Abstract
Land plant evolution has been marked by numerous genetic innovations, including novel catalytic reactions. Plants produce various carboxyl methyl esters using carboxylic acids as substrates, both of which are involved in diverse biological processes. The biosynthesis of methyl esters is catalyzed by SABATH methyltransferases, and understanding of this family has broadened in recent years. Meanwhile, the enzymes catalyzing demethylation—known as methylesterases (MESs)—have received less attention. Here, we present a comprehensive review of the plant MES family, focusing on known biochemical and biological functions, and evolution in the plant kingdom. Thirty-two MES genes have been biochemically characterized, with substrates including methyl esters of plant hormones and several other specialized metabolites. One characterized member demonstrates non-esterase activity, indicating functional diversity in this family. MES genes regulate biological processes, including biotic and abiotic defense, as well as germination and root development. While MES genes are absent in green algae, they are ubiquitous among the land plants analyzed. Extant MES genes belong to three groups of deep origin, implying ancient gene duplication and functional divergence. Two of these groups have yet to have any characterized members. Much remains to be uncovered about the enzymatic functions, biological roles, and evolution of the MES family.
Keywords: demethylation, α/β hydrolase, methyl esters, defense
1. Introduction to the Methylesterase Family
Methylation and demethylation are opposing biochemical reactions that occur in DNA, proteins, polysaccharides, and diverse small-molecule metabolites [1,2]. As such, they are important in many biological processes [3,4,5]. Among the small-molecule reactants and products of these reactions in plants are carboxylic acids and their respective methyl esters [6,7]. One protein family that catalyzes the methylation of carboxylic acids to form methyl esters is the SABATH methyltransferase family [4]. The known substrates of the SABATH family include plant hormones such as indole-3-acetic acid (IAA), gibberellic acid (GA), salicylic acid (SA), and jasmonic acid (JA). Methylation of these important metabolites regulates their availability. Their products, the respective methyl esters, may have new biological functions that differ from their substrates [4]. Since the establishment of the SABATH family in 2003, much progress has been made in discovering new catalytic and biological functions of SABATH genes [4,8]. Equally exciting was the discovery of the enzyme family that catalyzes the reverse reaction, namely demethylation of carboxylic acid methyl esters. This new protein family has been denoted the methylesterase (MES) family [9]. In this article, we provide a comprehensive synthesis of the studies of the MES family, especially their biochemical and biological functions. We also performed phylogenetic analysis to understand the evolution of the MES family in land plants.
2. MES Family History: Discovery and Relatedness to Other Enzyme Families
The first member of the plant MES family to be identified was polyneuridine aldehyde esterase (PNAE) from Rauvolfia serpentina, with the gene being reported in 2000 [10]. Following this, a gene encoding a methyl jasmonate esterase was discovered in tomato (SlMJE) in 2004 [11]. SlMJE was found to share significant sequence homology with PNAE [11]. The breakthrough discovery came with the realization that salicylic acid binding protein 2 from Nicotiana tabacum (NtSABP2) functions as methyl salicylate esterase [12]. It was found that NtSABP2, SlMJE, and PNAE all belong to the same protein family [12]. The name of the methylesterase (MES) family was coined when this protein family was investigated in Arabidopsis thaliana [9], to distinguish it from other families of similar function.
While the MES family was named to separate it from other families, this name can also lead to confusion with other methylesterases. Pectin methylesterases (PMEs) are a separate family of enzymes that have esterase activity with methylated pectin subunits, allowing them to alter pectin structure [5,13]. PMEs are prominent in plants but also exist in fungi and bacteria, especially in plant pathogens [5]. Meanwhile, the MES family is currently limited to the plant kingdom. Although they share the methylesterase name, MESs are not closely related to PMEs. They have distinct structures and catalytic mechanisms, leading to their respective esterase activities [13]. PMEs belong to the carbohydrate esterase family, while MESs are a part of the α/β hydrolase superfamily, one of the largest groups of enzymes that exist in all domains of life [13,14].
Members of the α/β hydrolase superfamily share a conserved catalytic triad, typically composed of either Ser-His-Asp or Ser-His-Glu [15,16]. Most members also share an oxyanion hole, which is known to stabilize reaction intermediates [16]. Despite these shared structures, members of the α/β hydrolase superfamily have diverse origins and functions [17]. Carboxylesterases (CXEs), another group within this superfamily, are also responsible for hydrolysis of carboxylesters [7]. Previously, the MES family was classified as a subcategory of CXE due to functional similarity between these groups [7]. Later, other studies showed that MESs are phylogenetically distinct from CXEs, indicating that they are not part of the same evolutionary lineage [9,18]. Furthermore, CXE enzymes can utilize a wide range of ester substrates [19,20]. MES members, on the other hand, are more limited in their known substrates, with most utilizing methyl esters of important plant hormones [9,11,21]. However, enzymatic diversity exists in this family, even among currently characterized members (Table 1). In the next section, we will describe in detail the known biochemical functions of the MES family.
Table 1.
List of all biochemically characterized MES enzymes with known substrates.
| Enzyme | Species | Major Substrate a | Reference |
|---|---|---|---|
| AtMES1 | Arabidopsis thaliana | MeSA | Vlot et al. 2008 [22] |
| AtMES7 | Arabidopsis thaliana | MeSA | Gao et al. 2021 [23] |
| AtMES9 | Arabidopsis thaliana | MeSA | Vlot et al. 2008 [22] |
| BnMES34 | Brassica napus | MeSA | Jia et al. 2024a [24] |
| CsMES1 | Citrus sinensis | MeSA | Lima Silva et al. 2019 [25] |
| FvMES2 | Fragaria vesca | MeSA | Jia et al. 2024b [26] |
| GmSABP2-1 | Glycine max | MeSA | Lin et al. 2024 [27] |
| LcSABP | Lycium chinense | MeSA | Li et al. 2019 [28] |
| PtSABP2-1 | Populus trichocarpa | MeSA | Zhao et al. 2009 [29] |
| PtSABP2-2 | Populus trichocarpa | MeSA | Zhao et al. 2009 [29] |
| PvMES1 | Phaseolus vulgaris | MeSA | Xue et al. 2021 [30] |
| NtSABP2 | Nicotiana tabacum | MeSA | Forouhar et al. 2005 [12] |
| SlMES1 | Solanum lycopersicum | MeSA | Frick et al. 2023 [31] |
| SlMES2 | Solanum lycopersicum | MeSA | Frick et al. 2023 [31] |
| SlMES3 | Solanum lycopersicum | MeSA | Frick et al. 2023 [31] |
| SlMES4 | Solanum lycopersicum | MeSA | Frick et al. 2023 [31] |
| StMES1 | Solanum tuberosum | MeSA | Manosalva et al. 2010 [32] |
| PpMES2 | Prunus persica | MeSA/MeJA | Cao et al. 2019 [18] |
| AtMES10 | Arabidopsis thaliana | MeJA | Koo et al. 2013 [33] |
| PpMES1 | Prunus persica | MeJA | Cao et al. 2019 [18] |
| SlMJE1 | Solanum lycopersicum | MeJA | Stuhlfelder et al. 2004 [11] |
| NaMJE | Nicotiana attenuata | MeJA | Wu et al. 2008 [34] |
| VvMES5 | Vitis vinifera | MeJA | Zhao et al. 2016 [35] |
| OeMES1 | Olea europaea | MeJA, MeIAA | Volk et al. 2019 [36] |
| OeMES2 | Olea europaea | MeJA, MeIAA | Volk et al. 2019 [36] |
| AtMES17 | Arabidopsis thaliana | MeIAA | Yang et al. 2008 [9] |
| AtMES2 | Arabidopsis thaliana | MeNA | Wu et al. 2018 [37] |
| AtMES16 | Arabidopsis thaliana | FCCs | Crist et al. 2012 [38] |
| ShMKS1 | Solanum habrochaites | 3-keto acids | Fridman et al. 2005 [39] |
| PNAE | Rauvolfia serpentina | PNA | Dogru et al. 2000 [10] |
| EAME1 | Olea europaea | Secoiridoids | Volk et al. 2019 [36] |
| EAME2 | Olea europaea | Secoiridoids | Volk et al. 2019 [36] |
a methyl salicylate (MeSA), methyl jasmonate (MeJA), methyl indole-3-acetate (MeIAA), methyl nicotinate (MeNA), fluorescent chlorophyll catabolites (FCCs), 3-keto acids, polyneuridine aldehyde (PNA), secoiridoids.
3. Known Enzymatic Functions of MES Members
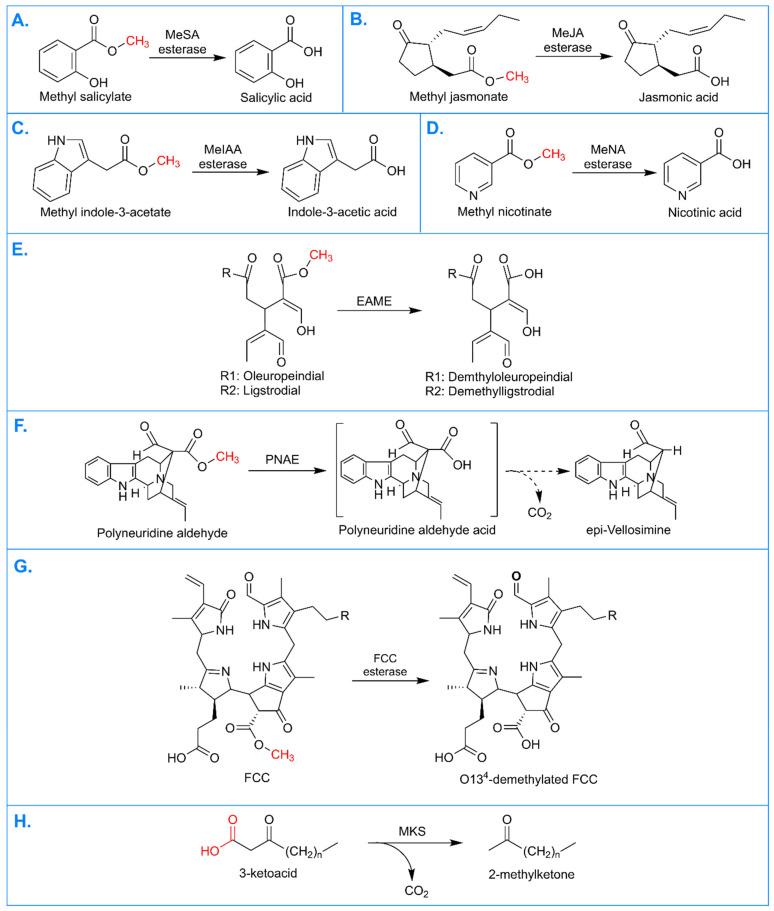
MES enzymes are currently known to utilize three methylated plant hormones as substrates, including methyl esters of SA, JA, and IAA (Figure 1A–C). Methyl salicylate (MeSA) esterases are by far the most represented among characterized members, with 18 known enzymes utilizing MeSA as their major substrate (Table 1). This class of enzymes—including NtSABP2—demethylates MeSA into SA, which is known to be crucial for signaling pathways [40]. Similarly, methyl jasmonate esterases (MJEs) demethylate methyl jasmonate (MeJA) into JA, another important signaling hormone [41]. Eight MES members have been found to have MJE activity (Table 1). Finally, three genes are identified as having esterase activity with methyl IAA (MeIAA) (Table 1). These genes demethylate MeIAA into IAA, one of the most well-studied plant hormones [42]. It is important to note that several genes show in vitro activity with multiple hormone substrates (Table 1). This highlights the importance of in vivo testing for determining the true biological function of MES enzymes.
Figure 1.
Biochemical reactions catalyzed by MES enzymes. The site of reaction for each substrate is indicated in red. (A) Reaction catalyzed by methyl salicylate (MeSA) esterase. (B) Reaction catalyzed by methyl jasmonate (MeJA) esterase. (C) Reaction catalyzed by methyl indole-3-acetate (MeIAA) esterase. (D) Reaction catalyzed by methyl nicotinate (MeNA) esterase. (E) Reaction catalyzed by elenolic acid methylesterase (EAME). (F) Reaction catalyzed by polyneuridine aldehyde esterase (PNAE). (G) Reaction catalyzed by fluorescent chlorophyll catabolite (FCC) esterase. (H) Reaction catalyzed by methyl ketone synthase (MKS).
MES enzymes can also favor substrates that are not hormones but maintain structural similarity to hormones. AtMES2 from Arabidopsis is known to utilize methyl nicotinate (MeNA) as a substrate, the structure of which is nearly identical to that of MeSA (Figure 1D) [37]. Demethylation of MeNA allows the recycling of nicotinate (NA) for an alternative biosynthesis pathway to maintain nicotinamide adenine dinucleotide (NAD) levels [37]. This is a crucial function in all plants; thus, it is likely that homologs of AtMES2 may be found across plant taxa.
Some MES members are also known to utilize specialized products to generate secondary metabolites. This includes elenolic acid methylesterases 1 and 2 (EAME1 and EAME2), which utilize secoiridoids as their substrates, including oleuropeindial and ligstrodial (Figure 1E). These enzymes are involved in the production of secoiridoid-derived polyphenols such as oleuropein [36]. Intriguingly, researchers also found that EAME1 and EAME2 can convert MeJA and MeIAA into ethyl JA and ethyl IAA, respectively, in the presence of ethanol [36]. This transesterification capability is not currently known to exist in any other MES enzymes. It is unclear whether this in vitro activity mirrors a true native function, but it indicates an interesting area of further study for novel MES capabilities. PNAE, the first characterized MES member, utilizes polyneuridine aldehyde as its substrate, which is structurally related to secoiridoids. PNAE converts polyneuridine aldehyde into polyneuridine aldehyde acid, an intermediate that is then nonenzymatically converted to epi-vellosimine (Figure 1F). This leads to downstream production of ajmaline, a compound of interest from Rauvolfia serpentina [10].
MES reactions with more complex substrates are also possible. AtMES16 from Arabidopsis has been found to break down fluorescent chlorophyll catabolites (FCCs) in the chloroplast, an important step in chlorophyll degradation [38]. AtMES16 was shown to demethylate FCCs at oxygen O134, generating O134-demethylated FCCs (Figure 1G). This demethylation was shown to be important for subsequent degradation steps [38]. FCCs are currently the largest known substrate of an MES enzyme, demonstrating the possibilities of this family. Chlorophyll degradation is important in all plants, though there are differences in the specific steps of FCC breakdown [38]. Thus, homologs of AtMES16 are likely to be found in other species but may not be conserved across all taxa. For example, the radish enzyme RsPPD has been shown to have similar functionality and, furthermore, shares sequence similarity with AtMES16 [38,43]. RsPPD has not been confirmed as a member of MES, but further study of this enzyme and other possible AtMES16 homologs can expand the understanding of specialized MES functions.
Finally, the enzyme ShMKS1 from the wild tomato species Solanum habrochaites has been found to be involved in methylketone synthesis [39]. Notably, the typical Ser-His-Asp triad is not found in this enzyme but is replaced by an Ala-His-Asn triad [44]. This change leads to unique enzymatic activity compared to other MES members. ShMKS1 catalyzes the final step of methylketone biosynthesis by converting 3-keto acids to 2-methylketones through decarboxylation (Figure 1H). This demonstrates that some MES members may have evolved new, non-esterase functions. Consideration of currently known MES reactions is useful in identifying activities of new members. Equally helpful is an understanding of MES biological functions, which will be discussed in the following section.
4. MES Biological Functions: Defense and Development
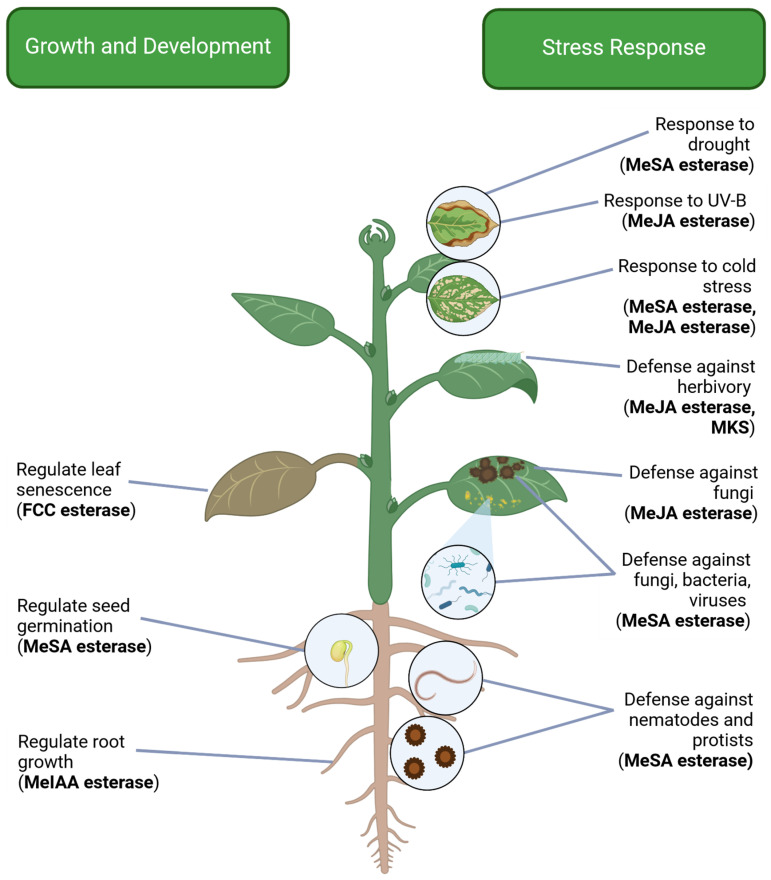
The majority of known MES members demethylate a specific phytohormone, which is important for the regulation of hormone concentrations and their respective processes. However, other MES members are involved in enzymatic reactions with other plant compounds, which can have a variety of functions throughout plant tissues and life stages. Currently, most biologically characterized MES genes are found to be involved in the stress response, though a few also factor into developmental processes (Figure 2). Understanding what is known about the biological roles of MES enzymes can lead to better knowledge of their origins.
Figure 2.
Known biological functions of MES enzymes in plants based on in vivo genetic studies. Created with BioRender. https://BioRender.com/v52h282 (accessed on 12 November 2024).
4.1. MeSA Esterases: Pathogen Defense, Drought Response, and Seed Germination
Being sessile organisms, plants must rely on chemical signaling pathways to defend themselves against stress and pathogens. The plant hormone salicylic acid (SA) has long been known to be crucial for defense against viral, bacterial, and fungal pathogens through the systemic acquired resistance (SAR) pathway [45,46,47]. Methylation of SA into MeSA is a crucial part of this signaling process [40]. Because MeSA can be transported more easily and become airborne, it serves as a mobile form to induce resistance pathways in uninfected tissues as well as neighboring plants [48]. Methylation of SA is performed by SAMTs and BSMTs, well-known members of the SABATH family [49,50]. However, once MeSA arrives in its target tissues, it must be demethylated back into SA to induce the SAR pathway.
Following its initial discovery, NtSABP2 was found to be involved in disease resistance by catalyzing the conversion of MeSA into SA [12,21]. Confirming the importance of MeSA in SAR signaling, it was shown that overexpression of NtSABP2 leads to reduced MeSA and impaired SAR infection response [40]. However, in a more recent study, overexpressing NtSABP2 in transgenic citrus fruit was found to enhance resistance to bacterial infection [51]. This is in line with research on SAMT genes, where modulation of SA:MeSA ratios can either hamper or enhance disease resistance in different cases [52,53]. This highlights the importance of understanding these genes and their involvement in immune pathways to correctly predict resulting phenotypes.
The NtSABP2 sequence has also been used to identify various MES genes in other species, many of which have been confirmed to have MeSA esterase activity (Table 1). Several of these have also been demonstrated to be directly involved in pathogen response(Figure 2). AtMES1, -7, and -9 from Arabidopsis were shown to be activated in response to infection with Pseudomonas syringae, and knockdown of these genes leads to inhibition of SAR pathways [22]. Similarly, CsMES1 from sweet orange was shown to be important for protection against citrus canker. CsMES1 transcripts were upregulated during pathogen infection; meanwhile, inhibition of this gene leads to increased canker formation [25]. In addition to bacterial pathogens, SA is also involved in responses to fungal pathogens. StMES1 from potato was found to be necessary for the SAR response to the potato blight fungus Phytophthora infestans [32]. Suppression of StMES1 led to compromised SAR responses against the fungus. PvMES1 from the common bean has similarly been shown to control SA signaling in response to Fusarium oxysporum [30]. It was found that overexpression of PvMES1 enhanced resistance, while silencing it led to increased susceptibility [30]. Likewise, FvMES2 from strawberry is an MeSA esterase involved in the fungal response. The gene was found to be upregulated in response to Botrytis cinerea infection, which was also highly correlated with SA signaling [26]. Meanwhile, strawberries overexpressing FvMES2 show increased resistance to B. cinerea [26]. MeSA esterases are also involved in specialized infection responses to protists and parasitic animals. The Brassica napus member BnMES34 is an MeSA esterase that controls responses to the protist pathogen Plasmodiophora brassicae [24]. It was found that heterologous expression of BnMES34 in Arabidopsis confers resistance to the clubroot disease caused by P. brassicae [24]. Finally, MeSA esterases can also respond to infection by nematodes. GmSABP2-1 from soybean was recently found to have MeSA activity, which can confer enhanced resistance to soybean cyst nematode [27]. This is in line with previous research in SAMTs, confirming that MeSA regulation is important during nematode infection [54].
Although pathogen response is perhaps the most discussed role of SA, it is not the only function of this hormone. Supporting this, some MeSA esterases have been found to have activities in other areas of stress response and plant development (Figure 2). LcSABP, an NtSABP2 ortholog from Lycium chinense, was found to enhance drought tolerance in transgenic tobacco [28]. It was found that this tolerance was conferred through an SA-dependent pathway, leading to increased production of reactive oxygen species and stress-responsive genes [28]. Similarly, AtMES9 was recently found to support an SA-mediated pathway during cold stress conditions [55]. Cold stress was shown to upregulate AtMES9 and other SA biosynthesis genes to maintain SA concentrations [55]. In another example of diverse regulation, AtMES7 was shown to modulate seed germination in Arabidopsis [23]. AtMES7 controls seed SA levels under normal and salt stress conditions, leading to varied germination responses [23]. As noted previously, AtMES7 and -9 were originally shown to activate expression in response to pathogen infection [22]. The more recent analyses demonstrate the possibility of MES genes regulating multiple processes at different times or in diverse tissues. This highlights the importance of in-depth biological function determination. Many MeSA-utilizing MES enzymes are known, but their functions are not necessarily limited to SAR pathogen response. Studying these members under the lens of other processes, such as abiotic stress or plant development, can lead to a better understanding of their in planta biological roles.
4.2. MeJA Esterases: Biotic and Abiotic Stress Response
Jasmonic acid (JA) is an established plant hormone that plays roles in an array of plant processes, including germination, growth, fruit ripening, and resistance to biotic and abiotic stresses [41,56,57]. Methyl jasmonate (MeJA) is a derivative of JA and is produced by JAMT of the SABATH family [58]. Similar to MeSA, MeJA serves as a mobile transport form to elicit responses throughout the plant or to neighboring plants [59,60]. MeJA is also considered an inactive form and must be demethylated back to JA for the regulation of biological processes [34]. This requires the action of an MeJA esterase (MJE), which is part of the MES family. These enzymes can serve various biological roles, notably in response to biotic and abiotic stresses (Figure 2).
SlMJE1, the first categorized MJE, was discovered in tomato and has been found to be important for the response to fungal pathogens [11,61]. Interestingly, neither overexpression nor RNAi knockdown of SlMJE1 shows consistent changes in levels of MeJA or other jasmonate derivatives [61]. Despite this, both conditions lead to increased susceptibility to the fungal pathogen Sclerotinia sclerotiorum [61]. Further study of this gene can help clarify how it regulates plant response to the fungus. On the other hand, JA is particularly known for its role in response to herbivory. NaMJE from Nicotiana attenuata is known to be involved in insect resistance. It was found that the conversion of MeJA back to JA is specifically required for resistance to the tobacco hornworm Manduca sexta [34]. In addition to biotic stress, JA signaling can also be involved in the abiotic stress response [56]. VvMJE1 from grapevine was shown to have MJE activity specifically activated by abiotic stresses, including cold stress and UV-B treatment [35]. Because JA can be involved in various plant responses, it is important to characterize the biological functions of MJE members. Several other enzymes with MJE activity have been identified, but their in vivo functions remain unknown (Table 1).
4.3. MeIAA Esterases: Regulation of Auxin-Mediated Development
Auxins are a class of plant hormones that are crucial for proper growth and development. Indole-3-acetic acid (IAA), the most common auxin, regulates diverse processes including cell elongation and differentiation, organ development, and phototropism [42,62]. Plants are known to utilize various IAA conjugates to regulate auxin levels, some of which are considered irreversible intermediates in auxin degradation [62,63]. Methyl IAA (MeIAA), on the other hand, has been used in experiments as a surrogate for IAA for many years, and is now known to be reversible to IAA through the action of an MES enzyme in Arabidopsis, AtMES17 [9,64]. It was found that AtMES17 mutants had altered responses to exogenous MeIAA—but not exogenous IAA—in regulating root growth [9] (Figure 2). This demonstrates the importance of demethylating MeIAA for proper auxin regulation. It is likely that this enzyme could regulate IAA in other parts of the plant as well, but this requires further study. Additionally, AtMES17 is the only enzyme that has been experimentally characterized as an MeIAA esterase in vivo, though other putative MeIAA esterases have been identified (Table 1). A number of recent studies have bioinformatically identified MES members from different species, including possible homologs of AtMES17 [18,24,35]. Whether these are true MeIAA esterases remains an open question. However, due to the importance of IAA, it is likely that MeIAA esterases will be well conserved across plant taxa. Further characterization of AtMES17 and other MeIAA esterases will be necessary to elucidate their native functions.
4.4. AtMES2: NAD Recycling
Nicotinamide adenine dinucleotide (NAD) is a crucial coenzyme in plants as well as in other organisms [65,66]. Because of its importance in many diverse processes, it is necessary to maintain adequate levels of NAD throughout the plant. NAD can be synthesized either via a de novo pathway, or it can be recycled from other derivatives [65]. The NAD salvage pathway begins with nicotinate (NA), but NA is known to be toxic to plants at high concentrations [67]. Due to this toxicity, it is converted into various other forms, including methyl nicotinate (MeNA), by an N-methyltransferase [68]. It was recently found that MeNA serves as a long-distance transport form of NA, which can later be demethylated by AtMES2 from Arabidopsis and recycled into NAD [37]. This is reminiscent of the roles of MeSA and MeJA and illustrates one of the reasons that methylated products are common in plants. This recycling process via MeNA can allow plants quick access to NAD, while ensuring that NA toxicity does not cause cellular damage. Furthermore, it is well established that abiotic stresses lead to increased degradation of NAD [65]. Supporting this, it was found that AtMES2 is suppressed under abiotic stress inducers, indicating that it may be involved in stress adaptation [37]. However, its true biological roles are not yet fully understood. Further study of this gene, as well as identification of homologs in other species, will expand understanding of NAD regulation by MES family members.
4.5. AtMES16: Chlorophyll Degradation During Leaf Senescence
Chlorophyll is the most abundant pigment on Earth and is clearly fundamental to plant survival [69]. Despite its importance, chlorophyll can also be toxic to cells and must be degraded during processes such as fruit ripening and leaf senescence [69]. This requires first the conversion of chlorophylls a and b into fluorescent chlorophyll catabolites (FCCs) [70]. These FCCs are then demethylated before passing through subsequent breakdown steps, and this demethylation is performed by AtMES16 in Arabidopsis [38]. This enzyme was shown to be important for chlorophyll degradation during leaf senescence, and mutants were seen to have increased retention of FCCs [38] (Figure 2). AtMES16 may be related to the previously identified radish enzyme RsPPD, which is known to demethylate pheophorbide, another member of the chlorophyll degradation pathway [43]. RsPPD has not been identified as a member of MES, but due to its homology and functional similarity to AtMES16, further investigation of this enzyme as a possible MES member is warranted. Furthermore, it is likely that MES members performing chlorophyll degradation may be found across plant taxa. The study of such proteins may reveal new roles in controlling leaf senescence and fruit ripening.
4.6. PNAE, EAME1, EAME2, and ShMKS1: Putative Defense and Stress Responses
PNAE from Rauvolfia serpentina was the first MES member to be functionally characterized [10]. Despite many years of study on this enzyme, its native biological functions remain unknown. However, examining the pathways it is involved in may hint at its possible roles. PNAE catalyzes the conversion of polyneuridine aldehyde into epi-vellosimine, an important precursor for ajmaline biosynthesis. Ajmaline is anthropologically significant due to its long history of use as a cardiovascular drug [71]. In plants, ajmaline has been found to be upregulated after treatment with MeJA, indicating a possible role in stress response or other areas of JA signaling [72]. Epi-vellosimine is also a precursor to the sarpagine-type alkaloids, making PNAE a catalyst in this pathway as well. Sarpagines are also thought to serve defensive functions similar to ajmaline and other alkaloids [73,74]. Further study of PNAE may reveal its biological role, perhaps in plant defense or other functions.
EAME1 and EAME2 were identified from olive based on their homology to PNAE and found to be involved in secoiridoid biosynthesis [36]. Oleuropein is the most common secoiridoid in olives, found in high levels in both leaves and fruits [75,76]. It is thought to serve a defense role against herbivory by activating protein denaturation mechanisms and decreasing nutritional value upon tissue damage [77]. Similar to PNAE, the true biological functions of these MESs are unconfirmed. Future analysis of EAME1 and -2 may reveal the importance of these enzymes.
Finally, ShMKS1 is known to be involved in the biosynthesis of methylketones in wild tomato [39]. Wild tomato plants are known to produce higher levels of methylketones than their domesticated relatives, which confers an advantage against herbivory [78,79]. Overexpression of methylketone biosynthesis pathway genes, including ShMKS1 in Arabidopsis and tobacco, demonstrated heterologous production of methylketones but also led to severe growth defects [80]. Meanwhile, targeted expression in cultivated tomatoes showed only a slight increase in methylketone synthesis [80]. Further biological characterization of MES enzymes is crucial to clarify the diversity in roles served by this family.
5. MES Family Evolution in Land Plants
Though the MES family has been known for over two decades, it has received relatively less attention than some other protein families. Currently, all characterized members are from a small number of angiosperms, limiting the ability to study the evolution of this family. The SABATH family, which serves as the inverse of MES, has three times as many characterized members, including several from gymnosperms and non-seed plants [81,82,83,84]. This allows for greater understanding of the family and how it arose and evolved across plant taxa [4]. To gain better insights into this evolution for the MES family, a phylogenetic analysis was performed using bioinformatically identified MES genes from selected sequenced plants. The findings from this analysis can help bring attention to the importance of this family across plant lineages and demonstrate the origins of MES enzymes.
5.1. MES Phylogenetic Analysis
A total of twenty-three species were selected for MES identification and phylogenetic analysis, including two green algae (Mesotaenium endlicherianum and Spirogloea muscicola), three mosses (Ceratodon purpureus, Physcomitrella patens, and Sphagnum fallax), two liverworts (Marchantia polymorpha and Ricciocarpos natans), two hornworts (Anthoceros agrestis and Anthoceros angustus), two lycophytes (Diphasiastrum complanatum and Selaginella moellendorffii), four ferns (Adiantum capillus-veneris, Alsophila spinulosa, Ceratopteris richardii, and Marsilea vestita), two gymnosperms (Picea abies and Thuja plicata), and six angiosperms (Amborella trichopoda, Arabidopsis thaliana, Nymphaea colorata, Oryza sativa, Populus trichocarpa, and Zea mays). Annotated protein sequences of the 23 species were retrieved from public databases, including NCBI (https://www.ncbi.nlm.nih.gov, accessed on 1 September 2024) and Phytozome (https://phytozome-next.jgi.doe.gov, accessed on 1 September 2024), and searched using BLASTp [85] with known MESs (Table 1) as queries. A total of 448 MES genes were identified. Most notably, no MES members were identified in either algal species, indicating the family may have evolved specifically in embryophytes or may be limited to only certain algae. All 21 selected land plant species exhibit MES members ranging from three to twenty-eight per species. There is no obvious trend of family expansion, as most taxa represent both the low and high ends of this range in different species. This indicates that MES enzymes have continued to duplicate and serve important purposes in all lineages. Of the 448 MES proteins, 274 have an annotated protein length of 200 to 400 amino acids, which were used in subsequent phylogenetic analysis.
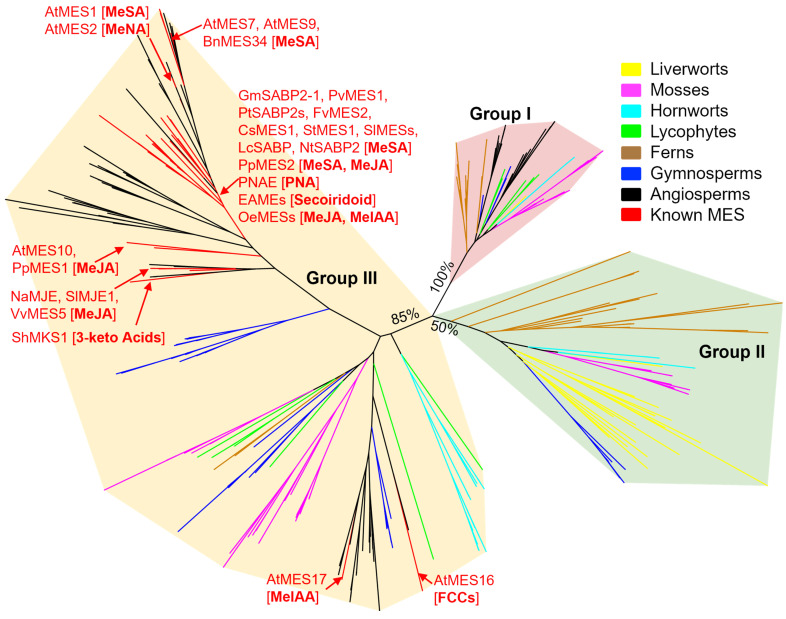
Phylogenetic analysis of the 274 putative MES proteins identified from selected plants and the 32 functionally characterized MES proteins from flowering plants was performed. Different schemes of grouping (e.g., three groups or five groups) of MES proteins in the phylogenetic tree could be proposed. In this article, we preferred the clustering of three groups (denoted Groups I, II, and III) (Figure 3) with the consideration of the phylogeny of the major lineages of land plants. Group I, with a robust 100% bootstrap value, contains 66 putative MES members representing all 21 selected plant genomes, making it the most well-conserved MES clade in land plants. Group II, with a bootstrap value of 50%, contains 65 putative MES members from 13 selected plant genomes, but is absent in the surveyed lyiccophytes and angiosperms. Group III, with a strong bootstrap value of 85%, contains 143 putative MES members across 19 of the 21 surveyed plant genomes, being absent only in liverworts. The lack of putative MES members from selected plants in Groups II and III indicates potential gene loss in specific lineages, including liverworts, lycophytes, and angiosperms.
Figure 3.
Phylogenetic analysis of MES proteins from 21 genomes representing various land plant taxa, plus all characterized MESs. Branches are color-coded as follows: liverworts, yellow; mosses, magenta; hornworts, cyan; lycophytes, green; ferns, brown; gymnosperms, blue; angiosperms, black; known MESs, red. Characterized MES members are indicated as red labels, with their substrates in brackets. The phylogenetic tree was constructed with RAxML (version 1.1.0) using the best-fit model LG + G4 + F with 1000 bootstrap replicates. Bootstrap support for the major groups is indicated. The tree was visualized and further annotated using iTOL (version 6.9.1).
The presence of mosses, hornworts, ferns, and gymnosperms in all three MES groups—each with multiple genes per species—suggests that the MES family underwent significant duplication and diversification early in the evolution of land plants, prior to the divergence of these groups (Figure 3). This broad distribution across different lineages indicates that the core MES functions were established early and may have been conserved throughout evolutionary history. In contrast, liverworts are represented in Groups I and II but are absent in Group III. It has been previously hypothesized that liverworts diverged earlier than mosses and hornworts [86]. If this were the case, it could imply that the MES family originally consisted of two genes, with a third evolving after the divergence of liverworts. However, this model of land plant evolution is highly debatable, and many alternative hypotheses exist [86]. For example, it could be the case that there were three ancestral MES genes, but Group III was later lost in liverworts. Notably, the loss of Group II in lycophytes and angiosperms—as well as the potential loss of Group III in liverworts—suggests that some MES genes may have become redundant or non-essential in certain lineages, indicating functional divergence that occurred during MES evolution. Exploring the evolution of known MES functions can provide key insights into the origins of this enzyme family.
5.2. Enzymatic Evolution of MES Functions
Many enzyme families are proposed to arise through catalytic promiscuity, wherein ancestral enzymes had the ability to utilize noncanonical substrates, which are sometimes positively selected for and later become preferred substrates [87]. This mechanism of enzymatic evolution has been demonstrated in many families, including several belonging to the α/β hydrolase superfamily [88,89,90]. The hydroxynitrile lyases (HNLs), which were once grouped together with MES under CXE class II, are now thought to have diverged from MES enzymes around 100 million years ago through promiscuity [90]. NtSABP2 was shown to switch from esterase to HNL activity following only a two-amino-acid alteration to its primary sequence [91]. Further supporting this, AtMJE was demonstrated to have HNL activity in its native form, suggesting that even modern enzymes can exhibit promiscuity [92]. Based on the current understanding of catalytic evolution, it is expected that ancestral enzymes would have demonstrated more promiscuity than their modern counterparts [87]. It therefore stands to reason that the MES ancestors may have had a core functionality for essential plant functions, while also demonstrating alternative substrate utilization that would have later led to the evolution of new enzymes through duplication and divergence. With this in mind, it is useful to explore modern functions of MES enzymes and make inferences as to how these functions may have evolved from ancestral enzymes.
All characterized MES proteins fall into Group III, with the majority of characterized members clustering together in the largest angiosperm subgroup, including MeSA esterases, MJEs, PNAE, ShMKS1, and others (Figure 3). It appears that these functions arose from an expansion in angiosperms and gymnosperms, which is not seen in other lineages. This could mean these functions are unique to these lineages or that unrelated enzymes could serve these roles in other taxa. It is known that SA-mediated defense signaling extends to bryophytes, such as the moss Physcomitrella patens [93]. There has also been one bryophyte SAMT gene identified, namely CsSAMT from the liverwort Conocephalum salebrosum [84]. This indicates that methylation of SA takes place in liverworts and possibly other bryophytes, suggesting a need for an esterase to demethylate MeSA back to its active form. However, if such an enzyme exists, it appears not to be closely related to known MeSA esterases from angiosperms.
Similarly, it is not well understood to what extent JA and MeJA are involved in signaling processes in nonvascular plants. One recent study in the liverwort Marchantia polymorpha found that it contained at least some of the core components for the JA signaling pathway [94]. On the other hand, it is known that 12-oxo-phytodienoic acid (OPDA), the precursor to JA, can act in place of JA as a signaling molecule [95]. This has been shown to take place in P. patens, where OPDA was shown to respond to fungal infection and wounding [96,97]. If OPDA is used by bryophytes in place of JA, it is unclear if an MES gene would be required for this function. Further study of JA signaling in non-angiosperm plants is required to clarify the need for MJEs in other lineages.
The only characterized MES members that lie outside the aforementioned subgroup—but still within Group III—are AtMES16 and AtMES17. As previously discussed, AtMES16 serves to break down chlorophyll during leaf senescence [38,43]. Because of the importance of chlorophyll to all plants, it is sensible that regulating it through degradation would be an early-evolved plant function. However, as mentioned previously, it is unclear how well-conserved chlorophyll degradation pathways are across plant taxa [38]. The other outlier, AtMES17, is an esterase of MeIAA [9]. IAA is known to be crucial for growth and development throughout plants, including mosses and liverworts [98]. MeIAA is known in many plants as a reversible storage form and one of the ways that auxin levels are regulated [99]. However, it is unknown if MeIAA is a storage form utilized within nonvascular plants and therefore unclear if they would require an MeIAA esterase. There are many unknown MES members from across taxa contained within Group III, clustering with AtMES16 and AtMES17. Some of these may be homologs of these known members, but others may serve different functions. Overall, this subgroup appears to be an ancestral branch to the angiosperm group containing all other characterized MESs. This indicates that AtMES16 and AtMES17 could represent earlier functions in MES evolution. Notably, several MES enzymes demonstrate in vitro activity with MeIAA at high concentrations [12,29,36]. This could be representative of their ancestral activities, which have been mostly lost but still function under specific conditions. Further research on the members within this group can elucidate their functions and better our understanding of MES evolution.
While there is much to be discovered regarding Group III, there is even more work to be done in Groups I and II (Figure 3). These groups have no characterized MES members, leaving great doubt as to their biological and enzymatic functions. Members of Group II are found in all taxa except for lycophytes and angiosperms. It appears that this is a well-conserved group dating back to the origin of land plants, but it was somehow lost during the evolution of lycophytes and flowering plants. It could be that these members serve functions that are no longer needed in these lineages. Alternatively, it may be that these functions are served by more distantly related members, such as those in the expanded angiosperm subgroup within Group III. Group I, meanwhile, is represented in all taxa of land plants, and thus may be expected to serve a fundamental role. This could be an MES role that arose early in evolutionary history, and therefore understanding the functions within this group may hint at ancestral MES functions. Another possibility that must be considered is that Groups I and II may not be true MES enzymes at all. As mentioned previously, it is likely that the MES family evolved from other enzymes through catalytic promiscuity [87,90]. Members in Groups I and II, therefore, could represent ancestral roles from which MESs later evolved, or alternatively could be separate branches that evolved from the same ancestor as the MES family, leading to enzymatically unrelated functions. Because there are no characterized members within these groups, it is impossible to say whether they are true MES enzymes. It is apparent that focusing on a subset of enzyme activities, as well as limiting analysis to angiosperms, has hindered a broader understanding of this family. Further study of enzymes within these unknown groups is crucial for the understanding of the family as a whole and could lead to the discovery of new functions or even new gene families.
6. Conclusions and Future Directions
The origin and diversification of land plants were enabled by vast genetic innovations, among which are the ability of land plants to produce diverse metabolites and regulate their concentrations [100,101]. One example is the biosynthesis of diverse methyl esters of carboxylic acids by SABATH methyltransferases [4]. It is intriguing that land plants have also evolved enzymes—namely methylesterases (MESs)—to catalyze the reverse reaction: demethylation of methyl esters to convert them back to carboxylic acids. A number of conclusions can be drawn based on the current understanding of this family. First, MESs form a small gene family within land plants, as discussed in Section 5.1. Second, the majority of known MES enzymes catalyze demethylation of carboxyl methyl esters, with exceptions. Some members have evolved new catalytic activities, e.g., the decarboxylation activity of ShMKS1 [39]. Third, MES genes appear to be involved in diverse biological processes, ranging from phytohormone regulation to the biosynthesis of secondary metabolites (Figure 2). Fourth, MES genes appear to be specific to land plants, suggesting their origin in the common ancestor of land plants after the divergence from green algae. Phylogenetic analysis also implies that the common ancestor of land plants contained either two or three copies of MES genes, leading to the three extant groups seen today (Figure 3).
While significant progress has been made in our understanding of the MES family in the last two decades, much remains to be uncovered. So far, only a small number of MES genes from a very limited number of plant species have been studied (Table 1). Furthermore, within the present phylogenetic analysis, all functionally characterized members are in Group III (Figure 3). It will be highly informative to determine biochemical and biological functions of the members from Groups I and II. Members of Group I appear to be conserved across taxa. It is therefore interesting to ask whether the MES genes in this group have conserved functions. It will also be useful to look at known methylated products in consideration of possible MES substrates. For example, the SABATH methyltransferase family has nearly 20 known substrates, with only three of these currently being represented by respective MES enzymes [4]. Focusing on these unrepresented substrates could help identify new MES functions, particularly for substrates that are known to be demethylated for proper function. It may also be the case that some MES members have evolved new roles, not directly related to esterase activity. This is already seen with ShMKS1 and is likely to be true for some other members as well, particularly the more divergent they are from known enzymes. If similar instances exist for other MESs, alternative methodologies and substrate analysis will be required to identify their activity. Functional elucidation of the new members of the MES family in Groups I and II will also establish a proper context to address the question of functional evolution in the MES family. Novel catalytic activities may inspire new structural studies, as three-dimensional structures and reaction mechanisms have been solved for only a few members of the MES family [12,44,102,103]. As more MES structures become available, in silico tools can be employed to guide further research. Software such as AlphaFold3 can predict MES structures and functions based on known members, allowing for deeper insights into family evolution [104,105]. AlphaFold can also be used to redesign enzymes with altered activity [106,107], permitting researchers to leverage the natural diversity in the MES family. As additional MES functions become known, the extent of this engineering potential will expand as well. The MES family is already known to be important in defense and development processes. Continuing to identify novel members of this family and characterize their functions will further highlight its importance and help discern its evolutionary origins.
Acknowledgments
We thank the High Performance and Scientific Computing group at the University of Tennessee for the computing resources used for this project.
Author Contributions
F.C. and J.-G.C. conceived the project. T.A.C. developed the original draft. W.W. performed phylogenetic analysis. All authors contributed to the revisions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries may be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was supported by a University of Tennessee AgResearch SPRINT award and by the Center for Bioenergy Innovation (CBI), U.S. Department of Energy, Office of Science, Biological and Environmental Research Program under Award Number ERKP886. Oak Ridge National Laboratory is managed by UT-Battelle, LLC. for the U.S. DOE under Contract Number DE-AC05-00OR22725.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Bauerle M.R., Schwalm E.L., Booker S.J. Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J. Biol. Chem. 2015;290:3995–4002. doi: 10.1074/jbc.R114.607044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu D., Gao Y.H., Yao X.S., Gao H. Recent advances in dissecting the demethylation reactions in natural product biosynthesis. Curr. Opin. Chem. Biol. 2020;59:47–53. doi: 10.1016/j.cbpa.2020.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Deleris A., Halter T., Navarro L. DNA Methylation and Demethylation in Plant Immunity. Annu. Rev. Phytopathol. 2016;54:579–603. doi: 10.1146/annurev-phyto-080615-100308. [DOI] [PubMed] [Google Scholar]
- 4.Wang W., Guo H., Bowman J.L., Chen F. Plant SABATH Methyltransferases: Diverse Functions, Unusual Reaction Mechanisms and Complex Evolution. Crit. Rev. Plant Sci. 2024;43:291–312. doi: 10.1080/07352689.2024.2335016. [DOI] [Google Scholar]
- 5.Kohli P., Kalia M., Gupta R. Pectin Methylesterases: A Review. J. Bioprocess. Biotech. 2015;5:227. doi: 10.4172/2155-9821.1000227. [DOI] [Google Scholar]
- 6.Noel J.P., Dixon R.A., Pichersky E., Zubieta C., Ferrer J.-L. Recent Advances in Phytochemistry. Volume 37. Elsevier; Amsterdam, The Netherlands: 2003. Chapter two Structural, functional, and evolutionary basis for methylation of plant small molecules; pp. 37–58. [Google Scholar]
- 7.Gershater M.C., Edwards R. Regulating biological activity in plants with carboxylesterases. Plant Sci. 2007;173:579–588. doi: 10.1016/j.plantsci.2007.08.008. [DOI] [Google Scholar]
- 8.D’Auria J.C., Chen F., Pichersky E. The SABATH family of MTs in Arabidopsis thaliana and other plant species. Recent Adv. Phytochem. 2003;37:253–284. [Google Scholar]
- 9.Yang Y., Xu R., Ma C.J., Vlot A.C., Klessig D.F., Pichersky E. Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol. 2008;147:1034–1045. doi: 10.1104/pp.108.118224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dogru E., Warzecha H., Seibel F., Haebel S., Lottspeich F., Stockigt J. The gene encoding polyneuridine aldehyde esterase of monoterpenoid indole alkaloid biosynthesis in plants is an ortholog of the alpha/betahydrolase super family. Eur. J. Biochem. 2000;267:1397–1406. doi: 10.1046/j.1432-1327.2000.01136.x. [DOI] [PubMed] [Google Scholar]
- 11.Stuhlfelder C., Mueller M.J., Warzecha H. Cloning and expression of a tomato cDNA encoding a methyl jasmonate cleaving esterase. Eur. J. Biochem. 2004;271:2976–2983. doi: 10.1111/j.1432-1033.2004.04227.x. [DOI] [PubMed] [Google Scholar]
- 12.Forouhar F., Yang Y., Kumar D., Chen Y., Fridman E., Park S.W., Chiang Y., Acton T.B., Montelione G.T., Pichersky E., et al. Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. USA. 2005;102:1773–1778. doi: 10.1073/pnas.0409227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markovic O., Janecek S. Pectin methylesterases: Sequence-structural features and phylogenetic relationships. Carbohydr. Res. 2004;339:2281–2295. doi: 10.1016/j.carres.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Nardini M., Dijkstra B.W. Alpha/beta hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 1999;9:732–737. doi: 10.1016/S0959-440X(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 15.Oakeshott J.G., Claudianos C., Russell R.J., Robin G.C. Carboxyl/cholinesterases: A case study of the evolution of a successful multigene family. Bioessays. 1999;21:1031–1042. doi: 10.1002/(SICI)1521-1878(199912)22:1<1031::AID-BIES7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 16.Ollis D.L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S.M., Harel M., Remington S.J., Silman I., Schrag J., et al. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 17.Mindrebo J.T., Nartey C.M., Seto Y., Burkart M.D., Noel J.P. Unveiling the functional diversity of the alpha/beta hydrolase superfamily in the plant kingdom. Curr. Opin. Struct. Biol. 2016;41:233–246. doi: 10.1016/j.sbi.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Cao X., Duan W., Wei C., Chen K., Grierson D., Zhang B. Genome-Wide Identification and Functional Analysis of Carboxylesterase and Methylesterase Gene Families in Peach (Prunus persica L. Batsch) Front. Plant. Sci. 2019;10:1511. doi: 10.3389/fpls.2019.01511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cummins I., Landrum M., Steel P.G., Edwards R. Structure activity studies with xenobiotic substrates using carboxylesterases isolated from Arabidopsis thaliana. Phytochemistry. 2007;68:811–818. doi: 10.1016/j.phytochem.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Ileperuma N.R., Marshall S.D., Squire C.J., Baker H.M., Oakeshott J.G., Russell R.J., Plummer K.M., Newcomb R.D., Baker E.N. High-resolution crystal structure of plant carboxylesterase AeCXE1, from Actinidia eriantha, and its complex with a high-affinity inhibitor paraoxon. Biochemistry. 2007;46:1851–1859. doi: 10.1021/bi062046w. [DOI] [PubMed] [Google Scholar]
- 21.Kumar D., Klessig D.F. High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc. Natl. Acad. Sci. USA. 2003;100:16101–16106. doi: 10.1073/pnas.0307162100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vlot A.C., Liu P.P., Cameron R.K., Park S.W., Yang Y., Kumar D., Zhou F., Padukkavidana T., Gustafsson C., Pichersky E., et al. Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 2008;56:445–456. doi: 10.1111/j.1365-313X.2008.03618.x. [DOI] [PubMed] [Google Scholar]
- 23.Gao W., Liu Y., Huang J., Chen Y., Chen C., Lu L., Zhao H., Men S., Zhang X. MES7 Modulates Seed Germination via Regulating Salicylic Acid Content in Arabidopsis. Plants. 2021;10:903. doi: 10.3390/plants10050903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia R., Yu L., Chen J., Hu L., Cao S., Dong X., Ma Q., Wang Y. Molecular evolution of methylesterase family genes and the BnMES34 is a positive regulator of Plasmodiophora brassicae stress response in Arabidopsis. Int. J. Biol. Macromol. 2024;260:129333. doi: 10.1016/j.ijbiomac.2024.129333. [DOI] [PubMed] [Google Scholar]
- 25.Lima Silva C.C., Shimo H.M., de Felicio R., Mercaldi G.F., Rocco S.A., Benedetti C.E. Structure-function relationship of a citrus salicylate methylesterase and role of salicylic acid in citrus canker resistance. Sci. Rep. 2019;9:3901. doi: 10.1038/s41598-019-40552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia R., Xing K., Tian L., Dong X., Yu L., Shen X., Wang Y. Analysis of Methylesterase Gene Family in Fragaria vesca Unveils Novel Insights into the Role of FvMES2 in Methyl Salicylate-Mediated Resistance against Strawberry Gray Mold. J. Agric. Food. Chem. 2024;72:11392–11404. doi: 10.1021/acs.jafc.4c01447. [DOI] [PubMed] [Google Scholar]
- 27.Lin J., Wang W., Mazarei M., Zhao N., Chen X., Pantalone V.R., Hewezi T., Stewart C.N., Jr., Chen F. GmSABP2-1 encodes methyl salicylate esterase and functions in soybean defense against soybean cyst nematode. Plant Cell Rep. 2024;43:138. doi: 10.1007/s00299-024-03224-9. [DOI] [PubMed] [Google Scholar]
- 28.Li Q., Wang G., Guan C., Yang D., Wang Y., Zhang Y., Ji J., Jin C., An T. Overexpression of LcSABP, an Orthologous Gene for Salicylic Acid Binding Protein 2, Enhances Drought Stress Tolerance in Transgenic Tobacco. Front. Plant. Sci. 2019;10:200. doi: 10.3389/fpls.2019.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao N., Guan J., Forouhar F., Tschaplinski T.J., Cheng Z.M., Tong L., Chen F. Two poplar methyl salicylate esterases display comparable biochemical properties but divergent expression patterns. Phytochemistry. 2009;70:32–39. doi: 10.1016/j.phytochem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Xue R., Feng M., Chen J., Ge W., Blair M.W. A methyl esterase 1 (PvMES1) promotes the salicylic acid pathway and enhances Fusarium wilt resistance in common beans. Theor. Appl. Genet. 2021;134:2379–2398. doi: 10.1007/s00122-021-03830-1. [DOI] [PubMed] [Google Scholar]
- 31.Frick E.M., Sapkota M., Pereira L., Wang Y., Hermanns A., Giovannoni J.J., van der Knaap E., Tieman D.M., Klee H.J. A family of methyl esterases converts methyl salicylate to salicylic acid in ripening tomato fruit. Plant Physiol. 2023;191:110–124. doi: 10.1093/plphys/kiac509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manosalva P.M., Park S.W., Forouhar F., Tong L., Fry W.E., Klessig D.F. Methyl esterase 1 (StMES1) is required for systemic acquired resistance in potato. Mol. Plant Microbe Interact. 2010;23:1151–1163. doi: 10.1094/MPMI-23-9-1151. [DOI] [PubMed] [Google Scholar]
- 33.Koo Y.J., Yoon E.S., Seo J.S., Kim J.-K., Choi Y.D. Characterization of a methyl jasmonate specific esterase in Arabidopsis. J. Korean Soc. Appl. Biol. Chem. 2013;56:27–33. doi: 10.1007/s13765-012-2201-7. [DOI] [Google Scholar]
- 34.Wu J., Wang L., Baldwin I.T. Methyl jasmonate-elicited herbivore resistance: Does MeJA function as a signal without being hydrolyzed to JA? Planta. 2008;227:1161–1168. doi: 10.1007/s00425-008-0690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao N., Lin H., Lan S., Jia Q., Chen X., Guo H., Chen F. VvMJE1 of the grapevine (Vitis vinifera) VvMES methylesterase family encodes for methyl jasmonate esterase and has a role in stress response. Plant Physiol. Biochem. 2016;102:125–132. doi: 10.1016/j.plaphy.2016.02.027. [DOI] [PubMed] [Google Scholar]
- 36.Volk J., Sarafeddinov A., Unver T., Marx S., Tretzel J., Zotzel J., Warzecha H. Two novel methylesterases from Olea europaea contribute to the catabolism of oleoside-type secoiridoid esters. Planta. 2019;250:2083–2097. doi: 10.1007/s00425-019-03286-0. [DOI] [PubMed] [Google Scholar]
- 37.Wu R., Zhang F., Liu L., Li W., Pichersky E., Wang G. MeNA, Controlled by Reversible Methylation of Nicotinate, Is an NAD Precursor that Undergoes Long-Distance Transport in Arabidopsis. Mol. Plant. 2018;11:1264–1277. doi: 10.1016/j.molp.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Christ B., Schelbert S., Aubry S., Sussenbacher I., Muller T., Krautler B., Hortensteiner S. MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant Physiol. 2012;158:628–641. doi: 10.1104/pp.111.188870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fridman E., Wang J., Iijima Y., Froehlich J.E., Gang D.R., Ohlrogge J., Pichersky E. Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell. 2005;17:1252–1267. doi: 10.1105/tpc.104.029736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S.W., Kaimoyo E., Kumar D., Mosher S., Klessig D.F. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science. 2007;318:113–116. doi: 10.1126/science.1147113. [DOI] [PubMed] [Google Scholar]
- 41.Creelman R.A., Mullet J.E. Biosynthesis and Action of Jasmonates in Plants. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1997;48:355–381. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- 42.Teale W.D., Paponov I.A., Palme K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki Y., Amano T., Shioi Y. Characterization and cloning of the chlorophyll-degrading enzyme pheophorbidase from cotyledons of radish. Plant Physiol. 2006;140:716–725. doi: 10.1104/pp.105.071290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auldridge M.E., Guo Y., Austin M.B., Ramsey J., Fridman E., Pichersky E., Noel J.P. Emergent decarboxylase activity and attenuation of alpha/beta-hydrolase activity during the evolution of methylketone biosynthesis in tomato. Plant Cell. 2012;24:1596–1607. doi: 10.1105/tpc.111.093997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White R.F. Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology. 1979;99:410–412. doi: 10.1016/0042-6822(79)90019-9. [DOI] [PubMed] [Google Scholar]
- 46.Rasmussen J.B., Hammerschmidt R., Zook M.N. Systemic Induction of Salicylic Acid Accumulation in Cucumber after Inoculation with Pseudomonas syringae pv syringae. Plant Physiol. 1991;97:1342–1347. doi: 10.1104/pp.97.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaffney T., Friedrich L., Vernooij B., Negrotto D., Nye G., Uknes S., Ward E., Kessmann H., Ryals J. Requirement of salicylic Acid for the induction of systemic acquired resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 48.Shulaev V., Silverman P., Raskin I. Airborne signalling by methyl salicylate in plant pathogen resistance. Nature. 1997;385:718–721. doi: 10.1038/385718a0. [DOI] [Google Scholar]
- 49.Tieman D., Zeigler M., Schmelz E., Taylor M.G., Rushing S., Jones J.B., Klee H.J. Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J. 2010;62:113–123. doi: 10.1111/j.1365-313X.2010.04128.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhao N., Guan J., Ferrer J.L., Engle N., Chern M., Ronald P., Tschaplinski T.J., Chen F. Biosynthesis and emission of insect-induced methyl salicylate and methyl benzoate from rice. Plant Physiol. Biochem. 2010;48:279–287. doi: 10.1016/j.plaphy.2010.01.023. [DOI] [PubMed] [Google Scholar]
- 51.Soares J.M., Weber K.C., Qiu W., Mahmoud L.M., Grosser J.W., Dutt M. Overexpression of the salicylic acid binding protein 2 (SABP2) from tobacco enhances tolerance against Huanglongbing in transgenic citrus. Plant Cell Rep. 2022;41:2305–2320. doi: 10.1007/s00299-022-02922-6. [DOI] [PubMed] [Google Scholar]
- 52.Koo Y.J., Kim M.A., Kim E.H., Song J.T., Jung C., Moon J.K., Kim J.H., Seo H.S., Song S.I., Kim J.K., et al. Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol. Biol. 2007;64:1–15. doi: 10.1007/s11103-006-9123-x. [DOI] [PubMed] [Google Scholar]
- 53.Zou X., Zhao K., Liu Y., Du M., Zheng L., Wang S., Xu L., Peng A., He Y., Long Q., et al. Overexpression of Salicylic Acid Carboxyl Methyltransferase (CsSAMT1) Enhances Tolerance to Huanglongbing Disease in Wanjincheng Orange (Citrus sinensis (L.) Osbeck) Int. J. Mol. Sci. 2021;22:2803. doi: 10.3390/ijms22062803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin J., Mazarei M., Zhao N., Zhu J.J., Zhuang X., Liu W., Pantalone V.R., Arelli P.R., Stewart C.N., Jr., Chen F. Overexpression of a soybean salicylic acid methyltransferase gene confers resistance to soybean cyst nematode. Plant Biotechnol. J. 2013;11:1135–1145. doi: 10.1111/pbi.12108. [DOI] [PubMed] [Google Scholar]
- 55.Wu Z., Han S., Zhou H., Tuang Z.K., Wang Y., Jin Y., Shi H., Yang W. Cold stress activates disease resistance in Arabidopsis thaliana through a salicylic acid dependent pathway. Plant Cell Environ. 2019;42:2645–2663. doi: 10.1111/pce.13579. [DOI] [PubMed] [Google Scholar]
- 56.Yu X., Zhang W., Zhang Y., Zhang X., Lang D., Zhang X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019;46:197–212. doi: 10.1071/FP18106. [DOI] [PubMed] [Google Scholar]
- 57.Linkies A., Leubner-Metzger G. Beyond gibberellins and abscisic acid: How ethylene and jasmonates control seed germination. Plant Cell Rep. 2012;31:253–270. doi: 10.1007/s00299-011-1180-1. [DOI] [PubMed] [Google Scholar]
- 58.Seo H.S., Song J.T., Cheong J.J., Lee Y.H., Lee Y.W., Hwang I., Lee J.S., Choi Y.D. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamogami S., Noge K., Abe M., Agrawal G.K., Rakwal R. Methyl jasmonate is transported to distal leaves via vascular process metabolizing itself into JA-Ile and triggering VOCs emission as defensive metabolites. Plant Signal. Behav. 2012;7:1378–1381. doi: 10.4161/psb.21762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baldwin I.T., Halitschke R., Paschold A., von Dahl C.C., Preston C.A. Volatile signaling in plant-plant interactions: “talking trees” in the genomics era. Science. 2006;311:812–815. doi: 10.1126/science.1118446. [DOI] [PubMed] [Google Scholar]
- 61.Findling S., Fekete A., Warzecha H., Krischke M., Brandt H., Blume E., Mueller M.J., Berger S. Manipulation of methyl jasmonate esterase activity renders tomato more susceptible to Sclerotinia sclerotiorum. Funct. Plant. Biol. 2014;41:133–143. doi: 10.1071/FP13103. [DOI] [PubMed] [Google Scholar]
- 62.Woodward A.W., Bartel B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ljun K., Hull A.K., Kowalczyk M., Marchant A., Celenza J., Cohen J.D., Sandberg G. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 2002;49:249–272. doi: 10.1023/A:1015298812300. [DOI] [PubMed] [Google Scholar]
- 64.Zimmerman P., Hitchcock A. Comparative effectiveness of acids, esters and salts as growth substances and methods of evaluating them. Contr. Boyce Thompson Inst. 1937;8:337–350. [Google Scholar]
- 65.Noctor G., Queval G., Gakiere B. NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 2006;57:1603–1620. doi: 10.1093/jxb/erj202. [DOI] [PubMed] [Google Scholar]
- 66.Lin H. Nicotinamide adenine dinucleotide: Beyond a redox coenzyme. Org. Biomol. Chem. 2007;5:2541–2554. doi: 10.1039/b706887e. [DOI] [PubMed] [Google Scholar]
- 67.Wang G., Pichersky E. Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J. 2007;49:1020–1029. doi: 10.1111/j.1365-313X.2006.03013.x. [DOI] [PubMed] [Google Scholar]
- 68.Li W., Zhang F., Wu R., Jia L., Li G., Guo Y., Liu C., Wang G. A Novel N-Methyltransferase in Arabidopsis Appears to Feed a Conserved Pathway for Nicotinate Detoxification among Land Plants and Is Associated with Lignin Biosynthesis. Plant Physiol. 2017;174:1492–1504. doi: 10.1104/pp.17.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hortensteiner S., Krautler B. Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta. 2011;1807:977–988. doi: 10.1016/j.bbabio.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 70.Mühlecker W., Ongania K.H., Kräutler B., Matile P., Hörtensteiner S. Tracking Down Chlorophyll Breakdown in Plants: Elucidation of the Constitution of a “Fluorescent” Chlorophyll Catabolite. Angew Chem. Int. Ed. 2003;36:401–404. doi: 10.1002/anie.199704011. [DOI] [Google Scholar]
- 71.Obayashi K., Nagasawa K., Mandel W.J., Vyden J.K., Parmley W.W. Cardiovascular effects of ajmaline. Am. Heart J. 1976;92:487–496. doi: 10.1016/S0002-8703(76)80049-X. [DOI] [PubMed] [Google Scholar]
- 72.Ruiz-May E., Galaz-Avalos R.M., Loyola-Vargas V.M. Differential secretion and accumulation of terpene indole alkaloids in hairy roots of Catharanthus roseus treated with methyl jasmonate. Mol. Biotechnol. 2009;41:278–285. doi: 10.1007/s12033-008-9111-2. [DOI] [PubMed] [Google Scholar]
- 73.Namjoshi O.A., Cook J.M. Sarpagine and Related Alkaloids. Alkaloids Chem. Biol. 2016;76:63–169. doi: 10.1016/bs.alkal.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsuura H.N., Fett-Neto A.G. Plant Toxins. Volume 2. Springer Science & Business Media; Dordrecht, The Netherlands: 2015. Plant alkaloids: Main features, toxicity, and mechanisms of action; pp. 1–15. [Google Scholar]
- 75.Amiot M.J., Fleuriet A., Macheix J.J. Importance and evolution of phenolic compounds in olive during growth and maturation. J. Agric. Food Chem. 1986;34:823–826. doi: 10.1021/jf00071a014. [DOI] [Google Scholar]
- 76.Le Tutour B., Guedon D. Antioxidative activities of Olea europaea leaves and related phenolic compounds. Phytochemistry. 1992;31:1173–1178. doi: 10.1016/0031-9422(92)80255-D. [DOI] [Google Scholar]
- 77.Konno K., Hirayama C., Yasui H., Nakamura M. Enzymatic activation of oleuropein: A protein crosslinker used as a chemical defense in the privet tree. Proc. Natl. Acad. Sci. USA. 1999;96:9159–9164. doi: 10.1073/pnas.96.16.9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams W.G., Kennedy G.G., Yamamoto R.T., Thacker J.D., Bordner J. 2-Tridecanone: A Naturally Occurring Insecticide from the Wild Tomato Lycopersicon hirsutum f. glabratum. Science. 1980;207:888–889. doi: 10.1126/science.207.4433.888. [DOI] [PubMed] [Google Scholar]
- 79.Kennedy G.G. Tomato, pests, parasitoids, and predators: Tritrophic interactions involving the genus Lycopersicon. Annu. Rev. Entomol. 2003;48:51–72. doi: 10.1146/annurev.ento.48.091801.112733. [DOI] [PubMed] [Google Scholar]
- 80.Yu G., Pichersky E. Heterologous expression of methylketone synthase1 and methylketone synthase2 leads to production of methylketones and myristic acid in transgenic plants. Plant Physiol. 2014;164:612–622. doi: 10.1104/pp.113.228502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang C., Chaiprasongsuk M., Chanderbali A.S., Chen X., Fu J., Soltis D.E., Chen F. Origin and evolution of a gibberellin-deactivating enzyme GAMT. Plant Direct. 2020;4:e00287. doi: 10.1002/pld3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaiprasongsuk M., Zhang C., Qian P., Chen X., Li G., Trigiano R.N., Guo H., Chen F. Biochemical characterization in Norway spruce (Picea abies) of SABATH methyltransferases that methylate phytohormones. Phytochemistry. 2018;149:146–154. doi: 10.1016/j.phytochem.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 83.Zhao N., Ferrer J.L., Moon H.S., Kapteyn J., Zhuang X., Hasebe M., Stewart C.N., Jr., Gang D.R., Chen F. A SABATH Methyltransferase from the moss Physcomitrella patens catalyzes S-methylation of thiols and has a role in detoxification. Phytochemistry. 2012;81:31–41. doi: 10.1016/j.phytochem.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 84.Zhang C., Chen X., Crandall-Stotler B., Qian P., Kollner T.G., Guo H., Chen F. Biosynthesis of methyl (E)-cinnamate in the liverwort Conocephalum salebrosum and evolution of cinnamic acid methyltransferase. Phytochemistry. 2019;164:50–59. doi: 10.1016/j.phytochem.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 85.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Simpson M.G. Plant Systematics. Elsevier; Amsterdam, The Netherlands: 2019. Evolution and diversity of green and land plants; pp. 55–74. [Google Scholar]
- 87.O’Brien P.J., Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 88.Marchot P., Chatonnet A. Enzymatic activity and protein interactions in alpha/beta hydrolase fold proteins: Moonlighting versus promiscuity. Protein Pept. Lett. 2012;19:132–143. doi: 10.2174/092986612799080284. [DOI] [PubMed] [Google Scholar]
- 89.Li C., Hassler M., Bugg T.D. Catalytic promiscuity in the alpha/beta-hydrolase superfamily: Hydroxamic acid formation, C--C bond formation, ester and thioester hydrolysis in the C–C hydrolase family. Chembiochem. 2008;9:71–76. doi: 10.1002/cbic.200700428. [DOI] [PubMed] [Google Scholar]
- 90.Devamani T., Rauwerdink A.M., Lunzer M., Jones B.J., Mooney J.L., Tan M.A., Zhang Z.J., Xu J.H., Dean A.M., Kazlauskas R.J. Catalytic Promiscuity of Ancestral Esterases and Hydroxynitrile Lyases. J. Am. Chem. Soc. 2016;138:1046–1056. doi: 10.1021/jacs.5b12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Padhi S.K., Fujii R., Legatt G.A., Fossum S.L., Berchtold R., Kazlauskas R.J. Switching from an esterase to a hydroxynitrile lyase mechanism requires only two amino acid substitutions. Chem. Biol. 2010;17:863–871. doi: 10.1016/j.chembiol.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 92.Andexer J., von Langermann J., Mell A., Bocola M., Kragl U., Eggert T., Pohl M. An R-selective hydroxynitrile lyase from Arabidopsis thaliana with an alpha/beta-hydrolase fold. Angew Chem. Int. Edit. Engl. 2007;46:8679–8681. doi: 10.1002/anie.200701455. [DOI] [PubMed] [Google Scholar]
- 93.Peng Y., Sun T., Zhang Y. Perception of Salicylic Acid in Physcomitrella patens. Front. Plant Sci. 2017;8:2145. doi: 10.3389/fpls.2017.02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Monte I., Franco-Zorrilla J.M., Garcia-Casado G., Zamarreno A.M., Garcia-Mina J.M., Nishihama R., Kohchi T., Solano R. A Single JAZ Repressor Controls the Jasmonate Pathway in Marchantia polymorpha. Mol. Plant. 2019;12:185–198. doi: 10.1016/j.molp.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 95.Dave A., Graham I.A. Oxylipin Signaling: A Distinct Role for the Jasmonic Acid Precursor cis-(+)-12-Oxo-Phytodienoic Acid (cis-OPDA) Front. Plant. Sci. 2012;3:42. doi: 10.3389/fpls.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ponce De Leon I., Schmelz E.A., Gaggero C., Castro A., Alvarez A., Montesano M. Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 2012;13:960–974. doi: 10.1111/j.1364-3703.2012.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luo W., Komatsu S., Abe T., Matsuura H., Takahashi K. Comparative Proteomic Analysis of Wild-Type Physcomitrella patens and an OPDA-Deficient Physcomitrella patens Mutant with Disrupted PpAOS1 and PpAOS2 Genes after Wounding. Int. J. Mol. Sci. 2020;21:1417. doi: 10.3390/ijms21041417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ester Sztein A., Cohen J.D., de la Fuente I.G., Cooke T.J. Auxin metabolism in mosses and liverworts. Am. J. Bot. 1999;86:1544–1555. doi: 10.2307/2656792. [DOI] [PubMed] [Google Scholar]
- 99.Casanova-Saez R., Mateo-Bonmati E., Ljung K. Auxin Metabolism in Plants. Cold Spring Harb. Perspect. Biol. 2021;13:a039867. doi: 10.1101/cshperspect.a039867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ishizaki K. Evolution of land plants: Insights from molecular studies on basal lineages. Biosci. Biotechnol. Biochem. 2017;81:73–80. doi: 10.1080/09168451.2016.1224641. [DOI] [PubMed] [Google Scholar]
- 101.Maeda H.A., Fernie A.R. Evolutionary History of Plant Metabolism. Annu. Rev. Plant. Biol. 2021;72:185–216. doi: 10.1146/annurev-arplant-080620-031054. [DOI] [PubMed] [Google Scholar]
- 102.Li H., Pu H. Crystal structure of methylesterase family member 16 (MES16) from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2016;474:226–231. doi: 10.1016/j.bbrc.2016.04.115. [DOI] [PubMed] [Google Scholar]
- 103.Yang L., Hill M., Wang M., Panjikar S., Stockigt J. Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids. Angew Chem. Int. Ed. Engl. 2009;48:5211–5213. doi: 10.1002/anie.200900150. [DOI] [PubMed] [Google Scholar]
- 104.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Duan Y., Tang H., Yu X. Phylogenetic and AlphaFold predicted structure analyses provide insights for A1 aspartic protease family classification in Arabidopsis. Front. Plant Sci. 2023;14:1072168. doi: 10.3389/fpls.2023.1072168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xia Y., Sun G., Xiao J., He X., Jiang H., Zhang Z., Zhang Q., Li K., Zhang S., Shi X., et al. AlphaFold-guided redesign of a plant pectin methylesterase inhibitor for broad-spectrum disease resistance. Mol. Plant. 2024;17:1344–1368. doi: 10.1016/j.molp.2024.07.008. [DOI] [PubMed] [Google Scholar]
- 107.Pinto G.P., Corbella M., Demkiv A.O., Kamerlin S.C.L. Exploiting enzyme evolution for computational protein design. Trends. Biochem. Sci. 2022;47:375–389. doi: 10.1016/j.tibs.2021.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries may be directed to the corresponding author.