Abstract
The integration of machine learning (ML) into material manufacturing has driven advancements in optimizing biopolymer production processes. ML techniques, applied across various stages of biopolymer production, enable the analysis of complex data generated throughout production, identifying patterns and insights not easily observed through traditional methods. As sustainable alternatives to petrochemical-based plastics, biopolymers present unique challenges due to their reliance on variable bio-based feedstocks and complex processing conditions. This review systematically summarizes the current applications of ML techniques in biopolymer production, aiming to provide a comprehensive reference for future research while highlighting the potential of ML to enhance efficiency, reduce costs, and improve product quality. This review also shows the role of ML algorithms, including supervised, unsupervised, and deep learning algorithms, in optimizing biopolymer manufacturing processes.
Keywords: ML, biopolymers, process optimization, materials science
1. Introduction
Biopolymers have garnered significant attention as sustainable alternatives to petroleum-based plastics due to their biodegradability and reduced environmental impact [1]. With the increasing awareness of environmental issues, industries are actively seeking ways to reduce plastic waste [2,3], and biopolymers have emerged as a key solution [4]. They are derived from renewable biological sources such as plants [5], algae [6], and microorganisms [7], offering a promising path to reduce the reliance on fossil fuels and mitigate the growing problem of plastic pollution. Their biodegradability is an advantage, as it ensures that these materials can break down into non-toxic components in the environment, contributing to a more sustainable and circular economy. As a result, biopolymers are finding expanding applications across various sectors, such as packaging [8], agriculture [9], automotive [10], textiles [11], and biomedical fields [12], where environmental sustainability and product performance are equally important.
The growing demand for biopolymers [13,14] has prompted the need for efficient, scalable manufacturing processes. However, the production of biopolymers presents several challenges that need to be addressed to fully capitalize on their potential. The manufacturing process involves complex biochemical reactions, including fermentation [15,16,17,18,19,20,21,22,23], polymerization [7,24,25,26,27,28,29,30,31,32], and extraction [33,34,35,36,37,38,39,40,41,42], which are highly sensitive to variations in raw material quality, environmental conditions, and process parameters. Even minor fluctuations in temperature [43,44,45,46,47], pH levels [48,49,50,51,52], or nutrient concentrations [9,53,54,55,56] during fermentation can significantly affect the yield and properties of the final product. Similarly, inconsistencies in raw materials [57,58,59,60,61]—such as variations in the composition of feedstocks or impurities—can disrupt polymerization and extraction processes, leading to inefficiencies, increased waste, and deviations from desired product specifications.
These fluctuations not only pose challenges for maintaining consistent product quality but also hinder scalability, as small-scale optimization techniques often fail to account for the variability encountered in large-scale operations [62]. Furthermore, the growing emphasis on sustainability compounds these challenges. Manufacturers must ensure that fluctuations do not compromise the eco-friendliness of the process, requiring continuous monitoring, precise control systems, and innovations in workflow optimization to minimize resource consumption, reduce emissions, and manage waste effectively [63,64,65,66,67,68,69,70,71,72]. Balancing these technical and environmental demands is crucial to achieving both production efficiency and long-term sustainability goals.
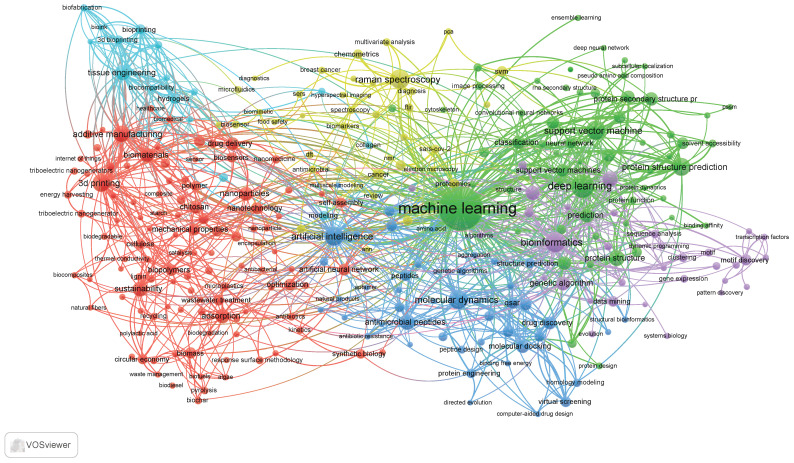
Machine learning (ML) offers promising solutions to these challenges by enabling real-time data processing [73], predictive modeling [74], and optimization of production workflows [75]. The integration of ML in biopolymer manufacturing can facilitate more precise control over the various stages of production, improving the consistency and quality of the final product. ML-driven predictive models can anticipate potential variations in raw material quality or environmental conditions [76,77], allowing manufacturers to adjust their processes proactively to maintain efficiency and sustainability. Moreover, ML can help to optimize energy consumption [78], reduce waste [79], and streamline logistics [80], further enhancing the overall sustainability of biopolymer production. ML algorithms, for instance, can analyze vast datasets from production lines to identify patterns and suggest process improvements, while ML-powered sensors [81] can monitor real-time performance metrics to ensure optimal operating conditions. A keyword map (Figure 1) was created using VOSviewer to analyze the scientific literature on the application of ML in biopolymers. This visualization highlights key topics, research directions, and their interconnections, providing insights into current trends and promising areas of study.
Figure 1.
Keyword map from articles on the application of ML in biopolymers.
ML has already made a significant impact in fields such as materials science and chemical engineering, and its transformative potential in biopolymers is becoming increasingly evident. For example, in materials science, ML has been used to predict material properties [82,83,84,85,86,87,88,89,90], optimize the design of alloys [84,90,91,92,93,94,95,96,97,98], and accelerate the discovery of novel materials [99,100,101,102,103,104,105,106,107,108]. Building on tools like ab initio calculations, density functional theory (DFT), and quantum chemistry databases, ML integrates computational and experimental approaches, facilitating breakthroughs in catalyst design, energy materials, and drug discovery [109].
The application of ML to biopolymers faces several challenges. Data quality and availability are key issues, as biopolymer systems involve complex variables and often suffer from sparse, noisy, or incomplete datasets, leading to overfitting or unreliable predictions [72,110,111,112,113,114]. Additionally, the nonlinear and multifactorial nature of biopolymer processes (e.g., fermentation, polymerization, extraction) complicates modeling with existing ML algorithms, which may assume simpler linear relationships [115,116,117,118]. Interpretability [119,120,121,122,123,124,125,126,127,128] is another challenge, as many ML models, especially deep learning, are seen as “black boxes”, making it difficult to understand the reasons behind predictions. This lack of transparency hinders trust and limits adoption in industrial settings where clear explanations are essential. ML models also struggle with scalability and generalization [128,129,130,131,132,133,134,135,136,137]; those trained on small-scale data often fail to perform well in larger, more variable real-world environments, restricting their broader application. Finally, the computational requirements [138,139,140,141,142,143,144,145,146,147] of advanced ML models can be a barrier for researchers and small manufacturers lacking access to high-performance computing resources, further complicating real-time model deployment in biopolymer production.
Recent reviews on biopolymers and the application of ML underscore the growing need for innovative approaches to address challenges associated with variable feedstock properties and complex manufacturing processes. These studies highlight the potential of ML to optimize key production stages, enhance sustainability, and improve the quality of final products across various industries.
In tissue engineering, significant advancements over the past 30 years have positioned additive manufacturing (AM) as a tool for replacing and regenerating injured tissues. AM, particularly 3D printing, is recognized as an effective method for synthesizing conducting polymer-based materials in applications such as flexible electronics, bioelectronics, and energy storage [148]. Das et al. [149] focus on gelatin methacryloyl (GelMA) as a versatile biomaterial for 3D bioprinting, discussing strategies to optimize its rheological, mechanical, and biophysical properties. Their review also explores the potential integration of artificial intelligence (AI) and ML to predict printability and functionality for clinically relevant applications, emphasizing the transformative role of computational tools in advancing tissue engineering solutions.
Similarly, the rising global prevalence of diabetes calls for innovative solutions, with hydrogel-based systems emerging as promising alternatives for non-invasive diagnosis, management, and treatment. Rahmani et al. [150] review the potential of hydrogels, their integration with Internet of Things (IoT) and ML technologies, and their role in advancing personalized and proactive diabetes care. These technologies offer new opportunities to improve patient outcomes through continuous monitoring and tailored therapeutic approaches.
In the food industry, consumer demand for safe and high-quality meat has driven the development of anthocyanin-based materials for real-time freshness monitoring. Xiong et al. [151] review recent advances in anthocyanin-based films, hydrogels, aerogels, and colorimetric sensor arrays. Their work highlights the potential of these materials to provide intuitive color signals indicating spoilage, while also addressing challenges related to sensitivity, stability, and integration with technologies like smartphones and ML for enhanced performance.
Furthermore, the integration of ML with algae-derived biopolymers, such as alginate and carrageenan, is transforming 3D printing by enabling sustainable and efficient additive manufacturing processes. Bin et al. [152] discuss advancements and challenges in this field, emphasizing ML’s role in optimizing material selection, predictive modeling, and quality control. Their findings demonstrate how the combination of ML and algae-based biopolymers can enhance mechanical properties and expand applications, particularly in areas such as bone tissue engineering.
The existing reviews highlight ML’s potential in biopolymer applications but overlook its specific role in optimizing manufacturing processes. A focused review is needed to address how ML can tackle challenges like feedstock variability, process efficiency, and product quality in biopolymer production.
This review aims to provide an overview of ML’s applications in the biopolymer manufacturing industry, outlining the potential of various ML techniques in improving production efficiency and sustainability. It explores key ML methodologies such as supervised and unsupervised learning, and how they are being utilized to address specific challenges in biopolymer production. Additionally, the review considers the future outlook of ML in the biopolymer industry, examining the potential for further innovations that could drive cost reductions, enhance material properties, and facilitate the development of new, advanced biopolymers that meet the demands of an evolving global market. To provide a clear structure, this review is organized as follows. It begins with an overview of biopolymer manufacturing (Section 2), covering key stages such as feedstock selection (Section 2.1), fermentation (Section 2.2), polymerization and extraction (Section 2.3), and quality control with post-processing techniques (Section 2.4). The discussion then shifts to the role of ML techniques in process optimization (Section 3), detailing approaches such as supervised learning (Section 3.1), unsupervised learning (Section 3.2), and neural networks with deep learning applications (Section 3.3). Challenges associated with integrating ML into biopolymer research are outlined in Section 4, followed by an exploration of future directions for advancing ML-driven solutions in this field (Section 5).
2. Overview of Biopolymer Manufacturing
Biopolymers are polymers derived from natural sources, including plants [153,154], animals [155,156], and microorganisms [7,157]. Unlike conventional petroleum-based plastics, biopolymers are sourced from renewable materials [158,159,160], which makes them highly attractive in the push for sustainable materials. They are biodegradable or compostable in many cases, reducing environmental impact and waste accumulation. Common types of biopolymers include polyhydroxyalkanoates (PHAs) [161,162], polylactic acid (PLA) [163,164], and starch-based polymers, each with unique properties suitable for various industrial applications. For instance, PHAs are microbial biopolymers that can be customized for diverse applications, while PLA, derived from fermented plant sugars, is widely used in packaging due to its favorable mechanical properties and biodegradability. Starch-based polymers, produced from plant-based starch, are often employed in food packaging and agricultural films.
ML is increasingly applied in biopolymer manufacturing to optimize the design, synthesis, and processing of biopolymers across various industries. By analyzing complex datasets, ML techniques can predict and enhance the functional properties of biopolymers, such as biodegradability, mechanical strength, and stability, enabling more efficient production processes, material selection, and product customization for applications in food, pharmaceuticals, packaging, and biomedical fields.
The production process of biopolymers typically involves several key stages that require careful control and optimization to ensure the quality and sustainability of the final product.
2.1. Feedstock Selection
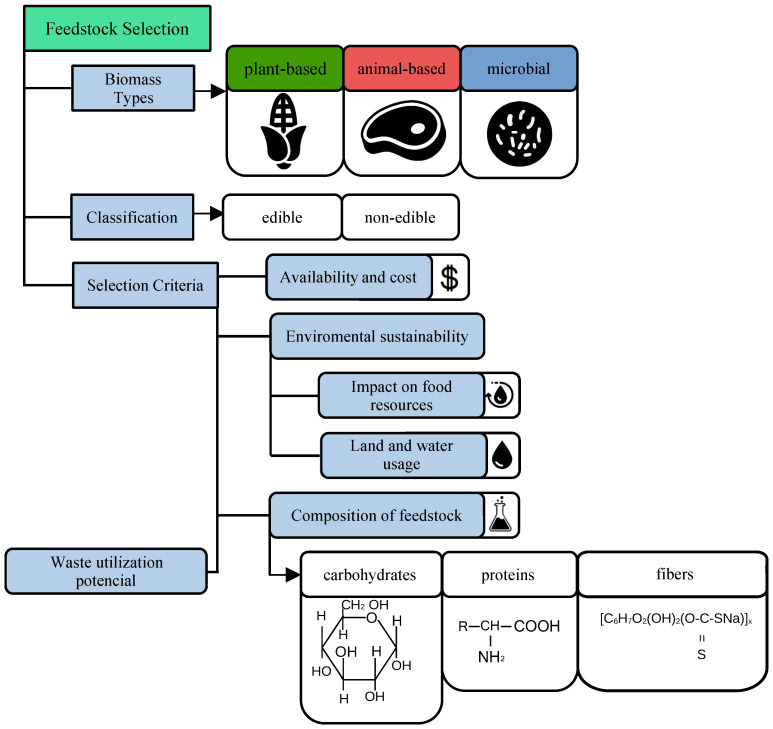
The use of traditional food-based feedstocks like corn starch [165] or sugarcane [166] has been widely adopted due to their high availability and consistent quality, making them suitable for the large-scale production of biopolymers such as polylactic acid (PLA). However, these feedstocks can compete with food crops for arable land and water resources, raising concerns about their impact on food security and land use [167]. This has led to an increased interest in using non-food biomass as an alternative, including agricultural residues (such as corn stover, wheat straw, or rice husks), forestry waste, and industrial by-products. These materials offer a more sustainable option, as they do not compete directly with food production and often represent waste streams that would otherwise go unused or be disposed of.
Using non-food biomass can reduce the overall environmental impact of biopolymer production [168]. For instance, agricultural residues that are left over after crop harvesting, such as straw or husks, can be collected and processed into valuable feedstocks for biopolymer production without requiring additional land or resources [169]. Similarly, industrial by-products, such as lignin from paper production or glycerol from biodiesel manufacturing, can be repurposed, reducing waste and improving the circularity of industrial processes [170]. This not only minimizes the carbon footprint associated with raw material extraction but also enhances the sustainability of biopolymer production by diverting waste from landfills or incineration.
However, the quality of these alternative feedstocks can vary significantly due to factors such as geographic location [171], climate [39], and farming practices [64]. For example, agricultural residues from different regions or seasons may have different moisture content, fiber composition, or levels of impurities, all of which can affect the efficiency of fermentation and polymerization processes [172]. Climate conditions such as drought or excessive rainfall can also alter the chemical composition of plants, leading to fluctuations in the availability of sugars or starches that are essential for biopolymer production [173]. In regions with inconsistent farming practices or where crop management varies, the feedstock may require additional pretreatment steps, such as cleaning or fractionation, to ensure it meets the quality standards necessary for biopolymer synthesis [174].
Furthermore, the supply chain logistics of sourcing feedstock from agricultural waste or non-food biomass can be complex [175]. Collecting, transporting, and storing large quantities of biomass often present logistical challenges due to the bulkiness and seasonal availability of these materials. In some cases, the energy required to process and transport low-density biomass can offset the environmental benefits of using renewable feedstocks [176]. To address these challenges, advancements in biorefining technologies are being developed to improve the conversion efficiency of various feedstocks into polymer precursors and optimize the overall sustainability of the production process [177]. Figure 2 is a visual representation of the key factors related to biopolymer feedstock selection and sustainability consideration.
Figure 2.
Biopolymer feedstock selection and sustainability considerations.
2.2. Fermentation
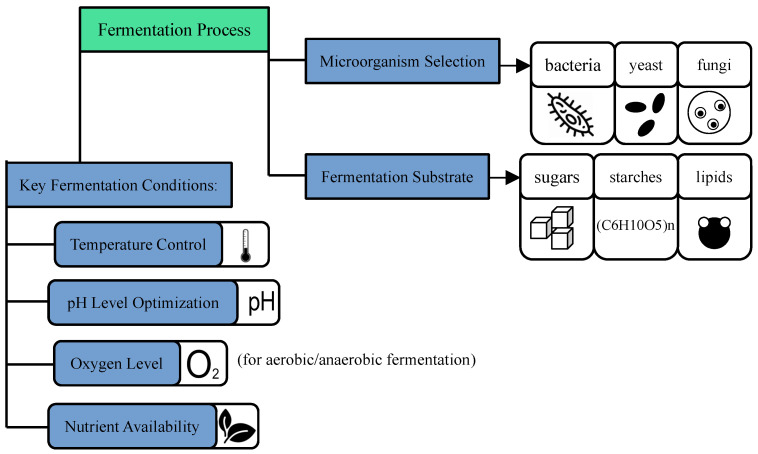
In the fermentation stage, microorganisms such as bacteria [178], yeast [179], or fungi [180] convert renewable feedstocks into polymer precursors, which are the building blocks of biopolymers. This bioconversion process is highly dependent on the metabolic capabilities of the selected microorganisms and the specific fermentation conditions. The microorganisms used in fermentation act as biocatalysts, breaking down the feedstock—typically composed of sugars, starches, or lipids—into simpler molecules such as organic acids or alcohols. These precursor molecules are then used in subsequent steps to produce various biopolymers [181,182].
For example, in the production of polylactic acid (PLA), specific strains of lactic acid bacteria (e.g., Lactobacillus spp.) are employed to ferment sugars derived from feedstocks such as corn, sugarcane, or other carbohydrates [183]. During fermentation, these bacteria metabolize the sugars and convert them into lactic acid, a key precursor for PLA production [183]. Once lactic acid is produced, it undergoes polymerization to form PLA, a biodegradable thermoplastic that is widely used in packaging, disposable cutlery, and biomedical applications.
Similarly, polyhydroxyalkanoates (PHAs) are another class of biopolymers produced through microbial fermentation. In PHA production, bacteria such as Cupriavidus necator or Ralstonia eutropha are typically used to ferment sugars, lipids, or other organic substrates [184,185,186]. Under conditions of nutrient limitation (such as nitrogen or phosphorus deficiency), these bacteria accumulate PHAs intracellularly as carbon and energy storage compounds. The PHAs are then harvested from the bacterial cells and processed into biodegradable plastics with a range of applications, from packaging materials to medical devices.
The selection of the microorganism strain is perhaps the most important factor in fermentation. Different strains have varying metabolic pathways, growth rates, and tolerance to environmental conditions. For instance, some bacterial strains may be more efficient at converting specific feedstocks into lactic acid or PHAs, while others may be more resistant to by-products that could inhibit fermentation [187]. Genetic engineering and synthetic biological techniques have also enabled the development of engineered microbial strains with enhanced fermentation capabilities [188]. These engineered strains can exhibit higher productivity, increased substrate versatility, and reduced by-product formation, all of which contribute to a more efficient fermentation process.
The composition and quality of the feedstock used in fermentation affect the efficiency of microbial activity [189]. Feedstocks rich in fermentable sugars, such as glucose, fructose, or sucrose, are often preferred for high-yield lactic acid or PHA production. However, feedstocks can vary in their composition, particularly when derived from agricultural residues or industrial by-products [190]. Complex feedstocks may contain inhibitors such as lignin, phenolic compounds, or heavy metals, which can slow down microbial growth or reduce the yield of polymer precursors [191]. To address these challenges, pretreatment processes such as enzymatic hydrolysis, acid or alkali treatments, or steam explosion may be required to break down complex biomass into fermentable sugars, enhancing the efficiency of fermentation [192].
The fermentation environment impacts the determination of the productivity of microorganisms. Maintaining optimal conditions for temperature [193], pH [194], and oxygen levels (in the case of aerobic fermentation) is essential for maximizing microbial growth and metabolic activity [195]. Each microorganism strain has a specific range of temperature and pH where it performs optimally. For instance, the Lactobacillus species used in lactic acid production typically thrive at moderately acidic pH values (around pH 5–6) and temperatures ranging from 30 to 40 °C [196,197]. Deviations from these optimal conditions can slow down fermentation, reduce yields, or lead to the formation of unwanted by-products such as acetic acid or ethanol. In some cases, the process may need to be aerobic [198] (with oxygen) or anaerobic [199] (without oxygen), depending on the metabolic pathway of the microorganism. Therefore, precise control over these variables is crucial for achieving efficient biopolymer precursor production.
The duration of fermentation also affects the final yield of polymer precursors. Longer fermentation times may lead to higher yields but can increase operational costs and energy consumption [200]. Conversely, shorter fermentation times can reduce costs but may result in lower yields if microbial growth and metabolism are not fully optimized [201]. Batch, fed-batch, and continuous fermentation processes are commonly used to balance yield and efficiency. In a batch fermentation, the feedstock is added at the beginning, and the process runs until completion [202]. Fed-batch fermentation allows for the gradual addition of feedstock, enabling better control over microbial growth and product formation [203]. Continuous fermentation, on the other hand, enables the constant input of feedstock and removal of products, allowing for more consistent yields and higher productivity over time [204].
During fermentation, unwanted by-products can form due to suboptimal conditions or microbial metabolic pathways [205]. These by-products, such as acetic acid, ethanol, or hydrogen sulfide, can inhibit microbial growth, lower the yield of polymer precursors, and increase the need for costly purification steps. By optimizing the fermentation conditions—such as adjusting nutrient concentrations, pH levels, or oxygen availability—manufacturers can minimize by-product formation and improve the efficiency of precursor production. The metabolic engineering of microorganisms is also being used to reprogram metabolic pathways, reducing the production of by-products and improving the conversion efficiency of feedstock into biopolymer precursors [206].
Given the complexity of microbial fermentation, maintaining optimal conditions is crucial for maximizing the yield of polymer precursors and minimizing the formation of by-products. Advances in bioprocess monitoring and control technologies are helping to improve the precision of fermentation processes. For instance, real-time sensors and monitoring systems can track variables such as temperature, pH, oxygen levels, and microbial growth rates [207]. These systems are increasingly integrated with ML algorithms, which can analyze large datasets in real-time and predict optimal conditions for maximizing yield. ML can also adjust fermentation parameters dynamically based on real-time data, improving process stability and ensuring a consistent production of high-quality polymer precursors [208]. Figure 3 is a visual representation of the key factors influencing fermentation in biopolymer production.
Figure 3.
Key factors influencing fermentation in biopolymer production.
2.3. Polymerization and Extraction
After the production of polymer precursors via fermentation, the next stage in biopolymer manufacturing involves polymerization and extraction. These processes transform the monomeric precursors, such as lactic acid or hydroxyalkanoates, into long-chain polymers that exhibit the desired mechanical and thermal properties [184]. The choice of polymerization method, extraction techniques, and subsequent purification affects the final properties of the biopolymer, as well as the overall sustainability and cost-effectiveness of the manufacturing process [209,210,211].
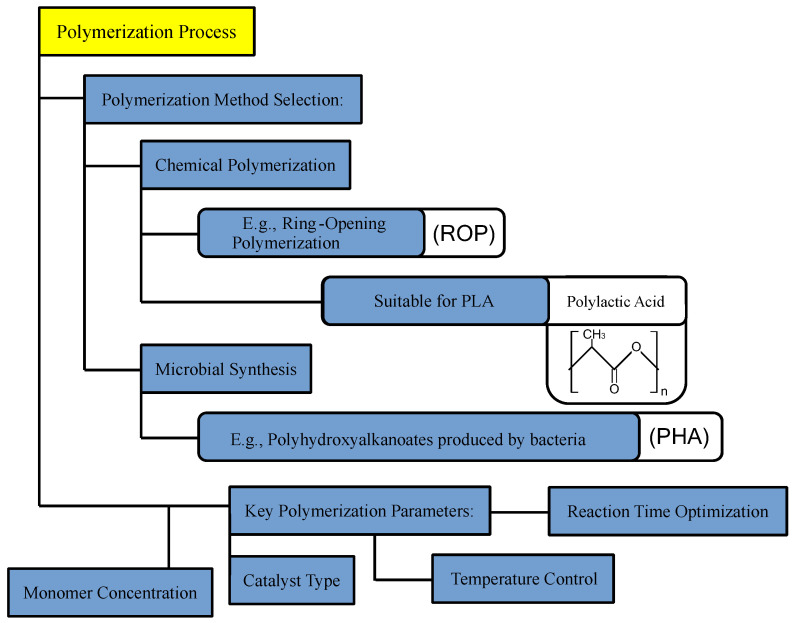
Polymerization is the chemical process by which monomer molecules are joined to form long-chain polymers [212,213,214]. In biopolymer production, this step can involve different techniques depending on the specific biopolymer being synthesized. The two primary methods for polymerizing biopolymers like polylactic acid (PLA) [215,216,217] and polyhydroxyalkanoates (PHAs) [218,219,220] are chemical polymerization and microbial synthesis, respectively.
In the production of polylactic acids (PLAs), the precursor lactic acid undergoes two main types of polymerization.
Lactic acid monomers are linked together via condensation reactions, during which water is released as a by-product [221]. This method is typically used for creating low-to-medium-molecular-weight PLAs. However, it is often less efficient for achieving high-molecular-weight polymers, which are needed for certain applications that require greater mechanical strength.
To produce higher-quality PLAs, the lactic acid is first converted into lactide, a cyclic dimer, which then undergoes ring-opening polymerization. In this process, a catalyst (often a metal-based catalyst) initiates the opening of the lactide ring, allowing it to polymerize into long PLA chains. ROP is the preferred method for producing high-molecular-weight PLAs due to its ability to create strong, durable polymers with controlled molecular architecture [222,223]. The temperature, catalyst type, and reaction time must be carefully optimized to achieve the desired polymer properties, such as molecular weight, crystallinity, and thermal stability.
Unlike PLAs, polyhydroxyalkanoates (PHAs) are synthesized directly by microorganisms through a biological polymerization process. In nutrient-limited conditions (e.g., limited nitrogen or phosphorus), certain bacterial strains such as Cupriavidus necator or Ralstonia eutropha store carbon and energy in the form of intracellular PHA granules [224]. These granules, consisting of long-chain PHA polymers, are stored inside the bacterial cells as reserve materials. The composition and properties of the PHA polymer can vary depending on the microorganism and the carbon source used in fermentation. For instance, the production of poly(3-hydroxybutyrate) (PHB), the most common PHA, is optimized by adjusting the feedstock and fermentation conditions [225]. After microbial synthesis, the PHA polymer needs to be extracted from the cells through physical or chemical processes.
Once the polymerization stage is complete, the next challenge is to extract the biopolymer from the reaction medium or, in the case of PHAs, from the microbial cells themselves [226]. Extraction techniques vary depending on the biopolymer and production process, but the goals are to isolate the polymer with high purity and ensure that its properties are not degraded during extraction [35].
For PLAs, the polymerized material needs to be separated from the reaction medium and any residual catalyst [227]. This is typically performed using filtration, precipitation, or solvent-based methods, depending on the production scale and the intended use of the PLA. The extracted PLA is then purified to remove any unreacted monomers or by-products. For commercial-scale production, processes such as melt filtration or solvent precipitation are often employed to produce high-purity PLA granules. After purification, the polymer is typically dried to remove any moisture, which can degrade the polymer during storage or further processing [228,229].
For PHAs, the extraction process is more complex because the polymer is stored inside microbial cells [219]. Several methods can be used for extracting PHAs. Organic solvents like chloroform, acetone, or methylene chloride can be used to dissolve the PHA granules from the bacterial cells [230]. This is a widely used method, but it can be energy-intensive and may involve toxic solvents, raising concerns about environmental safety and waste disposal. Another method involves the mechanical disruption of the bacterial cells, using techniques such as high-pressure homogenization, sonication, or bead milling, to physically break open the cells and release the PHA granules [35,231,232]. Once released, the PHA is recovered using filtration or centrifugation. More environmentally friendly methods involve enzymatic degradation of the non-PHA cellular components [233], or aqueous-based extraction [234], where the cell material is solubilized in water under specific conditions, leaving the PHA granules intact. These methods are still being optimized for industrial-scale applications but offer the advantage of reducing the need for toxic solvents.
Once extracted, the biopolymers undergo purification to remove impurities, unreacted monomers, residual solvents, and other by-products that could affect their performance or safety [26]. The specific purification methods depend on the type of polymer and the extraction process used. For example, PLA purification often involves solvent evaporation or crystallization techniques, while PHA purification may require additional washing and drying steps to remove residual bacterial cell fragments or solvents [235].
The purified polymers may then undergo post-processing to tailor their physical properties for specific applications [236]. This can include adjusting their molecular weight, crystallinity, or blending with other additives to enhance properties such as flexibility, impact resistance, or thermal stability. For instance, PLA might be blended with plasticizers to improve its flexibility for use in packaging, while PHAs might be blended with other biopolymers to enhance their durability for use in medical devices or agricultural films [237,238].
The extraction and purification steps in biopolymer production are often energy-intensive, involving processes like solvent evaporation, filtration, and drying, which require energy inputs [239]. For example, Kavitake et al. [240] focused on the extraction, purification, and characterization of an exopolysaccharide (EPS) from Enterococcus hirae OL616073, a strain isolated from Indian fermented food. The EPS was purified using ion exchange and size exclusion chromatography, yielding two major fractions with molecular masses of 7.7 × 104 and 6.5 × 104 Da. Structural analysis by FTIR, HPTLC, GC-MS, and NMR revealed that the EPS is a homopolysaccharide composed of glucose with -(1 → 6) and -(1 → 3) glycosidic linkages. The EPS demonstrated excellent physico-functional properties, including high water solubility, oil holding capacity, emulsifying activity, and shear-thinning rheology. These findings suggest that this EPS could be a promising functional biopolymer for applications in the food and pharmaceutical industries. If not carefully managed, these processes can offset the environmental benefits of using renewable feedstocks and biodegradable materials. The use of organic solvents in extraction, in particular, poses environmental challenges due to the potential for solvent emissions, hazardous waste, and energy consumption in solvent recovery or disposal.
To ensure that the environmental advantages of biopolymers are maintained, manufacturers are focusing on developing more energy-efficient and environmentally friendly extraction methods. For example, there is growing interest in using supercritical CO2 extraction as an alternative to organic solvents [241,242]. Supercritical CO2 is a non-toxic, non-flammable solvent that can be used at relatively low temperatures, reducing energy consumption and eliminating hazardous solvent waste [243]. The extraction of rare and precious metals from waste is becoming essential as mineral resources deplete, with supercritical CO2 extraction emerging as a promising, eco-friendly solution. Supercritical CO2 has unique properties, such as liquid-like solubility and gas-like mass transfer, allowing it to penetrate porous materials and extract metals effectively [244]. This method can be optimized by adjusting temperature and pressure to selectively dissolve and recover target metals while reducing unwanted by-products and solvent waste. Studies demonstrate high efficiency in extracting metals like copper, lead, and rare earth elements using supercritical CO2 combined with modifiers. Compared to conventional hydrometallurgical and pyrometallurgical methods, supercritical CO2 offers enhanced sustainability and purity. By improving process parameters and integrating advanced techniques, this technology has the potential to revolutionize waste recycling and metal recovery. Similarly, advances in biorefining technologies aim to integrate biopolymer production with other bio-based processes to minimize energy use and maximize resource efficiency [245]. Figure 4 is a diagram illustrating the stages and key steps involved in polymerization and extraction during biopolymer production.
Figure 4.
Stages and key steps involved in polymerization and extraction during biopolymer production.
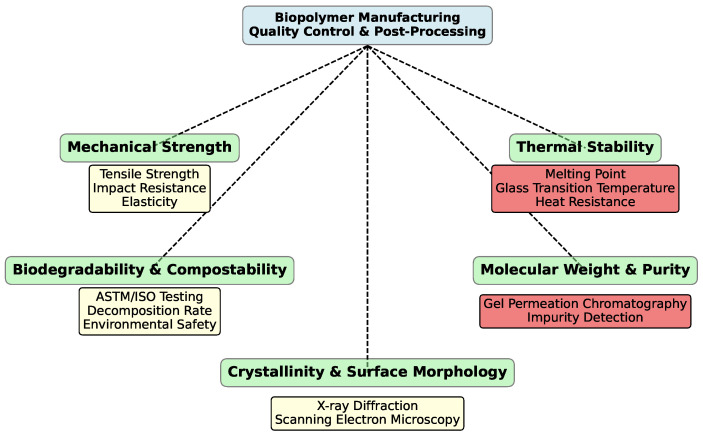
2.4. Quality Control and Post-Processing
After the polymerization stage, biopolymers undergo quality control to ensure that they meet the desired specifications for mechanical strength, thermal stability, and biodegradability [55,246]. Any inconsistencies in the polymer’s properties—such as molecular weight, crystallinity, or impurity levels—can affect the final product’s usability, durability, and biodegradability [247]. Quality control typically involves testing for mechanical properties, thermal stability, molecular weight, purity, and crystallinity. For example, mechanical strength is tested through tensile tests to ensure that the material can withstand the required stress for applications like packaging or automotive components [248,249]. Thermal stability, tested using differential scanning calorimetry (DSC) or thermogravimetric analysis (TGA), ensures that the polymer can endure temperature variations without degrading [250,251]. Molecular weight and purity are evaluated using techniques like gel permeation chromatography (GPC) or nuclear magnetic resonance (NMR), while crystallinity, which affects the polymer’s flexibility and transparency, is assessed using X-ray diffraction (XRD) or scanning electron microscopy (SEM) [252].
Biodegradability is a defining feature of biopolymers, and thorough testing ensures compliance with environmental standards such as ASTM D6400 [253,254,255] or EN 13432 [256,257]. These tests simulate industrial composting conditions to confirm that the material breaks down within a specified time frame and does not leave toxic residues. Ensuring biodegradability under the right conditions is crucial, especially for products marketed as compostable or environmentally friendly.
After passing quality control, biopolymers often undergo post-processing to enhance their properties for specific applications. Post-processing may include blending the biopolymer with additives such as plasticizers, which improve flexibility [258], or UV stabilizers [259], which protects the material from degradation when exposed to sunlight. For example, PLA is often brittle, so it is commonly blended with plasticizers like glycerol or oligomers to make it more flexible for use in packaging films [238]. PHAs, which are more thermally stable, might require impact modifiers to improve their toughness for applications like medical devices or textiles.
In addition to additives, biopolymers can be blended with other polymers, including both biopolymers and conventional plastics, to optimize their properties [209]. For example, blending a PLA with polycaprolactone (PCL) or polybutylene adipate-co-terephthalate (PBAT) improves its flexibility and biodegradability [260,261]. Surface treatments, such as plasma treatment or corona discharge, may be applied to enhance adhesion, printability, or barrier performance, making the material more suitable for packaging or medical applications. In medical contexts, surface treatments can also enhance biocompatibility, ensuring that the polymer is safe for use in implants or drug delivery systems.
Thermal and mechanical post-processing methods are also employed to further modify the polymer’s properties. Thermal treatments, such as annealing, can increase crystallinity in polymers like PLA, improving heat resistance and mechanical strength [262]. Techniques like extrusion, injection molding, and 3D printing are used to shape biopolymers into final products. During these processes, temperature and processing conditions must be carefully controlled to prevent polymer degradation. For instance, in injection molding, the temperature needs to be optimized to avoid thermal degradation, while blown film extrusion is often used for creating biodegradable packaging films [263].
Fungal pathogens are a significant threat to agricultural crops, reducing both the quantity and quality of yields. Usmanova et al. [264] developed innovative seed-coating formulations using biopolymers [209,265], such as polyhydroxyalkanoate (PHA) and pullulan, along with beneficial microorganisms for enhanced plant protection. The microbial strains used (e.g., Pseudomonas flavescens and Bacillus aerophilus) demonstrated key agricultural properties, including phytohormone production, antifungal activity, and salt tolerance. Pullulan, synthesized by Aureobasidium pullulans C7, exhibited ideal viscosity characteristics for seed coating, transitioning from Newtonian to pseudoplastic behavior at higher concentrations. Seed coatings combining microbial inoculants and polymers improved barley growth under phytopathogenic stress, enhancing germination rates, root and stem lengths, and photosynthetic pigment content. This approach highlights the potential of biopolymers and microbial strains to mitigate crop losses and promote sustainable agriculture.
Lastly, ensuring the environmental performance of biopolymers through biodegradability testing and regulatory compliance is crucial. Products must pass standardized tests, such as ISO 17088 [254,256,266] or ASTM D6400 [267,268,269], to ensure they decompose safely in industrial composting environments. These tests assess the rate of degradation, environmental safety, and the material’s ability to disintegrate under specific conditions. The goal is to confirm that biopolymers do not leave harmful residues and degrade within the expected time frames, reinforcing their role as sustainable alternatives to petroleum-based plastics.
The complexity and variability of raw materials, combined with fluctuating environmental and operational conditions, require a robust system for process monitoring and optimization. Traditional process control methods may struggle to keep up with these dynamic factors, leading to inconsistencies in product quality or inefficiencies in production. ML techniques can analyze vast amounts of data generated throughout the production process, enabling real-time monitoring and control.
For example, ML algorithms can be trained to predict the outcomes of fermentation based on the feedstock’s composition, environmental conditions, and microorganism strains, allowing for more precise control of the process [270,271]. Similarly, ML-driven predictive models can forecast fluctuations in raw material quality or identify optimal processing parameters for polymerization and extraction, ensuring consistent quality and minimizing waste [272,273]. Additionally, ML can enable the development of smart manufacturing systems that automatically adjust processing conditions in real-time to maximize efficiency and product performance, reducing the need for manual intervention.
In the realm of quality control, ML-powered systems can detect defects or inconsistencies in the biopolymer’s properties early in the production process, allowing for timely corrections before the final product is manufactured [274]. This can improve yield and reduce resource consumption, making the entire production process more sustainable. Moreover, ML can assist in optimizing the supply chain by predicting demand, optimizing inventory management, and minimizing transportation costs, further enhancing the sustainability of biopolymer manufacturing.
Biopolymer-bound soil composites (BSCs) are innovative, cement-free construction materials utilizing binders like starch, protein, and lignin. While they offer sustainable alternatives for diverse applications [275], their production presents challenges such as internal defects, improper mixing, and compaction issues. Traditional quality control methods, like vision or acoustic techniques, are often inefficient, as they focus on isolated issues and cannot monitor the unique strength gain process during desiccation. To address these gaps, the BioSys system, suggested by Miao et al. [276], employs vibration-based, non-destructive testing to evaluate BSC quality through impulse hammer-generated signals and accelerometer-recorded responses. BioSys utilizes ML models, achieving up to 99% accuracy in detecting defects, 100% accuracy in identifying improper compaction, and a 5% mean absolute percentage error (MAPE) in predicting strength gain. This system’s ability to simultaneously detect multiple defects, monitor compaction, and assess desiccation makes it a powerful tool for scaling the sustainable production of high-quality BSC materials.
Figure 5 is a diagram illustrating the stages and key steps involved in quality control during biopolymer production.
Figure 5.
Quality control during biopolymer production.
3. ML Techniques in Process Optimization
The integration of ML techniques in process optimization offers transformative potential for the manufacturing industry, particularly in the realm of biopolymer production. ML-driven tools enable a more precise control over production parameters, real-time adjustments, and better quality assurance. Below, we explore several ML methodologies that are driving innovations in this field.
ML is extensively utilized in process industries due to its ability to analyze vast datasets and make data-driven predictions. For biopolymer manufacturing, ML can be used for monitoring fermentation conditions, predicting product yields, and optimizing complex multi-step processes like extraction and purification [277]. The ability of ML models to continuously learn from production data makes them a vital tool for improving efficiency, minimizing waste, and ensuring consistent product quality [278].
3.1. Supervised Learning
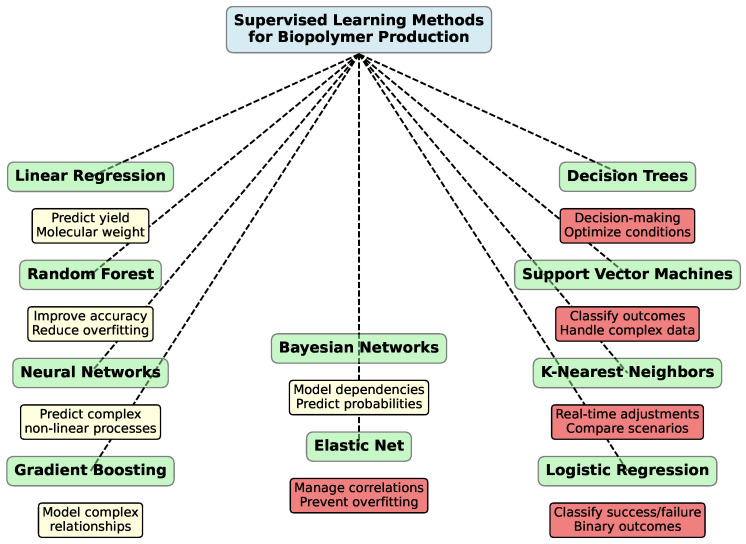
Supervised learning [279,280,281,282] is particularly useful in biopolymer production because it leverages labeled datasets to train models that can predict future outcomes. Figure 6 presents key supervised learning methods that assist in various aspects of biopolymer production, including predicting yields, molecular weight, and other characteristics of the final product.
Figure 6.
Overview of supervised learning methods applied in biopolymer production.
In biopolymer processes, this technique can forecast fermentation yields, molecular weight distribution, or viscosity of the final product based on key input parameters such as temperature, pH, feedstock composition, or microbial strain. For instance, predictive models trained on historical data can be employed to recommend adjustments to fermentation conditions in real-time, ensuring optimal performance under varying environmental conditions.
Moman et al. [283] addressed the computational prediction of ligand–biopolymer affinities, emphasizing ML’s role in modern drug discovery. Their work proposes using a nonparametric model of effective radii of atom descriptors, computable for the entire Periodic Table, which, when integrated with MLalgorithms, provides competitive predictive performance. The research involved querying the Protein Data Bank (PDB) [284,285,286] for relevant protein–ligand structures, converting affinity data into a usable format, and cleaning PDB files through automated scripts. The dataset was split into training (60%), validation (20%), and testing (20%) sets across multiple random splits to enhance robustness. ML models, specifically RandomForestClassifier [287,288] and RandomForestRegressor [289,290] from Scikit-learn, were utilized for classification and regression tasks. The final structure–activity database comprises 4703 biopolymer–ligand complexes, forming a valuable resource for predicting ligand affinities.
Biodegradable starch films are promising options for food packaging. Kathuria et al. [291] suggested using the k-Nearest Neighbor [292,293] (KNN) algorithm to classify these films based on parameters such as thickness, water vapor permeability (WVP), tensile strength (TS), and transparency. Twelve films from various botanical starch sources were produced via the casting method, resulting in a database of thirty-six samples. The 5% cassava starch formulation emerged as the best, with WVP 1.21 × 10−10 g · m−1 · s−1 · Pa−1, TS 2.34 MPa, thickness 0.193 mm, and water activity (Aw) 0.408. The KNN and principal component analysis effectively classified and selected optimal biodegradable starch films.
The automotive industry seeks cost-effective, renewable materials. Bejagam et al. [294] explored the use of wheat straw as a filler in polypropylene for automotive applications, aiming to meet mechanical property standards set by conventional fillers like glass fiber. Biocomposites [295,296,297] were created by varying the weight percentages of wheat straw and polypropylene through extrusion. The molded products underwent mechanical testing. Predictive models for the biocomposite properties were developed using Polynomial Regression, ANNs, and Support Vector Machines (SVMs). Results indicated that SVMs yielded the best predictive model, followed by ANNs and polynomial regression.
Xing et al. [298] compared an ANN [299,300] and an SVM [301,302,303] for predicting the molecular weight of polycaprolactone (PCL) synthesized via enzymatic catalysis. The study optimized synthesis parameters using a D-optimal design and employed ML techniques to predict the output molecular weight of biopolymers. The biocomposites were created by varying the weight percentages of ϵ-caprolactone and toluene, with mechanical testing performed on the molded products. Both the ANN and SVM were evaluated for prediction accuracy and the SVM was revealed to be the superior method in this context. Experimental data collection involved temperature, time, monomer/solvent ratios, and mixing speed, demonstrating the SVM’s effectiveness in handling the polymerization problem’s characteristics.
Pullulan is a biodegradable hydrogel biopolymer with applications in food, medicine, and cosmetics. Saber et al. [304] utilized the endophytic fungus Aureobasidium pullulans (accession number OP924554) for pullulan biosynthesis [305,306,307]. The fermentation process was optimized using Taguchi’s approach [308,309,310] and a decision tree [311,312,313] learning algorithm, which identified key variables affecting pullulan production. The decision tree model successfully reduced sucrose content by 33% without compromising pullulan yield. Optimal nutritional conditions were established as sucrose (60 or 40 g/L), K2HPO4 (6.0 g/L), NaCl (1.5 g/L), MgSO4 (0.3 g/L), and yeast extract (1.0 g/L) at pH 5.5, with a short incubation time of 48 h, achieving a pullulan yield of 7.23%. The structure of pullulan was confirmed through FT-IR [314,315,316] and -NMR [317,318,319] spectroscopy. This study marked the first application of Taguchi and decision tree methodologies for optimizing pullulan production, paving the way for further research on using ML to enhance fermentation processes.
Berger et al. [320] evaluated the conversion of orange peels into biodegradable polymers using a decision tree method to identify optimal production variables. The study analyzed factors such as the particle size of orange peel powder, starch types, cooling methods, and dehydration processes. The decision tree approach allowed for the efficient organization and analysis of these variables, leading to the identification of optimal conditions: a particle size of 250 μm, a 100% corn starch ratio, cooling at room temperature, and effective dehydration. The use of a decision tree model facilitated a structured exploration of the best combinations of ingredients and methods for producing high-quality bioplastics [321], demonstrating its effectiveness in optimizing the biopolymer production process.
The depletion of fossil fuels and rising plastic pollution necessitate sustainable alternatives like polyhydroxyalkanoates (PHAs). Bejagam et al. [294] employed ML to predict the melting temperature (Tm) of various PHA homo- and copolymers using a curated dataset of experimental Tm values, molecular weights, and polydispersity indices. Descriptors of polymer topology and charge/polarity were utilized to develop predictive ML models. This approach, integrated with a glass transition temperature (Tg) prediction model and an evolutionary algorithm, facilitated multiobjective optimization in polymer design.
Patnode et al. [322] developed bioplastic films using soy protein, zein, and plant oil-based monomer (POBM) latexes as sustainable alternatives to petrochemical-based food packaging. By leveraging the film-forming ability of soy protein, the strength of zein, and the plasticizing and hydrophobizing effects of POBM-latexes, strong, flexible, and moisture-resistant bioplastic films, termed proteoposites, were created. ML models with >85% accuracy were used to predict and optimize the bioplastics’ properties, confirming experimental outcomes. These proteoposite films show promise as biodegradable, high-performance packaging materials.
Biopolymer-based soil treatment (BPST) [323,324,325] is gaining traction in sustainable geotechnical engineering due to its low carbon footprint and effective ground improvement properties. Lee et al. [326] employed a decision tree ML model to predict the unconfined compressive strength (UCS) of BPST, achieving a high accuracy of R2 > 0.99. Their analysis revealed that biopolymer and water contents were critical factors influencing UCS. The model utilized data from eight published studies on BPST, focusing on various features that affect strength, including biopolymer type, soil type, and water content.
Bohar et al. [327] integrated ML and additive manufacturing to predict and optimize the mechanical strength of FDM-printed PEEK components, critical for aerospace, biomedical, and automotive industries. Key process parameters—infill density, layer height, printing speed, and infill pattern—were analyzed experimentally. Support Vector Regression (SVR) and Random Forest Regression (RFR) models achieved accurate tensile strength predictions, with deviations under 5%. Using a genetic algorithm (GA), the optimized parameters yielded a maximum tensile strength of 66.17 MPa. Microstructural analysis validated the results, demonstrating the potential of ML-driven optimization for high-performance 3D printing.
Ergun et al. [328] explored xanthan gum as a foam material for insulation and packaging, a novel application for this natural biopolymer. Foams were produced using varying ratios of xanthan gum and cellulose fiber in a 5% citric acid medium. Results showed that xanthan gum significantly influenced foam properties, with densities ranging from 49.42 to 172.2 kg/m3, compressive moduli from 235.25 to 1257.52 KPa, and flexural moduli from 1939.76 to 12,736.39 KPa. Five ML models were applied to predict material properties, with the generalized regression neural network (R2 > 0.97) achieving the best accuracy, optimizing the process while reducing experimental efforts.
Ergun et al. [329] investigated the use of guar gum-based foams for insulation applications, focusing on their properties and predicting them through regression analysis. The foams were produced by mixing guar gum, cellulose, and boric acid in varying proportions and drying the mixture. The resulting foams exhibited desirable properties such as low density, low thermal conductivity, and good mechanical strength. Regression models, including Multiple Linear Regression (MLR) and Gaussian Process Regression (GP), were used to estimate the foam’s density, compression modulus, and thermal conductivity. The GP model achieved high prediction accuracy (R2 up to 0.99), indicating that guar gum significantly influenced the foam’s properties.
Lofgren et al. [330] explored the optimization of the AquaSolv omni biorefinery for lignin using Bayesian optimization, an ML technique that enhances sample efficiency and guides data collection. The process links biorefinery conditions, such as hydrothermal pretreatment severity and temperature, with lignin’s structural features, analyzed through 2D nuclear magnetic resonance spectroscopy. By applying Pareto front analysis, the optimal processing conditions were identified to maximize lignin yield and preserve β-O-4 linkages for efficient depolymerization into platform chemicals. The research highlighted ML’s potential in advancing sustainable chemical processing for targeted applications.
Ifran et al. [331] developed an ML model using Gaussian Process Regression to predict nutrient release time from biopolymer-coated controlled-release fertilizers (CRFs). The model incorporates parameters like diffusion coefficient, coating thickness, and size distribution. With an R2 of 1 and an RMSE of 0.003, the model accurately predicted nutrient release, helping to optimize CRF performance in precision farming. It can be used to analyze and improve the release profiles of various biopolymer-coated CRFs.
Champa et al. [332] enhanced the mechanical properties of poly[(butylene succinate)-co-adipate] (PBSA) using functionalized single-walled carbon nanotubes (SWCNTs). Different SWCNT loadings were incorporated into PBSA via solution casting and optimized ultrasonication. The nanocomposites showed significant improvements in stiffness due to the superior reinforcing ability of SWCNTs. Four machine learning models—Polynomial Regression, Support Vector Machines, Gradient Boosting, and Artificial Neural Networks—were used to predict mechanical properties such as Young’s modulus, tensile strength, elongation at break, and impact strength, with R2 values ranging from 0.69 to 0.99. The study offers a promising approach to modeling and optimizing polymeric nanocomposites for various industrial applications.
For clarity, Table 1 summarizes the results of studies that utilized different supervised learning models for analyzing biopolymers. The table includes information on specific materials, applied models, obtained results, and limitations of the research.
Table 1.
Summary of supervised learning applications in biopolymer research.
| Reference | Focus | Material | Applied Model | Results | Limitations |
|---|---|---|---|---|---|
| Lee et al. (2023) [326] | Predicting ligand–biopolymer affinities | Biopolymer–ligand complexes | Random Forest Classifier, Random Forest Regressor | Achieved competitive predictive performance using 4703 complexes; dataset split into training, validation, and testing. | Limited to the structures available in the Protein Data Bank (PDB). |
| Kathuria et al. (2022) [291] | Classification of biodegradable starch films | Biodegradable starch films | k-Nearest Neighbor (KNN) | Identified optimal film formulation with WVP 1.21 × 10−10, TS 2.34 MPa, thickness 0.193 mm. | Limited dataset of 12 films. |
| Bejagam et al. (2022) [294] | Biocomposites for automotive applications | Wheat straw-filled polypropylene | Polynomial Regression, Artificial Neural Networks (ANNs), SVM | SVM provided the best predictive model for mechanical properties; significant variation in composite properties noted. | Reliance on specific material formulations may limit broader applicability. |
| Xing et al. (2002) [298] | Predicting molecular weight of polycaprolactone | Polycaprolactone (PCL) | ANN, SVM | SVM was superior for predicting molecular weight based on synthesis parameters; confirmed effectiveness in polymerization. | Focused only on PCL and its synthesis parameters. |
| Saber et al. (2023) [304] | Optimizing pullulan biosynthesis | Pullulan (from Aureobasidium pullulans) | Decision Tree Learning, Taguchi Method | Achieved a pullulan yield of 7.23% with reduced sucrose; optimal conditions identified. | Specific to one strain of fungus; broader applicability needs exploration. |
| Berger et al. (2020) [320] | Converting orange peels into biodegradable polymers | Orange peels | Decision Tree | Identified optimal production conditions for bioplastics; effective analysis of production variables. | Limited to orange peel feedstock; may not apply to other materials. |
| Bejagam et al. (2022) [294] | Predicting melting temperatures of PHAs | Polyhydroxyalkanoates (PHAs) | ML models | Developed ML models predicting melting temperature and facilitating polymer design optimization. | Limited dataset for training models; may affect accuracy. |
| Bohar et al. [327] | ML and additive manufacturing for mechanical strength prediction in FDM-printed components | PEEK (Polyether ether ketone) | Support Vector Regression (SVR), Random Forest Regression (RFR), Genetic Algorithm (GA) | Accurate tensile strength prediction (deviation < 5%), optimized parameters (66.17 MPa tensile strength). | Limited to FDM-printed PEEK components. |
| Ergun et al. [328] | Xanthan gum-based foam for insulation and packaging | Xanthan gum, cellulose fiber | Generalized Regression Neural Network (GRNN), multiple ML models | Xanthan gum impacted foam properties, R2 > 0.97 for GRNN model, optimized foam properties. | Limited to foam properties and materials studied. |
| Ergun et al. [329] | Guar gum-based foam for insulation applications | Guar gum, cellulose, boric acid | Multiple Linear Regression (MLR), Gaussian Process Regression (GP) | High prediction accuracy (R2 up to 0.99), low density, low thermal conductivity, good mechanical strength. | Focused on limited biopolymer-based foam formulations. |
| Lofgren et al. [330] | Optimization of AquaSolv biorefinery for lignin | Lignin | Bayesian Optimization, Pareto Front Analysis | Maximized lignin yield and β-O-4 linkages, optimized biorefinery conditions. | Limited to lignin depolymerization and chemical processing. |
| Ifran et al. [331] | ML model for nutrient release prediction from CRFs | Biopolymer-coated controlled-release fertilizers | Gaussian Process Regression (GPR) | R2 = 1, RMSE = 0.003, accurate nutrient release time prediction for CRFs. | Focus on CRFs, not applicable to all fertilizer types. |
| Champa et al. [332] | Enhancing mechanical properties of PBSA with SWCNTs | PBSA, single-walled carbon nanotubes (SWCNTs) | Polynomial Regression (PR), Support Vector Machines (SVMs), Gradient Boosting (GB), Artificial Neural Network (ANN) | Significant improvement in stiffness, R2 values ranging from 0.69 to 0.99 for various mechanical properties. | Variability in model performance based on predicted property. |
3.2. Unsupervised Learning
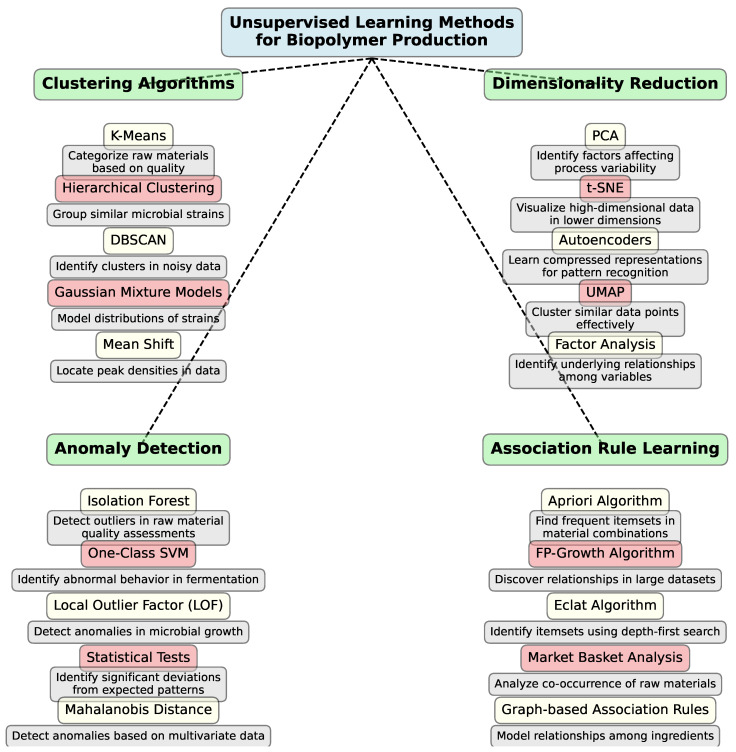
Unsupervised learning is beneficial for exploring and identifying hidden patterns in process data where no predefined labels exist. For example, clustering algorithms can categorize batches of raw materials based on their composition, quality, or suitability for biopolymer production. These techniques are also useful in analyzing microbial behavior during fermentation, where different strains might exhibit unique growth profiles, by grouping them based on similar characteristics or fermentation outcomes. Dimensionality reduction methods like Principal Component Analysis (PCA) can also uncover significant factors contributing to process variability, facilitating better control strategies. Figure 7 illustrates possible ways to apply unsupervised methods in biopolymer research.
Figure 7.
Unsupervised learning methods in biopolymer research with possible applications.
Lignin, the second most abundant biological polymer, has a complex structure and is primarily produced as a waste product in the pulp and paper industry, often underutilized. Understanding its structure is crucial for exploring potential applications. High-resolution nuclear magnetic resonance (NMR) spectroscopy is commonly used for dissolved lignin, but it cannot analyze insoluble technical lignins. Solid-state NMR spectroscopy offers a solution. Grishanovich et al. [333] introduced a method to classify the degree of lignin alteration using Hierarchical Cluster Analysis (HCA) on solid-state NMR spectra, addressing the lack of direct correlations between NMR spectra of dissolved and solid lignins.
Ireddy et al. [334] analyzed the surfaces of polyhydroxyalkanoate (PHA) films with varying monomer compositions using atomic force microscopy (AFM) and unsupervised ML algorithms. The aim was to classify films based on global attributes such as scan size, thickness, and monomer type. Their research benchmarked 12 widely used clustering algorithms through a hybrid approach, demonstrating the effectiveness of applying a one-dimensional (1D) Fourier Transform [335] (FT) on high-dimensional vectorized data for classification. Results indicated that the 1D FT produces the most accurate outcomes. The study also provided insights into individual algorithm performances and the impact of different data pools, alongside an early version of a tool designed for surface investigation using ML methods.
PLA is a bioresorbable polymer used in medical devices that require careful processing to avoid degradation. Mulrennan et al. [336] integrated in-process temperature, pressure, and NIR spectroscopy measurements with multivariate regression methods to predict the mechanical strength of extruded PLA products. Their work evaluated the feasibility of this method as an intelligent sensor for real-time quality analysis in compliance with medical device regulations. Their results indicated that combining NIR and conventional sensor data is essential for robust predictions across varying processing conditions. While partial least squares [337] (PLS) performed well, Random Forest (RF) and Support Vector Regression [338] (SVR) demonstrated superior reliability with a prior principal component dimension reduction step, suggesting that nonlinear methods may outperform traditional linear methods in predicting mechanical properties from complex sensor data.
DNA-binding proteins are crucial for genetic information processing but are often inefficiently identified by traditional methods. Zhang et al. [339] leveraged ML to extract and optimize four feature types: Reduced sequence and index-vectors (RS), Pseudo-amino acid components (PseAACS), Position-specific scoring matrix-Auto Cross Covariance Transform (PSSM-ACCT), and Position-specific scoring matrix-Discrete Wavelet Transform (PSSM-DWT). Using the LASSO method for dimension reduction, the optimized features were input into ensemble learning algorithms, achieving high accuracy rates of 86.98% and 88.9% in five-fold cross-validation with datasets PDB1075 and PDB594. The independent experiment showed an accuracy of 83.33%, indicating that the proposed methodology effectively predicts DNA-binding proteins.
The Kohonen self-organizing map [340] (SOM) was utilized by Qiao et al. [341] to map protein molecular surfaces, representing properties like shape and molecular electrostatics through 3D surface coordinates. This approach allows for visual comparisons of molecular features among proteins with similar topological or chemical characteristics. The SOM organizes input features onto a layered NN, creating globally ordered maps while preserving topological relationships and reducing dimensionality. The competitive learning process adjusts weights in the SOM, ensuring that neurons close in the network activate each other based on similar input, leading to global organization. This innovative method addresses the challenges of representing complex interrelationships in computational chemistry and biochemistry.
The consensus scaffolded mixture (CSM) position weight matrix model enhances the modeling of cis-regulatory elements by using overlapping components represented by multiple-position weight matrices (PWMs) linked to specific binding patterns. Jiang et al. [342] introduced a learning algorithm consisting of an initial structure learning phase based on frequent pattern mining, followed by refinement using the expectation maximization (EM) algorithm. In a case study of the transcription factor Leu3, CSM models aligned with conventional mixtures but demonstrated superior fitness via the Fermi-Dirac distribution. An analysis of predicted binding sites for 83 JASPAR transcription factors indicated that the CSM outperformed simple mixtures, context-specific independent (CSI) mixtures, and single PWM models in 83%, 84%, and 75% of the cases, respectively. A five-fold cross-validation across 46 TRANSFAC datasets confirmed the CSM model’s greater generality compared to other mixture models.
Motif discovery [343] in biological sequences is essential for understanding gene expression and regulation. Hasan et al. [344] reviewed the application of data mining techniques for motif discovery, noting a recent surge in interest despite limited prior usage compared to other algorithms. Various methodologies, including GYM, a program based on the a priori method [345], successfully identified helix-turn-helix motifs, improving detection rates without increasing false predictions. Challenges included the choice of training sets and minimum support thresholds. The modified prefix span method improved frequent pattern extraction by considering gaps, while BioPM utilized a prefix-projected method for efficient motif mining. Pushdown automata were employed for grammar-based motif extraction, and algorithms like informative motif mining and FP-growth enhanced performance by optimizing the search for biologically significant motifs.
Yousef et al. [346] explored how learning a suitable distance metric from labeled examples can significantly enhance k-Nearest Neighbor (kNN) classification performance. The proposed ensemble clustering kNN classifier [347] (EC-kNN) improved accuracy by defining distances based on co-clustering rather than solely geometric proximity. Through experiments involving seven plant microRNA species and eight feature selection methods, EC-kNN consistently outperformed traditional classifiers, including SVM. The EC-kNN approach also reduced data complexity by grouping points into equivalence classes, facilitating a novel data reduction technique complementary to methods like principal component analysis (PCA). The algorithm’s effectiveness was demonstrated through multiple runs and robust average results across different datasets.
Wei et al. [348] addressed the challenge of limited data in biochemistry, particularly in organic chemistry. To enhance modeling performance in the biopolymerization process, the authors proposed an ML approach that utilizes variational autoencoders and generative adversarial networks for data augmentation, mitigating overfitting. The Random Forest and ANN algorithms were employed for modeling. Results indicated that data augmentation significantly improves regression model performance, with the Random Forest model augmented by generative adversarial networks achieving the highest predictive accuracy—an R2 of 0.94 on the training set and 0.74 on the test set.
Lignin, an abundant biopolymer, presents substantial industrial potential, yet the limited molecular structure data restrict its applications. Eswaran et al. [349] introduced the Lignin Structural (LGS) Dataset, which features the molecular structures of milled wood lignin, emphasizing on two primary monomeric units (coniferyl and syringyl) and six prevalent interunit linkages. The dataset encompasses 60,000 newly generated lignin structures that accurately reflect experimental properties, achieving about 90% accuracy in matching literature data. The LGS dataset serves as a crucial resource for advancing lignin chemistry research, supporting computational simulations and predictive modeling.
Abreu et al. [350] investigated biohydrogen production from arabinose using four different anaerobic sludges across varying pH levels (4.5 to 8.0), with arabinose concentrations set at 30 g/L. The modified Gompertz equation was used to estimate production parameters, revealing that higher pH values led to greater hydrogen production across all sludges. Among the tested sludges, G2 (acclimated granular sludge) demonstrated the highest hydrogen yield and arabinose consumption. Granular sludges exhibited distinct behavior from suspended sludges, including shorter lag phases and varying fermentation pathways. A strong correlation (R2 = 0.973) between n-butyrate and ethanol percentages in G1 sludge suggested that ethanol/butyrate fermentation was predominant, while S1 showed a high correlation between n-butyrate and acetate (R2 = 0.980). The findings imply that granular sludge maintains efficiency across broader pH ranges, optimizing the hydrogen production of arabinose.
Fredricks et al. [351] highlighted the environmental concerns associated with non-degradable fossil-based plastics and advocates for biopolymers as sustainable alternatives. Biopolymers, synthesized by living organisms, offer desirable mechanical properties, compostability, and renewable sourcing. The paper discusses the hierarchical structure of three prominent biopolymer classes—cellulose, chitin, and protein beta-sheet structures—focusing on how their interaction networks contribute to mechanical strength. Various fabrication and processing techniques to develop macroscopic materials and composites from these biopolymers were reviewed. In addition, a novel approach that uses intact microorganisms or biological matter as building blocks for material construction was presented. The paper emphasizes the processing–structure–property relationships of biomatter-based materials and concludes with a perspective on the potential role of biopolymers in promoting a circular economy.
To provide a clear overview of key research in the fields of biopolymers and unsupervised learning, Table 2 summarizes the main studies. It highlights the research focus, materials used, applied models, results, and identified limitations.This summary facilitates a comparison between different approaches and models used in biopolymer analysis and related areas.
Table 2.
Summary of unsupervised learning applications in biopolymer research.
| Reference | Focus | Material | Applied Model | Results | Limitations |
|---|---|---|---|---|---|
| Grishanovich et al. (2024) [333] | Classifying the degree of lignin alteration using solid-state NMR spectroscopy. | Technical lignins | Hierarchical Cluster Analysis (HCA) on solid-state NMR spectra | Method effectively classifies lignin alterations, addressing gaps in correlating dissolved and solid lignins. | Limited to the accuracy of solid-state NMR and its analysis. |
| Ireddy et al. (2024) [334] | Analyzing PHA film surfaces using AFM and ML algorithms for classification. | Polyhydroxyalkanoate (PHA) films | Unsupervised ML algorithms; benchmarking 12 clustering algorithms; 1D Fourier Transform (FT) | The 1D FT yielded the highest accuracy for film classification. Insights provided on algorithm performance and data impact, along with a preliminary ML tool for surface investigation. | Focused on specific attributes; performance may vary with different film compositions. |
| Mulrennan et al. (2022) [336] | Predicting mechanical strength of PLA using real-time sensor data. | Polylactide (PLA) | Multivariate regression methods, including partial least squares (PLS), Random Forest (RF), SVR | Combining NIR and conventional sensor data enhanced predictions; RF and SVR showed superior reliability. Nonlinear methods outperformed linear methods. | Method complexity and need for real-time monitoring may limit applicability in practice. |
| Zhang et al. (2021) [339] | Identifying DNA-binding proteins using optimized features and ensemble learning. | DNA-binding proteins | ML algorithms, LASSO for feature selection, ensemble learning methods | Achieved high accuracy (86.98% and 88.9%) in five-fold cross-validation; effective prediction methodology. | Requires extensive dataset for robust validation; may not generalize to all protein types. |
| Qiao et al. (2001) [341] | Mapping protein molecular surfaces using SOM for visualization. | Protein molecular surfaces | Kohonen self-organizing map (SOM) | Provides a novel method for the visual comparison of molecular features, effectively addressing complex interrelationships in proteins. | SOM’s effectiveness may vary based on input feature quality. |
| Jiang et al. (2013) [342] | Improving cis-regulatory element modeling using the CSM model with PWMs. | Transcription factors | Consensus scaffolded mixture (CSM) position weight matrix model with EM algorithm | CSM model showed superior performance compared to other mixture models in 83% of cases, enhancing binding site prediction for transcription factors. | Limited to specific datasets; generalizability to other transcription factors may vary. |
| Hasan et al. (2014) [344] | Review of motif discovery methods in biological sequences. | Biological sequences | Various data mining techniques, including GYM, modified prefix span method, and grammar-based motif extraction | Identified methodologies improved motif detection rates while addressing training set and support threshold challenges. | Limited exploration of all possible algorithms; focus on recent developments. |
| Yousef et al. (2016) [346] | Enhancing kNN classification with a new distance metric learning approach. | Plant microRNA species | Ensemble clustering kNN classifier (EC-kNN) | EC-kNN consistently outperformed traditional classifiers, reducing data complexity and improving accuracy through co-clustering distance definitions. | Relies on labeled examples, limiting application to well-characterized datasets. |
| Wei et al. (2022) [348] | Addressing data limitations in biopolymerization modeling using ML. | Biopolymers | Variational autoencoders, generative adversarial networks (GANs), Random Forest (RF), ANN | Data augmentation improved regression model performance significantly, with RF achieving an R2 of 0.94 on the training set and 0.74 on the test set. | Dependence on quality of augmented data; may not fully replicate real-world variability. |
| Eswaran et al. (2022) [349] | Developing a structural dataset for lignin to facilitate computational research. | Milled wood lignin | Dataset creation and analysis | LGS dataset includes 60,000 structures with 90% accuracy in reflecting experimental properties, serving as a crucial resource for lignin chemistry research. | Limited by existing experimental data and the accuracy of generated structures. |
| Abreu et al. (2009) [350] | Investigating biohydrogen production from arabinose using anaerobic sludges. | Anaerobic sludges | Modified Gompertz equation for estimating hydrogen production parameters | Higher pH levels correlated with increased hydrogen production; G2 sludge showed the highest yields and efficiency. Strong correlations observed in fermentation pathways. | Specific to arabinose and pH conditions; results may not generalize to other substrates. |
| Fredricks et al. (2023) [351] | Analyzing biopolymers as sustainable alternatives to fossil-based plastics. | Cellulose, chitin, protein beta-sheet structures | Structural analysis and processing methods for biopolymers | Discusses mechanical properties, processing techniques, and the potential of biopolymers in promoting a circular economy. | Emphasis on selected biopolymer classes; further research needed for broader applicability. |
3.3. Neural Networks and Deep Learning
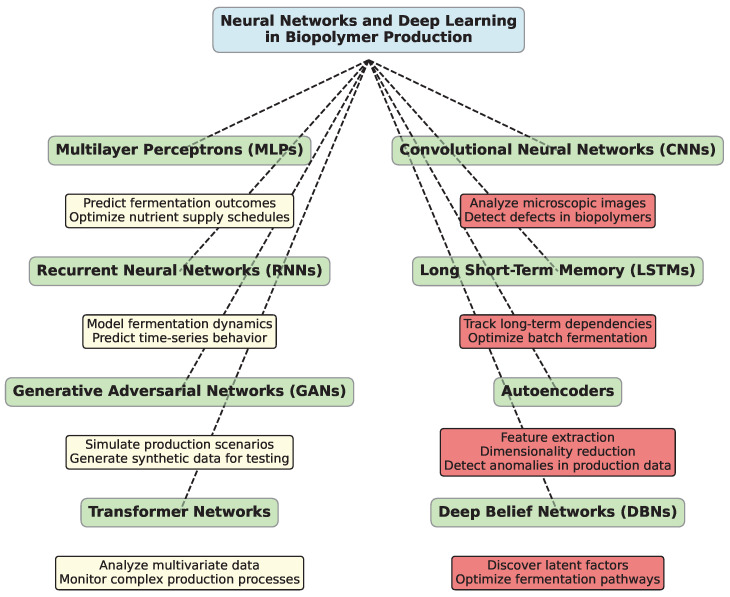
NNs, and more specifically deep learning architectures, handle highly complex, nonlinear relationships between variables within biopolymer production processes. These models excel in situations where traditional statistical models may fall short due to the sheer complexity of interactions, such as those observed in fermentation and polymer extraction. Deep learning can account for numerous variables and their interdependencies, enabling better control and optimization across different stages of the production pipeline. Figure 8 illustrates diagram with key NN architectures used in biopolymer production and their specific applications.
Figure 8.
Application of various NNs and deep learning architectures in biopolymer production.
Multilayer perceptrons [352,353,354,355] (MLPs) are a type of NN capable of approximating functions and making accurate predictions based on multiple input variables. In biopolymer manufacturing, MLPs can be used to predict fermentation outcomes based on real-time sensor data, such as pH, dissolved oxygen, and nutrient concentrations. These models can also optimize nutrient supply schedules to maximize microbial activity, thereby enhancing product yield and quality. Additionally, MLPs may assist in adaptive control systems that automatically adjust fermentation parameters during the process, leading to increased production efficiency.
Convolutional Neural Networks [356,357,358,359] (CNNs), though traditionally associated with image recognition, are finding innovative applications in biopolymer manufacturing, particularly in quality control. By analyzing microscopic images of biopolymers, CNNs can detect structural inconsistencies, contamination, or defects that may not be visible through conventional inspection methods. Furthermore, CNN-based systems could be employed in automated defect detection during the final stages of product refinement, ensuring only high-quality biopolymers reach the end users.
Recurrent Neural Networks [360,361,362,363] (RNNs) are specifically designed to handle sequential data, making them highly valuable for time-series prediction. In biopolymer production, RNNs can be employed to model fermentation dynamics by analyzing historical data from previous batches and forecasting future states. This capability enables real-time adjustments in fermentation parameters, reducing the likelihood of deviations and improving consistency in product quality.
Long short-term memory [364,365,366,367] (LSTM) networks are a specialized type of RNN designed to overcome the limitations of short-term memory in traditional RNNs. In biopolymer production, LSTMs can be applied to track long-term dependencies in complex processes, such as the progression of fermentation over extended periods. LSTMs are particularly useful in monitoring and predicting batch fermentation outcomes, optimizing nutrient input, and ensuring the stability of production processes over time.
Generative adversarial networks [368,369,370,371] (GANs) consist of two NNs (a generator and a discriminator) that work in opposition to improve the performance of both. In biopolymer production, GANs can be used to simulate the effects of different production parameters on product yield, aiding in process optimization [372,373]. Additionally, GANs can generate synthetic datasets that replicate the conditions of rare or expensive experiments, helping manufacturers explore different scenarios without conducting costly physical tests.
Autoencoders [374,375,376,377] are unsupervised learning architectures used for feature extraction and dimensionality reduction. In biopolymer manufacturing, autoencoders can be applied to compress large sets of sensor data collected during fermentation and extraction processes. This allows for a more efficient analysis of underlying patterns, leading to better control of key production variables. Autoencoders are also useful for anomaly detection, identifying irregularities in the data that could indicate process faults or contamination.
Transformer networks [378,379,380,381], originally developed for natural language processing, are gaining traction in industries requiring the analysis of long-range dependencies. In biopolymer production, transformers could be used to analyze multivariate time-series data, such as environmental conditions or equipment sensor data, and predict future states of the fermentation process. Their ability to handle large datasets and model complex relationships makes them highly applicable in optimizing batch production cycles.
Finally, Deep Belief Networks [382,383,384,385] (DBNs) are generative NNs that stack multiple layers of Restricted Boltzmann Machines (RBMs). They can learn to represent data hierarchically, making them useful for modeling complex relationships between variables in biopolymer production. DBNs can be applied to tasks such as optimizing fermentation pathways by discovering latent factors influencing microbial growth, leading to more efficient and controlled production.
Biological systems inspire materials science through their complex multiscale architectures. Combining ML with multiscale modeling provides insights into the structure–property–function relationships of biomaterials. Arevalo et al. [386] reviewed ML techniques—such as NNs and autoencoders—that are applied to predict and design biological materials, advocating for the integration of ML with physics-based models for high-throughput materials discovery.
Khare et al. [387] applied transformer models to predict the thermal stability of collagen triple helices based on amino acid sequences. They compared a small transformer model and a large pretrained ProtBERT model. Despite ProtBERT’s higher complexity, the small model achieved a nearly similar accuracy while using significantly fewer parameters. Both models showed good performance against experimental data, marking the first use of transformers for predicting biophysical properties from small data sets.
Bandyopadhyay et al. [388] presented a method to explore the conformational landscapes of mini-proteins and peptides using autoencoders. By projecting molecular dynamics simulations into a latent space, the method identifies key metastable states and predicts the folding behavior of complex proteins. The approach outperformed traditional dimensionality reduction techniques, offering a more optimized view of protein dynamics and folding pathways.
A generative model based on variational autoencoders was developed by Sadeghi et al. [389] to design DNA-stabilized silver nanoclusters (AgN-DNAs) with optimized fluorescence properties. This model allows for multiobjective property design, including the ability to generate AgN-DNAs with enhanced near-infrared emission for bioimaging. It also provides automatic feature extraction and reverse mapping from desired properties to DNA sequences, improving upon traditional models that require manual feature engineering.
Satteri et al.’s [390] review covers recent advancements in data-driven approaches for the inverse design of polymers with specific properties. It highlights three key strategies, all of which leverage materials data to explore chemical space efficiently: high-throughput virtual screening, global optimization, and generative models. The article discusses the challenges and opportunities in using these data-driven techniques to optimize polymer design.
ML techniques were applied by Baldizon et al. [391] to improve the classification of linear and circular DNA molecules in noisy data from solid-state nanopore experiments. Three methods—k-means clustering, principal component analysis with k-means, and long short-term memory (LSTM) models—were tested, with the LSTM model achieving the highest accuracy (80%), demonstrating its potential for better handling of noisy nanopore data.
Noor et al. [392] applied NNs, enhanced by bootstrap resampling, to predict the molecular weight of biopolymers produced in a batch reactor. The biopolymerization process, catalyzed by *Candida antarctica* lipase B, involved ε-caprolactone and toluene. NNs with a single hidden layer and trained with Levenberg–Marquardt optimization were used to model the process, using reaction temperature, time, and molecular weight as inputs. The model achieved accurate one-step-ahead predictions of biopolymer molecular weight, demonstrating its potential for controlling biopolymer quality.
Leal et al. [393] detailed the creation of a hydroxypropyl cellulose (HPC)-based sensor for estimating force. By mixing HPC with deionized water at varying concentrations, the sensor’s RGB color responses were analyzed, showing a correlation between HPC concentration and sensor sensitivity. A 63% HPC concentration yielded the highest sensitivity for red and green components, while a 57% concentration showed uniformity in sensitivity when force was applied at different positions. The sensor demonstrated sub-centimeter spatial resolution for force distribution assessment, and the integration of a CNN improved accuracy, achieving a mean squared error of 0.037.
Salma et al. [394] focused on predicting the drug release and skin permeation of Piroxicam (PX) topical films made from chitosan (CTS), xanthan gum (XG), and their carboxymethyl derivatives (CMXs). Using the solvent casting method with Tween 80 as a permeation enhancer, the films showed good physicochemical properties. Deep learning and ML models were employed to predict drug release and permeation rates. The optimal formulation (F8 based on CTS-CMX3) achieved a 99.97% drug release. The Deep Neural Network (DNN) emerged as the best predictive approach, demonstrating high accuracy with mean squared error values of 0.00098 for drug release and 0.00182 for permeation kinetics.
Araujo et al. [395] utilized thermogravimetric analysis to investigate chitosan’s thermal degradation under dynamic conditions, employing a multilayer perceptron (MLP) NN to quantify contributions from various kinetic models. The MLP architecture successfully approximated experimental data, showing the lowest residual error and determining activation energies ranging from 98.1 to 183.3 kJ/mol. The analysis revealed a relationship between activation energy increases and polymer dehydration, highlighting the MLP’s ability to capture complex thermal behavior during chitosan decomposition.
Wong et al. [396] discussed the biopolymerization of ε-caprolactone using the Novozyme 435 catalyst, varying reactor temperatures and impeller speeds. A multilayer feedforward neural network (FFNN) model was developed, comparing the performance of 11 training algorithms. Results indicated that the quasi-Newton and Levenberg–Marquardt algorithms outperformed others, achieving mean absolute percentage error (MAPE) values of 4.512%, 5.31%, and 3.21% for various molecular weight measures in the polycaprolactone biopolymerization process. This research identified effective training methods for estimating biopolymerization performance.
Laycock et al. [397] discussed the transition from traditional experimental methods to advanced computational approaches in the design and manufacture of biodegradable and bioderived polymeric materials. The Materials 4.0 framework integrates multiscale simulations, computational modeling, and artificial intelligence to model biopolymer structures, predict properties, and understand flow and processability. This holistic approach complements experimental techniques, facilitating the study of various biopolymeric materials, including biodegradable polyesters and polysaccharides. Furthermore, ML techniques were applied to optimize material properties and predict the effects of modifications and external factors. The article emphasizes the growing repository of computational modeling data that enhance design flexibility and processing options before costly experimental testing.
Kartal et al. [398] focused on the thermal degradation of biopolymeric structures in biomass—specifically hemicellulose, cellulose, and lignin. Given the complex structure of biomass, characterizing thermal degradation typically requires extensive experimental resources. The authors developed an ANN model to generate differential thermogravimetric analysis (DTG) curves for these biopolymers using proximate analysis results. Implemented with TensorFlow, the ANN model demonstrated excellent performance with R2 values exceeding 0.998, allowing for the estimation of thermal degradation at any temperature. This model enables immediate calculations of biopolymer fractions in degraded biomass, representing a novel advancement in the field.
Review [152] highlights the integration of ML with algae-derived biopolymers for enhancing 3D printing processes. It addresses the need for sustainable manufacturing solutions and discusses algae-based biopolymers like alginate and carrageenan, emphasizing their environmental advantages and technical challenges. The paper outlines how ML can optimize material selection, predictive modeling, and quality control, resulting in improved mechanical properties and printing parameter optimization [399,400,401]. Applications, such as Spirulina-based materials and carrageenan in bone tissue engineering, are highlighted. The article concludes that despite challenges, combining ML with algae-derived biopolymers has the potential to revolutionize sustainable additive manufacturing, with significant advancements in eco-friendly production techniques.
Asgharzadeh et al. [402] presented a deep learning method for segmenting biopolymer networks observed through confocal laser scanning microscopy (CLSM). The authors utilized an encoder–decoder network architecture, achieving a dice score of 0.88 for segmenting filamentous temperature-sensitive Z proteins from the chloroplasts of Physcomitrella patens. The segmentation process involved creating ground truth images through a semi-automated method, using adaptive local thresholding followed by expert modification. To enhance the dataset, 3D images were transformed into 2D slices, resulting in a training dataset of 15,015 images. The model was trained using a 5-fold cross-validation scheme, and performance was evaluated using the Intersection-over-Union (IoU) metric. The network, implemented in Keras and trained on an Nvidia GTX 1070 GPU, successfully produced segmented 3D images from the original CLSM data.
Leng et al. [403] discussed the development of an artificial fully connected neural network (FCNN) for modeling the behavior of representative volume elements (RVEs) in biopolymer gels, such as fibrin and collagen, which are important in tissue engineering. The FCNN was trained on data from 1100 fiber networks under biaxial deformations to predict strain energy derivatives. By incorporating constraints like the convexity of the strain energy function and symmetry of the Hessian, the FCNN was successfully integrated into the finite element software Abaqus as a user material subroutine (UMAT). The model outputs derivatives of strain energy in relation to deformation invariants, enhancing the simulation of biopolymer gels in nonlinear elasticity problems. The authors emphasized the potential for combining ML with computational mechanics to improve the modeling of biological materials with multiscale structures.
The growing environmental concerns over plastic pollution have heightened interest in producing biodegradable starch-based films. Nobrega et al. [404] emphasized the need for a comprehensive understanding of how various additives affect the properties of these films. Self-organizing maps (SOMs) were employed to analyze the mechanical and barrier properties of the films, highlighting the critical role of glycerol in films with low amounts of poly(butylene adipate-co-terephthalate) (PBAT) and its dependence on equilibrium relative humidity for water vapor permeability (WVP). The research utilized a multilayer perceptron model combined with a genetic algorithm to predict and optimize the properties of biodegradable films, achieving a high correlation between experimental and theoretical results with a maximum error of 24%. The authors suggested that further data are needed to enhance the model’s accuracy and ensure component compatibility.
Table 3 summarizes various studies that focus on the use of ML in the design and optimization of biopolymers. For each study, the key focus, materials investigated, applied models, results obtained, and limitations of the approaches are highlighted. This table illustrates the broad range of ML applications in materials science, emphasizing both the advancements made and the challenges that remain in this field.
Table 3.
Summary of deep learning applications in the design of biopolymers.
| Reference | Focus | Material | Dataset | Applied Model | Results | Limitations |
|---|---|---|---|---|---|---|
| Khare et al. [387] | Predicting thermal stability | Collagen triple helices | Amino acid sequences with experimental thermal stability data | Transformer models | Small transformer model achieved similar accuracy to larger ProtBERT while using fewer parameters; good performance against experimental data. | Limited to small datasets. |
| Bandyopadhyay et al. [388] | Exploring conformational landscapes | Mini-proteins and peptides | Molecular dynamics simulation data | Autoencoders | Method outperforms traditional techniques, providing optimized views of protein dynamics and folding pathways. | None specified. |
| Sadeghi et al. [389] | Designing DNA-stabilized silver nanoclusters | Silver nanoclusters (AgN-DNAs) | DNA sequences with fluorescence properties | Variational autoencoders | Enables multiobjective design for enhanced fluorescence properties and automatic feature extraction; improves on traditional manual engineering methods. | None specified. |
| Satteri et al. [390] | Inverse design of polymers | Polymers | Materials data with polymer properties | Data-driven approaches | Highlights strategies like high-throughput virtual screening and generative models; discusses optimization challenges. | Challenges in data-driven optimization discussed. |
| Baldizon et al. [391] | Classifying DNA molecules | Linear and circular DNA | Noisy data from solid-state nanopore experiments | LSTM models, PCA | LSTM achieved highest accuracy (80%) for noisy data classification from solid-state nanopore experiments. | Limited to noisy data context. |
| Noor et al. [392] | Predicting molecular weight of biopolymers | ε-caprolactone biopolymers | Reaction temperature, time, and molecular weight data | NNs | Accurate predictions of biopolymer molecular weight; demonstrated potential for controlling quality in biopolymerization processes. | Focused on a specific biopolymer process. |
| Leal et al. [393] | Force sensor development | Hydroxypropyl cellulose (HPC) | RGB color responses of HPC sensors under varying force and concentration | CNN | Achieved a mean squared error of 0.037; highest sensitivity noted at specific HPC concentrations. | None specified. |
| Salma et al. [394] | Drug release and skin permeation | Piroxicam films from chitosan and xanthan gum | Drug release and permeation data for various formulations | Deep learning, ML | DNN achieved high accuracy; optimal formulation reached 99.97% drug release. | None specified. |
| Araujo et al. [395] | Thermal degradation of chitosan | Chitosan | Thermogravimetric analysis data | Multilayer perceptron (MLP) | MLP effectively quantified contributions from various kinetic models; lowest residual error recorded. | None specified. |
| Wong et al. [396] | Biopolymerization performance | ε-caprolactone | Biopolymerization data with molecular weight measures | Multilayer feedforward NN | Identified effective training algorithms; MAPE values for various molecular weights. | None specified. |
| Laycock et al. [397] | Computational methods in biopolymer design | Biodegradable and bioderived polymers | Computational modeling data for polymeric materials | Multiscale simulations, AI | Integrated framework enhances design flexibility and predicts effects of modifications before testing. | None specified. |
| Kartal et al. [398] | Thermal degradation of biomass biopolymers | Hemicellulose, cellulose, lignin | Proximate analysis data and thermal degradation behavior | ANN | Excellent performance with R2 values over 0.998; allows for immediate calculations of biopolymer fractions in degraded biomass. | Complexity in biomass characterization remains. |
| Bin et al. [152] | ML in algae-derived biopolymers | Algae-based biopolymers | Material properties and 3D printing process parameters | ML | Highlights potential for sustainable manufacturing and improved mechanical properties; discusses applications in 3D printing. | Technical challenges in material properties optimization. |
| Asgharzadeh et al. [402] | Deep learning for confocal microscopy | Biopolymer networks | 3D confocal microscopy images of biopolymer networks (transformed to 2D slices) | Encoder-decoder network | Achieved a dice score of 0.88 in segmentation tasks; extensive training dataset created from 3D to 2D transformations. | None specified. |
| Leng et al. [403] | Modeling biopolymer gel behavior | Fibrin, collagen | Strain energy data from fiber networks under biaxial deformation | FCNN | Successfully predicts strain energy derivatives; integrated into finite element software for nonlinear elasticity problems. | None specified. |
| Nobrega et al. [404] | Biodegradable starch-based films | Starch-based films | Mechanical and barrier property data with additive effects | Self-organizing maps (SOMs), MLP | Achieved high correlation (max error 24%) in predicting mechanical and barrier properties; emphasizes role of glycerol. | Further data needed to improve model accuracy and compatibility. |
4. Challenges of Integrating ML in Biopolymer Research
Integrating ML into biopolymer research presents several challenges that limit its broader application and effectiveness. One of the primary issues is the limited availability of experimental data. In fields like biochemistry and organic chemistry, data collected from experiments often come in small quantities, making it difficult to train and validate robust models. For instance, in the biopolymerization process, small datasets can lead to overfitting, reducing the ability of models to make accurate predictions on new data. This data scarcity was exacerbated during the COVID-19 pandemic, which further restricted the ability to conduct experimental research.
Additionally, the molecular structures of biopolymers are complex and diverse, posing another major hurdle for accurate modeling. Structures such as lignin or polylactides contain various intermolecular interactions and bonds, making it challenging to mathematically represent these materials using conventional methods. Traditional ML models, like linear regression, often struggle to capture the nonlinear dependencies that characterize these systems. Therefore, more sophisticated modeling techniques and their integration with physical and chemical simulation methods are required.
Another key issue is the lack of high-quality labeled data, especially in biochemical processes. Automating the annotation of datasets is a significant effort, and without reliable labels, ML models cannot achieve high accuracy. Below are key comments based on reviewed papers:
Using variational autoencoders (VAEs) and generative adversarial networks (GANs) to synthesize new data from small experimental datasets can enhance model quality and mitigate the risk of overfitting. This approach has already been proven effective in certain biopolymer studies.
Applying nonlinear methods such as Random Forests, SVM, and NNs can significantly improve the prediction of biopolymer properties. These algorithms are particularly useful for handling data with complex molecular interactions.
ML in biopolymer research can benefit from closer integration with traditional computational chemistry methods, like molecular dynamics or quantum chemistry simulations. Combining knowledge from fundamental laws with ML capabilities will enable more accurate predictions.
Active learning algorithms can efficiently use small datasets by selecting the most informative experiments to prioritize data collection. This strategy can reduce the experimental workload required to train models.
As demonstrated by the Lignin Structural Dataset (LGS), the creation and publication of unique datasets for different biopolymers is importnant for advancing the field. These databases will support improved simulations, predictive models, and facilitate resource sharing among researchers.
5. Future Directions for Development
To successfully integrate ML into biopolymer research, it is essential to address current challenges and explore future development opportunities. In the future, improving the integration of ML into biopolymer research will aid in the development of new materials with targeted properties and optimize their production for sustainable use. These advancements will help overcome current limitations and open new avenues for innovation in biopolymer science.
Uncertainty quantification (UQ) helps account for variability in input data, measurement errors, and process instability [405]. It allows for assessing the accuracy of models and predictions, providing insights into the confidence of results. In biopolymers, UQ can be used to consider factors like composition, production conditions, and environmental influences on material properties. It can improve the prediction of material characteristics, optimize production processes, and increase the reliability of final products.
Explainable AI (XAI) [406] aims to make machine learning models more interpretable by providing insights into how decisions are made. It helps bridge the gap between model complexity and human understanding, making it easier to trust and validate model outputs. In biopolymer manufacturing, XAI can be applied to understand the relationship between raw materials, processing parameters, and the resulting properties of the biopolymer. This transparency can guide decision-making and improve the optimization of production processes.
Below are several key areas where ML can make a significant impact:
Grishanovich et al. (2024) [333] used Hierarchical Cluster Analysis (HCA) to classify lignin alterations using solid-state NMR spectra, addressing the gap between dissolved and solid lignins. Ireddy et al. (2024) [334] demonstrated that 1D Fourier Transform (FT) achieved high accuracy in classifying polyhydroxyalkanoate (PHA) films using unsupervised machine learning algorithms. Both approaches highlight the effectiveness of unsupervised techniques in classifying complex biopolymers. However, the accuracy of these models is constrained by the limitations of the underlying technologies (solid-state NMR and FT) and may vary with material composition. Future research should focus on integrating more advanced spectroscopic techniques or hybrid models to overcome these limitations and improve generalization across different biopolymer types.
Mulrennan et al. (2022) [336] combined near-infrared (NIR) and conventional sensor data with Random Forest (RF) and Support Vector Regression (SVR) models to predict the mechanical strength of polylactide (PLA). Similarly, Bejagam et al. (2022) [294] demonstrated that Support Vector Machines (SVMs) excelled in predicting the mechanical properties of wheat straw-filled polypropylene composites. Both studies show the superiority of nonlinear models like RF and SVM over traditional linear methods for material property prediction. However, the complexity of these models and the need for real-time data or specific formulations may limit practical applications. Future work could focus on simplifying these models for broader use and exploring their adaptability to different biopolymer formulations.
Zhang et al. (2021) [339] used ensemble learning for high-accuracy DNA-binding protein prediction, relying on feature selection with LASSO. Xing et al. (2002) [298] employed an SVM to predict the molecular weight of polycaprolactone (PCL), showing that SVM outperformed Artificial Neural Networks (ANNs) in this context. While these methods provide high accuracy, they require large, high-quality datasets for training and may not generalize well across different protein types or polymers. Future research could explore methods for data augmentation or transfer learning to expand these models’ applicability and robustness.
Qiao et al. (2001) [341] employed self-organizing maps (SOMs) for visualizing protein molecular surfaces, while Bandyopadhyay et al. (2021) [388] used autoencoders to predict protein dynamics and folding pathways. Both approaches highlight the importance of unsupervised learning in understanding complex biomolecular features. However, the effectiveness of an SOM depends on input feature quality, while autoencoders may struggle with very complex datasets. Future studies could integrate SOMs with deep learning-based feature extraction or enhance autoencoders by incorporating reinforcement learning to better model protein conformational landscapes.
Sadeghi et al. (2024) [389] used variational autoencoders (VAEs) for multiobjective optimization in the design of DNA-stabilized silver nanoclusters. Satteri et al. (2021) [390] emphasized the potential of data-driven models, such as generative models, for polymer design. Both studies demonstrate the power of data-driven techniques in optimizing material properties, but challenges remain in data quality and model generalizability. Future research should focus on improving model robustness and combining these techniques with traditional methods to achieve more accurate and versatile material design processes.
Kartal et al. (2023) [398] employed Artificial Neural Networks (ANNs) to predict the thermal degradation of biomass biopolymers with high accuracy, while Khare et al. (2022) [387] demonstrated the potential of small transformer models to predict the thermal stability of collagen triple helices. Both studies underline the importance of accurate prediction of biopolymer degradation, though the complexity of biomass and the limitations of smaller datasets in transformers may pose challenges. Future studies could integrate more advanced models, such as hybrid machine learning techniques, and explore the use of multi-modal datasets to improve prediction accuracy for biopolymer stability and degradation.
Ifran et al. (2020) [331] used Gaussian Process Regression (GPR) for accurate prediction of nutrient release in biopolymer-coated controlled-release fertilizers (CRFs), while Kathuria et al. (2022) [291] applied k-Nearest Neighbor (k-NN) models to optimize biodegradable starch film formulations. Both approaches show promise in predicting biopolymer properties, but their applicability may be limited by specific material conditions or dataset sizes. Future research should explore expanding these models to include a broader range of materials and applications and work towards integrating them with other predictive models for improved generalization.
Khare et al. (2022) [387] demonstrated that small transformer models can efficiently predict the thermal stability of biopolymer structures like collagen triple helices. These models provide a promising alternative to larger models such as ProtBERT, offering similar accuracy with fewer parameters. Future research could investigate the scalability of transformer models for larger, more complex datasets and explore their application to other biopolymer stability predictions.
Wei et al. (2022) [348] emphasized the importance of data augmentation in improving machine learning model performance for biopolymerization modeling. By enhancing the dataset, they were able to significantly boost prediction accuracy. Future research could focus on developing more robust data augmentation techniques and incorporating generative models, such as GANs, to handle real-world data variability and improve prediction reliability in biopolymer-related fields.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Naser A.Z., Deiab I., Darras B.M. Poly (lactic acid)(PLA) and polyhydroxyalkanoates (PHAs), green alternatives to petroleum-based plastics: A review. RSC Adv. 2021;11:17151–17196. doi: 10.1039/D1RA02390J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pilapitiya P.N.T., Ratnayake A.S. The world of plastic waste: A review. Clean. Mater. 2024;11:100220. doi: 10.1016/j.clema.2024.100220. [DOI] [Google Scholar]
- 3.Hahladakis J.N., Iacovidou E., Gerassimidou S. Environmental Materials and Waste. Elsevier; Amsterdam, The Netherlands: 2024. Plastic waste in a circular economy; pp. 99–134. [Google Scholar]
- 4.Frąckowiak P., Jędrzejczak E., Kaspryszyn F., Jesionowski T., Wysokowski M. Revolutionizing electrospinning: Sustainable solutions through deep eutectic solvents in biopolymer processing. J. Appl. Polym. Sci. 2024;141:e55864. doi: 10.1002/app.55864. [DOI] [Google Scholar]
- 5.Gholivand S., Tan T.B., Yusoff M.M., Qoms M.S., Wang Y., Liu Y., Nyam K.L., Tan C.P. Innovative microencapsulation of hemp seed oil using plant-based biopolymers: A comparative analysis of dehydration techniques on core stability, digestibility and release pattern. Food Hydrocoll. 2024;158:110683. doi: 10.1016/j.foodhyd.2024.110683. [DOI] [Google Scholar]
- 6.Joshi J.S., Langwald S.V., Ehrmann A., Sabantina L. Algae-Based Biopolymers for Batteries and Biofuel Applications in Comparison with Bacterial Biopolymers—A Review. Polymers. 2024;16:610. doi: 10.3390/polym16050610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saharan B.S., Kamal N., Badoni P., Kumar R., Saini M., Kumar D., Sharma D., Tyagi S., Ranga P., Parshad J., et al. Biopolymer and polymer precursor production by microorganisms: Applications and future prospects. J. Chem. Technol. Biotechnol. 2024;99:17–30. doi: 10.1002/jctb.7524. [DOI] [Google Scholar]
- 8.Jafarzadeh S., Nooshkam M., Zargar M., Garavand F., Ghosh S., Hadidi M., Forough M. Green synthesis of nanomaterials for smart biopolymer packaging: Challenges and outlooks. J. Nanostruct. Chem. 2024;14:113–136. doi: 10.1007/s40097-023-00527-3. [DOI] [Google Scholar]
- 9.Saberi Riseh R., Hassanisaadi M., Vatankhah M., Varma R.S., Thakur V.K. Nano/micro-structural supramolecular biopolymers: Innovative networks with the boundless potential in sustainable agriculture. Nano-Micro Lett. 2024;16:147. doi: 10.1007/s40820-024-01348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jineesh A., Parameswaranpillai J., Chavali M., Bahadur V., Prasad V., Hegde C., Bhansali P.R. Biopolymers. Elsevier; Amsterdam, The Netherlands: 2023. Automotive applications of biodegradable polymers; pp. 425–445. [Google Scholar]
- 11.Peng X., Umer M., Pervez M.N., Hasan K.F., Habib M.A., Islam M.S., Lin L., Xiong X., Naddeo V., Cai Y. Biopolymers-based microencapsulation technology for sustainable textiles development: A short review. Case Stud. Chem. Environ. Eng. 2023;7:100349. doi: 10.1016/j.cscee.2023.100349. [DOI] [Google Scholar]
- 12.Monia T. Sustainable natural biopolymers for biomedical applications. J. Thermoplast. Compos. Mater. 2024;37:2505–2524. doi: 10.1177/08927057231214468. [DOI] [Google Scholar]
- 13.Mehmood A., Raina N., Phakeenuya V., Wonganu B., Cheenkachorn K. The current status and market trend of polylactic acid as biopolymer: Awareness and needs for sustainable development. Mater. Today Proc. 2023;72:3049–3055. doi: 10.1016/j.matpr.2022.08.387. [DOI] [Google Scholar]
- 14.Westlake J.R., Tran M.W., Jiang Y., Zhang X., Burrows A.D., Xie M. Biodegradable biopolymers for active packaging: Demand, development and directions. Sustain. Food Technol. 2023;1:50–72. doi: 10.1039/D2FB00004K. [DOI] [Google Scholar]
- 15.Li F., Xie X., Xu X., Zou X. Water-soluble biopolymers calcium polymalate derived from fermentation broth of Aureobasidium pullulans markedly alleviates osteoporosis and fatigue. Int. J. Biol. Macromol. 2024;268:132013. doi: 10.1016/j.ijbiomac.2024.132013. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M., Zuo Z., Zhang X., Wang L. Food biopolymer behaviors in the digestive tract: Implications for nutrient delivery. Crit. Rev. Food Sci. Nutr. 2024;64:8709–8727. doi: 10.1080/10408398.2023.2202778. [DOI] [PubMed] [Google Scholar]
- 17.Lackner M., Mustakhimov I., García Martínez J.B., Pflügl S. Aerobic and anaerobic fermentation of gaseous and liquid one carbon feedstocks to produce food, feed, biopolymers and value-added products. Front. Bioeng. Biotechnol. 2024;12:1334864. doi: 10.3389/fbioe.2024.1334864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClements D.J. Composite hydrogels assembled from food-grade biopolymers: Fabrication, properties, and applications. Adv. Colloid Interface Sci. 2024;332:103278. doi: 10.1016/j.cis.2024.103278. [DOI] [PubMed] [Google Scholar]
- 19.Aquinas N., Bhat R.M., Selvaraj S. Submerged Fermentation and Kinetics of Newly Isolated Priestia megaterium for the Production of Biopolymer Curdlan. J. Polym. Environ. 2024;32:4683–4698. doi: 10.1007/s10924-024-03224-6. [DOI] [Google Scholar]
- 20.Khalfallah R., Mechmeche M., Ksontini H., Jmoui I., Hamdi M., Kachouri F. Novel Approach Coating by Edible Biopolymers of Sesame Fermented with Lactobacillus plantarum to Improve the Shelf Life of Strawberries and Preserve Their Nutritional Quality During Storage. J. Packag. Technol. Res. 2024;8:63–77. doi: 10.1007/s41783-024-00162-5. [DOI] [Google Scholar]
- 21.Hanifah A., Arfiathi, Mahardika M., Sumirat R., Nissa R.C., Nurhamiyah Y. Biomass Conversion and Sustainable Biorefinery: Towards Circular Bioeconomy. Springer; Singapore: 2024. Recent Updates on Biopolymers: Precursors, Process, Properties, Challenge, and Future Perspectives; pp. 19–42. [Google Scholar]
- 22.Rangel C., Lourenço N.D., Reis M.A., Carvalho G. Dynamics in the profile of biopolymers produced by mixed microbial cultures from ethanol-rich feedstocks. J. Environ. Chem. Eng. 2024;12:112609. doi: 10.1016/j.jece.2024.112609. [DOI] [Google Scholar]
- 23.Sinha S. An overview of biopolymer-derived packaging material. Polym. Renew. Resour. 2024;15:193–209. doi: 10.1177/20412479241226884. [DOI] [Google Scholar]
- 24.Abou-alfitooh S.A., El-Hoshoudy A. Eco-friendly modified biopolymers for enhancing oil production: A review. J. Polym. Environ. 2024;32:2457–2483. doi: 10.1007/s10924-023-03132-1. [DOI] [Google Scholar]
- 25.Khaydukova I.V., Ivannikova V.M., Zhidkov D.A., Belikov N.V., Peshkova M.A., Timashev P.S., Tsiganov D.I., Pushkarev A.V. Current State and Challenges of Tissue and Organ Cryopreservation in Biobanking. Int. J. Mol. Sci. 2024;25:11124. doi: 10.3390/ijms252011124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinaeva L.G., Noskov A.S. Biodegradable biopolymers: Real impact to environment pollution. Sci. Total. Environ. 2024;947:174445. doi: 10.1016/j.scitotenv.2024.174445. [DOI] [PubMed] [Google Scholar]
- 27.Opriș O., Mormile C., Lung I., Stegarescu A., Soran M.L., Soran A. An overview of biopolymers for drug delivery applications. Appl. Sci. 2024;14:1383. doi: 10.3390/app14041383. [DOI] [Google Scholar]
- 28.Hou X., Lin L., Li K., Jiang F., Qiao D., Zhang B., Xie F. Towards superior biopolymer gels by enabling interpenetrating network structures: A review on types, applications, and gelation strategies. Adv. Colloid Interface Sci. 2024;325:103113. doi: 10.1016/j.cis.2024.103113. [DOI] [PubMed] [Google Scholar]
- 29.Wahba M.I. A comprehensive review on genipin: An efficient natural cross-linker for biopolymers. Polym. Bull. 2024;81:14251–14305. doi: 10.1007/s00289-024-05406-7. [DOI] [Google Scholar]
- 30.Yang W., Yang W., Zeng J., Chen Y., Huang Y., Liu J., Gan J., Li T., Zhang H., Zhong L., et al. Biopolymer-based gel electrolytes for electrochemical energy Storage: Advances and prospects. Prog. Mater. Sci. 2024;144:101264. doi: 10.1016/j.pmatsci.2024.101264. [DOI] [Google Scholar]
- 31.Rahman A., Hasan K., Imran A.B. Bio-Based Polymers: Farm to Industry. Volume 3: Emerging Trends and Applications. ACS Publications; Washington, DC, USA: 2024. Biopolymers for Supercapacitors; pp. 57–80. [Google Scholar]
- 32.Mumtaz M., Hussain N., Ashraf M., Azam H.M.H., Iftikhar A. Applications of Biopolymers in Science, Biotechnology, and Engineering. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2024. Introduction to Biopolymers, Their Blend, IPN s, Gel, Composites, and Nanocomposites; pp. 1–29. [Google Scholar]
- 33.Werner J., Zgoła-Grześkowiak A., Grześkowiak T., Frankowski R. Biopolymers-based sorbents as a future green direction for solid phase (micro) extraction techniques. TrAC Trends Anal. Chem. 2024;173:117659. doi: 10.1016/j.trac.2024.117659. [DOI] [Google Scholar]
- 34.Hisham F., Akmal M.M., Ahmad F., Ahmad K., Samat N. Biopolymer chitosan: Potential sources, extraction methods, and emerging applications. Ain Shams Eng. J. 2024;15:102424. doi: 10.1016/j.asej.2023.102424. [DOI] [Google Scholar]
- 35.Abate T., Amabile C., Muñoz R., Chianese S., Musmarra D. Polyhydroxyalkanoate recovery overview: Properties, characterizations, and extraction strategies. Chemosphere. 2024;356:141950. doi: 10.1016/j.chemosphere.2024.141950. [DOI] [PubMed] [Google Scholar]
- 36.Durand K., Daassi R., Rodrigue D., Stevanovic T. Study of biopolymers and silica recovery from pre-hydrolyzed rice husks. Biomass Convers. Biorefinery. 2024:1–18. doi: 10.1007/s13399-024-05445-0. [DOI] [Google Scholar]
- 37.Ai J., Wang K., Fu Q., Dong T., Li L., Peng S., Wang D., Wang Q., Zhang W. Novel insights into the biopolymers transformation under wastewater sludge drying process at different temperatures in relation to drying behavior. Chem. Eng. J. 2024;486:150376. doi: 10.1016/j.cej.2024.150376. [DOI] [Google Scholar]
- 38.Ludka F.R., Klosowski A.B., Camargo G.A., Justo A.S., Andrade E.A., Beltrame F.L., Olivato J.B. Brewers’ spent grain extract as antioxidants in starch-based active biopolymers. Int. J. Food Sci. Technol. 2024;59:142–150. doi: 10.1111/ijfs.16785. [DOI] [Google Scholar]
- 39.Sudheshwar A., Vogel K., Nyström G., Malinverno N., Arnaudo M., Camacho C.E.G., Beloin-Saint-Pierre D., Hischier R., Som C. Unraveling the climate neutrality of wood derivatives and biopolymers. RSC Sustain. 2024;2:1487–1497. doi: 10.1039/D4SU00010B. [DOI] [Google Scholar]
- 40.Garnett M.T., Kumar H.K.S., Beckingham B.S., Alexander S.L. Extraction of cellulose from restaurant food waste. RSC Sustain. 2024;2:170–178. doi: 10.1039/D3SU00192J. [DOI] [Google Scholar]
- 41.Bhat S.G., Thivaharan V., Divyashree M. Sustainable Opportunities in the Downstream Processing of the Intracellular Biopolymer Polyhydroxyalkanoate. ChemBioEng Rev. 2024;11:79–94. doi: 10.1002/cben.202300040. [DOI] [Google Scholar]
- 42.List M., Maurischat J. Extraction of Protein-Based Biopolymers from Algae and Food Byproducts. 2024. [(accessed on 11 October 2024)]. Available online: https://opus4.kobv.de/opus4-rosenheim/frontdoor/index/index/docId/2739.
- 43.Dutta S., Ghosh A., Spakowitz A.J. Effect of local active fluctuations on structure and dynamics of flexible biopolymers. Soft Matter. 2024;20:1694–1701. doi: 10.1039/D3SM01491F. [DOI] [PubMed] [Google Scholar]
- 44.Shah Y.A., Bhatia S., Al-Harrasi A., Oz F., Khan M.H., Roy S., Esatbeyoglu T., Pratap-Singh A. Thermal properties of biopolymer films: Insights for sustainable food packaging applications. Food Eng. Rev. 2024:1–16. doi: 10.1007/s12393-024-09380-8. [DOI] [Google Scholar]
- 45.Abady M.M., Shawky A.M., Sakr F.A., Mohammed D.M., Goda E.S. Bio-Based Polymers: Farm to Industry. Volume 2: Current Trends and Applications. ACS Publications; Washington, DC, USA: 2024. Recent Advancements in Biosensors Using Biopolymers; pp. 81–112. [Google Scholar]
- 46.Chen K., Tian R., Jiang J., Xiao M., Wu K., Kuang Y., Deng P., Zhao X., Jiang F. Moisture loss inhibition with biopolymer films for preservation of fruits and vegetables: A review. Pt 1Int. J. Biol. Macromol. 2024;263:130337. doi: 10.1016/j.ijbiomac.2024.130337. [DOI] [PubMed] [Google Scholar]
- 47.Chiu I., Yang T. Biopolymer-based intelligent packaging integrated with natural colourimetric sensors for food safety and sustainability. Anal. Sci. Adv. 2024;5:e202300065. doi: 10.1002/ansa.202300065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawliszak P., Beheshti A., Møller A., Blencowe A., Beattie D.A., Krasowska M. Increasing surface hydrophilicity with biopolymers: A combined single bubble collision, QCM-D and AFM study. J. Colloid Interface Sci. 2024;667:393–402. doi: 10.1016/j.jcis.2024.04.073. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Li X., Gao N., Wang X., Sun S. Multivariable analysis of egg white protein-chitosan interaction: Influence of pH, temperature, biopolymers ratio, and ionic concentration. Food Chem. X. 2023;19:100817. doi: 10.1016/j.fochx.2023.100817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng J.P., Li S., Yan R., Wei C. Effect of pH value on effectiveness of biopolymer-based treatment of bauxite mine slurry. Physicochem. Probl. Miner. Process. 2023;59:167949. doi: 10.37190/ppmp/167949. [DOI] [Google Scholar]
- 51.Kumar Y., Bist Y., Thakur D., Nagar M., Saxena D.C. A review on the role of pH-sensitive natural pigments in biopolymers based intelligent food packaging films. Pt 1Int. J. Biol. Macromol. 2024;276:133869. doi: 10.1016/j.ijbiomac.2024.133869. [DOI] [PubMed] [Google Scholar]
- 52.Vegad U., Patel M., Khunt D., Zupančič O., Chauhan S., Paudel A. pH stimuli-responsive hydrogels from non-cellulosic biopolymers for drug delivery. Front. Bioeng. Biotechnol. 2023;11:1270364. doi: 10.3389/fbioe.2023.1270364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y., Ni J., Gu J., Liu S., Huang Y., Sadeghi H. Influence of biopolymer-vegetation interaction on soil hydro-mechanical properties under climate change: A review. Sci. Total. Environ. 2024;954:176535. doi: 10.1016/j.scitotenv.2024.176535. [DOI] [PubMed] [Google Scholar]
- 54.Schniete J.K., Brüser T., Horn M.A., Tschowri N. Specialized biopolymers: Versatile tools for microbial resilience. Curr. Opin. Microbiol. 2024;77:102405. doi: 10.1016/j.mib.2023.102405. [DOI] [PubMed] [Google Scholar]
- 55.Dey B., Prabhakar M.R., Jayaraman S., Gujjala L.K.S., Venugopal A.P., Balasubramanian P. Biopolymer-based solutions for enhanced safety and quality assurance: A review. Food Res. Int. 2024;191:114723. doi: 10.1016/j.foodres.2024.114723. [DOI] [PubMed] [Google Scholar]
- 56.Semenova M.G., Aslanova M.A., Galimova A.R., Fedulova L.V., Antipova A.S., Martirosova E.I., Zelikina D.V., Bero A.L., Utyanov D.A. Thermal stability and digestibility of a biopolymer system for the delivery of minor nutrients in enriched meat products. Theory and practice of meat processing. 2024;9:160–168. doi: 10.21323/2414-438X-2024-9-2-160-168. [DOI] [Google Scholar]
- 57.Gonçalves E.M., Silva M., Andrade L., Pinheiro J. From Fields to Films: Exploring Starch from Agriculture Raw Materials for Biopolymers in Sustainable Food Packaging. Agriculture. 2024;14:453. doi: 10.3390/agriculture14030453. [DOI] [Google Scholar]
- 58.da Silva Cazella P.H., de Souza M.V., Rodrigues F.R., da Silva S.A.M., Bispo R.A., De Araujo V.A., Christoforo A.L. Polyethylene terephthalate (PET) as a recycled raw material for particleboards produced from pinus wood and biopolymer resin. J. Clean. Prod. 2024;447:141460. doi: 10.1016/j.jclepro.2024.141460. [DOI] [Google Scholar]
- 59.Li Q., Wang F., Zhang Y., Shi M., Zhang Y., Yu H., Liu S., Li J., Tan S.C., Chen W. Biopolymers for hygroscopic material development. Adv. Mater. 2024;36:2209479. doi: 10.1002/adma.202209479. [DOI] [PubMed] [Google Scholar]
- 60.Liu M., Zhang X., Wei A., Li H., Zhang H., Zheng L., Xia N., Wang J. Protein-based active films: Raw materials, functions, and food applications. Compr. Rev. Food Sci. Food Saf. 2024;23:e13302. doi: 10.1111/1541-4337.13302. [DOI] [Google Scholar]
- 61.Wang K., Chi B.Y., Yang T.Y., Ren W.F., Gao X.J., Wang K.H., Sun R.C. Natural biopolymers derived kinematic and self-healing hydrogel coatings to continuously protect metallic zinc anodes. Electrochim. Acta. 2024;489:144238. doi: 10.1016/j.electacta.2024.144238. [DOI] [Google Scholar]
- 62.Guo X., Zhang Y., Li J., Hao Y., Ke H., Lv P., Wei Q. Wet Spinning Technology for Aerogel Fiber: Pioneering the Frontier of High-Performance and Multifunctional Materials. Adv. Fiber Mater. 2024:1–41. doi: 10.1007/s42765-024-00440-6. [DOI] [Google Scholar]
- 63.Sinha S. Bio-Based Superabsorbents: Recent Trends, Types, Applications And Recycling. Springer; Singapore: 2023. Synthesis of biopolymer based superabsorbent: An eco-friendly approach towards future sustainability; pp. 29–49. [Google Scholar]
- 64.TG Y.G., Ballupete Nagaraju S., Puttegowda M., Verma A., Rangappa S.M., Siengchin S. Biopolymer-based composites: An eco-friendly alternative from agricultural waste biomass. J. Compos. Sci. 2023;7:242. doi: 10.3390/jcs7060242. [DOI] [Google Scholar]
- 65.Sharma A., Jha A. Application of biopolymers in clothing and fashion. Int. J. Pharma Prof. Res. (IJPPR) 2023;14:1–21. doi: 10.48165/ijppronline.2023.14401. [DOI] [Google Scholar]
- 66.Liu B., Desai A.S., Sun X., Ren J., Pathan H.M., Dabir V., Ashok A., Hou H., Pan D., Guo X., et al. An overview of sustainable biopolymer composites in sensor manufacturing and smart cities. Adv. Compos. Hybrid Mater. 2024;7:146. doi: 10.1007/s42114-024-00938-y. [DOI] [Google Scholar]
- 67.Behrooznia Z., Nourmohammadi J. Polysaccharide-based materials as an eco-friendly alternative in biomedical, environmental, and food packaging. Giant. 2024;19:100301. doi: 10.1016/j.giant.2024.100301. [DOI] [Google Scholar]
- 68.Biswal T. Future perspectives of biopolymeric industry. Phys. Sci. Rev. 2024;9:2965–2988. doi: 10.1515/psr-2022-0192. [DOI] [Google Scholar]
- 69.Rahman M.Z., Rahman M., Mahbub T., Ashiquzzaman M., Sagadevan S., Hoque M.E. Advanced biopolymers for automobile and aviation engineering applications. J. Polym. Res. 2023;30:106. doi: 10.1007/s10965-023-03440-z. [DOI] [Google Scholar]
- 70.Khandeparkar A.S., Paul R., Sridhar A., Lakshmaiah V.V., Nagella P. Eco-friendly innovations in food packaging: A sustainable revolution. Sustain. Chem. Pharm. 2024;39:101579. doi: 10.1016/j.scp.2024.101579. [DOI] [Google Scholar]
- 71.Palaniappan S.K., Singh M.K., Rangappa S.M., Siengchin S. Eco-friendly Biocomposites: A Step Towards Achieving Sustainable Development Goals. Composites. 2023;7:7373. doi: 10.14416/j.asep.2024.02.003. [DOI] [Google Scholar]
- 72.Dhoundiyal S., Alam M.A., Kaur A., Maqsood S., Sharma S., Khan S.A. Biopolymers in the Textile Industry: Opportunities and Limitations. Springer; Singapore: 2024. Biopolymers in Sustainable Textile Dyeing and Printing; pp. 123–146. [Google Scholar]
- 73.Kanchetti D., Munirathnam R., Thakkar D. Integration of Machine Learning Algorithms with Cloud Computing for Real-Time Data Analysis. J. Res. Appl. Sci. Biotechnol. 2024;3:301–306. doi: 10.55544/jrasb.3.2.46. [DOI] [Google Scholar]
- 74.Momeni M., Afkanpour M., Rakhshani S., Mehrabian A., Tabesh H. A prediction model based on artificial intelligence techniques for disintegration time and hardness of fast disintegrating tablets in pre-formulation tests. BMC Med. Inform. Decis. Mak. 2024;24:88. doi: 10.1186/s12911-024-02485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Piras G., Muzi F., Tiburcio V.A. Digital Management Methodology for Building Production Optimization through Digital Twin and Artificial Intelligence Integration. Buildings. 2024;14:2110. doi: 10.3390/buildings14072110. [DOI] [Google Scholar]
- 76.Ayhan H.M., Kır S. Ml-driven approaches to enhance inventory planning: Inoculant weight application in casting processes. Comput. Ind. Eng. 2024;193:110280. doi: 10.1016/j.cie.2024.110280. [DOI] [Google Scholar]
- 77.Strielkowski W., Vlasov A., Selivanov K., Muraviev K., Shakhnov V. Prospects and challenges of the machine learning and data-driven methods for the predictive analysis of power systems: A review. Energies. 2023;16:4025. doi: 10.3390/en16104025. [DOI] [Google Scholar]
- 78.Mhlanga D. Artificial intelligence and machine learning for energy consumption and production in emerging markets: A review. Energies. 2023;16:745. doi: 10.3390/en16020745. [DOI] [Google Scholar]
- 79.Aguiar M.I., Sousa A.F., Teixeira G., Tavares A.P., Ferreira A.M., Coutinho J.A. Enhancing plastic waste recycling: Evaluating the impact of additives on the enzymatic polymer degradation. Catal. Today. 2024;429:114492. doi: 10.1016/j.cattod.2023.114492. [DOI] [Google Scholar]
- 80.Nasrin T., Pourkamali-Anaraki F., Peterson A.M. Application of machine learning in polymer additive manufacturing: A review. J. Polym. Sci. 2024;62:2639–2669. doi: 10.1002/pol.20230649. [DOI] [Google Scholar]
- 81.Miao B.H., Dong Y., Wu Z.Y., Alemdar B.N., Zhang P., Kohler M.D., Noh H.Y. Integration of physics-based building model and sensor data to develop an adaptive digital twin; Proceedings of the 9th ACM International Conference on Systems for Energy-Efficient Buildings, Cities, and Transportation; Boston, MA, USA. 9–10 November 2022; pp. 282–283. [Google Scholar]
- 82.Prateek S., Garg R., Kumar Saxena K., Srivastav V., Vasudev H., Kumar N. Data-driven materials science: Application of ML for predicting band gap. Adv. Mater. Process. Technol. 2024;10:708–717. doi: 10.1080/2374068X.2023.2171666. [DOI] [Google Scholar]
- 83.Chew A.K., Sender M., Kaplan Z., Chandrasekaran A., Chief Elk J., Browning A.R., Kwak H.S., Halls M.D., Afzal M.A.F. Advancing material property prediction: Using physics-informed machine learning models for viscosity. J. Cheminform. 2024;16:31. doi: 10.1186/s13321-024-00820-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J., Zhou M., Wu H.H., Wang L., Zhang J., Wu N., Pan K., Liu G., Zhang Y., Han J., et al. Machine Learning-Assisted Property Prediction of Solid-State Electrolyte. Adv. Energy Mater. 2024;14:2304480. doi: 10.1002/aenm.202304480. [DOI] [Google Scholar]
- 85.Cao Z., Barati Farimani O., Ock J., Barati Farimani A. Machine Learning in Membrane Design: From Property Prediction to AI-Guided Optimization. Nano Lett. 2024;24:2953–2960. doi: 10.1021/acs.nanolett.3c05137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kazemi-Khasragh E., Blázquez J.P.F., Gómez D.G., González C., Haranczyk M. Facilitating polymer property prediction with machine learning and group interaction modelling methods. Int. J. Solids Struct. 2024;286:112547. doi: 10.1016/j.ijsolstr.2023.112547. [DOI] [Google Scholar]
- 87.Yu T., Wang C., Yang H., Li F. Machine learning in metal-ion battery research: Advancing material prediction, characterization, and status evaluation. J. Energy Chem. 2024;90:191–204. doi: 10.1016/j.jechem.2023.10.049. [DOI] [Google Scholar]
- 88.Watpade A.D., Thakor S., Jain P., Mohapatra P.P., Vaja C.R., Joshi A., Shah D.V., Islam M.T. Comparative analysis of machine learning models for predicting dielectric properties in MoS2 nanofiller-reinforced epoxy composites. Ain Shams Eng. J. 2024;15:102754. doi: 10.1016/j.asej.2024.102754. [DOI] [Google Scholar]
- 89.Min Y., Ming X., Peihong C., Yang S., Zhang H., Lingfeng W., Liucheng Z., Yinghong L., Wanlin G. Machine learning for predicting fatigue properties of additively manufactured materials. Chin. J. Aeronaut. 2024;37:1–22. [Google Scholar]
- 90.Kalhor P., Jung N., Bräse S., Wöll C., Tsotsalas M., Friederich P. Functional material systems enabled by automated data extraction and machine learning. Adv. Funct. Mater. 2024;34:2302630. doi: 10.1002/adfm.202302630. [DOI] [Google Scholar]
- 91.Mi X., Dai L., Jing X., She J., Holmedal B., Tang A., Pan F. Accelerated design of high-performance Mg-Mn-based magnesium alloys based on novel bayesian optimization. J. Magnes. Alloy. 2024;12:750–766. doi: 10.1016/j.jma.2024.01.005. [DOI] [Google Scholar]
- 92.Liu S., Yang C. Machine learning design for high-entropy alloys: Models and algorithms. Metals. 2024;14:235. doi: 10.3390/met14020235. [DOI] [Google Scholar]
- 93.Sharma V., Misra J.P., Singhal S. Machine learning algorithms based advanced optimization of wire-EDM parameters: An experimental investigation into titanium alloy. Int. J. Interact. Des. Manuf. (IJIDeM) 2024;18:2855–2868. doi: 10.1007/s12008-023-01348-y. [DOI] [Google Scholar]
- 94.Padhy S.P., Chaudhary V., Lim Y.F., Zhu R., Thway M., Hippalgaonkar K., Ramanujan R.V. Experimentally validated inverse design of multi-property Fe-Co-Ni alloys. iScience. 2024;27:109723. doi: 10.1016/j.isci.2024.109723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou H.r., Yang H., Li H.q., Ma Y.c., Yu S., Shi J., Cheng J.c., Gao P., Yu B., Miao Z.q., et al. Advancements in machine learning for material design and process optimization in the field of additive manufacturing. China Foundry. 2024;21:101–115. doi: 10.1007/s41230-024-3145-3. [DOI] [Google Scholar]
- 96.Zhao S., Jiang B., Song K., Liu X., Wang W., Si D., Zhang J., Chen X., Zhou C., Liu P., et al. Machine learning assisted design of high-entropy alloys with ultra-high microhardness and unexpected low density. Mater. Des. 2024;238:112634. doi: 10.1016/j.matdes.2024.112634. [DOI] [Google Scholar]
- 97.Wu Y., Shang Z., Lu T., Zhou W., Li M., Lu W. Target-directed discovery for low melting point alloys via inverse design strategy. J. Alloys Compd. 2024;971:172664. doi: 10.1016/j.jallcom.2023.172664. [DOI] [Google Scholar]
- 98.Shi Y., Liu X., Lan S., Gao N., Yin S., Guo W., Fan Z., Wang K. Machine-learning assisted design of as-cast NiCoFeCrAlTi multi-principal element alloys with tensile yield strength over 1.35 GPa. Intermetallics. 2024;166:108170. doi: 10.1016/j.intermet.2023.108170. [DOI] [Google Scholar]
- 99.Lian Z., Ma Y., Li M., Lu W., Zhou W. Discovery Precision: An effective metric for evaluating performance of machine learning model for explorative materials discovery. Comput. Mater. Sci. 2024;233:112738. doi: 10.1016/j.commatsci.2023.112738. [DOI] [Google Scholar]
- 100.Cheetham A.K., Seshadri R. Artificial intelligence driving materials discovery? perspective on the article: Scaling deep learning for materials discovery. Chem. Mater. 2024;36:3490–3495. doi: 10.1021/acs.chemmater.4c00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu Y., Wang C.F., Ju M.G., Jia Q., Zhou Q., Lu S., Gao X., Zhang Y., Wang J. Universal machine learning aided synthesis approach of two-dimensional perovskites in a typical laboratory. Nat. Commun. 2024;15:138. doi: 10.1038/s41467-023-44236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Talukder M.J., Alshami A.S., Tayyebi A., Ismail N., Yu X. Membrane science meets machine learning: Future and potential use in assisting membrane material design and fabrication. Sep. Purif. Rev. 2024;53:216–229. doi: 10.1080/15422119.2023.2212295. [DOI] [Google Scholar]
- 103.Barcin T., Yucel M.A., Ersan R.H., Alagoz M.A., Dogen A., Burmaoglu S., Algul O. Deep learning approach to the discovery of novel bisbenzazole derivatives for antimicrobial effect. J. Mol. Struct. 2024;1295:136668. doi: 10.1016/j.molstruc.2023.136668. [DOI] [Google Scholar]
- 104.Mican J., Da’san MM J., Liu W., Weber G., Mazurenko S., Bornscheuer U.T., Damborsky J., Wei R., Bednar D. Exploring new galaxies: Perspectives on the discovery of novel PET-degrading enzymes. Appl. Catal. B Environ. 2024;342:123404. doi: 10.1016/j.apcatb.2023.123404. [DOI] [Google Scholar]
- 105.Song J., Lee J., Kim N., Min K. Artificial intelligence in the design of innovative metamaterials: A comprehensive review. Int. J. Precis. Eng. Manuf. 2024;25:225–244. doi: 10.1007/s12541-023-00857-w. [DOI] [Google Scholar]
- 106.Zhuang J., Midgley A.C., Wei Y., Liu Q., Kong D., Huang X. Machine-Learning-Assisted Nanozyme Design: Lessons from Materials and Engineered Enzymes. Adv. Mater. 2024;36:2210848. doi: 10.1002/adma.202210848. [DOI] [PubMed] [Google Scholar]
- 107.Lu B., Xia Y., Ren Y., Xie M., Zhou L., Vinai G., Morton S.A., Wee A.T., van der Wiel W.G., Zhang W., et al. When Machine Learning Meets 2D Materials: A Review. Adv. Sci. 2024;11:2305277. doi: 10.1002/advs.202305277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang J., Li F.Z., Arnold F.H. Opportunities and challenges for machine learning-assisted enzyme engineering. ACS Cent. Sci. 2024;10:226–241. doi: 10.1021/acscentsci.3c01275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Butler K.T., Davies D.W., Cartwright H., Isayev O., Walsh A. Machine learning for molecular and materials science. Nature. 2018;559:547–555. doi: 10.1038/s41586-018-0337-2. [DOI] [PubMed] [Google Scholar]
- 110.Ibarra-Pérez D., Faba S., Hernández-Muñoz V., Smith C., Galotto M.J., Garmulewicz A. Predicting the composition and mechanical properties of seaweed bioplastics from the scientific literature: A machine learning approach for modeling sparse data. Appl. Sci. 2023;13:11841. doi: 10.3390/app132111841. [DOI] [Google Scholar]
- 111.Patel R.A., Webb M.A. Data-driven design of polymer-based biomaterials: High-throughput simulation, experimentation, and machine learning. ACS Appl. Bio Mater. 2023;7:510–527. doi: 10.1021/acsabm.2c00962. [DOI] [PubMed] [Google Scholar]
- 112.Ahmed E., Mulay P., Ramirez C., Tirado-Mansilla G., Cheong E., Gormley A.J. Mapping biomaterial complexity by machine learning. Tissue Eng. Part A. 2024;30:662–680. doi: 10.1089/ten.tea.2024.0067. [DOI] [PubMed] [Google Scholar]
- 113.Stuart S., Watchorn J., Gu F.X. Sizing up feature descriptors for macromolecular machine learning with polymeric biomaterials. Npj Comput. Mater. 2023;9:102. doi: 10.1038/s41524-023-01040-5. [DOI] [Google Scholar]
- 114.Haarlemmer G., Matricon L., Roubaud A. Computer Aided Chemical Engineering. Volume 53. Elsevier; Amsterdam, The Netherlands: 2024. Hydrothermal liquefaction data for use in machine learning models; pp. 3169–3174. [Google Scholar]
- 115.Babatunde Q.O., Son D.G., Kim S.Y., Byun Y.H. Effect of Curing Condition and Solvent Content on Mechanical Properties of Zein-Biopolymer-Treated Soil. Sustainability. 2023;15:12048. doi: 10.3390/su151512048. [DOI] [Google Scholar]
- 116.Karkoszka M., Rok J., Wrześniok D. Melanin Biopolymers in Pharmacology and Medicine—Skin Pigmentation Disorders, Implications for Drug Action, Adverse Effects and Therapy. Pharmaceuticals. 2024;17:521. doi: 10.3390/ph17040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vázquez V., Giorgi V., Bonfiglio F., Menéndez P., Gioia L., Ovsejevi K. Lignocellulosic residues from bioethanol production: A novel source of biopolymers for laccase immobilization. RSC Adv. 2023;13:13463–13471. doi: 10.1039/D3RA01520C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Garduño-Juárez R., Tovar-Anaya D.O., Perez-Aguilar J.M., Lozano-Aguirre Beltran L.F., Zubillaga R.A., Alvarez-Perez M.A., Villarreal-Ramirez E. Molecular dynamic simulations for biopolymers with biomedical applications. Polymers. 2024;16:1864. doi: 10.3390/polym16131864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nasarian E., Alizadehsani R., Acharya U.R., Tsui K.L. Designing interpretable ML system to enhance trust in healthcare: A systematic review to proposed responsible clinician-AI-collaboration framework. Inf. Fusion. 2024;108:102412. doi: 10.1016/j.inffus.2024.102412. [DOI] [Google Scholar]
- 120.Räz T. ML interpretability: Simple isn’t easy. Stud. Hist. Philos. Sci. 2024;103:159–167. doi: 10.1016/j.shpsa.2023.12.007. [DOI] [PubMed] [Google Scholar]
- 121.Ciobanu-Caraus O., Aicher A., Kernbach J.M., Regli L., Serra C., Staartjes V.E. A critical moment in machine learning in medicine: On reproducible and interpretable learning. Acta Neurochir. 2024;166:14. doi: 10.1007/s00701-024-05892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Khayretdinova M., Zakharov I., Pshonkovskaya P., Adamovich T., Kiryasov A., Zhdanov A., Shovkun A. Prediction of brain sex from EEG: Using large-scale heterogeneous dataset for developing a highly accurate and interpretable ML model. NeuroImage. 2024;285:120495. doi: 10.1016/j.neuroimage.2023.120495. [DOI] [PubMed] [Google Scholar]
- 123.Jiang S., Sweet L.b., Blougouras G., Brenning A., Li W., Reichstein M., Denzler J., Shangguan W., Yu G., Huang F., et al. How interpretable machine learning can benefit process understanding in the geosciences. Earth’s Futur. 2024;12:e2024EF004540. doi: 10.1029/2024EF004540. [DOI] [Google Scholar]
- 124.Antonini A.S., Tanzola J., Asiain L., Ferracutti G.R., Castro S.M., Bjerg E.A., Ganuza M.L. Machine Learning model interpretability using SHAP values: Application to Igneous Rock Classification task. Appl. Comput. Geosci. 2024;23:100178. doi: 10.1016/j.acags.2024.100178. [DOI] [Google Scholar]
- 125.Ma D., Bortnik J., Ma Q., Hua M., Chu X. Machine learning interpretability of outer radiation belt enhancement and depletion events. Geophys. Res. Lett. 2024;51:e2023GL106049. doi: 10.1029/2023GL106049. [DOI] [Google Scholar]
- 126.Kobayashi K., Alam S.B. Explainable, interpretable, and trustworthy AI for an intelligent digital twin: A case study on remaining useful life. Eng. Appl. Artif. Intell. 2024;129:107620. doi: 10.1016/j.engappai.2023.107620. [DOI] [Google Scholar]
- 127.Zheng J.X., Li X., Zhu J., Guan S.Y., Zhang S.X., Wang W.M. Interpretable machine learning for predicting chronic kidney disease progression risk. Digit. Health. 2024;10:20552076231224225. doi: 10.1177/20552076231224225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nadizar G., Rovito L., De Lorenzo A., Medvet E., Virgolin M. An analysis of the ingredients for learning interpretable symbolic regression models with human-in-the-loop and genetic programming. ACM Trans. Evol. Learn. Optim. 2024;4:1–30. doi: 10.1145/3643688. [DOI] [Google Scholar]
- 129.Zou X., Perlaza S.M., Esnaola I., Altman E. Generalization analysis of machine learning algorithms via the worst-case data-generating probability measure. Proc. Aaai Conf. Artif. Intell. 2024;38:17271–17279. doi: 10.1609/aaai.v38i15.29674. [DOI] [Google Scholar]
- 130.Trivedi A. Addressing the Fallacy of Generalizing Caste Racism among Hindus: An AI/ML Approach to Deconstructing Misconceptions. Int. J. Multidiscip. Innov. Res. Methodol. 2024;3:50–63. [Google Scholar]
- 131.Ispirova G., Eftimov T., Džeroski S., Seljak B.K. MsGEN: Measuring generalization of nutrient value prediction across different recipe datasets. Expert Syst. Appl. 2024;237:121507. doi: 10.1016/j.eswa.2023.121507. [DOI] [Google Scholar]
- 132.Gil-Fuster E., Eisert J., Bravo-Prieto C. Understanding quantum machine learning also requires rethinking generalization. Nat. Commun. 2024;15:2277. doi: 10.1038/s41467-024-45882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Anagnostopoulos S.J., Toscano J.D., Stergiopulos N., Karniadakis G.E. Learning in PINNs: Phase transition, total diffusion, and generalization. arXiv. 20242403.18494 [Google Scholar]
- 134.Wang Y., Patel S., Ortner C. A theoretical case study of the generalization of machine-learned potentials. Comput. Methods Appl. Mech. Eng. 2024;422:116831. doi: 10.1016/j.cma.2024.116831. [DOI] [Google Scholar]
- 135.Alsaggaf I.A., Aloufi S.F., Baharith L.A. A new generalization of the inverse generalized Weibull distribution with different methods of estimation and applications in medicine and engineering. Symmetry. 2024;16:1002. doi: 10.3390/sym16081002. [DOI] [Google Scholar]
- 136.Corso G., Deng A., Fry B., Polizzi N., Barzilay R., Jaakkola T. Deep confident steps to new pockets: Strategies for docking generalization. arXiv. 20242402.18396 [Google Scholar]
- 137.Mora A., Bujari A., Bellavista P. Enhancing generalization in federated learning with heterogeneous data: A comparative literature review. Futur. Gener. Comput. Syst. 2024;157:1–15. doi: 10.1016/j.future.2024.03.027. [DOI] [Google Scholar]
- 138.Ramírez J.G.C., Islam M.M. Navigating the Terrain: Scaling Challenges and Opportunities in AI/ML Infrastructure. J. Artif. Intell. Gen. Sci. (JAIGS) 2024;2:241–250. [Google Scholar]
- 139.Singla A., Malhotra T. Challenges and Opportunities in Scaling AI/ML Pipelines. J. Sci. Technol. 2024;5:1–21. [Google Scholar]
- 140.Darzi S., Yavuz A.A. PQC meets ML or AI: Exploring the Synergy of Machine Learning and Post-quantum Cryptography. TechRxiv. 2024 doi: 10.36227/techrxiv.170785010.08081159/v1. [DOI] [Google Scholar]
- 141.Rane N., Mallick S., Kaya O., Rane J. Applied Machine Learning and Deep Learning: Architectures and Techniques. Deep Science Publishing; Palo Alto, CA, USA: 2024. From challenges to implementation and acceptance: Addressing key barriers in artificial intelligence, machine learning, and deep learning; pp. 153–166. [Google Scholar]
- 142.Patil D., Rane N., Desai P., Rane J. Trustworthy Artificial Intelligence in Industry and Society. Deep Science Publishing; Palo Alto, CA, USA: 2024. Machine learning and deep learning: Methods, techniques, applications, challenges, and future research opportunities; pp. 28–81. [Google Scholar]
- 143.Bianchi P., Monbaliu J.C.M. Revisiting the Paradigm of Reaction Optimization in Flow with a Priori Computational Reaction Intelligence. Angew. Chem. 2024;136:e202311526. doi: 10.1002/ange.202311526. [DOI] [PubMed] [Google Scholar]
- 144.Prakash S., Malaiyappan J.N.A., Thirunavukkarasu K., Devan M. Achieving regulatory compliance in cloud computing through ML. AIJMR-Adv. Int. J. Multidiscip. Res. 2024;2:1038. [Google Scholar]
- 145.Choudhury A., Ghose M., Islam A. Machine learning-based computation offloading in multi-access edge computing: A survey. J. Syst. Archit. 2024;148:103090. doi: 10.1016/j.sysarc.2024.103090. [DOI] [Google Scholar]
- 146.Rane J., Mallick S., Kaya O., Rane N. Future Research Opportunities for Artificial Intelligence in Industry 4.0 and 5. Deep Science Publishing; Palo Alto, CA, USA: 2024. Artificial intelligence, machine learning, and deep learning in cloud, edge, and quantum computing: A review of trends, challenges, and future directions; p. 2. [Google Scholar]
- 147.Safdar M., Paul P.P., Lamouche G., Wood G., Zimmermann M., Hannesen F., Bescond C., Wanjara P., Zhao Y.F. Fundamental requirements of a machine learning operations platform for industrial metal additive manufacturing. Comput. Ind. 2024;154:104037. doi: 10.1016/j.compind.2023.104037. [DOI] [Google Scholar]
- 148.Hussain A., Jabeen N., Tabassum A., Ali J. 3D Printed Conducting Polymers. CRC Press; Boca Raton, FL, USA: 2024. 3D-Printed Conducting Polymers for Solid Oxide Fuel Cells; pp. 179–195. [Google Scholar]
- 149.Das S., Jegadeesan J.T., Basu B. Gelatin Methacryloyl (GelMA)-Based Biomaterial Inks: Process Science for 3D/4D Printing and Current Status. Biomacromolecules. 2024;25:2156–2221. doi: 10.1021/acs.biomac.3c01271. [DOI] [PubMed] [Google Scholar]
- 150.Asl Z.R., Rezaee K., Ansari M., Zare F., Roknabadi M.H.A. A review of biopolymer-based hydrogels and IoT integration for enhanced diabetes diagnosis, management, and treatment. Int. J. Biol. Macromol. 2024;280:135988. doi: 10.1016/j.ijbiomac.2024.135988. [DOI] [PubMed] [Google Scholar]
- 151.Xiong G., Zhou X., Zhang C., Xu X. A comprehensive review of intelligent packaging materials based on biopolymers: Role of anthocyanins, type and properties of materials, and their application in monitoring meat freshness. Pt 6Int. J. Biol. Macromol. 2024;282:137462. doi: 10.1016/j.ijbiomac.2024.137462. [DOI] [PubMed] [Google Scholar]
- 152.Bin Abu Sofian A.D.A., Lim H.R., Chew K.W., Show P.L. Advancing 3D Printing through Integration of Machine Learning with Algae-Based Biopolymers. ChemBioEng Rev. 2024;11:406–425. doi: 10.1002/cben.202300054. [DOI] [Google Scholar]
- 153.Singh N.K., Baranwal J., Pati S., Barse B., Khan R.H., Kumar A. Application of plant products in the synthesis and functionalisation of biopolymers. Int. J. Biol. Macromol. 2023;237:124174. doi: 10.1016/j.ijbiomac.2023.124174. [DOI] [PubMed] [Google Scholar]
- 154.Silva F.C.O., Malaisamy A., Cahú T.B., de Araújo M.I.F., Soares P.A.G., Vieira A.T., dos Santos Correia M.T. Polysaccharides from exudate gums of plants and interactions with the intestinal microbiota: A review of vegetal biopolymers and prediction of their prebiotic potential. Pt 2Int. J. Biol. Macromol. 2023;254:127715. doi: 10.1016/j.ijbiomac.2023.127715. [DOI] [PubMed] [Google Scholar]
- 155.Bose I., Nousheen, Roy S., Yaduvanshi P., Sharma S., Chandel V., Biswas D. Unveiling the potential of marine biopolymers: Sources, classification, and diverse food applications. Materials. 2023;16:4840. doi: 10.3390/ma16134840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rahman S., Gogoi J., Dubey S., Chowdhury D. Animal derived biopolymers for food packaging applications: A review. Int. J. Biol. Macromol. 2024;255:128197. doi: 10.1016/j.ijbiomac.2023.128197. [DOI] [PubMed] [Google Scholar]
- 157.Chaari M., Smaoui S. Pullulan as a biopolymer from microorganisms: Role in food packaging. Curr. Food Sci. Technol. Rep. 2024;2:149–156. doi: 10.1007/s43555-024-00023-x. [DOI] [Google Scholar]
- 158.Weyhrich C.W., Petrova S.P., Edgar K.J., Long T.E. Renewed interest in biopolymer composites: Incorporation of renewable, plant-sourced fibers. Green Chem. 2023;25:106–129. doi: 10.1039/D2GC03384D. [DOI] [Google Scholar]
- 159.Kumar D.P., Nair A.S., Balakrishnan P., Gopi S. Handbook of Biopolymers. Springer; Singapore: 2023. Biopolymers from renewable sources; pp. 27–56. [Google Scholar]
- 160.Vasile C., Baican M. Lignins as promising renewable biopolymers and bioactive compounds for high-performance materials. Polymers. 2023;15:3177. doi: 10.3390/polym15153177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ladhari S., Vu N.N., Boisvert C., Saidi A., Nguyen-Tri P. Recent development of polyhydroxyalkanoates (PHA)-based materials for antibacterial applications: A review. ACS Appl. Bio Mater. 2023;6:1398–1430. doi: 10.1021/acsabm.3c00078. [DOI] [PubMed] [Google Scholar]
- 162.Mukherjee A., Koller M. Microbial polyHydroxyAlkanoate (PHA) biopolymers—Intrinsically natural. Bioengineering. 2023;10:855. doi: 10.3390/bioengineering10070855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Molinari G., Parlanti P., Aliotta L., Lazzeri A., Gemmi M. TEM morphological analysis of biopolymers: The case of Poly (Lactic Acid)(PLA) Mater. Today Commun. 2024;38:107868. doi: 10.1016/j.mtcomm.2023.107868. [DOI] [Google Scholar]
- 164.Alhulaybi Z.A. Fabrication and Characterization of Poly (lactic acid)-Based Biopolymer for Surgical Sutures. ChemEngineering. 2023;7:98. doi: 10.3390/chemengineering7050098. [DOI] [Google Scholar]
- 165.Koch M., Spierling S., Venkatachalam V., Endres H.J., Owsianiak M., Vea E.B., Daffert C., Neureiter M., Fritz I. Comparative assessment of environmental impacts of 1st generation (corn feedstock) and 3rd generation (carbon dioxide feedstock) PHA production pathways using life cycle assessment. Sci. Total Environ. 2023;863:160991. doi: 10.1016/j.scitotenv.2022.160991. [DOI] [PubMed] [Google Scholar]
- 166.Churam T., Usubharatana P., Phungrassami H. Sustainable production of carboxymethyl cellulose: A biopolymer alternative from sugarcane (Saccharum officinarum L.) leaves. Sustainability. 2024;16:2352. doi: 10.3390/su16062352. [DOI] [Google Scholar]
- 167.Govoni C., D’Odorico P., Pinotti L., Rulli M.C. Preserving global land and water resources through the replacement of livestock feed crops with agricultural by-products. Nat. Food. 2023;4:1047–1057. doi: 10.1038/s43016-023-00884-w. [DOI] [PubMed] [Google Scholar]
- 168.Mehmood M.A., Amin M., Haider M.N., Malik S., Malik H.A., Alam M.A., Xu J., Alessa A.H., Khan A.Z., Boopathy R. Wastewater-Grown Algal Biomass as Carbon-neutral, Renewable, and Low Water Footprint Feedstock for Clean Energy and Bioplastics. Curr. Pollut. Rep. 2024;10:172–188. doi: 10.1007/s40726-024-00294-x. [DOI] [Google Scholar]
- 169.Phiri R., Rangappa S.M., Siengchin S., Oladijo O.P., Dhakal H.N. Development of sustainable biopolymer-based composites for lightweight applications from agricultural waste biomass: A review. Adv. Ind. Eng. Polym. Res. 2023;6:436–450. doi: 10.1016/j.aiepr.2023.04.004. [DOI] [Google Scholar]
- 170.Sreeharsha R.V., Dubey N., Mohan S.V. Orienting biodiesel production towards sustainability and circularity by tailoring the feedstock and processes. J. Clean. Prod. 2023;414:137526. doi: 10.1016/j.jclepro.2023.137526. [DOI] [Google Scholar]
- 171.Ivankin A.N., Zarubina A.N., Borisova O.A. Bacteriostatic Paper–Polymer Composites Based on Styrene. Polym. Sci. Ser. D. 2024;17:719–724. doi: 10.1134/S1995421224701235. [DOI] [Google Scholar]
- 172.Bibi F., Ilyas N., Saeed M., Shabir S., Shati A.A., Alfaifi M.Y., Amesho K.T., Chowdhury S., Sayyed R.Z. Innovative production o f value-added products using agro-industrial wastes via solid-state fermentation. Environ. Sci. Pollut. Res. 2023;30:125197–125213. doi: 10.1007/s11356-023-28765-6. [DOI] [PubMed] [Google Scholar]
- 173.Low K.E., Tingley J.P., Klassen L., King M.L., Xing X., Watt C., Hoover S.E., Gorzelak M., Abbott D.W. Carbohydrate flow through agricultural ecosystems: Implications for synthesis and microbial conversion of carbohydrates. Biotechnol. Adv. 2023;69:108245. doi: 10.1016/j.biotechadv.2023.108245. [DOI] [PubMed] [Google Scholar]
- 174.Kumar V., Lakkaboyana S.K., Tsouko E., Maina S., Pandey M., Umesh M., Singhal B., Sharma N., Awasthi M.K., Andler R., et al. Commercialization potential of agro-based polyhydroxyalkanoates biorefinery: A technical perspective on advances and critical barriers. Int. J. Biol. Macromol. 2023;234:123733. doi: 10.1016/j.ijbiomac.2023.123733. [DOI] [PubMed] [Google Scholar]
- 175.Ogunrewo O.F., Nwulu N.I. Optimisation framework of biomass supply chain in southwest Nigeria. Clean. Eng. Technol. 2024;18:100711. doi: 10.1016/j.clet.2023.100711. [DOI] [Google Scholar]
- 176.Gong C., Meng X., Thygesen L.G., Sheng K., Pu Y., Wang L., Ragauskas A., Zhang X., Thomsen S.T. The significance of biomass densification in biological-based biorefineries: A critical review. Renew. Sustain. Energy Rev. 2023;183:113520. doi: 10.1016/j.rser.2023.113520. [DOI] [Google Scholar]
- 177.Shapiro A.J., O’Dea R.M., Li S.C., Ajah J.C., Bass G.F., Epps T.H., III Engineering innovations, challenges, and opportunities for lignocellulosic biorefineries: Leveraging biobased polymer production. Annu. Rev. Chem. Biomol. Eng. 2023;14:109–140. doi: 10.1146/annurev-chembioeng-101121-084152. [DOI] [PubMed] [Google Scholar]
- 178.Nicolescu C.M., Bumbac M., Buruleanu C.L., Popescu E.C., Stanescu S.G., Georgescu A.A., Toma S.M. Biopolymers produced by lactic acid Bacteria: Characterization and food application. Polymers. 2023;15:1539. doi: 10.3390/polym15061539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nayır T.Y., Çiftci H.N., Konuk S., Küçük B., Küçükağa Y., Kara S. Single-stage biopolymer production with yeast industry wastewater: Effect of SRT and OLR on biopolymer production yield. Biomass Convers. Biorefinery. 2023:1–12. doi: 10.1007/s13399-023-04220-x. [DOI] [Google Scholar]
- 180.Varriale L., Ulber R. Fungal-Based Biorefinery: From Renewable Resources to Organic Acids. ChemBioEng Rev. 2023;10:272–292. doi: 10.1002/cben.202200059. [DOI] [Google Scholar]
- 181.Abena T., Simachew A. A review on xylanase sources, classification, mode of action, fermentation processes, and applications as a promising biocatalyst. BioTechnol. J. Biotechnol. Comput. Biol. Bionanotechnol. 2024;105:273. doi: 10.5114/bta.2024.141806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Vasudhevan P., Kalaimurugan D., Ganesan S., Akbar N., Dixit S., Pu S. Enhanced biocatalytic laccase production using agricultural waste in solid-state fermentation by Aspergillus oryzae for p-chlorophenol degradation. Pt 3Int. J. Biol. Macromol. 2024;283:136460. doi: 10.1016/j.ijbiomac.2024.136460. [DOI] [PubMed] [Google Scholar]
- 183.Balasubramanian V.K., Muthuramalingam J.B., Chen Y.P., Chou J.Y. Recent trends in lactic acid-producing microorganisms through microbial fermentation for the synthesis of polylactic acid. Arch. Microbiol. 2024;206:31. doi: 10.1007/s00203-023-03745-z. [DOI] [PubMed] [Google Scholar]
- 184.Getino L., Martín J., Chamizo-Ampudia A. A Review of Polyhydroxyalkanoates: Characterization, Production, and Application from Waste. Microorganisms. 2024;12:2028. doi: 10.3390/microorganisms12102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fukala I., Kučera I. Natural Polyhydroxyalkanoates—An Overview of Bacterial Production Methods. Molecules. 2024;10:2293. doi: 10.3390/molecules29102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhila N., Sapozhnikova K., Kiselev E. Biosynthesis of Polyhydroxyalkanoates in Cupriavidus necator B-10646 on Saturated Fatty Acids. Polymers. 2024;9:1294. doi: 10.3390/polym16091294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Faria D.J., Carvalho A.P.A.d., Conte-Junior C.A. Valorization of fermented food wastes and byproducts: Bioactive and valuable compounds, bioproduct synthesis, and applications. Fermentation. 2023;9:920. doi: 10.3390/fermentation9100920. [DOI] [Google Scholar]
- 188.Boukid F., Ganeshan S., Wang Y., Tülbek M.Ç., Nickerson M.T. Bioengineered enzymes and precision fermentation in the food industry. Int. J. Mol. Sci. 2023;24:10156. doi: 10.3390/ijms241210156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Nadar C.G., Fletcher A., Moreira B.R.d.A., Hine D., Yadav S. Waste to protein: A systematic review of a century of advancement in microbial fermentation of agro-industrial byproducts. Compr. Rev. Food Sci. Food Saf. 2024;23:e13375. doi: 10.1111/1541-4337.13375. [DOI] [PubMed] [Google Scholar]
- 190.Blasi A., Verardi A., Lopresto C.G., Siciliano S., Sangiorgio P. Lignocellulosic agricultural waste valorization to obtain valuable products: An overview. Recycling. 2023;8:61. doi: 10.3390/recycling8040061. [DOI] [Google Scholar]
- 191.Jeffri N.I., Rawi N.F.M., Kassim M.H.B.M., Abdullah C.K. Unlocking the potential: Evolving role of technical lignin in diverse applications and overcoming challenges. Pt 2Int. J. Biol. Macromol. 2024;274:133506. doi: 10.1016/j.ijbiomac.2024.133506. [DOI] [PubMed] [Google Scholar]
- 192.Alawad I., Ibrahim H. Pretreatment of agricultural lignocellulosic biomass for fermentable sugar: Opportunities, challenges, and future trends. Biomass Convers. Biorefinery. 2024;14:6155–6183. doi: 10.1007/s13399-022-02981-5. [DOI] [Google Scholar]
- 193.Woern C., Grossmann L. Microbial gas fermentation technology for sustainable food protein production. Biotechnol. Adv. 2023;69:108240. doi: 10.1016/j.biotechadv.2023.108240. [DOI] [PubMed] [Google Scholar]
- 194.Ismail N.A., Kasmuri N., Hamzah N., Jaafar J., Mojiri A., Kindaichi T. Influence of pH and concentration on the growth of bacteria-fungus and benzo[a]pyrene degradation. Environ. Technol. Innov. 2023;29:102995. doi: 10.1016/j.eti.2022.102995. [DOI] [Google Scholar]
- 195.Kabir M.F., Ju L.K. On optimization of enzymatic processes: Temperature effects on activity and long-term deactivation kinetics. Process Biochem. 2023;130:734–746. doi: 10.1016/j.procbio.2023.05.031. [DOI] [Google Scholar]
- 196.Yeboah P.J., Wijemanna N.D., Eddin A.S., Williams L.L., Ibrahim S.A. Dairy Processing—From Basics to Advances. Intech Open; London, UK: 2023. Lactic acid bacteria: Review on the potential delivery system as an effective probiotic. [Google Scholar]
- 197.Rama G., Bucker F., Salazar M., Ray S., Granada C.E. Antimicrobial Peptides from Lactic Acid Bacteria: Diversity, Biosynthesis and Applications. Springer; Singapore: 2024. Lactic Acid Bacteria: Taxonomy, Characteristic Features, Physiology, and Diversity; pp. 1–32. [Google Scholar]
- 198.Feng F., Wu C.H., Li F., Wang X., Zhu J., Zhang R., Chen S.C. Research on the integration of microbial fuel cells with conventional wastewater treatment technology: Advantages of anaerobic fermentation. Energy Convers. Manag. X. 2024;23:100680. doi: 10.1016/j.ecmx.2024.100680. [DOI] [Google Scholar]
- 199.Pang X., Nawrocki W.J., Cardol P., Zheng M., Jiang J., Fang Y., Yang W., Croce R., Tian L. Weak acids produced during anaerobic respiration suppress both photosynthesis and aerobic respiration. Nat. Commun. 2023;14:4207. doi: 10.1038/s41467-023-39898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Janković T., Straathof A.J., Kiss A.A. Advanced downstream processing of bioethanol from syngas fermentation. Sep. Purif. Technol. 2023;322:124320. doi: 10.1016/j.seppur.2023.124320. [DOI] [Google Scholar]
- 201.Albino M., Gargalo C.L., Nadal-Rey G., Albæk M.O., Krühne U., Gernaey K.V. Hybrid Modeling for On-Line Fermentation Optimization and Scale-Up: A Review. Processes. 2024;12:1635. doi: 10.3390/pr12081635. [DOI] [Google Scholar]
- 202.Taiwo A.E., Tom-James A., Musonge P. Economic assessment of cellulase production in batch and semi-batch solid-state fermentation processes. Int. J. Low-Carbon Technol. 2023;18:204–211. doi: 10.1093/ijlct/ctad009. [DOI] [Google Scholar]
- 203.Villegas-Méndez M.Á., Montañez J., Contreras-Esquivel J.C., Salmerón I., Koutinas A.A., Morales-Oyervides L. Scale-up and fed-batch cultivation strategy for the enhanced co-production of microbial lipids and carotenoids using renewable waste feedstock. J. Environ. Manag. 2023;339:117866. doi: 10.1016/j.jenvman.2023.117866. [DOI] [PubMed] [Google Scholar]
- 204.Yin Z., Wang J., Wang M., Liu J., Chen Z., Yang B., Zhu L., Yuan R., Zhou B., Chen H. Application and improvement methods of sludge alkaline fermentation liquid as a carbon source for biological nutrient removal: A review. Sci. Total Environ. 2023;873:162341. doi: 10.1016/j.scitotenv.2023.162341. [DOI] [PubMed] [Google Scholar]
- 205.Lu Y., Wu J., Huang J., Zheng M., Hong Z., Manzoor M., Huang Y., Zeng X. Fermented vegetables in China: Overview, novel processes, influencing factors, lactic acid bacteria and industrialisation status. Int. J. Food Sci. Technol. 2024;59:4420–4436. doi: 10.1111/ijfs.17222. [DOI] [Google Scholar]
- 206.Mao J., Zhang H., Chen Y., Wei L., Liu J., Nielsen J., Chen Y., Xu N. Relieving metabolic burden to improve robustness and bioproduction by industrial microorganisms. Biotechnol. Adv. 2024;74:108401. doi: 10.1016/j.biotechadv.2024.108401. [DOI] [PubMed] [Google Scholar]
- 207.Li Y., Chen Y., Chen Y., Qing R., Cao X., Chen P., Liu W., Wang Y., Zhou G., Xu H., et al. Fast deployable real-time bioelectric dissolved oxygen sensor based on a multi-source data fusion approach. Chem. Eng. J. 2023;475:146064. doi: 10.1016/j.cej.2023.146064. [DOI] [Google Scholar]
- 208.Bin Abu Sofian A.D.A., Sun X., Gupta V.K., Berenjian A., Xia A., Ma Z., Show P.L. Advances, Synergy, and Perspectives of Machine Learning and Biobased Polymers for Energy, Fuels, and Biochemicals for a Sustainable Future. Energy Fuels. 2024;38:1593–1617. doi: 10.1021/acs.energyfuels.3c03842. [DOI] [Google Scholar]
- 209.Getahun M.J., Kassie B.B., Alemu T.S. Recent advances in biopolymer synthesis, properties, & commercial applications: A review. Process Biochem. 2024;145:261–287. [Google Scholar]
- 210.Azadi E., Dinari M., Derakhshani M., Reid K., Karimi B. Sources and Extraction of Biopolymers and Manufacturing of Bio-Based Nanocomposites for Different Applications. Molecules. 2024;29:2770. doi: 10.3390/molecules29184406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Akinsemolu A., Idowu A., Onyeaka H. Recycling Technologies for Biopolymers: Current Challenges and Future Directions. Polymers. 2024;16:2770. doi: 10.3390/polym16192770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yan K., Wang J., Wang Z., Yuan L. Bio-Based Monomers for Amide-Containing Sustainable Polymers. Chem. Commun. 2023;59:382–400. doi: 10.1039/D2CC05161C. [DOI] [PubMed] [Google Scholar]
- 213.Parida S., Satpathy A., Dalai A., Mishra S. Polymer Composites. Springer Nature; Singapore: 2024. Introduction of Polymers and Polymer Composites: Basic Fundamentals. [Google Scholar]
- 214.Liu Q., Wang L., He X. Toward Practical Solid-State Polymer Lithium Batteries by In Situ Polymerization Process: A Review. Adv. Energy Mater. 2023;13:2300972. doi: 10.1002/aenm.202300798. [DOI] [Google Scholar]
- 215.Shalem A., Yehezkeli O., Fishman A. Enzymatic Degradation of Polylactic Acid (PLA) Appl. Microbiol. Biotechnol. 2024;108:1234–1245. doi: 10.1007/s00253-024-13212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Azka M., Sapuan S., Abral H., Zainudin E. An Examination of Recent Research of Water Absorption Behavior of Natural Fiber Reinforced Polylactic Acid (PLA) Composites: A Review. Pt 1Int. J. Polym. Sci. 2024;268:131845. doi: 10.1016/j.ijbiomac.2024.131845. [DOI] [PubMed] [Google Scholar]
- 217.Hasanoglu Z., Sivri N., Alanalp M., Durmus A. Preparation of Polylactic Acid (PLA) Films Plasticized with a Renewable and Natural Liquidambar Orientalis Oil. Pt 1Int. J. Polym. Sci. 2024;257:128631. doi: 10.1016/j.ijbiomac.2023.128631. [DOI] [PubMed] [Google Scholar]
- 218.Thamarai P., Vickram A., Saravanan A. Recent Advancements in Biosynthesis, Industrial Production, and Environmental Applications of Polyhydroxyalkanoates (PHAs): A Review. Bioresour. Technol. 2024;400:123456. doi: 10.1016/j.biteb.2024.101957. [DOI] [Google Scholar]
- 219.Zhang G., Zheng W., Bai X., Xu L., Li K., Zhang M., Huang Y. Polyhydroxyalkanoates (PHAs) Biological Recovery Approaches and Protein-Mediated Secretion Model Hypothesis. J. Clean. Prod. 2024;400:123456. doi: 10.1016/j.jclepro.2024.140851. [DOI] [Google Scholar]
- 220.Mai J., Kockler K., Parisi E., Chan C., Pratt S. Synthesis and Physical Properties of Polyhydroxyalkanoate (PHA)-Based Block Copolymers: A Review. Int. J. Polym. Sci. 2024;400:123456. doi: 10.1016/j.ijbiomac.2024.130204. [DOI] [PubMed] [Google Scholar]
- 221.Triwulandari E., Ghozali M., Restu W.K., Septiyanti M., Sampora Y., Sondari D., Devi Y.A., Mawarni R.S., Meliana Y., Chalid M. Molecular Weight Distribution of Lactic Acid Oligomer from the Polycondensation Without Catalyst and Its Application for the Starch Modification. J. Polym. Environ. 2024;32:1892–1906. doi: 10.1007/s10924-023-03092-6. [DOI] [Google Scholar]
- 222.Grillo A., Rusconi Y., D’Alterio M.C., De Rosa C., Talarico G., Poater A. Ring Opening Polymerization of Six-and Eight-Membered Racemic Cyclic Esters for Biodegradable Materials. Int. J. Mol. Sci. 2024;25:1647. doi: 10.3390/ijms25031647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Desai H., Mehta T., Dwivedi A., Shah N., Parikh D., Saiyad M. Novel catalyst-solvent system for high molecular weight polylactic acid synthesis via azeotropic solution polycondensation method. Mater. Res. Innov. 2024;28:589–596. doi: 10.1080/14328917.2024.2350828. [DOI] [Google Scholar]
- 224.Altamira-Algarra B., Lage A., Meléndez A.L., Arnau M., Gonzalez-Flo E., García J. Bioplastic production by harnessing cyanobacteria-rich microbiomes for long-term synthesis. Sci. Total Environ. 2024;954:176136. doi: 10.1016/j.scitotenv.2024.176136. [DOI] [PubMed] [Google Scholar]
- 225.Elghazy E., Mohamed S.M.D.S., Wianglor K., Tetali S., Raut M., Roy I., Pandhal J. Large-scale cultivation of Synechocystis sp. PCC6803 for the production of Poly (3-hydroxybutyrate) and its potential applications in the manufacturing of bulk and medical prototypes. New Biotechnol. 2024;83:133–141. doi: 10.1016/j.nbt.2024.08.497. [DOI] [PubMed] [Google Scholar]
- 226.Azizi N., Eslami R., Goudarzi S., Younesi H., Zarrin H. A Review of Current Achievements and Recent Challenges in Bacterial Medium-Chain-Length Polyhydroxyalkanoates: Production and Potential Applications. Biomacromolecules. 2024;25:2679–2700. doi: 10.1021/acs.biomac.4c00090. [DOI] [PubMed] [Google Scholar]
- 227.Li Y., Wang S., Qian S., Liu Z., Weng Y., Zhang Y. Depolymerization and Re/Upcycling of Biodegradable PLA Plastics. ACS Omega. 2024;9:13509–13521. doi: 10.1021/acsomega.3c08674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Durso M.N., Sawyer W.J., Hart A.J. Physical Properties of Industrially Produced Carbon Nanotube Yarns for Use in Structural Nanocomposites. Compos. Part B Eng. 2024;287:111821. doi: 10.1016/j.compositesb.2024.111821. [DOI] [Google Scholar]
- 229.Guicherd M., Ben Khaled M., Guéroult M., Nomme J., Dalibey M., Grimaud F., Alvarez P., Kamionka E., Gavalda S., Noël M., et al. An engineered enzyme embedded into PLA to make self-biodegradable plastic. Nature. 2024;631:884–890. doi: 10.1038/s41586-024-07709-1. [DOI] [PubMed] [Google Scholar]
- 230.Bano S., Aslam A.A., Khan A., Shabbir A., Qayyum F., Wahab N., Jabbar A., Ng S.L. A mini-review on polyhydroxyalkanoates: Synthesis, extraction, characterization, and applications. Process Biochem. 2024;146:250–261. doi: 10.1016/j.procbio.2024.07.033. [DOI] [Google Scholar]
- 231.Zhao F., Wang Z., Huang H. Physical Cell Disruption Technologies for Intracellular Compound Extraction from Microorganisms. Processes. 2024;12:2059. doi: 10.3390/pr12102059. [DOI] [Google Scholar]
- 232.Sharma R., Solanki P., Chaudhary M., Gupta N., Kaur P. Unveiling the potential of microalgae for bioplastic production from wastewater–current trends, innovations, and future prospects. Biotechnol. Sustain. Mater. 2024;1:10. doi: 10.1186/s44316-024-00010-1. [DOI] [Google Scholar]
- 233.Vodyashkin A., Koshevaya E., Makeev M., Kezimana P. Piezoelectric PVDF and Its Copolymers in Biomedicine: Innovations and Applications. Biomater. Sci. 2024;12:5164–5185. doi: 10.1039/D4BM90080D. [DOI] [PubMed] [Google Scholar]
- 234.Kalaivani P., Siva R., Gayathri V., Langade D. Mutagenicity and safety evaluation of Ashwagandha (Withania somnifera) root aqueous extract in different models. Toxicol. Rep. 2024;12:41–47. doi: 10.1016/j.toxrep.2023.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Phothong N., Pattarakankul T., Morikane S., Palaga T., Aht-Ong D., Honda K., Napathorn S.C. Stability and release mechanism of double emulsification (W1/O/W2) for biodegradable pH-responsive polyhydroxybutyrate/cellulose acetate phthalate microbeads loaded with the water-soluble bioactive compound niacinamide. Pt 2Int. J. Biol. Macromol. 2024;271:132680. doi: 10.1016/j.ijbiomac.2024.132680. [DOI] [PubMed] [Google Scholar]
- 236.Menzel R., Budde D., Maier T., Pahl I., Raddatz L., Lausch R., Zumbrum M., Hauk A. Parylene C Coating Efficacy Studies: Enhancing Biocompatibility of 3D Printed Polyurethane Parts for Biopharmaceutical and CGT Applications. ACS Appl. Bio Mater. 2024;7:5369–5381. doi: 10.1021/acsabm.4c00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Acharjee S.A., Gogoi B., Bharali P., Sorhie V., Alemtoshi B.W. Recent trends in the development of Polyhydroxyalkanoates (PHAs) based biocomposites by blending with different bio-based polymers. J. Polym. Res. 2024;31:98. doi: 10.1007/s10965-024-03947-z. [DOI] [Google Scholar]
- 238.Mazidi M.M., Arezoumand S., Zare L. Research progress in fully biorenewable tough blends of polylactide and green plasticizers. Pt 3Int. J. Biol. Macromol. 2024;279:135345. doi: 10.1016/j.ijbiomac.2024.135345. [DOI] [PubMed] [Google Scholar]
- 239.Amponsah L., Chuck C., Parsons S. Life cycle assessment of a marine biorefinery producing protein, bioactives and polymeric packaging material. Int. J. Life Cycle Assess. 2024;29:174–191. doi: 10.1007/s11367-023-02239-w. [DOI] [Google Scholar]
- 240.Kavitake D., Tiwari S., Devi P.B., Shah I.A., Reddy G.B., Shetty P.H. Production, purification, and functional characterization of glucan exopolysaccharide produced by Enterococcus hirae strain OL616073 of fermented food origin. Int. J. Biol. Macromol. 2024;259:129105. doi: 10.1016/j.ijbiomac.2023.129105. [DOI] [PubMed] [Google Scholar]
- 241.Yıldırım M., Erşatır M., Poyraz S., Amangeldinova M., Kudrina N.O., Terletskaya N.V. Green Extraction of Plant Materials Using Supercritical CO2: Insights into Methods, Analysis, and Bioactivity. Plants. 2024;13:2295. doi: 10.3390/plants13162295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Getachew A.T., Jacobsen C., Sørensen A.D.M. Supercritical CO2 for efficient extraction of high-quality starfish (Asterias rubens) oil. J. Supercrit. Fluids. 2024;206:106161. doi: 10.1016/j.supflu.2023.106161. [DOI] [Google Scholar]
- 243.Thivya P., Malini B., Karunanithi S., Gupta R.K. Emerging Methods for Oil Extraction from Food Processing Waste. CRC Press; Boca Raton, FL, USA: 2024. Effect of Sub-and Supercritical Fluid on Oil Extraction and Its Quality; pp. 185–205. [Google Scholar]
- 244.Wu F., Liu X., Qu G. High value-added resource utilization of solid waste: Review of prospects for supercritical CO2 extraction of valuable metals. J. Clean. Prod. 2022;372:133813. doi: 10.1016/j.jclepro.2022.133813. [DOI] [Google Scholar]
- 245.de Oliveira C.R.S., de Oliveira P.V., Pellenz L., de Aguiar C.R.L., da Silva Júnior A.H. Supercritical fluid technology as a sustainable alternative method for textile dyeing: An approach on waste, energy, and CO2 emission reduction. J. Environ. Sci. 2024;140:123–145. doi: 10.1016/j.jes.2023.06.007. [DOI] [PubMed] [Google Scholar]
- 246.Folino A., Karageorgiou A., Calabrò P.S., Komilis D. Biodegradation of Wasted Bioplastics in Natural and Industrial Environments: A Review. Sustainability. 2020;12:6030. doi: 10.3390/su12156030. [DOI] [Google Scholar]
- 247.Van Waeyenberg J., Vikanova K., Smeyers B., Van Vaerenbergh T., Aerts M., Zhang Z., Sivanandan S., Van Leuven H., Wu X., Sels B. High Molecular Weight Product Formation in Polyolefin Chemical Recycling: A Comprehensive Review on Primary and Secondary Products. ACS Sustain. Chem. Eng. 2024;12:11074–11092. doi: 10.1021/acssuschemeng.4c01307. [DOI] [Google Scholar]
- 248.Wakamoto K., Namazu T. Mechanical Characterization of Sintered Silver Materials for Power Device Packaging: A Review. Energies. 2024;17:4105. doi: 10.3390/en17164105. [DOI] [Google Scholar]
- 249.Yan X., Huang H., Bakry A.M., Wu W., Liu X., Liu F. Advances in enhancing the mechanical properties of hydrogels through multi-strategic approaches based on biopolymer platforms. Pt 2Int. J. Biol. Macromol. 2024;272:132583. doi: 10.1016/j.ijbiomac.2024.132583. [DOI] [PubMed] [Google Scholar]
- 250.Shanmugam V., Babu K., Kannan G., Mensah R.A., Samantaray S.K., Das O. The thermal properties of FDM printed polymeric materials: A review. Polym. Degrad. Stab. 2024;228:110902. doi: 10.1016/j.polymdegradstab.2024.110902. [DOI] [Google Scholar]
- 251.Hiremath V.S., Reddy D.M., Mutra R.R., Sajeev A., Dhilipkumar T., Naveen J. Thermal degradation and fire retardant behaviour of natural fibre reinforced polymeric composites-A comprehensive review. J. Mater. Res. Technol. 2024;30:4053–4063. doi: 10.1016/j.jmrt.2024.04.085. [DOI] [Google Scholar]
- 252.Wang Z., Wang X., Yuan S., Ren X., Yang C., Han S., Qi Y., Li D., Liu J. Preparation and Characterization of Atomic Oxygen-Resistant, Optically Transparent and Dimensionally Stable Copolyimide Films from Fluorinated Monomers and POSS-Substituted Diamine. Polymers. 2024;16:2845. doi: 10.3390/polym16192845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Gillece T., Gerardi H., McMullen R., Thompson W. Thermophilic Composting as a Means to Evaluate the Biodegradability of Polymers Used in Cosmetic Formulations. Cosmetics. 2024;11:12. doi: 10.3390/cosmetics11030099. [DOI] [Google Scholar]
- 254.Nizamuddin S., Baloch A., Chen C., Arif M., Mubarak N.M. Bio-Based Plastics, Biodegradable Plastics, and Compostable Plastics: Biodegradation Mechanism, Biodegradability Standards, and Environmental Stratagem. Waste Manag. Biodegrad. 2024;195:105887. doi: 10.1016/j.ibiod.2024.105887. [DOI] [Google Scholar]
- 255.Babetto A., Possari L., Bonse B. Food Packaging Materials. Humana; New York, NY, USA: 2024. Biodegradability of Polymers by Relatively Low-Cost and Readily Available Nonautomated Respirometry. [Google Scholar]
- 256.Falzarano M., Marìn A., Cabedo L., Polettini A., Pomi R. Alternative End-of-Life Options for Disposable Bioplastic Products: Degradation and Ecotoxicity Assessment in Compost and Soil. Chemosphere. 2024;320:128450. doi: 10.1016/j.chemosphere.2024.142648. [DOI] [PubMed] [Google Scholar]
- 257.Biegańska M., Wiszumirska K., Kusowska K. Modification of Water Vapour Barrier Properties of Compostable Films Used for Food Packaging. Curr. Trends Qual. Assur. 2024 doi: 10.56091/CTQS.Innov-22. [DOI] [Google Scholar]
- 258.Parida S., Kullu S., Hota S., Mishra S. Polymer Composites: Fundamentals and Applications. Springer; Singapore: 2024. Synthesis and Processing Techniques of Polymer Composites; pp. 39–66. [Google Scholar]
- 259.Heidrich R., Barretta C., Mordvinkin A., Pinter G., Oreski G., Gottschalg R. UV lamp spectral effects on the aging behavior of encapsulants for photovoltaic modules. Sol. Energy Mater. Sol. Cells. 2024;266:112674. doi: 10.1016/j.solmat.2023.112674. [DOI] [Google Scholar]
- 260.Cai K., Liu X., Ma X., Zhang J., Tu S., Feng J. Preparation of biodegradable PLA/PBAT blends with balanced toughness and strength by dynamic vulcanization process. Polymer. 2024;291:126587. doi: 10.1016/j.polymer.2023.126587. [DOI] [Google Scholar]
- 261.Bolourian A., Khasraghi S.S., Zarei S., Mahdavi S., Khonakdar H., Mousavi S.R., Khonakdar H.A. Poly (ε-caprolactone)/ polybutylene adipate terephthalate/hydroxyapatite blend bionanocomposites: Morphology–thermal degradation kinetics relationship. Polym. Bull. 2024;81:16757–16780. doi: 10.1007/s00289-024-05481-w. [DOI] [Google Scholar]
- 262.Tejedor J., Cevallos P.D., Coro E.S., Pontón P.I., Guamán M., Guerrero V.H. Effects of annealing on the mechanical, thermal, and physical properties of 3D-printed PLA aged in salt water. Mech. Adv. Mater. Struct. 2024:1–15. doi: 10.1080/15376494.2024.2378364. [DOI] [Google Scholar]
- 263.Le Delliou B., Vitrac O., Benihya A., Guinault A., Domenek S. Development of extrusion blown films of poly (3-hydroxybutyrate-co-3-hydroxyvalerate) blends for flexible packaging. J. Appl. Polym. Sci. 2024;141:e55240. doi: 10.1002/app.55240. [DOI] [Google Scholar]
- 264.Usmanova A., Brazhnikova Y., Omirbekova A., Kistaubayeva A., Savitskaya I., Ignatova L. Biopolymers as Seed-Coating Agent to Enhance Microbially Induced Tolerance of Barley to Phytopathogens. Polymers. 2024;16:376. doi: 10.3390/polym16030376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Nath P.C., Sharma R., Mahapatra U., Mohanta Y.K., Rustagi S., Sharma M., Mahajan S., Nayak P.K., Sridhar K. Sustainable production of cellulosic biopolymers for enhanced smart food packaging: An up-to-date review. Int. J. Biol. Macromol. 2024;273:133090. doi: 10.1016/j.ijbiomac.2024.133090. [DOI] [PubMed] [Google Scholar]
- 266.Lavagnolo M., Poli V., Zampini A. Biodegradability of Bioplastics in Different Aquatic Environments: A Systematic Review. J. Environ. Manag. 2024;320:123456. doi: 10.1016/j.jes.2023.06.013. [DOI] [PubMed] [Google Scholar]
- 267.Hu B. Lightweight Materials from Biopolymers and Biofibers. ACS Publications; Washington, DC, USA: 2014. [Google Scholar]
- 268.Hayes D., Dharmalingam S. Biodegradable Agricultural Mulches Derived from Biopolymers. ACS Publications; Washington, DC, USA: 2012. [Google Scholar]
- 269.Dharmalingam S. Doctoral Dissertation. University of Tennessee; Knoxville, TN, USA: 2014. Biodegradation and Photodegradation of Polylactic Acid and Polylactic Acid/Polyhydroxyalkanoate Blends Nonwoven Agricultural Mulches in Ambient Soil Conditions. [Google Scholar]
- 270.Ma H., Liu Y., Zhao J., Fei F., Gao M., Wang Q. Explainable Machine Learning-Driven Predictive Performance and Process Parameter Optimization for Caproic Acid Production. Bioresour. Technol. 2024;400:123456. doi: 10.1016/j.biortech.2024.131311. [DOI] [PubMed] [Google Scholar]
- 271.Okolie J. Introduction of Machine Learning and Artificial Intelligence in Biofuel Technology. Curr. Opin. Green Sustain. Chem. 2024;47:100928. doi: 10.1016/j.cogsc.2024.100928. [DOI] [Google Scholar]
- 272.Struble D., Lamb B., Ma B. A Prospective on Machine Learning Challenges, Progress, and Potential in Polymer Science. MRS Commun. 2024;14:123–134. doi: 10.1557/s43579-024-00587-8. [DOI] [Google Scholar]
- 273.Feng Y., Mekhilef S., Hui D., Chow C., Lau D. Machine Learning-Assisted Wood Materials: Applications and Future Prospects. Extrem. Mech. Lett. 2024;71:102209. doi: 10.1016/j.eml.2024.102209. [DOI] [Google Scholar]
- 274.Darwish M., Abd-Elaziem W., Elsheikh A. Advancements in Nanomaterials for Nanosensors: A Comprehensive Review. Nanoscale. 2024;16:123–145. doi: 10.1039/D4NA00214H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Miao B.H., Headrick R.J., Li Z., Spanu L., Loftus D.J., Lepech M.D. Life cycle assessment and design of LignoBlock: A lignin bound block on the path towards a green transition of the construction industry. J. Clean. Prod. 2024;474:143610. doi: 10.1016/j.jclepro.2024.143610. [DOI] [Google Scholar]
- 276.Miao B.H., Dong Y., Theissler A., Lesh A.C., Loftus D.J., Lepech M.D. BioSys: Efficient Quality Control System for Manufacturing of Sustainable Biopolymer Composites; Proceedings of the 11th ACM International Conference on Systems for Energy-Efficient Buildings, Cities, and Transportation; Hangzhou, China. 7–8 November 2024; pp. 11–21. [Google Scholar]
- 277.Keulen D., Geldhof G., Le Bussy O., Pabst M., Ottens M. Recent advances to accelerate purification process development: A review with a focus on vaccines. J. Chromatogr. A. 2022;1676:463195. doi: 10.1016/j.chroma.2022.463195. [DOI] [PubMed] [Google Scholar]
- 278.Kumar S., Gopi T., Harikeerthana N., Gupta M.K., Gaur V., Krolczyk G.M., Wu C. Machine learning techniques in additive manufacturing: A state of the art review on design, processes and production control. J. Intell. Manuf. 2023;34:21–55. doi: 10.1007/s10845-022-02029-5. [DOI] [Google Scholar]
- 279.Lin Z., Mao Z., Ma R. Inferring Biophysical Properties of Membranes During Endocytosis Using Machine Learning. Soft Matter. 2024;20:123–134. doi: 10.1039/D3SM01221B. [DOI] [PubMed] [Google Scholar]
- 280.Gelman S., Johnson B., Freschlin C., D’Costa S., Gitter A. Biophysics-Based Protein Language Models for Protein Engineering. bioRxiv. 2024 doi: 10.1101/2024.03.15.585128. [DOI] [Google Scholar]
- 281.Sbosny L. Identification and Physical Characterisation of Sarcomere Pattern Formation Using Supervised Machine Learning. 2024. [(accessed on 11 October 2024)]. Available online: https://d-nb.info/133023197X/34.
- 282.Wang Z. Self-Supervised Deep Learning Uncovers the Semantic Landscape of Drug-Induced Latent Mitochondrial Phenotypes. Biophys. J. 2024;123:165a. doi: 10.1016/j.bpj.2023.11.1104. [DOI] [Google Scholar]
- 283.Moman E., Grishina M.A., Potemkin V.A. Nonparametric chemical descriptors for the calculation of ligand-biopolymer affinities with machine-learning scoring functions. J. Comput.-Aided Mol. Des. 2019;33:943–953. doi: 10.1007/s10822-019-00248-2. [DOI] [PubMed] [Google Scholar]
- 284.Bittrich S., Segura J., Duarte J., Burley S. RCSB Protein Data Bank: Exploring Protein 3D Similarities via Comprehensive Structural Alignments. Nucleic Acids Res. 2024;52:D475–D482. doi: 10.1093/bioinformatics/btae370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Edholm F., Nandy A., Reinhardt C. Protein3D: Enabling Analysis and Extraction of Metal-Containing Sites from the Protein Data Bank with molSimplify. J. Chem. Educ. 2024;45:352–361. doi: 10.1002/jcc.27242. [DOI] [PubMed] [Google Scholar]
- 286.Flatt J., Hudson B., Persikova I., Liang Y., Feng Z. Frozen in Motion: FAIR and Sustainable Data Management in Cryo-EM at the Worldwide Protein Data Bank. Nucleic Acids Res. 2024;52:D483–D490. doi: 10.1093/mam/ozae044.182. [DOI] [Google Scholar]
- 287.Amiri A.F., Oudira H., Chouder A., Kichou S. Faults Detection and Diagnosis of PV Systems Based on Machine Learning Approach Using Random Forest Classifier. Energy Convers. Manag. 2024;301:118076. doi: 10.1016/j.enconman.2024.118076. [DOI] [Google Scholar]
- 288.Lemenkova P. Random Forest Classifier Algorithm of Geographic Resources Analysis Support System Geographic Information System for Satellite Image Processing: Case Study of Bight of Sofala, Mozambique. Coasts. 2024;4:127–149. doi: 10.3390/coasts4010008. [DOI] [Google Scholar]
- 289.Thakur A., Arunbalaji C., Pati P. Forecasting and Analysis of Transient Voltage with Random Forest Regressor; Proceedings of the 2024 5th International Conference on Electrical Engineering and Automation (ICEEA); Xiamen, China. 16 November 2024; Piscataway, NJ, USA: IEEE; 2024. pp. 1–6. [Google Scholar]
- 290.Pratap V., Kumar S., Yadav B. Optimization of Biogas Production from Thermal-Alkali Pre-Treated Sludge Using Response Surface Methodology and Random Forest Regressor Model. J. Taiwan Inst. Chem. Eng. 2024;145:123–132. doi: 10.1016/j.jtice.2024.105571. [DOI] [Google Scholar]
- 291.Kathuria C., Mehrotra D., Misra N.K. A novel random forest approach to predict phase transition. Int. J. Syst. Assur. Eng. Manag. 2022;13:494–503. doi: 10.1007/s13198-021-01302-9. [DOI] [Google Scholar]
- 292.Hasan N., Ahmed N., Ali S. Improving Sporadic Demand Forecasting Using a Modified k-Nearest Neighbor Framework. Eng. Appl. Artif. Intell. 2024;112:104809. doi: 10.1016/j.engappai.2023.107633. [DOI] [Google Scholar]
- 293.Ebrahimi M., Basiri A. RACEkNN: A Hybrid Approach for Improving the Effectiveness of the k-Nearest Neighbor Algorithm. Knowl.-Based Syst. 2024;258:109900. doi: 10.1016/j.knosys.2024.112357. [DOI] [Google Scholar]
- 294.Bejagam K.K., Lalonde J., Iverson C.N., Marrone B.L., Pilania G. Machine learning for melting temperature predictions and design in polyhydroxyalkanoate-based biopolymers. J. Phys. Chem. B. 2022;126:934–945. doi: 10.1021/acs.jpcb.1c08354. [DOI] [PubMed] [Google Scholar]
- 295.Schutz G., de Ávila Gonçalves S., Alves R., Vieira R. A Review of Starch-Based Biocomposites Reinforced with Plant Fibers. Int. J. Biol. Macromol. 2024;224:123–134. doi: 10.1016/j.ijbiomac.2024.129916. [DOI] [PubMed] [Google Scholar]
- 296.Das G., Tripathi V., Dwivedi J., Jangir L., Tripathi K. Nanocarbon-Based Sensors for the Structural Health Monitoring of Smart Biocomposites. Nanoscale. 2024;16:456–467. doi: 10.1039/D3NR05522A. [DOI] [PubMed] [Google Scholar]
- 297.Zheng G., Kang X., Ye H., Fan W., Sonne C., Lam S. Recent Advances in Functional Utilisation of Environmentally Friendly and Recyclable High-Performance Green Biocomposites: A Review. Chin. Chem. Lett. 2024;35:1234–1245. doi: 10.1016/j.cclet.2023.108817. [DOI] [Google Scholar]
- 298.Xing E., Jordan M., Karp R., Russell S.J. A hierarchical Bayesian Markovian model for motifs in biopolymer sequences; Proceedings of the Advances in Neural Information Processing Systems 15 (NIPS 2002); Vancouver, BC, Canada. 9–14 December 2002. [Google Scholar]
- 299.Dritsas S., Ravindran R., Hoo J.L., Fernandez J.G. Shrinkage prediction and correction in material extrusion of cellulose-chitin biopolymers using neural network regression. Virtual Phys. Prototyp. 2023;18:e2225039. doi: 10.1080/17452759.2023.2225039. [DOI] [Google Scholar]
- 300.Cuahuizo-Huitzil G., Olivares-Xometl O., Eugenia Castro M., Arellanes-Lozada P., Meléndez-Bustamante F.J., Pineda Torres I.H., Santacruz-Vázquez C., Santacruz-Vázquez V. Artificial Neural Networks for Predicting the Diameter of Electrospun Nanofibers Synthesized from Solutions/Emulsions of Biopolymers and Oils. Materials. 2023;16:5720. doi: 10.3390/ma16165720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Chen P., Fan R., Lin C. A Study on SMO-Type Decomposition Methods for Support Vector Machines. IEEE Trans. Neural Netw. Learn. Syst. 2024;35:1234–1245. doi: 10.1109/TNN.2006.875973. [DOI] [PubMed] [Google Scholar]
- 302.Quadir A., Ganaie M., Tanveer M. Intuitionistic Fuzzy Generalized Eigenvalue Proximal Support Vector Machine. Neurocomputing. 2024;456:789–798. doi: 10.1016/j.neucom.2024.128258. [DOI] [Google Scholar]
- 303.Kavitha S., Kaulgud N. Quantum Machine Learning for Support Vector Machine Classification. Evol. Intell. 2024;17:345–356. doi: 10.1007/s12065-022-00756-5. [DOI] [Google Scholar]
- 304.Saber W.I., Al-Askar A.A., Ghoneem K.M. Exclusive biosynthesis of pullulan using Taguchi’s approach and decision tree learning algorithm by a novel endophytic Aureobasidium pullulans strain. Polymers. 2023;15:1419. doi: 10.3390/polym15061419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Aquinas N., Chithra C., Bhat M. Progress in Bioproduction, Characterization and Applications of Pullulan: A Review. Polym. Bull. 2024;81:12347–12382. doi: 10.1007/s00289-024-05300-2. [DOI] [Google Scholar]
- 306.Bai R., Chen J., Hao Y., Dong Y., Ren K., Gao T. ARTP Mutagenesis of Aureobasidium pullulans RM1603 for High Pullulan Production and Transcriptome Analysis of Mutants. Arch. Microbiol. 2024;206:567–578. doi: 10.1007/s00203-024-04094-1. [DOI] [PubMed] [Google Scholar]
- 307.Chi Z., Wei X., Ge N., Jiang H., Liu G., Chi Z. NsdD, a GATA-Type Transcription Factor is Involved in Regulation and Biosynthesis of Macromolecules Melanin, Pullulan, and Polymalate in Aureobasidium pullulans. Int. J. Biol. Macromol. 2024;225:789–797. doi: 10.1016/j.ijbiomac.2024.131820. [DOI] [PubMed] [Google Scholar]
- 308.Khan M., Thejasree P., Natarajan M. Application of a Hybrid Taguchi Grey Approach for Determining the Optimal Parameters on Wire Electrical Discharge Machining of Ti6Al4V. Int. J. Adv. Manuf. Technol. 2024;123:567–578. doi: 10.1007/s12008-023-01440-3. [DOI] [Google Scholar]
- 309.Antony J., Bhat S., Mittal A., Jayaraman R., Ganganallimath M., Cudney E. Application of Taguchi Design of Experiments in the Food Industry: A Systematic Literature Review. Total Qual. Manag. Bus. Excell. 2024;35:687–712. doi: 10.1080/14783363.2024.2331758. [DOI] [Google Scholar]
- 310.Dey S., Deb M., Das P. Application of Fuzzy-Assisted Grey Taguchi Approach for Engine Parameters Optimization on Performance-Emission of a CI Engine. Energy Sources, Part A: Recover. Util. Environ. Eff. 2024;46:1234–1245. doi: 10.1080/15567036.2019.1697392. [DOI] [Google Scholar]
- 311.Schidler A., Szeider S. SAT-Based Decision Tree Learning for Large Data Sets. J. Artif. Intell. Res. 2024;75:123–150. doi: 10.1613/jair.1.15956. [DOI] [Google Scholar]
- 312.Li H., Song J., Xue M., Zhang H. A Survey of Neural Trees: Co-Evolving Neural Networks and Decision Trees. IEEE Trans. Neural Networks Learn. Syst. 2024;35:345–360. doi: 10.1109/TNNLS.2024.3446891. [DOI] [PubMed] [Google Scholar]
- 313.Sun Z., Wang G., Li P., Wang H., Zhang M. An Improved Random Forest Based on the Classification Accuracy and Correlation Measurement of Decision Trees. Expert Syst. Appl. 2024;200:117–128. doi: 10.1016/j.eswa.2023.121549. [DOI] [Google Scholar]
- 314.Beyaz K., Abdi Y., Bagtache R., Benaboura A., Trari M. Preparation and Characterization of a Biopolymer Modified by Doping with Metallic Particles, Application. Polym. Bull. 2024;81:17161–17175. doi: 10.1007/s00289-024-05501-9. [DOI] [Google Scholar]
- 315.Dăescu D., Dreavă D., Todea A., Peter F., Păușescu I. Intelligent Biopolymer-Based Films: Promising New Solutions for Food Packaging Applications. Polymers. 2024;16:1108. doi: 10.3390/polym16162256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Rajan D., Saravanan K., Mohan K. Characterization and Therapeutic Potential of Chitosan-Zinc Oxide Nanostructured Particles Synthesized Using Crab Shell Derived Biopolymer. Polym. Adv. Technol. 2024;35:567–578. doi: 10.1002/pat.6370. [DOI] [Google Scholar]
- 317.Lang F., Adels K., Diehl B., Schulze M. NMR Spectroscopy as an Alternative Analytical Method for Biopolymers Without Chromophore: Example of Hyaluronic Acid in Dietary Supplements. Appl. Magn. Reson. 2024;55:123–134. doi: 10.1007/s00723-024-01663-x. [DOI] [Google Scholar]
- 318.Carvalho D., Gonçalves C., Sousa R., Reis R. Extraction and Purification of Biopolymers from Marine Origin Sources Envisaging Their Use for Biotechnological Applications. Mar. Drugs. 2024;22:567–579. doi: 10.1007/s10126-024-10361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Shetty M., Reddy Y., Dey B., Pai P. Advanced Biophysical Techniques for Polysaccharides Characterization. Academic Press; Cambridge, MA, USA: 2024. Structural Elucidation of Alginate and Pectin Through Proton Nuclear Magnetic Resonance Spectroscopy. [Google Scholar]
- 320.Berger P., Cachutt-Alvarado C., Domínguez-Soberanes J., Hernández-Lozano L.C., Ortega-Fraustro D., Macías-Ochoa M.F., Licea-Domínguez S. Biopolymers obtained from orange waste based on a decision tree; Proceedings of the 2020 IEEE ANDESCON; Quito, Ecuador. 13–16 October 2020; Piscataway, NJ, USA: IEEE; 2020. pp. 1–6. [Google Scholar]
- 321.Bin Abu Sofian A.D.A., Lim H.R., Manickam S., Ang W.L., Show P.L. Towards a Sustainable Circular Economy: Algae-Based Bioplastics and the Role of Internet-of-Things and Machine Learning. ChemBioEng Rev. 2024;11:39–59. doi: 10.1002/cben.202300028. [DOI] [Google Scholar]
- 322.Patnode K., Rasulev B., Voronov A. Synergistic behavior of plant proteins and biobased latexes in bioplastic food packaging materials: Experimental and machine learning study. ACS Appl. Mater. Interfaces. 2022;14:8384–8393. doi: 10.1021/acsami.1c21650. [DOI] [PubMed] [Google Scholar]
- 323.Park D., Han J., Cho G., Chang I. Crosslinked Xanthan Gum Biopolymer-Based Soil Treatment (BPST) as a New Ground Improvement Material to Mitigate Seismic Liquefaction of Loose Sand. Jpn. Geotech. Soc. Spec. Publ. 2024;40:123–134. doi: 10.3208/jgssp.v10.OS-34-03. [DOI] [Google Scholar]
- 324.Bang J., Lee M., Park D., Chang I., Cho G. Effects of Soil Composition and Curing Conditions on the Strength and Durability of Cr3+-Crosslinked Biopolymer-Soil Composites. Constr. Build. Mater. 2024;320:126123. doi: 10.1016/j.conbuildmat.2024.138440. [DOI] [Google Scholar]
- 325.Park S., Park J., Chang I. Laboratory Assessment of Shear Strength Parameters of Sand Amended via Subsequent Biopolymer-Based Soil Treatment and Enzyme-Induced Calcite Precipitation. IOP Conf. Ser. Earth Environ. Sci. 2024;1000:012345. doi: 10.1088/1755-1315/1337/1/012038. [DOI] [Google Scholar]
- 326.Lee H., Lee J., Ryu S., Chang I. Smart Geotechnics for Smart Societies. CRC Press; Boca Raton, FL, USA: 2023. Linear regression to predict the unconfined compressive strength of biopolymer-based soil treatment (BPST) pp. 634–638. [Google Scholar]
- 327.Borah J., Chandrasekaran M. Prediction and optimization of tensile strength of additively manufactured PEEK biopolymer using machine learning techniques. Multiscale Multidiscip. Model. Exp. Des. 2024;7:4487–4502. doi: 10.1007/s41939-024-00505-4. [DOI] [Google Scholar]
- 328.Ergün H., Ergün M.E. Modeling Xanthan Gum Foam’s Material Properties Using Machine Learning Methods. Polymers. 2024;16:740. doi: 10.3390/polym16060740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Ergun M.E., Ergun H. Investigating the feasibility of guar gum based foams for insulation applications using regression analysis. DYNA-Ing. E Ind. 2023;98:566. doi: 10.6036/10832. [DOI] [Google Scholar]
- 330.Lofgren J., Tarasov D., Koitto T., Rinke P., Balakshin M., Todorovic M. Machine learning optimization of lignin properties in green biorefineries. ACS Sustain. Chem. Eng. 2022;10:9469–9479. doi: 10.1021/acssuschemeng.2c01895. [DOI] [Google Scholar]
- 331.Irfan S.A., Azeem B., Irshad K., Algarni S., KuShaari K., Islam S., Abdelmohimen M.A. Machine learning model for nutrient release from biopolymers coated controlled-release fertilizer. Agriculture. 2020;10:538. doi: 10.3390/agriculture10110538. [DOI] [Google Scholar]
- 332.Champa-Bujaico E., Díez-Pascual A.M., Garcia-Diaz P., Sessini V., Mosquera M.E. Machine learning algorithms to optimize the properties of bio-based poly (butylene succinate-co-butylene adipate) nanocomposites with carbon nanotubes. Ind. Crop. Prod. 2024;219:119018. doi: 10.1016/j.indcrop.2024.119018. [DOI] [Google Scholar]
- 333.Grishanovich I.A., Sypalova Y.A., Shestakov S.L., Kozhevnikov A.Y. The Application of Hierarchical Cluster Analysis to Lignins Classification Based on Data of High-Resolution NMR and Solid-State NMR Spectra on 13C Nuclei. Appl. Magn. Reson. 2024;55:827–838. doi: 10.1007/s00723-024-01686-4. [DOI] [Google Scholar]
- 334.Ireddy A.T., Ghorabe F.D., Shishatskaya E.I., Ryltseva G.A., Dudaev A.E., Kozodaev D.A., Nosonovsky M., Skorb E.V., Zun P.S. Benchmarking Unsupervised Clustering Algorithms for Atomic Force Microscopy Data on Polyhydroxyalkanoate Films. ACS Omega. 2024;9:21595–21611. doi: 10.1021/acsomega.4c02502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Xu H., Qin Y., Hu G., Tsang H.K. Scalable integrated two-dimensional Fourier-transform spectrometry. Nat. Commun. 2024;15:436. doi: 10.1038/s41467-023-44518-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Mulrennan K., Munir N., Creedon L., Donovan J., Lyons J.G., McAfee M. NIR-based intelligent sensing of product yield stress for high-value bioresorbable polymer processing. Sensors. 2022;22:2835. doi: 10.3390/s22082835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Helmer M., Warrington S., Mohammadi-Nejad A.R., Ji J.L., Howell A., Rosand B., Anticevic A., Sotiropoulos S.N., Murray J.D. On the stability of canonical correlation analysis and partial least squares with application to brain-behavior associations. Commun. Biol. 2024;7:217. doi: 10.1038/s42003-024-05869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Liao Z., Dai S., Kuosmanen T. Convex support vector regression. Eur. J. Oper. Res. 2024;313:858–870. doi: 10.1016/j.ejor.2023.05.009. [DOI] [Google Scholar]
- 339.Zhang S., Zhu F., Yu Q., Zhu X. Identifying DNA-binding proteins based on multi-features and LASSO feature selection. Biopolymers. 2021;112:e23419. doi: 10.1002/bip.23419. [DOI] [PubMed] [Google Scholar]
- 340.Faye D., Kaly F., Dieng A.L., Wane D., Fall C.M.N., Mignot J., Gaye A.T. Regionalization of the Onset and Offset of the Rainy Season in Senegal Using Kohonen Self-Organizing Maps. Atmosphere. 2024;15:378. doi: 10.3390/atmos15030378. [DOI] [Google Scholar]
- 341.Qiao X.B., Jiang B., Hou T.J., Xu X.J. Representation of molecular electrostatic potentials of biopolymer by self-organizing feature map. Chin. J. Chem. 2001;19:1172–1178. doi: 10.1002/cjoc.20010191203. [DOI] [Google Scholar]
- 342.Jiang H., Zhao Y., Chen W., Zheng W., Zhang X. Improving cis-regulatory elements modeling by consensus scaffolded mixture models. Sci. China Inf. Sci. 2013;56:1–11. doi: 10.1007/s11432-011-4374-9. [DOI] [Google Scholar]
- 343.Mohammadi R., Neamatollahi P., Moradi M., Naghibzadeh M., Savadi A. Efficient Motif Discovery in Protein Sequences Using a Branch and Bound Algorithm. IEEE J. Biomed. Health Inform. 2024;28:2437–2444. doi: 10.1109/JBHI.2024.3355964. [DOI] [PubMed] [Google Scholar]
- 344.Hasan R., Uddin J. Data Mining Techniques for Informative Motif Discovery. Int. J. Comput. Appl. 2014;88:21–24. doi: 10.5120/15405-3901. [DOI] [Google Scholar]
- 345.Radhitya M.L., Widiantari N.K.M., Asana M.D.P., Wijaya B.K., Sudipa I.G.I. Product Layout Analysis Based on Consumer Purchasing Patterns Using Apriori Algorithm. J. Comput. Networks, Archit. High Perform. Comput. 2024;6:1701–1711. doi: 10.47709/cnahpc.v6i3.4400. [DOI] [Google Scholar]
- 346.Yousef M., Khalifa W., AbdAllah L. Ensemble clustering classification applied to competing SVM and one-class classifiers exemplified by plant MicroRNAs data. J. Integr. Bioinform. 2016;13:11–21. doi: 10.1515/jib-2016-304. [DOI] [PubMed] [Google Scholar]
- 347.Verma B., Rahman A. Cluster-oriented ensemble classifier: Impact of multicluster characterization on ensemble classifier learning. IEEE Trans. Knowl. Data Eng. 2011;24:605–618. doi: 10.1109/TKDE.2011.28. [DOI] [Google Scholar]
- 348.Wei S., Chen Z., Arumugasamy S.K., Chew I.M.L. Data augmentation and machine learning techniques for control strategy development in bio-polymerization process. Environ. Sci. Ecotechnol. 2022;11:100172. doi: 10.1016/j.ese.2022.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Eswaran S.c.d., Subramaniam S., Sanyal U., Rallo R., Zhang X. Molecular structural dataset of lignin macromolecule elucidating experimental structural compositions. Sci. Data. 2022;9:647. doi: 10.1038/s41597-022-01709-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Abreu A., Danko A., Costa J., Ferreira E., Alves M. Inoculum type response to different pHs on biohydrogen production from L-arabinose, a component of hemicellulosic biopolymers. Int. J. Hydrog. Energy. 2009;34:1744–1751. doi: 10.1016/j.ijhydene.2008.12.020. [DOI] [Google Scholar]
- 351.Fredricks J.L., Jimenez A.M., Grandgeorge P., Meidl R., Law E., Fan J., Roumeli E. Hierarchical biopolymer-based materials and composites. J. Polym. Sci. 2023;61:2585–2632. doi: 10.1002/pol.20230126. [DOI] [Google Scholar]
- 352.Jamali A., Roy S.K., Hong D., Atkinson P.M., Ghamisi P. Spatial Gated Multi-Layer Perceptron for Land Use and Land Cover Mapping. IEEE Geosci. Remote. Sens. Lett. 2024;21:5502105. doi: 10.1109/LGRS.2024.3354175. [DOI] [Google Scholar]
- 353.Zhang W., Shen X., Zhang H., Yin Z., Sun J., Zhang X., Zou L. Feature importance measure of a multilayer perceptron based on the presingle-connection layer. Knowl. Inf. Syst. 2024;66:511–533. doi: 10.1007/s10115-023-01959-7. [DOI] [Google Scholar]
- 354.Lu K., Gu Z., Qi F., Sun C., Guo H., Sun L. CMLP-Net: A convolution-multilayer perceptron network for EEG-based emotion recognition. Biomed. Signal Process. Control. 2024;96:106620. doi: 10.1016/j.bspc.2024.106620. [DOI] [Google Scholar]
- 355.Naseer A., Jalal A. Multimodal Objects Categorization by Fusing GMM and Multi-layer Perceptron; Proceedings of the 2024 5th International Conference on Advancements in Computational Sciences (ICACS); Lahore, Pakistan. 19–20 February 2024; Piscataway, NJ, USA: IEEE; 2024. pp. 1–7. [Google Scholar]
- 356.Siddique M.N.I., Shafiullah M., Mekhilef S., Pota H., Abido M. Fault classification and location of a PMU-equipped active distribution network using deep convolution neural network (CNN) Electr. Power Syst. Res. 2024;229:110178. doi: 10.1016/j.epsr.2024.110178. [DOI] [Google Scholar]
- 357.Rybacki P., Niemann J., Derouiche S., Chetehouna S., Boulaares I., Seghir N.M., Diatta J., Osuch A. Convolutional Neural Network (CNN) Model for the Classification of Varieties of Date Palm Fruits (Phoenix dactylifera L.) Sensors. 2024;24:558. doi: 10.3390/s24020558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Sarswat P.K., Singh R.S., Pathapati S.V.S.H. Real time electronic-waste classification algorithms using the computer vision based on convolutional neural network (cnn): Enhanced environmental incentives. Resour. Conserv. Recycl. 2024;207:107651. doi: 10.1016/j.resconrec.2024.107651. [DOI] [Google Scholar]
- 359.Lee B., Lee J., Lee J.O., Hwang Y., Bahn H.K., Park I., Jheon S., Lee D.S. Breath analysis system with convolutional neural network (CNN) for early detection of lung cancer. Sensors Actuators B Chem. 2024;409:135578. doi: 10.1016/j.snb.2024.135578. [DOI] [Google Scholar]
- 360.Lu M., Xu X. TRNN: An efficient time-series recurrent neural network for stock price prediction. Inf. Sci. 2024;657:119951. doi: 10.1016/j.ins.2023.119951. [DOI] [Google Scholar]
- 361.Zhang C., Zhou Z., Wu R. Analyzing and Predicting Financial Time Series Data Using Recurrent Neural Networks. J. Ind. Eng. Appl. Sci. 2024;2:15–21. [Google Scholar]
- 362.Shan F., He X., Armaghani D.J., Sheng D. Effects of data smoothing and recurrent neural network (RNN) algorithms for real-time forecasting of tunnel boring machine (TBM) performance. J. Rock Mech. Geotech. Eng. 2024;16:1538–1551. doi: 10.1016/j.jrmge.2023.06.015. [DOI] [Google Scholar]
- 363.Yeh Y.T., Hsiao W.Y., Yang Y.H. Hyper recurrent neural network: Condition mechanisms for black-box audio effect modeling. arXiv. 20242408.04829 [Google Scholar]
- 364.Ruhani B., Moghaddas S.A., Kheradmand A. Hydrogen production via renewable-based energy system: Thermoeconomic assessment and Long Short-Term Memory (LSTM) optimization approach. Int. J. Hydrog. Energy. 2024;52:505–519. doi: 10.1016/j.ijhydene.2023.03.456. [DOI] [Google Scholar]
- 365.Wu X., Du Z., Ma R., Zhang X., Yang D., Liu H., Zhang Y. Qualitative and quantitative studies of phthalates in extra virgin olive oil (EVOO) by surface-enhanced Raman spectroscopy (SERS) combined with long short term memory (LSTM) neural network. Food Chem. 2024;433:137300. doi: 10.1016/j.foodchem.2023.137300. [DOI] [PubMed] [Google Scholar]
- 366.Salamattalab M.M., Zonoozi M.H., Molavi-Arabshahi M. Innovative approach for predicting biogas production from large-scale anaerobic digester using long-short term memory (LSTM) coupled with genetic algorithm (GA) Waste Manag. 2024;175:30–41. doi: 10.1016/j.wasman.2023.12.046. [DOI] [PubMed] [Google Scholar]
- 367.Kratzert F., Gauch M., Klotz D., Nearing G. HESS Opinions: Never train a Long Short-Term Memory (LSTM) network on a single basin. Hydrol. Earth Syst. Sci. 2024;28:4187–4201. doi: 10.5194/hess-28-4187-2024. [DOI] [Google Scholar]
- 368.Koh A.J.H., Tan S.Y., Nasrudin M.F. A systematic literature review of generative adversarial networks (GANs) in 3D avatar reconstruction from 2D images. Multimed. Tools Appl. 2024;83:68813–68853. doi: 10.1007/s11042-024-18665-3. [DOI] [Google Scholar]
- 369.Wang Y., Zhang Q., Wang G.G., Cheng H. The application of evolutionary computation in generative adversarial networks (GANs): A systematic literature survey. Artif. Intell. Rev. 2024;57:182. doi: 10.1007/s10462-024-10818-y. [DOI] [Google Scholar]
- 370.Showrov A.A., Aziz M.T., Nabil H.R., Jim J.R., Kabir M.M., Mridha M., Asai N., Shin J. Generative Adversarial Networks (GANs) in Medical Imaging: Advancements, Applications and Challenges. IEEE Access. 2024;12:35728–35753. doi: 10.1109/ACCESS.2024.3370848. [DOI] [Google Scholar]
- 371.Chakraborty T., KS U.R., Naik S.M., Panja M., Manvitha B. Ten years of generative adversarial nets (GANs): A survey of the state-of-the-art. Mach. Learn. Sci. Technol. 2024;5:011001. doi: 10.1088/2632-2153/ad1f77. [DOI] [Google Scholar]
- 372.Kolosnitsyn A., Khamisov O., Semenkin E., Nelyub V. Application of the Parabola Method in Nonconvex Optimization. Algorithms. 2024;17:107. doi: 10.3390/a17030107. [DOI] [Google Scholar]
- 373.Stanovov V., Semenkin E. Adaptation of the Scaling Factor Based on the Success Rate in Differential Evolution. Mathematics. 2024;12:516. doi: 10.3390/math12040516. [DOI] [Google Scholar]
- 374.Gao L., la Tour T.D., Tillman H., Goh G., Troll R., Radford A., Sutskever I., Leike J., Wu J. Scaling and evaluating sparse autoencoders. arXiv. 20242406.04093 [Google Scholar]
- 375.Bengesi S., El-Sayed H., Sarker M.K., Houkpati Y., Irungu J., Oladunni T. Advancements in Generative AI: A Comprehensive Review of GANs, GPT, Autoencoders, Diffusion Model, and Transformers. IEEE Access. 2024;12:69812–69837. doi: 10.1109/ACCESS.2024.3397775. [DOI] [Google Scholar]
- 376.Baur M., Fesl B., Utschick W. Leveraging variational autoencoders for parameterized MMSE estimation. IEEE Trans. Signal Process. 2024;72:3731–3744. doi: 10.1109/TSP.2024.3439097. [DOI] [Google Scholar]
- 377.Berahmand K., Daneshfar F., Salehi E.S., Li Y., Xu Y. Autoencoders and their applications in machine learning: A survey. Artif. Intell. Rev. 2024;57:28. doi: 10.1007/s10462-023-10662-6. [DOI] [Google Scholar]
- 378.Xu R., Dong X.M., Li W., Peng J., Sun W., Xu Y. DBCTNet: Double branch convolution-transformer network for hyperspectral image classification. IEEE Trans. Geosci. Remote. Sens. 2024;62:5509915. doi: 10.1109/TGRS.2024.3368141. [DOI] [Google Scholar]
- 379.Alam W., Tayara H., Chong K.T. Unlocking the therapeutic potential of drug combinations through synergy prediction using graph transformer networks. Comput. Biol. Med. 2024;170:108007. doi: 10.1016/j.compbiomed.2024.108007. [DOI] [PubMed] [Google Scholar]
- 380.Volk A.A., Epps R.W., Ethier J.G., Baldwin L.A. Modeling Multi-Step Scientific Processes with Graph Transformer Networks. arXiv. 20242408.05425 [Google Scholar]
- 381.Bhatia S., Richie R. Transformer networks of human conceptual knowledge. Psychol. Rev. 2024;131:271. doi: 10.1037/rev0000319. [DOI] [PubMed] [Google Scholar]
- 382.Nguyen D.A., Nguyen V.B., Jang A. Ultrahigh-porosity Ranunculus-like MgO adsorbent coupled with predictive deep belief networks: A transformative method for phosphorus treatment. Water Res. 2024;249:120930. doi: 10.1016/j.watres.2023.120930. [DOI] [PubMed] [Google Scholar]
- 383.Sarangi S., Dash P.K., Bisoi R. Short-term prediction of wind power using an improved kernel based optimized deep belief network. Energy Convers. Manag. 2024;316:118821. doi: 10.1016/j.enconman.2024.118821. [DOI] [Google Scholar]
- 384.Shukla A.K., Muhuri P.K. A novel deep belief network architecture with interval type-2 fuzzy set based uncertain parameters towards enhanced learning. Fuzzy Sets Syst. 2024;477:108744. doi: 10.1016/j.fss.2023.108744. [DOI] [Google Scholar]
- 385.Meng S., Shi Z., Li G., Peng M., Liu L., Zheng H., Zhou C. A novel deep learning framework for landslide susceptibility assessment using improved deep belief networks with the intelligent optimization algorithm. Comput. Geotech. 2024;167:106106. doi: 10.1016/j.compgeo.2024.106106. [DOI] [Google Scholar]
- 386.Arevalo S.E., Buehler M.J. Learning from nature by leveraging integrative biomateriomics modeling toward adaptive and functional materials. MRS Bull. 2023;48:1140–1153. doi: 10.1557/s43577-023-00610-8. [DOI] [Google Scholar]
- 387.Khare E., Gonzalez-Obeso C., Kaplan D.L., Buehler M.J. CollagenTransformer: End-to-end transformer model to predict thermal stability of collagen triple helices using an NLP approach. ACS Biomater. Sci. Eng. 2022;8:4301–4310. doi: 10.1021/acsbiomaterials.2c00737. [DOI] [PubMed] [Google Scholar]
- 388.Bandyopadhyay S., Mondal J. A deep autoencoder framework for discovery of metastable ensembles in biomacromolecules. J. Chem. Phys. 2021;155:114106. doi: 10.1063/5.0059965. [DOI] [PubMed] [Google Scholar]
- 389.Sadeghi E., Mastracco P., Gonzàlez-Rosell A., Copp S.M., Bogdanov P. Multi-Objective Design of DNA-Stabilized Nanoclusters Using Variational Autoencoders With Automatic Feature Extraction. ACS Nano. 2024;18:26997–27008. doi: 10.1021/acsnano.4c09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Sattari K., Xie Y., Lin J. Data-driven algorithms for inverse design of polymers. Soft Matter. 2021;17:7607–7622. doi: 10.1039/D1SM00725D. [DOI] [PubMed] [Google Scholar]
- 391.Baldizon T.B.S.G. Master’s Thesis. California State University; Long Beach, CA, USA: 2024. Determining Biopolymer Topology via Nanopore Translocation and Machine Learning. [Google Scholar]
- 392.Noor R.M., Ahmad Z. Biopolycaprolactone molecular weight prediction based on neural network technique in a batch reactor; Proceedings of the 2012 7th IEEE Conference on Industrial Electronics and Applications (ICIEA); Singapore. 18–20 July 2012; Piscataway, NJ, USA: IEEE; 2012. pp. 1412–1416. [Google Scholar]
- 393.Leal-Junior A., Rocha H., Almeida P.L., Marques C. Force estimation with sustainable hydroxypropyl cellulose sensor using convolutional neural network. [(accessed on 11 October 2024)];IEEE Sensors J. 2024 24:1366–1373. doi: 10.1109/JSEN.2023.3332659. Available online: https://ieeexplore.ieee.org/abstract/document/10335606. [DOI] [Google Scholar]
- 394.Salma H., Melha Y.M., Sonia L., Hamza H., Salim N. Efficient prediction of in vitro piroxicam release and diffusion from topical films based on biopolymers using deep learning models and generative adversarial networks. J. Pharm. Sci. 2021;110:2531–2543. doi: 10.1016/j.xphs.2021.01.032. [DOI] [PubMed] [Google Scholar]
- 395.Araujo N.R., Sebastiao R.C., Freitas-Marques M.B., da Nova Mussel W., Yoshida M.I., Virtuoso L.S. Multilayer perceptron neural network applied to TG dynamic data of biopolymer chitosan–a robust tool to study the kinetics of solid thermal decomposition. Therm. Sci. Eng. Prog. 2022;36:101490. doi: 10.1016/j.tsep.2022.101490. [DOI] [Google Scholar]
- 396.Wong Y.J., Arumugasamy S.K., Jewaratnam J. Performance comparison of feedforward neural network training algorithms in modeling for synthesis of polycaprolactone via biopolymerization. Clean Technol. Environ. Policy. 2018;20:1971–1986. doi: 10.1007/s10098-018-1577-4. [DOI] [Google Scholar]
- 397.Laycock B.G., Chan C.M., Halley P.J. A review of computational approaches used in the modelling, design, and manufacturing of biodegradable and biobased polymers. Prog. Polym. Sci. 2024;157:101874. doi: 10.1016/j.progpolymsci.2024.101874. [DOI] [Google Scholar]
- 398.Kartal F., Dalbudak Y., Özveren U. Prediction of thermal degradation of biopolymers in biomass under pyrolysis atmosphere by means of machine learning. Renew. Energy. 2023;204:774–787. doi: 10.1016/j.renene.2023.01.017. [DOI] [Google Scholar]
- 399.Khamisov O.O., Khamisov O.V., Ganchev T.D., Semenkin E.S. A Method for Transforming Non-Convex Optimization Problem to Distributed Form. Mathematics. 2024;12:2796. doi: 10.3390/math12172796. [DOI] [Google Scholar]
- 400.Khamisov O., Semenkin E., Nelyub V. Allocation of Starting Points in Global Optimization Problems. Mathematics. 2024;12:606. doi: 10.3390/math12040606. [DOI] [Google Scholar]
- 401.Stanovov V., Kazakovtsev L., Semenkin E. Hyper-Heuristic Approach for Tuning Parameter Adaptation in Differential Evolution. Axioms. 2024;13:59. doi: 10.3390/axioms13010059. [DOI] [Google Scholar]
- 402.Asgharzadeh P., Birkhold A.I., Özdemir B., Reski R., Röhrle O. Biopolymer segmentation from CLSM microscopy images using a convolutional neural network. PAMM. 2021;20:e202000188. doi: 10.1002/pamm.202000188. [DOI] [Google Scholar]
- 403.Leng Y., Tac V., Calve S., Tepole A.B. Predicting the mechanical properties of biopolymer gels using neural networks trained on discrete fiber network data. Comput. Methods Appl. Mech. Eng. 2021;387:114160. doi: 10.1016/j.cma.2021.114160. [DOI] [Google Scholar]
- 404.Nobrega M.M., Bona E., Yamashita F. An artificial neural network model for the prediction of mechanical and barrier properties of biodegradable films. Mater. Sci. Eng. C. 2013;33:4331–4336. doi: 10.1016/j.msec.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 405.Zhang J., Yin J., Wang R. Basic framework and main methods of uncertainty quantification. Math. Probl. Eng. 2020;2020:6068203. doi: 10.1155/2020/6068203. [DOI] [Google Scholar]
- 406.Xu F., Uszkoreit H., Du Y., Fan W., Zhao D., Zhu J. Explainable AI: A brief survey on history, research areas, approaches and challenges; Proceedings of the Natural Language Processing and Chinese Computing: 8th CCF International Conference, NLPCC 2019; Dunhuang, China. 9–14 October 2019; Cham, Switzerland: Springer; 2019. pp. 563–574. Proceedings, part II 8. [Google Scholar]