Abstract
Antiapoptotic Bcl‐2 family proteins are involved in myeloma cell survival. To date, their expression in multiple myeloma (MM) patients has mostly been analyzed at the RNA level. In the present study, we quantified for the first time the protein expression of the Bcl2‐family members using a capillary electrophoresis immunoassay in 120 newly diagnosed MM patients, aged ≤65 years, treated in the context of the PETHEMA/GEM2012 study. We found that the pattern of expression of Bcl‐2 family proteins was highly heterogeneous among patients. Although cases with t(11;14) had significantly higher levels of Bcl‐2/Bcl‐xL and Bcl‐2+Bim+Bax/Bcl‐xL ratios than those without t(11;14), the presence of this translocation was not synonymous with such high levels of expression. Conversely, some patients with other genetic alterations also showed higher levels of those ratios. Survival analysis revealed that the high expression of Bad and Puma proteins was associated with significantly longer overall survival (p = 0.001 and p < 0.001, respectively). Bcl‐2 protein ratios predicting sensitivity to venetoclax in vitro were also able to distinguish patients with shorter time to progression after triplet‐based induction therapy and ASCT. This is the first study to assess the expression of the most important Bcl‐2 family proteins by a quantitative method in a large set of MM patients according to their cytogenetic abnormalities. We shed light on the impact of these proteins on MM prognosis, which could help to consider the levels of proteins involved in apoptosis in the development of new therapeutic strategies.
INTRODUCTION
Evasion of apoptotic mechanisms through the deregulation of Bcl‐2 family proteins may play an important role in the resistance of multiple myeloma (MM) to various therapeutic schemes. The balance between proapoptotic and antiapoptotic proteins is essential to ensure the apoptotic threshold is reached. A dependency on prosurvival proteins has been described in MM. In particular, plasma cells are characterized by Mcl‐1 dependency, 1 , 2 which enables them to evade apoptosis. However, the antiapoptotic dependencies differ between patients. 3 , 4 Some studies have associated the presence of t(11;14) with Bcl‐2 dependency, 5 and 1q21 amplification with a high level of Mcl‐1 expression. 6 Consequently, the different expression patterns of antiapoptotic proteins may influence the response to treatments.
Therapies targeting the Bcl‐2 protein, such as venetoclax, have been developed. The efficacy of Bcl‐2 inhibitors may be related to a particular expression of Bcl‐2 family proteins in MM cells. In fact, although the expression of these proteins is highly heterogeneous in MM cell lines, different ratios between the expression of some of the genes encoding them are known to predict sensitivity to venetoclax. 4 , 7 , 8 , 9 , 10 Additionally, the initial study evaluating venetoclax sensitivity in human MM cell lines (HMCLs) and MM samples revealed that MM cells carrying the t(11;14) translocation were more sensitive than those without it. 7 Hence, this translocation was considered a strong predictive biomarker for venetoclax sensitivity in MM. However, subsequent preclinical studies demonstrated that other cell lines lacking the translocation were also sensitive to venetoclax. 2 Additionally, other factors involved in the venetoclax response have been recently unveiled. Thus, the expression of B cell markers 5 , 11 , 12 and functional approaches, such as the use of ex vivo sensitivity 13 , 14 could help to identify venetoclax‐sensitive patients.
Taken together, these findings indicate that there are no accurate biomarkers for identifying MM patients who are sensitive to venetoclax, although the expression of BCL‐2 family members is considered the most robust set of biomarkers. To date, their expression in MM patients has most often been analyzed at the RNA level, particularly using quantitative polymerase chain reaction. With the aim of enabling the quantification of these potential biomarkers of response to venetoclax in clinical practice using samples from patients with MM, the determination of Bcl‐2, Bcl‐xL, and Mcl‐1 by immunohistochemistry has been proposed. 8
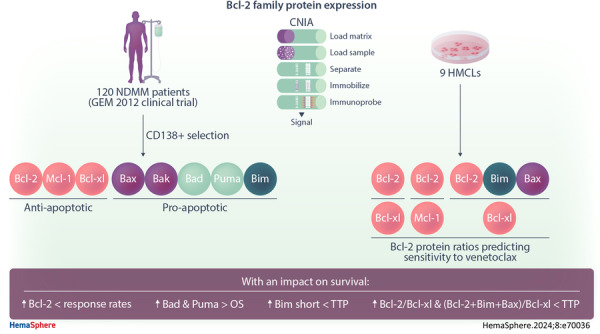
These circumstances prompted us to elaborate a profile of Bcl‐2 family protein expression, including Mcl‐1, Bcl‐2, Bcl‐xL, Bak, Bax, Bad, Puma and Bim, quantifying each of them by capillary nanoimmunoassay (CNIA) in a set of myeloma cell samples from newly diagnosed MM patients (NDMM) who were homogeneously treated and for whom information about clinical features, cytogenetic abnormalities and survival was available. This study builds, for the first time, a comprehensive protein expression profile of the Bcl‐2 family members in a large set of MM patients. This profile reveals the abundance of both antiapoptotic and proapoptotic proteins, depending on genetic abnormalities, and their impact on the clinical course of patients. Furthermore, we conducted in vitro assays using HMCLs to determine whether the expression ratios between Bcl‐2 family members, previously associated with response to venetoclax when analyzed at the RNA level, exhibited similar associations when examined in terms of protein expression.
MATERIALS AND METHODS
Primary samples
The study included 120 protein samples from NDMM patients enrolled in the GEM2012 clinical trial (NCT01916252), which involved bortezomib, lenalidomide, and dexamethasone (VRD) induction followed by autologous stem cell transplantation (ASCT) and consolidation treatment with VRD. Patients with at least minimal response were included in the maintenance trial GEM2014 (NCT02406144). Details of the GEM2012 trial and sample processing have been reported elsewhere. 15 , 16 , 17 Results of fluorescence in situ hybridization (FISH) studies to detect IGH rearrangements (t(11;14, t(4;14) and t(14;16)), 17p and 1p deletions, and 1q gains were available for all patients. 18 In this clinical trial, measurable residual disease (MRD) was assessed by next‐generation flow (NGF) cytometry. 19 Baseline characteristics of patients for whom data were available are summarized in Supporting Information S1: Table 1. This cohort of patients was representative of the entire GEM2012 trial dataset. 15
Human MM cell lines
The HMCLs H929, MM.1S, and U266 were acquired from the ATCC (American Type Culture Collection); KMS12‐BM, KMS12‐PE, JJN‐3, MOLP‐8, and SK‐MM‐2 were obtained from DMSZ (Deutsche Sammlung von Mikroorganismen and Zellkulturen); and KMS28‐BM were procured from the JCRB (Japanese Collection of Research Bioresources) cell bank.
FISH studies to detect IGH rearrangements, 17p and 1p deletions, and 1q gains were assessed for all HMCLs. Cells were routinely checked for the presence of mycoplasma with a MycoAlert kit (Lonza). Cell line identity was confirmed by STR analysis before starting the experiments using a PowerPlex 16 HS System kit (Promega) and online STR matching analysis (www.dsmz.de/fp/cgi-bin/str.html).
Reagents and cell viability assays
Venetoclax (ABT‐199) was purchased from Selleckchem (Catalog No. S8048). All HMCLs were treated with increasing concentrations of venetoclax (1 nM–5 µM), and cell viability was evaluated using the 3‐(4,5‐dimethylthiazol‐2‐yl)−2,5‐diphenyltetrazolium bromide (MTT) colorimetric assay (Sigma‐Aldrich), as described previously. 20 , 21
Protein extraction and capillary electrophoresis immunoassay
Proteins were extracted at the same time as genomic DNA and RNA and were purified by ice‐cold acetone precipitation, as previously described. 17 CNIA was performed using the WES system (ProteinSimple) according to the manufacturer's protocols and as previously outlined. 16 , 17 , 22 The primary antibodies used in this study were as follows: Bcl‐2 (sc‐7382), Bcl‐xL (54H6 Rabbit mAb #2764), Mcl‐1 (D35A5 Rabbit mAb #5453), Bax (D2E11 Rabbit mAb #5023), Bak (D4E4 Rabbit mAb #12105), Bim (C34C5 Rabbit mAb #2933), Puma (D30C10 Rabbit mAb #12450), Bad (D24A9 Rabbit mAb #9239), and Gapdh (14C10 Rabbit mAb #2118) proteins, the latter being used as the endogenous control.
All protein data were analyzed and quantified with Compass™ software (ProteinSimple) based on measurements of chemiluminescence peaks. Protein expression was reported as values relative to Gapdh, the endogenous control.
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics (version 28) and R (version 4.3.0). Statistical significance was concluded for values of p < 0.05. We analyzed the association between clinical parameters and levels of protein expression. Pearson's χ 2 test or Fisher's exact test were used to examine group differences of categorical variables. Differences in the levels of protein expression between groups were assessed with the two‐sided Student's t‐test or the Mann–Whitney U test, depending on whether the continuous variables were normally or nonnormally distributed, respectively, normality having been evaluated beforehand with the Shapiro–Wilk test.
We used Cutoff Finder software (http://molpath.charite.de/cutoff) to estimate the optimal cutoff values for each protein in the survival analysis, requiring a minimum of 20 samples per group. The endpoints for this analysis were time to progression (TTP), defined as the time from diagnosis to disease progression/relapse, and overall survival (OS), which refers to time from diagnosis until the date of death. Survival curves were plotted using the Kaplan–Meier method, and survival in the groups was compared using the log‐rank test. A multivariable Cox proportional hazards regression model was performed for the multivariable analysis. The contribution of each variable to the model was evaluated in terms of the proportion of the χ 2 value accumulated.
RESULTS
Expression of Bcl‐2 family proteins
Levels of expression of the antiapoptotic (Mcl‐1, Bcl‐2, Bcl‐xL) and proapoptotic (Bak, Bax, Bad, Puma, and Bim) proteins from the Bcl‐2 family were analyzed in 120 NDMM samples using CNIA (Figure 1A). As the different isoforms of Mcl‐1, Puma, and Bim proteins could be distinguished by molecular weight, we were able to note that the long isoforms were the most frequently expressed (Supporting Information S1: Figure 1).
Figure 1.
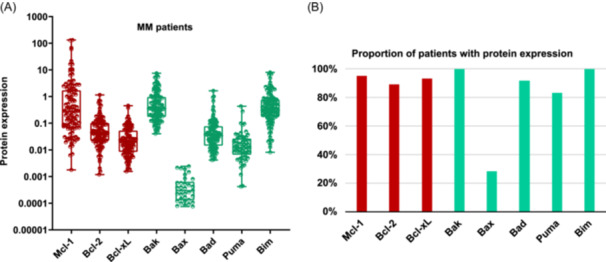
Bcl‐2 family protein expression in MM samples. (A) The expression of each protein was assessed by CNIA and normalized with respect to Gapdh expression in each case. (B) Percentage of patients with the presence of each protein. The antiapoptotic proteins (Mcl‐1, Bcl‐2, and Bcl‐xL) are represented in red, while the proapoptotic ones (Bak, Bax, Bad, Puma, and Bim) are shown in green. Only the patients with protein expression are shown.
All the proteins were present in more than 80% of patients, except for the effector proapoptotic protein Bax, which was found in fewer than 30% of patients. Bak, Bim, and Mcl‐1 exhibited the highest levels of expression, being present in almost 100% of patients. In contrast, Bax protein showed the lowest expression levels (Figure 1A,B).
Association between Bcl‐2 family proteins, and clinical features and cytogenetic abnormalities
Patients with low expression levels of the Bim protein showed a significantly higher incidence of plasmacytomas (p = 0.019). No associations were identified between protein expression and the other clinical parameters analyzed.
Exploring the relationship between the Bcl‐2 family protein levels and cytogenetic abnormalities, we found that Bak protein expression was significantly higher in patients with t(14;16) (p = 0.028), while Bcl‐xL expression was lower in patients with t(11;14) (p = 0.014 (Figure 2A). There was no association between the other cytogenetic abnormalities studied and any of the proteins tested (Supporting Information S1: Table 2).
Figure 2.
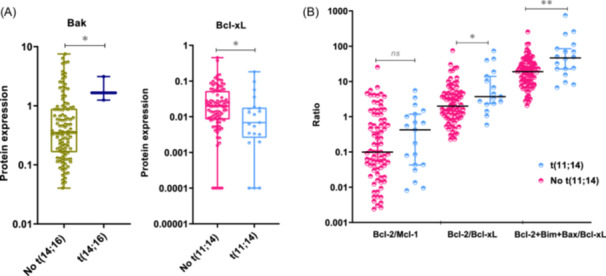
Association between proteins and cytogenetic abnormalities. (A) Expression level of Bak‐1 and Bcl‐xL according to the presence of t(14;16) and t(11;14), respectively. (B) Expression level of the Bcl‐2/Mcl‐1, Bcl‐2/Bcl‐xL, and Bcl‐2+Bim+Bax/Bcl‐xL ratios, according to the presence or absence of t(11;14). The statistically significant differences between groups were determined by the Mann–Whitney U test. *p < 0.05; **p < 0.01: ns, non‐statistically significant.
Previous studies have reported a great heterogeneity in the messenger RNA (mRNA) expression of genes coding for Bcl‐2 family proteins, with no individual gene serving as a reliable biomarker of venetoclax response. However, ratios based on the expression of some of these genes (BCL2/MCL1, BCL2/BCL2L1, and BCL2+BCL2L11+BAX/BCL2L1) do have a predictive value for the response to venetoclax. Additionally, the MM patients with high BCL2/MCL1 and BCL2/BCL2L1 gene expression ratios have been described to be enriched in cases with t(11;14), 7 , 9 , 23 and high BCL2+BCL2L11+BAX/BCL2L1 ratio have been associated with CCND1 MM molecular subgroup. 10 This prompted us to examine, for the first time, the association between these three reported ratios at the protein level and the most common cytogenetic abnormalities. We found that patients with t(11;14) had higher Bcl‐2/Bcl‐xL and Bcl‐2+Bim+Bax/Bcl‐xL ratios than those without t(11;14) (p = 0.022, p = 0.003, respectively) (Figure 2B). However, no statistically significant differences were observed between the ratios of expression in the group of patients with t(11;14) and each of the other cytogenetic groups considered separately (Figure 3). Within the t(11;14) group, some patients exhibited low expression level ratios, while other patients with cytogenetic abnormalities other than t(11;14) had expression levels higher than those of the third quartile of the t(11;14) group (Figure 3).
Figure 3.
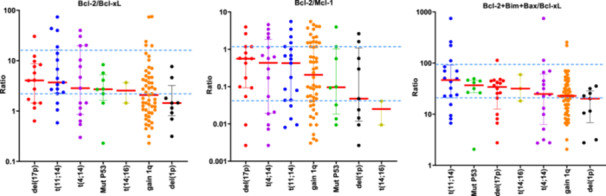
Distribution of the Bcl‐2/Mcl‐1, Bcl‐2/Bcl‐xL, and Bcl‐2+Bim+Bax/Bcl‐xL protein expression ratios according to the cytogenetic abnormalities del(17p), t(4;14), t(11;14), 1q gain, TP53 mutation, del(1p), t(14;16). The first and third quartiles of the protein expression ratios in the group of patients with t(11;14) are indicated with a blue dashed line. Patients with more than one alteration are represented more than once.
Bcl‐2 family protein expression in relation to in vitro sensitivity to venetoclax
To evaluate the expression pattern of Bcl‐2 family proteins in HMCLs and its association with venetoclax sensitivity, we analyzed Bcl‐2 family protein expression in nine HMCLs: KMS12‐PE, KMS12‐BM, U266, SK‐MM‐2, and MOLP‐8,which bear t(11;14); KMS28‐BM and H929 carrying t(4;14); MM.1S and JJN‐3 with t(14;16). As with patients, the proteins with the highest expression were Bak, Bim, and Mcl‐1, and the lowest was Bax (Supporting Information S1: Figure 2).
We conducted in vitro drug sensitivity assays to confirm the responsiveness or resistance to venetoclax of nine HMCLs previously reported by other authors. The HMCLs were treated with increasing concentrations of venetoclax for 48 h. The viability of HMCLs after exposure to 1 µM of venetoclax for 48 h was approximately 100% for U266, H929, MM.1S, and MOLP‐8, so they were categorized as resistant cell lines. Among the sensitive HMCLs, we distinguished three groups based on the degree of sensitivity: high sensitivity for KM12‐PE and SK‐MM‐2, intermediate sensitivity for KMS12‐BM and KMS28‐BM, and low sensitivity for JJN‐3. U266 and MOLP‐8 were both resistant to venetoclax in spite of harboring t(11;14). Conversely, other HMCLs without t(11;14), such as KMS28‐BM and JJN‐3, were sensitive to venetoclax (Figure 4).
Figure 4.
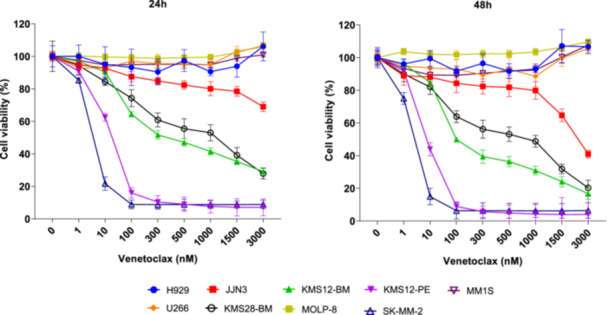
Antimyeloma activity of venetoclax in in vitro studies on human multiple myeloma cell lines (HMCLs). The HMCLs were treated with increasing concentrations of venetoclax for 24 and 48 h. Cell viability was assessed by 3‐(4,5‐dimethylthiazol‐2‐yl)−2,5‐diphenyltetrazolium bromide (MTT) assay. The means ± standard deviation of three independent experiments are represented.
After confirming that the selected set of HMCLs displayed a range of responses to venetoclax, spanning from highly sensitive to resistant, we investigated whether the observed variations in response were associated with the expression of Bcl‐2 family proteins. We observed that the group of HMCLs sensitive to venetoclax had higher Bcl‐2 and Puma expression levels than the resistant ones (p = 0.036) (Supporting Information S1: Figure 3A). Regarding the ratios predicting response to venetoclax, the group containing the sensitive cell lines showed higher levels of the three ratios than the group that included the resistant cell lines (p = 0.036) (Supporting Information S1: Figure 3B). When we considered the HCMLs individually, SK‐MM‐2 and KMS12‐PE were the HMCLs with the highest Bcl‐2/Mcl‐1, Bcl‐2/Bcl‐xL, and Bcl‐2+Bim+Bax/Bcl‐xL expression ratios (Table 1, Supporting Information S1: Figure 3C).
Table 1.
Sensitivity to venetoclax in HMCLs.
| Human MM cell lines | Sensitivity to venetoclax | IGH tx | TP53 status | Ratio Bcl2/Mcl1 | Ratio Bcl2/Bcl‐xl | Ratio Bcl2+Bim+Bax/Bcl‐XL |
|---|---|---|---|---|---|---|
| SK‐MM‐2 | Yes | t(11;14) | DH | 6.39 | 56.98 | 412.12 |
| KMS12‐PE | Yes | t(11;14) | DH | 0.67 | 18.86 | 122.4 |
| KMS12‐BM | Yes | t(11;14) | DH | 0.33 | 5.21 | 32.21 |
| KMS28‐BM | Yes | t(4;14) | DH | 0.6 | 4.36 | 26.68 |
| JJN‐3 | Yes | t(14;16) | No expression | 0.26 | 3.5 | 23.33 |
| U266 | No | t(11;14) | DH | 0.14 | 0.47 | 5.57 |
| H929 | No | t(4,14) | WT | 0.03 | 0.31 | 2.24 |
| MM.1S | No | t(14;16) | WT | 0.12 | 0.21 | 3.22 |
| MOLP‐8 | No | t(11;14) | WT | 0 | 0 | 18.45 |
Abbreviations: DH, TP53 double hit; WT, TP53 wild type.
 HMCLs sensitive to venetoclax.
HMCLs sensitive to venetoclax.
 HMCLs with low sensitivity to venetoclax.
HMCLs with low sensitivity to venetoclax.
 HMCLs resistant to venetoclax.
HMCLs resistant to venetoclax.
We did not find any association between sensitivity to venetoclax and the presence of t(11;14). With respect to TP53 status, all the highly sensitive HMCLs presented double hit TP53 (DH‐TP53) (4/4), while those with TP53 wild type were resistant (3/3) (Table 1). The group of HMCLs with DH‐TP53 had higher levels of Bcl‐2 and Bim (p = 0.032 and 0.016, respectively), as well as higher Bcl‐2/Mcl‐1 and Bcl‐2/Bcl‐xL ratios than the group of HMCLs without DH‐TP53 (p = 0.032). We found no other statistically significant differences between the levels of Bcl‐2 family proteins or their ratios and the other cytogenetic abnormalities present in MM cell lines.
Impact of Bcl‐2 family protein expression on response and survival
We examined the association between Bcl‐2 family protein expression at the time of diagnosis and the response at three different time points in the treatment scheme: after induction therapy, upon ASCT, and after consolidation. Within the group of patients who achieved a very good response or better after both induction and ASCT, Bcl‐2 expression was significantly lower than in the group of patients with partial response or worse (p = 0.001 and 0.028, respectively) (Figure 5A).
Figure 5.
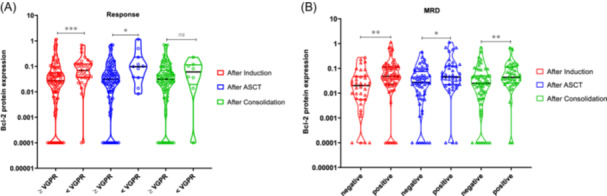
Association between Bcl‐2 protein expression analyzed at diagnosis and the response at three different time points in the treatment scheme (GEM2012 protocol). The response to treatment was assessed by IMWG response criteria (A) and minimal residual disease (MRD) by flow cytometry (B), at three points: after induction (in red), after autologous stem cell transplantation (ASCT) (in blue), and after consolidation (in green). The statistically significant differences between groups were determined by the Mann–Whitney U test. *p < 0.05, **p < 0.01, ***p < 0.001.
The link between high levels of Bcl‐2 expression and poorer response was confirmed when evaluating the MRD data at the same three assessment times. Our observations revealed that patients who were MRD‐positive after induction therapy, ASCT, and consolidation had significantly higher levels of Bcl‐2 protein expression at diagnosis than those who were MRD‐negative (p = 0.002, 0.033, and 0.007, respectively) (Figure 5B).
In the survival analysis, high levels of the proapoptotic proteins Bad and Puma were associated with longer OS (p = 0.001 and < 0.001, respectively) (Figure 6A). Conversely, patients with high levels of Bim expression had a shorter TTP than those with low levels (p = 0.046) (Figure 6B). Since the expression of four distinct protein isoforms of Bim could be distinguished, we evaluated their impact on survival and found that the negative effect on TTP was at the expense of the short isoform of Bim (BimS) (p = 0.016) (Supporting Information S1: Figure 4). For the antiapoptotic proteins, elevated levels of Bcl‐2 expression had a negative impact on TTP (p = 0.019) (Figure 6B). The levels of the remaining proteins were not significantly associated with survival.
Figure 6.
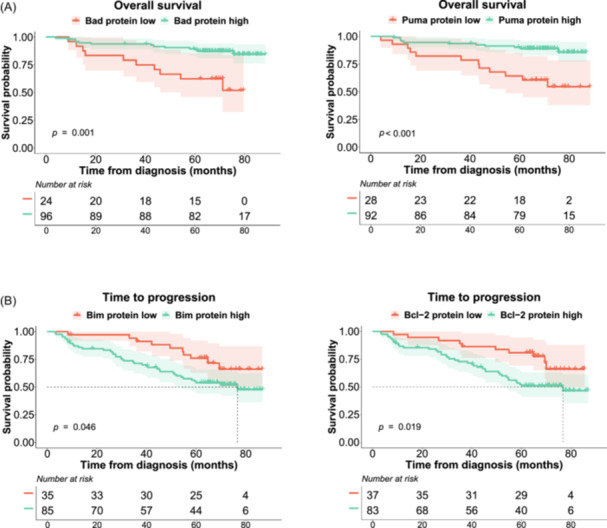
Kaplan–Meier curves for Bad, Puma, Bim, and Bcl‐2. Probability of overall survival (OS) (A) and survival without progression (time to progression [TTP]) (B) of MM patients by protein expression. The log‐rank test p values are shown.
Based on the observation that Bcl‐2 ratios seem to identify responders to venetoclax, and given that venetoclax has its own distinctive mechanism of action, we hypothesized that these ratios had prognostic implications in patients treated with conventional drugs, such as those included in the GEM2012 trial. We found that patients with higher Bcl‐2/Bcl‐xL and Bcl‐2+Bim+Bax/Bcl‐xL expression ratios had shorter TTP (p = 0.004 and < 0.001, respectively). Furthermore, this negative effect remained on OS for the Bcl‐2+Bim+Bax/Bcl‐xL ratio (p = 0.036) (Supporting Information S1: Figure 5).
In the multivariable models, we included conventional clinical variables such as the presence of plasmacytomas, LDH, and ISS score, as well as the high cytogenetic risk abnormalities and all the analyzed proteins, except for Bak due to its high collinearity. We observed that the presence of high cytogenetic risk and high levels of Puma expression retained their respective negative and positive impacts on OS (HR = 9.6, p < 0.001, and HR = 0.085, p < 0.001, respectively). In the multivariate Cox model for TTP, high cytogenetic risk and high levels of Bim remained as independent prognostic factors (HR = 3.1, p < 0.001, and HR = 2.5, p < 0.001, respectively) (Figure 7).
Figure 7.
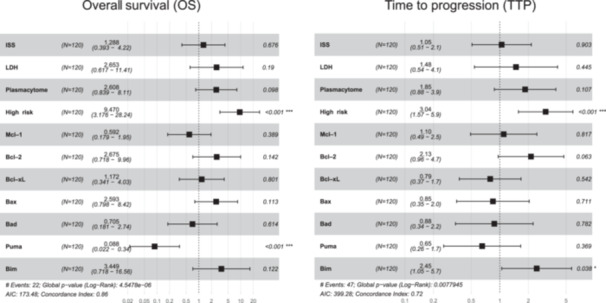
Multivariable analysis of OS and TTP. Forest plot of multivariable Cox proportional hazards regression models accounting for each potential risk factor associated with OS and TTP of MM: age, ISS III versus I/II, LDH high level, plasmacytoma, FISH risk (high cytogenetic risk, including del17p, t(4;14) and/or t(14;16), vs. standard risk), and the expression level of the studied proteins. For each factor, the hazard ratios and their 95% confidence intervals are shown. AIC, Akaike information criterion; FISH: fluorescence in situ hybridization; ISS, International Staging System; LDH, lactate dehydrogenase; OS, overall survival; TTP, time to progression.
DISCUSSION
The apoptosis pathway plays a central role in the survival of MM cells and their ability to acquire drug resistance. In this study, we quantified the expression of Bcl‐2 family proteins, including the antiapoptotic proteins Mcl‐1, Bcl‐2, and Bcl‐xL, and the proapoptotic proteins Bak, Bax, Bad, Puma, and Bim, in a cohort of 120 NDMM and transplant‐eligible homogeneously treated patients. Most studies assessing the expression of the Bcl‐2 family do so at the RNA level, and the few that have considered proteins have only tested the antiapoptotic proteins Bcl‐2, Bcl‐xL, and Mcl‐1 using immunohistochemistry. 8
We found the pattern of expression of Bcl‐2 proteins to be moderately heterogeneous among patients, as previously noted from analyses of gene expression. 24 , 25 We also did not find an imbalance in favor of the overexpression of antiapoptotic proteins in MM patients.
Since MM cells with t(11;14) are known to be more dependent on the antiapoptotic protein Bcl‐2, 5 we paid special attention to exploring relationships between a variety of cytogenetic alterations and the expression of eight Bcl‐2 family proteins. We were unable to find any association between the presence of t(11;14) and overexpression of Bcl‐2 protein, although we did detect low levels of the antiapoptotic protein Bcl‐xL, as previously described in the CCND1 molecular subgroup. 25 Additionally, expression levels of Bak protein were higher in patients with t(14;16), as was found when analyzing mRNA in the MAF molecular subgroup. 25 However, the limited number of patients with this translocation prevents definitive conclusions from being drawn.
The Bcl‐2 inhibitors have demonstrated their effect on hematological neoplasms and are approved to treat patients with chronic lymphocytic leukemia and acute myeloid leukemia. In MM, the ratios between gene expression of Bcl‐2 members, such as BCL2/BCL2L1, BCL2/MCL1 9 , and BCL2+BCL2L11+BAX/BCL2L1 10 have been correlated with the response to venetoclax. The early‐phase trials using venetoclax in MM patients revealed a significantly higher response rate in patients with t(11;14). This could be related to the unique apoptosis profile attributed to this translocation, which is characterized by the expression of high levels of BCL2 relative to BCL‐XL and MCL‐1. This led to the design of phase 3 trials specifically targeting patients with this translocation. 26 Although we confirmed in our study that patients with t(11;14) had higher protein expression for Bcl‐2/Bcl‐xL and Bcl‐2+Bim+Bax/Bcl‐xL ratios than patients without this translocation, we found considerable variation in these ratios across different cytogenetic groups. Thus, some patients with t(11;14) showed very low ratios, while a subgroup of patients with other cytogenetic abnormalities had higher ratios than those of the group with t(11;14).
In vitro venetoclax sensitivity assays performed in this work confirmed the sensitivity to venetoclax of SK‐MM‐2, KMS12‐PE, KMS12‐BM, and KMS28‐BM, while H929, U266, and MOLP‐8 were resistant, consistent with previous reports. 2 , 4 The group of HMCLs sensitive to venetoclax had higher expression levels of the Bcl‐2 protein ratios than the resistant ones. Therefore, we validated what had previously been described by others at the mRNA level 7 , 8 , 10 and protein level. 11 It should be noted that the HMCLs with t(11;14) that were sensitive to venetoclax were those with high Bcl‐2 protein ratios. Similarly, the HMCLs without that translocation that responded to venetoclax also had high ratios. These data confirm the evidence that t(11;14) is not an adequate biomarker for predicting response to venetoclax, while Bcl‐2 family protein ratios may function better as predictive biomarkers. Previous studies have shown that ratios based on antiapoptotic proteins (Bcl‐2, Bcl‐xl, and Mcl‐1) are predictive of response to venetoclax, which aligns with our findings. However, our work introduces a novel aspect by incorporating the quantification of five proapoptotic proteins, thereby detecting the association between high levels of Puma protein with sensitivity to venetoclax.
The influence of Bcl‐2 proteins on the response and survival of a cohort of MM patients treated as part of the GEM2012 trial was also investigated in this study. We observed that high levels of Bcl‐2 protein defined a group of MM patients with significantly poorer responses to VRD induction followed by ASCT and posttransplant consolidation (2 cycles of VRD), estimated by conventional techniques and NGF‐based MRD assessment. These results translated into a significantly shorter TTP for this group of patients. Moreover, high Bcl‐2/Mcl‐1, Bcl‐2/Bcl‐xL, and Bcl‐2+Bim+Bax/Bcl‐xL ratios were also associated with significantly shorter TTP. Considering these results, elevated Bcl‐2‐based ratios would not only identify a group of MM patients likely to respond to venetoclax but also patients who would have shorter survival if treated with VDR and ASCT.
High levels of Bad and Puma proteins also had a positive impact on the outcome of MM patients treated in the GEM2012 trial, being associated with significantly longer OS. Moreover, this association was retained for Puma in the multivariable analysis along with age, high LDH, and del(17p), which are well‐known prognostic factors in MM. 27 , 28 , 29 These results are in accordance with those of previous research in other neoplasms. 30 , 31 , 32 , 33 , 34 Recently, loss of Puma has been associated with venetoclax resistance by an epigenetic mechanism. 35 The independent prognostic value of Puma protein expression in NDMM patients treated with VRD and ASCT warrants validation in other independent studies.
We also found that high levels of the proapoptotic protein Bim had a significantly negative impact on the TTP of MM patients. Even though this result may seem counterintuitive, some studies have found an association between high levels of BIM with shorter PFS in non‐small cell lung cancer patients treated with gefitinib 36 or erlotinib, 37 and with shorter OS in breast cancer patients. 38 To shed more light on this matter, we analyzed the protein isoforms of Bim and confirmed that the negative prognostic impact was due to the short isoform (Bims), which has been considered the most potent apoptosis inductor, 39 as it binds directly to Bax without binding Bcl‐2 or Mcl‐1. 40 The other Bcl‐2 proteins analyzed did not show any impact on survival, including Mcl‐1, which is located on chromosome 1q21. However, we did not find any association between the expression of the Mcl‐1 protein and the presence of 1q gain.
Overall, our study provides valuable insights into Bcl‐2 family protein expression in patients with MM according to cytogenetic abnormalities, supporting the idea that t(11;14) is unlikely to be a consistent biomarker of venetoclax efficacy. Conversely, our results emphasize the value of quantifying Bcl‐2 protein members for identifying MM patients with short survival after receiving treatment with conventional triplet induction followed by ASCT, some of whom might benefit from incorporating venetoclax into their therapeutic approach.
Our results provide a comprehensive profile of Bcl‐2 family protein expression that could be helpful in understanding MM outcomes. We found that the high levels of Bcl‐2/Bcl‐xL and Bcl‐2+Bim+Bax/Bcl‐xL ratios were not exclusive to patients with t(11;14). This observation in MM patients was consistent with the in vitro sensitivity experiments to venetoclax, which showed that the sensitive MM cell lines exhibited high ratios of Bcl‐2 family proteins regardless of the presence of the t(11;14). Finally, the survival analysis showed that high expression of Bad and Puma proteins was associated with significantly longer OS of MM patients treated in the GEM2012 trial.
AUTHOR CONTRIBUTIONS
Cristina De Ramón designed the study, extracted and analyzed data, and wrote the manuscript. Elizabeta A. Rojas performed CNIA experiments, analyzed data, and prepared the figures. Irena Misiewicz‐Krzeminska was responsible for designing the CNIA protocol and supervised the data analysis. Ignacio J. Cardona‐Benavides participated in the preparation of figures and data analysis. Myriam Cuadrado contributed to interpreting results and creating tables. Isabel Isidro assisted with CNIA experiments. María‐José Calasanz and Manuela Fernandez reviewed the FISH results. Noemi Puig, M. Teresa Cedena, and Bruno Paiva were responsible for analyzing MRD. Ramón García‐Sanz, Laura Rosiñol, Joaquín Martínez‐López, Joan Bladé, Juan J. Lahuerta, Jesús F. San Miguel, and María V. Mateos provided patient samples and clinical data and were responsible for obtaining informed consent from patients. Luis A. Corchete analyzed data, supervised the whole study, and provided feedback on the manuscript. Norma C. Gutiérrez conceived the idea and designed the study, contributed to data interpretation, supervised the whole study and provided funding. All authors critically reviewed and approved the manuscript.
CONFLICT OF INTEREST STATEMENT
Cristina De Ramón has received travel grants from Beigene. Ramón García‐Sanz has received personal fees from Amgen, Janssen, Takeda, and Pfizer, honoraria from Pharmacyclics, research funding from Hospira, and travel accommodation from Celgene, unconnected with the submitted work. Bruno Paiva has received research funding from Bristol Myers Squibb/Celgene, Roche, and Sanofi and has served as a consultant or advisor for Adaptive, Amgen, Bristol Myers Squibb, Celgene, GlaxoSmithKline, Janssen, Kite, Roche, Sanofi, and Takeda. Noemi Puig has received honoraria from Amgen, Celgene, Janssen, Takeda, and The Binding Site, funding from Celgene, Janssen, Amgen, and Takeda, and travel grants from Amgen Celgene, Janssen, and Takeda. Laura Rosiñol has received consulting fees from Amgen, BMS/Celgene, Sanofi, Janssen, Takeda, GSK, and Karyofarm. Joaquín Martínez‐López has received honoraria and consulting fees from BMS/Celgene, Incyte, Janssen, Novartis, Sanofi, and Roche. Joan Bladé has received honoraria from Celgene, Janssen, and Amgen. Jesús F. San Miguel has consulting and advisory roles with Amgen, Bristol‐Myers Squibb, Celgene, Janssen, MSD, Novartis, Takeda, and Roche. María V. Mateos has served on speakers bureaus and advisory boards for AbbVie, Adaptive, Amgen, Celgene, GlaxoSmithKline, Janssen, Mundipharma, Oncopeptides, PharmaMar, Roche, Seattle Genetics, and Takeda. Norma C. Gutiérrez has received honoraria from Janssen and Amgen, and travel grants from Gilead. All the other authors have no conflicts of interest to disclose.
FUNDING
This study was funded by the Instituto de Salud Carlos III and co‐financed by FEDER (PI19/00674, PI23/00319); by the Asociación Española Contra el Cancer (AECC) (Proyectos Estratégicos: PROYE20047GUTI); and by the Gerencia Regional de Salud, Junta de Castilla y León grants (GRS2058/A/19, GRS2873/A1/2023). IJCB was supported by a fellowship from the Instituto de Salud Carlos III (contract PFIS‐2020: FI20/00226). CDR was supported by a fellowship from the AECC (CL‐JUN18010DERA); EARR was supported by the Consejería de Educación de Castilla y León and FEDER funds. The WES platform was acquired thanks to the INNOCAMPUS program (CEI10‐1‐0010).
Supporting information
Supporting information.
ACKNOWLEDGMENTS
The authors thank Vanesa Gutiérrez and Rut García for their technical assistance with MM cell purification and FISH analysis, and Phil Mason for his help in reviewing the English language of the manuscript.
DATA AVAILABILITY STATEMENT
Data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Gupta VA, Matulis SM, Conage‐Pough JE, et al. Bone marrow microenvironment‐derived signals induce Mcl‐1 dependence in multiple myeloma. Blood. 2017;129(14):1969‐1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gong JN, Khong T, Segal D, et al. Hierarchy for targeting prosurvival BCL2 family proteins in multiple myeloma: pivotal role of MCL1. Blood. 2016;128(14):1834‐1844. [DOI] [PubMed] [Google Scholar]
- 3. Slomp A, Peperzak V. Role and regulation of pro‐survival BCL‐2 proteins in multiple myeloma. Front Oncol. 2018;8:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gomez‐Bougie P, Maiga S, Tessoulin B, et al. BH3‐mimetic toolkit guides the respective use of BCL2 and MCL1 BH3‐mimetics in myeloma treatment. Blood. 2018;132(25):2656‐2669. [DOI] [PubMed] [Google Scholar]
- 5. Kitadate A, Terao T, Narita K, et al. Multiple myeloma with t(11;14)‐associated immature phenotype has lower CD38 expression and higher BCL2 dependence. Cancer Sci. 2021;112(9):3645‐3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Slomp A, Moesbergen LM, Gong J, et al. Multiple myeloma with 1q21 amplification is highly sensitive to MCL‐1 targeting. Blood Adv. 2019;3(24):4202‐4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Touzeau C, Dousset C, Le Gouill S, et al. The Bcl‐2 specific BH3 mimetic ABT‐199: a promising targeted therapy for t(11;14) multiple myeloma. Leukemia. 2014;28(1):210‐212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Punnoose EA, Leverson JD, Peale F, et al. Expression profile of BCL‐2, BCL‐XL, and MCL‐1 predicts pharmacological response to the BCL‐2 selective antagonist venetoclax in multiple myeloma models. Mol Cancer Ther. 2016;15(5):1132‐1144. [DOI] [PubMed] [Google Scholar]
- 9. Kumar S, Kaufman JL, Gasparetto C, et al. Efficacy of venetoclax as targeted therapy for relapsed/refractory t(11;14) multiple myeloma. Blood. 2017;130(22):2401‐2409. [DOI] [PubMed] [Google Scholar]
- 10. Tessoulin B, Papin A, Gomez‐Bougie P, et al. BCL2‐family dysregulation in B‐cell malignancies: from gene expression regulation to a targeted therapy biomarker. Front Oncol. 2019;8:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta VA, Barwick BG, Matulis SM, et al. Venetoclax sensitivity in multiple myeloma is associated with B‐cell gene expression. Blood. 2021;137(26):3604‐3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leblay N, Ahn S, Tilmont R, et al. Integrated epigenetic and transcriptional single‐cell analysis of t(11;14) multiple myeloma and its BCL2 dependency. Blood. 2024. Jan 4;143(1):42‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matulis SM, Gupta VA, Neri P, et al. Functional profiling of venetoclax sensitivity can predict clinical response in multiple myeloma. Leukemia. 2019;33(5):1291‐1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta VA, Matulis SM, Barwick BG, et al. Venetoclax ex vivo functional profiling predicts improved progression‐free survival. Blood Cancer J. 2022;12(8):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosiñol L, Oriol A, Rios R, et al. Bortezomib, lenalidomide, and dexamethasone as induction therapy prior to autologous transplant in multiple myeloma. Blood. 2019;134(16):1337‐1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Misiewicz‐Krzeminska I, Corchete LA, Rojas EA, et al. A novel nano‐immunoassay method for quantification of proteins from CD138‐purified myeloma cells: biological and clinical utility. Haematologica. 2018;103(5):880‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Misiewicz‐Krzeminska I, Isidro I, Gutiérrez N. Capillary nano‐immunoassay for quantification of proteins from CD138‐purified myeloma cells. Bio‐protocol. 2019;9(12):e3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. López‐Corral L, Gutiérrez NC, Vidriales MB, et al. The progression from MGUS to smoldering myeloma and eventually to multiple myeloma involves a clonal expansion of genetically abnormal plasma cells. Clin Cancer Res. 2011;17(7):1692‐1700. [DOI] [PubMed] [Google Scholar]
- 19. Paiva B, Puig N, Cedena MT, et al. Measurable residual disease by next‐generation flow cytometry in multiple myeloma. J Clin Oncol. 2020;38(8):784‐792. [DOI] [PubMed] [Google Scholar]
- 20. Misiewicz‐Krzeminska I, Sarasquete ME, Quwaider D, et al. Restoration of microRNA‐214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica. 2013;98(4):640‐648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rojas EA, Corchete LA, San‐Segundo L, et al. Amiloride, an old diuretic drug, is a potential therapeutic agent for multiple myeloma. Clin Cancer Res. 2017;23(21):6602‐6615. [DOI] [PubMed] [Google Scholar]
- 22. Misiewicz‐Krzeminska I, de Ramón C, Corchete LA, et al. Quantitative expression of Ikaros, IRF4, and PSMD10 proteins predicts survival in VRD‐treated patients with multiple myeloma. Blood Adv. 2020;4(23):6023‐6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cleynen A, Samur M, Perrot A, et al. Variable BCL2/BCL2L1 ratio in multiple myeloma with t(11;14). Blood. 2018;132(26):2778‐2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jourdan M, Reme T, Goldschmidt H, et al. Gene expression of anti‐ and pro‐apoptotic proteins in malignant and normal plasma cells. Br J Haematol. 2009;145(1):45‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gomez‐Bougie P, Amiot M. Apoptotic machinery diversity in multiple myeloma molecular subtypes. Front Immunol. 2013;4:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumar SK, Harrison SJ, Cavo M, et al. Venetoclax or placebo in combination with bortezomib and dexamethasone in patients with relapsed or refractory multiple myeloma (BELLINI): a randomised, double‐blind, multicentre, phase 3 trial. Lancet Oncol. 2020;21(12):1630‐1642. [DOI] [PubMed] [Google Scholar]
- 27. Abdallah NH, Smith AN, Geyer S, et al. Conditional survival in multiple myeloma and impact of prognostic factors over time. Blood Cancer J. 2023;13(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Palumbo A, Avet‐Loiseau H, Oliva S, et al. Revised international staging system for multiple myeloma: a report from international myeloma working group. J Clin Oncol. 2015;33(26):2863‐2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. D'Agostino M, Cairns DA, Lahuerta JJ, et al. Second revision of the international staging system (R2‐ISS) for overall survival in multiple myeloma: a European Myeloma Network (EMN) report within the HARMONY project. J Clin Oncol. 2022;40(29):3406‐3418. [DOI] [PubMed] [Google Scholar]
- 30. Sinicrope FA, Rego RL, Foster NR, et al. Proapoptotic Bad and Bid protein expression predict survival in stages II and III colon cancers. Clin Cancer Res. 2008;14(13):4128‐4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mann J, Githaka JM, Buckland TW, et al. Non‐canonical BAD activity regulates breast cancer cell and tumor growth via 14‐3‐3 binding and mitochondrial metabolism. Oncogene. 2019;38(18):3325‐3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sinicrope FA, Rego RL, Okumura K, et al. Prognostic impact of bim, puma, and noxa expression in human colon carcinomas. Clin Cancer Res. 2008;14(18):5810‐5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karst AM, Dai DL, Martinka M, Li G. PUMA expression is significantly reduced in human cutaneous melanomas. Oncogene. 2005;24(6):1111‐1116. [DOI] [PubMed] [Google Scholar]
- 34. Liu Z, Yan C, Xiao Y, et al. Expression and inhibitory effects of p53‐upregulated modulator of apoptosis in gallbladder carcinoma. Oncol Lett. 2021;21(3):234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thomalla D, Beckmann L, Grimm C, et al. Deregulation and epigenetic modification of BCL2‐family genes cause resistance to venetoclax in hematologic malignancies. Blood. 2022;140(20):2113‐2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chu CY, Lin CY, Lin CC, et al. Effect of BIM expression on the prognostic value of PD‐L1 in advanced non‐small cell lung cancer patients treated with EGFR‐TKIs. Sci Rep. 2023;13(1):3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karachaliou N, Codony‐Servat J, Teixidó C, et al. BIM and mTOR expression levels predict outcome to erlotinib in EGFR‐mutant non‐small‐cell lung cancer. Sci Rep. 2015;5:17499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maimaiti Y, Dong L, Aili A, Maimaitiaili M, Huang T, Abudureyimu K. Bim may be a poor prognostic biomarker in breast cancer patients especially in those with luminal A tumors. Cancer Biomarkers. 2017;19(4):411‐418. [DOI] [PubMed] [Google Scholar]
- 39. O'Connor L. Bim: a novel member of the Bcl‐2 family that promotes apoptosis. EMBO J. 1998;17(2):384‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marani M, Tenev T, Hancock D, Downward J, Lemoine NR. Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol Cell Biol. 2002;22(11):3577‐3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
Data that support the findings of this study are available from the corresponding author upon reasonable request.


