Abstract
Precision medicine has the potential to provide more accurate diagnosis, appropriate treatment and timely prevention strategies by considering patients’ biological makeup. However, this cannot be realized without integrating clinical and omics data in a data-sharing framework that achieves large sample sizes. Systems that integrate clinical and genetic data from multiple sources are scarce due to their distinct data types, interoperability, security and data ownership issues. Here we present a secure framework that allows immutable storage, querying and analysis of clinical and genetic data using blockchain technology. Our platform allows clinical and genetic data to be harmonized by combining them under a unified framework. It supports combined genotype–phenotype queries and analysis, gives institutions control of their data and provides immutable user access logs, improving transparency into how and when health information is used. We demonstrate the value of our framework for precision medicine by creating genotype–phenotype cohorts and examining relationships within them. We show that combining data across institutions using our secure platform increases statistical power for rare disease analysis. By offering an integrated, secure and decentralized framework, we aim to enhance reproducibility and encourage broader participation from communities and patients in data sharing.
Subject terms: Translational research, Research management
A secure framework that harmonizes storage and querying of clinical and genetic data using blockchain technology was developed to support combined genotype–phenotype queries, improving transparency into how and when health information is used.
Main
The goal of precision medicine is to individualize medical treatment for patients based on their characteristics, including genetics, physiology and environment. Given the potential to improve patient outcomes and reduce healthcare costs, it has become a national research agenda in the United States and elsewhere1. However, its potential cannot be realized without unifying clinical and genetic data to improve understanding of clinical observations within their genetic context2–5. Although some progress has been achieved, such as the All Of Us research project, the UK Biobank and the eMERGE research network, several key hurdles remain3,6. For example, integrated data systems that harmonize distinct clinical and omics data formats are lacking, leading to missed opportunities2,5. Furthermore, larger sample sizes and diverse populations are necessary when attempting to link genotypes to diseases, as biobanks tend to contain small numbers of patients from disease cohorts. Thus, achieving large sample sizes is only possible by aggregating data across institutions in a systematic way1. This requires a framework for uniformly processing and analyzing clinical and genetic data from multiple sources3. Importantly, a critical barrier to addressing these needs is a lack of robust tools to ensure data integration while maintaining security4. An ideal platform for storage and analysis of clinical and genetic data would enable unified data storage of multidomain data, protect from loss and manipulation, provide appropriate and controlled access to researchers and record usage logs.
Linking clinical information (for example, electronic health records (EHRs)) with genetic data across multiple sites presents several logistical challenges. First, they are stored under different file formats in separate databases. This increases technical requirements as users must learn distinct tools to parse different data types. There are also different data security requirements, as EHRs are considered protected health information and cannot be easily shared, even in anonymized form3,7. This often results in the creation of multiple databases for different domains, with a heavy analysis load required for linking them. Although initiatives to integrate clinical and genetic data are ongoing (for example, HL7 FHIR Genomics8), they do not address issues related to data security in multisite settings8,9. As such, concerns relating to data ownership, cost and dissemination procedures remain unresolved. Blockchain, a distributed ledger technology, can be an infrastructure solution that overcomes a number of these logistical challenges due to its properties of security, immutability and decentralization10. Furthermore, the inherent flexibility of blockchain technology makes it readily compatible with and complementary to efforts related to data standardization and harmonization.
Despite the fact that the implementation of blockchain technology in science is in its infancy, there are numerous blockchain-based solutions for clinical or genetic data sharing10–12. Blockchain has been used for next-generation sequencing data indexing and querying13, omics data access logging14,15, pharmacogenomics data querying16, patient-controlled data sharing17,18, coordinating EHR information19,20, infectious disease reporting21 and trustworthy machine learning22. However, most of these solutions do not enable the integration of genetic and clinical data. Moreover, most of them rely on storing the data outside the blockchain with only hashed references stored ‘on chain’22,23. When data are stored outside the blockchain, several limitations exist. First, the integrity of data stored outside the network is not guaranteed by blockchain’s immutability, posing a risk of tampering. Second, access cannot be as strictly controlled or audited when data can be accessed via methods outside of the network. Importantly, data contributors do not retain sovereignty with the ability to directly audit how their data are used. Finally, storing data ‘on chain’ offers the ability to perform computation directly on the network. This streamlines processing, as data are easily co-queryable, and also ensures stronger oversight on what computation is performed by researchers.
Here we present a decentralized data-sharing platform (PrecisionChain) using blockchain technology that unifies clinical and genetic data storage, retrieval and analysis. The platform works as a consortium network across multiple participating institutions, each with write and read access10,13. PrecisionChain was built using the free version of blockchain application programming interface (API) MultiChain, a well-maintained enterprise blockchain24. Although MultiChain provides data structures called ‘streams’ for data storage, the vanilla implementation does not allow for multimodal data harmonization and was shown to be still inefficient with performance overheads and high data storage cost13. Therefore, we created a data model and indexing schema that harmonizes clinical and genetic data storage, enables multimodal querying with low latency and contains an end-to-end analysis pipeline25. We included a proof-of-concept implementation with an accessible front end to showcase functionality for building cohorts and identifying genotype–phenotype relationships in a simulated network. We also showed that we can accurately reproduce existing association studies using UK Biobank data. We further assessed the utility of our framework within the context of a rare disease by analyzing genetic and clinical data of patients with amyotrophic lateral sclerosis (ALS) using data from 26 institutions in the New York Genome Center (NYGC) ALS Consortium.
Results
PrecisionChain enables efficient indexing of multimodal data
We envisage PrecisionChain to be used by a consortium of biomedical institutions that share genetic and clinical data for research purposes (Fig. 1a). We developed a data model and indexing schemes to enable simultaneous querying of clinical and genetic data. To overcome challenges related to blockchain technology, specifically transaction latency and lack of data structures for flexible querying, we developed an efficient data encoding mechanism and sparse indexing schema on top of MultiChain’s ‘data stream’ feature25.
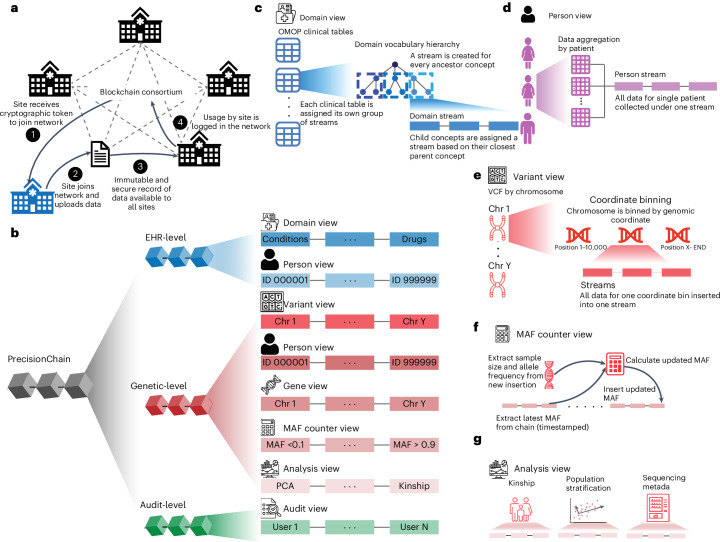
Fig. 1. Conceptual framework.
a, Consortium network. Network is made up of biomedical institutions. All sites share data on a decentralized blockchain platform maintained by all nodes. New joining institutions are verified via cryptographic tokens. Once joined, they can upload new data and access existing data. b, High-level indexing. Indexing of data into three levels: EHR, Genetic and Audit. EHR and Genetic levels are further divided into Domain and Person views and Variant, Person, Gene, MAF counter and Analysis views, respectively. Each view is made up of multiple streams with streams organized by property. c, Indexing of Domain view is by OMOP clinical table. Within each clinical table, we index streams using the OMOP vocabulary hierarchy. d, Indexing of Person views is by person ID with all data for a patient under one stream. This is done for both clinical and genetic data. e, Indexing of Variant view is by chromosome and genomic coordinates. f, MAF counter is organized by MAF range. MAF calculation occurs at every insertion. g, Analysis view records metadata to harmonize sequencing data, assess relatedness among samples and conduct population stratification.
We indexed data into three levels: clinical (EHR), genetics and access logs (Fig. 1 and Extended Data Fig. 1a–d). Within each level, we organized the data into views. We further used additional nesting within each view to enable efficient and flexible access to the data. We then created a mapping stream that records how data have been indexed and how to retrieve information under all views. This speeds up query time and allows us to efficiently store multimodal data under one network.
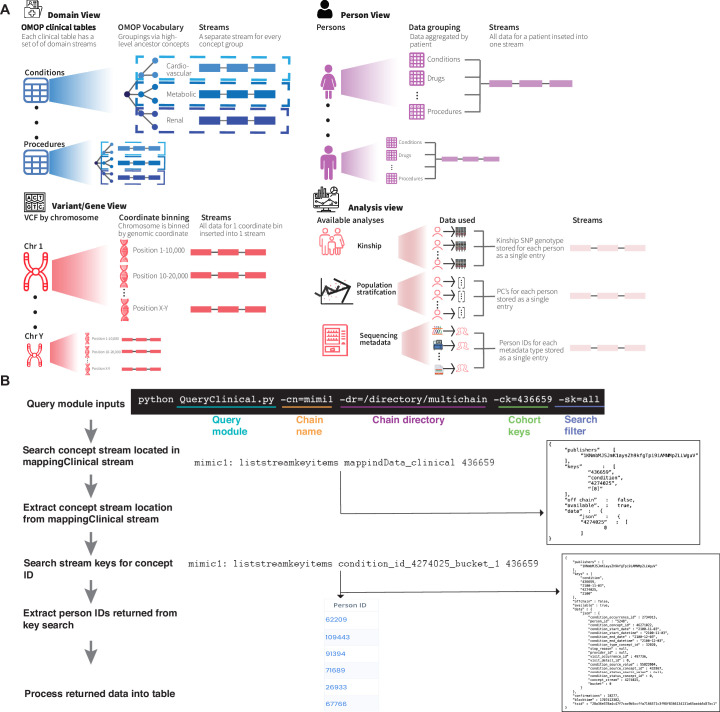
Extended Data Fig. 1. Indexing and Querying in PrecisionChain.
(A) Indexing in Domain view is done by clinical table and then OMOP vocabulary hierarchy. Each clinical domain has its own exclusive set of streams. Concepts are grouped by ancestor concept using the vocabulary hierarchy. Each ancestor group gets its own stream. Indexing in Person view is by person (person ID). All data (clinical or genetic) for a patient are inserted into the same stream. For clinical this is irrespective of domain and for variant this is irrespective of genomic coordinate bin. Note within a single stream, multiple patient data can be inserted. Indexing in Variant/Gene view is by genomic coordinate bin. All variants/genes within a set of continuous genomic coordinates are added to a single stream. Indexing in Analysis is by analysis type. Data for kinship and population stratification is stored per sample and data for sequencing metadata is stored by metadata type. (B) Flowchart of query process. User inputs required fields into the query module. The mapping stream is searched for the location of the stream holding data for that concept ID. The stream location is extracted from the mapping stream and the concept ID is searched in that stream. Person IDs returned from the stream search are retrieved and processed into a table. If additional search filters are added, these are processed on the returned data.
At the EHR level, we used the standardized vocabularies of the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) format (see Methods for details), which supports integration of clinical data from multiple sources26,27. OMOP is made up of concepts that represent some unique clinical information (for example, a specific medication or diagnosis)27. Under the EHR level, we developed two separate indexing schemes, which we call Domain view and Person view. The Domain view is indexed by concept type (for example, diagnosis, medications, etc.). If one queries the network with a concept (for example, diabetes diagnosis), the network will return all the patients with that record. The Person view is organized by patient ID—that is, each data stream contains the entire medical record of a patient to allow quick querying of single patients’ medical records. In both views, each entry contains several keys that can be used for specifying queries, including patient ID, concept ID, concept type, date of record (that is, when the clinical event occurred) and concept value or information (for example, laboratory value, medication dose, etc.). Combining multiple keys in a query enables cohort creation.
At the genetic level, we indexed data from variant call format (VCF) files to store and query genetic variants. We developed five sub-indexing schemes called Variant view, Person view, Gene view, Minor Allele Frequency (MAF) counter and Analysis view. Within the Variant view, we log all genetic variants into streams binned and indexed by their genomic coordinate. Users can extract patients’ genotypes for any variant using its genomic location. Patient view inserts all variant data for a patient into one stream, enabling fast retrieval of a patient’s genome. Gene view records the variants associated with specific genes, biological information about the gene and clinical and other annotations from ClinVar28, variant effect predictor (VEP) score29 and combined annotation-dependent depletion (CADD) score30. In doing so, gene view acts as a comprehensive repository of a variant’s known biological function and clinical impact. MAF counter is the most dynamic view of this level. It records the MAF values of the variants and is automatically updated as more patients are added to the network. We overcame the issues with immutability of stored data by timestamping the MAF values. System uses the MAF value with the most recent timestamp. In Analysis view, we recorded information necessary to conduct analysis, including sequencing and genotype calling metadata, principal components (PCs) for population stratification and variants needed to calculate kinship among samples.
A key aspect of controlled access is secure storage and query of audit logs to check for potential misuse. At the access logs level, whenever a researcher subscribes to the network or performs any kind of query or analysis, the information is automatically logged with a timestamp and the user’s wallet address. This creates an immutable record of use. Notably, the record is highly granular and can be searched in multiple ways. First, it is possible to determine exactly which records were viewed by a specific user. Second, it can return all user queries that returned a specific data point. The former is key for Health Insurance Portability and Accountability Act (HIPAA) audits, and the latter allows contributors to determine exactly how their data have been used. One key benefit of PrecisionChain is that the audit logs (and any other data on the chain) are immutable. This makes unauthorized alterations virtually impossible and ensures that any attempt to tamper with records is easily detectable.
One of the unique aspects of PrecisionChain is the ability to perform multimodal queries on the data. Query modules take advantage of the nested indexing scheme and mapping streams to efficiently retrieve information used for building cohorts and examining relationships (Fig. 2a). We provide the following queries in real time: (1) domain queries, such as pulling all patient IDs for individuals diagnosed with a particular disease; (2) patient queries, such as pulling all laboratory results for a single patient; (3) clinical cohort creation, based on any combination of clinical concepts, concept values, demographics and date ranges; (4) genetic variant queries, such as pulling all patient IDs that have a specific disease-causing variant; (5) patient variant queries, such as pulling the genome of a patient; (6) MAF queries, such as pulling all patients who have rare or common variants (that is, querying variants with a MAF threshold); (7) gene queries, such as pulling the IDs of all patients who have a disease-causing variant associated with a specific gene of interest; (8) variant annotation queries, such as retrieving all patients with variants annotated as pathogenic; (9) genetic data harmonization queries, such as extracting all samples aligned and processed using the same analysis pipeline; (10) kinship assessment queries, such as determining relatedness between samples in a cohort; (11) genetic cohort creation based on any combination of genetic variants, MAFs and sequencing metadata; (12) combination EHR and genetic cohort creation using both clinical and genetic logic gates (for example, all the patients with variant X and disease Y; Fig. 2b); and (13) combination EHR and genetic queries to identify genotype–phenotype relationships within a cohort (for example, presence of rare variants in a particular gene for patients with disease X compared to controls; Fig. 2c). See Extended Data Fig. 1e, Extended Data Table 1, Table 1 and Methods for more details on indexing and querying. Note that, due to the immutable nature of blockchain technology, the entries cannot be altered. Therefore, our system was designed to return queries with the latest timestamp if multiple entries exist.
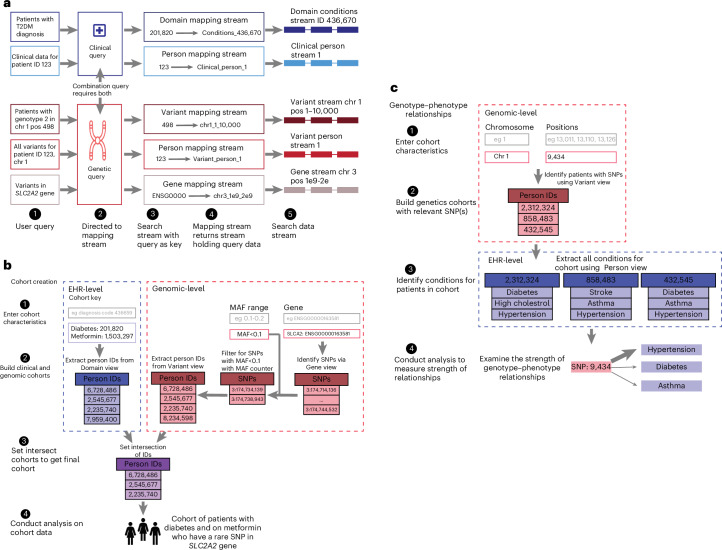
Fig. 2. Indexing and analysis on PrecisionChain.
a, Mapping stream indexing. Based on the users’ query, search keys are directed to the appropriate stream. A mapping stream is created for every view. Entries in the mapping stream follow a Key:Value structure (Key is the user’s input; Value is the stream where the data are stored). b, Cohort creation. Users input desired clinical characteristics, genes of interest and a MAF filter into the search function. Using the EHR-level ‘Domain view’, patient IDs for those that meet clinical criteria are identified. Using the Genetic-level ‘Gene, MAF counter and Variant views’, the appropriate variants are identified, and patient IDs with those variants are extracted. A set intersection of the two cohorts is done to create a final cohort, which can be analyzed further. c, Genotype–phenotype relationships. Users input variants of interest into the search function. Using the Genetic-level ‘Variant view’, IDs for patients with that variant(s) are extracted. All diagnoses for each patient are retrieved using the EHR-level ‘Person view’. The strength of relationship between each SNP and condition can be examined. ‘Gene view’ can give further information on what genes are carrying the variants, linking the clinical information to detailed genetic information (chr, chromosome; pos, position).
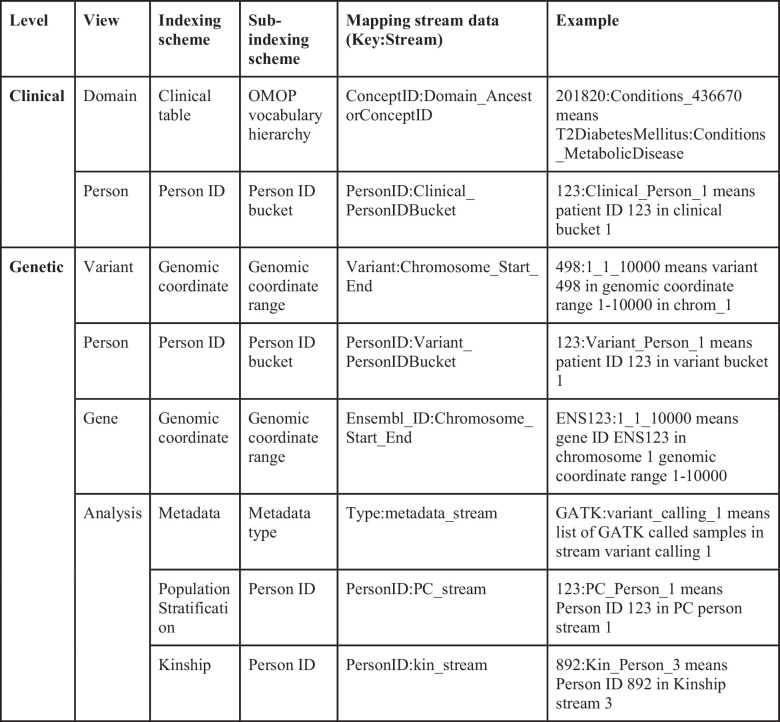
Extended Data Table 1.
How the mapping stream is structured to record the indexing structure of each view
Table 1.
Query modules available on the platform
| Query domain | Query name | Description | Example |
|---|---|---|---|
| Clinical | queryDomain | Query based on OMOP concept ID with date and/or value | Build cohort of patients diagnosed with diabetes and on metformin since 2015 |
| queryPerson | Query based on person ID(s) with date and/or concept and/ or value | Find all medications taken by patient(s) before 2018 | |
| Genetic | queryVariant | Query based on genomic coordinate and genotype with MAF and sequencing/technical analysis metadata filter | Find all patients with rare variants (MAF < 0.05) in chromosome 8 with variants called using GATK |
| queryPerson | Query based on person ID(s) and genomic location with MAF filter | Find all variants with MAF < 0.1 for specific patient(s) in chromosomes 11 and 12 | |
| queryVariantGene | Query based on variants associated with particular gene | Find all patients with rare variants (MAF < 0.05) in gene BRCA1 | |
| queryVariantAnnotations | Query based on variants with a particular clinical annotation | Find all patients with rare variants (MAF < 0.05) that are ClinVar annotated in chromosome 6 | |
| Analysis | querySamplePCA | Query based on person IDs that returns population stratification PC scores | Retrieve the top 20 PC scores for specific patient(s) |
| queryKinship | Query based on person IDs that determines relatedness between samples | Find level of relatedness between specific patient(s) in the network | |
| queryMetadata | Query based on sequencing and technical analysis metadata | Find all patients sequenced on Illumina NovaSeq, aligned using HISAT2, and variant called with GATK | |
| Combination | queryClinicalVariant | Query based on clinical cohort definition and gene of interest | Find all patients with rare variants in the SOD1 gene and a specific age of onset for ALS |
| queryVariantClinical | Query based on variants of interest returning patient clinical characteristics | Find all diseases for patients with a particular rare variant |
PrecisionChain provides security while being scalable
To showcase the value of our framework for precise cohort building and analysis, we simulated a data-sharing network consisting of clinical and genetic data for 12,000 synthetic patients. For clinical data, we used the Synthea patient generator to create EHR data in OMOP CDM format (Methods). Synthea has been shown to produce comprehensive and realistic longitudinal healthcare records that accurately simulate real-world datasets31,32. For genetic data, we simulated samples using 1000 Genomes Project (1000GP) individuals as reference (Methods)33,34. To reflect the technical considerations of genetic data sharing, we also assigned sequencing metadata to each sample (for example, sequencing machine, sequencing coverage, alignment pipeline and variant calling pipeline). We used the simulated network to showcase network functionality and the utility of combining genetics and clinical data in one infrastructure. For example, it is possible to build a cohort of patients who have a diagnosis of diabetes, take metformin and have a rare variant in the SLC2A2 gene, which is known to influence metformin response35. In this example, the availability of genetic and clinical information allows for more targeted cohort creation. See Table 1 for a full list of available query types.
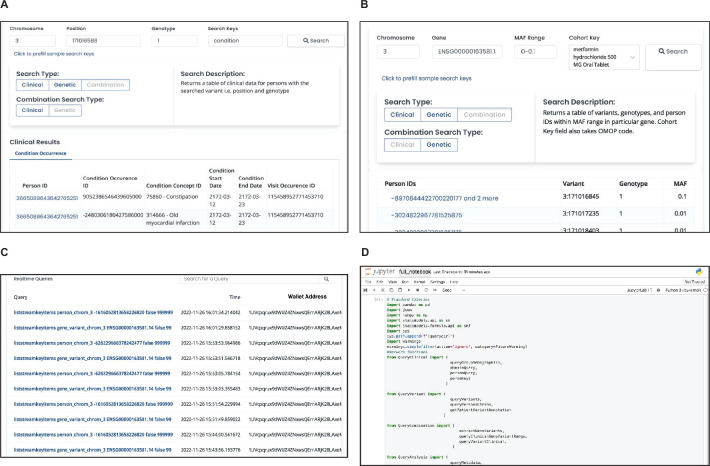
A major challenge with blockchain technology adoption is that end-users are not experts in distributed systems or cryptography. We developed a user-friendly front-end graphical user interface (GUI) for researchers to access the network, query data and run analysis via an interactive Jupyter Notebook (https://precisionchain.g2lab.org; username: test@test.com, password: test-ME). Researchers sign into the system with their username, and, in the back end, the blockchain recognizes the wallet addresses associated with the username and grants access. Users see drop-down menus and search windows in the front end, and all executions are performed over the blockchain network in the back end. For analysis, users can leverage one of the predefined functions in our query and analysis scripts or build new ones using the MultiChain command line interface (CLI) (Extended Data Fig. 2). We hope to increase adoption by abstracting away knowledge of blockchain algorithms while users are querying and analyzing data.
Extended Data Fig. 2. GUI.
(A) Combination Clinical Query. Users create a cohort using clinical and genetic data. In this example, a user is querying for patients with variants (MAF 0.0-0.1) in the SLC2A2 gene and are prescribed Metformin (SLC2A2 gene is known to influence metformin response). Variant level information for the cohort is returned. Clicking on the patient ID’s loads further demographic information (B) Combination Genetic Query. Extract clinical data for patients who have a specific variant of interest. In this example diagnosis information for patients with heterozygous genotype at position 3:17101658 (SLC2A2 gene) is returned. Clinical relationships with this variant can now be examined. (C) Administrative view. Administrators can view time-stamped logs of all queries conducted, filtering by user, query type and date. Information viewed is dependent on a user’s access level. (D) Analysis workbook. Users can leverage network functionality to build cohorts and conduct analysis that replicates traditional GWAS workflow.
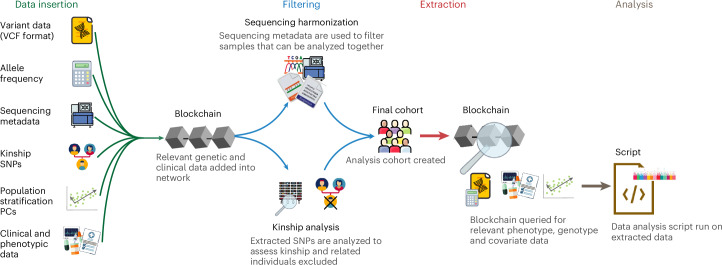
We then developed an ‘on chain’ analysis pipeline that can be used for a number of association studies for both common and truly rare genetic diseases, including genome-wide association studies (GWASs) and the classification of variants of uncertain significance (VUS) (see Methods for details). To illustrate the network’s analysis capabilities, we provide an example script to conduct a GWAS (can also be spinned as a Jupyter Notebook in the GUI), which uses built-in functionality to perform sample quality checks, harmonize genetic data from multiple sources, build analysis cohorts and extract relevant genetic, phenotypic and covariate information (Fig. 3). We provide security by enabling all analysis scripts to be run on the nodes of PrecisionChain to ensure that users cannot download patient data locally. Key to this analysis pipeline is the Analysis view, which facilitates genetic data harmonization, assessment of relatedness between samples and population stratification. For genetic data harmonization, we recorded technical metadata, such as sequencing machine, sequencing protocol, coverage, alignment pipeline and variant calling pipeline. Users can filter for samples based on any combination of metadata, enabling post hoc correction for batch effects. Although this approach mitigates much of the bias36,37, it does not completely eliminate all bias arising from analyzing data derived from different sources or variant calling pipelines. This issue is not unique to decentralized platforms but is a major challenge that large-scale genetics studies face. The research community suggested solutions, such as whole-genome sequencing (WGS) data processing standards that allow different groups to produce functionally equivalent results36 or iterative joint genotyping38. Owing to PrecisionChain’s modular build, these solutions can readily be adopted.
Fig. 3. Analysis pipeline and data insertion.
All relevant data are first inserted into the chain, including genetic and clinical data, sequencing metadata and population stratification PCs. Variant data are passed through a QC script before insertion. Filtering. Sequencing metadata are queried and filtered to extract patients who can be analyzed together. Patient relatedness is also assessed, and only unrelated samples are included in the final cohort. Extraction. Relevant phenotype, genotype and covariate information for the cohort is retrieved. Analysis. The data are analyzed and results are returned to the user.
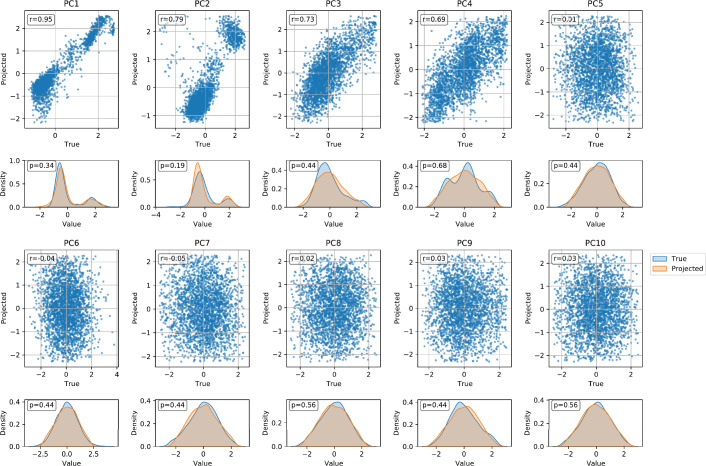
We used the National Center for Biotechnology Information (NCBI)’s Genetic Relationship and Fingerprinting (GRAF) protocol used in the database of Genotypes and Phenotypes (dbGaP)’s processing pipeline for kinship assessment39. We stored the kinship single-nucleotide polymorphisms (SNPs) used by GRAF in the kinship stream. For a given query of person IDs, one can extract the patients’ kinship SNP genotypes and calculate the kinship coefficient. For population stratification, we projected every patient’s genotype onto the PC loadings from the 1000GP and stored the top 20 PC scores for each patient. Although we use 1000GPʼs PCs to showcase this functionality, any reference population panel can be used. Notably, we calculated the PCs using an unbiased estimator shown to limit shrinkage bias40. Note that, for the purpose of a GWAS, these population stratification covariates are nuisance parameters—that is, their exact values are not essential as long as they correct for the population stratification (see Extended Data Fig. 3 for an empirical comparison)41.
Extended Data Fig. 3. Comparison of the top 10 actual and 1000GP projected PCs.
For each PC, we show a scatter plot and kernel density estimate (KDE) plot. In the scatter plots, the actual PC values are plotted on the x-axis and the projected PC values are plotted on the y-axis. The Pearson correlation coefficient is shown in the top left corner of each scatter plot. In the KDE plots, the true PC distribution is shown in blue and the projected PC distribution is shown in orange. The p-value of a two-sided Kolmogorov-Smirnov test comparing the two distributions is shown in the top left corner of each KDE plot. No multiple hypothesis correction was needed.
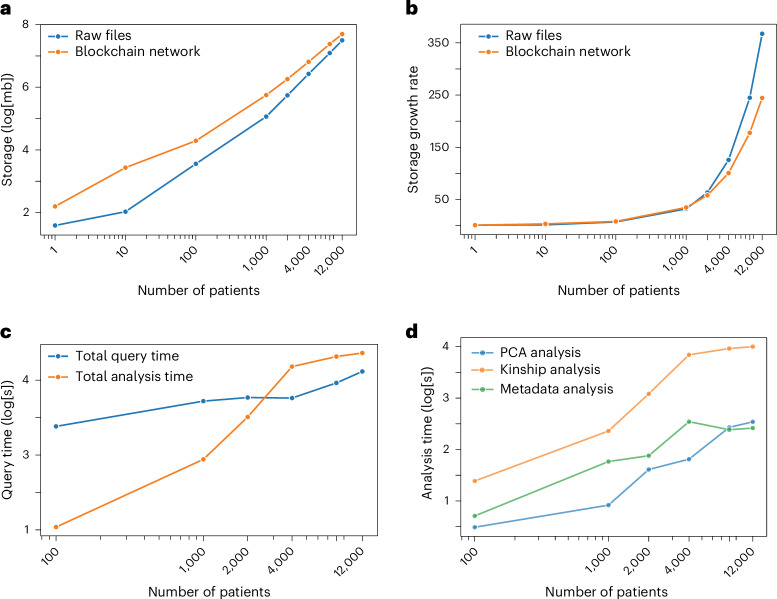
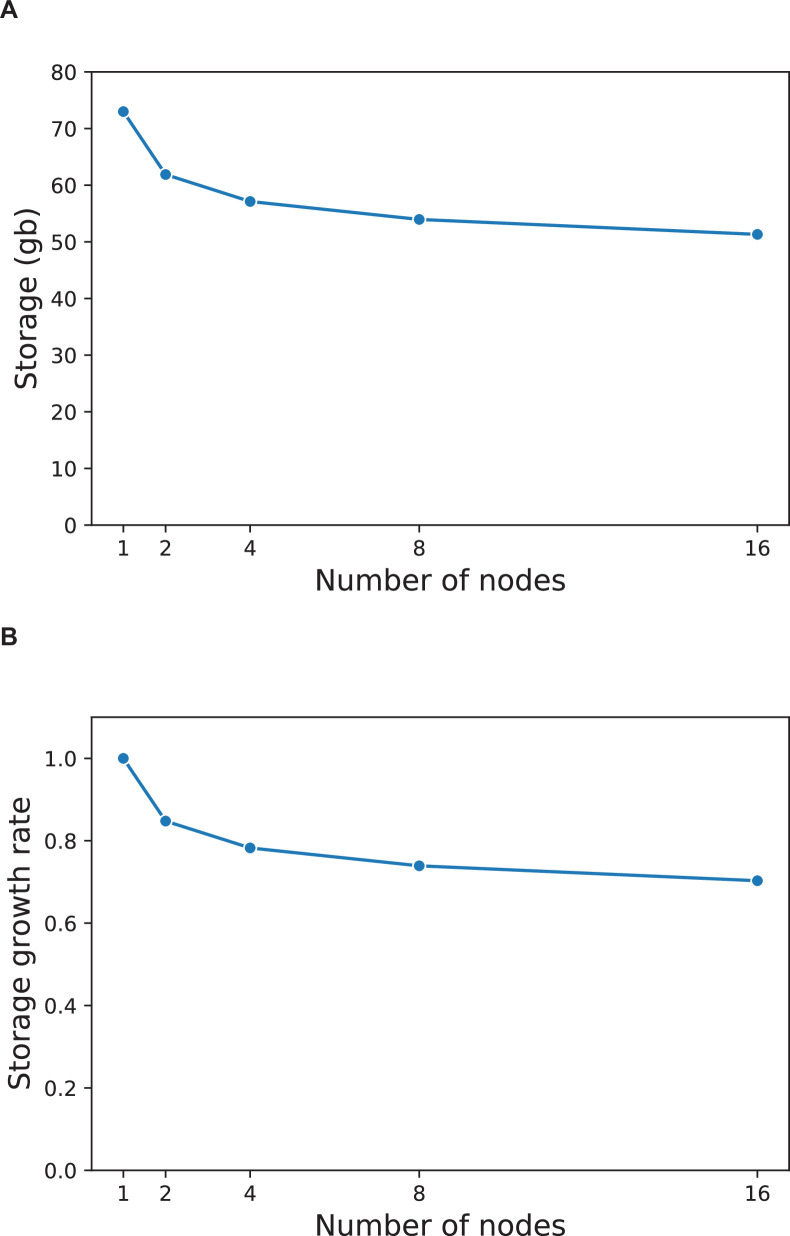
Blockchain technology has several inherent challenges, including large storage requirements, high latency and energy inefficiency. We addressed the storage requirements by organizing entries for both genetic and EHR data by clinical concept, genomic coordinate or person ID. This allows us to consolidate multiple related data points into a single entry and minimizes the storage overhead associated with making a unique entry. Figure 4a shows the total storage requirements (log[mb]) for the raw files and a single-node blockchain network with sample sizes between one and 12,000 patients. Total storage grows sublinearly for the blockchain network but remains higher than the raw files at all sample sizes, as expected. Figure 4b shows the growth rate in storage requirements, with values presented as a ratio to the storage costs of a network with a single patient. The growth of data storage within the blockchain network is slower compared to that of the raw files. Compared to a blockchain network with a single patient, a network with 12,000 patients requires only 245 times the storage. Next, we examined storage requirements as the number of nodes in the network increases. Extended Data Fig. 4 shows how storage costs of a network with 100 patients varies as the number of nodes increases from one to 16. We found that, as the number of nodes in the network increases, total data storage requirements at each node decrease. This is due to MultiChain’s inherent stream indexing property, which requires only the nodes pushing data onto the stream to hold full data copies and allows other nodes to store hashed references of the data. This process maintains data integrity on the blockchain without necessitating every node to store all the data42.
Fig. 4. Scalability.
a, Total data storage. Total data storage requirements (log[mb]) for the raw files and blockchain network at 100, 1,000, 2,000, 4,000, 8,000 and 12,000 patients. b, Storage growth rate. Growth rate in network storage requirements. Values are expressed as a ratio to storage requirements of a single patient network (baseline). c, Query time by query type (in log[s]). Query times broken down by query type. d, Analysis time by analysis type (in log[s]). Analysis times are broken down by analysis type.
Extended Data Fig. 4. Data Storage in PrecisionChain.
(A) Per node data storage. Data storage requirements (gb) for nodes in a 1, 2, 4, 8, and 16 node network with 100 patients. (B) Per node storage growth rate. Growth rate in network storage requirements. Values expressed as a ratio to storage requirements of a single node network (baseline).
To overcome high latency, we implemented an efficient indexing structure. Under each view, we binned the data into fixed sizes to create separate hash tables (that is, streams) from each bin, which allows the upper bound of query times to be proportional to the size of these hash tables. We then used a mapping stream to direct each query to the appropriate stream (Fig. 2a). We showed that query times increase linearly with the size of the stream rather than the full network. In Fig. 4c,d, we show the query and analysis times by network size. We found that, for most queries, query times remain constant after 4,000 patients, which is the point at which streams reach their maximum size. We showed that our platform’s query time is around 61 s (6.7 s per query), and analysis time is around 79 s (26.3 s per analysis). Note that, as the size of the network increases, query times may still increase as there is additional latency independent of stream size, including number of streams, size of the mapping stream and blockchain consensus mechanism. However, our empirical evaluations show that latency is dominated by stream size (Fig. 4c).
We addressed energy inefficiency by using a proof-of-authority (PoA) consensus mechanism whereby any institution with write access can insert data into the blockchain without approval from other nodes. This differs from proof-of-work (PoW), as used in Bitcoin, where 51% consensus is needed10. PoA drastically reduces the computational work on the network by a factor of 108 when compared to a public PoW network43. PoA is only possible within networks consisting of trusted entities. In our design, the hospitals and institutional nodes, which possess ‘write’ permissions, are the trusted entities, whereas the researchers with ‘query’ permission may not fall into this category and do not need to be fully trusted. Strict verification and access controls can be enforced on researchers. Although this verification requirement exists for any data-sharing network, the benefit of PrecisionChain is that institutions retain data sovereignty, setting their own access criteria and tracking use via the audit trail, allowing easier tracing of malicious actors.
Another challenge is the availability of the data on a blockchain to all nodes, which may not be desirable due to privacy concerns. To mitigate this, we propose a selective data masking system, which is composed of encryption of selected data, creating streams that contain sensitive data open only to select users and restrictions on querying. In addition, limits on user access can be implemented to minimize risks from excessive data exposure. These are available features in the API that can be readily adopted into PrecisionChain. See the Methods for the proposed selective data masking and user access controls.
PrecisionChain can identify genotype–phenotype insights
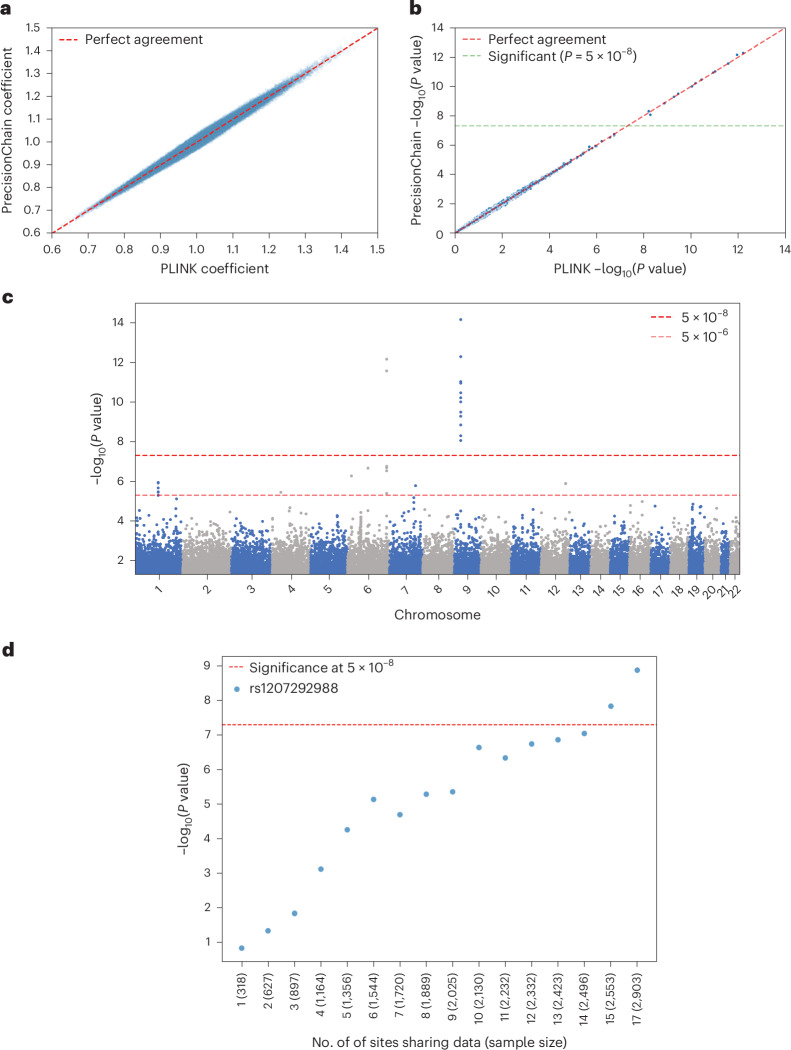
We replicated an existing GWAS from the UK Biobank dataset44 by storing data and performing computations on PrecisionChain. This study sought to identify genetic variants associated with coronary artery disease (CAD) among patients with type II diabetes mellitus (T2DM). We used this study because of its use of complex phenotyping algorithms and the large sample size involved45. We showed that we closely match the cohort size of the original study (Extended Data Table 2) and are able to streamline the cohort creation process (that is, rule-based phenotyping) by leveraging our indexing structure. Notably, we successfully replicated all of the significant lead variants identified in the original study, including rs74617384 (original odds ratio (OR) = 1.38, P = 3.2 × 10−12 versus our OR = 1.37, P = 2.6 × 10−12) and rs10811652 (original OR = 1.19, P = 6 × 10−11 versus our OR = 1.20, P = 5.3 × 10−13). We next showed that PrecisionChain minimizes the need to query multiple databases with distinct file formats (Extended Data Table 3). We also compared β coefficients and P values obtained performing GWAS on PrecisionChain compared to using the standard software, PLINK46 (Fig. 5a,b). We showed that there is excellent agreement between the two methods (Pearson correlation > 0.98, P < 0.05). For more details, see Methods and Extended Data Fig. 5a,b.
Extended Data Table 2.
Comparison of cohort sizes from our implementation and the original analysis
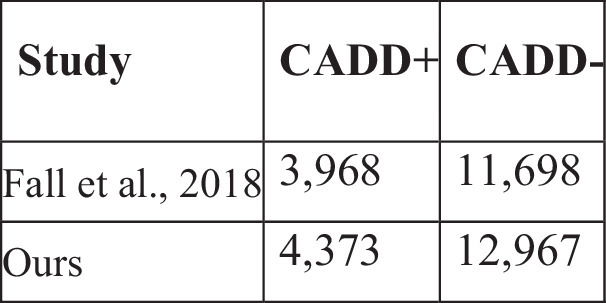
Extended Data Table 3.
Comparative analysis of GWAS using standard techniques, including querying VCFs and OMOP CDMs versus PrecisionChain approach
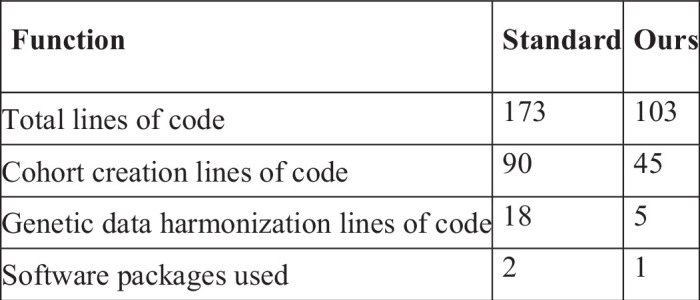
Fig. 5. Analysis results.
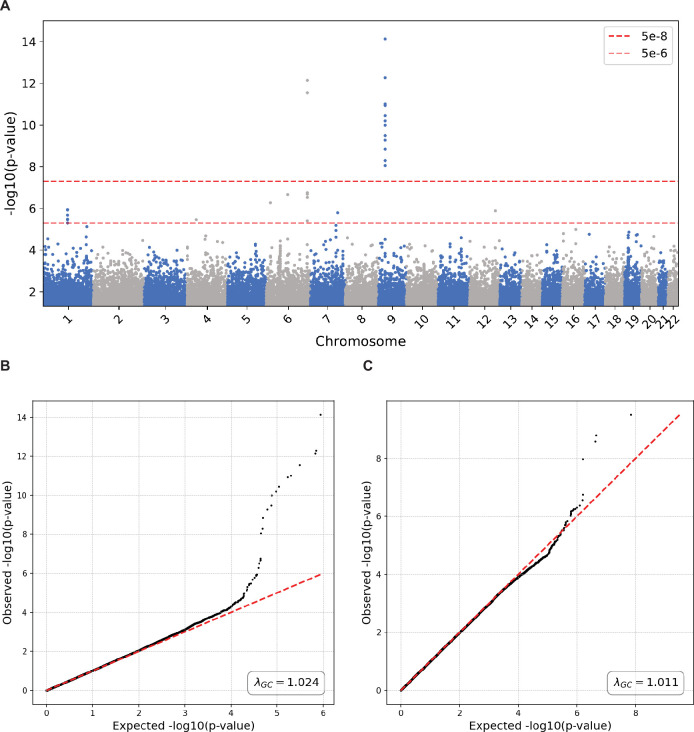
a, Effect size coefficient agreement between UK Biobank (UKBB) GWAS results from PLINK and PrecisionChain. Two-sided t-statistics were used. No multiple hypothesis testing was involved. b, P value agreement between UKBB GWAS results from PLINK and PrecisionChain. Two-sided t-statistics were used. No multiple hypothesis testing was involved. c, Manhattan plot for variants with P < 5 × 10−2 in ALS GWAS. Variants are ordered by genomic coordinates. y axis is the −log(P value). Dashed lines represent the significance line (5 × 10−8) and the suggestive line (5 × 10−6). Two-sided t-statistics were used with standard GWAS Bonferroni correction for multiple hypothesis testing with a cutoff of P < 5 × 10−8. d, Statistical strength of signal by the number of sites included in the ALS GWAS. Plot showing the signal strength (−log(P value)) for variant as a function of the number of sites (and sample size in parentheses) participating in the network. This variant is located on the FAM230C gene. Variant is labeled by rsID. Two-sided t-statistics were used with standard GWAS Bonferoni correction for multiple hypothesis testing with a cutoff of P < 5 × 10−8.
Extended Data Fig. 5. GWAS on PrecisionChain network.
(A) Manhattan plot for variants with p < 5e-2 in UKBB GWAS. In the original study two loci were found to be significant, 6q25 and 9p21. Two-sided t-statistics were used with standard GWAS Bonferroni correction for multiple hypothesis testing with a cut-off p < 5e-8. (B) QQ plot for all variants in UKBB GWAS. Lambda inflation factor=1.024. For GWAS p-values two-sided t-statistics were used with standard GWAS Bonferroni correction for multiple hypothesis testing with a cut-off p < 5e-8. (C) QQ plot for all variants in ALS GWAS. Lambda Inflation Factor = 1.011. For GWAS p-values two-sided t-statistics were used with standard GWAS Bonferroni correction for multiple hypothesis testing with a cut-off p < 5e-8.
We next assessed the utility of our data-sharing infrastructure to support discovery of genotype–phenotype relationships in a rare disease, ALS. We used data from the NYGC ALS Consortium that consists of 26 institutions and 4,734 patients. We conducted a GWAS to find significant associations between genotypes and site of onset (bulbar versus limb) and repeated this analysis using a replication cohort from the Genomic Translation for ALS Care (GTAC) dataset, which contains 1,340 patients collected from multiple sites47.
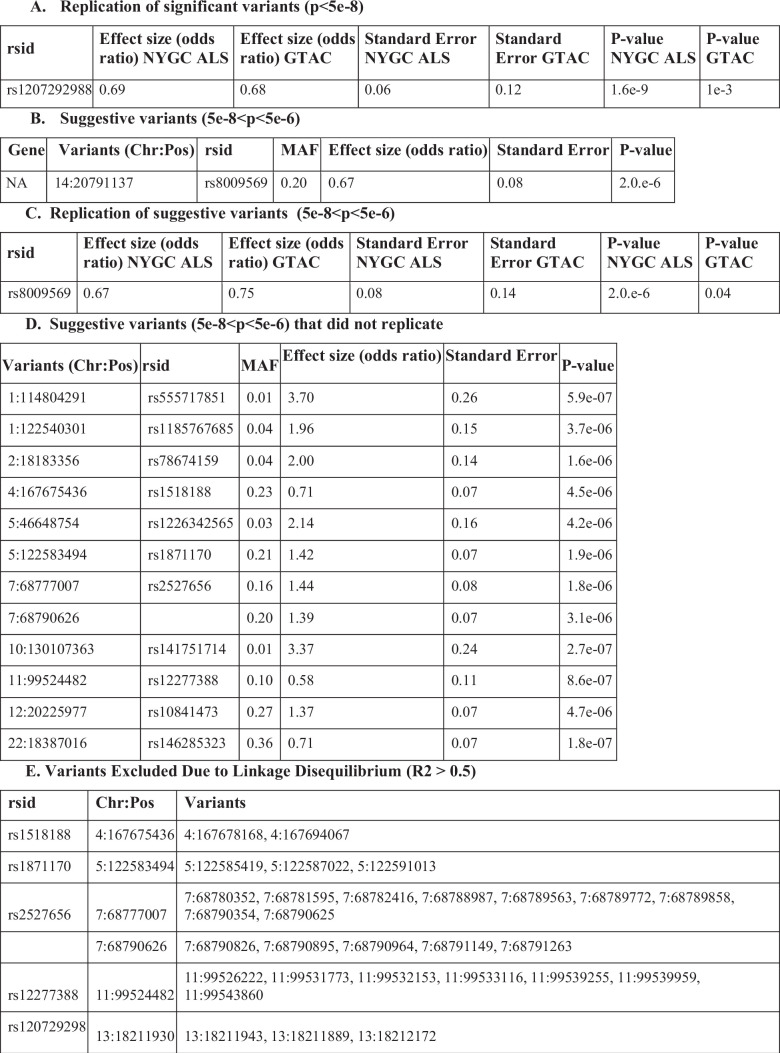
We identified one locus, 13q11 (lead SNP rs1207292988), associated with site of onset at a significance threshold of P < 5 × 10−8 that was successfully replicated on the GTAC dataset (Fig. 5c, Extended Data Fig. 5c and Extended Data Table 4). Extended Data Table 4 also contains details of the additional significant variants in 13q11 that were pruned for being in linkage disequilibrium (LD) (R2 > 0.5). The significant and replicated variant is located in the FAM230C gene. Although FAM230C, a long non-coding RNA (lncRNA), has not been previously implicated in ALS, there is growing evidence for the role lncRNAs in ALS48. For details on all significant and suggestive variants, categorized by replication status on the GTAC dataset, see Extended Data Table 4. We again compared results to a central GWAS conducted using PLINK and found an excellent agreement (Pearson correlation > 0.99) for both effect size coefficients and P values (Extended Data Fig. 6).
Extended Data Table 4.
ALS GWAS replication results
a, Comparison of significant variant results from original (NYGC ALS) and replication (GTAC) GWAS datasets. b, Suggestive variants that replicated and may be associated with site of onset in ALS. c, Comparison of suggestive variant results from original (NYGC ALS) and replication (GTAC) GWAS datasets. d, Suggestive variants that did not replicate and may be associated with site of onset in ALS. e, Variants pruned for LD. GWAS P values were calculated by two-sided t-statistics with standard GWAS Bonferroni correction for multiple hypothesis testing with a cutoff of P < 5 × 10−8.
Extended Data Fig. 6. GWAS comparison between PrecisionChain and PLINK.
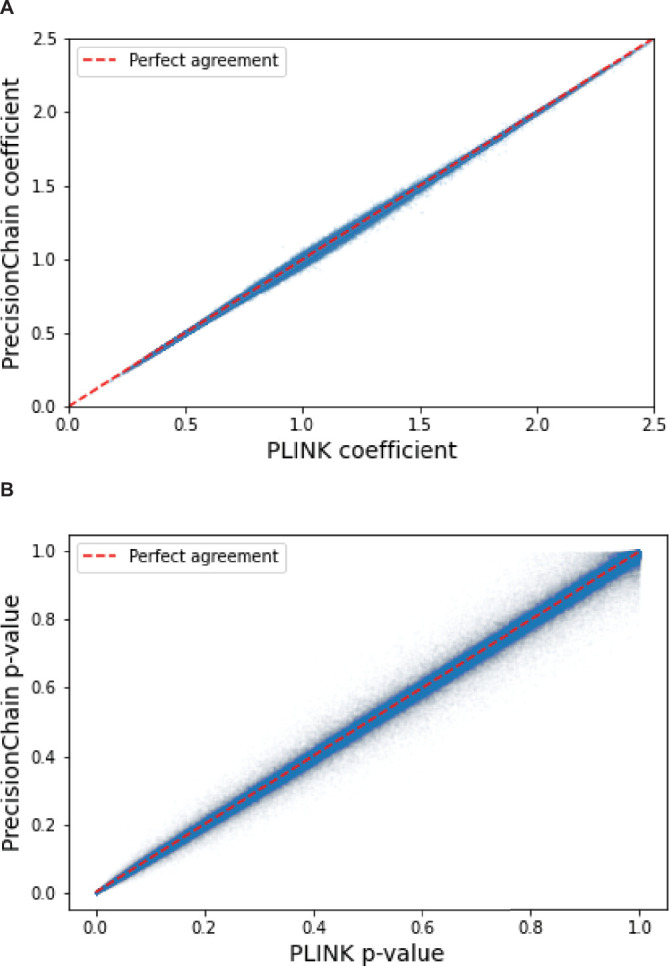
(A) Effect size coefficient agreement between ALS GWAS results from PLINK and PrecisionChain. Two-sided t-statistics were used. There is no multiple hypothesis testing involved. (B) P-value agreement between ALS GWAS results from PLINK and PrecisionChain. Two-sided t-statistics were used. There is no multiple hypothesis testing involved.
To assess the importance of a data-sharing network, we repeated ALS GWAS by varying the number of sites included in the network. We ordered sites by sample size contribution and iteratively added them to the analysis. This allowed us to determine the minimum number of sites required to meet our significance threshold. Note that we consolidated all sites with fewer than 50 patients into one bucket called ‘other’. We showed that a significant P value can be achieved only after data from all sites are included. We observed a linear relationship between the number of sites and −log10(P value), indicating that patients from all sites contribute to the results (Fig. 5d). This highlights the need for a formal data-sharing infrastructure in precision medicine, especially in the context of rare diseases.
Discussion
We present PrecisionChain, a data-sharing platform using a consortium blockchain infrastructure. PrecisionChain can harmonize genetic and clinical data, natively enable genotype–phenotype queries, record user access in granular auditable logs and support end-to-end robust association and phenotyping analysis. It achieves these while also storing all data on the blockchain (see Methods for the benefits of storing data ‘on chain’). We think that PrecisionChain can be used by precision medicine initiative networks to store and share data in a secure and decentralized manner. To achieve this framework, we developed three key innovations: a unified data model for clinical and genetic data, an efficient indexing system enabling low-latency multimodal queries and an end-to-end analysis pipeline specifically designed for research within a decentralized network.
Despite the adoption of blockchain technologies being in its infancy, we think that it can be a solution that overcomes many limitations of current data-sharing frameworks. A blockchain has inherent security safeguards and can cryptographically ensure tightly controlled access, tamper resistance and record usage10,11. These safeguards enable data provenance, increase transparency and enhance trust11,13,15. We enable this not only for storage of the data but also for performing computations ‘on chain’. This helps sovereignty and protection of the use of sensitive health data, particularly of marginalized groups49,50. We extend the inherent safeguards by introducing a granular audit system that can reliably track use. It provides a log of user activities, including records of when specific data were accessed with exact timestamps and which queries returned a specific data point. Access to this detailed and immutable log can support security audits necessary for regulatory compliance (for example, HIPAA compliance) as well as investigations into data misuse. The audit logs can be used to implement stricter access and verification controls on users and also allow greater supervision from patients and communities into how their data are stored and used for research purposes. This can help operationalize equitable data sharing, a key priority of any precision medicine research program49,51.
The flexible nature of the ledger technology allows a wide range of data types from different sources to be stored under one network. This reduces the processing tools needed by researchers to examine clinical and genetic data together. An added benefit of this flexibility is that data collection can be extended to capture patient-reported and clinical trial outcomes data, in effect making the network a research repository for any healthcare-related data52. By enabling conversion of the widely adopted file formats for clinical and genetic data, we hope to further promote semantic interoperability across health systems—a major barrier in biomedical informatics53. In addition, it opens the possibility of integrating the architecture directly into clinical practice, with all data collected on a patient available to any relevant institution and provider, irrespective of geographic location, in a more secure manner.
Data quality poses an inherent challenge in distributed networks. We proactively address this by integrating established pre-processing and data insertion safeguards with innovative blockchain-based solutions for data extraction. For clinical data, we employ Observational Health Data Sciences and Informatics (OHDSI)’s suite of tools to ensure that quality standards are met, which can be customized and automated at the point of insertion. For genetic data, we implemented a standard quality control (QC) process, address site-specific sequencing quality issues and embed advanced filtering techniques within the platform. Given the flexible nature of PrecisionChain, it is possible to integrate additional checks from external databases, such as the Genome Aggregation Database (gnomAD). Additionally, the immutable audit trail can help to identify and understand the sources of data quality issues, expose hidden patterns affecting data quality and facilitate targeted interventions.
Despite the many benefits of blockchain, it is still a nascent technology. Storing and querying large-scale data remains challenging due to inherent storage redundancy, transaction latency and a lack of data structures for flexible querying10,54. Although the former two ensure security guarantees, they also increase computational overhead. We think that decentralized control and security safeguards make this an acceptable tradeoff, especially given the increasing compute power available to institutions. In designing PrecisionChain, we balanced optimizing for both storage and query efficiency. We prioritized querying efficiency with a nested indexing system, as this has a greater impact on network functionality and user experience. Compared to a traditional system, our network offers more flexible multimodal queries but is less storage efficient. However, this deeper indexing can also slow down data insertion. We anticipate insertion to be planned on a monthly or quarterly basis once data have been transformed and quality checked. This update cadence is in line with standard institutional research data warehouse practices and can minimize QC concerns55,56. As our system has a slower increase in storage costs, we anticipate that the gap in costs should decline as the network grows. On-chain analysis also adds to the overhead. Thus, PrecisionChain strategically divides tasks between off-chain and on-chain processing. On-chain processing is reserved for tasks requiring data from multiple sites.
The data on a blockchain are available to all nodes, which may not be desirable due to privacy concerns. Although our platform was designed to be used in trusted consortium settings, we also propose a data masking system that can be readily adopted into our framework (see Methods, ‘Selective data masking’). In addition, limits on user access can be implemented to minimize risks from excessive data exposure (see Methods, ‘User access controls’). Moreover, through the platform’s control parameters, patient and research communities can exert direct control over data access and query rights for their community. For example, it is possible to manage data access via cryptographic tokens assigned to users’ wallets, revoking them after a certain time or after a specific use has been achieved.
Overall, PrecisionChain lays the groundwork for a decentralized multimodal data-sharing and analysis infrastructure. Although other decentralized platforms exist, they focus on either clinical or genetic data and not on a combination (see Methods for existing blockchain solutions)11,57–60. A unique advantage of PrecisionChain is the potential to implement trustless QC mechanisms and have a transparent analysis workflow. This allows for ‘methods-oriented’ research where workflows and findings can be easily verified36. In unifying multimodal data, we anticipate a growth in the discovery of more genotype–phenotype relationships that will translate into improved care. We hope that by enabling secure multimodal data sharing with decentralized control, we can encourage more institutions and communities to participate in collaborative biomedical research.
Methods
The authors confirm that research conducted in this study complies with all relevant ethical regulations. The NYGC ALS Consortium samples presented in this work were acquired through various institutional review board (IRB) protocols from member sites transferred to the NYGC in accordance with all applicable foreign, domestic, federal, state and local laws and regulations for processing, sequencing and analysis. The Biomedical Research Alliance of New York (BRANY) IRB serves as the central ethics oversight body for the NYGC ALS Consortium. Note that the BRANY IRB determined that the request for waiver of informed consent satisfies the waiver criteria set forth in 45 CFR 46.116(d).
We designed PrecisionChain using the blockchain API MultiChain, a framework previously used for biomedical applications24,61. MultiChain’s ‘data streams’ feature makes it possible for a blockchain to be used as a general purpose database as it enables high-level indexing of the data. The data published in every stream are stored by all nodes in the network. Each data stream consists of a list of items. Each item in the stream contains the following information as a JSON object: a publisher (string), Key:Value pairs (from one to 256 ASCII characters, excluding whitespace and single/double quotes) (string), data (hex string), a transaction ID (string), blocktime (integer) and confirmations (integer). When data need to be queried or streamed, they can be retrieved by searches using the Key:Value pairs. Publishing an item to a data stream constitutes a transaction (see below for a primer in MultiChain).
PrecisionChain is a permissioned blockchain where network access is limited to consortium members, and each joining node requires a token from validators (see below for a primer in blockchain). As it is a semi-private blockchain, we use a PoA consensus mechanism, whereby any trusted node can validate transactions (for example, data insertion and token access). Because all data insertion is made public to the consortium members, validating nodes are incentivized to maintain their reputation via accurate and timely data sharing. Once verified and joined, an institution can subscribe to and query the network as well as contribute data. Auditing is set up by querying the audit logs such that institutions receive a summary of their own usage and usage of their contributed data at regular intervals, but they are also able to query the usage in real time using audit logs level. We used the free version of MultiChain version 2.3.3 and Python version 3.6.13 in all implementations. VCF files are analyzed using BCFtools version version 1.9. Plaintext GWAS, including QC, was performed using PLINK version 1.90.
We developed three modules: buildChain, which creates a new chain; insertData, which inserts data into the streams and contains createStream, which creates the indexing structure for efficient data storage; and queryData, which enables multimodal queries.
Module 1: buildChain
buildChain initializes PrecisionChain, including runtime parameters and the access rights for all nodes joining from initialization (that is, read and write access). The default runtime parameters and node rights can be changed before initializing PrecisionChain.
Module 2: insertData
insertData creates the streams that are hierarchically indexed and inserts the data into the appropriate stream. For all views, we first created a mapping stream. The mapping stream records how data in the view have been indexed, including what is contained within each stream. insertData has nine submodules: createStream-Clinical, createStream-Variant, createStream-StructuralVariant, createStream-GTF, insertData-Clinical-Domain, insertData-Clinical-Person, insertData-Variant, insertData-Variant-Person and insertData-GTF. To ensure query efficiency, we fixed the maximum size of each stream. Once a stream exceeds this size, a second stream is created for additional data. Streams are then labeled with a bucket number to record the order of insertion.
Mapping streams
Extended Data Table 1 describes how we record indexing in the mapping stream for each view. The mapping stream has a Key:Value pair for each entry. The Key is the entity being queried (that is, variant or concept ID), and the Value is the name of the stream holding that information. This mapping is automatically assigned during insertion. Once a variant or concept is added to a certain stream, all instances of that entity will be added to the same stream.
EHR level
We inserted all standardized clinical data tables included in the OMOP CDM to the blockchain network. The OMOP CDM harmonizes disparate coding systems into a standardized vocabulary, increasing interoperability and supporting systematic analysis across many sites26,27,62. We took an approach that maximizes the efficiency of querying these data. We created two views: Domain and Person. Domain view contains data streams organized by concepts. Concept IDs are binned using the OMOP vocabulary hierarchy, with a group of related concepts assigned a separate stream. To achieve this, we selected a number of high-level OMOP concepts as ancestor concepts (for example, cardiovascular system, endocrinology system, etc.) and created a stream for each. Then, every remaining (child) concept is assigned to a unique ancestor concept (stream). This assignment is based on the first ancestor concept that subsumes the child concept in the OMOP vocabulary. To accommodate multiple inheritance, if a child concept belonged to multiple ancestor concepts, it was assigned the closest ancestor, and this assignment was recorded in the mapping stream. If one of the child’s other concept ancestors is queried, the mapping stream directs the query to the appropriate stream. This means that the assignment to an ancestor stream is purely for indexing purposes. As an illustrative example, imagine that ‘Hepatic failure’ (code 4245975) is queried. First, all of its descendants are extracted from the vocabulary, including ‘Hepatorenal syndrome’ (code 196455), which is stored under a different concept ancestor. The mapping stream is queried for ‘Hepatorenal syndrome’, and the concept ancestor ‘Disorder of kidney and/or ureter’ (code 404838287) is returned. The stream ‘Conditions_04838287’ is then searched to retrieve the relevant data for ‘Hepatorenal syndrome’. Using the vocabulary hierarchy ensures that related concepts are grouped together. On average, this would limit the number of streams searched in a single query, improving query times. In the patient view, we created a stream for a group of patients bucketed by patient ID ranges. Each stream includes the entire medical record of a patient in the stream, thereby enabling fast querying of all records from the same patient. As each stream contains data entries of different domains, it can be viewed similarly to a ‘noSQL’ database in its flexibility. Mapping stream keeps a record of which stream contains each patient.
Genetic level
We inserted genetic variants (SNPs, small insertions and deletions (indels) and structural variants (SVs)) and the information on the genes that overlap with these mutations. The genetic variant data are in VCF format, and the information on genes is in general transfer format (GTF). Before insertion, variant data are passed through a QC script to ensure that all inserted variants are suitable for analysis. We included five views: Variant, Person, Gene, Analysis and MAF counter. A mapping stream that records high-level indexing is also added. The mapping stream includes the stream that each variant, patient or gene is stored. Within the Variant view, we logged all genetic variants in the population into streams indexed by their genomic coordinate. That is, the genome is divided into discrete bins with each bin corresponding to a specific genomic coordinate range, and a stream is created for each bin. The exact genomic coordinate of the variant, alternative and reference allele and the genotype are included as keys for every entry, such that they are queryable. The data field for each entry includes the patients (person IDs) carrying the genotype in each entry. As such, each variant may have multiple entries, one for each genotype (genotype = 0, 1 and 2). Patient view creates a stream per patient and includes all heterozygous and homozygous alternative allele variants (genotype = 1 and 2) for the patient. To reduce storage requirements, homozygous reference alleles (genotype = 0) for a patient are not stored but are automatically recreated when queried because any variant in the network that was not logged for a specific patient is a homozygous reference for that patient (if the sequencing technology assayed that variant). Gene view stores the structural annotation of the variants, such as whether they overlap with different parts of a gene (for example, exons, introns and untranslated regions (UTRs)). Gene view also includes clinical and biological annotations from ClinVar, VEP and CADD, providing information on the pathogenicity and functional impact of the variants. Analysis view records information related to genetic analysis, including sequencing metadata, population stratification PCs and variants needed to calculate kinship among samples. The MAF counter records the MAF values of the variants and is automatically updated as more patients are added to the network. Because the blockchain is immutable, a new item with the updated MAF values is pushed with each insertion. During the insertion, our algorithm checks the timestamp of the existing entries to determine which MAF stream item is the most recent and accurate. It then extracts the relevant information (that is, sample size and allele frequency for each variant) and combines this with data from the current insertion to determine the new MAF. This is then inserted into the MAF counter stream.
Access logs
We created an access log view, a queryable stream that stores information on each subscription to the network and any query run on the network. An entry is inserted into the stream after an activity on the network. By inserting this into the blockchain, we created an immutable log on the blockchain that can be interrogated. We indexed and stored the type of query, the type of data (EHR or genetic), timestamp of the activity and user wallet ID. Each transaction (that is, query) is stored as a single item. The access log can be queried in real time, acting as an alert system for any potential misuse.
Submodule
createStream-Clinical
Creates streams for clinical data using the OMOP CDM format. For the domain view, each data table in the OMOP clinical tables database is a domain type for which streams are created. Each domain type has its own unique set of streams. Streams are created using a predefined stream structure based on ancestor concepts in the OMOP vocabulary. We selected a number of high-level concepts as ancestor concepts. These represent a broad clinical category (for example, cardiovascular system, endocrinology system, etc.). We assigned approximately 200 concepts as ancestor concepts, and, for every ancestor concept, we created a stream. Each stream follows the naming convention DOMAIN_ANCESTORCONCEPTID. For example, CONDITIONS_436670 represents all metabolic disease concepts, including diabetes (436670 is the concept ID for metabolic diseases). All known concepts are either covered under an ancestor concept ID or placed under the stream DOMAIN_0 (if no ancestor is found in the hierarchy). For the patient view, a stream is created for each group of patient IDs with the format CLINICAL_PERSON_IDBUCKET. For example, CLINICAL_PERSON_1 is the stream for ID bucket 1 (the mapping stream also contains a list of person IDs included in bucket 1). Using the full range of patient IDs (that is, minimum and maximum possible ID value), we create 20 uniformly sized buckets. Note that this range can be calculated automatically using the data or inputted by the user.
createStream-Variants
Creates streams based on genomic coordinates of genetic variants. This follows a predefined structure of CHROMOSOME_START_END. For example, 9_1_3000 includes all variants between positions 1 and 3,000 in chromosome 9. Person view streams are created in the same way as in createStream-Clinical.
createStream-GTF
Creates streams based on genomic coordinates of the genes. This follows a predefined structure of GTF_CHROMOSOME_START_END. For example, GTF_9_1_3000 includes all gene annotations between positions 1 and 3,000 on chromosome 9.
insertData-Clinical-Domain
Inserts tabular clinical data using the OMOP CDM format in the Domain view. Clinical data are added to the clinical domain streams (created via createStream-Clinical). Concept IDs are added to the mapping stream for all unique concept IDs. Each clinical table is a unique domain and has an exclusive set of streams to which data are added. To index concepts, we assigned every child concept (not an ancestor concept) to a unique ancestor concept (stream). This assignment is recorded in the mapping stream and is based on the OMOP vocabulary, so child concepts are assigned to ancestor concepts that subsume them in the hierarchy. Note that a predefined set of ancestor concepts is used to create the streams. Each entry in a stream represents a row of tabular data. This functionality allows querying with the following keys: Concept ID, Person ID, Year, Month-Year, Day-Month-Year and Value (if applicable).
insertData-Clinical-Person
Inserts tabular clinical data using the OMOP CDM format in the Person view. Clinical data are added to the clinical person streams, and patients (person IDs) are inserted into the mapping stream. Each person stream contains all medical record information for a patient irrespective of domain. This functionality allows querying with the following keys: Concept ID, Concept TYPE (the domain of the concept (for example, medication)), Person ID, Year, Month-Year, Day-Month-Year and Value (if applicable).
insertData-Variant
Inserts variant data using the VCF file format. Genetic variants are added to the variant streams, and the coordinates of these variants are added to the mapping stream. Each entry is a unique variant–genotype combination. That is, one genetic variant can have multiple entries, one for each genotype (for example, if, for a given variant, patients can have one of three genotypes (that is, 0, 1 or 2), then three separate entries are made). This functionality allows querying with the following keys: position, reference allele, alternative allele, genotype, sample size and MAF. Data for each entry then contain all patients (person IDs) with that particular variant–genotype combination. With each insertion, MAF is recalculated and inserted into the MAF counter stream.
insertData-Variant-Person
Inserts variant data using the VCF file format. Genetic variants are added to the Variant-Person streams, and patients (person IDs) carrying these mutations are added to the mapping stream. Each entry contains all genetic variants and associated information for a patient across a range of genomic coordinates (for example, Chr 1, Positions 1–300). Keys include person ID and genomic position start and end. Data for each entry are then a JSON entry with the genotype of the genetic variant. Note that only homozygous and heterozygous alternative alleles are stored to save space. With each insertion, MAF is re-calculated and inserted into the MAF counter stream.
insertData-GTF
Inserts details of genes overlapping with the genetic variants using GTF file format. Data are added to the GTF streams, and the gene IDs are added to the mapping stream. Each entry is a row from the GTF file and contains information for a particular gene. Keys include gene ID and gene feature (for example, intron, exon, transcript, etc.). Data are a JSON object and include gene start, gene end, gene type and strand.
insertData-Analysis
Inserts data necessary for genetic analysis. This includes sequencing and technical analysis metadata, SNPs used to calculate kinship using NCBI’s GRAF protocols and sample PCs for population stratification. Sequencing metadata entries are grouped by metadata such that all patients with a certain metadata are included in the same entry (for example, all patients sequenced by Oxford Nanopore PromethION are grouped under a single entry). Keys for the entry include the metadata type (for example, sequencing machine, variant calling pipeline, etc.) and the specific metadata itself (for example, Illumina NovaSeq 6000 and Genome Analysis Toolkit (GATK)). Data for kinship and population stratification are added per sample. This means that each patient will have a unique entry with their specific data. Data can then be extracted using sample IDs.
Module 3: queryData
We developed the queryData module to extract information for downstream analysis. To support efficient and multimodal querying, we leveraged our indexing schema and mapping streams for this module. By indexing all data types into distinct but related streams and using mapping streams to identify the appropriate streams, we reduced the query time significantly and enabled combination queries15. Our query module uses the Key:Value property of stream items to retrieve data from a chain based on a range of defined keys, including concept IDs, concept values, person IDs, dates, genomic locations, genotype, MAF, sequencing metadata and genes involved. When a user queries the chain, they first specify the query type (clinical, genetic or combined); our query module then finds the correct streams/bins based on the query information. This is achieved through querying the mapping streams, which contain a record of the data stored in each stream. Once the appropriate stream is identified, the module extracts the data, performs computation if necessary and returns the relevant information (see Extended Data Fig. 1e for more details). Table 1 describes all the available query functionalities. Overall, queries can be clinical (queryDomain, queryPerson), genetic (queryVariant, queryVariantPerson, queryVariantGene) or a combination of the two (queryClinicalVariant, queryVariantClinical). To increase flexibility and speed of the queries, we included multiple views with distinct indexes optimized for querying.
Three main query types are possible in the query module: clinical, genetic and combination. Multiple key searches are possible in each query using any of the keys included with an entry. For all queries, the mapping stream is first checked to determine the relevant streams to search. After each query, a call is made to the audit module to record the activity.
queryDomain
This functionality allows for a clinical query based on OMOP concept ID, date of concept ID occurrence and/or concept ID value. For each concept ID in the query, the mapping stream is searched to check for the appropriate stream, where data for that concept are stored. If the concept subsumes child concepts, these are also identified in the mapping stream and searched. Then, the relevant streams are searched with the queries, and the relevant patients (person IDs) are returned. It is possible to create cohorts by specifying multiple concept IDs, values and date ranges.
queryPerson
This functionality allows for a clinical query based on person ID, concept type (for example, diagnosis or test), concept ID, date of concept ID occurrence and/or concept ID value. The mapping stream is searched to determine which stream holds the patient’s data. All clinical data in that stream that meet the criteria (for example, person ID, concept type, date and value) can be returned. It is possible to specify a particular concept ID or concept type to return (known as the searchKey). If this is provided, then only data of that type or ID are returned.
queryVariant
This functionality allows for a genetic query based on the variant of interest’s genomic coordinate, genotype and MAF (optional). For each variant included in the query, the mapping stream is searched to determine the appropriate stream that holds data on that coordinate. The genomic coordinate and genotype are then searched in that stream, and patients (person IDs) with the variants of interest are returned. If a MAF range is inputted, variants outside that MAF range are filtered out.
queryVariantPerson
This functionality allows for a genetic query based on person ID, genetic coordinates (optional) and a MAF filter (optional). The person mapping stream is searched to identify which stream contains the person’s genetic data. Once found, that stream is then searched for the queried genetic coordinates, and the relevant variants are returned. If a MAF range is inputted, variants outside that MAF range are filtered out.
queryVariantGene
This functionality allows for a genetic query based on a gene of interest, genotype and MAF range (optional). For each gene (based on gene ID) included in the query, the mapping stream is searched to determine the appropriate stream to check. That stream is searched for variants that are associated with the gene. For each variant, patients (person IDs) with the queried genotype are returned. If a MAF range is inputted, variants outside that MAF range are filtered out.
queryAnalysis
This functionality enables cohort creation suitable for genetic analysis. This includes harmonizing samples across sites by accounting for differences in sequencing metadata, removing related samples from the cohort and extracting population stratification PCs. Metadata queries return all patient IDs that meet the search criteria. The criteria can include any number of metadata (for example, all patients with WGS on Illumina machines with at least 30–60× coverage depth and that were aligned using BWA63, and variants were called using GATK64). Kinship queries accept a list of patients and return a matrix with the pairwise kinship coefficients between all patients. To do this, all kinship SNPs for the cohort are extracted, and the kinship coefficient is then calculated similarly to GRAF39. GRAF is a tool developed by the NCBI specifically to support processing in dbGaP, a large database with phenotype and genotype data collected from multiple sources. It enables kinship calculation on any subset of samples given that they contain a limited number (10,000) of independent and highly informative SNPs39. Population stratification accepts a list of patient IDs and returns their PC scores. A user-defined value k is used to determine how many PC scores should be returned per patient.
queryClinicalVariant
This functionality allows for a ‘combination’ query based on clinical characteristics, genes of interest, genotype (optional) and MAF range (optional). It returns a cohort (person IDs) of patients with their clinical characteristics and relevant genetic variants. First, a clinical domain query is completed that returns a list of person IDs with the desired clinical characteristics (see ‘queryDomain’). Second, a genetic query is completed that returns person IDs for patients with relevant variants in the gene of interest (that is, variants within a MAF range or certain genotype (see ‘queryVariantGene’)). A set intersection of the two cohorts is done to create a final cohort with both clinical and genetic characteristics.
queryVariantClinical
This functionality allows for a ‘combination’ query based on variants and clinical characteristics of interest. First, patients (person IDs) with the queried genetic variants are returned (see ‘queryVariant’). These IDs are then searched using the ClinicalPerson module to extract relevant clinical characteristics for each person (see ‘queryClinicalPerson’). The characteristics are aggregated together as summary information.
Blockchain
Blockchain was initially proposed in 2008 as the cryptocurrency Bitcoin but now has a range of uses65. This is because blockchain has several desirable properties, including decentralization, security and immutability. Blockchains are made up of append-only data blocks that are shared among nodes in a decentralized, distributed network. Transactions are appended to the blockchain as new blocks. Each block is cryptographically connected to the previous block via a header, creating a verifiable chain with a deterministic order. As such, once a block is added to the chain, it is immutable, as any attempt to change a block alters the header and so disrupts the chain. To ensure ‘trustless’ maintenance of the network, there is a consensus mechanism before data are added to the chain. Most widely used is PoW, where participating nodes compete to solve a computationally difficult problem with the winner gaining rights to append the next block. Other mechanisms are proof-of-stake (PoS) and PoA. Public blockchain networks typically make use of PoW or PoS, which are suitable for large networks with unknown and untrusted users. Conversely, private blockchain networks include only individuals who are known, and so PoA can be used. The specific benefits of blockchain for biomedical research applications include decentralized management, immutability of data, transparent data provenance, no single point of failure and security and privacy.
MultiChain
Several blockchain platforms are available to develop a clinical/genetic data-sharing platform. Kuo et al.61 produced a detailed review of the options and identified Ethereum, Hyperledger and MultiChain as the most appropriate. MultiChain is a popular blockchain API used for biomedical applications as it can create permissioned networks that can be used as consortium blockchains65. This is preferred as biomedical data are sensitive and should be shared only with a set of individuals or institutions. MultiChain is a fork of the Bitcoin Blockchain that provides features such as permission management and improved data indexing. The MultiChain ‘streams’ feature makes it particularly good at indexing. Streams are append-only, on-chain lists of data. They have Key:Value retrieval capability, which makes storage and query functionality easy. Furthermore, it is possible to have multiple keys, enabling complex logic gates for a given data query.
OMOP CDM
The OMOP CDM is an open community data standard that aims to standardize the structure of clinical data across different sources27. By standardizing different clinical databases, it is possible to combine and analyze them together. This is achieved by transforming clinical databases into a common format (that is, data model) with a common representation (that is, terminologies, vocabularies and coding schemes). One of the major elements in the data model are the clinical data tables. These contain the key data elements extracted from the EHR related to a patientʼs clinical characteristics, including diagnoses, measurements, observations, notes, visits, procedures and devices. A common representation is achieved using the OMOP vocabulary, which standardizes the medical terms used. It contains records, or concepts, uniquely identifying each fundamental unit of meaning that is used to express clinical information27. Each concept, or piece of clinical information, has a unique ID that is used as its identifier in the data table. Concepts can represent broad categories (such as ‘Cardiovascular disease’), detailed clinical elements (‘Myocardial infarction of the anterolateral wall’) or modifying characteristics and attributes that define concepts (severity of a disease, associated morphology, etc.). Concepts are derived from national or international vocabularies, such as Standard Nomenclature of Medicine (SNOMED), RxNorm and Logical Observation Identifiers Names and Codes (LOINC). Concepts are grouped into a hierarchy with detailed terms subsumed by broader terms. The OMOP CDM is used by many existing research networks, including OHDSI, which has over 2,000 collaborators from 74 countries62. The main aim of the conversion to OMOP is to support secondary analysis of observational clinical data. In places where the data cannot be shared, analytic code can be shared and executed at a local site’s OMOP instance, preserving privacy. It is worth noting that conversion to OMOP has been shown to facilitate faster, more efficient and accurate analysis across sites and it is particularly helpful for rare diseases66,67. It is increasingly being adopted by existing research networks, such as the UK Biobank and the All of Us Research Program68. The recording of EHR to OMOP CDM has already been done by over 200 health systems covering over 800 million unique patients. The typical process involves leveraging local data experts and resources made available by OHDSI, including conversion scripts and QC tools. A multidiscplinary team of data engineers, informaticians and clinicians at an institution works on mapping EHR data elements to standardized terminologies.
OMOP conversion
EHR data are converted to OMOP CDM using extract, transform, load (ETL) guidelines and data standards by OHDSI27. Although the full pipeline will be different for each institution, the general steps are well established and many open-source tools and scripts exist to automate the process. Generally, the process requires expertise from clinicians, who understand the source data; informaticians, who understand the OMOP CDM; and data engineers, who can implement the ETL logic. The steps can be broken down into the following. (1) Analyzing the dataset to understand the structure of the tables, fields and values. This can be done using the open-source software ‘White Rabbit’, which can automatically scan the data and generate reports69. (2) Defining the ETL logic from the source data to OMOP CDM. The open-source tool ‘Rabbit-in-a-hat’ from ‘White Rabbit’ can be used for connecting source data to CDM tables and columns, completing field-to-field mappings and identifying value transformations. (3) Creating a mapping from source to OMOP codes. This step is optional and only necessary if the source codes have not been previously mapped to OMOP. In most cases, this is not necessary. The ‘Usagi’ tool can be used to support this process70. (4) Implementing the ETL. The exact pipeline will differ by institution, and a wide variety of tools have been used successfully (for example, SQL builders, SAS, C#, Java and Kettle). Several validated ETLs are publicly available and can be used71,72. (5) Validating the ETL. This is an iterative and ongoing process that requires unit testing, manual review and replication of existing OHDSI studies. Full details can be found in The Book of OHDSI, chapter 6 (ref. 27). It is worth highlighting that OHDSI has many communities, resources and established conventions that adopters of the OMOP CDM can use for support.
Generating synthetic patient population
Synthea
Synthea is an open-source software project that generates synthetic patient data to model real-world populations33. The synthetic patients generated by Synthea have complete medical histories, including medications, procedures, physician visits and other healthcare interactions. The data are generated based on modules that simulate different diseases to create a comprehensive and realistic longitudinal healthcare record for each synthetic patient. Synthea aims to provide high-quality and realistic synthetic data for analysis. Previous studies showed that Synthea produces accurate and valid simulations of real-world datasets31,32. We use Synthea OMOP, which creates datasets using standard OHDSI guidelines.
1000GP dataset
The 1000GP is a detailed dataset of human genetic variation, containing sequencing data for 2,504 genomes34. We simulate genetic data using MAF values observed in 1000GP data as a baseline.
Creating the population
For the patient population in the publicly available demonstration network (accessed via https://precisionchain.g2lab.org/), we combined synthetically generated clinical data using Synthea and synthetically generated genetic data that are based on the 1000GP dataset. For every patient, we randomly assigned an ID that can be used across both their clinical and genetic data. Note that this population is essentially random, and any analysis and results are for demonstration purposes only.
Genetic data pre-processing
Before insertion, we implemented an automated QC step for all genetic samples. This ensures that genetic data shared on the network are suitable for downstream analysis. First, we filtered the data based on sample call rate (<5% genotypes missing) and genotype call rate (<5% samples missing genotype). We also tested for Hardy–Weinberg equilibrium (HWE) (<1 × 10−6) and LD (50-SNP window size, 5-SNP step size and 0.5 R2 threshold) in that cohort. Any removed SNPs are then recorded on the network such that, when researchers combine data from multiple cohorts, they can track which SNPs have been removed. Pre-processing steps before insertion are done using PLINK46. Additionally, every sample undergoes population stratification projection using the 1000GP as a reference. This is achieved by using the method outlined by Prive et al.40 to limit shrinkage bias. Importantly, we noted that, for the purpose of a GWAS, these population stratification covariates are nuisance parameters41. We empirically evaluated the projected scores (Extended Data Fig. 3).
GWAS on the blockchain
The analysis follows a similar workflow as a GWAS on traditional infrastructure. However, we replace many of the steps with functionality from the network. The full script can be viewed on GitHub. First, using the Analysis view, we harmonize genetic data using technical sequencing metadata. In the provided script, we select for patients with the same reference genome (HG38), sequencing machine (Illumina, NovaSeq 6000), alignment pipeline (BWA) and variant calling pipeline (GATK) used. Next, for all included samples, we extract data on genetic ancestry, filtering for white Europeans (>80%). We then assess relatedness between all samples and filter for unrelated samples. Next, we retrieve necessary covariates, such as population stratification PCs and sequencing sites. Using the ClinicalPerson view, we then extract the site of onset (phenotype) and gender (covariate) for all samples in the cohort. Note that the site of onset phenotype is based on the patient having the OMOP concept code for ALS (4317965) with attribute site (4022057) bulbar or limb. Patients with other sites recorded were excluded. More advanced logic can be implemented for complex phenotypes. The final cohort is constructed of patients who have all relevant data. Next, we extract genotype data per variant. At this stage, it is possible to conduct additional MAF filtering or HWE testing if necessary. Note that this was already done during insertion. We then run a logistic regression model to test for genetic associations with age of onset, adjusting for PCs, site of sequencing and gender. This final analysis step is conducted using a standard software package. To protect data security, the analysis is done on the blockchain node with users accessing only the results. This is similar to how analysis is run on All of Us73. In the future, it is possible to have the analysis itself run on the network as a smart contract such that no data ever leave the blockchain.
Rare genetic disease analysis
The platform is well suited to support research into truly rare diseases, including the reclassification of VUS. First, our platform facilitates the secure sharing of patient clinical and genetic data among many institutions, enabling the creation of larger and more comprehensive datasets of patients with rare genetic diseases. Moreover, the platform also contains a repository of all annotations associated with a variant, simplifying the tracking of VUS updates. A key strength of this platform is the multimodal queries, which allow researchers to simply build patient cohorts based on both clinical presentations and the presence of a specific VUS. This functionality, coupled with the integrated analysis pipeline, could support association analyses that uncover potential links between a VUS and a particular disease. Should new links be identified and verified, the annotation stream can be updated. Notably, a user-level alert system can be implemented via the audit stream, enabling physicians and researchers to subscribe to updates for specific variants and stay informed about any reclassifications. This workflow reduces technical barriers associated with rare disease research and ensures that the latest knowledge is rapidly disseminated. Our platform offers two key benefits for such analysis: (1) it allows institutions to contribute even a few samples while retaining control over their data, thereby increasing sample sizes; and (2) it serves as a self-contained repository that integrates genetic, clinical and variant annotation data, along with the cohort-building and analysis functionality needed to classify a VUS.
GWAS projection using the 1000GP
To validate the projection of samples onto the 1000GP PC loadings, we compared the top 10 PCs from the 1000GP projection to those from the NYGC ALS dataset (Extended Data Fig. 3). Note that we use samples from 1000GP with European ancestry. This is valid as our study only uses samples with European ancestry. We use Pearson correlation coefficients to assess their linear relationship and the Kolmogorov–Smirnov test to determine distributional similarity. Our analysis shows high correlation for PCs 1–4, with their distributions aligning significantly (P > 0.05). In contrast, PCs 5–10 do not show a strong relationship. This is likely due the top four PCs accounting for 79% of the explained variance from the top 10 PCs.
Blockchain GWAS implementation accuracy
Extended Data Fig. 5 shows the QQ plot for both (UK Biobank and ALS) GWASs that we performed. The QQ plot represents the deviation of the observed P values from the null hypothesis. Notably, we observe that the P values closely match the expected distribution up to approximately P = 1 × 10−6, after which they rise above the expected line, suggesting that we have detected a true association74.
To validate our distributed blockchain-based GWAS implementation, we directly compared our results against a centralized analysis conducted on PLINK. Extended Data Fig. 6 compares the coefficients and P values obtained from the two methods. We observe a very high agreement (Pearson correlation coefficients > 0.99). This high concordance confirms the reliability and accuracy of our blockchain-powered GWAS.
Findings on UK Biobank GWAS
The original study identifies two significant loci, which we successfully replicate. We found two lead SNPs, rs74617384 and rs10811652, presented by the original authors, showing closely matching results. For rs74617384, we found an effect size of 1.37 versus the original study that found 1.38, and we found a P value of 2.8 × 10−12 versus the original study that found 3.2 × 10−12. For rs10811652, we found an effect size of 1.20 versus the original study that found 1.19, and we found a P value of 5.3 × 10−13 versus the original study that found 6 × 10−11. Figure 5 compares the coefficients and P values obtained from conducting the analysis on the blockchain versus using PLINK. We observe a very high agreement between the two methods (Pearson correlation > 0.98). Note that we cannot compare all of the results from the original study directly as the study released information on only the lead variants. However, visual comparison of the Manhattan plots suggest high concordance, as all significant loci were identified with no new loci found (Extended Data Fig. 5).
Comparing GWAS on the blockchain versus standard software
We demonstrate the efficiency advantages of our blockchain-based GWAS implementation compared to standard software tools on our repository at gwas_comparison/gwas_showcase.py. The script compares the steps required to run the GWAS using standard statistical software and using our blockchain implementation. Extended Data Table 3 summarizes the key findings. Our blockchain approach substantially decreases the code needed for clinical cohort generation and genetic data harmonization. Furthermore, we can conduct the GWAS using a single software solution, in contrast to the multiple tools typically required.
Selective data masking
To mitigate the privacy leak from nodes having access to all the data, we outline a potential strategy to selectively mask data. Although this system has not been implemented in the current platform, it uses features already present in the MultiChain API and is readily implemented in our framework. Below, we present the main steps:
Create key management streams: The MultiChain API generates public and private encryption keys for every user. A public key stream can be created to store user public keys for verification.
Selectively mask data: Data contributors can indicate whether they want to mask their data at the field level (specific data points) or stream level (entire streams). The decision to mask data is made during data upload by the data contributor.
Provide token-based access: For each masked entry or stream, the data contributor would assign tokens that control data access. These tokens would be encrypted and uploaded to an access stream. Encryption is done via the public keys of users who have been granted access.
Restricted querying: When a user queries the platform, encrypted data that the user does not have access to would not be returned.
User access controls
Although our system was developed for trusted users in a consortium setting, to mitigate the risk of privacy leakage from querying all of the data we suggest several complementary user access controls. First, query limitations can be implemented with restrictions tightly controlled by the audit trail. This can involve imposing limits on the number or type of queries that a user can perform within a given timeframe. Alternatively, limits based on data exposure could be imposed. These would be based on the percentage of patients queried or the overall amount of data accessed, preventing any single user from obtaining an excessive volume of sensitive information. These measures mirror the access controls found in centralized systems but with the added benefit of the blockchain’s immutable audit trail, which provides a transparent and tamper-proof record of all user activity that can be verified by all users. Second, as detailed in the ‘Selective data masking’ subsection, access to specific data streams or individual data entries can be tightly controlled through encryption and access tokens. This granular approach ensures that users have access only to the information that they are authorized to view. Finally, an institutional accountability mechanism can be established, using the audit trail to trigger alerts if a user exceeds predefined usage limits. Both the data-contributing institution and the user’s home institution would be notified, enabling action to address potential misuse according to their established protocols.
Existing blockchain solutions
A number of blockchain-based platforms are used for EHR and genetic data sharing. However, few attempt to unify both modalities in one data-sharing infrastructure. One such platform that has both EHR and genetic data is the Cancer Gene Trust (CGT)23. CGT was developed in 2019 and is described as a decentralized network that makes somatic mutation and clinical data about a patient publicly available23. It shares cancer registry data, including EHR, genetic and imaging data. However, for each, only a limited sample of data that are typically held in a cancer registry is available. All data are stored ‘off chain’ in Interplanetary File System (IFPS), a peer-to-peer distributed file sharing system, and raw sequencing data are stored locally. Our platform differs from CGT in that it stores clinical and genetic data ‘on chain’ to maximize data security and is capable of storing a wider range of clinical data.
Other platforms that focus solely on genomic data sharing include SAMChain13, CrypDist54, Zenome75, Nebula Genomic76 and Encrypgen/Gene-Chain77. Apart from SAMChain13, other solutions store data ‘off chain’ and retain pointers on the blockchain. Because data are not stored in the blockchain, they lose the benefit of secure and immutable data storage. CrypDist creates a custom blockchain using Java to store links to genomic data files that are then stored in the cloud. Zenome uses Ethereum smart contracts to support access to genomic data files. Users are incentivized to share by receiving ‘ZNA tokens’, a cryptocurrency. Nebula Genomics also uses Smart Contracts to communicate between nodes in the network and survey participants, and to facilitate data access permissions.
Platforms that focus on EHR data are numerous. Some of the notable ones are FHIRChain11, NotaryChain57, HIEChain58, mHealthChain59 and ImageChain60. FHIRChain aims to develop apps built on a blockchain (dApps) that make use of the Fast Healthcare Interoperability Resources (FHIR) data exchange protocol. All data are stored ‘off chain’ with URL links stored on the blockchain for data exchange. NotaryChain aims to notarize all research requests but adds the query and a hash of the returned values to the blockchain. Although this ensures integrity of queried data, no direct data-sharing component is facilitated by the blockchain. HIEChain is similar to FHIRChain in its approach, providing only links to the data on the blockchain. mHealthChain examined adding patient-inputted data from a mobile app onto the chain, but no indexing and querying system was put in place to ensure efficient storage and retrieval of the data. Finally, ImageChain was developed to share imaging data via a blockchain. Again, only a reference to the underlying data is kept ‘on chain’.
Storing metadata versus the data itself on the blockchain
The difference between storing metadata and the data itself on the blockchain is discussed in Gursoy et al.13. In summary, the benefit of security and immutability is limited only to data stored on the blockchain. As such, if only metadata are stored, underlying data that are kept in a centralized system remain vulnerable to manipulation and loss. However, if all data are stored, then all data are secured. Furthermore, access and computations can be more tightly controlled and audited if the data are ‘on chain’.
ALS consortium data analysis and replication in a different cohort
We used data from the NYGC ALS consortium, which has WGS data for 4,734 patients with ALS collected from 48 different sites. Approximately 1,964 (~42%) of 4,734 samples used from the NYGC ALS Consortium data are female. Average age of symptom onset for these patients was approximately 58 years. Clinical data for all patients were taken from NYGC ALS Consortium records and converted into OMOP format before insertion into the network. Data mostly included information specific to ALS and not the full health records for the patient. We limited our analysis to patients with European genetic ancestry (>80%), known gender and known site of onset. This leaves 2,903 patients—802 with bulbar ALS and 2,101 with limb ALS. We first performed QC pre-processing to remove individuals or variants with more than 5% missingness, variants with a MAF < 0.01, an HWE exact test P < 0.001 and LD pruning at R2 > 0.5. For each variant, we conducted a logistic regression adjusting for sex, site of sample collection and population stratification (top 10 PCs). We used a threshold of P < 5 × 10−8 for statistical significance.
For replication, we used data from the Columbia University GTAC dataset (dbGaP accession number phs002973.v1.p1), which has WGS data for 1,340 patients with ALS collected from many different sites. Note that only 250 patients were recruited at Columbia University. We followed similar cohort creation and data pre-processing steps in the replication GTAC dataset and were left with 916 patients (208 with bulbar onset and 708 with limb onset). To determine whether a variant had been replicated, we used an adjusted P < 5 × 10−2 and an effect size OR within 2 s.e. of each other. We adjusted the P value using Bonferroni correction, accounting for multiple testing across the three variants identified as significant in the NYGC dataset. This adjusted threshold for significance is justified, as we test variants already found to be significant in the primary NYGC dataset. Furthermore, the smaller sample size reduces our power to detect small P values. We think that this approach is prudent as we ensure that the effect size is concordant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41591-024-03239-5.
Supplementary information
Acknowledgements
This work has been supported by National Institutes of Health grants R00HG010909 and R35GM147004 to G.G. We thank Y. Shen, J. Morris, H. Phatnani and M. Harms for fruitful discussions.
All NYGC ALS Consortium activities are supported by the ALS Association (19-SI-459) and the Tow Foundation. We thank the Target ALS Human Postmortem Tissue Core, the New York Genome Center for Genomics of Neurodegenerative Disease, the ALS Association and the Tow Foundation.
We would like to acknowledge sample and data contributions from the ‘Genomic Translation for ALS Care’ project investigators and funding sources: principal investigator M. Harms (Columbia University) and site investigators S. Appel (Houston Methodist); R. Baloh (Cedars-Sinai); R. Bedlack (Duke University); S. Chandran (University of Edinburgh); L. Foster (University of Colorado); S. Gibson (University of Utah); D. Goldstein (Columbia University); S. Goutman (University of Michigan); C. Karam (Oregon Health Sciences); D. Lacomis (University of Pittsburgh); G. Manousakis (University of Minnesota); T. Miller (Washington University in St. Louis); C. Moreno (Columbia University); S. Pa (University of Edinburgh); D. Sareen (Cedars-Sinai); Z. Simmons (Pennsylvania State University); and L. Wang (Univeristy of Washington). Funding was provided by the ALS Association (National), the ALS Association (Greater New York Chapter) and Biogen.
Extended data
Author contributions
A.E. and G.G. conceived and designed the study. G.G. directed the study. A.E. developed the framework. A.E. and U.B. wrote the code. A.E., with guidance from G.G., analyzed the results. N.E. provided guidance and access to synthetic clinical data. K.N. and N.E. provided guidance on clinical data storage and analysis. K.N. provided guidance on OMOP CDM conversion and clinical data quality control. The NYGC ALS Consortium performed data collection. A.E. and G.G. wrote the manuscript. All authors reviewed, edited and approved the manuscript.
Peer review
Peer review information
Nature Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Lorenzo Righetto, in collaboration with the Nature Medicine team.
Data availability
The individual-level data from the NYGC ALS Consortium are available to authorized investigators through the dbGaP via accession code phs003067. Data dictionaries and variable summaries are available on the dbGaP public FTP site: https://ftp.ncbi.nlm.nih.gov/dbgap/studies/phs003067/phs003067.v1.p1. Public summary-level phenotype data may be browsed at the dbGaP study report page: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003067.v1.p1.
Subject Sample Telemetry Report (SSTR) for subject and sample IDs, consents, summary counts, processing status and molecular and sequence sample uses is available on the SSTR site: https://www.ncbi.nlm.nih.gov/gap/sstr/report/phs003067.v1.p1.
The data are available under General Research and Health, Medical, Biomedical data use limitations.
GTAC data can be accessed via dbGaP accession code phs002973.v1.p1.
Genetic data for the publicly accessible blockchain network are shared via the International Genome Sample Resource (1000 Genomes Project) and can be accessed through https://www.internationalgenome.org/data-portal/data-collection/30x-grch38.
UK Biobank data are available to authorized investigators through the UK Biobank portal.
Code availability
The most up-to-date code can be found at https://github.com/G2Lab/PrecisionChain/ and on Zenodo at 10.5281/zenodo.10067135. A demonstration of our user-friendly front end can be accessed via https://precisionchain.g2lab.org/ (use username: test@test.com and password: test-ME).
Competing interests
A patent application has been filed by Columbia University with A.E. and G.G listed as inventors (application number: 18/419,923; status of application: pending; specific aspect of manuscript covered in patent application: blockchain-based harmonization of clinical and genetic data). All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Gamze Gürsoy, Email: gamze.gursoy@columbia.edu.
NYGC ALS Consortium:
Hemali Phatnani, Justin Kwan, Dhruv Sareen, James R. Broach, Zachary Simmons, Ximena Arcila-Londono, Edward B. Lee, Vivianna M. Van Deerlin, Neil A. Shneider, Ernest Fraenkel, Lyle W. Ostrow, Frank Baas, Noah Zaitlen, James D. Berry, Andrea Malaspina, Pietro Fratta, Gregory A. Cox, Leslie M. Thompson, Steve Finkbeiner, Efthimios Dardiotis, Timothy M. Miller, Siddharthan Chandran, Suvankar Pal, Eran Hornstein, Daniel J. MacGowan, Terry Heiman-Patterson, Molly G. Hammell, Nikolaos A. Patsopoulos, Joshua Dubnau, Avindra Nath, Robert Bowser, Matt Harms, Eleonora Aronica, Mary Poss, Jennifer Phillips-Cremins, John Crary, Nazem Atassi, Dale J. Lange, Darius J. Adams, Leonidas Stefanis, Marc Gotkine, Robert H. Baloh, Suma Babu, Towfique Raj, Sabrina Paganoni, Ophir Shalem, Colin Smith, Bin Zhang, Brent Harris, Iris Broce, Vivian Drory, John Ravits, Corey McMillan, Vilas Menon, Lani Wu, Steven Altschuler, Yossef Lerner, Rita Sattler, Kendall Van Keuren-Jensen, Orit Rozenblatt-Rosen, Kerstin Lindblad-Toh, Katharine Nicholson, and Peter Gregersen
Extended data
is available for this paper at 10.1038/s41591-024-03239-5.
Supplementary information
The online version contains supplementary material available at 10.1038/s41591-024-03239-5.
References
- 1.Ginsburg, G. S. & Phillips, K. A. Precision medicine: from science to value. Health Aff. (Millwood)37, 694–701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward, R. & Ginsburg, G. S. Local and global challenges in the clinical implementation of precision medicine. In Genomic and Precision Medicine: Foundations, Translation, and Implementation 3rd edn (eds Ginsburg G. S. & Willard, H. F.) 105–117 (Academic Press, 2016).
- 3.Precision Cancer Medicine: Challenges and Opportunities (eds Roychowdhury, S. & Van Allen, E. M.) (Springer, 2020).
- 4.Acosta, J. N., Falcone, G. J., Rajpurkar, P. & Topol, E. J. Multimodal biomedical AI. Nat. Med.28, 1773–1784 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Haendel, M. A., Chute, C. G. & Robinson, P. N. Classification, ontology, and precision medicine. N. Engl. J. Med.379, 1452–1462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez, A. H., Gebo, K. A. & Harris, P. A. Progress with the All of Us research program: opening access for researchers. JAMA325, 2441–2442 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Hulsen, T. et al. From big data to precision medicine. Front. Med.6, 34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alterovitz, G. et al. FHIR Genomics: enabling standardization for precision medicine use cases. NPJ Genom. Med.5, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolin, R. H. et al. Introducing HL7 FHIR Genomics Operations: a developer-friendly approach to genomics-EHR integration. J. Am. Med. Inform. Assoc.30, 485–493 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo, T.-T., Kim, H.-E. & Ohno-Machado, L. Blockchain distributed ledger technologies for biomedical and health care applications. J. Am. Med. Inform. Assoc.24, 1211–1220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, P., White, J., Schmidt, D. C., Lenz, G. & Rosenbloom, S. T. FHIRChain: applying blockchain to securely and scalably share clinical data. Comput. Struct. Biotechnol. J.16, 267–278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubovitskaya, A., Xu, Z., Ryu, S., Schumacher, M. & Wang, F. Secure and trustable electronic medical records sharing using blockchain. AMIA Annu. Symp. Proc.2017, 650–659 (2017). [PMC free article] [PubMed] [Google Scholar]
- 13.Gürsoy, G. et al. Storing and analyzing a genome on a blockchain. Genome Biol.23, 134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattengale, N. D. & Hudson, C. M. Decentralized genomics audit logging via permissioned blockchain ledgering. BMC Med. Genomics13, 102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gürsoy, G., Bjornson, R., Green, M. E. & Gerstein, M. Using blockchain to log genome dataset access: efficient storage and query. BMC Med. Genomics13, 78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gürsoy, G., Brannon, C. M. & Gerstein, M. Using Ethereum blockchain to store and query pharmacogenomics data via smart contracts. BMC Med. Genomics13, 74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rai, B. K. PcBEHR: patient-controlled blockchain enabled electronic health records for healthcare 4.0. Health Serv. Outcomes Res. Methodol.23, 80–102 (2022). [Google Scholar]
- 18.Albalwy, F., Brass, A. & Davies, A. A blockchain-based dynamic consent architecture to support clinical genomic data sharing (ConsentChain): proof-of-concept study. JMIR Med. Inf.9, e27816 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chelladurai, U. & Pandian, S. A novel blockchain based electronic health record automation system for healthcare. J. Ambient Intell. Humaniz. Comput.13, 693–703 (2022). [Google Scholar]
- 20.Hajian, A., Prybutok, V. R. & Chang, H.-C. An empirical study for blockchain-based information sharing systems in electronic health records: a mediation perspective. Comput. Hum. Behav.138, 107471 (2023). [Google Scholar]
- 21.Kuo, T.-T. et al. Blockchain-enabled immutable, distributed, and highly available clinical research activity logging system for federated COVID-19 data analysis from multiple institutions. J. Am. Med. Inform. Assoc.30, 1167–1178 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Passerat-Palmbach, J. et al. Blockchain-orchestrated machine learning for privacy preserving federated learning in electronic health data. 2020 IEEE International Conference on Blockchain (Blockchain) 10.1109/blockchain50366.2020.00080 (IEEE, 2020).
- 23.Glicksberg, B. S. et al. Blockchain-authenticated sharing of genomic and clinical outcomes data of patients with cancer: a prospective cohort study. J. Med. Internet Res.22, e16810 (2020). [DOI] [PMC free article] [PubMed]
- 24.Greenspan, G. MultiChain Private Blockchain—White Paper. https://www.multichain.com/download/MultiChain-White-Paper.pdf (2015).
- 25.MultiChain data streams. https://www.multichain.com/developers/data-streams/
- 26.Voss, E. A. et al. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J. Am. Med. Inform. Assoc.22, 553–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Book of OHDSI. Observational Health Data Sciences and Informatics. https://ohdsi.github.io/TheBookOfOhdsi/ (2021).
- 28.Landrum, M. J. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res.44, D862–D868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol.17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res.47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meeker, D., Kallem, C., Heras, Y., Garcia, S. & Thompson, C. Case report: evaluation of an open-source synthetic data platform for simulation studies. JAMIA Open5, ooac067 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prasanna, A et al. Synthetic health data can augment community research efforts to better inform the public during emerging pandemics. Preprint at medRxiv 10.1101/2023.12.11.23298687 (2023).
- 33.Walonoski, J. et al. Synthea: an approach, method, and software mechanism for generating synthetic patients and the synthetic electronic health care record. J. Am. Med. Inform. Assoc.25, 230–238 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.1000 Genomes Project Consortiumet al. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, K. et al. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat. Genet.48, 1055–1059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Regier, A. A. et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun.9, 4038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellrott, K. et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst.6, 271–281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behera, S. et al. Comprehensive and accurate genome analysis at scale using DRAGEN accelerated algorithms. Preprint at bioRxiv10.1101/2024.01.02.573821 (2024).
- 39.Jin, Y., Schäffer, A. A., Sherry, S. T. & Feolo, M. Quickly identifying identical and closely related subjects in large databases using genotype data. PLoS ONE12, e0179106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Privé, F., Aschard, H., Ziyatdinov, A. & Blum, M. G. B. Efficient analysis of large-scale genome-wide data with two R packages: bigstatsr and bigsnpr. Bioinformatics34, 2781–2787 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, W., Chen, H., Jiang, X. & Harmanci, A. Federated generalized linear mixed models for collaborative genome-wide association studies. iScience26, 107227 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenspan, G. Scaling blockchains with off-chain data. https://www.multichain.com/blog/2018/06/scaling-blockchains-off-chain-data/ (2018).
- 43.Sedlmeir, J., Buhl, H. U., Fridgen, G. & Keller, R. The energy consumption of blockchain technology: beyond myth. Bus. Inf. Syst. Eng.62, 599–608 (2020).
- 44.Fall, T., Gustafsson, S., Orho-Melander, M. & Ingelsson, E. Genome-wide association study of coronary artery disease among individuals with diabetes: the UK Biobank. Diabetologia61, 2174–2179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eastwood, S. V. et al. Algorithms for the capture and adjudication of prevalent and incident diabetes in UK Biobank. PLoS ONE11, e0162388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet.81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harms, M. & Goldstein, D. Genomic Translation for ALS Care (GTAC)—WGS. dbGaP. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002973.v1.p1
- 48.Gagliardi, S. et al. Long non coding RNAs and ALS: still much to do. Noncoding RNA Res.3, 226–231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mackey, T. K. et al. Establishing a blockchain-enabled Indigenous Data Sovereignty framework for genomic data. Cell185, 2626–2631 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Zhang, X. & Poslad, S. Blockchain support for flexible queries with granular access control to electronic medical records (EMR). 2018 IEEE International Conference on Communications (ICC). 10.1109/icc.2018.8422883 (IEEE, 2018).
- 51.Shabani, M. Blockchain-based platforms for genomic data sharing: a de-centralized approach in response to the governance problems? J. Am. Med. Inform. Assoc.26, 76–80 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, D. R., Bhattacharya, S. & Butte, A. J. Prototype of running clinical trials in an untrustworthy environment using blockchain. Nat. Commun.10, 917 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker, J. et al. The value of health care information exchange and interoperability. Health Aff. (Millwood)10.1377/hlthaff.w5.10 (2005). [DOI] [PubMed]
- 54.Ozercan, H. I., Ileri, A. M., Ayday, E. & Alkan, C. Realizing the potential of blockchain technologies in genomics. Genome Res.28, 1255–1263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Post, A. R., Ai, M., Kalsanka Pai, A., Overcash, M. & Stephens, D. S. Architecting the data loading process for an i2b2 research data warehouse: full reload versus incremental updating. AMIA Annu. Symp. Proc.2017, 1411–1420 (2017). [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch, K. E. et al. Incrementally transforming electronic medical records into the observational medical outcomes partnership common data model: a multidimensional quality assurance approach. Appl. Clin. Inform.10, 794–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kleinaki, A.-S., Mytis-Gkometh, P., Drosatos, G., Efraimidis, P. S. & Kaldoudi, E. A blockchain-based notarization service for biomedical knowledge retrieval. Comput. Struct. Biotechnol. J.16, 288–297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhuang, Y. et al. A patient-centric health information exchange framework using blockchain technology. IEEE J. Biomed. Health Inf.24, 2169–2176 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Taralunga, D. D. & Florea, B. C. A blockchain-enabled framework for mHealth systems. Sensors (Basel)21, 2828 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koptyra, K. & Ogiela, M. R. Imagechain—application of blockchain technology for images. Sensors (Basel)21, 82 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuo, T.-T., Zavaleta Rojas, H. & Ohno-Machado, L. Comparison of blockchain platforms: a systematic review and healthcare examples. J. Am. Med. Inform. Assoc.26, 462–478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reinecke, I., Zoch, M., Reich, C., Sedlmayr, M. & Bathelt, F. The usage of OHDSI OMOP—a scoping review. Stud. Health Technol. Inform.283, 95–103 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res.20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakamoto, S. Bitcoin: A Peer-To-Peer Electronic Cash System. https://bitcoin.org/bitcoin.pdf (2008).
- 66.Zoch, M. et al. Adaption of the OMOP CDM for rare diseases. Stud. Health Technol. Inform.281, 138–142 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Hripcsak, G. et al. Facilitating phenotype transfer using a common data model. J. Biomed. Inform.96, 103253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramirez, A. H. et al. The All of Us Research Program: data quality, utility, and diversity. Patterns (N Y)3, 100570 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.OHDSI/WhiteRabbit. https://github.com/OHDSI/WhiteRabbit
- 70.OHDSI/Usagi.https://github.com/OHDSI/Usagi
- 71.A package supporting the conversion from Synthea CSV to OMOP CDM. https://ohdsi.github.io/ETL-Synthea/
- 72.OHDSI/ETL-CDMBuilder. https://github.com/OHDSI/ETL-CDMBuilder
- 73.All of Us Research Program Investigators et al. The ‘All of Us’ Research Program. N. Engl. J. Med. 381, 668–676 (2019). [DOI] [PMC free article] [PubMed]
- 74.Mezey, J. Basics of genome-wide association study (GWAS) analysis. https://physiology.med.cornell.edu/people/banfelder/qbio/resources_2013/2013_1_Mezey.pdf (2013).
- 75.Kulemin, N., Popov, S. & Gorbachev, A. Y. The Zenome Project: Whitepaper blockchain-based genomic ecosystem. https://zenome.io/download/whitepaper.pdf
- 76.Grishin, D. et al. Accelerating genomic data generation and facilitating genomic data access using decentralization, privacy-preserving technologies and equitable compensation. Blockchain in Healthcare Today. 10.30953/bhty.v1.34 (2018).
- 77.EncrypGen. http://encrypgen.com/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The individual-level data from the NYGC ALS Consortium are available to authorized investigators through the dbGaP via accession code phs003067. Data dictionaries and variable summaries are available on the dbGaP public FTP site: https://ftp.ncbi.nlm.nih.gov/dbgap/studies/phs003067/phs003067.v1.p1. Public summary-level phenotype data may be browsed at the dbGaP study report page: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003067.v1.p1.
Subject Sample Telemetry Report (SSTR) for subject and sample IDs, consents, summary counts, processing status and molecular and sequence sample uses is available on the SSTR site: https://www.ncbi.nlm.nih.gov/gap/sstr/report/phs003067.v1.p1.
The data are available under General Research and Health, Medical, Biomedical data use limitations.
GTAC data can be accessed via dbGaP accession code phs002973.v1.p1.
Genetic data for the publicly accessible blockchain network are shared via the International Genome Sample Resource (1000 Genomes Project) and can be accessed through https://www.internationalgenome.org/data-portal/data-collection/30x-grch38.
UK Biobank data are available to authorized investigators through the UK Biobank portal.
The most up-to-date code can be found at https://github.com/G2Lab/PrecisionChain/ and on Zenodo at 10.5281/zenodo.10067135. A demonstration of our user-friendly front end can be accessed via https://precisionchain.g2lab.org/ (use username: test@test.com and password: test-ME).