Abstract
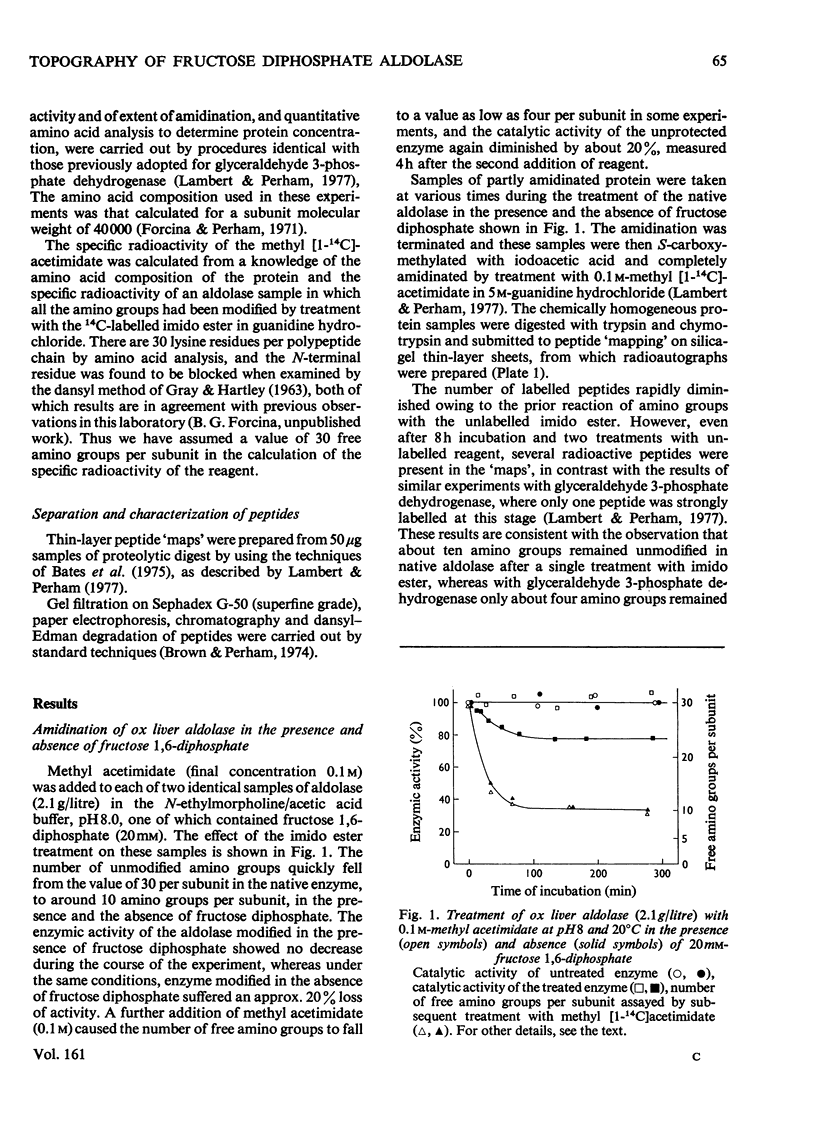
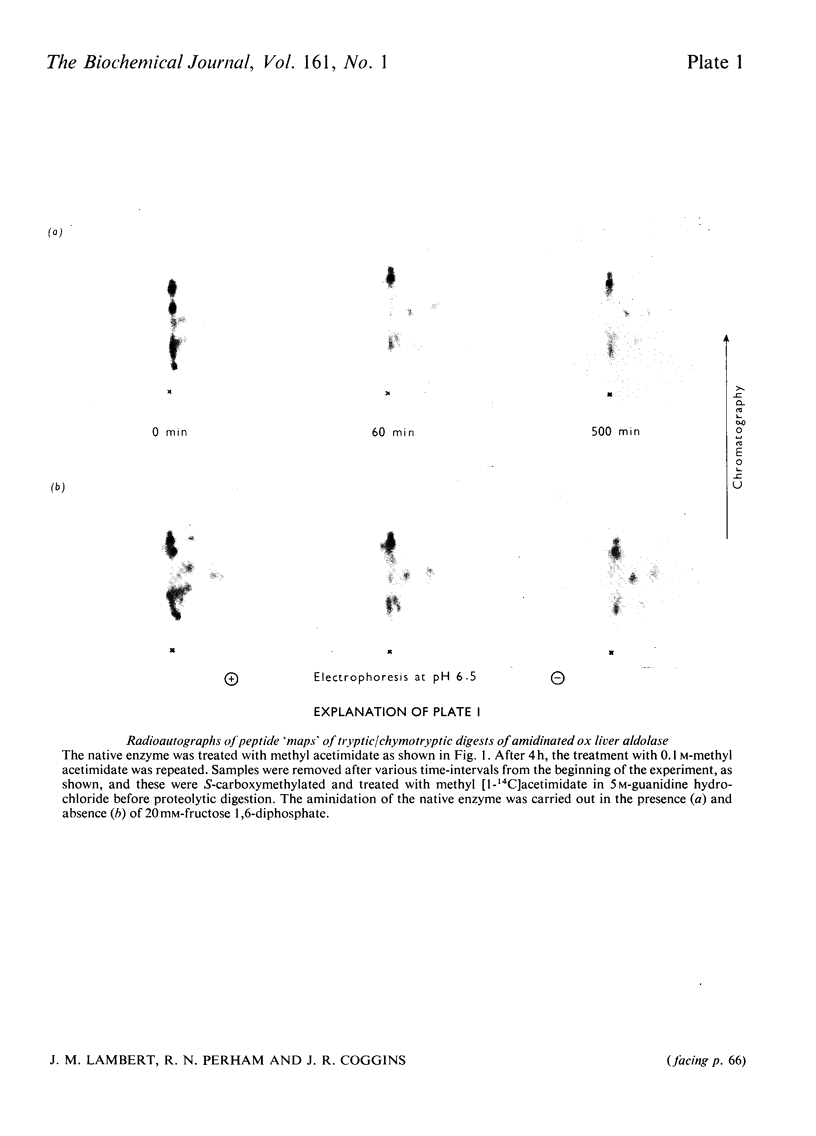
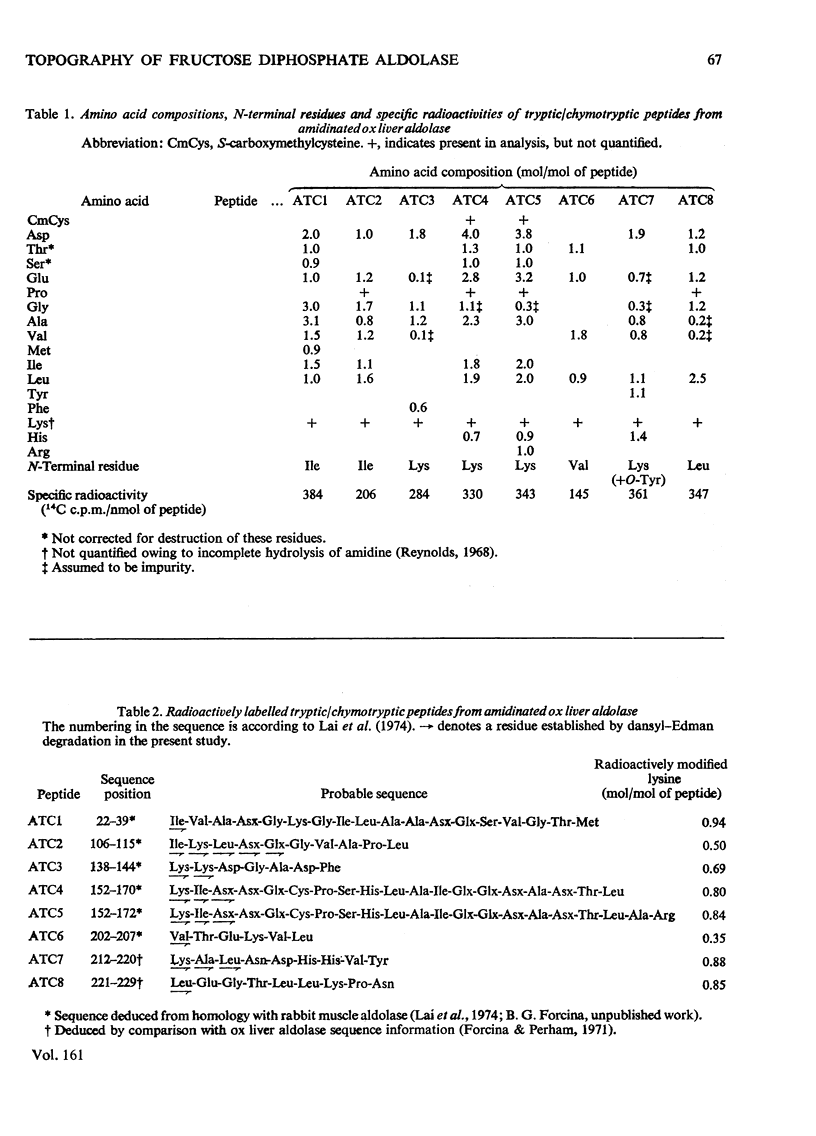
1. Treatment with methyl acetimidate was used to probe the topography of the tetrameric fructose 1,6-diphosphate aldolase from ox liver. A single treatment with imido ester in the presence or absence of 20mM-fructose 1,6-diphosphate caused the number of amino groups in the enzyme to fall to approx. 30% of the starting number (assumed to be 30 per subunit). The catalytic activity of the aldolase modified in the presence of fructose 1,6-diphosphate was unaffected, whereas that of the enzyme modified in the absence of substrate fell by about 20%. 2. Use of methyl [1-14C]acetimidate and small-scale methods of protein chemistry showed that the amino group of lysine-27 (the numbering is that of the highly homologous rabbit muscle enzyme) is essentially unavailable for amidination in the native enzyme and is therefore predicted to be buried in a hydrophobic environment, probably in the form of an ion-pair with a negatively charged side-chain carboxyl group. All the other lysine residues that reacted poorly with methyl acetimidate in the native enzyme (a total of 7) were found to be within the primary structure bounded by lysine-107 and lysine-227. An important member of this group of lysine residues displaying aberrant reactivity is lysine-227, which is known to form an imine with the substrate as part of the catalytic mechanism of the enzyme. 3. The results of the amidination experiments can be correlated in an interesting way with previous studies of thiol-group modification in the aldolases. Taken together, and arguing in part by analogy with the results of identical experiments with glyceraldehyde 3-phosphate dehydrogenases where the three-dimensional structure is known [Lambert & Perham (1977) Biochem. 4. 161. 49-62], they suggest that the region of primary structure from residues 107-227 may form the whole or part of a three-dimensional structural feature, perhaps a folding domain. A three-dimensional structure deduced from X-ray-crystallographic analysis will be needed to interpret these findings more closely. 4. The amino groups of lysine residues are commonly thought to reside at the 'surface' of protein structures. The patterns of specific lysine residues in glyceraldehyde 3-phosphate dehydrogenases and in aldolases that have been found to react poorly with methyl acetimidate in the native enzymes can be attributed to intramolecular ionic interactions deep in hydrophobic pockets and at the protein 'surface'. Such ionic interactions may contribute significantly to the stability of a given protein.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anai M., Lai C. Y., Horecker B. L. The pyridoxal phosphate-binding site of rabbit muscle aldolase. Arch Biochem Biophys. 1973 Jun;156(2):712–719. doi: 10.1016/0003-9861(73)90324-x. [DOI] [PubMed] [Google Scholar]
- Anderson P. J., Kaplan H. Reactivity of the active-centre lysine residue of rabbit muscle aldolase. Biochem J. 1974 Feb;137(2):181–184. doi: 10.1042/bj1370181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. J., Perham R. N. The reactivity of thiol groups and the subunit structure of aldolase. Biochem J. 1970 Apr;117(2):291–298. doi: 10.1042/bj1170291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. J. The number, location, and reactivity of the cysteine residues of sturgeon muscle aldolase. Can J Biochem. 1972 Feb;50(2):111–119. doi: 10.1139/o72-014. [DOI] [PubMed] [Google Scholar]
- BLOSTEIN R., RUTTER W. J. COMPARATIVE STUDIES OF LIVER AND MUSCLE ALDOLASE. II. IMMUNOCHEMICAL AND CHROMATOGRAPHIC DIFFERENTIATION. J Biol Chem. 1963 Oct;238:3280–3285. [PubMed] [Google Scholar]
- Bates D. L., Perham R. N., Coggins J. R. Methods for obtaining peptide maps of proteins on a subnanomole scale. Anal Biochem. 1975 Sep;68(1):175–184. doi: 10.1016/0003-2697(75)90692-2. [DOI] [PubMed] [Google Scholar]
- Brown J. P., Perham R. N. An amino acid sequence in the active site of lipoamide dehydrogenase from pig heart. Biochem J. 1974 Mar;137(3):505–512. doi: 10.1042/bj1370505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagles P. A., Johnson L. N., Joynson M. A., McMurray C. H., Gutfreund H. Subunit structure of aldolase: chemical and crystallographic evidence. J Mol Biol. 1969 Nov 14;45(3):533–544. doi: 10.1016/0022-2836(69)90310-6. [DOI] [PubMed] [Google Scholar]
- Forcina B. G., Perham R. N. Amino acid sequence homology between muscle and liver aldolases. FEBS Lett. 1971 Oct 15;18(1):59–63. doi: 10.1016/0014-5793(71)80406-4. [DOI] [PubMed] [Google Scholar]
- Gibbons I., Anderson P. J., Perham R. N. Amino acid sequence homology in the active site of rabbit and sturgeon muscle aldolases. FEBS Lett. 1970 Sep 18;10(1):49–53. doi: 10.1016/0014-5793(70)80413-6. [DOI] [PubMed] [Google Scholar]
- Gibbons I., Perham R. N. Kinetic and molecular properties of citraconyl-aldolase. The reversible denaturation and hybridization of the native and modified enzymes. Biochem J. 1974 May;139(2):331–342. doi: 10.1042/bj1390331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons I., Perham R. N. The reaction of aldolase with 2-methylmaleic anhydride. Biochem J. 1970 Mar;116(5):843–849. doi: 10.1042/bj1160843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidner E. G., Weber B. H., Eisenberg D. Subunit structure of aldolase. Science. 1971 Feb 19;171(3972):677–679. doi: 10.1126/science.171.3972.677. [DOI] [PubMed] [Google Scholar]
- Lai C. Y., Nakai N., Chang D. Amino acid sequence of rabbit muscle aldolase and the structure of the active center. Science. 1974 Mar;183(130):1204–1206. doi: 10.1126/science.183.4130.1204. [DOI] [PubMed] [Google Scholar]
- Lambert J. M., Perham R. N. A comparison of the glyceraldehyde 3-phosphate dehydrogenase from ox muscle and liver. FEBS Lett. 1974 Apr 1;40(2):305–308. doi: 10.1016/0014-5793(74)80250-4. [DOI] [PubMed] [Google Scholar]
- Lambert J. M., Perham R. N. Folding domains and intramolecular ionic interactions of lysine residues in glyceraldehyde 3-phosphate dehydrogenase. Biochem J. 1977 Jan 1;161(1):49–62. doi: 10.1042/bj1610049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler A. H., Cusic M. E., Jr Aldolase reaction with sugar diphosphates. Science. 1967 Mar 3;155(3766):1101–1103. doi: 10.1126/science.155.3766.1101. [DOI] [PubMed] [Google Scholar]
- Penhoet E. E., Kochman M., Rutter W. J. Ioslation of fructose diphosphate aldolases A, B, and C. Biochemistry. 1969 Nov;8(11):4391–4395. doi: 10.1021/bi00839a025. [DOI] [PubMed] [Google Scholar]
- Perham R. N., Anderson P. J. The reactivity of thiol groups in aldolase. Biochem Soc Symp. 1970;31:49–58. [PubMed] [Google Scholar]
- RUTTER W. J. EVOLUTION OF ALDOLASE. Fed Proc. 1964 Nov-Dec;23:1248–1257. [PubMed] [Google Scholar]
- Reynolds J. H. Acetimidation of bovine pancreatic ribonuclease A. Biochemistry. 1968 Sep;7(9):3131–3135. doi: 10.1021/bi00849a016. [DOI] [PubMed] [Google Scholar]
- Sajgó M., Hajós G. The amino acid sequence of rabbit muscle aldolase. Acta Biochim Biophys Acad Sci Hung. 1974;9(3):239–241. [PubMed] [Google Scholar]
- Saunders G. C., Weber B. H. Investigation of the role of lysine in the subunit contact regions of rabbit muscle aldolase. Arch Biochem Biophys. 1975 Jun;168(2):525–530. doi: 10.1016/0003-9861(75)90283-0. [DOI] [PubMed] [Google Scholar]
- Schmidt D. E., Jr, Westheimer F. H. PK of the lysine amino group at the active site of acetoacetate decarboxylase. Biochemistry. 1971 Mar 30;10(7):1249–1253. doi: 10.1021/bi00783a023. [DOI] [PubMed] [Google Scholar]
- Shapiro S., Enser M., Pugh E., Horecker B. L. The effect of pyridoxal phosphate on rabbit muscle aldolase. Arch Biochem Biophys. 1968 Nov;128(2):554–562. doi: 10.1016/0003-9861(68)90062-3. [DOI] [PubMed] [Google Scholar]
- Steinman H. M., Richards F. M. Participation of cysteinyl residues in the structure and function of muscle aldolase. Characterization of mixed disulfide derivatives. Biochemistry. 1970 Oct 27;9(22):4360–4372. doi: 10.1021/bi00824a017. [DOI] [PubMed] [Google Scholar]
- Szajáni B., Sajgó M., Biszku E., Friedrich P., Szabolcsi G. Identification of a cysteinyl residue involved in the activity of rabbit muscle aldolase. Eur J Biochem. 1970 Jul;15(1):171–178. doi: 10.1111/j.1432-1033.1970.tb00992.x. [DOI] [PubMed] [Google Scholar]



