Abstract
Mikania micrantha (“mile-a-minute” weed) is a global invasive alien weed that can cause severe damage to agroforestry ecosystems and significant agricultural losses worldwide. Although chemical, manual, or mechanical control methods are widely used to control M. micrantha, RNA interference (RNAi)-based biocontrol methods have rarely been reported for this species. The MONOPTEROS (MP) gene, encoding an auxin response factor, plays an essential role in embryonic root initiation in Arabidopsis thaliana. In this study, we identified the MP gene from M. micrantha via orthologous gene analysis. A total of 37 MP orthologous genes was identified in 4 plants, including 9 MP candidate genes in M. micrantha, 13 in Helianthus annuus, 6 in Chrysanthemum nankingense, and 9 in Lactuca sativa. Phylogenetic analysis revealed that an MP candidate gene in M. micrantha (Mm01G000655, named MmMP) was clustered into one clade with the MP gene in A. thaliana (AtMP). In addition, both MmMP and AtMP contain a B3-DNA binding domain that is shared by transcription factors that regulate plant embryogenesis. To study gene function, dsRNA against MmMP (dsMmMP) was applied to the roots of M. micrantha. Compared with those of the controls, the expression of MmMP was reduced by 43.3%, 22.1%, and 26.2% on the first, third, and fifth days after dsMmMP treatment, respectively. The dsMmMP-treated plants presented several morphological defects, mostly in the roots. Compared with water-treated plants, the dsMmMP-treated plants presented reduced developmental parameters, including root length, number of adventitious roots, root fresh and dry weights, plant height, and aboveground biomass. Additionally, safety assessment suggested that this dsMmMP treatment did not silence MP genes from non-target plants, including rice and tomato; nor did it inhibit root growth in those species. Collectively, these results suggest that MmMP plays an important role in root development in M. micrantha and provides a potential target for the development of species-specific RNAi-based herbicides.
Keywords: Mikania micrantha, invasive weed, MONOPTEROS gene, root development, RNA interference, RNAi-based herbicides
1. Introduction
Mikania micrantha is an extremely fast-growing invasive alien weed of the Asteraceae family that is native to Central and South America [1]. M. micrantha has the advantages of a strong ability to reproduce, fast asexual reproduction, and rapid nutritional growth. It is listed as one of the 100 most invasive species worldwide by the International Union for Conservation of Nature (IUCN) [2].and is also known as a “plant killer” [3,4]. M. micrantha is present in tropical Asia, parts of Papua New Guinea, islands in the Indian and Pacific Oceans, and Florida in the United States [4,5]. M. micrantha poses an immense threat to native plant species and wreaks havoc by climbing and winding around other plants to block their sunlight reception and photosynthesis, eventually resulting in the death of other plants [1,6]. As an invasive weed in cash crop fields and forests, the invasion of M. micrantha has caused economic losses of up to 4,000 hectares related to forest and crop production [7,8,9]. It also leads to a loss of genetic and species diversity, reduced soil and food web stability, and altered nutrient cycles [6].
Currently, the main control technologies for M. micrantha include chemical herbicide control, manual or mechanical eradication, and biological control [10,11,12]. Chemical herbicides are currently the most common control method for M. micrantha, given their fast effects and ease of operation. For example, glyphosate, paraquat, glufosinate, and 2,4-D have obvious control effects on M. micrantha [11,13]. However, studies have reported that the application of chemical herbicides to M. micrantha results in damage to surrounding plants, leading to reduced resistance of the native plant community to sub-invasions by M. micrantha [14,15]. On the other hand, manual or mechanical eradication can significantly reduce the coverage area of M. micrantha in a short period of time. However, this method is only effective in small areas and is not an effective means for large-scale control [16,17]. In recent years, researchers have focused on developing biological control technologies that use pathogenic bacteria and insects to control M. micrantha, such as Actinote thalia pyrrha, Actinote anteas, Brevipalpus phoenicis, and Pachypeltis micranthus [18,19]. These parasitic agents are not highly specific since they also harm other plants and are therefore not ideal for field applications [6,20].
RNA interference (RNAi) is a posttranscriptional gene-silencing technique [21]. The principle of RNAi is to induce the degradation of homologous gene sequences in organisms via the complementary pairing of double-stranded RNA (dsRNA) bases [22]. RNAi-based technologies show pronounced potential for use in agriculture, particularly for the management of destructive insect pests [23,24,25,26]. For example, a greater than 80% knockdown rate was observed by silencing the expression of the target gene V-type proton ATPase subunit D (ATPD) in the control of soybean aphids (Aphis glycines) [27]. There have been several relevant reports on the application of dsRNA to plant roots. Majidiani et al. evaluated the absorption of dsRNA by plant roots and found that long dsRNA molecules can also be absorbed and cause gene silencing in target insects that feed on leaves [27,28]. Konstantin et al. analyzed the importance of physiological conditions and different dsRNA application methods (brush spreading, spraying, infiltration, inoculation, needle injection, and pipetting) for the suppression of the neomycin phosphotransferase II (NPTII) transgene in Arabidopsis thaliana [29]. Jiang et al. applied a mixture of G2/dsSTM or G2/dsWER to roots, which led to a dramatic reduction in the expression of STM and WER in the entire seedling. The specific RNA interference of STM and WER mediated by G2-delivered dsRNA resulted in reduced SAM size and increased lateral roots, respectively [30]. Minsu et al. used dsRNA to treat A. thaliana by dipping and spraying, both of which can induce the inhibition of target gene expression after the application of target gene-specific exogenous dsRNA [31].
RNAi has been reported as a promising molecular control strategy, particularly in weed management. For example, the application of RNAi to manage glyphosate-resistant weeds has emerged as a new technology, providing a novel approach for weed control [32]. For the application of RNAi technology in M. micrantha, to our knowledge, only one study has reported the use of three types of RNAi molecules (double-stranded RNA, RNAi nanomicrosphere, and short hairpin RNA) to inhibit the gene expression of chlorophyll a/b-binding proteins in M. micrantha, resulting in yellow leaves and eventual wilt [33]. Previous studies have shown that MP (MONOPTEROS, MP), also known as auxin response factor 5 (ARF5), is an auxin response factor that plays a key role in embryonic root initiation in A. thaliana [34,35]. Mutants with complete loss of MP function die at the embryo stage due to their inability to form roots [36,37,38].
With the rapid development of molecular biology and the emergence of advanced sequencing technologies, the genome of M. micrantha has been sequenced and assembled [39]. On the basis of this genetic resource, in this study, we identified the MP gene from M. micrantha (MmMP). Owing to the biological characteristics of the rooting ability of each stem node in M. micrantha, we focused on developing an RNAi-based approach to knock down MmMP expression and therefore inhibit root development. The morphological phenotypes of the roots of M. micrantha were subsequently measured to evaluate the inhibition of growth. These findings revealed the functional role of MmMP in regulating root growth, making it a potential target for the development of RNAi-mediated herbicides.
2. Results
2.1. Identification of the MP Gene in M. micrantha
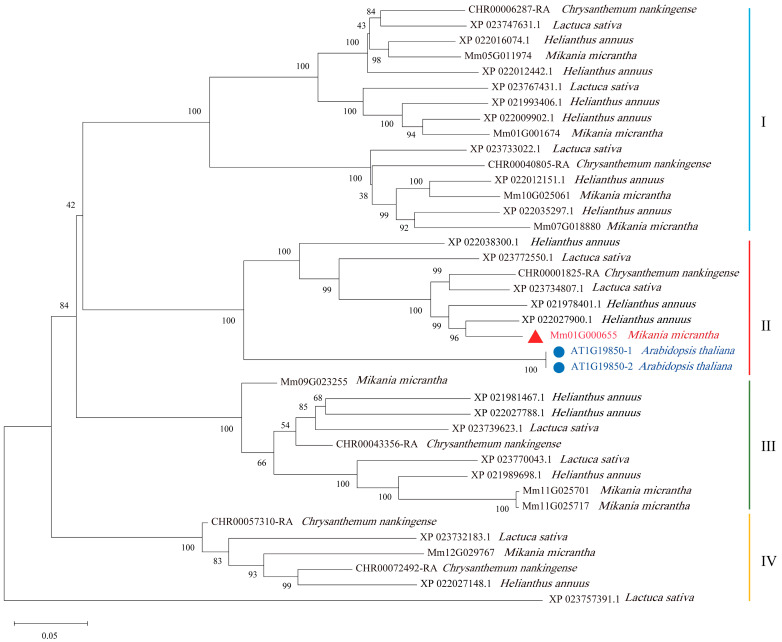
To identify the orthologs of MONOPTEROS (MP) in the four plants, we used the amino acid sequence of MP (AT1G19850) in A. thaliana for alignment with the protein sequences of M. micrantha, Chrysanthemum nankingense, Helianthus annuus, and Lactuca sativa. A total of 37 MP orthologous genes (Supplementary Table S3), including 9 MP candidate genes in M. micrantha, 13 in H. annuus, 6 in C. nankingense, and 9 in L. sativa, were used to construct the phylogenetic tree (Figure 1). The results revealed that these proteins in these five species clustered into four clades (I, II, III, and IV). One gene (Mm01G000655) in M. micrantha, three genes (XP_0220279001, XP_021978401.1, and XP_0220383001) in H. annuus, one gene (CHR00001825-RA) in C. nankingense, and two genes (XP_023772550.1 and XP_023734807.1) in L. sativa were clustered in the same clade as two copies of MP in A. thaliana (named AT1G19850-1 and AT1G19850-2), suggesting that these orthologous genes may perform the same function (Figure 1).
Figure 1.
Phylogenetic analysis of MP genes from M. micrantha and four other species, namely, A. thaliana, C. nankingense, H. annuus, and L. sativa. I~IV represent the MP proteins of the five species divided into four independent clades. The tree was constructed via the neighbor-joining method based on the protein sequence alignments. The numbers on the branches represent bootstrap values obtained from 1000 replicates. The branch lengths are proportional to the percentage of sequence difference (scale bar, 0.05% difference). The red triangle represents the M. micrantha gene Mm01G000655; the blue circles represent two protein-encoding genes in A. thaliana.
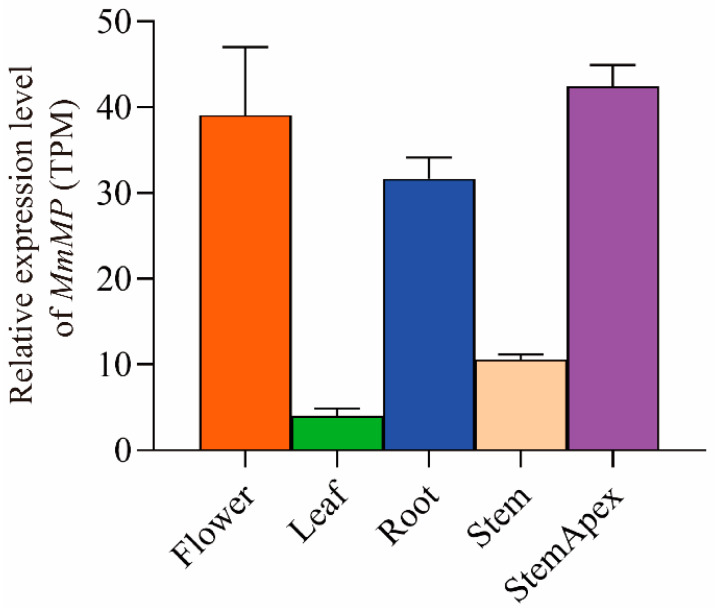
In addition, the amino acid sequence of the Mm01G000655 gene was highly conserved with those of the MP genes (AT1G19850-1 and AT1G19850-2) in A. thaliana (Supplementary Figure S1 and Supplementary Table S4). Further domain analysis revealed that Mm01G000655 has typical auxin response factor (ARF) structural characteristics, i.e., one plant-specific B3-DNA binding domain, one auxin-encoding domain, and one AUX-IAA domain, which is consistent with the structure of the MP gene in A. thaliana (Supplementary Figure S1). Therefore, Mm01G000655 (named MmMP) of M. micrantha may perform the same function as MP in A. thaliana. Transcriptome analysis of different tissues revealed that the highest expression of MmMP was detected in the roots (TPM = 31.7), flowers (TPM = 39.1), and stem tips (TPM = 42.4), followed by the stems (TPM = 10.6) and finally the leaves (TPM = 4.0) (Supplementary Table S5). The expression level in the roots was 7.9 and 2.9 times greater than that in the leaves and stems, respectively (Figure 2).
Figure 2.
TPM values of candidate gene expression in different tissues. The values represent the means ± SE derived from five biological replicates.
2.2. Regulation of MP Gene Expression by dsRNA in M. micrantha Roots
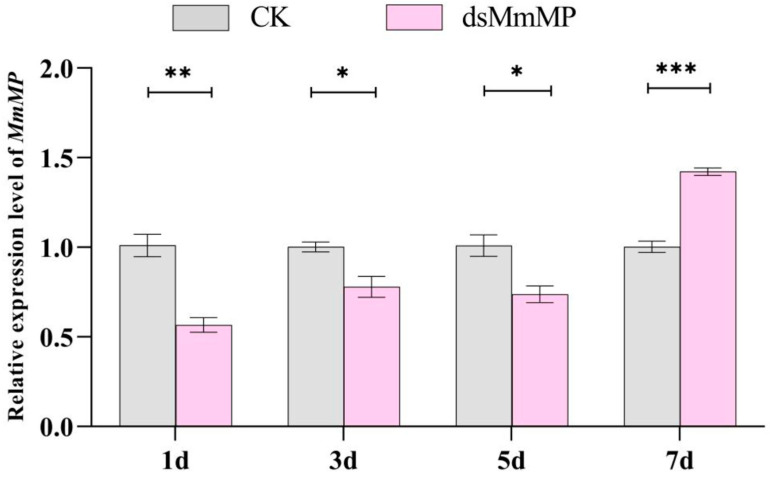
qRT–PCR was used to determine the effects of silencing via dsMmMP. Compared with that in the water-treated (CK) plants, MmMP expression in the dsMmMP-treated plants was inhibited by 43.3%, 22.1%, and 26.2% on the first (1 d), third (3 d), and fifth (5 d) days after dsRNA delivery, respectively (Figure 3). To further verify the inhibitory effect of dsRNA on the expression of target genes, we selected dsEGFP as a negative control. Compared with negative controls, dsMmMP significantly inhibited the expression of target genes, and the gene expression returned to normal levels by the seventh day (7d) (Supplementary Figure S2).
Figure 3.
The abundance of MmMP transcripts was measured at different time points after the roots were soaked in water (CK) or dsMmMP. Each bar represents the mean ± SE derived from three biological replicates. A t-test was used to determine significant differences in plant-silencing efficiency in this study (* p < 0.05; ** p < 0.01; *** p < 0.001). CK indicates treatment with water, and dsMmMP indicates double-stranded RNA of the M. micrantha MmMP gene.
2.3. Effects of MmMP Gene Silencing on the Root Growth of M. micrantha
To determine whether MmMP gene silencing affects the growth and development of M. micrantha, the morphological phenotypes were observed after 15 days of treatment. Compared with the plants in the CK treatment, those in the dsMmMP treatment exhibited significantly inhibited root growth (Figure 4).
Figure 4.
Effects of dsMmMP application on the phenotype of M. micrantha. Phenotypic changes in M. micrantha roots after continuous treatment for 15 days. CK, water control; dsMmMP, double-stranded RNA of the M. micrantha MmMP gene.
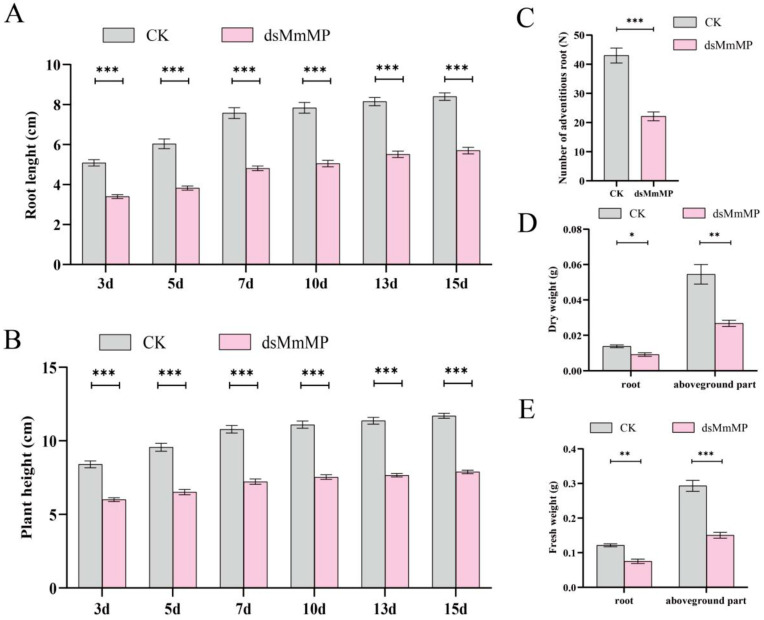
We subsequently measured the morphological phenotypes for 15 consecutive days under the dsRNA treatments. Compared with the CK plants, the plants treated with dsMmMP presented significantly shorter plant heights and lengths of the longest adventitious roots at the third, fifth, seventh, tenth, thirteenth, and fifteenth days. The average length of the longest adventitious root was inhibited by more than 30.1% under the dsMmMP treatment (Figure 5A). The average plant height was significantly inhibited by dsMmMP, representing an approximately 30.4% reduction (Figure 5B). After 15 days of treatment, the number of adventitious roots on average decreased by approximately 48.5% (Figure 5C). In addition, the fresh weights of the aboveground and belowground parts under the dsMmMP treatment significantly decreased by approximately 38.2% and 48.7%, respectively (Figure 5D). The average aboveground and belowground dry weights also decreased by 33.6% and 50.9%, respectively (Figure 5E).
Figure 5.
Statistical analysis of morphological indicators of M. micrantha. Statistical analysis of the length of the longest adventitious root (A), plant height (B), number of all adventitious roots (C), dry weights of all adventitious roots and aboveground parts (D), and fresh weights of all adventitious roots and fresh weights of the aboveground parts (E) after 15 d of continuous treatment. Each bar represents the mean and standard error for n = 8 biological replicates. A t-test was used to determine significant differences in plant-silencing efficiency in this study (* p < 0.05; ** p < 0.01; *** p < 0.001). CK, water control; dsMmMP, double-stranded RNA of the M. micrantha MmMP gene.
2.4. Safety Assessment for DsMmMP
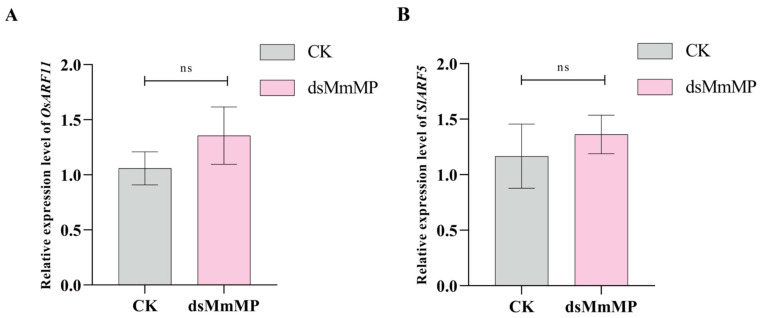
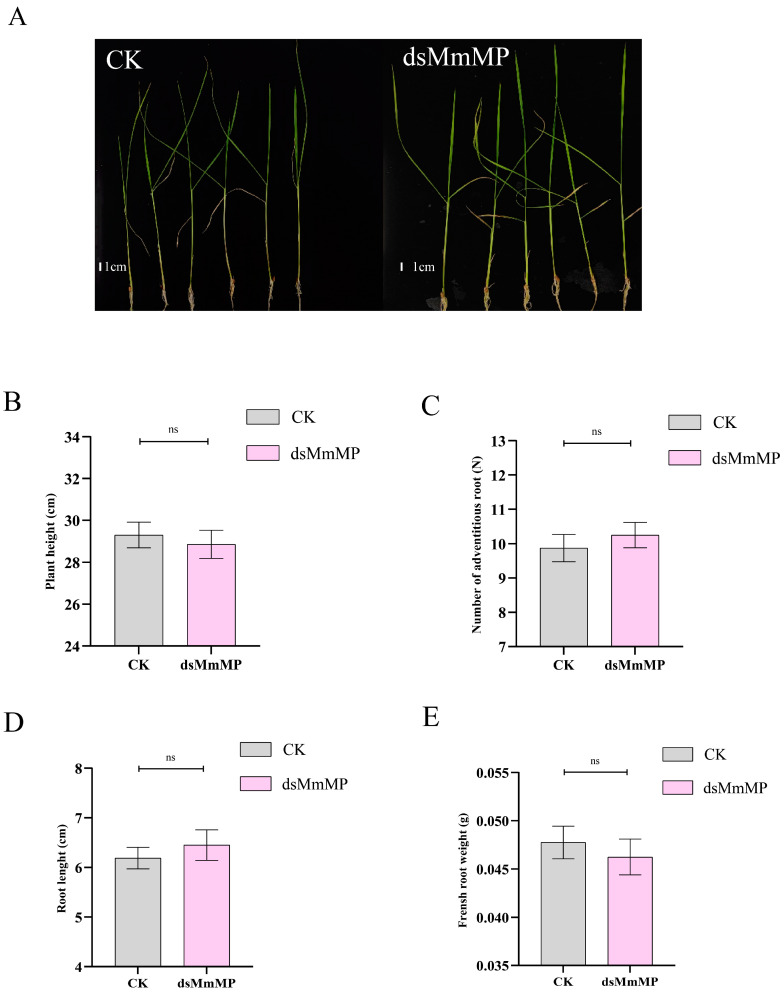
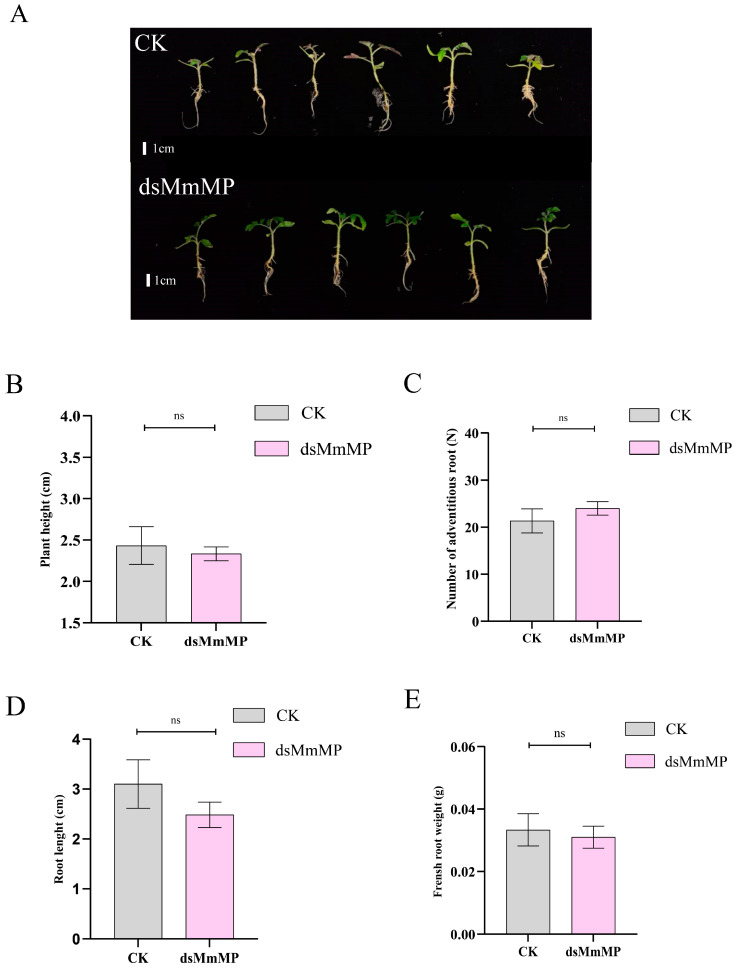
To determine whether dsMmMP also targets the MONOPTEROS genes in rice (OsARF11) and tomato (SlARF5), we verified gene expression after dsMmMP and water treatment via qRT–PCR. The results revealed that the gene expression of OsARF11 (Figure 6A) or SlARF5 (Figure 6B) 3 d after dsMmMP treatment was not significantly different from that after-water treatment. This might be explained by the lower identity of MmMP to OsARF11 (44.04%) and SlARF5 (60.37%) at the amino acid level (Supplementary Figure S3). In addition, the nucleotide-binding region of dsMmMP also presented low identity compared with that of OsARF11 (4.93%; Supplementary Figure S4A) or SlARF5 (13.65%; Supplementary Figure S4B). Therefore, rice and tomato MP genes are not targeted by dsMmMP. To further investigate whether treatment with dsMmMP affects the root growth of rice and tomato, the morphological phenotypes were examined 15 days after treatment. The results suggested that the plant height, root length, number of adventitious roots, and fresh weight of roots of rice were not significantly different between the CK and dsMmMP treatments (Figure 7A–E). Moreover, these parameters in tomato did not significantly differ between the CK and dsMmMP treatments (Figure 8A–E). Consequently, dsMmMP does not seem to inhibit the root growth of rice or tomato.
Figure 6.
Effects of dsMmMP on MP gene expression in non-target plants. The expression of the MP gene from rice (OsARF11) or tomato (SlARF5) was measured 3 d after dsMmMP and water treatment. (A) Rice (B) Tomato Each bar chart represents the mean and standard error of n = 3 biological replicates, and the data were analyzed via one-way ANOVA to identify significant differences in plant-inhibition efficiency and multiple Tukey comparisons (p < 0.05). “ns” indicates no significant difference between the CK and dsMmMP treatments. CK, water control; dsMmMP, double-stranded RNA of the M. micrantha MmMP gene.
Figure 7.
Effects of dsMmMP on the growth of O. sativa. (A) Image of a whole plant. (B) Plant height. (C) Number of adventitious roots. (D) Length of the longest adventitious root. (E) Fresh weight of all adventitious roots. Each bar represents the mean and standard error for n = 8 biological replicates, and the data were analyzed via one-way ANOVA to identify significant differences in plant-inhibition efficiency and multiple Tukey comparisons (p < 0.05). “ns” represents no significant difference between the CK and dsMmMP treatments. CK, water control; dsMmMP, double-stranded RNA of the M. micrantha MmMP gene.
Figure 8.
Effects of dsMmMP on the growth of S. lycopersicum. (A) Image of a whole plant. (B) Plant height. (C) Number of adventitious roots. (D) Length of the longest adventitious root. (E) Fresh weight of all adventitious roots. Each bar represents the mean and standard error for n = 6 biological replicates, and the data were analyzed via one-way ANOVA to identify significant differences in plant-inhibition efficiency and multiple Tukey comparisons (p < 0.05). “ns” indicates no significant difference between the CK and dsMmMP treatments. CK, water control; dsMmMP, double-stranded RNA of the M. micrantha MmMP gene.
3. Discussion
Sequence-specific knockdown of mRNA transcripts via RNAi provides countless opportunities for weed control. As a destructive invasive weed, M. micrantha has become a worldwide problem. Therefore, strong and effective biological strategies are urgently needed for the management of M. micrantha. RNAi is a conserved posttranscriptional gene-silencing process [40] that has been widely used in plant gene functional identification, quality improvement, and resistance research [41,42,43]. Previous studies have indicated that foliar application of dsRNAs encoding specific genes of plant pathogens triggered RNAi-mediated silencing of the target genes [43,44]. To data, the gene functional researches were major based on dsRNA-expressing transgenic plants and host-induced gene silencing [41,43]. However, few studies have reported the use of RNAi technology to control weeds. Recently, the importance of physiological conditions (plant age, time of day, soil moisture, high salinity, heat, and cold stresses) and different dsRNA application means (brush spreading, spraying, infiltration, inoculation, needle injection, and pipetting) for suppression of target gene were analyzed in A. thaliana, indicating that dsRNAs can suppress the expression of certain genes within an organism [29,45]. To select the gene that best suppresses M. micrantha, we chose a gene encoding an auxin-responsive protein, MP, as the target and designed the corresponding RNAi molecule. The use of RNAi technology to inhibit root development can effectively control the spread of M. micrantha, providing a theoretical basis for targeted control techniques that suppress the growth and development of M. micrantha through RNAi technology.
M. micrantha possesses strong reproductive capabilities, being able to reproduce sexually and spread through the production of a large number of seeds, as well as propagate through its strong asexual reproduction abilities. Therefore, the strong adventitious root formation at the nodes of M. micrantha stems is one of the main reasons for its ecological harm [1]. To improve the effectiveness of RNAi, it is particularly important to identify key target genes. This study focused on the inhibition of root growth and investigated the inhibitory effects of RNAi on root growth genes in M. micrantha, with the goal of biological control of this invasive weed. Previous studies have shown that the MP (MONOPTEROS, MP) gene is an auxin response factor that plays a key role in embryonic root initiation in A. thaliana [46,47]. Mutations resulting in complete loss of MP function result in embryo death due to the inability of A. thaliana to form root tips [38,48]. In this study, on the basis of genomics and transcriptome analysis, the MmMP gene was identified in M. micrantha. Further analysis revealed that the MmMP gene contained a conserved domain compared with the MP gene of A. thaliana, including a plant-specific B3-DNA binding domain, an auxin-resp, and an AUX-IAA [49,50]. We speculate that MmMP in M. micrantha has the same function as MP in A. thaliana. These findings reveal that the MmMP candidate gene may be important for further studies to inhibit the growth of M. micrantha.
At present, only a limited number of reports exist on the direct application of dsRNA to the exterior surface of plants, which can result in the silencing of endogenous target genes in the plant [51]. In this study, we analyzed the efficacy of target gene silencing in M. micrantha by soaking the roots of M. micrantha in dsRNA solution. The qRT–PCR results revealed that the expression levels of the MmMP gene were significantly reduced, indicating that RNAi was successful. Although the expression of the target gene was inhibited, the interference efficiency was not particularly high, which was speculated to be due to the relatively low concentration (0.3 μg/μL) applied, and increasing the dsRNA concentration could be considered in subsequent experiments. On the other hand, the relatively low efficiency might be because the applied dsRNA was not efficiently delivered inside the plant cell since it was taken up by the root and then transported through the xylem in woody plants [52]. To trigger intensive and global RNAi effects, they may need to be delivered inside the plant cell, either by mechanical damage to the cell wall [52] or through the use of nanoparticles [53] for efficient dsRNA delivery. Specifically, numerous studies have indicated that combining certain nanomaterials could significantly improve RNAi efficiency in some insects [54,55,56]. Moreover, studies have shown that, in plants, the application of dsRNA mixed with G2 nanoparticles to the root tip of A. thaliana markedly decreases the transcript levels of the target genes. Plants treated with G2/dsRNA exhibited slow growth, while dsRNA treatment alone did not lead to these effects [30]. In this study, we found that target gene expression in M. micrantha was significantly suppressed within 5 days after treatment, while the dsRNA may have lost its silencing effect on the seventh day. To improve the effects and duration of dsRNA action in plants, the use of nanoparticles may greatly facilitate the development of RNAi-based herbicides for M. micrantha and other weed pests.
Importantly, the results of the present study collectively revealed that dsMmMP had relatively strong and persistent inhibitory effects on plant development, verifying the gene function of MmMP. Phenotypic measurements revealed that the aboveground and belowground growth of M. micrantha was significantly inhibited after MmMP gene expression was reduced. Further safety assessment revealed that the nucleotide-binding region of dsMmMP shares low identity with those of OsARF11 (4.93%) and SlARF5 (13.65%), leading to be not significantly different between the water and dsMmMP treatments in rice and tomato, suggesting that the dsMmMp has an appropriate safety profile for important crops. On the other hand, the phylogenetic tree analysis of MP genes among Asteraceae plants, A. thaliana, rice, tomato, and M. micrantha indicated that the MP genes in M. micrantha are more distantly related to those in non-target plants such as rice and tomato (Supplementary Figure S5). Consequently, this further demonstrates the safety of the dsMmMP gene specific to M. micrantha against non-target plants. With respect to the study of root genes, our RNAi formulation can be applied through root irrigation, which is expected to make the application of RNAi biological herbicides possible, but the effectiveness of field control still needs to be further confirmed. Here, while we have demonstrated the safety of two non-target plants, the level of evidence may not be sufficient. In the future, we should continue to use a greater number of non-target plants to confirm the safety of off-target effects of dsRNA formulations and to verify the feasibility of using dsRNA-based herbicides to control invasive weeds. The development and use of RNAi bioherbicides can provide an efficient, safe, economical, and environmentally friendly method to effectively control M. micrantha in the field, which is highly important for advancing research on biological control techniques for M. micrantha.
4. Materials and Methods
4.1. Plant Material and Growth Conditions
M. micrantha seeds were collected from the farm of the Chinese Academy of Agricultural Sciences, Dapeng New District, Shenzhen City, Guangdong Province, China. M. micrantha seeds were cultured in plastic Petri dishes with wet filter paper under greenhouse conditions (a greenhouse with daily cycles of 16 h/light and 8 h/dark at 25 °C and a relative humidity of approximately 80%) for approximately 1 week until they took root and germinated. The plants were subsequently transplanted into Hoagland liquid medium (KNO3: 506 mg/L; NH4NO3: 80 mg/L; KH2PO4: 136 mg/L; MgSO4: 241 mg/L; FeNaEDTA: 36.7 mg/L; KI: 0.83 mg/L; H3BO3: 6.2 mg/L; MnSO4: 22.3 mg/L; Na2MoO4: 0.25 mg/L; ZnSO4: 8.6 mg/L; CuSO4: 0.025 mg/L; CoCl2: 0.025 mg/L; Ca(NO3)2: 945 mg/L; pH = 5.8) and grown in a greenhouse under constant illumination until the fourth leaf stage.
4.2. Identification of the M. micrantha MP Gene
The amino acid sequence of the MONOPTEROS (MP) (Gene ID: AT1G19850) gene in A. thaliana was downloaded from NCBI. We subsequently used the protein sequence of MP in A. thaliana to align to the protein sequences of four plants (M. micrantha, Chrysanthemum nankingense, Helianthus annuus, and Lactuca sativa; the species genome information is shown in Supplementary Table S1 via BLASTP with an E-value < 10−20. Candidate MP genes from these four plants were used to construct a phylogenetic tree via the neighbor–joining method in MEGA (version 11.0.13) with parameters of P-distance modeling, partial deletion of gaps, and 1000 bootstrap replicates [57,58,59]. Multiple sequence alignments of amino acid sequences were performed via ClustalW (version 2.1).
The transcriptome data for different tissues of M. micrantha, including flowers, leaves, roots, stems, and stem apices, were downloaded from NCBI (SRR8857616-SRR8857640) and used for subsequent gene expression analysis. To eliminate errors generated in sequencing, unreliable reads were filtered according to the following standards: (1) a 50% base Phred quality score was less than 20%; and (2) a read contained more than 10% ambiguous residues (‘N’). After filtering, the clean reads were mapped to the reference genome of M. micrantha via HISAT2 v2.1.0 with the default parameters [60]. Read counts were calculated with HTSeq-count v2.1.0 [61] via the results from SAMtools v1.1.3 [62], and transcripts per million (TPM) values were then calculated for every gene in the samples. GraphPad Prism 8.4.3 (Home-GraphPad) was used to map the expression of the M. micrantha candidate genes in different tissues.
4.3. RNA Extraction and Gene Cloning
Total RNA was extracted from M. micrantha leaves via a Plant Total RNA Isolation Kit Plus (FOREGENE, Chengdu, China) following the manufacturer’s protocols. The first strand of complementary DNA (cDNA) was synthesized from 1 μg of total RNA via the Hifair® III 1st Strand cDNA Synthesis Kit (Yeasen Biotech, Shanghai, China). For amplification of the cDNA sequence of MmMP, PCR primers were designed via Primer Premier 5 (Supplementary Table S2). The amplification procedure was 94 °C for 5 min, followed by 35 cycles of 94 °C for 10 s, 55 °C for 20 s, and 10 s at 72 °C, with a final elongation for 7 min at 72 °C. The expected product was cloned and inserted into the pMD18-T vector (TaKaRa, Dalian, China) and transformed into E. coli DH5α-competent cells (Yeasen Biotech, Shanghai, China). The plasmid was extracted and identified via sequencing.
4.4. Synthesis of dsRNA
A plasmid was used as a template to amplify dsRNA target gene sequences. The T7 RNA polymerase promoter (TAATACGACTCACTATAGGG) was added to the 5′ end of the dsMmMP primer (Supplementary Table S2). The amplified PCR products were purified and used as DNA templates, and dsRNA was synthesized using a T7 RNAi Transcription Kit (Vazyme, Nanjing, China). In addition, the dsRNA of enhanced green fluorescent protein (EGFP), a nonendogenous gene, was synthesized and used as a negative control (Supplementary Table S2). The synthesized dsRNA was collected via ethanol precipitation. Purified dsRNA was examined by agarose gel electrophoresis to determine its integrity and then stored at −20 °C before use.
4.5. Delivery of dsRNA and Phenotypic Analysis
dsRNA was applied to the roots of M. micrantha by soaking. The dsRNA application method was as follows: plant roots were immersed in 2.5 mL glass bottles containing 0.3 μg/μL dsRNA solution, and the treated plants were cultured at 25 °C and 80% relative humidity. The experiment was divided into a treatment group (dsMmMP) and two control groups (CK, water control; dsEGFP, negative control). Add 0.3 μg/μL dsRNA on the first day of treatment and not continuing to add it later. To determine RNAi efficiency, total RNA was extracted from the roots of plants treated with dsMmMP, dsEGFP, and CK on the first, third, fifth, and seventh days. The expression of MmMp gene in these days was detected. In addition, to determine the plant phenotypic effect, one plant was taken as a biological replicate, and each treatment had at least eight biological replicates. In the phenotypic measurement experiment, we added 0.3 μg/μL dsRNA every other day; the length of the longest adventitious root and the plant height were measured at 3 d, 5 d, 7 d, 10 d, 13 d, and 15 d after treatment. The number of adventitious roots and the fresh and dry root weights of all adventitious roots were measured at 15 d after treatment.
4.6. Gene Expression Analysis by Quantitative Real-Time PCR (qRT–PCR)
To reduce DNA contamination, a 5 × gDNA Digester Mix (Hifair® III 1st Strand cDNA Synthesis Kit) was used to remove gDNA contamination from the reverse transcription products. qPCR was performed via a CFX96 real-time PCR system (Bio-Rad, Hercules, CA, USA) with three biological replicates. Two internal controls, GAPDH (AGI: AT1G13440) and UBQ10 (AGI: AT4G05320), were selected from previous studies [63] as relevant reference genes for qRT–PCR in M. micrantha. The gene expression levels were calculated via the 2−ΔΔCt method [64]. The sequences of the gene-specific primers used for qPCR are listed in Supplementary Table S2.
4.7. Statistical Analysis
The experimental data were calculated as the mean ± standard error (SE), and one-way ANOVA was used to determine significant differences in plant-silencing efficiency. Tukey’s multiple comparisons and independent samples t-tests were performed in this study (p < 0.05), and the statistical analyses were performed in IBM SPSS Statistics version 29.0.1.0.
4.8. Safety Assessment of DsMmMP
Rice and tomato seeds were cultured in plastic Petri dishes with wet filter paper under greenhouse conditions (a greenhouse with daily cycles of 16 h/light and 8 h/dark at 25 °C and a relative humidity of approximately 80%) for approximately 1 week until they took root and germinated. The plants were subsequently transplanted into Hoagland liquid medium and grown in a light-constant greenhouse.
In accordance with previous studies, the MP genes of rice (OsARF11, GenBank: LOC4337309; Sims et al., 2021) [65] and tomato (SlARF5, GenBank: HM195248.1; Wu et al., 2011) [66] were downloaded from the National Center for Biotechnology Information (NCBI) database. The amino acid sequence alignments were performed via DNAMAN software (version 8; Lynnon Corporation, San Ramon, CA, USA). The alignments of the dsMmMP binding sequence of M. micrantha and the MP gene of rice or tomato were also performed via DNAMAN software.
DsMmMP was applied to the roots of rice and tomato plants by soaking. The dsRNA application method was the same as that described previously. After 3 d, the MP gene expression in the root tissues of rice (OsARF11) and tomato (SlARF5) plants was determined via qRT–PCR (Supplementary Table S2). The gene expression levels were calculated via the 2−ΔΔCt method [63,64]. The morphological phenotypes, including plant height, root number, root length, and root fresh weight, were measured after dsMmMP treatment for 15 d. This phenotypic analysis was conducted with at least eight biological replicates for each treatment.
5. Conclusions
In this study, an RNA interference system was successfully constructed for the invasive weed M. micrantha. We identified a gene encoding the MmMP auxin response protein of M. micrantha and verified the function of this gene, which is involved in root growth and development, via RNAi-mediated technology. These results suggest that MmMP plays an important role in root development in M. micrantha and therefore provides a potential target for the development of RNAi-based herbicides that might contribute to the biological control of invasive weeds.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms252312678/s1.
Author Contributions
B.L., F.W., and W.Q. conceived and designed the entire research project; B.L., Q.W., and Y.Y. provided experimental technical guidance; Z.O., Y.Z., K.W., and G.Z. provided bioinformatics analysis; Z.O. performed the experiment and analyzed the data; Z.O., B.L., X.Q., Y.Y., and Q.W. wrote and revised the manuscript. All authors contributed to this article and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Funding Statement
The work was funded by the National Natural Science Foundation of China (32072490), the National Key Research and Development Program of China (2021YFC2600100 and 2021YFC2600101), and the Agricultural Science and Technology Innovation Program. Shenzhen Science and Technology Program (KCXFZ20230731093259009). This project benefited from the collaboration between the Department of Insect Biotechnology in Plant Protection of the Justus-Liebig-University Giessen and the Agricultural Genomics Institute at Shenzhen of the Chinese Academy of Agricultural Sciences supported by the SINO-German Mobility Program (M-0050), DFG-NSFC, Germany-China.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Day M.D., Clements D.R., Gile C., Senaratne W.K.A.D., Shen S., Weston L.A., Zhang F. Biology and Impacts of Pacific Islands Invasive Species. 13. Mikania micrantha Kunth (Asteraceae) Pac. Sci. 2016;70:257–285. [Google Scholar]
- 2.Lowe S., Browne M., Boudjelas S., de Poorter M. 100 of the World’s Worst Invasive Alien Species. IUCN; Gland, Switzerland: 2000. [Google Scholar]
- 3.Li M.G., Zhang W.Y., Liao W.B., Wang B.S., Zan Q.J. The History and Status of the Study on Mikania micrantha. Ecol. Sci. 2000;3:41–45. [Google Scholar]
- 4.Zhang L.Y., Ye W.H., Cao H.L., Feng H.L. Mikania micrantha H. B. K. in China-an overview. Weed Res. 2004;44:42–49. doi: 10.1111/j.1365-3180.2003.00371.x. [DOI] [Google Scholar]
- 5.Manrique V., Diaz R., Cuda J.P., Overholt W.A. Suitability of a New Plant Invader as a Target for Biological Control in Florida. Invasive Plant Sci. Manag. 2011;4:1–10. doi: 10.1614/IPSM-D-10-00040.1. [DOI] [Google Scholar]
- 6.Li R.Y., Lin G.Q. Occurrence and control of exotic pest Mikania micrantha. J. Agric. Disaster Catastrophol. 2012;2:11–13+77. [Google Scholar]
- 7.Day M.D., Kawi A., Kurika K., Dewhurst C.F., Waisale S., Saul-Maora J., Fidelis J., Bokosou J., Moxon J., Orapa W., et al. Mikania micrantha Kunth (Asteraceae) (Mile-a-Minute): Its Distribution and Physical and Socioeconomic Impacts in Papua New Guinea. Pac. Sci. 2012;66:213–223. doi: 10.2984/66.2.8. [DOI] [Google Scholar]
- 8.XiaoQing Z., Huang Z., Si H., QiJie Z. Analysis of ecological-economic loss caused by weed Mikania micrantha on Neilingding Island, Shenzhen, China. J. Trop. Subtrop. Bot. 2004;12:167–170. [Google Scholar]
- 9.Macanawai A.R., Day M.D., Tumaneng-Diete T., Adkins S.W. Impact of Mikania micrantha on crop production systems in Viti Levu, Fiji. Pak. J. Weed Sci. Res. 2012;18:357–365. [Google Scholar]
- 10.Macanawai A.R., Day M.D., Adkins S.W. Effects of Age, Length, and Pattern of Burial on Survival of Mikania micrantha Stem Sections. Pac. Sci. 2015;69:95–102. doi: 10.2984/69.1.7. [DOI] [Google Scholar]
- 11.Li L.Z., Huang H.J. Research progress on prevention and control of Mikania micrantha. J. Zhongkai Univ. Agric. Eng. 2018;31:66–71. [Google Scholar]
- 12.Day M.D., Riding N. Host specificity of Puccinia spegazzinii (Pucciniales: Pucciniaceae), a biological control agent for Mikania micrantha (Asteraceae) in Australia. Biocontrol Sci. Technol. 2018;29:19–27. doi: 10.1080/09583157.2018.1520807. [DOI] [Google Scholar]
- 13.Xu S.E., Gao Y.B., Ling X.P., Liu J.F., Yu H.B., Liang W.S. Occurrence, control and research progress of Mikania micrantha in Guangdong. Guangdong For. Sci. Technol. 2013;29:83–89. [Google Scholar]
- 14.Huang D.G., Zhou X.Y., Wang Y.J., Liang W.B. The chemical control of Mikania micrantha in the country park of Hong Kong. J. S. China Norm. Univ. Nat. Sci. Ed. 2007;1:109–114+131. [Google Scholar]
- 15.Xu G.F., Yue Y., Shen S.C., Guo J., Jing G.M., Zhang F.D. Zhang Y H. Evaluation of the Controlling Methods on Inhibiting the Secondary Invasion of Mikania micrantha H.B.K. J. Ecol. Environ. 2017;26:911–918. [Google Scholar]
- 16.Ling C.X., Liao Q.W., Zeng L.M. Review of research on Mikania micrantha. Guangxi For. Sci. 2003;2:60–65. doi: 10.19692/j.cnki.gfs.2003.02.002. [DOI] [Google Scholar]
- 17.Mo J.B. Occurrence status and control measures of Mikania micrantha. Jiangxi Agric. 2020;12:93–94. [Google Scholar]
- 18.Cheng R.P., Xu Q.H., Li X.C. Application of Brevipalpus phoenici to the Controlling of Mikania micransa. J. Cent. South Univ. 2003;23:89–93. [Google Scholar]
- 19.Ze S.Z., Su E.G., Yan Z.L. The control effect of Pachypeltis micranthus on Mikania micrantha. J. West China For. Sci. 2013;42:46–52. [Google Scholar]
- 20.Xu X.W., Ze S.Z., Yang B., Ji M. Research status and prospect of distribution, biological control and resource utilization of Mikania micrantha. TAS. 2014;34:75–84. [Google Scholar]
- 21.Fire A., Xu S.Q., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 22.Rosa C., Kuo Y., Wuriyanghan H., Falk B.W. RNA Interference Mechanisms and Applications in Plant Pathology. Annu. Rev. Phytopathol. 2018;56:581–610. doi: 10.1146/annurev-phyto-080417-050044. [DOI] [PubMed] [Google Scholar]
- 23.Lundgren J.G., Duan J.J. RNAi-Based Insecticidal Crops: Potential Effects on Nontarget Species. Bioscience. 2013;63:657–665. doi: 10.1525/bio.2013.63.8.8. [DOI] [Google Scholar]
- 24.Mao Y., Cai W., Wang J., Hong G., Tao X., Wang L., Huang Y., Chen X. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y., Park Y., Gao X., Zhang X., Yao J., Pang Y., Jiang H., Zhu K.Y. Cholinergic and non-cholinergic functions of two acetylcholinesterase genes revealed by gene-silencing in Tribolium castaneum. Sci. Rep. 2012;2:288. doi: 10.1038/srep00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau S., Schwarzacher T., Othman R.Y., Harikrishna J.A. dsRNA silencing of an R2R3-MYB transcription factor affects flower cell shape in a Dendrobium hybrid. BMC Plant Biol. 2015;15:194. doi: 10.1186/s12870-015-0577-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan S., Qian J., Cai C., Ma Z., Li J., Yin M., Ren B., Shen J. Spray method application of transdermal dsRNA delivery system for efficient gene silencing and pest control on soybean aphid Aphis glycines. J. Pest Sci. 2019;93:449–459. doi: 10.1007/s10340-019-01157-x. [DOI] [Google Scholar]
- 28.Majidiani S., PourAbad R.F., Laudani F., Campolo O., Zappalà L., Rahmani S., Mohammadi S.A., Palmeri V. RNAi in Tuta absoluta management: Effects of injection and root delivery of dsRNAs. J. Pest Sci. 2019;92:1409–1419. doi: 10.1007/s10340-019-01097-6. [DOI] [Google Scholar]
- 29.Kiselev K.V., Suprun A.R., Aleynova O.A., Ogneva Z.V., Dubrovina A.S. Physiological Conditions and dsRNA Application Approaches for Exogenously induced RNA Interference in Arabidopsis thaliana. Plants. 2021;10:264. doi: 10.3390/plants10020264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang L., Ding L., He B., Shen J., Xu Z., Yin M., Zhang X. Systemic gene silencing in plants triggered by fluorescent nanoparticle-delivered double-stranded RNA. Nanoscale. 2014;6:9965–9969. doi: 10.1039/C4NR03481C. [DOI] [PubMed] [Google Scholar]
- 31.Park M., Um T.Y., Jang G., Choi Y.D., Shin C. Targeted gene suppression through double-stranded RNA application using easy-to-use methods in Arabidopsis thaliana. Appl. Biol. Chem. 2022;65:4. doi: 10.1186/s13765-022-00675-0. [DOI] [Google Scholar]
- 32.Chen L., Su S.Q. Status of glyphosate-resistant weeds and implications for their management by RNA interference. World Pestic. 2013;35:30–33. [Google Scholar]
- 33.Mai J., Liao L., Ling R., Guo X., Lin J., Mo B., Chen W., Yu Y. Study on RNAi-based herbicide for Mikania micrantha. Synth. Syst. Biotechnol. 2021;6:437–445. doi: 10.1016/j.synbio.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dastidar M.G.S.A. ARF5/MONOPTEROS directly regulates miR390 expression in the Arabidopsis thaliana primary root meristem. Plant Direct. 2019;2:e001162019. doi: 10.1002/pld3.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q., Nan W., Zhang H.M. Research progress in the regulation of ARF5/MONOPTEROS (MP) Acta Bot. Boreali-Occident. Sin. 2016;36:197–203. [Google Scholar]
- 36.Hardtke C.S. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aida M., Vernoux T., Furutani M., Traas J., Tasaka M. Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development. 2002;129:3965–3974. doi: 10.1242/dev.129.17.3965. [DOI] [PubMed] [Google Scholar]
- 38.Bhatia N., Bozorg B., Larsson A., Ohno C., Jönsson H., Heisler M.G. Auxin Acts through MONOPTEROS to Regulate Plant Cell Polarity and Pattern Phyllotaxis. Curr. Biol. 2016;26:3202–3208. doi: 10.1016/j.cub.2016.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B., Yan J., Li W.H., Yin L.J., Li P., Yu H.X., Xing L.S., Cai M.L., Wang H.C., Zhao M.X., et al. Mikania micrantha genome provides insights into the molecular mechanism of rapid growth. Nat. Commun. 2020;11:340. doi: 10.1038/s41467-019-13926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamthan A., Chaudhuri A., Kamthan M., Datta A. Small RNAs in plants: Recent development and application for crop improvement. Front. Plant Sci. 2015;6:208. doi: 10.3389/fpls.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saurabh S., Vidyarthi A.S., Prasad D. RNA interference: Concept to reality in crop improvement. Planta. 2014;239:543–564. doi: 10.1007/s00425-013-2019-5. [DOI] [PubMed] [Google Scholar]
- 42.Borges F., Martienssen R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morozov S.Y., Solovyev A.G., Kalinina N.O., Taliansky M.E. Double-Stranded RNAs in Plant Protection Against Pathogenic Organisms and Viruses in Agriculture. Acta Naturae. 2019;11:13–21. doi: 10.32607/20758251-2019-11-4-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramegowda V., Mysore K.S., Senthil-Kumar M. Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front. Plant Sci. 2014;5:323. doi: 10.3389/fpls.2014.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh A., Gautam V., Singh S., Sarkar Das S., Verma S., Mishra V., Mukherjee S., Sarkar A.K. Plant small RNAs: Advancement in the understanding of biogenesis and role in plant development. Planta. 2018;248:545–558. doi: 10.1007/s00425-018-2927-5. [DOI] [PubMed] [Google Scholar]
- 46.Krogan N.T., Ckurshumova W., Marcos D., Caragea A.E., Berleth T. Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 2012;194:391–401. doi: 10.1111/j.1469-8137.2012.04064.x. [DOI] [PubMed] [Google Scholar]
- 47.Schuetz M., Fidanza M., Mattsson J. Identification of Auxin Response Factor-Encoding Genes Expressed in Distinct Phases of Leaf Vein Development and with Overlapping Functions in Leaf Formation. Plants. 2019;8:242. doi: 10.3390/plants8070242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berleth T., Jürgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development. 1993;118:575–587. doi: 10.1242/dev.118.2.575. [DOI] [Google Scholar]
- 49.Liu Y., Dong Z. The Function and Structure of Plant B3 Domain Transcription Factor. MPB. 2017;15:1868–1873. [Google Scholar]
- 50.Li Y., Han S., Qi Y. Advances in structure and function of auxin response factor in plants. J. Integr. Plant Biol. 2022;65:617–632. doi: 10.1111/jipb.13392. [DOI] [PubMed] [Google Scholar]
- 51.Das P.R., Sherif S.M. Application of Exogenous dsRNAs-induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 2020;11:946. doi: 10.3389/fpls.2020.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalakouras A., Jarausch W., Buchholz G., Bassler A., Braun M., Manthey T., Krczal G., Wassenegger M. Delivery of Hairpin RNAs and Small RNAs Into Woody and Herbaceous Plants by Trunk Injection and Petiole Absorption. Front. Plant Sci. 2018;9:1253. doi: 10.3389/fpls.2018.01253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yong J., Wu M., Zhang R., Bi S., Mann C.W.G., Mitter N., Carroll B.J., Xu Z.P. Clay nanoparticles efficiently deliver small interfering RNA to intact plant leaf cells. Plant Physiol. 2022;190:2187–2202. doi: 10.1093/plphys/kiac430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Y., Hu Y., Yan S., Zhou H., Song D., Yin M., Shen J. A polymer/detergent formulation improves dsRNA penetration through the body wall and RNAi-induced mortality in the soybean aphid Aphis glycines. Pest Manag. Sci. 2019;75:1993–1999. doi: 10.1002/ps.5313. [DOI] [PubMed] [Google Scholar]
- 55.Zhou C., Zhu X., Zhu F., Zhang H., Han Z., Yang R., Wang K. Effects of nanoparticle carriers on the RNAi efficiency in the small brown plant hopper, Laodelphax striatellus (Hemiptera: Delphacidae) Acta Entomol. Sin. 2021;64:1153–1160. [Google Scholar]
- 56.Guo Y., Fan Y., Teng Z., Wang L., Tan X., Wan F., Zhou H. Efficacy of RNA interference using nanocarrier-based transdermal dsRNA delivery system in the woolly apple aphid, Eriosoma lanigerum. Arch. Insect Biochem. Physiol. 2022;110:e21888. doi: 10.1002/arch.21888. [DOI] [PubMed] [Google Scholar]
- 57.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 58.Nei M., Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; New York, NY, USA: 2000. pp. 3–16. [Google Scholar]
- 59.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 65.Sims K., Abedi-Samakush F., Szulc N., Macias H.M.G., Mattsson J. OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development. Int. J. Mol. Sci. 2021;22:4089. doi: 10.3390/ijms22084089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu J., Wang F., Cheng L., Kong F., Peng Z., Liu S., Yu X., Lu G. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 2011;30:2059–2073. doi: 10.1007/s00299-011-1113-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article and Supplementary Materials.