Abstract
Phenylalanine (Phe) is a potentially limiting amino acid for lactating cows. The mechanism by which Phe regulates milk protein synthesis remains unclear. The present study elucidates the mechanisms by which phenylalanine affects milk protein synthesis, amino acid utilization, and related signaling pathways in bovine mammary epithelial cells (BMECs). The BMECs were treated with five concentrations (0, 0.22, 0.44, 0.88, 1.76 mM, and serum free). Rapamycin inhibitors and RNA interference (RNAi) were used to inhibit the phosphorylation of the mammalian target of rapamycin (mTOR) signaling pathway and the expression of relevant amino acid transporters, respectively. The results showed that 4×Phe (0.88 mM) significantly increased (p < 0.05) both the mRNA and protein expression of α-casein (CSN1S1), β-casein (CSN2), and κ-casein (CSN3), as well as L-type amino acid transporter-1 (LAT1) mRNA expression. Protein expression and modification assays of mTOR-related proteins showed that 4×Phe could increase (p < 0.05) the expression of α-casein and eukaryotic initiation factor 4E-binding protein-1 (4EBP1) and tended to increase the expression of ribosomal protein S6 protein kinase (S6K1, p = 0.054). The general control nonderepressible 2 (GCN2) signaling pathway factor, eukaryotic initiation factor 2 (eIF2α), was downregulated by 4×Phe treatment (p < 0.05). The rapamycin inhibition test showed that Phe regulated casein synthesis via the mTOR signaling pathway. RNAi experiments showed that LAT1 mediated the entry of Phe into cells. Moreover, 4×Phe treatment tended to decrease (0.05 < p < 0.10) the consumption of valine, leucine, histidine, tyrosine, cysteine, alanine, asparagine, and serine in the medium. Collectively, phenylalanine enhanced α-casein synthesis by regulating the phosphorylation of 4EBP1 and eIF2α and promoting the formation of the mTOR-centered casein translation initiation complex.
Keywords: phenylalanine, milk protein synthesis, amino acid utilization, bovine mammary epithelial cells, mTOR
1. Introduction
Milk contains essential nutrients, especially high-quality protein, and can fight both viral infection and malnutrition in mothers, infants, children, adolescents, and elderly individuals [1,2,3]. The synthesis of milk protein in the mammary gland requires an adequate supply of energy (adenosine triphosphate) and amino acids (AAs) [4,5]. Lysine (Lys) and methionine (Met) play major limiting roles in dairy production [6], while the next potentially limiting amino acids are phenylalanine (Phe), histidine (His), and arginine (Arg) [7,8]. These AAs can also activate the translational machinery via the signaling pathway and affect the expression of milk protein genes [9,10]. The mammalian target of rapamycin (mTOR) pathway is closely related to milk protein synthesis and provides alternatives for the AA-mediated regulation of milk protein synthesis in bovine mammary epithelial cells (BMECs) [11,12]. The general control nonderepressible 2 (GCN2) signaling pathway is another signaling pathway that senses abundant AA and regulates protein synthesis [13], which senses the absence of one or more amino acids by direct binding to uncharged cognate transport RNAs [14,15]. Eukaryotic initiation factor 2α (eIF2α) is an important downstream factor, and its phosphorylation level directly reflects the activity of the signaling pathway [16]. The above signaling pathways are important for the amino acid regulation of milk protein synthesis [17], but whether Phe affects these pathways has not been reported.
Phe has been shown to affect milk protein production [18]. It has been reported that the mammary gland of dairy cows has a specific requirement for Phe, which increases milk protein production [19]. The supplemented Phe was extracted by the mammary gland in amounts equal to its secretion in milk protein [10,20], which indicates that Phe is almost exclusively utilized to support milk protein production. However, the mechanism by which Phe regulates protein synthesis and AA-mediated cellular signaling in BMECs is still unclear. Based on our previous study, the Phe regulates the synthesis of digestive protein via the mTOR signaling pathway in pancreatic acinar cells [21]. We hypothesized that Phe affects the potential cellular signals to regulate α-casein synthesis. The objective of the present study was to assess the effect of L-Phe on the expression of amino acid transporters as well as on the metabolism of L-Phe in bovine mammary epithelial cells, which produce casein, in an attempt to further elucidate the mechanism by which Phe regulates casein synthesis.
2. Results
2.1. BMEC Viability and PAH Activity
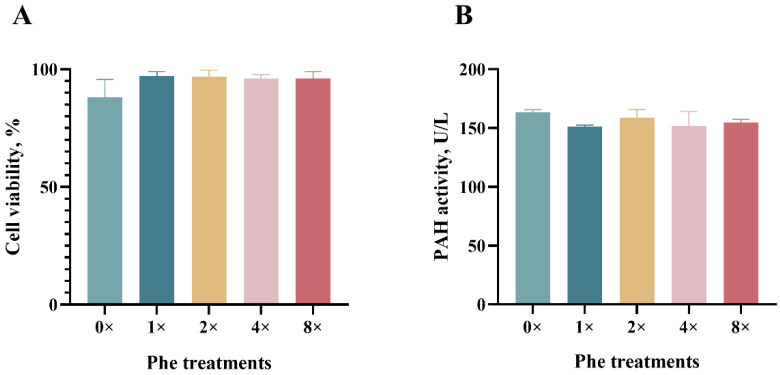
There was no significant difference in cell viability among the treatment media (p > 0.05, Figure 1A). The PAH activity did not differ among the treatment groups (p > 0.05, Figure 1B).
Figure 1.
Cell viability and intracellular phenylalanine hydroxylase activity in bovine epithelial cells treated with various concentrations of phenylalanine. (A) represents the cell viability. (B) represents intracellular phenylalanine hydroxylase (PHA) activity. The error bars represent the SDs (n = 3).
2.2. Casein mRNA Expression and Protein Synthesis
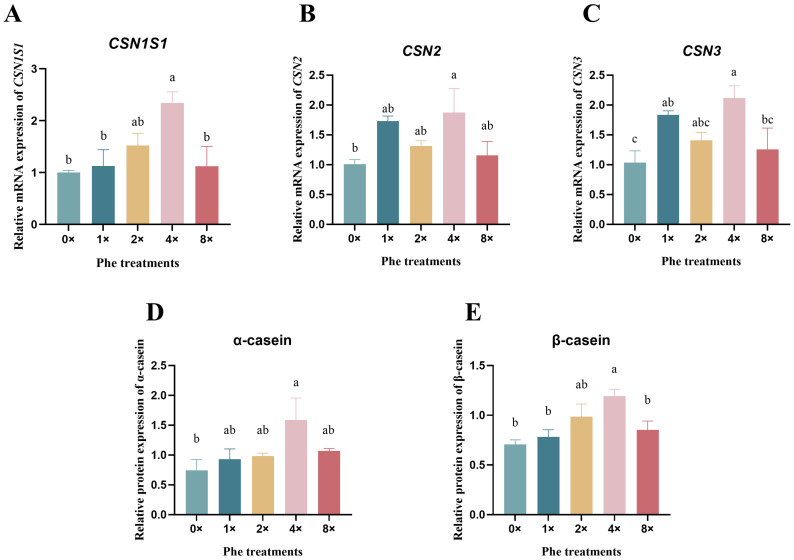
The mRNA expression levels of CSN1S1 (Figure 2A), CSN2 (Figure 2B), and CSN3 (Figure 2C) in the 4×Phe group were the greatest among all the treatments (p < 0.05). Compared with those in the 0×Phe group, both the α-casein (Figure 2D) and β-casein (Figure 2E) protein expression levels in the 4×Phe group increased (p < 0.05).
Figure 2.
The relative expression of casein mRNA and proteins in bovine mammary epithelial cells treated with various concentrations of phenylalanine. (A–C) represent the mRNA expression of CSN1S1, CSN2, and CSN3, respectively. (D,E) represent the protein expression of α-casein and β-casein. The error bars represent the SDs (n = 3). Different letters indicate significant differences (p < 0.05) (CSN1S1, α-casein; CSN2, β-casein; CSN3, κ-casein).
2.3. The Expression of Amino Acid Transporters
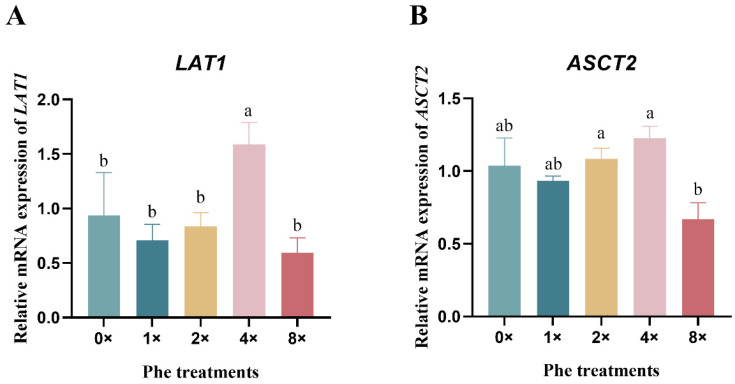
The LAT1 mRNA expression in the 4×Phe group increased (Figure 3A, p < 0.05). Compared with the 2×Phe and 4×Phe groups, the 8×Phe group exhibited reduced expression of ASCT2 mRNA (Figure 3B, p < 0.05).
Figure 3.
The relative expression of amino acid transporter mRNAs in bovine mammary epithelial cells treated with various concentrations of phenylalanine. (A) represents LAT1, and (B) represents ASCT2. The error bars represent the SDs (n = 3). Different letters indicate significant differences (p < 0.05) (LAT1, L-type amino acid transporter 1; ASCT2, sodium-dependent neutral amino acid transporter type 2).
2.4. Consumption of Amino Acids by Mammary Epithelial Cells
The results showed that the 4×Phe treatment tended to decrease (0.05< p < 0.10, Table 1) the consumption of Val, Leu, His, Tyr, Cys, Ala, Asp, and Ser. There was no significant difference in the other AAs among the treatments (p > 0.05).
Table 1.
The consumption of 16 types of amino acids in the medium of bovine mammary epithelial cells.
| Item/μg | Initial Mass | 0×Phe | 1×Phe | 2×Phe | 4×Phe | 8×Phe | SEM 1 | p Value |
|---|---|---|---|---|---|---|---|---|
| Lys | 273.8 | 144.0 | 142.5 | 143.9 | 144.3 | 142.2 | 0.57 | 0.126 |
| Phe | - | 0.0 | 69.1 | 104.8 | 278.8 | 155.9 | 0.41 | 0.007 |
| Thr | 160.4 | 105.7 | 104.9 | 105.6 | 104.2 | 104.6 | 0.34 | 0.108 |
| Met | 51.72 | 34.1 | 33.9 | 34.1 | 34.4 | 33.9 | 0.09 | 0.105 |
| Val | 158.6 | 104.5 | 103.9 | 104.4 | 102.2 | 103.7 | 0.30 | 0.092 |
| Leu | 177.2 | 116.4 | 116.2 | 116.7 | 114.0 | 115.9 | 0.34 | 0.069 |
| Ile | 163.4 | 108.9 | 108.8 | 108.9 | 109.4 | 108.5 | 0.07 | 0.443 |
| His | 94.4 | 46.2 | 46.1 | 46.2 | 45.5 | 46.0 | 0.11 | 0.068 |
| Arg | 442.5 | 241.0 | 238.5 | 240.6 | 239.7 | 237.9 | 0.86 | 0.118 |
| Gly | 53.55 | 37.0 | 36.7 | 37.0 | 37.0 | 36.6 | 0.12 | 0.138 |
| Tyr | 167.4 | 68.5 | 68.1 | 68.5 | 66.6 | 67.9 | 0.26 | 0.097 |
| Cys | 93.9 | 47.5 | 46.9 | 47.29 | 45.85 | 46.11 | 0.23 | 0.052 |
| Ala | 13.5 | 8.7 | 8.7 | 8.7 | 8.5 | 8.6 | 0.04 | 0.084 |
| Asp | 20.0 | 12.9 | 12.8 | 12.9 | 12.3 | 12.7 | 0.09 | 0.096 |
| Glu | 22.1 | 14.4 | 14.4 | 14.5 | 14.2 | 14.3 | 0.06 | 0.634 |
| Ser | 78.8 | 51.8 | 51.3 | 51.7 | 50.4 | 51.2 | 0.19 | 0.097 |
1 SEM, standard error of the mean.
2.5. Mesurement of Phosphorylation Levels of mTOR and GCN2 Signaling Pathway Markers
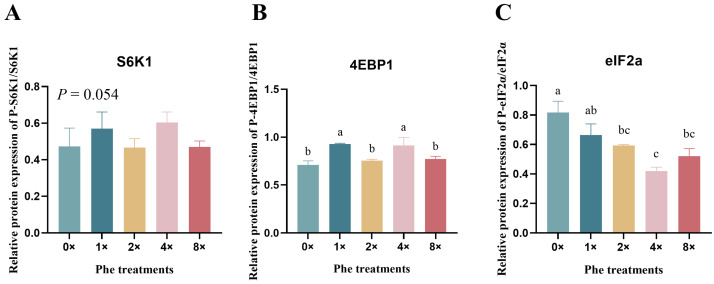
We detected the ratio of phosphorylated to total mTOR and GCN2 signaling pathway factors in BMECs treated with various concentrations of Phe (Figure 4). The phosphorylation level of S6K1 tended to increase in the 4×Phe group (Figure 4A, p = 0.054). The expression of 4EBP1 was upregulated by 4×Phe and 1×Phe compared with the other treatments (Figure 4B, p < 0.05). In contrast, the phosphorylation of eIF2α was significantly lower in the 4×Phe group than in the 0×Phe group (Figure 4C, p < 0.05).
Figure 4.
The phosphorylation levels of mTOR and GCN2 signaling pathway factors in bovine mammary epithelial cells treated with various concentrations of phenylalanine. (A–C) represent the phosphorylation levels of S6K, 4EBP1, and eIF2α, respectively. The error bars represent the SDs (n = 3). Different letters indicate significant differences (p < 0.05) (S6K1, ribosomal protein S6 protein kinase; 4EBP1, eukaryotic initiation factor 4E-binding protein 1; eIF2α, eukaryotic initiation factor 2α).
2.6. Relationships Among Casein Synthesis, Signaling Protein Phosphorylation or Expression, and AA Transporter
The results showed that α-casein tended to be negatively correlated with S6K1 phosphorylation (p < 0.05), while β-casein was positively correlated with eIF2 phosphorylation (p < 0.05, Table 2).
Table 2.
Correlations among casein synthesis, signaling protein phosphorylation or expression, and amino acid transporter.
| Dependent Variable | Independent Variable | Pearson Correlation | p Value |
|---|---|---|---|
| α-casein | β-casein | 0.36 | 0.43 |
| S6K1 | −0.69 | 0.08 | |
| 4EBP1 | 0.18 | 0.70 | |
| eIF2a | 0.51 | 0.24 | |
| ASCT2 | −0.24 | 0.61 | |
| LAT1 | −0.50 | 0.25 | |
| β-casein | S6K1 | −0.45 | 0.31 |
| 4EBP1 | 0.16 | 0.72 | |
| eIF2a | 0.75 | 0.05 | |
| ASCT2 | −0.01 | 0.98 | |
| LAT1 | 0.09 | 0.85 | |
| S6K1 | 4EBP1 | −0.06 | 0.90 |
| eIF2a | −0.69 | 0.08 | |
| ASCT2 | −0.21 | 0.65 | |
| LAT1 | 0.20 | 0.67 | |
| 4EBP1 | eIF2a | −0.12 | 0.79 |
| ASCT2 | −0.65 | 0.11 | |
| LAT1 | −0.85 | 0.02 | |
| eIF2a | ASCT2 | 0.30 | 0.52 |
| LAT1 | 0.11 | 0.81 | |
| ASCT2 | LAT1 | 0.68 | 0.09 |
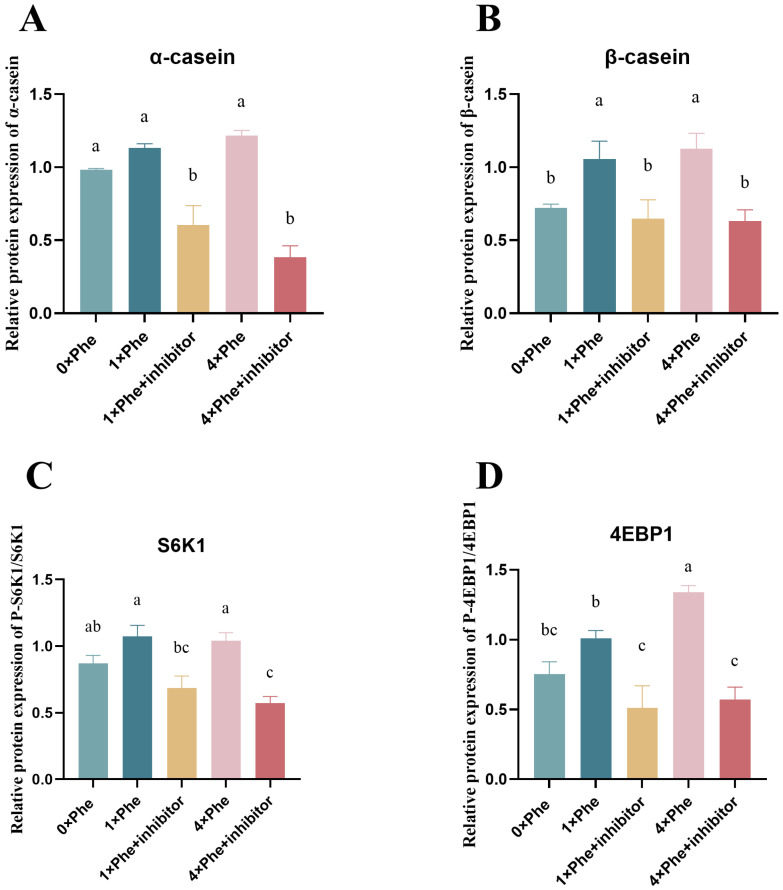
2.7. Rapamycin Inhibitor Expreriment
The effects of inhibition by mTOR phosphorylation on casein synthesis and phosphorylation of key downstream factors are shown in Figure 5. When treated with the rapamycin inhibitor, the protein expression of α- and β-casein was significantly reduced (Figure 5A,B, p < 0.05). The phosphorylation level of S6K1 and 4EBP1 was significantly decreased in the 1×Phe + inhibitor and 4×Phe + inhibitor treatment groups (Figure 5C,D, p < 0.05).
Figure 5.
Inhibition of mTOR downregulated S6K1 and 4EBP1 phosphorylation and casein synthesis. (A,B) represent the protein synthesis of α-casein and β-casein, respectively. (C,D) show the phosphorylation level of S6K1 and 4EBP1. The error bars represent the SDs (n = 3). Different letters indicate significant differences (p < 0.05) (S6K1, ribosomal protein S6 protein kinase; 4EBP1, eukaryotic initiation factor 4E-binding protein 1).
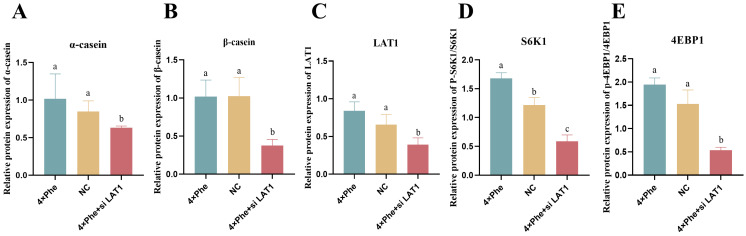
2.8. Inhibition of LAT1 Decreased Casein Synthesis and mTOR Signaling Pathway Phosphorylation
When the cells were treated with siLAT1, the protein abundances of α-casein and β-casein were significantly reduced (Figure 6A,B, p < 0.05). The protein expression of LAT1 decreased significantly in the 4×Phe + siLAT1 group (Figure 6C, p < 0.05). The phosphorylation levels of S6K1 and 4EBP1 were significantly decreased (Figure 6D,E, p < 0.05) after LAT1 inhibition.
Figure 6.
Inhibition of LAT1 downregulated S6K1 and 4EBP1 phosphorylation and casein synthesis. (A,B) represent the synthesis of α-casein and β-casein, respectively. (C) represents the expression of LAT1. (D,E) show the phosphorylation level of S6K1 and 4EBP1, respectively. The error bars represent the SDs (n = 3). Different letters indicate significant differences (p < 0.05) (LAT1, L-type amino acid transporter 1; S6K1, ribosomal protein S6 protein kinase; 4EBP1, eukaryotic initiation factor 4E-binding protein 1).
3. Discussion
3.1. Phe Affects Casein Synthesis
The current work elucidates the mechanisms by which phenylalanine affects milk protein synthesis, amino acid utilization, and related signaling pathways in vitro using BMECs. The cell viability was determined in order to measure the adverse effects of the medium on cell growth [22]. In our study, the viability of BMECs was not affected by the addition of Phe, which was consistent with the findings of the previous study [23].
Understanding the regulatory effects of single AA on milk protein synthesis is of great significance in improving the lactating protein and AA requirement models [24]. Lysine and methionine are considered the first and second limiting AAs in corn–soybean meal-based diets [25], while in grass silage-based diets, histidine tends to be the first limiting AA [26]. Therefore, many studies have focused on these three types of AAs on dairy cows [27,28]. Earlier studies showed that branch-chain amino acids (BCAAs) can regulate protein synthesis in dairy cow production [29,30,31]. Moreover, feeding rumen-protected arginine to dairy cows could increase the average daily milk yield [32]. It was also reported that arginine activates the synthesis of milk in sows [33]. The aromatic amino acids Phe and Trp also play important regulatory roles in protein synthesis [34]. The evidence showed that the digestive flow of Phe, on average, was lower than estimated [35]. Our previous study reported that Phe could enhance protein synthesis via the translation pathway in dairy calf pancreatic cells [21]. A recent study showed that when dairy cows got an abomasal infusion of ten EAAs without Phe, milk protein production was lower than that in the Phe-infused group [36], which indicated that Phe may be an important amino acid component that affects milk protein synthesis in dairy cows. The synthesis of milk protein in the mammary gland has specific requirements for Phe [19]; the evidence showed that the absence of Phe in AA infusions negatively affects milk and milk protein yield [8]. These results are consistent with our research, in which the Phe deprivation group negatively affected the gene expression involved in protein synthesis. However, previous research reported that when high milk-yield dairy cows received 7.5 g/cow/day of intestinally absorbable Phe, it was insufficient to support increased milk production because it was primarily used to support an increased body condition score [37]. Our study revealed a significant increase in α-casein synthesis in BMECs after treatment with 0.88 mM Phe in the medium. This result can be explained by differences between individual variations in animals and controlled laboratory environments.
3.2. Phe Affects AA Transporter and Cellular Metabolism
The AA uptake by mammary epithelial cells is conducted by different transporters located on the basolateral side of the plasma membrane in the mammary glands [38]. LAT1 (SLC7A5) is a sodium-independent amino acid exchanger that forms a complex with a larger surface glycoprotein, 4F2hc (SLC3A2), to transport several neutral, branched L-type amino acids, such as phenylalanine, tyrosine, and leucine [39]. LAT1 is the most abundant L-type AA transporter in the mammary gland [40]. In addition, LAT1 is involved in the transportation of EAAs, except for arginine, lysine, and threonine [41]. Previous research reported that L-Phe is one of several compounds that possesses a high LAT1 susceptibility, because the phenyl ring of L-phenylalanine provides a high susceptibility to the LAT1 protein [42]. Our results showed that LAT1 expression was positively influenced by the Phe concentration in the medium, which was consistent with the above findings. LAT1 is a key target for mTOR signaling pathway activation [43,44], and we also detected the phosphorylation of mTOR factors, which confirmed that they were positively correlated with LAT1 expression. ASCT2 can transport BCAAs, threonine, and some nonessential AAs to the cell interior [45]. ASCT2 expression showed no obvious change in the present study. This observation is consistent with a former study showing that Phe may not regulate the transport of other AAs into cells [23]. In addition, the high concentration (8×) of phenylalanine in the medium resulted in a decrease in the relative mRNA expression of LAT1 and ASCT2, probably because the addition of Phe inhibited the activity of amino acid transport carriers via a negative feedback mechanism [45].
The total absorption of individual amino acids and the uptake patterns in mammary epithelial cells are affected by the intracellular and extracellular amino acid concentrations and the number or activity of transporters and mammary cells [46,47]. In the present study, the uptake of Phe by mammary epithelial cells increased gradually as the Phe concentration increased, while the consumption of Val, Leu, His, Tyr, Cys, Ala, Asp, and Ser tended to decrease in the 4×Phe treatment group. Combined with the increase in casein synthesis, these results indicated that the AA utilization efficiency was greater in media supplemented with 0.88 mM Phe. Variations in concentrations of extracellular AAs can regulate the transcription, translation, posttranscriptional modifications, and epigenetic regulation of genes and proteins [48]. The results of the lower level of eIF2a phosphorylation indicated that the AA composition of the 4×Phe group was more balanced, which is also the reason for the above results.
PAH is the key rate-limiting enzyme for the conversion of Phe to Tyr [49], which is activated by Phe and then metabolizes excess phenylalanine in cells [50]. The PAH concentration in the mammary epithelial cells in this experiment did not significantly differ among the Phe treatments, especially in the group with a high Phe concentration. We speculated that the additional Phe was mainly involved in the synthesis of milk proteins. Studies of mammalian mammary tissues during lactation have shown that the uptake of Phe is usually balanced with its secretion in milk proteins [51]. When the Phe concentration is elevated, the high substrate concentration induces enzyme activation and promotes Phe catabolism, thereby maintaining the balance of Phe in the blood and providing the body with the required Tyr [49]. A previous publication reported that lactating goat mammary glands convert only 5–9% of the extracted Phe to Tyr, according to a stable isotope perspective [52]. These results suggest that PAH induces little conversion of Phe to Tyr in BMECs.
3.3. Phe Regulates Signaling Pathways Related to Casein Synthesis
We further measured the expression of mTORC1 signaling molecules and found that increasing the addition of Phe had a significant effect on the phosphorylation levels of 4EBP1 and S6K1. The mTORC1 signaling pathway controls the main protein synthesis process in mammalian cells [17,53]. When activated by AAs, mTOR in turn catalyzes the phosphorylation of S6K1 and 4EBP1 [54]. The S6K1 protein is a biomarker of mTOR and reflects the phosphorylation of the mTORC1 signaling pathway [55,56]. Phe regulates nucleocytosolic 26S proteasome translocation via mTOR [57], which indicates that Phe is closely related to the mTORC1 signaling pathway. Phe has been reported to regulate the transcription levels of fish target of rapamycin and S6K1 [58,59]. A recent study reported that 1.2 mM Phe in a medium could increase the relative mTOR or S6K1 expression and protein synthesis compared with those in the Phe-deprived group [23], which confirmed our results. A previous study reported that Trp, Phe, and Met had no effect on S6K1 phosphorylation in murine mammary epithelial cells [60]. In the present study, the addition of rapamycin resulted in a significant reduction in the abundance of α-casein and S6k1, indicating that Phe-induced α-casein synthesis via the mTOR pathway was inhibited. However, it has also been shown that the single use of rapamycin inhibitors did not completely inhibit mTORC1 activity and induced the feedback activation of its upstream signaling pathway [61]. Nevertheless, protein synthesis is a highly complex process that involves nutrients, energy status, and hormones [62]. The findings of the current study may help increase our understanding of the roles of Phe in casein synthesis in bovine mammary in vitro.
The GCN2 signaling pathway is sensitive to AA abundance in mammalian cells, which is activated by uncharged tRNA [63], and its biomarker is a downstream factor, eIF2α. GCN2 is activated when an essential amino acid is scarce or unbalanced and phosphorylates the α subunit of eIF2α [64]. The phosphorylation level of eIF2α negatively affects protein synthesis in mammalian cells [65]. In the present study, the phosphorylation level of eIF2α was highest in the 0×Phe group, indicating that the uncharged tRNA of Phe activated the GCN2 signaling pathway. The opposite was true in the 4×Phe group, which indicated that the balance of AA in the medium of the treatment group was more conducive to the synthesis of milk protein. The results of the casein synthesis confirmed these results.
4. Materials and Methods
4.1. Ethics Statement
The animal experiments in the present study were approved by the Institutional Animal Care and Use Committee at the Institute for Ruminant Research of Lanzhou University (CY-20211101).
4.2. Materials
In the present study, penicillin, hydrocortisone, L-Phe, and collagenases were bought from Sigma (Sigma, St. Louis, MO, USA). The fetal bovine serum (FBS) and Dulbecco’s Modified Eagle Medium/Nutrient Mixture F12 (DMEM/F12) were purchased from Invitrogen Company (Invitrogen, Carlsbad, CA, USA). qPCR reagents were purchased from Takara (Takara, Dalian, China). The primary mammary epithelial cells of dairy cows were used in this study.
4.3. Cell Culture and Treatment
Three dairy cattle (body weight 724.8 ± 20.88 kg; average lactation parity 3; mean ± SEM) at mid-lactation (lactation days 249 ± 19 d) were used for the BMEC isolation. After the cow was obtained, a 1 cm incision was made with a scalpel in the milking area, and the fascia tissue was stripped, the mammary tissue was exposed, and a small piece (0.5 cm3) of mammary tissue was quickly taken with scissors; the tissue was sutured after disinfection. The cells were isolated according to the method of a previous publication using the trypsin digestion technique [66]. Briefly, the mammary gland tissues were minced into 1 mm3 cubes and then digested in trypsin (0.25%) at 37 °C for 30 min and subsequently in collagenase I and collagenase II for 4 h at 37 °C.
Bovine mammary epithelial cells (BMECs) were cultured in DMEM/F12 (Cat No. SH30023.01, HyClone, Logan, UT, USA) supplemented with 10% FBS, 100 U/mL penicillin, 0.1 mg/mL streptomycin, and lactating hormones (5 μg/mL insulin, 1 μg/mL prolactin, and 1 μg/mL hydrocortisone) in a humidified 5% carbon dioxide incubator at 37 °C. BMECs purified from passages 5–10 were used for experimental assays. The scanning electron microscope image of mammary epithelial cells of dairy cow were shown in Figure S1.
When the cells were 80% confluent, they were cultured in 6-well plates (1 × 106 cells/well). To observe the effects of Phe on cells, the culture medium was changed to DMEM/F12 medium without FBS. The cells were divided into 5 groups: stripped Phe (0 mM), 1×Phe (0.22 mM, the normal plasma phenylalanine concentration of dairy cows), 2×Phe (0.44 mM), 4×Phe (0.88 mM), and 8×Phe (1.76 mM). After 24 h of incubation, the cells were harvested for further analysis. The experiment was repeated three times on three different days, and six technical replications per group were used on each day, so each experiment had a total of three replicates.
To explore whether mTORC1 signaling participated in the regulation of Phe, the cells were grown in 6-well plates (2 × 105 cells/well) to ~90% confluence and then incubated with 100 nM rapamycin or vehicle [0.02% (vol/vol) dimethyl sulfoxide]. After a 24 h incubation, the cells were collected for further analysis.
4.4. Cell Viability Assay
Cell viability was measured using the Cell Counting Kit-8 (CCK8, Cat No. K009-100, Zeta Life, San Francisco, CA, USA) with an Epoch Microplate Spectrophotometer (SynergyTM HTX, BioTek, Winooski, VT, USA), which referred to the previous study [67] and manufacturer’s protocol. Briefly, the procedure was divided into 4 steps: standard curve preparation, cell activity detection, and cell proliferation–toxicity detection and calculation.
4.5. Determination of Phenylalanine Hydroxylase Activity
The activity of phenylalanine hydroxylase (PAH) was measured using a commercial kit (ml060412, Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China) according to the manufacturer’s instructions. An Epoch Microplate Spectrophotometer (SynergyTM HTX, BioTek, Winooski, VT, USA) was used to detect the PAH activity. Briefly, the procedure included sample dilution, mixing, incubation (37 °C, 30 min), washing, enzymatic reaction, rewashing, color development, and determination.
4.6. Gene Expression by Real-Time Quantitative PCR (RT-qPCR)
The methods of total RNA extraction and RT-qPCR were performed according to the previous studies of our research group [68]. The total RNA of the cell sample was extracted using TRIzol reagent (Cat No. T9108, Takara, Osaka, Japan). The RNA quality and quantity were measured using a NanoDrop 1000® Spectrophotometer (Thermo Scientific, Waltham, MA, USA). The cDNA was synthesized using Evo M-MLV RT Premix (Cat No. 11706, Accurate Biology, Changsha, China). The RT-qPCR experiment was performed in a 96-well microwell plate using a CFX96 system (Bio-Rad, Herculus, CA, USA). The reactions were performed with a SYBR® Green Premix Pro Taq HS qPCR Kit (Cat No. 11701, Accurate Biology, Changsha, China), with β-actin, GAPDH, and 18S rRNA used as internal controls. The target genes included ASCT2 (sodium-dependent neutral amino acid transporter type 2), LAT1 (L-type amino acid transporter 1), CSN1S1 (α-casein), CSN2 (β-casein), and CSN3 (κ-casein), whose primers are shown in Table S1. The running program is as follows: 1 cycle of 95 °C for 30 s plus 40 cycles of amplification at 95 °C for 5 s and 60 °C for 30 s and after the melting curve of an additional 15 s at 95 °C, 1 min at 60 °C and 15 s at 95 °C.
4.7. Protein Expression and Modification Analysis
The methods of protein expression were according to our previous study [69]. Total cellular protein was extracted using RIPA lysis buffer (Cat No. R0020, Solabio, Beijing, China) containing phosphatase and protease inhibitors (Roche, Mannheim, Germany). Briefly, the protease and phosphatase inhibitors were mixed with RIPA solution and then added to the cell culture dish after the culture medium was removed and cleaned. The cells were scraped off with a scraper after being shaken on the ice to break the cells and were then transferred to a centrifuge tube for centrifugation. The supernatant was collected as the total protein of the cells. The protein concentration was measured using a PierceTM BCA assay kit (Thermo Fisher Scientific Inc., Rockford, IL, USA).
Equal amounts of protein extracts (20 μg protein each) were separated on 6–15% SDS-polyacrylamide gels (Bio-Rad, Richmond, CA, USA) and then transferred onto PVDF membranes (Cat No. ISEQ00010, 0.45 μm, Millipore, Bedford, MA, USA) in Tri-glycine buffer containing 20% methanol. The membranes were blocked and immunoblotted with a 1:1000 dilution of primary antibodies against β-tubulin, α-casein, β-casein, p70S6K, P-p70S6K, 4EBP1, P-4EBP1, eIF2α, and P-eIF2α. The target proteins were measured by a goat anti-rabbit IgG or anti-mouse IgG-conjugated secondary antibody (1:3000) with chemiluminescence (ECL) detection reagents (Bio-Rad, Hercules, CA, USA). The antibody information is referred to in Table S2. The Invitrogen iBright Imaging Systems (ThermoFisher Scientific, USA) and ImageJ (https://imagej.net/ij/) were used to detect the protein quantification.
4.8. Amino Acid Analysis in Culture Media
The methods of AA analysis were performed according to our previous study [70]. The sixteen AA concentrations in the medium were measured using an AA analyzer (A300 Advanced, MembraPure GmbH, Berlin, Germany). Briefly, the samples were made to react with sulfonyl salicylic acid and then centrifuged, diluted, and filtrated. The AA consumption was calculated as the amount of AA in the media before incubating the cells minus that after incubation.
4.9. Statistical Analyses
All data are presented as the means ± standard errors of the means (SEMs) from three independent experiments. The GLM procedure of the Statistical Analysis System 9.2 (SAS Inst. Inc. Cary, NC, USA) was used to perform all of the statistical analyses. The statistical model was as follows: Yi = μ + Ai + Bi, where Yi, μ, Ai, and Bi represent the dependent variable, overall mean, Phe treatment effect, and error term, respectively. Duncan’s test was performed for multiple comparisons of means. A significant difference was defined as p ≤ 0.05, and trends were defined as 0.05 < p ≤ 0.10. The relative expression of target gene mRNAs was calculated using the 2−ΔΔCT method, and the final result was calculated based on the geometric average of these three internal reference genes. The total protein expression levels were calculated as the ratio of the band intensity of β-actin or β-tubulin.
5. Conclusions
Overall, the present study suggested that phenylalanine regulates α-casein synthesis by promoting the LAT1–mTOR pathway in BMECs.
Abbreviations
Phenylalanine, Phe; bovine mammary epithelial cells, BMECs; the mammalian target of rapamycin, mTOR; α-casein, CSN1S1; β-casein, CSN2; κ-casein, CSN3; L-type amino acid transporter-1, LAT1; eukaryotic initiation factor 4E-binding protein-1, 4EBP1; eukaryotic initiation factor 2, eIF2α; amino acids, AAs; lysine, Lys; methionine, Met; isoleucine, Ile; threonine, Thr; histidine, His; arginine, Arg; chemiluminescence, ECL; phenylalanine hydroxylase, PAH; fetal bovine serum, FBS; Dulbecco’s Modified Eagle Medium/Nutrient Mixture F12, DMEM F12.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms252313135/s1.
Author Contributions
L.G. was involved in investigation, methodology, funding acquisition, writing—original draft, formal analysis, and data curation; C.Z. was involved in investigation, methodology, and writing—original draft; J.C. was involved in investigation, methodology, and writing—original draft; R.D. was involved in investigation, methodology, writing—reviewing and editing; F.L. was involved in data curation, supervision, and writing—reviewing and editing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
In the present study, the animal experiment was approved by the Institutional Animal Care and Use Committee and carried out strictly in compliance with the guidelines for the care and use of experimental animals at Lanzhou University (CY-20211101).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Material.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
Funding for this work was from the Science and Technology Program of Gansu Province [22YF7WA011, 23JRRA1078] and the Scientific Research Startup Foundation of Lanzhou University [561119219].
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Givens D.I. MILK Symposium review: The importance of milk and dairy foods in the diets of infants, adolescents, pregnant women, adults, and the elderly. J. Dairy Sci. 2020;103:9681–9699. doi: 10.3168/jds.2020-18296. [DOI] [PubMed] [Google Scholar]
- 2.Prasad A., Kothari N. Cow products: Boon to human health and food security. Trop. Anim. Health Prod. 2021;54:12. doi: 10.1007/s11250-021-03014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nili H., Bouzari M., Attaran H.R., Ghalegolab N., Rabani M., Mahmoudian A. Hyper-Immune Bovine Milk as an Immunological and Nutritional Supplement for COVID-19. Front. Nutr. 2022;9:868964. doi: 10.3389/fnut.2022.868964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pszczolkowski V.L., Halderson S.J., Meyer E.J., Lin A., Apelo S.I.A. Pharmacologic inhibition of mTORC1 mimics dietary protein restriction in a mouse model of lactation. J. Anim. Sci. Biotechnol. 2020;11:67. doi: 10.1186/s40104-020-00470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gross J.J. Limiting factors for milk production in dairy cows: Perspectives from physiology and nutrition. J. Anim. Sci. 2022;100:skac044. doi: 10.1093/jas/skac044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armentano L.E., Swain S.M., Ducharme G.A. Lactation response to ruminally protected methionine and lysine at two amounts of ruminally available nitrogen. J. Dairy Sci. 1993;76:2963–2969. doi: 10.3168/jds.S0022-0302(93)77635-3. [DOI] [PubMed] [Google Scholar]
- 7.Iroshan I.H., Lapierre H., Doepel L. The Effect of a Limited Supply of Phenylalanine, Threonine, and Tryptophan on Milk Yield and Composition. Wageningen Academic Publishers; Wageningen, The Netherlands: 2013. [DOI] [Google Scholar]
- 8.Seleem M.S., Wu Z.H., Xing C.Q., Zhang Y., Hanigan M.D., Bu D.P. Effects of rumen-encapsulated methionine and lysine supplementation and low dietary protein on nitrogen efficiency and lactation performance of dairy cows. J. Dairy Sci. 2024;107:2087–2098. doi: 10.3168/jds.2023-23404. [DOI] [PubMed] [Google Scholar]
- 9.Bionaz M., Hurley W., Loor J. Milk protein synthesis in the lactating mammary gland: Insights from transcriptomics analyses. Milk Protein. 2012;11:285–324. doi: 10.5772/2933. [DOI] [Google Scholar]
- 10.Wu Z., Heng J., Tian M., Song H., Chen F., Guan W., Zhang S. Amino acid transportation, sensing and signal transduction in the mammary gland: Key molecular signalling pathways in the regulation of milk synthesis. Nutr. Res. Rev. 2020;33:287–297. doi: 10.1017/S0954422420000074. [DOI] [PubMed] [Google Scholar]
- 11.Wang F., Shi H., Wang S., Wang Y., Cao Z., Li S. Amino acid metabolism in dairy cows and their regulation in milk synthesis. Curr. Drug Metab. 2019;20:36–45. doi: 10.2174/1389200219666180611084014. [DOI] [PubMed] [Google Scholar]
- 12.Appuhamy J.A., Knoebel N.A., Nayananjalie W.A., Escobar J., Hanigan M.D. Isoleucine and leucine independently regulate mTOR signaling and protein synthesis in MAC-T cells and bovine mammary tissue slices. J. Nutr. 2012;142:484–491. doi: 10.3945/jn.111.152595. [DOI] [PubMed] [Google Scholar]
- 13.Yuan W., Guo S., Gao J., Zhong M., Yan G., Wu W., Chao Y., Jiang Y. General control nonderepressible 2 (GCN2) kinase inhibits target of rapamycin complex 1 in response to amino acid starvation in Saccharomyces cerevisiae. J. Biol. Chem. 2017;292:2660–2669. doi: 10.1074/jbc.M116.772194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallinetti J., Harputlugil E., Mitchell J.R. Amino acid sensing in dietary-restriction-mediated longevity: Roles of signal-transducing kinases GCN2 and TOR. Biochem. J. 2013;449:1–10. doi: 10.1042/BJ20121098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosakamoto H., Okamoto N., Aikawa H., Sugiura Y., Suematsu M., Niwa R., Miura M., Obata F. Sensing of the non-essential amino acid tyrosine governs the response to protein restriction in Drosophila. Nat. Metab. 2022;4:944–959. doi: 10.1038/s42255-022-00608-7. [DOI] [PubMed] [Google Scholar]
- 16.Guo L., Liang Z., Zheng C., Liu B., Yin Q., Cao Y., Yao J. Leucine Affects α-Amylase Synthesis through PI3K/Akt-mTOR Signaling Pathways in Pancreatic Acinar Cells of Dairy Calves. J. Agric. Food Chem. 2018;66:5149–5156. doi: 10.1021/acs.jafc.8b01111. [DOI] [PubMed] [Google Scholar]
- 17.Osorio J.S., Lohakare J., Bionaz M. Biosynthesis of milk fat, protein, and lactose: Roles of transcriptional and posttranscriptional regulation. Physiol. Genom. 2016;48:231–256. doi: 10.1152/physiolgenomics.00016.2015. [DOI] [PubMed] [Google Scholar]
- 18.Bequette B.J., Backwell F.R.C., Kyle C.E., Calder A.G., Buchan V., Crompton L.A., France J., Macrae J.C. Vascular sources of phenylalanine, tyrosine, lysine, and methionine for casein synthesis in lactating goats. J. Dairy Sci. 1999;82:362–377. doi: 10.3168/jds.S0022-0302(99)75243-4. [DOI] [PubMed] [Google Scholar]
- 19.Schwab C.G., Satter L.D., Clay B. Response to lactating dairy cows to abomasal infusion of amino acids. J. Dairy Sci. 1976;59:1254–1270. doi: 10.3168/jds.S0022-0302(76)84354-8. [DOI] [PubMed] [Google Scholar]
- 20.Guinard J., Rulquin H. Effect of graded levels of duodenal infusions of casein on mammary uptake in lactating cows. 2. Individual amino acids. J. Dairy Sci. 1994;77:3304–3315. doi: 10.3168/jds.S0022-0302(94)77271-4. [DOI] [PubMed] [Google Scholar]
- 21.Guo L., Tian H., Shen J., Zheng C., Liu S., Cao Y., Cai C., Yao J. Phenylalanine regulates initiation of digestive enzyme mRNA translation in pancreatic acinar cells and tissue segments in dairy calves. Biosci. Rep. 2018;38:BSR20171189. doi: 10.1042/BSR20171189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansoorifar A., Koklu A., Ma S., Raj G.V., Beskok A. Electrical Impedance Measurements of Biological Cells in Response to External Stimuli. Anal. Chem. 2018;90:4320–4327. doi: 10.1021/acs.analchem.7b05392. [DOI] [PubMed] [Google Scholar]
- 23.Kim J., Lee J.E., Lee J.S., Park J.S., Moon J.O., Lee H.G. Phenylalanine and valine differentially stimulate milk protein synthetic and energy-mediated pathway in immortalized bovine mammary epithelial cells. J. Anim. Sci. Technol. 2020;62:263–275. doi: 10.5187/jast.2020.62.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Appuhamy J.A., Bell A.L., Nayananjalie W.A., Escobar J., Hanigan M.D. Essential amino acids regulate both initiation and elongation of mRNA translation independent of insulin in MAC-T cells and bovine mammary tissue slices. J. Nutr. 2011;141:1209–1215. doi: 10.3945/jn.110.136143. [DOI] [PubMed] [Google Scholar]
- 25.National Academies of Sciences, Engineering, and Medicine . Nutrient Requirements of Dairy Cattle: Eighth Revised Edition. National Academies Press; Washington, DC, USA: 2020. [DOI] [PubMed] [Google Scholar]
- 26.Vanhatalo A., Huhtanen P., Toivonen V., Varvikko T. Response of dairy cows fed grass silage diets to abomasal infusions of histidine alone or in combinations with methionine and lysine. J. Dairy Sci. 1999;82:2674–2685. doi: 10.3168/jds.S0022-0302(99)75524-4. [DOI] [PubMed] [Google Scholar]
- 27.Vyas D., Erdman R.A. Meta-analysis of milk protein yield responses to lysine and methionine supplementation. J. Dairy Sci. 2009;92:5011–5018. doi: 10.3168/jds.2008-1769. [DOI] [PubMed] [Google Scholar]
- 28.Lin X., Li S., Zou Y., Zhao F.Q., Liu J., Liu H. Lysine Stimulates Protein Synthesis by Promoting the Expression of ATB0,+ and Activating the mTOR Pathway in Bovine Mammary Epithelial Cells. J. Nutr. 2018;148:1426–1433. doi: 10.1093/jn/nxy140. [DOI] [PubMed] [Google Scholar]
- 29.Appuhamy J.A., Knapp J.R., Becvar O., Escobar J., Hanigan M.D. Effects of jugular-infused lysine, methionine, and branched-chain amino acids on milk protein synthesis in high-producing dairy cows. J. Dairy Sci. 2011;94:1952–1960. doi: 10.3168/jds.2010-3442. [DOI] [PubMed] [Google Scholar]
- 30.Liu G., Hanigan M., Lin X., Zhao K., Jiang F.G., White R.R., Wang Y., Hu Z.Y., Wang Z.H. Methionine, leucine, isoleucine, or threonine effects on mammary cell signaling and pup growth in lactating mice. J. Dairy Sci. 2017;100:4038–4050. doi: 10.3168/jds.2016-11973. [DOI] [PubMed] [Google Scholar]
- 31.Che L., Xu M., Gao K., Zhu C., Wang L., Yang X., Wen X., Xiao H., Jiang Z., Wu D. Valine increases milk fat synthesis in mammary gland of gilts through stimulating AKT/MTOR/SREBP1 pathway†. Biol. Reprod. 2019;101:126–137. doi: 10.1093/biolre/ioz065. [DOI] [PubMed] [Google Scholar]
- 32.Kirchgessner M., Maierhofer R., Schwarz F.J., Eidelsburger U. Effect of feeding protected arginine on food intake, milk yield and growth hormone and amino acid levels in blood plasma of cows during the summer feeding period with grass. Arch. Tierernahr. 1993;45:57–69. doi: 10.1080/17450399309386088. [DOI] [PubMed] [Google Scholar]
- 33.Mateo R.D., Wu G., Moon H.K., Carroll J.A., Kim S.W. Effects of dietary arginine supplementation during gestation and lactation on the performance of lactating primiparous sows and nursing piglets. J. Anim. Sci. 2008;86:827–835. doi: 10.2527/jas.2007-0371. [DOI] [PubMed] [Google Scholar]
- 34.Lapierre H., Lobley G.E., Doepel L., Raggio G., Rulquin H., Lemosquet S. Triennial Lactation Symposium: Mammary metabolism of amino acids in dairy cows. J. Anim. Sci. 2012;90:1708–1721. doi: 10.2527/jas.2011-4645. [DOI] [PubMed] [Google Scholar]
- 35.Doepel L., Pacheco D., Kennelly J.J., Hanigan M.D., López I.F., Lapierre H. Milk protein synthesis as a function of amino acid supply. J. Dairy Sci. 2004;87:1279–1297. doi: 10.3168/jds.S0022-0302(04)73278-6. [DOI] [PubMed] [Google Scholar]
- 36.Doepel L., Hewage I.I., Lapierre H. Milk protein yield and mammary metabolism are affected by phenylalanine deficiency but not by threonine or tryptophan deficiency. J. Dairy Sci. 2016;99:3144–3156. doi: 10.3168/jds.2015-10320. [DOI] [PubMed] [Google Scholar]
- 37.Swanepoel N., Robinson P., Erasmus L.J. Effects of ruminally protected methionine and/or phenylalanine on performance of high producing Holstein cows fed rations with very high levels of canola meal. Anim. Feed Sci. Technol. 2015;205:10–22. doi: 10.1016/j.anifeedsci.2015.04.002. [DOI] [Google Scholar]
- 38.Hou X., Song S., Xu Z., Shi Y., Yang Y., Zhang L., Cui Y., Wang C., Lin Y. Prolactin upregulates amino acids uptake in dairy cow mammary epithelial cells via LAT1. J. Dairy Sci. 2024;28:9948–9959. doi: 10.3168/jds.2024-24746. [DOI] [PubMed] [Google Scholar]
- 39.Huttunen J., Peltokangas S., Gynther M., Natunen T., Hiltunen M., Auriola S., Ruponen M., Vellonen K.S., Huttunen K.M. L-Type Amino Acid Transporter 1 (LAT1/Lat1)-Utilizing Prodrugs Can Improve the Delivery of Drugs into Neurons, Astrocytes and Microglia. Sci. Rep. 2019;9:12860. doi: 10.1038/s41598-019-49009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baumrucker C.R. Amino acid transport systems in bovine mammary tissue. J. Dairy Sci. 1985;68:2436–2451. doi: 10.3168/jds.S0022-0302(85)81119-X. [DOI] [PubMed] [Google Scholar]
- 41.Rosario F.J., Urschitz J., Powell T.L., Brown T.L., Jansson T. Overexpression of the LAT1 in primary human trophoblast cells increases the uptake of essential amino acids and activates mTOR signaling. Clin. Sci. 2023;137:1651–1664. doi: 10.1042/CS20230490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srisongkram T., Bahrami K., Järvinen J., Timonen J., Rautio J., Weerapreeyakul N. Development of Sesamol Carbamate-L-Phenylalanine Prodrug Targeting L-Type Amino Acid Transporter1 (LAT1) as a Potential Antiproliferative Agent against Melanoma. Int. J. Mol. Sci. 2022;23:8446. doi: 10.3390/ijms23158446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaira K., Oriuchi N., Takahashi T., Nakagawa K., Ohde Y., Okumura T., Murakami H., Shukuya T., Kenmotsu H., Naito T., et al. LAT1 expression is closely associated with hypoxic markers and mTOR in resected non-small cell lung cancer. Am. J. Transl. Res. 2011;3:468–478. [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez R.S., Salji M.J., Rushworth L., Ntala C., Rodriguez Blanco G., Hedley A., Clark W., Peixoto P., Hervouet E., Renaude E., et al. SLFN5 Regulates LAT1-Mediated mTOR Activation in Castration-Resistant Prostate Cancer. Cancer Res. 2021;81:3664–3678. doi: 10.1158/0008-5472.CAN-20-3694. [DOI] [PubMed] [Google Scholar]
- 45.Rius A.G., Appuhamy J.A., Cyriac J., Kirovski D., Becvar O., Escobar J., McGilliard M.L., Bequette B.J., Akers R.M., Hanigan M.D. Regulation of protein synthesis in mammary glands of lactating dairy cows by starch and amino acids. J. Dairy Sci. 2010;93:3114–3127. doi: 10.3168/jds.2009-2743. [DOI] [PubMed] [Google Scholar]
- 46.Shennan D.B., Boyd C.A. The functional and molecular entities underlying amino acid and peptide transport by the mammary gland under different physiological and pathological conditions. J. Mammary Gland Biol. Neoplasia. 2014;19:19–33. doi: 10.1007/s10911-013-9305-5. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y., Zhou Z., Peng J., Loor J.J. Methionine and valine activate the mammalian target of rapamycin complex 1 pathway through heterodimeric amino acid taste receptor (TAS1R1/TAS1R3) and intracellular Ca (2+) in bovine mammary epithelial cells. J. Dairy Sci. 2018;101:11354–11363. doi: 10.3168/jds.2018-14461. [DOI] [PubMed] [Google Scholar]
- 48.Sah N., Wu G., Bazer F.W. Regulation of gene expression by amino acids in animal cells. Adv. Exp. Med. Biol. 2021;1332:1–15. doi: 10.1007/978-3-030-74180-8_1. [DOI] [PubMed] [Google Scholar]
- 49.Berguig G.Y., Martin N.T., Creer A.Y., Xie L., Zhang L., Murphy R., Pacheco G., Bullens S., Olbertz J., Weng H.H. Of mice and men: Plasma phenylalanine reduction in PKU corrects neurotransmitter pathways in the brain. Mol. Genet. Metab. 2019;128:422–430. doi: 10.1016/j.ymgme.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Fitzpatrick P.F. Allosteric regulation of phenylalanine hydroxylase. Arch. Biochem. Biophys. 2012;519:194–201. doi: 10.1016/j.abb.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verbeke R., Roets E., Massart-Leën A.M., Peeters G. Metabolism of (U- 14 C)-L-threonine and (U- 14 C)-L-phenylalanine by the isolated perfused udder. J. Dairy Res. 1972;39:239–250. doi: 10.1017/S0022029900014072. [DOI] [PubMed] [Google Scholar]
- 52.Bequette B.J., Backwell F.R., Calder A.G., Metcalf J.A., Beever D.E., MacRae J.C., Lobley G.E. Application of a U-13C-labeled amino acid tracer in lactating dairy goats for simultaneous measurements of the flux of amino acids in plasma and the partition of amino acids to the mammary gland. J. Dairy Sci. 1997;80:2842–2853. doi: 10.3168/jds.S0022-0302(97)76249-0. [DOI] [PubMed] [Google Scholar]
- 53.Liu G.Y., Sabatini D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., Proud C.G. The mTOR pathway in the control of protein synthesis. Physiology. 2006;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 55.Dann S.G., Selvaraj A., Thomas G. mTOR Complex1-S6K1 signaling: At the crossroads of obesity, diabetes and cancer. Trends Mol. Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Magnuson B., Ekim B., Fingar D.C. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 57.Livneh I., Cohen-Kaplan V., Fabre B., Abramovitch I., Lulu C., Nataraj N.B., Lazar I., Ziv T., Yarden Y., Zohar Y., et al. Regulation of nucleo-cytosolic 26S proteasome translocation by aromatic amino acids via mTOR is essential for cell survival under stress. Mol. Cell. 2023;83:3333–3346.e3335. doi: 10.1016/j.molcel.2023.08.016. [DOI] [PubMed] [Google Scholar]
- 58.Feng L., Li W., Liu Y., Jiang W.D., Kuang S.Y., Wu P., Jiang J., Tang L., Tang W.N., Zhang Y.A., et al. Protective role of phenylalanine on the ROS-induced oxidative damage, apoptosis and tight junction damage via Nrf2, TOR and NF-κB signalling molecules in the gill of fish. Fish Shellfish Immunol. 2017;60:185–196. doi: 10.1016/j.fsi.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 59.Yang P., Wang H., Ma L., Yin H., Zhu Z., Liu C., Huang W., Zhou Z., Wu X., Taj S. The optimum dietary phenylalanine requirement of Hybrid Grouper (Epinephelusfuscoguttatus ♀ × Epinepheluslanceolatus ♂) Juveniles: Effects on growth performance, gut micromorphology, and antioxidation. Aquac. Nutr. 2023;2023:9155290. doi: 10.1155/2023/9155290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prizant R.L., Barash I. Negative effects of the amino acids Lys, His, and Thr on S6K1 phosphorylation in mammary epithelial cells. J. Cell Biochem. 2008;105:1038–1047. doi: 10.1002/jcb.21904. [DOI] [PubMed] [Google Scholar]
- 61.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 62.White J.P. Amino acid trafficking and skeletal muscle protein synthesis: A case of supply and demand. Front. Cell. Dev. Biol. 2021;9:656604. doi: 10.3389/fcell.2021.656604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A.G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6:269–279. doi: 10.1016/S1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 64.Berlanga J.J., Santoyo J., De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur. J. Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 65.Chen R., Rato C., Yan Y., Crespillo-Casado A., Clarke H.J., Harding H.P., Marciniak S.J., Read R.J., Ron D. G-actin provides substrate-specificity to eukaryotic initiation factor 2α holophosphatases. eLife. 2015;4:e04871. doi: 10.7554/eLife.04871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang C., Sun Y., Zhao F., Liu J., Liu H. Functional characterization of peptide transporters in bovine mammary epithelial cells. J. Agric. Food Chem. 2018;67:213–219. doi: 10.1021/acs.jafc.8b05637. [DOI] [PubMed] [Google Scholar]
- 67.Zhao Y., Tang J., Yang D., Tang C., Chen J. Staphylococcal enterotoxin M induced inflammation and impairment of bovine mammary epithelial cells. J. Dairy Sci. 2020;103:8350–8359. doi: 10.3168/jds.2019-17444. [DOI] [PubMed] [Google Scholar]
- 68.Xie T.T., Tian H., Du R.F., Li F., Guo L. Mitogen-activated protein kinase-interacting kinase 1 is involved in mTOR-mediated pancreatic exocrine function of bovine pancreatic acinar cells with leucine supply. Arch. Anim. Nutr. 2013;77:155–169. doi: 10.1080/1745039X.2023.2199839. [DOI] [PubMed] [Google Scholar]
- 69.Guo L., Yao J.H., Zheng C., Tian H.B., Liu Y.L., Liu S.M., Cai C.J., Xu X.R., Cao Y.C. Leucine regulates α-amylase and trypsin synthesis in dairy calf pancreatic tissue in vitro via the mammalian target of rapamycin signalling pathway. Animal. 2018;13:1899–1906. doi: 10.1017/S1751731118003683. [DOI] [PubMed] [Google Scholar]
- 70.Zheng C., Yao J., Guo L., Cao Y., Liang Z., Yang X., Cai C. Leucine-induced promotion of post-absorptive EAA utilization and hepatic gluconeogenesis contributes to protein synthesis in skeletal muscle of dairy calve. J. Anim. Physiol. Anim. Nutr. 2018;103:705–712. doi: 10.1111/jpn.13072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article and Supplementary Material.