Abstract
Primary sclerosing cholangitis (PSC) and Primary biliary cholangitis (PBC) are chronic inflammatory biliary diseases characterized by progressive damage of the bile ducts, resulting in hepatobiliary fibrosis and cirrhosis. Currently, specific biomarkers that allow to distinguish between PSC and PBC do not exist. In this study, we examined the salivary proteome by carrying out a comprehensive and non-invasive screening aimed at highlighting possible quali-quantitative protein deregulations that could be the starting point for the identification of effective biomarkers in future. Saliva samples collected from 6 PBC patients were analyzed using a liquid chromatography–tandem mass spectrometry technique, and the results were compared with those previously obtained in the PSC group. We identified 40 proteins as significantly deregulated in PSC patients compared to the PBC group. The Gene Ontology and pathway analyses highlighted that several proteins (e.g., small integral membrane protein 22, cofilin-1, macrophage-capping protein, plastin-2, and biliverdin reductase A) were linked to innate immune responses and actin cytoskeleton remodeling, which is a critical event in liver fibrosis and cancer progression. These findings provide new foundations for a deeper understanding of the pathophysiology of PSC and demonstrate that saliva is a suitable biological sample for obtaining proteomic fingerprints useful in the search for biomarkers capable of discriminating between the two cholestatic diseases.
Keywords: primary sclerosing cholangitis, primary biliary cholangitis, saliva, proteomics, LC-MS/MS, cytoskeleton
1. Introduction
Primary sclerosing cholangitis (PSC) and primary biliary cholangitis (PBC) are chronic liver diseases characterized by inflammation and progressive damage of the bile ducts, resulting in hepatobiliary fibrosis and cirrhosis. PSC is a rare disease more frequent in men than in women (ratio 2:1) and shows a peak incidence around 40 years [1]. On the other hand, PBC predominantly affects women (ratio 1:9) with a peak range between 60 and 79 years [2]. These cholestatic diseases share multiple clinical (e.g., fatigue, pruritus, and jaundice) and biochemical features (elevated levels of alkaline phosphatase (ALP) and gamma-glutamyl transferase (GGT)), making differential diagnosis a challenge [3]. Histological examinations can provide additional information, showing the typical ‘onion skin’ lesions in PSC [4], while PBC displays lymphocytic infiltrates and granulomas surrounding the small bile ducts [5]. However, liver biopsies are very invasive and risky, therefore they are unattractive diagnostic tools in clinical routine. Thus, reliable and non-invasive biomarkers that can accurately distinguish between PSC and PBC, or can assess disease activity and prognosis, represent an urgency for patients and a challenge for researchers and clinicians.
Recent advances in proteomics mass spectrometry (MS)-based analyses are promising in this area, as they can provide a comprehensive profile of the protein content within biological samples such as serum, bile, or saliva, showing the alterations in protein expression that accompany a particular disease [6]. This approach has occasionally been applied in PSC and PBC patients to identify discriminatory proteins towards healthy individuals and patients with liver and/or biliary tumors or autoimmune liver disease. For instance, bile, serum, and urine MS-based proteomics were investigated to discriminate PSC from cholangiocarcinoma (CCA) and benign biliary disorders [7,8,9,10] or to identify proteins with potential prognostic value [11]. MS techniques were also applied to identify serum proteins able to discriminate PBC patients from healthy subjects [12] or autoimmune hepatitis patients [13]. Over the last 10 years, human saliva has been investigated for diagnostic purposes because it reflects the general health status of an individual [14,15]. Many studies have shown that saliva is a useful biofluid for detecting oral diseases [16,17] and also systemic conditions, including heart disease [18,19], diabetes [20], and several types of cancer [21,22,23,24]. In the hepatic field, some evidence suggested that liver function parameters can be assessed in saliva [25] and proposed several metabolites to discriminate between healthy individuals and patients with liver cirrhosis or hepatocellular carcinoma [26]. Interestingly, salivary proteomics investigations were carried out to identify salivary protein differences between subjects affected by PBC and autoimmune hepatitis [27,28], and between PSC patients and healthy individuals [29]. In this scenario, our study is the first to have conducted a comparative salivary proteomic analysis between subjects with PSC and PBC, providing new foundations for a deeper understanding of the PSC pathophysiology, and therefore it might have relevant implications for improving diagnosis, prognosis, and treatment strategies.
2. Results
2.1. Salivary Proteomics of PSC Patients
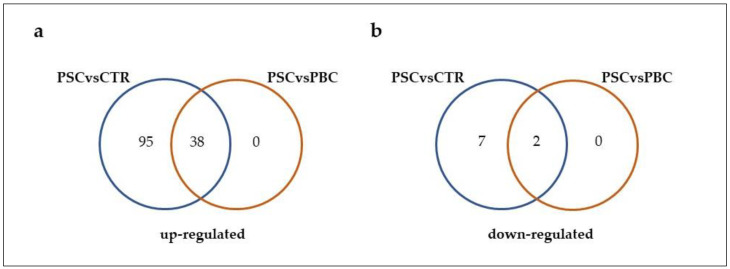
Saliva samples from PSC patients and healthy controls were compared, and, 142 proteins passed the requirement of having a Mann–Whitney p-value of less than 0.05 and log2FC ≤ 0.58 or log2FC ≥ 0.58 [29]. Among them, the expression levels of 40 proteins were also deregulated in PSC patients in respect to those in the PBC group (Figure 1 and Figure S1). As reported in Figure 1a, 38 proteins were up-regulated in PSC patients compared to both healthy individuals and PBC patients, and 2 proteins were down-regulated (Figure 1b) in PSC patients compared to both healthy and PBC subjects. In Table 1, we listed these proteins, and for each one, the log2FC and the p-value were reported. The protein with the highest FC was small integral membrane protein 22 (log2FC = 3.589). Multiple other proteins also displayed significant positive change in expression between the two groups, such as biliverdin reductase A (log2FC = 3.117), F-box only protein 50 (log2FC = 2.814), and glycogenin-1 (log2FC = 2.609). In addition, in the PSC group we detected a reduction in expression level for prosaposin and immunoglobulin lambda variable 3–10 when compared with those determined in PBC patients, with a log2FC of −0.779 and −0.615, respectively. In addition, 102 proteins showed significant change in their abundance in PSC subjects compared to healthy controls, but no change was identified with respect to the PBC group (Figure 1). In particular, we identified 7 down-regulated and 95 up-regulated proteins that are listed in Table S1. In Table 2, we listed the 7 down-regulated and the top 20 out of 95 up-regulated DEPs in PSC subjects compared to healthy controls, which showed no significant change compared to the PBC group, and for each one the log2FC and the p-value were reported. Among the down-regulated proteins, the serum amyloid P-component exhibited the lower FC (log2FC = −1.079), followed by Mucin-5B (log2FC = −0.980), Mucin-16 (log2FC = −0.936), Cystatin-A (log2FC = −0.812), Zinc-alpha-2-glycoprotein (log2FC = −0.781), WAP four-disulfide core domain protein 2 (log2FC = −0.700), and Immunoglobulin kappa constant (log2FC = −0.584). On the other hand, the protein that displayed the highest FC was Kallikrein-6 (log2FC = 5.136). Several other proteins also displayed up-regulation in the PSC group without significant change if compared to the PBC subjects, such as Protein S100-A7 (log2FC = 2.793), Aldo-keto reductase family 1 member B10 (log2FC = 2.513), Complement factor I (log2FC = 2.309), Low-affinity immunoglobulin gamma Fc region receptor III-A (log2FC = 2.231), and Osteoclast-stimulating factor 1 (log2FC = 2.218).
Figure 1.
Venn diagrams showing the number of up-regulated (a) and down-regulated (b) proteins in PSC patients compared to both healthy subjects and PBC patients. Each colored circle represents a different dataset, and areas of overlap indicate shared proteins. The cutoff criteria for statistical significance were defined based on log2FC ≤ 0.58 or log2FC ≥ 0.58, and p-value < 0.05.
Table 1.
List of 40 DEPs in PSC patients compared to both healthy and PBC subjects, which passed the cutoff log2FC ≤ 0.58 or log2FC ≥ 0.58 FC, and a Mann–Whitney p-value < 0.05.
| Protein ID | Protein Name | log2FC(PSCvsPBC) | p-Value |
|---|---|---|---|
| DOWN-REGULATED PROTEINS | |||
| P07602 | Prosaposin | −0.779 | 0.006 |
| A0A075B6K4 | Immunoglobulin lambda variable 3–10 | −0.615 | 0.004 |
| UP-REGULATED PROTEINS | |||
| K7EJ46 | Small integral membrane protein 22 | 3.589 | 0.002 |
| P53004 | Biliverdin reductase A | 3.117 | 0.0001 |
| Q6ZVX7 | F-box only protein 50 | 2.814 | 0.006 |
| P46976 | Glycogenin-1 | 2.609 | 0.001 |
| O15511 | Actin-related protein 2/3 complex subunit 5 | 2.370 | 0.017 |
| P47929 | Galectin-7 | 2.267 | 0.0001 |
| O75367 | Core histone macro-H2A.1 | 2.225 | 0.015 |
| P60842 | Eukaryotic initiation factor 4A-I | 2.096 | 0.013 |
| P49327 | Fatty acid synthase | 2.058 | 0.011 |
| P40121 | Macrophage-capping protein | 1.995 | 0.001 |
| P29350 | Tyrosine-protein phosphatase non-receptor type 6 | 1.968 | 0.008 |
| P49368 | T-complex protein 1 subunit gamma | 1.714 | 0.002 |
| P23526 | Adenosylhomocysteinase | 1.543 | 0.005 |
| Q96BQ1 | Protein FAM3D | 1.503 | 0.005 |
| P19823 | Inter-alpha-trypsin inhibitor heavy chain H2 | 1.464 | 0.015 |
| P23528 | Cofilin-1 | 1.413 | 0.001 |
| Q08431 | Lactadherin | 1.358 | 0.016 |
| P14550 | Aldo-keto reductase family 1 member A1 | 1.309 | 0.021 |
| P13639 | Elongation factor 2 | 1.195 | 0.0002 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | 1.190 | 0.042 |
| P53396 | ATP-citrate synthase | 1.183 | 0.041 |
| P51159 | Ras-related protein Rab-27A | 1.159 | 0.014 |
| P07900 | Heat shock protein HSP 90-alpha | 1.140 | 0.013 |
| P17900 | Ganglioside GM2 activator | 1.120 | 0.042 |
| P09211 | Glutathione S-transferase P | 1.102 | 0.009 |
| P16152 | Carbonyl reductase [NADPH] 1 | 1.051 | 0.039 |
| Q15080 | Neutrophil cytosol factor 4 | 1.012 | 0.045 |
| Q9UKR3 | Kallikrein-13 | 0.996 | 0.032 |
| Q13404 | Ubiquitin-conjugating enzyme E2 variant 1 | 0.960 | 0.0120 |
| P18206 | Vinculin | 0.953 | 0.038 |
| P09960 | Leukotriene A-4 hydrolase | 0.892 | 0.043 |
| Q92743 | Serine protease HTRA1 | 0.840 | 0.033 |
| P08238 | Heat shock protein HSP 90-beta | 0.825 | 0.020 |
| P51149 | Ras-related protein Rab-7a | 0.807 | 0.003 |
| P13489 | Ribonuclease inhibitor | 0.798 | 0.021 |
| P13796 | Plastin-2 | 0.716 | 0.045 |
| P43490 | Nicotinamide phosphoribosyltransferase | 0.654 | 0.021 |
| P52209 | 6-phosphogluconate dehydrogenase, decarboxylating | 0.625 | 0.004 |
Table 2.
List of the 7 down-regulated and the top 20 out of 95 up-regulated DEPs identified in PSC subjects compared to healthy controls, which showed no significant change compared to the PBC group. For each protein, the cutoff of log2FC ≤ 0.58 or log2FC ≥ 0.58, and a Mann–Whitney p-value < 0.05 were applied.
| Protein ID | Protein Name | log2FC(PSCvsControl) | p-Value |
|---|---|---|---|
| DOWN-REGULATED PROTEINS | |||
| P02743 | Serum amyloid P-component | −1.079 | 0.013 |
| Q9HC84 | Mucin-5B | −0.980 | 0.0002 |
| Q8WXI7 | Mucin-16 | −0.936 | 0.031 |
| P01040 | Cystatin-A | −0.812 | 0.034 |
| P25311 | Zinc-alpha-2-glycoprotein | −0.781 | 0.021 |
| Q14508 | WAP four-disulfide core domain protein 2 | −0.700 | 0.037 |
| P01834 | Immunoglobulin kappa constant | −0.584 | 0.048 |
| UP-REGULATED PROTEINS | |||
| Q92876 | Kallikrein-6 | 5.136 | 0.006 |
| P31151 | Protein S100-A7 | 2.793 | 0.003 |
| O60218 | Aldo-keto reductase family 1 member B10 | 2.513 | 0.048 |
| P05156 | Complement factor I | 2.309 | 0.017 |
| P08637 | Low affinity immunoglobulin gamma Fc region receptor III-A | 2.231 | 0.012 |
| Q92882 | Osteoclast-stimulating factor 1 | 2.218 | 0.009 |
| P19878 | Neutrophil cytosol factor 2 | 2.096 | 0.005 |
| O75923 | Dysferlin | 2.018 | 0.006 |
| Q96QK1 | Vacuolar protein sorting-associated protein 35 | 1.916 | 0.01 |
| Q14002 | Carcinoembryonic antigen-related cell adhesion molecule 7 | 1.897 | 0.045 |
| Q13393 | Phospholipase D1 | 1.865 | 0.001 |
| P53618 | Coatomer subunit beta | 1.822 | 0.02 |
| Q9BQR3 | Serine protease 27 | 1.771 | 0.015 |
| Q92839 | Hyaluronan synthase 1 | 1.757 | 0.023 |
| P30101 | Protein disulfide-isomerase A3 | 1.683 | 0.001 |
| Q6JEL2 | Kelch-like protein 10 | 1.678 | 0.018 |
| O43240 | Kallikrein-10 | 1.603 | 0.002 |
| Q8N6Q3 | CD177 antigen | 1.548 | 0.006 |
| Q9Y2X7 | ARF GTPase-activating protein GIT1 | 1.507 | 0.001 |
| Q14974 | Importin subunit beta-1 | 1.503 | 0.015 |
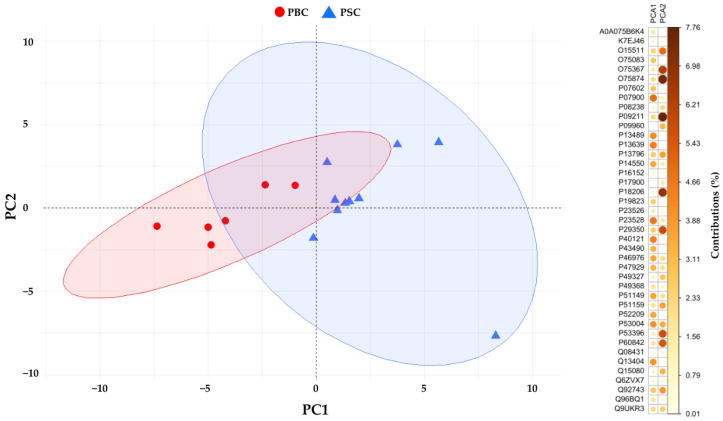
All the 40 identified DEPs were subjected to Principal Component Analysis (PCA), and the reduced dataset was used to investigate the usability of these proteins as more specific for PSC status (Figure 2). The first two principal components (PC1 and PC2) explain 38.4% and 17.1% of the data variance, respectively. Among the two components, the first one contains the highest percentage of variance between the data, and itis of interest in discriminating between the two groups of patients.
Figure 2.
PCA scores plot with 95% confidence ellipse of the first two principal components (PC) obtained by the analysis of proteins significantly different in abundance (p < 0.05) between PSC patients (blue triangles) and PBC patients (red circles). For each protein, the contribution in the calculation of the variability of the first two principal components is expressed as a percentage.
2.2. Gene Ontology Enrichment and Pathway Analysis
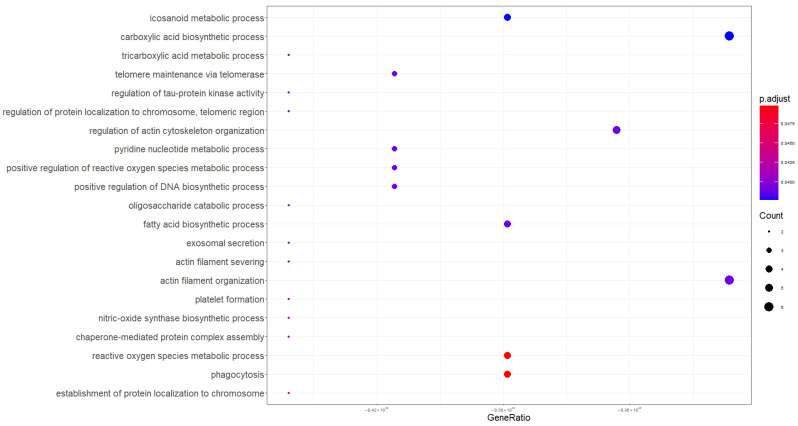
To obtain more biological insights, the GO enrichment analysis was performed using the clusterProfiler V.4.8.3 R package. Firstly, GO analysis was performed on the proteins listed in Table 1 to identify the changes in the BPs more tightly modulated by the PSC. As shown in Table 3 and Figure 3, the BPs were primarily enriched in biosynthetic and metabolic processes involving carboxylic acid, eicosanoids, fatty acids, and pyridine nucleotides. In addition, processes related to the organization of actin filaments, platelet formation, and the reactive oxygen species metabolic process also emerged. For deeper investigation, a pathway analysis was performed using g:Profiler, discovering significant enriched terms related to the innate immune responses and neutrophils degranulation (Figure 4). This analysis also highlighted two terms not statistically significant but biologically relevant, identified as metabolic reprogramming in colon cancer and glutathione metabolism. PSC and PBC are both chronic liver diseases involving bile ducts, sharing several overlapping clinical and serological features, though they are distinct pathologies. In order to highlight possible biological processes common to both diseases, a GO analysis was conducted on Table S1 proteins, which are deregulated in the PSC compared to the control group but not significantly different in expression compared to subjects with PBC. As shown in Table 4, GO enrichment analysis revealed that the protein-related BPs were markedly concentrated in inflammatory responses, phagocytosis, and cellular redox homeostasis (GO:0072593, GO:1990748, GO:0042744).
Table 3.
GO enrichment analysis of the DEPs identified in PSC patients compared to both healthy control and PBC subjects. The GO terms belonging to BPs are listed, and for each one the significance level (p-adjust < 0.05) and the gene list were reported.
| ID | Description | p-Adjust | Gene ID |
|---|---|---|---|
| GO:0046394 | carboxylic acid biosynthetic process | 0.0387 | GSTP1/LTA4H/AKR1A1/CBR1/FASN/ACLY |
| GO:0006690 | eicosanoid metabolic process | 0.0377 | CES2/GSTP1/LTA4H/CBR1 |
| GO:0007015 | actin filament organization | 0.0390 | ARPC5/WDR1/RNH1/LCP1/CFL1/CAPG |
| GO:0032956 | regulation of actin cytoskeleton organization | 0.0390 | SMIM22/ARPC5/WDR1/RNH1/CAPG |
| GO:0006633 | fatty acid biosynthetic process | 0.0390 | GSTP1/CBR1/FASN/ACLY |
| GO:0007004 | telomere maintenance via telomerase | 0.0390 | HSP90AA1/HSP90AB1/CCT3 |
| GO:2000379 | positive regulation of reactive oxygen species metabolic process | 0.0390 | GSTP1/CBR1/RAB27A |
| GO:2000573 | positive regulation of DNA biosynthetic process | 0.0390 | HSP90AA1/HSP90AB1/CCT3 |
| GO:0019362 | pyridine nucleotide metabolic process | 0.0390 | IDH1/NAMPT/PGD |
| GO:0072350 | tricarboxylic acid metabolic process | 0.0390 | IDH1/ACLY |
| GO:1902947 | regulation of tau-protein kinase activity | 0.0390 | HSP90AA1/HSP90AB1 |
| GO:1904814 | regulation of protein localization to chromosome, telomeric region | 0.0390 | MACROH2A1/CCT3 |
| GO:0051014 | actin filament severing | 0.0390 | CFL1/CAPG |
| GO:0009313 | oligosaccharide catabolic process | 0.0390 | GM2A/PGD |
| GO:1990182 | exosomal secretion | 0.0390 | RAB7A/RAB27A |
| GO:0030220 | platelet formation | 0.0406 | WDR1/PTPN6 |
| GO:0051767 | nitric-oxide synthase biosynthetic process | 0.0406 | GSTP1/NAMPT |
| GO:0051131 | chaperone-mediated protein complex assembly | 0.0414 | HSP90AA1/HSP90AB1 |
| GO:0006909 | phagocytosis | 0.0498 | RAB7A/RAB27A/MFGE8/NCF4 |
| GO:0072593 | reactive oxygen species metabolic process | 0.0498 | GSTP1/CBR1/RAB27A/NCF4 |
| GO:0070199 | establishment of protein localization to chromosome | 0.0498 | MACROH2A1/CCT3 |
Figure 3.
Dot plot showing enrichment of Gene Ontology Biological Processes (PBs) for the DEPs identified in PSC patients compared to both healthy control and PBC subjects.
Figure 4.
Pathway analysis via the online tool g:Profiler. For each term, the statistical significance and the involved proteins are reported. * Terms not statistically significant but biologically relevant.
Table 4.
GO enrichment analysis of the DEPs identified in PSC patients compared to healthy control, which showed no significant change compared to the PBC group. The GO terms belonging to BPs are listed, and for each one the significance level (p-adjust < 0.05) and the gene list were reported.
| ID | Description | p-Adjust | Gene ID |
|---|---|---|---|
| GO:0006909 | phagocytosis | 0.0028 | DYSF/AHSG/ANXA3/CYBA/NCF2/ICAM3/ANXA11/ PTPRJ/PYCARD |
| GO:0072593 | reactive oxygen species metabolic process | 0.0028 | HBD/HBB/G6PD/CYBA/ARF4/NCF2/PRDX5/CD177/ DHRS4 |
| GO:0007596 | blood coagulation | 0.0308 | KNG1/HBB/SERPING1/SAA1/GNA13/FERMT3/TLN1 |
| GO:0006098 | pentose-phosphate shunt | 0.0308 | H6PD/G6PD/PGAM1 |
| GO:0006740 | NADPH regeneration | 0.0308 | H6PD/G6PD/PGAM1 |
| GO:0052547 | regulation of peptidase activity | 0.0329 | KNG1/AHSG/SERPING1/MMP9/CD44/PRDX5/PSME1/ ITIH4/PYCARD |
| GO:0002526 | acute inflammatory response | 0.0329 | IGHG1/AHSG/FCGR3A/SAA1/ITIH4 |
| GO:1990748 | cellular detoxification | 0.0329 | AKR1B10/HBD/HBB/PRDX6/PRDX5 |
| GO:0042060 | wound healing | 0.0347 | KNG1/HBB/SERPING1/ANXA6/SAA1/CD44/GNA13/ FERMT3/TLN1 |
| GO:0051156 | glucose 6-phosphate metabolic process | 0.0347 | H6PD/G6PD/PGAM1 |
| GO:0042744 | hydrogen peroxide catabolic process | 0.0383 | HBD/HBB/PRDX5 |
| GO:0050878 | regulation of body fluid levels | 0.0385 | KNG1/HBB/SERPING1/SAA1/CYBA/GNA13/FERMT3/ TLN1 |
| GO:0006958 | complement activation, classical pathway | 0.0428 | SERPING1/CFI/C7 |
| GO:0002437 | inflammatory response to antigenic stimulus | 0.0441 | IGHG1/FCGR3A/IL1RN/IL36A |
| GO:0030212 | hyaluronan metabolic process | 0.0471 | CD44/ITIH4/HAS1 |
3. Discussion
In this explorative study, we have analyzed and compared the saliva proteomic profiles of PBC patients with those obtained in our previous study on PSC subjects and healthy controls [29]. Comparative analysis showed 142 DEPs in the PSC group compared to healthy subjects, and among them, 102 proteins did not have significantly different levels with respect to the PBC group (Table 2), suggesting that these proteins are not good discriminators between the two cholestatic diseases. Interestingly, 40 of the 142 proteins might be more indicative of PSC, having shown significant deregulation compared to patients with PBC (Table 1). For these proteins we performed a GO analysis highlighting 7 proteins as significantly related to the actin filaments organization (Table 3; Figure 3). Among them, the most up-regulated protein was small integral membrane protein 22, with a log2FC = 3.589. Although no literature data reported information on its involvement in cholestatic diseases or related manifestations, Polycarpou-Schwarz and colleagues demonstrated that small integral membrane protein 22 is involved in actin cytoskeleton organization in the MCF7 breast cancer cell line, affecting cell proliferation, cell migration, and cell cycle progression [30]. Cofilin-1 is an actin-binding protein that regulates the cytoskeleton dynamics in hepatic stellate cells (HSCs), triggering the deposition of type I collagen during liver fibrosis [31]. Although the up-regulation of cofilin-1 was only detected in another salivary proteomics study on patients with PBC [27], two recent studies demonstrated the increase in cofilin-1 expression in hepatocellular carcinoma tissues [32,33]. Macrophage-capping protein is a calcium-sensitive member of the gelsolin family and is involved in the remodeling of the cytoskeleton actin filaments [34]. No clinical study has shown its deregulation in biological fluids or tissues from patients with PSC or PBC; however, the association of increased macrophage-capping protein expression with vascular invasion in patients with cholangiocarcinoma [35] and hepatocellular carcinoma tissues [36] was recently demonstrated. Another deregulated salivary protein related to cytoskeleton shaping was actin-related protein 2/3 complex subunit 5 that modulates the actin polymerization after the stimulation by nucleation-promoting factor [37]. Huang and colleagues highlighted higher actin-related protein 2/3 complex subunit 5 expression in hepatocellular carcinoma tissues and cells when compared with healthy liver tissues or normal liver cells [38]. Although our results are preliminary and supporting literature data are scarce, they seem to suggest the proteins discussed above as possible salivary indicators of liver fibrosis caused by actin cytoskeleton remodeling in HSCs. Furthermore, their up-regulation in hepatocellular carcinoma and cholangiocarcinoma would suggest that these proteins could be indicative of the progression of cholestatic disease towards a cancerous form. Among the proteins linked to cytoskeletal shaping, plastin-2 is the one with the lowest degree of deregulation in PSC patients compared to the PBC group (log2FC = 0.716). Tit-Oon and colleagues analyzed the secretoma of cholangiocarcinoma cells cultured in 3D, identifying plastin-2 as a secreted protein; protein not found in mixed hepatocarcinoma-cholangiocarcinoma cell culture [39]. Joshi and colleagues have recently demonstrated the connection between plastin-2 and the NLRP3 inflammasome activation in macrophages using a mouse model of lung fibrosis [40]. It is well known that increased NLRP3 inflammasome expression was a key event in HSC activation and in cholangiocytes switch to a reactive phenotype, contributing to the development of liver and biliary fibrosis [41,42,43]. Consistent results were observed in liver sections of patients affected by PSC [41] and in a Mdr2-knockout mouse model [44]. These findings are very interesting as they would suggest a more pronounced relationship of plastin-2 with the biliary tissue rather than the liver parenchyma, corroborating its salivary up-regulation as related to biliary fibrosis in PSC, in which macrophages play a major role [45]. Considering the secretion of plastin-2 by cholangiocarcinoma cells, our results could suggest this protein as a salivary indicator of a probable evolution towards cancer. Although biliverdin reductase A did not emerge as being significantly associated with any biological process in our GO analysis, we believe it may have considerable clinical relevance. Very little is known about the role of biliverdin reductase A in the pathogenesis and progression of cholestatic diseases; however, according to our results, recent studies highlighted its up-regulation in serum samples from PSC patients [46]. Biliverdin reductase A is an enzyme that converts biliverdin into bilirubin, which possesses cytoprotective properties in response to oxidative stress and regulates the innate immune responses [47,48,49]. Recently, Weaver and colleagues suggested that biliverdin reductase A may prevent HSCs fibrogenesis by antagonizing the toll-like receptor 4 (TLR4) signaling pathway [50]. Indeed, TLR4 signaling plays a key role in activated HSCs, which represent the major fibrogenic cell type during liver fibrosis [43,51]. In addition, increased biliverdin reductase A expression was detected on the plasma membrane of macrophages after lipopolysaccharide treatment, used to initiate an inflammatory response [52]. Collectively, this evidence would attribute to biliverdin reductase A a key role in the activation of HSCs by actively contributing to the restraint of the fibrotic process. In this perspective, the increased salivary expression of biliverdin reductase A in patients with PSC could be indicative of its cytoprotective activity against liver fibrosis. In this study, we demonstrated that PSC significantly alters the salivary proteome compared to PBC and healthy controls, identifying multiple cytoskeleton-related proteins that might represent a specific signature of the PSC. To corroborate our results, all the 40 DEPs were subjected to PCA, and the reduced dataset was used to investigate if these proteins were more specific for PSC status (Figure 2). Our analysis had shown that the expression of four proteins (biliverdin reductase A, macrophage-capping protein, heat shock protein HSP 90-beta, and cofilin-1) out of the 40 salivary DEPs might be more indicative of PSC than the PBC group. These data would seem to support the results of the pathway analysis (Figure 4) that showed a greater involvement of the innate immune system in PSC than in PBC subjects. Indeed, cofilin, macrophage-capping protein, and HSP90 are proteins whose role in modulating the innate immune response by macrophages is well documented [53,54,55,56]. In addition, previous studies showed a higher number of macrophages in liver biopsies from patients with PSC than in those from PBC or other liver diseases [57,58]. This phenomenon could underlie the greater involvement of innate immune system responses in patients with PSC than in the other groups analyzed in this study, suggesting that the salivary proteins macrophage-capping protein, cofilin-1, and heat shock protein HSP 90-beta could be indicators of the macrophage involvement in the pathophysiology of PSC. Since some of the proteins discussed (e.g., actin-related protein 2/3 complex subunit 5, cofilin-1,plastin-2 and macrophage-capping protein) go into post-translational modifications (PMTs), such as phosphorylation and glycosylation, that regulate their activity, distribution, and stability [59,60,61,62,63,64], further MS-based PTM analyses are needed to clarify the role that might have in the onset and progression of PSC [65,66]. Certainly, the most impactful limitation of this study is related to the small patient cohort that should be addressed in larger population studies to confirm the peculiar phenomena observed. However, this preliminary exploratory study provides important information for a deeper understanding of the pathophysiology of PSC, demonstrating that saliva is a suitable biological sample for obtaining proteomic fingerprints useful in the search for biomarkers that can effectively discriminate between the two cholestatic diseases.
4. Materials and Methods
4.1. Reagents
All the reagents and materials used for sample collection, processing and analysis in liquid chromatography–tandem mass spectrometry (LC-MS/MS) were detailed in our previous article [67].
4.2. Patient Characteristics
Six female patients with confirmed PBC were subsequently recruited at the University Hospital of Pisa (Hepatology Unit and Gastroenterology Unit), according to the established inclusion criteria. The patient group was in a range of 52–70 years (median age of 60.5 years). A saliva sample was obtained from each subject as previously described [29,68]. At the time of saliva collection, patients showed a median value for alanine aminotransferase (ALT), aspartate aminotransferase (AST), and total bilirubin of 17.00 U/L (10–48), 18.00 U/L (18–42), and 0.36 mg/dL (0.22–0.49), respectively. The median GGT, ALP, albumin, and gamma globulin values were 79 U/L (12–161), 123 U/L (84–350), 4.35 g/L (3.8–5.0), and 19.7% (13.9–24.6), respectively. A total of 2 patients out of 6 were positive for anti-nuclear antibodies (ANA) and anti-mitochondrial antibodies (AMA). Among them, three patients were positive for ANA and P-ANCA, and just one patient was positive for anti-smooth muscle antibodies (ASMA). A total of 6 males and 4 females were enrolled for both the PSC group and healthy individuals, aged between 18 and 70 years (median age of 49 years) and 27–65 years (median age of 41 years), respectively.
The study obtained the approval of the Comitato Etico Area Vasta Nord Ovest (Protocol code 57532, 30 October 2019, Pisa, Italy). All the patients signed an informed consent to participate in the study.
4.3. Saliva Collection, Processing, and LC-MS/MS Analysis
Spontaneous saliva from each PBC patient was collected and enriched in extracellular vesicles by applying a differential centrifugation isolation protocol; then the proteins were extracted and quantified according to our previous papers [29,68]. A total of 100 µg of proteins for each sample were subsequently reduced, alkylated, and digested. After that, the peptide mixtures were centrifuged, desalted, resuspended in CH3CN/0.1% HCOOH (ratio 5/95) to achieve a final peptide concentration of 2 µg/µL, and analyzed by LC-MS/MS as previously reported [29]. Sample analysis was performed according to our previous paper [29] using a micro-HPLC (Eksigent Ekspert microLC 200, AB Sciex, Concord, ON, Canada) coupled with a Triple TOF 5600 mass spectrometer (AB Sciex, Concord, ON, Canada). The protein quantification was achieved through Sequential Window Acquisition of all Theoretical fragment ion Mass Spectra (SWATH-MS) methodology, as previously reported [29].
4.4. Data Processing and Statistical Analysis
SWATH raw files were processed using the free universal software DIA-NN (version 1.8) as previously described [29], quantifying 733 proteins. This dataset was compared with that obtained from our previous salivary proteomic analysis on patients with PSC and healthy controls [29]. For each protein, we compared the fold change (FC) calculated as the ratio between the mean of abundance in PSC patients and the PBC group with those obtained as the ratio between the mean of abundance in PSC patients and the healthy control group. Proteins were considered significantly differentially expressed (DEPs) when FC ≤ 1/1.5 or FC ≥ 1.5, and p-value < 0.05 (determined using the non-parametric Wilcoxon test).
The DEPs were also subjected to Gene Ontology (GO) over-representation analysis, which was performed using the clusterProfiler package in R (version 4.8.3) and the OrgDb annotation database, Genome-wide annotation for Human (version 3.17.0). Significant GO terms were identified using a p-value threshold of 0.05, with False Discovery Rate (FDR) correction applied via the Benjamini–Hochberg method. The pathway analysis was performed using the g:Profiler tool [69]. Principal component analysis (PCA) was performed using the FactoMineR package implemented in R (version 2.11). To highlight the variables that most contributed to each dimension, the corrplot package (version 0.92) was used.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29235783/s1, Table S1: List of down and up-regulated proteins identified in PSC patients compared to healthy control which showed no significant change compared to the PBC group. The cutoff log2FC ≤ 0.58 or log2FC ≥ 0.58 FC, and a Mann–Whitney p-value < 0.05 were applied; Figure S1: Volcano plot of the significantly DEPs in PSC patients compared to both healthy and PBC subjects. The red dots represent the 38 up-regulated proteins, blue dots mean the 2 down-regulated proteins, and gray ones are non-significantly altered proteins. The x-axis represents the protein difference reported as log2FC, and the y-axis corresponds to the −log10(p-value).
Author Contributions
Conceptualization, methodology, data curation, writing—original draft preparation, writing—review and editing, E.C.; methodology, writing—review and editing, F.N.; data curation, writing—review and editing, A.M.; methodology, writing—review and editing, B.C.; writing—review and editing, M.B.; writing—review and editing, M.R.B.; supervision, writing—review and editing, A.C.; resources, project administration, funding acquisition, writing—review and editing, S.R. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Comitato Etico Area Vasta Nord Ovest (protocol code 57532, 30 October 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data will be made available on request.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the AIRCS—Italian Association for Sclerosing Cholangitis.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Boonstra K., Beuers U., Ponsioen C.Y. Epidemiology of Primary Sclerosing Cholangitis and Primary Biliary Cirrhosis: A Systematic Review. J. Hepatol. 2012;56:1181–1188. doi: 10.1016/j.jhep.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 2.Trivella J., John B.V., Levy C. Primary Biliary Cholangitis: Epidemiology, Prognosis, and Treatment. Hepatol. Commun. 2023;7:e0179. doi: 10.1097/HC9.0000000000000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das D.D., Sharma N., Chawla P.A. Current Trends and Future Perspectives in the Treatment of PBC and PSC: A Review. Health Sci. Rev. 2022;5:100065. doi: 10.1016/j.hsr.2022.100065. [DOI] [Google Scholar]
- 4.Karlsen T.H., Folseraas T., Thorburn D., Vesterhus M. Primary Sclerosing Cholangitis—A Comprehensive Review. J. Hepatol. 2017;67:1298–1323. doi: 10.1016/j.jhep.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 5.Mironova M., Gopalakrishna H., Rodriguez Franco G., Holland S.M., Koh C., Kleiner D.E., Heller T. Granulomatous Liver Diseases. Hepatol. Commun. 2024;8:e0392. doi: 10.1097/HC9.0000000000000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birhanu A.G. Mass Spectrometry-Based Proteomics as an Emerging Tool in Clinical Laboratories. Clin. Proteom. 2023;20:32. doi: 10.1186/s12014-023-09424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lankisch T.O., Metzger J., Negm A.A., Vosskuhl K., Schiffer E., Siwy J., Weismüller T.J., Schneider A.S., Thedieck K., Baumeister R., et al. Bile Proteomic Profiles Differentiate Cholangiocarcinoma from Primary Sclerosing Cholangitis and Choledocholithiasis. Hepatology. 2011;53:875–884. doi: 10.1002/hep.24103. [DOI] [PubMed] [Google Scholar]
- 8.Navaneethan U., Lourdusamy V., Gk Venkatesh P., Willard B., Sanaka M.R., Parsi M.A. Bile Proteomics for Differentiation of Malignant from Benign Biliary Strictures: A Pilot Study. Gastroenterol. Rep. 2015;3:136–143. doi: 10.1093/gastro/gou066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzger J., Negm A.A., Plentz R.R., Weismüller T.J., Wedemeyer J., Karlsen T.H., Dakna M., Mullen W., Mischak H., Manns M.P., et al. Urine Proteomic Analysis Differentiates Cholangiocarcinoma from Primary Sclerosing Cholangitis and Other Benign Biliary Disorders. Gut. 2013;62:122–130. doi: 10.1136/gutjnl-2012-302047. [DOI] [PubMed] [Google Scholar]
- 10.Kan M., Chiba T., Konno R., Kouchi Y., Mishima T., Kawashima Y., Kishimoto T., Ohtsuka M., Ohara O., Kato N. Bile Proteome Analysis by High-Precision Mass Spectrometry to Examine Novel Biomarkers of Primary Sclerosing Cholangitis. J. Hepato Biliary Pancreat. 2023;30:914–923. doi: 10.1002/jhbp.1299. [DOI] [PubMed] [Google Scholar]
- 11.Vesterhus M., Holm A., Hov J.R., Nygård S., Schrumpf E., Melum E., Thorbjørnsen L.W., Paulsen V., Lundin K., Dale I., et al. Novel Serum and Bile Protein Markers Predict Primary Sclerosing Cholangitis Disease Severity and Prognosis. J. Hepatol. 2017;66:1214–1222. doi: 10.1016/j.jhep.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Li Y., Tang R., Leung P.S.C., Gershwin M.E., Ma X. Proteomics in Primary Biliary Cholangitis. Methods Mol. Biol. 2019;1981:163–173. doi: 10.1007/978-1-4939-9420-5_11. [DOI] [PubMed] [Google Scholar]
- 13.Bowlus C.L., Seeley E.H., Roder J., Grigorieva J., Roder H., Caprioli R.M., Gershwin M. In Situ Mass Spectrometry of Autoimmune Liver Diseases. Cell Mol. Immunol. 2011;8:237–242. doi: 10.1038/cmi.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaffe T., Cooper-White J., Beyerlein P., Kostner K., Punyadeera C. Diagnostic Potential of Saliva: Current State and Future Applications. Clin. Chem. 2011;57:675–687. doi: 10.1373/clinchem.2010.153767. [DOI] [PubMed] [Google Scholar]
- 15.Cui Y., Yang M., Zhu J., Zhang H., Duan Z., Wang S., Liao Z., Liu W. Developments in Diagnostic Applications of Saliva in Human Organ Diseases. Med. Nov. Technol. Devices. 2022;13:100115. doi: 10.1016/j.medntd.2022.100115. [DOI] [Google Scholar]
- 16.Punyadeera C., Slowey P.D. Nanobiomaterials in Clinical Dentistry. Elsevier; Amsterdam, The Netherlands: 2019. Saliva as an Emerging Biofluid for Clinical Diagnosis and Applications of MEMS/NEMS in Salivary Diagnostics; pp. 543–565. [Google Scholar]
- 17.Tang K.D., Baeten K., Kenny L., Frazer I.H., Scheper G., Punyadeera C. Unlocking the Potential of Saliva-Based Test to Detect HPV-16-Driven Oropharyngeal Cancer. Cancers. 2019;11:473. doi: 10.3390/cancers11040473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., Karunathilaka N., Senanayake S., Subramaniam V.N., Chan W., Kostner K., Fraser J., Atherton J.J., Punyadeera C. The Potential Prognostic Utility of Salivary Galectin-3 Concentrations in Heart Failure. Clin. Res. Cardiol. 2020;109:685–692. doi: 10.1007/s00392-019-01557-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., Walsh T., Atherton J.J., Kostner K., Schulz B., Punyadeera C. Identification and Validation of a Salivary Protein Panel to Detect Heart Failure Early. Theranostics. 2017;7:4350–4358. doi: 10.7150/thno.21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aitken J.P., Ortiz C., Morales-Bozo I., Rojas-Alcayaga G., Baeza M., Beltran C., Escobar A. α-2-Macroglobulin in Saliva Is Associated with Glycemic Control in Patients with Type 2 Diabetes Mellitus. Dis. Markers. 2015;2015:128653. doi: 10.1155/2015/128653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assad D., Mascarenhas E., Normando A.G., Chardin H., Barra G., Pratesi R., Nóbrega Y.K., Acevedo A.C., Guerra E. Correlation between Salivary and Serum CA15-3 Concentrations in Patients with Breast Cancer. Mol. Clin. Oncol. 2020;13:155–161. doi: 10.3892/mco.2020.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding F., Sun K., Sun N., Jiang Q., Cao M., Wu Z. iTRAQ-Based Proteomics Reveals SOD2 as a Potential Salivary Biomarker in Liver Cancer. Int. J. Biol. Markers. 2019;34:221–231. doi: 10.1177/1724600819841619. [DOI] [PubMed] [Google Scholar]
- 23.Wu Z.-Z., Wang J.-G., Zhang X.-L. Diagnostic Model of Saliva Protein Finger Print Analysis of Patients with Gastric Cancer. WJG. 2009;15:865. doi: 10.3748/wjg.15.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao H., Zhang L., Zhou H., Lee J.M., Garon E.B., Wong D.T.W. Proteomic Analysis of Human Saliva From Lung Cancer Patients Using Two-Dimensional Difference Gel Electrophoresis and Mass Spectrometry. Mol. Cell. Proteom. 2012;11:M111.012112. doi: 10.1074/mcp.M111.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Idkaidek N., Qawasmi H., Hanahen A., Abuqatouseh L., Hamadi S., Bustami M. Applicability of Saliva for Evaluation of Some Biochemical Parameters of Kidney and Liver Function in Healthy Individuals. mljgoums. 2020;14:1–6. doi: 10.29252/mlj.14.4.1. [DOI] [Google Scholar]
- 26.Hershberger C.E., Rodarte A.I., Siddiqi S., Moro A., Acevedo-Moreno L., Brown J.M., Allende D.S., Aucejo F., Rotroff D.M. Salivary Metabolites Are Promising Non-invasive Biomarkers of Hepatocellular Carcinoma and Chronic Liver Disease. Liver Cancer Int. 2021;2:33–44. doi: 10.1002/lci2.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guadalupi G., Contini C., Iavarone F., Castagnola M., Messana I., Faa G., Onali S., Chessa L., Vitorino R., Amado F., et al. Combined Salivary Proteome Profiling and Machine Learning Analysis Provides Insight into Molecular Signature for Autoimmune Liver Diseases Classification. IJMS. 2023;24:12207. doi: 10.3390/ijms241512207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olianas A., Guadalupi G., Cabras T., Contini C., Serrao S., Iavarone F., Castagnola M., Messana I., Onali S., Chessa L., et al. Top-Down Proteomics Detection of Potential Salivary Biomarkers for Autoimmune Liver Diseases Classification. Int. J. Mol. Sci. 2023;24:959. doi: 10.3390/ijms24020959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceccherini E., Michelucci E., Signore G., Coco B., Zari M., Bellini M., Brunetto M.R., Cecchettini A., Rocchiccioli S. The Clinical Utility of the Saliva Proteome in Rare Diseases: A Pilot Study for Biomarker Discovery in Primary Sclerosing Cholangitis. J. Clin. Med. 2024;13:544. doi: 10.3390/jcm13020544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polycarpou-Schwarz M., Groß M., Mestdagh P., Schott J., Grund S.E., Hildenbrand C., Rom J., Aulmann S., Sinn H.-P., Vandesompele J., et al. The Cancer-Associated Microprotein CASIMO1 Controls Cell Proliferation and Interacts with Squalene Epoxidase Modulating Lipid Droplet Formation. Oncogene. 2018;37:4750–4768. doi: 10.1038/s41388-018-0281-5. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y., Duan J., Ji W., Liu C., Li X., Wu Q., Gao C., Su C. A Novel Matrine Derivative, WM130, Inhibits Activation and Movement of Human Hepatic Stellate LX-2 Cells by Targeting Cofilin 1. Cytotechnology. 2022;74:613–622. doi: 10.1007/s10616-022-00548-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao P.-H., Hsu H.-H., Chen T.-S., Chen M.-C., Day C.-H., Tu C.-C., Lin Y.-M., Tsai F.-J., Kuo W.-W., Huang C.-Y. Phosphorylation of Cofilin-1 by ERK Confers HDAC Inhibitor Resistance in Hepatocellular Carcinoma Cells via Decreased ROS-Mediated Mitochondria Injury. Oncogene. 2017;36:1978–1990. doi: 10.1038/onc.2016.357. [DOI] [PubMed] [Google Scholar]
- 33.Yao B., Li Y., Chen T., Niu Y., Wang Y., Yang Y., Wei X., Liu Q., Tu K. Hypoxia-induced Cofilin 1 Promotes Hepatocellular Carcinoma Progression by Regulating the PLD1/AKT Pathway. Clin. Transl. Med. 2021;11:e366. doi: 10.1002/ctm2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Impe K., De Corte V., Eichinger L., Bruyneel E., Mareel M., Vandekerckhove J., Gettemans J. The Nucleo-Cytoplasmic Actin-Binding Protein CapG Lacks a Nuclear Export Sequence Present in Structurally Related Proteins. J. Biol. Chem. 2003;278:17945–17952. doi: 10.1074/jbc.M209946200. [DOI] [PubMed] [Google Scholar]
- 35.Morofuji N., Ojima H., Onaya H., Okusaka T., Shimada K., Sakamoto Y., Esaki M., Nara S., Kosuge T., Asahina D., et al. Macrophage-Capping Protein as a Tissue Biomarker for Prediction of Response to Gemcitabine Treatment and Prognosis in Cholangiocarcinoma. J. Proteom. 2012;75:1577–1589. doi: 10.1016/j.jprot.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 36.Kimura K., Ojima H., Kubota D., Sakumoto M., Nakamura Y., Tomonaga T., Kosuge T., Kondo T. Proteomic Identification of the Macrophage-Capping Protein as a Protein Contributing to the Malignant Features of Hepatocellular Carcinoma. J. Proteom. 2013;78:362–373. doi: 10.1016/j.jprot.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Welch M.D., DePace A.H., Verma S., Iwamatsu A., Mitchison T.J. The Human Arp2/3 Complex Is Composed of Evolutionarily Conserved Subunits and Is Localized to Cellular Regions of Dynamic Actin Filament Assembly. J. Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S., Sun L., Hou P., Liu K., Wu J. A Comprehensively Prognostic and Immunological Analysis of Actin-Related Protein 2/3 Complex Subunit 5 in Pan-Cancer and Identification in Hepatocellular Carcinoma. Front. Immunol. 2022;13:944898. doi: 10.3389/fimmu.2022.944898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tit-Oon P., Chokchaichamnankit D., Khongmanee A., Sawangareetrakul P., Svasti J., Srisomsap C. Comparative Secretome Analysis of Cholangiocarcinoma Cell Line in Three-Dimensional Culture. Int. J. Oncol. 2014;45:2108–2116. doi: 10.3892/ijo.2014.2636. [DOI] [PubMed] [Google Scholar]
- 40.Joshi H., Almgren-Bell A., Anaya E.P., Todd E.M., Van Dyken S.J., Seth A., McIntire K.M., Singamaneni S., Sutterwala F., Morley S.C. L-Plastin Enhances NLRP3 Inflammasome Assembly and Bleomycin-Induced Lung Fibrosis. Cell Rep. 2022;38:110507. doi: 10.1016/j.celrep.2022.110507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maroni L., Agostinelli L., Saccomanno S., Pinto C., Giordano D.M., Rychlicki C., De Minicis S., Trozzi L., Banales J.M., Melum E., et al. Nlrp3 Activation Induces Il-18 Synthesis and Affects the Epithelial Barrier Function in Reactive Cholangiocytes. Am. J. Pathol. 2017;187:366–376. doi: 10.1016/j.ajpath.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Sun M., Zhang Y., Guo A., Xia Z., Peng L. Progress in the Correlation Between Inflammasome NLRP3 and Liver Fibrosis. J. Clin. Transl. Hepatol. 2024;12:191–200. doi: 10.14218/JCTH.2023.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ceccherini E., Cecchettini A., Morales M.A., Rocchiccioli S. The Potentiality of Herbal Remedies in Primary Sclerosing Cholangitis: From In Vitro to Clinical Studies. Front. Pharmacol. 2020;11:813. doi: 10.3389/fphar.2020.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao L., Schneider K.M., Galvez E.J.C., Frissen M., Marschall H.-U., Su H., Hatting M., Wahlström A., Haybaeck J., Puchas P., et al. Intestinal Dysbiosis Augments Liver Disease Progression via NLRP3 in a Murine Model of Primary Sclerosing Cholangitis. Gut. 2019;68:1477–1492. doi: 10.1136/gutjnl-2018-316670. [DOI] [PubMed] [Google Scholar]
- 45.Trussoni C.E., LaRusso N.F. Macrophages Make a Difference in Cholestatic Liver Diseases—But How? J. Hepatol. 2023;79:1349–1351. doi: 10.1016/j.jhep.2023.09.022. [DOI] [PubMed] [Google Scholar]
- 46.Holm M., Joenväärä S., Saraswat M., Tohmola T., Saarela T., Tenca A., Arola J., Renkonen R., Färkkilä M. Quantitative Bile and Serum Proteomics for the Screening and Differential Diagnosis of Primary Sclerosing Cholangitis. PLoS ONE. 2022;17:e0272810. doi: 10.1371/journal.pone.0272810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barañano D.E., Rao M., Ferris C.D., Snyder S.H. Biliverdin Reductase: A Major Physiologic Cytoprotectant. Proc. Natl. Acad. Sci. USA. 2002;99:16093–16098. doi: 10.1073/pnas.252626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miralem T., Hu Z., Torno M.D., Lelli K.M., Maines M.D. Small Interference RNA-Mediated Gene Silencing of Human Biliverdin Reductase, but Not That of Heme Oxygenase-1, Attenuates Arsenite-Mediated Induction of the Oxygenase and Increases Apoptosis in 293A Kidney Cells. J. Biol. Chem. 2005;280:17084–17092. doi: 10.1074/jbc.M413121200. [DOI] [PubMed] [Google Scholar]
- 49.Wegiel B., Otterbein L.E. Go Green: The Anti-Inflammatory Effects of Biliverdin Reductase. Front. Pharmacol. 2012;3:47. doi: 10.3389/fphar.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weaver L., Hamoud A., Stec D.E., Hinds T.D. Biliverdin Reductase and Bilirubin in Hepatic Disease. Am. J. Physiol. -Gastrointest. Liver Physiol. 2018;314:G668–G676. doi: 10.1152/ajpgi.00026.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang C.-Y., Yuan W.-G., He P., Lei J.-H., Wang C.-X. Liver Fibrosis and Hepatic Stellate Cells: Etiology, Pathological Hallmarks and Therapeutic Targets. WJG. 2016;22:10512. doi: 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wegiel B., Baty C.J., Gallo D., Csizmadia E., Scott J.R., Akhavan A., Chin B.Y., Kaczmarek E., Alam J., Bach F.H., et al. Cell Surface Biliverdin Reductase Mediates Biliverdin-Induced Anti-Inflammatory Effects via Phosphatidylinositol 3-Kinase and Akt. J. Biol. Chem. 2009;284:21369–21378. doi: 10.1074/jbc.M109.027433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jönsson F., Gurniak C.B., Fleischer B., Kirfel G., Witke W. Immunological Responses and Actin Dynamics in Macrophages Are Controlled by N-Cofilin but Are Independent from ADF. PLoS ONE. 2012;7:e36034. doi: 10.1371/journal.pone.0036034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silacci P., Mazzolai L., Gauci C., Stergiopulos N., Yin H.L., Hayoz D. Gelsolin Superfamily Proteins: Key Regulators of Cellular Functions. CMLS Cell. Mol. Life Sci. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PubMed] [Google Scholar]
- 55.Bzowska M., Nogieć A., Bania K., Zygmunt M., Zarębski M., Dobrucki J., Guzik K. Involvement of Cell Surface 90 kDa Heat Shock Protein (HSP90) in Pattern Recognition by Human Monocyte-Derived Macrophages. J. Leukoc. Biol. 2017;102:763–774. doi: 10.1189/jlb.2MA0117-019R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byrd C.A., Bornmann W., Erdjument-Bromage H., Tempst P., Pavletich N., Rosen N., Nathan C.F., Ding A. Heat Shock Protein 90 Mediates Macrophage Activation by Taxol and Bacterial Lipopolysaccharide. Proc. Natl. Acad. Sci. USA. 1999;96:5645–5650. doi: 10.1073/pnas.96.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cameron R.G., Blendis L.M., Neuman M.G. Accumulation of Macrophages in Primary Sclerosing Cholangitis. Clin. Biochem. 2001;34:195–201. doi: 10.1016/S0009-9120(01)00215-6. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y.-Y., Arndtz K., Webb G., Corrigan M., Akiror S., Liaskou E., Woodward P., Adams D.H., Weston C.J., Hirschfield G.M. Intrahepatic Macrophage Populations in the Pathophysiology of Primary Sclerosing Cholangitis. JHEP Rep. 2019;1:369–376. doi: 10.1016/j.jhepr.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C., Morley S.C., Donermeyer D., Peng I., Lee W.P., Devoss J., Danilenko D.M., Lin Z., Zhang J., Zhou J., et al. Actin-Bundling Protein L-Plastin Regulates T Cell Activation. J. Immunol. 2010;185:7487–7497. doi: 10.4049/jimmunol.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hagi A., Hirata H., Shinomiya H. Analysis of a Bacterial Lipopolysaccharide-Activated Serine Kinase That Phosphorylates P65/L-Plastin in Macrophages. Microbiol. Immunol. 2006;50:331–335. doi: 10.1111/j.1348-0421.2006.tb03801.x. [DOI] [PubMed] [Google Scholar]
- 61.Freeley M., O’Dowd F., Paul T., Kashanin D., Davies A., Kelleher D., Long A. L-Plastin Regulates Polarization and Migration in Chemokine-Stimulated Human T Lymphocytes. J. Immunol. 2012;188:6357–6370. doi: 10.4049/jimmunol.1103242. [DOI] [PubMed] [Google Scholar]
- 62.De Clercq S., Zwaenepoel O., Martens E., Vandekerckhove J., Guillabert A., Gettemans J. Nanobody-Induced Perturbation of LFA-1/L-Plastin Phosphorylation Impairs MTOC Docking, Immune Synapse Formation and T Cell Activation. Cell. Mol. Life Sci. 2013;70:909–922. doi: 10.1007/s00018-012-1169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anaya E.P., Lin X., Todd E.M., Szasz T.P., Morley S.C. Novel Mouse Model Reveals That Serine Phosphorylation of L-Plastin Is Essential for Effective Splenic Clearance of Pneumococcus. J. Immunol. 2021;206:2135–2145. doi: 10.4049/jimmunol.2000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wabnitz G.H., Michalke F., Stober C., Kirchgessner H., Jahraus B., Van Den Boomen D.J.H., Samstag Y. L-plastin Phosphorylation: A Novel Target for the Immunosuppressive Drug Dexamethasone in Primary Human T Cells. Eur. J. Immunol. 2011;41:3157–3169. doi: 10.1002/eji.201041366. [DOI] [PubMed] [Google Scholar]
- 65.Finamore F., Ucciferri N., Signore G., Cecchettini A., Ceccherini E., Vitiello M., Poliseno L., Rocchiccioli S. Proteomics Pipeline for Phosphoenrichment and Its Application on a Human Melanoma Cell Model. Talanta. 2020;220:121381. doi: 10.1016/j.talanta.2020.121381. [DOI] [PubMed] [Google Scholar]
- 66.Illiano A., Pinto G., Melchiorre C., Carpentieri A., Faraco V., Amoresano A. Protein Glycosylation Investigated by Mass Spectrometry: An Overview. Cells. 2020;9:1986. doi: 10.3390/cells9091986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ceccherini E., Signore G., Tedeschi L., Vozzi F., Di Giorgi N., Michelucci E., Cecchettini A., Rocchiccioli S. Proteomic Modulation in TGF-β-Treated Cholangiocytes Induced by Curcumin Nanoparticles. Int. J. Mol. Sci. 2023;24:10481. doi: 10.3390/ijms241310481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Giorgi N., Cecchettini A., Michelucci E., Signore G., Ceccherini E., Ferro F., Elefante E., Tani C., Baldini C., Rocchiccioli S. Salivary Proteomics Markers for Preclinical Sjögren’s Syndrome: A Pilot Study. Biomolecules. 2022;12:738. doi: 10.3390/biom12060738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolberg L., Raudvere U., Kuzmin I., Adler P., Vilo J., Peterson H. G:Profiler—Interoperable Web Service for Functional Enrichment Analysis and Gene Identifier Mapping (2023 Update) Nucleic Acids Res. 2023;51:W207–W212. doi: 10.1093/nar/gkad347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.