Abstract
Eukaryotes carry three types of structural maintenance of chromosome (SMC) protein complexes, condensin, cohesin, and SMC5/6, which are ATP-dependent motor proteins that remodel the genome via DNA loop extrusion (LE). SMCs modulate DNA supercoiling but remains incompletely understood how this is achieved. Using a single-molecule magnetic tweezers assay that directly measures how much twist is induced by individual SMCs in each LE step, we demonstrate that all three SMC complexes induce the same large negative twist (i.e., linking number change of ~−0.6 at each LE step) into the extruded loop, independent of step size and DNA tension. Using ATP hydrolysis mutants and nonhydrolyzable ATP analogs, we find that ATP binding is the twist-inducing event during the ATPase cycle, coinciding with the force-generating LE step. The fact that all three eukaryotic SMC proteins induce the same amount of twist indicates a common DNA-LE mechanism among these SMC complexes.
Measuring single DNA loop extrusion steps of individual eukaryotic SMCs revealed that ATP binding induces a DNA twist of −0.6.
INTRODUCTION
SMC (structural maintenance of chromosomes) protein complexes are essential in all organisms to ensure proper chromosome organization. In eukaryotes, three SMC protein complexes exist: cohesin, condensin, and SMC5/6 (1). These SMC complexes share a common architecture (fig. S1A) where two Smc subunits heterodimerize at their hinge and an intrinsically disordered kleisin subunit connects their adenosine triphosphatase (ATPase) domains, thus forming a ring structure. Cohesin and condensin associate with two HAWK (HEAT repeat–containing proteins associated with kleisins) subunits on the kleisin, while SMC5/6 carries two KITE (kleisin interacting winged-helix tandem element) on its kleisin and a third, Nse2, on the coiled coil of Smc5 (2). The three SMCs participate in a large range of diverse biological processes in vivo (2). From S-phase until metaphase, cohesin topologically embraces the two replicated sister chromatids (3, 4), while condensin compacts the chromatids via DNA loop extrusion in mitosis (5, 6) for subsequent chromosome segregation. During interphase, cohesin also folds genomic DNA into loops (7–9), which has been implicated in the regulation of transcription (10), recombination (11, 12), and the local separation of sister chromatids (13). SMC5/6 predominantly contributes to DNA repair (14), but its exact role remains elusive. Despite their varying roles, all three eukaryotic SMC complexes are able to extrude DNA into loops (5, 8, 15–18) via an adenosine triphosphate (ATP)–dependent DNA loop extrusion mechanism that remains incompletely understood (19).
SMCs have been reported to change the amount of DNA supercoiling. Early biochemical reconstitutions of condensin from Xenopus egg extracts demonstrated that condensin generates positive DNA supercoiling (20–22), i.e., an additional twist deposited in DNA in the same right-handed direction as the double helix. Supercoiling is ubiquitously present in cells and contributes to the packaging of chromosomes and the regulation of genetic processes. Recent data have shown that condensin (23) and SMC5/6 (24) complexes are able to loop already supercoiled DNA and that cohesin also actively induces DNA supercoiling using ATP (20, 23, 25, 26). Condensin dosage compensation complex (DCC) has been associated with local negative supercoils at its high-occupancy binding sites of the X chromosome in Caenorhabditis elegans (27). Likely, DNA supercoiling and DNA loop extrusion stem from the same conformational changes during the ATPase cycle, suggesting that also SMC5/6 should exhibit a DNA supercoiling activity as well. Notably, DNA loop extrusion by condensin (28) and cohesin (29) occurs in large discrete steps of up to hundred base pairs each, which are generated by ATP binding to the SMC complex (28). It remains unknown how SMC proteins induce DNA twist, whether or how this mechanism is related to the loop extrusion mechanism, and whether the DNA twist directionality and degree of twist are comparable across the SMC family.
Here, we use a high-resolution magnetic tweezers (MT) assay to quantitatively examine, at the single-molecule level, the twist induced into DNA during loop extrusion by all three eukaryotic SMC complexes: human cohesin, yeast condensin, and yeast SMC5/6. We find that all three SMC complexes twist DNA negatively in each DNA loop extrusion step, irrespective of the loop extrusion step size and the applied DNA tension. Notably, a large linking number change (ΔLk, ~−0.6) was resolved in each step of DNA loop extrusion by individual SMCs on single DNA molecules. Using ATP hydrolysis mutants and nonhydrolyzable ATP analogs, we show that ATP binding is not only the DNA extrusion step but also the twist-generating event of the ATPase cycle. Notably, all three SMC complexes quantitatively induced a comparable amount of negative twist in the extruded DNA loop, indicating a conserved mechanism shared by all eukaryotic SMCs to generate supercoils that is an intrinsic property of the DNA loop extrusion cycle.
RESULTS
SMC complexes induce a negative DNA twist at individual DNA loop extrusion steps
With the objective to probe the degree of DNA twist induced by SMCs during single DNA loop extrusion steps, we turned to MT as this single-molecule technique has been proven to be able to resolve both single loop extrusion steps of yeast condensin and human cohesin (28, 29), as well as minute changes in DNA twist (30–33). Torsionally constrained 3.6-kbp-long double-stranded DNA (dsDNA) molecules were tethered between a superparamagnetic 1-μm bead and an anti-digoxygenin–functionalized glass surface via handles on the end of DNA molecules containing either multiple biotin or digoxygenin labels, respectively (Methods). A pair of permanent magnets was mounted above the flow cell which allowed to exert a calibrated force on the surface-tethered DNA molecules (Fig. 1A) (34). Here, we applied a force of 0.3 pN, at which individual SMC-mediated loop extrusion steps can be measured (28, 29). Rotation of the magnet pair within the plane of the surface yielded a corresponding rotation of the magnetized bead. Since the attached DNA molecule is torsionally constrained, its linking number Lk changes proportionally with the rotation of the bead. The DNA extension is at its maximum when no rotation is applied, i.e., when ΔLk = 0. Applying a low number of positive (negative) rotations overwinds (underwinds) and twists the DNA. Beyond a buckling point (35), twist is converted into writhe which absorbs the applied turns into plectonemic supercoils, resulting in the shortening of the DNA end-to-end extension (30, 31, 33, 36). Monitoring the DNA end-to-end extension versus positive and negative rotations thus generates a rotation curve which is symmetric around zero rotations (ΔLk = 0) at low forces [≤0.6 pN, i.e., where DNA remains in its B-form (30, 31, 33, 36)] (Fig. 1A). Rotation curves were measured for all DNA molecules in the field of view as a reference before the introduction of SMCs into the flow chamber.
Fig. 1. Measuring twist induced by single DNA loop extrusion steps by use of MT.
(A) Assay schematic (top) and corresponding experimental data (bottom) of the DNA end-to-end extension as a function of magnet rotation for a torsionally constrained 3.6-kbp DNA molecule. At a constant force of 0.3 pN, the DNA extension is maximal when no external rotations are applied. Upon applying positive/negative rotations, the over/underwinding of the DNA changes the linking number Lk and leads to the formation of plectonemic supercoils, which, at this low force, are symmetric for both positive and negative coiling. Solid line is a Gaussian fit to the rotation curve data. (B) Assay schematic (top) and experimental data (bottom) which depict a representative trajectory of human cohesin (100 pM human cohesin, 250 pM NIPBL-Mau2) showing the step-wise DNA loop extrusion in the presence of 1 mM ATP at 0.3 pN. Black line shows fit result from a step-finding algorithm. During loop extrusion, human cohesin induces negative supercoils (−ΔLk) into the extruded DNA loop, while compensatory, positive supercoils (+ΔLk) form in the DNA molecule outside the loop. (C) Assay schematic (top) and experimental data (bottom) of DNA extension as a function of magnet rotation, similar to (A), conducted directly after the DNA loop extrusion experiment at 0.3 pN. The rotation curve after loop extrusion (red; line depicts Gaussian fit to the data) shows that the maximum DNA extension was shifted to negative magnet rotation compared to the initial rotation curve (black), which was caused by the uncoiling of the positive complementary supercoils (+ΔLk) that were formed in the DNA molecule outside the DNA loop. The degree of negative supercoils (−ΔLk) that are generated by human cohesin residing in the loop is thus directly measured from the shift of the rotation-curve maxima. Black arrows in the trajectories depict single steps. (D) Same as (A) to (C), for a trace showing only a single loop extrusion step (20 pM human cohesin with 50 pM NIPBL-Mau2 and 1 mM ATP). A lower amount of induced negative supercoils (−ΔLk) is observed, compared to (A) to (C).
The MT assay allows to resolve both the steps and the twist induced by individual SMCs during their loop extrusion activity. In a first series of experiments, human cohesin and ATP were flushed into the flow chamber, while a high force of 7 pN was applied to the tethers to prevent compaction of the DNA molecules during flush-in by SMC-mediated DNA loop extrusion. When the flow was stopped and the force was quickly lowered to 0.3 pN, a stepwise decrease in the DNA end-to-end extension was observed (Fig. 1B). These can be attributed to discrete DNA loop extrusion steps as previously demonstrated for yeast condensin and human cohesin (28, 29). Directly after monitoring the steps for 10 min, another rotation curve was acquired at the same force (Fig. 1C). The peak of the rotation curve after loop extrusion (Fig. 1C, red curve) was systematically shifted to a lower bead height and to a negative number of rotations, compared to the reference rotation curve acquired before the extrusion of DNA by SMCs (Fig. 1, A and C, black line). As the SMC reeled DNA into the loop, the DNA tether length decreased, explaining the decrease of the bead height (28, 29). The frequency of DNA loop extrusion steps (which reflects the loop extrusion speed) observed in MT experiments at 0.3 pN (28, 29) was observed to be considerably lower than the loop extrusion speed measured in single-molecule fluorescence experiments that was deduced at DNA tensions close to 0 pN [at the time that SMC complexes just started loop extrusion and when the loop extrusion rate was measured (5, 8, 29, 37)]. Notably, these MT experiments are inevitably carried out at forces at or above the low stalling force of these SMC complexes (28, 29) because MTs intrinsically need to be operated at nonzero (practically >0.2 pN) forces to allow for both the trapping of the magnetic bead and achieving a reasonable signal-to-noise ratio, where these molecular motor proteins only occasionally make a few steps (28).
The shift of the rotation curve signals the twist that was inserted into the extruded loop by the SMC. To obtain the maximum DNA extension in the final rotation curve (Fig. 1C, red curve), negative rotations have to be externally applied to the DNA tether (outside the extruded loop) to remove the writhe. In other words, the DNA tether outside the loop was positively twisted after loop extrusion. Since the overall linking number of the entire torsionally constrained DNA molecule was conserved, the positive linking number change (ΔLk > 0) that was measured in the nonlooped DNA tether compensates the negative linking number change (ΔLk < 0) that was generated by the SMC inside the extruded loop (Fig. 1C). In this way, our assay thus directly measures, at the single-molecule level, the induced twist that is generated into the extruded loop through one or more steps of the SMC. The magnitude of the induced twist ΔLk is quantified as the difference of the peak positions of Gaussian fits to the rotation curves before and after loop extrusion (Methods), and it can be correlated to the number of downward steps that was observed in the time trace. A small but still negative shift between the rotation curves before and after the induction of loop extrusion could even be resolved for DNA molecules that showed only a single downward step (Fig. 1D). This observation is a direct demonstration that single SMC molecules negatively twist DNA, concomitant with individual loop extrusion steps.
ATP binding induces an equal amount of twist in each loop extrusion step, irrespective of the step size and DNA tension
Using MT, we previously determined that ATP binding (i.e., not ATP hydrolysis) is the step-generating event during the DNA loop extrusion cycle of yeast condensin (28). To test whether this is also the case for cohesin, as well as to assess whether the ATP binding step is involved in DNA twisting, we used adenylyl-imidodiphosphate (AMP-PNP), a nonhydrolyzable ATP analog, as well as an ATP hydrolysis (EQ/EQ)–deficient mutant during cohesin-mediated loop extrusion to obtain trajectories with a single loop extrusion step. The DNA end-to-end extension exhibited only a single downward step (e.g., as in Fig. 1D) or repeated downward-reverse step combinations (fig. S2A) for human cohesin under these conditions. No subsequent consecutive downward steps were observed, confirming that ATP binding is the step-generating event for human cohesin. Downward-reverse step combinations represent single DNA loop extrusion steps which were followed by a reversal, possibly upon dissociation of DNA from one of the binding sites before the reeled-in DNA was merged with the existing loop. Reversals were found to not contribute to the induced DNA twist (i.e., yielding ΔLk = 0). The change in DNA linking number ΔLk for traces with a single downward step (Fig. 2A) was −0.58 ± 0.16 (mean ± SD, N = 37).
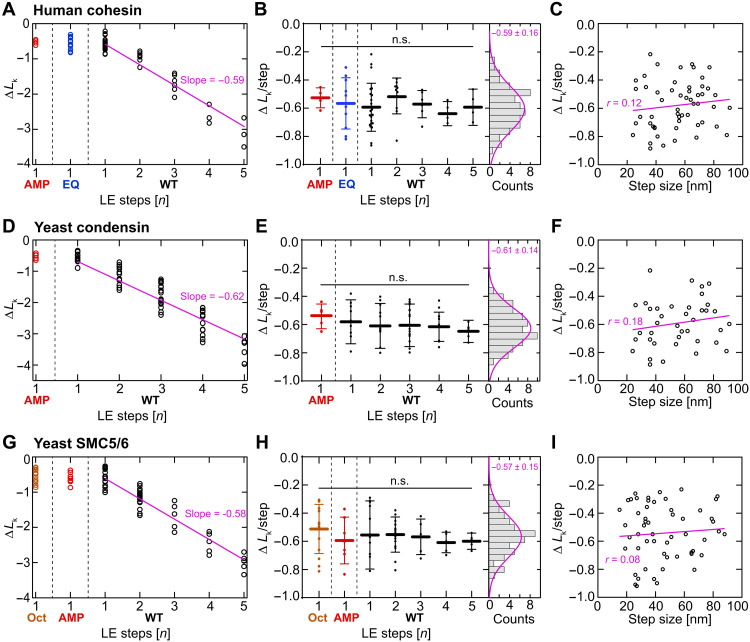
Fig. 2. Induced DNA twist is constant for each loop extrusion step and step size independent for all eukaryotic SMC complexes.
(A) Linking number change ΔLk versus the number of steps for WT human cohesin in the presence of AMP-PNP (red, N = 5), for EQ/EQ mutant of human cohesin in the presence of ATP (blue, N = 10), and for WT human cohesin in the presence of ATP (black, N = 63). The pink line is a linear fit without offset. (B) Same data as in (A) but displaying the linking number change per step [i.e., ΔLk divided by the number of loop extrusion (LE) steps]. Histogram on the right represents all data points (AMP-PNP, EQ, and WT) and is fitted by a Gaussian with −0.59 ± 0.16 ΔLk/step (mean ± SD). Statistical significance was assessed using a one-way analysis of variance (ANOVA) with a significance level α = 0.05 [95% confidence interval; not significant (n.s.) = P > 0.05]. (C) Linking number change versus the measured step size, for traces with only a single step (pooled from AMP-PNP, EQ, and WT experiments, N = 54). Linear fit and Pearson’s correlation coefficient are shown in pink. (D to F) Same as (A) to (C) but for yeast condensin [N = 5 for AMP-PNP, N = 64 for WT, and N = 39 in (F)]. (G to I) Same as (A) to (C) but for the yeast SMC5/6 hexamer [N = 15 for the SMC5/6 octamer (SMC5/6 with Nse5-Nse6), N = 7 for AMP-PNP, N = 56 for WT, and N = 55 in (I)].
By contrast, traces of wild-type (WT) human cohesin in the presence of ATP often exhibited several consecutive downward steps (Fig. 1B and fig. S2A), similar to previous MT experiments on condensin (28). We quantitatively estimated whether these consecutive steps were performed by a single SMC complex that made multiple subsequent steps or, alternatively, by multiple independent SMCs that bound at the same DNA, each performing only one step. We found that, in most cases, the multiple steps were caused by a single SMC that performed multiple loop extrusion cycles and not by several SMCs, each performing only one step. We concluded this based on a statistical assessment of our experimental data that considered which of these scenarios is more likely to occur (see Supplementary Methods): Using the fraction of “dead traces” per experiment (i.e., traces that did not exhibit any downward step or reversal activity), we calculated the expected number of SMCs per DNA molecule via a Poisson distribution under the assumption of independent DNA binding (i.e., noncooperative binding at the very low concentrations used here), and we compared this with the observed number of steps per tether (see Supplementary Methods and fig. S2D). The analysis showed that SMCs underwent multiple loop extrusion cycles in 64 to 71% or 77 to 86% of the experiments with traces of more than two or three steps, respectively (Supplementary Methods and fig. S2, D and E). In summary, for the majority of our single-molecule MT traces, our statistics analysis indicates that we probed single SMC complexes performing multiple DNA loop extrusion steps.
The change in linking number ΔLk after cohesin-mediated loop extrusion was found to scale linearly with the number of loop extrusion steps (Fig. 2A). Division of the final linking number change by the number of loop extrusion steps in the trace showed that each step induced, on average, a DNA twist of ΔLk = −0.59 ± 0.16 (mean ± SD, N = 78; Fig. 2B). Notably, the linking number change was independent of the size of the loop extrusion step (Fig. 2C), as similar ~−0.6 values were found for small (~20 nm) steps and for large (~100 nm) steps. In addition, loop extrusion experiments with yeast condensin were performed at different applied forces, ranging from 0.2 to 1.5 pN. The data (fig. S2F) show that the twist generation is also independent of the tension acting on the DNA. These results suggest that DNA twist is generated by local conformational changes within the SMC complex, independent of the DNA tension and the amount of DNA that is fed into the loop.
All eukaryotic SMCs induce ΔLk = −0.6 per step into the extruded DNA loop
All three eukaryotic SMC complexes—cohesin, condensin, and SMC5/6—share the ability to extrude DNA into loops, and they do so with very similar characteristics, viz., at a comparable extrusion rate and up to a comparable subpiconewton stalling force (5, 8, 15, 16, 29). We therefore examined whether their ability to twist DNA is also shared. To answer this question, we repeated MT experiments described above for human cohesin with budding yeast condensin (Fig. 2, D to F; figs. S1B and S2B; and Methods) and budding yeast SMC5/6 [Fig. 2, G to I; figs. S1C and S2C; Methods; and (37)]. We then analyzed whether individual loop extrusion steps can be observed using torsionally unconstrained DNA and measured the size of these steps (Methods). At an applied force of 0.2 pN, we found that the median step size of SMC5/6 is ~30 nm, while cohesin and condensin take steps of median size of ~40 nm (fig. S1D). The step size decreased as the DNA tension is increased, a phenomenon observed for all eukaryotic SMC complexes [fig. S1D and (28, 29)].
MT experiments on yeast condensin and SMC5/6 yielded very similar results as for human cohesin. In the presence of AMP-PNP, WT yeast condensin and SMC5/6 showed only a single downward step and downward-reverse step combinations (fig. S2, B and C), while in the presence of ATP, multiple consecutive downward steps were observed (fig. S2, B and C). The SMC5/6 hexamer can be converted to an octameric version by the addition of the cofactor Nse5/6 (38, 39). Octameric SMC5/6 does not loop DNA (16, 37) but promotes salt-stable loading of SMC5/6 onto DNA, which is indicative of topological loading (16, 38). Loop extrusion traces of octameric SMC5/6 exhibited a single downward step or downward-reverse step combinations (but never multiple consecutive downward steps) (fig. S2C), similar to traces of WT SMCs in the presence of a nonhydrolyzable AMP-PNP or an ATP hydrolysis–deficient mutant [fig. S2C and (28), respectively]. This suggests that Nse5/6 blocks ATP hydrolysis but not ATP binding to SMC5/6, consistent with an earlier report that ATP binding but not hydrolysis is required to topologically load SMC5/6 onto DNA (38).
As for cohesin, the linking number change induced by condensin and SMC5/6 hexamer was negative and scaled with the amount of loop extrusion steps, while the linking number change per step remained constant at a value of ΔLk = −0.61 ± 0.14 and −0.57 ± 0.15, respectively (mean ± SD; Fig. 2, E and H). These values were also independent of the size of the loop extrusion steps (Fig. 2, F and I). Notably, all three SMC complexes thus induced a comparable amount of DNA twist per step of ΔLk = −0.59 ± 0.02 (mean ± SD across the three SMCs; Fig. 3A).
Fig. 3. Eukaryotic SMC complexes induce comparable negative DNA twist per loop extrusion step.
(A) Induced twist per step for all three eukaryotic SMC complexes, where all values of the changes in the linking number ΔLk per DNA loop extrusion step were pooled from Fig. 2 (B, E, and H) (N = 59 for cohesin, N = 64 for yeast condensin, and N = 56 for SMC5/6). Statistical significance was assessed using a one-way ANOVA with a significance level α = 0.05 (95% confidence interval; n.s. = P > 0.05). (B) Potential mechanism yielding a negative DNA twist of ΔLk ~−0.6 based on the reel-and-seal model (19). DNA is nontopologically held by kleisin and HEAT-A (step 1). Upon ATP binding, clamping of DNA on top of the engaged ATPase heads temporarily forces a small loop of DNA pseudo-topologically into the Smc lumen (step 2a). Rotation of HEAT-A (if bound to DNA in the apo as well as the engaged state) induces a negative twist of about 60°, yielding ΔLk ~−0.17 (fig. S3, D and E), into the loop [see inset for twist direction (44)]. Spontaneous rotation of the coiled coils with hinge-bound DNA by about 180° (53, 54, 56) induces a further linking number change of ΔLk ~−0.5 into the loop (step 2b).
DISCUSSION
We here presented a high-resolution MT assay on single torsionally constrained DNA molecules that directly measures how much twist is induced by an individual SMC in each loop extrusion step. The data provide direct evidence that all eukaryotic SMC complexes negatively twist DNA by the same amount at every loop extrusion step, irrespective of the DNA tension and the amount of DNA that is reeled into the DNA loop. The linking number change of ΔLk ~−0.6 is generated upon ATP binding and is found to be a conserved value across the eukaryotic SMC family.
Previous deletion and mutation studies with bulk plasmid assays suggested that SMC proteins induce positive supercoiling of genomic DNA in vivo in eukaryotes (40) and in prokaryotes (41, 42). Positive DNA supercoiling by Xenopus 13S condensin was also reconstituted in vitro (20). Recently, however, it was discovered that two modes of supercoiling can be observed in these plasmid assays for condensin (25) and cohesin (26), depending on the molar ratio of SMC to plasmid as well as the protein and DNA concentrations (25). In these studies, it appears that condensin and cohesin twist DNA positively when it is in large excess to DNA, whereas negative twist is induced at low protein-DNA ratios and concentrations (25, 26). Our results are in agreement with the negative supercoiling induced at a low concentration of condensin (25) and cohesin (26). In these studies, it remained unclear what constitutes the change in twist direction induced by these SMCs on a molecular level.
Kimura and Hirano (20) noted that ATP hydrolysis is necessary for the supercoiling reaction to occur. By contrast, for conditions where ATP hydrolysis was inhibited, we observed that SMCs generate at most one step with ~−0.6 turns (Fig. 2, A, D, and G, and fig. S2)—which shows that SMCs induce negative twist into DNA upon ATP binding alone. Possibly, the topoisomerases used in the experiments in (20) were unable to catalyze sufficient changes in the linking number to be resolved in these bulk assays. In contrast, Martínez-García et al. (25) found that the incubation of the supercoiling reaction with ATP and subsequent incubation with an excess of AMP-PNP increase the amount of topo I–fixed supercoils twofold compared to an incubation with ATP alone. From the average levels of induced supercoiling, the authors estimated that a negative twist of ΔLk ~−0.8 is induced upon ATP binding which is reduced to ΔLk ~−0.4 upon ATP hydrolysis. While the induction of negative supercoils during each loop extrusion step aligns with our observation, the ΔLk values differ from our direct single-molecule measurements, as we observed that ATP binding alone induces a twist of ΔLk ~−0.6 which remains unchanged by subsequent ATP hydrolysis. Possibly, condensin is stabilized on DNA in the ATP-bound state in the plasmid assay, preventing the loss of condensin-mediated supercoils upon loop dissociation before topo I could have permanently induced a linking number change. The discrepancy between the estimates of ΔLk per ATPase cycle by Martínez-García et al. (25) and our results may be explainable by the assumptions that had to be made in the bulk assay to assess the supercoiling activity of condensin. Specifically, all condensin molecules were assumed to be active, and topo I was assumed to be 100% efficient in removing the generated supercoils. In contrast, our single-molecule experiments directly measure the induced twist on the single-step level during loop extrusion by a single SMC complex, without any treatment by topoisomerases. Martínez-García et al. (25) further concluded that each condensin molecule only transiently bends DNA during each loop extrusion step, whereas our measurements suggest that SMCs are able to accumulate negatively supercoiled DNA over multiple loop extrusion steps.
When we converted the SMC5/6 hexamer to a SMC5/6 octamer, we observed that the octamer can only induce DNA twist in a single step (inducing a linking number change of ΔLk = −0.51 ± 0.17, mean ± SD) despite its inability to hydrolyze ATP (38, 43) or extrude DNA loops [Fig. 2G, fig. S2C, and (16, 37)], suggesting that the SMC5/6 octamer is able to bind ATP. Since the SMC5/6 octamer has been shown to topologically embrace DNA (16, 39), it is possible that the ATP binding–induced power stroke by the SMC5/6 octamer serves as an intermediate to the topological loading reaction of SMC complexes onto DNA. Current models of DNA loop extrusion postulate that a transient DNA loop is inserted pseudo-topologically into the SMC lumen upon ATP binding to the complex due to a DNA clamping onto the engaged Smc ATPase heads (19, 39, 44–51) that causes the power stroke (46). Transient hinge opening in this state would lead to a topological entrapment of the DNA duplex [see fig. S3 (H to J) for such a potential pathway to topological entrapment].
Since DNA twisting occurs concomitantly with loop extrusion, we conclude that DNA twisting by ~−0.6 turns in each step is inherent to the ATP-driven DNA loop extrusion cycle of SMC complexes. While the DNA loop extrusion mechanism still remains incompletely resolved, future models have to quantitatively consider the twist of −0.6 turns induced into the loop upon ATP binding. On the basis of previous structural data, we can point to a few elements that may explain the mechanism driving the induced negative supercoiling (Fig. 3B) in a reel-and-seal model (19) with a twist. Upon ATP binding, the HEAT-A subunit [accessory subunit I, Nipped-B-like protein (NIPBL)-Mau2 for human cohesin, in fig. S1A] undergoes a rigid body rotation about 60° (ΔLk, ~−0.17) which forms a positively charged channel to clamp DNA onto the ATPase heads (Fig. 3B, steps 1 to 2a, and fig. S3, A and B) (44, 52). While the DNA trajectory within SMC complexes in the apo state has so far not been resolved, continuous DNA binding to HEAT-A in both states may direct DNA to the binding site formed by the engaged ATPase heads and thus induce ΔLk ~−0.17 into the extruded loop. Upon head engagement, the coiled coils unfold (53) and may spontaneously intertwine as has been observed by molecular dynamic simulations (fig. S3C) (54), cryo–electron microscopy (cryo-EM) (fig. S3D) (55), negative staining electron microscopy (fig. S3E) (56), and high-speed atomic force microscopy (fig. S3F) (53). DNA binding to the hinge has been proposed to be essential for DNA loop extrusion (53). If intertwining of the coiled coils occurs after DNA is bound at both the hinge and the ATPase heads, then this would induce a negative linking number change of about −0.5 into the DNA loop. Such a mechanism is supported by the finding that a mutant, which is defective in DNA binding at the hinge (53), shows reduced supercoiling activity (26). These two contributions, the rigid body rotation of HEAT-A and the spontaneous intertwining of the Smc coiled coils, therefore may additively contribute an induced linking number of −0.6 to −0.7, which is close to the observed negative linking number change of approximately −0.6 upon ATP binding. Subsequent ATP hydrolysis disengages the ATPase heads and transfers the accumulated twist into the extruded loop, clearing the way for the next cycle of DNA loop extrusion (fig. S3E). We note that the above model is consistent with the absence of a force dependence of the induced twist that we measured (fig. S2F): Since twist is induced by conformational changes in the SMC complex associated with the local clamping of DNA onto the ATPase heads upon ATP binding, one would not expect a dependence on DNA tension. This contrast the force dependence of the step size (fig. S1D) as the amount of DNA that is reeled into the loop in each step can be variable in this picture.
What is the impact of SMC-induced DNA supercoiling on the genome? DNA binding of CCCTC-binding factor (CTCF) demarcates topologically associated domains (TADs) via an orientational interaction of CTCF with cohesin (29, 57–63). The two- to threefold higher contact frequency within TADs than across TADs (64–67) might be partially explained by the barrier function of CTCF on loop progression. However, it is also plausible that supercoiling accumulates within a subset of (sub-)TADs (68) which could account for the increased intra-TAD contact frequency (67). While the accumulation of supercoils within TADs has so far been thought to be due to transcription-mediated supercoiling (67, 69, 70), this could also be the result of cohesin-mediated loop extrusion itself. The accumulation of supercoiling within TADs can contribute to their insulating property (71) as well as to the facilitation of long-range contacts between genomic loci within a TAD, e.g., promoter-enhancer contacts (72). The latter could explain how cohesin may drive transcription of enhancer-controlled developmental genes even in the absence of loop anchors positioned at enhancer-promoter pairs (10). SMCs preferentially bind and loop onto positively over negatively supercoiled DNA (23, 24, 73), which could prevent SMCs to load onto already extruded (negatively supercoiled) loops. This bias could contribute to the appearance of characteristic CTCF-dependent TAD corner dots in high-throughput chromosome conformation capture (Hi-C) maps (59, 74, 75) since those depend on a fraction of TADs whose DNA is completely contained in SMC-mediated loops (76) as observed in vivo (77, 78). Similarly, extrusion of the entire chromosomes without gaps between loop anchors is instrumental to compact metazoan mitotic chromosomes for sister chromatid segregation (76), which could also be mediated by the preferential recruitment to positively supercoiled, nonlooped DNA. Similarly, the SMC-mediated positive supercoiling of genomic regions outside loops might direct topo II to decatenate sister chromatids before relaxing supercoils (40). If bacterial SMC complexes induce a similar amount of twist as their eukaryotic relatives, then this could contribute substantially to the observed supercoiling density of many bacteria in vivo [supercoiling density σ = ~−0.06 (79, 80)] since we observed that SMCs induce supercoiling densities of −0.04 to −0.06 in the extruded DNA [for example, σ ~−0.6/150 bp*10.5/bp ~−0.04, given a typical step size of ~150 bp; fig. S1D and (28)].
Our observation that all eukaryotic SMC complexes twist DNA negatively at every loop extrusion step establishes DNA supercoiling as an integral part of the DNA loop extrusion mechanism. The measured quantitative loop extrusion–induced DNA twist of −0.59 ± 0.02 (mean ± SD) will help modeling and polymer simulations to capture yet finer structures of genomes (67) and will inform refined models of DNA loop extrusion with the handedness and amount of induced twist upon ATP binding.
METHODS
Protein expression and purification
WT and EQ/EQ human cohesin, as well as NIPBL-Mau2, were expressed in and purified from Sf9 insect cells as described previously (8). Saccharomyces cerevisiae condensin was expressed in and purified from S. cerevisiae as described previously (5). S. cerevisiae SMC5/6 was expressed in and purified from Escherichia coli as described previously (39).
Synthesis of torsionally constrained dsDNA
Linear, torsionally constrained dsDNA constructs were synthesized on the basis of either a 3.4-kbp fragment of the pRL-SV40 plasmid (Promega, USA), digested with Bam HI and Xba I, or a 10-kb fragment from pSuperCos-Lambda1,2, digested with Xho I (dig-handle), Eco RI–HF (bio-handle), and Bam HI–HF. Afterward, the fragment was enzymatically ligated to 600-bp dsDNA handles that contained multiple digoxigenins on one end of the dsDNA construct and multiple biotins at the other end of the dsDNA construct, as previously described (81, 82). To synthesize these handles, a 1.2-kb fragment from pBluescript SK+ (Stratagene, USA) was amplified by polymerase chain reaction in the presence of biotin–16–deoxyuridine triphosphate (dUTP) (Roche, Switzerland) or digoxigenin-11-dUTP (Roche, Switzerland) in a 1:5 ratio, using the forward primer 5′-GACCGAGATAGGGTTGAGTG and the reverse primer 5′-CAGGGTCGGAACAGGAGAGC. Before enzymatic ligation via T4 DNA ligase (New England Biolabs, UK) overnight, the handle fragment was digested with either Bam HI or Xba I for a 3.6-kbp fragment and Xho I or Eco RI–HF for a 10-kbp fragment. The final dsDNA constructs were cleaned up from the access of handles and other DNA fragments by running on a 1% agarose gel and extract the dsDNA construct using a gel purification kit (A9282, Promega).
Magnetic tweezers
The MT used in this study was described previously (28, 83). Briefly, light transmitted through the sample was collected by a 50× oil-immersion objective (CFI Plan 50XH, Achromat; 50×; numerical aperture = 0.9; Nikon) and projected onto a 4-megapixel complementary metal-oxide semiconductor camera (4M60, Falcon2; Teledyne DALSA) with a sampling frequency of 60 Hz. The applied magnetic field was generated by a pair of vertically aligned permanent neodymium-iron-boron magnets (Supermagnete GmbH, Germany), separated by a distance of 1 mm, and suspended on a motorized stage (M-126.PD2; Physik Instrumente) above the flow cell. In addition, the magnet pair could be rotated around the illumination axis by an applied dc servo step motor (C-150.PD; Physik Instrumente). Image processing of the collected light allowed to track the real-time position of both surface-attached reference beads and superparamagnetic beads coupled to the torsionally constrained dsDNA construct in three dimensions over time. Bead x, y, z position tracking was achieved with a spatial resolution of ~2 nm (28) using a cross-correlation algorithm realized with custom-written software in LabVIEW (2011, National Instruments Corporation) (84). The software determined the bead positions with spectral corrections to correct for camera blur and aliasing.
Measurement of DNA loop extrusion–induced DNA twist
The flow cell preparation for the MT experiments used in this study has been described previously in detail (28, 83). Briefly, polystyrene reference beads (Polysciences Europe) of 1.5 μm in diameter were diluted 1:1500 in phosphate-buffered saline (PBS) buffer (pH 7.4) and adhered to 1 M KOH-treated surface of the flow cell channel as fiducial markers. Next, digoxigenin sheep antibody Fab fragments (0.5 mg/ml; Roche, Switzerland) in PBS buffer were incubated for 1 hour within the flow cell channel, following incubation for 2 hours of bovine serum albumin (10 mg/ml; New England Biolabs, UK), diluted in PBS buffer. For each experiment, 1 pM of the torsionally constrained dsDNA construct was incubated in PBS buffer for 20 min in the flow cell channel. After washing with 500 ml of PBS buffer, the addition of 100-μl streptavidin-coated superparamagnetic beads with a diameter of 1 μm (diluted 1:400 in PBS buffer; MyOne #65601 Dynabeads, Invitrogen/Life Technologies) for 5 min resulted in the attachment of the beads to biotinylated dsDNA constructs. Non-attached beads were washed out with PBS buffer. Before conducting the force-extension experiments, we assessed whether dsDNA tethers were singly tethered by applying a high force (5 pN) and 20 negative rotations, and whether they were torsionally constrained by applying 20 rotations to each direction at a low force (0.3 pN). Only single and torsionally constrained DNA tethers were analyzed. All experiments were conducted at room temperature (22°C).
For assessing the twist generated during the DNA loop extrusion experiments, a reference rotation curve was first acquired at 0.3 pN with negative and positive rotations of each 12 turns, with a rotation speed of 0.25 turns s−1. Subsequently, the SMC proteins (cohesin WT: 20 to 100 pM; cohesin EQ/EQ: 10 to 20 pM; condensin: 0.4 to 1 nM; SMC5/6: 3 to 6 nM) in the presence of 1 mM ATP or 1 mM AMP-PNP (Sigma-Aldrich, The Netherlands)—as indicated in the Results section—were inserted into the flow cell channel while applying 7 pN to the dsDNA tethers. We initially varied the protein concentration to assess whether the results would depend on protein concentrations. However, this was not the case, and data obtained from experiments under varying protein concentrations were therefore pooled. The force was then quickly lowered to a constant force of 0.3 pN (unless indicated otherwise; see fig. S2F) to record the DNA loop extrusion steps via the change in bead Z-position (28, 29) for 10 min, which is a time where the DNA loop extrusion activity of most SMCs ceased. To assess the degree of DNA twist that was introduced by each SMC during DNA loop extrusion, another rotation curve was recorded at 0.3 pN with negative and positive rotations of 12 turns (25 turns for 10-kbp DNA) with a rotation speed of 0.25 turns s−1).
Modeling of the experimentally determined cryo-EM map of apo S. cerevisiae cohesin
A full pseudo-atomic model of S. cerevisiae cohesin in the apo state (in the absence of ATP and DNA) was fitted into the cryo-EM map EMD-12880 using UCSF ChimeraX (85). The lower part containing the ATPase heads of Smc1 and Smc3, kleisin, as well as Scc2 was taken from Protein Data Bank (PDB) entry 6ZZ6 (50), and the folded coiled coils were taken from PDB entry 7OGT (86) as performed previously (87). The EM maps and protein structures in Fig. 3B and fig. S3 were depicted using UCSF ChimeraX (85).
Quantification and statistical analyses
MT datasets were processed and analyzed using custom-written Igor v6.37–based scripts. From our raw data, we removed traces that showed surface-adhered magnetic beads as well as dsDNA tethers where the DNA-bead attachment points were close (<100 nm) to the magnetic equator of the magnetic beads using a previously described method (88). Tethers that detached from the surface during the measurement were also rejected from further analysis. All traces and rotation curves resulting from experiments conducted at identical conditions for each SMC were pooled and filtered to 1 Hz (moving average) for extrusion step identification and before Gaussian fitting of the rotations curves.
DNA loop extrusion step sizes were extracted from the change in bead Z-positions in the traces using a previously described methodology and a stepfinder algorithm (28, 89). The induced DNA twist during loop extrusion was determined by fitting a Gaussian to the bead Z-positions as a function of applied turns. The peak position of the fitted Gaussian before DNA loop extrusion Cbefore was subtracted from the peak position determined after the DNA loop extrusion experiments Cafter which reflect the induced DNA twists ΔLk = ΔTw = Cafter − Cbefore in the unit of turns for each trace.
The statistical analyses comparing the DNA twist results for each SMC and condition were conducted using one-way analysis of variance (ANOVA) with a significance level α = 0.05 (95% confidence interval).
Acknowledgments
We thank J. van der Torre and S. Gruber for discussion and M. Taschner for providing purified SMC5/6 proteins.
Funding: Research in the laboratory of C.D. was funded by ERC Advanced Grant 883684 (DNA looping), NWO grant OCENW.GROOT.2019.012, and the BaSyC program. Research in the laboratory of J.-M.P. was supported by Boehringer Ingelheim, the Austrian Research Promotion Agency (headquarter grant FFG-FO999902549), the European Research Council under the European Union’s Horizon 2020 research and innovation program GA no. 101020558, the Human Frontier Science Program (grant RGP0057/2018), and the Vienna Science and Technology Fund (grant LS19-029).
Author contributions: R.J., R.B., and C.D. designed experiments; R.J. and R.B. conducted single-molecule experiments, and R.J. analyzed the single-molecule data; R.B. made structure predictions; I.F.D. purified human cohesin; R.B., R.J., and C.D. wrote the manuscript; J.-M.P. and C.D. acquired funding; C.D. supervised the work.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The plasmids for proteins can be provided by J.-M.P. pending scientific review and a completed material transfer agreement. Requests for the plasmids should be submitted to J.-M.P. at peters@imp.ac.at.
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S3
References
REFERENCES AND NOTES
- 1.Harvey S. H., Krien M. J., O’Connell M. J., Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases. Genome Biol. 3, reviews3003.1 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim E., Barth R., Dekker C., Looping the genome with SMC complexes. Annu. Rev. Biochem. 92, 15–41 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Haering C. H., Farcas A. M., Arumugam P., Metson J., Nasmyth K., The cohesin ring concatenates sister DNA molecules. Nature 454, 297–301 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Peters J.-M., Nishiyama T., Sister chromatid cohesion. Cold Spring Harb. Perspect. Biol. 4, a011130–a011130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganji M., Shaltiel I. A., Bisht S., Kim E., Kalichava A., Haering C. H., Dekker C., Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibcus J. H., Samejima K., Goloborodko A., Samejima I., Naumova N., Nuebler J., Kanemaki M. T., Xie L., Paulson J. R., Earnshaw W. C., Mirny L. A., Dekker J., A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nora E. P., Caccianini L., Fudenberg G., So K., Kameswaran V., Nagle A., Uebersohn A., Hajj B., Saux A. L., Coulon A., Mirny L. A., Pollard K. S., Dahan M., Bruneau B. G., Molecular basis of CTCF binding polarity in genome folding. Nat. Commun. 11, 5612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson I. F., Bauer B., Goetz D., Tang W., Wutz G., Peters J. M., DNA loop extrusion by human cohesin. Science 366, 1338–1345 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Davidson I. F., Peters J. M., Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 22, 445–464 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Horsfield J. A., Full circle: A brief history of cohesin and the regulation of gene expression. FEBS J. 290, 1670–1687 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Piazza A., Bordelet H., Dumont A., Thierry A., Savocco J., Girard F., Koszul R., Cohesin regulates homology search during recombinational DNA repair. Nat. Cell Biol. 23, 1176–1186 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Litwin I., Pilarczyk E., Wysocki R., The emerging role of cohesin in the DNA damage response. Genes 9, 581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batty P., Langer C. C., Takács Z., Tang W., Blaukopf C., Peters J., Gerlich D. W., Cohesin-mediated DNA loop extrusion resolves sister chromatids in G2 phase. EMBO J. 42, e113475 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fousteri M. I., A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 19, 1691–1702 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y., Shi Z., Zhang H., Finkelstein I. J., Yu H., Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pradhan B., Kanno T., Umeda Igarashi M., Loke M. S., Baaske M. D., Wong J. S. K., Jeppsson K., Björkegren C., Kim E., The Smc5/6 complex is a DNA loop-extruding motor. Nature 616, 843–848 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golfier S., Quail T., Kimura H., Brugués J., Cohesin and condensin extrude DNA loops in a cell-cycle dependent manner. eLife 9, e53885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong M., Cutts E. E., Pan D., Beuron F., Kaliyappan T., Xue C., Morris E. P., Musacchio A., Vannini A., Greene E. C., Human condensin I and II drive extensive ATP-dependent compaction of nucleosome-bound DNA. Mol. Cell 79, 99–114.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekker C., Haering C. H., Peters J.-M., Rowland B. D., How do molecular motors fold the genome? Science 382, 646–648 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Kimura K., Hirano T., ATP-dependent positive supercoiling of DNA by 13S condensin: A biochemical implication for chromosome condensation. Cell 90, 625–634 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Kimura K., Rybenkov V. V., Crisona N. J., Hirano T., Cozzarelli N. R., 13S condensin actively reconfigures DNA by introducing global positive writhe: Implications for chromosome condensation. Cell 98, 239–248 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Hagstrom K. A., Holmes V. F., Cozzarelli N. R., Meyer B. J., C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16, 729–742 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E., Gonzalez A. M., Pradhan B., van der Torre J., Dekker C., Condensin-driven loop extrusion on supercoiled DNA. Nat. Struct. Mol. Biol. 29, 719–727 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Jeppsson K., Pradhan B., Sutani T., Sakata T., Umeda Igarashi M., Berta D. G., Kanno T., Nakato R., Shirahige K., Kim E., Björkegren C., Loop-extruding Smc5/6 organizes transcription-induced positive DNA supercoils. Mol. Cell 84, 867–882.e5 (2024). [DOI] [PubMed] [Google Scholar]
- 25.Martínez-García B., Dyson S., Segura J., Ayats A., Cutts E. E., Gutierrez-Escribano P., Aragón L., Roca J., Condensin pinches a short negatively supercoiled DNA loop during each round of ATP usage. EMBO J. 42, e111913 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.I. F. Davidson, R. Barth, S. Horn, R. Janissen, K. Nagasaka, G. Wutz, R. R. Stocsits, B. Bauer, C. Dekker, J.-M. Peters, Cohesin supercoils DNA during loop extrusion. bioRxiv 586228 [Preprint] (2024). 10.1101/2024.03.22.586228 [DOI]
- 27.Krassovsky K., Ghosh R. P., Meyer B. J., Genome-wide profiling reveals functional interplay of DNA sequence composition, transcriptional activity, and nucleosome positioning in driving DNA supercoiling and helix destabilization in C. elegans. Genome Res. 31, 1187–1202 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu J.-K., Rah S.-H., Janissen R., Kerssemakers J. W. J., Bonato A., Michieletto D., Dekker C., Condensin extrudes DNA loops in steps up to hundreds of base pairs that are generated by ATP binding events. Nucleic Acids Res. 50, 820–832 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson I. F., Barth R., Zaczek M., Van Der Torre J., Tang W., Nagasaka K., Janissen R., Kerssemakers J., Wutz G., Dekker C., Peters J.-M., CTCF is a DNA-tension-dependent barrier to cohesin-mediated loop extrusion. Nature 616, 822–827 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dohnalová H., Seifert M., Matoušková E., Klein M., Papini F. S., Lipfert J., Dulin D., Lankaš F., Temperature-dependent twist of double-stranded RNA probed by magnetic tweezer experiments and molecular dynamics simulations. J. Phys. Chem. B 128, 664–675 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strick T. R., Allemand J.-F., Bensimon D., Bensimon A., Croquette V., The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Mosconi F., Allemand J. F., Bensimon D., Croquette V., Measurement of the torque on a single stretched and twisted DNA using magnetic tweezers. Phys. Rev. Lett. 102, 078301 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Vlijm R., Mashaghi A., Bernard S., Modesti M., Dekker C., Experimental phase diagram of negatively supercoiled DNA measured by magnetic tweezers and fluorescence. Nanoscale 7, 3205–3216 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Ostrofet E., Papini F. S., Dulin D., Correction-free force calibration for magnetic tweezers experiments. Sci. Rep. 8, 15920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forth S., Deufel C., Sheinin M. Y., Daniels B., Sethna J. P., Wang M. D., Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys. Rev. Lett. 100, 148301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lionnet T., Dawid A., Bigot S., Barre F.-X., Saleh O. A., Heslot F., Allemand J.-F., Bensimon D., Croquette V., DNA mechanics as a tool to probe helicase and translocase activity. Nucleic Acids Res. 34, 4232–4244 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R. Barth, I. F. Davidson, J. Van Der Torre, M. Taschner, S. Gruber, J.-M. Peters, C. Dekker, SMC motor proteins extrude DNA asymmetrically and contain a direction switch. bioRxiv 572892 [Preprint] (2023). 10.1101/2023.12.21.572892. [DOI]
- 38.Taschner M., Basquin J., Steigenberger B., Schäfer I. B., Soh Y., Basquin C., Lorentzen E., Räschle M., Scheltema R. A., Gruber S., Nse5/6 inhibits the Smc5/6 ATPase and modulates DNA substrate binding. EMBO J. 40, e107807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taschner M., Gruber S., DNA segment capture by Smc5/6 holocomplexes. Nat. Struct. Mol. Biol. 30, 619–628 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baxter J., Sen N., López Martínez V., Monturus De Carandini M. E., Schvartzman J. B., Diffley J. F. X., Aragón L., Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 331, 1328–1332 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Lindow J. C., Britton R. A., Grossman A. D., Structural maintenance of chromosomes protein of Bacillus subtilis affects supercoiling in vivo. J. Bacteriol. 184, 5317–5322 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawitzke J. A., Austin S., Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 97, 1671–1676 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hallett S. T., Schellenberger P., Zhou L., Beuron F., Morris E., Murray J. M., Oliver A. W., Nse5/6 is a negative regulator of the ATPase activity of the Smc5/6 complex. Nucleic Acids Res. 49, 4534–4549 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Z., Gao H., Bai X. C., Yu H., Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science 368, 1454–1459 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Shaltiel I. A., Datta S., Lecomte L., Hassler M., Kschonsak M., Bravo S., Stober C., Ormanns J., Eustermann S., Haering C. H., A hold-and-feed mechanism drives directional DNA loop extrusion by condensin. Science 376, 1087–1094 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Nomidis S. K., Carlon E., Gruber S., Marko J. F., DNA tension-modulated translocation and loop extrusion by SMC complexes revealed by molecular dynamics simulations. Nucleic Acids Res. 50, 4974–4987 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marko J. F., De Los Rios P., Barducci A., Gruber S., DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. Nucleic Acids Res. 47, 6956–6972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diebold-Durand M. L., Lee H., Ruiz Avila L. B., Noh H., Shin H. C., Im H., Bock F. P., Bürmann F., Durand A., Basfeld A., Ham S., Basquin J., Oh B. H., Gruber S., Structure of full-length SMC and rearrangements required for chromosome organization. Mol. Cell 67, 334–347.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee B. G., Rhodes J., Löwe J., Clamping of DNA shuts the condensin neck gate. Proc. Natl. Acad. Sci. U.S.A. 119, e2120006119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collier J. E., Lee B. G., Roig M. B., Yatskevich S., Petela N. J., Metson J., Voulgaris M., Llamazares A. G., Löwe J., Nasmyth K. A., Transport of DNA within cohesin involves clamping on top of engaged heads by SCC2 and entrapment within the ring by SCC3. eLife 9, e59560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Y., Li S., Ser Z., Kuang H., Than T., Guan D., Zhao X., Patel D. J., Cryo-EM structure of DNA-bound Smc5/6 reveals DNA clamping enabled by multi-subunit conformational changes. Proc. Natl. Acad. Sci. U.S.A. 119, e2202799119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higashi T. L., Eickhoff P., Sousa J. S., Locke J., Nans A., Flynn H. R., Snijders A. P., Papageorgiou G., O’Reilly N., Chen Z. A., O’Reilly F. J., Rappsilber J., Costa A., Uhlmann F., A structure-based mechanism for DNA entry into the cohesin ring. Mol. Cell 79, 917–933.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauer B. W., Davidson I. F., Canena D., Wutz G., Tang W., Litos G., Horn S., Hinterdorfer P., Peters J. M., Cohesin mediates DNA loop extrusion by a “swing and clamp” mechanism. Cell 184, 5448–5464.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krepel D., Davtyan A., Schafer N. P., Wolynes P. G., Onuchic J. N., Braiding topology and the energy landscape of chromosome organization proteins. Proc. Natl. Acad. Sci. U.S.A. 117, 1468–1477 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Q., Zhang J., Haluska C., Zhang X., Wang L., Liu G., Wang Z., Jin D., Cheng T., Wang H., Tian Y., Wang X., Sun L., Zhao X., Chen Z., Wang L., Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms. Nat. Struct. Mol. Biol. 31, 1532–1542 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hons M. T., in ‘t Veld P. J. H., Kaesler J., Rombaut P., Schleiffer A., Herzog F., Stark H., Peters J.-M., Topology and structure of an engineered human cohesin complex bound to Pds5B. Nat. Commun. 7, 12523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nora E. P., Goloborodko A., Valton A. L., Gibcus J. H., Uebersohn A., Abdennur N., Dekker J., Mirny L. A., Bruneau B. G., Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wutz G., Várnai C., Nagasaka K., Cisneros D. A., Stocsits R. R., Tang W., Schoenfelder S., Jessberger G., Muhar M., Hossain M. J., Walther N., Koch B., Kueblbeck M., Ellenberg J., Zuber J., Fraser P., Peters J., Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao S. S. P., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., Sanborn A. L., Machol I., Omer A. D., Lander E. S., Aiden E. L., A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., Ren B., Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y., Haarhuis J. H. I., Sedeño Cacciatore Á., Oldenkamp R., van Ruiten M. S., Willems L., Teunissen H., Muir K. W., de Wit E., Rowland B. D., Panne D., The structural basis for cohesin–CTCF-anchored loops. Nature 578, 472–476 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., Mirny L. A., Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanborn A. L., Rao S. S. P., Huang S. C., Durand N. C., Huntley M. H., Jewett A. I., Bochkov I. D., Chinnappan D., Cutkosky A., Li J., Geeting K. P., Gnirke A., Melnikov A., McKenna D., Stamenova E. K., Lander E. S., Aiden E. L., Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 112, E6456–E6465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finn E. H., Pegoraro G., Brandão H. B., Valton A. L., Oomen M. E., Dekker J., Mirny L., Misteli T., Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176, 1502–1515.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansen A. S., Cattoglio C., Darzacq X., Tjian R., Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus 9, 20–32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang L. H., Ghosh S., Noordermeer D., TADs and their borders: Free movement or building a wall? J. Mol. Biol. 432, 643–652 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Racko D., Benedetti F., Dorier J., Stasiak A., Are TADs supercoiled? Nucleic Acids Res. 47, 521–532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naughton C., Avlonitis N., Corless S., Prendergast J. G., Mati I. K., Eijk P. P., Cockroft S. L., Bradley M., Ylstra B., Gilbert N., Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 20, 387–395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Racko D., Benedetti F., Dorier J., Stasiak A., Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res. 46, 1648–1660 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janissen R., Barth R., Polinder M., van der Torre J., Dekker C., Single-molecule visualization of twin-supercoiled domains generated during transcription. Nucleic Acids Res. 52, 1677–1687 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benedetti F., Dorier J., Burnier Y., Stasiak A., Models that include supercoiling of topological domains reproduce several known features of interphase chromosomes. Nucleic Acids Res. 42, 2848–2855 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benedetti F., Dorier J., Stasiak A., Effects of supercoiling on enhancer–promoter contacts. Nucleic Acids Res. 42, 10425–10432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diman A., Panis G., Castrogiovanni C., Prados J., Baechler B., Strubin M., Human Smc5/6 recognises transcription-generated positive DNA supercoils. Nat. Commun. 15, 7805 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang Z., Luo O. J., Li X., Zheng M., Zhu J. J., Szalaj P., Trzaskoma P., Magalska A., Wlodarczyk J., Ruszczycki B., Michalski P., Piecuch E., Wang P., Wang D., Tian S. Z., Penrad-Mobayed M., Sachs L. M., Ruan X., Wei C. L., Liu E. T., Wilczynski G. M., Plewczynski D., Li G., Ruan Y., CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163, 1611–1627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Wit E., Vos E. S. M., Holwerda S. J. B., Valdes-Quezada C., Verstegen M. J. A. M., Teunissen H., Splinter E., Wijchers P. J., Krijger P. H. L., de Laat W., CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Banigan E. J., van den Berg A. A., Brandão H. B., Marko J. F., Mirny L. A., Chromosome organization by one-sided and two-sided loop extrusion. eLife 9, e53558 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gabriele M., Brandão H. B., Grosse-Holz S., Jha A., Dailey G. M., Cattoglio C., Hsieh T. H. S., Mirny L., Zechner C., Hansen A. S., Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging. Science 376, 496–501 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mach P., Kos P. I., Zhan Y., Cramard J., Gaudin S., Tünnermann J., Marchi E., Eglinger J., Zuin J., Kryzhanovska M., Smallwood S., Gelman L., Roth G., Nora E. P., Tiana G., Giorgetti L., Cohesin and CTCF control the dynamics of chromosome folding. Nat. Genet. 54, 1907–1918 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bauer W. R., Structure and reactions of closed duplex DNA. Annu. Rev. Biophys. Bioeng. 7, 287–313 (1978). [DOI] [PubMed] [Google Scholar]
- 80.Zechiedrich E. L., Khodursky A. B., Bachellier S., Schneider R., Chen D., Lilley D. M. J., Cozzarelli N. R., Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 275, 8103–8113 (2000). [DOI] [PubMed] [Google Scholar]
- 81.Lipfert J., Kerssemakers J. W. J., Jager T., Dekker N. H., Magnetic torque tweezers: Measuring torsional stiffness in DNA and RecA-DNA filaments. Nat. Methods 7, 977–980 (2010). [DOI] [PubMed] [Google Scholar]
- 82.Janissen R., Berghuis B. A., Dulin D., Wink M., van Laar T., Dekker N. H., Invincible DNA tethers: Covalent DNA anchoring for enhanced temporal and force stability in magnetic tweezers experiments. Nucleic Acids Res. 42, e137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janissen R., Eslami-Mossallam B., Artsimovitch I., Depken M., Dekker N. H., High-throughput single-molecule experiments reveal heterogeneity, state switching, and three interconnected pause states in transcription. Cell Rep. 39, 110749 (2022). [DOI] [PubMed] [Google Scholar]
- 84.Cnossen J. P., Dulin D., Dekker N. H., An optimized software framework for real-time, high-throughput tracking of spherical beads. Rev. Sci. Instrum. 85, 103712 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Petela N. J., Llamazares A. G., Dixon S., Hu B., Lee B. G., Metson J., Seo H., Ferrer-Harding A., Voulgaris M., Gligoris T., Collier J., Oh B. H., Löwe J., Nasmyth K. A., Folding of cohesin’s coiled coil is important for scc2/4-induced association with chromosomes. eLife 10, e67268 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Srinivasan M., Scheinost J. C., Petela N. J., Gligoris T. G., Wissler M., Ogushi S., Collier J. E., Voulgaris M., Kurze A., Chan K. L., Hu B., Costanzo V., Nasmyth K. A., The cohesin ring uses its hinge to organize DNA using non-topological as well as topological mechanisms. Cell 173, 1508–1519.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klaue D., Seidel R., Torsional stiffness of single superparamagnetic microspheres in an external magnetic field. Phys. Rev. Lett. 102, 028302 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Loeff L., Kerssemakers J. W. J., Joo C., Dekker C., AutoStepfinder: A fast and automated step detection method for single-molecule analysis. Patterns 2, 100256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ryu J. K., Bouchoux C., Liu H. W., Kim E., Minamino M., de Groot R., Katan A. J., Bonato A., Marenduzzo D., Michieletto D., Uhlmann F., Dekker C., Bridging-induced phase separation induced by cohesin SMC protein complexes. Sci. Adv. 7, eabe5905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cliff N., Dominance statistics: Ordinal analyses to answer ordinal questions. Psychol. Bull. 114, 494–509 (1993). [Google Scholar]
- 92.M. Hess, J. Kromrey, “Robust confidence intervals for effect sizes: A comparative study of Cohen’s d and Cliff’s delta under non-normality and heterogeneous variances” in American Educational Research Association Annual Meeting (2004).
- 93.Collier J. E., Nasmyth K. A., DNA passes through cohesin’s hinge as well as its Smc3–kleisin interface. eLife 11, e80310 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gruber S., Arumugam P., Katou Y., Kuglitsch D., Helmhart W., Shirahige K., Nasmyth K., Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell 127, 523–537 (2006). [DOI] [PubMed] [Google Scholar]
- 95.Buheitel J., Stemmann O., Prophase pathway-dependent removal of cohesin from human chromosomes requires opening of the Smc3–Scc1 gate. EMBO J. 32, 666–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nasmyth K., Cohesin: A catenase with separate entry and exit gates? Nat. Cell Biol. 13, 1170–1177 (2011). [DOI] [PubMed] [Google Scholar]
- 97.Nagasaka K., Davidson I. F., Stocsits R. R., Tang W., Wutz G., Batty P., Panarotto M., Litos G., Schleiffer A., Gerlich D. W., Peters J.-M., Cohesin mediates DNA loop extrusion and sister chromatid cohesion by distinct mechanisms. Mol. Cell 83, 3049–3063.e6 (2023). [DOI] [PubMed] [Google Scholar]
- 98.Wilhelm L., Bürmann F., Minnen A., Shin H. C., Toseland C. P., Oh B. H., Gruber S., SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife 4, e06659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Çamdere G. Ö., Carlborg K. K., Koshland D., Intermediate step of cohesin’s ATPase cycle allows cohesin to entrap DNA. Proc. Natl. Acad. Sci. U.S.A. 115, 9732–9737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Minamino M., Higashi T. L., Bouchoux C., Uhlmann F., Topological in vitro loading of the budding yeast cohesin ring onto DNA. Life Sci. Alliance 1, e201800143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murayama Y., Uhlmann F., Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505, 367–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hassler M., Shaltiel I. A., Kschonsak M., Simon B., Merkel F., Thärichen L., Bailey H. J., Macošek J., Bravo S., Metz J., Hennig J., Haering C. H., Structural basis of an asymmetric condensin ATPase Cycle. Mol. Cell 74, 1175–1188.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S3
References