Abstract
To overcome their limited genetic capacity, numerous viruses encode multifunctional proteins. The birnavirus VP3 protein plays key roles during infection, including scaffolding of the viral capsid during morphogenesis, recruitment, and regulation of the viral RNA polymerase, shielding of the double-stranded RNA genome and targeting of host endosomes for genome replication, and immune evasion. The dimeric form of VP3 is critical for these functions. In previous work, we determined the X-ray structure of the central domains (D2–D3) of VP3 from the infectious bursal disease virus (IBDV). However, the structure and function of the IBDV VP3 N-terminal domain (D1) could not be determined at that time. Using integrated structural biology approaches and functional cell assays, here we characterize the IBDV VP3 D1 domain, unveiling its unexplored roles in virion stability and infection. The X-ray structure of D1 shows that this domain folds in four α-helices arranged in parallel dimers, which are essential for maintaining the dimeric arrangement of the full-length protein. Combining small-angle X-ray scattering analyses with molecular dynamics simulations allowed us to build a structural model for the D1–D3 domains. This model consists of an elongated structure with high flexibility in the D2–D3 connection, keeping D1 as the only driver of VP3 dimerization. Using reverse genetics tools, we show that the obliteration of D1 domain prevents the VP3 scaffold function during capsid assembly and severely impacts IBDV infection. Altogether, our study elucidates the structure of the VP3 D1 domain and reveals its role in VP3 protein dimerization and IBDV infection.
Keywords: IBDV, dsRNA-binding protein, moonlighting proteins, scaffolding protein, viral replication
Significant Statement.
The infectious bursal disease virus (IBDV) is a major threat for poultry worldwide. IBDV outbreaks are arduous to control and bring devastating economic losses for farmers. Despite its relevance, the molecular mechanisms behind IBDV infection and pathogenesis remain poorly understood. VP3 is a structural viral protein in charge of multiple crucial functions, most of which depend on its capacity to oligomerize. In this work, we characterized the structure and function of the aminoterminal domain (D1) of IBDV VP3, revealing its key role in VP3 dimerization. Our data also demonstrate that the absence of D1 results in the lack of virus replication and progeny recovery, shedding light on the role for this domain in IBDV infection and pathogenesis.
Introduction
Birnaviruses are a family of small nonenveloped double-stranded RNA (dsRNA) viruses that infect a diverse range of hosts, including birds, fishes, insects, and rotifers (1). The infectious bursal disease virus (IBDV) stands out as the prototype of the Avibirnavirus genus, and it is the cause of the Gumboro disease, a highly contagious and immunosuppressive infection of domestic chicks worldwide (2). IBDV is very resistant to decontamination and despite the implementation of rigorous vaccination programs, outbreaks remain as one of the leading causes of severe economic losses within the poultry industry (3–5).
The dsRNA genome of IBDV comprises two segments, A and B, with segment A containing two overlapping open reading frames (ORFs). The largest ORF encodes a polyprotein that is cleaved during translation, generating the capsid protein precursor (pVP2), the serine-protease VP4, and the moonlighting protein VP3. The small ORF encodes the VP5 protein involved in virus egress (6). Segment B has a single ORF encoding the viral RNA-dependent RNA polymerase (RdRP) VP1 (7). VP3 is a 32-kDa polypeptide that plays pivotal roles during the viral replication cycle: acting as scaffolding protein during capsid morphogenesis (8–10), recruiting the viral RdRP to the virus particle and regulating its activity (11–14), shielding of the dsRNA genome (15, 16), controlling the host innate antiviral responses triggered by dsRNA (17–20), and targeting host proteins and endosomes for genome replication (21–23). The delicate coordination of VP3 activities throughout the viral replication cycle may involve one or more of the following elements: (i) the presence of intrinsically disordered regions; (ii) conformational changes, or (iii) variations in its oligomeric state, as it has been previously reported that VP3 oligomerization via its flexible C-terminal tail was essential for viral capsid formation (9).
We had previously solved the crystal structure of the central domains of IBDV VP3 (D2–D3) (24), providing insights into VP3 functions. However, the structure and functional relevance of the VP3 N-terminal moiety (containing D1 domain), which appears to be implicated in homotypic and host protein interactions (25–27), remains largely unknown.
In this study, we have performed a multidisciplinary analysis, encompassing biophysical, biochemical, and cellular strategies to characterize the structure and function of the D1 domain of IBDV VP3. Our results show that D1 folds in four α-helices arranged as a parallel dimer that is crucial for maintaining the dimeric state of the VP3 protein in solution. Importantly, D1 dimerization is essential for capsid assembly, targeting of endosomes and virus viability. Altogether, our findings shed new light on the structural and functional characteristics of VP3 and identified the D1 domain as an attractive target candidate for antiviral design.
Results
Structure of the VP3 N-terminal domain
We started by seeking high-resolution structural information of the core VP3 protein. To this end, we produced a recombinant construct that contained the D1–D2–D3 domains of IBDV VP3 (residues 1–221) in Escherichia coli (Fig. 1A and B). The VP3 C-terminal tail (residues 222–257) was removed because it compromised the solubility and stability of the full-length protein (24), possibly because it lacks secondary structure elements, as predicted by AlphaFold3 (AF3) (28) (Figs. 2A, B and S1A). The His-tagged construct was purified and subjected to high-throughput crystallization screenings. A mixture of trigonal and monoclinic crystals appeared after 2 months in the same crystallization drop. In addition, new orthorhombic P212121 crystals were obtained in different conditions (see Materials and methods). X-ray data were collected for the three crystal forms that diffracted to 1.46, 1.34, and 3.5 Å resolution, respectively, for the P21, P3221, and P212121 crystals (Table S1). During initial data analysis, we realized that the construct was cleaved during the crystallization process and monoclinic P21 and trigonal P3221 crystals corresponded to the D1 domain, whereas the P212121 crystals were from D2 to D3.
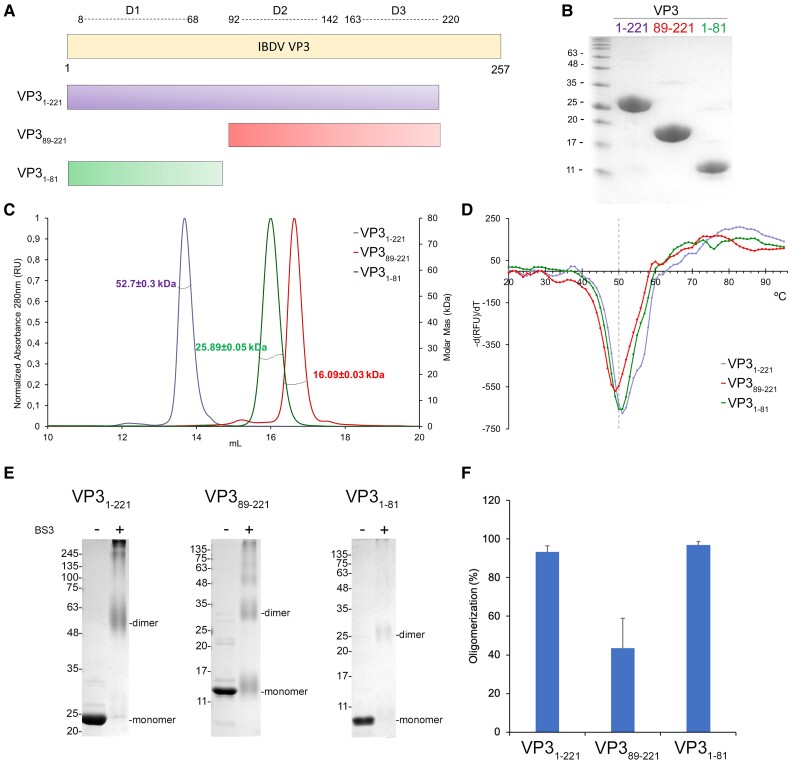
Fig. 1.
Characterization of IBDV VP3 constructs. A) Schematic representation of VP3 (residues 1 to 257), the domain definition (D1–D3), and the recombinant protein constructs expressed in E. coli. B) Coomassie-stained SDS–PAGE gels loaded with the recombinant purified proteins used in this study. C) SEC-MALS profiles of VP3 constructs, in buffer 50 mM Tris pH 8, 500 mM NaCl, 5 mM DTT. The lines intercepting each peak correspond to the measured molecular masses shown in the right axis (VP31–221: 52 ± 0.3 kDa; VP31–81: 20 ± 0.2 kDa; VP389–221: 16.1 ± 0.03 kDa). The theoretical MWs for each construct monomer are 24.8, 13.9, and 11.0 kDa, respectively. D) Plot of the first derivative of fluorescence signal obtained in DSF experiments to measure the VP3 constructs stability at increasing temperatures (VP31–81, green; VP389–221, red; VP31–221, violet). Dashed line is indicating 50 °C. E) SDS–PAGE of the different VP3 constructs crosslinked with BS3. F) Bar plot of the quantification by densitometric analysis of SDS–PAGE gel showing that BS3 covalently stabilizes dimers and higher MW species of constructs VP31–221 (93.3 ± 3.2%), VP31–81 (96.9 ± 1.7%), and VP389–221 (43.5 ± 15.4%).
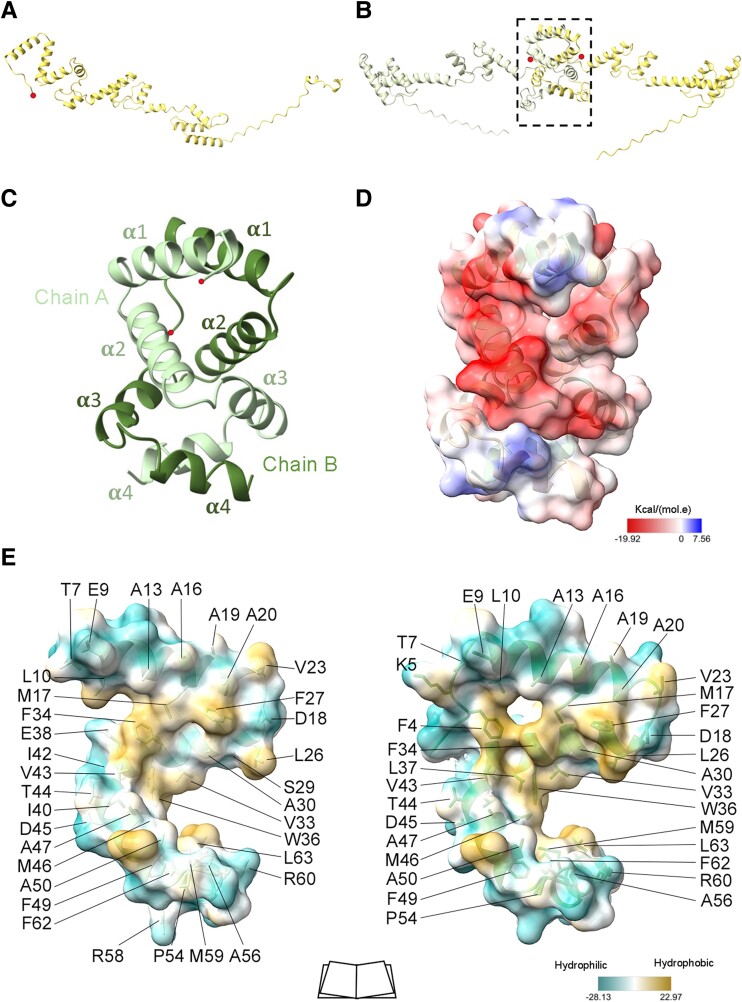
Fig. 2.
Structure of the IBDV VP3 aminoterminal domain. A) Ribbon representation of the monomeric structure of full-length VP3 (residues 1–257) predicted by AF3. The red dot highlights the aminoterminal end. B) As in A, but for the prediction of dimeric VP3. The aminoterminal domain (D1) is highlighted with the dashed lines box. C) Ribbon diagram of the structure of the VP3 D1 domain. Chains A and B of the D1 dimer are represented, with the secondary structural elements (α1 to α4) explicitly labeled and the aminoterminal ends depicted as red dots. D) Electrostatic potential of the protein surface showing from red to blue the negative to positive potential respectively. E) D1 monomer–monomer interactions, the molecular lipophilicity potential of protein surface is represented, from dark cyan to dark goldenrod colors, showing the hydrophilic to hydrophobic residues, respectively. Amino acids involved in monomer–monomer contacts are labeled and lateral chains showed with stick representation.
The structure of the D2–D3 crystals was solved by molecular replacement (MR), using the structure of the equivalent VP3 domains previously solved (PDB ID: 2R18 (24)) as a search model (Table S1). As expected, the D2–D3 polypeptide adopts the same extended fold, consisting of two α-helical modules connected by a long hinge, in line with the previously determined structure (24) (Fig. S2). This conformation is conserved in the eight molecules, contained in the asymmetric unit, that were organized in four head-to-tail dimers (Fig. S2A and B). Comparisons between the nine independent molecules from the asymmetric units of the two crystals compared, showed rmsd of 0.68 ± 0.06 Å for the superimposition of all Cα atoms (residues 92–220; Fig. S2C and D).
The D1 structure in P3221 crystal was solved by ab initio methods using ARCIMBOLDO software (29). Initial maps allowed unambiguous tracing of the first 66 residues of VP3 (Fig. S3A). The D1 polypeptide was folded into four α-helices (α1, spanning residues from P8 to V23; α2, residues from P25 to N40; α3, residues from V43 to D53; and α4, residues from H57 to N65) that were arranged forming an S-like structure and organized in parallel dimers (Fig. 2C and D). The D1 dimer was mainly stabilized through hydrophobic contacts, connecting helices α1-α1, α2-α2, and α4-α4 of the two neighboring molecules, burying the hydrophobic faces of each α-helix in the dimerization interface (Figs. 2E and S3). Furthermore, helices α2 and α4 of one molecule contacted helix α3 of its partner (Fig. 2C). This compact D1–D1 interface has a buried surface of ∼1,940 Å2, representing ∼34% of the domain surface involved in this interaction.
Calculations of the electrostatic potential of D1 dimers show a predominantly negative surface at physiological pH (Fig. 2D). This is in contrast to the extensive positively charged regions observed in the VP3 central domains D2 and D3 (24), which were shown to be essential for dsRNA binding (18).
The structure of the P21 crystals was solved by MR using the P3221 structure as a search model (Table S1). The asymmetric unit of these crystals contained two D1 dimers oriented in opposite directions (Fig. S3B). The individual D1 chains were almost identical (rmsd = 0.3 Å for the superimposition Cα atoms of residues 7–68). D1 dimers were also similar, with rmsd of 0.8 Å (Cα superimposition of residues 7–68, chain A). In addition, AF3 predicted the same structure (rmsd = 0.48 Å, Cα alignment of residues 10–66), with high confidence for IBDV D1 in the context of the full-length VP3 protein (Fig. S1A and D).
VP3 N-terminal domain drives dimerization
To investigate the role of the D1 domain in the context of the VP3 quaternary structure, we analyzed three different constructs, each containing the three, two, or one domain (1–221, 89–221, 1–81, respectively; Fig. 1A and B). Size exclusion chromatography (SEC) coupled to multiangle light scattering (MALS) analyses showed that the purified VP31–221 eluted in a predominant peak of 52.7 ± 0.3 kDa (98.8% of total sample). This mass matched with the expected size of a protein dimer (calculated monomer weight = 27.6 kDa; Fig. 1C). Conversely, the fragment containing the central domains of VP3 (D2–D3), in the construct VP389–221, was predominantly observed as a protein monomer (97.2% of total sample) with a mass of 16.09 ± 0.03 kDa. Nonetheless, as observed for the previous construct, a small proportion (2.8%) of a higher molecular weight (MW) oligomer (42 ± 1 kDa) was also detected (Fig. 1C). In contrast, the first 81 residues of VP3, encompassing the D1 domain, eluted as a clear peak of 25.89 ± 0.05 kDa compatible with a dimer (calculated monomer weight = 12.9 kDa). These results indicate that VP3 dimerization is promoted by its N-terminal D1 domain.
Stability analyses of the constructs, performed by differential scanning fluorimetry (DSF) showed that the three VP3 polypeptides were stable proteins, displaying small differences in melting temperature (Tm; Fig. 1D). VP389–221 showed the lowest Tm = 49.0 °C, while VP31–81 and VP31–221 showed a Tm of 50.1 and 51.2 °C, respectively (Fig. 1D).
Crosslinking experiments, using bis-(sulfosuccinimidyl)suberate (BS3) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis, showed that BS3 covalently stabilized 93.2 ± 3.3% of the protein sample of VP31–221 construct in dimers and higher MW species (Fig. 1E and F). Similar results were obtained for the VP31–81 construct, with 96.9 ± 1.7% of the band corresponding to the dimeric form of the domain (Fig. 1E and F). However, the VP389–221 construct showed a notable decrease in dimerization/oligomerization capacity (43.5 ± 15.5%). Similar results were obtained using glutaraldehyde as a crosslinker (Fig. S4), reinforcing the importance of the D1 domain in VP3 dimerization. Indeed, AF3 predicted with high confidence that the full-length protein formed a dimer through D1 contacts (Fig. S1A and D).
Small-angle X-ray scattering data show the high flexibility of VP3 dimers in solution
To better understand the structural dynamics of VP3 dimers in solution, we characterized the three VP3 constructs by small-angle X-ray scattering (SAXS). The SAXS profiles are shown in Fig. 3A with the Guinier plots [log I(s) vs. s2] shown in the insets. The radii of gyration, Rg, of the VP31–81, VP389–221, and VP31–221 constructs were 2.43 ± 0.01, 2.54 ± 0.02, and 3.86 ± 0.02 nm, respectively. The corresponding MWs, determined using Bayesian estimation (30) from four concentration-independent methods (Porod invariant (31), SAXSMoW (32), volume of correlation (33), and size and shape (34)), were 21.2, 14.8, and 46.7 kDa, respectively (Table S2). In line with the data obtained by SEC-MALS analysis, the MWs corresponded to homodimeric forms for VP31–81 and VP31–221, while VP389–221 appeared as a monomer (calculated monomer weight = 17.8 kDa). The pair-wise distance distribution, P(r), analyses for VP31–221 and VP389–221 were typical of elongated flexible proteins, with maximum intramolecular distances of 17.0 and 10.0 nm, respectively. In accordance with these observations, their Kratky plots indicated that the VP31–221 and VP389–221 fragments contain disordered regions. To further understand the structure of these constructs in solution, we analyzed the SAXS data in terms of ensembles.
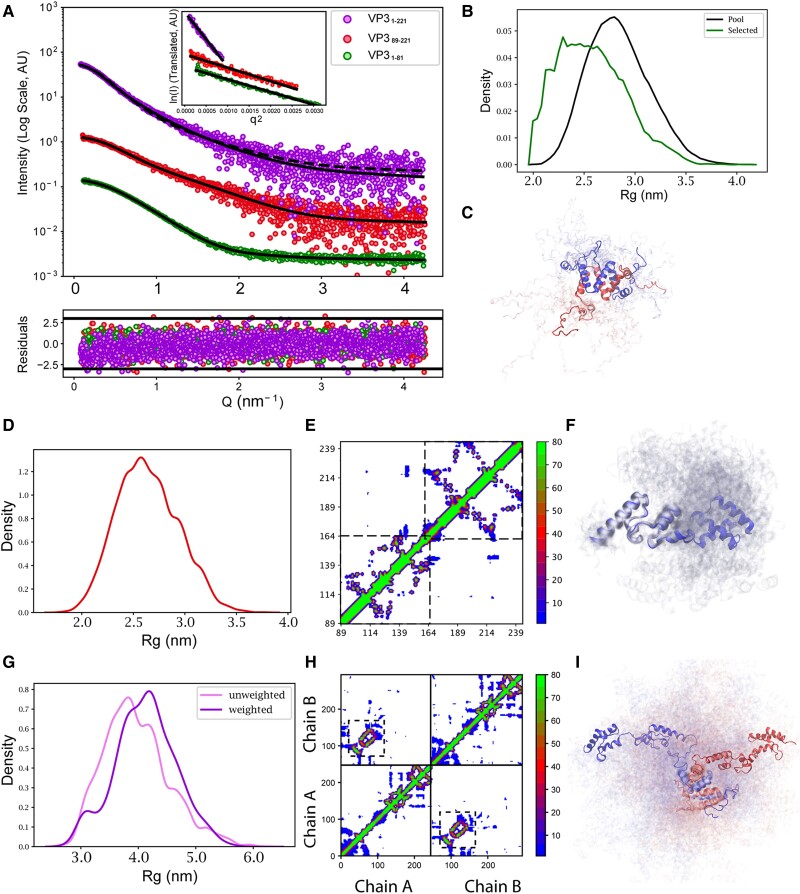
Fig. 3.
SAXS analyses of the IBDV VP3 constructs. A) Scattering intensity as a function of the momentum transfer Q measured for VP31–81 (green), VP389–221 (red), and VP31–221 (purple). The intensities are displaced along the y-axis for better inspection. Black lines correspond to the structural fitting of the data using different approaches (see main text). In the case of VP31–221, comparison with the simulation (dashed line) is shown together with the fitting after the Bayesian inference reweighting (solid line). The point-by-point residuals of each fit are shown at the bottom of the panel with the same color code. The inset displays the Guinier region for each one of the experimental profiles. B) Radius of gyration (Rg) distribution of the EOM-derived subensembles (green) of VP31–81 compared with that of the initial pool of conformations computed with RanCh (black). C) Subensemble of 20 conformations of VP31–81 derived from the EOM fit. The two chains of the dimer are colored in two different colors. D) Rg distribution of the conformations from the trajectory computed for VP389–221 using all-atom MD simulations, which describe the data without the need for further optimization. The broad distribution of Rg values indicates the important degree of flexibility of the protein. E) Contact map (percentage of conformations with Cα–Cα distances below 8 Å) computed from the MD trajectory of VP389–221. The His-tag is not displayed. The two squares indicate the position of the folded domains in this construct, highlighting the presence of transient interdomain interactions. F) Ensemble of 20 structures randomly selected from the trajectory representing the conformational sampling of VP389–221. G) Rg distribution of the reweighted ensemble using the Bayesian Inference approach (purple) of VP31–221 compared with that of the initial CG trajectory computed with Martini3 (pink). H) Contact map (percentage of conformations with Cα–Cα distances below 8 Å) computed from the trajectory of VP31–221. The His-tag is not displayed. Solid lines indicate the two chains of the dimer. Off-diagonal signals indicate interchain interactions within the ensemble with the dashed-line squares indicating the dimerization interface. I) Ensemble of 20 structures randomly selected from the reweighted trajectory representing the conformational sampling of VP31–221. The two chains of the dimer are colored in different colors.
For VP31–81, we generated 10,000 structures of the flexible loops using RanCh (35) while keeping the arrangement of the dimer intact, as seen in the crystal structure. The resulting pool was filtered using the ensemble optimization method (EOM) to get a subensemble that collectively fits the SAXS data (30). An excellent alignment with the SAXS data (χ2 = 1.02) was obtained (Fig. 3A), further supporting the dimeric form of VP31–81 in solution. The Rg distribution of EOM-selected pool exhibited compaction compared with the initial pool, suggesting some interactions between the flexible loops (Fig. 3B and C).
The VP389–221 ensemble was generated using all-atom molecular dynamics (MD) simulations. Four 800 ns trajectories were simulated using a 99SB-disp force field. Then, 20,000 equally spaced frames were extracted from the trajectories and added the flexible N-terminal tail using RanCh (35). The average theoretical SAXS curves resulting from this ensemble had a complete alignment with the one obtained experimentally (χ2 = 0.97), and no further EOM optimization was required (Fig. 3A). The contact map calculated from the combined trajectories further supports that the two subdomains retain their tertiary structures (Fig. 3E and F). Interestingly, despite retaining an important degree of flexibility, the contact map indicates that this construct presents transient interdomain contacts. To quantify them, we clustered the trajectory in terms of the interresidue contacts and found three prominent clusters (Fig. S5). The largest cluster consists of structures with no contact between the two domains of VP389–221. The two smaller clusters involve contacts between residues 115–118 (ATPEWV) and 175–180 (GAPGQA), and between residues 89–90 (GP) and 173–179 (IYGAPGQ) of the wild-type (WT) protein, equivalent to 50–55, 110–115, 22–25, and 108–114 sequences of the recombinant protein construct, respectively. Importantly, those residues correspond to hydrophobic regions of the protein.
In order to generate an ensemble of VP31–221, which is a large dimeric particle, coarse-grained MD simulations were performed using Martini3 force field (36). As standard Martini parameters are known to give overtly compact structures for multidomain proteins, and following previous studies (37), three trajectories were generated by rescaling the solute–water interactions by 2, 4, and 6%, and their individual agreements evaluated with the experimental SAXS profile. The simulation with rescaling factor of 4% displayed a correct agreement with the experimental SAXS data (χ2 = 2.1) and very similar Rg (3.90 vs. 3.86 nm; Fig. 3A, dashed line). Reweighting this trajectory using Bayesian inference approach (38) led to a substantial improvement to the SAXS data (χ2 = 1.1; Fig. 3A, solid line) with an Rg that is slightly more compact than the initial pool (Figs. 3G and S6). The contact map of the reweighted ensemble showed interactions between the D1 and D2–D3 domains of each monomer as well as between the D2–D3 domains of the two monomers of the dimer (Fig. 3H). These transient interdomain interactions rationalize the abovementioned enhanced compactness of the optimized ensemble.
Taken together, the biophysical and biochemical characterizations indicate that D1 drives the dimerization of IBDV VP3.
D1 domain is crucial for IBDV viability
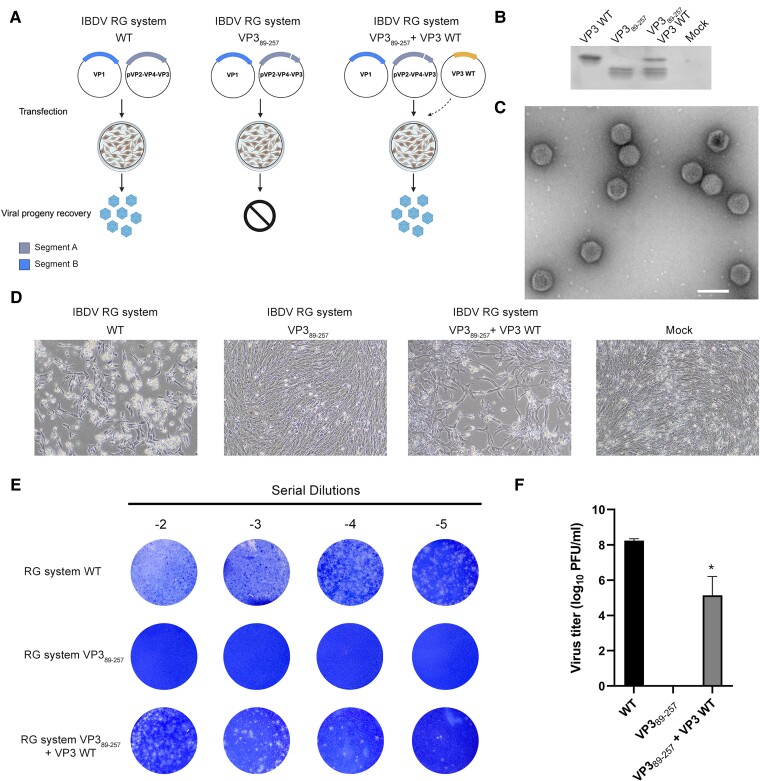
To investigate the role of the aminoterminal moiety of VP3 in the context of a viral infection, comprehensive reverse genetic (RG) systems, each expressing the viral genomic segments under the polymerase II promoter (WT or VP389–257), were developed (Fig. 4A). Upon co-transfection of both WT plasmids, viral proteins were successfully synthesized, as confirmed by western blot analysis (Fig. 4B). Subsequently, a substantial amount of viruses, capable of inducing cytopathic effects, was produced in avian cell cultures (cell line QM7), 96 h posttransfection (Fig. 4C and D). The plaque assay confirmed the recovery of infectious viral particles in the cell supernatant after transfection, reaching an average viral titer of 1.73 ± 0.5 × 108 PFU/mL (Fig. 4E and F). However, when the region of VP3 containing D1 was truncated, no cytopathic effect was observed posttransfection despite the expression of the viral proteins (Fig. 4B and D). More importantly, the plaque assay confirmed the lack of infectious particles in the supernatants of cells that were transfected with the truncated RG system (Fig. 4E and F). The latter phenotype was effectively reversed by co-transfection with a plasmid containing the full-length VP3 sequence, leading to the recovery of the cytopathic effects, as observed in cells transfected with both WT genomic segments (Fig. 4B and D). Indeed, viable viruses were detected in those supernatants, albeit the viral titers were significantly lower than those obtained with the WT plasmids for the RG system (Fig. 4E and F). Transfection efficiency of WT VP3 plasmid directly impacts in the virus recovery.
Fig. 4.
The D1 of IBDV VP3 is essential for the virus life cycle. A) Schematic representation of the RG system encoding each of the IBDV genomic segments in plasmids (pCAGEN-SegA, pCAGEN-SegB) and the experimental set-up. Both plasmids were transfected in QM7 cells and virus production was analyzed by monolayer visualization and staining. Genomic Segment A with a deletion of the N-terminal domain (pCAGEN SegA VP389–257) was co-transfected with pCDNA3-VP3 encoding the full-length WT protein “Created in BioRender. Gimenez, M. (2024) BioRender.com/i89z961”. B) Western blot analysis of the transfected cells with the RG plasmids. C) Purified IBDV virus produced with the RG system. D) Bright field micrographs of QM7 monolayers 96 h posttransfection. E) Titration of extracellular viruses of cells transfected with the RG systems using the plaque assay, as described in Materials and methods. Images are representative of three independent trials. F) Bar graph shows the mean viral titers of three independent trials expressed in PFU/mL. Error bars show SD. *P < 0.05.
Altogether, these results demonstrate the essential role that VP3 D1 plays in the intricate machinery of IBDV.
D1 is essential for capsid assembly
To understand how VP3 N-terminal domain deletion affected IBDV production, two main functions of this protein were evaluated: binding to dsRNA (18, 39) and scaffolding in capsid morphogenesis (10, 40, 41). To investigate the role of the VP3 D1 in dsRNA binding, an electrophoretic mobility shift assay (EMSA) was performed with a short dsRNA oligonucleotide (28 bp). The assay indicated that the N-terminal moiety of VP3 containing D1 was not able to bind the oligonucleotide even in a 120-fold protein excess (Fig. S7). In contrast, constructs containing D2–D3 (VP389–221) and D1–D2–D3 (VP31–221) were able to shift dsRNA mobility, showing direct binding to the short dsRNA (Fig. S7). Strikingly, at identical molar concentration, the dsRNA shift in samples containing D2–D3 was less than the observed for the D1–D2–D3 construct counterpart, suggesting that D1 can contribute to dsRNA binding to some extent (Fig. S7).
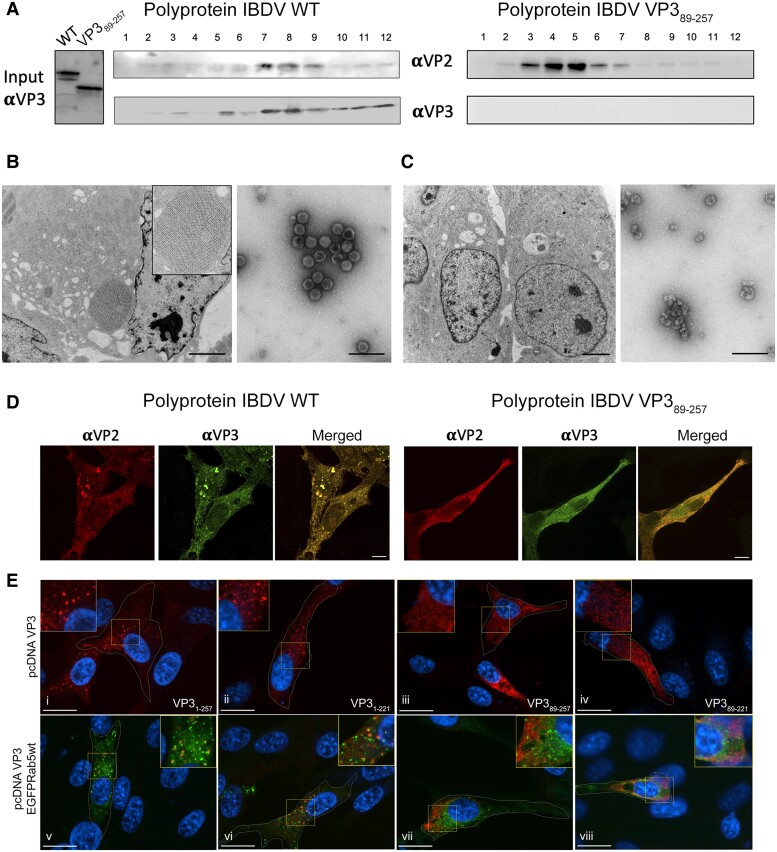
Previous characterization of birnavirus capsid morphogenesis showed that procapsid assembly is assisted by a dual scaffolding role of VP3 (40). Assessing the capacity of the polyprotein (pVP2-VP4-VP3) to form icosahedral capsids with T = 13 geometry is a direct indicator of the scaffolding function of VP3. Thus, in the context of the host cells, transfection of QM7 avian cells with the WT RG system resulted in the occurrence of typical cytoplasmatic paracrystals, characterized by tightly packed T = 13 IBDV capsids (Fig. 5B). In contrast, in cells transfected with RG system VP389–257, the absence of D1 resulted in the complete lack of any similar structures within the cytoplasm of the cells (Fig. 5C). To confirm these findings, the WT or VP389–257 versions of IBDV polyprotein were expressed (see Materials and methods and SI Appendix, Section 1), and the resulting virus-like particles (VLPs) were purified and analyzed by negative staining EM (Fig. 5A and B). Expression of the WT polyprotein yielded the typical icosahedral capsids of ∼70 nm, as previously described (42), similar to authentic virions (Figs. 4C and 5B). In contrast, after expression of the VP389–257 polyprotein, the icosahedral VLPs were replaced by amorphous assemblies of VP2 (Fig. 5A and C), and no VP3 was detected in gradient fractions (Fig. 5A). Most of the mutant VP3 protein remains in the soluble fraction of the first purification step and therefore detached from VP2 structures purified in the sucrose gradient.
Fig. 5.
The IBDV VP3 D1 domain is crucial for capsid morphogenesis and for VP3 targeting of EEs. A) Western blot analysis of VP3 in input samples and purification (left panel), fractions of purification gradient (fractions 1 to 12 from top to bottom in gradient) for WT (central panel), and VP389–257 (right panel) polyprotein. B) Left panel, TEM of a QM7 cell transfected with the RG system WT. The inset depicts a typical cytoplasmatic paracrystalline array of IBDV viruses. Scale bar represents 2.5 µm. Right panel, electron microscopy images of purified WT VLPs expressed with recombinant vaccinia virus (fraction 8 of the gradient showed in A). Scale bar represents 200 nm. C) Left panel, TEM of QM7 cells transfected with the RG system VP389-257. Right panel, electron microscopy images of sample corresponding to fraction 5 of the gradient shown in A. D) Immunofluorescence labeling showing the subcellular distribution of IBDV VP3 and VP2 in QM7 cells transfected with RG WT or VP389–257 plasmids (segments A). Scale bars represent 5 µm E) Immunofluorescence labeling showing the subcellular distribution of the above indicated VP3 constructs in QM7 cells co-expressing EGFP-Rab5 WT. Scale bars represent 10 µm.
As a scaffolding protein, VP3 must interact with pVP2 and later with VP2 to accomplish its function. Therefore, the subcellular distribution of both proteins was expected to be similar. Indeed, the immunofluorescence labeling of both proteins in QM7 cells transfected with the WT RG plasmid for segment A, showed that the capsid protein VP2 overlapped with VP3 (Fig. 5D). Both proteins were distributed throughout the cytoplasm, generally in fine granules, but some accumulations of both fluorescent signals were observed in large aggregates (Fig. 5D), as previously described (9). In contrast, these aggregates were not observed when the RG plasmid VP389–257 was expressed (Fig. 5D), and both proteins were diffusely distributed with scarce overlapping.
Taken together, these results indicate that D1 is essential for the scaffolding function of VP3.
D1 domain is necessary for endosomal targeting
It was previously demonstrated that both viral and recombinant IBDV VP3 presented in clusters on the outer leaflet of early endosomes (EEs) through their association to phosphatidylinositol 3-phosphate [PtdIns(3)P] (22, 23, 43). This was suggested to be related to the oligomeric capacity of VP3 (23). Thus, the dimerization of VP3 was hypothesized to be a necessary intermediate step for protein oligomerization and for VP3 targeting of EEs (22, 23, 43, 44). To test this hypothesis, immunofluorescence against VP3 was performed in QM7 cells ectopically co-expressing variants of VP3 and EGFP-Rab5 GTPase, the latter representing a hallmark of EEs. In agreement with previous findings, the full-length VP31–257 showed a punctate cytoplasmic distribution (Fig. 5Ei) (22, 23, 43, 44). A similar distribution was observed with construct VP31–221, which lacks the C-terminal tail (Fig. 5Eii). The puncta patterns were not altered by co-expression of EGFP-Rab5 GTPase, indeed both proteins showed a marked overlapping with Rab5, indicating their co-occurrence on EEs membranes (Fig. 5Ev and vi). On the other hand, the expression of mutants VP389–257 and VP389–221, lacking the D1 domain, exhibited a fine-grained cytoplasmic and diffuse distribution not associated with the EEs marker Rab5 (Fig. 5Eiii, iv, vii, and viii). These results support that VP3 dimerization and posterior oligomerization produced VP3 accumulation on EEs membranes independent of its C-terminal tail.
Discussion
In this study, we performed a comprehensive structural, biochemical, and cellular characterization of the N-terminal domain of IBDV VP3, demonstrating that it is an independent dimerization domain, crucial for virus viability.
The homotypic interaction of this protein region was suggested in a previous study, based on the analysis of sequential VP3 deletion mutants in yeast two hybrid assays (25). Here, we provide the structural basis underlying those results. The compact dimeric arrangement of the four α-helices forming this domain (Figs. 2C and S3B) is maintained through multiple interactions involving hydrophobic side chains, as observed in the crystal structures (Fig. 2E). The dimeric assembly is retained in the largest protein construct (VP31–221, containing D1–D2–D3), but is lost in the absence of D1 (Fig. 1C–F). Structural data obtained in solution by SAXS, and also supported by SEC-MALS experiments, confirm the role of D1 as the dimerization domain of IBDV VP3.
SAXS data along with MD simulations allowed building a more detailed structural model for the VP3 polypeptide in solution, highlighting the dynamic nature of this protein. The linker connecting D1 with D2 and D3 is intrinsically disordered, and the connection between domains D2 and D3 also displays extensive flexibility. Notably, intramolecular contacts between D2 and D3 have been identified, inducing an important population of compact conformations. This compact structure is in equilibrium with a more extended one, closely resembling the crystal structure of the D2–D3 domains previously elucidated (24) and equivalent to that solved here for the P212121 crystals (Fig. S2). Similarly, the VP31–221 construct is explained as an ensemble of multiple conformations, featuring the dimeric arrangement of the N-terminal domain, followed by multiple open and closed conformations of the remaining domains in solution (Fig. 3G–I). The most extended conformation of this protein exposes all the electropositive patches involved in RNA binding per dimeric VP3 molecule (18, 24). This conformation is likely adopted by the full-length protein during binding to genomic dsRNA, in the formation of the RNP complexes, where no dsRNA compaction was observed (15). It has been described that one VP3 molecule could cover 26 bp (∼73 Å) (15), a distribution that would be compatible with an extended conformation of dimeric VP3s on the dsRNA surface. In this configuration, electropositive patches in D2 and D3 of each protein monomer could face the nucleic acid phosphate backbone. In this context, the role of the D1 domain of VP3 would be to facilitate access to the two dsRNA-binding sites in the same molecule, rather than the direct participation of the domain in the binding as indicated in the EMSA experiments (Fig. S7).
Removal of the N-terminal end from the full-length protein severely impairs other VP3 functions, highlighting the importance of its multimeric state for protein function. In particular, truncation of the D1 moiety prevents viral replication, as evidenced by the absence of intracellular viruses, the lack of infective particles in cell culture supernatants, as well as a lack of VLP recovery (Figs. 4D–F and 5B, C).
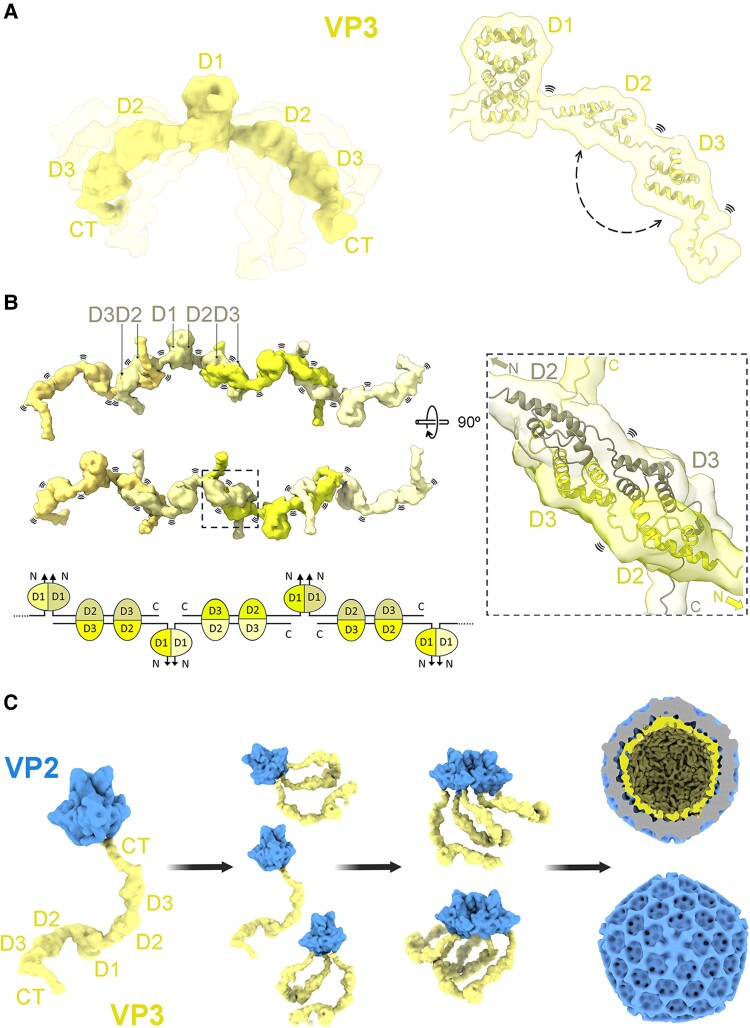
VP3 is known to act as a scaffolding protein, forming an inner shell beneath the viral VP2 capsid (9, 45). Multiple interactions occur between VP3 and pVP2 or VP2 during the IBDV morphogenesis, facilitating the modulation of the structural polymorphism and subsequent capsid stabilization (9, 10, 46). The oligomeric state of VP3 is a key determinant of this function (41, 45). Consequently, disruption of the dimerization capacity of VP3—seen as an indispensable state for oligomerization—inhibits capsid assembly, demonstrating that interactions between the C-terminal of VP3 and pVP2 are not sufficient for capsid formation. In fact, the interaction of the C-terminal tail of VP3 with VP2 (10) is compatible with the VP3 dimerization through its D1 domain, as seen in the crystal structures shown in this work. In this scenario, domains D2 and D3 are able to interact with each other, both forming self-interactions, as indicated by SAXS data (Fig. S5). These formed intermolecular contacts can be seen in the crystal structures of the D2–D3 domains (24) (Fig. S2), and are suggested by crosslinking experiments (Figs. 1E, F and S4). This interplay of interactions would result in the formation of flexible VP3 multimers, significantly increasing the possibilities of size and shape explored by the VP3 quaternary structure, that together with the VP2–VP3 interactions mediated by the VP3 C-terminus, function as scaffolding protein in capsid morphogenesis (Fig. 6).
Fig. 6.
IBDV VP3 contacts and capsid assembly model. A) Full-length VP3 dimeric model generated by AlphaFold (28) combined with crystal structures (PDB ID: 2R18). Protein flexibility is depicted in semitransparent. Regions involved in interdomain contacts are indicated with a dashed arrow and flexible regions with the symbol (right panel). B) Model for VP3 multimerization mediated by D1 and by the contacts between D2 and D3 of neighbor dimeric molecules, mimicking the antiparallel arrangement observed in D2–D3 crystals (24 and P212121 structure of this work), forming highly flexible elongated oligomers. The dashed box in the right panel is a magnification of the selected region of the model. The bottom panel shows a schematic representation of the model. C) Contribution of VP3 dimerization to the IBDV capsid assembly. Model of the dimeric VP3 interacting with VP2 trimers. While one or two VP3 C-terminal tails of one VP3 dimer are interacting with one or two VP2 trimers, the N-terminal domains of VP3 are maintaining its dimeric state. At the same time, some C-terminal tails or the middle domains (D2–D3) of a neighbor VP2–VP3 complex can establish contacts with the first complex, pulling from VP2 and guiding the capsid formation as a scaffold protein.
Consistent with this model, we did not observe the accumulation of VP3 on EEs (Fig. 5E), supporting the hypothesis that VP3 oligomerizes on PtdIns(3)P-enriched endosomes (22, 23, 43).
In addition to our findings, previous studies have also highlighted the importance of VP3 D1 domain in modulating IBDV replication. For instance, residue H28 is essential for binding to the chicken elongation factor 1α, thereby enhancing virus replication (26). Additionally, residues in the aminoterminal domain of VP3 bind Beclin1, inhibiting autophagy, a natural defense response of host cells (27).
The VP3 D1 domain sequence is relatively well-conserved across related pathogens within the Birnaviridae family (Fig. S8) (24). In particular, the strictly conserved G90 (IBDV numbering), located in the D1–D2 linker, could be one of the residues responsible for the high flexibility observed between D1 and D2D3 domains in birnavirus VP3 proteins (Figs. 3I, S1, and S8). In addition, some residues involved in the stabilization of the D1 dimeric interface, such as W36 in IBDV, or the residues L31 and D53, participating in the positioning of the D1 helices α1-α2 and α3-α4, are also strictly conserved. Earlier research highlighted the importance of the equivalent domain in infectious pancreatic necrosis virus (IPNV), a pathogen of great concern in the aquaculture industry (47). Using a set of deletion mutants, it was demonstrated that the aminoterminal end (residues 1–101) was involved in IPNV VP3 self-interactions as determined by a yeast two-hybrid approach (47). This region overlaps with the IBDV VP3 D1. Indeed, the first 42 residues of IPNV VP3 are not necessary for dsRNA binding (47), but successive truncations starting from residue 81 progressively reduce the binding until prevented. In line with these results, we demonstrate that the IBDV VP389–221 construct has a lower dsRNA binding capacity in comparison with VP31–221 (Fig. S7). Even with limited sequence similarity, other distantly related members of the Birnaviridae family, such as the Entomobirnavirus Drosophila X virus (DXV), show a closely related dimeric structure for its VP3 D1 domain (48) (Fig. S8C). The structures solved between residues 43–90 were three α-helixes that overlap with α-helix 2 to 4 in IBDV VP3 (rmsd = 2.23 Å, Cα alignment of residues 45–85 for DXV and 25–65 for IBDV). These observations suggest that both the helical structure and the ability of D1 to form compact dimers are common features of birnaviruses. This is also suggested by the structural predictions with AF3 for the full-length protein (Fig. S1). Interactions with other VP3 partners, like the viral polymerase VP1 and the dsRNA, in which VP3 structure could change, are not considered in this work.
Collectively, our findings emphasize the critical role of the VP3 D1 domain in the IBDV life cycle and highlight the potential of this small domain for the design of antiviral therapies not only against IBDV but also against other economically significant pathogens of the Birnaviridae family.
Materials and methods
Recombinant plasmids and cloning
Details of plasmids construction are given in SI Appendix, Section 1.
Protein expression and purification
All protein constructs for structural and biophysical analyses were expressed in E. coli BL21 (DE3) at 17 °C overnight. After lysis in 50 mM Tris-HCl pH 8, 500 mM NaCl, 1 mM DTT (buffer A) adding 10 µg/mL lysozyme, 10 µg/mL DNase; 10 µg/mL RNase; 4 mM MgCl2 and centrifugation, recombinant proteins were purified by His-trap and Superdex 200 10/300 column and stored in buffer A at −80 °C. Details of protein purification are summarized in SI Appendix, Section 1.
Crystallization data collection
Crystallization screenings were performed using a nanoliter-drop crystallization robot in 96-well Greiner plates. Protein solution of VP31–222 (30 mg/mL) crystallized in 1.5 M NaCl, 0.1 M sodium acetate pH 4.5 (P21 and P3221 crystals) and 0.8 M LiSO4, 0.05 M sodium cacodylate pH 6 (P212121 crystals). All crystals were flash-frozen in liquid nitrogen and protected with 20% (v/v) glycerol. More details are summarized in SI Appendix, Section 1.
Structure solution and refinement
The structure in P3221 space group was solved by de novo phasing method using ARCIMBOLDO software (49). The P21 structure was solved by MR with MOLREP (50), using one D1 monomer from the trigonal structure as a search model containing four copies of the polypeptide chain. The P212121 crystal was solved by MR using PDB ID: 2R18 (24) as a search model. All structure refinements were performed by alternating cycles of automatic refinement in Phenix (51) and manual model rebuilding using Coot (52). Data collection and refinement statistics are displayed in Table S1. Additional details are summarized in SI Appendix, Section 1.
Size exclusion chromatography MALS
The purified proteins were subjected to SEC using a Superdex 200 10/300 GL column in buffer A and monitored by an UV detector, a differential refractometer for refractive index (OPTI-rEx; Wyatt Corp.) and a static MALS detector (Dawn-Heleos; Wyatt Corp.). ASTRA 7 software (Wyatt Corp.) was used for data collection and analysis. Additional details are summarized in SI Appendix, Section 1.
Differential scanning fluorimetry
The stability of VP3 constructs was measured in 25 µL mixture reaction containing 2 µM protein, 5X SYPRO orange by DSF in 96-well PCR plates using iCycler instrument coupled to an IQ5 multicolor real-time PCR detection system (BioRad). Fluorescence intensity in each well was measured via the SYBR/6-carboxifluorescein (FAM) filter during a temperature ramp from 20 to 95 °C. Additional details are summarized in SI Appendix, Section 1.
Crosslinking experiments
Crosslinking experiments were performed in 100 mM phosphate buffer pH 8, 500 mM NaCl using 120 and 24 µM of each VP3 construct in the presence of 12 μM of BS3 in 20 μL. Reactions were incubated 30 min at 25 °C. Alternatively, glutaraldehyde crosslinking was employed. Coomassie-stained gels resolving crosslinked samples were analyzed with GelAnalyzer 23.1.1 software. Additional details are summarized in SI Appendix, Section 1.
Electrophoretic mobility shift assay
A 28-bp dsRNA labeled with cyanine5.5 (Cy5.5; 8.3 nM constant final concentration) was mixed with variable protein concentrations (1 µM, 500 nM, and 50 nM) for each construct in 20 mM Tris pH 8 20 mM NaCl, 5% glycerol (24 µL final concentration), incubated during 10 min at room temperature (RT) and resolved in 15% polyacrylamide gels. Cy5.5 signal was registered with Odyssey infrared imaging system. Additional details are summarized in SI Appendix, Section 1.
RGs and IBDV production
IBDV recombinants were generated, as previously described (53), using a modified version of the RGs system of Qi et al. (54). Transfection of QM7 cells with plasmids pCAGEN-SegA and pCAGEN-SegB containing IBDV genomic segments A and B, respectively, produces WT IBDV virus (53). Additional details are summarized in SI Appendix, Section 1.
Negative staining and transmission electron microscope images
Purified viruses or VLPs were placed on carbon-coated copper grids and stained with 2% (w/v) uranyl acetate. All grids were visualized in Jeol JEM 1010 80 kV transmission electron microscope (TEM) with a Megaview 1kx1k CCD digital camera. Additional details are summarized in SI Appendix, Section 1.
Western blot and antibodies
See details in SI Appendix, Section 1.
Indirect immunofluorescence in cells and confocal microscope imaging
QM7 cells were seeded onto 12-mm glass coverslips and transfected or co-transfected employing FuGENE HD, following the manufacturer's instructions. After 12 h posttransfection, cells were fixed with 4% paraformaldehyde and permeabilized with a solution of 0.05% saponin containing 0.2% bovine serum albumin for 20 min at RT. Anti-VP3 or anti-VP2 antibodies were incubated for 90 min followed by a 90-min incubation with the corresponding secondary antibodies. Images were acquired with a Leica DMI6000B spinning-disc confocal microscope and analyzed by Volocity 6.1 software (PerkinElmer). Additional details are summarized in SI Appendix, Section 1.
SAXS experiments
The SAXS data for VP31–81 were collected at ESRF at variable protein concentrations in batch mode. The SAXS data for VP389–221 were collected at PETRAIII in SEC-SAXS mode while the data for VP31–221 were collected in batch mode at variable protein concentrations. The SEC-SAXS data were analyzed using CHROMIXS (55) from ATSAS suite (56). The ensemble of 10,000 structures for VP31–81 was generated using RanCh, and the subensemble to fit the SAXS data was selected using GAJOE (35, 57). Additional details are summarized found in SI Appendix, Section 1 and Table S2.
MD simulations
The MD simulations for VP389–221 were carried out using a 99SB-disp force field (58). The coarse-grained MD simulations for VP31–221 were carried out using Martini3 (36) force field. Ten microseconds long simulations rescaling the water–protein interactions for 2, 4, and 6% (37). Twenty thousand structures collected during the simulations were back mapped to all-atom structures (59), and the SAXS profiles were calculated using CRYSOL (60). The ensemble obtained with solute–solvent interaction scaling of 4% was further reweighted to fit the SAXS data using BioEN approach (38). Additional details are given in SI Appendix, Section 1.
Supplementary Material
Acknowledgments
The authors thank the Center for Neurobiology of Stress, and in particular Durga Achayra, at the University of Toronto Scarborough for the use of the imaging facility and the assistance during preparation and analysis of TEM samples. The authors thank Serene Moussaoui from the University of Toronto Scarborough for proofreading this manuscript. The authors thank the Platform for Automated Crystallization (PAC, Join IBMB-CSIC/IRB-Barcelona) and its staff members for the crystallization experiments.
Contributor Information
Diego Sebastian Ferrero, Institut de Biologia Molecular de Barcelona, CSIC, Parc Científic de Barcelona, Baldiri i Reixac 15, 08028 Barcelona, Spain.
María Cecilia Gimenez, Department of Biological Sciences, University of Toronto at Scarborough, Toronto, ON M1C 1A4, Canada.
Amin Sagar, Centre de Biologie Structurale (CBS), Université de Montpellier, INSERM and CNRS, 34090 Montpellier, France.
Javier María Rodríguez, Department of Structure of Macromolecules, Centro Nacional de Biotecnología (CNB-CSIC), C Darwin, 3, 28049 Madrid, Spain.
José R Castón, Department of Structure of Macromolecules, Centro Nacional de Biotecnología (CNB-CSIC), C Darwin, 3, 28049 Madrid, Spain.
Mauricio R Terebiznik, Department of Biological Sciences, University of Toronto at Scarborough, Toronto, ON M1C 1A4, Canada; Department of Cell and Systems Biology, University of Toronto, Toronto, ON M5S 3G5, Canada.
Pau Bernadó, Centre de Biologie Structurale (CBS), Université de Montpellier, INSERM and CNRS, 34090 Montpellier, France.
Nuria Verdaguer, Institut de Biologia Molecular de Barcelona, CSIC, Parc Científic de Barcelona, Baldiri i Reixac 15, 08028 Barcelona, Spain.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
The work at IBMB was funded by the Spanish Ministry of Science and Innovation (PID2020-117976GB-I00). The work at the CBS was funded by Labex EpiGenMed, an “Investissements d’avenir” program (ANR-10-LABX-12-01). The work at the CNB was supported by grants from the Spanish Ministry of Science and Innovation (PID2020-113287RB-I00) and the Comunidad Autónoma de Madrid (P2018/NMT-4389) to J.R.C. Mauricio R. Terebiznik funding to this project was provided by the Discovery Grants programs RGPIN-2018-05734 and RGPAS2018-522692 from the Natural Sciences and Engineering Research Council of Canada (NSERC). X-ray data were collected at ESRF, beamline ID29 (ESRF, Grenoble, France), and XALOC (ALBA, Cerdanyola del Vallés, Spain). SAXS data were collected at EMBL-bioSAXS P12 Beamline at Petra III storage ring (Hamburg) and BM29 BioSAXS beamline at ESRF (Grenoble). Financial support was provided by Instruct-ERIC (PID6902) for access to Hamburg synchrotron. The CBS is a member of France-BioImaging (FBI) and the French Infrastructure for Integrated Structural Biology (FRISBI), two national infrastructures supported by the French National Research Agency (ANR-10-INBS-04-01 and ANR-10-INBS-05, respectively).
Data Availability
All the structures determined in this study have been deposited in the Protein Data Bank with IDs: 9EQN, 9EQO, 9EQP. The SAXS data for protein constructs have been deposited at the Small Angle Scattering Biological Data Bank with accession codes: SASDUS6, SASDUR7, and SASDUS7.
References
- 1. Delmas B, et al. 2019. ICTV virus taxonomy profile: birnaviridae. J Gen Virol. 100:5–6. [DOI] [PubMed] [Google Scholar]
- 2. Rehman ZU, Meng C, Umar S, Munir M, Ding C. 2016. Interaction of infectious bursal disease virus with the immune system of poultry. Worlds Poult Sci J. 72:805–820. [Google Scholar]
- 3. Mahgoub HA. 2012. An overview of infectious bursal disease. Arch Virol. 157:2047–2057. [DOI] [PubMed] [Google Scholar]
- 4. Zachar T, et al. 2016. A 5-year study of the incidence and economic impact of variant infectious bursal disease viruses on broiler production in Saskatchewan, Canada. Can J Vet Res. 80:255–261. [PMC free article] [PubMed] [Google Scholar]
- 5. Jordan AB, Gongora V, Hartley D, Oura C. 2018. A review of eight high-priority, economically important viral pathogens of poultry within the Caribbean region. Vet Sci. 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mundt E, Beyer J, Müller H. 1995. Identification of a novel viral protein in infectious bursal disease virus-infected cells. J Gen Virol. 76:437–443. [DOI] [PubMed] [Google Scholar]
- 7. von Einem UI, et al. 2004. VP1 of infectious bursal disease virus is an RNA-dependent RNA polymerase. J Gen Virol. 85:2221–2229. [DOI] [PubMed] [Google Scholar]
- 8. Oña A, et al. 2004. The C-terminal domain of the pVP2 precursor is essential for the interaction between VP2 and VP3, the capsid polypeptides of infectious bursal disease virus. Virology. 322:135–142. [DOI] [PubMed] [Google Scholar]
- 9. Maraver A, et al. 2003. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J Virol. 77:6438–6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saugar I, et al. 2010. Electrostatic interactions between capsid and scaffolding proteins mediate the structural polymorphism of a double-stranded RNA virus. J Biol Chem. 285:3643–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lombardo E, et al. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J Virol. 73:6973–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tacken MGJ, Peeters BPH, Thomas AAM, Rottier PJM, Boot HJ. 2002. Infectious bursal disease virus capsid protein VP3 interacts both with VP1, the RNA-dependent RNA polymerase, and with viral double-stranded RNA. J Virol. 76:11301–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garriga D, et al. 2007. Activation mechanism of a noncanonical RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 104:20540–20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferrero D, Garriga D, Navarro A, Rodríguez JF, Verdaguer N. 2015. Infectious bursal disease virus VP3 upregulates VP1-mediated RNA-dependent RNA replication. J Virol. 89:11165–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luque D, et al. 2009. Infectious bursal disease virus: ribonucleoprotein complexes of a double-stranded RNA virus. J Mol Biol. 386:891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luque D, et al. 2009. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc Natl Acad Sci U S A. 106:2148–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Busnadiego I, Maestre AM, Rodríguez D, Rodríguez JF. 2012. The infectious bursal disease virus RNA-binding VP3 polypeptide inhibits PKR-mediated apoptosis. PLoS One. 7:e46768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Valli A, et al. 2012. The VP3 factor from viruses of birnaviridae family suppresses RNA silencing by binding both long and small RNA duplexes. PLoS One. 7:e45957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Cleef KWR, et al. 2014. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res. 42:8732–8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye C, et al. 2014. Inhibition of antiviral innate immunity by birnavirus VP3 protein via blockage of viral double-stranded RNA binding to the host cytoplasmic RNA detector MDA5. J Virol. 88:11154–11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma ST, et al. 2021. Mass spectrometry-based proteomic analysis of potential infectious bursal disease virus VP3-interacting proteins in chicken embryo fibroblasts cells. Virus Genes. 57:194–204. [DOI] [PubMed] [Google Scholar]
- 22. Gimenez MC, Zanetti FA, Terebiznik MR, Colombo MI, Delgui LR. 2018. Infectious bursal disease virus hijacks endosomal membranes as the scaffolding structure for viral replication. J Virol. 92:e01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gimenez MC, et al. 2021. Phosphatidylinositol 3-phosphate mediates the establishment of infectious bursal disease virus replication complexes in association with early endosomes. J Virol. 95:e02313-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Casañas A, et al. 2008. Structural insights into the multifunctional protein VP3 of birnaviruses. Structure. 16:29–37. [DOI] [PubMed] [Google Scholar]
- 25. Tacken MGJ, et al. 2003. Homotypic interactions of the infectious bursal disease virus proteins VP3, pVP2, VP4, and VP5: mapping of the interacting domains. Virology. 312:306–319. [DOI] [PubMed] [Google Scholar]
- 26. Yang B, et al. 2020. Chicken eEF1α is a critical factor for the polymerase complex activity of very virulent infectious bursal disease virus. Viruses. 12:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y, et al. 2020. Binding of avibirnavirus VP3 to the PIK3C3-PDPK1 complex inhibits autophagy by activating the AKT-MTOR pathway. Autophagy. 16:1697–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abramson J, et al. 2024. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 630:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodríguez D, et al. 2012. Practical structure solution with ARCIMBOLDO. Acta Crystallogr. D68:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hajizadeh NR, Franke D, Jeffries CM, Svergun DI. 2018. Consensus Bayesian assessment of protein molecular mass from solution X-ray scattering data. Sci Rep. 8:7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Porod G. 1951. Die Röntgenkleinwinkelstreuung von dichtgepackten kolloiden Systemen - I. Teil. Kolloid Zeitschrift. 124:83–114. [Google Scholar]
- 32. Fischer H, De Oliveira Neto M, Napolitano HB, Polikarpov I, Craievich AF. 2010. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J Appl Crystallogr. 43:101–109. [Google Scholar]
- 33. Rambo RP, Tainer JA. 2013. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 496:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Franke D, Jeffries CM, Svergun DI. 2018. Data mining in structural biology: machine learning methods for data analysis of biological macromolecules in solution. Biophys J. 114:2485–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. 2007. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 129:5656–5664. [DOI] [PubMed] [Google Scholar]
- 36. Souza PCT, et al. 2021. Martini 3: a general purpose force field for coarse-grained molecular dynamics. Nat Methods. 18:382–388. [DOI] [PubMed] [Google Scholar]
- 37. Thomasen FE, Pesce F, Roesgaard MA, Tesei G, Lindorff-Larsen K. 2022. Improving martini 3 for disordered and multidomain proteins. J Chem Theory Comput. 18:2033–2041. [DOI] [PubMed] [Google Scholar]
- 38. Köfinger J, et al. 2019. Efficient ensemble refinement by reweighting. J Chem Theory Comput. 15:3390–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kochan G, Gonzalez D, Rodriguez JF, Bionostra SL. 2003. Characterization of the RNA-binding activity of VP3, a major structural protein of infectious bursal disease virus. Arch Virol. 148:723–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luque D, et al. 2007. Infectious bursal disease virus capsid assembly and maturation by structural rearrangements of a transient molecular switch. J Virol. 81:6869–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mertens MJ, et al. 2015. A protein with simultaneous capsid scaffolding and dsRNA-binding activities enhances the birnavirus capsid mechanical stability. Sci Rep. 5:13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saugar I, et al. 2008. Structural polymorphism of the major capsid protein of a double-stranded RNA virus: an amphipathic α helix as a molecular switch. Structure. 13:1007–1017. [DOI] [PubMed] [Google Scholar]
- 43. Gimenez MC, et al. 2022. Rab1b-GBF1-ARF1 secretory pathway axis is required for birnavirus replication. J Virol. 96:e0200521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Delgui LR, Rodríguez JF, Colombo MI. 2013. The endosomal pathway and the Golgi complex are involved in the infectious bursal disease virus life cycle. J Virol. 87:8993–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mata CP, et al. 2018. The RNA-binding protein of a double-stranded RNA virus acts like a scaffold protein. J Virol. 92:e00968-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Irigoyen N, Castón JR, Rodríguez JF. 2012. Host proteolytic activity is necessary for infectious bursal disease virus capsid protein assembly. J Biol Chem. 287:24473–24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pedersen T, Skjesol A, Jørgensen JB. 2007. VP3, a structural protein of infectious pancreatic necrosis virus, interacts with RNA-dependent RNA polymerase VP1 and with double-stranded RNA. J Virol. 81:6652–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferrero DS, et al. 2021. Structure and double-stranded RNA-binding activity of the birnavirus Drosophila X virus VP3 protein. J Virol. 95:e02166-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodríguez D, et al. 2009. Crystallographic ab initio protein structure solution below atomic resolution. Nat Methods. 6:651–653. [DOI] [PubMed] [Google Scholar]
- 50. Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 66:22–25. [DOI] [PubMed] [Google Scholar]
- 51. Liebschner D, et al. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Biol Crystallogr. D75:861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zanetti FA, et al. 2024. Birnaviral hijacking of endosomal membranes. eLife. 13:RP97261. [Google Scholar]
- 54. Qi X, et al. 2007. An improved method for infectious bursal disease virus rescue using RNA polymerase II system. J Virol Methods. 142:81–88. [DOI] [PubMed] [Google Scholar]
- 55. Panjkovich A, Svergun DI. 2018. CHROMIXS: automatic and interactive analysis of chromatography-coupled small-angle X-ray scattering data. Bioinformatics. 34:1944–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Manalastas-Cantos K, et al. 2021. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J Appl Crystallogr. 54:343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tria G, Mertens HDT, Kachala M, Svergun DI. 2015. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2:207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Robustelli P, Piana S, Shaw DE. 2018. Developing a molecular dynamics force field for both folded and disordered protein states. Proc Natl Acad Sci U S A. 115:E4758–E4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wassenaar TA, Pluhackova K, Böckmann RA, Marrink SJ, Tieleman DP. 2014. Going backward: a flexible geometric approach to reverse transformation from coarse grained to atomistic models. J Chem Theory Comput. 10:676–690. [DOI] [PubMed] [Google Scholar]
- 60. Svergun D, Barberato C, Koch MHJ. 1995. CRYSOL– a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 28:768–773. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the structures determined in this study have been deposited in the Protein Data Bank with IDs: 9EQN, 9EQO, 9EQP. The SAXS data for protein constructs have been deposited at the Small Angle Scattering Biological Data Bank with accession codes: SASDUS6, SASDUR7, and SASDUS7.