Abstract
Protein structure prediction via artificial intelligence/machine learning (AI/ML) approaches has sparked substantial research interest in structural biology and adjacent disciplines. More recently, AlphaFold2 (AF2) has been adapted for the prediction of multiple structural conformations in addition to single-state structures. This novel avenue of research has focused on proteins (typically 50 residues in length or greater), while multi-conformation prediction of shorter peptides has not yet been explored in this context. Here, we report AF2-based structural conformation prediction of a total of 557 peptides (ranging in length from 10 to 40 residues) for a benchmark dataset with corresponding nuclear magnetic resonance (NMR)-determined conformational ensembles. De novo structure predictions were accompanied by structural comparison analyses to assess prediction accuracy. We found that the prediction of conformational ensembles for peptides with AF2 varied in accuracy versus NMR data, with average root-mean-square deviation (RMSD) among structured regions under 2.5 Å and average root-mean-square fluctuation (RMSF) differences under 1.5 Å. Our results reveal notable capabilities of AF2-based structural conformation prediction for peptides but also underscore the necessity for interpretation discretion.
INTRODUCTION
AlphaFold2 (AF2) is an artificial intelligence/machine learning (AI/ML) model capable of predicting the 3-dimensional (3D) structures of proteins from amino acid sequence alone with accuracy comparable to lower-resolution experimentally determined protein structures [1, 2]. AF2 was trained on multiple sequence alignment (MSA) of metagenomic sequencing data in combination with protein structures determined by X-ray crystallography and cryo-electron microscopy (cryoEM) from the Protein Data Bank (PDB) [3]. Since its release, AF2 has garnered widespread use for various applications across the biological sciences and was recognized with a share of the 2024 Nobel Prize in Chemistry [4, 5]. Recently, AF2 has been adapted for predicting the structures of multiple protein conformations, going beyond the original scope of single static structure prediction [6–9]. This is accomplished by using multiple random seeds, i.e., stochastic prediction initializations, and subsampling of the input MSA, resulting in an ensemble of predicted structures [8]. The prediction of multiple conformations with AF2 has been demonstrated to align with experimentally determined conformational ensemble data for certain proteins as determined by nuclear magnetic resonance (NMR) spectroscopy [6, 7], which is notable considering that the training data for AF2 did not contain NMR structures [1]. However, AF2-based structural conformation prediction is a novel area of research, and—as with the prediction of static structures—accuracy is expected to vary and should be considered with discretion [8]. Furthermore, the focus of this research has been on regular-sized proteins, while prediction of structural conformations for peptides in this context has not yet been explored.
McDonald et al. [10] benchmarked the structure prediction capabilities of AF2 for a rich dataset of 588 peptides with NMR-determined 3D structures in the PDB. These peptides range from 10 to 40 amino acid residues in length, and were categorized into various structural groupings, including α-helical membrane-associated peptides (AH MP), α-helical soluble peptides (AH SL), β-hairpin peptides (BHPIN), disulfide-rich peptides (DSRP), mixed secondary structure membrane-associated peptides (MIX MP), and mixed secondary structure soluble peptides (MIX SL). While the benchmark study comprehensively evaluated the accuracy of conventional (individual) structure prediction capabilities of AF2, the more recent approach for predicting multiple structural conformations was not explored. Herein, we report AF2-based conformational ensemble prediction of peptide structures from the aforementioned dataset using multiple random seeds and MSA subsampling. The original dataset was filtered to include only peptides for which the NMR-determined ensemble contained five or more structural conformers for a total of n = 557 peptides to assess conformational variability. High-throughput prediction and computational analyses with various metrics, including root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and MSA sequence depth comparisons, were performed. Our analyses revealed that AF2-based conformational ensemble prediction of peptides varied in accuracy compared to NMR data, with overall RMSD under 2.5 Å and overall RMSF differences under 1.5 Å. These results highlight notable capabilities of predicting peptide structural conformations with AF2 yet underscore the need for discretion in prediction interpretation.
RESULTS
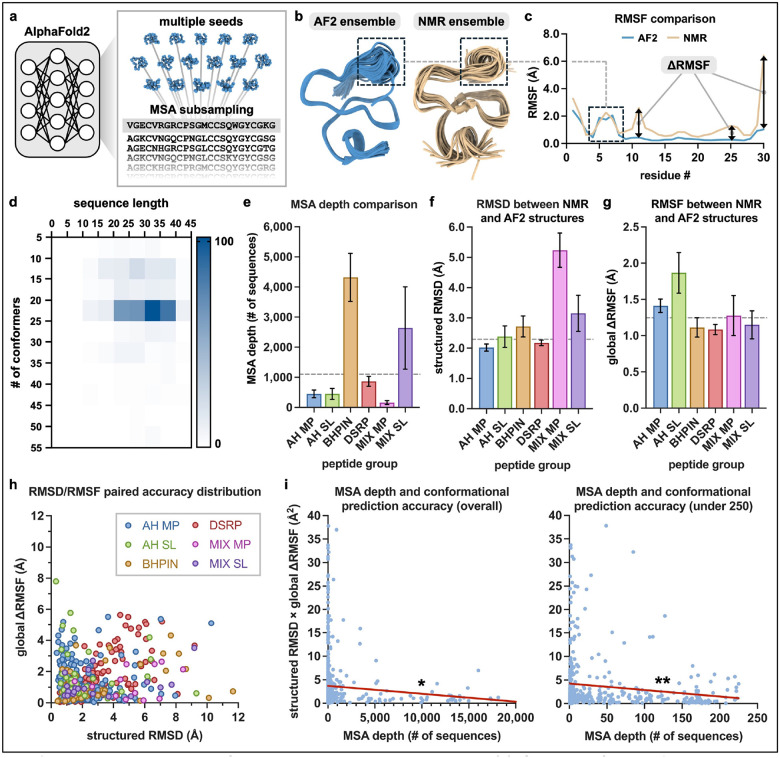
The ColabFold [8] implementation of AF2 was used for structure prediction, with multiple random seeds and MSA subsampling (Fig. 1a). The number of seeds was set to 16, with five structures per seed (n = 80 structures per prediction), and MSA subsampling parameters were set to 16 for number of sequence cluster points and 32 for extra sequences (see Methods for additional information). An example of a prediction and its assessment is shown in Figs. 1b and 1c for an antimicrobial peptide derived from Amaranthus caudatus (PDB ID 1ZUV) [11], which highlights an area of conformational variability (Fig. 1b) plus per-residue RMSF differences (ΔRMSF) versus the NMR-determined conformational ensemble (Fig. 1c). Predictions were run for 557 peptides from the McDonald et al. dataset, filtered to include only peptides with NMR ensembles consisting of five or more conformational structures (Fig. 1d). The six peptide categorizations of (1) AH MP, (2) AH SL, (3) BHPIN, (4) DSRP, (5) MIX MP, and (6) MIX SL from the original dataset were also adopted in the current study. The average depth of MSAs generated by ColabFold (via MMseqs2) [8, 12] was 1,102 sequences across all peptides (Fig. 1e). Alpha carbon (Cα) comparative analysis of structured regions, as previously annotated [10], between the AF2-predicted and NMR-determined conformational ensembles of the peptides was performed to calculate RMSD, which was found to be 2.294 Å overall across all peptides (Fig. 1f). To gain insight into conformational similarity between AF2-predicted and NMR-determined conformational ensembles in terms of structural flexibility, RMSF differences were assessed using a global ΔRMSF metric (eqn. 1), where i is the residue position, nr is the number of residues, and ΔRMSF (Fig. 1c) is the per-residue difference between the individual RMSF values of two conformational ensembles. The global ΔRMSF was found to be 1.247 Å overall across all peptides (Fig. 1g).
Fig. 1: Peptide structural conformation prediction and analysis.
(a) Overview of the AF2-based multiple conformation prediction approach with multiple seeds and MSA subsampling. (b) AF2-predicted and NMR-determined conformational ensembles of an antimicrobial peptide (PDB ID 1ZUV). (c) Comparisons of the Cα RMSF values of the conformational ensembles from (b). A flexible region present in both ensembles is outlined with a dashed box. Examples of individual ΔRMSF values are also indicated with arrows. (d) Heatmap of the n = 557 peptide NMR dataset displaying sequence length versus number of conformational structures per ensemble, with the scale bar representing the number of PDB entries. (e-g) Among the six different peptide groupings: (e) MSA depth (number of sequences within the MSA) for each peptide, (f) structured region Cα RMSD between NMR-determined and AF2-predicted conformational ensembles, and (g) Cα global ΔRMSF (eqn. 1) between NMR-determined and AF2-predicted conformational ensembles. Data represented as means ± SEM, with dashed lines indicating overall mean. AH MP n = 172 peptides; AH SL n = 37 peptides; BHPIN = 56 peptides; DSRP n = 261 peptides; MIX MP n = 12 peptides; MIX SL n = 19 peptides. (h) Distribution of structured region RMSD paired with global ΔRMSF between AF2-predicted and NMR-determined conformational ensembles among the six different peptide groupings. (i) MSA depth versus the product of structured region RMSD and global ΔRMSF between NMR-determined and AF2-predicted conformational ensembles across all peptides considering the entire MSA depth range (left graph) or peptides with an MSA depth less than 250 sequences (right graph). Pearson correlation R2 = 0.0076 (entire MSA depth, n = 557 peptides) and R2 = 0.021 (MSA depth < 250, n = 415 peptides), * p < 0.05, ** p < 0.005. Lines determined by linear regression are depicted in red. Two data-points were not graphically represented for eligibility and space preservation, as described in the Methods section.
| eqn. 1 |
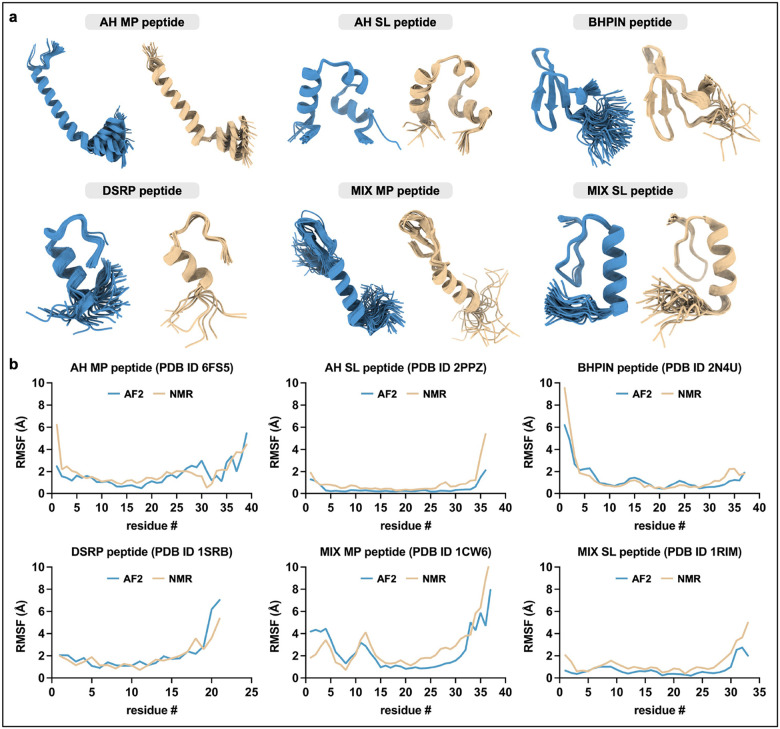
While certain trends appear to be present across the peptide groupings among the RMSD, RMSF, and MSA depth metrics (Fig. 1e–g), it is important to consider that the number of peptides across these groups vary substantially—for example, the DSRP group consists of 261 peptides while the MIX MP group consists of only 12 peptides. With this in consideration, the BHPIN and MIX SL groups had the greatest MSA depth. Additionally, predictions for the DSRP peptide group appear to have been the most accurate in terms of having the lowest of both structured region RMSD and global ΔRMSF, i.e. when considering both metrics, while predictions for the MIX MP and AH SL peptide groups were the least accurate with respect to structured region RMSD and global ΔRMSF, respectively (Figs. 1f and 1g). The distribution plotted for the structured region RMSD metric paired with the global ΔRMSF metric for each peptide did not indicate any observable differences among the six groupings, but overall revealed that 54.2% and 73.8% of peptides had values of less than 2 Å and 3 Å for both metrics, respectively (Fig. 1h). Comparison of MSA depth with the structured region RMSD and global ΔRMSF metrics in combination (treated here as the product of the two metrics) indicates a weak inverse correlation, both for peptides across the entire MSA depth range (R2 = 0.0076) and for peptides with an MSA depth of < 250 sequences (R2 = 0.021) (Fig. 1i), suggesting that any effect that greater MSA depth may have on improving prediction accuracy in this context is minimal but may be beneficial. Examples of AF2-predicted conformational ensembles compared to NMR-based conformational ensembles are shown in Fig. 2, with structural depictions for each of the six peptide groupings (Fig. 2a) and corresponding RMSF comparisons (Fig. 2b). The results presented here provide insights into AF2-based peptide conformational ensemble prediction, which is generally favorable yet considerably variable in terms of RMSD and RMSF-based accuracy metrics, and may moderately benefit from increased MSA depth.
Fig. 2: Exemplary peptide structural conformation predictions.
(a) AF2-predicted (blue) and NMR-determined (beige) conformational ensembles of exemplary peptides from each of the six peptide groupings: AH MP (PDB ID 6FS5) [13], AH SL (PDB ID 2PPZ) [14], BHPIN (PDB ID 2N4U) [15], DSRP (PDB ID 1SRB) [16], MIX MP (PDB ID 1CW6) [17], and MIX SL (PDB ID 1RIM) [18]. (b) Cα RMSF comparisons between AF2-predicted (blue lines) and NMR-determined (beige lines) conformational ensembles corresponding to the peptides depicted in (a). Global ΔRMSF values for each peptide are as follows: AH MP, 0.607 Å; AH SL, 0.455 Å; BHPIN, 0.433 Å; DSRP, 0.506 Å; MIX MP, 1.038 Å; MIX SL, 0.561 Å. Cα RMSD values for each peptide are as follows: AH MP, 1.759 Å; AH SL, 2.152 Å; BHPIN, 1.212 Å; DSRP, 2.621 Å; MIX MP, 2.633 Å; MIX SL, 1.410 Å.
DISCUSSION
In the current work, we utilized AF2 for the prediction of peptide conformational ensembles by multiple seed initialization and MSA subsampling. This approach was applied to predict structural conformations of a dataset of 557 peptides categorized into six distinct structural groups, originating from a previously curated peptide dataset with NMR-determined conformational ensembles [10]. We performed structural analyses to compare the AF2-predicted and NMR-determined ensembles using metrics of RMSD of structured regions and global ΔRMSF, with the former serving to measure rigid structure prediction accuracy and the latter for assessing structural flexibility prediction accuracy. The predicted conformations were often structurally similar to NMR-determined conformations, with over 50% of the predictions differing by no more than 2 Å for both RMSD and RMSF metrics, and nearly 75% differing by less than 3 Å. Additionally, we found that greater MSA depth may contribute to increased prediction accuracy, albeit to a very limited extent.
There are, however, various limitations that merit consideration. Firstly, while NMR is arguably the most conformationally sensitive experimental structure determination approach, it does not necessarily capture the breadth of conformational space, especially when an NMR-determined ensemble consists of a limited number of structural conformers. While time-resolved X-ray crystallography [19] and cryoEM [20] are also conformationally sensitive, these experimental approaches yield even fewer diverse structural conformers. Secondly, RMSD and RMSF are useful metrics both independently and in combination but only partially characterize structural differences between conformational ensembles. As such conformational structure prediction approaches continue to evolve, more comprehensive evaluation metrics must be formulated. Another challenge is that shorter peptide sequences generally have far fewer homologous sequences available for assembling MSAs. However, the number of annotated protein-coding sequences continues to increase [21], and the discovery of widespread noncanonical open reading frames may also contribute to protein-coding sequence diversity [22]. It is also important to consider that while NMR structures were not included for AF2 model training [1], corresponding X-ray crystallography or cryoEM structures with identical or highly similar sequences from the PDB that may have been included in the training data of AF2 would be expected to influence prediction accuracy— though it is unclear how such structures might inform conformational variability.
In conclusion, AI/ML-based structural conformation prediction holds promise as a novel avenue for the prediction of peptide conformational ensembles. AF2 serves as a capable model in this regard, and other models are likely to be developed/adapted for this purpose. However, the novelty of such approaches and the current variability in terms of prediction accuracy underscore the necessity for discretion and careful interpretation of results. Further research is needed to better characterize conformational ensemble predictions and to advance the current state of the methodology.
METHODS
AF2 Predictions
Predictions were performed using the ColabFold [8] implementation of AF2 in a Google Colab Jupyter Notebook environment with Hardware Accelerator set to T4 GPU. The num_seeds parameter was adjusted to 16 and the max_msa parameter was set to 16:32, while all other parameters were left unmodified from their default settings, i.e. num_relax: 0; template_mode: none; pair_mode: unpaired_paired; msa_mode: mmseqs2_uniref_env; num_recycles: 3; recycle_early_stop_tolerance: auto; use_dropout: unselected. For each input sequence, five structures were computed per seed for a total of n = 80 structures.
Structural Analyses
RMSD and RMSF values were computed using custom implementations of Python code available online from the CHARMM-GUI lecture series [23, 24]. Whole-structure alignments were performed for both RMSD and RMSF calculations. For RMSD determination, each structure in a given AF2-predicted conformational ensemble was compared to each structure in the corresponding NMR-determined conformational ensemble, and the average of all comparisons was used as the final RMSD value for each peptide. RMSD calculations included only Cα atoms of structured regions, as annotated in the original peptide dataset by McDonald et al. for regions with secondary structures and stretches with multiple disulfide bonds [10]. For RMSF determination, the individual structures in a given AF2-predicted conformational ensemble were aligned to the top-ranked (highest pLDDT score) structure within the ensemble, while the individual structures of the NMR-determined conformational ensembles were aligned to the initial structure in the PDB entry. Per-residue RMSF values were calculated based on Cα atoms for each residue across all structures within a given conformational ensemble.
Statistical Analyses and Graphical Representation
Statistical analyses of standard error of the mean, Pearson correlation, and linear regression, as well as graphical representations shown in Figs. 1 and 2, were performed using GraphPad Prism 10. In Fig. 2i, for the graph depicting entire MSA depth range (left graph), two data-points (PDB ID 2RR0 with MSA depth of 23,764 and RMSD × ΔRMSF product of 1.635 Å2; and PDB ID 6A8Y with MSA depth of 436 and RMSD × ΔRMSF product of 52.410 Å2) are not shown on the graph but were not excluded from statistical analyses.
ACKNOWLEDGEMENTS
This work was supported by the Levy-Longenbaugh Donor-Advised Fund (to R.P. and W.A.). RCSB Protein Data Bank is jointly funded by the National Science Foundation (DBI-2321666, PI: S.K.B.), the US Department of Energy (DE-SC0019749, PI: S.K.B.), and the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, and the National Institute of General Medical Sciences of the National Institutes of Health (R01GM157729, PI: S.K.B.). The annotated peptide dataset curated by McDonald et al. [10] was a foundational aspect of this work. AF2 predictions were performed using ColabFold, an open-source platform developed by the research group of Dr. Martin Steinegger at Seoul National University. Molecular analyses were performed with custom implementations of Python code available online from the CHARMM-GUI lecture series developed by the research group of Dr. Wonpil Im at Lehigh University. Molecular graphics were visualized with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from the National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases [25].
Footnotes
COMPETING INTERESTS
A.M.I. is a founder and partner of North Horizon, which is engaged in the development of artificial intelligence-based software. C.M. declares no competing interests. S.K.B. declares no competing interests. R.P. is a founder and equity shareholder of PhageNova Bio. R.P. is Chief Scientific Officer and a paid consultant of PhageNova Bio. R.P. is a founder and equity shareholder of MBrace Therapeutics. R.P. serves as a paid consultant for MBrace Therapeutics. R.P. has Sponsored Research Agreements (SRAs) in place with PhageNova Bio and with MBrace Therapeutics. These arrangements are managed in accordance with the established institutional conflict-of-interest policies of Rutgers, The State University of New Jersey. This study falls outside of the scope of these SRAs. W.A. is a founder and equity shareholder of PhageNova Bio. W.A. is a founder and equity shareholder of MBrace Therapeutics. W.A. serves as a paid consultant for MBrace Therapeutics. W.A. has Sponsored Research Agreements (SRAs) in place with PhageNova Bio and with MBrace Therapeutics. These arrangements are managed in accordance with the established institutional conflict-of-interest policies of Rutgers, The State University of New Jersey. This study falls outside of the scope of these SRAs.
REFERENCES
- 1.Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. Aug 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. Critical assessment of methods of protein structure prediction (CASP)-Round XIV. Proteins. Dec 2021;89(12):1607–1617. doi: 10.1002/prot.26237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burley SK, Berman HM. Open-access data: A cornerstone for artificial intelligence approaches to protein structure prediction. Structure. Jun 3 2021;29(6):515–520. doi: 10.1016/j.str.2021.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovalevskiy O, Mateos-Garcia J, Tunyasuvunakool K. AlphaFold two years on: Validation and impact. Proc Natl Acad Sci U S A. Aug 20 2024;121(34):e2315002121. doi: 10.1073/pnas.2315002121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Press release: Nobel Prize in Chemistry 2024. 2024. https://www.nobelprize.org/prizes/chemistry/2024/press-release/
- 6.Wayment-Steele HK, Ojoawo A, Otten R, et al. Predicting multiple conformations via sequence clustering and AlphaFold2. Nature. Jan 2024;625(7996):832–839. doi: 10.1038/s41586-023-06832-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monteiro da Silva G, Cui JY, Dalgarno DC, Lisi GP, Rubenstein BM. High-throughput prediction of protein conformational distributions with subsampled AlphaFold2. Nat Commun. Mar 27 2024;15(1):2464. doi: 10.1038/s41467-024-46715-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim G, Lee S, Levy Karin E, et al. Easy and accurate protein structure prediction using ColabFold. Nat Protoc. Oct 14 2024;doi: 10.1038/s41596-024-01060-5 [DOI] [PubMed] [Google Scholar]
- 9.Del Alamo D, Sala D, McHaourab HS, Meiler J. Sampling alternative conformational states of transporters and receptors with AlphaFold2. Elife. Mar 3 2022;11 doi: 10.7554/eLife.75751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald EF, Jones T, Plate L, Meiler J, Gulsevin A. Benchmarking AlphaFold2 on peptide structure prediction. Structure. Jan 5 2023;31(1):111–119 e2. doi: 10.1016/j.str.2022.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chavez MI, Andreu C, Vidal P, et al. On the importance of carbohydrate-aromatic interactions for the molecular recognition of oligosaccharides by proteins: NMR studies of the structure and binding affinity of AcAMP2-like peptides with non-natural naphthyl and fluoroaromatic residues. Chemistry. Nov 18 2005;11(23):7060–74. doi: 10.1002/chem.200500367 [DOI] [PubMed] [Google Scholar]
- 12.Steinegger M, Soding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. Nov 2017;35(11):1026–1028. doi: 10.1038/nbt.3988 [DOI] [PubMed] [Google Scholar]
- 13.Mercurio FA, Scaloni A, Caira S, Leone M. The antimicrobial peptides casocidins I and II: Solution structural studies in water and different membrane-mimetic environments. Peptides. Apr 2019;114:50–58. doi: 10.1016/j.peptides.2018.09.004 [DOI] [PubMed] [Google Scholar]
- 14.Gronwald W, Hohm T, Hoffmann D. Evolutionary Pareto-optimization of stably folding peptides. BMC Bioinformatics. Feb 19 2008;9:109. doi: 10.1186/1471-2105-9-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maisuradze GG, Medina J, Kachlishvili K, et al. Preventing fibril formation of a protein by selective mutation. Proc Natl Acad Sci U S A. Nov 3 2015;112(44):13549–54. doi: 10.1073/pnas.1518298112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atkins AR, Martin RC, Smith R. 1H NMR studies of sarafotoxin SRTb, a nonselective endothelin receptor agonist, and IRL 1620, an ETB receptor-specific agonist. Biochemistry. Feb 14 1995;34(6):2026–33. doi: 10.1021/bi00006a024 [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Henz ME, Gallagher NL, et al. Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria. Biochemistry. Nov 23 1999;38(47):15438–47. doi: 10.1021/bi991351x [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Liu Z, Androphy E, Chen J, Baleja JD. Design and characterization of helical peptides that inhibit the E6 protein of papillomavirus. Biochemistry. Jun 15 2004;43(23):7421–31. doi: 10.1021/bi049552a [DOI] [PubMed] [Google Scholar]
- 19.Branden G, Neutze R. Advances and challenges in time-resolved macromolecular crystallography. Science. Aug 27 2021;373(6558)doi: 10.1126/science.aba0954 [DOI] [PubMed] [Google Scholar]
- 20.Amann SJ, Keihsler D, Bodrug T, Brown NG, Haselbach D. Frozen in time: analyzing molecular dynamics with time-resolved cryo-EM. Structure. Jan 5 2023;31(1):4–19. doi: 10.1016/j.str.2022.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. Jan 6 2023;51(D1):D523–D531. doi: 10.1093/nar/gkac1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright BW, Yi Z, Weissman JS, Chen J. The dark proteome: translation from noncanonical open reading frames. Trends Cell Biol. Mar 2022;32(3):243–258. doi: 10.1016/j.tcb.2021.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Im Lab, Lehigh University. CHARMM-GUI lecture series: RMSD Between Two Structures. https://www.charmm-gui.org/?doc=lecture&module=scientific&lesson=11
- 24.Im Lab, Lehigh University. CHARMM-GUI lecture series: Average and RMSF from Multiple Structures. https://www.charmm-gui.org/?doc=lecture&module=scientific&lesson=12
- 25.Meng EC, Goddard TD, Pettersen EF, et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. Nov 2023;32(11):e4792. doi: 10.1002/pro.4792 [DOI] [PMC free article] [PubMed] [Google Scholar]