was stirred overnight. Then, DCM was evaporated, and the product was precipitated in cold MeOH thrice. Last, the residue was collected and dried under HV to obtain PLGA-NHS (90% yield).
Polymer 2 (0.27 g, 0.129 mmol, 3 equiv) was taken in a round-bottom flask. N,N-diisopropylethylamine (DIPEA) (0.04 ml, 0.216 mmol, 5.0 equiv) and dry chloroform (10 ml) were added and stirred at RT for 10 min followed by the addition of PLGA-NHS (0.3 g, 0.043 mmol) and stirred at RT overnight. Then, the reaction mixture was heated at 45°C for 6 hours, and the chloroform was removed. The residue was washed with cold MeOH thrice (−20°C), and the residue was collected by centrifugation. The final product was dried under HV to obtain PLGA-PEG-SO3Na (polymer 3) as a white solid (70% yield).
1H NMR (400 MHz; CDCl3; 25°C): δ (ppm) = 5.35 to 5.10 [m, 92H, CH, PLGA backbone], 4.95 to 4.55 (m, 184H, CH2, PLGA backbone), 3.76 to 3.53 (m, 180H, PEG backbone), 1.66 to 1.50 (m, 276H, CH3, PLGA backbone).
Synthesis of PLGA-PEG-GLY-(OSO3Na)2 (polymer 8)
Synthesis of compound A
Solketal (0.5 g, 3.78 mmol, 1 equiv) was taken in a flame-dried round-bottom flask, and dry DCM (8 ml) was added under argon. Then carbonyldiimidazol (0.79 g, 4.9 mmol, 1.3 equiv) was dissolved in dry DCM (3 ml) and added to the above reaction mixture instantly. The reaction mixture was stirred at RT for 3 hours and washed with brine. The organic layer was dried with sodium sulfate, and a flash column was performed using ethyl acetate/hexane. Pure product A (oil) was obtained with 20% ethyl acetate/hexane (yield 70%).
1H NMR (400 MHz; CDCl3; 25°C): δ (ppm) = 8.15 (s, 1H, -N-CH=N-CH), 7.44 [s, 1H, -N(CH)-CH=CH-], 7.07 (s, 1H, -N(CH)-CH=CH-), 4.55 to 4.35 (m, 3H, -O-CH2-CH-), 4.2 to 4.1 (m, 1H, -O-CH2-CH-CH2), 3.9 to 3.8 (m, 1H, -O-CH2-CH-CH2), 1.4 (s, 3H, -CH3), 1.3 (s, 3H, -CH3).
Synthesis of NH2-PEG-acetal (polymer 4)
PEG-bis-NH2 (polymer 1) (1.0 g, 0.5 mmol, 1 equiv) was taken in a round-bottom flask equipped with a magnetic stir bar. Then, 18 ml of dry chloroform and DIPEA (0.13 ml, 0.75 mmol, 1.5 equiv) were added under argon, and the reaction mixture was stirred for 5 min at RT. Then, compound A (0.178 g, 0.6 mmol, 1.2 equiv) dissolved in dry chloroform (2 ml) was added dropwise to the reaction mixture and stirred overnight at RT.
Then, the chloroform was evaporated, and the reaction mixture was loaded on a MeOH-based LH20 (Sephadex) size exclusion column. Fractions that contained the product (identified by coloring with iodine) were unified, the organic solvents were evaporated to dryness, and the product was dried under HV. Polymer 4 was obtained as a white solid in quantitative yield.
1H NMR (400 MHz; CDCl3; 25°C): δ (ppm) = 5.45 (s, 1H, CH2-NH-CO2-), 4.35 to 4.05 (m, 5H, -CO2-CH2-, CH-CH2-O-, and CH2-CH-O-CH of the solketal group), 4.0 to 3.25 (m, 180H, PEG backbone), 2.94 (t, J = 5.04 Hz, 2H, -CH2-CH2-NH-CO2-), 2.8 (t, J = 5.35 Hz, 2H, -CH2-CH2-NH2), 1.4 (s, 3H, -CH3), 1.3 (s, 3H, -CH3).
Synthesis of NH2-PEG-GLY-(OH)2 (polymer 5)
Polymer 4 was dissolved in 4 ml of dry DCM, and 3 ml of TFA was added dropwise to the solution and stirred overnight. TFA and DCM were evaporated, and the reaction mixture was loaded on a MeOH-based LH20 (Sephadex) size exclusion column. Fractions that contained the product (identified by coloring with iodine) were unified, the organic solvents were evaporated to dryness, and the product was dried under HV. Polymer 5 was obtained as a white solid in quantitative yield.
1H NMR (400 MHz; CDCl3; 25°C): δ (ppm) = 5.94 (s, 1H, CH2-NH-CO2-), 4.4 to 4.1 [m, 2H, -NH-CO-CH2-CH (OH)-CH2-OH], 4.0 to 3.1 (m, 180H, PEG backbone, -CH-CH2-OH from glycerol and -CH2-CH2-NH-), 3.2 to 3.05 (m, 2H, -CH2-CH2-NH2).
ESI-MS spectra (fig. S10) of polymer 1 and polymer 5 showed Mp (peak with high intensity) values that were found to be at 1823.1 and 1940.9 g/mol, respectively. Thus, the Mp peak shifted to higher mass/charge ratio (m/z) by 118 g/mol, which corresponds to one glycerol unit.
Further, all the masses in the distribution can be explained by the following equations
| (3) |
| (4) |
Synthesis of Acetyl-PLGA-OH (polymer 6)
PLGA (Resomer RG 502 H, Sigma-Aldrich) (0.5 g, 0.072 mmol, 1 equiv) was taken in a round-bottom flask equipped with a magnetic stir bar. Dry DCM (8 ml) and triethylamine (0.06 ml, 0.432 mmol, 6 equiv) were added and stirred at RT for 10 min. Then, acetyl chloride (0.03 ml, 0.486 mmol, 4.5 equiv) was added at 0°C and stirred at RT overnight. Afterward, DCM was removed, and the product was precipitated in cold MeOH thrice (−20°C). The residue was collected and dried under HV to obtain polymer 6 as a white solid (93% yield).
1H NMR (400 MHz; CDCl3; 25°C): δ (ppm) = 5.5 to 5.1 (m, 92H, CH, PLGA backbone), 5.0 to 4.0 (m, 184H, CH2, PLGA backbone), 2.2 (s, 3H, CH3-COO-), 1.7 to 1.3 (m, 276H, CH3, PLGA backbone).
Synthesis of PLGA-PEG-GLY-(OH)2 (polymer 7)
Polymer 6 (1.0 g, 0.143 mmol, 1 equiv) was taken in a round-bottom flask. Then, NHS (0.083 g, 0.715 mmol, 5.0 equiv) and dry DCM (10 ml) were added and stirred at RT. Then, EDC (0.11 g, 0.572 mmol, 4 equiv) was added and stirred overnight. DCM was removed, and the product was precipitated in cold MeOH thrice (−20°C). The residue was collected by centrifugation and dried under HV to obtain NHS ester of polymer 6 as a white solid in quantitative yield.
Polymer 5 (0.27 g, 0.129 mmol, 3 equiv) was taken in a round-bottom flask. Then, DIPEA (0.04 ml, 0.216 mmol, 5.0 equiv) and dry chloroform (10 ml) were added and stirred at RT for 10 min. Then, the NHS ester of polymer 6 (0.3 g, 0.043 mmol, 1 equiv) was added and stirred at RT overnight. Next, the reaction mixture was heated at 45°C for 6 hours, and chloroform was removed. The residue was washed with cold MeOH thrice (−20°C) and collected by centrifugation. Then, it was dried under HV to obtain polymer 7 as a white solid (79% yield).
1H NMR (400 MHz; CDCl3; 25°C): δ (ppm) = 5.5 to 5.1 (m, 92H, CH, PLGA backbone), 5.0 to 4.0 (m, 184H, CH2, PLGA backbone), 4.0 to 3.5 (m, 180H, CH2, PEG backbone), 2.16 (s, 3H, CH3), 1.7 to 1.5 (m, 276H, CH3, PLGA backbone).
Synthesis of PLGA-PEG-GLY-(OSO3Na)2 (polymer 8)
Polymer 7 (0.236 g, 0.052 mmol, 1 equiv) was dissolved in 5 ml of dry dimethylformamide (DMF) and heated to 65°C. Then, the SO3-DMF complex (80 mg, 0.524 mmol, 10 equiv) was dissolved in 3 ml of dry DMF and added dropwise to the solution of 7 under argon. Next, the reaction mixture was heated to 65°C for 24 hours and stirred at RT for another 24 hours. Then, DMF was removed using HV and aqueous NaHCO3 solution was added and stirred for a few minutes. The product was dialyzed in MilliQ water keeping the NaCl solution outside for 24 hours and then in MilliQ water respectively for another 48 hours. The solution present inside the dialysis was lyophilized to obtain polymer 8 as a white solid of 0.17 g (72% yield).
1H NMR (400 MHz; CDCl3; 25°C): δ (ppm) = 5.5 to 5.1 (m, 92H, CH, PLGA backbone), 5.0 to 4.0 (m, 184H, CH2, PLGA backbone), 4.0 to 3.5 (m, 180H, PEG backbone), 2.2 (s, 3H, CH3), 1.7 to 1.5 (m, 276H, CH3, PLGA backbone).
Optimized synthesis of PLGA-PEG-GLY-(OSO3Na)2 (polymer 12)
Commercially available polymer 9 HO-PLGA12K-PEG2K-allyl was first characterized by 1H-NMR and SEC analysis (fig. S13, B and G).
1H NMR (400 MHz, CDCl3): δ (ppm) = 5.97 to 5.84 (m, 1H, CH2 = CH-CH2-O-), 5.33 to 5.06 [m, 83H, -O-CH(CH3)-CO-, PLGA backbone], 4.95 to 4.55 (m, 100H, -O-CH2-CO-, PLGA backbone), 4.09 to 3.92 (m, 2H, CH2 = CH-CH2-O-), 3.85 to 3.43 (m, 181H, -O-CH2-CH2-O-, PEG backbone), 1.78 to 1.38 [m, 250H, -O-CH(CH3)-CO-, PLGA backbone]. SEC (DMF + 25 mM NH4Ac, PEG standards calibration): Mn = 3.8 kDa, Ð = 2.2.
Synthesis of AcO-PLGA-PEG-allyl (polymer 10)
According to fig. S13A, polymer 9 (1.95 g, Mw 11 kDa, 1 equiv) was dissolved in DCM (10 ml per 1 g of polymer). Et3N (240 μl, 10 equiv) and acetyl-Cl (250 μl, 20 equiv) were added, and the reaction was left to stir overnight at RT. The reaction mixture was diluted with DCM, washed with brine, and then with saturated ammonium chloride solution twice. The organic layer was separated and dried and then the organic solvent was evaporated to dryness and the product was dried under high vacuum. The product was obtained as a white solid in quantitative yield (1.85 g).
1H NMR (400 MHz, CDCl3): δ (ppm) = 5.98 to 5.84 (m, 1H, CH2 = CH-CH2-O-), 5.38 to 5.06 [m, 83H, -O-CH(CH3)-CO-, PLGA backbone and CH2 = CH-CH2-O-], 4.97 to 4.51 (m, 100H, -O-CH2-CO-, PLGA backbone), 4.01 (dt, J = 5.6, 1.5 Hz, 2H, CH2 = CH-CH2-O-), 3.88 to 3.40 (m, 181H, -O-CH2-CH2-O-, PEG backbone), 2.06 (s, 3H, -O-CO-CH3), 1.77 to 1.35 [m, 250H, -O-CH(CH3)-CO-, PLGA backbone]. SEC (DMF + 25 mM NH4Ac, PEG standards calibration): Mn = 4.3 kDa, Ð = 2.0.
Synthesis of AcO-PLGA-PEG-glycol (polymer 11)
Polymer 10 (1.80 g, 1 equiv), 1-thioglycerol (430 μl, 30 equiv), and DMPA (13 mg, 0.3 equiv; 1 mol% concerning the thiol) were dissolved in DMF (3 ml per 1 g of hybrid). The solution was purged with nitrogen for 30 min and then stirred under UV light (365 nm) for 2 hours. Then, the reaction mixture was dissolved in a minimal amount of DCM, and the product was precipitated using a slow addition of ether (150 ml). The obtained solid was filtered, washed with ether, and lastly dried under a high vacuum. The product was obtained as a white solid in quantitative yield (1.80 g).
1H NMR (400 MHz, CDCl3): δ (ppm) = 5.39 to 5.05 [m, 83H, -O-CH(CH3)-CO-, PLGA backbone], 4.98 to 4.51 (m, 100H, -O-CH2-CO-, PLGA backbone), 3.88 to 3.40 (m, 186H, -O-CH2-CH2-O-, PEG backbone and glycol), 2.73 to 2.52 (m, 4H, -CH2-S-CH2-), 2.13 (s, 3H, -O-CO-CH3), 1.93 to 1.76 (m, 2H, -O-CH2-CH2-CH2-S-), 1.77 to 1.35 [m, 250H, -O-CH(CH3)-CO-, PLGA backbone]. SEC (DMF + 25 mM NH4Ac, PEG standards calibration): Mn = 5.0 kDa, Ð = 1.8.
Synthesis of AcO-PLGA-PEG-(OSO3Na)2 (polymer 12)
Polymer 11 (980 mg, 1 equiv) was dissolved in dry DMF (5 ml per 1 g), and SO3-DMF complex (270 mg, 20 equiv) was added to the solution. Then, the reaction mixture was heated to 65°C for 2 hours. The solvent was evaporated, and the mixture was quenched by the addition of 5 ml of aqueous NaHCO3 and further diluted with 50 ml of water. The aqueous mixture was left to cool overnight, and the polymer was precipitated and separated from the solution using a centrifuge. The white solid was then resuspended in water, cooled, and centrifuged. This step was repeated twice more. The precipitate was lastly resuspended in water and lyophilized to obtain the final product in 83% yield (810 mg). Because of interactions of the sulfonates with the column, this polymer could not be analyzed by SEC.
1H NMR (400 MHz, CDCl3): δ (ppm) = 5.32 to 5.05 [m, 120H, -O-CH(CH3)-CO-, PLGA backbone], 4.96 to 4.53 (m, 137H, -O-CH2-CO-, PLGA backbone), 3.86 to 3.41 (m, 186H, -O-CH2-CH2-O-, PEG backbone and glycol), 2.82 to 2.65 (m, 4H, -CH2-S-CH2-, partially falls under water), 2.12 (s, 3H, -O-CO-CH3), 1.93 to 1.81 (m, 2H, -O-CH2-CH2-CH2-S-), 1.77 to 1.34 [m, 360H, -O-CH(CH3)-CO, PLGA backbone].
The analyzed NMR of the product (polymer 12), along with the lower yield, indicated a fractional precipitation that produced a separation of the hybrids’ population; while hybrids containing the PLGA block with higher molecular weight (MW) were precipitated under the said conditions, the lower MW PLGA block remained in the solution due to its relative increased solubility in water. The separated aqueous supernatant was centrifuged once more, using dialysis tubes, to estimate the amount of polymer as well as its Mn. The white solid was resuspended in water and lyophilized and then analyzed by 1H-NMR. The weight ratio of the two samples (83 and 15%) correlated well with the calculated MW of the PLGA block (12 and 4 kDa).
1H NMR (400 MHz, CDCl3): δ (ppm) = 5.28 to 5.05 [m, 39H, -O-CH(CH3)-CO-, PLGA backbone], 4.96 to 4.53 (m, 52H, -O-CH2-CO-, PLGA backbone), 3.86 to 3.41 (m, 186H, -O-CH2-CH2-O-, PEG backbone and glycol), 2.82 to 2.65 (m, 4H, -CH2-S-CH2-), 2.12 (s, 3H, -O-CO-CH3), 1.93 to 1.78 (m, 2H, -O-CH2-CH2-CH2-S-), 1.77 to 1.36 [m, 114H, -O-CH(CH3)-CO-, PLGA backbone].
Size exclusion chromatography
The polymers were characterized using Malvern Viscotek GPCmax equipped with 2xPSS GRAM 1000 Å. After a needle wash with DMF, the polymer was run with a mobile phase composed of DMF + 25 mM ammonium acetate (NH4Ac) with a flow rate of 0.5 ml/min with a runtime of 60 min. Fifty microliters from a sample amphiphiles solution (20 mg/ml) in the mobile phase was injected, and before that, the polymer solution was filtered with a 0.45 μM PTFE syringe filter. Column temperature: 50°C; Detector: Viscotek VE3580 RI detector (PEG standards were used for calibration, and the Mn and D values of the amphiphiles were calculated accordingly).
X-ray photoelectron spectroscopy
To determine the PLGA-PEG-Glycerol-(SO3)2 NP elemental composition and to determine that the sulfate groups are present on the surface of the nanoparticles, XPS, a surface-sensitive and quantitative analysis, was performed. Samples were prepared by depositing 60 μl of freshly prepared PLGA-PEG-Glycerol-(SO3)2 NPs in silica wafers. The samples were covered with a petri dish to avoid any contamination for 72 hours, until the complete evaporation of the drop. Before the spectral acquisition, surface cleaning was performed via sputtering with a 1000 Cluster Ar ion beam at an energy of 4 keV. The sputtering duration was set to 60 s, an interval selected to effectively remove surface contaminants without significantly altering the inherent surface chemistry of the sample. The analyses used an x-ray beam with a diameter of 650 μm under ultrahigh vacuum conditions. The XPS system was equipped with a monochromatic Al Kα x-ray source to excite the electrons. Spectra were acquired using pass energy appropriate for high-resolution scans, with the exact value determined based on the equipment’s specifications and the information depth required. Charge compensation was achieved using a dual-beam system. This system employs both low-energy electrons and a low-energy argon ion beam concurrently to neutralize the positive charge that can accumulate during XPS analysis. All measurements were performed at RT, and the data were calibrated against the C 1s peak at 284.8 eV if required, to compensate for any charging effects that were not neutralized by the dual beam system.
Internalization of PLGA-Cy5 NPs into 3D spheroids
D4M.3A, WM115, and EMT6 3D spheroids were prepared by seeding the cells (1000 cells per spheroid for WM115 cells and 500 cells per spheroid for D4M.3A cells) in an ultralow attachment round-bottom 96-well plate (Corning) and allowing the cells to form 3D spheroids for 96 hours. Next, the medium was replaced with a fresh medium containing 1 μM Cy5-labeled targeted and nontargeted NPs. For NP internalization following treatments with P-selectin inhibitor, the 3D spheroids were treated with P-selectin inhibitor (0.1, 1, and 10 μM) for 1 hour before adding the NPs. The plate was then placed in the IncuCyte Live cell analysis system (Essen Bioscience) and the Cy5 fluorescence intensity within the 3D spheroids (bright field) was monitored for 20 to 24 hours.
To image the internalization of PLGA-PEG-GLY-(OSO3Na)2 NPs into the core of D4M.3A spheroids, the spheroids were prepared and treated as described above, in the presence or absence of pretreatment with 10 μM P-selectin inhibitor for 1 hour at 37°C. Then, after 24 hours of incubation with 1 μM Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs, the spheroids were fixed with 4% PFA, mounted on a slide with ProLong Gold Antifade Mountant with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen), and imaged with BioTek Cytation C10, 20× magnification. The picture was taken from the middle of the Z-stack.
NPs binding to immobilized P-selectin
Recombinant human P-selectin (1 ng/μl, 50 μl per well) was incubated in a Nunc-Immuno 96-microwell plate (Sigma-Aldrich) at 4°C, overnight. Then, the wells were blocked with 3% skim milk solution (200 μl per well) for 1 hour, washed three times with 200 μl of PBS, and incubated with Cy5-labeled PLGA-PEG-GLY-(OSO3Na)2 NPs or PLGA-PEG NPs for 15 min at three different concentrations: 500, 300, and 150 μg/ml. Next, the wells were washed three times with 200 μl of PBS, and the Cy5 fluorescence signal was measured by a SpectraMax M5e plate reader.
Ethics statement
Animals were housed in the Tel Aviv University animal facility. All experiments received ethical approval from the Animal Care and Use Committee (IACUC) of Tel Aviv University (approval nos. 01-18-027 and 01-21-005) and were conducted under NIH guidelines.
Animal studies
To evaluate the antitumor activity of drug-loaded NPs compared to free drugs, 7- to 8-week-old male C57BL/6 mice (Envigo CRS, Israel) were inoculated subcutaneously with 1 × 106 D4M.3A cells. The tumors were allowed to be established for 10 days (tumor size ~180 mm3), and then the mice were treated with DBF, TRM, or their combination as drug-loaded NPs or free drugs. Treatments were administered intravenously at 3 mg/kg DBF and 0.3 mg/kg TRM. Control groups were treated with PBS, Blank (empty) NPs, and vehicle [0.01% DMSO, PEG 200 (83 mg/ml), and Tween 80 (14 mg/ml) in PBS]. Mice were treated QOD, and the tumors were measured by a digital caliper according to the formula: width2 × length × 0.52. Body weight and tumor size were monitored every other day. After the seventh treatment, the mice were euthanized, perfused with PBS and 4% formaldehyde, and the tumors were harvested. The tissues were then embedded in an OCT compound and frozen sectioned into 5-μm-thick sections. Immunostaining of the frozen sections was performed using the BOND RX automated IHC stainer as described above. The slides were stained with rabbit anti-mouse phospho-Braf/phospho-MEK1/2/phospho-Erk1/2 for 1 hour, followed by staining with secondary antibody goat anti-rabbit Alexa Fluor 647. Nuclei were stained with Hoechst fluorescent dye (1:5000) for an additional 10 min. The tissues were fixed and mounted on a glass microscope slide with a glass coverslip using ProLong Gold antifade reagent (Invitrogen).
For drug dosing assessment, 1 × 106 D4M.3A cells were inoculated subcutaneously into 7- to 8-week-old C57BL/6 mice (Envigo CRS, Israel) and allowed to form tumors for 10 days (tumor size ~70 mm3). Mice were then randomized into six groups and treated with intravenous injections of either free or drug-loaded NPs at 1 or 2 mg/kg DBF and 0.1 or 0.2 mg/kg TRM, QOD. The body weight and tumor volume were monitored every other day, and the mice were euthanized when tumor volume reached >1000 mm3 or when mice lost more than 20% of their initial (day 0) body weight. Blood samples were drawn from the submandibular vein following the ninth treatment for a complete blood count.
An additional efficacy study was performed with P-selectin–targeted NPs. D4M.3A cells (1 × 106 cells per mouse) were subcutaneously injected into 6- to 8-week-old male C57BL/6 mice (Envigo CRS, Israel). Following 10 days (tumor size ~50 mm3), the mice were randomized into five groups and were intravenously administered with P-selectin–targeted or nontargeted NPs or with the combination of free drugs, at 1 mg/kg DBF and 0.1 mg/kg TRM. When the first treated group reached tumor size >1000 mm3, all treatments were discontinued. Body weight and tumor size were monitored every other day, and mice were euthanized when tumor volume > 1000 mm3 or when they lost more than 20% of their initial (day 0) body weight. Blood samples were drawn from the submandibular vein following the eighth treatment for biochemistry analysis. In addition, for immunohistochemical staining, D4M.3A cells (1 × 106 cells per mouse) were subcutaneously injected into 6- to 8-week-old male C57BL/6 mice (Envigo CRS, Israel). Once the tumor size reached ~50 mm3, the mice were randomized into five groups (four to five mice per group). The mice were treated with eight doses of P-selectin–targeted or nontargeted NPs or with the combination of free drugs, at 1 mg/kg DBF and 0.1 mg/kg TRM, QOD, and then euthanized, perfused with PBS and 4% formaldehyde, and the tumors were harvested. The tissues were embedded in an OCT compound and cryo-sectioned into 5-μm-thick sections. Tumor sections were stained with rabbit anti-mouse phospho-Erk1/2/Ki67/cleaved caspase 3, followed by the secondary antibody, goat anti-rabbit Alexa Fluor 647. Nuclei were stained with Hoechst fluorescent dye (1:5000). All fluorescence images were captured using a fluorescence microscope (Evos FL Auto, Life Technologies) at 40× magnification.
To evaluate the antitumor activity of compound-loaded NPs compared to free compounds in the BC model, 6- to 7-week-old female BALB/c (Envigo CRS, Israel) mice were inoculated into the mammary fat pad with 0.2 × 106 EMT6 cells. The tumors were allowed to establish for 15 days and then the mice were treated with talazoparib and SM56 as compound-loaded NPs (nontargeted and P-selectin–targeted) or free compounds or the corresponding controls. Treatments were administered intravenously at 3.33 mg/kg SM56 and 0.33 mg/kg talazoparib. Control groups were treated with saline, Blank (empty) NPs, and vehicle (0.2% DMSO). Mice were treated 3 days/week, and the tumors were measured by a digital caliper according to the following formula: width2 × length × 0.52. The body weight and the tumor volume were monitored 3 days/week, and the mice were euthanized if they lost more than 20% of their initial body weight. After the sixth treatment, the mice were euthanized.
dSTORM imaging of PD-L1 after exposure of EMT6 to talazoparib
EMT6 cells were seeded in Ibidi μ-slide eight-well glass-bottom chambered coverslips at a density of 5000 cells per well. After 24 hours of incubation, cells were treated with talazoparib for 48 hours (0, 10, and 50 mM). After 48 hours, cells were washed with PBS and fixed using 3.7% formaldehyde solution for 10 min at RT. After fixation, cells were washed thrice with PBS and incubated with 5% BSA solution overnight at 4°C. Antibody staining with anti–PD-L1 (Mouse PD-L1 Alexa Fluor 647 conjugated) or control antibody (Rabbit IgG Alexa Fluor 647 conjugated) was performed for 2 hours at RT at 1 μg/ml antibody dilution in PBS containing 5% BSA (150 μl per well). Subsequently, cells were washed thrice with PBS and post-fixated using 1% formaldehyde solution for 10 min at RT. Last, cells were washed thrice with PBS and stored at 4°C before imaging. Before dSTORM imaging, cells were incubated with WGA-488 (0.5 μg/ml) for 10 min at RT to visualize the cell outline. Cells were washed thrice with PBS and incubated with STORM buffer [5% w/v glucose, 100 mM cysteamine, glucose oxidase (0.5 mg/ml), catalase (40 μg/ml) in PBS]. Before STORM imaging, a low-resolution Total Internal Reflection Fluorescence (TIRF) image was acquired at 5% 647 laser power and 2% 488 laser power. For dSTORM imaging, cells were acquired for 20,000 frames at 16-ms exposure time and 100% 647 laser power. Between 6 and 10 cells were imaged for each condition. dSTORM images were analyzed with the Nikon NIS elements software (version 5.21.01). dSTORM localizations were detected using Gaussian fitting, with a minimum intensity threshold height of 150 for the 647 channel. Molecules detected in five consecutive frames were considered as a single blinking event, while molecules detected in more than five frames were discarded. Drift correction was performed in the NIS elements software, based on an autocorrelation function. The resulting x-y coordinates of the detected localizations were imported and run through a custom MATLAB script to quantify the localization density per cell. Each cell was manually selected by drawing a region of interest (ROI) around the cell contour (detected in the 488 channel). The resulting localization densities per cell were plotted in a box plot using Origin 2020 software.
Murine splenocytes isolation and coculture with EMT6 mCherry-labeled 3D spheroids
Splenocytes were isolated from the spleen of adult Balb/c mice and cultured in RPMI supplemented with 10% FBS, 1% Hepes buffer, 1% sodium pyruvate, and 0.1% β-mercaptoethanol. The splenocytes were activated with anti-CD3 (100 ng/ml; Ultra-LEAF Purified anti-mouse CD3 antibody; BioLegend no. 100340), anti-CD28 (10 ng/ml; Ultra-LEAF Purified anti-mouse CD28; BioLegend no. 102116) and E. coli–derived human IL-2 protein (10 ng/ml; R&D Systems no. 202-IL-500) for 48 hours. All cells were grown at 37°C in 5% CO2. In parallel, EMT6 mCherry-labeled 3D spheroids were prepared by seeding the cells (500 cells per well per spheroid) in an ultralow attachment round-bottom 96-well plate (Corning) and allowing them to form 3D spheroids for 72 hours. After the 3D spheroids’ complete formation, the medium was replaced with a fresh medium containing activated splenocytes (cancer cell:splenocytes = 1:100 ratio) and talazoparib and/or SM56. The plate was placed in the Incucyte Live cell analysis system (Essen Bioscience), and the treatment efficacy was assessed by measuring the mCherry fluorescence intensity from the 3D spheroid for 72 hours.
Statistical analysis
Data were expressed as mean ± SD for in vitro assays and as mean ± SEM for in vivo assays. The sample size (N) for each statistical analysis was added for each experiment. Statistical significance was determined using an analysis of variance (ANOVA). P < 0.05 was considered statistically significant. All statistical measurements were two sided. The Kaplan-Meier curve was created to assess the survival of mice in vivo. The software used for statistical analysis was GraphPad Prism 8.
Acknowledgments
We thank the Sackler Cellular and Molecular Imaging Center (SCMIC) for the imaging services, and special thanks to S. Lichtenstein and Y. Zilberstein for their kind and professional assistance. LC-MS analysis and subsequent quantification of the entitled molecules were performed by V. Galperin and researcher J. Lepsky from the BLAVATNIK CENTER for Drug Discovery at Tel Aviv University, which was supported by L. Blavatnik and the Blavatnik Family Foundation.
Funding: This work was supported by European Research Council (ERC) Advanced Grant 835227, 3DBrainStrom (R.S.-F.); ERC Proof of Concept (PoC) Grant 862580, 3DCanPredict (R.S.-F.); the Israel Science Foundation 1969/18 (R.S.-F.); the Israel Cancer Research Fund PROF-18-682 (R.S.-F.); the Melanoma Research Alliance (MRA) Established Investigator Award 615808 (R.S.-F.); the Morris Kahn Foundation 590219 (R.S.-F.); the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 765497, THERACAT (R.S.-F., R.J.A., and L.A.); the Israel Science Foundation 1553/18 (R.J.A.); the ADAMA Center for Novel Delivery Systems in Crop Protection (S.T.); and the Zuckerman-CHE STEM Leadership Program and Marie Skłodowska-Curie Actions (MSCA) fellowships (R.B.).
Author contributions: Conceptualization: S.K.-M., D.R.A., P.D., H.F.F., R.J.A., and R.S.-F. Methodology: S.K.-M., D.R.A., P.D., R.Kl., S.T., Y.E., M.G.B., R.Kh., S.P., G.S.-K., E.Y., L.W., R.B., S.L., A.S., L.A., R.J.A., I.B., H.F.F., and R.S.-F. Investigation: S.K.-M., D.R.A., P.D., R.Kl., S.T., Y.E., M.G.B., R.Kh., S.P., G.S.-K., E.Y., L.W., R.B., and S.L. Visualization: S.K.-M., D.R.A., P.D., H.F.F., R.J.A., L.A., and R.S.-F. Resources: S.K.-M., D.R.A., P.D., M.G.B., and R.S.-F. Validation: S.K.-M., D.R.A., P.D., M.G.B., H.F.F., and R.S.-F. Formal analysis: S.K.-M., D.R.A., and R.S.-F. Project administration: S.K.-M., R.J.A., and R.S.-F. Funding acquisition: R.S.-F. Data curation: R.S.-F. Supervision: S.K.-M., R.J.A., and R.S.-F. Writing—original draft: S.K.-M., D.R.A., P.D., S.T., and R.S.-F. Writing—review and editing: S.K.-M., D.R.A., P.D., R.Kl., M.G.B., S.P., H.F.F., L.A., R.J.A., and R.S.-F.
Competing interests: R.S.-F. is a board director at Teva Pharmaceutical Industries Ltd. and receives unrelated research funding from Merck KGaA. R.S.-F. is a cofounder and officer with an equity interest in Selectin Therapeutics Inc. R.S.-F., S.K.-M., and P.D. are inventors of a related patent, “P-selectin targeted nanoparticles and uses thereof” PCT patent application no. PCT/IL2024/050131 filed on 2 February 2024, by Ramot at TAU. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S22
REFERENCES AND NOTES
- 1.Danhier F., Ansorena E., Silva J. M., Coco R., Le Breton A., Préat V., PLGA-based nanoparticles: An overview of biomedical applications. J. Control. Release 161, 505–522 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Matsumura Y., Maeda H., A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 46, 6387–6392 (1986). [PubMed] [Google Scholar]
- 3.Monsky W. L., Fukumura D., Gohongi T., Ancukiewcz M., Weich H. A., Torchilin V. P., Yuan F., Jain R. K., Augmentation of transvascular transport of macromolecules and nanoparticles in tumors using vascular endothelial growth factor. Cancer Res. 59, 4129–4135 (1999). [PubMed] [Google Scholar]
- 4.Schroeder A., Heller D. A., Winslow M. M., Dahlman J. E., Pratt G. W., Langer R., Jacks T., Anderson D. G., Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer 12, 39–50 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Prabhakar U., Maeda H., Jain R. K., Sevick-Muraca E. M., Zamboni W., Farokhzad O. C., Barry S. T., Gabizon A., Grodzinski P., Blakey D. C., Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Cancer Res. 73, 2412–2417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferber S., Tiram G., Sousa-Herves A., Eldar-Boock A., Krivitsky A., Scomparin A., Yeini E., Ofek P., Ben-Shushan D., Vossen L. I., Licha K., Grossman R., Ram Z., Henkin J., Ruppin E., Auslander N., Haag R., Calderón M., Satchi-Fainaro R., Co-targeting the tumor endothelium and P-selectin-expressing glioblastoma cells leads to a remarkable therapeutic outcome. eLife 6, e25281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shamay Y., Elkabets M., Li H., Shah J., Brook S., Wang F., Adler K., Baut E., Scaltriti M., Jena P. V., Gardner E. E., Poirier J. T., Rudin C. M., Baselga J., Haimovitz-Friedman A., Heller D. A., P-selectin is a nanotherapeutic delivery target in the tumor microenvironment. Sci. Transl. Med. 8, 345ra387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solhi L., Sun H. S., Daswani S. H., Springate C. M. K., Brumer H., Controlled sulfation of poly (vinyl alcohol) for biological and technical applications using response surface methodology. Mol. Syst. Des. Eng. 5, 1671–1678 (2020). [Google Scholar]
- 9.Chen M., Geng J.-G., P-selectin mediates adhesion of leukocytes, platelets, and cancer cells in inflammation, thrombosis, and cancer growth and metastasis. Arch. Immunol. Ther. Exp. (Warsz.) 54, 75–84 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Vestweber D., Blanks J. E., Mechanisms that regulate the function of the selectins and their ligands. Physiol. Rev. 79, 181–213 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Yeini E., Ofek P., Pozzi S., Albeck N., Ben-Shushan D., Tiram G., Golan S., Kleiner R., Sheinin R., Israeli Dangoor S., Reich-Zeliger S., Grossman R., Ram Z., Brem H., Hyde T. M., Magod P., Friedmann-Morvinski D., Madi A., Satchi-Fainaro R., P-selectin axis plays a key role in microglia immunophenotype and glioblastoma progression. Nat. Commun. 12, 1912 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwamura T., Caffrey T. C., Kitamura N., Yamanari H., Setoguchi T., Hollingsworth M. A., P-selectin expression in a metastatic pancreatic tumor cell line (SUIT-2). Cancer Res. 57, 1206–1212 (1997). [PubMed] [Google Scholar]
- 13.Hoos A., Protsyuk D., Borsig L., Metastatic growth progression caused by PSGL-1–mediated recruitment of monocytes to metastatic sites. Cancer Res. 74, 695–704 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Luke J. J., Flaherty K. T., Ribas A., Long G. V., Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 14, 463–482 (2017). [DOI] [PubMed] [Google Scholar]
- 15.GlaxoSmithKline, Tafinlar® (dabrafenib capsules): US prescribing information (GlaxoSmithKline) (2013); www.accessdata.fda.gov/drugsatfda_docs/label/2022/202806s022lbl.pdf.
- 16.Novartis, Mekinist (trametinib) tablets, for oral use (2013); www.accessdata.fda.gov/drugsatfda_docs/label/2022/204114s024lbl.pdf.
- 17.FDA, FDA grants accelerated approval to dabrafenib in combination with trametinib for unresectable or metastatic solid tumours with BRAF V600E mutation (2022); https://fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-dabrafenib-combination-trametinib-unresectable-or-metastatic-solid.
- 18.Robert C., Grob J. J., Stroyakovskiy D., Karaszewska B., Hauschild A., Levchenko E., Chiarion Sileni V., Schachter J., Garbe C., Bondarenko I., Gogas H., Mandalá M., Haanen J. B. A. G., Lebbé C., Mackiewicz A., Rutkowski P., Nathan P. D., Ribas A., Davies M. A., Flaherty K. T., Burgess P., Tan M., Gasal E., Voi M., Schadendorf D., Long G. V., Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N. Engl. J. Med. 381, 626–636 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Infante J. R., Fecher L. A., Falchook G. S., Nallapareddy S., Gordon M. S., Becerra C., DeMarini D. J., Cox D. S., Xu Y., Morris S. R., Peddareddigari V. G. R., Le N. T., Hart L., Bendell J. C., Eckhardt G., Kurzrock R., Flaherty K., Burris H. A. III, Messersmith W. A., Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: A phase 1 dose-escalation trial. Lancet Oncol. 13, 773–781 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Falchook G. S., Long G. V., Kurzrock R., Kim K. B., Arkenau T. H., Brown M. P., Hamid O., Infante J. R., Millward M., Pavlick A. C., O’Day S. J., Blackman S. C., Curtis C. M., Lebowitz P., Ma B., Ouellet D., Kefford R. F., Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: A phase 1 dose-escalation trial. Lancet 379, 1893–1901 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falchook G. S., Long G. V., Kurzrock R., Kim K. B., Arkenau H. T., Brown M. P., Hamid O., Infante J. R., Millward M., Pavlick A., Chin M. T., O'Day S. J., Blackman S. C., Curtis C. M., Lebowitz P., Ma B., Ouellet D., Kefford R. F., Dose selection, pharmacokinetics, and pharmacodynamics of BRAF inhibitor dabrafenib (GSK2118436). Clin. Cancer Res. 20, 4449–4458 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Bianchini G., Balko J. M., Mayer I. A., Sanders M. E., Gianni L., Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 13, 674–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Talhouet S., Peron J., Vuilleumier A., Friedlaender A., Viassolo V., Ayme A., Bodmer A., Treilleux I., Lang N., Tille J.-C., Chappuis P. O., Buisson A., Giraud S., Lasset C., Bonadona V., Trédan O., Labidi-Galy S. I., Publisher correction: Clinical outcome of breast cancer in carriers of BRCA1 and BRCA2 mutations according to molecular subtypes. Sci. Rep. 10, 19248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litton J. K., Rugo H. S., Ettl J., Hurvitz S. A., Gonçalves A., Lee K. H., Fehrenbacher L., Yerushalmi R., Mina L. A., Martin M., Roché H., Im Y. H., Quek R. G. W., Markova D., Tudor I. C., Hannah A. L., Eiermann W., Blum J. L., Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 379, 753–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.US Food and Drug Administration, “FDA approves talazoparib for gBRCAm HER2-negative locally advanced or metastatic breast cancer” (2018); https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-talazoparib-gbrcam-her2-negative-locally-advanced-or-metastatic-breast-cancer.
- 26.Turk A. A., Wisinski K. B., PARP inhibitors in breast cancer: Bringing synthetic lethality to the bedside. Cancer 124, 2498–2506 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang A., Garraway L. A., Ashworth A., Weber B., Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 19, 23–38 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Lord C. J., Ashworth A., Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat. Med. 19, 1381–1388 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Sato H., Niimi A., Yasuhara T., Permata T. B. M., Hagiwara Y., Isono M., Nuryadi E., Sekine R., Oike T., Kakoti S., Yoshimoto Y., Held K. D., Suzuki Y., Kono K., Miyagawa K., Nakano T., Shibata A., DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 8, 1751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiao S., Xia W., Yamaguchi H., Wei Y., Chen M.-K., Hsu J.-M., Hsu J. L., Yu W.-H., du Y., Lee H.-H., Li C.-W., Chou C.-K., Lim S.-O., Chang S.-S., Litton J., Arun B., Hortobagyi G. N., Hung M.-C., PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin. Cancer Res. 23, 3711–3720 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lampert E. J., Zimmer A., Padget M., Cimino-Mathews A., Nair J. R., Liu Y., Swisher E. M., Hodge J. W., Nixon A. B., Nichols E., Bagheri M. H., Levy E., Radke M. R., Lipkowitz S., Annunziata C. M., Taube J. M., Steinberg S. M., Lee J.-M., Combination of PARP inhibitor olaparib, and PD-L1 inhibitor durvalumab, in recurrent ovarian cancer: A proof-of-concept phase II study. Clin. Cancer Res. 26, 4268–4279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmer A. S., Nichols E., Cimino-Mathews A., Peer C., Cao L., Lee M. J., Kohn E. C., Annunziata C. M., Lipkowitz S., Trepel J. B., Sharma R., Mikkilineni L., Gatti-Mays M., Figg W. D., Houston N. D., Lee J. M., A phase I study of the PD-L1 inhibitor, durvalumab, in combination with a PARP inhibitor, olaparib, and a VEGFR1–3 inhibitor, cediranib, in recurrent women's cancers with biomarker analyses. J. Immunother. Cancer 7, 197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pantelidou C., Sonzogni O., de Oliveria Taveira M., Mehta A. K., Kothari A., Wang D., Visal T., Li M. K., Pinto J., Castrillon J. A., Cheney E. M., Bouwman P., Jonkers J., Rottenberg S., Guerriero J. L., Wulf G. M., Shapiro G. I., PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao P., Li L., Jiang X., Li Q., Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J. Hematol. Oncol. 12, 54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou T.-C., Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70, 440–446 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Sperduto P. W., Mesko S., Li J., Cagney D., Aizer A., Lin N. U., Nesbit E., Kruser T. J., Chan J., Braunstein S., Lee J., Kirkpatrick J. P., Breen W., Brown P. D., Shi D., Shih H. A., Soliman H., Sahgal A., Shanley R., Sperduto W. A., Lou E., Everett A., Boggs D. H., Masucci L., Roberge D., Remick J., Plichta K., Buatti J. M., Jain S., Gaspar L. E., Wu C. C., Wang T. J. C., Bryant J., Chuong M., An Y., Chiang V., Nakano T., Aoyama H., Mehta M. P., Survival in patients with brain metastases: Summary report on the updated diagnosis-specific graded prognostic assessment and definition of the eligibility quotient. J. Clin. Oncol. 38, 3773–3784 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies M. A., Liu P., McIntyre S., Kim K. B., Papadopoulos N., Hwu W. J., Hwu P., Bedikian A., Prognostic factors for survival in melanoma patients with brain metastases. Cancer 117, 1687–1696 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Colombino M., Capone M., Lissia A., Cossu A., Rubino C., de Giorgi V., Massi D., Fonsatti E., Staibano S., Nappi O., Pagani E., Casula M., Manca A., Sini M. C., Franco R., Botti G., Caracò C., Mozzillo N., Ascierto P. A., Palmieri G., BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol. 30, 2522–2529 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Pisarevsky E., Blau R., Epshtein Y., Ben-Shushan D., Eldar-Boock A., Tiram G., Koshrovski-Michael S., Scomparin A., Pozzi S., Krivitsky A., Shenbach-Koltin G., Yeini E., Fridrich L., White R., Satchi-Fainaro R., Rational design of polyglutamic acid delivering an optimized combination of drugs targeting mutated BRAF and MEK in melanoma. Adv. Ther. 3, 2000028 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King A. J., Arnone M. R., Bleam M. R., Moss K. G., Yang J., Fedorowicz K. E., Smitheman K. N., Erhardt J. A., Hughes-Earle A., Kane-Carson L. S., Sinnamon R. H., Qi H., Rheault T. R., Uehling D. E., Laquerre S. G., Dabrafenib; preclinical characterization, increased efficacy when combined with trametinib, while BRAF/MEK tool combination reduced skin lesions. PLOS ONE 8, e67583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acúrcio R. C., Pozzi S., Carreira B., Pojo M., Gómez-Cebrián N., Casimiro S., Fernandes A., Barateiro A., Farricha V., Brito J., Leandro A. P., Salvador J. A. R., Graça L., Puchades-Carrasco L., Costa L., Satchi-Fainaro R., Guedes R. C., Florindo H. F., Therapeutic targeting of PD-1/PD-L1 blockade by novel small-molecule inhibitors recruits cytotoxic T cells into solid tumor microenvironment. J. Immunother. Cancer 10, e004695 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dangoor S. I., Hadad O., Satchi-Fainaro R., Abstract 2850: Astrocyte-breast cancer interactions facilitate brain metastasis. Cancer Res. 81, 2850 (2021). [Google Scholar]
- 43.Weinhart M., Gröger D., Enders S., Riese S. B., Dernedde J., Kainthan R. K., Brooks D. E., Haag R., The role of dimension in multivalent binding events: Structure–activity relationship of dendritic polyglycerol sulfate binding to L-selectin in correlation with size and surface charge density. Macromol. Biosci. 11, 1088–1098 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Paulus F., Schulze R., Steinhilber D., Zieringer M., Steinke I., Welker P., Licha K., Wedepohl S., Dernedde J., Haag R., The effect of polyglycerol sulfate branching on inflammatory processes. Macromol. Biosci. 14, 643–654 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Hoyle C. E., Bowman C. N., Thiol-ene click chemistry. Angew. Chem. Int. Ed. Engl. 49, 1540–1573 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Tylawsky D. E., Kiguchi H., Vaynshteyn J., Gerwin J., Shah J., Islam T., Boyer J. A., Boué D. R., Snuderl M., Greenblatt M. B., Shamay Y., Raju G. P., Heller D. A., P-selectin-targeted nanocarriers induce active crossing of the blood–brain barrier via caveolin-1-dependent transcytosis. Nat. Mater. 22, 391–399 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flaherty K. T., Infante J. R., Daud A., Gonzalez R., Kefford R. F., Sosman J., Hamid O., Schuchter L., Cebon J., Ibrahim N., Kudchadkar R., Burris H. A. III, Falchook G., Algazi A., Lewis K., Long G. V., Puzanov I., Lebowitz P., Singh A., Little S., Sun P., Allred A., Ouellet D., Kim K. B., Patel K., Weber J., Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 367, 1694–1703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long G. V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J., Garbe C., Jouary T., Hauschild A., Grob J. J., Chiarion Sileni V., Lebbe C., Mandalà M., Millward M., Arance A., Bondarenko I., Haanen J. B. A. G., Hansson J., Utikal J., Ferraresi V., Kovalenko N., Mohr P., Probachai V., Schadendorf D., Nathan P., Robert C., Ribas A., DeMarini D. J., Irani J. G., Casey M., Ouellet D., Martin A.-M., Le N., Patel K., Flaherty K., Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 371, 1877–1888 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Davies M. A., Saiag P., Robert C., Grob J. J., Flaherty K. T., Arance A., Chiarion-Sileni V., Thomas L., Lesimple T., Mortier L., Moschos S. J., Hogg D., Márquez-Rodas I., del Vecchio M., Lebbé C., Meyer N., Zhang Y., Huang Y., Mookerjee B., Long G. V., Dabrafenib plus trametinib in patients with BRAFV600-mutant melanoma brain metastases (COMBI-MB): A multicentre, multicohort, open-label, phase 2 trial. Lancet Oncol. 18, 863–873 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Long G. V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J., Garbe C., Jouary T., Hauschild A., Grob J. J., Chiarion-Sileni V., Lebbe C., Mandalà M., Millward M., Arance A., Bondarenko I., Haanen J. B. A. G., Hansson J., Utikal J., Ferraresi V., Kovalenko N., Mohr P., Probachai V., Schadendorf D., Nathan P., Robert C., Ribas A., DeMarini D. J., Irani J. G., Swann S., Legos J. J., Jin F., Mookerjee B., Flaherty K., Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: A multicentre, double-blind, phase 3 randomised controlled trial. Lancet 386, 444–451 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Atkinson V. G., Quaglino P., Aglietta M., Vecchio M. D., Depenni R., Consoli F., Bafaloukos D., Ferrucci P. F., Tulyte S., Krajsová I., Ascierto P. A., Gueli R., Arance A., Gogas H., Banerjee H., Saliba T., de Jong E., Neyns B., A retrospective analysis of dabrafenib and/or dabrafenib plus trametinib combination in patients with metastatic melanoma to characterize patients with long-term benefit in the individual patient program (DESCRIBE III). Cancer 13, 2466 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markovsky E., Baabur-Cohen H., Satchi-Fainaro R., Anticancer polymeric nanomedicine bearing synergistic drug combination is superior to a mixture of individually-conjugated drugs. J. Control. Release 187, 145–157 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Stroock A. D., Dertinger S. K. W., Ajdari A., Mezić I., Stone H. A., Whitesides G. M., Chaotic mixer for microchannels. Science 295, 647–651 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Operti M. C., Dölen Y., Keulen J., van Dinther E. A. W., Figdor C. G., Tagit O., Microfluidics-assisted size tuning and biological evaluation of PLGA particles. Pharmaceutics 11, 590 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang J., Islam W., Maeda H., Exploiting the dynamics of the EPR effect and strategies to improve the therapeutic effects of nanomedicines by using EPR effect enhancers. Adv. Drug Deliv. Rev. 157, 142–160 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Mincu R. I., Mahabadi A. A., Michel L., Mrotzek S. M., Schadendorf D., Rassaf T., Totzeck M., Cardiovascular adverse events associated with BRAF and MEK inhibitors: A systematic review and meta-analysis. JAMA Netw. Open 2, e198890 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cherri M., Ferraro M., Mohammadifar E., Quaas E., Achazi K., Ludwig K., Grötzinger C., Schirner M., Haag R., Biodegradable dendritic polyglycerol sulfate for the delivery and tumor accumulation of cytostatic anticancer drugs. ACS Biomater Sci. Eng. 7, 2569–2579 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Gilmartin A. G., Bleam M. R., Groy A., Moss K. G., Minthorn E. A., Kulkarni S. G., Rominger C. M., Erskine S., Fisher K. E., Yang J., Zappacosta F., Annan R., Sutton D., Laquerre S. G., GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 17, 989–1000 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Cheng Y., Tian H., Current development status of MEK inhibitors. Molecules 22, 1551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jafari M., Sriram V., Xu Z., Harris G. M., Lee J. Y., Fucoidan-doxorubicin nanoparticles targeting P-selectin for effective breast cancer therapy. Carbohydr. Polym. 249, 116837 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Braatz D., Dimde M., Ma G., Zhong Y., Tully M., Grötzinger C., Zhang Y., Mavroskoufis A., Schirner M., Zhong Z., Ballauff M., Haag R., Toolbox of biodegradable dendritic (poly glycerol sulfate)-SS-poly(ester) micelles for cancer treatment: Stability, drug release, and tumor targeting. Biomacromolecules 22, 2625–2640 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Achazi K., Haag R., Ballauff M., Dernedde J., Kizhakkedathu J. N., Maysinger D., Multhaup G., Understanding the interaction of polyelectrolyte architectures with proteins and biosystems. Angew. Chem. Int. Ed. Engl. 60, 3882–3904 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boreham A., Pikkemaat J., Volz P., Brodwolf R., Kuehne C., Licha K., Haag R., Dernedde J., Alexiev U., Detecting and quantifying biomolecular interactions of a dendritic polyglycerol sulfate nanoparticle using fluorescence lifetime measurements. Molecules 21, E22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cunha L., Grenha A., Sulfated seaweed polysaccharides as multifunctional materials in drug delivery applications. Mar. Drugs 14, 42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang M., Yang C., Yan X., Sung J., Garg P., Merlin D., Highly biocompatible functionalized layer-by-layer ginger lipid nano vectors targeting P-selectin for delivery of doxorubicin to treat colon cancer. Adv. Ther. 2, 1900129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barber L. J., Sandhu S., Chen L., Campbell J., Kozarewa I., Fenwick K., Assiotis I., Rodrigues D. N., Reis-Filho J. S., Moreno V., Mateo J., Molife L. R., de Bono J., Kaye S., Lord C. J., Ashworth A., Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J. Pathol. 229, 422–429 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Norquist B., Wurz K. A., Pennil C. C., Garcia R., Gross J., Sakai W., Karlan B. Y., Taniguchi T., Swisher E. M., Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J. Clin. Oncol. 29, 3008–3015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cruz C., Castroviejo-Bermejo M., Gutiérrez-Enríquez S., Llop-Guevara A., Ibrahim Y. H., Gris-Oliver A., Bonache S., Morancho B., Bruna A., Rueda O. M., Lai Z., Polanska U. M., Jones G. N., Kristel P., de Bustos L., Guzman M., Rodríguez O., Grueso J., Montalban G., Caratú G., Mancuso F., Fasani R., Jiménez J., Howat W. J., Dougherty B., Vivancos A., Nuciforo P., Serres-Créixams X., Rubio I. T., Oaknin A., Cadogan E., Barrett J. C., Caldas C., Baselga J., Saura C., Cortés J., Arribas J., Jonkers J., Díez O., O’Connor M. J., Balmaña J., Serra V., RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann. Oncol. 29, 1203–1210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller R. E., El-Shakankery K. H., Lee J.-Y., PARP inhibitors in ovarian cancer: Overcoming resistance with combination strategies. J. Gynecol. Oncol. 33, e44 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galluzzi L., Buqué A., Kepp O., Zitvogel L., Kroemer G., Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell 28, 690–714 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Lee E. K., Konstantinopoulos P. A., PARP inhibition and immune modulation: Scientific rationale and perspectives for the treatment of gynecologic cancers. Ther. Adv. Med. Oncol. 12, 1758835920944116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jenkins M. H., Steinberg S. M., Alexander M. P., Fisher J. L., Ernstoff M. S., Turk M. J., Mullins D. W., Brinckerhoff C. E., Multiple murine BRafV600E melanoma cell lines with sensitivity to PLX4032. Pigment Cell Melanoma Res. 27, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cruz-Munoz W., Man S., Xu P., Kerbel R. S., Development of a preclinical model of spontaneous human melanoma central nervous system metastasis. Cancer Res. 68, 4500–4505 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Timmins N. E., Nielsen L. K., Generation of multicellular tumor spheroids by the hanging-drop method. Methods Mol. Med. 140, 141–151 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Jung M. E., Kim W. J., Practical syntheses of dyes for difference gel electrophoresis. Bioorg. Med. Chem. 14, 92–97 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Redy O., Kisin-Finfer E., Sella E., Shabat D., A simple FRET-based modular design for diagnostic probes. Org. Biomol. Chem. 10, 710–715 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Han D. K., Lee N. Y., Park K. D., Kim Y. H., I. Cho H., Min B. G., Heparin-like anticoagulant activity of sulphonated poly(ethylene oxide) and sulphonated poly(ethylene oxide) grafted polyurethane. Biomaterials 16, 467–471 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S22