Abstract
Bitter taste perception is crucial for animal survival. By detecting potentially harmful substances, such as plant secondary metabolites, as bitter, animals can avoid ingesting toxic compounds. In vertebrates, this function is mediated by taste receptors type 2 (T2Rs), a family of G protein-coupled receptors (GPCRs) expressed on taste buds. Given their vital roles, T2Rs have undergone significant selective pressures throughout vertebrate evolution, leading to frequent gene duplications and deletions, functional changes, and intrapopulation differentiation across various lineages. Recent advancements in genomic and functional research have uncovered the repertoires and functions of bitter taste receptors in a wide range of vertebrate species, shedding light on their evolution in relation to dietary habits and other ecological factors. This review summarizes recent research on bitter taste receptors and explores the mechanisms driving the diversity of these receptors from the perspective of vertebrate ecology and evolution.
Keywords: bitter taste perception, G protein-coupled receptors, molecular evolution
1. Introduction
Vertebrates use a variety of sensory systems, such as taste, smell, vision, touch, and hearing, to obtain the environmental cues essential for their survival. Among them, taste is crucial for evaluating the nutritional value and potential toxicity of food before ingestion. Different species rely on diverse food sources, resulting in significant variation in taste stimuli detection among species. Consequently, taste receptors exhibit remarkable diversity in gene repertoire, receptive breadth, and sensitivity to tastants, often correlating with an animal’s ecology, including habitat, dietary preferences, and feeding strategies. Among the basic tastes (sweet, umami, bitter, sour, and salty), the T2R receptor family, which is encoded by Tas2r genes, was identified as bitter taste receptors around the year 2000 [1,2,3]. Since then, the gene repertoires and functions of these receptors have been analyzed in various species. In particular, the sequencing of numerous vertebrate genomes over the past decade has significantly advanced the research on taste receptors in various non-model animals, including those that are difficult to access or endangered, thereby enhancing our understanding of the evolution and diversity of taste receptors in vertebrates. In this review, we highlight recent achievements in bitter taste receptor research from an ecological and evolutionary perspective, mainly focusing on taste function.
2. T2R Receptors: Phylogeny and Evolutionary Origins
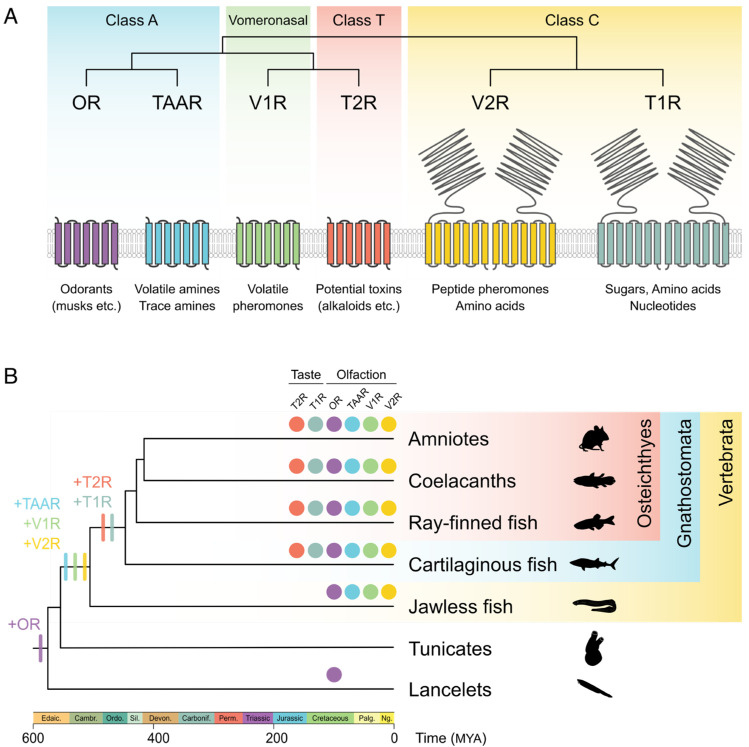
Bitter taste is one of the chemical senses (taste and olfaction) and is considered to be an important sense for vertebrates to detect potential toxins in food, as all toxic compounds do not always exhibit bitterness [4]. Bitterness is mediated by a GPCR family member, T2Rs [1,2,3]. T2Rs are mainly expressed in taste bud cells on sensory organs, such as the tongue and oral palate, and detect potentially harmful substances with diverse structures [5,6]. Chemical senses other than bitterness are also mainly mediated by GPCRs similar to T2Rs. Vertebrates have six major GPCR families responsible for taste and olfaction (Figure 1A). Taste receptors include taste receptor type 1 (T1Rs), encoded by Tas1r genes, as well as T2Rs, while odorant receptors are classified into olfactory receptors (ORs), trace amine-associated receptors (TAARs), and vomeronasal receptor type 1 and type 2 (V1Rs and V2Rs, respectively). T2Rs detect potentially harmful substances, whereas T1Rs detect nutrient-related molecules such as sugars, amino acids, and nucleotides as sweet or umami taste. T2Rs have a small N-terminus extracellular domain similar to ORs, TAARs, and V1Rs, unlike T1Rs and V2Rs, and their phylogenetically closest relative is the V1R family (Figure 1A).
The evolutionary origins are different among chemosensory genes (Figure 1B). Odorant receptors have earlier origins than taste receptors. OR genes were already present in the common ancestor of Chordata, and the origins of other odorant receptor genes were traced back to the common ancestor of vertebrates [7,8,9,10]. Although OR-like genes, encoding class A GPCRs, are also reported in invertebrates such as Echinodermata (e.g., sea urchins and starfish) and Cnidaria (e.g., hydras and sea anemones), their orthology with vertebrate-type ORs is questionable [10,11,12,13,14,15]. In taste receptor genes, Tas1rs originated in the common ancestor of jawed vertebrates (Gnathostomata) [16,17], whereas it has been believed that the evolutionary origin of the Tas2rs traced back to the common ancestor of bony vertebrates (Osteichthyes), due to the absence of Tas2rs in cartilaginous fish (Chondrichthyes) and jawless fish (Cyclostomata) genomes [16,18]. However, several research groups have recently identified Tas2rs in a subclass of cartilaginous fish, Elasmobranchii (sharks, rays, skates, sawfish, and their close relatives) [8,19,20]. Consequently, the evolutionary origin of the Tas2r gene family is now assumed to be the common ancestor of jawed vertebrates, similar to the Tas1r gene family (Figure 1B) [8,17]. To date, no Tas2r orthologs have been found in jawless fish [8,20], although lampreys, a lineage of jawless fish, have a well-developed gustatory system that includes taste buds [21]. Thus, it remains an open question whether ancient jawless fish had Tas2r orthologs and subsequently lost them or whether they possess unknown genes that function as bitter receptor genes instead of Tas2rs.
Figure 1.
Classification and origins of T2Rs in chemosensory GPCR families. (A) A schematic illustration of the phylogeny, structures, and ligands of chemosensory GPCR families. The GPCR family classification and phylogeny refer to GPCRdb and several literature sources [22,23,24,25]. (B) The evolutionary origins of chemosensory GPCR families in Chordata. The animal silhouettes are retrieved from Phylopic (https://www.phylopic.org/, accessed on 28 May 2024).
3. Evolution of Tas2r Gene Repertoires
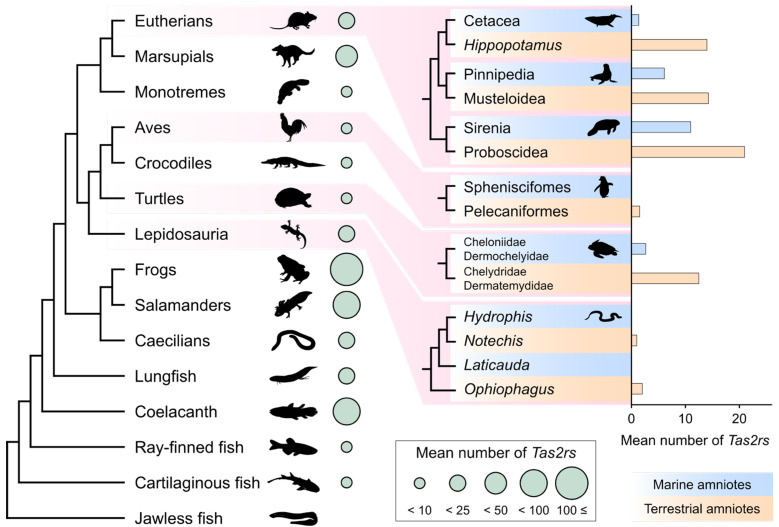
The evolution of the vertebrate Tas2r repertoire has been studied in a vast number of species (over 1500 species as of 2024) [8,19,20,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47]. Generally, the receptors were identified from genome assemblies via BLAST-based gene mining. In many of these studies, intact genes were defined as those hit sequences that are predicted to encode proteins with a seven-transmembrane topology and form a monophyletic clade with known Tas2rs in a phylogenetic tree. This definition is primarily adopted in the present study. The number of Tas2r genes varies widely among vertebrates, ranging from zero (e.g., some cetaceans, penguins, and sea snakes) to over two hundred in certain frogs (Figure 2 and Figure 3) [8,32,41,42,45]. Cartilaginous fish and ray-finned fish generally have small Tas2r repertoires, whereas diversification and expansion of the gene family occurred in the lineage leading to tetrapods [8]. The Tas2r gene family has experienced drastic birth-and-death evolution [8,26,48]. Consequently, there are many lineage-specific Tas2r subtypes [8,26]. For example, there is no orthologous Tas2rs between “fish” (cartilaginous fish, ray-finned fish, and lobe-finned fish) and amniotes (mammals, reptiles, and birds), whereas some amphibians (e.g., salamanders and caecilians) have both amniote-specific Tas2r subtypes and Tas2r subtypes shared with coelacanths [8,28]. Although this relationship is supported by several studies [8,20,26,28], the unified classification of Tas2r subtypes across vertebrates and the phylogenetic relationships among the subtypes are not fully established. This is because the topology of the Tas2r gene tree is prone to change depending on the input dataset [8,20,28]. For example, the true topology near the stem root of the Tas2r gene tree [8,20,28] and whether there is a Tas2r subtype globally shared among ray-finned fish, lobe-finned fish, and amphibians [8,28] are topics for further research.
Figure 2.
Variation of Tas2r gene number in vertebrates. The mean number of Tas2rs in major vertebrate lineages is represented as different sized circles. The inset bar plot shows that the comparisons of the mean number of Tas2rs between marine and terrestrial amniotes are represented as bar plots. Data were retrieved from Policarpo et al. (The number of complete Tas2rs with a predicted seven-transmembrane topology from the whole-genome assembly with > 80% BUSCO gene completeness) [8]. The animal silhouettes are obtained from PhyloPic (https://www.phylopic.org/, accessed on 28 May 2024). The silhouettes of the Tasmanian devil and platypus drawn by Sarah Werning are reused under the CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/). The silhouette of Hydrophis curtus drawn by Christina Zdenek and the manatee drawn by Flurin Leugger are reused under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
In vertebrates, a general trend shows that herbivores have larger Tas2r repertoires than carnivores [26]. The number of Tas2rs in amniotes is also influenced by feeding patterns and living environments. Species that swallow their food whole, having less opportunity to sense the chemicals from their prey, tend to have fewer Tas2rs [32,35,45,46]. The transition from land to marine environments also leads to a reduction in Tas2r repertoires, as observed in other chemoreceptor genes (see the inset of Figure 2) [8,32,44,45,46,49]. Furthermore, the number of Tas2rs is associated with other chemical sensing modalities [8]. Niimura et al. found that gains or losses of Tas2rs occurred synchronously with those of ORs, V1Rs, and V2Rs during Hystricomorph rodent evolution, suggesting no trade-off among different chemical sensing modalities [50]. It should be noted that such trends in the Tas2r repertoire related to vertebrate ecology or phenotypes are not consistently observed across all vertebrate classes or orders. Therefore, it is crucial to identify the specific periods in phylogenetic trees when Tas2r genes were gained or lost and to individually estimate the causes of these changes [8,29,41,50]. Hereafter, we overview the characteristics of the receptor repertoire in each vertebrate lineage.
3.1. Cartilaginous, Ray-Finned, and Lobe-Finned Fish
Cartilaginous fish have the smallest repertoires (ranging from zero to one) among vertebrates. Their Tas2rs consist of a single, unique ortholog, which is considered one of the earliest-emerged Tas2r orthologs [8,19,20]. This gene is highly conserved with a low frequency of gene duplication, and its presence or absence is not correlated with diet [20].
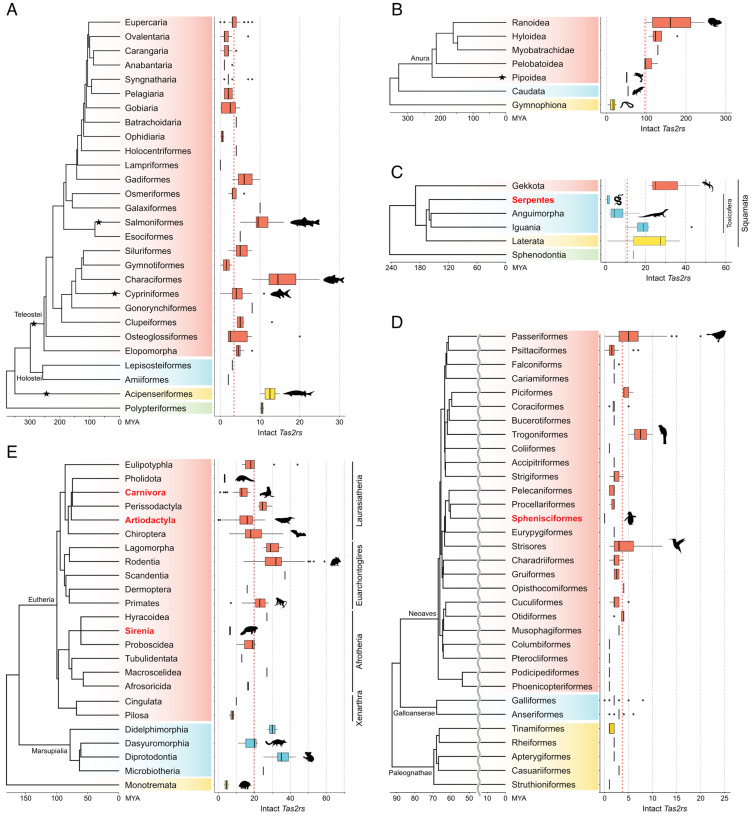
Ray-finned fish also have small repertoires, not correlated with any ecological traits such as diet, temperature preference, water habitat, or living depth [8]. Ray-finned fish receptors are classified into three or four clades, with varying degrees of gene gains or losses between clades [8,36]. Some lineages have experienced massive expansion of Tas2rs (Figure 3A). In particular, Characiformes has one of the largest Tas2r repertoires [8]. For example, the Mexican tetra (Astyanax mexicanus) has over twenty Tas2rs, roughly the same number as humans [8,36]. This species has two morphs: cave and surface forms. Cave populations have an increased number of taste buds, enhanced chemosensory capabilities, and improved food detection in dark environments [51,52]. The cave form has 21 Tas2rs, whereas the surface form has 25 [8,36]. Given that gene mining methodologies and genome assembly quality affect gene identification, further detailed investigation using the same gene mining method using high-quality genome assembly of two morphs and population data is required to understand the differences in bitter taste between two morphs. For additional details on this species’ taste study, please refer to another review [51]. Ray-finned fish have experienced lineage-specific whole-genome duplication (WGD) events in a few lineages, including Acipenseriformes (sturgeons and paddlefish), the stem of Teleostei, Salmoniformes, and carps (a subclade of Cypriniformes). Since carps, Salmoniformes, and Acipenseriformes have a relatively larger number of Tas2rs (Figure 3A), WGD might be a factor affecting Tas2r numbers in ray-finned fish.
The Tas2r repertoire of lobe-finned fish differs largely between coelacanths (Coelacanthiformes) and lungfish (Dipnoi). The West Indian Ocean coelacanth (Latimeria chalumnae) has the second largest Tas2r repertoire among vertebrates, following batrachians (frogs and salamanders) [8,28,37,38]. Coelacanth Tas2rs expanded through species-specific gene duplication, likely due to accumulated repeat elements surrounding Tas2r loci [37]. Of the coelacanth Tas2rs, Tas2r01 is orthologous to ray-finned fish Tas2r1, the only known clear orthologous gene pair between the ray-finned fish and lobe-finned lineages [8,28,37]. This gene is believed to be one of the basal Tas2r orthologs. Conversely, lungfish have less than half the Tas2rs of the coelacanth [8]. Since both lungfish and tetrapods lack Tas2rs orthologous to coelacanth Tas2r01, this ortholog was likely lost in the common ancestor of lungfish and tetrapods.
3.2. Amphibians
Amphibians exhibit remarkable variations in Tas2r gene repertoires (Figure 2 and Figure 3B). Batrachians have the largest repertoires among vertebrates, with the Japanese wrinkled frog (Glandirana rugosa) having 247 Tas2rs [8,41,42]. In contrast, caecilians (Gymnophiona) have relatively smaller repertoires than batrachians, ranging from 3 to 25 [8,41]. Many caecilians, unlike batrachians, are highly adapted to subterranean environments and are carnivorous, likely leading to reduced exposure to potential toxins and thus a reduced importance of bitter taste [41]. Anuran (frog) Tas2rs frequently expand in a lineage-specific manner [41]. In some frogs, many clusters of Tas2rs are observed on certain chromosomes, with particularly large clusters tending to be located near chromosome ends [41]. On the other hand, allotetraploid Xenopus frogs (African clawed frog, X. laevis, and Marsabit clawed frog, X. borealis) have almost the same number of Tas2rs as their diploid relative, the tropical clawed frog (X. tropicalis) [8,41]. Given these pieces of evidence, one of the most likely mechanisms for the massive expansion of batrachian Tas2rs is tandem duplication near chromosome ends, where nonhomologous recombination frequently occurs, rather than large-scale mutations such as WGD. It is so far unclear whether repeat elements are involved in Tas2r duplication of batrachians, as in the coelacanth [37]. To understand the expansion mechanisms in more detail, a larger comparative genomic analysis between batrachians and other vertebrates is required. Furthermore, the ecological and physiological significance of batrachian Tas2r expansion also remains controversial. Many batrachians are sit-and-wait or ambush predators, suggesting the biological importance of bitter taste lies more in the ability to detect and reject inedible or potentially harmful prey once captured rather than in detecting prey [53]. Additionally, many batrachians inhabit both aquatic and terrestrial environments, leading to encounters with a larger variety of harmful substances. Anurans, in particular, experience an ontogenetic dietary shift from herbivory or detritivory to carnivory or insectivory through metamorphosis. Hao et al. demonstrated that a subset of Tas2rs is differentially expressed in the oral cavity between tadpole and adult American bullfrogs (Lithobates catesbeianus), suggesting differential use of Tas2rs between life stages [42]. However, only a fraction of 180 bullfrog Tas2rs show differential expression in the oral cavity during metamorphosis. Therefore, there may be other potential drivers for Tas2r expansion beyond taste-related functions, such as extra-oral roles.
3.3. Birds and Reptiles
Avian Tas2r repertoires are generally small, with some lineages, such as Passeriformes (perching birds), Trogoniformes (trogons), and Strisores (nighthawk, sparrows, and hummingbirds), experiencing significant expansions of Tas2rs, up to 20 genes (Figure 2 and Figure 3D) [8]. Conversely, Sphenisciformes (penguins), which lack taste buds on their tongues, have lost all Tas2rs [8,39,45]. Possible causes of this loss include swallowing food whole and marine adaptation (both interrelated), while the previously suggested association with their life in cold environments is questionable, as it has not been observed in other cold-adapted birds and mammals [8,45]. The number of intact Tas2rs correlates with dietary preference in birds; herbivorous and insectivorous species have more Tas2rs than carnivorous species [39]. A recent large-scale analysis indicated a correlation between Tas2r number and migratory behavior, though the dietary association was not observed in that study [8].
Non-avian reptiles, including turtles, crocodilians, lizards, and snakes, generally have small Tas2r repertoires, except for some lizards (Figure 2 and Figure 3C). The number of Tas2rs in Lepidosauria (lizards and snakes) is highly variable (Figure 3C). Many lizards possess more than twenty Tas2rs, with the Japanese gecko (Gekko japonicus; Gekkota) having nearly 50 Tas2rs [8,35]. In contrast, snakes (Serpentes) and Varanus monitor lizards (Anguimorpha), which have taste-bud-less tongues, have a minimal Tas2r repertoire, ranging from zero to three [8,35,40]. In squamate reptiles, herbivorous and insectivorous species have more Tas2rs than carnivorous species, indicating dietary association [35]. Additionally, foraging patterns may influence Tas2r repertoires [35]. Snakes and monitor lizards, which swallow their prey whole, have small Tas2r repertoires similar to cetaceans and penguins (though marine adaptation may also play a role) [35,44,45,46]. Sea turtles and sea snakes have reduced Tas2r repertoires compared to their terrestrial relatives, exemplifying genetic convergence relevant to marine adaptation (see the inset of Figure 2) [8]. Notably, sea snakes have completely lost the few remaining Tas2rs found in terrestrial snakes [8], suggesting that both swallowing feeding and marine adaptation contribute to the reduction of Tas2r repertoires in snakes. This raises the possibility that for most terrestrial snakes, the few remaining Tas2rs may be crucial for terrestrial life.
3.4. Mammals
Mammals have medium-sized Tas2r repertoires, ranging from 0 (some cetaceans) to 59 (African woodland thicket rat, Grammomys surdaster) (Figure 2 and Figure 3E) [8,26,29,32,50]. Monotremes (platypus and echidna) have one of the smallest numbers of Tas2rs in non-fully aquatic mammals, whereas therians (eutherians and marsupials) have an average of ten or more. In marsupials, Diprotodontia (e.g., koalas, possums, and kangaroos) have the largest number of Tas2rs, followed by Didelphimorphia (opossums), Microbiotheria (monito del monte), and Dasyuromorphia (e.g., quolls and Tasmanian devils). In eutherians, Euarchontoglires (e.g., rodents, rabbits, and primates) have the largest Tas2r repertoire, followed by Afrotheria (e.g., elephants and tenrecs) and Laurasiatheria (e.g., ruminants, bats, and Carnivora). Xenarthra (e.g., sloths and armadillos) have the smallest Tas2r repertoire, 10 or fewer genes, among four major eutherian lineages. A recent large-scale study showed that the mammalian Tas2r repertoire is larger in omnivorous species, followed by herbivorous and carnivorous species [8]. This trend differs from the vertebrate-wide trend inferred in the earlier study [26], although in a mammalian lineage, Laurasiatheria, the trend matches the vertebrate-wide pattern [32]. Swallowing behavior is also associated with Tas2r reduction in mammals, such as cetaceans and pinnipeds [32].
Massive reductions in Tas2r repertoires have independently occurred in many mammalian lineages (Figure 3E). For example, pangolins (Pholidota), a Laurasiatherian lineage, has only a few Tas2rs, likely due to dietary specialization to ants and termites (myrmecophagy) [8,32]. Similar reductions occurred convergently in marsupial and monotreme myrmecophagous species, like the numbat (Myrmecobius fasciatus; Dasyuromorphia) and the short-beaked echidna (Tachyglossus aculeatus) [43,54]. Sanguivorous vampire bats also have reduced Tas2rs [55,56]. Colobine monkeys, specialized folivores, reduced the number of Tas2rs after diverging from their omnivorous cercopithecine relatives, which is thought to be associated with high tolerance to toxic compounds through mechanisms like foregut fermentation by symbiotic microbiota [47]. In relation to marine adaptation, convergent reductions in Tas2r repertoires are also found in marine mammals, including sirenians, pinnipeds, and cetaceans (see the inset of Figure 2) [8,32,44,46,49]. Conversely, massive expansions of receptors are also found in many mammalian lineages (Figure 3E). For instance, the koala (Phascolarctos cinereus) has a large Tas2r repertoire including tandemly duplicated Tas2r41 and Tas2r705 [31,57]. Some of these duplicates are sensitive to analogs of eucalyptus toxins, potentially aiding in the detection of eucalyptus toxins and the selection of edible eucalyptus leaves [31,57]. In primates, the common ancestor of hominoids and cercopithecoids independently experienced a massive expansion of Tas2rs, possibly related to increased folivory with increased body size (i.e., switching of protein sources from insects to leaves) [29].
Figure 3.
Variation of Tas2r gene number within representative vertebrate clades. The intact Tas2r gene number for (A) ray-finned fish, (B) amphibians, (C) birds, (D) Lepidosaurians, and (E) mammals is shown as boxplots with the mean Tas2r number for each clade. Phylogenetic trees were retrieved from previous studies (ray-finned fish, amphibians, and birds) [58,59,60] or the TimeTree 5 database (Lepidosaurians and mammals) [61]. Stars indicate whole-genome duplication events within the branches. Taxonomic groups in red include species fully adapted to aquatic environments. The taxonomic clades mentioned in the main text are marked with animal silhouettes. Animal silhouettes are obtained from PhyloPic (https://www.phylopic.org/, accessed on 28 May 2024). The silhouettes of the koala and numbat drawn by Sarah Werning and the silhouette of the vampire bat drawn by Roberto Díaz Sibaja are reused under CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/). The silhouette of the manatee drawn by Flurin Leugger are reused under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
4. Functional Evolution of T2Rs
4.1. Ancestral Functions of T2Rs
The frequent expansion and contraction of Tas2rs and the great diversity of agonist repertoires make it challenging to identify the ancestral function of T2Rs. However, the early-emerged orthologs or functionally conserved orthologs between distantly related species provide insight into ancestral T2R functions. For instance, a distinct one-to-one ortholog conserved between lobe-finned and ray-finned lineages, namely ray-finned fish T2R1 and coelacanth T2R01, have the same agonists, chloroquine and denatonium benzoate [28]. Cartilaginous fish-specific T2R orthologs also respond to these substances, suggesting that these agonists may be reflected in the function of T2R founders [19,20]. Additionally, endogenous steroids, such as bile acids and steroid hormones, are known as agonists of T2Rs in diverse vertebrates from ray-finned fish to mammals [28,62,63]. Shark receptors also recognize bile acids, suggesting the bifunctional nature of T2R founders to endogenous and exogenous compounds [19].
4.2. Relationships Between the Tuning Breadth and Repertoire Size of T2Rs
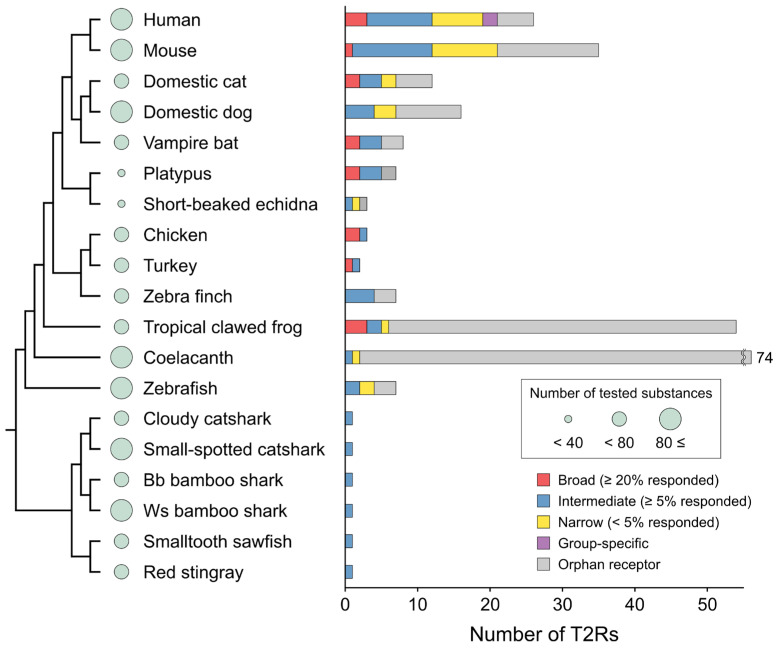
T2Rs from many vertebrates have been functionally characterized using bitter compound panels [5,6,19,20,27,28,30,42,56,57,64,65,66,67,68,69,70] (see the BitterDB database (https://bitterdb.agri.huji.ac.il/dbbitter.php) [71,72] for humans, mice, cats, and chickens and Supplementary Table S1 for other species). These data on the tuning breadth of receptors indicate an intriguing relationship with Tas2r repertoire size (Figure 4). Even species with fewer receptors, such as chickens and turkeys, have broadly tuned receptors, suggesting a wider receptive range than expected from the size of Tas2r repertoires [27,28]. Conversely, the receptive range in the dietary specialized echidna is narrower than in the platypus, aligning with a reduction in the Tas2r repertoire [57]. Species with many receptors, such as humans and mice, also have a few broadly tuned receptors, while many receptors are intermediate or narrowly tuned [73]. Consequently, the size of the Tas2r repertoire does not always reflect the receptive range of bitter substances. To fully understand the animals’ bitter taste space, analyses of both receptor repertoire and tuning breadth are essential. However, it should be noted that bitter compound panels used in most studies are biased toward compounds known as bitter to humans. Further investigation with panels of ecologically relevant substances, such as defense substances of potential prey, would be worthwhile to examine the significance of bitter sense in each animal.
Figure 4.
The tuning breadth of vertebrate T2Rs. The receptive ranges of representative vertebrate T2Rs are classified into “broad”, “intermediate”, “narrow”, and “group-specific” based on the ratio of agonists in the total number of substances tested in the screening studies, which included in total over 20 substances except for a receptor of vampire bats (19 substances) [5,6,19,20,27,28,56,57,64,65,66,67,73] and represented in bar plots. The number of tested substances is shown as the size of circles next to the species names. Bb bamboo shark and Ws bamboo shark indicate the brownbanded bamboo shark and whitespotted bamboo shark, respectively.
4.3. Functional Diversity of Orthologous T2Rs Among Species
The interspecific variation of T2R properties is observed in ligand sensitivity and specificity, and receptive ranges. Comprehensive comparisons between human and mouse T2Rs revealed that even one-to-one orthologs generally have distinct agonist profiles [6]. Similarly, T2R1 has achieved differential agonist spectra among bats [69]. In contrast, there is also evidence for the conservative ligand specificity of T2Rs among distantly related species. For instance, eutherian T2R16 and its marsupial ortholog (T2R705) respond to β-glucosides and their analogs, one of the major plant secondary metabolites, with high specificity in many species [5,30,57,74,75,76,77]. Furthermore, monotreme T2Rs in the same orthologous group as T2R16/T2R705 respond to β-glucosides, suggesting this bitter sensing trait was retained from the common ancestor of mammals [57]. Sensitivity to metal ions in T2R7 is conserved between humans and vampire bats [70,78,79], and zebrafish T2R1 and its coelacanth ortholog T2R01 have the same agonist property mentioned above [28].
The functional divergence in agonist sensitivity of each T2R receptor contributes to species-specific bitter taste space. For example, T2R16 orthologs are known as receptors for β-glucosides and their analogs, but agonist sensitivity and selectivity frequently change. Reduced sensitivity to β-glucosides is independently observed in many primate lineages, with complete loss in marmosets, tamarins, and galagos [74,75,80]. In some lemurs, specific β-glucosides, that act as agonists in other primates, act as inverse agonists [77]. Behavioral tests corroborate some of these functional reductions, such as in macaques and lemurs [74,77]. Moreover, bamboo lemurs (Hapalemur and Prolemur), which rely on cyanogenic bamboo, have receptors that are less sensitive to β-glucosides [80]. This reduced sensitivity is inferred to be an advantageous trait for ingesting bamboo containing cyanogenic glucosides [80]. Conversely, enhanced sensitivity to β-glucosides is observed in some primates, such as the white-faced saki (Pithecia pithecia) and certain human haplotypes, which independently increased sensitivity to β-glucosides through the convergent mutation at the same amino acid position [75]. The T2R38 ortholog known as a receptor for phenylthiocarbamide (PTC), also exhibits functional differences between species. T2R38 orthologs respond to PTC in humans and domestic cats, but not in mice and domestic dogs [6,64,65,81]. Even within primates, humans, chimpanzees, and macaques have PTC-sensitive T2R38 [81,82,83], whereas folivorous colobine monkeys lost or reduced the PTC sensitivity of T2R38 through lineage-specific mutations [84,85]. This receptor phenotype corresponds to behavioral avoidance of PTC, hypothesized as an adaptation related to leaf-eating behavior [84,85].
Gene duplication is another major mechanism for functional divergence in T2Rs. Studies of highly duplicated T2Rs in hummingbirds (T2R1) and Myotis bats (T2R16) have demonstrated functional divergence among duplicated T2Rs [30,68]. Some receptor duplicates retain agonists shared with the no-duplicated ortholog in close relatives and acquire novel agonists, while others lose these shared agonists. This diversification may be related to the hummingbirds’ unique nectar diet and the ability of Myotis species to adapt to diverse environments across all continents except Antarctica [30,68].
Besides interspecific comparisons of T2R orthologs, ancestral sequence reconstruction is a useful approach to identify the key residues and evolutionary trajectory of receptor function. Studies on platyrrhines and strepsirrhines identified the evolutionary time points of functional changes by comparisons between extant species receptors and reconstructed ancestral receptors [80,86]. Reconstruction of intact sequences of pseudogenized T2Rs in modern humans revealed that functional redundancy could cause pseudogenization of receptors [87]. Because T2Rs form a multigene family, estimating accurate ancestral sequences prior to massive gene duplications can be challenging, but ancestral reconstruction is a powerful tool for understanding the evolutionary history of individual receptors at the functional level.
5. Intraspecific Variation in T2Rs and Agonist Sensitivity
Intraspecific variations of Tas2r repertoires and agonist sensitivity provide individual differences in bitter perception. These variations are numerous in human populations and can result in phenotypic differences in bitter perception [81,88,89,90]. It has long been debated how this diversity in human TAS2Rs was formed, with several probable causes, such as natural selection or demographic history being suggested [88,91,92,93,94,95,96]. A recent comprehensive genomic survey, using data from the 1000 Genomes Project, demonstrated that selective pressure on most TAS2Rs has relaxed during recent human evolution [97]. Genetic variations in Tas2rs are also observed in non-human animals, influencing taste perception or food selection [83,98,99,100,101,102,103,104]. For example, the polymorphism of three linked amino acid positions (P49A, A262V, and V296I) in T2R38 determines whether it is PTC-sensitive (PAV) or not (AVI) in humans [81,105]. Different polymorphic sites lead to similar phenotypic variations in non-human primates. Haplotypes with a loss of start codon are found in chimpanzees and Japanese macaques, while haplotypes with a premature stop codon or polymorphic sites leading to a loss of PTC sensitivity are present in several species of Sulawesi macaques [82,83,99,106]. These genetic variations correspond well to differences in aversive behavior toward PTC. T2Rs are also functionally differentiated among wild populations. In neighboring populations of the blind mole rat (Spalax galili) inhabiting contrasting soil environments (basalt and chalk), several T2R haplotypes enriched in the basalt population have a higher sensitivity to bitterants than those enriched in the chalk population [103]. This suggests that functional differentiation of T2Rs could aid in optimal food selection from the different food resources [103]. In giant pandas, the functional differentiation of T2R20, a quercitrin receptor, is associated with different quercitrin contents in dietary bamboo among populations [104]. In chimpanzees, most Tas2r haplotypes are specific to each of four subspecies, potentially linked to subspecies-specific dietary repertoires [98,107]. Further studies are needed to clarify the relationship between the functional differentiation of bitter taste receptors and diet in wild animal populations, as research on this topic is less extensive compared to human populations.
6. Extra-Oral Expressions of T2Rs in Vertebrates
6.1. Diverse Functions of Extra-Oral T2Rs
Although T2R expression outside oral tissues was already known in one of the earliest studies [3], subsequent studies have revealed that T2R expression is widely distributed in various extra-oral tissues such as the brain, muscles, cardiovascular system, skin, adipose tissues, immune cells, respiratory tract, gastrointestinal tract, and reproductive organs [108,109,110,111]. Human and rodent studies have revealed that the extra-oral receptors act as local chemoreceptors and are involved in important physiological functions, including not only protection from pathogens and harmful substances similar to their role in the oral cavity but also in metabolic regulatory pathways and the reproductive system.
For instance, T2Rs in the cardiac muscles and smooth muscles of various organs, such as the airway, gastrointestinal tract, vessel, uterus, and bladder, are involved in muscular relaxation (and contraction in some organs) [112,113,114,115,116,117,118,119,120,121,122,123]. In the respiratory tract, nasal tuft cells (known as solitary chemosensory cells) [124,125,126,127,128,129], tracheal tuft cells (known as brush cells) [130,131,132,133], and airway ciliated cells [126,134,135,136,137,138] have T2Rs responding to bitterants and/or bacterial quorum-sensing molecules (QSMs). These receptors induce bacterial clearance systems such as neurogenic inflammation, the production of antimicrobial molecules (nitric oxide and β-defensins), and the upregulation of mucociliary clearance [130,131,132,133,139,140,141]. In the gastrointestinal tract, T2Rs are found in specific epithelial cells, including enteroendocrine cells (EECs), tuft cells, goblet cells, and Paneth cells [142,143,144,145,146,147,148,149,150]. EEC T2R signals can modulate the secretion of distinct gut hormones (ghrelin, cholecystokinin, glucagon-like peptide-1, etc.) depending on the gut segments or cell types [150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165], which regulate food digestion, nutrient absorption, and metabolic homeostasis. Analogous to respiratory tracts, mouse T2Rs in intestinal tuft cells initiate a type 2 innate immune response to helminth infection [166]. Human T2Rs in goblet and Paneth cells can regulate the mRNA expression of antimicrobial peptides, mucins, and chemokines, potentially maintaining intestinal homeostasis [167]. Interestingly, T2Rs are also found in a variety of immune cells (e.g., lymphocytes, myeloid cells, neutrophils, leucocytes, and macrophages) as well as epithelial immune-related cells in the airways and intestines [168,169,170,171,172,173]. T2Rs detect QSMs in human macrophages, leading to enhanced bacterial phagocytosis [172,174]. Furthermore, T2Rs are also expressed in both male and female reproductive organs, such as the testis, sperm, and ovary, in humans and mice [116,175,176,177,178,179,180,181,182,183,184]. Ablation of testicular T2R105-expressing cells leads to loss of spermatids and infertility in mice [176]. Sperm T2Rs may be involved in chemotaxis by detection of progesterone or chemokines derived from cumulus–oocyte complex because progesterone is a T2R agonist [6,177,184]. Ovarian T2Rs may be involved in progesterone production [185].
T2R-expressing cell types, T2R subtypes, and functions in extra-oral tissues of humans and rodents are summarized in the tables of several recent review articles [109,110,111,186]. Please see the recent specific reviews for more details on the extra-oral functions of T2Rs and their inter- and intra-cellular mechanisms (e.g., overall review [109,174], smooth muscle and cardiovascular system [186,187], respiratory tract [124,139], gastrointestinal tract [111,151,188], and reproductive system [184,189]).
6.2. Interspecific Differences of Extra-Oral T2Rs
The recent research advances in extra-oral T2R functions are illuminating the broad roles in mammalian physiology. Many studies have demonstrated that stimulation of T2Rs by certain bitterants, such as denatonium benzoate, elicits similar physiological responses in a variety of organs between humans and rodents, whereas the chemical response properties of T2R-expressing cells may have species differences. This is because the receptor repertoire, tuning breadth, and agonist sensitivity are largely different between species. Indeed, comparing Tas2r expression in the gastrointestinal tract and muscle between humans and rodents, the expression patterns are often different even between one-to-one orthologs [151,187]. In the upper airways, QSMs, particularly acyl-homoserine lactones, can stimulate mouse tuft cells but do not appear to stimulate human cells [124]. These differences may be associated with species-specific habitats because the respiratory and gastrointestinal tracts are interfaces to external environments; hence, it is worthwhile to study them in the framework of evolution and environmental adaptation. In particular, it is important to study non-human primates, which have a similar Tas2r repertoire to humans, to clarify the specificities and evolution of humans and for application to human clinical practice. For instance, although not an exact match, the overall expression pattern of intestinal Tas2rs is very similar between humans and macaque monkeys [147,148], and further studies both in vivo and in vitro (primary or organoid culture) are expected in the future. It is also interesting to note that easily evolved T2Rs are used in internal organs that are less directly exposed to external stimuli, such as the brain, ovary, and muscle. Given the possibility that vertebrate T2Rs have potentially endogenous agonists, such as bioactive steroids [6,19,28,62,63], these internal functions may be important to understand the evolutionary ancestry of T2Rs.
Although the various physiological roles of extra-oral T2Rs have been identified in model rodents and humans, the functional conservation and diversity of extra-oral T2Rs across vertebrates remain poorly understood. This is because novel functions have recently been elucidated in rodents and humans, while research on non-mammalian vertebrates remains limited. Despite the limited number of studies, the expression of Tas2rs is observed in a variety of extra-oral tissues of cartilaginous fish, ray-finned fish, and birds [66,190,191,192,193,194,195]. This suggests that the extra-oral function of T2Rs developed as early as their oral function. However, it is so far difficult to compare gene expression patterns and putative functions among species because of the limited number of analyzed organs and few cellular level data. Even in zebrafish, the Tas2r expression across the entire body has not been fully analyzed; thus, it is an important direction to accumulate comparative transcriptomic information focusing on taste receptors and their signaling molecules, including other model animals (e.g., medaka fish and Xenopus frogs). Furthermore, CRISPR/Cas-based gene editing would enable in vivo analyses of the physiological functions of extra-oral taste receptors even in non-model animals, which will accelerate our understanding of evolution in extra-oral T2R physiology.
7. Structural Features of T2Rs in the GPCR Superfamily
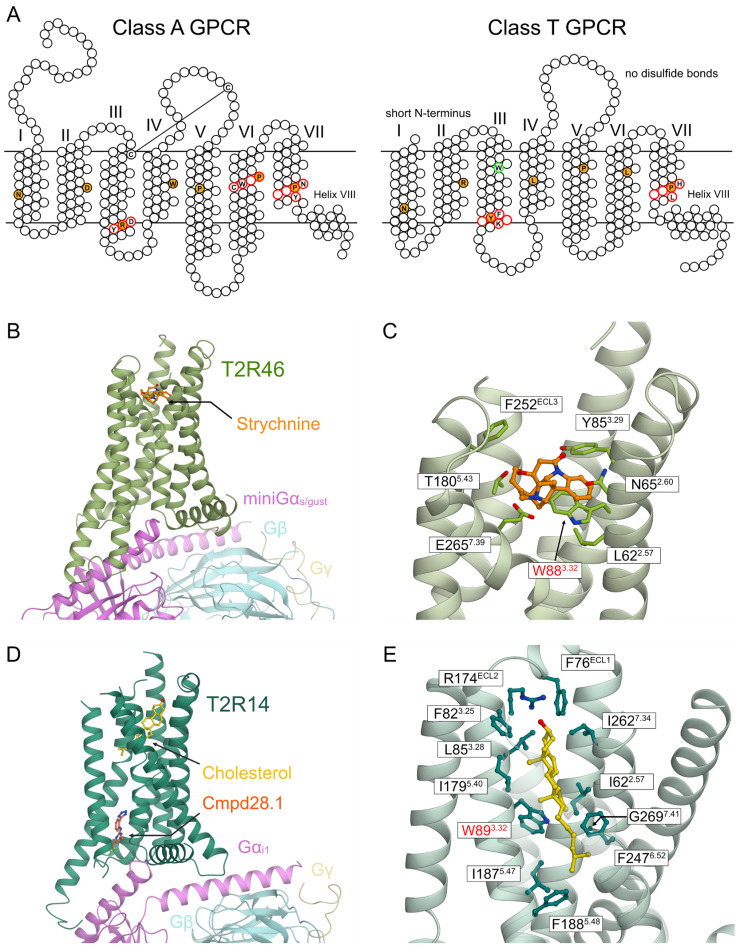
In recent years, significant progress has also been achieved in the structural biology of bitter taste receptors. T2Rs have the typical GPCR motif; seven transmembrane domains with an extracellular N-terminus and an intracellular C-terminus. GPCRs are generally categorized into class A, B, C, and F depending on the shape of the N-terminus domain [1,2,3,196]. While T2Rs were previously classified as class A GPCRs due to their lack of an extracellular N-terminus domain, their unique amino acid sequences distinguish them from this class. Thus, the T2R family was classified into a distinct GPCR class, namely class T, in 2019 [197]. T2Rs differ from class A GPCRs in two major structural features (Figure 5A). First, the N-termini of most T2Rs are very short or almost nonexistent. This characteristic often makes it difficult to properly express T2Rs in a cell culture system; hence, N-terminus signal sequences of other membrane proteins such as rat somatostatin receptor 3 are often added to the native T2Rs to improve cell surface expression in vitro [198,199]. Second, the T2R family lacks a disulfide bond between transmembrane (TM) III and extracellular loop 2. The disulfide bond contributes to receptor stability and limits the width or variety of the ligand binding pocket; hence, this feature may allow the binding pocket of T2Rs to flexibly accept a variety of substances.
Figure 5.
Three-dimensional structures of T2Rs. (A) The snake plots of class A and T GPCRs. The residues filled by orange indicate X.50 residues (generic residue numbering), most conserved residues of each transmembrane domain for each GPCR class. Residues colored in red and green indicate conserved sequence motifs and a key residue for ligand binding, respectively. (B) The overall structure of T2R46 complexed with strychnine retrieved from the PDB (7XP6). (C) The binding mode of strychnine to T2R46 retrieved from the PDB (7XP6). (D) The overall structure of T2R14 complexed with cholesterol and cmpd28.1 (an analog of fulfenamic acid) retrieved from the PDB (8VY7). (E) The binding mode of cholesterol to T2R14 retrieved from the PDB (8VY7). The illustrations were drawn using CueMol2 (http://www.cuemol.org/, downloaded on 24 May 2024).
For a comparison of mutational effects, ligand interactions, and structural motifs, the transmembrane residues of GPCRs are numbered based on a specific numbering system. In Figure 5A, residues filled in orange represent X.50 residue based on the generic residue numbering in which the most conserved residue in each TM helix is numbered as 50 [200]. For instance, N1.50 in the class A GPCR indicates the most conserved residue in TM I. Furthermore, each class of GPCRs has well-conserved sequence motifs (Figure 5A). For example, class A GPCRs have motifs such as D3.49R3.50Y3.51, C6.47W6.48xP6.50, and N7.49P7.50xxY7.53 [201]. In contrast, T2Rs have activation-related motifs, F3.49Y3.50xxK3.53 and H7.49S/P7.50xxL7.53, which correspond to the class A motifs, D3.49R3.50Y3.51 and N7.49P7.50xxY7.53, respectively [201].
In 2022, Xu et al. reported the first experimental structure of human T2R46 complexed with and without strychnine by cryo-electron microscopy (cryo-EM) technology (Figure 5B,C) [202]. Furthermore, cryo-EM structures of human T2R14 complexed with cholesterol, flufenamic acid, and aristolochic acid were reported in 2024 (Figure 5D,E) [203,204,205]. These studies revealed the distinct 3D structures of T2Rs compared to any other GPCRs. The primary (orthosteric) binding site of T2Rs is located at the extracellular side of the transmembrane domain. The highly conserved tryptophan residue at position 3.32 (found in 84% of human T2Rs, shown as a green circle in Figure 5A), which is in the orthosteric binding site and forms CH−π or π−π interactions with ligands, plays a critical role in ligand binding for T2R14 and T2R46 (Figure 5C,E) [202,203,204,205]. As W3.32 also largely affects agonist sensitivity in the other T2Rs [206,207], it may generally contribute to the ligand binding of T2Rs by the same mechanism as T2R14 and T2R46. The experimental structure of T2R14 complexed with ligands demonstrates that this receptor has two or three distinct binding sites (Figure 5D) [203,204,205]. The most extracellular site is the orthosteric binding site, whereas the other intracellular sites are the allosteric binding sites. In particular, the molecule bound in the intracellular part interacts with the coupled G protein alpha subunit. These multiple binding sites may contribute to the broad receptive breadth range in T2R14. T2R14 and T2R46 are broadly tuned T2Rs; therefore, the experimental structures of narrowly tuned or group-specific T2Rs, such as T2R38 or T2R16, would be aid in understanding the mechanisms regulating the receptive ranges of ligands.
8. Conclusions
Recent advances in genomic and functional studies of taste receptors have highlighted the remarkable diversity of vertebrate bitter taste receptors. Both gene repertoires and protein functions are associated with vertebrate diets, although not always observed in all vertebrate taxa. In particular, many examples of such associations have been found in mammals and birds. Therefore, cooperation is required between sequence-based and protein function-based analyses to deeply understand the evolution of taste receptor functions associated with dietary variation. To identify the taste evolution involved in phenotypic and physiological changes in vertebrate evolution, it is also essential to test various species with appropriate outgroups (and sometimes the reconstructed ancestral sequences) and identify the key amino acid changes on the phylogenetic tree with high temporal resolution. Moreover, deeply understanding the genetic basis of dietary shift requires exploring the evolutionary coordination of taste receptors with other diet-related proteins, such as digestive/detoxification enzymes.
While comparative studies on bitter taste receptor evolution have primarily focused on the relation to feeding behavior and taste perception, T2Rs also play crucial roles in the chemosensory cells of various extra-oral organs, including the brain, airways, and gastrointestinal and reproductive organs. While the function of these ectopic taste receptors has been intensively studied in mammalian models, the evolutionary origins and conservation of each ectopic function remain poorly understood. These functions may also contribute to the T2R evolution in both aspects of gene number and receptive ranges of substances, hence the comparative evolutionary study on such ectopic functions using a variety of species can shed light on the deeper mechanisms of bitter taste receptor evolution.
Acknowledgments
We thank the members of Ishimaru lab at Meiji University for the valuable discussion.
Abbreviations
| T2R | Taste receptor type 2 (bitter taste receptor) protein |
| Tas2r/TAS2R | Taste receptor type 2 (bitter taste receptor) gene |
| T1R | Taste receptor type 1 (sweet/umami taste receptor) protein |
| Tas1r/TAS1R | Taste receptor type 1 (sweet/umami taste receptor) gene |
| OR | Olfactory receptor (protein in upright or gene in italic) |
| TAAR | Trace amine-associated receptor (protein in upright or gene in italic) |
| V1R | Vomeronasal receptor type 1 (protein in upright or gene in italic) |
| V2R | Vomeronasal receptor type 2 (protein in upright or gene in italic) |
| GPCR | G protein-coupled receptor |
| TM | Transmembrane |
| QSM | Quorum-sensing molecule |
| EEC | Enteroendocrine cell |
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms252312654/s1.
Author Contributions
Conceptualization, A.I.; writing—original draft, A.I.; writing—review and editing, A.I., T.N., and Y.T.; funding acquisition, A.I. and Y.T. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by JSPS KAKENHI (Nos. 22K15176 and 23KJ1994 for A.I.; 23H02168 for Y.T.) and the Lotte Shigemitsu Prize (Y.T.).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Adler E., Hoon M.A., Mueller K.L., Chandrashekar J., Ryba N.J.P., Zuker C.S. A Novel Family of Mammalian Taste Receptors. Cell. 2000;100:693–702. doi: 10.1016/S0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 2.Chandrashekar J., Mueller K.L., Hoon M.A., Adler E., Feng L., Guo W., Zuker C.S., Ryba N.J.P. T2Rs Function as Bitter Taste Receptors. Cell. 2000;100:703–711. doi: 10.1016/S0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 3.Matsunami H., Montmayeur J.-P., Buck L.B. A Family of Candidate Taste Receptors in Human and Mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 4.Nissim I., Dagan-Wiener A., Niv M.Y. The Taste of Toxicity: A Quantitative Analysis of Bitter and Toxic Molecules. IUBMB Life. 2017;69:938–946. doi: 10.1002/iub.1694. [DOI] [PubMed] [Google Scholar]
- 5.Meyerhof W., Batram C., Kuhn C., Brockhoff A., Chudoba E., Bufe B., Appendino G., Behrens M. The Molecular Receptive Ranges of Human TAS2R Bitter Taste Receptors. Chem. Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 6.Lossow K., Hübner S., Roudnitzky N., Slack J.P., Pollastro F., Behrens M., Meyerhof W. Comprehensive Analysis of Mouse Bitter Taste Receptors Reveals Different Molecular Receptive Ranges for Orthologous Receptors in Mice and Humans. J. Biol. Chem. 2016;291:15358–15377. doi: 10.1074/jbc.M116.718544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowatschew D., Korsching S.I. Lamprey Possess Both V1R and V2R Olfactory Receptors, but Only V1Rs Are Expressed in Olfactory Sensory Neurons. Chem. Senses. 2022;47:bjac007. doi: 10.1093/chemse/bjac007. [DOI] [PubMed] [Google Scholar]
- 8.Policarpo M., Baldwin M.W., Casane D., Salzburger W. Diversity and Evolution of the Vertebrate Chemoreceptor Gene Repertoire. Nat. Commun. 2024;15:1421. doi: 10.1038/s41467-024-45500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churcher A.M., Taylor J.S. Amphioxus (Branchiostoma Floridae) Has Orthologs of Vertebrate Odorant Receptors. BMC Evol. Biol. 2009;9:242. doi: 10.1186/1471-2148-9-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niimura Y. On the Origin and Evolution of Vertebrate Olfactory Receptor Genes: Comparative Genome Analysis Among 23 Chordate Species. Genome Biol. Evol. 2009;1:34–44. doi: 10.1093/gbe/evp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churcher A.M., Taylor J.S. The Antiquity of Chordate Odorant Receptors Is Revealed by the Discovery of Orthologs in the Cnidarian Nematostella Vectensis. Genome Biol. Evol. 2011;3:36–43. doi: 10.1093/gbe/evq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niimura Y. Olfactory Receptor Multigene Family in Vertebrates: From the Viewpoint of Evolutionary Genomics. Curr. Genom. 2012;13:103–114. doi: 10.2174/138920212799860706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marquet N., Cardoso J.C.R., Louro B., Fernandes S.A., Silva S.C., Canário A.V.M. Holothurians Have a Reduced GPCR and Odorant Receptor-like Repertoire Compared to Other Echinoderms. Sci. Rep. 2020;10:3348. doi: 10.1038/s41598-020-60167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall M.R., Kocot K.M., Baughman K.W., Fernandez-Valverde S.L., Gauthier M.E.A., Hatleberg W.L., Krishnan A., McDougall C., Motti C.A., Shoguchi E., et al. The Crown-of-Thorns Starfish Genome as a Guide for Biocontrol of This Coral Reef Pest. Nature. 2017;544:231–234. doi: 10.1038/nature22033. [DOI] [PubMed] [Google Scholar]
- 15.Krishnan A., Almén M.S., Fredriksson R., Schiöth H.B. Remarkable Similarities between the Hemichordate (Saccoglossus kowalevskii) and Vertebrate GPCR Repertoire. Gene. 2013;526:122–133. doi: 10.1016/j.gene.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Sharma K., Syed A.S., Ferrando S., Mazan S., Korsching S.I. The Chemosensory Receptor Repertoire of a True Shark Is Dominated by a Single Olfactory Receptor Family. Genome Biol. Evol. 2019;11:398–405. doi: 10.1093/gbe/evz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishihara H., Toda Y., Kuramoto T., Kamohara K., Goto A., Hoshino K., Okada S., Kuraku S., Okabe M., Ishimaru Y. A Vertebrate-Wide Catalogue of T1R Receptors Reveals Diversity in Taste Perception. Nat. Ecol. Evol. 2024;8:111–120. doi: 10.1038/s41559-023-02258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldwin M.W., Ko M.-C. Functional Evolution of Vertebrate Sensory Receptors. Horm. Behav. 2020;124:104771. doi: 10.1016/j.yhbeh.2020.104771. [DOI] [PubMed] [Google Scholar]
- 19.Behrens M., Lang T., Korsching S.I. A Singular Shark Bitter Taste Receptor Provides Insights into the Evolution of Bitter Taste Perception. Proc. Natl. Acad. Sci. USA. 2023;120:e2310347120. doi: 10.1073/pnas.2310347120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoigawa A., Toda Y., Kuraku S., Ishimaru Y. Evolutionary Origins of Bitter Taste Receptors in Jawed Vertebrates. Curr. Biol. 2024;34:R271–R272. doi: 10.1016/j.cub.2024.02.024. [DOI] [PubMed] [Google Scholar]
- 21.Barreiro-Iglesias A., Anadón R., Rodicio M.C. The Gustatory System of Lampreys. Brain Behav. Evol. 2010;75:241–250. doi: 10.1159/000315151. [DOI] [PubMed] [Google Scholar]
- 22.Cvicek V., Iii W.A.G., Abrol R. Structure-Based Sequence Alignment of the Transmembrane Domains of All Human GPCRs: Phylogenetic, Structural and Functional Implications. PLOS Comput. Biol. 2016;12:e1004805. doi: 10.1371/journal.pcbi.1004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji Y., Zhang Z., Hu Y. The Repertoire of G-Protein-Coupled Receptors in Xenopus Tropicalis. BMC Genom. 2009;10:263. doi: 10.1186/1471-2164-10-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gloriam D.E., Fredriksson R., Schiöth H.B. The G Protein-Coupled Receptor Subset of the Rat Genome. BMC Genom. 2007;8:338. doi: 10.1186/1471-2164-8-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pándy-Szekeres G., Caroli J., Mamyrbekov A., Kermani A.A., Keserű G.M., Kooistra A.J., Gloriam D.E. GPCRdb in 2023: State-Specific Structure Models Using AlphaFold2 and New Ligand Resources. Nucleic Acids Res. 2023;51:D395–D402. doi: 10.1093/nar/gkac1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D., Zhang J. Diet Shapes the Evolution of the Vertebrate Bitter Taste Receptor Gene Repertoire. Mol. Biol. Evol. 2014;31:303–309. doi: 10.1093/molbev/mst219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behrens M., Korsching S.I., Meyerhof W. Tuning Properties of Avian and Frog Bitter Taste Receptors Dynamically Fit Gene Repertoire Sizes. Mol. Biol. Evol. 2014;31:3216–3227. doi: 10.1093/molbev/msu254. [DOI] [PubMed] [Google Scholar]
- 28.Behrens M., Di Pizio A., Redel U., Meyerhof W., Korsching S.I. At the Root of T2R Gene Evolution: Recognition Profiles of Coelacanth and Zebrafish Bitter Receptors. Genome Biol. Evol. 2021;13:evaa264. doi: 10.1093/gbe/evaa264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayakawa T., Suzuki-Hashido N., Matsui A., Go Y. Frequent Expansions of the Bitter Taste Receptor Gene Repertoire during Evolution of Mammals in the Euarchontoglires Clade. Mol. Biol. Evol. 2014;31:2018–2031. doi: 10.1093/molbev/msu144. [DOI] [PubMed] [Google Scholar]
- 30.Jiao H., Wang Y., Zhang L., Jiang P., Zhao H. Lineage-Specific Duplication and Adaptive Evolution of Bitter Taste Receptor Genes in Bats. Mol. Ecol. 2018;27:4475–4488. doi: 10.1111/mec.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson R.N., O’Meally D., Chen Z., Etherington G.J., Ho S.Y.W., Nash W.J., Grueber C.E., Cheng Y., Whittington C.M., Dennison S., et al. Adaptation and Conservation Insights from the Koala Genome. Nat. Genet. 2018;50:1102–1111. doi: 10.1038/s41588-018-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., Liu G., Hailer F., Orozco-terWengel P., Tan X., Tian J., Yan Z., Zhang B., Li M. Dietary Specialization Drives Multiple Independent Losses and Gains in the Bitter Taste Gene Repertoire of Laurasiatherian Mammals. Front. Zool. 2016;13:28. doi: 10.1186/s12983-016-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang S., Wu X., Chen J., Zhang H., Zhong H., Wei Q., Yan J., Li H., Liu G., Sha W., et al. The Repertoire of Bitter Taste Receptor Genes in Canids. Amino Acids. 2017;49:1159–1167. doi: 10.1007/s00726-017-2422-5. [DOI] [PubMed] [Google Scholar]
- 34.Shang S., Zhang H., Wu X., Chen J., Zhong H., Wei Q., Zhao C., Yan J., Chen Y., Tang X., et al. The Repertoire of Bitter Taste Receptor Genes in Ovalentaria Fish. Environ. Biol. Fish. 2017;100:1489–1496. doi: 10.1007/s10641-017-0659-1. [DOI] [Google Scholar]
- 35.Zhong H., Shang S., Zhang H., Chen J., Wu X., Zhang H. Characterization and Phylogeny of Bitter Taste Receptor Genes (Tas2r) in Squamata. Genetica. 2019;147:131–139. doi: 10.1007/s10709-019-00056-4. [DOI] [PubMed] [Google Scholar]
- 36.Shiriagin V., Korsching S.I. Massive Expansion of Bitter Taste Receptors in Blind Cavefish, Astyanax Mexicanus. Chem. Senses. 2019;44:23–32. doi: 10.1093/chemse/bjy062. [DOI] [PubMed] [Google Scholar]
- 37.Syed A.S., Korsching S.I. Positive Darwinian Selection in the Singularly Large Taste Receptor Gene Family of an ‘Ancient’ Fish, Latimeria Chalumnae. BMC Genom. 2014;15:650. doi: 10.1186/1471-2164-15-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picone B., Hesse U., Panji S., Van Heusden P., Jonas M., Christoffels A. Taste and Odorant Receptors of the Coelacanth—A Gene Repertoire in Transition. J. Exp. Zool. B Mol. Dev. Evol. 2014;322:403–414. doi: 10.1002/jez.b.22531. [DOI] [PubMed] [Google Scholar]
- 39.Wang K., Zhao H. Birds Generally Carry a Small Repertoire of Bitter Taste Receptor Genes. Genome Biol. Evol. 2015;7:2705–2715. doi: 10.1093/gbe/evv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong H., Shang S., Wu X., Chen J., Zhu W., Yan J., Li H., Zhang H. Genomic Evidence of Bitter Taste in Snakes and Phylogenetic Analysis of Bitter Taste Receptor Genes in Reptiles. PeerJ. 2017;5:e3708. doi: 10.7717/peerj.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong H., Huang J., Shang S., Yuan B. Evolutionary Insights into Umami, Sweet, and Bitter Taste Receptors in Amphibians. Ecol. Evol. 2021;11:18011–18025. doi: 10.1002/ece3.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao X., Jiao H., Zou D., Li Q., Yuan X., Liao W., Jiang P., Zhao H. Evolution of Bitter Receptor Genes and Ontogenetic Dietary Shift in a Frog. Proc. Natl. Acad. Sci. USA. 2023;120:e2218183120. doi: 10.1073/pnas.2218183120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y., Shearwin-Whyatt L., Li J., Song Z., Hayakawa T., Stevens D., Fenelon J.C., Peel E., Cheng Y., Pajpach F., et al. Platypus and Echidna Genomes Reveal Mammalian Biology and Evolution. Nature. 2021;592:756–762. doi: 10.1038/s41586-020-03039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kishida T., Thewissen J., Hayakawa T., Imai H., Agata K. Aquatic Adaptation and the Evolution of Smell and Taste in Whales. Zool. Lett. 2015;1:9. doi: 10.1186/s40851-014-0002-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H., Li J., Zhang J. Molecular Evidence for the Loss of Three Basic Tastes in Penguins. Curr. Biol. 2015;25:R141–R142. doi: 10.1016/j.cub.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu K., Zhou X., Xu S., Sun D., Ren W., Zhou K., Yang G. The Loss of Taste Genes in Cetaceans. BMC Evol. Biol. 2014;14:218. doi: 10.1186/s12862-014-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou M., Akhtar M.S., Hayashi M., Ashino R., Matsumoto-Oda A., Hayakawa T., Ishida T., Melin A.D., Imai H., Kawamura S. Reduction of Bitter Taste Receptor Gene Family in Folivorous Colobine Primates Relative to Omnivorous Cercopithecine Primates. Primates. 2024;65:311–331. doi: 10.1007/s10329-024-01124-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Go Y. Lineage-Specific Expansions and Contractions of the Bitter Taste Receptor Gene Repertoire in Vertebrates. Mol. Biol. Evol. 2006;23:964–972. doi: 10.1093/molbev/msj106. [DOI] [PubMed] [Google Scholar]
- 49.Feng P., Zheng J., Rossiter S.J., Wang D., Zhao H. Massive Losses of Taste Receptor Genes in Toothed and Baleen Whales. Genome Biol. Evol. 2014;6:1254–1265. doi: 10.1093/gbe/evu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niimura Y., Biswa B.B., Kishida T., Toyoda A., Fujiwara K., Ito M., Touhara K., Inoue-Murayama M., Jenkins S.H., Adenyo C., et al. Synchronized Expansion and Contraction of Olfactory, Vomeronasal, and Taste Receptor Gene Families in Hystricomorph Rodents. Mol. Biol. Evol. 2024;41:msae071. doi: 10.1093/molbev/msae071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berning D., Gross J.B. The Constructive Evolution of Taste in Astyanax Cavefish: A Review. Front. Ecol. Evol. 2023;11:1177532. doi: 10.3389/fevo.2023.1177532. [DOI] [Google Scholar]
- 52.Berning D., Heerema H., Gross J.B. The Spatiotemporal and Genetic Architecture of Extraoral Taste Buds in Astyanax Cavefish. Commun. Biol. 2024;7:951. doi: 10.1038/s42003-024-06635-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barlow L.A. The Biology of Amphibian Taste. In: Heatwole H., Dawley E.M., editors. Amphibian Biology, Volume 3: Sensory Perception. Surrey Beatty & Sons Pty Ltd.; Chipping Norton, NSW, Australia: 1998. pp. 743–782. [Google Scholar]
- 54.Peel E., Silver L., Brandies P., Hayakawa T., Belov K., Hogg C.J., Silver L., Brandies P., Hayakawa T., Belov K., et al. Genome Assembly of the Numbat (Myrmecobius fasciatus), the Only Termitivorous Marsupial. Gigabyte. 2022;2022:gigabyte47. doi: 10.46471/gigabyte.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong W., Zhao H. Vampire Bats Exhibit Evolutionary Reduction of Bitter Taste Receptor Genes Common to Other Bats. Proc. R. Soc. B. 2014;281:20141079. doi: 10.1098/rspb.2014.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Q., Jiao H., Wang Y., Norbu N., Zhao H. Molecular Evolution and Deorphanization of Bitter Taste Receptors in a Vampire Bat. Integr. Zool. 2021;16:659–669. doi: 10.1111/1749-4877.12509. [DOI] [PubMed] [Google Scholar]
- 57.Itoigawa A., Hayakawa T., Zhou Y., Manning A.D., Zhang G., Grutzner F., Imai H. Functional Diversity and Evolution of Bitter Taste Receptors in Egg-Laying Mammals. Mol. Biol. Evol. 2022;39:msac107. doi: 10.1093/molbev/msac107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes L.C., Ortí G., Huang Y., Sun Y., Baldwin C.C., Thompson A.W., Arcila D., Betancur-R R., Li C., Becker L., et al. Comprehensive Phylogeny of Ray-Finned Fishes (Actinopterygii) Based on Transcriptomic and Genomic Data. Proc. Natl. Acad. Sci. USA. 2018;115:6249–6254. doi: 10.1073/pnas.1719358115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stiller J., Feng S., Chowdhury A.-A., Rivas-González I., Duchêne D.A., Fang Q., Deng Y., Kozlov A., Stamatakis A., Claramunt S., et al. Complexity of Avian Evolution Revealed by Family-Level Genomes. Nature. 2024;629:851–860. doi: 10.1038/s41586-024-07323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hime P.M., Lemmon A.R., Lemmon E.C.M., Prendini E., Brown J.M., Thomson R.C., Kratovil J.D., Noonan B.P., Pyron R.A., Peloso P.L.V., et al. Phylogenomics Reveals Ancient Gene Tree Discordance in the Amphibian Tree of Life. Syst. Biol. 2021;70:49–66. doi: 10.1093/sysbio/syaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S., Suleski M., Craig J.M., Kasprowicz A.E., Sanderford M., Li M., Stecher G., Hedges S.B. TimeTree 5: An Expanded Resource for Species Divergence Times. Mol. Biol. Evol. 2022;39:msac174. doi: 10.1093/molbev/msac174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ziegler F., Steuer A., Di Pizio A., Behrens M. Physiological Activation of Human and Mouse Bitter Taste Receptors by Bile Acids. Commun. Biol. 2023;6:612. doi: 10.1038/s42003-023-04971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaefer S., Ziegler F., Lang T., Steuer A., Di Pizio A., Behrens M. Membrane-Bound Chemoreception of Bitter Bile Acids and Peptides Is Mediated by the Same Subset of Bitter Taste Receptors. Cell. Mol. Life Sci. 2024;81:217. doi: 10.1007/s00018-024-05202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lei W., Ravoninjohary A., Li X., Margolskee R.F., Reed D.R., Beauchamp G.K., Jiang P. Functional Analyses of Bitter Taste Receptors in Domestic Cats (Felis catus) PLoS ONE. 2015;10:e0139670. doi: 10.1371/journal.pone.0139670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibbs M., Winnig M., Riva I., Dunlop N., Waller D., Klebansky B., Logan D.W., Briddon S.J., Holliday N.D., McGrane S.J. Bitter Taste Sensitivity in Domestic Dogs (Canis familiaris) and Its Relevance to Bitter Deterrents of Ingestion. PLoS ONE. 2022;17:e0277607. doi: 10.1371/journal.pone.0277607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shimizu T., Kubozono T., Asaoka R., Toda Y., Ishimaru Y. Expression Profiles and Functional Characterization of Common Carp (Cyprinus carpio) T2Rs. Biochem. Biophys. Rep. 2021;28:101123. doi: 10.1016/j.bbrep.2021.101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar P., Redel U., Lang T., Korsching S.I., Behrens M. Bitter Taste Receptors of the Zebra Finch (Taeniopygia guttata) Front. Physiol. 2023;14:1233711. doi: 10.3389/fphys.2023.1233711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y., Jiao H., Jiang P., Zhao H. Functional Divergence of Bitter Taste Receptors in a Nectar-Feeding Bird. Biol. Lett. 2019;15:20190461. doi: 10.1098/rsbl.2019.0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Y., Li Y., Hu H., Jiao H., Zhao H. Genomic and Functional Insights into Dietary Diversification in New World Leaf-Nosed Bats (Phyllostomidae) J. Syst. Evol. 2024;62:928–941. doi: 10.1111/jse.13059. [DOI] [Google Scholar]
- 70.Ziegler F., Behrens M. Bitter Taste Receptors of the Common Vampire Bat Are Functional and Show Conserved Responses to Metal Ions in Vitro. Proc. R. Soc. B. 2021;288:20210418. doi: 10.1098/rspb.2021.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiener A., Shudler M., Levit A., Niv M.Y. BitterDB: A Database of Bitter Compounds. Nucleic Acids Res. 2012;40:D413–D419. doi: 10.1093/nar/gkr755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dagan-Wiener A., Di Pizio A., Nissim I., Bahia M.S., Dubovski N., Margulis E., Niv M.Y. BitterDB: Taste Ligands and Receptors Database in 2019. Nucleic Acids Res. 2019;47:D1179–D1185. doi: 10.1093/nar/gky974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Behrens M., Meyerhof W. Vertebrate Bitter Taste Receptors: Keys for Survival in Changing Environments. J. Agric. Food Chem. 2018;66:2204–2213. doi: 10.1021/acs.jafc.6b04835. [DOI] [PubMed] [Google Scholar]
- 74.Imai H., Suzuki N., Ishimaru Y., Sakurai T., Yin L., Pan W., Abe K., Misaka T., Hirai H. Functional Diversity of Bitter Taste Receptor TAS2R16 in Primates. Biol. Lett. 2012;8:652–656. doi: 10.1098/rsbl.2011.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang H., Yang S., Fan F., Li Y., Dai S., Zhou X., Steiner C.C., Coppedge B., Roos C., Cai X., et al. A New World Monkey Resembles Human in Bitter Taste Receptor Evolution and Function via a Single Parallel Amino Acid Substitution. Mol. Biol. Evol. 2021;38:5472–5479. doi: 10.1093/molbev/msab263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bufe B., Hofmann T., Krautwurst D., Raguse J.-D., Meyerhof W. The Human TAS2R16 Receptor Mediates Bitter Taste in Response to β-Glucopyranosides. Nat. Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 77.Itoigawa A., Hayakawa T., Suzuki-Hashido N., Imai H. A Natural Point Mutation in the Bitter Taste Receptor TAS2R16 Causes Inverse Agonism of Arbutin in Lemur Gustation. Proc. R. Soc. B. 2019;286:20190884. doi: 10.1098/rspb.2019.0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y., Zajac A.L., Lei W., Christensen C.M., Margolskee R.F., Bouysset C., Golebiowski J., Zhao H., Fiorucci S., Jiang P. Metal Ions Activate the Human Taste Receptor TAS2R7. Chem. Senses. 2019;44:339–347. doi: 10.1093/chemse/bjz024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Behrens M., Redel U., Blank K., Meyerhof W. The Human Bitter Taste Receptor TAS2R7 Facilitates the Detection of Bitter Salts. Biochem. Biophys. Res. Commun. 2019;512:877–881. doi: 10.1016/j.bbrc.2019.03.139. [DOI] [PubMed] [Google Scholar]
- 80.Itoigawa A., Fierro F., Chaney M.E., Lauterbur M.E., Hayakawa T., Tosi A.J., Niv M.Y., Imai H. Lowered Sensitivity of Bitter Taste Receptors to β-Glucosides in Bamboo Lemurs: An Instance of Parallel and Adaptive Functional Decline in TAS2R16? Proc. R. Soc. B. 2021;288:20210346. doi: 10.1098/rspb.2021.0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bufe B., Breslin P.A.S., Kuhn C., Reed D.R., Tharp C.D., Slack J.P., Kim U.-K., Drayna D., Meyerhof W. The Molecular Basis of Individual Differences in Phenylthiocarbamide and Propylthiouracil Bitterness Perception. Curr. Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wooding S., Bufe B., Grassi C., Howard M.T., Stone A.C., Vazquez M., Dunn D.M., Meyerhof W., Weiss R.B., Bamshad M.J. Independent Evolution of Bitter-Taste Sensitivity in Humans and Chimpanzees. Nature. 2006;440:930–934. doi: 10.1038/nature04655. [DOI] [PubMed] [Google Scholar]
- 83.Suzuki-Hashido N., Hayakawa T., Matsui A., Go Y., Ishimaru Y., Misaka T., Abe K., Hirai H., Satta Y., Imai H. Rapid Expansion of Phenylthiocarbamide Non-Tasters among Japanese Macaques. PLoS ONE. 2015;10:e0132016. doi: 10.1371/journal.pone.0132016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Purba L.H.P.S., Widayati K.A., Tsutsui K., Suzuki-Hashido N., Hayakawa T., Nila S., Suryobroto B., Imai H. Functional Characterization of the TAS2R38 Bitter Taste Receptor for Phenylthiocarbamide in Colobine Monkeys. Biol. Lett. 2017;13:20160834. doi: 10.1098/rsbl.2016.0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Purba L.H.P.S., Widayati K.A., Suzuki-Hashido N., Itoigawa A., Hayakawa T., Nila S., Juliandi B., Suryobroto B., Imai H. Evolution of the Bitter Taste Receptor TAS2R38 in Colobines. Primates. 2020;61:485–494. doi: 10.1007/s10329-020-00799-1. [DOI] [PubMed] [Google Scholar]
- 86.Tsutsui K., Otoh M., Sakurai K., Suzuki-Hashido N., Hayakawa T., Misaka T., Ishimaru Y., Aureli F., Melin A.D., Kawamura S., et al. Variation in Ligand Responses of the Bitter Taste Receptors TAS2R1 and TAS2R4 among New World Monkeys. BMC Evol. Biol. 2016;16:208. doi: 10.1186/s12862-016-0783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Risso D., Behrens M., Sainz E., Meyerhof W., Drayna D. Probing the Evolutionary History of Human Bitter Taste Receptor Pseudogenes by Restoring Their Function. Mol. Biol. Evol. 2017;34:1587–1595. doi: 10.1093/molbev/msx097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Soranzo N., Bufe B., Sabeti P.C., Wilson J.F., Weale M.E., Marguerie R., Meyerhof W., Goldstein D.B. Positive Selection on a High-Sensitivity Allele of the Human Bitter-Taste Receptor TAS2R16. Curr. Biol. 2005;15:1257–1265. doi: 10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 89.Roudnitzky N., Bufe B., Thalmann S., Kuhn C., Gunn H.C., Xing C., Crider B.P., Behrens M., Meyerhof W., Wooding S.P. Genomic, Genetic and Functional Dissection of Bitter Taste Responses to Artificial Sweeteners. Hum. Mol. Genet. 2011;20:3437–3449. doi: 10.1093/hmg/ddr252. [DOI] [PubMed] [Google Scholar]
- 90.Pronin A.N., Xu H., Tang H., Zhang L., Li Q., Li X. Specific Alleles of Bitter Receptor Genes Influence Human Sensitivity to the Bitterness of Aloin and Saccharin. Curr. Biol. 2007;17:1403–1408. doi: 10.1016/j.cub.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 91.Wooding S., Kim U., Bamshad M.J., Larsen J., Jorde L.B., Drayna D. Natural Selection and Molecular Evolution in PTC, a Bitter-Taste Receptor Gene. Am. J. Hum. Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X., Thomas S.D., Zhang J. Relaxation of Selective Constraint and Loss of Function in the Evolution of Human Bitter Taste Receptor Genes. Hum. Mol. Genet. 2004;13:2671–2678. doi: 10.1093/hmg/ddh289. [DOI] [PubMed] [Google Scholar]
- 93.Campbell M.C., Ranciaro A., Zinshteyn D., Rawlings-Goss R., Hirbo J., Thompson S., Woldemeskel D., Froment A., Rucker J.B., Omar S.A., et al. Origin and Differential Selection of Allelic Variation at TAS2R16 Associated with Salicin Bitter Taste Sensitivity in Africa. Mol. Biol. Evol. 2014;31:288–302. doi: 10.1093/molbev/mst211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kulichová I., Mouterde M., Mokhtar M.G., Diallo I., Tříska P., Diallo Y.M., Hofmanová Z., Poloni E.S., Černý V. Demographic History Was a Formative Mechanism of the Genetic Structure for the Taste Receptor TAS2R16 in Human Populations Inhabiting Africa’s Sahel/Savannah Belt. Am. J. Biol. Anthr. 2022;177:540–555. doi: 10.1002/ajpa.24448. [DOI] [PubMed] [Google Scholar]
- 95.Risso D.S., Mezzavilla M., Pagani L., Robino A., Morini G., Tofanelli S., Carrai M., Campa D., Barale R., Caradonna F., et al. Global Diversity in the TAS2R38 Bitter Taste Receptor: Revisiting a Classic Evolutionary PROPosal. Sci. Rep. 2016;6:25506. doi: 10.1038/srep25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Valente C., Alvarez L., Marques P.I., Gusmão L., Amorim A., Seixas S., João Prata M. Genes from the TAS1R and TAS2R Families of Taste Receptors: Looking for Signatures of Their Adaptive Role in Human Evolution. Genome Biol. Evol. 2018;10:1139–1152. doi: 10.1093/gbe/evy071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wooding S.P., Ramirez V.A. Global Population Genetics and Diversity in the TAS2R Bitter Taste Receptor Family. Front. Genet. 2022;13:952299. doi: 10.3389/fgene.2022.952299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hayakawa T., Sugawara T., Go Y., Udono T., Hirai H., Imai H. Eco-Geographical Diversification of Bitter Taste Receptor Genes (TAS2Rs) among Subspecies of Chimpanzees (Pan Troglodytes) PLoS ONE. 2012;7:e43277. doi: 10.1371/journal.pone.0043277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Widayati K.A., Yan X., Suzuki-Hashido N., Itoigawa A., Purba L.H.P.S., Fahri F., Terai Y., Suryobroto B., Imai H. Functional Divergence of the Bitter Receptor TAS2R38 in Sulawesi Macaques. Ecol. Evol. 2019;9:10387–10403. doi: 10.1002/ece3.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.SU Y., LI D., GAUR U., WANG Y., WU N., CHEN B., XU Z., YIN H., HU Y., ZHU Q. Genetic Diversity of Bitter Taste Receptor Gene Family in Sichuan Domestic and Tibetan Chicken Populations. J. Genet. 2016;95:675–681. doi: 10.1007/s12041-016-0684-4. [DOI] [PubMed] [Google Scholar]
- 101.Zhao F., Zhang T., Xie J., Zhang S., Nevo E., Su J., Lin G. Genetic Variation in Bitter Taste Receptor Genes Influences the Foraging Behavior of Plateau Zokor (Eospalax baileyi) Ecol. Evol. 2016;6:2359–2367. doi: 10.1002/ece3.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Davenport K.M., Taylor J.B., Henslee D., Southerland C., Yelich J., Ellison M.J., Murdoch B.M. Variation in Type Two Taste Receptor Genes Is Associated with Bitter Tasting Phenylthiocarbamide Consumption in Mature Targhee and Rambouillet Rams. Transl. Anim. Sci. 2021;5:txab142. doi: 10.1093/tas/txab142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiao H., Wang Q., Wang B.-J., Li K., Lövy M., Nevo E., Li Q., Su W., Jiang P., Zhao H. Local Adaptation of Bitter Taste and Ecological Speciation in a Wild Mammal. Mol. Biol. Evol. 2021;38:4562–4572. doi: 10.1093/molbev/msab205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hu X., Wang G., Shan L., Sun S., Hu Y., Wei F. TAS2R20 Variants Confer Dietary Adaptation to High-Quercitrin Bamboo Leaves in Qinling Giant Pandas. Ecol. Evol. 2020;10:5913–5921. doi: 10.1002/ece3.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim U., Jorgenson E., Coon H., Leppert M., Risch N., Drayna D. Positional Cloning of the Human Quantitative Trait Locus Underlying Taste Sensitivity to Phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 106.Suzuki N., Sugawara T., Matsui A., Go Y., Hirai H., Imai H. Identification of Non-Taster Japanese Macaques for a Specific Bitter Taste. Primates. 2010;51:285–289. doi: 10.1007/s10329-010-0209-3. [DOI] [PubMed] [Google Scholar]
- 107.Sugawara T., Go Y., Udono T., Morimura N., Tomonaga M., Hirai H., Imai H. Diversification of Bitter Taste Receptor Gene Family in Western Chimpanzees. Mol. Biol. Evol. 2011;28:921–931. doi: 10.1093/molbev/msq279. [DOI] [PubMed] [Google Scholar]
- 108.Gilca M., Dragos D. Extraoral Taste Receptor Discovery: New Light on Ayurvedic Pharmacology. Evid.-Based Complement. Altern. Med. 2017;2017:5435831. doi: 10.1155/2017/5435831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tuzim K., Korolczuk A. An Update on Extra-Oral Bitter Taste Receptors. J. Transl. Med. 2021;19:440. doi: 10.1186/s12967-021-03067-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Behrens M. International Union of Basic and Clinical Pharmacology. CXVII: Taste 2 Receptors: Structures, Functions, Activators and Blockers. Pharmacol. Rev. 2024;76:1063–1088. doi: 10.1124/pharmrev.123.001140. [DOI] [PubMed] [Google Scholar]
- 111.Wang Q., Liszt K.I., Depoortere I. Extra-Oral Bitter Taste Receptors: New Targets against Obesity? Peptides. 2020;127:170284. doi: 10.1016/j.peptides.2020.170284. [DOI] [PubMed] [Google Scholar]
- 112.Deshpande D.A., Wang W.C.H., McIlmoyle E.L., Robinett K.S., Schillinger R.M., An S.S., Sham J.S.K., Liggett S.B. Bitter Taste Receptors on Airway Smooth Muscle Bronchodilate by Localized Calcium Signaling and Reverse Obstruction. Nat. Med. 2010;16:1299–1304. doi: 10.1038/nm.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhai K., Yang Z., Zhu X., Nyirimigabo E., Mi Y., Wang Y., Liu Q., Man L., Wu S., Jin J., et al. Activation of Bitter Taste Receptors (Tas2rs) Relaxes Detrusor Smooth Muscle and Suppresses Overactive Bladder Symptoms. Oncotarget. 2016;7:21156–21167. doi: 10.18632/oncotarget.8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Upadhyaya J.D., Singh N., Sikarwar A.S., Chakraborty R., Pydi S.P., Bhullar R.P., Dakshinamurti S., Chelikani P. Dextromethorphan Mediated Bitter Taste Receptor Activation in the Pulmonary Circuit Causes Vasoconstriction. PLoS ONE. 2014;9:e110373. doi: 10.1371/journal.pone.0110373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Avau B., Rotondo A., Thijs T., Andrews C.N., Janssen P., Tack J., Depoortere I. Targeting Extra-Oral Bitter Taste Receptors Modulates Gastrointestinal Motility with Effects on Satiation. Sci. Rep. 2015;5:15985. doi: 10.1038/srep15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zheng K., Lu P., Delpapa E., Bellve K., Deng R., Condon J.C., Fogarty K., Lifshitz L.M., Simas T.A.M., Shi F., et al. Bitter Taste Receptors as Targets for Tocolytics in Preterm Labor Therapy. FASEB J. 2017;31:4037–4052. doi: 10.1096/fj.201601323RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Manson M.L., Säfholm J., Al-Ameri M., Bergman P., Orre A.-C., Swärd K., James A., Dahlén S.-E., Adner M. Bitter Taste Receptor Agonists Mediate Relaxation of Human and Rodent Vascular Smooth Muscle. Eur. J. Pharmacol. 2014;740:302–311. doi: 10.1016/j.ejphar.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 118.Chen J.-G., Ping N.-N., Liang D., Li M.-Y., Mi Y.-N., Li S., Cao L., Cai Y., Cao Y.-X. The Expression of Bitter Taste Receptors in Mesenteric, Cerebral and Omental Arteries. Life Sci. 2017;170:16–24. doi: 10.1016/j.lfs.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 119.Foster S.R., Porrello E.R., Purdue B., Chan H.-W., Voigt A., Frenzel S., Hannan R.D., Moritz K.M., Simmons D.G., Molenaar P., et al. Expression, Regulation and Putative Nutrient-Sensing Function of Taste GPCRs in the Heart. PLoS ONE. 2013;8:e64579. doi: 10.1371/journal.pone.0064579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Woo J.-A.A., Castaño M., Kee T.R., Lee J., Koziol-White C.J., An S.S., Kim D., Kang D.E., Liggett S.B. A Par3/LIM Kinase/Cofilin Pathway Mediates Human Airway Smooth Muscle Relaxation by TAS2R14. Am. J. Respir. Cell Mol. Biol. 2023;68:417–429. doi: 10.1165/rcmb.2022-0303OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou Y.-W., Sun J., Wang Y., Chen C.-P., Tao T., Ma M., Chen X., Zhang X.-N., Yang L.-Y., Zhang Z.-L., et al. Tas2R Activation Relaxes Airway Smooth Muscle by Release of Gαt Targeting on AChR Signaling. Proc. Natl. Acad. Sci. USA. 2022;119:e2121513119. doi: 10.1073/pnas.2121513119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Conaway Jr S., Huang W., Hernandez-Lara M.A., Kane M.A., Penn R.B., Deshpande D.A. Molecular Mechanism of Bitter Taste Receptor Agonist-Mediated Relaxation of Airway Smooth Muscle. FASEB J. 2024;38:e23842. doi: 10.1096/fj.202400452R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim D., Woo J.A., Geffken E., An S.S., Liggett S.B. Coupling of Airway Smooth Muscle Bitter Taste Receptors to Intracellular Signaling and Relaxation Is via Gαi1,2,3. Am. J. Respir. Cell Mol. Biol. 2017;56:762–771. doi: 10.1165/rcmb.2016-0373OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carey R.M., Lee R.J. Taste Receptors in Upper Airway Innate Immunity. Nutrients. 2019;11:2017. doi: 10.3390/nu11092017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tizzano M., Gulbransen B.D., Vandenbeuch A., Clapp T.R., Herman J.P., Sibhatu H.M., Churchill M.E.A., Silver W.L., Kinnamon S.C., Finger T.E. Nasal Chemosensory Cells Use Bitter Taste Signaling to Detect Irritants and Bacterial Signals. Proc. Natl. Acad. Sci. USA. 2010;107:3210–3215. doi: 10.1073/pnas.0911934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen J., Larson E.D., Anderson C.B., Agarwal P., Frank D.N., Kinnamon S.C., Ramakrishnan V.R. Expression of Bitter Taste Receptors and Solitary Chemosensory Cell Markers in the Human Sinonasal Cavity. Chem. Senses. 2019;44:483–495. doi: 10.1093/chemse/bjz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ualiyeva S., Hallen N., Kanaoka Y., Ledderose C., Matsumoto I., Junger W.G., Barrett N.A., Bankova L.G. Airway Brush Cells Generate Cysteinyl Leukotrienes through the ATP Sensor P2Y2. Sci. Immunol. 2020;5:eaax7224. doi: 10.1126/sciimmunol.aax7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Saunders C.J., Christensen M., Finger T.E., Tizzano M. Cholinergic Neurotransmission Links Solitary Chemosensory Cells to Nasal Inflammation. Proc. Natl. Acad. Sci. USA. 2014;111:6075–6080. doi: 10.1073/pnas.1402251111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee R.J., Kofonow J.M., Rosen P.L., Siebert A.P., Chen B., Doghramji L., Xiong G., Adappa N.D., Palmer J.N., Kennedy D.W., et al. Bitter and Sweet Taste Receptors Regulate Human Upper Respiratory Innate Immunity. J. Clin. Investig. 2014;124:1393–1405. doi: 10.1172/JCI72094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hollenhorst M.I., Jurastow I., Nandigama R., Appenzeller S., Li L., Vogel J., Wiederhold S., Althaus M., Empting M., Altmüller J., et al. Tracheal Brush Cells Release Acetylcholine in Response to Bitter Tastants for Paracrine and Autocrine Signaling. FASEB J. 2020;34:316–332. doi: 10.1096/fj.201901314RR. [DOI] [PubMed] [Google Scholar]
- 131.Krasteva G., Canning B.J., Papadakis T., Kummer W. Cholinergic Brush Cells in the Trachea Mediate Respiratory Responses to Quorum Sensing Molecules. Life Sci. 2012;91:992–996. doi: 10.1016/j.lfs.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 132.Perniss A., Liu S., Boonen B., Keshavarz M., Ruppert A.-L., Timm T., Pfeil U., Soultanova A., Kusumakshi S., Delventhal L., et al. Chemosensory Cell-Derived Acetylcholine Drives Tracheal Mucociliary Clearance in Response to Virulence-Associated Formyl Peptides. Immunity. 2020;52:683–699.e11. doi: 10.1016/j.immuni.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 133.Hollenhorst M.I., Nandigama R., Evers S.B., Gamayun I., Wadood N.A., Salah A., Pieper M., Wyatt A., Stukalov A., Gebhardt A., et al. Bitter Taste Signaling in Tracheal Epithelial Brush Cells Elicits Innate Immune Responses to Bacterial Infection. J. Clin. Investig. 2022;132:e150951. doi: 10.1172/JCI150951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee R.J., Xiong G., Kofonow J.M., Chen B., Lysenko A., Jiang P., Abraham V., Doghramji L., Adappa N.D., Palmer J.N., et al. T2R38 Taste Receptor Polymorphisms Underlie Susceptibility to Upper Respiratory Infection. J. Clin. Investig. 2012;122:4145–4159. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shah A.S., Ben-Shahar Y., Moninger T.O., Kline J.N., Welsh M.J. Motile Cilia of Human Airway Epithelia Are Chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yan C.H., Hahn S., McMahon D., Bonislawski D., Kennedy D.W., Adappa N.D., Palmer J.N., Jiang P., Lee R.J., Cohen N.A. Nitric Oxide Production Is Stimulated by Bitter Taste Receptors Ubiquitously Expressed in the Sinonasal Cavity. Am. J. Rhinol. Allergy. 2017;31:85–92. doi: 10.2500/ajra.2017.31.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hariri B.M., McMahon D.B., Chen B., Freund J.R., Mansfield C.J., Doghramji L.J., Adappa N.D., Palmer J.N., Kennedy D.W., Reed D.R., et al. Flavones Modulate Respiratory Epithelial Innate Immunity: Anti-Inflammatory Effects and Activation of the T2R14 Receptor. J. Biol. Chem. 2017;292:8484–8497. doi: 10.1074/jbc.M116.771949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee R.J., Chen B., Redding K.M., Margolskee R.F., Cohen N.A. Mouse Nasal Epithelial Innate Immune Responses to Pseudomonas Aeruginosa Quorum-Sensing Molecules Require Taste Signaling Components. Innate Immun. 2014;20:606–617. doi: 10.1177/1753425913503386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hollenhorst M.I., Krasteva-Christ G. Chemosensory Cells in the Respiratory Tract as Crucial Regulators of Innate Immune Responses. J. Physiol. 2023;601:1555–1572. doi: 10.1113/JP282307. [DOI] [PubMed] [Google Scholar]
- 140.Montoro D.T., Haber A.L., Biton M., Vinarsky V., Lin B., Birket S.E., Yuan F., Chen S., Leung H.M., Villoria J., et al. A Revised Airway Epithelial Hierarchy Includes CFTR-Expressing Ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nadjsombati M.S., McGinty J.W., Lyons-Cohen M.R., Jaffe J.B., DiPeso L., Schneider C., Miller C.N., Pollack J.L., Gowda G.A.N., Fontana M.F., et al. Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity. 2018;49:33–41.e7. doi: 10.1016/j.immuni.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Prandi S., Bromke M., Hübner S., Voigt A., Boehm U., Meyerhof W., Behrens M. A Subset of Mouse Colonic Goblet Cells Expresses the Bitter Taste Receptor Tas2r131. PLoS ONE. 2013;8:e82820. doi: 10.1371/journal.pone.0082820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Prandi S., Voigt A., Meyerhof W., Behrens M. Expression Profiling of Tas2r Genes Reveals a Complex Pattern along the Mouse GI Tract and the Presence of Tas2r131 in a Subset of Intestinal Paneth Cells. Cell. Mol. Life Sci. 2018;75:49–65. doi: 10.1007/s00018-017-2621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vegezzi G., Anselmi L., Huynh J., Barocelli E., Rozengurt E., Raybould H., Sternini C. Diet-Induced Regulation of Bitter Taste Receptor Subtypes in the Mouse Gastrointestinal Tract. PLoS ONE. 2014;9:e107732. doi: 10.1371/journal.pone.0107732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Latorre R., Huynh J., Mazzoni M., Gupta A., Bonora E., Clavenzani P., Chang L., Mayer E.A., Giorgio R.D., Sternini C. Expression of the Bitter Taste Receptor, T2R38, in Enteroendocrine Cells of the Colonic Mucosa of Overweight/Obese vs. Lean Subjects. PLoS ONE. 2016;11:e0147468. doi: 10.1371/journal.pone.0147468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gonda S., Matsumura S., Saito S., Go Y., Imai H. Expression of Taste Signal Transduction Molecules in the Caecum of Common Marmosets. Biol. Lett. 2013;9:20130409. doi: 10.1098/rsbl.2013.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Imai H., Hakukawa M., Hayashi M., Iwatsuki K., Masuda K. Expression of Bitter Taste Receptors in the Intestinal Cells of Non-Human Primates. Int. J. Mol. Sci. 2020;21:902. doi: 10.3390/ijms21030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hayashi M., Inaba A., Hakukawa M., Iwatsuki K., Imai H., Masuda K. Expression of TAS2R14 in the Intestinal Endocrine Cells of Non-Human Primates. Genes. Genom. 2021;43:259–267. doi: 10.1007/s13258-021-01054-7. [DOI] [PubMed] [Google Scholar]
- 149.Wu S.V., Rozengurt N., Yang M., Young S.H., Sinnett-Smith J., Rozengurt E. Expression of Bitter Taste Receptors of the T2R Family in the Gastrointestinal Tract and Enteroendocrine STC-1 Cells. Proc. Natl. Acad. Sci. USA. 2002;99:2392–2397. doi: 10.1073/pnas.042617699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dotson C.D., Zhang L., Xu H., Shin Y.-K., Vigues S., Ott S.H., Elson A.E.T., Choi H.J., Shaw H., Egan J.M., et al. Bitter Taste Receptors Influence Glucose Homeostasis. PLoS ONE. 2008;3:e3974. doi: 10.1371/journal.pone.0003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Descamps-Solà M., Vilalta A., Jalsevac F., Blay M.T., Rodríguez-Gallego E., Pinent M., Beltrán-Debón R., Terra X., Ardévol A. Bitter Taste Receptors along the Gastrointestinal Tract: Comparison between Humans and Rodents. Front. Nutr. 2023;10:1215889. doi: 10.3389/fnut.2023.1215889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xie C., Wang X., Young R.L., Horowitz M., Rayner C.K., Wu T. Role of Intestinal Bitter Sensing in Enteroendocrine Hormone Secretion and Metabolic Control. Front. Endocrinol. 2018;9:576. doi: 10.3389/fendo.2018.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Janssen S., Laermans J., Verhulst P.-J., Thijs T., Tack J., Depoortere I. Bitter Taste Receptors and α-Gustducin Regulate the Secretion of Ghrelin with Functional Effects on Food Intake and Gastric Emptying. Proc. Natl. Acad. Sci. USA. 2011;108:2094–2099. doi: 10.1073/pnas.1011508108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Avau B., Thijs T., Laermans J., Tack J., Depoortere I. Endocrine and Smooth Muscle Responses of the Bitter Agonist, Denatonium Benzoate, in the Stomach. Regul. Pept. 2012;177:S15. doi: 10.1016/j.regpep.2012.05.019. [DOI] [Google Scholar]
- 155.Deloose E., Janssen P., Corsetti M., Biesiekierski J., Masuy I., Rotondo A., Van Oudenhove L., Depoortere I., Tack J. Intragastric Infusion of Denatonium Benzoate Attenuates Interdigestive Gastric Motility and Hunger Scores in Healthy Female Volunteers1, 2, 3. Am. J. Clin. Nutr. 2017;105:580–588. doi: 10.3945/ajcn.116.138297. [DOI] [PubMed] [Google Scholar]
- 156.Deloose E., Corsetti M., Van Oudenhove L., Depoortere I., Tack J. Intragastric Infusion of the Bitter Tastant Quinine Suppresses Hormone Release and Antral Motility during the Fasting State in Healthy Female Volunteers. Neurogastroenterol. Motil. 2018;30:e13171. doi: 10.1111/nmo.13171. [DOI] [PubMed] [Google Scholar]
- 157.Chen M.C., Wu S.V., Reeve J.R., Rozengurt E. Bitter Stimuli Induce Ca2+ Signaling and CCK Release in Enteroendocrine STC-1 Cells: Role of L-Type Voltage-Sensitive Ca2+ Channels. Am. J. Physiol.-Cell Physiol. 2006;291:C726–C739. doi: 10.1152/ajpcell.00003.2006. [DOI] [PubMed] [Google Scholar]
- 158.Jeon T.-I., Zhu B., Larson J.L., Osborne T.F. SREBP-2 Regulates Gut Peptide Secretion through Intestinal Bitter Taste Receptor Signaling in Mice. J. Clin. Investig. 2008;118:3693–3700. doi: 10.1172/JCI36461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jeon T.-I., Seo Y.-K., Osborne T.F. Gut Bitter Taste Receptor Signalling Induces ABCB1 through a Mechanism Involving CCK. Biochem. J. 2011;438:33–37. doi: 10.1042/BJ20110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Le Nevé B., Foltz M., Daniel H., Gouka R. The Steroid Glycoside H.g.-12 from Hoodia Gordonii Activates the Human Bitter Receptor TAS2R14 and Induces CCK Release from HuTu-80 Cells. Am. J. Physiol.-Gastrointest. Liver Physiol. 2010;299:G1368–G1375. doi: 10.1152/ajpgi.00135.2010. [DOI] [PubMed] [Google Scholar]
- 161.Kim K.-S., Egan J.M., Jang H.-J. Denatonium Induces Secretion of Glucagon-like Peptide-1 through Activation of Bitter Taste Receptor Pathways. Diabetologia. 2014;57:2117–2125. doi: 10.1007/s00125-014-3326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Park J., Kim K.-S., Kim K.-H., Lee I.-S., Jeong H., Kim Y., Jang H.-J. GLP-1 Secretion Is Stimulated by 1,10-Phenanthroline via Colocalized T2R5 Signal Transduction in Human Enteroendocrine L Cell. Biochem. Biophys. Res. Commun. 2015;468:306–311. doi: 10.1016/j.bbrc.2015.10.107. [DOI] [PubMed] [Google Scholar]
- 163.Yue X., Liang J., Gu F., Du D., Chen F. Berberine Activates Bitter Taste Responses of Enteroendocrine STC-1 Cells. Mol. Cell Biochem. 2018;447:21–32. doi: 10.1007/s11010-018-3290-3. [DOI] [PubMed] [Google Scholar]
- 164.Yu Y., Hao G., Zhang Q., Hua W., Wang M., Zhou W., Zong S., Huang M., Wen X. Berberine Induces GLP-1 Secretion through Activation of Bitter Taste Receptor Pathways. Biochem. Pharmacol. 2015;97:173–177. doi: 10.1016/j.bcp.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 165.Pham H., Hui H., Morvaridi S., Cai J., Zhang S., Tan J., Wu V., Levin N., Knudsen B., Goddard W.A., et al. A Bitter Pill for Type 2 Diabetes? The Activation of Bitter Taste Receptor TAS2R38 Can Stimulate GLP-1 Release from Enteroendocrine L-Cells. Biochem. Biophys. Res. Commun. 2016;475:295–300. doi: 10.1016/j.bbrc.2016.04.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Luo X.-C., Chen Z.-H., Xue J.-B., Zhao D.-X., Lu C., Li Y.-H., Li S.-M., Du Y.-W., Liu Q., Wang P., et al. Infection by the Parasitic Helminth Trichinella Spiralis Activates a Tas2r-Mediated Signaling Pathway in Intestinal Tuft Cells. Proc. Natl. Acad. Sci. USA. 2019;116:5564–5569. doi: 10.1073/pnas.1812901116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liszt K.I., Wang Q., Farhadipour M., Segers A., Thijs T., Nys L., Deleus E., der Schueren B.V., Gerner C., Neuditschko B., et al. Human Intestinal Bitter Taste Receptors Regulate Innate Immune Responses and Metabolic Regulators in Obesity. J. Clin. Investig. 2022;132:e144828. doi: 10.1172/JCI144828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tran H.T.T., Herz C., Ruf P., Stetter R., Lamy E. Human T2R38 Bitter Taste Receptor Expression in Resting and Activated Lymphocytes. Front. Immunol. 2018;9:2949. doi: 10.3389/fimmu.2018.02949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Maurer S., Wabnitz G.H., Kahle N.A., Stegmaier S., Prior B., Giese T., Gaida M.M., Samstag Y., Hänsch G.M. Tasting Pseudomonas Aeruginosa Biofilms: Human Neutrophils Express the Bitter Receptor T2R38 as Sensor for the Quorum Sensing Molecule N-(3-Oxododecanoyl)-l-Homoserine Lactone. Front. Immunol. 2015;6:369. doi: 10.3389/fimmu.2015.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Grassin-Delyle S., Salvator H., Mantov N., Abrial C., Brollo M., Faisy C., Naline E., Couderc L.-J., Devillier P. Bitter Taste Receptors (TAS2Rs) in Human Lung Macrophages: Receptor Expression and Inhibitory Effects of TAS2R Agonists. Front. Physiol. 2019;10:1267. doi: 10.3389/fphys.2019.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kobayashi D., Watarai T., Ozawa M., Kanda Y., Saika F., Kiguchi N., Takeuchi A., Ikawa M., Matsuzaki S., Katakai T. Tas2R Signaling Enhances Mouse Neutrophil Migration via a ROCK-Dependent Pathway. Front. Immunol. 2022;13:973880. doi: 10.3389/fimmu.2022.973880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gopallawa I., Freund J.R., Lee R.J. Bitter Taste Receptors Stimulate Phagocytosis in Human Macrophages through Calcium, Nitric Oxide, and Cyclic-GMP Signaling. Cell. Mol. Life Sci. 2021;78:271–286. doi: 10.1007/s00018-020-03494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Malki A., Fiedler J., Fricke K., Ballweg I., Pfaffl M.W., Krautwurst D. Class I Odorant Receptors, TAS1R and TAS2R Taste Receptors, Are Markers for Subpopulations of Circulating Leukocytes. J. Leukoc. Biol. 2015;97:533–545. doi: 10.1189/jlb.2A0714-331RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kouakou Y.I., Lee R.J. Interkingdom Detection of Bacterial Quorum-Sensing Molecules by Mammalian Taste Receptors. Microorganisms. 2023;11:1295. doi: 10.3390/microorganisms11051295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Voigt A., Hübner S., Lossow K., Hermans-Borgmeyer I., Boehm U., Meyerhof W. Genetic Labeling of Tas1r1 and Tas2r131 Taste Receptor Cells in Mice. Chem. Senses. 2012;37:897–911. doi: 10.1093/chemse/bjs082. [DOI] [PubMed] [Google Scholar]
- 176.Li F., Zhou M. Depletion of Bitter Taste Transduction Leads to Massive Spermatid Loss in Transgenic Mice. Mol. Hum. Reprod. 2012;18:289–297. doi: 10.1093/molehr/gas005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xu J., Cao J., Iguchi N., Riethmacher D., Huang L. Functional Characterization of Bitter-Taste Receptors Expressed in Mammalian Testis. Mol. Hum. Reprod. 2013;19:17–28. doi: 10.1093/molehr/gas040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Governini L., Semplici B., Pavone V., Crifasi L., Marrocco C., De Leo V., Arlt E., Gudermann T., Boekhoff I., Luddi A., et al. Expression of Taste Receptor 2 Subtypes in Human Testis and Sperm. J. Clin. Med. 2020;9:264. doi: 10.3390/jcm9010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wu S., Xue P., Grayson N., Bland J.S., Wolfe A. Bitter Taste Receptor Ligand Improves Metabolic and Reproductive Functions in a Murine Model of PCOS. Endocrinology. 2019;160:143–155. doi: 10.1210/en.2018-00711. [DOI] [PubMed] [Google Scholar]
- 180.Liu S., Lu S., Xu R., Atzberger A., Günther S., Wettschureck N., Offermanns S. Members of Bitter Taste Receptor Cluster Tas2r143/Tas2r135/Tas2r126 Are Expressed in the Epithelium of Murine Airways and Other Non-Gustatory Tissues. Front. Physiol. 2017;8:849. doi: 10.3389/fphys.2017.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Semplici B., Luongo F.P., Passaponti S., Landi C., Governini L., Morgante G., De Leo V., Piomboni P., Luddi A. Bitter Taste Receptors Expression in Human Granulosa and Cumulus Cells: New Perspectives in Female Fertility. Cells. 2021;10:3127. doi: 10.3390/cells10113127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wölfle U., Elsholz F.A., Kersten A., Haarhaus B., Schumacher U., Schempp C.M. Expression and Functional Activity of the Human Bitter Taste Receptor TAS2R38 in Human Placental Tissues and JEG-3 Cells. Molecules. 2016;21:306. doi: 10.3390/molecules21030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Taher S., Borja Y., Cabanela L., Costers V.J., Carson-Marino M., Bailes J.C., Dhar B., Beckworth M.T., Rabaglino M.B., Post Uiterweer E.D., et al. Cholecystokinin, Gastrin, Cholecystokinin/Gastrin Receptors, and Bitter Taste Receptor TAS2R14: Trophoblast Expression and Signaling. Am. J. Physiol. -Regul. Integr. Comp. Physiol. 2019;316:R628–R639. doi: 10.1152/ajpregu.00153.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lu P., Moore Simas T.A., Delpapa E., ZhuGe R. Bitter Taste Receptors in the Reproductive System: Function and Therapeutic Implications. J. Cell. Physiol. 2024;239:e31179. doi: 10.1002/jcp.31179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jiang J., Liu S., Qi L., Wei Q., Shi F. Activation of Ovarian Taste Receptors Inhibits Progesterone Production Potentially via NO/cGMP and Apoptotic Signaling. Endocrinology. 2021;162:bqaa240. doi: 10.1210/endocr/bqaa240. [DOI] [PubMed] [Google Scholar]
- 186.Bloxham C.J., Foster S.R., Thomas W.G. A Bitter Taste in Your Heart. Front. Physiol. 2020;11:431. doi: 10.3389/fphys.2020.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Luo M., Ni K., Jin Y., Yu Z., Deng L. Toward the Identification of Extra-Oral TAS2R Agonists as Drug Agents for Muscle Relaxation Therapies via Bioinformatics-Aided Screening of Bitter Compounds in Traditional Chinese Medicine. Front. Physiol. 2019;10:861. doi: 10.3389/fphys.2019.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Behrens M., Lang T. Extra-Oral Taste Receptors—Function, Disease, and Perspectives. Front. Nutr. 2022;9:881177. doi: 10.3389/fnut.2022.881177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Luddi A., Governini L., Wilmskötter D., Gudermann T., Boekhoff I., Piomboni P. Taste Receptors: New Players in Sperm Biology. Int. J. Mol. Sci. 2019;20:967. doi: 10.3390/ijms20040967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kong S., Dong C., Lv H., Chen L., Zhang J., Pu F., Li X., Xu P. Genome Wide Identification of Taste receptor Genes in Common Carp (Cyprinus carpio) and Phylogenetic Analysis in Teleost. Gene. 2018;678:65–72. doi: 10.1016/j.gene.2018.07.078. [DOI] [PubMed] [Google Scholar]
- 191.Hamdard E., Lv Z., Jiang J., Wei Q., Shi Z., Malyar R.M., Yu D., Shi F. Responsiveness Expressions of Bitter Taste Receptors Against Denatonium Benzoate and Genistein in the Heart, Spleen, Lung, Kidney, and Bursa Fabricius of Chinese Fast Yellow Chicken. Animals. 2019;9:532. doi: 10.3390/ani9080532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Birdal G., D’Gama P.P., Jurisch-Yaksi N., Korsching S.I. Expression of Taste Sentinels, T1R, T2R, and PLCβ2, on the Passageway for Olfactory Signals in Zebrafish | Chemical Senses | Oxford Academic. Chem. Senses. 2023;48:bjad040. doi: 10.1093/chemse/bjad040. [DOI] [PubMed] [Google Scholar]
- 193.Mayeur H., Leyhr J., Mulley J., Leurs N., Michel L., Sharma K., Lagadec R., Aury J.-M., Osborne O.G., Mulhair P., et al. The Sensory Shark: High-Quality Morphological, Genomic and Transcriptomic Data for the Small-Spotted Catshark Scyliorhinus Canicula Reveal the Molecular Bases of Sensory Organ Evolution in Jawed Vertebrates. bioRxiv. 2024 doi: 10.1101/2024.05.23.595469. [DOI] [PubMed] [Google Scholar]
- 194.Cheled-Shoval S.L., Druyan S., Uni Z. Bitter, Sweet and Umami Taste Receptors and Downstream Signaling Effectors: Expression in Embryonic and Growing Chicken Gastrointestinal Tract. Poult. Sci. 2015;94:1928–1941. doi: 10.3382/ps/pev152. [DOI] [PubMed] [Google Scholar]
- 195.Liu X., Yu Y., Qin D., Song Z., Huang Z., Meng K., Cao J., Xu F., Cheng G., Ji W., et al. Expression Analysis of Taste Receptor Genes (T1R1, T1R3, and T2R4) in Response to Bacterial, Viral and Parasitic Infection in Rainbow Trout, Oncorhynchus mykiss. Fish. Shellfish. Immunol. 2020;101:176–185. doi: 10.1016/j.fsi.2020.03.055. [DOI] [PubMed] [Google Scholar]
- 196.Wu H., Wang C., Gregory K.J., Han G.W., Cho H.P., Xia Y., Niswender C.M., Katritch V., Meiler J., Cherezov V., et al. Structure of a Class C GPCR Metabotropic Glutamate Receptor 1 Bound to an Allosteric Modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Munk C., Mutt E., Isberg V., Nikolajsen L.F., Bibbe J.M., Flock T., Hanson M.A., Stevens R.C., Deupi X., Gloriam D.E. An Online Resource for GPCR Structure Determination and Analysis. Nat. Methods. 2019;16:151–162. doi: 10.1038/s41592-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ammon C., Schäfer J., Kreuzer O.J., Meyerhof W. Presence of a Plasma Membrane Targeting Sequence in the Amino-Terminal Region of the Rat Somatostatin Receptor 3. Arch. Physiol. Biochem. 2002;110:137–145. doi: 10.1076/apab.110.1.137.908. [DOI] [PubMed] [Google Scholar]
- 199.Tan S.M., Seetoh W.-G. Construction of a Bioluminescence-Based Assay for Bitter Taste Receptors (TAS2Rs) Sci. Rep. 2022;12:17658. doi: 10.1038/s41598-022-21678-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Isberg V., de Graaf C., Bortolato A., Cherezov V., Katritch V., Marshall F.H., Mordalski S., Pin J.-P., Stevens R.C., Vriend G., et al. Generic GPCR Residue Numbers—Aligning Topology Maps While Minding the Gaps. Trends Pharmacol. Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Di Pizio A., Levit A., Slutzki M., Behrens M., Karaman R., Niv M.Y. Comparing Class A GPCRs to Bitter Taste Receptors: Structural Motifs, Ligand Interactions and Agonist-to-Antagonist Ratios. In: Shukla A.K., editor. Methods in Cell Biology. Volume 132. Academic Press; Cambridge, MA, USA: 2016. pp. 401–427. G Protein-Coupled Receptors. [DOI] [PubMed] [Google Scholar]
- 202.Xu W., Wu L., Liu S., Liu X., Cao X., Zhou C., Zhang J., Fu Y., Guo Y., Wu Y., et al. Structural Basis for Strychnine Activation of Human Bitter Taste Receptor TAS2R46. Science. 2022;377:1298–1304. doi: 10.1126/science.abo1633. [DOI] [PubMed] [Google Scholar]
- 203.Peri L., Matzov D., Huxley D.R., Rainish A., Fierro F., Sapir L., Pfeiffer T., Waterloo L., Huebner H., Weikert D., et al. Intracellular Binding Pocket Revealed in the Human Bitter Taste Receptor TAS2R14. bioRxiv. 2024 doi: 10.1101/2024.04.10.588278. [DOI] [Google Scholar]
- 204.Kim Y., Gumpper R.H., Liu Y., Kocak D.D., Xiong Y., Cao C., Deng Z., Krumm B.E., Jain M.K., Zhang S., et al. Bitter Taste Receptor Activation by Cholesterol and an Intracellular Tastant. Nature. 2024;628:664–671. doi: 10.1038/s41586-024-07253-y. [DOI] [PubMed] [Google Scholar]
- 205.Hu X., Ao W., Gao M., Wu L., Pei Y., Liu S., Wu Y., Zhao F., Sun Q., Liu J., et al. Bitter Taste TAS2R14 Activation by Intracellular Tastants and Cholesterol. Nature. 2024;631:459–466. doi: 10.1038/s41586-024-07569-9. [DOI] [PubMed] [Google Scholar]
- 206.Born S., Levit A., Niv M.Y., Meyerhof W., Behrens M. The Human Bitter Taste Receptor TAS2R10 Is Tailored to Accommodate Numerous Diverse Ligands. J. Neurosci. 2013;33:201–213. doi: 10.1523/JNEUROSCI.3248-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Thomas A., Sulli C., Davidson E., Berdougo E., Phillips M., Puffer B.A., Paes C., Doranz B.J., Rucker J.B. The Bitter Taste Receptor TAS2R16 Achieves High Specificity and Accommodates Diverse Glycoside Ligands by Using a Two-Faced Binding Pocket. Sci. Rep. 2017;7:7753. doi: 10.1038/s41598-017-07256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.