Abstract
Objectives: To investigate the role of the long non-coding RNA VCAN antisense RNA 1 (VCAN-AS1) in gastric cancer (GC) progression and elucidate its underlying molecular mechanisms, focusing on its interaction with ELAV-like RNA-binding protein 1 (ELAVL1, known as HuR) and its effect on F11 receptor (F11R) expression. Methods: VCAN-AS1 expression levels in GC tissues and cell lines were measured using real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR). In vitro and in vivo experiments, including cell counting kit-8 (CCK-8) assay, colony formation assay. Transwell migration assay, and a xenograft tumor model, were performed to evaluate VCAN-AS1’s function in GC progression. RNA immunoprecipitation (RIP) and RNA pull-down assays were used to confirm the interaction between VCAN-AS1 and HuR. Results: VCAN-AS1 expression was significantly elevated in GC tissues and cell lines, with higher expression levels linked to poorer prognosis in GC patients. Functional assays demonstrated that VCAN-AS1 knockdown suppressed GC cell proliferation and migration. RIP and RNA pull-down experiments confirmed a specific interaction between VCAN-AS1 and HuR. Additionally, VCAN-AS1 regulated F11R expression in a HuR-dependent manner, and rescue experiments confirmed that F11R contributed to VCAN-AS1’s oncogenic role in GC. Conclusions: These findings suggest that VCAN-AS1 facilitates GC progression through the HuR/F11R pathway, offering new insights into GC pathogenesis and identifying VCAN-AS1 as a potential therapeutic target for GC treatment.
Keywords: VCAN-AS1, gastric cancer, HuR, F11R
Introduction
Gastric cancer (GC) is one of the most prevalent and deadly cancers worldwide, ranking as the fifth most common cancer and the third leading cause of cancer-related deaths [1,2]. Despite progress in diagnostic and therapeutic strategies, the prognosis for GC remains poor due to late-stage diagnosis, frequent metastasis, and high recurrence rates [3-5]. Understanding the molecular mechanisms underlying GC progression is crucial for developing more effective diagnostic and therapeutic approaches. Recent studies have highlighted the significant roles of non-coding RNAs in various biological processes and their potential as novel biomarkers and therapeutic targets in cancer [6-8].
Long non-coding RNAs (lncRNAs), which are non-coding RNAs longer than 200 nucleotides, have emerged as key regulators of gene expression at transcriptional, post-transcriptional, and epigenetic levels [9-12]. They are involved in diverse physiological and pathological processes, including cell proliferation, differentiation, and apoptosis [13,14]. In cancer, lncRNAs can act as oncogenes or tumor suppressors, influencing tumor growth, invasion, metastasis, and resistance to therapy [15-17]. Given their versatile roles, lncRNAs represent a promising research area for understanding cancer biology and developing new therapeutic strategies.
VCAN antisense RNA 1 (VCAN-AS1) is a lncRNA that has recently gained attention for its potential role in cancer progression. Previous studies have reported upregulation of VCAN-AS1 in various cancers, including breast cancer and GC, where it is associated with poor prognosis [18-21]. VCAN-AS1 is believed to regulate cancer progression through different mechanisms, such as interacting with proteins or functioning as a competitive endogenous RNA (ceRNA) [18,19]. For example, it has been shown to promote breast cancer progression by modulating the miR-106a-5p/signal transducer and activator of transcription 3 (STAT3) signaling pathway [19]. Emerging evidence also indicates that VCAN-AS1 is involved in other diseases, such as diabetic kidney disease, where it plays a central role in a lncRNA-associated ceRNA network [22].
However, the role and mechanisms of VCAN-AS1 in GC remain largely unexplored. This study aims to investigate the role of VCAN-AS1 in GC progression and clarify the molecular mechanisms involved. Our findings indicate that VCAN-AS1 promotes GC progression by interacting with Human antigen R (HuR, also known as ELAVL1) and enhancing the stability of F11 receptor (F11R) mRNA. These insights into VCAN-AS1’s function in GC may help identify new therapeutic targets for improving the management of this deadly disease.
Materials and methods
Bioinformatic analysis
Expression levels of VCAN-AS1 in gastric cancer (GC) tissues and adjacent normal tissues were obtained from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases. Survival analysis, including overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI), was performed using the Kaplan-Meier method and log-rank test based on data from TCGA database [23,24]. The interaction between VCAN-AS1 and HuR was predicted using the RNA-Protein Interaction Prediction (RPISeq) tool as previously reported [25].
Human tissues
GC tissues and adjacent normal tissues were collected from patients who underwent surgical resection at Suzhou Municipal Hospital between March 2023 and June 2024, with written informed consent from all participants. Tissue samples were immediately snap-frozen in liquid nitrogen and stored in liquid nitrogen until RNA extraction. The study was approved by the Ethics Committee of Suzhou Municipal Hospital.
Cell culture and transfection
Human GC cell lines AGS, HGC27, Hs746T, and NCI-N87, along with the normal gastric epithelial cell line GES-1, were obtained from the Chinese Academy of Cell Collection (Shanghai, China). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37°C in a humidified atmosphere containing 5% CO2. For transfection, siRNAs targeting VCAN-AS1 and HuR, a negative control (NC) siRNA, and sh-VCAN-AS1 vectors were purchased from GenePharma (Shanghai, China). The pcDNA3.1-F11R overexpression plasmid (OE-F11R) and empty vector were obtained from Sangon (Shanghai, China). Cells were transfected with siRNAs or plasmids using Lipofectamine 2000 (Invitrogen, CA, USA) according to the manufacturer’s instructions. Cells were harvested for subsequent experiments 48 hours post-transfection. The siRNA sequences were as follows: si-VCAN-AS1 #1, 5’-AUGUUUUCCUUGGCUUUUGGA-3’; si-VCAN-AS1 #2, 5’-UGGCUUUUGGAUUAAUACAAU-3’. Sequences targeting HuR and the NC siRNA were used as previously described [25].
RNA extraction and real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples and cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. The quality and concentration of the RNA were assessed with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). cDNA synthesis was performed using the PrimeScript RT reagent kit (Vazyme, Nanjing, China) with gDNA Eraser to remove genomic DNA contamination. RT-qPCR was conducted using SYBR Green Master Mix (Vazyme) on an Applied Biosystems 7500 system. The relative expression levels of VCAN-AS1 and F11R were calculated using the 2^-ΔΔCt method, with 18 s rRNA as the internal control. The primer sequences were listed as follows: VCAN-AS1, forward 5’-AATGCCACATCACAGCTGAC-3’ and reverse 5’-AGAGCCACCAACATACTTGACA-3’. The primers for 18s rRNA and F11R amplification were reported previously [25].
Cell counting kit-8 (CCK-8) assay, colony formation assay, and Transwell assay
Cell viability was assessed with the CCK-8 assay (Beyotime, Shanghai, China) as previously described [26,27]. Briefly, transfected cells were seeded in 96-well plates at a density of 2×103 cells per well and incubated for 24, 48, 72, and 96 hours. CCK-8 reagent was added, and the absorbance at 450 nm was measured using a microplate reader (Bio-Rad). For the colony formation assay, transfected cells were seeded in 6-well plates at 500 cells per well and cultured for two weeks. Colonies were then fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and counted under a microscope. Cell migration was evaluated using Transwell chambers (Corning) as previously reported [28,29]. Cells (1×105) were placed in the upper chamber with serum-free medium, while the lower chamber contained medium with 10% FBS. After 48 hours, cells on the lower membrane surface were fixed, stained, and counted under a microscope (Axioskop 2 plus, Zeiss, Germany).
Xenograft model
The tumorigenic potential of VCAN-AS1 knockdown was evaluated in vivo using a xenograft mouse model. Four-week-old female BALB/c nude mice were purchased from Vital River Laboratory (Beijing, China) and housed under specific pathogen-free conditions. The si-VCAN-AS1-1 sequence was cloned into an LV-2N/Puro lentiviral vector (GenePharma). Lentiviral vectors and packaging plasmids were co-transfected into HEK-293T cells (ATCC, USA), and the collected viruses were used to infect AGS cells, which were then selected for puromycin resistance for two weeks. AGS cells stably expressing sh-VCAN-AS1 or control shRNA were subcutaneously injected on both flanks of each mouse (1×107 cells per mouse). Tumor growth was measured every three days, and tumor volume was calculated as V = 0.5 × length × width2. Mice were euthanized 12 days after injection using carbon dioxide exposure followed by cervical dislocation, and tumors were excised and weighed. All animal experiments were approved by the Institutional Animal Care and Use Committee of Nanjing Medical University.
RNA pull-down
Biotin-labeled RNA probes targeting VCAN-AS1 and a control probe were synthesized by Ribobio (Guangzhou, China). RNA pull-down assays were carried out using the Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific) as previously described [30]. Briefly, biotinylated RNA probes were incubated with cell lysates from AGS and HGC27 cells at 4°C for 4 hours with gentle rotation. RNA-protein complexes were then captured by adding streptavidin magnetic beads to the mixture, followed by incubation at 4°C for 2 hours with rotation. The bound proteins were eluted, separated by SDS-PAGE, and identified through Western blotting.
RNA immunoprecipitation (RIP)
RIP assays were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) as previously described [31,32]. AGS and HGC27 cell lysates were incubated with HuR antibodies (Proteintech, Chicago, IL, USA) or control IgG at 4°C overnight with rotation. RNA-protein complexes were captured using Protein A/G beads, followed by 2 hours of incubation at 4°C with rotation. Co-precipitated RNAs were extracted with TRIzol reagent, and the presence of VCAN-AS1 and F11R was detected by RT-qPCR.
Western blotting
Total protein was extracted from cells using RIPA lysis buffer (Beyotime) supplemented with protease inhibitors, as described previously [33,34]. Protein concentrations were measured with the BCA Protein Assay Kit (Beyotime). Equal amounts of protein were separated via SDS-PAGE and transferred onto PVDF membranes (Millipore). Membranes were blocked with 5% non-fat milk in TBST and incubated overnight at 4°C with primary antibodies against HuR (1:1000, Proteintech) and Tubulin (1:5000, Beyotime). After washing, membranes were incubated with HRP-conjugated secondary antibodies (1:1000, Thermo Fisher Scientific), and the immunoreactive bands were visualized using the ECL detection system (Tanon, Shanghai, China).
Statistical analysis
Data were analyzed with GraphPad Prism software (version 8.0). Quantitative results are presented as mean ± standard deviation (SD) from at least three independent experiments. Differences between groups were assessed using Student’s t-test or one-way ANOVA followed by Tukey’s post-hoc test. A p-value < 0.05 was considered statistically significant.
Results
VCAN-AS1 is upregulated in GC tissues and cells, and associated with poor prognosis
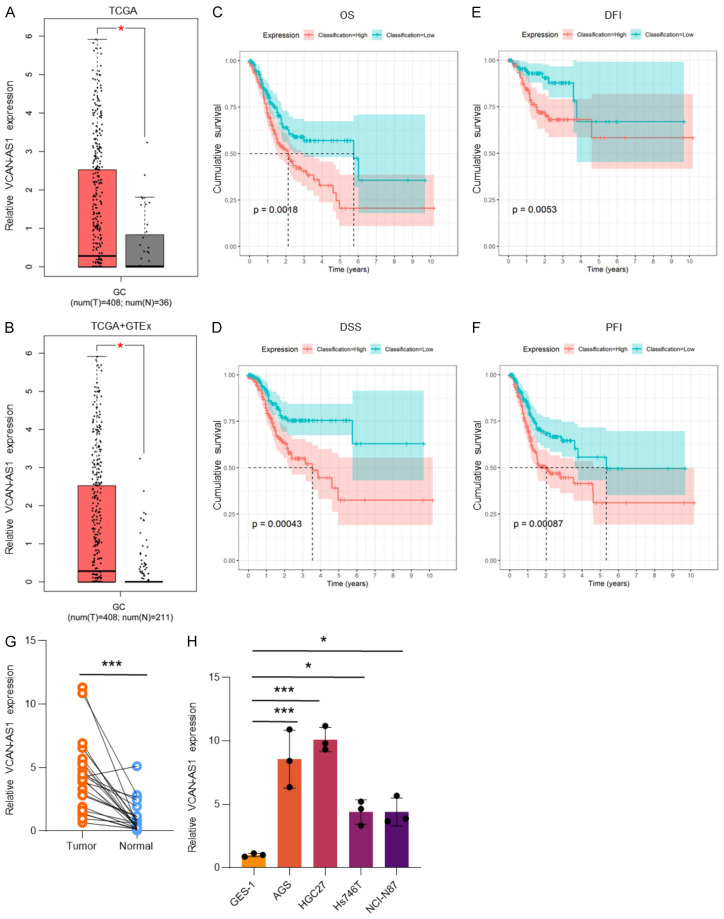
Analysis of the TCGA and GTEx databases showed that VCAN-AS1 was significantly upregulated in GC tissues compared to normal tissues (Figure 1A and 1B). Higher expression levels of VCAN-AS1 in GC tissues were correlated with worse patient outcomes, including OS (Figure 1C), DSS (Figure 1D), DFI (Figure 1E), and PFI (Figure 1F). Further validation by RT-qPCR confirmed elevated VCAN-AS1 expression in both GC tissues (Figure 1G) and cell lines (Figure 1H). Among the cell lines tested, AGS and HGC27 showed the highest VCAN-AS1 expression and were thus selected for subsequent experiments.
Figure 1.
VCAN-AS1 is overexpressed in GC tissues and cell lines, and associated with poor prognosis. (A, B) Relative expression of VCAN-AS1 in TCGA database (A) and TCGA+GTEx databases (B). (C-F) Kaplan-Meier analysis of overall survival (OS, C), disease-specific survival (DSS, D), disease-free interval (DFI, E), and progression-free interval (PFI, F) in GC patients. (G) Relative expression of VCAN-AS1 in 24 paired GC tissues and adjacent tissues. (H) Relative expression of VCAN-AS1 in normal gastric epithelial cell line GES-1 and GC cell lines AGS, HGC27, Hs746T, and NCI-N87. *P < 0.05, ***P < 0.001.
Knockdown of VCAN-AS1 inhibits GC cell proliferation and migration in vitro
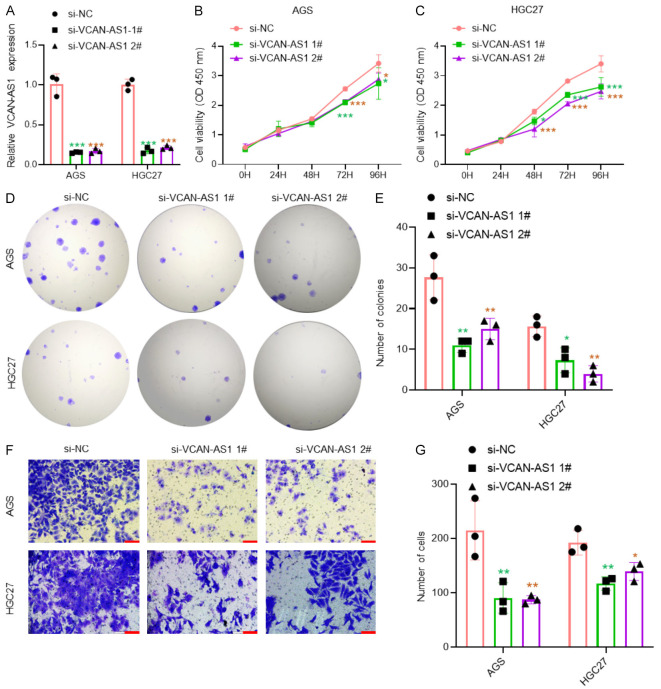
To investigate the role of VCAN-AS1 in GC, two specific siRNAs were designed to knock down VCAN-AS1 expression in AGS and HGC27 cells, with RT-qPCR confirming effective knockdown (Figure 2A). Functional assays, including CCK-8, colony formation, and Transwell assays, demonstrated that silencing VCAN-AS1 significantly reduced the proliferation (Figure 2B-E) and migration (Figure 2F, 2G) of GC cells.
Figure 2.
Silencing VCAN-AS1 inhibits proliferation and migration in GC cells. A. RT-qPCR analysis of VCAN-AS1 expression in AGS and HGC27 cells transfected with si-NC and si-VCAN-AS1 for 48 hours. B, C. CCK-8 assays in AGS and HGC27 cells transfected with si-NC and si-VCAN-AS1 for 48 hours. n=6 per group. D, E. Colony formation assays in AGS and HGC27 cells transfected with si-NC and si-VCAN-AS1 for 48 hours. n=3 per group. F, G. Transwell assays in AGS and HGC27 cells transfected with si-NC and si-VCAN-AS1 for 48 hours. n=3 per group. Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
VCAN-AS1 knockdown suppresses tumor growth in vivo
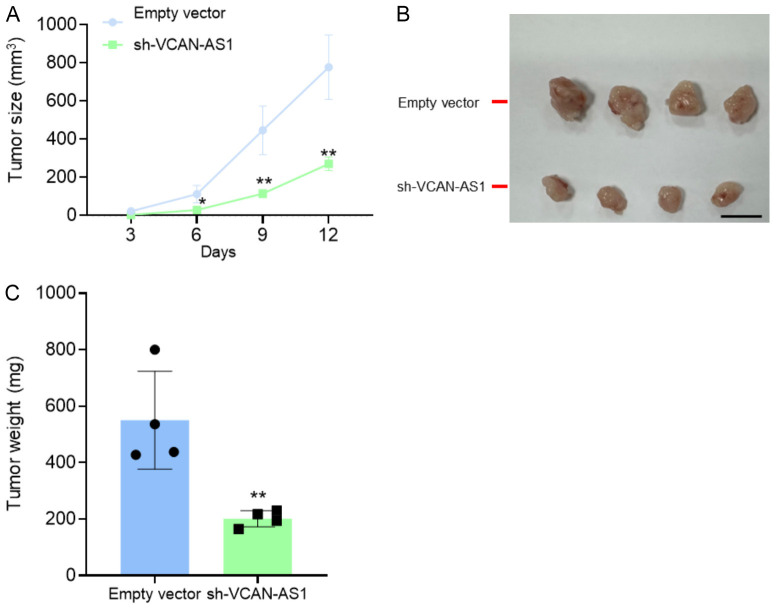
The effect of VCAN-AS1 knockdown on tumor growth was further evaluated in a subcutaneous xenograft model using nude mice. AGS cells stably transfected with sh-VCAN-AS1 or an empty vector were injected into the mice. Tumors derived from VCAN-AS1-knockdown cells exhibited significantly slower growth and lower tumor weight compared to the control group (Figure 3A-C). These findings indicate that silencing VCAN-AS1 inhibits GC cell growth in vivo.
Figure 3.
Silencing VCAN-AS1 inhibits tumor growth in vivo. A total of 4 nude mice were injected subcutaneously with stably transfected AGS cells with empty vector or sh-VCAN-AS1. A. Tumor volumes were assayed every 3 days. B, C. Tumors were collected and weighted. n=4 per group. Scale bar: 1 cm. *P < 0.05, **P < 0.01.
VCAN-AS1 interacts with HuR and regulates F11R expression
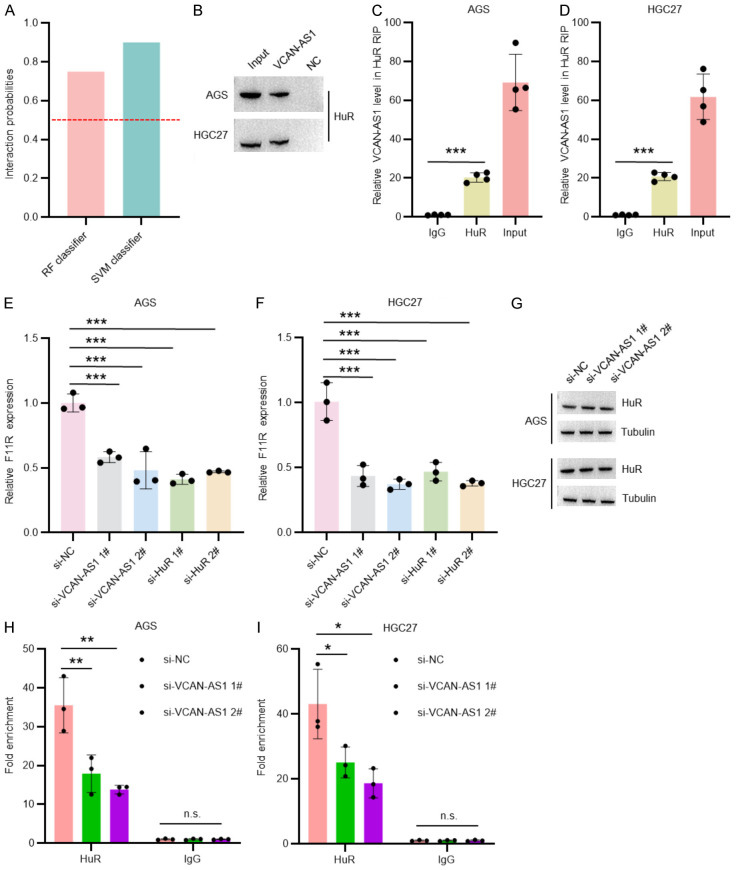
Using RPISeq software, we predicted a potential interaction between VCAN-AS1 and the RNA-binding protein HuR (Figure 4A). This interaction was confirmed by RNA pull-down (Figure 4B) and RNA immunoprecipitation (RIP) assays (Figure 4C and 4D). Previous studies have shown that HuR binds to the 3’-UTR of F11R mRNA, stabilizing it and promoting its oncogenic role in GC [25,35]. Our results revealed that knockdown of either VCAN-AS1 or HuR significantly reduced F11R mRNA expression in GC cells (Figure 4E and 4F). Notably, silencing VCAN-AS1 did not alter HuR protein levels but impaired HuR’s ability to bind F11R mRNA (Figure 4G-I). These findings suggest that VCAN-AS1 participates in the HuR-F11R regulatory axis involved in GC progression.
Figure 4.
VCAN-AS1 interacts with HuR and affects F11R mRNA expression. A. VCAN-AS1-HuR interactions were predicted by RPISeq. B. Western blot analysis of RNA pull-down products based on an anti-HuR antibody in AGS and HGC27 cells. C, D. RIP assays of VCAN-AS1 binding to HuR in AGS and HGC27 cells. n=3 per group. E, F. RT-qPCR analysis of F11R expression in AGS and HGC27 cells transfected with si-NC, si-VCAN-AS1, and si-HuR for 48 hours. n=3 per group. G. Western blot analysis of HuR expression in AGS and HGC27 cells transfected with si-NC and si-VCAN-AS1 for 48 hours. H, I. RIP assays of F11R binding to HuR in AGS and HGC27 cells transfected with si-NC and si-VCAN-AS1 for 48 hours. n=3 per group. *P < 0.05, **P < 0.01, ***P < 0.001.
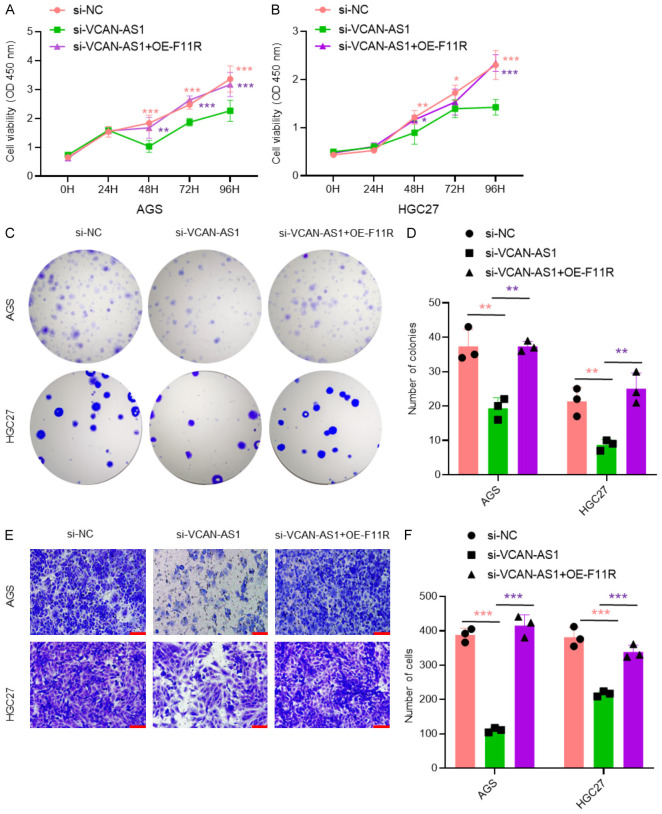
F11R overexpression rescues the effects of VCAN-AS1 knockdown on GC cell proliferation and migration
To further clarify the mechanism by which VCAN-AS1 promotes GC progression, we performed rescue experiments by overexpressing F11R in VCAN-AS1-knockdown cells. The results showed that F11R overexpression effectively restored the reduced proliferation (Figure 5A-D) and migration (Figure 5E and 5F) caused by VCAN-AS1 knockdown, indicating that VCAN-AS1 promotes GC cell proliferation and migration through F11R regulation.
Figure 5.
F11R is a major downstream of VCAN-AS1/HuR axis. A, B. CCK-8 assays in AGS and HGC27 cells transfected with si-NC, si-VCAN-AS1, and si-VCAN-AS1+OE-F11R for 48 hours. n=6 per group. C, D. Colony formation assays in AGS and HGC27 cells transfected with si-NC, si-VCAN-AS1, and si-VCAN-AS1+OE-F11R for 48 hours. n=3 per group. E, F. Transwell assays in AGS and HGC27 cells transfected with si-NC, si-VCAN-AS1, and si-VCAN-AS1+OE-F11R for 48 hours. n=3 per group. Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
This study investigated the role of the long non-coding RNA (lncRNA) VCAN-AS1 in GC and uncovered its involvement in GC progression. Our results demonstrate that VCAN-AS1 is significantly upregulated in GC tissues and cell lines, correlating with poor prognosis. Both in vitro and in vivo knockdown of VCAN-AS1 suppressed tumor growth. Mechanistically, we identified that VCAN-AS1 interacts with HuR, which in turn enhances the stability of F11R mRNA, promoting GC progression. These findings provide new insights into the molecular pathways of GC involving lncRNAs, suggesting the VCAN-AS1-HuR-F11R axis as a potential therapeutic target for GC.
Furthermore, identifying VCAN-AS1 as a critical regulator of GC progression may offer a novel biomarker for predicting GC prognosis. Unlike previous studies focusing primarily on protein-coding genes in GC biology [36,37], our work expands the understanding of GC carcinogenesis, emphasizing the significant role of lncRNAs in cancer regulation [38-40]. A common mechanism through which lncRNAs function as competing endogenous RNAs (ceRNAs), binding to microRNAs (miRNAs) and preventing them from interacting with their target mRNAs. For example, lncRNA PCED1B-AS1 has been shown to promote GC metastasis and epithelial-mesenchymal transition by sponging miR-3681-3p, leading to the upregulation of MAP2K7 [41]. Similarly, LINC01140, in conjunction with miR-140-5p and FGF9, forms a lncRNA-miRNA-mRNA axis that promotes GC aggressiveness [42].
In addition to acting as ceRNAs, accumulating evidence indicates that many lncRNAs exert their oncogenic roles by directly binding to proteins. For instance, lncRNA HOXA11-AS has been shown to facilitate GC progression by scaffolding the epigenetic factors EZH2, LSD1, and DNMT1 [43]. Similarly, lncRNA NRSN2-AS1 plays a crucial role in ovarian cancer malignancy through interaction with PTK2, thereby activating β-catenin signaling [30]. In this study, we found that VCAN-AS1 directly interacts with the RNA-binding protein HuR, which enhances the stability of F11R mRNA, thereby promoting its oncogenic function in GC. This novel mechanism illustrates the diverse functions of lncRNAs and their complex roles in cancer biology.
HuR is a well-established RNA-binding protein known for its function in stabilizing target mRNAs by binding to AU-rich elements in their 3’-untranslated regions (3’-UTRs) [44,45]. Previous research has shown that HuR stabilizes TIMM44 mRNA, promoting ovarian cancer proliferation [46], and enhances the stability of CDK3 mRNA in breast cancer, contributing to cancer cell growth [47]. In GC, HuR has been reported to interact with lincRNAs such as LINC00707 and DEPDC1-AS1, leading to the stabilization of F11R mRNA and driving GC progression [25,35]. Our findings demonstrate that HuR interacts with VCAN-AS1, which in turn stabilizes F11R mRNA, uncovering a new layer of regulatory control in GC progression.
In conclusion, our findings emphasize the significance of VCAN-AS1 in GC progression and suggest new avenues for developing targeted therapies aimed at the VCAN-AS1-HuR-F11R axis.
Acknowledgements
We would like to thank Walgenron Bio-Pharm Co., Ltd. (Shenzhen, China) for language editing. This work was supported by the Suzhou Youth Science and Technology Project (KJXW2022025 to W.X.), the Suzhou Key Medical Disease Project (LCZX202111 to X.G.), and the Suzhou Medical Key Discipline Gastrointestinal Surgery (SZXK202109 to X.G.).
Disclosure of conflict of interest
None.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Xuan J, Li F, Li J, Gong C, Li J, Mo Z, Jin Q. The role of senescence genes in the treatment, prognosis, and tumor microenvironment of gastric cancer. Am J Transl Res. 2023;15:6926–6938. [PMC free article] [PubMed] [Google Scholar]
- 3.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 4.Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020;21:4012. doi: 10.3390/ijms21114012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocken C. Predictive biomarkers in gastric cancer. J Cancer Res Clin Oncol. 2023;149:467–481. doi: 10.1007/s00432-022-04408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Ding W, Yu W, Zhang Y, Ao X, Wang J. Long non-coding RNAs: biogenesis, functions, and clinical significance in gastric cancer. Mol Ther Oncolytics. 2021;23:458–476. doi: 10.1016/j.omto.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma S, Zhou M, Xu Y, Gu X, Zou M, Abudushalamu G, Yao Y, Fan X, Wu G. Clinical application and detection techniques of liquid biopsy in gastric cancer. Mol Cancer. 2023;22:7. doi: 10.1186/s12943-023-01715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan H, Zhang S, Zhang J, Zhu L, Chen Y, Yang H, Chen Y, An Y, Liu B. Long non-coding RNAs in gastric cancer: new emerging biological functions and therapeutic implications. Theranostics. 2020;10:8880–8902. doi: 10.7150/thno.47548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan JH, Liao KQ, Yao L, Chen JL, Xiong LF, Tao XZ. LncRNA AL645608.3 mediates malignant progression of acute myeloid leukemia. Am J Transl Res. 2024;16:342–355. doi: 10.62347/TXKA6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 12.Crespi M. Long non-coding RNAs reveal new regulatory mechanisms controlling gene expression. C R Biol. 2023;345:15–39. doi: 10.5802/crbiol.106. [DOI] [PubMed] [Google Scholar]
- 13.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 14.Zong Y, Wang X, Cui B, Xiong X, Wu A, Lin C, Zhang Y. Decoding the regulatory roles of non-coding RNAs in cellular metabolism and disease. Mol Ther. 2023;31:1562–1576. doi: 10.1016/j.ymthe.2023.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65:625–639. doi: 10.1042/EBC20200032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benacka R, Szaboova D, Gulasova Z, Hertelyova Z, Radonak J. Non-coding RNAs in human cancer and other diseases: overview of the diagnostic potential. Int J Mol Sci. 2023;24:16213. doi: 10.3390/ijms242216213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saw PE, Xu X, Chen J, Song EW. Non-coding RNAs: the new central dogma of cancer biology. Sci China Life Sci. 2021;64:22–50. doi: 10.1007/s11427-020-1700-9. [DOI] [PubMed] [Google Scholar]
- 18.Feng L, Li J, Li F, Li H, Bei S, Zhang X, Yang Z. Long noncoding RNA VCAN-AS1 contributes to the progression of gastric cancer via regulating p53 expression. J Cell Physiol. 2020;235:4388–4398. doi: 10.1002/jcp.29315. [DOI] [PubMed] [Google Scholar]
- 19.Du P, Luo K, Li G, Zhu J, Xiao Q, Li Y, Zhang X. Long non-coding RNA VCAN-AS1 promotes the malignant behaviors of breast cancer by regulating the miR-106a-5p-mediated STAT3/HIF-1alpha pathway. Bioengineered. 2021;12:5028–5044. doi: 10.1080/21655979.2021.1960774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Ding Y, Wu Y, Wang X. Identification of the complex regulatory relationships related to gastric cancer from lncRNA-miRNA-mRNA network. J Cell Biochem. 2020;121:876–887. doi: 10.1002/jcb.29332. [DOI] [PubMed] [Google Scholar]
- 21.Liao Y, Cao W, Zhang K, Zhou Y, Xu X, Zhao X, Yang X, Wang J, Zhao S, Zhang S, Yang L, Liu D, Tian Y, Wu W. Bioinformatic and integrated analysis identifies an lncRNA-miRNA-mRNA interaction mechanism in gastric adenocarcinoma. Genes Genomics. 2021;43:613–622. doi: 10.1007/s13258-021-01086-z. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Tan J, Xu C, Wu H, Zhang Y, Xiong Y, Yi C. Identification and construction of lncRNA-associated ceRNA network in diabetic kidney disease. Medicine (Baltimore) 2021;100:e26062. doi: 10.1097/MD.0000000000026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zia T, Bangfan L, Nadeem A, Hussain A, Abdel-Maksoud MA, Zakri AM, Bashir MK, Ali M, Jabeen N, Jamil M, Al-Qahtani WH, Almanaa TN. Comprehensive multi-level expression profiling of key biomarkers in breast cancer patients. Am J Transl Res. 2023;15:6058–6070. [PMC free article] [PubMed] [Google Scholar]
- 24.Ding XS, Hua YC, Han BX, An J, Zhou LL, Xu WR, Shi H, Zheng XX, Shi WW, Li XY. The prognostic value of cancer stage-associated genes in clear cell renal cell carcinoma. Am J Transl Res. 2023;15:5145–5158. [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Wang J, Xu J, Li S, Zhang R, Shen C, Xie M, Zheng B, Gu M. Long non-coding RNA DEPDC1-AS1 promotes proliferation and migration of human gastric cancer cells HGC-27 via the human antigen R-F11R pathway. J Int Med Res. 2022;50:3000605221093135. doi: 10.1177/03000605221093135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Li J, Qian C, Qiu F, Shen Q, Tong R, Yang Q, Xu J, Zheng B, Lv J, Hou J. LINC00624/TEX10/NF-kappaB axis promotes proliferation and migration of human prostate cancer cells. Biochem Biophys Res Commun. 2022;601:1–8. doi: 10.1016/j.bbrc.2022.02.078. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, Wu Y, Lin M, Wang G, Liu J, Xie M, Zheng B, Shen C, Shen J. BMI1 promotes osteosarcoma proliferation and metastasis by repressing the transcription of SIK1. Cancer Cell Int. 2022;22:136. doi: 10.1186/s12935-022-02552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue J, Wu T, Huang C, Shu M, Shen C, Zheng B, Lv J. Identification of proline-rich protein 11 as a major regulator in mouse spermatogonia maintenance via an increase in BMI1 protein stability. Mol Biol Rep. 2022;49:9555–9564. doi: 10.1007/s11033-022-07846-8. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y, Shen C, Wu T, Huang X, Li H, Zheng B. Syntaxin binding protein 2 in sertoli cells regulates spermatogonial stem cell maintenance through directly interacting with connexin 43 in the testes of neonatal mice. Mol Biol Rep. 2022;49:7557–7566. doi: 10.1007/s11033-022-07564-1. [DOI] [PubMed] [Google Scholar]
- 30.Wu YB, Li SY, Liu JY, Xue JJ, Xu JF, Chen T, Cao TY, Zhou H, Wu TT, Dong CL, Qian WF, Qiao LW, Hou SY, Wang T, Shen C. Long non-coding RNA NRSN2-AS1 promotes ovarian cancer progression through targeting PTK2/beta-catenin pathway. Cell Death Dis. 2023;14:696. doi: 10.1038/s41419-023-06214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou JY, Liu JY, Tao Y, Chen C, Liu SL. LINC01526 promotes proliferation and metastasis of gastric cancer by interacting with TARBP2 to induce GNG7 mRNA decay. Cancers (Basel) 2022;14:4940. doi: 10.3390/cancers14194940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L, Li S, Xu J, Shen C, Qian Q. AGAP2-AS1/BRD7/c-Myc signaling axis promotes skin cutaneous melanoma progression. Am J Transl Res. 2023;15:350–362. [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, Wang T, Zhao Z, Liu S, Shen C, Li H, Liu M, Zheng B, Yu J, Huang X. Retinoic acid induced protein 14 (Rai14) is dispensable for mouse spermatogenesis. PeerJ. 2021;9:e10847. doi: 10.7717/peerj.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen C, Zhang K, Yu J, Guo Y, Gao T, Liu Y, Zhang X, Chen X, Yu Y, Cheng H, Zheng A, Li H, Huang X, Ding X, Zheng B. Stromal interaction molecule 1 is required for neonatal testicular development in mice. Biochem Biophys Res Commun. 2018;504:909–915. doi: 10.1016/j.bbrc.2018.09.044. [DOI] [PubMed] [Google Scholar]
- 35.Xie M, Ma T, Xue J, Ma H, Sun M, Zhang Z, Liu M, Liu Y, Ju S, Wang Z, De W. The long intergenic non-protein coding RNA 707 promotes proliferation and metastasis of gastric cancer by interacting with mRNA stabilizing protein HuR. Cancer Lett. 2019;443:67–79. doi: 10.1016/j.canlet.2018.11.032. [DOI] [PubMed] [Google Scholar]
- 36.Lei ZN, Teng QX, Tian Q, Chen W, Xie Y, Wu K, Zeng Q, Zeng L, Pan Y, Chen ZS, He Y. Signaling pathways and therapeutic interventions in gastric cancer. Signal Transduct Target Ther. 2022;7:358. doi: 10.1038/s41392-022-01190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou W, Zhao Y, Zhu H. Predictive biomarkers for immunotherapy in gastric cancer: current status and emerging prospects. Int J Mol Sci. 2023;24:15321. doi: 10.3390/ijms242015321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, Wang C, Yang Y, Xu R, Li Z. LncRNA and its role in gastric cancer immunotherapy. Front Cell Dev Biol. 2023;11:1052942. doi: 10.3389/fcell.2023.1052942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, Liu J, Xu Y, Shen Y, Yang M. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19:62. doi: 10.1186/s12943-020-01185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie S, Chang Y, Jin H, Yang F, Xu Y, Yan X, Lin A, Shu Q, Zhou T. Non-coding RNAs in gastric cancer. Cancer Lett. 2020;493:55–70. doi: 10.1016/j.canlet.2020.06.022. [DOI] [PubMed] [Google Scholar]
- 41.Cao J, Yang Y, Duan B, Zhang H, Xu Q, Han J, Lu B. LncRNA PCED1B-AS1 mediates miR-3681-3p/MAP2K7 axis to promote metastasis, invasion and EMT in gastric cancer. Biol Direct. 2024;19:34. doi: 10.1186/s13062-024-00468-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh J, Narayan G, Dixit VK. The long intergenic non-coding RNA LINC01140 modulates gastric cancer phenotypes and cancer cell lines aggressiveness. Dig Liver Dis. 2024;56:1776–1783. doi: 10.1016/j.dld.2024.03.006. [DOI] [PubMed] [Google Scholar]
- 43.Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 44.Lachiondo-Ortega S, Delgado TC, Banos-Jaime B, Velazquez-Cruz A, Diaz-Moreno I, Martinez-Chantar ML. Hu Antigen R (HuR) protein structure, function and regulation in hepatobiliary tumors. Cancers (Basel) 2022;14:2666. doi: 10.3390/cancers14112666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X, Xu L. The RNA-binding protein HuR in human cancer: a friend or foe? Adv Drug Deliv Rev. 2022;184:114179. doi: 10.1016/j.addr.2022.114179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X, Li Y, Ding Y, Zhang H, Ding N, Lu M. HuR promotes ovarian cancer cell proliferation by regulating TIMM44 mRNA stability. Cell Biochem Biophys. 2020;78:447–453. doi: 10.1007/s12013-020-00939-w. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Huang A, Zhang A, Zhou C. HuR promotes breast cancer cell proliferation and survival via binding to CDK3 mRNA. Biomed Pharmacother. 2017;91:788–795. doi: 10.1016/j.biopha.2017.04.063. [DOI] [PubMed] [Google Scholar]