Abstract
Objectives: Lung adenocarcinoma (LUAD) is a prevalent form of non-small cell lung cancer with high morbidity and mortality rates. Identifying molecular markers and therapeutic targets is crucial for improving LUAD diagnosis and treatment. Pituitary tumor-transforming gene (PTTG) family members, PTTG1, PTTG2, and PTTG3P, have been linked to several malignancies; however, it is unclear how these genes relate to LUAD. The current retrospective study investigates the diagnostic, prognostic, and therapeutic importance of PTTG genes in LUAD. Methods: We used detailed in silico and in vitro experiments involving cell lines for experimental design. Results: Using RT-qPCR, we documented that PTTG1, PTTG2, and PTTG3P are significantly up-regulated in LUAD cell lines compared to controls, with PTTG2 showing the highest diagnostic potential (AUC = 1.0). Promoter methylation analysis revealed hypomethylation in LUAD samples, particularly for PTTG3P, further supporting its diagnostic relevance. Immunohistochemical staining confirmed overexpression of PTTG1 and PTTG2 proteins in LUAD tissue samples. The mutational analysis highlighted PTTG2 as the most frequently mutated gene within the PTTG family in LUAD samples, with mutations primarily being missense types. Survival analysis demonstrated that high expression of PTTG genes is associated with poorer overall survival, indicating their prognostic value. Gene enrichment analysis suggested that PTTG genes are involved in critical cancer-related pathways and cellular processes. Functional assays following siRNA-mediated knockdown of PTTG genes in A549 cells showed a significant reduction in cell proliferation. Lastly, drug sensitivity analysis revealed strong correlations between PTTG1/PTTG3P expression and resistance to various anticancer drugs. Conclusion: The findings of the current study highlight the potential of PTTG genes as diagnostic biomarkers, prognostic indicators, and therapeutic targets in LUAD.
Keywords: PTTG family genes, LUAD, biomarker, therapeutic target
Introduction
Lung adenocarcinoma (LUAD) is the most prevalent form of lung cancer, representing a significant global health burden [1,2]. It contributes significantly to the mortality and morbidity caused by cancer globally [2]. In 2023, despite advancements in diagnostic and therapeutic approaches, LUAD remains a leading cause of cancer-related deaths worldwide. It accounts for approximately 40% of all lung cancer cases, with a notable increase in incidence among nonsmokers and younger populations [3]. LUAD arises primarily from the epithelial cells lining the alveoli and bronchioles of the lung and is characterized by its aggressive nature, early metastasis, and resistance to conventional therapies [4,5]. The increasing incidence of LUAD, particularly among nonsmokers, emphasizes the urgent need for effective diagnostic, prognostic, and therapeutic strategies [6].
Several studies have demonstrated the critical role that genetic and molecular variables play in the development of LUAD, including changes to tumor suppressor genes and oncogenes [7,8]. Among the numerous molecular targets under investigation, the Pituitary Tumor Transforming Gene (PTTG) family has garnered significant attention [9]. The PTTG family, comprising PTTG1 (PTTG) and its homolog PTTG2 (securin), plays a crucial role in cell cycle regulation, chromosomal stability, and genomic integrity [10]. These genes have been implicated in the progression of various cancers, including breast, prostate, and thyroid cancers, where their overexpression is often associated with poor prognosis and increased tumor aggressiveness [11].
The role of PTTG genes in the context of LUAD has not been fully investigated. PTTG1 can boost oncogenic processes by promoting cell proliferation, suppressing apoptosis, and enabling chromosomal instability, as previous investigations in various cancer types have shown [12,13]. Additionally, the up-regulation of PTTG1 has been correlated with advanced tumor stage, lymph node metastasis, and reduced survival rates in several cancers [14-16]. However, the exact mechanisms through which PTTG genes influence the development and progression of LUAD remain inadequately understood, and there is a paucity of data on their diagnostic and therapeutic potential in this context.
This study aims to fill this gap by comprehensively analyzing the expression patterns, prognostic implications, and potential therapeutic targets of PTTG family genes in LUAD. We aim to clarify the function of PTTG genes in LUAD pathogenesis and investigate their potential as diagnostic and therapeutic targeting biomarkers by a thorough analysis of genomic databases and experimental models. Our research may lead to the development of innovative diagnostic techniques and therapeutic approaches, which would eventually improve the clinical results for LUAD patients.
Methodology
Cell culture
For the current retrospective study, 10 LUAD cell lines, A549, H1299, H1975, HCC827, H1650, H2228, H358, H460, PC-9, Calu-3, and 5 normal control lung cell lines, BEAS-2B, WI-38, MRC-5, NHLF, and CCD-19Lu were purchased from the American Type Culture Collection (ATCC). All LUAD cell lines were cultured in RPMI-1640 medium, supplemented with 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin, at 37°C in a 5% CO2 atmosphere. While, BEAS-2B cells were grown in Bronchial Epithelial Cell Growth Medium (BEGM), WI-38, MRC-5, and CCD-19Lu were cultured in EMEM with 10% FBS and 1% penicillin-streptomycin. Lastly, NHLF cells were maintained in a Fibroblast Growth Medium (FGM) with 10% FBS and 1% penicillin-streptomycin.
Nucleic acid extraction
DNA was extracted using the organic method [17] while RNA was extracted using the TRIzol method [18].
Expression analysis of PTTG genes and other key genes
We used RT-qPCR to examine the expression of four other genes in this section of the study in addition to the PTTG genes, which are EGFR, KRAS, TP53, and MMP9. These genes were chosen because they are essential for vital processes connected to cancer, including invasion, migration, survival, and proliferation of cells. To create complementary DNA (cDNA), the RNA was reverse-transcribed using the PrimeScriptTM RT reagent kit that included a gDNA Eraser (RR047A, TaKaRa, China).
Using a CFX96 real-time PCR detection system, RT-qPCR was carried out using TB Green® Premix Ex TaqTM II (RR820A, TaKaRa, China). The 2-ΔΔCt technique was utilized to determine the relative mRNA expression levels of PTTG1, PTTG2, and PTTG3P in LUAD cells. The expression levels were then normalized to the endogenous control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The following is a list of the primers utilized in this investigation.
GAPDH-F: 5’-ACCCACTCCTCCACCTTTGAC-3’, GAPDH-R: 5’-CTGTTGCTGTAGCCAAATTCG-3’; PTTG1-F: 5’-GCTTTGGGAACTGTCAACAGAGC-3’, PTTG1-R: 5’-CTGGATAGGCATCATCTGAGGC-3’; PTTG2-F: 5’-CTTTGGGCACTGTCAACAGAGC-3’, PTTG2-R: 5’-TCTGGATAGGCGTCATCTGAGG-3’; PTTG3P-F: 5’-CTGCCTGAAGAGCACCAGATTG-3’, PTTG3P-R: 5’-CATGGTGGAGAGGGCATCTTCA-3’; EGFR-F: 5’-AACACCCTGGTCTGGAAGTACG-3’, EGFR-R: 5’-TCGTTGGACAGCCTTCAAGACC-3’; KRAS-F: 5’-CAGTAGACACAAAACAGGCTCAG-3’, KRAS-R: 5’-TGTCGGATCTCCCTCACCAATG-3’; TP53-F: 5’-CCTCAGCATCTTATCCGAGTGG-3’, TP53-R: 5’-TGGATGGTGGTACAGTCAGAGC-3’; MMP9-F: 5’-GCCACTACTGTGCCTTTGAGTC-3’, MMP9-R: 5’-CCCTCAGAGAATCGCCAGTACT-3’.
Bisulfite sequencing analysis
The first step of bisulfite sequencing involved the fragmentation of 1 µg of total DNA into fragments of approximately 200-300 bp using the Covarias sonication system (Covarias, Woburn, MA, USA). After that, a variety of enzymes, including T4 DNA polymerase, Klenow Fragment, and T4 polynucleotide kinase, helped the DNA fragments go through several activities, including blunt end repair and phosphorylation. The repaired fragments were then subject to 3’ adenylation using Klenow Fragment (3’-5’ exo-), followed by ligation with adapters. These adapters contained 5’-methylcytosine in place of 5’-cytosine and index sequences, and the ligation was executed using T4 DNA Ligase. After the construction of libraries, quantification was performed utilizing a Qubit fluorometer with the Quant-iT dsDNA HS Assay Kit (Invitrogen, Carlsbad, CA, USA). The prepared libraries were subsequently dispatched to Beijing Genomic Institute (BGI), China, for targeted bisulfite sequencing. Once the sequencing was completed, the methylation data was subjected to a normalization process, resulting in the generation of beta values.
Immunohistochemistry (IHC)
All collaborating patients provided written consent for the collection of four tissue samples (three LUAD and one normal control), including permission for the results to be published. The study involved LUAD patients who underwent tumor surgical resection at Taizhou Central Hospital, China. The tissue samples were stored at -80°C until further analysis. This study adhered to the Declaration of Helsinki guidelines and received approval from the Medical Research Ethics Committee of Taizhou University.
Tissue samples were subjected to formalin fixation, paraffin embedding, and subsequent sectioning into 3-μm-thick slices. The sections were subjected to heat-induced antigen retrieval by boiling in sodium citrate buffer after deparaffinization and endogenous catalase neutralization. Following this, non-specific binding was blocked with a 1-hour incubation in 5% normal goat serum. The sections were then exposed to anti-PTTG1 and anti-PTTG2 antibodies (diluted 1:50; CAT#: TA322776 and CAT#: TA324611) overnight at 4°C, and antibody binding was detected using the avidin-biotin-peroxidase method. The sections were then counterstained with hematoxylin, and the results were assessed independently by two skilled investigators.
Expression analysis of PTTG genes across The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets
UALCAN (https://ualcan.path.uab.edu/) is a user-friendly web portal providing a comprehensive analysis of cancer transcriptome data from The Cancer Genome Atlas (TCGA) [19]. It offers accessible tools for exploring gene expression, survival analysis, and clinicopathological correlation across various cancer types, aiding researchers in uncovering potential biomarkers and therapeutic targets. In this study, UALCAN was used to analyze PTTG gene expression across the TCGA LUAD cohort.
Moreover, we obtained the standardized matrix profile (*series matrix.txt) of GSE10072, a microarray dataset containing expression profiles from 58 LUAD and 49 control tissue samples sourced from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) [20]. The GEO2R tool combined with the Limma package made it easier to analyze the GSE10072 dataset. Differentially expressed genes (DEGs) were identified by using GEO2R, a t-test, and the Benjamin-Hochberg technique to obtain P-values and false discovery rates (FDR). DEG screening adhered strictly to established criteria: P < 0.05 and logFC > 1.
Promoter methylation analysis of PTTG genes across TCGA cohort
MEXPRESS (https://mexpress.ugent.be/) is a user-friendly web tool offering an interactive exploration of DNA methylation and gene expression data across various cancers [21]. It integrates datasets from TCGA, allowing researchers to visualize correlations, conduct differential analysis, and identify potential biomarkers. MEXPRESS enhances understanding of epigenetic mechanisms in cancer progression and therapy response. In the current study, the MEXPRESS database was used to analyze promoter methylation levels of PTTG genes across the TCGA LUAD cohort.
Mutational analysis of PTTG genes
cBioPortal (https://www.cbioportal.org/) is an open-access platform that enables visualization and analysis of complex cancer genomics data [22]. It facilitates the investigation of genetic changes, survival rates, and possible treatment targets across a range of cancer types by integrating molecular profiling results from many cancer research. cBioPortal is used by researchers to learn more about the biology of cancer and to guide customized care strategies. The cBioPortal database was used in this study to analyze PTTG gene mutations in LUAD.
Survival analysis and prognostic model development
The Kaplan Meier (KM) plotter tool (https://kmplot.com/analysis/) is a web-based application that allows researchers to perform survival analysis using gene expression data [23]. It compiles information from open databases such as TCGA and GEO, allowing researchers to investigate the relationship between gene expression levels and patient survival outcomes across a range of cancer types. Prognostic biomarkers and prospective treatment targets can be found with the use of the KM plotter. The KM plotter tool was utilized in our work to do a survival analysis of PTTG genes in LUAD.
Furthermore, to develop and validate the prognostic model of PTTG genes, gene expression data from GSE29016 was used as the training dataset, while six independent datasets (GSE29013, GSE26939, GSE19188, GSE14814, GSE13213, and GSE11969) served as validation datasets. Initially, candidate prognostic genes were identified through univariate Cox regression analysis in the training dataset. A multivariate Cox proportional hazards model was then constructed using these selected genes, estimating the hazard ratios (HR) for each gene to indicate the risk associated with their expression levels. Model parameters were optimized using cross-validation within the training dataset to prevent overfitting. The model was internally validated via bootstrapping and further externally validated on the six independent datasets by calculating HRs and concordance indices (C-index) to assess predictive power.
Gene enrichment analysis
The DAVID (https://david.ncifcrf.gov/) tool is a web-based resource used for functional annotation and enrichment analysis of gene lists [24]. The identification of enriched biological terminology, such as Gene Ontology (GO) concepts and pathways, inside gene sets facilitates the interpretation of biological relevance by researchers. Using gene lists produced from experimental data, DAVID makes it easier to comprehend the biological pathways and underlying processes that are connected to those lists. The PTTG gene enrichment study in LUAD was conducted using this database.
Drug sensitivity analysis
GSCA (https://david.ncifcrf.gov/) is a bioinformatics tool developed by the Guo Lab, designed for comprehensive analysis of gene sets in the context of cancer [25]. By combining multi-omics data from different kinds of cancer, it enables researchers to investigate relationships between gene sets and clinical outcomes. The drug sensitivity analysis of the PTTG genes in LUAD was conducted in our study using GSCA.
Knockdown of PTTG genes in A549 cells
siRNAs targeting PTTG genes were obtained from OBiO Company. A549 LUAD cell lines were transfected with these siRNAs using INTERFERin Transfection Reagent (from France) to silence PTTG family genes, including PTTG1, PTTG2, and PTTG3P.
Cell Counting Kit-8 assay
Cell proliferation in A549 LUAD cells was assessed using the Cell Counting Kit-8 (CCK-8, APExBIO, USA). Initially, 3×10^3 cells were seeded into 96-well plates following transfection. Subsequently, plates were incubated at 37°C for different durations (0, 24, 48, and 72 hours). Following the addition of CCK-8 reagent to each well, the optical absorbance at 450 nm was measured to assess the vitality of the cells.
Colony formation assay
A549 LUAD cells that had been transfected were cultivated in 6-well plates and kept in DMEM that had been enhanced with 10% FBS and 1% Penicillin-Streptomycin. Following seeding, the media was substituted with DMEM that included 10% FBS and 500 μg/mL G418 (Thermo Fisher Scientific, Catalog #11811031) to identify transfected cells that were effectively propagated. To create colonies, the cells were allowed to develop for 10-14 days, with medium changes occurring every 3 days. The colonies were then counted after being fixed with 4% paraformaldehyde and stained with Crystal Violet Staining Solution (Thermo Fisher Scientific, Catalog #C3886). The outcomes showed a noteworthy decrease in colony formation in the siRNA-transfected cells in contrast to the control cells, underscoring the cooperative function of PTTG1, PTTG2, and PTTG3P in fostering cellular growth and viability.
Statistics
This study employed the t-test to compare group differences for normally distributed variables. The diagnostic potential of PTTG gene expressions was assessed using receiver operating characteristic (ROC) curves, with the area under the curve (AUC) as the diagnostic metric. Survival analyses utilized the Cox proportional hazards model, Kaplan-Meier analysis, and log-rank test. Correlations were evaluated using Spearman’s or Pearson’s test. All statistical tests were two-sided, with significance defined as P < 0.05. Statistical analyses were conducted using R software (version 4.0.2).
Results
Expression landscape of PTTG genes in LUAD cell lines
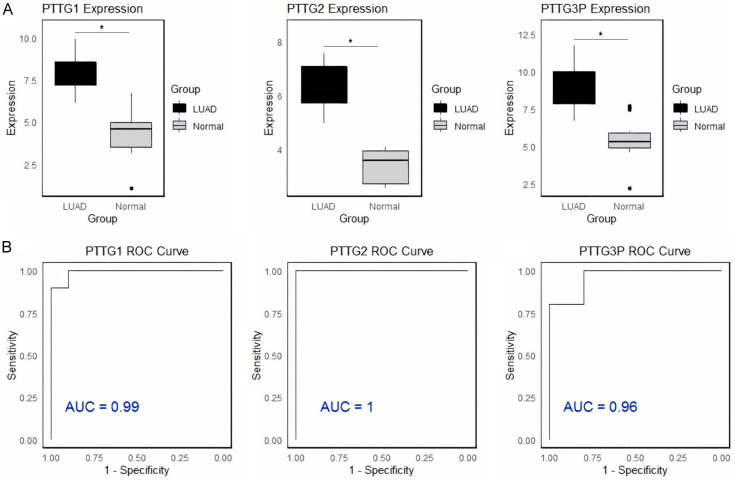
Firstly, the expression of PTTG genes in LUAD and control cell lines was documented via the RT-qPCR. Data on PTTG1, PTTG2, and PTTG3P gene expression levels in LUAD and control cell lines are shown in Figure 1, together with ROC curves illustrating these genes’ diagnostic efficacy. Figure 1A shows that the expression levels of PTTG1, PTTG2, and PTTG3P are significantly (p-value < 0.05) higher in LUAD cell lines compared to control cell lines, indicating up-regulation in LUAD. Figure 1B illustrates the evaluations of the diagnostic accuracy of these genes, with ROC curves indicating AUC values of 0.99 for PTTG1, 1.0 for PTTG2, and 0.96 for PTTG3P. The high AUC value for PTTG3P suggests it is a highly effective biomarker for distinguishing LUAD from control samples, followed by PTTG1 and PTTG2. Overall, PTTG3P shows the greatest potential as a diagnostic biomarker for LUAD.
Figure 1.
Expression and diagnostic performance of pituitary tumor-transforming gene (PTTG) genes in lung adenocarcinoma (LUAD) and control cell lines. A. RT-qPCR analysis of PTTG1, PTTG2, and PTTG3P gene expression levels in LUAD (orange) and control (green) cell lines. B. Receiver operating characteristic (ROC) curves assessing the diagnostic accuracy of PTTG1, PTTG2, and PTTG3P expression levels. The area under the curve (AUC) values are 0.88 for PTTG1, 0.77 for PTTG2, and 0.98 for PTTG3P, demonstrating that PTTG3P has the highest diagnostic potential for distinguishing LUAD from control samples, followed by PTTG1 and PTTG2. *P-value < 0.05.
Promoter methylation landscape of PTTG genes
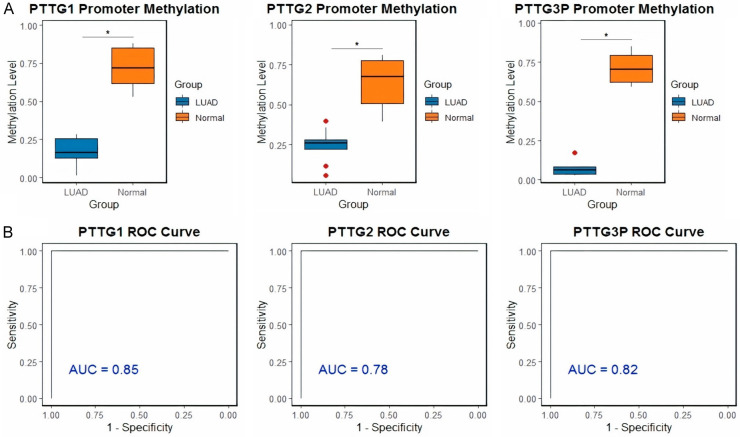
Promoter methylation analysis of PTTG in LUAD and control cell lines was conducted via bisulfite sequencing. In Figure 2A, box plots of beta values indicate that the methylation levels of PTTG1, PTTG2, and PTTG3P are lower in LUAD cell lines compared to control cell lines, suggesting hypomethylation in the cancerous samples. Additionally, ROC curves assessing the methylation levels for these genes’ diagnostic accuracy are displayed in Figure 2B. AUC values for PTTG1, PTTG2, and PTTG3P are 0.85, 0.78, and 0.82, respectively, suggesting that all three genes have excellent discriminative ability between persons with LUAD and those without (Figure 2B). Among these, PTTG3P has the highest AUC, suggesting it is the most effective biomarker for distinguishing between LUAD and normal tissues based on methylation status. These findings demonstrate the PTTG gene family methylation’s potential as a LUAD diagnostic tool.
Figure 2.
Promoter methylation analysis and diagnostic performance of pituitary tumor-transforming gene (PTTG) genes in lung adenocarcinoma (LUAD) and control cell lines. A. Box plots of beta values obtained from bisulfite sequencing indicate the promoter methylation levels of PTTG1, PTTG2, and PTTG3P genes in LUAD (red) and control (blue) cell lines. B. Receiver operating characteristic (ROC) curves evaluating the diagnostic accuracy of promoter methylation levels for PTTG1, PTTG2, and PTTG3P. The areas under the curve (AUC) values are 0.81 for PTTG1, 0.84 for PTTG2, and 0.88 for PTTG3P, demonstrating good discriminative power between LUAD and normal individuals, with PTTG3P showing the highest diagnostic potential. *P-value < 0.05.
Expression landscape of EGFR, KRAS, TP53, and MMP9 in LUAD cell lines
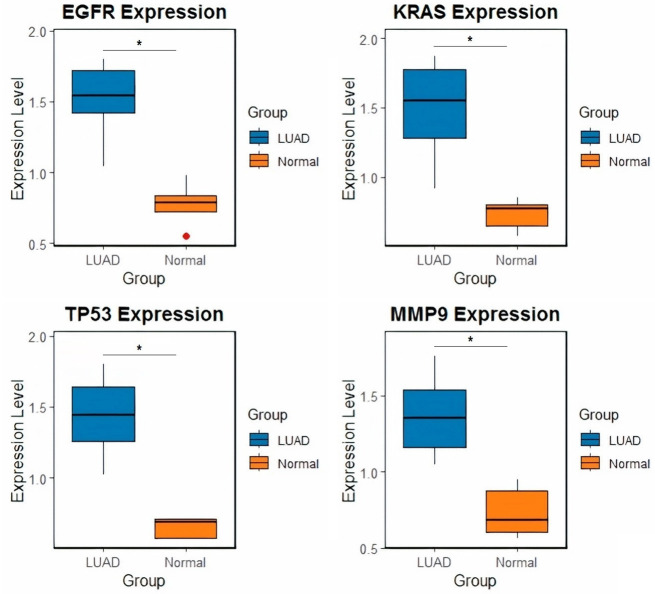
EGFR, KRAS, TP53, and MMP9 expression levels are substantially (p-value < 0.05) higher in LUAD cell lines than in normal control cell lines, according to the findings of the RT-qPCR study (Figure 3). These genes may be important in the initiation and development of this kind of cancer, based on the increased levels of LUAD.
Figure 3.
Differential gene expression of EGFR, KRAS, TP53, and MMP9 in lung adenocarcinoma (LUAD) and normal control cell lines. Box plots representing the expression levels of four genes (EGFR, KRAS, TP53, and MMP9) in LUAD and normal control cell lines as determined by Reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The blue boxes represent LUAD samples, while the orange boxes represent normal control samples. *P-value < 0.05.
Immunohistochemical staining of PTTG1 and PTTG2 proteins
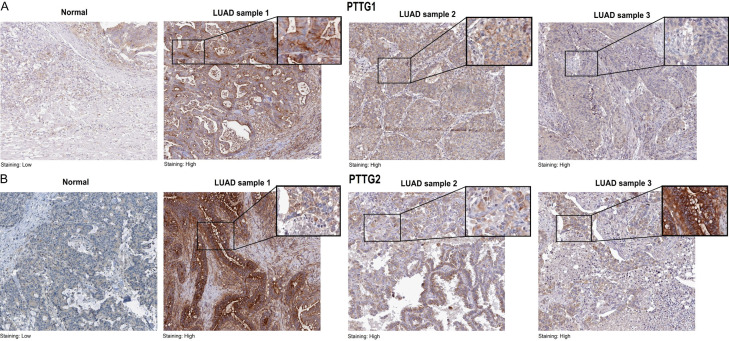
The immunohistochemistry (IHC) technique was used to evaluate the protein expression of PTTG1 and PTTG2 proteins in LUAD and control tissue samples. PTTG1 and PTTG2 protein expression levels in LUAD and control tissue samples are compared using immunohistochemical pictures shown in Figure 4. In the normal tissue, the staining is low, indicating minimal or baseline expression of the PTTG1 and PTTG2 proteins (Figure 4A, 4B). In contrast, the LUAD tissues exhibit high staining intensity, reflecting a significant overexpression of these proteins (Figure 4A, 4B). The observed variation in expression indicates a significant up-regulation of PTTG1 and PTTG2 proteins in LUAD compared to normal tissue, which may indicate their possible involvement in the development and advancement of LUAD tumors.
Figure 4.
Immunohistochemical staining of PTTG1 and PTTG2 proteins expression in normal and lung adenocarcinoma (LUAD) tissue samples. A. Representative images of PTTG1. B. Representative images of PTTG1.
Expression validation of the PTTG gene using additional TCGA and GEO cohorts of LUAD patients
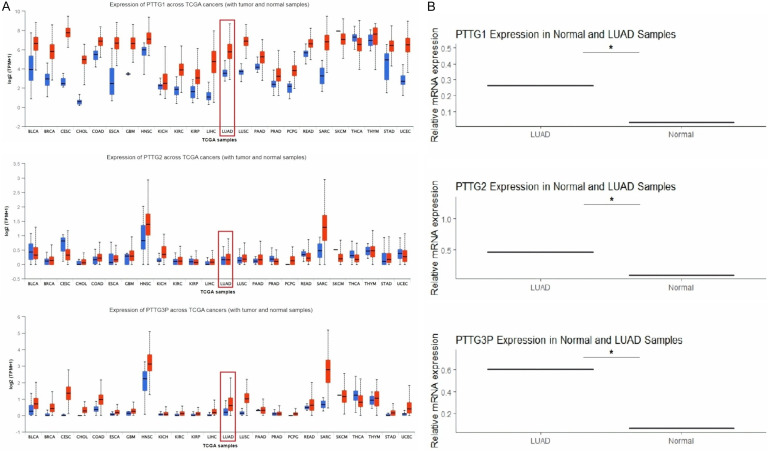
The expression of PTTG genes was further validated on two independent TCGA and GEO expression datasets. Figure 5 illustrates the expression levels of PTTG1, PTTG2, and PTTG3P in LUAD samples across two datasets: TCGA from the UALCAN database and the GSE10072 dataset from the GEO database. Figure 5A displays box plots from the UALCAN database, demonstrating a substantial (p-value < 0.05) up-regulation of all three genes (PTTG1, PTTG2, and PTTG3P) in LUAD tumor tissues compared to normal samples. Figure 5B reinforces these findings with bar graphs from the GSE10072 dataset, which also display a significant increase in the relative mRNA expression of PTTG1, PTTG2, and PTTG3P in LUAD samples compared to normal samples. The consistency of these results across two independent datasets suggests a strong association of these genes with LUAD, indicating their potential role in the disease’s pathogenesis or progression.
Figure 5.
Comparative analysis of pituitary tumor-transforming gene (PTTG) family gene expression in The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets. A. Expression validation of PTTG1, PTTG2, and PTTG3P across LUAD TCGA dataset using the UALCAN platform. B. Expression analysis of PTTG1, PTTG2, and PTTG3P in normal and LUAD samples using GSE10072 expression dataset from the GEO database. *P-value < 0.05.
Promoter methylation validation of the PTTG gene using an additional TCGA cohort of LUAD patients
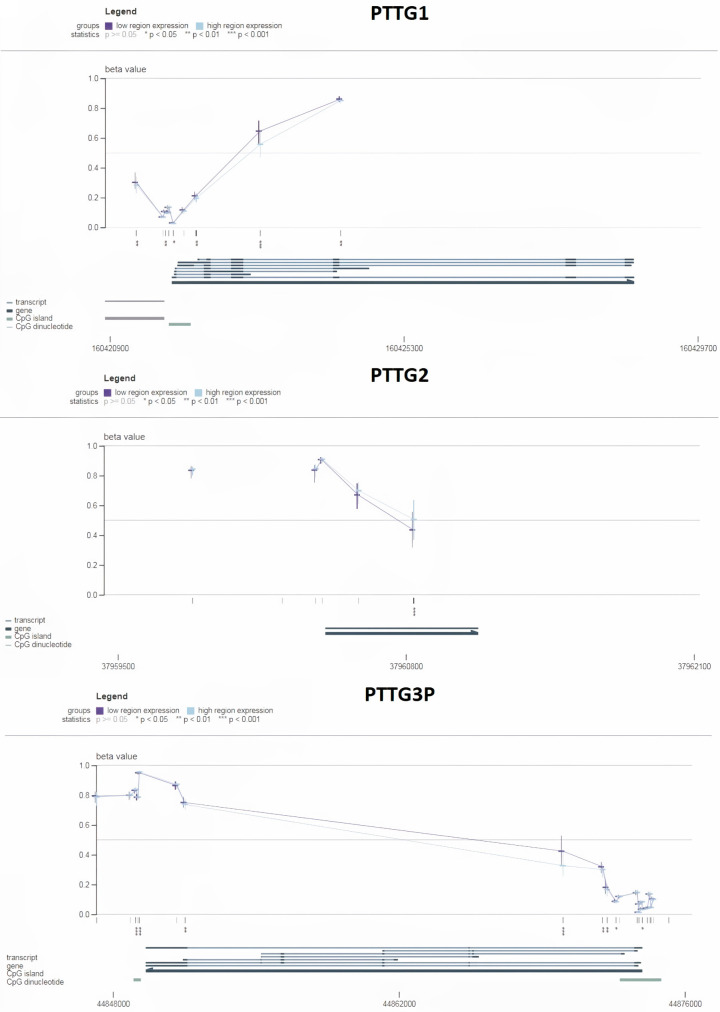
Next, the validation of DNA methylation levels in the promoter regions of PTTG1, PTTG2, and PTTG3P was done using the MEXPRESS database. Figure 6 shows the degree of methylation as a beta value, with values closer to 0 denoting lesser methylation and values closer to 1 denoting more methylation. For PTTG1, the plot reveals a general decrease in beta values from high to low-expression groups, suggesting lower methylation in the high-expression group (Figure 6). For PTTG2, there is a clear trend of decreasing beta values from low to high-expression groups, indicating lower methylation in the high-expression group (Figure 6). This implies that methylation and expression of PTTG2 are inversely correlated. Similar to PTTG2, PTTG3P exhibits a similar pattern, with lower beta values in the group with high expression (Figure 6), demonstrating the inverse link between methylation and expression. Overall, these results suggest that the expression levels of PTTG1, PTTG2, and PTTG3P are likely regulated by DNA methylation, where lower methylation is associated with higher gene expression.
Figure 6.
Promoter methylation analysis of PTTG1, PTTG2, and PTTG3P genes in lung adenocarcinoma (LUAD) samples using MEXPRESS database. Each graph shows beta values indicating methylation levels across different regions of the respective gene promoters. P-value < 0.05.
Mutational analysis of PTTG genes
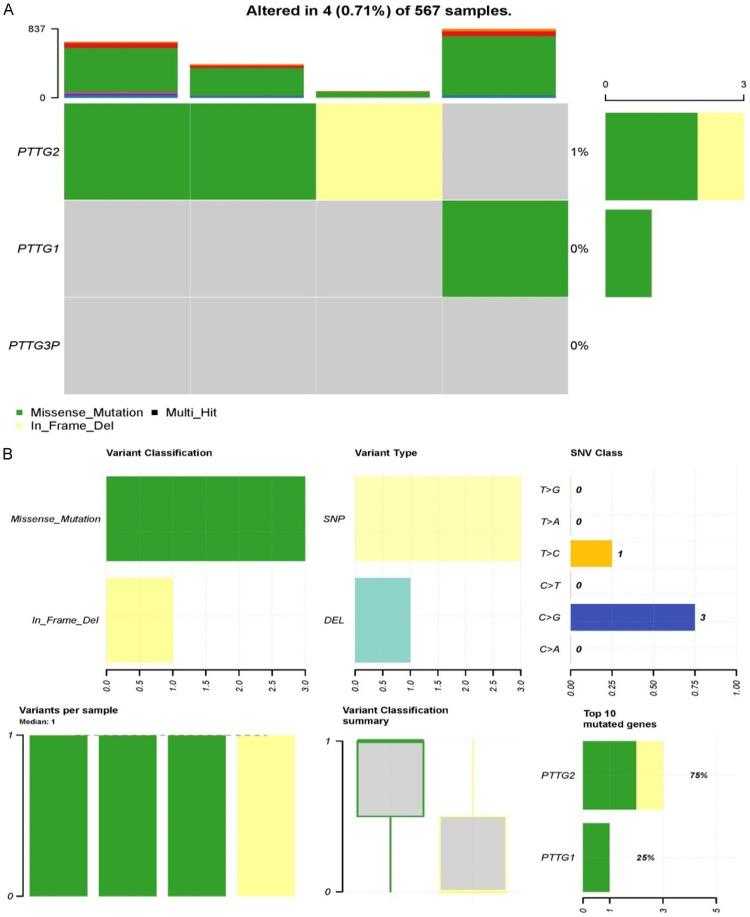
Mutational analysis of PTTG genes was conducted via the cBioPortal platform. Figure 7 presents mutation data for PTTG1, PTTG2, and PTTG3P in a dataset of 567 samples, indicating that alterations were observed in 4 samples (0.71%). The heatmap highlights that PTTG2 is the most frequently mutated gene among the three, with a mutation rate of 1%, while PTTG1 and PTTG3P show no mutations (Figure 7A). The two main kinds of mutations found in PTTG2 are in-frame deletions (yellow) and missense mutations (green). According to the categorization summary, single nucleotide polymorphisms (SNPs) are the most common variation type, and missense mutations account for the bulk of alterations (Figure 7B). Within the SNP class, the most common single nucleotide variant (SNV) is C>G (three occurrences), followed by a single C>T change (Figure 7B). The variant classification summary box plot indicates that each of the four samples has at least one mutation, with a median of one variant per sample. This suggests that PTTG2 mutations, especially missense mutations and in-frame deletions, could be relevant in the context of the studied condition, while PTTG1 and PTTG3P do not appear to be commonly mutated in LUAD samples.
Figure 7.
Mutational landscape of pituitary tumor-transforming gene (PTTG) family genes in 567 lung adenocarcinoma (LUAD) samples. A. Frequencies of genetic mutations in PTTG gene family (PTTG1, PTTG2, PTTG3P) in 567 samples. B. Detailed summery of the observed genetic alterations.
Survival analysis and prognostic model development
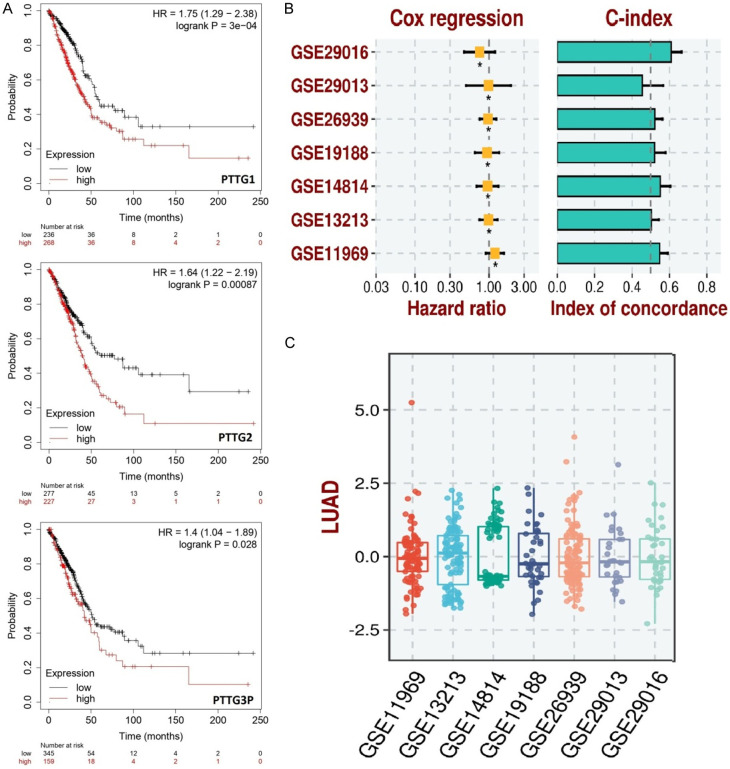
Survival analysis of PTTG genes in LUAD patients was conducted using the KM plotter tool. The Kaplan-Meier survival curves for the PTTG1, PTTG2, and PTTG3P genes are displayed in Figure 8A. The expression levels of each gene are divided into high and low groups, demonstrating that higher expression levels are linked to lower overall survival. Specifically, PTTG1 (HR = 1.75, logrank P = 3e-04), PTTG2 (HR = 1.64, logrank P = 0.00087), and PTTG3P (HR = 1.4, logrank P = 0.028) are all significantly correlated with reduced survival probability over time (Figure 8A). Additionally, Figure 8B provides an overview of the concordance index (C-index) and Cox regression findings for seven distinct datasets, demonstrating the prognostic model’s resilience across various datasets. The hazard ratios for each dataset are all above 1, indicating that higher expression consistently predicts worse outcomes (Figure 8B). The C-index values range from approximately 0.4 to 0.8, suggesting moderate to good predictive power (Figure 8B). Box plots of PTTG gene expression levels from the same seven datasets are shown in Figure 8C. These plots indicate variance but similar patterns, confirming the general finding that PTTG gene expression is a significant prognostic factor in LUAD.
Figure 8.
Survival analysis and prognostic model evaluation of pituitary tumor-transforming gene (PTTG) family genes in lung adenocarcinoma (LUAD). A. This panel displays Kaplan-Meier survival curves for PTTG1, PTTG2, and PTTG3P, using the KM plotter tool in LUAD patients. B. This panel shows the results of Cox regression analysis and the concordance index (C-index) for seven gene expression datasets (GSE29016, GSE29013, GSE26939, GSE19188, GSE14814, GSE13213, and GSE11969). C. This panel shows the distribution of prognostic scores for LUAD across the seven gene expression datasets (GSE29016, GSE29013, GSE26939, GSE19188, GSE14814, GSE13213, and GSE11969). *P-value < 0.05.
Gene enrichment analysis of PTTG genes
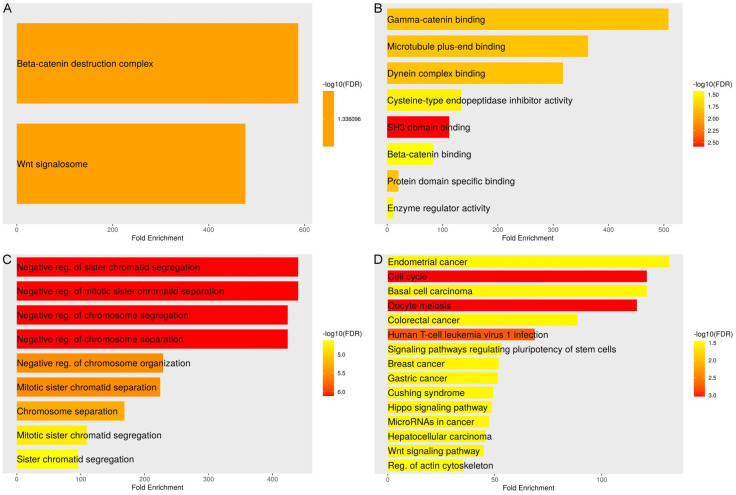
Gene enrichment analysis of PTTG genes was conducted via the DAVID tool. Concerning cellular components (CC), Figure 9A illustrates that the PTTG genes are significantly overexpressed in the “Wnt signalosome” and the “Beta-catenin destruction complex”. The high fold enrichment values and the significant -log10 (FDR) scores indicate strong associations with these cellular structures, suggesting that PTTG genes play crucial roles in these complexes. The molecular functions (MF) enriched by PTTG genes are illustrated in Figure 9B. Key functions include “Gamma-catenin binding”, “Microtubule plus-end binding”, “Dynein complex binding”, “Cysteine-type endopeptidase inhibitor activity”, “SH3 domain binding”, and “Beta-catenin binding”. The fold enrichment values and the varying -log10 (FDR) scores, with the highest enrichment observed in SH3 domain binding, imply significant involvement in these molecular interactions and activities. The PTTG gene-related biological processes (BP) are shown in Figure 9C. The three most important processes are “sister chromatid segregation, chromosome organization, and negative regulation of chromosome segregation”. This suggests that PTTG genes are critical regulators in maintaining chromosomal stability and proper segregation during cell division, as evidenced by the high fold enrichment and strong -log10 (FDR) scores. An understanding of the pathways enriched by PTTG genes is shown in Figure 9D. These pathways include “Endometrial cancer”, “Cell cycle”, “Basal cell carcinoma”, “Oocyte meiosis”, “Colorectal cancer”, “Human T-cell leukemia virus 1 infection”, “Signaling pathways regulating pluripotency of stem cells”, “Breast cancer”, “Gastric cancer”, “Cushing syndrome”, “Hippo signaling pathway”, “MicroRNAs in cancer”, “Hepatocellular carcinoma”, “Wnt signaling pathway”, and “Regulation of actin cytoskeleton”. The high -log10 (FDR) scores and considerable fold enrichment for these pathways suggest that PTTG genes are extensively engaged in cell cycle control, signaling processes, and a variety of cancer-related pathways. Overall, the enrichment analysis suggests that PTTG genes are integral to various cellular processes, particularly those related to cell division, signal transduction, and cancer pathways.
Figure 9.
Functional enrichment analysis of pituitary tumor-transforming gene (PTTG) genes. A. Illustrates the significant enrichment of PTTG genes in cellular components (CC). B. Focuses on molecular functions (MF) of PTTG genes. C. Details biological processes (BP) of PTTG genes. D. Identifies pathways associated with PTTG genes.
Cell Counting Kit-8 and colony formation assays
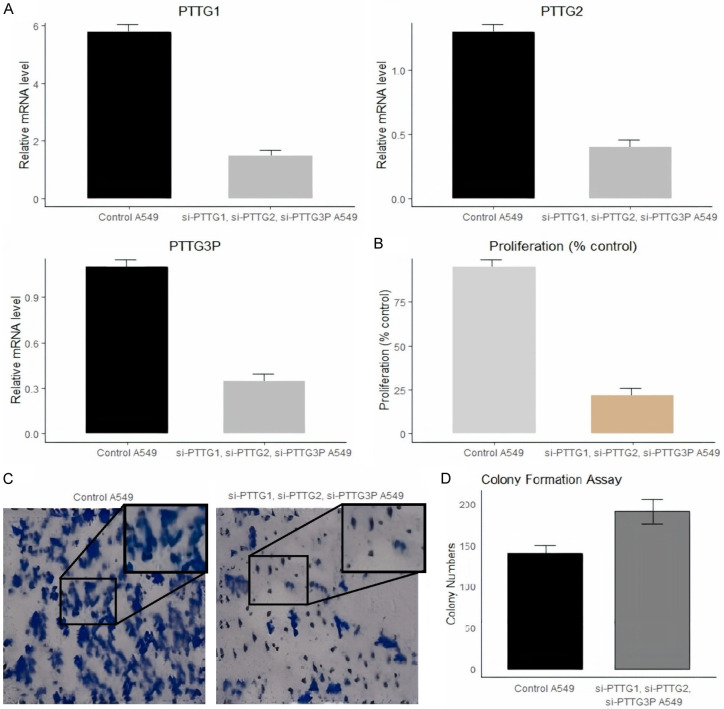
The PTTG1, PTTG2, and PTTG3P genes synergistically regulate processes such as cell cycle regulation, tumor development, and other cellular functions. To examine their combined impact on several parameters, siRNA was used to concurrently silence all three genes in A549 cells. The silencing efficiency was evaluated via RT-qPCR. As shown in Figure 10A, there was a significant reduction in the expression levels of PTTG1, PTTG2, and PTTG3P in the transfected A549 cells compared to the control A549 cells. Further analysis using a CCK-8 assay revealed a marked decrease in cellular proliferation in cells with silenced PTTG1, PTTG2, and PTTG3P, compared to the control A2058 cells (Figure 10B). The treated group, on the other hand, has fewer and smaller colonies, as indicated by Figure 10C’s visual comparison of colony formation between control and treated cells calculated in Figure 10D.
Figure 10.
Effects of PTTG1, PTTG2, and PTTG3P knockdown on mRNA levels and proliferation of A549 cells. A. Relative mRNA levels of PTTG1, PTTG2, and PTTG3P in A549 cells. The bar graphs depict the mRNA expression levels of PTTG1, PTTG2, and PTTG3P in control A549 cells and A549 cells treated with siRNAs targeting PTTG1, PTTG2, and PTTG3P. B. Proliferation of A549 cells post-siRNA treatment. C. Images from the colony formation assay depicting control A549 cells and siRNA-treated A549 cells. D. Quantification of colony numbers from the colony formation assay.
Drug sensitivity analysis of PTTG genes
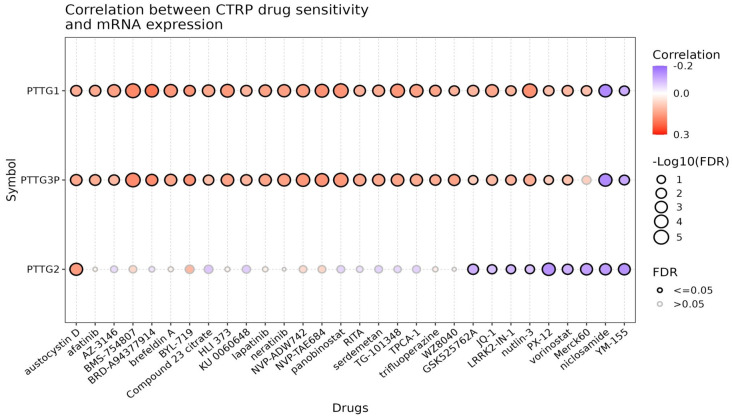
Drug sensitivity analysis of PTTG genes was conducted using the GSCA database. Figure 11 illustrates the correlation between the expression levels of PTTG genes (PTTG1, PTTG2, PTTG3P) and drug sensitivity, as derived from the CTRP dataset. PTTG1 and PTTG3P show strong positive correlations (drug resistance) with sensitivity to a broad range of drugs, indicated by red-colored dots with significant correlations denoted by large dots with thick black borders (FDR ≤ 0.05). Drugs such as Afatinib, AZD3146, BRD-A94377914, Compound 23_citrate, KU006373, and Lapatinib exhibit notable positive correlations (drug resistance) with these genes. Conversely, PTTG2 displays a more varied correlation profile, with many correlations being non-significant (smaller dots with grey borders, FDR > 0.05). While some drugs like Austocystin D, BMS-754807, and Brefeldin A show some level of correlation with PTTG2, these are generally weaker and less significant. Interestingly, a few drugs, such as IPI-145 and Nutlin-3, exhibit a slight negative correlation (drug sensitivity) with PTTG2 expression, but these correlations are also weak and typically non-significant. Overall, this research indicates that PTTG1 and PTTG3P influence drug sensitivity more significantly than PTTG2, suggesting that they may play a role in regulating drug response in cancer treatment.
Figure 11.
Correlation analysis between drug sensitivity data from the Cancer Therapeutics Response Portal (CTRP) and mRNA expression levels of PTTG1, PTTG2, and PTTG3P in lung adenocarcinoma (LUAD). Each circle represents a drug, with the color indicating the correlation coefficient between mRNA expression and drug sensitivity (red for positive correlation, blue for negative correlation). The size of the circle corresponds to the -log10 (FDR) value, with larger circles indicating more significant correlations. P-value < 0.05.
Discussion
Lung adenocarcinoma (LUAD) is the most prevalent subtype of non-small cell lung cancer (NSCLC), characterized by its aggressive nature and poor prognosis [26,27]. The five-year survival rate is still poor despite therapy breakthroughs, highlighting the critical need for new biomarkers and therapeutic targets [28,29]. The pituitary tumor-transforming gene (PTTG) family, comprising PTTG1, PTTG2, and PTTG3P, has been implicated in various malignancies, but their roles in LUAD remain to be fully elucidated [16,30]. To gain a fresh understanding of the diagnostic and prognostic implications of PTTG genes, the current research examines their expression, promoter methylation, and possible clinical relevance in LUAD.
Our findings demonstrate significant up-regulation of PTTG1, PTTG2, and PTTG3P in LUAD cell lines and tissue samples compared to normal controls. This is consistent with other studies that found PTTG1 to be an oncogene in a variety of malignancies, such as thyroid, colorectal, and breast cancers, where it stimulates the growth and metastasis of cells [31-33]. PTTG2 and PTTG3P, though less studied, have also shown elevated expression in specific cancers. Our study is the first to collectively analyze these genes in LUAD, highlighting their potential as biomarkers. PTTG1, acting as a securin, inhibits separase to prevent premature sister chromatid separation, but its overexpression can lead to aneuploidy and genomic instability by disrupting the G2/M transition of the cell cycle [34]. Additionally, it hinders DNA repair processes by interacting with proteins such as p53, which in turn promotes the buildup of DNA damage and mutagenesis [34]. Furthermore, PTTG1 promotes invasion and increases tumor blood supply by upregulating vascular endothelial growth factor (VEGF) [12]. PTTG2, though less studied, is believed to function similarly to PTTG1, contributing to genomic instability and altered cell cycle progression [35,36]. PTTG3P, a pseudogene, may act through competing endogenous RNA (ceRNA) mechanisms, sponging microRNAs that regulate oncogenic pathways, thereby indirectly promoting tumorigenesis [37]. Overall, the abnormal cellular homeostasis caused by the over-expression of these PTTG genes results in unchecked cell proliferation, genomic instability, and increased tumor development and dissemination.
Moreover, it appears that epigenetic dysregulation is a factor in the upregulation of PTTG1, PTTG2, and PTTG3P promoters, which are hypomethylated in LUAD cell lines and tissue samples. Previous research has demonstrated that oncogenes in cancer can be activated by promoter hypomethylation [38,39]. Mutational analysis revealed low mutation frequencies for PTTG genes in LUAD, with PTTG2 showing the highest rate of 1%. While mutations in PTTG genes are rare, their overexpression driven by other mechanisms, such as hypomethylation, might be more critical in LUAD pathogenesis. This contrasts with genes like TP53, where mutations are a primary driver [40]. Kaplan-Meier survival analysis and Cox regression models consistently showed that high expression of PTTG genes is associated with poorer overall survival in LUAD patients. This conclusion is especially noteworthy for PTTG1 and PTTG2, which is consistent with other research that found a negative correlation between PTTG1 expression and colorectal cancer outcomes [41]. Our prognostic model, which has been confirmed on several datasets, shows that PTTG genes have the potential to be reliable prognostic indicators.
Silencing PTTG genes significantly reduced cell proliferation in LUAD cell lines, indicating their potential as therapeutic targets. Drug sensitivity investigation revealed a correlation between the expression levels of PTTG1 and PTTG3P and resistance to several anticancer medicines, indicating that targeting these genes may improve treatment outcomes. Studies examining the interplay between drugs and genes in cancer therapy have revealed similar results [42-46].
This study has several advantages and limitations that merit consideration. One of the primary advantages is the comprehensive analysis of PTTG1, PTTG2, and PTTG3P gene expression, methylation, and protein levels across multiple datasets and experimental platforms, providing robust and reproducible results. The results are more valid since data from patient samples and cell lines are combined, and a variety of analytical techniques, including RT-qPCR, bisulfite sequencing, and immunohistochemistry, are employed. Moreover, the reliability of the prognostic models created is improved by the study’s utilization of independent validation cohorts. However, there are limitations, including the relatively small sample size in some analyses, which may affect the generalizability of the results. The study is also limited by its retrospective nature and reliance on publicly available datasets, which may introduce selection bias. Moreover, while the findings suggest mechanistic roles for the PTTG genes in LUAD, further in vivo studies and functional assays are needed to confirm these mechanisms.
Conclusion
In conclusion, our thorough investigation shows that PTTG genes are highly hypomethylated and up-regulated in LUAD and that high expression levels are associated with a poor prognosis. These genes constitute important options for use as therapeutic targets as well as prognostic and diagnostic biomarkers. Our results contribute to the increasing amount of data regarding the involvement of PTTG genes in cancer and lay the groundwork for further investigations into their mechanisms and possible therapeutic applications in LUAD.
Acknowledgements
This work was supported by the Application research of 3D printed personalized guideplate in acetabular reconstruction of total hip arthroplasty. This work was supported by the Study on the interaction network of FHL1 and MMP-2 involved in the inhibition of invasion and metastasis of lung cancer cells, Taizhou Science and Technology Program, Project No. 22ywa18.
Disclosure of conflict of interest
None.
References
- 1.Wang C, Shao J, Song L, Ren P, Liu D, Li W. Persistent increase and improved survival of stage I lung cancer based on a large-scale real-world sample of 26,226 cases. Chin Med J (Engl) 2023;136:1937–1948. doi: 10.1097/CM9.0000000000002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill W, Lim EL, Weeden CE, Lee C, Augustine M, Chen K, Kuan FC, Marongiu F, Evans EJ Jr, Moore DA, Rodrigues FS, Pich O, Bakker B, Cha H, Myers R, van Maldegem F, Boumelha J, Veeriah S, Rowan A, Naceur-Lombardelli C, Karasaki T, Sivakumar M, De S, Caswell DR, Nagano A, Black JRM, Martínez-Ruiz C, Ryu MH, Huff RD, Li S, Favé MJ, Magness A, Suárez-Bonnet A, Priestnall SL, Lüchtenborg M, Lavelle K, Pethick J, Hardy S, McRonald FE, Lin MH, Troccoli CI, Ghosh M, Miller YE, Merrick DT, Keith RL, Al Bakir M, Bailey C, Hill MS, Saal LH, Chen Y, George AM, Abbosh C, Kanu N, Lee SH, McGranahan N, Berg CD, Sasieni P, Houlston R, Turnbull C, Lam S, Awadalla P, Grönroos E, Downward J, Jacks T, Carlsten C, Malanchi I, Hackshaw A, Litchfield K TRACERx Consortium. DeGregori J, Jamal-Hanjani M, Swanton C. Lung adenocarcinoma promotion by air pollutants. Nature. 2023;616:159–167. doi: 10.1038/s41586-023-05874-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou X, Ji L, Ma Y, Tian G, Lv K, Yang J. Intratumoral microbiota-host interactions shape the variability of lung adenocarcinoma and lung squamous cell carcinoma in recurrence and metastasis. Microbiol Spectr. 2023;11:e0373822. doi: 10.1128/spectrum.03738-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinjab A, Rahal Z, Kadara H. Cell-by-Cell: unlocking lung cancer pathogenesis. Cancers (Basel) 2022;14:3424. doi: 10.3390/cancers14143424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saab S, Zalzale H, Rahal Z, Khalifeh Y, Sinjab A, Kadara H. Insights into lung cancer immune-based biology, prevention, and treatment. Front Immunol. 2020;11:159. doi: 10.3389/fimmu.2020.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Y, Kelava L, Kiss I. MiRNAs in lung adenocarcinoma: role, diagnosis, prognosis, and therapy. Int J Mol Sci. 2023;24:13302. doi: 10.3390/ijms241713302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi YX, Sheng DQ, Cheng L, Song XY. Current landscape of epigenetics in lung cancer: focus on the mechanism and application. J Oncol. 2019;2019:8107318. doi: 10.1155/2019/8107318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papadimitriou E, Chatzellis E, Dimitriadi A, Kaltsas GA, Theocharis S, Alexandraki KI. Prognostic biomarkers in pituitary tumours: a systematic review. touchREV Endocrinol. 2023;19:42–53. doi: 10.17925/EE.2023.19.2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlotides G, Eigler T, Melmed S. Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocr Rev. 2007;28:165–186. doi: 10.1210/er.2006-0042. [DOI] [PubMed] [Google Scholar]
- 11.Smith VE, Franklyn JA, McCabe CJ. Pituitary tumor-transforming gene and its binding factor in endocrine cancer. Expert Rev Mol Med. 2010;12:e38. doi: 10.1017/S1462399410001699. [DOI] [PubMed] [Google Scholar]
- 12.Cui L, Ren T, Zhao H, Chen S, Zheng M, Gao X, Feng D, Yang L, Jin X, Zhuo R. Suppression of PTTG1 inhibits cell angiogenesis, migration and invasion in glioma cells. Med Oncol. 2020;37:73. doi: 10.1007/s12032-020-01398-2. [DOI] [PubMed] [Google Scholar]
- 13.Noll JE, Vandyke K, Hewett DR, Mrozik KM, Bala RJ, Williams SA, Kok CH, Zannettino AC. PTTG1 expression is associated with hyperproliferative disease and poor prognosis in multiple myeloma. J Hematol Oncol. 2015;8:106. doi: 10.1186/s13045-015-0209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang W, Wu X, Huang C, Wang M, Zhao X, Luo G, Li Y, Jiang G, Xiao X, Zeng F. PTTG1 regulated by miR-146a-3p promotes bladder cancer migration, invasion, metastasis and growth. Oncotarget. 2017;8:664–678. doi: 10.18632/oncotarget.13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren Q, Jin B. The clinical value and biological function of PTTG1 in colorectal cancer. Biomed Pharmacother. 2017;89:108–115. doi: 10.1016/j.biopha.2017.01.115. [DOI] [PubMed] [Google Scholar]
- 16.Grzechowiak I, Graś J, Szymańska D, Biernacka M, Guglas K, Poter P, Mackiewicz A, Kolenda T. The oncogenic roles of PTTG1 and PTTG2 genes and pseudogene PTTG3P in head and neck squamous cell carcinomas. Diagnostics (Basel) 2020;10:606. doi: 10.3390/diagnostics10080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta N. DNA extraction and polymerase chain reaction. J Cytol. 2019;36:116–117. doi: 10.4103/JOC.JOC_110_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent) Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5439. pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- 19.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang G, Cho M, Wang X. OncoDB: an interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 2022;50:D1334–D1339. doi: 10.1093/nar/gkab970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lánczky A, Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res. 2021;23:e27633. doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu CJ, Hu FF, Xie GY, Miao YR, Li XW, Zeng Y, Guo AY. GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief Bioinform. 2023;24:bbac558. doi: 10.1093/bib/bbac558. [DOI] [PubMed] [Google Scholar]
- 26.Lu C, Bera K, Wang X, Prasanna P, Xu J, Janowczyk A, Beig N, Yang M, Fu P, Lewis J, Choi H, Schmid RA, Berezowska S, Schalper K, Rimm D, Velcheti V, Madabhushi A. A prognostic model for overall survival of patients with early-stage non-small cell lung cancer: a multicentre, retrospective study. Lancet Digit Health. 2020;2:e594–e606. doi: 10.1016/s2589-7500(20)30225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sial N, Rehman JU, Saeed S, Ahmad M, Hameed Y, Atif M, Rehman A, Asif R, Ahmed H, Hussain MS, Khan MR, Ambreen A, Ambreen A. Integrative analysis reveals methylenetetrahydrofolate dehydrogenase 1-like as an independent shared diagnostic and prognostic biomarker in five different human cancers. Biosci Rep. 2022;42:BSR20211783. doi: 10.1042/BSR20211783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melocchi V, Dama E, Mazzarelli F, Cuttano R, Colangelo T, Di Candia L, Lugli E, Veronesi G, Pelosi G, Ferretti GM, Taurchini M, Graziano P, Bianchi F. Aggressive early-stage lung adenocarcinoma is characterized by epithelial cell plasticity with acquirement of stem-like traits and immune evasion phenotype. Oncogene. 2021;40:4980–4991. doi: 10.1038/s41388-021-01909-z. [DOI] [PubMed] [Google Scholar]
- 29.Ullah L, Hameed Y, Ejaz S, Raashid A, Iqbal J, Ullah I, Ejaz SA. Detection of novel infiltrating ductal carcinoma-associated BReast CAncer gene 2 mutations which alter the deoxyribonucleic acid-binding ability of BReast CAncer gene 2 protein. J Cancer Res Ther. 2020;16:1402–1407. doi: 10.4103/jcrt.JCRT_861_19. [DOI] [PubMed] [Google Scholar]
- 30.Shih JH, Chen HY, Lin SC, Yeh YC, Shen R, Lang YD, Wu DC, Chen CY, Chen RH, Chou TY, Jou YS. Integrative analyses of noncoding RNAs reveal the potential mechanisms augmenting tumor malignancy in lung adenocarcinoma. Nucleic Acids Res. 2020;48:1175–1191. doi: 10.1093/nar/gkz1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y, Guo J, Zhou J, Lu J, Chen Q, Zhang C, Qing C, Koeffler HP, Tong Y. FoxM1 transactivates PTTG1 and promotes colorectal cancer cell migration and invasion. BMC Med Genomics. 2015;8:49. doi: 10.1186/s12920-015-0126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou C, Tong Y, Wawrowsky K, Melmed S. PTTG acts as a STAT3 target gene for colorectal cancer cell growth and motility. Oncogene. 2014;33:851–861. doi: 10.1038/onc.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read ML, Seed RI, Fong JC, Modasia B, Ryan GA, Watkins RJ, Gagliano T, Smith VE, Stratford AL, Kwan PK, Sharma N, Dixon OM, Watkinson JC, Boelaert K, Franklyn JA, Turnell AS, McCabe CJ. The PTTG1-binding factor (PBF/PTTG1IP) regulates p53 activity in thyroid cells. Endocrinology. 2014;155:1222–1234. doi: 10.1210/en.2013-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy D, Ferreira MCMR, Reichert CO, de Almeida LV, Brocardo G, Lage LAPC, Culler HF, Nukui Y, Bydlowski SP, Pereira J. Cell cycle changes, DNA ploidy, and PTTG1 gene expression in HTLV-1 patients. Front Microbiol. 2020;11:1778. doi: 10.3389/fmicb.2020.01778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panguluri SK, Yeakel C, Kakar SS. PTTG: an important target gene for ovarian cancer therapy. J Ovarian Res. 2008;1:6. doi: 10.1186/1757-2215-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakachi I, Helfrich BA, Spillman MA, Mickler EA, Olson CJ, Rice JL, Coldren CD, Heasley LE, Geraci MW, Stearman RS. PTTG1 levels are predictive of saracatinib sensitivity in ovarian cancer cell lines. Clin Transl Sci. 2016;9:293–301. doi: 10.1111/cts.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, Tang J, Zhang H, Kong F, Zhu H, Li P, Li Z, Kong X, Wang K. A novel lncRNA PTTG3P/miR-132/212-3p/FoxM1 feedback loop facilitates tumorigenesis and metastasis of pancreatic cancer. Cell Death Discov. 2020;6:136. doi: 10.1038/s41420-020-00360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Sun Y, Yang J, Wu D, Yu S, Liu J, Hu T, Luo J, Zhou H. DNMT1-targeting remodeling global DNA hypomethylation for enhanced tumor suppression and circumvented toxicity in oral squamous cell carcinoma. Mol Cancer. 2024;23:104. doi: 10.1186/s12943-024-01993-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nejati-Koshki K, Roberts CT, Babaei G, Rastegar M. The epigenetic reader methyl-CpG-binding protein 2 (MeCP2) is an emerging oncogene in cancer biology. Cancers (Basel) 2023;15:2683. doi: 10.3390/cancers15102683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandey R, Johnson N, Cooke L, Johnson B, Chen Y, Pandey M, Chandler J, Mahadevan D. TP53 mutations as a driver of metastasis signaling in advanced cancer patients. Cancers (Basel) 2021;13:597. doi: 10.3390/cancers13040597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao F, Chen YY, Wang HC. GLI1 and PTTG1 expression in colorectal carcinoma patients undergoing radical surgery and their correlation with lymph node metastasis. World J Gastrointest Surg. 2024;16:1328–1335. doi: 10.4240/wjgs.v16.i5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neary B, Zhou J, Qiu P. Identifying gene expression patterns associated with drug-specific survival in cancer patients. Sci Rep. 2021;11:5004. doi: 10.1038/s41598-021-84211-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cecchin E, De Mattia E, Ecca F, Toffoli G. Host genetic profiling to increase drug safety in colorectal cancer from discovery to implementation. Drug Resist Updat. 2018;39:18–40. doi: 10.1016/j.drup.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Xu W, Li H, Hameed Y, Abdel-Maksoud MA, Almutairi SM, Mubarak A, Aufy M, Alturaiki W, Alshalani AJ, Mahmoud AM, Li C. Elucidating the clinical and immunological value of m6A regulator-mediated methylation modification patterns in adrenocortical carcinoma. Oncol Res. 2023;31:819–831. doi: 10.32604/or.2023.029414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu H, Umair M, Khan SA, Sani AI, Iqbal S, Khalid F, Sultan R, Abdel-Maksoud MA, Mubarak A, Dawoud TM, Malik A, Saleh IA, Al Amri AA, Algarzae NK, Kodous AS, Hameed Y. CDCA8, a mitosis-related gene, as a prospective pan-cancer biomarker: implications for survival prognosis and oncogenic immunology. Am J Transl Res. 2024;16:432–445. doi: 10.62347/WSEF7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdel-Maksoud MA, Ullah S, Nadeem A, Shaikh A, Zia MK, Zakri AM, Almanaa TN, Alfuraydi AA, Mubarak A, Hameed Y. Unlocking the diagnostic, prognostic roles, and immune implications of BAX gene expression in pan-cancer analysis. Am J Transl Res. 2024;16:63–74. doi: 10.62347/TWOY1681. [DOI] [PMC free article] [PubMed] [Google Scholar]