Abstract
Objectives: This study aims to elucidate the role of cAMP responsive element binding protein 3 like 4 (CREB3L4) in the pathogenesis of lung adenocarcinoma (LUAD) and to provide new insights and approaches for its effective treatment. An analysis was conducted on the expression and prognostic implications of CREB3L4 in LUAD. Methods: Potential downstream target genes regulated by CREB3L4 were identified through chromatin immunoprecipitation assay sequencing and mRNA sequencing analyses, and the regulatory relationship, mechanism, and prognostic significance of the identified target gene in LUAD were subsequently confirmed. Moreover, immune microenvironment analysis, the identification of immunotherapy targets, and chemotherapy drug sensitivity analyses were performed for LUAD with different levels of CREB3L4 expression. Results: CREB3L4 is upregulated in LUAD and is significantly associated with tumor staging and poor prognosis, influencing cell proliferation and migration. Comprehensive analysis through chromatin immunoprecipitation assay sequencing and mRNA sequencing highlighted RAS and EF-hand domain containing (RASEF) as a potential target gene under the regulation of CREB3L4, which was found to be overexpressed in LUAD and linked to tumor staging, as well as cell proliferation and migration. Notably, the knockdown of CREB3L4 markedly reduced RASEF promoter-driven luciferase activity. The aberrant expression of CREB3L4 in LUAD was intricately related to the complex tumor microenvironment and immune therapy targets, including PD-L1, CTLA-4, CD28, CD80, and demonstrated increased sensitivity to chemotherapy drugs such as osimertinib, gefitinib, and afatinib. Conclusions: These findings provide preliminary evidence for the involvement of the CREB3L4/RASEF signaling pathway in LUAD pathogenesis and suggest its potential as a novel biomarker for accurate diagnosis and targeted therapy.
Keywords: Lung adenocarcinoma, CREB3L4, RASEF, signaling pathway, biomarker
Introduction
Recent research findings indicate that lung cancer is the deadliest form of cancer worldwide [1]. There has been an increase in the incidence of lung adenocarcinoma (LUAD), which now surpasses squamous cell carcinoma as the dominant histological type within non-small cell lung carcinoma [2,3]. Understanding the pathogenesis of lung cancer, identifying novel molecular markers, and finding therapeutic targets remain crucial challenges in global research. Significant progress has been made in this field, with the ongoing identification of genes and potential therapeutic targets linked to lung cancer’s initiation and progression [4-6]. Moreover, considerable advances have been made in targeted lung cancer therapy [7,8]. Located in the southeast of Yunnan Province, China, the city of Xuanwei has one of the highest incidence and mortality rates for lung cancer both in China and globally [9]. Lung cancer in Xuanwei is predominantly a highly malignant and poor-prognosis LUAD type. Due to distinctive incidence characteristics [10,11], it is classified as a LUAD subtype. Comprehensive research, including studies by our group, has focused on the pathogenesis of lung cancer in XuanWei (LCXW), attributed to its unique incidence features [12-19]. The JT cells used in this study are a subtype of LUAD cells, previously established by our team, and are representative of LCXW cells.
cAMP responsive element binding protein 3 like 4 (CREB3L4), located on 1q21.3, encodes a transcriptional activation factor that plays crucial roles in various biological pathways, including protein processing, transcription regulation, signal transduction, cellular homeostasis, and small molecule transport [20]. CREB3L4 plays a pivotal role in the pathogenesis and progression of various malignancies [21-23]. However, research on the biological functions and molecular mechanisms of CREB3L4 in tumorigenesis is currently limited. Notably, no literature reports were found on the correlation between CREB3L4 and lung cancer during our review. This report aims to bridge the gap in understanding the association between CREB3L4 and lung cancer, providing new insights and references for further research into CREB3L4’s role in other tumor types.
RAS and EF-hand domain containing (RASEF), a member of the Rab family of GTPases, is involved in the regulation of membrane traffic and may potentially act as a tumor suppressor. Recent studies have shown a significant association between increased RASEF expression and improved prognosis in colorectal cancer [24]. Additionally, RASEF has been identified as a tumor suppressor gene in uveal melanoma through epigenetic regulation [25]. However, our study indicates that RASEF exhibits distinct expression patterns in LUAD compared to other tumors, suggesting potential differential roles in various malignancies. Further investigation is needed to elucidate the specific mechanisms and clinical significance of RASEF in LUAD, offering new perspectives for personalized therapy.
The biological function and molecular mechanism of the novel CREB3L4/RASEF pathway axis in the initiation and progression of LUAD were thoroughly examined at molecular, cellular, tissue, and clinical levels. Analyses were conducted on tumor-associated lymphocyte infiltration, immune therapy targets, and chemotherapy drug sensitivity related to CREB3L4. The goal is to develop a theoretical framework to enhance understanding of LUAD pathogenesis and identify novel therapeutic targets.
Materials and methods
Collection of cell lines, clinical specimens, and related data
In this study, human bronchial epithelial cells BEAS-2B and 16HBE, along with the LUAD cell line A549, were obtained from the American Type Culture Collection (ATCC) in Virginia, USA. The LCXW cell line JT was developed by the Department of Clinical Laboratory, First Affiliated Hospital of Kunming Medical University.
Patients were recruited from the First Affiliated Hospital of Kunming Medical University (Kunming, China). Cancer tissues identified as LCXW and adjacent non-tumor tissues were collected post-surgery, processed, and stored in RNAlater, then preserved in liquid nitrogen. Following approval from the Ethics Committee of the First Affiliated Hospital of Kunming Medical University, patient data including gender, age, smoking history, tumor lymph node metastasis, and staging were obtained from medical records. Regular follow-ups were conducted every six months through telephone, WeChat, email, etc., to gather data on survival and disease progression. All sample collection, research testing, and data gathering received ethical approval from the institution’s review board, ensuring participant confidentiality and adherence to national laws and regulations.
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University. The ethical certification materials are provided in the supplementary materials, and the approval date: June 28, 2022.
Real-time quantitative polymerase chain reaction
Total RNA from cells and tissues in each experimental group was extracted using the Trizol method (Tiangen, Beijing, China). High-purity and integrity mRNA was reverse-transcribed to cDNA using a PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Kusatsu, Japan). Primers for CREB3L4, RASEF, and the reference gene β-actin were designed and synthesized: CREB3L4, 5’-CTGCCCTGTCAAACCCTGTT-3’ (forward primer) and 5’-GCTTGTTACGGATTTTCCTCCT-3’ (reverse primer); RASEF, 5’-GGCTGACATTCGTGACACTG-3’ (forward primer) and 5’-GGAATTGGTCCCGGTTAGAT-3’ (reverse primer); PCNA, 5’-TGGAGAACTTGGAAATGGAAA-3’ (forward primer) and 5’-GAACTGGTTCATTCATCTCTATGG-3’ (reverse primer); Ki-67, 5’-TCCTTTGGTGGGCACCTAAGACCTG-3’ (forward primer) and 5’-TGATGGTTGAGGTCGTTCCTTGATG-3’ (reverse primer); β-actin, 5’-GTGGCCGAGGACTTTGATTG-3’ (forward primer) and 5’-CCTGTAACAACGCATCTCATATT-3’ (reverse primer). RT-qPCR detection was performed using SuperReal PreMix Plus (SYBR Green) (Tiangen, Beijing, China).
Western-blotting and immunohistochemistry
Cells were lysed using RIPA Lysis Buffer (Beyotime, Shanghai, China), and protein concentration was determined. Proteins were denatured, and SDS-PAGE was prepared using the Color PAGE Gel Rapid Preparation Kit (Epizyme, Shanghai, China). After electrophoresis, proteins were transferred to membranes, blocked, and incubated with primary antibodies: human RASEF at 1:500 (Novus, Littleton, Colorado, USA), CREB3L4 at 1:1000 (abbexa, Cambridge, UK), and GAPDH at 1:5000 (Immunoway, Plano, USA). The HRP-conjugated Goat Anti-Mouse IgG (H+L) secondary antibody (Servicebio, Wuhan, China) was used at 1:3000. Target proteins were visualized and analyzed post-development.
The organization and adjacent non-tumor tissues were fixed in formaldehyde and embedded in paraffin for tissue sectioning. After antigen retrieval, the primary antibody was diluted to 1:200 for mouse anti-human RASEF (Novus, Littleton, Colorado, USA) and 1:250 for mouse anti-human CREB3L4 (abbexa, Cambridge, UK), then applied and incubated overnight at 4°C. The second antibody, HRP-conjugated Goat Anti-Mouse IgG (H+L) (Servicebio, Wuhan, China), was used at a 1:300 dilution and incubated at room temperature, followed by PBS washes. A color development solution was applied and incubated, then the sections were rinsed with distilled water to observe positive staining under a microscope. The sections were washed again, counterstained with hematoxylin-eosin, and subjected to a dehydration process using an ethanol gradient (80%→95%→95%→100%→100%), cleared in xylene, mounted with neutral resin on coverslips, air-dried, and imaged microscopically.
EdU cell proliferation assay and cell scratch test
The BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 555 (Beyotime, Shanghai, China) was used for the proliferation assay. Plates were treated with 1× EdU working solution at 37°C and incubated for 2 hours. After labeling, the medium was replaced with 100 μL of fixative and incubated at room temperature for 15 minutes. A permeabilization step was conducted with 100 μL of permeabilization solution for 10-15 minutes at room temperature. ClickAdditive Solution was prepared and added (100 μL per well), followed by a 30-minute incubation in darkness at room temperature. Cells were washed thrice, then stained with 100 μL of 1× Hoechst33342 for 10 minutes in darkness for fluorescence detection using an inverted fluorescence microscope.
For the scratch test, precise vertical scratches were made with a pipette tip in PBS, followed by washing to remove detached cells before adding complete medium. Images were taken at 0 and 48 hours to calculate the cell migration rate: (area of scratch at zero hours - area of scratch at forty-eight hours)/area of scratch at zero hours.
Dual-luciferase reporter assay
The relationship between the enrichment peak of SLC38A2 and the transcription start site (TSS) in chromatin immunoprecipitation assay sequencing (CHIP-seq) data was analyzed. Visualization of the promoter’s differential enrichment peak on the UCSC website determined the sequence location for the target gene promoter synthesis. A significant enrichment peak for RASEF was identified at chr9: 85678051-85678200. The Shanghai GM Company synthesized the antisense complementary sequence for this RASEF promoter region, which was then cloned into the PGL3-basic vector. Experiments were conducted using the Dual-Glo® Luciferase Assay System (Promega, Wisconsin, USA). 293T cells were grown in antibiotic-free medium before transfection, with a transfection complex DNA to Attractene ratio of 1:3. The plates were incubated at 37°C with 5% CO2, and dual luciferase activity was measured after 48 hours using an inverted fluorescence microscope.
Cell transfection
We constructed sh-CREB3L4-1, sh-CREB3L4-2, and control sh-NC plasmid using the pHBLV-U6-MCS-CMV-ZsGreen-PGK-PURO vector. Lentiviral packaging of each plasmid was conducted using the Lenti-Pac HIV Expression Packaging Kit (Genecopoeia, Rockville, USA), followed by cell transfection and selection of stable cell lines using puromycin (Solarbio, Beijing, China). Additionally, we utilized siRNA duplexes to inhibit RASEF expression, with sequences: siRNA-RASEF, 5’-CTTCATCCGTGAGATCAGA-3’; siRNA-NC, 5’-TTCTCCGAACGTGTCACGT-3’. Transfection was performed using Lipofectamine 2000 (Thermo Fisher, Massachusetts, USA) at a 1:3 ratio.
Sequencing and data analysis of CHIP-seq and mRNA-seq
CloudSeq Biotech (Shanghai, China) provided high-throughput sequencing services. Chromatin immunoprecipitation followed the method outlined by Wamstad et al. [26]. ChIP DNA yield was quantified using the Quant IT fluorescence assay (Life Technologies, State of California, USA), and ChIP reaction efficiencies were assessed by qPCR. Illumina sequencing libraries were prepared with the NEBNext® Ultra™ DNA Library Prep Kit (New England Biolabs, Massachusetts, USA) and quality-checked using the Agilent 2100 Bioanalyzer (Agilent, State of California, USA), followed by 150 base paired-end sequencing on an Illumina Hiseq sequencer. For mRNA-seq, total RNA was processed to deplete rRNAs using the Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA, USA), with RNA libraries constructed using the TruSeq Stranded Total RNA Library Prep Kit (Illumina, San Diego, CA, USA). Library quality and quantity were verified using the BioAnalyzer 2100 system before sequencing. Data were compared and analyzed against the human reference genome (UCSC hg19) [27-31].
Data collection for bioinformatics analysis
Transcriptome RNA sequencing data (FPKM normalization) for LUAD patients, along with their clinical pathological features, were downloaded from The Cancer Genome Atlas (TCGA) Data Portal, which included data on 522 patients and 594 samples (535 cancer samples and 59 normal samples) (accessed on February 26, 2022). The survival analysis of target genes was performed using the GEPIA 2 platform (http://gepia2.cancer-pku.cn/#index). Gene expression visualization in various cell types was conducted using the TISCH 2 platform (http://tisch.comp-genomics.org/search-gene/) for tumor microenvironment-related single-cell sequencing data.
Statistical analysis
Statistical analysis of LUAD samples from the TCGA database were conducted using R language version 4.3.3 and the SPSS 22.0 software package. The normality of data was assessed using the Kolmogorov-Smirnov (K-S) test. For normally distributed metric data, the t-test was used for comparisons between two groups, while the Wilcoxon rank-sum test was applied for non-normally distributed data. Chi-square test was used for differential analysis of IHC results. The clinical relevance of target genes was analyzed using the Kruskal-Wallis rank sum test. The Log-Rank test was employed to assess the relationship between target gene expression levels and survival rates, with the Spearman correlation coefficient used for gene expression correlation analysis. A p-value <0.05 was considered statistically significant in all analyses.
Results
Expression, clinical significance, and cellular biological functions of CREB3L4 in LUAD
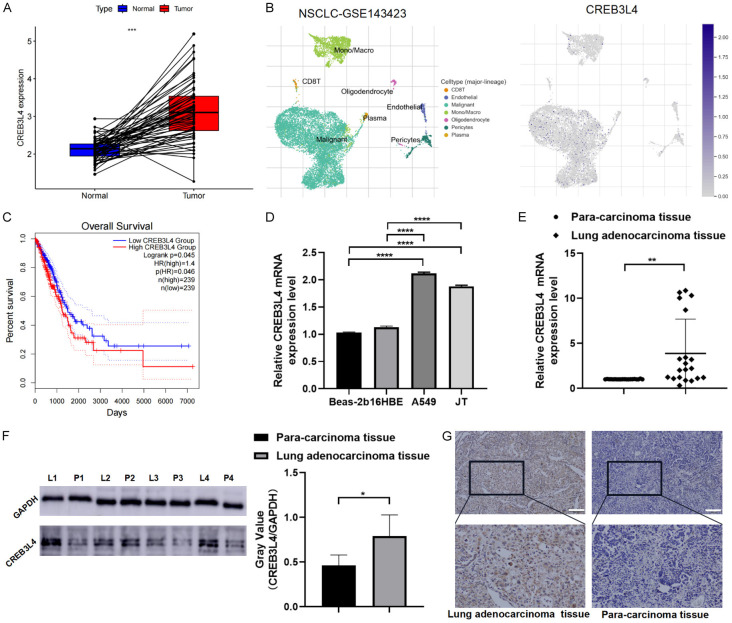
Analysis of the TCGA dataset revealed that CREB3L4 mRNA expression was significantly higher in LUAD samples compared to adjacent normal tissues, as illustrated in 59 paired samples (Figure 1A). Single-cell analysis from the GSE143423 dataset showed CREB3L4 predominantly expressed in malignant cells within the tumor immune microenvironment (TIME) (Figure 1B). Survival analysis indicated that the high expression group of CREB3L4 (HEG-CREB3L4) correlates with poor prognosis (Figure 1C). Further validation in normal human bronchial epithelial cells (BEAS-2B and 16HBE) and LUAD cells (A549 and LCXW cell line JT) confirmed elevated CREB3L4 levels in cancer cells (P<0.05) (Figure 1D).
Figure 1.
Validation and clinical relevance of cAMP responsive element binding protein 3 like 4 (CREB3L4) expression. A. CREB3L4 expression in paired cancer tissues (TCGA database); B. CREB3L4 expression across different immune phenotype-related single-cell atlases (GSE143423); C. Survival analysis based on CREB3L4 expression (TCGA database); D. CREB3L4 mRNA expression in various cell lines; E. RT-qPCR analysis of CREB3L4 in 20 pairs of cancer tissues; F. Western blot analysis of CREB3L4 in 4 pairs of cancer tissues (samples from the same experiment processed in parallel); G. IHC analysis of CREB3L4 in cancer tissues. L: lung adenocarcinoma tissue, P: para-carcinoma tissue. *P<0.05, **P<0.01, ****P<0.0001. Scale bar: 200 µm.
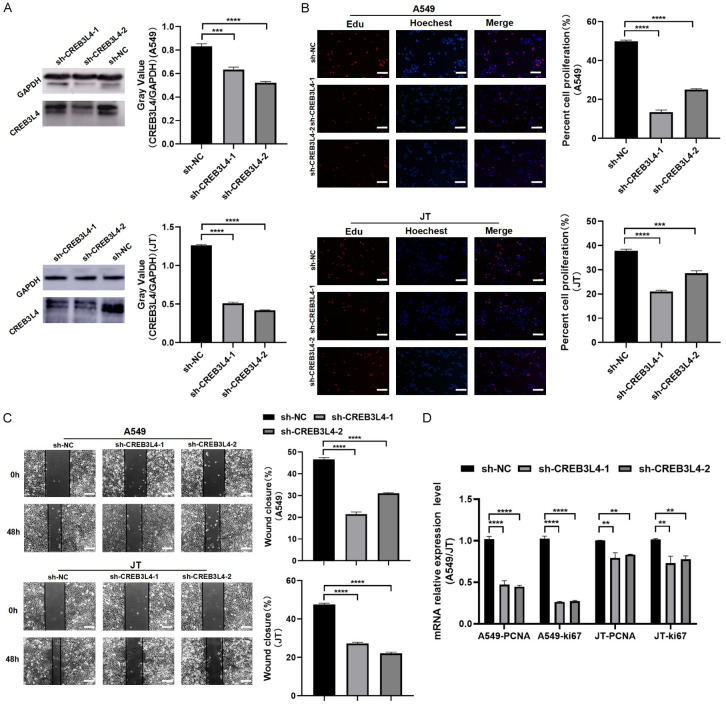
We analyzed 20 pairs of LCXW cancer tissues and adjacent non-tumor tissues from the First Affiliated Hospital of Kunming Medical University for transcription level validation, with 4 pairs assessed for protein levels. These analyses consistently showed higher CREB3L4 expression in cancer tissues (P<0.05) (Figure 1E, 1F). In 98 pairs of tissues analyzed via immunohistochemistry (IHC), CREB3L4 immunostaining appeared as yellow-brown granules, mainly in the cytoplasm, with some nuclear positivity (Figure 1G). IHC scores significantly differed between cancerous and adjacent tissues, indicating increased CREB3L4 expression in cancer (χ2=17.251, P<0.01) (Table 1). When stratifying the 98 patients by clinical characteristics, the non-parametric Kruskal-Wallis rank sum test showed a significant rise in CREB3L4 expression detected in stage III+IV lung cancer tissues compared to stages I+II (χ2=8.233, P=0.004) and a significantly higher CREB3L4 levels in patients with lymph node metastasis stages N1-3 compared to the N0 stage (χ2=4.009, P=0.045) (Table 2). We employed carrier-based short hairpin RNA to synthesize specific CREB3L4 shRNA and corresponding control plasmids. After lentiviral infection and puromycin screening, stable knockdown LUAD cell models for sh-CREB3L4-1 and sh-CREB3L4-2 were established, along with a control sh-NC cell model (Figure 2A). EDU assay results showed a significant reduction in the proliferation of A549 and JT cells in the knockdown group (P<0.05) (Figure 2B). Similarly, scratch assay results demonstrated a significant decrease in migration ability of A549 and JT cells in the knockdown group compared to the control (P<0.05) (Figure 2C). Moreover, the expression levels of proliferation markers PCNA and Ki-67 were significantly reduced in the knockdown group relative to the control group (P<0.05) (Figure 2D).
Table 1.
Expression of cAMP responsive element binding protein 3 like 4 (CREB3L4) in lung adenocarcinoma and para-carcinoma tissues
| Groups | Number | CREB3L4 | Positive (%) | χ2 | P | |
|---|---|---|---|---|---|---|
|
| ||||||
| - | + | |||||
| Lung cancer tissue | 98 | 38 | 60 | 61.22% | 17.251 | <0.01 |
| Para-carcinoma tissue | 98 | 67 | 31 | 31.63% | ||
Table 2.
Clinical correlation analysis of CREB3L4
| Groups | Number | CREB3L4 | χ2 | P | Mean Rank | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| - | + | ++ | +++ | ||||||
| Gender | Male | 50 | 2 | 2 | 30 | 16 | 0.070 | 0.792 | 50.14 |
| Female | 48 | 1 | 3 | 30 | 14 | 48.83 | |||
| Age | <55 | 70 | 4 | 2 | 46 | 18 | 2.270 | 0.132 | 47.14 |
| ≥55 | 28 | 1 | 1 | 14 | 12 | 55.39 | |||
| Smoking | Yes | 34 | 6 | 7 | 7 | 14 | 0.012 | 0.913 | 49.10 |
| No | 64 | 7 | 1 | 42 | 14 | 49.71 | |||
| FIGO staging | I+II | 86 | 2 | 6 | 56 | 22 | 8.233 | 0.004* | 46.85 |
| III+IV | 12 | 0 | 0 | 4 | 8 | 68.50 | |||
| Lymphatic Metastasis | N1~3 | 36 | 0 | 0 | 22 | 14 | 4.009 | 0.045* | 56.00 |
| N0 | 62 | 3 | 5 | 38 | 16 | 45.73 | |||
P<0.05.
Figure 2.
Biological functions of CREB3L4. A. Western blot analysis post-CREB3L4 knockdown in lung adenocarcinoma (LUAD) cells (samples from the same experiment processed in parallel); B. EDU assay results for knockdown and control groups in LUAD cells; C. Wound healing assay results for knockdown and control groups in LUAD cells; D. Proliferation marker RT-qPCR analysis for the knockdown and control groups in LUAD cells. sh-CREBL34-1: Group 1 with CREB3L4 knockdown. sh-CREBL34-2: Group 2 with CREB3L4 knockdown. sh-NC: Knockdown control group. **P<0.01, ***P<0.001, ****P<0.0001. Scale bar: 200 µm.
Differential enrichment peak annotation in CHIP and differential mRNA expression following aberrant CREB3L4 expression, with Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genome (KEGG) pathway analysis
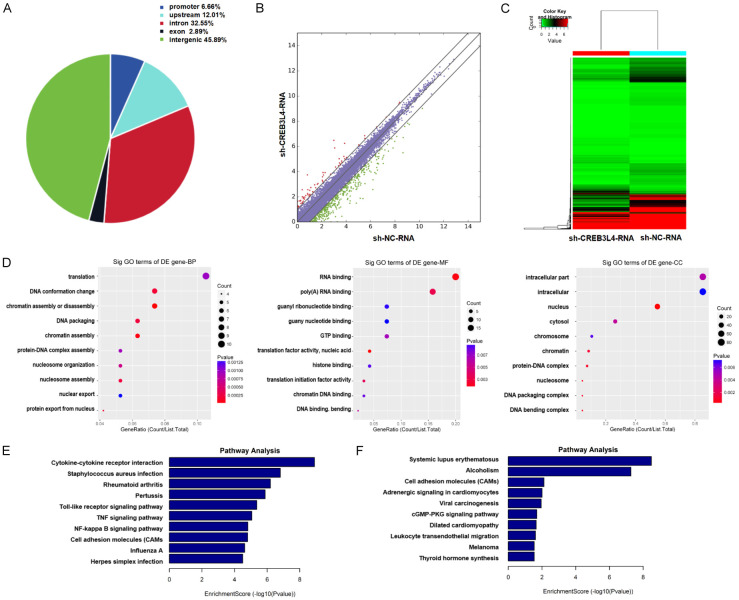
In stable JT cells with sh-CREB3L4-2 and shNC, ChIP-seq identified 7675 enriched peaks showing consistent changes (5323 up-regulated and 2352 down-regulated) in the sh-CREB3L4-chip_vs_sh-NC-chip comparison (Supplementary Table 1). Among these, there were 511 “promoter” peaks, 922 “upstream” peaks, 2498 “intron” peaks, 222 “exon” peaks, and 3522 “intergenic” peaks. The distribution of enriched peaks across various genomic features was visualized (Figure 3A). In the mRNA-seq analysis for the sh-CREB3L4-RNA versus sh-NC-RNA group, 2319 mRNA species were detected, with 518 up-regulated and 1801 down-regulated transcripts (Figure 3B, 3C; Supplementary Table 2).
Figure 3.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) analysis results from chromatin immunoprecipitation assay sequencing and mRNA sequencing. A. Distribution of enriched peaks across genomic features in the sh-CREB3L4-chip vs. sh-NC-chip group; B. Scatter plot showing differentially expressed genes in the sh-CREB3L4-RNA vs. sh-NC-RNA group; C. Heatmap illustrating differentially expressed genes in the sh-CREB3L4-RNA vs. sh-NC-RNA group; D. Top ten GeneRatio-related genes in biological processes (BP), molecular functions (MF), and cellular components (CC) in the downregulated sh-CREB3L4-chip vs. sh-NC-chip group; E. Signaling pathways associated with the top ten enrichment score-related genes in the downregulated sh-CREB3L4-RNA vs. sh-NC-RNA group; F. Signaling pathways associated with the top ten enrichment score-related genes in the upregulated sh-CREB3L4-RNA_vs_sh-NC-RNA. sh-CREB3L4: CREB3L4 knockdown group. sh-NC: Knockdown control group.
Given CREB3L4’s affiliation with the CREB/ATF transcription factor family, this study aims to clarify how CREB3L4 influences cancer progression via regulation of target gene transcription. Specifically, GO functional and KEGG pathway analyses will focus on genes linked to differentially enriched peaks within the promoter region (TSS-2000 to TSS+2000) as identified by ChIP-seq.
The GO analysis of the down-regulated group in sh-CREBL4-chip_vs_sh-NC-chip, reflecting CREB3L4’s binding to target genes, indicated that CREB3L4 is involved in molecular functions and biological processes including protein-DNA complex assembly, chromatin DNA binding, and DNA conformational changes (Figure 3D). Notably, CREB3L4 was predominantly linked to translation initiation factor activity and the process of translation, suggesting a potential interactive mechanism between the transcription factor CREB3L4 and post-transcriptional translation, which merits further exploration.
Differentially expressed genes in the sh-CREB3L4-RNA_vs_sh-NC-RNA down-regulated group, indicating positive regulation, and the up-regulated group, indicating negative regulation, were significantly enriched in cancer-associated pathways, including cytokine-cytokine receptor interaction, Toll-like receptor signaling pathway, TNF signaling pathway, and NF-κB signaling pathway (Figure 3E, 3F). These findings underscore the critical role of CREB3L4 in elucidating the mechanisms of tumorigenesis.
Combined analysis of CHIP-seq and mRNA-seq to identify potential downstream targets of CREB3L4
Leveraging CREB3L4’s role as a transcription factor, we undertook a thorough analysis of ChIP-seq and mRNA-seq data to pinpoint possible downstream targets regulated by CREB3L4. This process involved searching for sequences within gene regulatory regions where CREB3L4 might bind and evaluating the mRNA expression levels of these potential targets. Our integrated analysis identified several likely candidates under CREB3L4’s positive regulation, notably RASEF, which showed decreased expression in both the sh-CREB3L4-chip versus sh-NC-chip group and the sh-CREB3L4-RNA versus sh-NC-RNA group. This pattern indicates that CREB3L4 may enhance RASEF expression (Table 3).
Table 3.
Partial candidate target genes potentially regulated by CREB3L4 identified through the joint analysis of chromatin immunoprecipitation assay sequencing (CHIP-seq) and mRNA sequencing (mRNA-seq)
| GeneSymbol | Regulation: shCREB3L4_vs_shNC | CHIP-seq results | mRNA-seq results | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Peak_classification | Fold change | P-value | FDR | P-value | FDR | ||
| RASEF | Down | Promoter | -164.3 | 0.0000 | 0.0055 | 0.0572 | 0.9996 |
| ACTG2 | Down | Promoter | -164.3 | 0.0000 | 0.0055 | 0.0015 | 0.228 |
| HIST1H2AB | Down | Promoter | -164.3 | 0.0000 | 0.0055 | 1 | 1 |
| CACNG7 | Down | Promoter | -164.3 | 0.0000 | 0.0055 | 0.0081 | 0.5828 |
| DPEP1 | Down | Promoter | -156.1 | 0.0000 | 0.0071 | 0.2513 | 0.9996 |
| ITPR2 | Down | Promoter | -156.1 | 0.0000 | 0.0071 | 0.0125 | 0.8371 |
| PCDHA1 | Down | Promoter | -148 | 0.0000 | 0.0086 | 1 | 1 |
| GPCPD1 | Down | Promoter | -22.34 | 0.0000 | 0.0002 | 0.0219 | 0.8883 |
Expression and clinical significance of RASEF in LUAD, its cellular functions, and regulatory mechanism with CREB3L4
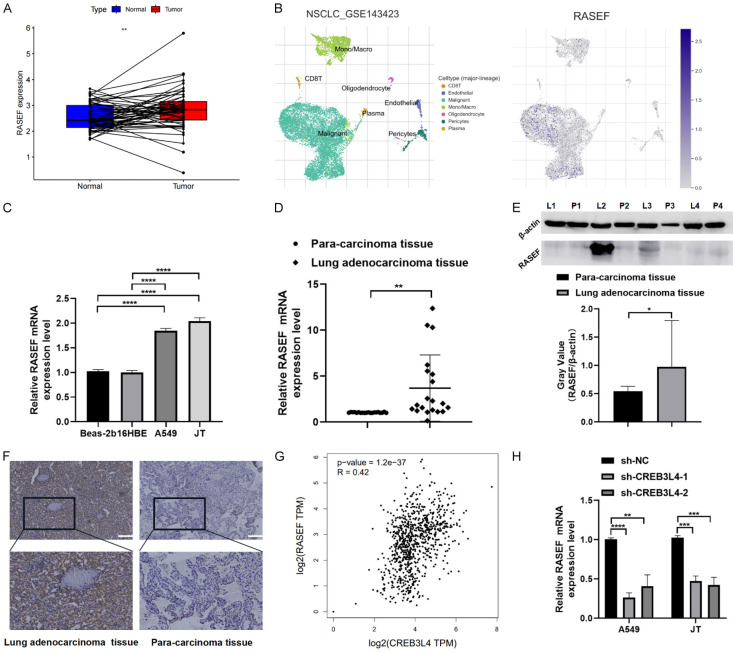
TCGA database analysis indicated that RASEF expression is significantly higher in LUAD tissues than in adjacent normal tissues (P<0.05) (Figure 4A). Single-cell analysis within the tumor microenvironment (GSE143423) showed predominant RASEF distribution in malignant cells (Figure 4B). RASEF validation in BEAS-2B, 16HBE, A549, and JT cell lines confirmed its elevated expression in LUAD cells compared to normal cells (Figure 4C). RT-qPCR results, using the 2-ΔΔCt method, demonstrated statistically significant upregulation of RASEF mRNA in cancer tissues (P<0.05) (Figure 4D). Protein level differences in RASEF were assessed in 4 pairs of tissues, using β-actin as a reference, showing higher expression in cancerous tissues (P<0.05) (Figure 4E; Table 4). The immunostaining for RASEF manifested as yellow-brown granules, predominantly showing cytoplasmic positivity with some nuclear expression (Figure 4F). To examine the correlation between increased RASEF expression in LUAD and clinical patient data, we employed a non-parametric Kruskal-Wallis rank sum test. This analysis of 98 pairs of lung cancer tissue samples and their clinical characteristics found no significant association between RASEF expression and patient gender, age, smoking status, or TNM staging (P>0.05). Notably, a significant rise in RASEF expression was detected in stage III+IV lung cancer tissues compared to stages I+II (χ2=10.270, P=0.001) (Table 5), indicating that RASEF expression correlates with advanced tumor staging in LUAD.
Figure 4.
Expression validation and clinical relevance analysis of RAS and EF-hand domain containing (RASEF). A. RASEF expression in paired cancer tissues (TCGA database); B. RASEF expression across different immune phenotype-related single-cell atlases (GSE143423); C. RASEF expression in various cell lines; D. RT-qPCR analysis of RASEF in 20 pairs of cancer tissues; E. Western blot analysis of RASEF in 4 pairs of cancer tissues (samples processed in parallel); F. IHC analysis of RASEF in cancer tissues. Correlation validation between RASEF and CREB3L4 expression levels. G. Correlation analysis between CREB3L4 and RASEF (TCGA and GTEx databases); H. mRNA expression levels of RASEF in LUAD cells with CREB3L4 knockdown vs. control group. L: lung adenocarcinoma tissue, P: adjacent non-cancerous tissue. sh-CREBL34-1: Group 1 with knocked-down CREBL34. sh-CREBL34-2: Group 2 with knocked-down CREBL34. sh-NC: Knockdown control group. **P<0.01, ***P<0.001, ****P<0.0001. Scale bar: 200 µm.
Table 4.
Expression of RAS and EF-hand domain containing (RASEF) in lung adenocarcinoma and para-carcinoma tissues
| Groups | Number | RASEF | Positive (%) | χ2 | P | |
|---|---|---|---|---|---|---|
|
| ||||||
| - | + | |||||
| Lung cancer tissue | 98 | 37 | 61 | 62.24% | 16.027 | <0.01 |
| Para-carcinoma tissue | 98 | 65 | 33 | 33.67% | ||
Table 5.
Clinical correlation analysis of RASEF
| Groups | Number | RASEF | χ2 | P | Mean Rank | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| - | + | ++ | +++ | ||||||
| Gender | Male | 50 | 14 | 14 | 13 | 9 | 0.414 | 0.520 | 47.75 |
| Female | 48 | 12 | 11 | 15 | 10 | 51.32 | |||
| Age | <55 | 70 | 24 | 18 | 18 | 10 | 1.968 | 0.161 | 47.04 |
| ≥55 | 28 | 6 | 7 | 9 | 6 | 55.64 | |||
| Smoking | Yes | 34 | 8 | 8 | 6 | 12 | 0.353 | 0.552 | 51.76 |
| No | 64 | 16 | 13 | 21 | 14 | 48.30 | |||
| FIGO staging | I+II | 86 | 17 | 34 | 18 | 17 | 10.270 | 0.001* | 46.20 |
| III+IV | 12 | 1 | 1 | 1 | 9 | 73.17 | |||
| Lymphatic Metastasis | N1~3 | 36 | 9 | 11 | 8 | 8 | 0.001 | 0.973 | 49.63 |
| N0 | 62 | 17 | 14 | 20 | 11 | 49.43 | |||
P<0.05.
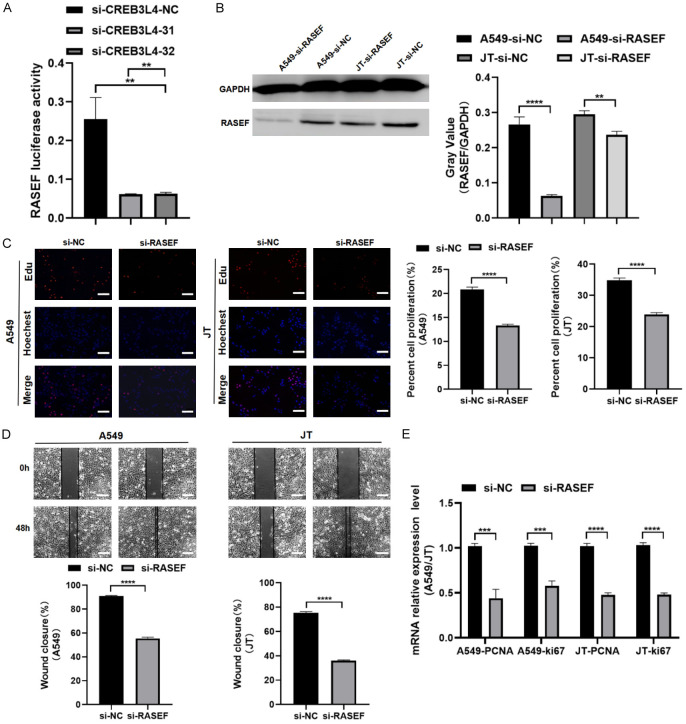
The analysis using the TCGA and GTEx databases on the “GEPIA 2” platform indicated a significant positive correlation between CREB3L4 and RASEF (Spearman correlation coefficient R=0.42, P<0.05) (Figure 4G). Knockdown of CREB3L4 resulted in a decrease in RASEF mRNA levels within cells (P<0.05) (Figure 4H). Furthermore, the ratio of firefly to renilla luciferase indicated that CREB3L4 knockdown significantly reduced luciferase activity driven by the RASEF promoter, showing statistically significant differences (P<0.05) (Figure 5A).
Figure 5.
Interaction validation between RASEF and CREB3L4. A. Fluorescence enzyme activity of the RASEF reporter gene in CREB3L4 knockdown and control groups. Biological functions of RASEF. B. Western blot results for RASEF in LUAD cells post-interference (samples from the same experiment processed in parallel); C. EDU assay results in LUAD cells post-RASEF interference compared to the control group; D. Wound healing assay results in LUAD cells post-RASEF interference compared to the control group; E. RT-qPCR analysis of proliferation markers in LUAD cells post-RASEF interference compared to the control group. sh-CREBL34-1: Group 1 with knocked-down CREBL34. sh-CREBL34-2: Group 2 with knocked-down CREBL34. sh-NC: Control group with knocked-down. si-RASEF: RASEF interference group. si-NC: interference control group. **P<0.01, ***P<0.001, ****P<0.0001. Scale bar: 200 µm.
Using small interfering RNA (siRNA) to transiently reduce RASEF expression in A549 and JT cells (Figure 5B), EDU assays showed a marked decrease in proliferation in the knockdown group (P<0.05) (Figure 5C), and scratch assays demonstrated significantly reduced migration compared to the control group (P<0.05) (Figure 5D). Furthermore, the levels of proliferation markers PCNA and Ki-67 were significantly lower in the knockdown group than in the control group (P<0.05) (Figure 5E). These results suggest that CREB3L4 directly binds to the RASEF promoter region, enhancing its expression and subsequently affecting cell proliferation and migration in LUAD cells.
Differential expression genes related to RASEF and their GO and KEGG analysis
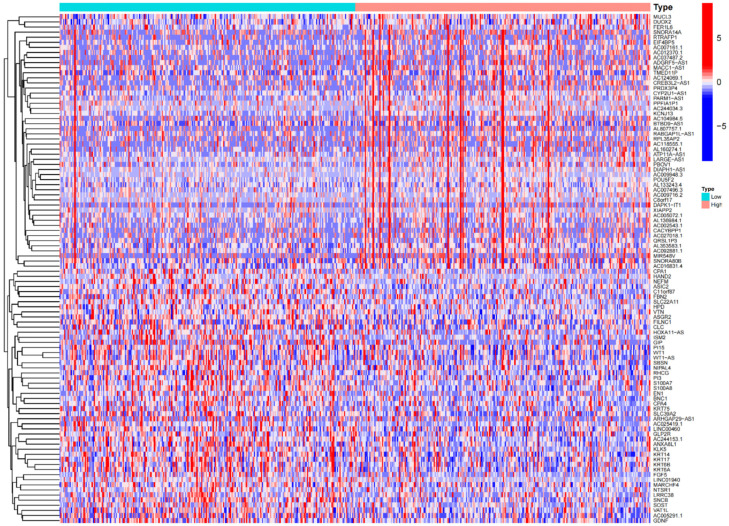
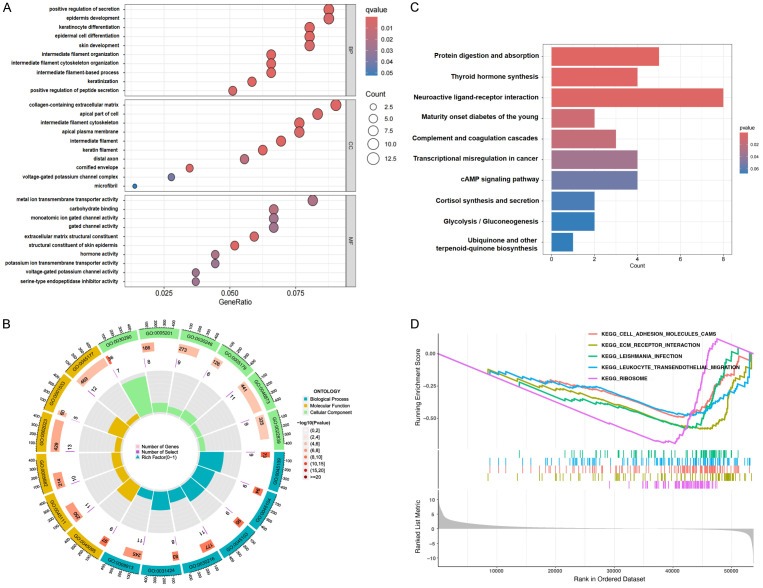
The limma package in R was used to identify differentially expressed genes (DEGs) associated with RASEF, uncovering 696 genes with statistically significant changes. A heatmap of these DEGs is shown in Figure 6. Following this, GO, KEGG, and GSEA enrichment analyses were performed, yielding significant enrichment in 53 GO terms and 6 KEGG pathways. The top 10 terms for biological processes (BP), molecular functions (MF), and cellular components (CC) are illustrated (Figure 7A, 7B). Notable enrichments included “positive regulation of secretion” in BP, “metal ion transmembrane transporter activity”, “carbohydrate binding”, and “serine-type endopeptidase inhibitor activity” in MF, and “collagen-containing extracellular matrix” and “voltage-gated potassium channel complex” in CC. The top 10 KEGG pathways with the smallest p-values highlighted RASEF’s involvement in processes such as protein digestion and absorption, thyroid hormone synthesis, transcriptional misregulation in cancer, and the cAMP signaling pathway (Figure 7C, 7D). These bioinformatics analyses reveal RASEF’s potential biological functions and its involvement in various biological processes and pathways related to cancer and other diseases, underscoring its pivotal role in LUAD progression and providing a substantial basis for further research.
Figure 6.
Heatmap displaying a selection of RASEF-related differentially expressed genes.
Figure 7.
GO and KEGG analysis of RASEF-related differentially expressed genes. A and B. Top-ranked BP, MF, CC functions involved in RASEF-related differentially expressed genes; C and D. Top-ranked KEGG pathways associated with RASEF-related differentially expressed genes.
Evaluation of the efficacy of immunotherapy and chemotherapy in CREB3L4-related LUAD
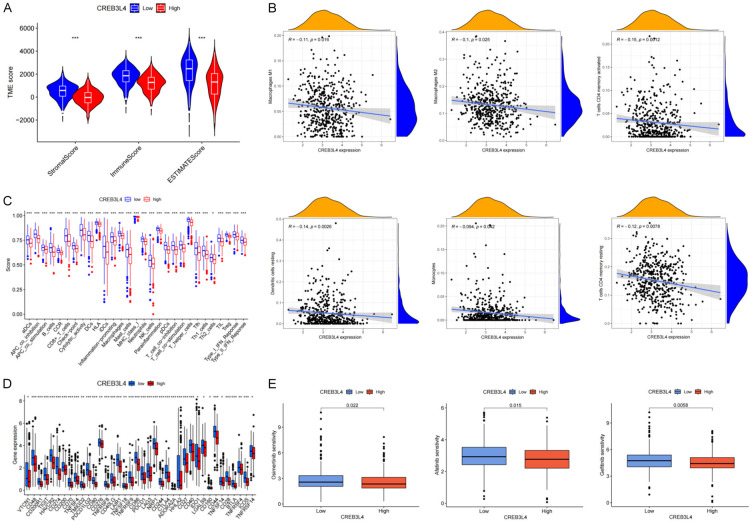
The ESTIMATE algorithm analysis, shown in Figure 8A, indicated that the low expression group of CREB3L4 (LEG-CREB3L4) had higher immune, matrix, and ESTIMATE scores, suggesting that LUAD patients with LEG-CREB3L4 might have a better prognosis than those with high expression of CREB3L4 (HEG-CREB3L4), aligning with previous survival curve analyses. Furthermore, LEG-CREB3L4 was associated with increased infiltration of various immune cells, including myeloid dendritic cells, M2 macrophages, M1 macrophages, neutrophils, CD4+ T cells, and CD8+ T cells (Figure 8B).
Figure 8.
Analysis of immune microenvironment, immune targets, and chemotherapy drug sensitivity in CREB3L4-related LUAD. A. Violin plot of immune score, stromal score, and estimate score in CREB3L4-related LUAD; B. Immune cell infiltration in CREB3L4-related LUAD; C. Immunological function evaluation in CREB3L4-related LUAD; D. Analysis of immune checkpoints in CREB3L4-related LUAD; E. Sensitivity analysis of chemotherapy drugs in CREB3L4-related LUAD. *P<0.05, **P<0.01, ***P<0.001.
Single sample gene set enrichment analysis (ssGSEA) scores revealed greater immune cell infiltration in LEG-CREB3L4 samples, with elevated immune scores for co-stimulation of antigen-presenting cells, checkpoint, cytolytic activity, human leukocyte antigen, inflammation promoting, major histocompatibility complex class I, parainflammation, T cell co-stimulation, and Type I interferon response, consistent with the observed poor prognosis in HEG-CREB3L4 patients (Figure 8C).
Additionally, a significant correlation was found between LEG-CREB3L4 and higher expression of immunotherapy targets like PD-L1, CD28, CD80, and CTLA-4 (Figure 8D), suggesting that LEG-CREB3L4 patients might respond better to immune checkpoint therapy. Given that chemotherapy is a primary treatment for advanced LUAD, we assessed the drug sensitivity for common chemotherapy drugs. The results showed that drugs such as Osimertinib, gefitinib, and Afatinib were more effective in the LEG-CREB3L4 group (Figure 8E), indicating potential enhanced therapeutic outcomes for patients with LEG-CREB3L4 in CREB3L4-associated LUAD treatment.
Discussion
The advent and progression of omics technologies have overcome the limitations of traditional research methods, providing a high-throughput, systematic, and in-depth approaches for dissecting the pathogenesis of complex diseases like tumors. This advancement offers unprecedented opportunities to identify molecular markers and therapeutic targets. In our quest to pinpoint essential genes involved in the development and progression of LUAD, our team undertook a multi-omics analysis of 8 LCXW cases and their corresponding adjacent non-cancerous tissues. We established a DNA methylation-CNA-miRNA-mRNA co-expression network, centering on differentially expressed mRNAs. Through this network, we identified several core genes significantly upregulated in the LUAD samples, notably CREB3L4 [32], indicating its potential significance in research. Given the current scarcity of literature on the relationship between CREB3L4 and LUAD progression, our study provides a thorough analysis to bridge this knowledge gap.
The validation results showed a significant increase in CREB3L4 expression in LUAD, correlating with lymph node metastasis and poor patient prognosis. The elevated levels of CREB3L4, a novel transcription factor, were found to enhance LUAD cell proliferation and migration, laying a solid foundation for further mechanistic studies. Through combined CHIP-seq and mRNA-seq analysis, we identified several potential CREB3L4-regulated target genes, including RASEF. We opted to delve deeper into RASEF’s role. Subsequent validation confirmed RASEF’s significant upregulation in LUAD, linked to tumor staging, and its involvement in cell proliferation and migration, highlighting RASEF’s crucial role in LUAD progression. Additionally, RASEF expression was positively correlated with CREB3L4, and luciferase reporter assays indicated CREB3L4’s binding to the RASEF promoter, enhancing its transcriptional activity. These results suggest the CREB3L4/RASEF signaling pathway as a promising avenue for further investigation into LUAD’s pathogenesis.
Initially aimed at exploring CREB3L4’s unique expression and functions in LCXW with specific disease features, our comparative analysis across molecular, cellular, and tissue levels found no significant differences between LCXW and LUAD. Thus, we expanded our focus to include CREB3L4’s role in LUAD pathogenesis.
The gene RASEF is posited to potentially serve as a tumor suppressor [24,25]. However, our findings reveal that RASEF assumes an activating role in LUAD progression, diverging from its expression patterns in other cancer types. This variability in expression across different cancers could be due to factors such as tissue specificity, mutations, epigenetic changes, and abnormal regulatory mechanisms, necessitating further studies to clarify their specific molecular mechanisms.
To fully understand the biological functions of organisms, it is critical to extend research beyond in vitro studies and lung cancer samples. Future research should include the development of in vivo animal models related to CREB3L4 and its target genes, providing data that better reflect clinical scenarios and offering valuable insights for subsequent clinical research.
Prior to validating clinical samples, we conducted bioinformatics analyses using R language. Yet, the clinical relevance findings for CREB3L4 diverged from the actual validation results, likely due to the small sample size and the high heterogeneity of the samples. It is thus crucial to increase the sample size and enhance sample uniformity to ensure the accuracy of experimental results.
The TIME is crucial in tumor initiation, progression, and treatment response. The immune and stromal scores, calculated using the ESTIMATE algorithm, assess the tumor’s immune and stromal components. Our analysis revealed that the LEG-CREB3L4 group had higher immune and stromal scores than the high-expression group, correlating with a superior Estimate score. This suggests that LUAD patients with LEG-CREB3L4 have a notably better prognosis compared to those with HEG-CREB3L4, consistent with our survival curve analysis.
Despite advancements in LUAD treatment, the complexity of TIME often hampers the effectiveness of targeted immunotherapies. The immune cell landscape within TIME includes neutrophils, eosinophils, and basophils involved in acute inflammation, macrophages, natural killer (NK) cells, and dendritic cells (DCs) integral to innate immunity [33,34]. Various subgroups such as helper T cell 1 (Th1), Th2, Th17 and regulatory T cell (Treg) produced through CD4+ T cell differentiation. Research has shown that M1 macrophages enhance the anti-tumor response of Th1 cells and counteract regulatory immune cells’ suppressive functions [35,36]. DCs, upon activation by external stimuli, migrate to lymphoid organs to initiate B or T cell responses, thereby contributing to adaptive immune anti-tumor activity [37,38]. NK cells, key players in innate immunity, help control infections and tumor growth [39,40]. B cells support T cell-mediated responses through antibody production, cytokine release, and antigen presentation, enhancing anti-tumor effects [41,42]. In our study, we noted significant infiltration of anti-tumor immune cells, including M1 macrophages, DCs, NK cells, and CD4+ and CD8+ T cells, in the TIME of LEG-CREB3L4 LUAD patients. This suggests that patients with LEG-CREB3L4 LUAD may have an enhanced response to immunotherapy, correlating with their improved prognosis. Understanding the dynamics and regulatory mechanisms of the tumor immune microenvironment is crucial for developing new cancer therapies and improving patient outcomes.
Immunotherapy has become a critical advancement in treating LUAD. Particularly, anti-PD-1/PD-L1 inhibitors, as key immune checkpoint inhibitors, have significantly advanced LUAD treatment [43-47]. Our study showed a notable association between high LEG-CREB3L4 expression and immune therapy targets, including PD-L1, CD28, CD80, and CTLA-4, indicating that patients with higher LEG-CREB3L4 expression may respond better to immunotherapy targeting these checkpoints compared to those with lower expression levels. The application of inhibitors against these immune checkpoints could be crucial in treating patients with CREB3L4-associated LUAD. Despite the progress in targeted immunotherapy, challenges such as treatment resistance and maintaining long-term therapeutic effects remain. Future research should aim to develop optimization strategies for immunotherapy in LUAD, aiming to improve patient outcomes and survival rates.
Chemotherapy remains a cornerstone in LUAD management, with significant advancements in recent years enhancing its efficacy. Traditional agents like cisplatin, paclitaxel, and pemetrexed have been standard treatments, but their effectiveness and tolerability are limited. The introduction of new chemotherapy agents, especially targeted therapies such as Gefitinib, Afatinib, and Osimertinib, which focus on EGFR mutations [48-56], and Alectinib and Crizotinib, targeting ALK fusion genes, has sparked renewed hope in LUAD therapy. Our findings indicate that Osimertinib, gefitinib, and Afatinib show increased sensitivity in LEG-CREB3L4 groups, highlighting their potential in treating CREB3L4-associated LUAD and suggesting a link between EGFR mutations and CREB3L4 expression. Additionally, the exploration of combination chemotherapy regimens and novel agents targeting the PI3K/AKT/mTOR and KRAS pathways opens new avenues for LUAD treatment, emphasizing the promise of using Osimertinib, gefitinib, Afatinib, and other agents to target the CREB3L4/RASEF signaling pathway. These developments in chemotherapy research provide broader treatment options for LUAD patients, laying the groundwork for personalized and precision medicine. Despite these advances, current prediction methods for chemotherapy efficacy still have limitations, calling for ongoing research and improvement.
Conclusions
In conclusion, the pathogenesis and progression of LUAD involves a range of molecular and cellular aberrations, leading to changes in the expression of specific markers, including the transcription factor CREB3L4, which is the focus of our investigation. We have observed notable dysregulation of CREB3L4 in LUAD, with initial evidence indicating its role in modulating the disease’s development and progression via its interaction with RASEF. Evaluating CREB3L4 expression in LUAD patients may enhance early detection and improve prognostic accuracy. Moreover, personalizing immunotherapeutic and chemotherapeutic treatments based on CREB3L4 expression could provide a more targeted approach to LUAD management. This study marks the initial identification and reporting of the oncogene CREB3L4 in LUAD. Our findings not only expand our understanding of CREB3L4’s function in LUAD but also offer new perspectives for a deeper comprehension of the disease’s onset and progression.
Acknowledgements
We would like to thank the Department of Pathology and Thoracic Surgery Department of the First Affiliated Hospital of Kunming Medical University for providing LCXW tissue specimens, and we are grateful to Dr. Ma-Li Ju from the Laboratory Department of the First Affiliated Hospital of Kunming Medical University for screening and constructing JT cells. This research was funded by Yunnan Provincial Department of Science and Technology-Kunming Medical University Joint Special Fund for Applied Basic Research (Key Project), grant number 202301AY070001-005; The leader of the “535” high-level talent discipline at the First Affiliated Hospital of Kunming Medical University, grant number 2023535D08; The “High-level Talent Training Support Program” for young top-notch talents in Yunnan Province, grant number YNWR-QNBJ-2019-159; Yunnan Provincial Department of Science and Technology-Basic Research Program-General Project, grant number 202101AT070223; Funding Major Science and Technology Projects of Yunnan Privince/First Affiliated Hospital of Kunming Medical University, grant number 2023zdpy05; and National Natural Science Foundation Regional Science Fund project, grant number 81660388.
Disclosure of conflict of interest
None.
Supplementary Table 1
Supplementary Table 2
References
- 1.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Adams SJ, Stone E, Baldwin DR, Vliegenthart R, Lee P, Fintelmann FJ. Lung cancer screening. Lancet. 2023;401:390–408. doi: 10.1016/S0140-6736(22)01694-4. [DOI] [PubMed] [Google Scholar]
- 3.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, Geisinger K, Hirsch FR, Ishikawa Y, Kerr KM, Noguchi M, Pelosi G, Powell CA, Tsao MS, Wistuba I WHO Panel. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Ma J, Shao J, Zhang S, Liu Z, Yu Y, Li W. Predicting EGFR and PD-L1 status in NSCLC patients using multitask AI system based on CT images. Front Immunol. 2022;13:813072. doi: 10.3389/fimmu.2022.813072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y, Liu X, Qi X, Liu X, Peng F, Li H, Fu H, Pei S, Chen L, Chi X, Zhang L, Zhu X, Song Y, Wang Y, Meng S, Jiang T, Shao S. PDIA6 modulates apoptosis and autophagy of non-small cell lung cancer cells via the MAP4K1/JNK signaling pathway. EBioMedicine. 2019;42:311–325. doi: 10.1016/j.ebiom.2019.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H, Du Z, Li Z, Liu X, Li X, Zhang X, Ma J. MUC1-EGFR crosstalk with IL-6 by activating NF-κB and MAPK pathways to regulate the stemness and paclitaxel-resistance of lung adenocarcinoma. Ann Med. 2024;56:2313671. doi: 10.1080/07853890.2024.2313671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou C, Wang Z, Sun Y, Cao L, Ma Z, Wu R, Yu Y, Yao W, Chang J, Chen J, Zhuang W, Cui J, Chen X, Lu Y, Shen H, Wang J, Li P, Qin M, Lu D, Yang J. Sugemalimab versus placebo, in combination with platinum-based chemotherapy, as first-line treatment of metastatic non-small-cell lung cancer (GEMSTONE-302): interim and final analyses of a double-blind, randomised, phase 3 clinical trial. Lancet Oncol. 2022;23:220–233. doi: 10.1016/S1470-2045(21)00650-1. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, Fang J, Hao X, Zhang S, Liu Y, Wang L, Chen J, Hu Y, Hang X, Li J, Liu C, Zhang Y, Wang Z, Hu Y, Gu K, Huang J, Zhang L, Shan J, Ouyang W, Zhao Y, Zhuang W, Yu Y, Zhao J, Zhang H, Lu P, Li W, Si M, Ge M, Geng H. Safety and activity of WX-0593 (Iruplinalkib) in patients with ALK- or ROS1-rearranged advanced non-small cell lung cancer: a phase 1 dose-escalation and dose-expansion trial. Signal Transduct Target Ther. 2022;7:25. doi: 10.1038/s41392-021-00841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin H, Ning B, Li J, Zhao G, Huang Y, Tian L. Temporal trend of mortality from major cancers in Xuanwei, China. Front Med. 2015;9:487–495. doi: 10.1007/s11684-015-0413-z. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Guo W, Ran J, Tang R, Lin H, Chen X, Ning B, Li J, Zhou Y, Chen LC, Tian L, Huang Y. Five-year lung cancer mortality risk analysis and topography in Xuan Wei: a spatiotemporal correlation analysis. BMC Public Health. 2019;19:173. doi: 10.1186/s12889-019-6490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Sun X, Ren H, Wan X, Huang H, Ma X, Ning B, Zou X, Hu W, Yang G. The mortality patterns of lung cancer between 1990 and 2013 in Xuanwei, China. Lung Cancer. 2015;90:155–160. doi: 10.1016/j.lungcan.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, Wang X, Huang Y, Chen Y, Zhao G, Yao Q, Jin C, Huang Y, Liu X, Li G. Down-regulated SOX4 expression suppresses cell proliferation, metastasis and induces apoptosis in Xuanwei female lung cancer patients. J Cell Biochem. 2015;116:1007–1018. doi: 10.1002/jcb.25055. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Yang JL, Xue ZZ, Cai QC, Hou C, Li HJ, Zhao LX, Zhang Y, Gao CW, Cong L, Wang TZ, Chen DM, Li GS, Luo SQ, Yao Q, Yang CJ, Zhu QS, Cao CH. Effects and mechanism of microRNA-218 against lung cancer. Mol Med Rep. 2021;23:28. doi: 10.3892/mmr.2020.11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Hou C, Zhao LX, Cai QC, Zhang Y, Li DL, Tang Y, Liu HY, Liu YY, Zhang YY, Yang YK, Gao CW, Yao Q, Zhu QS, Cao CH. The association of microRNA-34a with high incidence and metastasis of lung cancer in Gejiu and Xuanwei Yunnan. Front Oncol. 2021;11:619346. doi: 10.3389/fonc.2021.619346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan Y, Li WX, Wang Y, Zhao Y, Shen J, Deng CJ, Li Q, Chen R, Liu X, Zhang YL. Integrated analysis of lncRNAs and mRNAs identifies a potential driver lncRNA FENDRR in lung cancer in Xuanwei, China. Nutr Cancer. 2021;73:983–995. doi: 10.1080/01635581.2020.1779323. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Zhou X, Deng CJ, Zhao Y, Shen J, Wang Y, Zhang YL. Comprehensive analyses of T-UCR expression profiles and exploration of the efficacy of uc.63- and uc.280+ as biomarkers for lung cancer in Xuanwei, China. Pathol Res Pract. 2020;216:152978. doi: 10.1016/j.prp.2020.152978. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Lu LJ, Duan Y, Liu X, Mao Y, Chen Y, Zhang YL. Analysis of circular RNA expression profiles of lung cancer in Xuanwei, China. J Clin Lab Anal. 2020;34:e23521. doi: 10.1002/jcla.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv L, Liu Z, Liu Y, Zhang W, Jiang L, Li T, Lu X, Lei X, Liang W, Lin J. Distinct EGFR mutation pattern in patients with non-small cell lung cancer in Xuanwei region of China: a systematic review and meta-analysis. Front Oncol. 2020;10:519073. doi: 10.3389/fonc.2020.519073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Duan Y, Meng QH, Gong R, Guo C, Zhao Y, Zhang Y. Integrated analysis of DNA methylation profiling and gene expression profiling identifies novel markers in lung cancer in Xuanwei, China. PLoS One. 2018;13:e0203155. doi: 10.1371/journal.pone.0203155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben Aicha S, Lessard J, Pelletier M, Fournier A, Calvo E, Labrie C. Transcriptional profiling of genes that are regulated by the endoplasmic reticulum-bound transcription factor AIbZIP/CREB3L4 in prostate cells. Physiol Genomics. 2007;31:295–305. doi: 10.1152/physiolgenomics.00097.2007. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Z, Shi B, Zhang Y, Yu T, Cheng Y, Zhu J, Zhang G, Zhong M, Hu S, Ma X. CREB3L4 promotes hepatocellular carcinoma progression and decreases sorafenib chemosensitivity by promoting RHEB-mTORC1 signaling pathway. iScience. 2024;27:108843. doi: 10.1016/j.isci.2024.108843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TH, Park JM, Kim MY, Ahn YH. The role of CREB3L4 in the proliferation of prostate cancer cells. Sci Rep. 2017;7:45300. doi: 10.1038/srep45300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pu Q, Lu L, Dong K, Geng WW, Lv YR, Gao HD. The novel transcription factor CREB3L4 contributes to the progression of human breast carcinoma. J Mammary Gland Biol Neoplasia. 2020;25:37–50. doi: 10.1007/s10911-020-09443-6. [DOI] [PubMed] [Google Scholar]
- 24.Yu X, Fang Z, Li G, Zhang S, Liu M, Wang Y. High RASEF expression is associated with a significantly better prognosis in colorectal cancer. Int J Clin Exp Pathol. 2018;11:4276–4282. [PMC free article] [PubMed] [Google Scholar]
- 25.Maat W, Beiboer SH, Jager MJ, Luyten GP, Gruis NA, van der Velden PA. Epigenetic regulation identifies RASEF as a tumor-suppressor gene in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49:1291–1298. doi: 10.1167/iovs.07-1135. [DOI] [PubMed] [Google Scholar]
- 26.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 28.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Xue Q, Pan G, Meng QH, Tuo X, Cai X, Chen Z, Li Y, Huang T, Duan X, Duan Y. Integrated analysis of genome-wide copy number alterations and gene expression profiling of lung cancer in Xuanwei, China. PLoS One. 2017;12:e0169098. doi: 10.1371/journal.pone.0169098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Song C, Tian Y, Yang X. Single-cell RNA sequencing in lung cancer: revealing phenotype shaping of stromal cells in the microenvironment. Front Immunol. 2022;12:802080. doi: 10.3389/fimmu.2021.802080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, McGraw T, Mittal V. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19:9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Y, Yu Y, Wang X, Zhang T. Tumor-associated macrophages in tumor immunity. Front Immunol. 2020;11:583084. doi: 10.3389/fimmu.2020.583084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen D, Zhang X, Li Z, Zhu B. Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages. Theranostics. 2021;11:1016–1030. doi: 10.7150/thno.51777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardner A, Ruffell B. Dendritic cells and cancer immunity. Trends Immunol. 2016;37:855–865. doi: 10.1016/j.it.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen M, Giladi A, Barboy O, Hamon P, Li B, Zada M, Gurevich-Shapiro A, Beccaria CG, David E, Maier BB, Buckup M, Kamer I, Deczkowska A, Le Berichel J, Bar J, Iannacone M, Tanay A, Merad M, Amit I. The interaction of CD4+ helper T cells with dendritic cells shapes the tumor microenvironment and immune checkpoint blockade response. Nat Cancer. 2022;3:303–317. doi: 10.1038/s43018-022-00338-5. [DOI] [PubMed] [Google Scholar]
- 39.Wu SY, Fu T, Jiang YZ, Shao ZM. Natural killer cells in cancer biology and therapy. Mol Cancer. 2020;19:120. doi: 10.1186/s12943-020-01238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17:1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 41.Hao D, Han G, Sinjab A, Gomez-Bolanos LI, Lazcano R, Serrano A, Hernandez SD, Dai E, Cao X, Hu J, Dang M, Wang R, Chu Y, Song X, Zhang J, Parra ER, Wargo JA, Swisher SG, Cascone T, Sepesi B, Futreal AP, Li M, Dubinett SM, Fujimoto J, Solis Soto LM, Wistuba II, Stevenson CS, Spira A, Shalapour S, Kadara H, Wang L. The single-cell immunogenomic landscape of B and plasma cells in early-stage lung adenocarcinoma. Cancer Discov. 2022;12:2626–2645. doi: 10.1158/2159-8290.CD-21-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song P, Li W, Wu X, Qian Z, Ying J, Gao S, He J. Integrated analysis of single-cell and bulk RNA-sequencing identifies a signature based on B cell marker genes to predict prognosis and immunotherapy response in lung adenocarcinoma. Cancer Immunol Immunother. 2022;71:2341–2354. doi: 10.1007/s00262-022-03143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, Kiialainen A, Hanhart J, Schill C, Hess C, Savic Prince S, Wiese M, Lardinois D, Ho PC, Klein C, Karanikas V, Mertz KD, Schumacher TN, Zippelius A. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ajona D, Ortiz-Espinosa S, Moreno H, Lozano T, Pajares MJ, Agorreta J, Bértolo C, Lasarte JJ, Vicent S, Hoehlig K, Vater A, Lecanda F, Montuenga LM, Pio R. A combined PD-1/C5a blockade synergistically protects against lung cancer growth and metastasis. Cancer Discov. 2017;7:694–703. doi: 10.1158/2159-8290.CD-16-1184. [DOI] [PubMed] [Google Scholar]
- 45.Creelan BC, Wang C, Teer JK, Toloza EM, Yao J, Kim S, Landin AM, Mullinax JE, Saller JJ, Saltos AN, Noyes DR, Montoya LB, Curry W, Pilon-Thomas SA, Chiappori AA, Tanvetyanon T, Kaye FJ, Thompson ZJ, Yoder SJ, Fang B, Koomen JM, Sarnaik AA, Chen DT, Conejo-Garcia JR, Haura EB, Antonia SJ. Tumor-infiltrating lymphocyte treatment for anti-PD-1-resistant metastatic lung cancer: a phase 1 trial. Nat Med. 2021;27:1410–1418. doi: 10.1038/s41591-021-01462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng L, Wang Y, Liu F, Qiu X, Zhang X, Fang C, Qian X, Li Y. Peripheral blood markers predictive of outcome and immune-related adverse events in advanced non-small cell lung cancer treated with PD-1 inhibitors. Cancer Immunol Immunother. 2020;69:1813–1822. doi: 10.1007/s00262-020-02585-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B, Hu X, Feng K, Gao R, Xue Z, Zhang S, Zhang Y, Corse E, Hu Y, Han W, Zhang Z. Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat Cancer. 2022;3:108–121. doi: 10.1038/s43018-021-00292-8. [DOI] [PubMed] [Google Scholar]
- 48.Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu DT, Saijo N, Duffield EL, Rukazenkov Y, Speake G, Jiang H, Armour AA, To KF, Yang JC, Mok TS. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J. Clin. Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 49.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T North-East Japan Study Group. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 50.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, Asami K, Katakami N, Takada M, Yoshioka H, Shibata K, Kudoh S, Shimizu E, Saito H, Toyooka S, Nakagawa K, Fukuoka M West Japan Oncology Group. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 51.Sequist LV, Yang JC, Yamamoto N, O’Byrne K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, Su WC, Bennouna J, Kato T, Gorbunova V, Lee KH, Shah R, Massey D, Zazulina V, Shahidi M, Schuler M. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J. Clin. Oncol. 2013;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 52.Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, Li W, Hou M, Shi JH, Lee KY, Xu CR, Massey D, Kim M, Shi Y, Geater SL. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213–222. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 53.Park K, Tan EH, O’Byrne K, Zhang L, Boyer M, Mok T, Hirsh V, Yang JC, Lee KH, Lu S, Shi Y, Kim SW, Laskin J, Kim DW, Arvis CD, Kölbeck K, Laurie SA, Tsai CM, Shahidi M, Kim M, Massey D, Zazulina V, Paz-Ares L. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016;17:577–589. doi: 10.1016/S1470-2045(16)30033-X. [DOI] [PubMed] [Google Scholar]
- 54.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, Okamoto I, Zhou C, Cho BC, Cheng Y, Cho EK, Voon PJ, Planchard D, Su WC, Gray JE, Lee SM, Hodge R, Marotti M, Rukazenkov Y, Ramalingam SS FLAURA Investigators. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 55.Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, Zhou C, Reungwetwattana T, Cheng Y, Chewaskulyong B, Shah R, Cobo M, Lee KH, Cheema P, Tiseo M, John T, Lin MC, Imamura F, Kurata T, Todd A, Hodge R, Saggese M, Rukazenkov Y, Soria JC FLAURA Investigators. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med. 2020;382:41–50. doi: 10.1056/NEJMoa1913662. [DOI] [PubMed] [Google Scholar]
- 56.Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WS, Lee CK, Sebastian M, Templeton A, Mann H, Marotti M, Ghiorghiu S, Papadimitrakopoulou VA AURA3 Investigators. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.