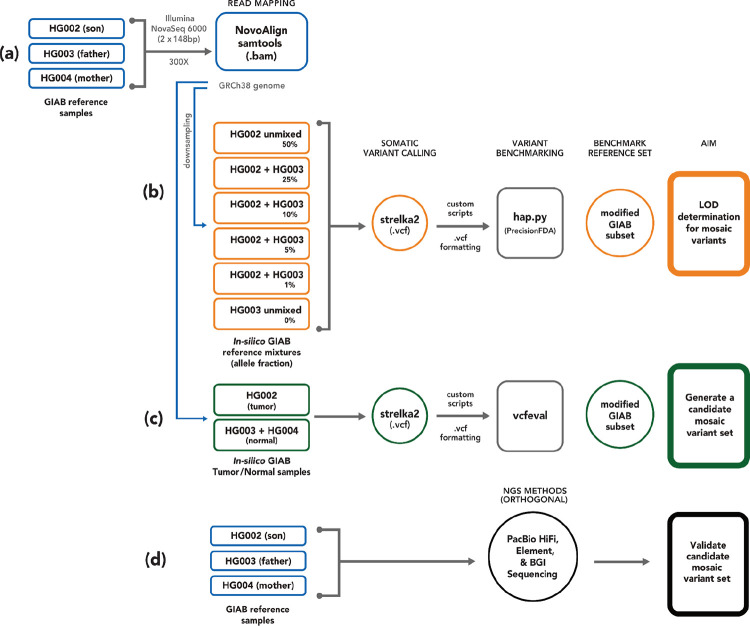
Figure 1 -.
Trio-based methodology using high coverage Illumina data, Strelka2 somatic caller, and orthogonal next generation sequencing datasets for candidate mosaic variant detection and validation in HG002. (A) AJ trio (NIST RM - HG002, HG003, and HG004) sequencing and reference mapping (GRCh38) were initially performed by Zook et al 2016. (B) In silico sample mixtures were created using HG002 and HG003, treating HG003 as normal and the mixtures as tumor, to determine the limit of detection for variant allele fraction. Strelka2 somatic calling and benchmarking with hap.py was conducted using the GIAB mixtures to estimate a limit of detection (LOD). (C) To identify potential mosaic and de novo variants, a tumor-normal Strelka2 somatic run, with HG002 (son) as tumor and HG003+HG004 (combined parents) as normal, was performed. (D) The Strelka2 callset was benchmarked against the GIAB v4.2.1 small variant benchmark with vcfeval to create a candidate variant set, and three orthogonal high-coverage short- and long-read sequencing technologies were used for validation.