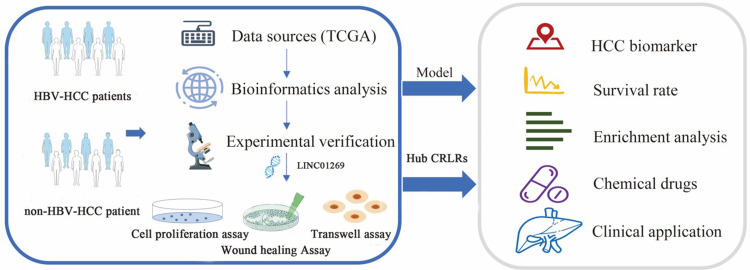
Abstract
Background
Hepatocellular carcinoma (HCC) triggered by Hepatitis B virus (HBV) remains a significant clinical challenge, necessitating novel therapeutic interventions. Copper ionophores, recognized for introducing an innovative type of programmed cell death termed cuproptosis, present promising potentials for cancer therapy. Nevertheless, The role of cuproptosis-related lncRNAs (CRLRs) in HBV-HCC has not been clearly elucidated.
Methods
This study utilised univariate Cox, least absolute shrinkage and selection operator (LASSO), and multivariable Cox regression analyses to establish a signature for CRLRs in HBV-HCC. This prognostic model was validated with an independent internal validation cohort, combined with clinical parameters, and used to construct a nomogram for patient survival predictions. Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) were employed to explore associated biological pathways. Additionally, a protein-protein interaction (PPI) network was developed, and implications for tumour mutational burden (TMB) and drug response were examined. A comprehensive bioinformatics analysis of these hub CRLRs was performed, followed by experimental validation through quantitative real-time PCR (qRT-PCR) and functional cellular assays.
Results
The nomogram showed high predictive accuracy for HBV-HCC patient survival. GO and GSEA analyses indicated that these lncRNAs are involved in pathways related to cancer and oestrogen metabolism. A PPI network consisting of 201 nodes and 568 edges was developed, and the TMB and drug response differed significantly between high- and low-risk groups. Analyses identified three hub CRLRs, SOS1-IT1, AC104695.3, and LINC01269, which were significantly differentially expressed in HCC tissues. In vitro, LINC01269 was found to enhance HCC cell proliferation, invasion, and migration.
Conclusion
The first systematic exploration of the roles of CRLRs in HBV-HCC demonstrates their critical involvement in the disease’s pathogenesis and possible therapeutic implication. The distinct expression patterns and significant biological pathways suggest that these lncRNAs may facilitate novel therapeutic targets.
Keywords: cuproptosis, HBV-HCC, proliferation, invasion, migration, LINC01269
Graphical Abstract
Introduction
Hepatocellular carcinoma (HCC) is the predominant type of primary liver cancer, contributing significantly to global cancer morbidity and mortality. With approximately 830,000 annual deaths, it is the third leading cause of cancer-related fatalities worldwide.1 Chronic hepatitis B virus (HBV) infection is a major risk factor for HCC,2 especially prevalent in Asia3 and Africa.4 Early-stage HBV-associated HCC (HBV-HCC) can be treated surgically,5 but advanced stages have limited treatment options, with systemic therapies like multikinase inhibitors offering minimal survival benefits.6,7 Recent attempts to utilise immune checkpoint inhibitors have also been hindered by varying patient responses8 and significant side effects.9 Consequently, there is an urgent need for novel therapeutic strategies that target specific molecular pathways involved in HBV-HCC progression. Recent discoveries have highlighted the role of cuproptosis10—a copper-dependent form of regulated cell death distinct from apoptosis and ferroptosis11—in cancer development. This has opened new avenues for research into cancer prognosis and treatment. Previous studies suggest that long non-coding RNAs (lncRNAs) are pivotal in regulating cell death pathways. However, the specific contribution of lncRNAs related to cuproptosis in HBV-HCC remains unexplored.
Emerging evidence indicates that cuproptosis-related lncRNAs (CRLRs) might play crucial roles in cancer biology, influencing tumour progression, recurrence, and response to immunotherapy. In a recent study by Wang et al,12 16 CRLRs were used to create a prognostic marker score. Enrichment analyses have shown that the biological functions of lncRNAs are associated with tumorigenesis. Wang et al13 developed a risk model based on 10 CRLRs that can be used to predict the prognosis of patients with gastric cancer. Xu et al14 used 11 CRLRs to construct prognostic prediction models, with good results in predicting the prognosis of patients with low-grade glioma. Han et al15 constructed a scoring system for CRLRs in a study of soft tissue sarcoma that can be used as an independent prognostic indicator; patients in the high-risk group had a worse prognosis than those in the low-risk group. Functional enrichment analysis and tumour immune microenvironment analysis have also indicated that CRLRs may play a role in the tumour immune microenvironment. At present, there is limited research on the potential role of CRLRs in the tumour immunoenvironment, yet some studies have further explored this aspect. For instance, Guo et al16 discovered that in high-risk breast cancer patients, key pathways are enriched in immune response and cell activation, with the low-risk group showing higher ESTIMATE scores and improved responses to immunotherapy due to elevated CD8+ T cell infiltration and TMB status. Similarly, Zhang et al17 identified differences in immune-related pathways between high- and low-risk groups in breast cancer. Xu et al18 ‘s study also found significant associations between CRLR characteristics and immune responses in colorectal cancer, demonstrating notable differences in immune function, cell scores, and checkpoint expression between risk groups.
To date, there have been no studies on cuproptosis in HBV-HCC, and it remains unclear whether cuproptosis-related genes are associated with the prognosis of patients with HBV-HCC; furthermore, the potential role of cuproptosis-related genes in HBV-HCC is unknown. Effective and innovative prognostic models are needed to improve the feasibility of targeted therapy for HBV-HCC. Given that few HBV-HCC-related lncRNAs have been identified, some effective indicators related to cuproptosis to predict the prognosis of HBV-HCC patients may provide potential new targets for the future development of HBV-HCC nucleic acid therapeutics from a lncRNA perspective.
Materials and Methods
Data Collection
The RNA sequencing data (RNA-seq) of HBV-HCC patients were obtained from TCGA database (https://portal.gdc.cancer.gov/), and after excluding patients with incomplete survival follow-up data or with a survival time of 0, a total of six normal samples and 102 hBV-HCC samples were obtained. HBV-HCC mutation data were downloaded from the TCGA database. Complete clinical data of the corresponding patients were subsequently collected from the cBio Portal database (http://www.cbioportal.org/). The current study follows TCGA data access policies and publication guidelines. In this study, sixteen cases of fresh tissue samples of HCC tumors and adjacent cancer were obtained from Jiangsu Provincial Cancer Hospital. Our study has been performed in accordance with the Declaration of Helsinki, and approved by the ethics committee of Jurong hospital affiliated to Jiangsu University. We confirm that all methods were performed in accordance with the relevant guidelines and regulations. Informed consent was obtained from all subjects involved in the study.
Identification of Cuproptosis-Associated lncRNAs with Prognostic Significance in HBV-HCC
A total of 19 genes (Supplementary Table 1) were identified through a review literature and summarization of cuproptosis-related genes. The TCGA transcriptional expression profiling data were classified into lncRNAs or mRNAs using Perl. The expression levels of cuproptosis-related mRNAs and lncRNAs were extracted using the ‘limma’ package in R software to construct a cuproptosis-related mRNA‒lncRNA co-expression network (filtering criterion: 0.4 for the correlation coefficient and p < 0.001 for the p-value of the correlation test). Then, HBV-HCC CRLRs were identified. The co-expression results were visualized by generating Sankey diagrams using the ‘ggplot2’ and ‘ggalluvial’ packages in R software.
Construction of a Risk Scoring Model Based on Prognostic CRLRs in HCC
Prognosis-related CRLRs were screened by univariate COX regression (P < 0.01), and to reduce overfitting during the modeling process, we then performed least absolute shrinkage and selection operator (LASSO) Cox regression analysis,19 employing the R package glmnet to perform ten-fold cross-validation on the obtained data, implementing 1000 cycles, to identify prognostic CRLRs. To construct a prognostic risk model, a multivariable Cox regression analysis was performed using the Akaike Information Criterion (AIC) to identify CRLRs with independent prognostic significance. The risk score was defined as the sum of the product of the expression level and the coefficient of each gene: risk score = (expression level of gene A × coefficient of gene A) + (expression level of gene B × coefficient of gene B), etc. Heatmaps of correlations between CRLRs and cuproptosis-related mRNAs in the model were constructed using the ‘reshape2’ and ‘tidyverse’ packages in R. Using the risk score for HBV-HCC patients, the patients were divided into a high-risk group and a low-risk group, and The Kaplan-Meier survival analysis20 was utilised to compare the two groups, employing the ‘Survival’ and ‘Survminer’ packages. Furthermore, the ‘pheatmap’ package was used to create a distribution of CRLR risk scores in the prediction model, determined the survival status of each patient, and construct a heatmap of the risk of expression of CRLRs for each sample. Validation was conducted using an internal validation cohort, with data for the internal validation set obtained through the ‘boot’ package in R software using Bootstrap resampling with replacement, a method recommended for the internal validation of prognostic models.21–24 Then, univariate and multivariate COX regression analyses were performed for combinations of clinical features to determine whether the 3-CRLR model was an independent prognostic indicator for HCC. The validity of this prognostic model was evaluated by constructing multiple receiver operating characteristics (ROCs) and time-dependent ROCs using the “Survival”, “Survminer”, “time ROC”, “dplyr”, “rms”, and ‘pec’ packages, and the predictive ability of the model was evaluated using the C-index. Finally, PCA was performed using the ‘limma’ and ‘scatterplot3d’ packages to obtain 3-dimensional distribution patterns of risk samples classified under different models, followed by an investigation of whether the lncRNAs involved in the model construction could better differentiate between high- and low-risk groups of patients with HBV-HCC.
Construction of Clinical Nomograms Based on Clinical Features
To construct clinical nomograms that predict overall survival (OS) at 1, 3, and 5 years, we employed several R packages, including “survival”, “regplot”, and “rms”. These tools facilitated the integration of both high-risk population data and clinicopathological features, allowing for the development of a comprehensive prognostic model. The ‘survival’ package was instrumental in analysing event time data, specifically tailored to capture OS rates across different time frames. Meanwhile, the ‘regplot’ package provided advanced plotting capabilities, enabling us to visually represent the regression models effectively. The ‘rms’ package offered robust support for regression modelling strategies, crucial for the calibration and validation of our nomograms. To ensure the reliability and accuracy of our predictions, we also generated calibration curves. These curves play a crucial role in assessing how well the nomogram-predicted probabilities of OS align with actual outcomes. By depicting the relationship between observed survival rates and nomogram predictions, the calibration curves provide visual validation of the model’s consistency and predictive accuracy.
Enrichment Analysis and PPI Network Analysis
Differentially expressed genes (DEGs) between the high- and low-risk groups were identified using the “limma” package and filtered by setting the absolute value of logFC > 1 and false discovery rate (FDR) < 0.05. The identified differentially expressed genes (DEGs) were subjected to Gene Ontology (GO) analysis to provide annotations and classifications for genes based on biological processes (BP), molecular functions (MF), and cellular components (CC). This analysis was conducted using the “clusterProfiler”, “org.Hs.eg.db”, “enrichplot”, and “ComplexHeatmap” packages. Additionally, GSEA was utilised to identify biological functions and pathways significantly enriched in the high- and low-risk groups, with a false discovery rate (FDR) of less than 0.05; the following gene sets were used: c2.all.v2022.1.Hs.symbols.gmt, c5.all.v2022.1.Hs.symbols.gmt and h.all.v2022.1.Hs.symbols.gmt. To investigate the potential protein-protein interactions among the DEGs associated with risk, we constructed a Protein-Protein Interaction (PPI)25 network utilising the STRING database (https://string-db.org/). During the construction of the network, we focused on maintaining a high level of confidence in the interaction data. Therefore, only those interaction pairs with a confidence score exceeding 0.4 were deemed meaningful and retained for further analysis. Within this network, genes with a connection number equal to or greater than five were identified as hub genes, indicating their potential significance in the biological pathways or processes being studied.
Tumour Mutational Burden (TMB) Analysis
For the analysis of TMB, we extracted detailed genomic data, including gene information, mutation locations, and mutation types, using Perl software. This process allowed us to comprehensively assess the mutational landscape. Following data extraction, the ‘maftools’ package26 was employed to identify the 15 genes exhibiting the highest mutation frequencies, which are of particular interest due to their potential impact on tumour behaviour and patient prognosis. To visualise these mutations, we generated TMB waterfall plots that distinctly represent the mutation profiles across both high-risk and low-risk groups. Additionally, to further explore and present the variability between the two groups, we constructed a violin plot using the “ggpubr” package in R. This visual representation provides an intuitive comparison of TMB distributions, illustrating the central tendencies and variabilities effectively. Subsequently, we conducted a survival analysis based on the TMB data using the same R package. This analysis aimed to evaluate the prognostic significance of TMB in relation to patient survival.
Drug Sensitivity Prediction
To assess the potential impact of specific features on predicting clinical responses to HBV-HCC treatment, we conducted drug sensitivity predictions using R software. This involved utilising the “limma”, “oncoPredict”, and “parallel” packages to estimate and evaluate the half-maximal inhibitory concentration (IC50) values for various chemotherapy and targeted drugs across both high- and low-risk patient groups. The IC50 values serve as a critical metric for determining the efficacy of a drug, reflecting the concentration required to inhibit cell viability by 50%. Maeser et al27 developed an R package called oncopredict to predict drug responses in tumour patients. First, we utilized the oncopredict package to score the drug sensitivity for each sample. Subsequently, the limma and ggpubr packages were employed to predict the differential drug sensitivity between the high-risk and low-risk groups. We employed the Wilcoxon signed-rank test, a non-parametric statistical test, ensuring a rigorous evaluation of the data with a significance threshold set at p < 0.01. A lower IC50 value indicates higher sensitivity to the drug, which can guide the clinical selection of drugs for patients.
Analysis of Risk CRLRs in the Model
To comprehensively analyse the survival data and assess how different factors impact survival time, we stratified the expression levels of three critical genes into high- and low-expression groups based on their median values. This stratification allowed us to conduct Kaplan-Meier survival analysis, which is instrumental in evaluating the prognostic value of selected lncRNA genes in HBV-HCC patients. By comparing survival curves between these groups, we could determine the influence of gene expression levels on patient outcomes. Following the survival analysis, we utilised the “limma”, “plyr”, and “reshape2” packages in R to perform a pan-cancer differential analysis of CRLRs that demonstrated prognostic significance in the K-M analysis. This step was crucial to explore the expression patterns of CRLRs across various cancer types and identify potential prognostic biomarkers. We then performed Pearson correlation analysis using the “limma” and “tidyverse” packages to identify genes significantly correlated with the CRLRs, applying a stringent filtering criterion of correlation coefficients greater than 0.5 and p-values less than 0.001. The resulting correlations were visualised through heatmaps created with the “ComplexHeatmap” package, providing an intuitive view of gene coexpression networks. To further explore the biological implications of these findings, we assessed the biological functions and pathways associated with coexpression of hub CRLR risk genes using GO annotation and GSEA. The “clusterProfiler”, “org.Hs.eg.db”, “dplyr”, and “ComplexHeatmap” packages in R facilitated these analyses, with significance thresholds set at FDR < 0.05 and P < 0.05. This analysis helped identify critical biological processes and pathways linked to CRLR expression. Finally, to evaluate drug responsiveness, we estimated the drug sensitivity of each sample using the “limma”, “oncoPredict”, and “parallel” packages. We then analysed the differential expression, ROC curves, and time-dependent ROC curves for risk CRLRs to assess their potential clinical diagnostic value. This multi-faceted approach not only highlights the prognostic significance of CRLRs but also provides insights into their role in treatment outcomes.
Cell Culture and Transfection
The normal hepatic cell line LO2, along with HCC cell lines HepG2, Hep3B2.1–7, and HCC-LM3, were sourced from the Cell Bank of the Chinese Academy of Sciences in Shanghai, China. The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM), which was supplemented with 10% fetal bovine serum from Gibco to provide essential nutrients and growth factors. Culturing was performed at a stable temperature of 37°C in an atmosphere containing 5% CO2, ensuring conditions ideal for cell growth and replication. To investigate the role of LINC01269 in HCC, specific small interfering RNA (siRNA) sequences from GenePharma (China) were used to knock down its expression. HCC cells were transfected with siRNAs by LipofectamineTM3000 (Invitrogen, USA). This process involved preparing a complex of siRNA and Lipofectamine™ 3000, followed by its addition to the HCC-LM3 cells cultured in growth medium. The cells were then incubated under standard conditions to allow sufficient time for the siRNA to exert its gene-silencing effects, effectively reducing LINC01269 expression.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) Detection of the Expression Level of Prognostic CRLRs in Clinical Tumor Samples and Cell Lines
Total RNA was isolated using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Carlsbad, CA, USA) under the guidance of the manufacturer’s scheme. RNA concentration and purity were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Next, cDNA synthesis was done by reverse transcription of RNA (1 μg) with HiScript 1st Strand cDNA Synthesis Kit (Vazyme, China), and it was amplified using SYBR Green Kit (Vazyme). Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal reference comparison gene. The expression of GAPDH was normalized, and the 2−ΔΔCt method was used to calculate the expression of the related gene lncRNA. The primer sequences are as follows: AC104695.3 forward primer TCCCCAGAGTGAACTTTTGC reverse primer TGGCATTTATTTGTCCCTCA; LINC01269 forward primer ATTTGGCTCAGGCATTTGTC, reverse primer CGTTGGTGATGACCAGAAAA; SOS1-IT1 forward primer TTGTTGTCTTTGGGCCATCT REVERSE PRIMER CCCAGCACTTTGGAAGGTTA; GAPDH forward primer GCACCGTCAAGGCTGAGAAC, reverse primer TGGTGAAGACGCCAGTGGA.
Cell Proliferation Assay
The assessment of cell proliferation was conducted using the Cell Count Kit-8 (CCK-8) assay, which facilitates sensitive colorimetric quantification of viable cells. Cells were seeded at a density of 2000 cells per well in 96-well flat-bottomed plates (Corning Inc., Corning, NY, USA) and incubated overnight at 37°C in a 5% CO2 atmosphere. Following 24, 48, 72, and 96 hours of incubation, at specified post-treatment intervals, the cell culture medium was replaced with 100 μL of 10% CCK-8 reagent (Beyotime, China) prepared in serum-free DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). The plates were then incubated for an additional 2 hours at 37°C in the same incubator. To evaluate relative cell numbers under different conditions, the absorbance of the CCK-8 reaction product was measured at 450 nm using a SpectraMax M3 microplate reader (Molecular Devices, San Jose, CA, USA). Cells were seeded in 6-well plates and cultured at 500 cells per well for approximately 14 days. Cells were fixed with 4% paraformaldehyde for 20 minutes, gently washed once with PBS and the crystal violet dye was added to each well. After 20 minutes, excess crystal violet was washed with pure water, then photographed and counted.
Wound Healing Assay
A wound healing assay was conducted on HCC-LM3 cell lines under different treatment conditions. Each cell culture was dissociated into single-cell suspensions and prepared in medium at a density of 6 × 105 cells/mL. Following this, 1 mL of this cell suspension was introduced into each well of a six-well plate, combined with 1 mL of the cell culture medium, and subjected to incubation at 37°C for a duration of 12 hours. A sterile pipette tip was employed to incise a scratch in the cell monolayer, and images were captured at time zero (t = 0 h) using a microscope to determine the initial wound area. Further imaging occurred at 48 hours to monitor the wound closure attributable to cell migration in the scratched region. The wound area was subsequently assessed and computed using the ImageJ software.
Transwell Migration and Invasion
The migratory and invasive capabilities of cells under various experimental conditions were evaluated through transwell migration and invasion assays. Transwell inserts possessing 8 μm pores were placed into 24-well companion plates (Corning Inc., Corning, NY, USA). For invasion assays, these inserts were pre-coated with a 20% Matrigel matrix layer (Corning Matrigel Matrix, Corning Inc., Corning, NY, USA), which functioned as an extracellular matrix barrier to assess the invasive potential of the cells. Post-treatment, cells were harvested and introduced into the upper chambers of the transwell inserts in 200 μL of serum-free DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). The lower chambers were filled with DMEM supplemented with 10% fetal bovine serum (FBS) as a chemoattractant (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). The cells were then incubated for 24 hours under standard culture conditions at 37 °C with 5% CO2. Following incubation, cells that had not invaded were carefully removed from the upper surface of the transwell filters using cotton swabs (Puritan Medical Products, Guilford, ME, USA). Migrated or invaded cells on the underside of the inserts were fixed using a 4% paraformaldehyde solution (Sigma-Aldrich, St. Louis, MO, USA) and then stained with 0.4% crystal violet dye (Sigma-Aldrich, St. Louis, MO, USA).
Results
Identification of HBV-HCC CRLRs and Construction of a 3-Gene Signature
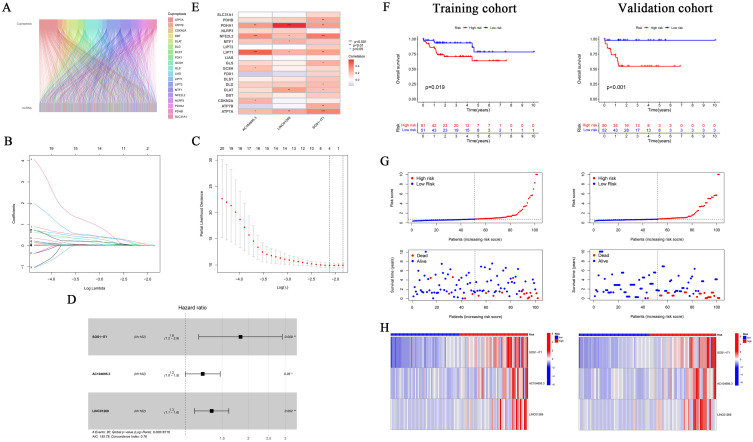
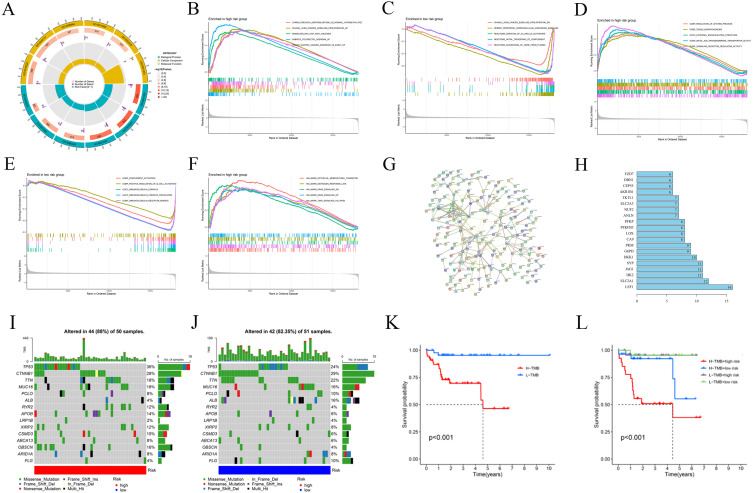
A total of 869 CRLRs were identified from 16,876 lncRNAs (filtering criteria: correlation coefficient > 0.4 and p < 0.001). The results were visualized using a Sankey diagram of co-expressed CRLRs and cuproptosis-related mRNAs (Figure 1A).
Figure 1.
Identification of Cuproptosis-associated lncRNA prognostic features in hepatocellular carcinoma (HBV-HCC). (A) Sankey relationship diagram of cuproptosis genes and cuproptosis-associated Long non-coding RNAs (lncRNAs). (B, C) Least absolute shrinkage and selection operator (LASSO) regression screened of cuproptosis-related lncRNAs at the minimum point of cross-validation. (D) The forest plot shows the results of the multivariate Cox regression analysis. (E) Correlation of lncRNAs with cuproptosis-related genes in risk models. (F) Kaplan–Meier survival curves of overall survival of HBV-HCC patients between low- and high-risk groups in the train and internal validation sets, respectively. (G) Distribution and overall survival status of patient risk scores in the train and internal validation sets. (H) The expression heatmap of 3 cuproptosis-associated lncRNAs in the train sets and validation sets. Significant differences are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
Through univariate regression analysis, 34 prognostic genes associated with cuproptosis were identified from 869 CRLRs (Supplementary Table 2). Next, LASSO regression analysis was used to screen risk genes, eventually resulting in 4 cuproptosis-related lncRNAs, namely, SOS1.IT1, AC145207.8, AC104695.3 and LINC01269 (Figure 1B and C). Further multivariate Cox regression analysis identified 3 robust cuproptosis-related lncRNAs with independent prognostic significance, namely, SOS1.IT1, AC104695.3 and LINC01269 (Figure 1D). The risk score was calculated as follows: risk score = expression value of SOS1.IT1 × (0.6069) + expression value of AC104695.3 × (0.1920) + expression value of LINC01269 × (0.2906). The correlation heatmap of the 3 selected lncRNAs and cuproptosis-related mRNAs is shown in Figure 1E.
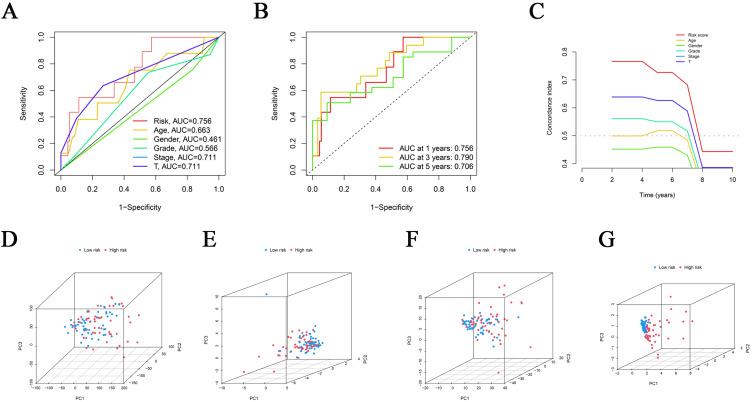
The two sets (the training set and the internal validation set) of data were comparable in terms of clinical features (P > 0.05, Supplementary Table 3). Next, Kaplan-Meier (K-M) survival analysis was used to determine the prognostic significance of the 3-CRLR model. The results showed that patients in the high-risk group in both the training set and the internal validation set had a shorter survival time, with statistically significant differences (p =0.019 0.05 and p < 0.001, respectively, Figure 1F). No significant differences in Progression Free Survival were observed between the training set and the internal validation set (Supplementary Figure 1). As seen in the risk survival status plot, the survival time of patients was inversely proportional to the risk score in both the training set and internal validation set (Figure 1G and H). We constructed a 3-CRLR model to predict the survival time of HBV-HCC patients. The 3-CRLR risk model had an area under the curve (AUC) of 0.756, which was higher than that of univariate models that included age, sex, tumour grade, Stage tumor-node-metastasis (TNM) and T stage, indicating that the 3-CRLR model predicted the survival time of HBV-HCC patients with higher accuracy than did the clinical univariate models (Figure 2A). The 1-, 3-, and 5-year AUCs for the constructed 3-CRLR model were 0.756, 0.790, and 0.706, respectively, indicating that the 3-CRLR model had good predictive accuracy for HBV-HCC patients (Figure 2B). In addition, the 3-CRLR model had the largest consistency index (C-index) value and thus outperformed other clinicopathological factors, indicating that the model was the best to predict the survival time of HBV-HCC patients (Figure 2C). Principal component analysis (PCA) indicated that lncRNAs included in the construction of the CRLR model (Figure 2G) could better differentiate between high- and low-risk groups of HBV-HCC patients than all gene (Figure 2D), cuproptosis-related mRNAs (Figure 2E) and CRLRs (Figure 2F), demonstrating the accuracy of the 3-CRLR model.
Figure 2.
Receiver operating characteristic curves (ROC), C-index and Principal Component Analysis (PCA). (A) Predictive accuracy of the risk model compared with clinicopathologic characteristics such as age, gender, stage and T; (B) Accuracy of the risk characteristic based on a whole-group prediction of 1-, 3-, and 5-years receiver operating characteristic curves. (C) C-index curve of the risk model. PCA plots depicted the distribution of samples based on the all genes of the prognostic signature (D), cuproptosis-related genes (E), cuproptosis-related lncRNAs (F), and the expression of three lncRNAs of the prognostic signature (G).
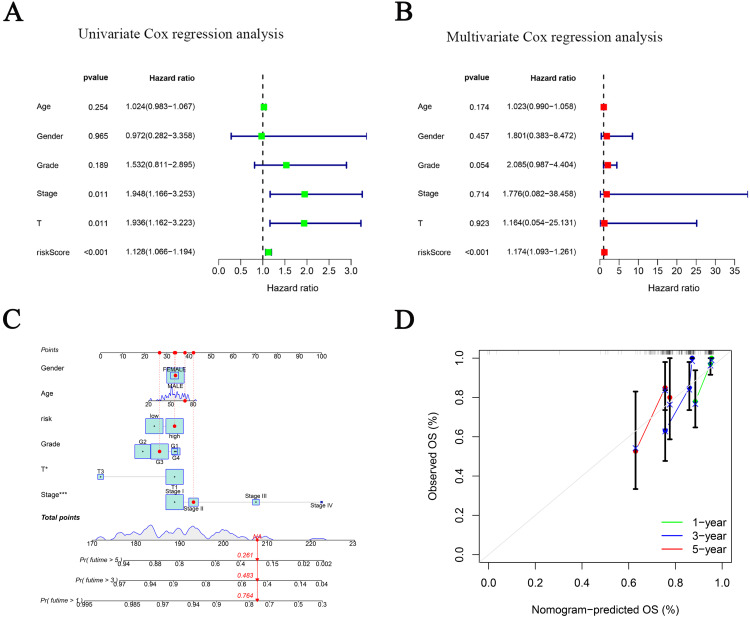
Univariate analysis was used to compare the model and clinical factors with the survival time and survival status of patients, respectively, to observe whether the model was correlated with prognosis. Multivariate analysis was used to assess the interactions between factors to determine whether the model could be used as a prognostic factor independent of other clinical traits. The results showed that the 3-CRLR risk model was associated with prognosis in both the univariate (Figure 3A) and multivariate analyses (Figure 3B) and was statistically significant (both P < 0.01), indicating that the 3-CRLR model can be used as an independent prognostic tool for HBV-HCC.
Figure 3.
Verification of the independent prognostic ability and clinical predictive ability of the 3 cuproptosis-related lncRNAs signature for HBV-HCC in The Cancer Genome Atlas (TCGA) and construction nomogram of cuproptosis-related lncRNAs prognostic signatures and clinicopathological characteristics. Forest plots of univariate (A) and multivariate (B) Cox regression analyses revealed that risk score could be an independent prognostic factor. (C) The nomogram for both clinical-pathological factors and risk score could predict the probability of survival based on the total points; (D) Calibration curves test the agreement between actual and predicted outcomes at 1, 3, and 5 years. Significant differences are indicated by ***P < 0.001.
Clinical Prediction by Nomogram
To provide a quantitative tool for clinical application, a nomogram with age, sex, pathological grade, N stage, T stage, Stage TNM, and risk score as indicators was developed and used to predict the survival time of HBV-HCC patients (Figure 3C). Calibration plots showed good agreement between the actual and predicted ratios for 1-, 3-, and 5-year overall survival (OS) (Figure 3D).
Risk DEGs Gene Ontology (GO) Analysis, Gene Set Enrichment Analysis (GSEA) and Protein-Protein Interaction (PPI) Network Analysis
GO analysis (Figure 4A) indicated that DEGs were significantly enriched in 54 BPs (FDR < 0.05), with the top 5 being “kidney development”, “renal system development”, “urogenital system development”, “anatomical structure homeostasis” and “response to hypoxia”. DEGs were significantly enriched in 9 CCs (FDR < 0.05), with the top 5 being “apical plasma membrane”, “apical part of cell”, “neuron to neuron synapse”, “postsynaptic density” and “asymmetric synapse”. However, DEGs were not enriched in MF, and thus, these DEGs may not function primarily in the extracellular matrix. GSEA (Figure 4B-F) results for the gene set c2.all.v2022.1.Hs.symbols.gmt, indicated the involvement of multiple tumour-related pathways in patients in the high-risk group, for example, carrilloreixach hepatoblastoma vs normal hypomethylated and up, Chiang liver cancer subclass proliferation up, and the involvement of multiple tumour, complement activation, and haeme clearance pathways in patients in the low-risk group, for example, Chiang liver cancer subclass proliferation dn, desert periportal hepatocellular carcinoma subclass up, and scavenging of heme from plasma. GSEA results for the gene set c5.all.v2022.1.Hs.symbols.gmt indicated the involvement of gobp regulation of system process, gobp tissue morphogenesis, and gomf signaling receptor regulator activity in patients in the high-risk group and the involvement of many bp, cc and mf pathways, such as gobp complement activation, gobp positive regulation of b cell activation, and gocc immunoglobulin complex, in patients in the low-risk group. GSEA results for the gene set h.all.v2022.1.Hs.symbols.gmt indicated the involvement of epithelial-mesenchymal transition, oestrogen metabolism, KRAS signalling, and NFKB/TNFA signalling pathways, such as hallmark epithelial mesenchymal transition, hallmark estrogen response late, hallmark kras signaling dn, hallmark kras signaling up, and hallmark tnfa signaling via nfkb, in patients in the high-risk group; in contrast, there was no significant enrichment in the low-risk group. The risk DEG PPI network constructed using STRING consisted of 201 nodes and 568 edges (Figure 4G). Based on a total of 31 hub genes (degree ≥ 5) in the network, the top 20 genes with the most connections were identified and the gene with the highest degree (degree = 16) was LEF1 (Figure 4H).
Figure 4.
Gene Ontology (GO) analysis, gene set enrichment analysis (GSEA), protein-protein interaction (PPI) network and Tumor mutation burden (TMB). (A) Gene Ontology (GO) analysis demonstrated the richness of molecular biological processes (BP), cellular components (CC), and molecular functions (MF). (B-F) GSEA revealed significant differences in the enrichment of c2.all.v2022.1.Hs.symbols.gmt, c5.all.v2022.1.Hs.symbols.gmt, and h.all.v2022.1.Hs.symbols.gmt in the TCGA HBV-HCC cohort. (G) PPI network of risk differentially expressed genes (DEGs). (H) Information about nodes in the PPI network. Tumor mutation burden (TMB) in the TCGA HBV-HCC cohort. Waterfall plots of somatic mutation characteristics in the two groups (I and J). K–M survival curves between the high- and low-TMB groups (K); K–M survival curves between the four groups (L).
Risk Score-Related Tumour Mutational Burden (TMB) Analysis
Analyses of HCC gene mutation profiles downloaded from the Cancer Genome Atlas (TCGA) database and we found the 15 genes with the highest mutation frequency. The high-risk group (Figure 4I) had high frequencies of TP53, CTNNB1, TTN, MUC16, RYR2, OBSCN and XIRP2 mutations, and the low-risk group (Figure 4J) had high frequencies of CTNN-B1, TP53, TTN, MUC16, ALB, FLG and ARID1A mutations in these 15 genes. The mutation frequencies of 8 genes in the low-risk group were lower than those in the high-risk group, and the mutation frequencies of 6 genes in the low-risk group were higher than those in the high-risk group. The top 15 genes with the highest mutation frequency were visualized using a waterfall plot. Subsequently, a differential analysis of TMB between the high- and low-risk groups was performed, and there was no significant difference between the 2 groups (Supplementary Figure 2). Furthermore, K-M analysis of the TMB indicated that patients with a low TMB had better OS outcomes (Figure 4K). Finally, K-M analysis of the TMB combined with risk score indicated that patients with a low TMB + low risk score and patients with a low TMB + high risk score had the best prognosis, followed by patients with a high TMB + low risk score; patients with a high TMB + high risk score had the worst prognosis (Figure 4L).
HBV-HCC Potential Drug Screening
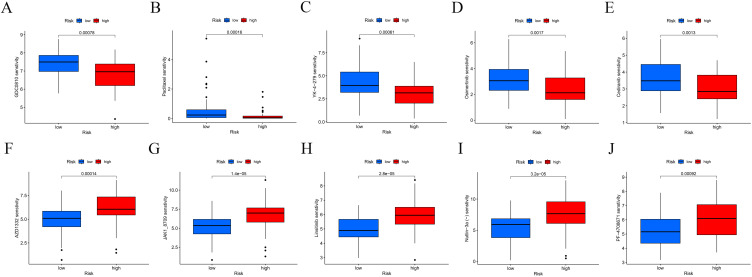
Drug sensitivity was compared between the 2 groups. The lower the half maximal inhibitory concentration (IC50) value, the higher was the drug sensitivity. Patients in the high-risk group were more sensitive to GDC0810 (Figure 5A), Paclitaxel (Figure 5B), YK-4-279 (Figure 5C), Osimertinib (Figure 5D) and Cediranib (Figure 5E). In addition, the high-risk group was more resistant to AZD1332 (Figure 5F), JAK1_8709 (Figure 5G), Linsitinib (Figure 5H), Nutlin-3a (-) (Figure 5I) and PF-4708671 (Figure 5J).
Figure 5.
Drug sensitivity analysis of low- and high-risk groups. The distribution of the half maximal inhibitory concentration (IC50) showed a significant difference of patients in the low- and high- risk groups among (A) GDC0810, (B) Paclitaxel, (C) YK-4-279, (D) Osimertinib, (E) Cediranib, (F)AZD1332, (G) JAK1_8709, (H) Linsitinib, (I) Nutlin-3a (-), and (J) PF-4708671.
Analysis of Risk Variables in the Model
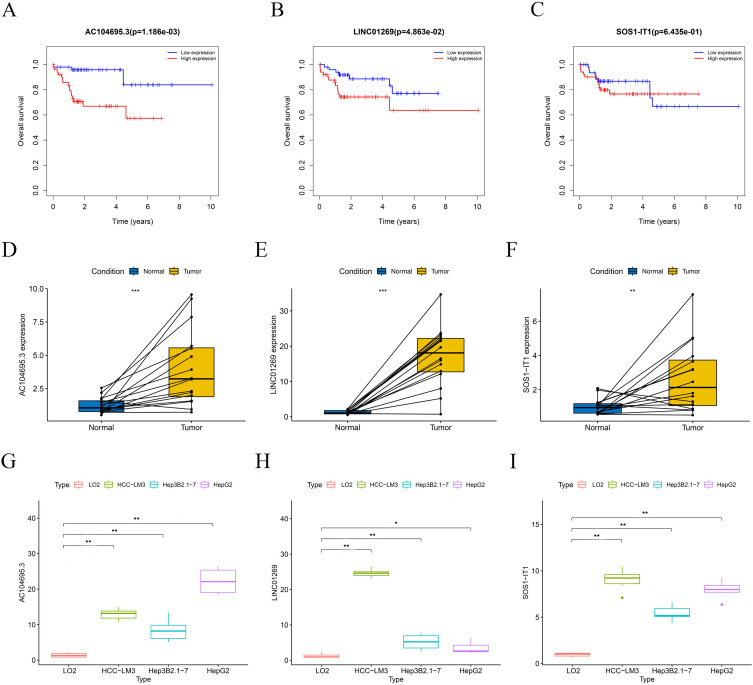
We explored the differential expression of AC104695.3 (Figure 6A), LINC01269 (Figure 6B) and SOS1-IT1 (Figure 6C) in pan-cancer data (including 32 other cancer types) in the TCGA database. The results indicate that the three hub CRLRs exhibit differential expression across various tumor tissues. Furthermore, the genes regulated by 3 hub CRLRs were identified by co-expression analysis. Supplementary Figure 3, 4 and 5 show the top 10 genes co-expressed with 3 hub CRLRs, respectively; Figure 6D–F are the corresponding correlation heatmaps. Spearman correlation coefficients were utilized to ascertain the associations between three prognostic CRLRs and 22 levels of immune cell infiltration. The NK cells resting (P=0.042) demonstrated a noteworthy inverse correlation with the expression of AC104695.3 (Figure 6G). As depicted in Figure 6I, T cells regulatory (Tregs) (P=0.002) exhibited a significant negative correlation with SOS1-IT1. However, as illustrated in Figure 6H, the expression of LINC01269 did not exhibit a substantial correlation with various types of immune cells.
Figure 6.
Differential expression, co-expressed genes in HBV-HCC patients, and immune infiltration. The differential expression of AC104695.3 (A) and LINC01269 (B) and SOS1-IT1(C) in pan-cancer data (including 32 other cancer types) in the TCGA database. The genes regulated by AC104695.3 (D) LINC01269 (E) and SOS1-IT1(F) were identified by co-expression analysis (corresponding correlation heatmaps). Correlation between the relative abundances of 22 immune cells and lncRNA AC104695.3 (G) LINC01269 (H) and SOS1-IT1(I) expression level. Significant differences are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
The GO analysis reveals that the genes co-expressed with the three hub CRLRs are enriched in different biological pathways (Supplementary Figure 6A-C). GSEA results for the gene sets c2.all.v2022.1.Hs.symbols.gmt, c5.all.v2022.1.Hs.symbols.gmt and h.all.v2022.1.Hs.symbols.gmt indicated significant enrichment of multiple tumour-associated pathways, complement pathways, and immune pathways in the AC104695.3 (Supplementary Figure 6D-F), LINC01269 (Supplementary Figure 6G-I) and SOS1-IT1 (Supplementary Figure 6J-L) high- and low-expression groups. As demonstrated in Supplementary Figures 7A-E, patients in the high expression group of AC104695.3 exhibit lower IC50 values compared to those in the low expression group. This indicates increased sensitivity to drugs such as AZD7762, GDC0810, Pevonedistat, Staurosporine, and WIKI4, suggesting that effective inhibition can be achieved with lower doses, potentially allowing for reduced medication intake while still achieving satisfactory therapeutic outcomes. Conversely, as shown in Supplementary Figures 7F-H, the high-expression group exhibits higher IC50 values, indicating a reduced sensitivity to JAK1_8709, Linsitinib, and PF-4708671. This suggests a certain degree of resistance in these patients, necessitating higher doses or alternative treatment strategies for effective management. Similarly, in the high expression group of LINC01269, presented in Supplementary Figures 8A-E, there is a lower IC50, indicating heightened sensitivity to AMG-319, Dasatinib, MK-8776, PD173074, and Savolitinib. However, in Supplementary Figures 8F-J, higher IC50 values suggest a level of resistance to Cytarabine, ERK_2440, Luminespib, PLX-4720, and Sabutoclax. Further analysis reveals that patients in the high expression group of SOS1-IT1 are likely more sensitive to Daporinad, GDC0810, I-BRD9, Paclitaxel, and YK-4-279 (Supplementary Figures 9A-E), but show resistance to AZD1332, AZD2014, JAK1_8709, JQ1, and Nutlin-3a (-) (Supplementary Figures 9F-J). The lower IC50 values in the high-expression groups may reflect the enhanced drug mechanism mediated by these lncRNA biomarkers. These findings provide valuable insights for personalised therapy, potentially guiding drug selection and dosage adjustments.
In addition, the expression levels of 3 hub CRLRs were significantly higher in specimens from the TCGA HBV-HCC cohort than in normal tissue (Supplementary Figure 10). The receiver operating characteristic curve (ROC) analyses suggest that the prediction of three HUB CRLRs is more accurate for patients with HBV-HCC (Supplementary Figure 10). The screened risk variables AC104695.3, LINC01269 and SOS1-IT1 were again subjected to K-M analysis, revealing that HBV-HCC patients with high AC104695.3 (Figure 7A) and LINC01269 (Figure 7B) expression had a shorter survival time and that high SOS1-IT1 (Figure 7C) expression was not associated with the prognosis of HBV-HCC patients.
Figure 7.
Hub cuproptosis-related lncRNA (CRLR) survival analysis in the model. K–M survival curves between the high- and low-expression groups of lncRNA AC104695.3 (A), LINC01269 (B) and lncRNA SOS1-IT1 (C). Validation of prognostic LncRNAs in the 3- CRLRs model in clinical HCC tumor samples (D - F). Expression levels of AC104695.3 (D), LINC01269 (E) and SOS1-IT1(F) in HCC cell lines HepG2, Hep3B2.1–7, HCC-LM3, and normal liver cell LO2(G - I). Significant differences are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
Validation of Signature‑related LncRNA Expression Levels
Validation with clinical tumor samples showed that, significantly higher expression levels of 3 hub CRLRs in the HCC than in the para-carcinoma normal tissues (Figure 7D - F). Validation results at the cellular level also provided support for this finding. The results showed that compared with LO2, the expression of 3 hub CRLRs increased in all three HCC cell lines (Figure 7G - I). These results suggest that these three CRLRs may play important roles in HCC.
LINC01269 Promote Cell Proliferation, Migration and Invasion in vitro
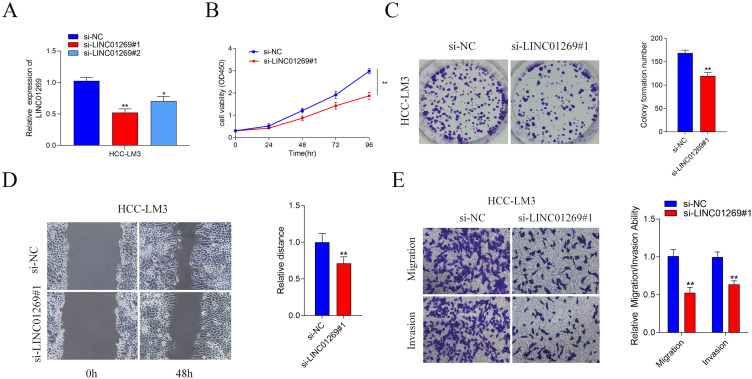
To estimate whether the expression of LINC01269 had the potential to affect the proliferation, migration, and invasion of HCC cells. Two small interfering RNAs (siRNAs) were created to knockdown LINC01269 expression, and qPCR was used to measure the relative expression of LINC01269 in HCC cells (Figure 8A). CCK-8 and colony formation assays showed that knockdown of LINC01269 inhibited the cell proliferation capacity (Figure 8B and C). We validated the influence of LINC01269 on cell metastasis. As shown in Figure 8D and E, knockdown of LINC01269 inhibited the migration of HCC cells in the Wound healing assay. Moreover, Transwell analysis demonstrated that the migration and invasion ability of HCC cells in si-LINC01269 group compared with the si-NC group.
Figure 8.
The effect of LINC01269 on HCC cells proliferation, migration and invasion. LINC01269 expression in HCC cells of different groups detected by qPCR (A) CCK-8 assay (B), Colony formation assay (C), Wound healing assay (D) and Transwell assay (E) of HCC cells transfected with si-NC and si-RNA. Significant differences are indicated by *P < 0.05 and **P < 0.01.
Discussion
Alpha-fetoprotein (AFP) is the most widely used serum biomarker for the diagnosis of HCC.28 However, only 60–70% of HCC patients have elevated serum AFP concentrations.29 Therefore, more sensitive, non-invasive biomarkers are needed for the better diagnosis of HCC. Copper is a dynamic signalling element with considerable influences on various processes, including lipolysis, cell proliferation, autophagy and neural activity.30,31 Excitingly, Tsvetkov et al10 found that copper ionophores induce a new form of cell death, one that is distinct from the classical apoptotic pathway. Cells that are highly dependent on mitochondrial respiration are more sensitive to copper-induced cell death, and the mitochondrial respiration pathway is necessary for copper ionophore-induced cell death. Copper-dependent death occurs through the direct binding of copper to lipidated components of the tricarboxylic acid (TCA) cycle. This leads to lipidated protein aggregation and the subsequent loss of iron-sulphur cluster proteins, resulting in proteotoxic stress and ultimately cell death.
A growing number of studies have explored the potential impact of cuproptosis genes on tumours. Lv et al32 showed that among melanoma patients, those with higher copper-related gene expression had a longer overall survival time than those with lower gene expression; in particular, the prognostic value of the LIPT1 gene in melanoma was identified, revealing the correlation of LIPT1 expression with immune infiltration in melanoma. Mo et al33 found that the cuproptosis lncRNA MIR31HG acts as an oncogene in lung adenocarcinoma to regulate TNFRSF21 by targeting miR-193a-3p. Therefore, it is of great significance to investigate the relationship between cuproptosis genes and tumours.
Based on the critical role of cuproptosis in cancer and the close interaction between cuproptosis and lncRNAs, TCGA transcriptome data were used in this study to analyse the potential mechanism and prognostic value of CRLRs in HBV-HCC, and 3 key CRLRs (SOS1-IT1, AC104695.3, LINC01269) were identified, thus leading to the development of a new prognostic risk model for HBV-HCC. Importantly, univariate and multivariate analyses indicated that the risk model was an independent prognostic factor for HBV-HCC. Next, a high-precision clinical prognostic nomogram was established to provide 1-, 3-, and 5-year OS predictions for patients with HBV-HCC, and the results indicated that the 3-CRLR model could be used to predict patient prognosis. Additionally, following the methodologies of previous researchers,34 we performed GO analysis on the identified DEGs, employing the clusterProfiler package for gene set enrichment analysis. DEGs were evaluated using an FDR-corrected p-value (adjusted P < 0.05) and an absolute log2 fold change (|log2 FC| ≥ 1). This similar analytical approach further corroborates the reliability of our study’s findings. The GO analysis results indicate that DEGs are enriched in biological functions such as “kidney development”, “renal system development”, and “apical plasma membrane” between the high-risk and low-risk groups. Similarly, we applied methods from previous studies to conduct GSEA.35 This was used to interpret the enriched biological terms that categorise prognostic risk scores between high-risk and low-risk groups in HBV-HCC patients, further supporting our conclusions. The GSEA results showed significant differences in the enrichment of various tumour-related signalling pathways, complement activation, haeme clearance, epithelial-mesenchymal transition, and oestrogen metabolism between the 2 groups. Considering that the TME plays an important role in tumourigenesis and progression, the TMB status of HBV-HCC was investigated, revealing that the TP53, CTNNB1, TTN, and MUC16 genes had high mutation frequencies in both the high-risk group and low-risk group. K-M analysis of the TMB combined with the risk score showed that patients with a high TMB + high risk score had the worst prognosis. The TMB analysis in our study is designed to provide an overview of the mutation landscape within HBV-HCC, highlighting the 15 genes with the highest mutation frequencies. This broad genetic analysis helps to characterise the tumour’s heterogeneity, which is paramount in understanding potential responses to therapies. While the TMB analysis provides an overarching view of mutation prevalence, the CRLRs in our model are selected for their direct correlation with clinical outcomes, representing a targeted approach in prognostication. The connection between TMB and the CRLR model lies in their complementary roles: TMB provides a macroscopic view of mutational diversity, which might influence tumour behaviour and therapeutic responses, whereas the CRLRs offer a microscopic, targeted perspective on prognostic evaluation and potential therapeutic targeting within the context of HBV-HCC. On this basis, sensitivity to chemotherapeutic drugs was investigated using the model. Eight drugs with significant differences in sensitivity were identified, namely, GDC0810, Paclitaxel, YK-4-279, AZD1332, JAK1_8709, Linsitinib, Nutlin-3a (-) and PF-4708671, providing reliable choices for clinical use. Among the high-risk patient group, there is a pronounced sensitivity to GDC-0810, Paclitaxel, YK-4-279, Osimertinib, and Cediranib. Notably, Paclitaxel, recognized as a microtubule stabilizer, possesses anticancer capabilities that transcend mere cell cycle inhibition. As a cornerstone of cancer treatment, cell cycle inhibitors—especially Taxanes—when used in tandem with anti-PD-1 therapies, have demonstrated the potential to disrupt HCC growth in murine models and extend survival periods. This underscores the immune-mediated tumour suppressive properties of cell cycle inhibitors and highlights their potential as adjunctive therapies in immunotherapy protocols.36 Cediranib, a broad-spectrum VEGFR tyrosine kinase inhibitor, has previously been shown in studies37 to have a high incidence of toxicity alongside preliminary antitumor efficacy at a daily dose of 30 mg in advanced HCC. Furthermore, other studies38 identify Osimertinib as a promising candidate for HCC treatment through dual targeting of tumour cells and angiogenesis. Currently, research on GDC-0810 and YK-4-279 in the context of HCC is mostly limited to bioinformatics with a paucity of clinical trials. Presently available sensitive drugs comprise targeted therapies and immune checkpoint inhibitors, which mainly target four critical receptors in HCC: VEGFR, PDGFR, EGFR, and FGFR. These drugs achieve effective treatment by inhibiting the activation of multiple pathways, blocking downstream signal transduction, and reducing tumour angiogenesis. Structural impacts of drugs such as Sorafenib, Cabozantinib, Lenvatinib, and Regorafenib on the corresponding HCC targets provide promising avenues for new drug development.39 Recent studies40 indicate that monotherapy with immune checkpoint inhibitors (ICIs) does not result in significant overall survival benefits for most advanced HCC patients. Consequently, research has shifted towards exploring potentially active combinations, like those involving ICIs with anti-angiogenic agents or two different ICIs. At present, the combination therapies of atezolizumab (anti-PD-L1) with bevacizumab (anti-VEGF), or durvalumab (anti-PD-L1) with tremelimumab (anti-CTLA-4), are considered the standard treatment for advanced HCC.41 The drugs identified as sensitive for high-risk HBV-HCC patients in our screening differ from those currently available due to their status as bioinformatically screened candidates, which inherently possess data dependency. Nonetheless, meticulous experimental and clinical validation is imperative to substantiate the therapeutic efficacy and value of these drug candidates for high-risk HBV-HCC populations.
The 3 lncRNA genes SOS1-IT1, AC104695.3 and LINC01269, identified as risk variables by the Cox-LASSO regression model, have been little studied, and there is a lack of research on their role in HBV-HCC. Previous studies have shown that the lncRNA SOS1-IT1 may affect endometrial carcinogenesis and progression by regulating autophagy processes42 and may serve a role in predicting the prognosis of patients with endometrial cancer.43 SOS1-IT1 expression is significantly increased in endometrial cancer under hypoxic conditions, and the hypoxia regulator HIF-1α can directly bind to the SOS1-IT1 promoter region and affect its expression level.44 In addition, SOS1-IT1 is associated with ivermectin targeting the lncRNA-EIF4A3-mRNA pathway in ovarian cancer.45 Studies on lncRNA AC104695.3 have shown that newly developed prognostic models that include AC104695.3 can predict the prognosis and immunotherapy response of patients with gastric adenocarcinoma and gastric cancer.46,47 Lv et al48 found that lncRNA LINC01269 was associated with genetic instability in breast cancer. The results of another study showed that LINC01269 can activate KRT80 and increase the therapeutic effect in head and neck squamous cell carcinoma.49
The results of the pan-cancer analysis showed that 3 hub CRLRs were significantly differentially expressed in a variety of cancers. The expression of 3 hub CRLRs was also significantly elevated in the TCGA HBV-HCC cohort. ROC analysis confirmed the 3 hub CRLRs expression results, suggesting that 3 hub CRLRs may play tumour-promoting roles in HBV-HCC and may serve as prognostic markers of cuproptosis with special significance in HBV-HCC. AC104695.3 was found to be involved in cell differentiation, development, regulation and protein transport, all of which are associated with tumorigenesis, invasion and metastasis.50 Through GSEA of lncRNA AC104695.3, several HCC-related signalling pathways, such as “desert periportal hepatocellular carcinoma subclass up”, “chiang liver cancer subclass polysomy7 up”, “liver cancer subclass proliferation dn” and “hsiao liver specific genes”, were identified, indicating its involvement in tumourigenesis and progression. In addition, lncRNA AC104695.3 was also associated with immune pathways, EMT and hypoxia. LINC01269 may play a role in glucose metabolism, in particular the pentose-phosphate shunt, NADPH regeneration, glucose 6-phosphate metabolic process, NADP metabolic process and monosaccharide binding. Tumour cells have a unique energy metabolism, and various glucose metabolic pathways play important roles in the regulation, metabolism and survival of cancer cells.51–54 The GSEA of LINC01269 also showed that it is associated with complement and immune pathways. SOS1-IT1 is also involved in RNA splicing and nuclear export pathways, and demonstrates enrichment in pathways associated with tumorigenesis, immunity, and cellular activation. Finally, the differences in drug sensitivity and resistance between three hub CRLRs high- and low-expression groups were explored. We also investigated the proliferation and migration of LINC01269 using CCK-8, Colony formation assay, Wound healing and Transwell assay. We found that knockdown of LINC01269 inhibited the proliferation and migration of HCC cells.
Effective and innovative prognostic models are needed to improve the feasibility of targeted therapy for HBV-HCC. A number of validated indicators associated with cuproptosis to predict the prognosis of HBV-HCC patients may provide potential options for the treatment of HBV-HCC. In this study, for the first time, a prognostic risk model was constructed using 3 CRLRs, and the model was able to predict the prognosis of patients with HBV-HCC and thus potentially guide clinical treatment. Our research identifies key biomarkers in HCC, which hold promising implications for clinical practice. The identification of specific CRLRs offers avenues for not only understanding the underlying pathology of HCC but also developing personalised medicine approaches. By integrating these CRLR markers into clinical settings, there is potential to enhance diagnosis precision and stratify patients based on their risk profiles, enabling tailored treatment strategies that could improve patient outcomes. Furthermore, our findings suggest that targeting these CRLRs could lead to new therapeutic interventions. The correlation of CRLR expression with treatment responses indicates that these markers could be employed to predict patient responsiveness to existing drugs, thereby informing treatment choices. Moreover, developing novel therapeutics aimed at modulating CRLR expression or function could lead to breakthrough treatments that specifically target tumour biology. We also recognise that the translation of these findings into clinical practice necessitates rigorous validation through clinical trials and real-world studies. Future research should focus on these aspects to confirm the utility and safety of CRLR-based interventions.
The risk model is feasible and valid. However, our study has some limitations. As the data and results of this study were based on TCGA transcriptomic, mutational and clinical data, the findings require additional prospective studies to validate the predictive value of this model. Second, although the findings of this study contribute to the understanding of the relationship of 3 CRLRs with HBV-HCC, research on their roles in cancer is still in its infancy. Due to the limited sample size and few relevant experimental studies, the functional mechanisms of 3 CRLRs need further experimental validation.
Conclusion
In conclusion, for the first time, the relationship between CRLRs and the prognosis of HBV-HCC patients was revealed. A new HBV-HCC prognosis prediction model based on CRLRs and a nomogram based on CRLR markers were developed, and potential sensitive drugs were screened. This information can be used for target selection and individualized immunotherapy for HBV-HCC. This study not only supports the clinical application of the 3-CRLR model for HBV-HCC patients but also provides useful suggestions for the research field of cuproptosis. We look forward to further advances in the study of cuproptosis in HBV-HCC in the near future, and on the basis of new knowledge gained, we will validate and improve the model to make it more accurate and effective.
Acknowledgments
We thank Zhe Zhang of the Jiangsu Cancer Hospital, Affiliated Cancer Hospital of Nanjing Medical University for providing HCC tissue specimens for this project. The authors gratefully thank the open-source provided by TCGA databases, as well as the developers of the R package.
Funding Statement
This study was supported by Project of Modern Hospital Management and Development Institute, Nanjing University, Aid project of Nanjing Drum Tower Hospital Health, Education & Research Foundation (project no. NDYG2021046), and Nursing Research Project at Nanjing Drum Tower Hospital (2021) (project no. ZSA491-3).
Abbreviations
AFP, alpha-fetoprotein; AIC, Akaike Information Criterion; AUC, area under curve; BP, biological process; C-index, consistency index; CC, cellular component; CCK-8, Cell Count Kit-8; CRLRs, cuproptosis-related lncRNAs; DEGs, differentially expressed genes; DMEM, Dulbecco’s Modified Eagle Medium; EMT, epithelial-mesenchymal transition; FBS, fetal bovine serum; FDR, false discovery rate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GO, Gene Ontology; GSEA, gene set enrichment analysis; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; IC50, half maximal inhibitory concentration; ICIs, immune checkpoint inhibitors; K-M, Kaplan-Meier; LASSO, least absolute shrinkage and selection operator; lncRNAs, Long noncoding RNAs; MF, molecular function; OS, overall survival; PCA, Principal component analysis; PPI, protein-protein interaction; qRT-PCR, quantitative real time polymerase chain reaction; RNA-seq, RNA sequencing; ROC, receiver operating characteristic curve; siRNA, small interfering RNA; TCA, tricarboxylic acid; TCGA, The Cancer Genome Atlas; TMB, tumour mutational burden; TNM, tumor-node-metastasis.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, L SR, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petruzziello A. Epidemiology of hepatitis B virus (HBV) and hepatitis C virus (HCV) related hepatocellular carcinoma. Open Virol J. 2018;12(1):26–32. doi: 10.2174/1874357901812010026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plummer M, de Martel C, Vignat J, et al. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4(9):e609–16. doi: 10.1016/S2214-109X(16)30143-7 [DOI] [PubMed] [Google Scholar]
- 5.M LJ, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19(3):151–172. doi: 10.1038/s41571-021-00573-2 [DOI] [PubMed] [Google Scholar]
- 6.Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised Phase 3 non-inferiority trial. Lancet. 2018;391(10126):1163–1173. doi: 10.1016/S0140-6736(18)30207-1 [DOI] [PubMed] [Google Scholar]
- 7.Bruix QS, Merle P, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389(10064):56–66. doi: 10.1016/S0140-6736(16)32453-9 [DOI] [PubMed] [Google Scholar]
- 8.Centanni M, Moes D, F TI, et al. Clinical pharmacokinetics and pharmacodynamics of immune checkpoint inhibitors. Clin Pharmacokinet. 2019;58(7):835–857. doi: 10.1007/s40262-019-00748-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, D GM, Li S, et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat Med. 2021;27(1):152–164. doi: 10.1038/s41591-020-1131-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;6586:1254–1261. doi: 10.1126/science.abf0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bock FJ, Tait SWG. Mitochondria as multifaceted regulators of cell death. Nat Rev Mol Cell Biol. 2020;21(2):85–100. doi: 10.1038/s41580-019-0173-8 [DOI] [PubMed] [Google Scholar]
- 12.Wang F, Lin H, Su Q, et al. Cuproptosis-related lncRNA predict prognosis and immune response of lung adenocarcinoma. World J Surg Oncol. 2022;20(1):275.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Hu Y, Liu S, et al. A newly established cuproptosis-associated long non-coding RNA signature for predicting prognosis and indicating immune microenvironment features in soft tissue sarcoma. J Oncol. 2022;2022:8489387. doi: 10.1155/2022/8489387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Liu K, Shen K, et al. A novel risk model construction and immune landscape analysis of gastric cancer based on cuproptosis-related long noncoding RNAs. Front Oncol. 2022;12:1015235. doi: 10.3389/fonc.2022.1015235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Wang C, Li S, et al. Prognosis and immune response of a cuproptosis-related lncRNA signature in low grade glioma. Front Genet. 2022;13:975419. doi: 10.3389/fgene.2022.975419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Q, Qiu P, Pan K, et al. Comprehensive analysis of cuproptosis-related long non-coding RNA signature and personalized therapeutic strategy of breast cancer patients. Front Oncol. 2022;12:1081089. doi: 10.3389/fonc.2022.1081089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Zhang Y, Bao J, et al. Cuproptosis combined with lncRNAs predicts the prognosis and immune microenvironment of breast cancer. Comput Math Methods Med. 2022;2022:5422698. doi: 10.1155/2022/5422698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu M, Mu J, Wang J, et al. Construction and validation of a cuproptosis-related lncRNA signature as a novel and robust prognostic model for colon adenocarcinoma. Front Oncol. 2022;12:961213. doi: 10.3389/fonc.2022.961213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tibshirani R. Regression shrinkage and selection via the lasso. J Royal 20 Statistical Society. 1996;58(58):267–288. doi: 10.1111/j.2517-6161.1996.tb02080.x [DOI] [Google Scholar]
- 20.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. doi: 10.1080/01621459.1958.10501452 [DOI] [Google Scholar]
- 21.Blackstone EH. Breaking down barriers: helpful breakthrough statistical methods you need to understand better. J Thorac Cardiovasc Surg. 2021;122(3):430–439. doi: 10.1067/mtc.2001.117536 [DOI] [PubMed] [Google Scholar]
- 22.Gu X, Li H, Sha L, et al. A prognostic model composed of four long noncoding RNAs predicts the overall survival of Asian patients with hepatocellular carcinoma. Cancer Med. 2020;9(16):5719–5730. doi: 10.1002/cam4.3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu X, Sha L, Zhang S, et al. Neutrophils and lymphocytes can help distinguish asymptomatic COVID-19 from moderate COVID-19. Front Cell Infect Microbiol. 2021;11:654272. doi: 10.3389/fcimb.2021.654272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sha L, Xu T, Ge X, et al. Predictors of death within 6 months of stroke onset: a model with Barthel index, platelet/lymphocyte ratio and serum albumin. Nurs Open. 2021;8(3):1380–1392. doi: 10.1002/nop2.754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeser D, Gruener RF, Huang RS. OncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22:bbab260. doi: 10.1093/bib/bbab260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A JA, Kulik LM, Sirlin CB, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American association for the study of liver diseases. Hepatology. 2018;68(2):723–750. doi: 10.1002/hep.29913 [DOI] [PubMed] [Google Scholar]
- 29.L LC, Rong Y, Chen H, et al. A logistic regression model for noninvasive prediction of AFP-negative hepatocellular carcinoma. Technol Cancer Res Treat. 2019;18:1533033819846632. doi: 10.1177/1533033819846632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang T, M PJ, A GA, et al. Copper is an essential regulator of the autophagic kinases ULK1/2 to drive lung adenocarcinoma. Nat Cell Biol. 2020;22(4):412–424. doi: 10.1038/s41556-020-0481-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao T, M AC, C CE, et al. Copper regulates rest-activity cycles through the locus coeruleus-norepinephrine system. Nat Chem Biol. 2018;14(7):655–663. doi: 10.1038/s41589-018-0062-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv H, Liu X, Zeng X, et al. Comprehensive analysis of cuproptosis-related genes in immune infiltration and prognosis in melanoma. Front Pharmacol. 2022;13:930041. doi: 10.3389/fphar.2022.930041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo X, Hu D, Yang P, et al. A novel cuproptosis-related prognostic lncRNA signature and lncRNA MIR31HG/miR-193a-3p/TNFRSF21 regulatory axis in lung adenocarcinoma. Front Oncol. 2022;12:927706. doi: 10.3389/fonc.2022.927706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song Q, Ruiz J, Xing F, et al. Single-cell sequencing reveals the landscape of the human brain metastatic microenvironment. Commun Biol. 2023;6:760. doi: 10.1038/s42003-023-05124-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouch RJ, Zhang J, Miller BC, et al. Distinct inflammatory Th17 subsets emerge in autoimmunity and infection. J Exp Med. 2023;220:e20221911. doi: 10.1084/jem.20221911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong PY, Chan CYK, Xue HDG, et al. Cell cycle inhibitors activate hypoxia-induced DDX41-STING pathway to mediate anti-tumor immune response in liver cancer. JCI Insight. 2024;9:e170532. doi: 10.1172/jci.insight.170532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu AX, Ancukiewicz M, Supko JG, et al. Efficacy, safety, pharmacokinetics, and biomarkers of cediranib monotherapy in advanced hepatocellular carcinoma: a Phase II study. Clin Cancer Res. 2013;19:1557–1566. doi: 10.1158/1078-0432.CCR-12-3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Q, He S, Zhan D. Osimertinib is a dual inhibitor of hepatocellular carcinoma and angiogenesis in an EGFR-independent manner, and synergizes with venetoclax. J Cancer Res Clin Oncol. 2023;149:10727–10735. doi: 10.1007/s00432-023-04926-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng Q, Huang Y, Zeng J, et al. Recent advancements in the small-molecule drugs for hepatocellular carcinoma (HCC): structure-activity relationships, pharmacological activities, and the clinical trials. Biomed Pharmacother. 2024;179:117343. doi: 10.1016/j.biopha.2024.117343 [DOI] [PubMed] [Google Scholar]
- 40.Rimassa L, Finn RS, Sangro B. Combination immunotherapy for hepatocellular carcinoma. J Hepatol. 2023;79:506–515. doi: 10.1016/j.jhep.2023.03.003 [DOI] [PubMed] [Google Scholar]
- 41.Childs A, Aidoo-Micah G, Maini MK, et al. Immunotherapy for hepatocellular carcinoma. JHEP Rep. 2024;6:101130. doi: 10.1016/j.jhepr.2024.101130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Zhang J, Liu Y, et al. An integrated autophagy-related long noncoding RNA signature as a prognostic biomarker for human endometrial cancer: a bioinformatics-based approach. Biomed Res Int;2020. 5717498. doi: 10.1155/2020/5717498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H, Cheng Y. Identification of autophagy-related long non-coding RNAs in endometrial cancer via comprehensive bioinformatics analysis. BMC Women’s Health. 2022;22(1):85. doi: 10.1186/s12905-022-01667-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Wan J, Feng Q, et al. Long non-coding RNA SOS1-IT1 promotes endometrial cancer progression by regulating hypoxia signaling pathway. J Cell Commun Signal. 2022;16(2):253–270. doi: 10.1007/s12079-021-00651-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li N, Zhan X. Anti-parasite drug ivermectin can suppress ovarian cancer by regulating lncRNA-EIF4A3-mRNA axes. EPMA J. 2020;11(2):289–309. doi: 10.1007/s13167-020-00209-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng C, Liu Y, He R, et al. Identification and validation of a novel cellular senescence-related lncRNA prognostic signature for predicting immunotherapy response in stomach adenocarcinoma. Front Genet. 2022;13:935056. doi: 10.3389/fgene.2022.935056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S, Li X, Tang C, et al. Inflammation-related long non-coding RNA signature predicts the prognosis of gastric carcinoma. Front Genet. 2021;12:736766. doi: 10.3389/fgene.2021.736766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv Z, Wang Q, Liu X, et al. Genetic instability-related lncRNAs predict prognosis and influence the immune microenvironment in breast cancer. Front Genet. 2022;13:926984. doi: 10.3389/fgene.2022.926984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y, Huang X, Zhang Z, et al. The long noncoding transcript HNSCAT1 activates KRT80 and triggers therapeutic efficacy in head and neck squamous cell carcinoma. Oxid Med Cell Longev. 2022;2022:4156966. doi: 10.1155/2022/4156966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen T, You Y, Jiang H, et al. Epithelial-mesenchymal transition (EMT): a biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol. 2017;232(12):3261–3272. doi: 10.1002/jcp.25797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39(8):347–354. doi: 10.1016/j.tibs.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Q JH, F LJ, Tian T, et al. NADPH homeostasis in cancer: functions, mechanisms and therapeutic implications. Signal Transduct Target Ther. 2020;5(1):231. doi: 10.1038/s41392-020-00326-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M RG, A PA, Szekely Z, et al. In cancer, all roads lead to NADPH. Pharmacol Ther. 2021;226:107864. doi: 10.1016/j.pharmthera.2021.107864 [DOI] [PubMed] [Google Scholar]
- 54.Ge T, Yang J, Zhou S, et al. The role of the pentose phosphate pathway in diabetes and cancer. Front Endocrinol. 2020;11:365. doi: 10.3389/fendo.2020.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.