Abstract
Human leukocyte antigen-G (HLA-G) is linked to the development of human malignancies via immune escape mechanisms. The chief variations for HLA-G were found in three prime untranslated regions (3’UTR). The current study aims to evaluate the distribution of HLA-G rs1063320 (G+3142 C>G), HLA-G*01:03, and HLA-G*01:05N polymorphisms with risk of acute lymphocytic leukemia (ALL) in Saudi Arabia. This case-control study analyzed 232 samples from 117 patients with ALL and 115 healthy controls (HCN) using the PCR-RFLP method. Associations between HLA-G and ALL risk were analyzed using allele contrasts. The HLAG rs1063320 G+3142 C>G polymorphism results showed a reduced risk of ALL in the dominant G/C model odds ratios (OR) = 0.34, 95% confidence interval (CI) = 0.12-0.98, P = 0.041), stratified by age. However, those stratified by gender, showed decreased risk of ALL in all genetic inheritance models tested: codominant model C/G versus G/G (OR = 0.24, 95% CI = 0.06-0.99), C/C versus G/G (OR = 0.01, 95% CI = 0.00-0.12), P = 0.0001), and dominant model C/G-C/C versus G/G (OR = 0.12, 95% CI = 0.03-0.47, P = 0.000004), and the recessive model C/C versus G/G-C/G (OR = 0.03, 95% CI = 0.00-0.24, P = 0.0001), log-additive (OR = 0.12, 95% CI = 0.04-0.35, P = 0.0001). Conversely, the G*01:03 allele was not found in ALL or HCN, whereas the G*01:05N allele showed polymorphic frequencies that were not significant. In conclusion, the HLA-G +3142 C>G polymorphism significantly decreased the prevalence of ALL stratified by gender and age polymorphisms in the risk of ALL in the pediatric Saudi population.
Keywords: Acute lymphocytic leukemia, HLA-G polymorphisms, genotyping, non-classical HLA , RFLP, Saudi Arabia
Introduction
Cancer ranks as the second most prevalent cause of death on a global scale after heart disease, thus it imposes a notable economic burden on both developed and developing nations. In addition, approximately 1,918,030 novel cancer cases and 609,360 deaths due to malignant tumors have occurred in the United States alone [1,2]. Cancer is becoming increasingly common in the Middle East at an astonishing rate. The top five cancers in children in the Arab World in 2020 were identical to those reported globally [3,4]. Childhood cancer ranks as the second most prevalent cause of mortality, and leukemia accounts for 28% of all cases [1]. More than 18,000 children in Arab countries are annually diagnosed with cancer [4]. Leukemia is one of the five most common cancers in Saudi Arabia [5].
The HLA-G molecule differs from HLA class I genes by having few alternative molecular codes. Although the coding region is the most conserved [6,7]. The HLA-G molecules function as immunological checkpoints [8], regulating immune responses by binding to receptors on immune cells, including the inhibitory receptors ILT2 and ILT4 [9]. HLA-G expression plays a vital role in preserving the tolerance between mother and fetus and may impact the development of many diseases, such as autoimmune disorders, malignancies, and viral infections [10,11]. The HLA-G rs1063320 SNP G +3142 C>G is located in the 3’-UTR [12]. The presence of G at position +3142 enhances its affinity for three microRNAs (miRNA-148a, miRNA-148b, and miRNA-152) and decreases HLA-G mRNA stability and production [13]. Several studies in different populations have investigated the association between corroborating the unfavorable role of G+3142 C>G and cancer risk, including cervico-vaginal cancer [14], papillomavirus [15], cervical squamous cell carcinoma [16], breast cancer [17,18], thyroid tumors [19], and colorectal cancer [20].
The HLA-G*01:05N allele is a well-known part of the human leukocyte antigen (HLA) system [21]. It is known for its part in controlling the immune system and its effects on many diseases. This allele is classified as a null variant due to a specific mutation that leads to a truncated protein product, significantly affecting its functionality [22]. HLA-G*01:05N is distinguished by the deletion of a single base, spliced HLAG4 isoforms, and full-length soluble isoforms HLA-G5 [21]. The HLA-G*01:05N null allele is distinguished by a single base-pair deletion in exon 3 [23], it blocks HLA-G1 and -G5 translation, in exon 5 in the codon 297 (TAG) [24]. The reduced expression of HLA-G associated with the *01:05N variant may cause altered immunological responses, possibly increasing the susceptibility to certain diseases or influencing transplant results [9,21].
Several investigations have examined the impact of HLA-G polymorphisms on the chance of developing leukemia; however, the results have been inconsistent [15,25]. In CLL, HLA-G expression is linked to impaired immunodeficiency in tumor cells [26]. In acute leukemia, soluble HLA-G levels are increased [27]. HLA-G expression has been identified as a prognostic marker in B-CLL [28-30], and ALL [31]. This study aimed to evaluate the genetic polymorphisms of HLA-G rs1063320 (G+3142 C>G), G*01:03, and G*01:05N alleles in patients of Saudi Arabia with ALL.
Materials and methods
Ethics statement
The ethics committee of King Khaled Hospital University (KKHU) in Riyadh, Saudi Arabia, gave its approval to this research project prior to its implementation. The ethical norms of King Saud University’s Institutional Review Board (IRB) were adhered to throughout the course of this investigation (IRB code: E-20-5346).
Participants
The participants in this study were 117 pediatric patients with ALL (male = 73 and female = 44), identified between April 2017 and March 2020. The healthy control (HCN) group consisted of 115 ethnically matched individuals (male = 77 and female = 38). As a retrospective study, control patients had no diseases linked to any form of cancer and no history of immune disorders.
Genomic DNA extraction
Placed in EDTA vials, three ml of blood was taken from individuals who were diagnosed with ALL and from HCN. Following the directions provided by the manufacturer, genomic DNA was extracted with the use of the DNeasy Blood & Tissue Kit which was manufactured by QIAGENE GmbH, Germany. In this investigation, both forward and reverse primers sequences were used, and they are included in Table 1.
Table 1.
PCR-RFLP for HLA-G rs1063320 (G+3142 C>G), G*01:03 and G*01:05N
| HLA-G | Primers | RE | Product size (bp) | Specificities |
|---|---|---|---|---|
| rs1063320 (G+3142 C>G) | F: 5’-CATGCTGAACTGCATTCCTTCC-3’ | BseSI | 460 | C |
| R: 5’-CTGGGCCGGAGTTACTCACT-3’ | 316+90 | G | ||
| G*01:03 (Exon 2) | F: 5’-TCCATGAGGTATTTCAGCGC/G-3’ | HinfI | 79+125+27 | G*01:03 |
| R: 5’-CTGGGCCGGAGTTACTCACT-3’ | 106+175 | All but G*01:03 | ||
| G*01:05N (Exon 3) | F: 5’-CACACCCTCCAGTGGATGAT-3’ | PpuMI | 276 | G*01:05N |
| R: 5’-GGTACCCGCGCGCTGCAGCA-3’ | 108+168 | All but G*01:05N |
F, forward; R, reverse; rs, reference SNP cluster ID; RE, restriction endonuclease; bp, base pair.
Genotyping for HLA-G
Genotyping of HLA-G involves the analysis of genetic variations within the HLA-G gene to understand its polymorphisms and their associations with ALL. Using the PCR-RFLP technique, the genotyping of HLA-G rs1063320 (G+3142 C>G) as well as two alleles (HLA-G*01:03 and HLA-G*01:05N) was carried out. In brief, the reaction mix for HLA-G +3142 C>G comprised of the following components: DNA (1 μL), 7.5 μL 2X (GoTaq® Master Mixes, Promega, USA), forward and reverse primers (0.5 μL), and nuclease-free water. The total volume of the reaction mix was 15 μL. The thermocycling settings were as follows: a three-minute initial denaturation at 95°C was followed by thirty seconds of denaturation at 94°C, thirty seconds of annealing at 57-65°C, forty-five seconds of elongation at 72°C, and finally five minutes of final elongation at 72°C.
The digestion products were separated on 2.5% agarose by gel electrophoresis (Bio-Rad Laboratories, GmbH, Munchen). The bands were stained with ethidium bromide (Sigma-Aldrich, US) to ensure that the primers were working and that alleles were present or absent, and they were visualized by a UV trans-illuminator (BioDocAnalyze, Biometra GmbH, Germany).
The specific enzymes used for digestion were BseSI (rs1063320), Hinf-I (G*01:03), and PpuM-I (G*01:05N). The procedures were conducted according to the instructions provided by the manufacturer (Thermo Fisher Scientific).
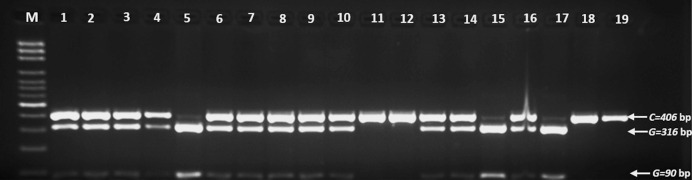
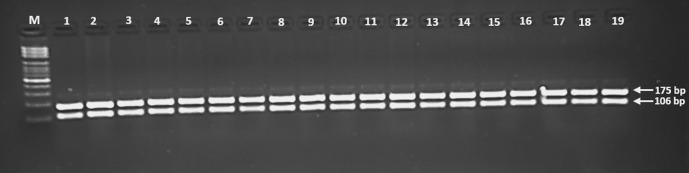
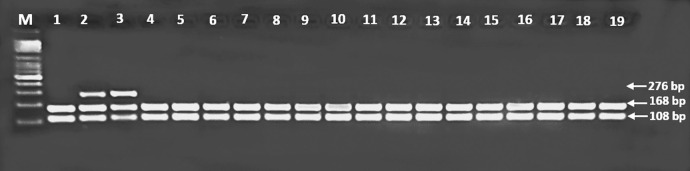
The digested products were separated by electrophoresis using 2.5% agarose gels. HLA-G rs1063320 (G+3142 C>G) was defined as a band of 406 bp. After digestion with BseS-I, three bands were observed at 406, 316, and 90 bp (Figure 1). In addition, for HLA-G*01:03, two bands of 106 and 175 bp were found after digestion with Hinf-I (Figure 2). While HLA-G*01:05N was defined by a 276 bp band after PpuMI digestion, three bands of 108, 168, and 276 bp were observed (Figure 3).
Figure 1.
A photograph of the PCR-RFLP products of HLA-G rs1063320 (G+3142 C>G); G allele was digested with BseS-I, three bands were produced: 406 bp C allele, 316 and 90 bp G allele; lanes 1-4 and 6-10, as well as 13, 14, and 16: CG; 5, 15, and 17: GG; 11, 12, 18, and 19: CC; Lane M: 100 bp DNA Marker.
Figure 2.
A photograph of the PCR-RFLP products of HLA-G*01:03 allele, digested with Hinf-I, shows two bands (106 and 175 bp); lanes 1-19 show the absence of HLA-G*01:03 allele; lane M: 100 bp DNA marker.
Figure 3.
A photograph of HLA-G*01:05N, PCR-RFLP products digested with PpuM-I; Lanes 2 and 3 show heterozygous HLA-G*01:05N allele; lanes 1 and 4-19 show homozygote, absence of HLA-G*01:05N allele; Lane M: 100 bp DNA marker.
Statistical analysis
In control subjects, the Hardy-Weinberg equilibrium (HWE) was tested for genotype frequencies. We used the two-tailed Fisher’s exact test; HLA-G (G+3142 C>G, G*01:03, and G*01:05N) was tested in terms of numerical values and percentages and was used to establish the correlation between alleles. To evaluate the risk of ALL, OR and 95% CI, and p-value ≤ 0.05 were determined.
The analysis methods utilized in this study effectively elucidated the relationship between HLA-G genetic variants and ALL susceptibility, by employing SNP genotyping, using SNPStats software [32], statistical modeling, and stratification by age and sex.
Results
In total, 232 patients were enrolled in this study, including 117 patients with ALL and 115 unrelated HCN. We investigated the HLA-G rs1063320 (G+3142 C>G) SNP and the variants of two alleles, G*01:03 and G*01:05N, in the 3’-UTR HLA-G polymorphic sites. The genotyping results for the G+3142 C>G polymorphism are shown in Figure 1. The G*01:03 allele was not present with ALL or HCN. Contrarily, the G*01:05N allele exhibited variable frequencies and distribution with ALL compared to HCN.
The allele frequencies of G+3142 C>G, G*01:03, and G*01:05N with ALL and in HCN are shown in Table 2. The G+3142 G SNP was less common with ALL (55.0%) than in those in the HCN group (63.3%). The G+3142 SNP was more prevalent with ALL (45.0%) than in those in the HCN group (36.7%). The results of G+3142 C>G are presented in Table 3. In addition, we conducted tests categorized by age, and the findings are shown in Table 4. In the dominant model, the SNP G+3142 C>G showed a substantial age-dependent difference. The SNP G+3142 C>G was found to reduce the risk of ALL in patients older than 18 years in the dominant group.
Table 2.
Frequency of HLA-G +3142 C>G SNP, G*01:03 and G*01:05N alleles in patients with ALL and HCN
| HLA-G alleles | HCN | ALL | X2 | OR | 95% CI | P-value | |
|---|---|---|---|---|---|---|---|
| N = 115 | N = 117 | ||||||
| Alleles (%) | |||||||
| G*01:03 | ACG | 230 (100.0%) | 234 (100.0%) | NS | NS | NS | NS |
| TCG | 0.0 (100.0%) | 0.0 (100.0%) | |||||
| G*01:05N | CTG | 223 (97.0%) | 230 (98.3%) | 0.89 | 1.80 | (0.52-6.25) | 0.38 |
| -TG | 7 (3.0%) | 4 (1.7%) | 0.55 | (0.16-1.92) | |||
ALL, acute lymphoblastic leukemia; HCN, healthy control; N, number of individuals; Allele, allele frequency; OR, odds ratio; CI, confidence interval; -TG, deletion C from codon 130 position 1597 in exon 3 in G*01:05N; TCG, Sequences at codon 31 encode a serine in G*01:03 and ACG encode a threonine in all but G*01:03. Bold letters indicate the positions of amino acid variability.
Table 3.
Genotypic association of the HLA-G +3142 C>G polymorphism in patients with ALL and HCN, displaying genetic models: codominant, dominant, recessive, over dominant, and log-additive
| Model | Allele/Genotype | ALL | HCN | OR (95% CI) | P-value | AIC |
|---|---|---|---|---|---|---|
| G+3142 | G | 77 (55.0%) | 105 (63.3%) | 0.71 (0.44-1.12) | 0.16 | |
| C | 63 (45.0%) | 61 (36.7) | 1.41 (0.89-2.23) | |||
| Codominant | G/G | 22 (31.4%) | 33 (39.8%) | 1.00 | 0.33 | 214.8 |
| C/G | 33 (47.1%) | 39 (47%) | 0.79 (0.39-1.60) | |||
| C/C | 15 (21.4%) | 11 (13.2%) | 0.49 (0.19-1.26) | |||
| Dominant | G/G | 22 (31.4%) | 33 (39.8%) | 1.00 | 0.28 | 213.8 |
| C/G-C/C | 48 (68.6%) | 50 (60.2%) | 0.69 (0.36-1.36) | |||
| Recessive | G/G-C/G | 55 (78.6%) | 72 (86.8%) | 1.00 | 0.18 | 213.2 |
| C/C | 15 (21.4%) | 11 (13.2%) | 0.56 (0.24-1.32) | |||
| Over dominant | G/G-C/C | 37 (52.9%) | 44 (53%) | 1.00 | 0.98 | 215 |
| C/G | 33 (47.1%) | 39 (47%) | 0.99 (0.53-1.88) | |||
| Log-additive | --- | --- | --- | 0.71 (0.45-1.13) | 0.15 | 212.9 |
AIC, Akaike information criterion.
Table 4.
Correlations between HLA-G +3142 C>G polymorphism and susceptibility in ALL patients stratified by age
| Model | Genotype | ALL (Age ≤ 18) | ALL (Age >18) | OR (95% CI) | P-value | AIC | BIC |
|---|---|---|---|---|---|---|---|
| Codominant | G/G | 8 (21.1%) | 14 (43.8%) | 1.00 | 0.11 | 98.2 | 104.9 |
| C/G | 20 (52.6%) | 13 (40.6%) | 0.37 (0.12-1.13) | ||||
| C/C | 10 (26.3%) | 5 (15.6%) | 0.29 (0.07-1.14) | ||||
| Dominant | G/G | 8 (21.1%) | 14 (43.8%) | 1.00 | 0.041 | 96.4 | 100.8 |
| C/G-C/C | 30 (79%) | 18 (56.2%) | 0.34 (0.12-0.98) | ||||
| Recessive | G/G-C/G | 28 (73.7%) | 27 (84.4%) | 1.00 | 0.27 | 99.3 | 103.8 |
| C/C | 10 (26.3%) | 5 (15.6%) | 0.52 (0.16-1.72) | ||||
| Over dominant | G/G-C/C | 18 (47.4%) | 19 (59.4%) | 1.00 | 0.32 | 99.5 | 104 |
| C/G | 20 (52.6%) | 13 (40.6%) | 0.62 (0.24-1.59) | ||||
| Log-additive | --- | --- | --- | 0.51 (0.26-1.02) | 0.051 | 96.7 | 101.2 |
BIC, Bayesian information criterion. The significant p-values are indicated in bold.
Table 5 shows our findings from testing the inheritance models for the genotype frequencies of the G+3142 C>G SNP and susceptibility in patients with ALL stratified by sex. The G+3142 C>G variant decreased the risk for ALL in males in the codominant model C/G VS G/G (OR = 0.24, 95% CI = 0.06-0.99) and C/C versus G/G (OR = 0.01, 95% CI = 0.00-0.12), P = 0.0001. In addition, the dominant model was C/G-C/C VS G/G (OR = 0.12, 95% CI = 0.03-0.47, P = 0.000004). The recessive model was C/C VS G/G-C/G (OR = 0.03, 95% CI = 0.00-0.24, P = 0.0001), and log-additive (OR = 0.12, 95% CI = 0.04-0.35, P = 0.0001).
Table 5.
Correlations between HLA-G +3142 C>G polymorphism and susceptibility in patients with ALL stratified by sex
| Model | Genotype | ALL Female | ALL Male | OR (95% CI) | P-value | AIC | BIC |
|---|---|---|---|---|---|---|---|
| Codominant | G/G | 3 (10%) | 19 (47.5%) | 1.00 | <0.0001 | 75.1 | 81.9 |
| C/G | 13 (43.3%) | 20 (50%) | 0.24 (0.06-0.99) | ||||
| C/C | 14 (46.7%) | 1 (2.5%) | 0.01 (0.00-0.12) | ||||
| Dominant | G/G | 3 (10%) | 19 (47.5%) | 1.00 | 5e-04 | 87.3 | 91.8 |
| C/G-C/C | 27 (90%) | 21 (52.5%) | 0.12 (0.03-0.47) | ||||
| Recessive | G/G-C/G | 16 (53.3%) | 39 (97.5%) | 1.00 | <0.0001 | 77.7 | 82.2 |
| C/C | 14 (46.7%) | 1 (2.5%) | 0.03 (0.00-0.24) | ||||
| Over dominant | G/G-C/C | 17 (56.7%) | 20 (50%) | 1.00 | 0.58 | 99.3 | 103.8 |
| C/G | 13 (43.3%) | 20 (50%) | 1.31 (0.50-3.39) | ||||
| Log-additive | --- | --- | --- | 0.12 (0.04-0.35) | <0.0001 | 74.7 | 79.2 |
The significant p-values are indicated in bold.
Discussion
HLA-G has been implicated in various pathological conditions, including cancer and autoimmune diseases. Its expression can be upregulated in tumors, contributing to immune evasion and tumor progression. Studies have shown that HLA-G can facilitate a tumor-driven immune escape mechanism, making it a potential target for therapeutic interventions in oncology.
This study looked at how variations in the HLA-G gene, specifically the HLA-G +3142 C>G polymorphism and two alleles, G01:03 and G01:05N, are linked to the risk of getting ALL in healthy, unrelated people. These findings provide notable and significant insights into the genetic determinants that may potentially impact ALL’s vulnerability.
This is the first study to investigate the effects of the G+3142 C>G SNP, G*01:03, and G*0105N allele polymorphisms on the risk of ALL in the pediatric Saudi population. The genotyping findings for the HLA-G +3142 C>G indicated that the G+3142 G allele was less frequent patients with ALL (55.0%) than that in HCN (63.3%), whereas the G+3142 C allele was more common with ALL (45.0%) than in HCN (36.7%). The p-values and ORs in Table 2 indicate that these differences were not statistically significant. This finding implies that the previously aforementioned polymorphism may not have a significant influence on the predisposition to ALL within the Saudi population. Results of this study were consistent with that of our previous studies where [33]. This frequency matches with the findings in other reports [17] and [34].
Our results showed a high risk between the G+3142 C>G and susceptibility in patients with ALL by age in the genetic dominant inheritance model only, which showed a decrease in the risk of ALL in the Saudi population. This was maintained only in the dominant model (C/G-C/C) after stratification by age ≤ 18 years (OR = 0.34, 95% CI = 0.12-0.98, P = 0.041). Conversely, our study results showed a significant association stratified by sex, and showed a protective effect against ALL in all inheritance models. The calculated OR suggests that the homozygous G/G and heterozygous C/G genotypes have a protective effect against ALL. Our findings suggest that the G+3142 C>G mutation may have a protective effect in patients with ALL. Therefore, the results of our study are consistent with another study [35]. This is also consistent with the findings of other researchers [36], who found that G+3142 G>C decreased the risk of rheumatoid arthritis in an Iranian patients. Another study [37] showed that none of the HLA-G variant locations studied were linked to susceptibility to AML. Contrarily, additional research [21] found that 3’-UTR HLA-G alleles are a risk for the development of CML. Another study [31] also mentioned that HLA-G expression in patients with ALL. Similarly, [38] it was observed that acute leukemia increases soluble HLA-G levels.
Saudi Arabia’s population exhibits substantial genetic diversity, influenced by its complex demographic structure, which includes a mix of native and immigrant populations. This study examined two HLA-G alleles, G*01:03 and G*01:05N. The lack of the G*01:03 allele in persons with acute lymphoblastic leukemia (ALL) or healthy controls (HCN) indicates that it is uncommon among the Saudi population. This discovery is consistent with broader genetic studies that emphasize the substantial variation and distinct allele frequencies observed in various populations, especially in locations characterized by high levels of consanguinity, like Saudi Arabia [39]. Consanguineous marriages are quite common in South Africa (about 57.7%), contributing to a unique genetic environment. This genetic background is critical for understanding the distribution of many alleles, including those of the HLA-G gene, which contains the G*01:03 variant [40-42].
Recent study data has shown that the G01:03 allele is not found in patients with inflammatory bowel disease (IBD) [43], suggesting a low prevalence in both populations. Other alleles, such as G01:04 and G01:05N, had polymorphism frequencies and were linked to a higher risk of disorders such as ulcerative colitis (UC) and Crohn’s disease (CD) [44]. The lack of G01:03 in both ALL and HCN populations shows that it may not have a substantial role in the genetic susceptibility to both diseases in the Saudi population.
In conclusion, this study provides evidence suggesting a potential correlation between the G+3142 C>G SNP and ALL susceptibility in the Saudi population. This association was further influenced by the stratification according to age and sex. The observed protective effect of this genetic variation seems to be more prominent among younger individuals and females. Nevertheless, further investigations using more extensive sample sizes and a broader range of populations are necessary to validate these discoveries and provide a comprehensive understanding of the genetic predisposition to ALL. Understanding the genetic elements that contribute to the risk of acute ALL may have implications for disease prevention and personalized treatment strategies in the future.
Acknowledgements
This work was supported by Research Supporting Project number (RSP 2024R75), at King Saud University, Riyadh, Saudi Arabia.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Alhakak A, Østergaard L, Butt JH, Vinther M, Philbert BT, Jacobsen PK, Yafasova A, Torp-Pedersen C, Køber L, Fosbøl EL, Mogensen UM, Weeke PE. Cause-specific death and risk factors of 1-year mortality after implantable cardioverter-defibrillator implantation: a nationwide study. Eur Heart J Qual Care Clin Outcomes. 2022;8:39–49. doi: 10.1093/ehjqcco/qcaa074. [DOI] [PubMed] [Google Scholar]
- 3.Al-Shamsi HO, Abu-Gheida IH, Iqbal F, Al-Awadhi A. Cancer in the Arab world. Springer Nature. 2022 [Google Scholar]
- 4.Zandaki Da, Sultan I. Pediatric Oncology in the Arab World. Cancer in the Arab World. Springer; 2022. pp. 409–425. [Google Scholar]
- 5.Alanezi N, Abdalhabib E, Alfayez A, Alsalman D, Alanezi F, Al-Rayes S, Alyousef S, AlNujaidi H, Al-Saif AK, Attar R. Knowledge and awareness of leukaemia and its risks among the population of Saudi Arabia. Inform Med Unlocked. 2022:100971. [Google Scholar]
- 6.Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res. 2014;2014:734068. doi: 10.1155/2014/734068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amodio G, Gregori S. HLA-G genotype/expression/disease association studies: success, hurdles, and perspectives. Front Immunol. 2020;11:1178. doi: 10.3389/fimmu.2020.01178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: an immune checkpoint molecule. Adv Immunol. 2015;127:33–144. doi: 10.1016/bs.ai.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Martín-Villa JM, Vaquero-Yuste C, Molina-Alejandre M, Juarez I, Suárez-Trujillo F, López-Nares A, Palacio-Gruber J, Barrera-Gutiérrez L, Fernández-Cruz E, Rodríguez-Sainz C, Arnaiz-Villena A. HLA-G: too much or too little? Role in cancer and autoimmune disease. Front Immunol. 2022;13:796054. doi: 10.3389/fimmu.2022.796054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhuang B, Shang J, Yao Y. HLA-G: an important mediator of maternal-fetal immune-tolerance. Front Immunol. 2021;12:744324. doi: 10.3389/fimmu.2021.744324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira LMR, Meissner TB, Tilburgs T, Strominger JL. HLA-G: at the interface of maternal-fetal tolerance. Trends Immunol. 2017;38:272–286. doi: 10.1016/j.it.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Martelli-Palomino G, Pancotto JA, Muniz YC, Mendes-Junior CT, Castelli EC, Massaro JD, Krawice-Radanne I, Poras I, Rebmann V, Carosella ED, Rouas-Freiss N, Moreau P, Donadi EA. Polymorphic sites at the 3’untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PLoS One. 2013;8:e71742. doi: 10.1371/journal.pone.0071742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manaster I, Goldman-Wohl D, Greenfield C, Nachmani D, Tsukerman P, Hamani Y, Yagel S, Mandelboim O. MiRNA-mediated control of HLA-G expression and function. PLoS One. 2012;7:e33395. doi: 10.1371/journal.pone.0033395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva ID, Muniz YC, Sousa MC, Silva KR, Castelli EC, Filho JC, Osta AP, Lima MI, Simões RT. HLA-G 3’UTR polymorphisms in high grade and invasive cervico-vaginal cancer. Hum Immunol. 2013;74:452–458. doi: 10.1016/j.humimm.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 15.Rizzo R, Audrito V, Vacca P, Rossi D, Brusa D, Stignani M, Bortolotti D, D’Arena G, Coscia M, Laurenti L, Forconi F, Gaidano G, Mingari MC, Moretta L, Malavasi F, Deaglio S. HLA-G is a component of the CLL escape repertoire to generate immune suppression: impact of HLA-G 14 bp (rs66554220) polymorphism. Haematologica. 2014;99:888–96. doi: 10.3324/haematol.2013.095281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang YC, Chang TY, Chen TC, Lin WS, Chang SC, Lee YJ. Human leucocyte antigen-G polymorphisms are associated with cervical squamous cell carcinoma risk in Taiwanese women. Eur J Cancer. 2014;50:469–474. doi: 10.1016/j.ejca.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Zidi I, Dziri O, Zidi N, Sebai R, Boujelebene N, Ben Hassine A, Ben Yahia H, Laaribi AB, Babay W, Rifi H, Mezlini A, Chelbi H. Association of HLA-G +3142 C>G polymorphism and breast cancer in Tunisian population. Immunol Res. 2016;64:961–968. doi: 10.1007/s12026-015-8782-6. [DOI] [PubMed] [Google Scholar]
- 18.Ouni N, Chaaben AB, Kablouti G, Ayari F, Douik H, Abaza H, Gara S, Elgaaied-Benammar A, Guemira F, Tamouza R. The impact of HLA-G 3’UTR polymorphisms in breast cancer in a Tunisian population. Immunol Invest. 2019;48:521–532. doi: 10.1080/08820139.2019.1569043. [DOI] [PubMed] [Google Scholar]
- 19.de Figueiredo-Feitosa NL, Martelli Palomino G, Ciliao Alves DC, Mendes Junior CT, Donadi EA, Maciel LM. HLA-G 3’ untranslated region polymorphic sites associated with increased HLA-G production are more frequent in patients exhibiting differentiated thyroid tumours. Clin Endocrinol (Oxf) 2017;86:597–605. doi: 10.1111/cen.13289. [DOI] [PubMed] [Google Scholar]
- 20.Dhouioui S, Laaribi AB, Boujelbene N, Jelassi R, Ben Salah H, Bellali H, Ouzari HI, Mezlini A, Zemni I, Chelbi H, Zidi I. Association of HLA-G 3’UTR polymorphisms and haplotypes with colorectal cancer susceptibility and prognosis. Hum Immunol. 2022;83:39–46. doi: 10.1016/j.humimm.2021.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Arnaiz-Villena A, Juarez I, Suarez-Trujillo F, López-Nares A, Vaquero C, Palacio-Gruber J, Martin-Villa JM. HLA-G: function, polymorphisms and pathology. Int J Immunogenet. 2021;48:172–192. doi: 10.1111/iji.12513. [DOI] [PubMed] [Google Scholar]
- 22.Vihinen M. Functional effects of protein variants. Biochimie. 2021;180:104–120. doi: 10.1016/j.biochi.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Le Discorde M, Moreau P, Sabatier P, Legeais JM, Carosella ED. Expression of HLA-G in human cornea, an immune-privileged tissue. Hum Immunol. 2003;64:1039–1044. doi: 10.1016/j.humimm.2003.08.346. [DOI] [PubMed] [Google Scholar]
- 24.Alizadeh N, Majidi J, Movassaghpoor A, Farzadi L, Mohammadian M, Baradaran B. Relation between HLA-G gene null allele (HLA-G* 0105n) and recurrent miscarriage. Shiraz E-Med J. 2015;16:e26471. [Google Scholar]
- 25.Klimkiewicz-Wojciechowska G, Grzybowska-Izydorczyk O, Borowiec M, Wyka K, Chmielewska M, Cebula-Obrzut B, Makuch-Lasica H, Robak T, Warzocha K, Mlynarski W, Lech-Maranda E. Polymorphisms of human leukocyte antigen-G gene and clinical outcome of patients with chronic lymphocytic leukemia. Blood. 2013;122:4151. [Google Scholar]
- 26.Nückel H, Rebmann V, Dürig J, Dührsen U, Grosse-Wilde H. HLA-G expression is associated with an unfavorable outcome and immunodeficiency in chronic lymphocytic leukemia. Blood. 2005;105:1694–1698. doi: 10.1182/blood-2004-08-3335. [DOI] [PubMed] [Google Scholar]
- 27.Gros F, Sebti Y, de Guibert S, Branger B, Bernard M, Fauchet R, Amiot L. Soluble HLA-G molecules are increased during acute leukemia, especially in subtypes affecting monocytic and lymphoid lineages. Neoplasia. 2006;8:223–230. doi: 10.1593/neo.05703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Attia MA, Nosair NA, Gawally A, Elnagar G, Elshafey EM. HLA-G expression as a prognostic indicator in B-cell chronic lymphocytic leukemia. Acta Haematol. 2009;13:53–58. doi: 10.1159/000353757. [DOI] [PubMed] [Google Scholar]
- 29.Giannopoulos K, Schmitt M, Kowal M, Własiuk P, Bojarska-Junak A, Roliński J, Dmoszyńska A. The significance of soluble HLA-G plasma levels as well as messenger HLA-G for B-cell chronic lymphocytic leukemia (B-CLL) Leuk Res. 2008;32:1815–1819. doi: 10.1016/j.leukres.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Erikci AA, Karagoz B, Ozyurt M, Ozturk A, Kilic S, Bilgi O. HLA-G expression in B chronic lymphocytic leukemia: a new prognostic marker? Hematology. 2009;14:101–105. doi: 10.1179/102453309X385197. [DOI] [PubMed] [Google Scholar]
- 31.Alkhouly N, Shehata I, Ahmed MB, Shehata H, Hassan S, Ibrahim T. HLA-G expression in acute lymphoblastic leukemia: a significant prognostic tumor biomarker. Med Oncolo. 2013;30:460. doi: 10.1007/s12032-013-0460-8. [DOI] [PubMed] [Google Scholar]
- 32.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 33.Al-Tamimi J, Al Omar SY, Al-Khulaifi F, Aljuaimlani A, Alharbi SA, Al-jurayyan A, Mansour L. Evaluation of the relationships between HLA-G 14 bp polymorphism and two acute leukemia in a saudi population. J King Saud Univ Sci. 2022;34:102139. [Google Scholar]
- 34.Eskandari-Nasab E, Hashemi M, Hasani SS, Omrani M, Taheri M, Mashhadi MA. Association between HLA-G 3’UTR 14-bp ins/del polymorphism and susceptibility to breast cancer. Cancer Biomark. 2013;13:253–259. doi: 10.3233/CBM-130364. [DOI] [PubMed] [Google Scholar]
- 35.Liu KS, Pan F, Mao XD, Liu C, Chen YJ. Biological functions of circular RNAs and their roles in occurrence of reproduction and gynecological diseases. Am J Transl Res. 2019;11:1–15. [PMC free article] [PubMed] [Google Scholar]
- 36.Fathi SE, Seleim H, Hammad M, Ezzeldin N, Pasha H. Association of human leukocyte antigen-g gene polymorphisms with rheumatoid arthritis: relationship with disease activity, severity and treatment response. Zagazig University Medical Journal. 2024;30:248–258. [Google Scholar]
- 37.Locafaro G, Amodio G, Tomasoni D, Tresoldi C, Ciceri F, Gregori S. HLA-G expression on blasts and tolerogenic cells in patients affected by acute myeloid leukemia. J Immunol Res. 2014;2014:636292. doi: 10.1155/2014/636292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cross-Najafi AA, Farag K, Isidan A, Li W, Zhang W, Lin Z, Walsh JR, Lopez K, Park Y, Higgins NG, Cooper DKC, Ekser B, Li P. Co-expression of HLA-E and HLA-G on genetically modified porcine endothelial cells attenuates human NK cell-mediated degranulation. Front Immunol. 2023;14:1217809. doi: 10.3389/fimmu.2023.1217809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreira JC, Alshamali F, Pereira L, Fernandes V. Characterization of Arabian Peninsula whole exomes: exploring high inbreeding features. bioRxiv. 2022 doi: 10.1016/j.isci.2022.105336. 2022.2002. 2022.4814611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khayat AM, Alshareef BG, Alharbi SF, AlZahrani MM, Alshangity BA, Tashkandi NF. Consanguineous marriage and its association with genetic disorders in Saudi Arabia: a review. Cureus. 2024;16:e53888. doi: 10.7759/cureus.53888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alanazi M, Pathan AA, Ajaj SA, Khan W, Shaik JP, Al Tassan N, Parine NR. DNA repair genes XRCC1, XRCC3, XPD, and OGG1 polymorphisms among the central region population of Saudi Arabia. Biol Res. 2013;46:161–167. doi: 10.4067/S0716-97602013000200007. [DOI] [PubMed] [Google Scholar]
- 42.Chentoufi AA, Uyar FA, Chentoufi HA, Alzahrani K, Paz M, Bahnassy A, Elyamany G, Elghazaly A. HLA diversity in Saudi population: high frequency of homozygous HLA alleles and haplotypes. Front Genet. 2022;13:898235. doi: 10.3389/fgene.2022.898235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashton JJ, Latham K, Beattie RM, Ennis S. Review article: the genetics of the human leucocyte antigen region in inflammatory bowel disease. Aliment Pharmacol Ther. 2019;50:885–900. doi: 10.1111/apt.15485. [DOI] [PubMed] [Google Scholar]
- 44.Abdul-Hussein SS, Ali EN, Zaki NH, Ad’hiah AH. Genetic polymorphism of HLA-G gene (G*01:03, G*01:04, and G*01:05N) in Iraqi patients with inflammatory bowel disease (ulcerative colitis and Crohn’s disease) Egypt J Med Hum Genet. 2021;22:1–10. [Google Scholar]