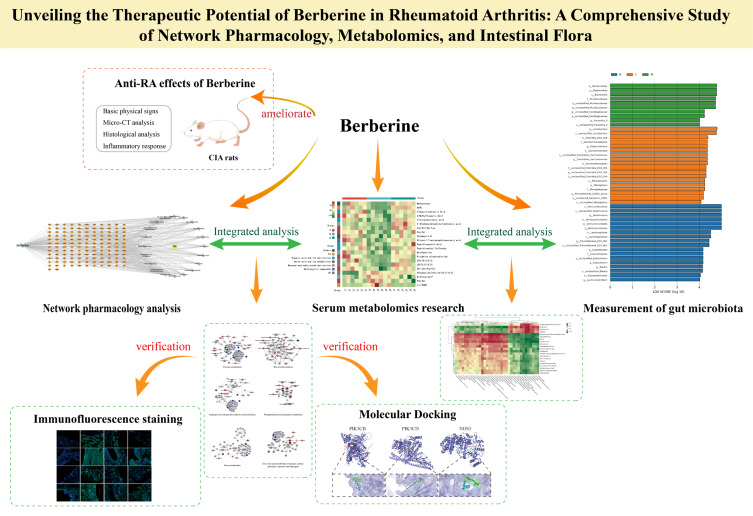
Abstract
Purpose
Rheumatoid arthritis (RA) is a chronic inflammatory autoimmune disease influenced by environmental triggers, including the commensal microbiota. Recent research has highlighted distinctive features of the gut microbiota in RA patients. This study investigates the therapeutic potential of berberine (BBR), a gut microbiota modulator known for its significant anti-RA effects, and elucidates the underlying mechanisms.
Methods
Utilizing the collagen-induced arthritis (CIA) rat model, we comprehensively evaluated the anti-rheumatoid arthritis effects of BBR in vivo through various indices, such as paw edema, arthritis index, ankle diameter, inflammatory cytokine levels, pathological conditions, and micro-CT analysis. Employing network pharmacology, we identified potential targets involved in RA alleviation by BBR. To analyze comprehensive metabolic profiles and identify underlying metabolic pathways, we conducted a serum-based widely targeted metabolomics analysis utilizing LC-MS technology. An integrated network encompassing metabolomics and network pharmacology data was constructed using Cytoscape. The potential therapeutic targets and signaling pathways of BBR in the management of RA were predicted using network pharmacology. Key targets and pathways were further validated by molecular docking and immunofluorescent staining, which integrated findings from serum metabolomics and network pharmacology analysis. Additionally, we analyzed the gut microbiota composition in rats employing 16S rDNA sequencing and investigated the effects of BBR on the microbiota of CIA rats through bioinformatics and statistical methods.
Results
Our results showed that BBR demonstrated significant efficacy in alleviating RA symptoms in CIA rats, as evidenced by improvements in paw redness and swelling, attenuation of bone and cartilage damage, reduction in synovial hyperplasia, inflammatory cell infiltration, and suppression of proinflammatory cytokines IL-1β, IL-6, IL-17A, and TNF-α. KEGG analysis highlighted the PI3K/AKT signaling pathway as a key mediator of BBR’s anti-RA effects. Metabolomics profiling via LC-MS revealed 22 potential biomarkers. Arginine and proline metabolism, cutin, suberine and wax biosynthesis, glycine, serine and threonine metabolism and taurine and hypotaurine metabolism are the most related pathways of BBR anti-RA. Molecular docking studies corroborated high affinities between BBR and key targets. Furthermore, 16S analysis demonstrated BBR’s capacity to modulate gut bacteria composition, including an increase in the abundance of Lachnoclostridium, Akkermansia, Blautia, Romboutsia, and Faecalibacterium genera, alongside a decrease in Prevotella_9 abundance in genus level. Integrated analysis underscored a strong correlation between serum microbiota and fecal metabolites.
Conclusion
Our findings elucidate the multifaceted mechanisms underlying BBR’s therapeutic efficacy in RA, involving inhibition of the PI3K/AKT pathway, modulation of intestinal flora, and regulation of host metabolites. These insights provide novel perspectives on BBR’s role in RA management.
Keywords: berberine, rheumatoid arthritis, gut microbiota, 16S rRNA sequencing, serum metabolism, network pharmacology
Graphical Abstract
Introduction
Rheumatoid arthritis (RA) is an autoimmune disorder distinguished by persistent synovitis, leading to localized bone and cartilage damage, along with various systemic manifestations. The quality of life for individuals with RA is significantly compromised due to joint stiffness and swelling, which can result in irreversible bone damage in milder instances and profound bone destruction or disability in severe cases.1 Despite extensive research, the exact etiology and pathology of RA remain elusive, and the quest for less toxic yet efficacious drugs remain ongoing, with no specific pharmacological breakthroughs to date.1 Despite the availability of numerous effective therapies for RA, remission rates remain below 50%, with a notable proportion of patients classified as refractory, underscoring the absence of a definitive cure for this condition.2
Emerging evidence underscores the intricate relationship between alterations in gut microbiota compositions, serum metabolites, and RA, known as the gut-joint axis.3–5 Coptis chinensis (CC) is known for its diverse pharmacological effects including antibacterial, anti-inflammatory, antioxidant, anti-tumor, antiarrhythmic properties.6 With over 130 chemical constituents, CC primarily contains alkaloids, coumarins, organic acids, and flavonoids. Extracts from CC, such as BBR, exhibit notable antibacterial and anti-inflammatory properties.7 BBR, an isoquinoline alkaloid with a history of over 400 years in traditional Chinese, Indian, and Middle Eastern medicine, has gained recent attention for its therapeutic potential, particularly in managing RA.8–10 Studies suggest that berberine effectively alleviates RA symptoms by reducing inflammatory cytokine levels, modulating intestinal flora, enhancing uric acid excretion, and mitigating inflammatory damage to joints and surrounding tissues.11 However, the specific mechanism underlying berberine’s anti-RA effects warrants further investigation.
Network pharmacology, primarily investigated through systems biology research methods, enhances comprehension of cellular and organ behaviors at the molecular level. It accelerates the identification of drug targets, discovery of new biomarkers, and construction of biological networks.12 Additionally, molecular docking simulation, matching a ligand (drug) to a target molecule (receptor) by generating various conformations, proves invaluable in drug discovery and molecular design.13,14 The successful integration of molecular docking simulation and network pharmacology offers a robust model for treating RA with BBR.
In this investigation, we established a rat model of RA by administering Freund’s Incomplete Adjuvant combined with Bovine Type II Collagen and assessed the establishment of the CIA model and the effectiveness of BBR in treating RA through parameters such as paw thickness, arthritis score, cytokine levels, histopathological examination, and micro-CT analysis. We utilized network pharmacology to identify the main targets, and potential mechanisms of BBR in treating RA. The predictive pathways identified through network pharmacology were validated using molecular docking and Immunofluorescence (IF) techniques. Leveraging LC-MS technology, we conducted serum metabolomics to identify endogenous metabolites and metabolic pathways. Furthermore, we investigated the composition of intestinal flora and serum metabolites, analyzing the correlation between serum metabolites and gut microbiota. By integrating the findings from network pharmacology, metabolomics, and gut microbiota analysis, we systematically elucidated the mechanisms underlying the anti-RA effects of BBR.
Materials and Methods
Materials
Freund’s Incomplete Adjuvant (F5506) were purchased from Sigma Biological Engineering Co., Ltd. Bovine Type II Collagen (20022) was gotten from Chondrex Co., Ltd. Hematoxylin-Eosin staining solution (G1120), Modified Safranin O-Fast Green Cartilage Staining Kit (G1371) and Tartrate-Resistant Acid Phosphatase (TRAP) Stain Kit (GP1492) were gotten from Beijing Solarbio Science Technology Co., Ltd. ELISA kits of TNF-α (MM-0180R1), IL-1β (MM-0922R2), IL-6 (MM-0190R1) and IL-17A (MM-70049R1) were purchased from Jiangsu Meimian Industrial Co., Ltd (Jiangsu, China). Antibodies against inhibitor of PIK3CB (ab151549) and second antibody, including Alexa Fluor 488 (ab150077 and ab150113) were purchased from Abcam Co., Ltd (Cambridge, UK), PIK3CD (12788-1-AP), AKT (60203-2-Ig), NOS3 (27,120-1-AP) were were got from Proteintech Co., Ltd (Wuhan, China). Berberine (purity ≥98%) was purchased from Zelang Pharmaceutical Technology Co., Ltd. (Nanjing, China). MTX (XW00590521) was got from Sinopharm Chemical Reagent Co., Ltd. and used as the positive control, TGuide S96 Magnetic Soil/Stool DNA Kit was purchased from Tiangen Biotech Co., Ltd (Beijing, China).
Animals
SPF-grade male Sprague-Dawley (SD) rats, weighing 200 ± 10 grams, were procured from Hubei Beinte Bio-Tech Co., Ltd. (Hubei, China) (certified under number SCXY (E) 2021–0027). These animals were housed in a well-regulated environment with a standard 12-hour light/dark cycle, maintained at a constant temperature of 24 ± 1°C and relative humidity of 50 ± 10%. Food and water were provided to the animals without restriction. All experimental procedures involving animals adhered strictly to the ethical guidelines approved by the Animal Experiments Ethics Committee of Hubei University of Chinese Medicine (Approval No.00298217).
CIA Induction and Drug Administration
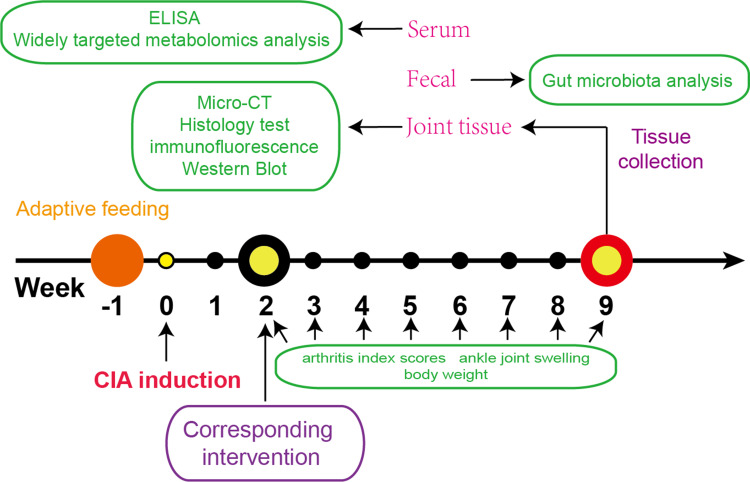
In this study, a collagen-induced arthritis (CIA) model was established in rats through the administration of type II collagen and Freund’s incomplete adjuvant following adaptive feeding.15 The rats were then categorized into three groups: model group; BBR group (treatment at a dosage of 200 mg/kg administered orally)16 and MTX group (treated with MTX at a dosage of 1 mg/kg once per week).17 Each group consisted of six rats, and all underwent a 7-week treatment period to evaluate the efficacy and effects of the respective interventions on arthritis progression. The experimental timeline was shown in Figure 1.
Figure 1.
Eighteen eight-week-old Sprague–Dawley rats received an type II collagen and Freund’s incomplete adjuvant to build RA model after seven days of adaptive feeding. Then, experimental groups underwent BBR and MTX treatment 14 days after the initial immunization, and the arthritis index scores, ankle joint swelling and body weight tested every week. After seven weeks of intervention, all rats were sacrificed. Serum were collected to conduct Elisa and widely targeted metabolomics analysis. Feces were collected to conduct gut microbiota analysis. Cartilage and synovium from ankle joint were collected to conduct histology test immunofluorescence labeling.
Measurement of Arthritis Index Scores, Ankle Joint Swelling, Body Weight and ELISA Assay
Throughout the study, various parameters were assessed to monitor arthritis progression. Body weight, paw thickness, and arthritis score were measured every 3 days from week 2 to week 9. Hind-paw thickness was quantified using a digital vernier scale. Arthritis severity was evaluated using a scoring system ranging from 0 to 4, with the cumulative score of both hind paws serving as the arthritis index, with a maximum value of 8.18 Importantly, all scoring procedures were conducted in a blinded manner to minimize bias and ensure the accuracy and reliability of the data collected.
In week 9, Blood sample was collected to estimate cytokine levels. Serum was obtained by centrifuging blood samples at 3500 rpm and 4 °C for 15 minutes. ELISA kits were employed to quantify levels of IL-1β, IL-6, IL-17A, and TNF-α in the serum, following the manufacturer’s recommendations.
Histopathological and Micro-CT Analysis
At week 9, the hind paws of rats were collected and the skin was removed, and they were then fixed in 4% paraformaldehyde. Tissue samples underwent decalcification using 10% EDTA for 8 weeks before being embedded in paraffin. Hematoxylin and eosin (H&E), safranin O-fast green, and tartrate-resistant acid phosphatase (TRAP) stains were conducted on 5-micrometer sagittal sections of the ankle joint to assess histopathological changes. The histological assessment of ankle joints was conducted in a blinded manner, adhering to the following criteria.19 Synovial Inflammation: 0 = No evidence of hyperemia or inflammation; 1 = Mild hyperemia accompanied by 2–4 layers of reactive synovial cells; 2 = Moderate hyperplasia, >4 layers of reactive synovial cells and scattered inflammatory cell infiltration; 3 = Severe hyperplasia, extensive inflammatory infiltration into the synovial space. Cartilage damage: 0 = Smooth joint surface without any erosion; 1 = Rough articular surface with minimal bone erosion; 2 = Ulcerated articular surface, significant cartilage erosion; 3 = Severe bone erosion with complete absence of staining in cartilage. Immunofluorescence staining was performed using primary antibodies targeting PIK3CB (1:100), PIK3CD (1:100), AKT (1:100), and NOS3 (1:100), followed by secondary labeling with Alexa Fluor 488 (1:500) and nuclear staining with DAPI. Immunofluorescence images were captured using a microscope system (Carl Zeiss, Axio Observer Z1), ensuring a minimum of 3 random replicates for each sample. CIA rats’ joint bone images were obtained using a Skyscan 1276 micro-CT scanner (Bruker microCT, Kontich, Belgium) and reconstructed using NRecon.
Targets Prediction, Pathway Enrichment Analysis and Network Construction
BBR targets were sourced from the TCMSP database,20 the SwissTargetPrediction database,21 and the PharmMapper server.22 Targets related to “Rheumatoid arthritis” were identified in the DisGeNET database,23 the Therapeutic Target database, the GeneCards database,23 and the OMIM database,24 and the retrieved gene targets were merged.
To elucidate the pathways through which BBR treats RA, the David database was utilized for GO biological processes pathway and KEGG enrichment analysis.25 The top 20 KEGG pathways were selected based on the order of p-values, arranged from smallest to largest. Detailed information is provided in Table S1.
Protein-Protein Interaction (PPI) network, drug-targets-pathways network and drug-reaction-enzyme-gene network was constructed by Cytoscape 3.10.2 software. Using MetScape plug-in to obtain the drug-reaction-enzyme-gene network.
Widely Targeted Metabolomics Analysis
Serum samples were thawed on ice from −80°C, vortexed for 10 seconds, then added to a 2 mL microcentrifuge tube along with 50μL of sample and 300μL of extraction solution (ACN: Methanol = 1:4, V/V). The mixture was vortexed for 3 minutes, centrifuged at 12000 rpm for 10 minutes at 4°C, and 200μL of the supernatant were collected and stored at −20°C for 30 minutes. 180μL of the supernatant was then analyzed by ultra-performance liquid chromatography (UPLC, ExionLC AD) and quadrupole-time of flight mass spectrometry (TripleTOF 6600, AB SCIEX).
UPLC conditions included a Waters ACQUITY UPLC HSS T3 C18 column (1.8μm, 2.1 mm×100 mm) maintained at 40°C, with a flow rate of 0.4 mL/min, and an injection volume ranging from 2μL to 5μL. The solvent system consisted of water (0.1% formic acid) and acetonitrile (0.1% formic acid), with a gradient program starting at 95:5 V/V at 0 minutes, transitioning to 10:90 V/V at 10.0 minutes, maintaining 10:90 V/V until 11.0 minutes, returning to 95:5 V/V at 11.1 minutes, and holding at 95:5 V/V until 14.0 minutes.
The mass spectrometry (MS) conditions were set as follow: electrospray ionization (ESI) temperature at 500°C; Ion spray voltage (IS) at 5500 V, positive mode and −4500 V, negative mode. Gas I (GSI) and gas II (GSII) at 50 psi each, curtain gas (CUR) at 25.0 psi. Collision gas (CAD) at high. A specific set of multiple reaction monitoring (MRM) transitions was monitored for each period based on eluted metabolites, ensuring accurate and comprehensive data acquisition and analysis.
Gut Microbiota Analysis
Fecal samples from colon were collected fresh and kept at −80 °C after rats were sacrificed. The TGuide S96 Magnetic Soil/Stool DNA Kit was used in this study to extract genomic DNA from rat feces. The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified using primer pairs 338F: 5’-ACTCCTACGGGAGGCAGCA-3’ and 806R: 5’-GGACTACHVGGGTWTCTAAT-3’. PCR products were visualized on agarose gel and subsequently purified using the Omega DNA purification kit. The purified PCR products underwent paired-end sequencing (2 × 250 bp) on the Illumina Novaseq 6000 platform. Sequences were then clustered into operational taxonomic units (OTUs) at a 97% similarity threshold using USEARCH (version 10.0). Taxonomy annotation of OTUs/ASVs was performed using the Naive Bayes classifier in QIIME2 with the SILVA database (release 138.1), confidence threshold was 70%.
Alpha diversity analysis was conducted using QIIME2 software to assess species diversity complexity in each sample. Principal coordinate analysis (PCoA) was utilized for beta diversity calculations. Bacterial abundance and diversity were compared using one-way analysis of variance. Linear discriminant analysis coupled with effect size (LEfSe) was applied to identify differentially abundant taxa. Finally, sequencing data were analyzed using the online platform BMKCloud (https://www.biocloud.net).
Statistical Analysis
GraphPad Prism (version 8.3.0) facilitated our statistical analysis, with results expressed as means ± SD. For data not conforming to normal distribution, we applied the Mann–Whitney test. To discern significant variances, ANOVA and subsequent Tukey’s test were employed. Serum metabolite-genera correlations hinged on Spearman’s method via the Metware cloud platform (https://cloud.metware.cn/). Only completed the correlation in those genera (P < 0.05) and metabolites (P < 0.05, VIP > 1) were found to be statistically significant between groups.
Results
Alleviating Effects of BBR on Physical Indices and Bone Damage in CIA Rats
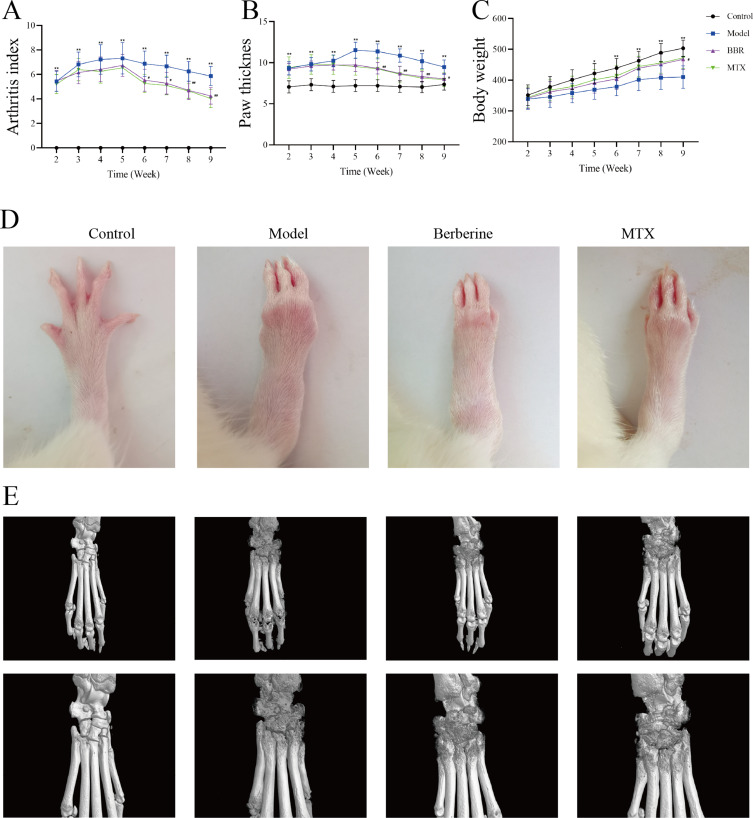
To investigate the effects of BBR on CFA-induced arthritis, we assessed arthritis index scores, ankle joint swelling, and body weight changes. In the model group, arthritis index scores markedly increased from 2th to 9th weeks post-primary immunization compared to the control group (P < 0.01). Treatment with BBR and Methotrexate (MTX) resulted in a significant reduction in arthritis index scores compared to the model group at the 6th week (P < 0.05, Figure 2A). Hind paw thickness notably increased in CIA rats compared to the control group (P < 0.01). Following treatment with BBR and MTX in CIA rats, paw thickness began to decrease from the 6th week (P < 0.01, Figure 2B). By the 5th week, the control group exhibited a significantly greater increase in body weight compared to the model groups (P < 0.05). However, by the 9th week, rats treated with BBR exhibited a notable increase in weight compared to the CIA model rats (P < 0.05, Figure 2C). These findings underscore the significant ameliorative effects of BBR on various symptoms associated with CFA-induced arthritis in rats.
Figure 2.
(A) The arthritis index of rats. (B) The paw thickness of rats. (C) The body weight changes of rats. Data were expressed as mean ± SD. *p < 0.05, **p < 0.01, compared with Control; #p < 0.05, ##p < 0.001 compared with Model. (D) The represented paw of rat in different treatment groups. (E) Represented image of Micro-CT analysis of joint destructions of rats.
The effects of BBR on joint damage in CIA rats were scrutinized through comprehensive morphological analysis and Micro-CT evaluations. As shown in Figure 2D, the morphological changes in paw appearance were compared among groups, revealing that while paws of control group rats appeared slim and flexible, those of model rats displayed evident RA symptoms, characterized by redness and swelling. Following treatment with BBR and MTX, these RA symptoms were alleviated to varying degrees. Micro-CT analysis of CIA rats displayed significant damage to bone structure, including erosion and the presence of irregular, coarse surfaces on the ankle and toe joints, indicating severe bone destruction, as shown in Figure 2E. After administration of BBR and MTX, there was a marked reduction in the severity of bone destruction, along with noticeable improvement in the roughness of the articular surfaces.
Effect of BBR on Histopathological Changes and Inflammatory Cytokines in CIA Rats
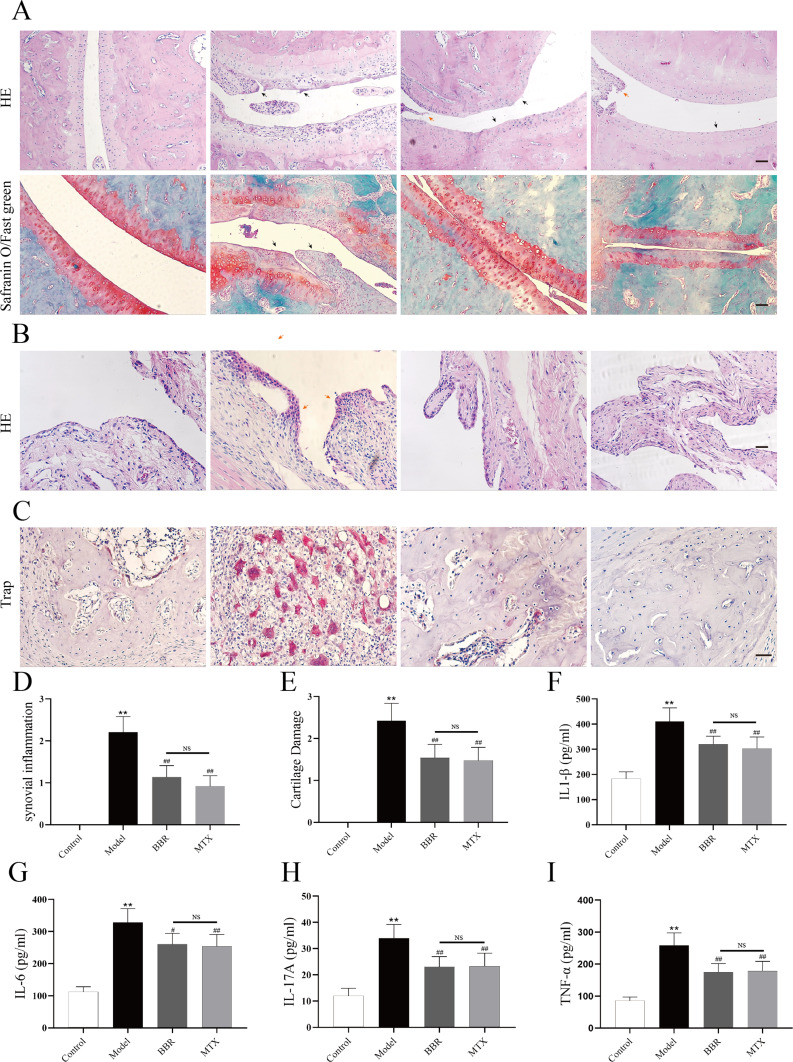
Results of histopathological alterations in ankle joints revealed that BBR modulated the pathological state of CIA rats. Representative images of H&E and Safranin O&Fast Green staining displayed cartilage damage, erosion, synovial hyperplasia, and inflammatory cell infiltration in model group rats compared to controls (Figure 3A and B). However, administration of MTX and BBR significantly ameliorated these pathological changes in RA. Histological assessment scores clearly demonstrated that BBR treatment effectively reduced cartilage damage and alleviated synovial inflammation in CIA rats, as depicted in Figure 3D and E. Furthermore, trap staining images (Figure 3C) indicated that the number of trap-positive cells elevated in the model group compared to controls. Treatment with MTX and BBR can decrease the number of trap-positive cells, suggesting BBR’s potential in ameliorating claw joint bone destruction in CIA rats.
Figure 3.
(A) and (B) Representative pathological images of ankle joints stained with HE and Safranin O/Fast green staining (100× magnification). Black arrow represents the cartilage damage and red arrow represents the Synovial inflammatory changes Scale bar = 100 μm. (C) Representative pathological images of subchondral bone stained with trap, Scale bar = 20 μm. (D and E) Histological evaluation score of cartilage damage and synovial inflammation (n = 6). **p < 0.01, compared with Control; ##p < 0.001 compared with Model; NS = no significant. (F–I) The serum concentrations of inflammatory cytokines IL-1β, IL-6, IL-17A, TNF-α. **p < 0.01, compared with Control; #p < 0.05, ##p < 0.001 compared with Model; NS = no significant.
The expression levels of IL-1β (Figure 3F), IL-6 (Figure 3G), IL-17A (Figure 3H), and TNF-α (Figure 3I) in serum was tested by Elisa. The expression of these cytokines in the model group was significantly elevated compared to the control group (P < 0.01). Treatment with BBR and MTX resulted in reduced levels of inflammatory cytokines compared to the model group (P < 0.05). These results showed that the CIA model was established successfully and both BBR and MTX effectively suppressed the inflammatory response.
The Combination Interaction of BBR and Target
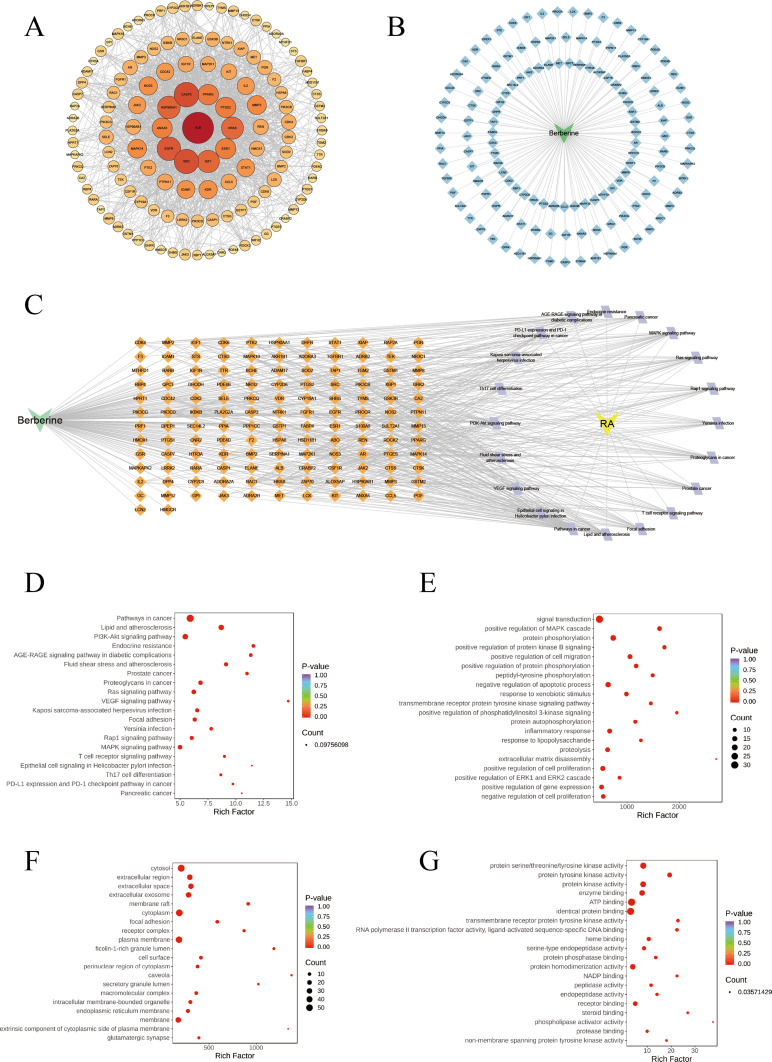
314 targets associated with BBR were found and labeled based on their gene symbols. Through database searches, 2298 genes relevant to RA were identified. Among these, 134 common target genes were input into the STRING database, applying a minimum required interaction score > 0.9 and specifying Homo sapiens as the species. The resulting protein-protein interaction (PPI) network was visually analyzed using Cytoscape 3.10.2 software, revealing 131 nodes and 1349 edges (Figure 4A). Through network analysis, nodes were color-coded and sized based on their degree to identify core targets. Key targets implicated in the anti-RA effects of BBR included ALB, CASP3, SRC, EGFR, HSP90AA1, and IGF1, based on their degree in the network, The details of these targets are shown in Table S2. Further investigation into the combined mechanisms of BBR for treating RA involved the construction of a drug-target network using 136 targets (Figure 4B). This analysis revealed that BBR interacts with multiple targets, resulting in 64 drug-target associations, highlighting its multifaceted approach in treating RA.
Figure 4.
(A) PPI network of BBR for the treatment of RA. (B) Drug-target network between the interaction network of 136 intersection genes (light blue diamond) and BBR. (C) Network of drug-targets-pathways. The Orange diamond represents related targets of top 20 KEGG signaling pathways, the lavender diamond represents top 20 KEGG signaling pathways. (D) Bubble chart of top 20 KEGG signaling pathways related to the effect of BBR treat RA. The results of GO enrichment analyzed by DAVID. (E) Biological process (BP), (F) Cell component (CC), (G) Molecular function (MF).
Effects of BBR on Pathways
The drug-targets-pathway network encompassing of 156 nodes (20 pathways, 134 targets, 1 drug, 1 disease) and 541 edges (Figure 4C). Notably, “Pathways in cancer” (degree = 47), PI3K/AKT signaling pathway (degree = 29), and Lipid and atherosclerosis (degree = 28), exhibited a relatively high number of target connections among others. The top three targets based on degrees were PIK3CB (degree = 18), PIK3CD (degree = 18), and HRAS (degree = 16).
We utilized DAVID 2021 to conduct GO function and KEGG analysis on the 134 candidate target genes, aiming to elucidate the molecular mechanism of BBR in treating RA. The KEGG pathway analysis revealed a strong correlation between several targets and the PI3K/AKT, MAPK, VEGF signaling pathways, all these pathways are intricately intertwined with inflammatory responses (Figure 4D). The GO evaluations were categorized into biological process (BP), cell component (CC), and molecular function (MF) terms. A total of 342 enrichment results in the BP category were identified, encompassing signal transduction, positive regulation of MAPK cascade, protein phosphorylation, and positive regulation of protein kinase B signaling (Figure 4E). Furthermore, 57 enrichment results were associated with CC, including cytosol, extracellular region, extracellular space, and extracellular exosome (Figure 4F). Additionally, 100 enrichment processes related to MF were identified, covering protein serine/threonine/tyrosine kinase activity, protein tyrosine kinase activity, protein kinase activity, and others (Figure 4G). Therefore, the PI3K/AKT signaling pathway was considered a key pathway in the anti-RA effects of BBR. These findings underscored the multifaceted nature of BBR’s treatment for RA, operating through multiple pathways.
Multivariate Data Analysis and Screening and Identification of Differential Metabolites
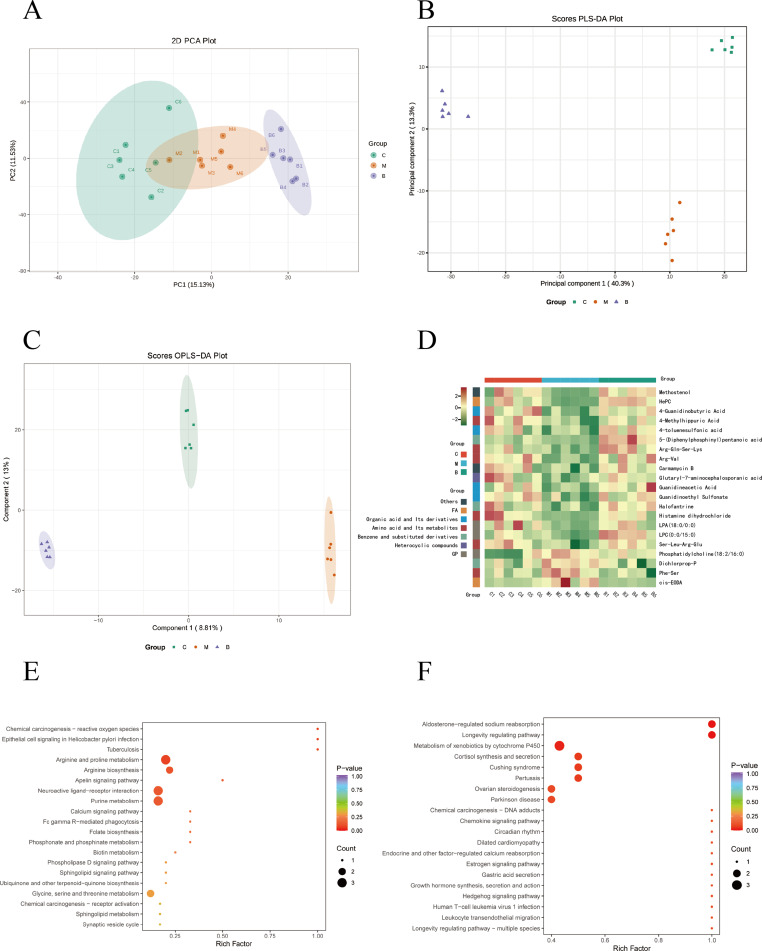
Initially, an unsupervised PCA model was employed to analyze all samples (Figure 5A). The PCA results effectively distinguished the control group from the model group, confirming the successful establishment of the RA model. Furthermore, the BBR group exhibited clear differentiation from the model group, indicating a discernible effect of BBR in treating RA. Subsequently, PLS-DA was utilized to extract variation information among groups to identify distinct compounds (Figure 5B). The PLS-DA score plots, demonstrated significant classification effects, with each group clearly separated. The supervised OPLS-DA model depicted the overall trends of all groups (Figure 5C).
Figure 5.
Metabolomics analysis of serum in rats. (A) PCA plot chart. PC1 represents the first principal component, PC2 represents the second principal component, and percentage represents the interpretation rate of this principal component to the data set. Each point in the diagram represents a sample, and samples from the same group are represented using the same color. (B) Partial Least Squares Discriminant Analysis (PLS-DA) plot of all groups. Data from different groups were considered significant when p < 0.05, as labeled by different lowercase letters. (C) OPLS-DA score chart. The horizontal coordinate indicates the predicted component score value, and the horizontal coordinate direction shows the gap between groups; the vertical coordinate indicates the orthogonal component score value, and the vertical coordinate direction shows the gap within groups; the percentage indicates the degree of explanation of the component to the dataset. (D), Hierarchical clustering analysis heatmap of 21 differential metabolites. (C) Control group, (M) Model group, (B) BBR group. (E and F) KEGG functional enrichment analysis of differential metabolites. Rich factor indicates the ratio of the number of differentially expressed metabolites in the corresponding pathway to the total number of metabolites annotated in the pathway. p value is closer to 0, indicating a more significant enrichment. The size of the dots in the graph represents the number of differentially significant metabolites enriched to the corresponding pathway.
Differential metabolites were identified based on VIP values (> 1.0) and T-test (p value < 0.05). This screening process yielded a total of 176 differential metabolites. Comparative analysis revealed that in the model group, 82 differential metabolites exhibited significant increases, while 94 showed noticeable decreases compared to the control group. However, treatment with BBR partially reversed the levels of these metabolites. Specifically, following BBR administration, 21 biomarkers (such as Deethylatrazine, Docosahexaenoic acid, Isoquinoline, etc.) exhibited a tendency to normalize. The heatmap analysis of these 21 differential metabolites across different groups illustrating a distinct cluster separation between the model and control groups (Figure 5D). Consequently, BBR demonstrated a capacity to mitigate metabolic disorders in CIA rats to some extent. The details of the differential metabolites are shown in Table S3.
Metabolic Pathways Analysis
The differentially expressed metabolites in the control and model groups were mainly enriched in arginine and proline metabolism, arginine biosynthesis, neuroactive ligand-receptor interaction and purine metabolism (Figure 5E). In addition, arginine and proline metabolism, cutin, suberine and wax biosynthesis, glycine, serine and threonine metabolism, taurine and hypotaurine metabolism were highly enriched in the model and BBR groups (Figure 5F).
Integrated Analysis of Metabolomics and Network Pharmacology
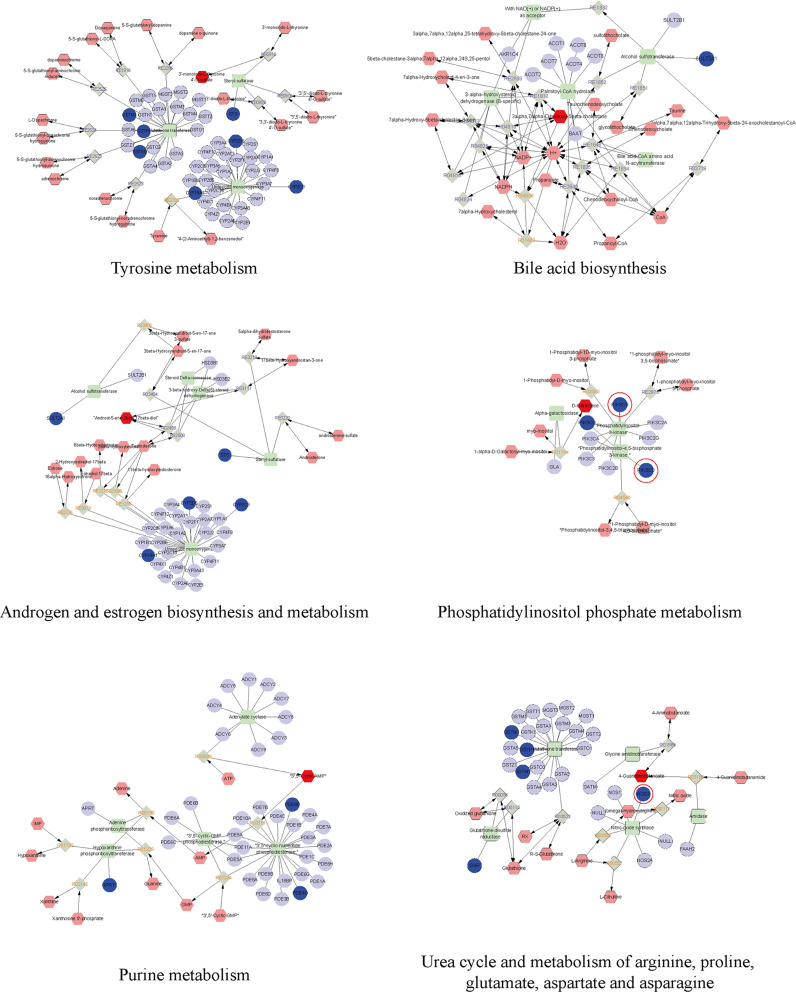
For a comprehensive understanding of BBR’s mechanisms in combating RA, we established an interaction network amalgamating metabolomics and network pharmacology (Figure 6). Differential metabolites were incorporated into the MetScape plugin within Cytoscape. Through correlating potential targets from network pharmacology with genes in MetScape analysis, 24 key targets were pinpointed, encompassing STS, GSTP1, NOS3, and others. The associated key metabolites included D-Galactose, 4-Guanidinobutanoate, and 3’-monoiodo-L-thyronine. Furthermore, the affected pathways encompassed tyrosine metabolism, phosphatidylinositol phosphate metabolism, androgen and estrogen biosynthesis and metabolism, bile acid biosynthesis, purine metabolism, and urea cycle and metabolism of arginine, proline, glutamate, aspartate, and asparagine. These pathways may contribute significantly to the therapeutic effects of BBR on RA. The screened targets were further matched with hub targets identified in the PPI pathway network; we identified NOS3. For subsequent validation, we focused on genes related to the PIK3-AKT pathway among the key genes, which are highlighted in red.
Figure 6.
The drug-reaction-enzyme-gene network networks of the key metabolites and targets. The red hexagons, grey diamonds, green round rectangle and blue circles represent the active drug, reactions, enzyme and genes, respectively. Key metabolites, proteins and genes were darkened. PIK3-AKT pathway relevant genes are circled in red.
Molecular Docking
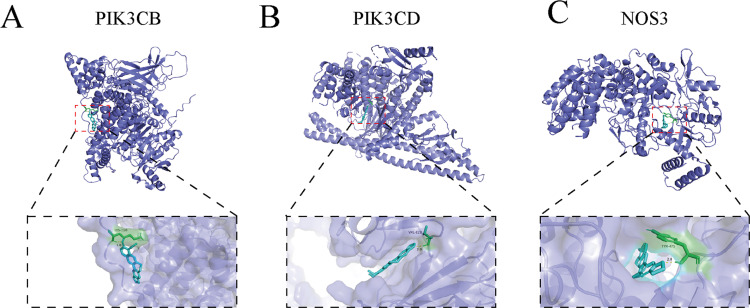
Drawing from the outcomes of network pharmacological analysis, our focus was on the core PI3K/AKT pathway, for which three key targets were selected for molecular docking investigations. Employing AutoDock Vina, we conducted molecular docking analyses between BBR and the three chosen targets: PI3KCB, PI3KCD, and NOS3. The PDB IDs of the receptor proteins for PIK3CD and NOS3 are 8bcy and 1m9q, respectively. As PI3KCB lacks PDB data, we utilized AlphaFold structure prediction (AF-P42338-F1). Our findings exhibited robust binding affinity between BBR and PI3KCB, with a binding energy of −5.62 kcal/mol (Figure 7A). Docking analyses unveiled hydrogen bonds formed between BBR and the LYS318 residues of the PI3KCB protein. Similarly, a strong binding affinity was observed between BBR and PI3KCD in the ligand binding domain of PI3KCD (binding energy of −6.44 kcal/mol), with hydrogen bonds formed between BBR and the GLN-116 residues of the PI3KCD (Figure 7B). Furthermore, BBR demonstrated favorable interaction with NOS3 (binding energy of −7.07 kcal/mol), forming hydrogen bonds with the TYR475 residues of the NOS3 (Figure 7C).
Figure 7.
Molecular docking simulation of compound-target binding. (A) MDT of BBR on PI3KCB. (B) MDT of BBR on PI3KCD. (C) MDT of BBR on NOS3.
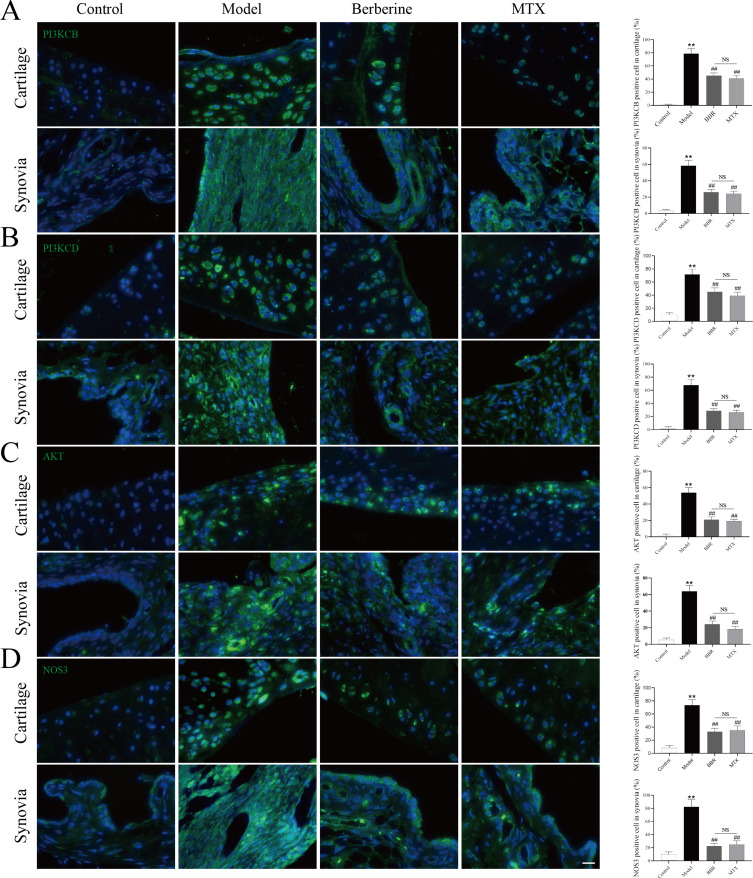
Effects of BBR on PI3K-AKT Signaling Pathways
The PI3K-AKT signaling pathway emerged as a pivotal route for RA treatment according to network pharmacology analysis. To delve deeper into the potential anti-RA mechanisms of BBR, we investigated its effects on PI3K-AKT signaling pathways (including PIK3CB, PIK3CD, AKT and NOS3), utilizing IF staining. The expression levels of PIK3CB, PIK3CD, AKT and NOS3 in the cartilage and synovial tissues of rats are depicted in Figure 8. Compared to the control group, the model group exhibited significantly heightened expression of PIK3CB, PIK3CD, AKT and NOS3 (P < 0.01). Following treatments with BBR and MTX in CIA rats, the expression of PIK3CB, PIK3CD, AKT and NOS3 markedly decreased compared to the model group (P < 0.01). Similarly, there was no significant difference observed between the BBR and MTX groups (Figure 8A–D).
Figure 8.
Immunofluorescence staining images for PIK3CB (A), PIK3CD (B), AKT (C) and NOS3 (D) in cartilage and synovia. Quantitative analysis of positive cells counting in cartilage and synovia was shown after immunofluorescence staining images (each group: n = 6, ANOVA). Scale bar = 20 μm. Values are mean ± SD. **P < 0.01: compared with the control group. ##P < 0.01: compared with the model group. Turkey’s post hoc tests were used for multiple comparisons. NS: no significant.
Effects of BBR on Gut Microbiota in CIA Rats
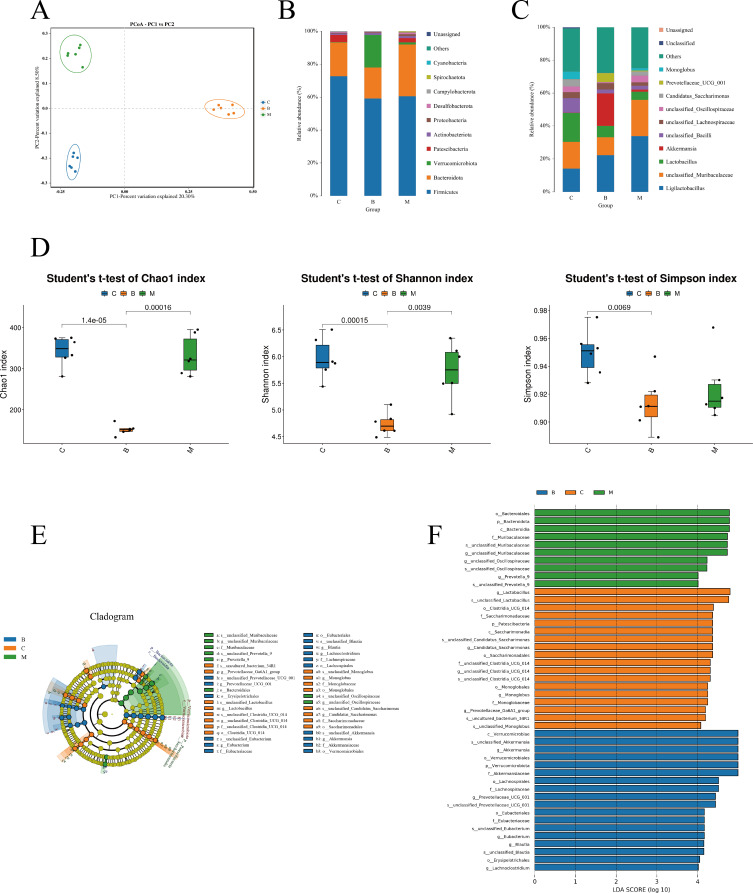
To investigate BBR’s impact on gut microbiota during RA treatment, 16S rRNA sequencing was employed. For β-diversity, there was a distinct separation in the distribution of microbiota composition among the three groups by PCoA analysis (Figure 9A). At phylum and genus levels, results of bacterial composition and relative abundance analysis showed that nine phyla with abundance exceeding 1%, among these, Firmicutes, Patescibacteria, and Bacteroidota were notably prevalent in each sample in each sample. Firmicutes emerged as the dominant phylum, representing approximately 70% in each group (Figure 9B). Bacteroidota and Proteobacteria followed Firmicutes in abundance. 11 genera with abundances of ≥1% were identified, with ligilactobacillus and unclassified_Muribaculaceae being the predominant genera across all three groups (Figure 9C). The other genera with high abundance in the control group were lactobacillus (17.61%), unclassified_Bacilli (8.82%), Candidatus_Saccharimonas (4.39%), and unclassified_Oscillospiraceae (3.49%). In the model group, ligilactobacillus (33.8%), lactobacillus (5%) and unclassified_bacilli (2.28%) evaluated remarkable compared with control group (P<0.05), We also found the abundance of unclassified_Muribaculaceae (10.8%) decreased (P<0.05) and akkermansia increased (P<0.05) in BRB group comparing with model group. Results of alpha diversity showed that, BBR significantly BBR significantly decreased the Chao1 and Shannon Index of gut microbiota in CIA rats.26,27 This finding supports the notion that BBR possesses natural antibacterial properties (Figure 9D). Linear Discriminate Analysis Effect Size (LEfSe) analysis was then conducted and the results indicated that the unclassified_Lactobacillus, Candidatus_Saccharimonas, unclassified_Clostridia_UCG_014 and Prevotellaceae_Ga6A1_group genus were enriched in the control group; in the CIA group, the unclassified_Muribaculaceae, unclassified_Oscillospiraceae and prevotella_9 genera were enriched; the Akkermansia, Prevotellaceae_UCG_001, Eubacterium, Blautia and Lachnoclostridium were enriched in the BRB groups (Figure 9E and F).
Figure 9.
(A) Beta diversity of the gut microbiota was assessed by principal coordinate analyses (PCoA). (B) The relative abundance of bacteria at the phylum level. (C) The relative abundance of bacteria at the genus level. (D)Alpha diversity indices including Chao1, Shannon and Simpson. (E) LEfSe classification evolutionary tree. (F) LEfSe linear discriminant analysis (LDA) score.
Correlation Analysis of the Widely Targeted Metabolomics and the Gut Microbiota
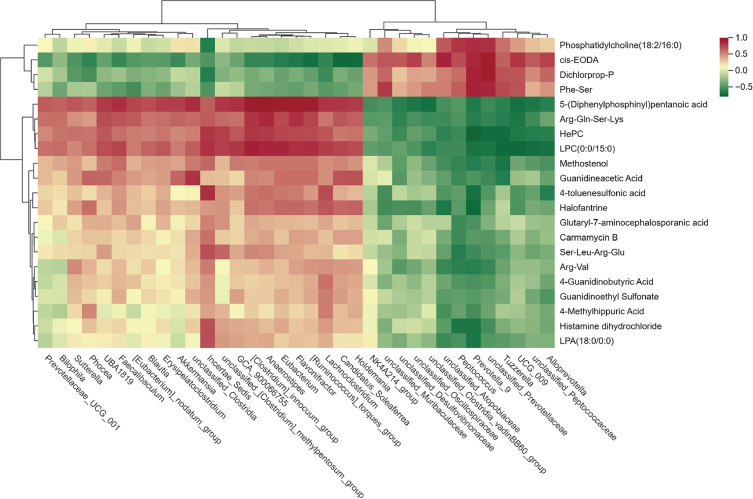
Spearman rank correlation coefficient analysis was employed to investigate the correlation between differential genera of intestinal flora and serum metabolites, which was then visualized in a heatmap chart. (Figure 10). The associations between 35 differential genera and 21 serum metabolites were investigated and identified numerous associations were existed. For instance,
Figure 10.
Spearman correlations between the gut microbiome and serum metabolites rats after BBR treatment. The magnitude of correlation was represented by color intensity (green, negative correlation; red, positive correlation).
Peptococcus, Prevotella_9 and unclassified_Prevotellaceae were negatively correlated with 4-Guanidinobutyric Acid. Incertae_Sedis was negatively, while Peptococcus, Prevotella_9, unclassified_Atopobiaceae and unclassified_Prevotellaceae were positively correlated with Phosphatidylcholine(18:2/16:0). Tuzzerella, unclassified_Prevotellaceae and Prevotella_9 were negatively, while Incertae_Sedis, [Clostridium]_innocuum_group and Anaerostipes, unclassified_[Clostridium]_methylpentosum_group were positively correlated with Hepc. Differential bacterial genera in terms of relative abundance and listed at Table S4.
Discussion
Metabolomics has become a widely employed tool for investigating disease mechanisms and intervention strategies. However, metabolomics studies typically only list potential metabolites and related pathways without delving into their direct relationships comprehensively. Due to the complexity and heterogeneity of metabolomics data, collaborative efforts are needed for data analysis and interpretation.28 The field of network pharmacology, grounded in systems biology, assesses the impact of drug polypharmacology on a molecular scale to foresee the interplay between natural substances and proteins, and to pinpoint key mechanisms.29,30 It further validates therapeutic regulation of metabolic networks and aids to identify of key targets and biomarkers.31 In our study, metabolomics analysis revealed that BBR administration reversed alterations in serum metabolites in CIA rats. Specifically, BBR significantly increased levels of Methostenol, HePC, 4-Guanidinobutyric Acid, 4-Methylhippuric Acid, 4-toluenesulfonic acid, 5-(Diphenylphosphinyl)pentanoic acid, Arg-Gln-Ser-Lys, Arg-Val, Carmamycin B, Glutaryl-7-aminocephalosporanic acid, Guanidineacetic Acid, Guanidinoethyl Sulfonate, Halofantrine, Histamine dihydrochloride, LPA(18:0/0:0), LPC(0:0/15:0), and Ser-Leu-Arg-Glu. Conversely, BBR led to a decrease in the abundance of Phosphatidylcholine (18:2/16:0), Dichlorprop-P, Phe-Ser, and cis-EODA in the serum of these rats. Guanidinoethyl sulfonate is the N-amidino derivative of taurine. TauCl, metabolite of Taurine exhibits anti-inflammatory and connective tissue protective effects, as demonstrated in rats with modified adjuvant-induced arthritis, where it notably down-regulated the generation of inflammatory mediators.32 Guanidinoacetic acid is the N-amidino derivative of glycine. Glycine has the protective effects to anti-oxidation and anti-inflammation and enhanced ferroptosis via SAM-related GPX4 promoter methylation to ameliorate CIA progression.33 Phe-Ser is a dipeptide that is the N-(L-phenylalanyl) derivative of L-serine. Serine metabolism is necessary for macrophage glutathione synthesis to support IL−1β cytokine production.34 Arg-Val is a dipeptide formed from L-arginine and L-valine residues while L-arginine is reported to ameliorate arthritis and bone erosion through metabolic reprogramming and perturbation of purine metabolism in osteoclasts.35 In RA patients, the levels of numerous amino acids are diminished to varying degrees, potentially indicating a dysregulation in amino acid metabolism that could play a pivotal role in the development of RA.36 Interventions with BBR may offer relief for RA patients by contributing to the amelioration of dysregulated amino acid (Guanidinoethyl sulfonate, Guanidinoacetic acid, Phe-Ser and Arg-Val) derivatives in serum, suggesting a potential mechanism for its therapeutic efficacy in RA management. Additionally, other metabolites such as Histamine dihydrochloride, LPA (18:0/0:0), and 4-guanidinobutyric acid may serve as biomarkers of RA remission after BBR intervention, although further studies are needed to elucidate their exact role in RA improvement.
By integrating metabolomics with network pharmacology, a comprehensive approach is established to identify core targets and mechanisms, offering a more precise understanding of the network of BBR action against RA. We identified 24 key targets (PIK3CB, PIK3CD, and NOS3, among others), 6 key metabolites (D-Galactose, 4-Guanidinobutanoate, 3’-monoiodo-L-thyronine, 3alpha,7alpha-Dihydroxy-5beta-cholestane, Androst-5-ene-3beta,17beta-diol, and 3’,5’-Cyclic AMP), and 6 related pathways (tyrosine metabolism, phosphatidylinositol phosphate metabolism, androgen and estrogen biosynthesis and metabolism, bile acid biosynthesis, purine metabolism, and urea cycle and metabolism of arginine, proline, glutamate, aspartate, and asparagine). This approach provides a method to verify the results of both approaches. The integrated analysis of metabolomics and network pharmacology revealed that the PI3K/AKT pathway plays a pivotal role in the treatment of RA. This pathway influences the mammalian target of rapamycin protein (mTOR), inhibiting fibroblast-like synoviocyte (FLS) autophagy, and exacerbating RA by promoting synovial cell proliferation.37 Furthermore, PI3K/AKT signaling not only contributes to abnormal FLS proliferation and synovial inflammation but also affects osteoclast differentiation and generation.38 Elevated PI3K expression in RA synovial tissue may regulate synovial fibroblasts, contributing to inflammatory erosive arthritis and TNF-α-mediated cartilage destruction. TNF-α prompts T cells to produce macrophage-colony stimulating factor, triggering osteoblasts to generate RANKL, ultimately leading to osteoclast formation.39 These osteoclasts migrate, causing bone and articular cartilage destruction, ultimately worsening RA progression.38 In our investigation, we conducted molecular docking and IF to validate the interaction between BBR and the PI3K/AKT pathway. The findings indicated that BBR tightly binds to PIK3CB, PIK3CD, and NOS3, and reduced the expression of PIK3CB, PIK3CD, AKT and NOS3. These results suggested that BBR may alleviate inflammation by inhibiting the PI3K/AKT pathway.
The concept of bacterial involvement in the onset and progression of RA traces its origins back to the 19th century when a correlation between RA and tuberculosis emerged, suggesting Mycobacterium infection as a potential cause.40,41 In recent times, the gut-joint-axis hypothesis gained traction, supported by observations that 20% of inflammatory bowel disease patients experience recurrent episodes of peripheral arthritis.42 The gut microbiota represents the largest microbial community within the human body, often referred to as the “invisible organ”. Its metabolic capacity underscores its crucial role in both host health and disease.43 While our data unveiled a novel target for BBR in CIA rats, the precise mechanism through which BBR guards against metabolic abnormalities remains elusive.3 Microbiota dysbiosis is a hallmark of RA patients,3 and well-established rodent arthritis models corroborate this finding.44 Moreover, specific alterations in gut bacterial abundance have been linked to clinical RA manifestations.45 For instance, diminished levels of Lactobacillus and heightened levels of Prevotella are recognized features of RA patients’ gut microbiota.46 This study observed similar distinctions in gut microbial compositions between control and RA rats. BBR could altered gut dysbiosis at the phylum level by downregulating Patescibacteria, Proteobacteria, Bacteroidota and upregulating Verrucomicrobiota. At the genus level, BBR increased the abundance of Akkermansia, Prevotellaceae_UCG_001, Eubacterium, Blautia, Lachnoclostridium, UBA1819, Erysipelatoclostridium, [Ruminococcus]_torques_group, Faecalibaculum, Romboutsia, Incertae_Sedis, Flavonifractor, unclassified_Clostridia, Anaerostipes, [Eubacterium]_nodatum_group, [Clostridium]_innocuum_group, Holdemania, unclassified_[Clostridium]_methylpentosum_group, Sutterella and decreased the abundances of Candidatus_Saccharimonas, unclassified_Oscillospiraceae, unclassified_Muribaculaceae, Christensenellaceae_R_7_group, Monoglobus, unclassified_Ruminococcaceae, unclassified_Desulfovibrionaceae, uncultured_rumen_bacterium, Candidatus_Soleaferrea, NK4A214_group, Colidextribacter, unclassified_Peptococcaceae, Prevotellaceae_Ga6A1_group, unclassified_Prevotellaceae, unclassified_Clostridia_vadinBB60_group, Tuzzerella, Prevotella_9, Papillibacter, unclassified_RF39, UCG_009, Alloprevotella, Peptococcus, Candidatus_Arthromitus, unclassified_Atopobiaceae, Rikenellaceae_RC9_gut_group in CIA rats. Hence, there is compelling evidence suggesting that BBR’s impact on RA may stem from its regulation of specific intestinal microorganisms. Among these, Akkermansia predominates in healthy populations, conferring protective effects on digestive health while bolstering the host’s immune system, metabolism, and gut barrier integrity.47 Conversely, Blautia can serve as an exogenous antigen, stimulating lymphocyte proliferation and cytokine activation, thus fostering inflammatory responses that contribute to cartilage injury and bone changes.48 Romboutsia and Faecalibacterium are associated with intestinal health, being abundant in healthy mucosa and reduced in oncogenic states such as colorectal cancer. Their decrease may signify a vulnerability to disease progression.49 Prevotella_9 exhibits a positive correlation with serum levels of antibodies to cyclic citrullinated peptide (ACPA) and rheumatoid factor (RF), suggesting a potential role in RA pathogenesis.50 Increasing its abundance could mitigate inflammation and benefit RA patients. Lachnoclostridium, an anaerobic bacterium found in the human intestinal flora, produces butyric acid, contributing to colorectal cancer prevention. It correlates positively with age and disease duration in RA patients, indicating a potential protective role.48 Based on our findings and existing literature, Akkermansia, Blautia, Romboutsia, Faecalibacterium, Prevotella_9, and Lactobacillus emerge as pivotal bacteria for RA relief. BBR intervention modulates the abundance of these bacteria, rebalancing the intestinal microecology, reducing inflammation, and thereby alleviating RA symptoms.
The gut microbiota plays a crucial role in host metabolism through its interaction with host signaling pathways. For instance, the heightened presence of Collinsella in the gut microbiota of RA patients correlates with increased levels of alpha-aminoadipic acid and asparagine.51 In our study, we observed a significant negative correlation between Guanidinoethyl sulfonate and the abundance of Alloprevotella. Guanidinoacetic acid showed positive correlations with Candidatus_Soleaferrea, Holdemania, and unclassified_Clostridia, while negatively correlating with Peptococcus, Prevotella_9, Tuzzerella, unclassified_Desulfovibrionaceae, and unclassified_Prevotellaceae. Phe-Ser exhibited positive correlations with Peptococcus, Prevotella_9, Tuzzerella, UCG_009, unclassified_Muribaculaceae, and unclassified_Prevotellaceae, but negative correlations with [Eubacterium]_nodatum_group and Incertae_Sedis. Arg-Val was negatively correlated with Peptococcus, Prevotella_9, unclassified_Atopobiaceae, and unclassified_Prevotellaceae. These host metabolites likely interact with the gut microbiota, yet the precise mechanism by which BBR influences this metabolism in CIA rats requires further investigation.
Conclusion
In summary, our comprehensive analysis, which combined network pharmacology, metabolomics, intestinal flora studies, and molecular biology techniques, has elucidated the pharmacological effects and molecular mechanisms by which BBR alleviates RA. The findings highlight the PI3K/AKT, MAPK, and VEGF signaling pathways—critical mediators of inflammation and immune response—as key targets of BBR. Molecular docking and experimental validation further confirmed the involvement of the PI3K/AKT pathway. Additionally, BBR was shown to modulate the intestinal microbiota. These results enhance our understanding of the molecular underpinnings of BBR’s anti-RA effects. However, further research, including in vitro studies, targeted metabolomics, fecal microbiota transplantation and fecal metabolomics analyses, is needed to fully validate these mechanisms.
Acknowledgments
Thanks to the Acupuncture and Orthopedics Laboratory of Hubei University of Chinese Medicine for providing the experimental platform for this study.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388(10055):2023–2038. doi: 10.1016/s0140-6736(16)30173-8 [DOI] [PubMed] [Google Scholar]
- 2.Bécède M, Alasti F, Gessl I, et al. Risk profiling for a refractory course of rheumatoid arthritis. Semin Arthritis Rheumatism. 2019;49(2):211–217. doi: 10.1016/j.semarthrit.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 3.Zaiss MM, Joyce Wu HJ, Mauro D, Schett G, Ciccia F. The gut-joint axis in rheumatoid arthritis. Nat Rev Rheumatol. 2021;17(4):224–237. doi: 10.1038/s41584-021-00585-3 [DOI] [PubMed] [Google Scholar]
- 4.Kyriachenko Y, Falalyeyeva T, Korotkyi O, Molochek N, Kobyliak N. Crosstalk between gut microbiota and antidiabetic drug action. World J Diabetes. 2019;10(3):154–168. doi: 10.4239/wjd.v10.i3.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. New Engl J Med. 2016;375(24):2369–2379. doi: 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- 6.Gai X, Liu S, Ren T, Liu Y. Advances in chemical constituents and pharmacological effects of coptis chinensis. Chin Herb Med. 2008;49(20):4919–4927. [Google Scholar]
- 7.Rui Z, Chang-Pei X, Jing-Jing Z, Hong-Jun Y. Research progress on chemical compositions of coptidis rhizoma and pharmacological effects of berberine. Zhongguo Zhong Yao Za Zhi. 2020;45(19):4561–4573. doi: 10.19540/j.cnki.cjcmm.20200527.202 [DOI] [PubMed] [Google Scholar]
- 8.Chang W, Chen L, Hatch GM. Berberine as a therapy for type 2 diabetes and its complications: from mechanism of action to clinical studies. Biochem Cell Biol. 2015;93(5):479–486. doi: 10.1139/bcb-2014-0107 [DOI] [PubMed] [Google Scholar]
- 9.Yin J, Ye J, Jia W. Effects and mechanisms of berberine in diabetes treatment. Acta Pharmaceutica Sinica B. 2012;2(4):327–334. doi: 10.1016/j.apsb.2012.06.003 [DOI] [Google Scholar]
- 10.Huang D-N, Wu -F-F, Zhang A-H, Sun H, Wang X-J. Efficacy of berberine in treatment of rheumatoid arthritis: from multiple targets to therapeutic potential. Pharmacol Res. 2021;169:105667. doi: 10.1016/j.phrs.2021.105667 [DOI] [PubMed] [Google Scholar]
- 11.Fan -X-X, Leung EL-H, Xie Y, et al. Suppression of lipogenesis via reactive oxygen species-AMPK signaling for treating malignant and proliferative diseases. Antioxid Redox Signal. 2018;28(5):339–357. doi: 10.1089/ars.2017.7090 [DOI] [PubMed] [Google Scholar]
- 12.Li L, Yang L, Yang L, et al. Network pharmacology: a bright guiding light on the way to explore the personalized precise medication of traditional Chinese medicine. ChinMed. 2023;18(1):146. doi: 10.1186/s13020-023-00853-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Zhao C, Zhang Y, et al. (1)H NMR-based metabolomics coupled with molecular docking reveal the anti-diabetic effects and potential active components of berberis vernae on type 2 diabetic rats. Front Pharmacol. 2020;11:932. doi: 10.3389/fphar.2020.00932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saikia S, Bordoloi M. Molecular docking: challenges, advances and its use in drug discovery perspective. Curr Drug Targets. 2019;20(5):501–521. doi: 10.2174/1389450119666181022153016 [DOI] [PubMed] [Google Scholar]
- 15.Liu C, He L, Wang J, et al. Anti-angiogenic effect of shikonin in rheumatoid arthritis by downregulating PI3K/AKT and MAPKs signaling pathways. J Ethnopharmacol. 2020;260:113039. doi: 10.1016/j.jep.2020.113039 [DOI] [PubMed] [Google Scholar]
- 16.Yue M, Tao Y, Fang Y, et al. The gut microbiota modulator berberine ameliorates collagen-induced arthritis in rats by facilitating the generation of butyrate and adjusting the intestinal hypoxia and nitrate supply. FASEB j. 2019;33(11):12311–12323. doi: 10.1096/fj.201900425RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan Y, Sun W, Yang J, et al. The protective effect of traditional Chinese medicine jinteng qingbi granules on rats with rheumatoid arthritis. Front Pharmacol. 2024;15:1327647. doi: 10.3389/fphar.2024.1327647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mateen S, Shahzad S, Ahmad S, et al. Cinnamaldehyde and eugenol attenuates collagen induced arthritis via reduction of free radicals and pro-inflammatory cytokines. Phytomedicine. 2019;53:70–78. doi: 10.1016/j.phymed.2018.09.004 [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Cheng Z, Zhong Y, et al. Extracorporeal photopheresis reduces inflammation and joint damage in a rheumatoid arthritis murine model. J Transl Med. 2024;22(1):305. doi: 10.1186/s12967-024-05105-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminf. 2014;6:13. doi: 10.1186/1758-2946-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–w364. doi: 10.1093/nar/gkz382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Shen Y, Wang S, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45(W1):W356–w360. doi: 10.1093/nar/gkx374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piñero J, Saüch J, Sanz F, Furlong LI. The DisGeNET Cytoscape app: exploring and visualizing disease genomics data. Comput Struct Biotechnol J. 2021;19:2960–2967. doi: 10.1016/j.csbj.2021.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amberger JS, Hamosh A. Searching online Mendelian inheritance in man (OMIM): a knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinform. 2017;58:1.2.1–1.2.12. doi: 10.1002/cpbi.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 26.Yao J, Kong W, Jiang J. Learning from berberine: treating chronic diseases through multiple targets. Sci China Life Sci. 2015;58(9):854–859. doi: 10.1007/s11427-013-4568-z [DOI] [PubMed] [Google Scholar]
- 27.Qiu S, Sun H, Zhang AH, et al. Natural alkaloids: basic aspects, biological roles, and future perspectives. Chinese J Nat Med. 2014;12(6):401–406. doi: 10.1016/s1875-5364(14)60063-7 [DOI] [PubMed] [Google Scholar]
- 28.Eicher T, Kinnebrew G, Patt A, et al. Metabolomics and multi-omics integration: a survey of computational methods and resources. Metabolites. 2020;10(5):202. doi: 10.3390/metabo10050202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong Y, Luo J, Tang T, et al. Exploring pharmacological mechanisms of Xuefu Zhuyu decoction in the treatment of traumatic brain injury via a network pharmacology approach. Evid Based Complementary Altern Med. 2018;2018:8916938. doi: 10.1155/2018/8916938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheng S, Wang J, Wang L, et al. Network pharmacology analyses of the antithrombotic pharmacological mechanism of Fufang Xueshuantong capsule with experimental support using disseminated intravascular coagulation rats. J Ethnopharmacol. 2014;154(3):735–744. doi: 10.1016/j.jep.2014.04.048 [DOI] [PubMed] [Google Scholar]
- 31.Yu H, Chen J, Xu X, et al. A systematic prediction of multiple drug-target interactions from chemical, genomic, and pharmacological data. PLoS One. 2012;7(5):e37608. doi: 10.1371/journal.pone.0037608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcinkiewicz J, Kontny E. Taurine and inflammatory diseases. Amino Acids. 2014;46(1):7–20. doi: 10.1007/s00726-012-1361-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling H, Li M, Yang C, et al. Glycine increased ferroptosis via SAM-mediated GPX4 promoter methylation in rheumatoid arthritis. Rheumatology. 2022;61(11):4521–4534. doi: 10.1093/rheumatology/keac069 [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez AE, Ducker GS, Billingham LK, et al. Serine metabolism supports macrophage IL-1β production. Cell Metab. 2019;29(4):1003–1011.e4. doi: 10.1016/j.cmet.2019.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao S, Li Y, Song R, et al. L-arginine metabolism inhibits arthritis and inflammatory bone loss. Ann Rheumatic Dis. 2024;83(1):72–87. doi: 10.1136/ard-2022-223626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Liu L, Luo J, Peng X. Metabolites from specific intestinal bacteria in vivo fermenting Lycium barbarum polysaccharide improve collagenous arthritis in rats. Int J Biol Macromol. 2023;226:1455–1467. doi: 10.1016/j.ijbiomac.2022.11.257 [DOI] [PubMed] [Google Scholar]
- 37.Qu Y, Wu J, Deng JX, et al. MicroRNA-126 affects rheumatoid arthritis synovial fibroblast proliferation and apoptosis by targeting PIK3R2 and regulating PI3K-AKT signal pathway. Oncotarget. 2016;7(45):74217–74226. doi: 10.18632/oncotarget.12487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S, Ma H, Zhang H, Deng C, Xin P. Recent advances on signaling pathways and their inhibitors in rheumatoid arthritis. Clin Immunol. 2021;230:108793. doi: 10.1016/j.clim.2021.108793 [DOI] [PubMed] [Google Scholar]
- 39.Hayer S, Pundt N, Peters MA, et al. PI3Kgamma regulates cartilage damage in chronic inflammatory arthritis. FASEB j. 2009;23(12):4288–4298. doi: 10.1096/fj.09-135160 [DOI] [PubMed] [Google Scholar]
- 40.Whitehouse MW. Rheumatoid arthritis and tuberculosis. Lancet. 1986;2(8508):688–689. doi: 10.1016/s0140-6736(86)90195-9 [DOI] [PubMed] [Google Scholar]
- 41.Holoshitz J, Klajman A, Drucker I, et al. T lymphocytes of rheumatoid arthritis patients show augmented reactivity to a fraction of mycobacteria cross-reactive with cartilage. Lancet. 1986;2(8502):305–309. doi: 10.1016/s0140-6736(86)90003-6 [DOI] [PubMed] [Google Scholar]
- 42.Jethwa H, Abraham S. The evidence for microbiome manipulation in inflammatory arthritis. Rheumatology. 2017;56(9):1452–1460. doi: 10.1093/rheumatology/kew374 [DOI] [PubMed] [Google Scholar]
- 43.Yadav M, Verma MK, Chauhan NS. A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol. 2018;200(2):203–217. doi: 10.1007/s00203-017-1459-x [DOI] [PubMed] [Google Scholar]
- 44.Rehaume LM, Mondot S, Aguirre de Cárcer D, et al. ZAP-70 genotype disrupts the relationship between microbiota and host, leading to spondyloarthritis and ileitis in SKG mice. Arthritis Rheumatol. 2014;66(10):2780–2792. doi: 10.1002/art.38773 [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Quezada SL, Martínez-Bonilla GE, Cruz-Castro A, et al. AB0607 effect of probiotic lactobacillus casei shirota on clinical manifestations and serum cytokines in patients with rheumatoid arthritis. Ann Rheum Dis. 2013;71(Suppl 3):673. doi: 10.1136/annrheumdis-2012-eular.607 [DOI] [Google Scholar]
- 46.Maeda Y, Takeda K. Role of gut microbiota in rheumatoid arthritis. J Clin Med. 2017;6(6):60. doi: 10.3390/jcm6060060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Effendi R, Anshory M, Kalim H, et al. Akkermansia muciniphila and faecalibacterium prausnitzii in immune-related diseases. Microorganisms. 2022;10(12):2382. doi: 10.3390/microorganisms10122382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Z, Wu Y, Luo Y, et al. Self-balance of intestinal flora in spouses of patients with rheumatoid arthritis. Front Med. 2020;7:538. doi: 10.3389/fmed.2020.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mangifesta M, Mancabelli L, Milani C, et al. Mucosal microbiota of intestinal polyps reveals putative biomarkers of colorectal cancer. Sci Rep. 2018;8(1):13974. doi: 10.1038/s41598-018-32413-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozhakhmetov S, Babenko D, Issilbayeva A, et al. Oral microbial signature of rheumatoid arthritis in female patients. J Clin Med. 2023;12(11):3694. doi: 10.3390/jcm12113694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J, Wright K, Davis JM, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome med. 2016;8(1):43. doi: 10.1186/s13073-016-0299-7 [DOI] [PMC free article] [PubMed] [Google Scholar]