Abstract
Gene model for the ortholog of Ecdysone-inducible gene L2 ( ImpL2 ) in the May 2017 (Princeton ASM75419v2/DsimGB2) Genome Assembly (GenBank Accession: GCA_000754195.3 ) of Drosophila simulans . This ortholog was characterized as part of a developing dataset to study the evolution of the Insulin/insulin-like growth factor signaling pathway (IIS) across the genus Drosophila using the Genomics Education Partnership gene annotation protocol for Course-based Undergraduate Research Experiences.
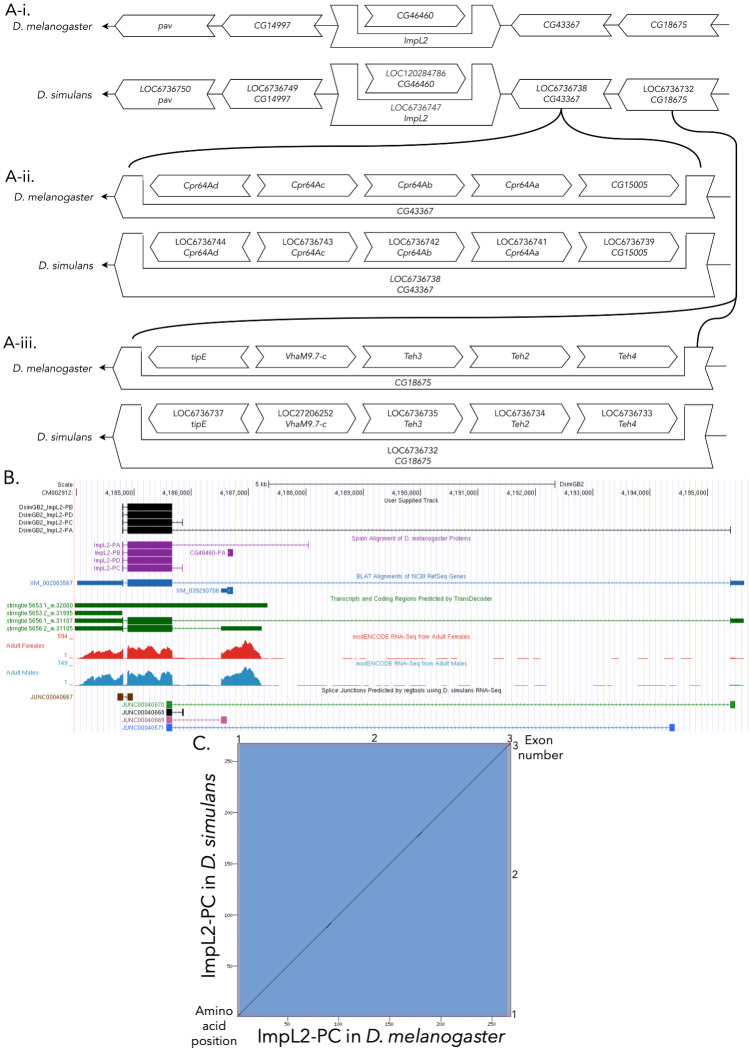
Figure 1. Genomic neighborhood and gene model for ImpL2 in D. simulans .
(A-i) Synteny of genomic neighborhood of ImpL2 in both D. melanogaster and D. simulans . Gene arrows pointing in the same direction as target gene in both D. simulans and D. melanogaster are on the same strand as the target gene; gene arrows pointing in the opposite direction are on the opposite strand. The thin underlying arrows pointing to the left indicate that ImpL2 is on the – strands. White arrows in D. simulans indicate the locus ID and the orthology to the corresponding gene in D. melanogaster . The gene names given in the D. simulans gene arrows indicate the orthologous gene in D. melanogaster , while the locus identifiers are specific to D. simulans . (A-ii) Nested genes within CG43367 , shown in the same format as figure A-i. (A-iii) Nested genes within CG18675 , shown in the same format as figure A-i . (B) Gene Model in UCSC Track Hub (Raney et al. 2014): Image from the Track Hub instance housing the gene model in D. simulans , Spaln of D. melanogaster Proteins (alignment of Refseq proteins from D. melanogaster ), BLAT alignments of NCBI RefSeq Genes (alignment of Refseq genes for D. simulans ), RNA-Seq from adult male and females (alignment of Illumina RNA-Seq reads from D. simulans ), and Transcripts and Coding Regions Predicted by TransDecoder and Splice Junctions Predicted by regtools using D. simulans RNA-Seq (Gravely et al. 2011; SRP006203 ). Splice junctions shown have a minimum read-depth of 9 with 1-9, 10-49, 50-99, 100-499, 500-999 supporting reads in black, blue, green, pink, and brown, respectively. The custom gene model (User Supplied Track) is indicated in black with CDSs depicted with boxes and introns with narrow lines (arrows indicate direction of transcription). (C) Dot Plot of ImpL2-PC in D. melanogaster ( x -axis) vs. the orthologous peptide in D. simulans ( y -axis) . Amino acid number is indicated along the left and bottom; CDS number is indicated along the top and right, and CDSs are also highlighted with alternating colors. Note that the first and last CDSs of Impl2 are very small, so their corresponding bands of color are very thin.
Description
|
This article reports a predicted gene model generated by undergraduate work using a structured gene model annotation protocol defined by the Genomics Education Partnership (GEP; thegep.org ) for Course-based Undergraduate Research Experience (CURE). The following information in this box may be repeated in other articles submitted by participants using the same GEP CURE protocol for annotating Drosophila species orthologs of Drosophila melanogaster genes in the insulin signaling pathway. "In this GEP CURE protocol students use web-based tools to manually annotate genes in non-model Drosophila species based on orthology to genes in the well-annotated model organism fruitfly Drosophila melanogaster . The GEP uses web-based tools to allow undergraduates to participate in course-based research by generating manual annotations of genes in non-model species (Rele et al., 2023) . Computational-based gene predictions in any organism are often improved by careful manual annotation and curation, allowing for more accurate analyses of gene and genome evolution (Mudge and Harrow 2016; Tello-Ruiz et al., 2019) . These models of orthologous genes across species, such as the one presented here, then provide a reliable basis for further evolutionary genomic analyses when made available to the scientific community.” (Myers et al., 2024) . “The particular gene ortholog described here was characterized as part of a developing dataset to study the evolution of the Insulin/insulin-like growth factor signaling pathway (IIS) across the genus Drosophila . The Insulin/insulin-like growth factor signaling pathway (IIS) is a highly conserved signaling pathway in animals and is central to mediating organismal responses to nutrients (Hietakangas and Cohen 2009; Grewal 2009) .” (Myers et al., 2024) . |
The model presented here is the ortholog of ImpL2 in the DsimGB2 assembly of D. simulans ( Drosophila 12 Genomes Consortium et al., 2007; GCA_000754195.3 ) and corresponds to the Gnomon Peptide ID ( XP_002083603.1 ) predicted model in D. simulans ( LOC6736747 ). This gene model is based on RNA-seq data from D. simulans (Gravely et al., 2011; SRP006203 ) and the ImpL2 in D. melanogaster from FB2022_02 (Larkin et al., 2021; Gramates et al., 2022; Jenkins et al., 2022; GCA_000001215.4 ).
Ecdysone-inducible gene L2 was originally known as I nducible m embrane-bound p olysomal transcript 2 (Natzle et al., 1986) , and is also known as Imaginal morphogenesis protein-Late 2, ImpL2 , Imp-L2 , or ImpL-2 (FBgn0001257). ImpL2 is a putative insulin-binding protein, and is thought to antagonize insulin pathway activity (Honegger et al., 2008; Alic et al., 2011) . ImpL2 function is essential in the development of the Drosophila nervous system during early embryogenesis (Garbe et al., 1993; Bader et al., 2013) . Impl2 is the sole fly homolog of mammalian IGFBPs [insulin-like growth factor binding proteins]. In fly tumor models, ImpL2 acts as a secreted wasting factor that contributes to loss of tissue mass by antagonizing system-wide IGF1 signaling (Figueroa-Clarevega and Bilder 2015; Kwon et al., 2015) . It was recently demonstrated that the growth-decreasing effects of ImpL2 in model Drosophila midgut tumors are diminished by upregulation of Wingless (Wg) signaling, although whether this is a direct mechanism remains to be established (Lee et al., 2021) .
D. simulans ( NCBI:txid7240) is part of the melanogaster species group within the subgenus Sophophora of the genus Drosophila (Sturtevant 1939; Bock and Wheeler 1972) . It was first described by Sturtevant (1919). D. simulans is a sibling species to D. melanogaster , thus extensively studied in the context of speciation genetics and evolutionary ecology (Powell 1997) . Historically, D. simulans was a tropical species native to sub-Saharan Africa (Lemeunier et al., 1986) where figs served as a primary host (Lachaise and Tsacas 1983) . However, D. simulans's range has expanded worldwide within the last century as a human commensal using a broad range of rotting fruits as breeding sites (https://www.taxodros.uzh.ch, accessed 1 Feb 2023).
Synteny
ImpL2 occurs on chromosome 3L in D. melanogaster. CG46460 is a dicistronic transcript that is nested within a UTR CDS of ImpL2 . ImpL2 is flanked upstream by pav and CG14997 and downstream by CG43367 and CG18675 . C pr64Ad , C pr64Ac , C pr64Ab , C pr64Aa , and CG15005 are nested within CG43367 . tipE , VhaM9.7-c , Teh3 , Teh2 , Teh4 are nested within CG18675 . We determined that the putative ortholog of ImpL2 is found on scaffold CM002912.1 in D. simulans ( GCA_000754195.3 ) with LOC6736747 ( XP_002083603.1 ) (via tblastn search with an e-value of 0.0 and percent identity of 99.25%). LOC120284786 ( XP_039149640.1 ), which corresponds to CG46460 in D. melanogaster with an e-value of 6e-116 and a percent identity of 100%, as determined by blastp, is nested within the ImpL2 ortholog in D. simulans . It is flanked upstream by LOC6736750 ( XP_016030374.1 ) and LOC6736749 ( XP_002083605.1 ) which correspond to pav and CG14997 , with e-values of 0.0 and 0.0, respectively, and percent identities of 99.55% and 97.81%, respectively, as determined by blastp. It is flanked downstream by LOC6736738 ( XP_016030369.1 ) and LOC6736732 ( XP_002083588.1 ) , which correspond to CG43367 and CG18675 in D. melanogaster with e-values of 0.0 and 0.0, respectively, and percent identities of 99.23% and 96.85%, respectively, as determined by blastp ( Figure 1A -i, Altschul et al., 1990). LOC6736744 ( XP_016030372.1 ), LOC6736743 ( XP_002083599.1 ), LOC6736742 ( XP_002083598.1 ), LOC6736741 ( XP_002083597.1 ), and LOC6736739 ( XP_039149637.1 ), which correspond to C pr64Ad , C pr64Ac , C pr64Ab , C pr64Aa , and CG15005 in D. melanogaster with e-values of 2e-65, 5e-69, 5e-54, 4e-48, and 0.0, respectively, and percent identities of 99.02%, 99.47%, 98.02%, 100%, and 96.94%, respectively, as determined by blastp, are nested within the CG43367 ortholog found at LOC6736738 ( XP_016030369.1 ; Figure 1A -ii, Altschul et al., 1990) . LOC6736737 ( XP_016030363.1 ), LOC27206252 ( XP_016030361.1 ), LOC6736735 ( XP_016030360.1 ), LOC6736734 ( XP_002083590.1 ), and LOC6736733 ( XP_039149638.1 ), which correspond to tipE , VhaM9.7-c , Teh3 , Teh2 , Teh4 in D. melanogaster with e-values of 0.0, 3e-45, 0.0, 0.0, and 0.0, respectively, and percent identities of 99.12%, 96.43%, 100%, 99.65%, and 98.66%, respectively, as determined by blastp, are nested within the CG18675 ortholog found at LOC6736732 ( XP_002083588.1 ; 1A-iii, Altschul et al., 1990). We believe this is the correct ortholog assignment for ImpL2 in D. simulans because local synteny is completely conserved and because all of the blastp results used to determine the orthology of neighboring genes are very high-quality matches.
Protein Model
ImpL2 in D. simulans has three unique protein coding isoforms ( Figure 1B ). ImpL2-PA and ImpL2-PC contain three CDSs, the first of which is different and the other two are the same. mRNAs ImpL2-RB and ImpL2-RD , which differ in their 3' UTRs, contain the same two CDSs. Relative to the ortholog in D. melanogaster, all isoforms are present in D. simulans and have the same respective number of CDSs . The sequence of ImpL2-PC in D. simulans has 99.3% identity similarity with ImpL2-PC in D. melanogaster as determined by blastp ( Figure 1C ). Note that the annotation for the first CDS of ImpL2-PA (Flybase ID: 1_294_0) d oes not align with the Spaln of D. melanogaster Proteins track prediction for the first CDS of the isoform (Flybase IDs from FB2022_03). The placement of this CDS in our model was determined based multiple lines of evidence including the BLAT alignments of NCBI RefSeq Genes, RNA-Seq from adult male and females, Transcripts and Coding Regions Predicted by TransDecoder, and Splice Junctions Predicted by regtools using D. simulans RNA-Seq, all of which support the CDS we selected. Additionally, the peptide sequence of this first CDS in D. melanogaster is MQ, which is identical to the sequence of the chosen orthologous placement in D. simulans , whereas the Spaln of D. melanogaster Proteins track prediction indicates a CDS with peptide sequence MS. This combined data provides evidence to support the determined location of the first CDS of ImpL2-PA (Flybase ID: 1_294_0) in D. simulans . The coordinates of the curated gene models can be found in NCBI at GenBank/BankIt using the accessions BK063005 , BK063006 , BK063007 , and BK063008 . These data are also available in Extended Data files below, which are archived in CaltechData.
Methods
Detailed methods including algorithms, database versions, and citations for the complete annotation process can be found in Rele et al. (2023). Briefly, students use the GEP instance of the UCSC Genome Browser v.435 ( https://gander.wustl.edu ; Kent WJ et al., 2002; Navarro Gonzalez et al., 2021) to examine the genomic neighborhood of their reference IIS gene in the D. melanogaster genome assembly (Aug. 2014; BDGP Release 6 + ISO1 MT/dm6). Students then retrieve the protein sequence for the D. melanogaster target gene for a given isoform and run it using tblastn against their target Drosophila species genome assembly ( Drosophila simulans ( GCA_000754195.3 - Graveley et al., 2010)) on the NCBI BLAST server ( https://blast.ncbi.nlm.nih.gov/Blast.cgi , Altschul et al., 1990) to identify potential orthologs. To validate the potential ortholog, students compare the local genomic neighborhood of their potential ortholog with the genomic neighborhood of their reference gene in D. melanogaster . This local synteny analysis includes at minimum the two upstream and downstream genes relative to their putative ortholog. They also explore other sets of genomic evidence using multiple alignment tracks in the Genome Browser, including BLAT alignments of RefSeq Genes, Spaln alignment of D. melanogaster proteins, multiple gene prediction tracks (e.g., GeMoMa, Geneid, Augustus), and modENCODE RNA-Seq from the target species. Genomic structure information (e.g., CDSs, CDS number and boundaries, number of isoforms) for the D. melanogaster reference gene is retrieved through the Gene Record Finder ( https://gander.wustl.edu/~wilson/dmelgenerecord/index.html ; Rele et al ., 2023). Approximate splice sites within the target gene are determined using tblastn using the CDSs from the D. melanogaste r reference gene. Coordinates of CDSs are then refined by examining aligned modENCODE RNA-Seq data, and by applying paradigms of molecular biology such as identifying canonical splice site sequences and ensuring the maintenance of an open reading frame across hypothesized splice sites. Students then confirm the biological validity of their target gene model using the Gene Model Checker ( https://gander.wustl.edu/~wilson/dmelgenerecord/index.html ; Rele et al., 2023), which compares the structure and translated sequence from their hypothesized target gene model against the D. melanogaster reference gene model. At least two independent models for each gene are generated by students under mentorship of their faculty course instructors. These models are then reconciled by a third independent researcher mentored by the project leaders to produce a final model like the one presented here. Note: comparison of 5' and 3' UTR sequence information is not included in this GEP CURE protocol.
Extended Data
Description: Model Data (FNA, FAA, GTF). Resource Type: Model. DOI: 10.22002/ffnsa-1r156
Acknowledgments
Acknowledgments
We would like to thank Wilson Leung who created and maintain the GEP technological infrastructure. We would also like to thank Sarah P. Crocker-Buta for helping us submit the model to the TPA database, and the constructive comments from the review panel. Thank you to FlyBase for providing the definitive database for Drosophila melanogaster gene models. FlyBase is supported by grants: NHGRI U41HG000739 and U24HG010859, UK Medical Research Council MR/W024233/1, NSF 2035515 and 2039324, BBSRC BB/T014008/1, and Wellcome Trust PLM13398.
Funding Statement
This material is based upon work supported by the National Science Foundation under Grant No. IUSE-1915544 to LKR and the National Institute of General Medical Sciences of the National Institutes of Health Award R25GM130517 to LKR. The Genomics Education Partnership is fully financed by Federal moneys. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Alic N, Hoddinott MP, Vinti G, Partridge L. Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging Cell. 2011 Feb 1;10(1):137–147. doi: 10.1111/j.1474-9726.2010.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bader R, Sarraf-Zadeh L, Peters M, Moderau N, Stocker H, Köhler K, Pankratz MJ, Hafen E. The IGFBP7 homolog Imp-L2 promotes insulin signaling in distinct neurons of the Drosophila brain. J Cell Sci. 2013 Apr 16;126(Pt 12):2571–2576. doi: 10.1242/jcs.120261. [DOI] [PubMed] [Google Scholar]
- Bock IR, Wheeler MR. 1972. The Drosophila melanogaster species group. Univ. Texas Publs Stud. Genet. 7(7213): 1-102. FBrf0024428
- Figueroa-Clarevega A, Bilder D. Malignant Drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Dev Cell. 2015 Apr 6;33(1):47–55. doi: 10.1016/j.devcel.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe JC, Yang E, Fristrom JW. IMP-L2: an essential secreted immunoglobulin family member implicated in neural and ectodermal development in Drosophila. Development. 1993 Dec 1;119(4):1237–1250. doi: 10.1242/dev.119.4.1237. [DOI] [PubMed] [Google Scholar]
- Gramates LS, Agapite J, Attrill H, Calvi BR, Crosby MA, Dos Santos G, Goodman JL, Goutte-Gattat D, Jenkins VK, Kaufman T, Larkin A, Matthews BB, Millburn G, Strelets VB, the FlyBase Consortium. Fly Base: a guided tour of highlighted features. Genetics. 2022 Apr 4;220(4) doi: 10.1093/genetics/iyac035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster. Nature. 2010 Dec 22;471(7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SS. Insulin/TOR signaling in growth and homeostasis: a view from the fly world. Int J Biochem Cell Biol. 2008 Oct 18;41(5):1006–1010. doi: 10.1016/j.biocel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu Rev Genet. 2009;43:389–410. doi: 10.1146/annurev-genet-102108-134815. [DOI] [PubMed] [Google Scholar]
- Honegger B, Galic M, Köhler K, Wittwer F, Brogiolo W, Hafen E, Stocker H. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J Biol. 2008 Apr 15;7(3):10–10. doi: 10.1186/jbiol72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Huang ZL, Yang JH, Xu YH, Sun JS, Zheng Q, Wei C, Song W, Yuan Z. Pancreatic cancer cell-derived IGFBP-3 contributes to muscle wasting. J Exp Clin Cancer Res. 2016 Mar 15;35:46–46. doi: 10.1186/s13046-016-0317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins VK, Larkin A, Thurmond J, FlyBase Consortium Using FlyBase: A Database of Drosophila Genes and Genetics. Methods Mol Biol. 2022;2540:1–34. doi: 10.1007/978-1-0716-2541-5_1. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002 Jun 1;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y, Song W, Droujinine IA, Hu Y, Asara JM, Perrimon N. Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev Cell. 2015 Apr 6;33(1):36–46. doi: 10.1016/j.devcel.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, Tsacas L. 1983. Breeding-sites of tropical African Drosophilids. Ashburner, Carson, Thompson, 1981-1986 3d: 221-332. FBrf0038884
- Larkin A, Marygold SJ, Antonazzo G, Attrill H, Dos Santos G, Garapati PV, Goodman JL, Gramates LS, Millburn G, Strelets VB, Tabone CJ, Thurmond J, FlyBase Consortium. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021 Jan 8;49(D1):D899–D907. doi: 10.1093/nar/gkaa1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Ng KG, Dombek KM, Eom DS, Kwon YV. Tumors overcome the action of the wasting factor ImpL2 by locally elevating Wnt/Wingless. Proc Natl Acad Sci U S A. 2021 Jun 8;118(23) doi: 10.1073/pnas.2020120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeunier F, David J, Tsacas L, Ashburner M. 1986. The melanogaster species group. Ashburner, Carson, Thompson, 1981-1986 e: 147-256. FBrf0043749.
- Mudge JM, Harrow J. The state of play in higher eukaryote gene annotation. Nat Rev Genet. 2016 Oct 24;17(12):758–772. doi: 10.1038/nrg.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A, Hoffmann A, Natysin M, Arsham AM, Stamm J, Thompson JS, Rele CP. 2024. Gene model for the ortholog of Myc in Drosophila ananassae . microPublication Biology, in press [DOI] [PMC free article] [PubMed]
- Natzle JE, Hammonds AS, Fristrom JW. Isolation of genes active during hormone-induced morphogenesis in Drosophila imaginal discs. J Biol Chem. 1986 Apr 25;261(12):5575–5583. [PubMed] [Google Scholar]
- Navarro Gonzalez J, Zweig AS, Speir ML, Schmelter D, Rosenbloom KR, Raney BJ, Powell CC, Nassar LR, Maulding ND, Lee CM, Lee BT, Hinrichs AS, Fyfe AC, Fernandes JD, Diekhans M, Clawson H, Casper J, Benet-Pagès A, Barber GP, Haussler D, Kuhn RM, Haeussler M, Kent WJ. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021 Jan 8;49(D1):D1046–D1057. doi: 10.1093/nar/gkaa1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR. 1997. Progress and prospects in evolutionary biology: the Drosophila model 1st edition. Oxford University Press. ISBN:9780195076912
- Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, Wang T, Nguyen N, Paten B, Zweig AS, Karolchik D, Kent WJ. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2013 Nov 13;30(7):1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rele Chinmay P., Sandlin Katie M., Leung Wilson, Reed Laura K. Manual annotation of Drosophila genes: a Genomics Education Partnership protocol. F1000Research. 2022 Dec 23;11:1579–1579. doi: 10.12688/f1000research.126839.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. 1919. A new species closely resembling to Drosophila melanogaster. Psyche 26: 488–500. FBrf0000977
- Sturtevant AH. On the Subdivision of the Genus Drosophila. Proc Natl Acad Sci U S A. 1939 Mar 1;25(3):137–141. doi: 10.1073/pnas.25.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello-Ruiz MK, Marco CF, Hsu FM, Khangura RS, Qiao P, Sapkota S, Stitzer MC, Wasikowski R, Wu H, Zhan J, Chougule K, Barone LC, Ghiban C, Muna D, Olson AC, Wang L, Ware D, Micklos DA. Double triage to identify poorly annotated genes in maize: The missing link in community curation. PLoS One. 2019 Oct 28;14(10):e0224086–e0224086. doi: 10.1371/journal.pone.0224086. [DOI] [PMC free article] [PubMed] [Google Scholar]