Abstract
Multinuclear NMR study has demonstrated that G-quadruplex adopted by d(G3T4G4) exhibits two cation binding sites between three of its G-quartets. Titration of tighter binding K+ ions into the solution of d(G3T4G4)2 folded in the presence of ions uncovered a mixed mono-K+-mono- form that represents intermediate in the conversion of into di-K+ form. Analogously, ions were found to replace Na+ ions inside d(G3T4G4)2 quadruplex. The preference of over Na+ ions for the two binding sites is considerably smaller than the preference of K+ over ions. The two cation binding sites within the G-quadruplex core differ to such a degree that ions bound to the site, which is closer to the edge-type loop, are always replaced first during titration by K+ ions. The second binding site is not taken up by K+ ion until K+ ion already resides at the first binding site. Quantitative analysis of concentrations of the three di-cation forms, which are in slow exchange on the NMR time scale, at 12 K+ ion concentrations afforded equilibrium binding constants. K+ ion binding to sites U and L within d(G3T4G4)2 is more favorable with respect to ions by Gibbs free energies of approximately −24 and −18 kJ mol−1 which includes differences in cation dehydration energies, respectively.
INTRODUCTION
Quadruplexes are stable structures adopted by DNA guanine-rich sequences that can be found in telomeres, immunoglobulin switch regions and gene promoter regions; these have also been implicated in association with human diseases, as therapeutic targets in drug design and in potential technical applications as nanomolecular devices (1–14). The basic structural motif of G-quadruplex is a G-quartet, which consists of four guanine bases held together in a coplanar arrangement by eight hydrogen bonds. G-quadruplexes differ in stoichiometry and orientation of strands. The major requirement for G-quadruplex formation is the presence of cations, which reduce repulsions of guanine carbonyl oxygen atoms and also promote stacking of G-quartets.
A fairly wide variety of cations is capable of inducing G-quadruplex formation (1,15). DNA oligonucleotides with double G-rich repeats like d(G4T4G4), d(G3T4G3) and d(G3T4G4) form dimeric fold-back G-quadruplex structures in the presence of Na+, K+ or ions (16–18). In contrast, d(G4T4G3) adopts a single G-quadruplex structure only in the presence of Na+ ions, whereas multiple structures form in the presence of K+ or ions (19). In the presence of Na+ ions, oligonucleotide consisting of four repeats of the human telomeric motif, d(TTAGGG), has been shown to fold into an intramolecular G-quadruplex composed of three G-quartets connected by one diagonal and two lateral TTA loops (20). In contrast, the crystal structure in the presence of K+ ions has demonstrated a propeller-type folding topology (21). Recent study by circular dichroism has shown that vertebrate telomere repeat, d(TTAGGG), sequences favor the formation of parallel-stranded structures with propeller-type topologies under conditions of high potassium and low sodium concentration as found in nuclei (22). Changeable cation concentration or gradients in cation concentration combined with the presence of different cations can lead to even more complicated situations where prediction of prevailing G-quadruplex species is difficult. Differences in binding properties of K+ and Na+ ions have long been noted and have been discussed in terms of metal ion switches inducing conformational changes from one type of G-quadruplex to another (23–25).
Solution and solid-state NMR (16–19,26–32), X-ray crystallography (33–35), computer simulations (36–39) as well as other spectroscopic methods have been applied to localize cations and evaluate cation-dependent structural changes (15,40–43). Further studies are however needed to understand subtle interrelationship between thermodynamic preference of cations for available binding sites, structural details that are cation specific and stability as well as folding pathways of G-quadruplexes. G-quadruplexes represent a good model system due to their strong coordination of metal ions. Recently, 15N-labeled ammonium ion has been used as a non-metallic substitute in combination with 2D NMR spectroscopy to demonstrate that cations are localized between two neighboring G-quartets in solution (44,45).
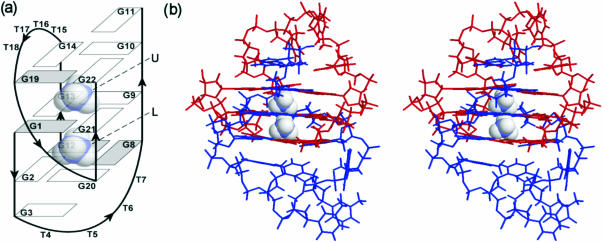
In our previous work, we have shown that d(G3T4G4)2 quadruplex, which consists of three G-quartet planes, two overhanging guanine residues and diagonal as well as edge-type loops (Figure 1) shows no variation with the nature of monovalent cation although the NMR spectra exhibit specific (de)shielding effects (18,46). The present solution NMR study demonstrates that two ions are localized between three G-quartet planes of the dimeric d(G3T4G4)2 structure (Figure 1). Monovalent cations in general stabilize G-quadruplexes in the following order: K+ > > Na+. The introduction of tighter binding cation into solution (e.g. K+) results in the replacement of weaker binding cation (e.g. or Na+). Progressive addition of tighter binding K+ ions was therefore expected to result in a replacement of ions at both binding sites of d(G3T4G4)2. The prediction as to the preference of K+ over ions for one of the two binding sites could not be made. One of the possibilities involved simultaneous replacement of both bound ions by K+. Other possibility could be that K+ preferentially binds over ions at one of the two binding sites. Titration experiments uncovered mixed mono-K+–mono- form of d(G3T4G4)2 which represents intermediate in the conversion of into di-K+ form. We herein show that K+ for ammonium ion replacement in d(G3T4G4)2 is a sequential process. Spectral differences amongst di-cation forms of d(G3T4G4)2 enabled their identification by NMR and evaluation of preference of the two cations for both binding sites in a quantitative way.
Figure 1.
(a) Schematic representation of topology of d(G3T4G4)2 G-quadruplex and its two cation binding sites labeled as U and L. The guanine bases are shown as rectangles, where filled rectangles represent syn nucleobases. (b) Stereo view of high resolution NMR structure (pdb = 1U64) of d(G3T4G4)2. Note a 180° rotation to the left with respect to (a). Strands G1–G11 and G12–G22 are colored blue and red, respectively.
MATERIALS AND METHODS
Sample preparation
DNA oligonucleotide d(G3T4G4) was synthesized on an Expedite 8909 synthesizer using phosphoramidite chemistry following the manufacturer's protocol and deprotected with concentrated aqueous ammonia. DNA was purified on 1.0 m Sephadex G15 column. Fractions containing only full-length oligonucleotide were pooled, lyophilized, redissolved in 1 ml H2O and extensively dialyzed against 10 mM LiCl. The DNA was then lyophilized and subsequently redissolved in 0.3 ml of 90% H2O/10% 2H2O. LiOH or HCl were added to adjust pH of the sample to 5.0. Oligonucleotide concentration was 3.5 mM in strand (1.75 mM in G-quadruplex) for experiments and 3.4 mM in strand for experiments. Aqueous solutions of KCl, NaCl or 15NH4Cl were titrated into the samples.
NMR spectroscopy
NMR data were collected on a Varian Unity Inova 600 MHz NMR spectrometer at 25°C. The standard and 15N-filtered 1H spectra were acquired using 15N decoupling. Both 2D NOESY (τm of 80 and 300 ms) and ROESY (τm of 140 ms) spectra were acquired in 10% 2H2O at 25°C using WATERGATE solvent suppression, 4096 complex points in the t2 dimension, 400 increments in the t1 dimension and a spectral width of 9.9 kHz. Spectra were 15N decoupled in both F1 and F2 dimensions.
RESULTS
Identification of mixed form of d(G3T4G4)2
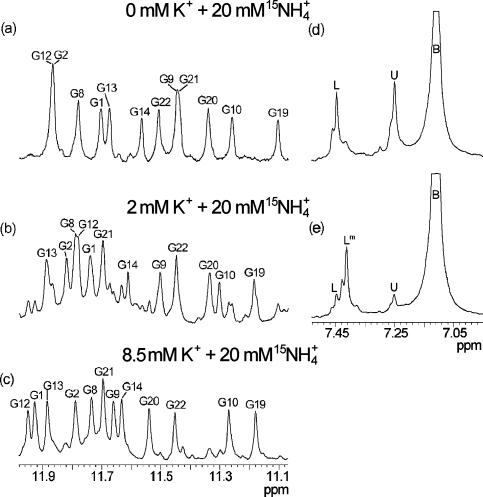
d(G3T4G4) has been folded into a G-quadruplex structure by titration of 15NH4Cl up to a 20 mM concentration that resulted in well-resolved imino (and other) resonances in 1H NMR spectrum (Figure 2a). NMR assignment and 3D structure determination have been described earlier (18,46). We herein examined the effect of gradual titration of K+ ions into the solution of d(G3T4G4)2 folded in the presence of 20 mM ions. New set of resonances appeared corresponding to a mixed form of G-quadruplex which was a major species (55%) when K+ ion concentration reached 2 mM (Figure 2b). The minor peaks in Figure 2b were assigned to and di-K+ forms (22.5% each). It is interesting to note that the percentage of mixed form of d(G3T4G4)2 in solution is dependent solely on the ratio between and K+ ion concentrations and not on their absolute concentrations. Further increase in the relative concentration of K+ ions resulted in the increased relative amount of di-K+ form, which became greatly predominant at 8.5 mM KCl and 20 mM 15NH4Cl (Figure 2c). The increase in K+ ion concentration up to 20 mM did not result in further changes in 1H NMR spectra, which were equivalent to the spectrum of d(G3T4G4)2 with only K+ ions present in solution (46). Although chemical shifts of imino protons for , di-K+ and mixed forms differ, their NOESY cross-peak patterns in the aromatic–anomeric and other spectral regions clearly confirm the formation of G-quadruplex structure with the same general fold as presented in Figure 1 (46).
Figure 2.
Standard and 15N-filtered 1D 1H NMR spectra of d(G3T4G4)2 at 25°C and pH 5.0 in 10% 2H2O in the presence of 20 mM 15NH4Cl and 0 (a and d), 2 (b and e) and 8.5 mM (c) KCl. Assignments of individual imino resonances are indicated (a–c). Labels U (δ7.25 p.p.m.), L (δ7.45 p.p.m.), Lm (δ7.41 p.p.m.) and B (δ7.11 p.p.m.) indicate ions at distinct binding sites within the architecture of d(G3T4G4)2 and in bulk solution, respectively. Downfield shoulders of the main peaks in (d) and (e) are due to isotopomers like 15NH3D+. The oligonucleotide concentration was 3.5 mM in strand.
We have also performed 15N-filtered 1H NMR experiments which in the presence of ions alone give rise to two resonances at δ7.25 and 7.45 p.p.m. corresponding to ions bound to U and L binding sites within the architecture of d(G3T4G4)2, respectively (Figures 1a and 2d). The largest peak at δ7.11 p.p.m. corresponds to ions that are found in bulk solution (Figure 2d and e). A decrease in intensity of U and L resonances was observed in the course of titration with K+ ions, which was in agreement with decrease in the amount of form. At the same time, a new resonance started to appear at δ7.41 p.p.m. corresponding to ion bound in a mixed form (Lm, Figure 2e). All three di-cation forms can be identified in equilibrium at given concentrations of KCl and 15NH4Cl and their relative ratios do not change even after several months in solution. Further increase in the concentration of K+ ions, however, resulted in disappearance of resonance at δ7.41 p.p.m., which is in agreement with the decreasing amount of mixed form and simultaneous increasing concentration of di-K+ form of d(G3T4G4)2. The mixed form has also been identified during reverse titration of ions into the solution of G-quadruplex, which was initially folded in the presence of K+ ions alone.
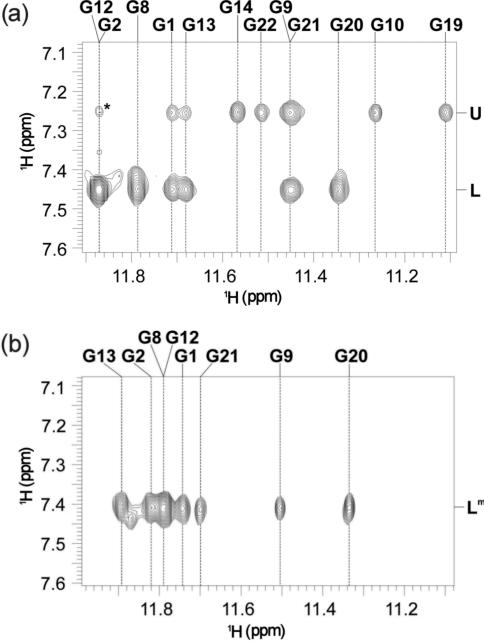
ROESY spectra shown in Figure 3 were used to assign and localize ammonium ions within d(G3T4G4)2 G-quadruplex. ions resonating at both δ7.25 (U) and 7.45 p.p.m. (L) show cross-peaks with imino protons of G1, G21, G9 and G13 residues (Figure 3a) that constitute the middle G-quartet (Figure 1a). In addition, ions at binding site U show correlations with imino protons of G19, G22, G10 and G14 residues (Figure 3a), which places them between the planes of the middle and the outer G-quartet that is adjacent to the edge-type loop consisting of T15–T18 (Figure 1a). The ions resonating at δ7.45 p.p.m., on the other hand, show additional cross-peaks with imino protons belonging to residues G2, G20, G8 and G12 (Figure 3a), and are therefore localized between the middle G-quartet and the outer G-quartet which is spanned by the diagonal loop (Figure 1a). ROESY spectrum shown in Figure 3b was used to localize ions at δ7.41 p.p.m. in the mixed form of d(G3T4G4)2. Eight ROE cross-peaks with imino protons of the two neighboring G-quartets (Figure 3b) offer unequivocal evidence that ions in the mixed form are residing between the planes of the middle G-quartet and the outer G-quartet that is spanned by the diagonal loop (i.e. at binding site L in Figure 1a).
Figure 3.
Part of ROESY spectra (τm =140 ms) of d(G3T4G4)2 at 25°C in the presence of 20 mM 15NH4Cl (a) showing cross-peaks between bound and nearby imino protons and in the presence of 20 mM 15NH4Cl and 2 mM KCl (b). Cross-peak designated with asterisk in (a) corresponds to G2(NH) and G12(H8) correlation.
Although exchange between di-K+ and mixed forms is slow on the NMR time scale respective chemical shift differences are large enough to identify and quantify both species only in the imino region of 1H NMR spectrum. ROESY experiments that would enable us to follow exchange between di-cation forms based on imino resonances do not allow separation of such exchange process from the exchange of protons between hydrogen-bonded sites. Anyhow, no cross-peaks have been observed above noise level in the imino region of ROESY spectra supposedly due to the opposite signs of correlation signals originating from dipolar interactions of spatially close imino protons and those from exchange processes. Chemical shift differences in other spectral regions were too small and thus cross-peaks were too close to the diagonal to allow the study of exchange of K+ cations between the di-cation forms of d(G3T4G4)2.
Determination of binding constants for two distinct binding sites
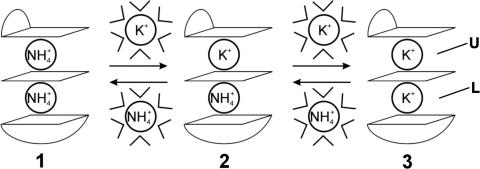
Two cation binding sites within d(G3T4G4)2 G-quadruplex differ to such a degree that ions bound to site U, which is closer to the edge-type loop, are always replaced first during titration by K+ ions (Figures 1 and 4). Furthermore, binding site L is not taken up by K+ ion until K+ ion already resides at binding site U.
Figure 4.
Cation displacement pathway amongst (1), mixed (2) and di-K+ (3) forms of d(G3T4G4)2. Three G-quartets shown schematically define two cation binding sites. The outer G-quartets are spanned by edge-type and diagonal loops. Cations have to be dehydrated to enter ion cavity of G-quadruplex.
We have then examined cationic preferences within G-quadruplex core in a quantitative way. The conversion from through mixed to di-K+ form (Figure 4) can be expressed by the two equilibrium constants, K1 and K2;
| 1 |
| 2 |
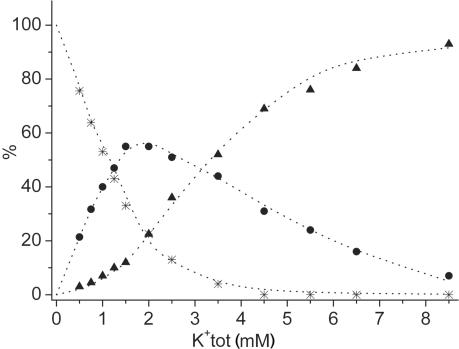
where 1, 2 and 3 correspond to the three di-cation forms defined in Figure 4. Integration of respective resonances in 1H NMR spectra enabled us to determine the concentrations of all three di-cation forms of d(G3T4G4)2 at 12 K+ ion concentrations, while the concentration of ions was constant at 20 mM (Figure 5). The concentrations of free K+ and ions in Equations 1 and 2 were calculated from the total concentration of inorganic salts and the amount of cations bound inside the three forms of d(G3T4G4)2 assuming that both of its binding sites are completely occupied by cations. Individual equilibrium constants K1 and K2 were calculated by the least-squares fitting procedure of concentrations of (1), mixed (2) and di-K+ (3) forms to Equations 1 and 2. The K1 and K2 values that best fitted the experimental data are 234 (±20) and 29 (±5), respectively (Figure 5). First K+ ion residing at binding site U stabilizes d(G3T4G4)2 G-quadruplex by apparent Gibbs free energy of −13.5 (±0.2) kJ mol−1 at 25°C with respect to form. Additional stabilization by the second K+ ion, which binds to binding site L is −8.3 (±0.4) kJ mol−1. It is noteworthy that dehydration of K+ ions with respect to ions is energetically more demanding process. The difference in the Gibbs free energies of their (de)hydration is 10 kJ mol−1 (47). The binding of K+ ions to binding sites U and L of d(G3T4G4)2 are thus more favorable with respect to ions by Gibbs free energies of approximately −24 and −18 kJ mol−1, which include differences in cation dehydration energies, respectively.
Figure 5.
Relative concentrations of the three di-cation forms of d(G3T4G4)2 as a function of total concentration of K+ ions. Stars, circles and triangles represent experimental relative concentrations of (1), mixed (2) and di-K+ (3) forms, respectively. Dotted curves represent the best fits of the experimental data to Equations 1 and 2. The maximum individual discrepancy between experimental and calculated fractions of 1–3 was 5 unit%, whereas root-mean-square deviation was 2 unit%.
Mixed form of d(G3T4G4)2
We furthermore examined the effect of addition of ions into the solution of d(G3T4G4)2 folded in the presence of 10 mM NaCl. At 3 mM and 10 mM Na+ ion concentrations, a single resonance was observed in 15N-filtered 1H spectrum at δ7.35 p.p.m., which demonstrated that ions selectively bind and replace Na+ ions at one of the two binding sites (see Figure S1, Supplementary Material). At 7.5 mM concentration of ions, a mixture of (δ7.25 and 7.45 p.p.m.), mixed (δ7.35 p.p.m.) and di-Na+ forms were present in solution. Increase in ion concentrations up to 40 mM resulted in the complete replacement of Na+ ions inside d(G3T4G4)2 and became the only species in solution. The resonances of mixed form were not resolved from their pure di-cation analogues in 1H NMR spectrum, which prevented localization of ammonium ions in the mixed form and determination of individual cation binding preferences. Additionally, signals in 1H NMR spectra became broader upon titration with ammonium chloride due to intermediate exchange on the NMR time scale between the three di-cation forms. This largely affected the quality of the spectra which did not allow assignment of resonances of individual forms and thus their quantitative evaluation at all concentrations of added ions that would enable us to determine equilibrium constants. We have nevertheless attempted to analyze concentrations of di-cation forms in the limited range from 3 to 12 mM (∼30% of , 25% of and 45% of di-Na+ forms) total concentration of ions. The limited number of six data points gave estimates of equilibrium constants K1 and K2 of 0.5 and 1.3, respectively. Note that these values should be considered as very rough estimates, as in the ion concentration range used, only approximately half of the di-Na+ forms have been transformed into ammonium forms, which greatly affects the fitting procedure.
DISCUSSION
Multinuclear NMR data have demonstrated that G-quadruplex adopted by d(G3T4G4) exhibits two cation binding sites U and L. The cation binding affinities and electrostatic potentials of these two binding sites are different as evident by their preferential binding of K+, Na+ and ions. The nature and preferential affinity of individual binding site for a given monovalent cation is influenced by different orientation of the loops and syn/anti alternation of the glycosidic torsion angles within individual G-quartets. The binding site U is characterized by syn-anti-anti-anti orientation of guanines in both G19-G22-G10-G14 and G1-G21-G9-G13 quartets (Figure 1a). In contrast, cation at binding site L is sandwiched between G1-G21-G9-G13 and G12-G8-G20-G2 quartets with syn-anti-anti-anti and syn-syn-anti-anti orientations, respectively, which affects directionality of hydrogen bonds and thus head-to-head versus head-to-tail nature of stacking interactions of guanine residues of the two G-quartets (Figure 1a). The characteristics of binding site U are defined by the outer G-quartet, which is spanned by the edge-type loop that consists of four thymine residues (T15–T18). Neighboring guanine residue G11 is stacked on the same G-quartet and in this way influences cation interactions at binding site U. In contrast, the outer G-quartet of binding site L is spanned by five residue diagonal loop consisting of four thymines T4–T7 and G3 (Figure 1a).
In general, monovalent cations stabilize G-quadruplexes in the following order: K+ > > Na+. Introduction of tighter binding cations results in replacement of cations with weaker stabilization energies. Several scenarios were expected when KCl was titrated into the solution of d(G3T4G4)2 folded in the presence of 15NH4Cl. First, ions at both sites could be replaced simultaneously. Second, K+ ions could take preference over ions at one of the two binding sites and subsequently at the remaining binding site. We however found that increase in K+ ion concentration first leads to replacement of ion at binding site U of d(G3T4G4)2 which is afterwards converted into di-K+ form. The slow exchange on the NMR time scale enabled us to demonstrate the existence of mixed form. In this form, the binding site U is occupied by K+ ion, whereas binding site L is still occupied by ion. The other two minor forms in solution were di-K+ and with the former becoming the only form as K+ ion concentration was increased further. Previous observations of related gradual transition from sodium to potassium forms of G-quadruplexes of d(G3T4G3)2 (16) and d(G4T4G4)2 (17) were interpreted by the assumed mixed Na+-K+ forms. No separate resonances could be observed due to fast exchange between cation forms on the NMR time scale (16,17). Existence of mixed cation forms is supported by the recent crystallographic study on d(TGGGGT)4 quadruplex which is stabilized by both Tl+ and Na+ ions in a single G-quadruplex (48). Quantitative analysis showed that binding of K+ ion to sites U and L within d(G3T4G4)2 is more favorable with respect to ions by Gibbs free energies of approximately −24 and −18 kJ mol−1 which includes differences in the cation dehydration energies, respectively. The comparison of equilibrium constants K1 and K2 for K+ binding to d(G3T4G4)2 suggests that cation binding is a process of negative cooperativity. It is however well established that it is very difficult to distinguish between negative cooperativity and binding to non-identical binding sites. The nature of the two cation binding sites of d(G3T4G4)2 quadruplex is determined by different orientations of the two loops and stacking interactions of non-identical G-quartets. It can be expected that consecutive binding or replacement of cations between pairs of G-quartets is a general feature of G-quadruplexes with alternating orientations of nucleotides within G-quartets and/or strand directionalities. On the other hand, the analysis of fraction saturation parameter (i.e. {[2]+[3]}/{[1]+[2]+[3]{) as a function of concentration of free K+ ions according to the Hill equation afforded a slope of 1.1. The calculated value of Hill coefficient >1, but less than the number of binding sites implies positive cooperativity of cation binding to d(G3T4G4)2.
Folding pathways of G-quadruplexes depend significantly on the oligonucleotide sequence and the nature of cations in solution. Folding process can proceed in a stepwise fashion where formation of a G-quartet is in the presence of alkali cations followed by stacking of neighboring G-quartet. Further stacking of additional G-quartets on such dimeric species leads to final G-quadruplex structure. Alternative mechanism results in a folded structure with all cation binding sites being occupied simultaneously. Demonstrations of both scenarios can be found in the literature. The thrombin binding aptamer, d(GGTTGGTGTGGTTGG), for example, forms G-quadruplex structures in the presence of K+ ions in a stepwise fashion (49). A chair-type G-quadruplex structure is formed at one molar equivalent of K+ ions per oligonucleotide. The binding of the second K+ ion primarily alters the structure of the loop while G-quadruplex remains in a chair-type structure (49). Tetrahymena telomere sequence d(T2G4) forms G-quadruplex monomer at low K+ ion concentration (50 mM) and dimer of co-axial G-quaduplexes at 300 mM K+ ion concentration (50). The presence of major and minor conformers of d(TAGGAGGT)4 in 100 mM KCl solution was interpreted by two and three bound K+ ions per G-quadruplex, respectively (51). Our NMR experiments have unequivocally shown that titration of K+, or Na+ ions into aqueous solution of d(G3T4G4) leads to immediate formation of asymmetric bimolecular G-quadruplex structure that consists of three G-quartet planes, two overhanging guanine residues and diagonal as well as an edge-type loops where both binding sites are occupied (18,46). In the process of titrations of unfolded d(G3T4G4) with ammonium chloride, two NMR resonances corresponding to ions bound at binding sites U and L appear simultaneously. There is no experimental evidence that folding of d(G3T4G4) proceeds in a stepwise fashion.
CONCLUSION
Some G-rich oligonucleotides fold into different G-quadruplex structures in the presence of Na+, K+ or other cations. Situation is even more complex in physiological systems where G-rich nucleotides get exposed to mixed ionic environments. Localization of cations interacting with macromolecules in solution is still elusive. G-quadruplexes represent a good model system due to their strong coordination of cations by four guanine carbonyl oxygen atoms per G-quartet. d(G3T4G4)2 is a unique system where K+ and di-cation forms are in slow exchange on the NMR time scale. Titration of tighter binding K+ ions into the solution of G-quadruplex folded in the presence of ions demonstrated formation of one of the two possible mixed forms of d(G3T4G4)2 G-quadruplex. Two cation binding sites within d(G3T4G4)2 differ to such a degree that ions bound to the site which is closer to the edge-type loop are always replaced first during titration by K+ ions. The second binding site is not taken up by K+ ion until K+ ion already resides at the upper binding site. Quantitative analysis afforded equilibrium binding constants of 234 and 29. K+ ion binding to sites U and L within d(G3T4G4)2 is more favorable with respect to ions by Gibbs free energies of approximately −24 and −18 kJ mol−1, which includes differences in cation dehydration energies, respectively. Taken altogether, mixed form of d(G3T4G4)2 presented here is to the best of our knowledge the first G-quadruplex structure identified in solution with two different cations bound inside G-quadruplex core. Understanding preference of different cations for binding sites presented here, thermodynamics of cation binding and dynamics of their replacement uncovers intriguing questions on polymorphism of nucleic acids and G-quadruplexes in particular.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Slovenian Research Agency (ARRS) and the Ministry of Higher Education, Science and Technology of the Republic of Slovenia (Grant Nos P1-0242-0104 and J1-6140-0104) for their financial support. Funding to pay the Open Access publication charges for this article was provided from the above grants.
Conflict of interest statement: None declared.
REFERENCES
- 1.Keniry M.A. Quadruplex structures in nucleic acids. Biopolymers. 2001;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 3.Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neidle S., Read M.A. G-quadruplexes as therapeutic targets. Biopolymers. 2001;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Shafer R.H., Smirnov I. Biological aspects of DNA/RNA quadruplexes. Biopolymers. 2001;56:209–227. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 7.Rezler E.M., Bearss D.J., Hurley L.H. Telomeres and telomerases as drug targets. Curr. Opin. Pharmacol. 2002;2:415–423. doi: 10.1016/s1471-4892(02)00182-0. [DOI] [PubMed] [Google Scholar]
- 8.Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 9.Arthanari H., Bolton P.H. Functional and dysfunctional roles of quadruplex DNA in cells. Chem. Biol. 2001;8:221–230. doi: 10.1016/s1074-5521(01)00007-2. [DOI] [PubMed] [Google Scholar]
- 10.Kim M.Y., Vankayalapati H., Kazuo S., Wierzba K., Hurley L.H. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 11.Seenisamy J., Rezler E.M., Powell T.J., Tye D., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 12.Alberti P., Mergny J.L. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armitage B.A. The impact of nucleic acid secondary structure on PNA hybridization. Drug Discov. Today. 2003;8:222–228. doi: 10.1016/s1359-6446(03)02611-4. [DOI] [PubMed] [Google Scholar]
- 14.da Silva M.W. Association of DNA quadruplexes through G : C : G : C tetrads. Solution structure of d(GCGGTGGAT) Biochemistry. 2003;42:14356–14365. doi: 10.1021/bi0355185. [DOI] [PubMed] [Google Scholar]
- 15.Hardin C.C., Perry A.G., White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2001;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 16.Hud N.V., Smith F.W., Anet F.A.L., Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 17.Schultze P., Hud N.V., Smith F.W., Feigon J. The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G4T4G4) Nucleic Acids Res. 1999;27:3018–3028. doi: 10.1093/nar/27.15.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crnugelj M., Sket P., Plavec J. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. [DOI] [PubMed] [Google Scholar]
- 19.Crnugelj M., Hud N.V., Plavec J. The solution structure of d(G4T4G3)2: a bimolecular G-quadruplex with a novel fold. J. Mol. Biol. 2002;320:911–924. doi: 10.1016/s0022-2836(02)00569-7. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 21.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 22.Rujan I.N., Meleney J.C., Bolton P.H. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sen D., Gilbert W. A sodium–potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 24.Hardin C.C., Henderson E., Watson T., Prosser J.K. Monovalent cation induced structural transitions in telomeric DNAs—G-DNA folding intermediates. Biochemistry. 1991;30:4460–4472. doi: 10.1021/bi00232a013. [DOI] [PubMed] [Google Scholar]
- 25.Miura T., Thomas G.J. Structural polymorphism of telomere DNA—interquadruplex and duplex–quadruplex conversions probed by raman-spectroscopy. Biochemistry. 1994;33:7848–7856. doi: 10.1021/bi00191a012. [DOI] [PubMed] [Google Scholar]
- 26.Wu G., Wong A. Direct detection of the bound sodium ions in self-assembled 5′-GMP gels: a solid-state 23Na NMR approach. Chem. Commun. 2001:2658–2659. [Google Scholar]
- 27.Wu G., Wong A., Gan Z.H., Davis J.T. Direct detection of potassium cations bound to G-quadruplex structures by solid-state 39K NMR at 19.6 T. J. Am. Chem. Soc. 2003;125:7182–7183. doi: 10.1021/ja0340634. [DOI] [PubMed] [Google Scholar]
- 28.Kettani A., Bouaziz S., Gorin A., Zhao H., Jones R.A., Patel D.J. Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol. 1998;282:619–636. doi: 10.1006/jmbi.1998.2030. [DOI] [PubMed] [Google Scholar]
- 29.Bouaziz S., Kettani A., Patel D.J. A K cation-induced conformational switch within a loop spanning segment of a DNA quadruplex containing G-G-G-C repeats. J. Mol. Biol. 1998;282:637–652. doi: 10.1006/jmbi.1998.2031. [DOI] [PubMed] [Google Scholar]
- 30.Strahan G.D., Keniry M.A., Shafer R.H. NMR structure refinement and dynamics of the K+-[d(G3T4G3)]2 quadruplex via particle mesh Ewald molecular dynamics simulations. Biophys. J. 1998;75:968–981. doi: 10.1016/S0006-3495(98)77585-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keniry M.A., Strahan G.D., Owen E.A., Shafer R.H. Solution structure of the Na+ form of the dimeric quadruplex [d(G3T4G3)]2. Eur. J. Biochem. 1995;233:631–643. doi: 10.1111/j.1432-1033.1995.631_2.x. [DOI] [PubMed] [Google Scholar]
- 32.Sket P., Crnugelj M., Kozminski W., Plavec J. ion movement inside d(G4T4G4)2 G-quadruplex is accelerated in the presence of smaller Na+ ions. Org. Biomol. Chem. 2004;2:1970–1973. doi: 10.1039/b406815g. [DOI] [PubMed] [Google Scholar]
- 33.Haider S., Parkinson G.N., Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 34.Phillips K., Dauter Z., Murchie A.I.H., Lilley D.M.J., Luisi B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 angstrom resolution. J. Mol. Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]
- 35.Horvath M.P., Schultz S.C. DNA G-quartets in a 1.86 angstrom resolution structure of an Oxytricha nova telomeric protein–DNA complex. J. Mol. Biol. 2001;310:367–377. doi: 10.1006/jmbi.2001.4766. [DOI] [PubMed] [Google Scholar]
- 36.Spackova N., Berger I., Sponer J. Nanosecond molecular dynamics simulations of parallel and antiparallel guanine quadruplex DNA molecules. J. Am. Chem. Soc. 1999;121:5519–5534. [Google Scholar]
- 37.Fadrna E., Spackova N., Stefl R., Koca J., Cheatham T.E., Sponer J. Molecular dynamics simulations of guanine quadruplex loops: advances and force field limitations. Biophys. J. 2004;87:227–242. doi: 10.1529/biophysj.103.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spackova N., Berger I., Sponer J. Structural dynamics and cation interactions of DNA quadruplex molecules containing mixed guanine/cytosine quartets revealed by large-scale MD simulations. J. Am. Chem. Soc. 2001;123:3295–3307. doi: 10.1021/ja002656y. [DOI] [PubMed] [Google Scholar]
- 39.Chowdhury S., Bansal M. G-quadruplex structure can be stable with only some coordination sites being occupied by cations: a six-nanosecond molecular dynamics study. J. Phys. Chem. B. 2001;105:7572–7578. [Google Scholar]
- 40.Miyoshi D., Nakao A., Toda T., Sugimoto N. Effect of divalent cations on antiparallel G-quartet structure of d(G4T4G4) FEBS Lett. 2001;496:128–133. doi: 10.1016/s0014-5793(01)02416-4. [DOI] [PubMed] [Google Scholar]
- 41.Davis J.T. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 42.Mergny J.L., De Cian A., Ghelab A., Sacca B., Lacroix L. Kinetics of tetramolecular quadruplex. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hazel P., Huppert J., Balasubramanian S., Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 44.Hud N.V., Schultze P., Feigon J. Ammonium ion as an NMR probe for monovalent cation coordination sites of DNA quadruplexes. J. Am. Chem. Soc. 1998;120:6403–6404. [Google Scholar]
- 45.Hud N.V., Schultze P., Sklenar V., Feigon J. Binding sites and dynamics of ammonium ions in a telomere repeat DNA quadruplex. J. Mol. Biol. 1999;285:233–243. doi: 10.1006/jmbi.1998.2327. [DOI] [PubMed] [Google Scholar]
- 46.Sket P., Cmugelj M., Plavec J. d(G3T4G4) forms unusual dimeric G-quadruplex structure with the same general fold in the presence of K+, Na+ or NH+4 ions. Bioorg. Med. Chem. 2004;12:5735–5744. doi: 10.1016/j.bmc.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Marcus Y. A simple empirical-model describing the thermodynamics of hydration of ions of widely varying charges, sizes, and shapes. Biophys. Chem. 1994;51:111–127. [Google Scholar]
- 48.Caceres A., Wright G., Gouyette C., Parkinson G., Subirana J.A. A thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+/Na+ ions. Nucleic Acids Res. 2004;32:1097–1102. doi: 10.1093/nar/gkh269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marathias V.M., Bolton P.H. Structures of the potassium-saturated 2:1 and intermediate 1:1 forms of a quadruplex DNA. Nucleic Acids Res. 2000;28:1969–1977. doi: 10.1093/nar/28.9.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y., Patel D.J. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry. 1992;31:8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- 51.Searle M.S., Williams H.E.L., Gallagher C.T., Grant R.J., Stevens M.F.G. Structure and K+ ion-dependent stability of a parallel-stranded DNA quadruplex containing a core A-tetrad. Org. Biomol. Chem. 2004;2:810–812. doi: 10.1039/b314559j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.