Abstract
Microorganisms that colonize the intestine communicate with the host in various ways and affect gut function and health. Extracellular vesicles (EVs), especially their encapsulated microRNAs (miRNAs), participate in the complex and precise regulation of microbiota–host interactions in the gut. These roles make miRNAs critically important for the prevention, diagnosis, and treatment of intestinal diseases. Here, we review the current knowledge on how different sources of EVs and miRNAs, including those from diets, gut microbes, and hosts, maintain gut microbial homeostasis and improve the intestinal barrier and immune function. We further highlight the roles of EVs and miRNAs in intestinal diseases, including diarrhea, inflammatory bowel disease, and colorectal cancer, thus providing a perspective for the application of EVs and miRNAs in these diseases.
Keywords: extracellular vesicles, inflammation, microbiota, microRNAs
1. Introduction
The intestine is colonized by thousands of microorganisms, and healthy gut microbial communities are beneficial for intestinal function and necessary to resist the invasion of external pathogens and harmful substances. Microbiota–host crosstalk refers to disease resistance, immune system homeostasis, and intestinal maturation [1]. However, the key regulatory mechanism of the intestine–microbiota crosstalk remains to be elucidated. Owing to advances in sequencing technology, multiple biogenic pathways and small RNA activities have been discovered over the past decades. Small RNAs, including microRNAs (miRNAs), siRNAs, and PIWI-associated RNAs, have been widely studied [2], with miRNAs being the most studied. MiRNAs are endogenous, non-coding, single-stranded RNAs derived from a conserved short hairpin structure consisting of approximately 22 base pairs. MiRNAs recognize post-transcriptional genes through base complementary pairing and guide genes to degradation or silencing [3]. Extracellular vesicles (EVs) are extracellular membrane-bound vesicles with diameters ranging from 40 to 100 nm that are widely present in various biological fluids. Different types of molecules can be present in EVs, such as proteins, lipids, and nucleic acids [4]. The prediction and early diagnosis of gastrointestinal diseases remain significant challenges in contemporary research. It is crucial to identify an efficient and practical approach to address these issues. Recently, numerous studies have focused on the roles of EVs and encapsulated miRNAs, suggesting their potential in advancing this field. They alter the composition of the gut microbiota and mediate the occurrence of intestinal diseases. EVs and miRNAs participate in the gut microbiota crosstalk and bind to key factors in important pathways, thereby regulating gene expression [5]. Moreover, the synergistic use of miRNAs in conjunction with traditional biomarkers has shown a significant increase in sensitivity compared to the use of individual biomarkers alone [6]. As a result, EVs and encapsulated miRNAs are increasingly being employed as biomarkers for disease prediction, diagnosis, and treatment.
Accordingly, this manuscript summarizes the underlying mechanisms of EVs and their encapsulated miRNAs in mediating gut–microbiota crosstalk, as well as their roles in intestinal diseases, providing a basis for using miRNAs to regulate host intestinal health.
2. Crosstalk Between Microbiota–Host in the Intestine
Recently, the role of the microbiota in mammalian intestinal health has received increasing attention. Many studies have proven that a microbiome is presented in the gut prior to birth, and many factors affect intestinal microbiota as the host matures [7,8]. The large intestine is the major site of microbial fermentation, whereas the small intestine contains microbiota related to immune functions [9,10]. The dynamic balance of the microbiota plays a substantial role in resisting intestinal diseases, such as diarrhea and inflammatory bowel diseases (IBD) [11]. Therefore, understanding how microbiota interacts with intestinal cells is beneficial for the prevention and treatment of diseases.
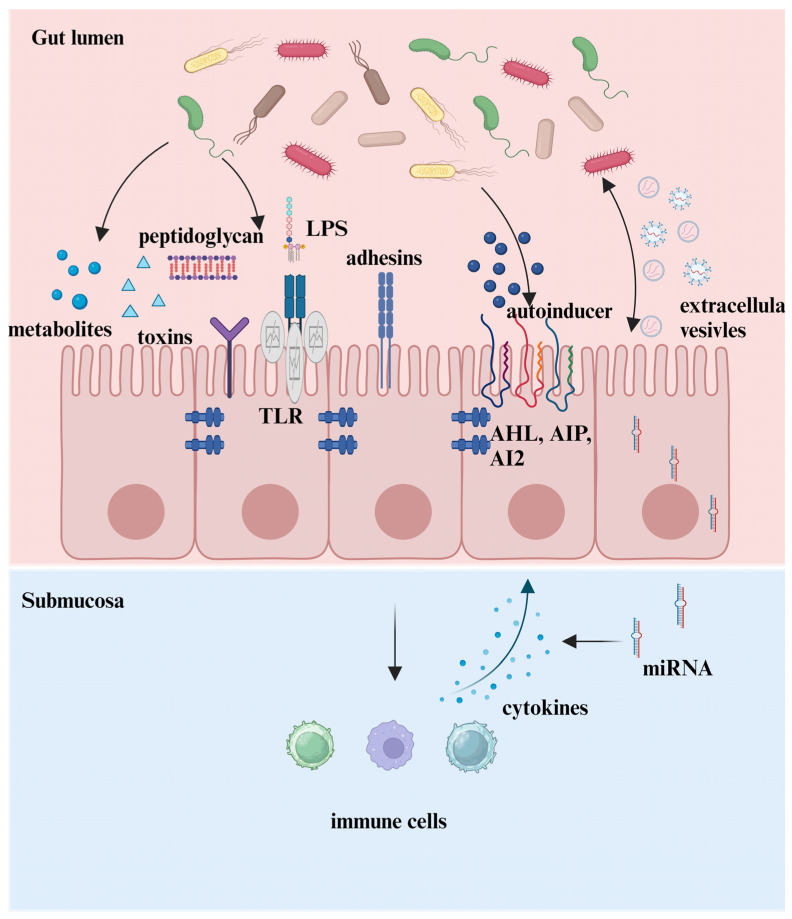
In general, crosstalk between the microbiota and intestine is mediated by microbial metabolites. Bacteriocin secreted by Lactobacillus can resist diarrhea by enhancing intestinal fluid absorption and decreasing intestinal fluid secretion [11], while the pathogenic effects of E. coli can be mitigated by bacteriocins secreted by commensal probiotic E. coli strains of human origin, both in vitro and in vivo [12]. The widely researched microbial metabolites, short-chain fatty acids (SCFAs), can maintain intestine barrier function by increasing the abundance of occludin and claudin-1 in the duodenum and ileum and decreasing the protein abundance of IL-1β in the colon, thus improving intestinal immune function [13]. Many receptors located on intestinal cells react with gut microbes and their metabolites. Lipopolysaccharides (LPS) from Gram-negative bacteria are recognized by toll-like receptor 4 (TLR4) and cluster of differentiation 14 (CD14) in epithelial cells, inducing the secretion of proinflammatory cytokines [14,15]. Peptidoglycan, found in microbial cell walls, can bind to peptidoglycan recognition proteins (PGLYRP1–PGLYRP4), which are expressed mainly in intestinal immune cells and epithelial cells, to exert antibacterial activity [16]. Bacterial adhesins, such as fimbriae F4 and F18, expressed in enterotoxigenic E. coli can bind to various receptors on epithelial cells, further activating the immune response and mediating crosstalk between microorganisms and the intestine [17].
Quorum sensing (QS) is a population density-dependent physiological behavior that regulates gene expression. Typically, when the concentration of the autoinducer released by bacteria reaches a certain threshold, it alters the expression of specific genes in bacteria [18]. The main signaling molecules adopted by different bacterial species are N-acyl-homoserine lactones, autoinducing peptides, and autoinducer 2 (AI-2) [19]. Quorum sensing is involved in a variety of biological processes and affects the spatial distribution of the intestinal flora [20]. Recently, numerous studies in quorum-sensing have been conducted. Pathogenic bacteria, such as E. coli, induce the death of intestinal epithelial cells by secreting AI-2 [21], and the activity of AI-2 decreases in the presence of Lactobacillus acidophilus strain 30SC cell extract [22]. The inhibition of AI-2 molecules also reduces the ability of lactobacilli to adhere to the host gut [23].
In addition to these mediators, which have been extensively studied and reviewed [24], the functions of EVs and their encapsulated miRNAs have become a new focus as advances in sequencing technology (Figure 1). An increasing number of publications have greatly expanded our understanding of their roles in the microbe–intestine crosstalk in recent years.
Figure 1.
The crosstalk between microbe–host in the intestine. Microbiotas interact with the host through five manners. (1) Microbiota-sourced metabolites. (2) Components of microbiota bind to receptors on epithelial cells. (3) Microbiota colonizes in the host by bacterial adhesins. (4) Microbiotas interact with the host via quorum sensing. (5) The microbiota and host interact bilaterally via EVs and their encapsulated miRNAs.
3. Extracellular Vesicles Mediate Microbiota–Intestine Crosstalk
EVs produced in the endosomes of eukaryotic cells can be classified based on the vesicular budding pathway from the cell plasma membrane. One is directly secreted from the cell membrane, defined as microvesicles and apoptotic bodies, and the other is exosomes, which primitively bud inward from the cell plasma membrane and form endosomes. These endosomes invaginate and form many small vesicles called multivesicular bodies (MVBs). Ultimately, MVBs fuse with the plasma membrane and release its intraluminal endosomal vesicles [4,25,26]. EVs are equipped with heterogeneous macromolecules such as proteins, nucleic acids, and lipids. Among these cargos, mRNAs and miRNAs carried by EVs play critical roles in cell–cell communication and have attracted the most attention [25]. As reported previously, the opportunities for different miRNAs to enter EVs vary. For example, miR-150 and miR-142-3p preferentially enter EVs [27]. EVs expression varies under different conditions. Four major underlying mechanisms for the sorting of miRNA into EVs have been reported [4]: (1) the neural sphingomyelinase 2-dependent pathway; (2) the miRNA-induced silencing complex-related pathway; (3) the 3′-end of the miRNA sequence-dependent pathway; and (4) the miRNA motif and sumoylated heterogeneous nuclear ribonucleoprotein-dependent pathway. These mechanisms emphasize the importance of specific exosomal miRNA sequences that can guide miRNAs into EVs. Usually, miRNAs encapsulated in EVs incorporate with their target genes in recipient cells to regulate their function [28]. Additionally, utilizing in vivo and in vitro models, tumor-secreted EVs, miR-21 and miR-29a, were found to act as glands binding to TLR, indicating that exosomal miRNAs can exert their functions as paracrine agonists [29].
The crosstalk between the intestine and microbiota relies heavily on the secretion and transportation of EVs. EVs are crucial carriers of bioactive molecules from the microbiota, capable of crossing the intestinal barrier to interact with host cells [1]. EVs have three fates after formation. Surface transmembrane proteins on EVs directly bind to receptors on target cells [30]. Second, the transported cargo is released into the target cells through endocytosis. The specific mechanisms are clathrin-dependent, caveolae-dependent, phagocytosis, macropinocytosis, and lipid raft-mediated uptake [31]. Third, the EVs develop into endosomes via endocytosis, mature into lysosomes, and undergo degradation [4].
Diet-derived EVs have a profound influence on the composition of the host intestinal microbes. Ginger-derived EVs increase the abundance of Lactobacillaceae and Bacteroidales and decrease the abundance of Clostridiaceae in the intestine; IL-22 is induced by ginger-derived EVs to improve intestinal barrier function and ameliorate colitis [32]. These results suggest that externally sourced EVs help maintain healthy gut function by improving microbiota–host cell interactions. Host cells can secrete EVs, which are considered the connecting kingdom for host and gut microbiota intercommunication. Notably, EVs secreted by host cells can deliver miRNAs to bacteria and bind to bacterial genes, affecting their abundance and phenotype [33]. Intestinal epithelial cells (IECs) secrete EVs with high levels of miR-130a and miR-30c expression to inhibit autophagy-related proteins, including ATG5 and ATG16L1, thereby favoring E. coli intracellular replication and colonization [34]. Circular RNAs of HIF1α encapsulated in EVs interact with the KH domain of IGF2BP3 in a m6A-modified manner, inhibiting the cell cycle and consequently suppressing pathological colonization in the intestine [35]. Importantly, the miRNAs contained in the IEC-derived EVs correlated with gut microbes. For instance, EVs containing miR-200b-3p, miR-200b-5p, and miR-26a-5p show a negative correlation with Dubosiella and Lactobacillus in the colon of an acute colitis model [36]. Treatment with the Bacillus amyloliquefaciens SC06 probiotic promotes miR-222 expression in EVs derived from porcine epithelial cell lines (IPEC-J2) and stimulates macrophage polarization to M1 [37].
Bacteria can also secrete bacterial extracellular vesicles (BEVs). BEVs are double-layered vesicles with diameters of 20–300 nm. Although BEVs differ from EVs, they can cross the intestinal barrier and enter cells via endocytosis, macropinocytosis, and endocytosis. Thus, BEVs are essential mediators in the gut microbiota crosstalk by transporting bacterial molecules, including LPS, peptidoglycans, lipids, proteins, nucleic acids, and even toxins, to host intestinal cells [38]. Small RNAs (sRNAs) are the dominant components of BEVs. These molecules play an undeniable role in the microbe–intestine crosstalk [39]. sRNA71 is abundant in Lactobacillus plantarum-derived EVs based on small RNA sequencing. It is validated that cellular protein expression is adjusted by sRNA71 by binding to the 3′-untranslated region (UTR) of target mRNAs in vitro, indicating its role in microbe–host interactions [40]. Unlike eukaryotes, BEV-encapsulated sRNA regulates host genes by relying on Hfq instead of the miRNA-induced silencing complex (RISC) [31]. Hfq is a hexameric protein conserved among Gram-negative and -positive bacteria that contains a portal structure, distal structure, lateral rim, and C-terminal domains. Usually, the portal structure binds to single-stranded sRNA, whereas the distal structure binds to mRNA [41]. BEVs are beneficial to intestinal bacterial colonization and proliferation by delivering adhesion factors and polysaccharide-degrading enzymes [42,43]. Additionally, BEVs are closely involved in regulating the host intestinal immune response and have intricate connections with various diseases. The connection between BEVs and immune function was confirmed as forty-eight Bacteroides thetaiotaomicron EV proteins were found to interact with host immune cells [44]. Unfortunately, some BEVs can impair the host innate immune response to promote bacterial colonization, such as Vibrio cholerae, which EVs can induce miR-146a expression to reduce the innate immune reactions in IEC and inhibit inflammation to facilitate host colonization [45]. However, BEVs derived from Gram-negative E. coli Nissle 1917 and its commensal, ECOR12, contain peptidoglycans, which can activate the NF-kB pathway in a NOD1-dependent manner in IEC [46]; BEVs derived from Fusobacterium nucleatum exhibit multiple outer membrane protein porin FomA, which can activate the NF-kB pathway through a TLR2-dependent manner in IEC [46]. The activation of these two pathways helps the host maintain immune homeostasis and resist invasion by exogenous pathogenic bacteria. Macrophages are key intermediaries in BEV-mediated inflammatory responses, which have a superior ability to maintain the gut barrier integrity [47]. EVs derived from Lactobacillus johnsonii modulate macrophage conversion to the M2 phenotype by suppressing the NLR family pyrin domain-containing 3 (NLRP3) signaling pathway in IEC, which promotes gut barrier repair [48]. BEVs derived from Limosilactobacillus mucosae profoundly alleviate diarrheal disease symptoms by regulating macrophage phenotypes in germ-free mice [49]. Bifidobacterium longum exerts anti-inflammatory effects by activating the immune system [50]. BEVs derived from Bifidobacterium longum are rich in anti-inflammatory proteins, including ABC transporters, quorum-sensing proteins, and extracellular solute-binding proteins. Notably, BEVs can induce secretion of the anti-inflammatory cytokine IL-10, which emphasizes their immunomodulatory effects [51]. However, certain pathogenic bacteria deliver toxins to their hosts, resulting in inflammation and proptosis. For instance, BEVs from Neisseria gonorrhoeae, uropathogenic E. coli, and Pseudomonas aeruginosa can activate macrophages to induce mitochondrial apoptosis and NLRP3 inflammasome activation [47]. In addition, amino acids can stimulate microorganisms to produce EVs, thereby alleviating host homeostatic dysfunction. Glycine promotes Bacteroides acidifaciens to secrete BEVs and alters their protein profile, considerably enhancing intestinal barrier repairment [52]. The complex interaction between BEVs and the intestine relies on bacteria, intestinal physiological conditions, and other substances, and the use of BEVs to maintain intestinal health and treat diseases is a long and arduous task (Table 1).
Table 1.
The mechanisms of BEVs mediating host–microbiota crosstalk.
| Microbiota | Encapsulated Substances | Role | References |
|---|---|---|---|
| Lactobacillus plantarum | sRNA71 | Modulate cellular protein expression | [40] |
| Bacteroides thetaiotaomicron | protein | Target various immune cells | [44] |
| E. coli Nissle 1917, Fusobacterium nucleatum | peptidoglycans, outer membrane protein | Activate NF-κB pathway | [46] |
| Limosilactobacillus mucosae, Lactobacillus johnsonii | / | Modulate the macrophage and intestinal epithelial barrier | [48,49] |
| Bifidobacterium longum | protein | Induce anti-inflammatory cytokines secretion | [51] |
4. MiRNAs Mediate Microbiota–Intestine Crosstalk
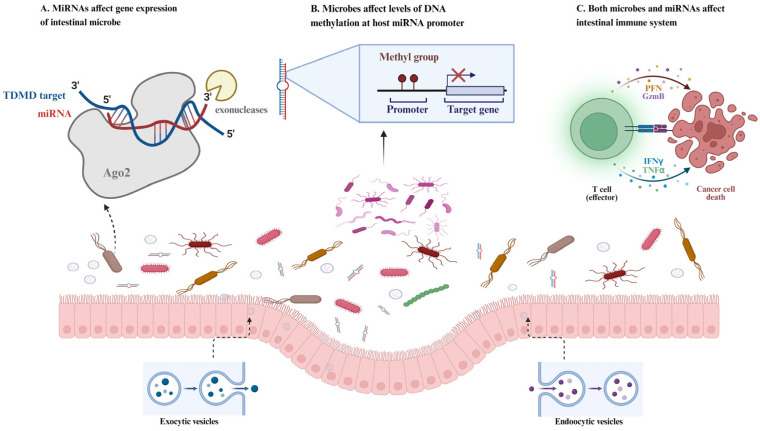
Intestinal epithelial and hopx-positive cells secrete various miRNAs and are the main sources of intestinal miRNAs [53]. Canonically, the degree of complementarity between miRNAs and their target mRNA determines their fate. Generally, when there is complete complementarity, the target mRNA is degraded, whereas, if the miRNA is not fully complementary to the target mRNA, translation is hindered [54]. Notably, when miRNAs interact with the gene promoter, 5′-UTRs, and coding sequences, they may also lead to activation of gene expression or other consequences (Figure 2) [55]. MiRNAs can be transported via EVs or present in EV-free forms associated with high-density lipoproteins or argonaute protein [56].
Figure 2.
The microbe–host crosstalk mediated by miRNAs in the intestine. (A) MiRNAs target key genes in microbiota to regulate their abundance and metabolism. (B) Microbes affect the level of DNA methylation at the host miRNA promoter to regulate the expression of miRNAs. (C) MiRNAs interact with microbiotas and then affect the intestinal immune system.
There are strong interactions between miRNAs and the intestinal microbiota. First, miRNAs mediate microbial metabolism. Succinate is an important metabolite of Prevotella, and its excessive accumulation in the colon usually causes diarrhea. One study found that ssc-miR-425-5p and ssc-miRNA-423-3p reduce the succinate concentration by targeting the gene of a key enzyme, fumarate reductase, thereby alleviating diarrhea [57]. In addition, miRNAs can directly alter the microbiota abundance to treat inflammation. MiRNA-142a-3p can promote the growth of Lactobacillus reuteri to prevent colitis induced by dextran sulfate sodium (DSS), and its potential targets are locus tags LREU_RS06530 and LREU_RS03575, which encode DNA polymerase I and primase, respectively [58]. Host miRNAs enter bacteria and specifically alter bacterial transcripts. When Fusobacterium nucleatum is co-cultured with human miR-515-5p, the ratio of F. nucleatum 16S rRNA/23S rRNA transcripts increased, whereas mutated miRNAs did not exhibit this effect [53]. Fecal miRNAs, which can influence suitable intestinal flora, are also popular in research. IEC-miRNA-deficient mice exhibit a completely different microbial composition from that of wild-type (WT) mice, and WT fecal miRNA transplantation restores this alteration. Moreover, WT fecal miRNA transplantation can alleviate the symptoms of, and has the potential to treat, colitis [53].
In turn, intestinal microbiota can regulate the expression of miRNAs, thereby altering the intestinal homeostasis. Compared to germ-free mice, specific pathogen-free mice have a more abundant miRNA profile [53]. Probiotic treatment alleviates macroscopic colonic damage in mice by regulating miRNA expression. Lactobacillus fermentum increases miR-159 and miR-143 expression, thereby preserving the intestinal barrier function [59]. Akkermansia muciniphila colonizing the intestinal mucosa is beneficial for maintaining the intestinal barrier and a healthy mucus layer, and a decrease in its abundance is correlated with the development of diseases such as IBD, appendicitis, and obesity [60]. A. muciniphila can upregulate the expression of miR-143 and miR-145, which promote IEC regeneration and barrier integrity [61]. Pathogenic bacteria can alter miRNA expression to improve cell survival, replication, and persistence [62]. Salmonella enterica is one of the main pathogenic bacteria that cause gastrointestinal disease, mortality, and substantial economic loss to the livestock industry [63]. After intranasal inoculation with S. enterica, 62 differentially expressed miRNAs were identified in vivo in whole blood. The expression of miR-214 decreased, whereas that of miR-331-3p increased. MiR-214 and miR-331-3p induce immune responses against S. enterica by targeting important immune sites, including SLC11A1, LILR-like, and VAV2 [63]. MiR-125a is a direct target of the major histocompatibility complex-class I component PSMB8, which is also decreased in mesenteric lymph nodes treated with S. enterica [64]. Other pathogenic bacteria, such as P. aeruginosa and Helicobacter pylori, also influence miRNA expression in the gastro-intestine [65,66]. Several studies have highlighted the potential mechanisms of action. Bacteria can regulate the levels of DNA methylation, a type of epigenetic modification, via the microbial metabolite SCFA [67]. Additionally, miRNAs respond to immune-related factors. For example, commensal bacteria downregulate miR-10a expression in dendritic cells by stimulating TLR ligands in a myeloid differentiation primary response gene 88 (MyD88)-dependent manner [68]. However, the exact mechanism by which intestinal microbiota affects miRNA expression needs to be further elucidated (Table 2).
Table 2.
The mechanism of EVs and encapsulated miRNAs mediating host–microbiota crosstalk.
| EVs and miRNAs | Microbiota | Role | References |
|---|---|---|---|
| Ginger-derived EVs | Lactobacillaceae, Bacteroidales, Clostridiaceae | Improve intestinal barrier function | [32] |
| miR-130a, miR-30c | E. coli | Inhibit autophagy-related proteins | [34] |
| miRNA-142a-3p | Lactobacillus reuteri | Regulating expression of DNA polymerase I and primase | [58] |
| miR-515-5p | Fusobacterium nucleatum | Directly increase microbiota 16S rRNA/23S rRNA transcripts | [53] |
| miR-159 and miR-143 | Lactobacillus fermentum | Preserving the intestinal barrier function | [59] |
| miR-143 and miR-145 | Akkermansia muciniphila | Promote IEC regeneration and barrier integrity | [61] |
| miR-214, miR-331-3p, miRNA-125a | Salmonella enterica | Induce immune response | [63,64] |
5. MiRNAs and Gut Microbiota Interactions in Intestinal Diseases
Research has focused on the role of EVs and miRNAs in disease prediction and diagnosis because of their sensitivity to alterations in the homeostasis. Using EVs and miRNAs to mediate intestinal–microbiota crosstalk is expected to provide new avenues for the treatment of currently intractable diseases and to maintain intestinal health. Furthermore, as exposed miRNAs are easy to degrade during long-distance transportation, EVs are a common and dependable carrier. The application of EVs and miRNAs in disease treatment involves two aspects. First, since EVs can be secreted by all cells, they may serve as a diagnostic marker for diseases. Specific miRNAs in EVs have diagnostic or prognostic potential for many diseases [69]. Second, EVs can deliver cargo to recipient cells over long distances, and marker proteins, such as CD63, on EVs facilitate their immune capture and enrichment; thus, many studies have reported their application in disease treatment [69,70]. EVs loaded with miRNAs have been used to treat various diseases in many animal models [71]. In the following discussion, we will focus on the application of EVs and their encapsulated miRNAs in the treatment of major intestinal diseases (Figure 3).
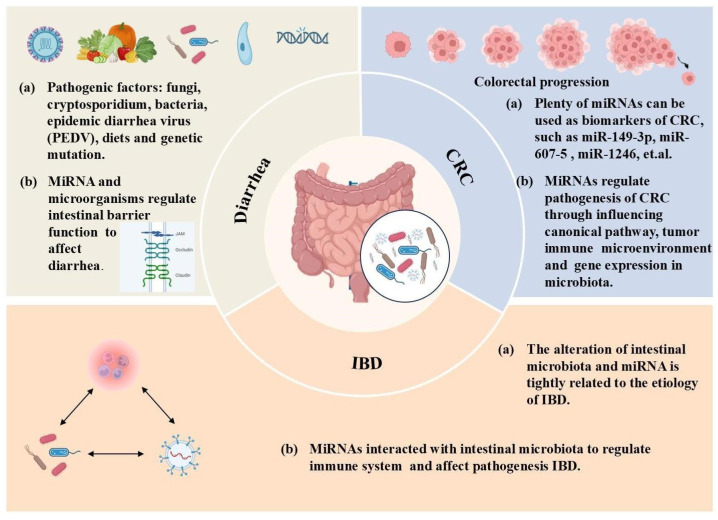
Figure 3.
The dominant function of extracellular vesicles and their encapsulated miRNAs in intestinal diseases. Interactions among microbiota, miRNAs, and the immune system involves the pathogenesis of IBD. MiRNAs interact with microbiota to affect intestinal barrier function in diarrhea. MiRNAs influence the canonical pathway, tumor immune environment, and gene expression in microbiota in CRC.
5.1. Inflammatory Bowel Diseases
The gastrointestinal tract is the chief immune organ of paramount importance for host health. Inflammation is a lethal threat to intestinal health and a critical part of the complex biological response when stimuli attack the intestine. Inflammation refers to a crucial consequence of the immune response when the host is attacked by pathogens; however, this process needs to be controlled in the case of oversecreted cytokines, which are detrimental to gut health and the immune system [72]. Moreover, inflammation is the cause of many diseases, such as necrotizing enterocolitis and diarrhea [73]. IBD is a common chronic gastrointestinal disease that mainly comprises ulcerative colitis (UC) and Crohn’s disease (CD). Germline genetics, the immune system, environmental factors, and intestinal microbiota are closely associated with IBD pathogenesis [74]. Among these factors, intestinal microbiota has recently received increasing attention. Patients with UC and CD have different intestinal microbiota compositions compared to that of healthy individuals. The abundance of Leuconostocaceae in CD and Ruminococcaceae in UC, which can produce SCFA, are decreased, whereas pro-inflammatory microbiota, such as Enterobacteriaceae in CD, is increased [75]. Therefore, adjusting the interaction between microbiota and intestine may be pivotal for the pathogenesis of IBD and could be a breakthrough point to cure IBD.
Recently, the role of EVs and miRNAs in the regulation of microbiota homeostasis and the development of intestinal inflammation was reported. EVs derived from host IEC, immune cells, and microbiota are both critical mediators in IBD, which would exert an immense role in the pathogeny and therapeutics of IBD. Exogenous miRNA transplantation can play a non-trivial role in shaping gut microbes. Ginger-derived miRNAs can promote more L. rhamnosus colonization in the intestinal lumen rather than mucosa, which is beneficial to restore colitis, thus promoting intestinal barrier function [32,76]. Dicer-specific knockout mice without the expression of miRNA-processing enzymes, exhibit exacerbated colitis. However, fecal miRNA presented within EV transplantation in wild-type mice restores the impaired intestinal flora and rescues colitis [53]. Specifically, oral administration of fecal miR-142-3p can increase the abundance of L. reuteri in the gut, which is beneficial for the recovery of colitis [58]. MiRNAs, separate or wrapped in EVs, such as miR-31, miR-215, miR-22, and miR-19b, can be used as biomarkers for IBD [77]. Specifically, some miRNAs have anti-inflammatory effects, and restoring their abundance is beneficial for attenuating inflammation. The expression of miR-10b in the intestinal villus upper cells and crypt cells decreased in a post-weaning piglet model as the intestinal inflammation intensified. Moreover, miR-10b knockout mice were more susceptible to DSS, indicating the anti-inflammatory potential of miR-10b [78]. MiR-143 and miR-145 couple with cAMP-responsive element-binding protein H (CREBH), which expression can be enhanced by A. muciniphila, to alleviate intestinal inflammation [61]. Researchers have also investigated the effects of exosomal miRNAs in breast milk on intestinal inflammation. They found that exosomal miR-4334 and miR-219 reduced intestinal inflammation, and miR-338 inhibited cell apoptosis induced by LPS. When co-transfected with these miRNAs, synergistic effects were observed in the prevention of apoptosis [79]. Notably, overexpression of certain miRNAs may aggravate inflammation. For example, a miR-30d inhibitor can attenuate inflammatory injury in IPEC-J2 cells caused by Clostridium perfringens beta2 (CPB2) toxin, which can cause necrotizing enterocolitis [73]. The regulatory function of EVs and miRNAs in IBD may occur through the induction of macrophage differentiation into the M1 pro-inflammatory phenotype, as indicated by EVs derived from Fusobacterium nucleatum [80].
The communication between miRNAs, EVs, and microbes affects the etiology of IBD. Probiotics can alleviate inflammation by regulating miRNA expression [81], whereas pathogenic bacteria disturb the miRNA expression profile and induce inflammation [62]. The effects of miRNAs on IBD are usually realized by regulating the intestinal immune system. The aryl hydrocarbon receptor (AhR) ligand is a product of tryptophan metabolism by intestinal microorganisms. AhR is an important component of intestinal immunity, and AhR ligands regulate T cell differentiation, which can be mediated by miRNA-132 [82]. MiR-31 can bind to 3′-UTRs of the genes encoding receptors for both IL-7 and IL-17 to relieve intestinal inflammation, and its expression is increased in colon tissues of patients with CD and UC [83]. miR-30c and miR-130a are targets of adherent-invasive E. coli, which prevents IEC autophagy by downregulating these two miRNAs and improving the survival of inflammatory cells [84]. The intestinal barrier is an essential part of the innate immune system, and a defective intestinal barrier is one of the pathogeneses of IBD [85]. Commensal bacteria can upregulate the expression of miR-21-5p, which increases the intestinal permeability by targeting ADP ribosylation factor 4 [86]. F. nucleatum-infected IEC-derived EVs delivered miR-129-2-3p to uninfected IEC, resulting in intestinal barrier dysfunction and inflammation [87]. Furthermore, pathogenic bacteria can inhibit autophagy-related gene expression in inflammatory cells by regulating the levels of miRNAs that promote the replication of inflammatory cells [88]. These studies suggest that unraveling the interaction mechanisms between miRNAs and gut microbes could provide new insights into the role of exosomal miRNAs in the prevention and treatment of intestinal inflammation.
5.2. Diarrhea
Diarrhea is a common clinical manifestation of various diseases caused by several factors, such as Cryptosporidium, fungi, bacteria, epidemic diarrhea virus (PEDV), diet, and genetic mutations [89,90,91,92,93]. Diarrhea can be chronic or acute, resulting in a series of adverse symptoms, including septicemia, peritonitis, electrolyte disorders, hypoalbuminemia, and anemia. EVs and their encapsulated miRNAs play critical roles in the diagnosis and treatment of diarrhea.
Diarrhea-predominant irritable bowel syndrome (IBS-D) is a chronic functional gastrointestinal disorder caused by genetic susceptibility, psychological factors, visceral hypersensitivity, increased mucosal permeability, and abnormal gut microbiology [94]. New insights into the pathogenesis of IBS-D suggest that EVs and miRNA-mediated signaling is essential. MiR-29a expression increased, while the tight junction proteins, ZO-1 and CLDN1, decreased in both in vivo and in vitro inflammatory models, indicating that miR-29a is involved in the pathogenesis of IBS-D by regulating intestinal barrier function [95]. MiR-16 can relieve IBS-D by inhibiting its target TLR4/NF-κB pathway [96]. In an IBS-D mouse model, miR-200a downregulates the expression of cannabinoid receptor 1 and serotonin transporters, thereby resulting in visceral hypersensitivity [97]. C. perfringens type C and E. coli F18 are widely studied pathogenic bacteria that cause severe diarrhea. Certain miRNAs, including miR-532-3p, miR-218-3p, and miR-500, are associated with diarrhea caused by these pathogens. These miRNAs participate in pathways related to diarrhea and regulate the expression of essential proteins involved [98,99,100]. For instance, miR-532-3p and miR-133 increase the susceptibility of C. perfringens type C by directly targeting NFATC4, a critical gene in the Wnt signaling pathway [100]. Additionally, long non-coding RNA (lncRNA) interact with miRNAs involved in the pathogenesis of diarrhea. MiR-122-5p is a diarrhea-related gene, and its overexpression aggravates C. perfringens type C-induced diarrheal injury. The ALDB-898 lncRNA completely bind to miR-122–5p and thereby mitigate diarrheal injury [101]. This suggests that miRNAs are promising therapeutic targets in IBS-D.
Diarrheal diseases pose a significant threat to livestock and cause serious economic loss [102]. EVs isolated from Vero cells infected with PEDV showed different miRNA expression levels compared to those of the control cells. Researchers have found that 80 miRNAs are upregulated and 35 miRNAs are downregulated in the EVs of infected cells [92], indicating that miRNAs are critically involved in diarrheal diseases and could be potential biomarkers for diarrhea prediction. MiRNAs can interact with the gut microbiota to regulate the occurrence of diarrhea. Host-sourced miR-425-5p and miR-423-3p could target the fumarate reductase (frd) gene in the Prevotella genus, and their reduction in the colons of piglets resulted in the overaccumulation of succinate by the Prevotella genus, further causing diarrhea [57]. Notably, BEVs alleviate diarrhea by modulating macrophage function. L. johnsonii-derived EVs ameliorate diarrheal symptoms by enhancing M2 macrophage polarization and promoting gut barrier homeostasis [48]. L. mucosae-derived EVs can also alleviate piglet diarrhea in a macrophage phenotype-dependent manner [49]. Exosomal ssc-miR-1343 inhibits FAM131C expression, thereby suppressing the innate immune response and reducing PEVD replication [103]. However, whether the miRNAs encapsulated in these EVs critically mediate their beneficial effects in diarrhea requires further elucidation.
Due to the close correlation between miRNA and diarrhea, experimental studies have been conducted to investigate the advantages of vector-delivered artificial miRNA against PEDV. The results showed that the transient expression of two artificial miRNAs, miR-349 and miR-1447, markedly decreased the expression of PEDV RNA and protein in African green monkey kidney cells [104]. Overall, vector-delivered miRNAs represent a novel therapeutic strategy for the treatment of diarrhea.
5.3. Colorectal Cancer
Colorectal cancer (CRC) is a common malignant tumor of the digestive tract with the second-highest mortality rate among cancers. Recently, as the incidence rate of CRC has increased [105], a variety of therapeutic approaches, such as surgery, radiation, and chemotherapy, have been applied in clinical practice. Phenotypic symptoms in the early stages of CRC are difficult to detect; therefore, EVs and their encapsulated miRNAs have been extensively researched as potential diagnostic and therapeutic biomarkers to promptly understand CRC progression. EVs play an important role in shaping the tumor microenvironment and act as critical participants in intercellular crosstalk by transmitting miRNAs from cancer or stromal cells to recipient cells to regulate the expression of oncogenes and tumor suppressor genes [106]. Several exosomal miRNAs, including miR-19a, miR-149-3p, miR-607-5, and miR-1246, are aberrantly expressed in patients with CRC compared to healthy individuals and those with other gastrointestinal diseases, implying the potential of miRNAs as biomarkers [107,108]. MiRNA dysregulation is related to survival after rectal cancer diagnosis. MiR-1 and miR-101-3p expression are associated with poor survival in patients with rectal cancer, indicating that the presence of miR-1 and miR-101-3p can be the basis for evaluating the prognosis of CRC [109]. MiRNAs regulate the pathogenesis and survival of CRC primarily by influencing the canonical pathway and tumor immune microenvironment [110]. MiR-34a binds to IL-6R, which ligand activates the oncogenic STAT3 transcription factor to relieve CRC progression [111]. Another miR-34 family member, miR-34a-5p, acts as a tumor suppressor by regulating DNA methylation [112]. MiR-342-5p could target placental growth factor (PGF), which participates in the activation of the mitogen-activated protein kinase (MAPK) pathway that regulates carcinogenesis, tumor cell invasion, and metastasis [113,114]. MiR-342-5p can reduce PGF expression, thus inhibiting CRC cell migration and invasion [115]. Additionally, the protein kinase B (AKT)/mammalian target of rapamycin (mTOR) pathway and Wnt/β-catenin pathway, which are both involved in the carcinogenic process of CRC, are pivotal targets of many miRNAs [116,117,118]. Furthermore, miRNAs play crucial roles in the host–intestinal microbiota crosstalk in CRC. F. prausnitzii promoted apoptosis and diminished the proliferation of CRC cells by producing butyrate. Butyrate can inhibit miR-92a transcription, thus increasing the p57 levels to suppress tumor activity [119]. Fecal microbiota transplantation (FMT) is a potential strategy to modulate the intestinal microbiome and regulate the immune system, making it a promising approach for CRC management [120]. As research on FMT progresses, it has been discovered that the mechanism also involves the transfer of miRNAs, as patients with CRC exhibit distinct fecal miRNA profiles compared to healthy individuals [107]. Mechanistically, fecal miRNAs influence the intestinal microbiota through two primary mechanisms. First, they can alter the composition and function of the microbiota, which, in turn, affects cytokine secretion. Second, they directly regulate the expression of components of the intestinal barrier, thereby influencing the microbiota [121]. Specifically, fecal miR-515-5p and miR-1226-5p can enter Fusobacterium nucleatum and Escherichia coli, promoting bacterial proliferation by regulating gene expression, which consequently exacerbates CRC [53,122,123]. These findings suggest that miRNAs can directly alter the expression of tumor-related genes and reshape the intestinal microbiota to interfere with CRC. However, our knowledge of the vast number of miRNAs and intestinal microbiota associated with CRC remains limited.
6. Conclusions
The exosomal miRNA–microbiota–host network is important for a variety of physiological processes. The microbiota can directly or indirectly influence intestinal health, and its dysregulation can be attributed to various intestinal diseases. As a pivotal mediator, exosomal miRNAs act as a bridge for communication between the gut and microorganisms. Additionally, EVs and encapsulated miRNAs are critical in gastrointestinal disorders, mediating crosstalk between the host and microbiota.
7. Perspective
Microorganisms and miRNAs are highly sensitive to alterations in each other’s abundance and are closely related to the pathology of intestinal diseases. Additionally, miRNAs are promising materials for designing disease-specific molecular signatures to achieve non-invasive screening tests because of their high sensitivity, specificity, and easy accessibility, which are of great help in the diagnosis, prognosis, and therapy of diseases. Due to the accuracy of nucleic acid pairing, miRNAs can precisely regulate the specific gene expression of microbes and their metabolism. Alterations in miRNA abundance can reflect disease progression in real time, enabling the adjustment of treatment strategies over time. Therefore, miRNAs and microbiota are increasingly valued as diagnostic and therapeutic tools for diseases.
However, there are still many practical issues regarding the application of miRNAs and microbiota in diseases that need to be addressed. First, we only discussed three diseases related to the miRNA–microbiota–host network. However, owing to the large population of both miRNAs and microorganisms, it is necessary to investigate the relationship between miRNAs, microbiota, and various diseases in populations of different age groups, countries, and dietary habits. Furthermore, the complex underlying mechanisms of miRNAs within cells and their roles in various diseases have yet to be fully elucidated. Second, biosensors may provide novel alternatives for miRNA analysis because of their high sensitivity. However, many problems still need to be solved, including their high cost, no portability, and high signal-to-noise ratio. Finally, some plant miRNAs have been identified as effective in treating animal gastrointestinal diseases. Therefore, whether miRNAs can achieve species cross-communication is a future research hotspot. Moreover, a multifunctional and accurate delivery system is required to ensure that exosomal miRNAs are delivered to target cells and microorganisms. The sophisticated sorting mechanisms of exosomal miRNAs are not comprehensively understood. The relationship between miRNAs and microorganisms is not one to one; therefore, how to accurately deliver miRNAs to microorganisms and regulate the abundance of target microorganisms remains to be explored. It is difficult to regulate specific miRNA expression in the target intestinal segment and precisely guide endogenous and exogenous miRNAs to intestinal lesions. Further studies are expected to improve EV–miRNA-based targeted therapies. These challenges have hindered the clinical application of EVs and miRNAs. The full potential of EVs and encapsulated miRNAs still requires further exploration.
Abbreviations
EVs, extracellular vesicles; miRNAs, microRNAs; IBD, inflammatory bowel disease; CREBH, cAMP-responsive element-binding protein H; CRC, colorectal cancer; SCFAs, short-chain fatty acids; LPS, lipopolysaccharides; TLR4, toll-like receptor 4; CD, cluster of differentiation; PGLYRP, peptidoglycan recognition proteins; IECs, intestinal epithelial cells; QS, Quorum sensing; AI-2, autoinducer 2; MVBs, multivesicular bodies; IPEC-J2, porcine epithelial cell lines; BEVs, bacteria can also secrete bacterial extracellular vesicles; sRNA, small RNA; UTR, untranslated region; RISC, miRNA-induced silencing complex; NLRP3, NLR family pyrin domain-containing 3; DSS, dextran sulfate sodium; WT, wild-type; MyD88, myeloid differentiation primary response gene 88; UC, ulcerative colitis; CD, Crohn’s disease; CPB2, Clostridium perfringens beta2; AhR, aryl hydrocarbon receptor; PEDV, epidemic diarrhea virus; IBS-D, diarrhea-predominant irritable bowel syndrome; lncRNA, long non-coding RNA; frd, fumarate reductase; PGF, placental growth factor; MAPK, mitogen-activated protein kinase; AKT, protein kinase B; mTOR, mammalian target of rapamycin.
Author Contributions
Writing—original draft, Y.Z.; Writing—review and editing, X.Z. and Y.Z.; Visualization, Y.Z.; Supervision, Y.Y.; Supervision, X.Z. All authors have read and agreed to the published version of the manuscript.
Informed Consent Statement
No applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Funding Statement
This work was supported by the National Natural Science Foundation of China [grant number 32341053]; the design and cultivation of the new breed in pigs with high-quality meat [grant number 2023ZD04046]; the Key Fundamental Research Program of Hunan Province [grant number 2024JC0007]; the Science and Technology Innovation Program of Hunan Province [grant number 2023RC1074]; and Youth Innovation Promotion Association Chinese Academic of Science.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Zhao X., Jiang L., Fang X., Guo Z., Wang X., Shi B., Meng Q. Host-microbiota interaction-mediated resistance to inflammatory bowel disease in pigs. Microbiome. 2022;10:115. doi: 10.1186/s40168-022-01303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peter J.O., Santos-Ortega Y., Flynt A. Guiding RNAi Design Through Characterization of Endogenous Small RNA Pathways. Methods Mol. Biol. 2022;2360:33–47. doi: 10.1007/978-1-0716-1633-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J., Li S., Li L., Li M., Guo C., Yao J., Mi S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Li C., Zhang B., Li Z., Zeng W., Luo R., Cao J., Cheng G., Fan S., He Q. Differential expression and correlation analysis of miRNA-mRNA profiles in swine testicular cells infected with porcine epidemic diarrhea virus. Sci. Rep. 2021;11:1868. doi: 10.1038/s41598-021-81189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boicean A., Boeras I., Birsan S., Ichim C., Todor S.B., Onisor D.M., Brusnic O., Bacila C., Dura H., Roman-Filip C., et al. In Pursuit of Novel Markers: Unraveling the Potential of miR-106, CEA and CA 19-9 in Gastric Adenocarcinoma Diagnosis and Staging. Int. J. Mol. Sci. 2024;25:7898. doi: 10.3390/ijms25147898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi Y., Tu Y., Zhang N., Wang S., Zhang F., Suen G., Shao D., Li S., Diao Q. Multiomics analysis reveals the presence of a microbiome in the gut of fetal lambs. Gut. 2021;70:853–864. doi: 10.1136/gutjnl-2020-320951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan L., Xia Y., Wang Y., Han D., Liu Y., Li J., Fu J., Wang L., Gan Z., Liu B., et al. Gut microbiota bridges dietary nutrients and host immunity. Sci. China Life Sci. 2023;66:2466–2514. doi: 10.1007/s11427-023-2346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao W., Wang Y., Liu S., Huang J., Zhai Z., He C., Ding J., Wang J., Wang H., Fan W., et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS ONE. 2015;10:e0117441. doi: 10.1371/journal.pone.0117441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adhikari B., Kim S.W., Kwon Y.M. Characterization of Microbiota Associated with Digesta and Mucosa in Different Regions of Gastrointestinal Tract of Nursery Pigs. Int. J. Mol. Sci. 2019;20:1630. doi: 10.3390/ijms20071630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J., Ma L., Nie Y., Chen J., Zheng W., Wang X., Xie C., Zheng Z., Wang Z., Yang T., et al. A Microbiota-Derived Bacteriocin Targets the Host to Confer Diarrhea Resistance in Early-Weaned Piglets. Cell Host Microbe. 2018;24:817–832.e818. doi: 10.1016/j.chom.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Hrala M., Bosák J., Micenková L., Křenová J., Lexa M., Pirková V., Tomáštíková Z., Koláčková I., Šmajs D. Escherichia coli Strains Producing Selected Bacteriocins Inhibit Porcine Enterotoxigenic Escherichia coli (ETEC) under both In Vitro and In Vivo Conditions. Appl. Environ. Microbiol. 2021;87:e0312120. doi: 10.1128/AEM.03121-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diao H., Jiao A.R., Yu B., Mao X.B., Chen D.W. Gastric infusion of short-chain fatty acids can improve intestinal barrier function in weaned piglets. Genes Nutr. 2019;14:4. doi: 10.1186/s12263-019-0626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens M., von der Weid P.-Y. Lipopolysaccharides modulate intestinal epithelial permeability and inflammation in a species-specific manner. Gut Microbes. 2020;11:421–432. doi: 10.1080/19490976.2019.1629235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huntley N.F., Nyachoti C.M., Patience J.F. Lipopolysaccharide immune stimulation but not β-mannanase supplementation affects maintenance energy requirements in young weaned pigs. J. Anim. Sci. Biotechnol. 2018;9:47. doi: 10.1186/s40104-018-0264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Royet J., Gupta D., Dziarski R. Peptidoglycan recognition proteins: Modulators of the microbiome and inflammation. Nat. Rev. Immunol. 2011;11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 17.Luise D., Lauridsen C., Bosi P., Trevisi P. Methodology and application of Escherichia coli F4 and F18 encoding infection models in post-weaning pigs. J. Anim. Sci. Biotechnol. 2019;10:53. doi: 10.1186/s40104-019-0352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abisado R.G., Benomar S., Klaus J.R., Dandekar A.A., Chandler J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio. 2018;9:e02331-17. doi: 10.1128/mBio.02331-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meroni G., Panelli S., Zuccotti G., Bandi C., Drago L., Pistone D. Probiotics as Therapeutic Tools against Pathogenic Biofilms: Have We Found the Perfect Weapon? Microbiol. Res. 2021;12:916–937. doi: 10.3390/microbiolres12040068. [DOI] [Google Scholar]
- 20.Wu L., Luo Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 2021;12:611413. doi: 10.3389/fmicb.2021.611413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J., Yin X., Yu H., Zhao L., Sabour P., Gong J. Involvement of quorum sensing and heat-stable enterotoxin a in cell damage caused by a porcine enterotoxigenic Escherichia coli strain. Infect. Immun. 2011;79:1688–1695. doi: 10.1128/IAI.01281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J., Kim J., Kim Y., Oh S., Song M., Choe J.H., Whang K.Y., Kim K.H., Oh S. Influences of quorum-quenching probiotic bacteria on the gut microbial community and immune function in weaning pigs. Anim. Sci. J. 2018;89:412–422. doi: 10.1111/asj.12954. [DOI] [PubMed] [Google Scholar]
- 23.Yeo S., Park H., Ji Y., Park S., Yang J., Lee J., Mathara J.M., Shin H., Holzapfel W. Influence of gastrointestinal stress on autoinducer-2 activity of two Lactobacillus species. FEMS Microbiol. Ecol. 2015;91:fiv065. doi: 10.1093/femsec/fiv065. [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Zhang Y., Xu J., Shi Q., Peng Y., Long C., Li L., Yin Y. Listening to enteric bacteria from the perspective of antibiotic alternatives in animal husbandry. Innov. Life. 2023;1:100022. doi: 10.59717/j.xinn-life.2023.100022. [DOI] [Google Scholar]
- 25.Lai J.J., Chau Z.L., Chen S.Y., Hill J.J., Korpany K.V., Liang N.W., Lin L.H., Lin Y.H., Liu J.K., Liu Y.C., et al. Exosome Processing and Characterization Approaches for Research and Technology Development. Adv. Sci. 2022;9:e2103222. doi: 10.1002/advs.202103222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle L.M., Wang M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells. 2019;8:727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldie B.J., Dun M.D., Lin M., Smith N.D., Verrills N.M., Dayas C.V., Cairns M.J. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014;42:9195–9208. doi: 10.1093/nar/gku594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Balkom B.W., de Jong O.G., Smits M., Brummelman J., den Ouden K., de Bree P.M., van Eijndhoven M.A., Pegtel D.M., Stoorvogel W., Würdinger T., et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. 2013;121:3997–4006. doi: 10.1182/blood-2013-02-478925. [DOI] [PubMed] [Google Scholar]
- 29.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J., et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munich S., Sobo-Vujanovic A., Buchser W.J., Beer-Stolz D., Vujanovic N.L. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology. 2012;1:1074–1083. doi: 10.4161/onci.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulcahy L.A., Pink R.C., Carter D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles. 2014;3:24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teng Y., Ren Y., Sayed M., Hu X., Lei C., Kumar A., Hutchins E., Mu J., Deng Z., Luo C., et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe. 2018;24:637–652.e638. doi: 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanton B.A. Extracellular Vesicles and Host-Pathogen Interactions: A Review of Inter-Kingdom Signaling by Small Noncoding RNA. Genes. 2021;12:1010. doi: 10.3390/genes12071010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larabi A., Dalmasso G., Delmas J., Barnich N., Nguyen H.T.T. Exosomes transfer miRNAs from cell-to-cell to inhibit autophagy during infection with Crohn’s disease-associated adherent-invasive E. coli. Gut Microbes. 2020;11:1677–1694. doi: 10.1080/19490976.2020.1771985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J., Gao Y., Liu F., Zhang Y., Li J., Ding L., Ren S., Yang J., Jiao J., Feng G., et al. m6A-modified exosome-derived circHIF1α binding to KH domain of IGF2BP3 mediates DNA damage and arrests G1/S transition phase to resists bacterial infection in bacteremia. J. Nanobiotechnol. 2024;22:654. doi: 10.1186/s12951-024-02932-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Q., Huang Z., Ma L., Yao J., Luo T., Zhao Y., Xiao Y., Jin Y. Extracellular vesicle miRNAs promote the intestinal microenvironment by interacting with microbes in colitis. Gut Microbes. 2022;14:2128604. doi: 10.1080/19490976.2022.2128604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X., Liu R., Zhou X., Zhang Z., Zhu T., Huang Y., Chai L., Wang Y., Zhao Z., Li W., et al. Characterization of exosomes derived from IPEC-J2 treated with probiotic Bacillus amyloliquefaciens SC06 and its regulation of macrophage functions. Front. Immunol. 2022;13:1033471. doi: 10.3389/fimmu.2022.1033471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Díaz-Garrido N., Badia J., Baldomà L. Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J. Extracell. Vesicles. 2021;10:e12161. doi: 10.1002/jev2.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joshi B., Singh B., Nadeem A., Askarian F., Wai S.N., Johannessen M., Hegstad K. Transcriptome Profiling of Staphylococcus aureus Associated Extracellular Vesicles Reveals Presence of Small RNA-Cargo. Front. Mol. Biosci. 2020;7:566207. doi: 10.3389/fmolb.2020.566207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu S., Zhao Z., Hao P., Qiu Y., Zhao M., Zhou G., Zhang C., Kang J., Li P. Biological Functions and Cross-Kingdom Host Gene Regulation of Small RNAs in Lactobacillus plantarum-Derived Extracellular Vesicles. Front. Microbiol. 2022;13:944361. doi: 10.3389/fmicb.2022.944361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watkins D., Arya D. Models of Hfq interactions with small non-coding RNA in Gram-negative and Gram-positive bacteria. Front. Cell. Infect. Microbiol. 2023;13:1282258. doi: 10.3389/fcimb.2023.1282258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishiyama K., Takaki T., Sugiyama M., Fukuda I., Aiso M., Mukai T., Odamaki T., Xiao J.-z., Osawa R., Okada N. Extracellular Vesicles Produced by Bifidobacterium longum Export Mucin-Binding Proteins. Appl. Environ. Microbiol. 2020;86:e01464-20. doi: 10.1128/AEM.01464-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valguarnera E., Scott N.E., Azimzadeh P., Feldman M.F. Surface Exposure and Packing of Lipoproteins into Outer Membrane Vesicles Are Coupled Processes in Bacteroides. mSphere. 2018;3:10–1128. doi: 10.1128/mSphere.00559-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gul L., Modos D., Fonseca S., Madgwick M., Thomas J.P., Sudhakar P., Booth C., Stentz R., Carding S.R., Korcsmaros T. Extracellular vesicles produced by the human commensal gut bacterium Bacteroides thetaiotaomicron affect host immune pathways in a cell-type specific manner that are altered in inflammatory bowel disease. J. Extracell. Vesicles. 2022;11:e12189. doi: 10.1002/jev2.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bitar A., Aung K.M., Wai S.N., Hammarström M.-L. Vibrio cholerae derived outer membrane vesicles modulate the inflammatory response of human intestinal epithelial cells by inducing microRNA-146a. Sci. Rep. 2019;9:7212. doi: 10.1038/s41598-019-43691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cañas M.A., Fábrega M.J., Giménez R., Badia J., Baldomà L. Outer Membrane Vesicles from Probiotic and Commensal Escherichia coli Activate NOD1-Mediated Immune Responses in Intestinal Epithelial Cells. Front. Microbiol. 2018;9:498. doi: 10.3389/fmicb.2018.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deo P., Chow S.H., Han M.-L., Speir M., Huang C., Schittenhelm R.B., Dhital S., Emery J., Li J., Kile B.T., et al. Mitochondrial dysfunction caused by outer membrane vesicles from Gram-negative bacteria activates intrinsic apoptosis and inflammation. Nat. Microbiol. 2020;5:1418–1427. doi: 10.1038/s41564-020-0773-2. [DOI] [PubMed] [Google Scholar]
- 48.Tao S., Fan J., Li J., Wu Z., Yao Y., Wang Z., Wu Y., Liu X., Xiao Y., Wei H. Extracellular vesicles derived from Lactobacillus johnsonii promote gut barrier homeostasis by enhancing M2 macrophage polarization. J. Adv. Res. 2024. Online ahead of print . [DOI] [PubMed]
- 49.Li J., Feng S., Wang Z., He J., Zhang Z., Zou H., Wu Z., Liu X., Wei H., Tao S. Limosilactobacillus mucosae-derived extracellular vesicles modulates macrophage phenotype and orchestrates gut homeostasis in a diarrheal piglet model. NPJ Biofilms Microbiomes. 2023;9:33. doi: 10.1038/s41522-023-00403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alessandri G., Ossiprandi M.C., MacSharry J., van Sinderen D., Ventura M. Bifidobacterial Dialogue With Its Human Host and Consequent Modulation of the Immune System. Front. Immunol. 2019;10:2348. doi: 10.3389/fimmu.2019.02348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandelbaum N., Zhang L., Carasso S., Ziv T., Lifshiz-Simon S., Davidovich I., Luz I., Berinstein E., Gefen T., Cooks T., et al. Extracellular vesicles of the Gram-positive gut symbiont Bifidobacterium longum induce immune-modulatory, anti-inflammatory effects. NPJ Biofilms Microbiomes. 2023;9:30. doi: 10.1038/s41522-023-00400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng C., Zhong Y., Zhang W., Wang Z., Xiao H., Zhang W., Xie J., Peng X., Luo J., Xu W. Chlorogenic Acid Ameliorates Post-Infectious Irritable Bowel Syndrome by Regulating Extracellular Vesicles of Gut Microbes. Adv. Sci. 2023;10:e2302798. doi: 10.1002/advs.202302798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu S., da Cunha A.P., Rezende R.M., Cialic R., Wei Z., Bry L., Comstock L.E., Gandhi R., Weiner H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe. 2016;19:32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sobolewski C., Calo N., Portius D., Foti M. MicroRNAs in Fatty Liver Disease. Semin. Liver Dis. 2015;35:012–025. doi: 10.1055/s-0034-1397345. [DOI] [PubMed] [Google Scholar]
- 55.Correia de Sousa M., Gjorgjieva M., Dolicka D., Sobolewski C., Foti M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019;20:6249. doi: 10.3390/ijms20246249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 57.Zhou X., Liu Y., Xiong X., Chen J., Tang W., He L., Zhang Z., Yin Y., Li F. Intestinal accumulation of microbiota-produced succinate caused by loss of microRNAs leads to diarrhea in weanling piglets. Gut Microbes. 2022;14:2091369. doi: 10.1080/19490976.2022.2091369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He L., Zhou X., Liu Y., Zhou L., Li F. Fecal miR-142a-3p from dextran sulfate sodium-challenge recovered mice prevents colitis by promoting the growth of Lactobacillus reuteri. Mol. Ther. J. Am. Soc. Gene Ther. 2022;30:388–399. doi: 10.1016/j.ymthe.2021.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodríguez-Nogales A., Algieri F., Garrido-Mesa J., Vezza T., Utrilla M.P., Chueca N., Garcia F., Olivares M., Rodríguez-Cabezas M.E., Gálvez J. Differential intestinal anti-inflammatory effects of Lactobacillus fermentum and Lactobacillus salivarius in DSS mouse colitis: Impact on microRNAs expression and microbiota composition. Mol. Nutr. Food Res. 2017;61 doi: 10.1002/mnfr.201700144. [DOI] [PubMed] [Google Scholar]
- 60.Belzer C., de Vos W.M. Microbes inside--from diversity to function: The case of Akkermansia. ISME J. 2012;6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wade H., Pan K., Duan Q., Kaluzny S., Pandey E., Fatumoju L., Saraswathi V., Wu R., Harris E.N., Su Q. Akkermansia muciniphila and its membrane protein ameliorates intestinal inflammatory stress and promotes epithelial wound healing via CREBH and miR-143/145. J. Biomed. Sci. 2023;30:38. doi: 10.1186/s12929-023-00935-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riahi Rad Z., Riahi Rad Z., Goudarzi H., Goudarzi M., Mahmoudi M., Yasbolaghi Sharahi J., Hashemi A. MicroRNAs in the interaction between host-bacterial pathogens: A new perspective. J. Cell. Physiol. 2021;236:6249–6270. doi: 10.1002/jcp.30333. [DOI] [PubMed] [Google Scholar]
- 63.Bao H., Kommadath A., Liang G., Sun X., Arantes A.S., Tuggle C.K., Bearson S.M., Plastow G.S., Stothard P., Guan L.L. Genome-wide whole blood microRNAome and transcriptome analyses reveal miRNA-mRNA regulated host response to foodborne pathogen Salmonella infection in swine. Sci. Rep. 2015;5:12620. doi: 10.1038/srep12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herrera-Uribe J., Zaldívar-López S., Aguilar C., Luque C., Bautista R., Carvajal A., Claros M.G., Garrido J.J. Regulatory role of microRNA in mesenteric lymph nodes after Salmonella Typhimurium infection. Vet. Res. 2018;49:9. doi: 10.1186/s13567-018-0506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang F., Liu J., Zou Y., Jiao Y., Huang Y., Fan L., Li X., Yu H., He C., Wei W., et al. MicroRNA-143-3p, up-regulated in H. pylori-positive gastric cancer, suppresses tumor growth, migration and invasion by directly targeting AKT2. Oncotarget. 2017;8:28711–28724. doi: 10.18632/oncotarget.15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roy B.C., Subramaniam D., Ahmed I., Jala V.R., Hester C.M., Greiner K.A., Haribabu B., Anant S., Umar S. Role of bacterial infection in the epigenetic regulation of Wnt antagonist WIF1 by PRC2 protein EZH2. Oncogene. 2015;34:4519–4530. doi: 10.1038/onc.2014.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du J., Zhang P., Luo J., Shen L., Zhang S., Gu H., He J., Wang L., Zhao X., Gan M., et al. Dietary betaine prevents obesity through gut microbiota-drived microRNA-378a family. Gut Microbes. 2021;13:1–19. doi: 10.1080/19490976.2020.1862612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xue X., Feng T., Yao S., Wolf K.J., Liu C.G., Liu X., Elson C.O., Cong Y. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J. Immunol. 2011;187:5879–5886. doi: 10.4049/jimmunol.1100535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanada M., Bachmann M.H., Hardy J.W., Frimannson D.O., Bronsart L., Wang A., Sylvester M.D., Schmidt T.L., Kaspar R.L., Butte M.J., et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. USA. 2015;112:E1433–E1442. doi: 10.1073/pnas.1418401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng L., Sharples R.A., Scicluna B.J., Hill A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lauridsen C. From oxidative stress to inflammation: Redox balance and immune system. Poult Sci. 2019;98:4240–4246. doi: 10.3382/ps/pey407. [DOI] [PubMed] [Google Scholar]
- 73.Xie K., Yang Q., Yan Z., Gao X., Huang X., Wang P., Zhang J., Yang J., Li J., Gun S. miR-30d Inhibition Protects IPEC-J2 Cells Against Clostridium perfringens Beta2 Toxin-Induced Inflammatory Injury. Front. Vet. Sci. 2022;9:909500. doi: 10.3389/fvets.2022.909500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kostic A.D., Xavier R.J., Gevers D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., Reyes J.A., Shah S.A., LeLeiko N., Snapper S.B., et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He S., Ran C., Qin C., Li S., Zhang H., de Vos W.M., Ringø E., Zhou Z. Anti-Infective Effect of Adhesive Probiotic Lactobacillus in Fish is Correlated With Their Spatial Distribution in the Intestinal Tissue. Sci. Rep. 2017;7:13195. doi: 10.1038/s41598-017-13466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu F., Zhang S., Dassopoulos T., Harris M.L., Bayless T.M., Meltzer S.J., Brant S.R., Kwon J.H. Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm. Bowel Dis. 2010;16:1729–1738. doi: 10.1002/ibd.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Z., Pi G., Song W., Li Y. Investigation of the Expression Pattern and Functional Role of miR-10b in Intestinal Inflammation. Animals. 2023;13:1236. doi: 10.3390/ani13071236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie M.Y., Hou L.J., Sun J.J., Zeng B., Xi Q.Y., Luo J.Y., Chen T., Zhang Y.L. Porcine Milk Exosome MiRNAs Attenuate LPS-Induced Apoptosis through Inhibiting TLR4/NF-κB and p53 Pathways in Intestinal Epithelial Cells. J. Agric. Food Chem. 2019;67:9477–9491. doi: 10.1021/acs.jafc.9b02925. [DOI] [PubMed] [Google Scholar]
- 80.Liu L., Liang L., Yang C., Zhou Y., Chen Y. Extracellular vesicles of Fusobacterium nucleatum compromise intestinal barrier through targeting RIPK1-mediated cell death pathway. Gut Microbes. 2021;13:1–20. doi: 10.1080/19490976.2021.1902718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oliveira E.C.S., Quaglio A.E.V., Magro D.O., Di Stasi L.C., Sassaki L.Y. Intestinal Microbiota and miRNA in IBD: A Narrative Review about Discoveries and Perspectives for the Future. Int. J. Mol. Sci. 2023;24:7176. doi: 10.3390/ijms24087176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abdulla O.A., Neamah W., Sultan M., Chatterjee S., Singh N., Nagarkatti M., Nagarkatti P. AhR Ligands Differentially Regulate miRNA-132 Which Targets HMGB1 and to Control the Differentiation of Tregs and Th-17 Cells During Delayed-Type Hypersensitivity Response. Front. Immunol. 2021;12:635903. doi: 10.3389/fimmu.2021.635903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tian Y., Xu J., Li Y., Zhao R., Du S., Lv C., Wu W., Liu R., Sheng X., Song Y., et al. MicroRNA-31 Reduces Inflammatory Signaling and Promotes Regeneration in Colon Epithelium, and Delivery of Mimics in Microspheres Reduces Colitis in Mice. Gastroenterology. 2019;156:2281–2296.e2286. doi: 10.1053/j.gastro.2019.02.023. [DOI] [PubMed] [Google Scholar]
- 84.Nguyen H.T., Dalmasso G., Müller S., Carrière J., Seibold F., Darfeuille-Michaud A. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology. 2014;146:508–519. doi: 10.1053/j.gastro.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 85.Michielan A., D’Incà R. Intestinal Permeability in Inflammatory Bowel Disease: Pathogenesis, Clinical Evaluation, and Therapy of Leaky Gut. Mediat. Inflamm. 2015;2015:628157. doi: 10.1155/2015/628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakata K., Sugi Y., Narabayashi H., Kobayakawa T., Nakanishi Y., Tsuda M., Hosono A., Kaminogawa S., Hanazawa S., Takahashi K. Commensal microbiota-induced microRNA modulates intestinal epithelial permeability through the small GTPase ARF4. J. Biol. Chem. 2017;292:15426–15433. doi: 10.1074/jbc.M117.788596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei S., Wu X., Chen M., Xiang Z., Li X., Zhang J., Dong W. Exosomal-miR-129-2-3p derived from Fusobacterium nucleatum-infected intestinal epithelial cells promotes experimental colitis through regulating TIMELESS-mediated cellular senescence pathway. Gut Microbes. 2023;15:2240035. doi: 10.1080/19490976.2023.2240035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Casado-Bedmar M., Viennois E. MicroRNA and Gut Microbiota: Tiny but Mighty-Novel Insights into Their Cross-talk in Inflammatory Bowel Disease Pathogenesis and Therapeutics. J. Crohns Colitis. 2022;16:992–1005. doi: 10.1093/ecco-jcc/jjab223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feng R., Niu Z., Zhang X., Hou W., Zhang Y., Jian F., Ning C., Zhang L., Zhang S., Wang R. Cryptosporidium parvum downregulates miR-181d in HCT-8 cells via the p50-dependent TLRs/NF-κB pathway. Vet. Parasitol. 2022;305:109710. doi: 10.1016/j.vetpar.2022.109710. [DOI] [PubMed] [Google Scholar]
- 90.Huang H., Liao D., Zhou G., He B., Pu R., Cui Y. MicroRNA-194-3p impacts autophagy and represses rotavirus replication via targeting silent information regulator 1. Virol. J. 2023;20:210. doi: 10.1186/s12985-023-02175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thiagarajah J.R., Donowitz M., Verkman A.S. Secretory diarrhoea: Mechanisms and emerging therapies. Nat. Rev. Gastroenterol. Hepatol. 2015;12:446–457. doi: 10.1038/nrgastro.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yin L., Shen X., Yin D., Wang J., Zhao R., Dai Y., Pan X. Characteristics of the MicroRNA Expression Profile of Exosomes Released by Vero Cells Infected with Porcine Epidemic Diarrhea Virus. Viruses. 2022;14:806. doi: 10.3390/v14040806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou X., He Y., Chen J., Xiong X., Yin J., Liang J., Peng C., Huang C., Guan G., Yin Y. Colonic phosphocholine is correlated with Candida tropicalis and promotes diarrhea and pathogen clearance. NPJ Biofilms Microbiomes. 2023;9:62. doi: 10.1038/s41522-023-00433-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gupta A. Peripheral mechanisms in irritable bowel syndrome. N. Engl. J. Med. 2013;368:578. doi: 10.1056/NEJMc1214185. [DOI] [PubMed] [Google Scholar]
- 95.Zhu H., Xiao X., Shi Y., Wu Y., Huang Y., Li D., Xiong F., He G., Chai Y., Tang H. Inhibition of miRNA-29a regulates intestinal barrier function in diarrhea-predominant irritable bowel syndrome by upregulating ZO-1 and CLDN1. Exp. Ther. Med. 2020;20:155. doi: 10.3892/etm.2020.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xi M., Zhao P., Li F., Bao H., Ding S., Ji L., Yan J. MicroRNA-16 inhibits the TLR4/NF-κB pathway and maintains tight junction integrity in irritable bowel syndrome with diarrhea. J. Biol. Chem. 2022;298:102461. doi: 10.1016/j.jbc.2022.102461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hou Q., Huang Y., Zhang C., Zhu S., Li P., Chen X., Hou Z., Liu F. MicroRNA-200a Targets Cannabinoid Receptor 1 and Serotonin Transporter to Increase Visceral Hyperalgesia in Diarrhea-predominant Irritable Bowel Syndrome Rats. J Neurogastroenterol. Motil. 2018;24:656–668. doi: 10.5056/jnm18037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang P., Huang X., Yan Z., Yang Q., Sun W., Gao X., Luo R., Gun S. Analyses of miRNA in the ileum of diarrheic piglets caused by Clostridium perfringens type C. Microb. Pathog. 2019;136:103699. doi: 10.1016/j.micpath.2019.103699. [DOI] [PubMed] [Google Scholar]
- 99.Wu Z., Qin W., Wu S., Zhu G., Bao W., Wu S. Identification of microRNAs regulating Escherichia coli F18 infection in Meishan weaned piglets. Biol. Direct. 2016;11:59. doi: 10.1186/s13062-016-0160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang P., Yang Q., Yan Z., Huang X., Gao X., Gun S. Identification of MicroRNAs Regulating Clostridium perfringens Type C Infection in the Spleen of Diarrheic Piglets. Curr. Issues Mol. Biol. 2023;45:3193–3207. doi: 10.3390/cimb45040208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gao X., Yang Q., Zhang S., Huang X., Yan Z., Wang P., Gun S. LncRNA ALDB-898 modulates intestinal epithelial cell damage caused by Clostridium perfringens type C in piglet by regulating ssc-miR-122-5p/OCLN signaling. Mol. Immunol. 2022;149:143–156. doi: 10.1016/j.molimm.2022.07.002. [DOI] [PubMed] [Google Scholar]
- 102.Duan C., Luo Y., Liang X., Wang X. A Review of Bioactive Compounds against Porcine Enteric Coronaviruses. Viruses. 2022;14:2217. doi: 10.3390/v14102217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qin W., Jiang J., Wu J., Xie Y., Wu Z., Sun M., Bao W. Exosomal ssc-miR-1343 targets FAM131C to regulate porcine epidemic diarrhea virus infection in pigs. Vet. Res. 2024;55:91. doi: 10.1186/s13567-024-01345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu T., Qian J., Shen Z., Shao H., Qian K., Jin W., Qin A. Vector-delivered artificial miRNA effectively inhibits Porcine epidemic diarrhea virus replication. Virol. J. 2023;20:164. doi: 10.1186/s12985-023-02129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Siegel R.L., Wagle N.S., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023;73:233–254. doi: 10.3322/caac.21772. [DOI] [PubMed] [Google Scholar]
- 106.Yang F., Ning Z., Ma L., Liu W., Shao C., Shu Y., Shen H. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol. Cancer. 2017;16:148. doi: 10.1186/s12943-017-0718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pardini B., Ferrero G., Tarallo S., Gallo G., Francavilla A., Licheri N., Trompetto M., Clerico G., Senore C., Peyre S., et al. A Fecal MicroRNA Signature by Small RNA Sequencing Accurately Distinguishes Colorectal Cancers: Results from a Multicenter Study. Gastroenterology. 2023;165:582–599.e588. doi: 10.1053/j.gastro.2023.05.037. [DOI] [PubMed] [Google Scholar]
- 108.Matsumura T., Sugimachi K., Iinuma H., Takahashi Y., Kurashige J., Sawada G., Ueda M., Uchi R., Ueo H., Takano Y., et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer. 2015;113:275–281. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Slattery M.L., Herrick J.S., Pellatt D.F., Mullany L.E., Stevens J.R., Wolff E., Hoffman M.D., Wolff R.K., Samowitz W. Site-specific associations between miRNA expression and survival in colorectal cancer cases. Oncotarget. 2016;7:60193–60205. doi: 10.18632/oncotarget.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Han L., Chen S., Luan Z., Fan M., Wang Y., Sun G., Dai G. Immune function of colon cancer associated miRNA and target genes. Front. Immunol. 2023;14:1203070. doi: 10.3389/fimmu.2023.1203070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rokavec M., Öner M.G., Li H., Jackstadt R., Jiang L., Lodygin D., Kaller M., Horst D., Ziegler P.K., Schwitalla S., et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ma Z.B., Kong X.L., Cui G., Ren C.C., Zhang Y.J., Fan S.J., Li Y.H. Expression and clinical significance of miRNA-34a in colorectal cancer. Asian Pac. J. Cancer Prev. 2014;15:9265–9270. doi: 10.7314/APJCP.2014.15.21.9265. [DOI] [PubMed] [Google Scholar]
- 113.Shain A.H., Joseph N.M., Yu R., Benhamida J., Liu S., Prow T., Ruben B., North J., Pincus L., Yeh I., et al. Genomic and Transcriptomic Analysis Reveals Incremental Disruption of Key Signaling Pathways during Melanoma Evolution. Cancer Cell. 2018;34:45–55.e44. doi: 10.1016/j.ccell.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tutuka C.S.A., Andrews M.C., Mariadason J.M., Ioannidis P., Hudson C., Cebon J., Behren A. PLX8394, a new generation BRAF inhibitor, selectively inhibits BRAF in colonic adenocarcinoma cells and prevents paradoxical MAPK pathway activation. Mol. Cancer. 2017;16:112. doi: 10.1186/s12943-017-0684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin C., Ma M., Zhang Y., Li L., Long F., Xie C., Xiao H., Liu T., Tian B., Yang K., et al. The N(6)-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol. Cancer. 2022;21:80. doi: 10.1186/s12943-022-01560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y., Lauriola M., Kim D., Francesconi M., D’Uva G., Shibata D., Malafa M.P., Yeatman T.J., Coppola D., Solmi R., et al. Adenomatous polyposis coli (APC) regulates miR17-92 cluster through β-catenin pathway in colorectal cancer. Oncogene. 2016;35:4558–4568. doi: 10.1038/onc.2015.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grillari J., Hackl M., Grillari-Voglauer R. miR-17-92 cluster: Ups and downs in cancer and aging. Biogerontology. 2010;11:501–506. doi: 10.1007/s10522-010-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Slattery M.L., Mullany L.E., Sakoda L.C., Samowitz W.S., Wolff R.K., Stevens J.R., Herrick J.S. Expression of Wnt-signaling pathway genes and their associations with miRNAs in colorectal cancer. Oncotarget. 2018;9:6075–6085. doi: 10.18632/oncotarget.23636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hu S., Liu L., Chang E.B., Wang J.Y., Raufman J.P. Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol. Cancer. 2015;14:180. doi: 10.1186/s12943-015-0450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brusnic O., Onisor D., Boicean A., Hasegan A., Ichim C., Guzun A., Chicea R., Todor S.B., Vintila B.I., Anderco P., et al. Fecal Microbiota Transplantation: Insights into Colon Carcinogenesis and Immune Regulation. J. Clin. Med. 2024;13:6578. doi: 10.3390/jcm13216578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Casado-Bedmar M., Roy M., Berthet L., Hugot J.P., Yang C., Manceau H., Peoc’h K., Chassaing B., Merlin D., Viennois E. Fecal let-7b and miR-21 directly modulate the intestinal microbiota, driving chronic inflammation. Gut Microbes. 2024;16:2394249. doi: 10.1080/19490976.2024.2394249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang Y., Weng W., Peng J., Hong L., Yang L., Toiyama Y., Gao R., Liu M., Yin M., Pan C., et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851–866.e824. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cougnoux A., Dalmasso G., Martinez R., Buc E., Delmas J., Gibold L., Sauvanet P., Darcha C., Déchelotte P., Bonnet M., et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63:1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.